Abstract
Purpose
To investigate the frequency of a founder mutation in NLRP7, L750V, in independent cohorts of Mexican patients with recurrent hydatidiform moles (RHMs).
Methods
Mutation analysis was performed by Sanger sequencing on DNA from 44 unrelated Mexican patients with RHMs and seven molar tissues from seven additional unrelated patients.
Results
L750V was present in homozygous or heterozygous state in 37 (86%) patients and was transmitted on the same haplotype to patients from different states of Mexico. We also identified a second founder mutation, c.2810+2T>G in eight (18.1%) patients, and a novel premature stop-codon mutation W653*.
Conclusion
Our data confirm the strong founder effect for L750V, which appears to be the most common mutation in NLRP7. We also report on six healthy live births to five patients with biallelic NLRP7 mutations, two from spontaneous conceptions and four from donated ovum and discuss our recommendations for DNA testing and genetic counseling.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-021-02132-1.
Keywords: Recurrent hydatidiform moles, Founder mutation
Introduction
Hydatidiform mole (HM) is an aberrant human pregnancy characterized by abnormal embryonic development and excessive proliferation of the trophoblast. Common HM is sporadic and affects 1 in every 600 pregnancies [1]. At the histopathological level, HM is classified as complete or partial. Complete hydatidiform moles (CHMs) are characterized by the absence of embryo and excessive proliferation of the trophoblast. Partial hydatidiform moles (PHMs) have moderate focal trophoblastic proliferation and may contain embryonic tissues. CHMs are androgenetic while PHMs are triploid dispermic [2]. Recurrent hydatidiform moles (RHMs) are defined by the occurrence of at least two molar pregnancies in the same patient and affect approximately 1-9.4% of women with a prior HM, depending on studies and populations [3–7]. Based on morphological analysis, RHMs may be classified as CHM or PHM.
Biallelic NLRP7 mutations are the major cause for RHMs (OMIM 231090) [8] and explain the genetic etiology of 55% of patients [9]. A second gene responsible for RHMs, KHDC3L, was identified in 2011 [10] and its biallelic mutations explain the etiology of 5% of patients with RHMs (hydatidiform mole, recurrent type 2 (OMIM 611687)) [9, 11]. Molar tissues from patients with mutations in NLRP7 or KHDC3L are diploid biparental. Both genes are components of the subcortical maternal complex, which is essential for epigenetic reprogramming of the oocyte genome and the activation of the embryonic genome [12–14]. Recently, biallelic mutations in three other genes, MEI1, TOP6BL (C11orf80), and REC114, with roles in meiotic double-strand break formation have been identified in patients with recurrent androgenetic complete hydatidiform moles, miscarriages, and infertility [15].
In 2013, our group analyzed NLRP7 mutations in 20 Mexican patients with RHMs and found that 17 of them have biallelic mutations in NLRP7 [16] and all the 17 patients had at least one copy of a previously reported mutation, c.2248C>G, p.Leu750Val (L750V) in two Mexican patients [17]. Furthermore, of the 17 patients, 12 were homozygous for L750V. These 12 patients were born in different parts of Mexico and all denied consanguinity between their parents. In addition, the L750V was found in a heterozygous state in 5% of control subjects from the general Mexican population [16]. These data suggested a strong founder effect for L750V in the Mexican population.
Founder mutations in NLRP7 have been reported in other populations, including the Indian [c.2078G>C, p.(Arg693Pro) and c.2738A>G, p.(Asn913Ser)] and Egyptian [c.-39-387_2129+265dup, p.(Glu710Aspfs*7)] populations [17–19]. However, the founder effect in the Mexican population appeared stronger because the same mutation was found in all the 17 patients with biallelic mutations we reported in Estrada et al. [16]. We therefore set up to analyze another independent cohort of 44 unrelated Mexican patients with RHMs, and seven molar conceptions from unrelated patients with RHMs. Thirty-one of these patients and the seven moles were recruited or retrieved from the Instituto Nacional de Perinatologia in Mexico City. We also reviewed the mutation analysis results of another cohort of 13 unrelated patients with RHMs of Mexican origin who were referred either from the USA or Mexico to the Research Institute of the McGill University Health Centre (RI-MUHC) for mutation analysis. Our data confirm our previous findings and highlight the strong founder effect for L750V in Mexico and its inheritance on the same haplotype to patients from various states. Our study also revealed a second founder mutation, c.2810+2T>G and a novel protein-truncating mutation in the Mexican population.
Material and methods
Patients with RHMs
The study was approved by the review boards of the Instituto Nacional de Perinatologia (INPer), study number: 212250-3220-11108-01-14 and McGill University (study number: A01-M07-03A). Patients with at least two HMs were referred from different hospitals in Mexico. A complete clinical evaluation including family and reproductive histories of the patients and their first-degree relatives was taken for all patients. When possible, sisters with RHMs and parents were invited to participate in the study. Written informed consents were obtained from all participants prior to obtaining venous blood samples. A total of 44 unrelated patients were included in this study, 31 were referred to INPer, and 13 were referred to the RI-MUHC. Archived formalin-fixed paraffin embedded (FFPE) molar tissues were retrieved from seven patients with RHMs from the INPer by screening the pathology department record for patients with RHMs.
