Abstract
In vitro fertilization has been regarded as a forefront solution in treating infertility for over four decades, yet its effectiveness has remained relatively low. This could be attributed to the lack of advancements for the method of observing and selecting the most viable embryos for implantation. The conventional morphological assessment of embryos exhibits inevitable drawbacks which include time- and effort-consuming, and imminent risks of bias associated with subjective assessments performed by individual embryologists. A combination of these disadvantages, undeterred by the introduction of the time-lapse incubator technology, has been considered as a prominent contributor to the less preferable success rate of IVF cycles. Nonetheless, a recent surge of AI-based solutions for tasks automation in IVF has been observed. An AI-powered assistant could improve the efficiency of performing certain tasks in addition to offering accurate algorithms that behave as baselines to minimize the subjectivity of the decision-making process. Through a comprehensive review, we have discovered multiple approaches of implementing deep learning technology, each with varying degrees of success, for constructing the automated systems in IVF which could evaluate and even annotate the developmental stages of an embryo.
Keywords: In vitro fertilization, Deep learning, Artificial intelligence, Embryo assessment, Embryo selection
Introduction
Approximately 13% of women and 10% of men had suffered from infertility [1]. While the percentages may appear to be insignificant, the sum could aggravate to a much more grave and critical concern when the world’s entire population is considered. Hence, multiple treatments have been developed and formulated along the years as therapeutic strategies for infertility, collectively referred to as assisted reproductive technology (ART) [2].
The implementation of ARTs has been evolving and improving along with the marginal increase in the number of patients undergoing the treatments, estimated at about 474 ART cycles per million of the population in the year 2011 [3]. In vitro fertilization (IVF) has prevailed as the most effective and commonly utilized type of ART, which comprises of Intracytoplasmic Sperm Injection IVF (IVF-ICSI) and Intracytoplasmic Morphologically selected Sperm Injection (IVF-IMSI) [4]. However, despite of the state-of-the-art insemination technology, the actual success rate of IVF cycles remained underwhelmingly low, evident by a recorded global pregnancy rate of only 24.0% in 2011 [3].
For an extensive period of time, improvements in IVF procedures have been particularly focused on the medical practices and laboratory techniques, yet little innovations are established for the process of embryo grading and subsequent embryo selection for transfer. Conventionally, embryo assessment is conducted under ×200–400 magnification by utilizing an inverted microscope equipped with a heated stage. Embryos are graded morphologically following a standardized scoring system based on several parameters such as embryo age, blastomere number, size regularity, and fragmentation percentage. During observation, the embryo culture has to therefore be withdrawn from the incubator, presenting risks of exposure to unstable environments which could presumably perturb the embryo development. To eliminate such risks, a safer and semi-automated incubator system, called the time-lapse (TL) system, has been adopted for IVF which permits a real-time observation of embryo culture during incubation. [5, 6].
The TL system integrates the functions of an incubator and an optical microscope by equipping the incubation chambers with internal microscope cameras. The system is operated through a software which authorizes the recording and annotation of embryo morphokinetics in real time, allowing a non-intrusive method for examining embryo culture. Effectively, TL also facilitates the gain of additional information concerning the patterns of embryo development that were previously undetected through the conventional embryo assessments [5]. Morphological evaluation of embryos through the TL system has evidently demonstrated efficacy in improving pregnancy and live birth rates, as compared to the conventional method [7].
A few TL systems are also equipped with prediction models or algorithms that could assist embryologists in selecting embryos with the highest implantation potential for transfer. Principally, such prediction models could be developed through images or videos captured using the conventional inverted microscopes [8] or TL incubators [9, 10]. The advanced visual information provided by the TL technology is, nonetheless, more preferable than that of the conventional microscopy because of the more informative and retrievable nature of TL data. As demonstrated by Regneir and colleagues, commercially available predictions models such as KIDscore have displayed significant predictive values for embryo implantation [11]. However, none of the existing prediction models have been generally accepted or applied in IVF clinics [12]. Furthermore, the effectiveness of the IVF prediction models is still currently on a prospective validation process. Two global manufactures of IVF products (Vitrolife, Sweeden, and Genea-Biomedx, Australia) have also accommodated versions of AI within their TL incubators (Embryoscope and Geri). The specific built-in algorithms would enable the TL incubators to hypothetically perform automated annotation and selection of embryos for transfer. Nevertheless, both systems are still considered experimental [66].
Similar disadvantages previously recognized in the conventional embryo assessment are also apparent in the TL system. Manual annotation of embryo culture yields subjective outcomes and consumes a considerable amount of time and effort, even with the application of a real-time visual documentation function in the form of videos [14]. In order to eradicate such complications, the future of embryo assessment has upheld an interest towards the development of an automated embryo grading system, either by utilizing TL-based data or even conventional microscope. That being said however the incorporation of an AI-based image analysis technique has allowed the possibility of constructing an automated system that could independently and automatically perform embryo assessments and annotations by analyzing the recorded time-lapse images and videos during incubation. In addition to simplifying the embryo grading methodology, such computerized system would accommodate specific and accurate algorithms that could minimize the risk of subjectivity. Furthermore, promising outcomes in utilizing AI, specifically the Convolutional Neural Network (CNN) algorithms, for embryo assessments have been reported and demonstrated to deliver a better performance than the conventional method [9].
Objective
Existing studies on the implementation of AI for the automation of embryo assessment are hereby reviewed. The purpose of this review is to examine just what method of AI-based technology have been implemented into ART technology, and to see how far said study have come, in addition to comparing the differences and similarity that may exist within each researches. However, the fact is that many other reviews already exist in the field, ranging from reviews that provides a broad overview of automated algorithm in reproductive data [13, 15] to reviews that specifically reviews machine learning algorithms implemented in ART technologies [16, 17], and even a dedicated webpage that compiles some of the existing AI technologies in ART [18]. In that respect, this review mainly differs from the other existing ones due to the focus towards researches that can specifically be categorized as computer vision, the selection criteria which is detailed in “Method” below. While the extent of AI-based systems has been described for other aspects of IVF, visual-based technology can still be considered new. This can be attributed due to the fact that TL technology, which is capable of providing advanced visual data, is still being gradually adopted in most clinics. This does not indicates that other reviews does not include computer vision-based technology, but instead to emphasize the fact that we attempt to take a closer and more detailed look at computer vision based researches in IVF as opposed to performing a more general overview of every AI-related researches in IVF. While this decrease the width of the area that the review covers, it instead offers more depth in the specific area that it covers.
