Abstract
In the present scenario of a major demand for new compounds with antimicrobial activity, bacteriocin and bacteriocin-like inhibitory substances (BLIS) are promising tools against deteriorating and pathogenic microorganisms, thus having potential applications in both the food industry and infectious disease control. In the present report, we describe the genetic and phenotypic characteristics of BLIS produced by Enterococcus faecium E86, a strain previously isolated and sequenced by our group, focusing on the structural genes of two bacteriocins identified: enterocin TW21 and enterocin P. Transcription of all four genes associated with the biosynthesis and immunity of enterocin P and enterocin TW21 were confirmed by RT-PCR. However, Sanger sequencing confirmed a truncation of the structural gene of enterocin TW21 due to one base pair deletion (A/T). Thus, although E. faecium E86 was shown to carry two bacteriocinogenic gene clusters, only one cluster encodes a functional bacteriocin, enterocin P. Enterocin P was able to inhibit different strains of Listeria monocytogenes and vancomycin-resistant enterococci (both Enterococcus faecalis and Enterococcus faecium), showing intense bacteriolytic activity, in most cases.
Keywords: Bacteriocin, Enterocin P, Inhibitory activity, Listeria monocytogenes, VRE
Introduction
Members of the genus Enterococcus are usually referred as ubiquitous bacteria, which are widespread in various habitats [1], and two major species, Enterococcus faecalis and Enterococcus faecium, are frequently found in food and clinical samples [2, 3]. Although these species have been recognized as important causes of challenging to treat healthcare-associated infections due to acquisition of antimicrobial resistance [4], in foods such as cheese, sausages, and fermented products, some enterococci may contribute to texture, taste, aroma, and food safety by producing diverse aromatic compounds, enzymes, and bacteriocins [2, 5]. Studies also suggest probiotic benefits of E. faecium isolated from naturally fermented foods [6, 7]. Accordingly, at least two strains of the genus have already been introduced in the market as probiotics: E. faecium SF68® (NCIMB 10415; Cerbios-Pharma SA, Barbengo, CH) and E. faecalis Symbioflor 1 (SymbioPharm, Herborn, DE).
Bacteriocins are proteinaceous multi-functional compounds ribosomally synthesized by prokaryotes, which classically have antimicrobial activity against microorganisms of the same species or species related to the bacteriocin-producing strain [8, 9]. Bacteriocinogenic clusters usually include genes encoding the bacteriocin precursor peptide, processing enzymes, and transport, regulatory, and immunity proteins. In some cases, such clusters also harbor genes involved in amino acid modification [10, 11]. Currently, bacteriocins can be classified in six different classes, according to their structure: class I, also called lantibiotics, usually composed of thermostable, small peptides (< 5 kDa) carrying post-translationally modified amino acids, such as lanthionine and β-methyllanthionine [12]; class II, thermostable, small peptides (≤ 10 kDa) that do not undergo extensive post-translational modifications [13]; class III, proteins larger than 25 kDa, mostly heat-sensitive [14]; class IV, circular and thermostable bacteriocins, presenting carboxy and amino termini that are linked by an amide bond type, after cleavage of the leader peptide [15, 16]; class V, circular or linear peptides, with a characteristic binding of the cysteine thiol group to an α-carbon, mediated by post-translational modifications [17, 18]; and class VI, also called thiopeptides, small macrocyclic peptides (≤ 5 kDa) with vast post-translational modifications resulting in a central nitrogen-containing six-membered ring [19, 20]. It is important to highlight that there is no consensus in the literature on the classification of this group of substances. Thus, other research groups have proposed different classifications [21, 22].
These compounds have many biotechnological applications, especially in the food industry as biopreservatives [23]. Studies have shown that bacteriocins increase the shelf life of foods and beverages and reduce the risk of transmission of pathogens through the production chain. These natural antimicrobial compounds allow the use of less chemical additives, which is a common requirement from consumers nowadays [13, 24, 25]. Besides that, bacteriocins may be applied to different products since they differ in aspects such as solubility, stability, and spectra of action [13, 26].
Bacteriocins produced by enterococci, commonly called enterocins, are usually stable over a wide range of pH and temperature and may have a very diverse spectrum of action, including other enterococci and lactic acid bacteria (LAB), as well as pathogenic species of the genera Listeria, Staphylococcus, and Clostridium, besides the species Clostridioides difficile [5, 27]. Bacteriocins of different classes have been isolated and characterized in Enterococcus spp., most of them belonging to class II, and produced by E. faecium or E. faecalis, from both clinical and food origin [13, 28].
Some examples of this group would be enterocin P and enterocin TW21. Enterocin P is a class IIa bacteriocin, and its producing strain, E. faecium P13, was isolated from a fermented sausage. Its genetic group comprises two genes that encode the bioactive peptide (entP) and its immunity protein (entiP). The mature peptide is composed of 44 residues and has a theoretical molecular mass of 4493 Da. Its spectrum of action is wide and includes bacteria belonging to the genera Enterococcus spp., Lactobacillus spp., Pediococcus spp., and some food pathogens (Bacillus cereus, L. monocytogenes, and Staphylococcus aureus) [29]. Enterocin TW21 also belongs to class IIa and has a genetic group composed of two genes: the structural one (entTW21) and other related to immunity (entiTW21) [30]. The mature peptide has 48 residues and a molecular mass of 5300.6 Da [31]. Its spectrum of action comprises species such as Lactobacillus sakei, E. faecium, C. perfringens, and L. monocytogenes, among others [32].