DNA extraction and mutation analysis
Genomic DNA was isolated from the patient venous peripheral blood. Sequence analysis was performed at the INPer (Mexico) first for exon 6 of NLRP7 to investigate the presence of the founder mutation L750V. Patients without biallelic mutations were screened for mutations in the other exons, 1 to 5 and 7 to 11, at the RI-MUHC (Montreal, Canada). Primer sequences and polymerase chain reaction (PCR) conditions were as previously described [17, 20] (Supplementary Table 1). PCR products were purified and directly sequenced in forward and reverse orientations using terminator dye in an ABI Prism 3130 (Applied Biosystems). All identified mutations were compared with the reference sequence NM_001127255.1 (http://fmf.igh.cnrs.fr/ISSAID/infevers/) and annotated according to the Human Genome Variation Society (HGVS) (http://varnomen.hgvs.org/). Sequence variant nomenclature is given according to the following references: NM_001127255.1 (cDNA), NG_008056.1 (genomic DNA), and NP_001120727.1 (protein). Patients who were negative for mutations in NLRP7 were analyzed for mutations in KHDC3L as previously described [21].
Parental contribution to the molar tissues
Sections of FFPE molar tissues were stained with hematoxylin and eosin. Chorionic villi were separated from maternal tissues under a stereomicroscope and used to extract DNA as previously described [9, 22]. Multiplex microsatellite DNA genotyping was performed using the Powerplex 16 HS System (Promega Corporation, Fitchburg, WI, USA), and analyzed as previously described [9, 22].
Results
During the study period, a total of 31 unrelated patients with RHMs were recruited and analyzed for mutations in NLRP7 (Table 1). Of these 31 patients, seven had a family history of RHMs and nine (29%) patients had gestational trophoblastic disease after one of their molar pregnancies. Mutation analysis on these 31 patients revealed biallelic NLRP7 mutations in 26 (83.8%) of them. Of these patients, seventeen were homozygous for L750V; five were compound heterozygous for L750V and c.2810+2T>G, another previously reported mutation in Mexican patients [17]; one patient was compound heterozygous for L750V and a large deletion in the promoter region, c.-6831_-39-1586del, that leads to the absence of transcripts from the allele carrying it [23]; one patient was compound heterozygous for L750V and c.2471+1G>A, p.Leu825* (L825*); one patient was compound heterozygous for L750V and a novel premature stop-codon mutation c.1959G>A, p.Trp653* (W653*); and one patient was homozygous for c.1168del p.Arg390Alafs*26 (R390Afs*26) [9]. Five patients (16.1%) did not have any pathogenic or likely pathogenic variant in NLRP7 and were screened for KHDC3L, but none of them had any mutation.
Table 1.
Recapitulation of data on 71 analyzed patients with RHMs from Mexico
| Case N. | Patient ID | Reproductive history (complications) | NLRP7 mutations | Complication | References |
|---|---|---|---|---|---|
| Mexican patients recruited in Mexico between 2013-2020 | |||||
| 1 | ACC | 6 PHM | L750V hom | ||
| 2 | CEA | 4 HM | L750V hom | ||
| 3 | BBL | 2 HM, END (preeclampsia), LB | L750V hom | GTD | |
| 4 | MMN (consanguinity) | 4 HM | L750Vhom | ||
| 5 | GHR* | 3 HM, MC | L750V hom | GTD | |
| 6 | VGDE* | 2 HM | L750V hom | ||
| VGLE (sister) | HM | L750V hom | |||
| 7 | DJEY* | 3 HM | L750V hom | ||
| DJER (sister) | 2 HM | L750V hom | |||
| DJEG (sister) | 2 HM | L750V hom | |||
| 8 | OLO* | 2 HM, MC | L750V hom | ||
| 9 | CLL | 2 HM | L750V hom | GTD | |
| 10 | RLMC | 2 HM | L750V hom | ||
| 11 | PQRM | 3 HM | L750V hom | ||
| 12 | PAF | 4 HM, MC | L750V hom | ||
| 13 | GGE* | 4 HM | L750V hom | GTD | |
| 14 | DSL | 3 PHM, MC | L750V hom | ||
| 15 | VOM | 3 HM | L750V hom | ||
| 16 | CRA | HM, 2 MC | L750V hom | GTD | |
| 17 | ABH | 5 CHM | L750V hom | GTD | |
| 18 | GEM | 2 PHM | L750V, c.2810+2T>G | ||
| 19 | MADM | HM, CHM, 2 MC | L750V, c.2810+2T>G | ||
| 20 | CR | 5 HM, LB | L750V, c.2810+2T>G | GTD | |
| 21 | PVI | 2 HM, MC | L750V, c.2810+2T>G | ||