Method
Specific criteria were established in considering the relevant papers that are included in this review, as follows:
Studies which utilized deep-learning based computer vision technology. Regardless of any other technology implemented alongside, the inclusion of a deep learning-based computer vision technology is necessary and sufficient.
Studies that applied said technology to automate an IVF related process, such as embryo annotation, embryo assessment, etc.. The visual data could be sourced from either the TL images/videos or traditional microscopic images.
The related research papers, 21 in total, were reviewed and compared in regard to the particulars of the associated technology, how it was implemented, and to what extent the technology was able to present efficiency for the respective IVF procedure. Comparisons were made to highlight the differences and similarities among the studies, and to determine the up-to-date progress on the application of the AI-based systems in IVF.
Deep Learning and computer vision
Deep Learning
Deep Learning (DL) is a concept evolved from Machine Learning (ML) technology within the AI superset, whose development has vastly improved the scopes of machine learning and artificial intelligence-related technology [19]. ML is a data processing technique that facilitates a machine’s capability to learn to do a specific task independently. Traditional ML has not been sufficient in achieving the desired level of task automation as it requires the behavior and the so-called feature extractor to be manually defined so that the ML system could work. Even then, the ML system is often incapable of functioning when a different and new set of data input is introduced [20, 21]. DL overcomes this obstacle by capitalizing on a technique called representation learning (RL). RL expands the construct of automation by authorizing a fully automated feature extraction process to operate from raw data. All in all, while ML requires a defined feature extractor, DL has gone a step further by automatically defining its feature extractor [22, 23]. RL is applied in the form of an artificial neural network, a computing system that is designed to mimic the functions of biological neurons. The architecture of this neural network consists of multiple layers and levels which regulate the execution of a particular process and its subsequent results. Hence, the designs of neural network architecture could differ depending on the specific task that is performed [21, 24–26].
Computer vision and CNN
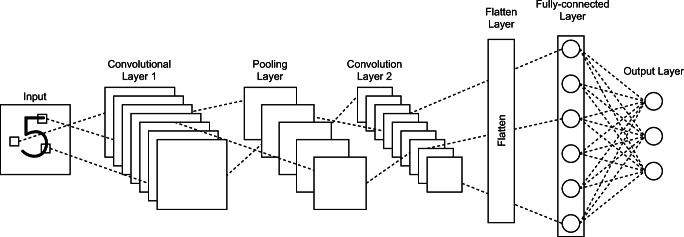
As established previously, the rapid advancement of DL has enabled the development of fully automated systems that are applicable for all sort of tasks in various fields [27, 28]. Among the most prominent applications of DL technology is for computer vision, which is a branch of technology that offers a machine the capability to process visual information, essentially giving them eyes to see and recognize vision sensory data such as images and videos [29]. Such concept of computer vision had a long history, but only with the recent advancement of DL did it see a rapid development for it to be ubiquitously utilized in a wide array of tasks. DL in computer vision is mostly driven by Convolutional Neural Network (CNN) [24], a specific type of DL that is designed to manage image-based data. Figure 1 displays an example of a CNN architecture. The exclusive convolutional layer, which distinguishes CNN from other neural networks, is in control of the image processing capability. The layer works on an image by segregating its unique features, step-by-step, that are subsequently used to predict an outcome, usually for the classification of the input image into a pre-defined class. Different types of CNN exist which differ mainly in the architectures of the layers [25, 30, 31].
Fig. 1.
Example of a convolutional neural network’s architecture
Architecture of CNN
The architecture of a neural network decides the behavior of a prediction model, which corresponds to the type of data it is able to process and how it would perform the task with the given data. Hence, deep learning model can be defined as the result of a CNN architecture developed through training [24, 26]. Additionally, retraining an existing model is possible so available architectures and training methods could be employed to create an entirely new model that can conduct an altogether different task. This technique is referred to as Transfer Learning which is often more advantageous than constructing an entirely new architecture or model. Favorable outcomes could be derived through transfer learning even with a minimal amount of training data, but the technique is not fault-free and not applicable for certain types of task and data [32].
Computer vision in IVF
Implementation of computer vision technology in IVF is a very specific yet diverse methodology. It often involves the processing of visual data, pertaining to the stages of embryonic development, which is derived from either the TL incubator system or conventional microscopy. Different information obtained from the particular types of images/videos highly influence the methods of CNN implementation. Varying types of microscope models used to capture the data also produce contrasting results which actively challenge the susceptibility of CNN to differences between images, adding another layer of variation to CNN implementation. In this review, the studies examined were categorized into four main subjects of discussion according to the IVF procedures that the CNN was implemented for (alternatively defined as CNN outputs).
Embryo development annotation
This section examines the task of automatically recognizing and subsequently annotating the developmental stage of an embryo at a given time. The papers included in this section did not attempt to predict the outcomes of IVF cycles and instead focused on analyzing the development of embryos. One particular method associated with such function is referred to as Cell Counting which, as the name indicates, is a process of quantifying the number of cells during a biological observation. Despite being a ubiquitous task, it is mostly done manually by examining a biological image to discern each cell individually or by observing for defined features of the cells, which challenges its accuracy due to the subjective nature of manual observations [15, 33].The manual annotation of embryonic development also bears the same risk of subjectivity. Cell counting in IVF is related to the evaluation of embryo morphokinetic attributes such as the physical characteristics of embryos (primarily in the regularity of blastomere shape), the time of cell division and the occurrence of abnormality such as multinucleation, all of which are said to influence the implantation potential of the related embryo [34] (Table 1).
Table 1.