In 2008, Miguel and collaborators [33] reported on an isolate, recovered from meat pie and identified as E. faecium E86, capable of producing a bacteriocin-like inhibitory substance. Afterward, Farias and collaborators (2019) [30] sequenced and assembled the genome of E. faecium E86 and found two bacteriocin gene clusters (encoding enterocin P and enterocin TW21), with the structural gene of TW21 appearing to have a premature stop codon due to a base pair deletion. Considering the potential of E. faecium E86 as a biopreservative producer, the present report describes genetic and phenotypic characteristics of the bacteriocins produced by this strain. Moreover, our results demonstrate the relevance of an accurate genetic characterization of bacteriocinogenic genes to assign the activity of bacteriocins.
Material and methods
Bacterial isolates and growth conditions
Strain E. faecium E86, the focus of the present study, was previously isolated from meat pie by our research group [33]. Listeria spp. [provided by the Listeria Collection (CLIST) of the Laboratory of Bacterial Zoonoses (Fiocruz)] and vancomycin-resistant enterococci (VRE; belonging to our bacterial collection) were used to evaluate the antimicrobial action of the enterocins produced by E. faecium E86. Listeria monocytogenes isolates (total of 25), belonging to the main serotypes involved in foodborne infections (1/2a, 1/2b, 1/2c, and 4b), were recovered from different ready-to-eat foods. Listeria innocua 2 was used as a control for the enterocin inhibitory activity test since this strain was shown to be highly susceptible to the products of E. faecium E86 [33]. Enterococcus isolates (total of 14) are representative of clinical samples of cases of infection or colonization in humans. E. faecalis ATCC 10100 was used as a van operon negative control. Listeria spp. and Enterococcus spp. isolates were grown in brain heart infusion [BHI (Difco, Detroit, USA)] and MRS [Man, Rogosa, and Sharpe (Difco)] culture media, respectively, whether or not added with agar [Merck, Darmstadt, DE; 1.5% (w/v) or 0.7% (w/v), depending on the need of the experiment].
Sequencing of the enterocin TW21 structural gene
Genomic DNA from E. faecium E86 was obtained as previously described [30] and subjected to polymerase chain reaction (PCR) for the amplification of the enterocin TW21 structural gene. PCR was carried out as recommended [34] by using an annealing temperature of 58 °C. The amplicons were purified by using the ExoSAP-IT enzyme (Promega, Madison, USA). Purified DNA (100 ng) and 5.0 pmol of each of the following primer oligonucleotides were used for the DNA sequencing employing the Sanger method: TW21Sanger-F (5’TAAAAAAGGGAGGCAATTATATGAA3’) and TW21 Sanger-R (5’TCAAAAAGTTTTTCTTTTTTATCTTCC3’). Sequencing was performed by Macrogen Inc. Both strands were sequenced. Nucleotide sequence analyses were performed using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) [35].
Transcriptional analysis of the genes involved in the biosynthesis of the bacteriocin(s) present in the E. faecium E86 genome
E. faecium E86 was grown in 5 mL of MRS for 6 and 10 h at 37 °C. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, DE) as described [36], with the use of 20 mg/mL lysozyme (Sigma-Aldrich, St. Louis, USA) and 5.000 U mutanolysin (Sigma-Aldrich) for cell disruption. The cDNA synthesis was performed with 500 ng of the DNase I-pre-treated RNA as substrate, using the Revert Aid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, USA). Pairs of primers for the genes involved in enterocin P (entP, 120 bp; entiP, 182 bp) and enterocin TW21 (entTW21, 138 bp; entiTW21, 274 bp) biosynthesis were applied to RT-PCR analyses, as described in Table 1. 16S rRNA-encoding genes were adopted as internal controls using the primers 16FX and 16RX [38]. The PCR reactions were carried out as described in the last item, except for the annealing temperature, which was 50 °C for the 16S gene primers and 55 °C for the other primers. Genomic DNA, RNA, and 16S rRNA cDNA were used as templates in these reactions as positive control, negative control, and internal control, respectively.
Table 1.
Oligonucleotide primers used in the RT-PCR experiments
| Genes | Oligonucleotides | Sequence (5’–3’) | tm (°C) | Used pairing temperature (°C) | Amplicon size expected (bp) | Reference |
|---|---|---|---|---|---|---|
| entP | entP F | TATGGTAATGGTGTTTATTGTAAT | 53 | 55 | 120 | [37] |
| entP R | ATGTCCCATACCTGCCAAAC | 53 | ||||
| entiP | entiP F | TAGCCACCCCAGAAATTAAA | 57 | 55 | 182 | This work |
| entiPR | TTCAGCAAGCAACTCCAATA | 57 | ||||
| entTW21 | entTW21 F | TGCTGCAACTTATTATGGAAA | 56 | 55 | 138 | This work |
| entTW21 R | ACTCCACCTAGCACTTTCGT | 56 | ||||
| entiTW21 | entiTW21 F | TTTGACTTAATTGCGAATGC | 56 | 55 | 274 | This work |
| entiTW21 R | GCATAGACATGGCACCATAA | 57 | ||||
| 16S rRNA | 16S primer 10 FX | GACTACCNGGGTATCTAATCC | 59 | 50 | 800 | [38] |
| 16S primer 804 RX | AGAGTTTGATCCTGGCTNAG | 58 |
tm, average denaturation temperature; bp, base pairs
Test of the antimicrobial activity of strain E. faecium E86 on solid medium
The antimicrobial activity of E. faecium E86 against 25 L. monocytogenes and 14 VRE isolates was tested by the agar-spot assay [39]. Ten microliters of the producing strain broth growth (7.0 log CFU) were inoculated as a spot on the surface of a MRS agar plate, which was then covered with 3 mL of BHI (L. monocytogenes) or MRS (VRE) soft agar containing 6.0 log CFU/mL of each target strain. Plates were incubated at 37 °C for 24 h, and the size of the growth inhibition zones was measured. These experiments were performed in triplicate. The strains L. innocua 2 and E. faecalis ATCC 10100 were also tested for comparison.
Partial purification of E. faecium E86 enterocin P
The enterocin P was partially purified by ammonium sulfate precipitation followed by cation exchange chromatography [40] from a 1-liter culture of E. faecium E86, grown in MRS, using optimized conditions for the bacteriocin production as previously described [33]. This enterocin preparation was dialyzed against ultrapure water and quantified by the agar diffusion assay to determine the arbitrary units per mL (AU/mL) [41], using 100 μL of twofold serial bacteriocin dilutions prepared in MRS broth and 100 μL (106 cells) of a L. innocua 2 suspension as the indicator microorganism. AU/mL represented the reciprocal of the highest bacteriocin dilution showing at least 50% inhibition of the bacterial growth, after incubation at 37 °C for 18 h, when compared with the control with no bacteriocin added, multiplied by 10. The final pH of the enterocin preparation was in the range of 5.5–6.0.
Activity kinetics of enterocin P against L. monocytogenes and VRE isolates
The activity kinetics of the partially purified enterocin P (640 AU/mL) was determined by the microtiter plate assay [42] using as target the following strains (OD600 of ~0.2): (i) two L. monocytogenes isolates displaying the highest and lowest sensitivity to the bacteriocin in the agar-spot assay, respectively, and (ii), for VRE, the most sensitive strains of each species (E. faecium and E. faecalis) included in the study, according to the agar-spot assay results. Strains L. innocua 2 and E. faecalis ATCC 10100 were also tested for comparison. The OD600 of the medium without inoculum was discounted for the final results. Also, at the beginning and at the end of the experiment (0 and 24 h), viable cell counts of the Listeria spp. and Enterococcus spp. strains were performed, respectively, in BHI and MRS agar plates, to evaluate the inhibitory effect of the bacteriocin. These experiments were performed in triplicate.
Results
Confirmation of the mutation in the entTW21 gene
Sanger sequencing confirmed the deletion of a base pair (A/T) in the enterocin TW21 encoding gene found in E. faecium 86 when compared with the nucleotide sequence of the gene already described in the literature (Fig. 1) [31]. Due to this fact, from now on enterocin P will be considered the sole bacteriocin responsible for the antimicrobial activity exerted by this strain.
Fig. 1.
Sequence alignments. a Alignment of the DNA sequences of the structural gene encoding enterocin TW21 in strain E. faecium E86 (E86; accession no. SIHT00000000.1) and in the reference strain (Ref.; accession no. JX880073.1) retrieved from the GenBank data base. The base pair deletion found in strain E86 is highlighted in yellow. The same sequence and consequently deletion were observed after sequencing, in the present study, the amplicon corresponding to the structural gene of enterocin TW21 found in strain E86. b Alignment of the amino acid sequences encoded by each gene. The arrow indicates the peptide bond that is cleaved during processing of the precursor peptide of enterocin TW21 to generate the mature and bioactive bacteriocin of 48 amino acids
Transcription analysis of bacteriocinogenic clusters
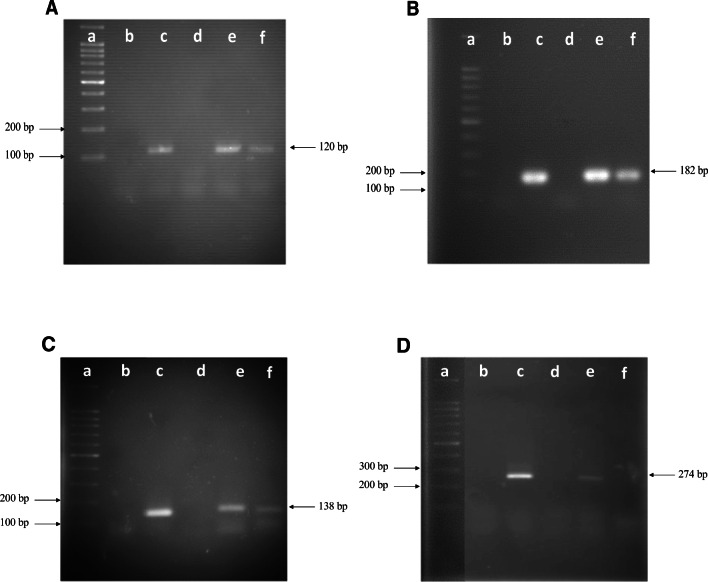
Results from RT-PCR experiments for entP, entiP, entTW21, and entiTW21 showed that these genes are expressed in E. faecium E86, since cDNA synthesized from RNA extracted within 6 h of cultivation resulted in amplicons with the expected size for each gene studied. In contrast, cDNA synthesized from the RNA extracted within 10 h of cultivation indicated the transcription of the entP, entiP, and entTW21 genes only. At this incubation time, no amplification of the entiTW21 gene was observed, which may be due either to its non-transcription or to the fact that there was no enough cDNA to be detected by the analysis. Importantly, for all genes tested, there was a weaker amplification at 10 h of growth in comparison to the amount detected in cultures after at 6 h of incubation (Fig. 2).