Comparison table of studies associated with the cell counting procedure
| Paper | AI Architecture | Input data type | Training data size | CNN function | Annotation limit | Overall accuracy |
|---|---|---|---|---|---|---|
| Khan et al. (2016) [33] | Custom CNN architecure [39] | Dark-field time-lapse image sequence | 265 sequence/ 148.993 frames | Automatically count number of cells | Up to 5 cells stage | ± 92.18% |
| Ng and McAuley et al. (2018) [35] | Modified ResNet model [40] | EmbryoScope time lapse videos | 1309 videos/ 191.449 frames | Automatically detect embryo developmental stage | Up to 4+ cells stage | ± 87% |
| Malmsten et al. (2019) [36] | InceptionV3 model [41] | Time-lapse microscopy image | 47.584 images | Predict/detect cell division | Up to 4-cell stage | ± 90.77% |
| Liu et al. (2019) [42] | Multi-task deep learning with dynamic programming (MTDL-DP) | Time-lapse microscopy video | Extracted 59.500 frames in total | Classify embryo development stages | From initialization (tStart) to 4+ cells (t4+) | ± 86% |
| Leahy et al. (2020) [37] | ResNeXt101 [43] | Embryoscope time-lapse images | 73 embryos with 23.850 labels | Multiple function pipeline, one of which is detecting embryo developmental stage | From start of incubation to finish | ± 87.9%% |
| Dirvanauskas et al. (2019) [38] | AlexNet [39] + 2nd classifier | Miri TL-captured images | 3000 images from 6 embryo sets + 600 images of embryo fragmentation | Classify the development stage of an embryo | Classify for 1 cell, 2 cells, 4 cells, 8 cells and No Embryo | ± 97.5%% |
| Lau et al. (2019) [44] | RPN + ResNet-50 [40] | EmbryoScope-captured videos | 1309 time-lapse videos extracted by frames | Locate cell and classify development stage | Classify for tStart, tPnf, t2, t3, t4, t4+ | ± 90% |
| Raudonis et al. (2019) [45] | Comparing VGG [46] and AlexNet [39] | Miri TL-captured images | 300 TL sequences for a totl of 114793 frames | Locate cell and classify development stage | Classify for 1 cell, 2 cells, 3 cells, 4 cells, > 4 cells | VGG: ± 93.6% AlexNet: ± 92.7% |
Multiple CNN-based methodologies have been investigated for the automation of cell counting which involved a computer vision technique called classification. Classification is a basic computer vision task, which instructs a machine to distinguish images and assign them to pre-defined labels/classes. In cell counting, this classification function enables an AI model to recognize the features and characteristics of each cellular developmental stage and perform a “pseudo counting”. In these types of research [33, 35–38], the AI models were not trained to recognize individual cells and perform actual cell counting, but instead trained to recognize the collective shape of each cell count/embryo stage.
In training an AI model, the quality of training data is most substantial. The paper by Khan [33] emphasized the advantage of the classification method, in that it required minimal preparation of training data by simply labelling the class of said data. However, Khan notably utilized training data captured using dark-field microscopy which had simpler features compared to that of the bright field microscopy.
McAuley [35], Malmsten [36], Liu [42], Leahy [37], Dirvanauskas [38], Lau [44] and Raudonis [45] all utilized training data captured using time-lapse technology equipped with bright field microscopy. Such data established the benefits of additional contextual information even though its sequential nature was not put to use instead only utilizing static images. Regardless, time-lapse as training data has offered a more detailed and complete observation of embryos compared to data gained via the traditional microscope.
Mcauley [35], Liu [42] and Lau [44] made full use of the image sequence feature, taking into account the monotonically non-decreasing nature of embryo development which does not exist in the regular CNN. To achieve this, a monotonicity rule is implemented via dynamic programming. While the particulars of the methods presented in the three studies differ considerably, the basics resided in imposing a rule that prevented classification for frame n to be a class lower than frame n-1.
Malmsten [36], Leahy [37], Dirvanauskas [38], and Raudonis [45] also adopted the time-lapse training data, but the proposed methods did not make use of its sequential characteristic. Leahy did, however, exploit the multiple focal points that were additionally captured through time-lapse observation. Consequently, it improved the cell counting results but did not offer the same temporal features that are provided by the time-lapse image sequences. Additionally, the research by Dirvanauskas also utilizes a slightly different method, namely in how it strips the CNN of its classification layer, and instead utilize a separate classifier (based on machine learning techniques such as decision tree, etc.) to perform the classification, the features output from the CNN as input for the classifier.
Embryonic cell detection and tracking
Relatively similar to cell counting, the purpose of embryonic cell detection and tracking is to authorize the automated annotation of embryo developments through images. The major difference, on the other hand, would be in the methods and types of data that are utilized. Studies which investigated embryonic cell detection and tracking did not only attempt to classify or detect the developmental stage of an embryo, but to also locate each individual cell within the given image or video. For that reason, the two functions would require a different set of neural network and type of labeled data (Table 2).
Table 2.
Comparison table of studies associated with the cell detection and tracking procedure
| Paper | AI Architecture | Input data type | Training data size | CNN function | Detection limit | Overall accuracy |
|---|---|---|---|---|---|---|
| Matusevičius et al. (2017) [47] | R-CNN model [48] | Light microscopy image | 700 embryo images | Detecting position and size of cells | Only locates cell without stage prediction | Size prediction min error at 11.92% and detection at 5.68% |
| Rad et al. (2018) [49] | U-Net [25] modified with residual dilation | Light microscopy image | 224 embryo images | Locating individual cell(by centroid) and counting total | Up to 5-cells stage | 88.2% |
| Rad et al. (2019) [50] | ResNet model [40] modified with a special encoding & decoding scheme | Light microscopy image | 176 embryo images | Locating individual cell(by centroid) and counting total | Up to 8-cells stagel | 86.1% |
| Kutlu and Avci (2019) [51] | Faster R-CNN model [52] | Mouse embryo microscopy image | 565 mouse-embryo images | Locating individual cell and counting | Up until 4-cells stage | Averages 95% |
| Leahy et al. (2020) [37] | Mask R-CNN architecture [53] | Embryoscope time-lapse images | 102 embryos labeled at 16,284 times with 8 or fewer cells; | Multiple function pipeline, one of which is locating and segmenting individual cell | Tracking on images that were previously detected to have 1-8 cells | 82.8% |
Multiple implementations of the cell detection function have been initiated. All of the studies discussed here adopted the same object detection function but different methods of execution [37, 47, 49, 50]. Furthermore, there also varying circumstances among the studies on whether the cell detection function is utilized as the final goal of the research, or as means for another objective.