Fig. 2.
Detection of the entP (120 bp; a), entiP (182 bp; b), entTW21 (138 bp; c) and entiTW21 (274 bp; d) genes by PCR and their transcription by RT-PCR. The analyses were done with cDNA obtained from the RNA extracted after 6 and 10 h of incubating E. faecium E86 strain cultures, and the amplicons were detected by electrophoresis on 1.8% agarose gels in TAE buffer, at 90 V for 1.5 h. a 100-bp DNA Ladder ready-to-use (Sinapse Inc). b blank (samples without nucleic acid); c positive control of the reaction (amplification of the gene from DNA); d negative control of the reaction (DNA of E. coli strain DH5α); e cDNA obtained from RNA extracted after 6 h of culturing E. faecium E86; f cDNA obtained from RNA extracted after 10 h of culturing E. faecium E86. The numbers to the left of the figure indicate the size of some DNA fragments present in the molecular size marker. The number to the right of each figure corresponds to the size of the expected amplicon
Sensitivity of L. monocytogenes and VRE to enterocin P
All L. monocytogenes and Enterococcus strains tested were sensitive to the bacteriocin expressed by E. faecium 86, according to the agar-spot assay (Tables 2 and 3). However, the inhibition capacity varied among the target strains, with inhibition halos ranging from 10.3 to 31 mm.
Table 2.
Listeria strains used in the present study and respective sizes of zones of inhibition produced by strain E. faecium E86
| Indicator strains | Serotypes | Inhibition zone (standard deviation)* |
|---|---|---|
| L. monocytogenes CLIST 3236 | 1/2b | 10.3 (0.6) |
| L. monocytogenes CLIST 480 | 4b | 10.3 (0.6) |
| L. monocytogenes CLIST 529 | 4b | 10.3 (0.6) |
| L. monocytogenes CLIST 3980 | 1/2a | 11 (0.0) |
| L. monocytogenes CLIST 654 | 1/2b | 11.7 (0.6) |
| L. monocytogenes CLIST 636 | 1/2b | 12 (0.0) |
| L. monocytogenes CLIST 2546 | 1/2a | 17 (1.0) |
| L. monocytogenes CLIST 1400 | 4b | 19 (0.0) |
| L. monocytogenes CLIST 3982 | 4b | 19.7 (0.6) |
| L. monocytogenes CLIST 653 | 1/2b | 20 (0.0) |
| L. monocytogenes CLIST 3652 | 1/2b | 20.3 (0.6) |
| L. monocytogenes CLIST 305 | 1/2c | 22 (0.0) |
| L. monocytogenes CLIST 484 | 1/2a | 22 (0.0) |
| L. monocytogenes CLIST 965 | 1/2b | 22 (0.0) |
| L. monocytogenes CLIST 3986 | 1/2c | 22 (0.0) |
| L. monocytogenes CLIST 3988 | 1/2c | 22 (0.6) |
| L. monocytogenes CLIST 3602 | 1/2c | 23 (0.0) |
| L. monocytogenes CLIST 2547 | 1/2a | 23 (0.0) |
| L. monocytogenes CLIST 3984 | 1/2b | 23 (0.0) |
| L. monocytogenes CLIST 2876 | 4b | 24 (0.0) |
| L. monocytogenes CLIST 2557 | 1/2b | 25 (0.0) |
| L. monocytogenes CLIST 1071 | 4b | 25 (0.0) |
| L. monocytogenes CLIST 3981 | 1/2b | 26 (0.0) |
| L. monocytogenes CLIST 1064 | 1/2c | 27 (0.0) |
| L. monocytogenes CLIST 2968 | 1/2b | 31 (0.0) |
| L. innocua 2 | - | 25 (0.0) |
*Results represent the approximate mean value of the diameters of the inhibition zones and the standard deviation in millimeters, obtained in three independent experiments.
Table 3.
Enterococcus strains used in the present study and respective sizes of zones of inhibition produced by strain E. faecium E86
| Indicator strains | Resistance genotype | Inhibition zone (standard deviation)* |
|---|---|---|
| E. faecium CL–6770 | vanA | 18 (1.0) |
| E. faecium CL–8527 | vanA | 18.3 (0.6) |
| E. faecium CL–720ª | vanA | 19 (1.0) |
| E. faecium CL–6179 | vanA | 21.3 (1.0) |
| E. faecium RS73 | vanA | 22 (0.0) |
| E. faecium RS70 | vanA | 22 (2.0) |
| E. faecium RS71 | vanA | 24 (0.0) |
| E. faecium CL–6258 | vanA | 25 (0.0) |
| E. faecalis RS72 | vanB | 17 (1.0) |
| E. faecalis CL–9325 | vanA | 17 (1.0) |
| E. faecalis CL–8187 | vanA | 18 (1.0) |
| E. faecalis CL–5241 | vanA | 18.7 (0.6) |
| E. faecalis CL–5865 | vanA | 20 (0.6) |
| E. faecalis CL–8286 | vanA | 20.3 (1.0) |
| E. faecalis ATCC 10100 | - | 19 (0.0) |
*Results represent the approximate mean value of the diameters of the inhibition zones and the standard deviation in millimeters, obtained in three independent experiments.
Bacteriolytic activity of enterocin P against L. monocytogenes and VRE
The mode of action test aimed to verify the bactericidal (bacteriolytic or not) or bacteriostatic activity of a partially purified enterocin P preparation. The results are shown in Fig. 3.
Fig. 3.
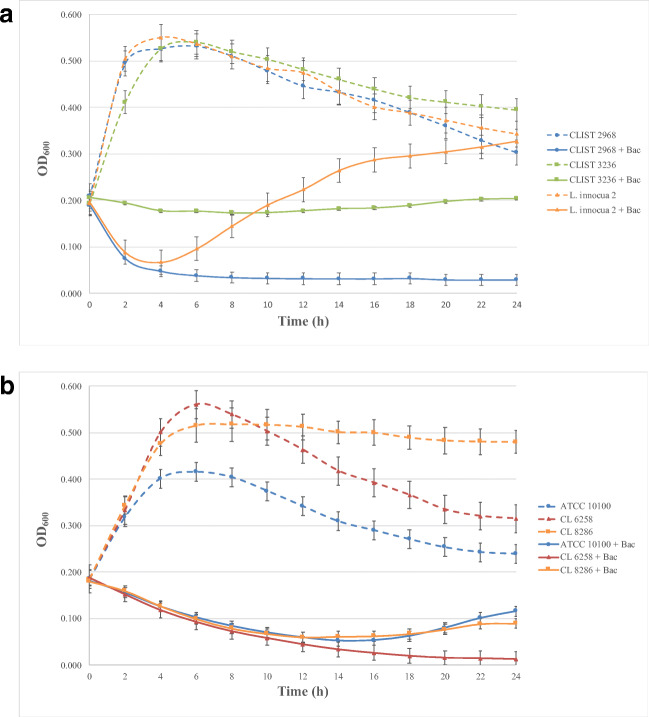
Mode of action of enterocin P (640 AU/mL) against Listeria strains (b) and Enterococcus strains (b), grown in BHI and MRS broth, respectively, for 24 h at 37 °C. The results represent the average and standard deviations of three independent experiments. Bac, bacteriocin
Against L. monocytogenes (Fig. 3a), enterocin P displayed a strong or weak bacteriolytic activity, depending on the isolate, within 4 h of incubation, after which the culture OD remained almost the same. The initial viable cell counts for the three strains were 1.0 ± 0.1 × 109 CFU/mL. L. monocytogenes CLIST 2968, which was the most sensitive L. monocytogenes strain evaluated in the agar-spot test, reached 1.8 ± 0.2 × 106 CFU/mL at the end of the experiment (24 h). In contrast, final counts for L. monocytogenes CLIST 3236, the less sensitive strain, reached 9.5 ± 0.5 × 108 CFU/mL. The bacteriocin also caused a sharp reduction in the culture OD of L. innocua 2 during the first 4 h of incubation, after which the strain resumed the growth, reaching viable cell counts of 2.0 ± 0.2 × 109 CFU/mL at 24 h of incubation.
In the presence of 640 AU/mL enterocin P, all enterococci showed a continuous growth reduction until 14 h of incubation, suggesting a bacteriolytic activity against these strains (Fig. 3b). The initial viable cell counts for the three strains were 1.1 ± 0.2 × 109 CFU/mL. After that, E. faecium CL-6258 counts remained low, with viable cell counts of 2.9 ± 5.0 × 103 CFU/mL at 24 h. Both E. faecalis strains tested, in contrast, started a slight regrowth after this time, and their viable cell counts at 24 h were close to 1.0 ± 1.5 × 108 CFU/mL.
Discussion and Conclusion
The demand for new compounds exhibiting antimicrobial activity has greatly motivated the study of bacteriocins and BLIS during the last decades aiming applications in both the food industry and the medical field. Some bacteriocins produced by Enterococcus spp., known as enterocins, can inhibit various human and animal pathogens, such as C. difficile, VRE, and L. monocytogenes, among others [5]. Early results from our group have shown that strain E. faecium 86 carries two bacteriocinogenic clusters [30].
However, DNA sequencing and in silico analyses of the entTW21 structural gene present in the genome of this strain have indicated an adenine deletion in the coding region of the gene. The presence of this mutation leads to a change in the reading frame for the translation event and to the formation of a premature termination codon (TGA; in nucleotides between positions 27 and 29 of the sequence presented in Fig. 1). As a consequence, a truncated peptide without its antimicrobial function is synthesized.
A preliminary analysis of the expression of the genes involved in bacteriocinogenic clusters (entP, entiP, entTW21, and entiTW21 genes) showed that all these genes are transcribed and, therefore, are expected to be translated. The presence of RBS regions preceding these genes corroborates the hypothesis of their possible translation [29–31]. The choice of 6 and 10 h of growth for this analysis was based on their correlation, respectively, to the log phase of the microorganism growth and to the apex of the antimicrobial activity detected [33]. Importantly, the transcription of the entiTW21 gene was no longer observed at 10 h. Such an event may have occurred as a result of cell autoregulation.
Therefore, genomic analysis of E. faecium E86 associated with molecular analysis lead to the conclusion that the enterocin TW21 precursor peptide is synthesized in a truncated manner and that the inhibitory action of this strain results from the activity of the other bacteriocin produced by it, which proved to be identical to enterocin P [29].