A preliminary research by [51] has explored the use of mouse embryo data in constructing a cell detection model. Their findings on the detection of embryo cells within images captured by the microscope camera have provided key hypotheses that could be adapted for future training of human embryo data.
Matusevicius [47] and Leahy [37] performed the detection of cells through a method known as segmentation, which operates by approximating the area that contains the features of the desired object and subsequently masking the predicted area. Matusevicius resorted to the use of a region-based bounding box technique to set a series of coordinates that form a box around the cells while Leahy [35] implemented a similar bounding box method with additional masking of the predicted area of cell(s). In particular, Leahy’s method consisted of multiple pipelines and is trained using images of one-cell to eight-cell embryos that were previously confirmed via the classification technique.
Rad [49, 50] actually defined the objective of their studies as cell counting, yet it is excluded from “Embryo development annotation” due to the method of implementation that was utilized. Rad presented the task as a regression function which automatically approximates the location of each individual cell for cell counting. For this reason, the method is deemed more similar to object detection in its input pipeline, rather than cell counting by classification.
IVF outcome prediction
This section is concerned with researches that have developed ML function categorized under prediction, specifically in regard to predicting the outcome of an IVF cycle or a specific IVF procedure. Theoretically, this section should cover a lot of different outputs however there was a relatively limited number of studies that are associated with the prediction function. This could be attributed to the aspect of predicting tentative outcomes, such as blastocyst formation, that is regarded as an unreliable indicator for clinical pregnancy [54].
The ultimate goal of an IVF cycle is to produce a clinical pregnancy that is followed by a live birth. Hence, the establishment of prediction models that could foresee such outcomes would very be useful in improving the quality of IVF programs [55]. Experimental tests of the prediction models have demonstrated promising efficacy but some doubts exist appertaining to the accuracy and practicality of the models for clinical use (Table 3).
Table 3.
Comparison table of studies associated with predicting IVF outcomes
| Paper | AI Architecture | Input data type | Training data size | CNN function | Prediction accuracy |
|---|---|---|---|---|---|
| Kan-Tor et al. (2020) [56] | DNN with Decision Tree classifier | Time-lapse videos captured using various incubator from various clinics | ± 6200 labeled embryo divided into train-validation with 28% separated as test | Predicting blastocysts formation | (AUC) Training: ± 0.83 Test across age group: ± 0.75 |
| Kan-Tor et al. (2020) [56] | DNN with Logistic Regression classifier | Time-lapse videos captured using various incubator from various clinics | ± 5500 labeled embryo separated into train-validation with 21% separated as test | Predicting embryo implantation potential | (AUC) ± 0.75 average |
| Kanakasabapathy et al. (2020) [55] | Xception [57] | 3469 recorded video from 543 patients | Training: 1190, Test: 748 | Predicting blastocysts formation | ± 71.87% |
Kan-Tor [56] and Kanakasabapathy [55] both attempted to perform predictions of blastocyst formation which typically occurs on day 5 or day 6 of embryo culture after insemination. A pregnancy would only ensue if an embryo develops to become a blastocyst. Transfer of blastocysts, on the other hand, does not entirely guarantee a clinical pregnancy, regardless of the quality of embryos. The prediction of blastocyst formation therefore serves to assist the selection of embryos for transfer but it is not capable of measuring the overall outcome of an IVF cycle. Kan-Tor and Kanakasabapathy both highlighted the benefits of this function during the decision-making process for transfer. In IVF, there exist two instance where a transfer can be made, either in day 3 or in day 5 post insemination. The benefit of blastocyst formation prediction comes into effect on the decision of day 3 transfer, as the formation of blastocysts only happens in either day 5 or day 6. Thus the implementation of this prediction technology can lend more confidence to the decision of day 3 transfer.
Additionally, Kan-Tor’s [56] paper attempted to predict embryo implantation potential by making use of Known Implantation Data (KID) labeling. KID refers to a data-labelling scheme that predicates the transfer statistic of an embryo and describes the relation between the embryo being transferred and its respective implantation outcome. In short, a positive label denotes a high implantation potential while a negative label predicts an implantation failure. Following a successful transfer, the next step is for the embryo to implant, which may or may not occur [14]. KID labelling is a score that calculates the occurence of implantation in respect to transfer. Thus a system that can predict based on KID labeling can also improve on transfer decision by providing a better picture of the expected implantation occurrence.
Embryo grading and selection
This section reviews studies which sought to create an AI system that is capable of assigning a grade to an embryo. As morphological characteristics of an embryo vary greatly across its developmental stages, the methods that are utilized to carry out the automated grading task would also vary. The results of embryo grading directly reflect the quality of the embryo and are consequently critical for the embryo transfer decision. One of the prevailing issues in IVF is the lack of correlation between the predicted and actual outcomes. Favorable circumstances may culminate in a pregnancy failure and, vice versa, unfavorable predictions could lead to a successful cycle. Regardless, the benchmark for assessing the performances of automated embryo grading and selection systems is established through comparison with the results of manual grading, as means to gauge the AI capability against that of the human embryologists [54, 55, 58–62] (Table 4).
Table 4.
Comparison table of studies associated with the embryo grading task
| Paper | AI Architecture | Input data type | Training data size | CNN function | Function limit | Prediction accuracy |
|---|---|---|---|---|---|---|
| Kragh et al. (2019) [58] | Xception CNN model [57] paired with custom RNN model | EmbryoScope time-lapse images | 8.664 embryo | Assigning ICM and TE grades | – | ICM ± 65%, TE ± 69% |
| Khosravi et al. (2019) [60] | Inception-V1 model [41] along with CHAID decision tree [64] | EmbryoScope time-lapse image | 12,001 time-lapse image up to seven focal depths | Classifying embryo into good and poor quality & predicting pregnancy probability | – | Grading precision: 95.7% |
| Chen et al. (2019) [59] | ResNet50 model [40] | Microscopic images | 171,239 static images from 16,201 embryos of 4,146 IVF cycles | Automatic grading based on Blastocysts Development, ICM, and TE grades | Only detected D5/D6 embryo images for prediction | Blastocyst: 96.24%, TE: 84.42%, ICM: 91.08% |
| VerMilyea et al. (2020) [54] | Ensemble of multiple ResNet models [40] and DenseNet modesl [31] | Optical light microscope images | 1667 microscopic images of embryo in Day 5 | Predicting embryo viability | Analysis only on Day 5 embryos | 64.3% prediction accuracy |
| Bormann et al. (2020) [61] | Xception [57] with Genetic Algorithm | Vitro-life Embryoscope time-lapse videos | 2440 static human embryo images recorded at 113hpi | Embryo selection based on embryo quality and implantation outcome | Only utilizes images from single timepoint even with time-lapse | Based on: embryo quality = 90.97% implantation outcome = 82.76% |
| Kanakasabapathy et al. (2020) [55] | Xception [57] + Genetic Algorithm | 3469 recorded video from 543 patients | Training: 1190, Validation: 511 | Embryo selection based on morphology quality | – | SET: ± 83.51% DET: ± 96.90% |
| Thirumalaraju et al. (2020) [62] | Multiple tested, Xception [57] deemed as best | Embryoscope time-lapse system | Training: 1188 images Validation: 510 images Additional 742 as test | Grading blastocysts | – | ± 90.90% |
| Silver et al. (2020) [63] | An implementation of CNN (no specific architecture were named) | Time-lapse videos | 8789 videos in total Additionally 272 were KID labeled | Embryo grading and implantation prediction | – | AUC score of ± 0.82% |
Among the studies, the two methods that are largely used to achieve grading automation include classification [59, 60, 62, 63] and combined stack [54, 58]. Classification-based grading is very similar to classification cell counting in technique, while combined stack here refers to a method which utilizes a function pipelines.
Chen [59], Khosravi [60], Thirumalaraju [62] and Silver [63] all applied the classification-based approach to automate the process of embryo grading. Analogous to the cell-counting classification function, the AI was trained to assign the input images or videos into the appropriate labels which interpret embryo traits and nomenclature in compliance with the current embryo grading system. Technically, there would be some differences in the architectures of the AI used, but the basic principle remains largely the same.
Chen utilized a grading system established by Gardner to evaluate three embryo morphological characteristics: Blastocysts with developmental ranking between 3-6, Inner Cell Mass (ICM), and Trophectoderm (TE) quality. A simpler grading scheme was adopted by Khosravi which merely distinguished between good and bad quality embryos. Additionally, Khosravi also developed a decision tree model that considered variables such as embryo grades and patient age as predictors of pregnancy potential, as an additional function separate from the grading (albeit it utilizes the output of the embryo grading). Thirumalaraju performed said classification by grouping the images into 5 different classes, which is then categorized again into two groups, as blastocysts and non-blastocysts. The paper focused on comparing the efficacy of 5 different CNN architectures, with 4 being pre-trained CNN that is re-trained utilizing transfer learning and 1 new multilayer CNN that is trained from scratch. Among the tested CNN, the Xception architecture outputs the best result, and thus was also chosen for the additional test of parameter tuning, to display the effect of different parameter setting on the CNN’s performance. Additionally Thirumalaraju also performs additional external test on the resulting models, utilizing data from different source that was captured with an entirely different method (the models were trained on data captured using Embryoscope and the external test utilized data captured with inverted bright-field microscope). Only the Xception model displayed a similar result on both external and internal testing, while all the other models performed worse on the external test, each to varying degrees. Silver employs CNN to perform grading on a number of pre-prepared time-lapse videos, over which half is graded by external embryologists, which aids in confirming/comparing the CNN’s performance to real embryologist’s grading. Additionally, Silver also attempted to predict embryo’s implantation probability, utilzing the same CNN (with only the head modified to fit with the desired output) on 272 videos with known implantation data. This implantation prediction also utilizes a panel of embryologists with which the CNN’s performance is compared.
Kragh [58], VerMilyea [54], Bormann [61] and Kanakasabapathy [55] employed a function pipeline which incorporated at least one CNN based function among others. This approach is more complex than the classification method as it involved the merging of multiple techniques together to construct a single process. Taking advantage of the time-lapse data, Kragh used a combination of classification and regression techniques (similar to those utilized in cell counting and cell detection) to perform embryo assessments according to the Gardner system. Likewise, Khosravi performed the classification function on the input time-lapse image sequences and subsequently used regression to integrate the classification results, as means of getting a more accurate interpretation of the embryo’s grade.
VerMilyea, on the other hand, attempted to directly predict the viability of embryos instead of performing quality grading. This was done by making use of Day 5 embryo images to train a pipeline which comprised of two different DL architectures. Bormann and Kanakasabapathy employed similar techniques, by using CNN to initially classify embryo images into the pre-determined labels and then utilizing Genetic Algorithm to perform a selection based on the classification results. The difference lies in where Kanakasabapathy only utilized labels of embryo quality based on morphology, Bormann utilized this technique two-fold first labelling based on embryo quality and second based on recorded implantation outcomes.
Additional papers
Within this section, is included a group of additional papers whose functions only tentatively fall under the selection criteria. Primarily, these papers may or may not employ deep-learning based computer vision technology, which is tentative due to a relatively obscure details in the method section. That being said however, these papers does indeed utilize neural network technology, which is trained using a combination of different data, of which a portion was extracted from images, utilizing the aformentioned methods that may or may not be deep-learning based computer vision. What is certain however, is the impressive result of the papers, achieved through the combination of image variables and tabular data. While the precise methods utilized is still relatively obscure on some parts, the result achieved is unquestionably impressive, which is the primary reason for their inclusion in this review despite being a tentative match with the decided criteria.