Regarding the spectrum of action, enterocin P expressed by E. faecium E86 was able to inhibit the growth of diverse L. monocytogenes and VRE strains, showing a typical spectrum of action of class IIa enterocins [5, 28, 43, 44]. In addition, the results consolidate the data found by Cintas et al. (1997) [29] and Miguel et al. (2008) [33] on the antimicrobial activity of enterocin P against these bacterial genera.
Finally, the mode of action of a given bacteriocin is an important property when it is considered to be applied as a biopreservative or an alternative drug in clinical settings. The mode of action of enterocin P on L. innocua 2 and L. monocytogenes CLIST 2968 showed a marked bactericidal (and bacteriolytic) activity as early as the first 4 h of the experiment. This suggests that the presence of virulence genes, which differentiates these species [45], is not related to their susceptibility to enterocin P. However, only a bacteriostatic activity was observed against L. monocytogenes CLIST 3236, and after 24 h of incubation, the viable cell count of this strain was equal to the initial counting. The result correlates to the lower sensitivity of this strain as detected by agar-spot assays. This phenomenon can be explained by four characteristics that may vary among representatives of this species: (1) a more positively charged cell wall, which occurs by the D-alanization of the teichoic and lipoteichoic acids; (2) a cell membrane with a more neutral charge; (3) increased membrane fluidity, which is caused by increased unsaturated phosphatidylglycerol and short acyl chains; and (4) a large reduction in gene mptA expression, which codes for the Man-PTS IIAB subunit. Also, differences in bacterial metabolism may lead to changes in the cell membrane and cell wall that directly influence susceptibility to antimicrobials [46].
The mode of action of enterocin P against representatives of the genus Enterococcus showed a marked bactericidal (and bacteriolytic) activity along at least 10 h of incubation. For the three strains tested, a reduction of more than 90% of viable cells was observed after 24 h of experiment. The occurrence of a vancomycin resistance phenotype (in E. faecalis CL-8286 and E. faecium CL-6258) was irrelevant to the performance of the bacteriocin in comparison to a vancomycin-susceptible reference strain (E. faecalis ATCC 10100). This observation is consistent with the fact that the full vancomycin resistance occurs by changing the terminal amino acids D-Ala-D-Ala of the peptidoglycan cell wall precursor, with no effects at the membrane level, where enterocin P acts [47]. It is important to highlight that the resumption of bacterial growth by many strains (Enterococcus spp. or Listeria spp.) was probably due to the depletion of enterocin. Future studies with purified bacteriocin may help to understand this phenomenon.
The molecular characterization of the bacteriocinogenic clusters present in E. faecium E86 strain performed in this work demonstrates the complexity associated with the study of bacteriocins. The simple detection, by PCR, of a bacteriocin structural gene is not enough to assume that this substance is produced and responsible for a given antimicrobial action. In the case of enterocin P, such activity varied among different Listeria isolates but was consistent against VRE strains. Moreover, the identification of the still little known TW21 enterocin opens the possibility of further studies regarding its structure, mechanism of action, and biotechnological application, if an intact version of the gene is assembled by heterologous expression. The study of this strain is an example of the potentialities of enterocin application, which may contribute to the development of new strategies to combat microorganisms in both the clinical and food conservation context. With an appropriate technological development, this bacteriocin has a potential against infections caused by Listeria and VRE; on the other hand, due to the low cost of its purification, it could easily be incorporated as dairy products preservative [25, 48].
Author contributions
FMF carried out the experiments and wrote the paper with the support of LMT, DCH, MCFB, MALM, and RRB. DCH and LMT provided isolates for testing and contributed to the discussion of the results. MCFB, MALM, and RRB supervised the design and implementation of the research.
Funding
FMF is the recipient of a PhD scholarship from Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ-Doutorado Nota 10; grant no. 200.322/2020). This work was partially supported by Coordenação de Aperfeiçoamento de Pessoal do Ensino Superior (CAPES, grant no. 001) and by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. pp. 5–64. [PubMed] [Google Scholar]
- 2.Vu J, Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol. 2011;6:1749–1757. doi: 10.1007/s11515-011-1167-x. [DOI] [Google Scholar]
- 3.Araújo TF, Ferreira CLDLF. The genus Enterococcus as probiotic: safety concerns. Braz Arch Biol Technol. 2013;56:457–466. doi: 10.1590/S1516-89132013000300014. [DOI] [Google Scholar]