The paper by Chavez-Badiola et al. [8] emphasized the creation of their algorithm, referred to as Embryo Ranking Intelligent Classification Algorithm (ERICA), touted as an automatic algorithm capable of assisting with embryo ploidy and implantation prediction. The algorithm is defined as a ``deep machine learning artificial intelligence algorithm” and is composed of two different modules. The first is defined as a pre-processing module which includes image augmentation and feature extractor, which generates an output in the form of multiple variables based on the input images. It is this module that’s defined to be a computer vision module, for the purpose of extracting features in the form of continuous variables from an image input. The module made use of various process, which includes convolution, albeit it appears to be done manually (in terms of custom designed filters), unlike CNN in which the convolution filters are automatically inferred by the algorithm. Following successful extraction of variables, the second module comes into play, which is the deep learning model designed to perform the automatic ranking of the embryo, utilizing a combination of image-extracted variables and the metadata of the related embryo. Evaluation of the algorithm is done through 4 different approaches: (1) Testing set, (2) Comparison against randomly assigned label), (3) Comparison against professional analysis, (4) Quantifying the ranking and selection result. This multiple evaluation approach allows for a more accurate assesment of the algorithm’s ability, in which same evaluation result between different approach can work to validate each other.
The paper by L. Bori et al. [65] define itself as developing an Artificial Neural Network (ANN) for predicting the likelihood of live birth, which utilize a combination of proteomic data and image-extracted variable similar to Chavez-Badiola et al.’s paper, albeit utilizing different methods. Where Chavez-Badiola et al. utilized convolution for feature extraction, L. Bori et al. utilized a more manual method that involves Hough transformation and the calculation of features of the various sections of the images, such as measurement of the area, number of pixels present in a specific segment, the binary patterns and texture analysis. In total, 33 variables were obtained through this method, which is expected to be representative of the characteristics of the related blastocyst. Following extraction, an additional check was performed to determine the correlation of the variables, which as a result reduces the number of variables to a total of 20. A collinearity analysis is then also performed for the proteomic data, which results in a different set of data utilized in each ANN attempt. The prediction model is trained utilizing genetic algorithm with each ANN acting as the individual of a population, which sees an increase in performance with each generation. Performance of the model is tested with two methods, ROC curve and Confusion Matrix.
Conclusion
The implementation of DL technology, particularly, for computer vision in IVF is quite extensive albeit few in practice. DL is still actively developing and existing researches have achieved variable degrees of success. Nevertheless, findings on the utilization of different AI functions that are discussed here have displayed favorable outcomes despite the presently limited research in the field. Undoubtedly, the prospective advancement of machine assistance could revolutionize the procedure for embryo assessment in IVF. This is reflected through the aforementioned studies which have formulated effective approaches in achieving machine automation for cell counting, cell detection, and even embryo grading that are on par with embryologist performances. Most of the papers reviewed here have only discussed the experimental results without actual implementation of AI in clinical cases. Hence, the number of clinical cases which have implemented IVF automation is minimal in comparison to the number of successful preliminary studies. A favorable experimental outcome, deduced from a controlled environment, may not translate very well during implementation in a real clinical case. While a CNN by itself is capable of performing a task, it behaves like a tool without a handle in which its applicability retains an absence of direction. Clinical implementation of CNN is therefore imperative especially if the technology is intended to be adopted by IVF practitioners. As of now, CNN technology in IVF is a fascinating advancement worthy of further research, but its current application for commercial and practical purposes is still lacking. The implementation of this automated technology will be a great boon for practicioners of IVF, and proving that AI is capable of automating a process as delicate and critical as IVF is a major step forward towards making AI-based technology more ubiquitous in the world.
Declarations
Conflict of interest
Not applicable
Ethical approval
Not applicable
Informed concern
Not applicable
Conflict of interest
Not applicable
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barbieri RL. Female Infertility. Eighth Edi. Amsterdam: Elsevier Inc.; 2019. [Google Scholar]
- 2.Zegers-Hochschild F, Adamson GD, De Mouzon J, Ishihara O, Mansour R, Nygren K, Sullivan E, Van Der Poel S. The International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary on ART terminology, 2009. Hum. Reprod. 2009;24(11):2683–2687. doi: 10.1093/humrep/dep343. [DOI] [PubMed] [Google Scholar]
- 3.Adamson GD, de Mouzon J, Chambers GM, Zegers-Hochschild F, Mansour R, Ishihara O, Banker M, Dyer S. International committee for monitoring assisted reproductive technology: world report on assisted reproductive technology, 2011. Fertil. Steril. 2018;110(6):1067–1080. doi: 10.1016/j.fertnstert.2018.06.039. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Nisenblat V, Yang P, Zhang X, Ma C. Reproductive outcomes in women with unicornuate uterus undergoing in vitro fertilization: A nested case-control retrospective study. Reprod. Biol. Endocrinol. 2018;16(1):1–8. doi: 10.1186/s12958-017-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castelló D, Motato Y, Basile N, Remohí J, Espejo-Catena M, Meseguer M. How much have we learned from time-lapse in clinical IVF? Mol. Hum. Reprod. 2016;22(10):719–727. doi: 10.1093/molehr/gaw056. [DOI] [PubMed] [Google Scholar]