- 4.Selleck EM, Van Type D, Gilmore MS (2019) Pathogenicity of Enterococci. Microbiol Spectr 7. 10.1128/microbiolspec.GPP3-0053-2018 [DOI] [PMC free article] [PubMed]
- 5.Franz CMAP, Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev. 2007;31:293–310. doi: 10.1111/j.1574-6976.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- 6.Zommiti M, Cambornel M, Maillot O, Barreau M, Sebei K, Feuilloley M, Ferchichi M, Connil N (2018) Evaluation of probiotic properties and safety of Enterococcus faecium isolated from artisanal Tunisian meat Dried Ossban”. Front Microbiol 9:1685. 10.3389/fmicb.2018.01685 [DOI] [PMC free article] [PubMed]
- 7.Žugić Petrović TD, Ilić PD, Grujović MŽ, Mladenović KG, Kocić-Tanackov SD, Čomić LR. Assessment of safety aspect and probiotic potential of autochthonous Enterococcus faecium strains isolated from spontaneous fermented sausage. Biotechnol Lett. 2020;42:1513–1525. doi: 10.1007/s10529-020-02874-5. [DOI] [PubMed] [Google Scholar]
- 8.Rea MC, Dobson A, O'sullivan O, Crispie F, Fouhy F, Cotter PD, Shanahan F, Kiely B, Hill C, Ross RP. Effect of broad and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A. 2011;108:4639–4464. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018;49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack RW, Tagg JG, Ray B (1995) Bacteriocins of Gram-positive bacteria. Microbiol Rev 59:171-200. PMC239359 [DOI] [PMC free article] [PubMed]
- 11.Bastos MCF, Ceotto H, Coelho MLV, Nascimento JS. Staphylococcal antimicrobial peptides: relevant properties and potential biotechnological applications. Curr Pharm Biotechnol. 2009;10:38–61. doi: 10.2174/138920109787048580. [DOI] [PubMed] [Google Scholar]
- 12.Bierbaum G, Sahl H-G. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 13.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro127. [DOI] [PubMed] [Google Scholar]
- 14.Heng NCK, Wescombe PA, Burton JP, Jack RW, Tagg JR. The diversity of bacteriocins in Gram-positive bacteria. In: Riley MA, Chavan MA, editors. Bacteriocins: ecology and evolution, 1sted. New York: Springer; 2007. pp. 45–83. [Google Scholar]
- 15.Gabrielsen C, Brede DA, Nes IF, Diep DB. Circular bacteriocins: biosynthesis and mode of action. Appl Environ Microbiol. 2014;80:6854–6862. doi: 10.1128/AEM.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez RH, Zendo T, Sonomoto K. Circular and leaderless bacteriocins: biosynthesis, mode of action, applications and prospects. Front Microbiol. 2018;9:2085. doi: 10.3389/fmicb.2018.02085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathur H, Rea MC, Cotter PD, Hill C, Ross RP. The sactibiotic subclass of bacteriocins: an update. Curr Protein Pept Sci. 2015;16:549–558. doi: 10.2174/1389203716666150515124831. [DOI] [PubMed] [Google Scholar]
- 18.Coelho MLV, Duarte AFS, Bastos MCF. Bacterial labionin-containing peptides and sactibiotics: unusual types of antimicrobial peptides with potential use in clinical settings (a review) Curr Top Med Chem. 2017;17:1–22. doi: 10.2174/1568026616666160930144809. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Q, Fang H, Liu W (2017) Post-translational modifications involved in the biosynthesis of thiopeptide antibiotics. Org Biomol Chem 15:3376–3390. 10.1039/c7ob00466d [DOI] [PubMed]
- 20.Bastos MCF, Farias FM, Fagundes PC, Coelho MLV. Staphylococcins: an update on antimicrobial peptides produced by staphylococci and their diverse potential applications. Appl Microbiol Biotechnol. 2020;104:10339–10368. doi: 10.1007/s00253-020-10946-9. [DOI] [PubMed] [Google Scholar]
- 21.Acedo JZ, Chiorean S, Vederas JC, van Belkum MJ. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol Rev. 2018;42:805–828. doi: 10.1093/femsre/fuy033. [DOI] [PubMed] [Google Scholar]
- 22.Zimina M, Babich O, Prosekov A, Sukhikh S, Ivanova S, Shevchenko M, Noskova S. Overview of global trends in classification, methods of preparation and application of bacteriocins. Antibiotics. 2020;9:553. doi: 10.3390/antibiotics9090553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Cuellar MR, Rodríguez-Hernández A-I, Chavarría-Hernández N. LAB bacteriocin applications in the last decade. Biotechnol Biotechnol Equip. 2016;30:1039–1050. doi: 10.1080/13102818.2016.1232605. [DOI] [Google Scholar]
- 24.Gálvez A, Abriouel H, López RL, Ben-Omar N. Bacteriocin-based strategies for food biopreservation. Int J Food Microbiol. 2007;120:51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Silva CC, Silva SP, Ribeiro SC. Application of bacteriocins and protective cultures in dairy food preservation. Front Microbiol. 2018;9:594. doi: 10.3389/fmicb.2018.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan K, Field D, Rea MC, Ross RP, Hill C, Cotter PD. Bacteriocins: novel solutions to age old spore-related problems? Front Microbiol. 2016;7:461. doi: 10.3389/fmicb.2016.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hachin H, Hammami R, Gingras H, Kourda R, Bergeron MG, Ben Hámida J, Ouellette M, Fliss I. Inhibition of MRSA and of Clostridium difficile by duracin 61A: synergy with bacteriocins and antibiotics. Future Microbiol. 2017;12:205–212. doi: 10.2217/fmb-2016-011. [DOI] [PubMed] [Google Scholar]
- 28.Nes IF, Diep DB, Ike Y. Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. pp. 637–657. [PubMed] [Google Scholar]
- 29.Cintas LM, Casaus P, Håvarstein LS, Hernandez PE, Nes IF (1997) Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol 63:4321–4330. 10.1128/AEM.63.11.4321-4330.1997 [DOI] [PMC free article] [PubMed]
- 30.Farias FM, Francisco MS, Santos INS, Marques-Bastos SLS, Miguel MAL, Albano RM, Bastos MCF. Draft genome sequence of Enterococcus faecium E86, a strain producing broad-spectrum antimicrobial peptides: description of a novel bacteriocin immunity protein and a novel sequence type. J Glob Antimicrob Resist. 2019;17:195–197. doi: 10.1016/j.jgar.2019.04.00. [DOI] [PubMed] [Google Scholar]
- 31.Chang SY, Chen YS, Pan SF, Lee YS, Chang CH, Yu B, Wu HC. Enterocin TW21, a novel bacteriocin from dochi-isolated Enterococcus faecium D081821. J Appl Microbiol. 2013;115:673–678. doi: 10.1111/jam.12265. [DOI] [PubMed] [Google Scholar]
- 32.Chen YS, Yanagida F, Srionnual S. Characteristics of bacteriocin-like inhibitory substances from dochi-isolated Enterococcus faecium D081821 and D081833. Lett Appl Microbiol. 2007;44:320–325. doi: 10.1111/j.1472-765X.2006.02058.x. [DOI] [PubMed] [Google Scholar]
- 33.Miguel MAL, Castro ÂCD, Leite SFG. Inhibition of vancomycin and high-level aminoglycoside-resistant enterococci strains and Listeria monocytogenes by bacteriocin-like substance produced by Enterococcus faecium E86. Curr Microbiol. 2008;57:429–436. doi: 10.1007/s00284-008-9224-7. [DOI] [PubMed] [Google Scholar]
- 34.Arcuri EF, Ângelo FF, Guimarães MFM, Talon R, Borges MF, Leroy S, Loiseau G, Lange CC, Andrade NJ, Montet D. Toxigenic status of Staphylococcus aureus isolated from bovine raw milk and Minas frescal cheese in Brazil. J Food Prot. 2010;73:2225–2231. doi: 10.4315/0362-028x-73.12.2225. [DOI] [PubMed] [Google Scholar]
- 35.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins D. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coelho MLV, Coutinho BG, Santos OCS, Nes IF, Bastos MCF. Immunity to the Staphylococcus aureus leaderless four-peptide bacteriocin aureocin A70 is conferred by AurI, an integral membrane protein. Res Microbiol. 2014;165:50–59. doi: 10.1016/j.resmic.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Shehata AA, Tarabees R, Basiouni S, Gamil M, Kamal AS, Krüger M. Phenotypic and genotypic characterization of bacteriocinogenic enterococci against Clostridium botulinum. Probiotics Antimicrob Prot. 2017;9:182–188. doi: 10.1007/s12602-016-9240-z. [DOI] [PubMed] [Google Scholar]
- 38.Dutka-Malen S, Evers S, Courvalin P (1995) Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 33:24-27. PMC227872 [DOI] [PMC free article] [PubMed]
- 39.Giambiagi-deMarval M, Mafra MA, Penido EGC, Bastos MCF. Distinct groups of plasmids correlated with bacteriocin production in Staphylococcus aureus. J Gen Microbiol. 1990;136:1591–1599. doi: 10.1099/00221287-136-8-1591. [DOI] [PubMed] [Google Scholar]
- 40.Ceotto H, Holo H, Costa KFS, Nascimento JS, Salehian S, Nes IF, Bastos MCF. Nukacin 3299, an anti-streptococci and anti-staphylococci bacteriocin produced by Staphylococcus simulans 3299. Vet Microbiol. 2010;146:124–131. doi: 10.1016/j.vetmic.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 41.Schillinger U, Geisen R, Holzapfel WH. Potential of antagonistic microorganisms and bacteriocins for the biological preservation of foods. Trends Food Sci Technol. 1996;71:58–64. doi: 10.1016/0924-2244(96)81256-8. [DOI] [Google Scholar]
- 42.Ceotto H, Brede D, Salehian Z, Nascimento JS, Fagundes PC, Nes IF, Bastos MCF. Aureocins 4185, bacteriocins produced by Staphylococcus aureus 4185: potential application in food preservation. Foodborne Pathog Dis. 2010;7:1255–1262. doi: 10.1089/fpd.2010.0578. [DOI] [PubMed] [Google Scholar]
- 43.Eguchi T, Kaminaka K, Shima J, Kawamoto S, Mori K, Choi SH, Doi K, Ohmomo S, Ogata S. Isolation and characterization of enterocin SE-K4 produced by thermophilic enterococci, Enterococcus faecalis K-4. Biosci Biotechnol Biochem. 2001;65:247–253. doi: 10.1271/bbb.65.247. [DOI] [PubMed] [Google Scholar]
- 44.Uteng M, Hauge HH, Markwick PR, Fimland G, Mantzilas D, Nissen-Meyer J, Muhle-Goll C. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and a sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry. 2003;42:11417–11426. doi: 10.1021/bi034572i. [DOI] [PubMed] [Google Scholar]
- 45.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskane O, Furtado MR, Wiedmann M. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vadyvaloo V, Arous S, Gravesen A, Hechard Y, Chauhan-Haubrock R, Hastings JW, Rautenbach M. Cell-surface alterations in class IIa bacteriocin-resistant Listeria monocytogenes strains. Microbiology. 2004;150:3025–3033. doi: 10.1099/mic.0.27059-0. [DOI] [PubMed] [Google Scholar]
- 47.Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti-Infect Ther. 2014;12:1221–1236. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng ZJ, Zarin MA, Lee CK, Tan JS. Application of bacteriocins in food preservation and infectious disease treatment for humans and livestock: a review. RSC Adv. 2020;10:38937–38964. doi: 10.1039/d0ra06161a. [DOI] [PMC free article] [PubMed] [Google Scholar]