- 6.Pribenszky C, Losonczi E, Molnár M, Lang Z, Mátyás S, Rajczy K, Molnár K, Kovács P, Nagy P, Conceicao J, Vajta G. Prediction of in-vitro developmental competence of early cleavage-stage mouse embryos with compact time-lapse equipment. Reprod. BioMed. Online. 2010;20(3):371–379. doi: 10.1016/j.rbmo.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Pribenszky C, Nilselid AM, Montag M. Time-lapse culture with morphokinetic embryo selection improves pregnancy and live birth chances and reduces early pregnancy loss: a meta-analysis. Reprod. BioMed. Online. 2017;35(5):511–520. doi: 10.1016/j.rbmo.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Chavez-Badiola A, Flores-Saiffe-Farías A, Mendizabal-Ruiz G, Drakeley AJ, Cohen J. Embryo ranking intelligent classification algorithm (ERICA): artificial intelligence clinical assistant predicting embryo ploidy and implantation. Reprod. BioMed. Online. 2020;41(4):585–593. doi: 10.1016/j.rbmo.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Bormann CL, Thirumalaraju P, Kanakasabapathy MK, Kandula H, Souter I, Dimitriadis I, Gupta R, Pooniwala R, Shafiee H. Consistency and objectivity of automated embryo assessments using deep neural networks. Fertil. Steril. 2020;113(4):781–787.e1. doi: 10.1016/j.fertnstert.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feyeux M, Reignier A, Mocaer M, Lammers J, Meistermann D, Barrière P, Paul-Gilloteaux P, David L, Fréour T. Development of automated annotation software for human embryo morphokinetics. Hum. Reprod. 2020;35(3):557–564. doi: 10.1093/humrep/deaa001. [DOI] [PubMed] [Google Scholar]
- 11.Reignier A, Girard JM, Lammers J, Chtourou S, Lefebvre T, Barriere P, Freour T. Performance of Day 5 KIDScore™morphokinetic prediction models of implantation and live birth after single blastocyst transfer. J. Assist. Reprod. Genet. 2019;36(11):2279–2285. doi: 10.1007/s10815-019-01567-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aparicio-Ruiz B, Romany L, Meseguer M. Selection of preimplantation embryos using time-lapse microscopy in in vitro fertilization: State of the technology and future directions. Birth Defects Res. 2018;110(8):648–653. doi: 10.1002/bdr2.1226. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez EI, Ferreira AS, Cecílio MHM, Chéles DS, de Souza RCM, Nogueira MFG, Rocha JC. Artificial intelligence in the IVF laboratory: overview through the application of different types of algorithms for the classification of reproductive data. J. Assist. Reprod. Genet. 2020;37(10):2359–2376. doi: 10.1007/s10815-020-01881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adolfsson E, Andershed AN. Morphology vs morphokinetics: A retrospective comparison of interobserver and intra-observer agreement between embryologists on blastocysts with known implantation outcome. Jornal Brasileiro de Reproducao Assistida. 2018;22(3):228–237. doi: 10.5935/1518-0557.20180042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filho ES, Noble JA, Wells D. A review on automatic analysis of human embryo microscope images. Open Biomed Eng Jo. 2010;4(1):170–177. doi: 10.2174/1874120701004010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raef B, Ferdousi R. A review of machine learning approaches in assisted reproductive technologies. Acta Informatica Medica. 2019;27(3):205–211. doi: 10.5455/aim.2019.27.205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patil AS. A review of soft computing used in assisted reproductive techniques (ART) International Journal of Engineering Trends and Applications (IJETA) 2015;2(3):88–93. [Google Scholar]
- 18.LLC. ARVAAN TECHNO-LAB. Reproai — Home.
- 19.Deng L, Yu D. Deep learning: methods and applications. Found Trends Signal Process. 2013;7(3-4):197–387. doi: 10.1561/2000000039. [DOI] [Google Scholar]
- 20.Du KL, Swamy MNS. 2014. Neural networks and statistical learning. 9781447155.
- 21.Schmidhuber J. Deep Learning in neural networks: An overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Najafabadi MM, Villanustre F, Khoshgoftaar TM, Seliya N, Wald R, Muharemagic E. Deep learning applications and challenges in big data analytics. J Big Data. 2015;2(1):1–21. doi: 10.1186/s40537-014-0007-7. [DOI] [Google Scholar]
- 23.Lecun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 24.O’Shea K, Nash R. 2015. An introduction to convolutional neural networks.
- 25.Navab N, Hornegger J, Wells WM, Frangi AF. U-Net: Convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2015;9351:234–241. [Google Scholar]
- 26.Albawi S, Mohammed TA, Al-Zawi S. Understanding of a convolutional neural network. Proceedings of 2017 international conference on engineering and technology, ICET 2017, 2018-Janua(April 2018)1–6; 2018.
- 27.Deng L, Hinton G, Kingsbury B. New types of deep neural network learning for speech recognition and related applications: An overview. ICASSP, IEEE international conference on acoustics, speech and signal processing - proceedings, pp 8599–8603; 2013.
- 28.Alex Graves AM, Hinton G. 2013. Speech recognition with deep recurrent neural networks , department of computer science, university of toronto, (3):6645–6649.
- 29.Szeliski R. Computer vision: algorithms and applications. Choice Rev Online. 2011;48(09):48–5140–48–5140. [Google Scholar]
- 30.Girshick R. Fast R-CNN. Proceedings of the IEEE international conference on computer vision, 2015 Inter:1440–1448; 2015.
- 31.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings - 30th IEEE conference on computer vision and pattern recognition, CVPR 2017, 2017-Janua:2261–2269; 2017.
- 32.Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 33.Khan A, Gould S, Salzmann M. 2016. Deep Convolutional Neural Networks for Human Embryonic Cell Counting, ECCV Workshops, 1–10.
- 34.Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remoh J. The use of morphokinetics as a predictor of embryo implantation. Hum. Reprod. 2011;26(10):2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 35.McAuley J, Ng NH, Lipton ZC, Gingold JA, Desai N. Predicting embryo morphokinetics in videos with late fusion nets & dynamic decoders. 6th international conference on learning representations, ICLR 2018 - workshop track proceedings; 2018. p. 1–4.
- 36.Malmsten J, Zaninovic N, Zhan Q, Rosenwaks Z, Shan J. Automated cell stage predictions in early mouse and human embryos using convolutional neural networks. 2019 IEEE EMBS international conference on biomedical and health informatics, BHI 2019 - Proceedings, 1–4; 2019.
- 37.Leahy BD, Jang WD, Yang HY, Struyven R, Wei D, Sun Z, Lee KR, Royston C, Cam L, Kalma Y, Azem F, Ben-Yosef D, Pfister H, Needleman D. 2020. Automated Measurements of Key Morphological Features of Human Embryos for IVF. [DOI] [PMC free article] [PubMed]
- 38.Dirvanauskas D, Maskeliunas R, Raudonis V, Damasevicius R. Embryo development stage prediction algorithm for automated time lapse incubators. Comput. Methods Programs Biomed. 2019;177:161–174. doi: 10.1016/j.cmpb.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 39.Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks Alex. Advances in neural information processing systems; 2012. p. 1097–1105.
- 40.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE computer society conference on computer vision and pattern recognition, 2016-Decem:770–778; 2016.
- 41.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. Proceedings of the IEEE computer society conference on computer vision and pattern recognition, 2016-Decem:2818– 2826; 2016.
- 42.Liu Z, Huang B, Cui Y, Xu Y, Zhang B, Zhu L, Wang Y, Jin L, Wu D. Multi-task deep learning with dynamic programming for embryo early development stage classification from time-lapse videos. IEEE Access. 2019;7:122153–122163. doi: 10.1109/ACCESS.2019.2937765. [DOI] [Google Scholar]
- 43.Xie S, Girshick R, Doll P. 2017. Aggregated residual transformations for deep neural networks arXiv:1611.05431v2. Cvpr.
- 44.Lau T, Ng N, Gingold J, Desai N, McAuley J, Lipton ZC. 2019. Embryo staging with weakly-supervised region selection and dynamically-decoded predictions. arXiv, 1–10.
- 45.Raudonis V, Paulauskaite-Taraseviciene A, Sutiene K, Jonaitis D. Towards the automation of early-stage human embryo development detection. BioMedical Eng Online. 2019;18(1):1–20. doi: 10.1186/s12938-019-0738-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 3rd International conference on learning representations, ICLR 2015 - conference track proceedings; 2015. p. 1–14.
- 47.Matusevičius A, Dirvanauskas D, Maskeliūnas R, Raudonis V. Embryo cell detection using regions with convolutional neural networks. CEUR Workshop Proceedings. 2017;1856:89–93. [Google Scholar]
- 48.Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. Proceedings of the IEEE computer society conference on computer vision and pattern recognition; 2014. p. 580– 587.
- 49.Rad RM, Saeedi P, Au J, Havelock J. Blastomere cell counting and centroid localization in microscopic images of human embryo. 2018 IEEE 20th international workshop on multimedia signal processing, MMSP 2018; 2018. p. 1–6.
- 50.Rad RM, Saeedi P, Au J, Member S. Cell-Net : embryonic cell counting and centroid localization via residual incremental atrous pyramid and progressive upsampling convolution. IEEE Access. 2019;PP(c):1. [Google Scholar]
- 51.Kutlu H, Avci E. Detection of cell division time and number of cell for in vitro fertilized (IVF ) embryos in time-lapse videos with deep learning techniques. The 6Th international conference on control & signal processing; 2019. p. 7–10.
- 52.Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(6):1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 53.Doll P, Girshick R, Ai F. Mask R-CNN ar.
- 54.VerMilyea M, Hall JMM, Diakiw SM, Johnston A, Nguyen T, Perugini D, Miller A, Picou A, Murphy AP, Perugini M. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Human Reprod (Oxford, England) 2020;35(4):770–784. doi: 10.1093/humrep/deaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanakasabapathy MK, Thirumalaraju P, Bormann CL, Gupta R, Pooniwala R, Kandula H, Souter I, Dimitriadis I, Shafiee H. 2020. Deep learning mediated single time-point image-based prediction of embryo developmental outcome at the cleavage stage. arXiv 1–31.
- 56.Kan-Tor Y, Zabari N, Erlich I, Szeskin A, Amitai T, Richter D, Or Y, Shoham Z, Hurwitz A, Har-Vardi I, Gavish M, Ben-Meir A, Buxboim A. Automated evaluation of human embryo blastulation and implantation potential using deep learning. Adv Intell Syst. 2020;2(10):2000080. doi: 10.1002/aisy.202000080. [DOI] [Google Scholar]
- 57.Chollet F. Xception: Deep learning with depthwise separable convolutions. Proceedings - 30th IEEE conference on computer vision and pattern recognition, CVPR 2017, 2017-Janua:1800–1807; 2017.
- 58.Kragh MF, Rimestad J, Berntsen J, Karstoft H. Automatic grading of human blastocysts from time-lapse imaging. Comput. Biol. Med. 2019;115(August):103494. doi: 10.1016/j.compbiomed.2019.103494. [DOI] [PubMed] [Google Scholar]
- 59.Chen T-J, Zheng W-L, Liu C-H, Huang I, Lai H-H, Liu M. Using deep learning with large dataset of microscope images to develop an automated embryo grading system. Fertility Reproduction. 2019;01(01):51–56. doi: 10.1142/S2661318219500051. [DOI] [Google Scholar]
- 60.Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, Sigaras A, Lavery S, Cooper LAD, Hickman C, Meseguer M, Rosenwaks Z, Elemento O, Zaninovic N, Hajirasouliha I. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. npj Digit Med. 2019;2(1):1–9. doi: 10.1038/s41746-019-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bormann CL, Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, Kandula H, Hariton E, Souter I, Dimitriadis I, Ramirez LB, Curchoe CL, Swain J, Boehnlein LM, Shafiee H. Performance of a deep learning based neural network in the selection of human blastocysts for implantation. eLife. 2020;9:1–14. doi: 10.7554/eLife.55301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thirumalaraju P, Kanakasabapathy MK, Bormann CL, Gupta R, Pooniwala R, Kandula H, Souter I, Dimitriadis I, Shafiee H. 2020. Evaluation of deep convolutional neural networks in classifying human embryo images based on their morphological quality. 1–21. [DOI] [PMC free article] [PubMed]
- 63.Silver DH, Feder M, Gold-Zamir Y, Polsky AL, Rosentraub S, Shachor E, Weinberger A, Mazur P, Zukin VD, Bronstein AM. 2020. Data-driven prediction of embryo implantation probability using IVF time-lapse imaging. arXiv, 1–6.
- 64.Kass GV. An exploratory technique for investigating large quantities of categorical data. Appl. Stat. 1980;29(2):119. doi: 10.2307/2986296. [DOI] [Google Scholar]
- 65.Bori L, Dominguez F, Fernandez EI, Gallego RD, Alegre L, Hickman C, Quiñonero A, Nogueira MFG, Rocha JC, Meseguer M. 2020. An artificial intelligence model based on the proteomic profile of euploid embryos and time-lapse images: a preliminary study, Reprod. BioMed. Online. [DOI] [PubMed]
- 66.Zaninovic N, Elemento O, Rosenwaks Z. Artificial intelligence: its applications in reproductive medicine and the assisted reproductive technologies. Fertil Steril. 2019;112(1):28–30. doi: 10.1016/j.fertnstert.2019.05.019. [DOI] [PubMed] [Google Scholar]