| 22 | RJG | 2 PHM, MC | L750V, c.2810+2T>G | ||
| 23 | HME | HM, CHM, 2 PHM | L750V, c.-6831_-39-1586del | GTD | Rezaei et al. [23] |
| 24 | RGR* | HM, 2 CHM, MC | L750V, c.2471+1G>A | ||
| 25 | LCMV | HM, 2 PHM | L750V, W653* | ||
| 26 | TGR* | 2 HM | R390Afs*26 hom | Nguyen et al. [9] | |
| 27 | GBNA | CHM, PHM | No mutation | ||
| 28 | VPA | 2 HM, LB | No mutation | ||
| 29 | QVSL | PHM (triploid dispermic), PHM revised to MC | No mutation | ||
| 30 | MCV | 2 CHM (2 androgenetic monospermic), MC | No mutation | GTD | |
| 31 | MTMC | 2 PHM, 3 MC, MC (92,XXYY) | No mutation | ||
| Screening for L750V in HM tissues from patients with RHMs received between 2003 and 2019 | |||||
| 32 | MTO | 2 HM (1 diploid biparental) | L750V hom | ||
| 33 | PFME* | 3 HM (1 diploid biparental) | L750V het | ||
| 34 | PSJ | 5 HM (1 diploid biparental), MC | L750V het | ||
| 35 | MGMJ | 3 HM (1 diploid biparental) | L750V het | ||
| 36 | CPE | MC, 3 HM (1 diploid biparental) | Negative for L750V | ||
| 37 | DCRN | MC, 3 HM (1 diploid biparental) | Negative for L750V | ||
| 38 | MAD | 2HM (1 androgenetic monospermic) | not screened | ||
| Patients of Mexican origin referred from various clinics and hospitals to the MUHC-RI between 2006 and 2020 | |||||
| 39 | 655 | 2 PHM, MC, PHM | L750V hom | Deveault et al. 2009; Nguyen et al. [29] | |
| 657 (sister) | PHM, CHM, HM | L750V hom | |||
| 30 | 733 | 2 HM, MC, 2 HM, IVF-PGT-HM, 5 HM, donated ovum-LB | L750V hom | Nguyen et al. [29] | |
| 41 | 908* | 4 HM (with 3 partners) | L750V hom | ||
| 42 | 1220* | 3 HM | L750V hom | ||
| 1224 (sister) | 3 HM, donated ova-2 LB | L750V hom | |||
| 1227 (sister) | HM | L750V hom | |||
| 43 | 1352 | 2 PHM, 3 HM | L750V hom | ||
| 44 | 1371* | 2 HM, CHM, 2 HM (with 3 partners) | L750V hom | ||
| 45 | 1878 | 5 HM, BO, donated ovum-LB | L750V hom | ||
| 46 | 1359 | 4 HM | L750V, c.-13413_2982-344del | Reddy et al. [24] | |
| 47 | 1243 | PHM, 8 MC, MC, PHM | L750V, c.2810+2T>G | Reddy et al. [24] | |
| 48 | 1674 | 2 HM, MC, HM, MC, PHM, MC | L750V, c.2810+2T>G | ||
| 49 | 1888* | MC, 2 HM, MC, HM | L750V, c.2471+1G>A | ||
| 1889 (sister | 2 HM, HAT | L750V, c.2471+1G>A | |||
| 50 | 1074 | MC, PHM, HM, 5 MC (2 after clomide), HM, CHM | Y872X, c.2810+2T>G | Nguyen et al. [29]; Reddy et al. [24] | |
| 51 | 1333 | 4 MC, 4 CHM (4 androgenetic monospermic) | Biallelic MEI1 mutations | Nguyen et al. [9] | |
HM, hydatidiform mole, which is used when the pathology report did not specify the classification; CHM, complete hydatidiform mole; PHM, partial hydatidiform mole; MC, miscarriage; END, early neonatal death; LB, live birth; GTD, gestational trophoblastic disease; BO, blighted ovum; IVF, in vitro fertilization; PGD, preimplantation genetic testing; HAT, total hysterectomy; hom, homozygous; het, heterozygous
In the light of the high frequency of L750V in the 31 patients, we screened the record of the Pathology Department of the Instituto Nacional de Perinatologia for cases of RHMs since 2003. We found seven archived FFPE molar tissues, from seven additional unrelated patients that were available for analysis. DNA extraction from the chorionic villi of these tissues and their genotyping demonstrated that six are diploid biparental and one is diploid androgenetic monospermic. We next tested the six biparental moles for the presence of the founder L750V mutation. We found that two molar tissues were negative for L750V, three were heterozygous for L750V, and one was homozygous for L750V (Table 1). The latter observation indicates that the father of the HM carries the L750V, known to be present in 5% of control subjects from the general Mexican population [16].
We next reviewed the results of all Mexican patients with RHMs who were referred from various hospitals and medical centers from the USA or Mexico to the RI-MUHC since 2006 for NLRP7 and KHDC3L mutation analyses. We found 13 unrelated patients, of them 12 had biallelic mutations in NLRP7 (Table 1). Seven were homozygous for L750V; one was compound heterozygous for L750V and another previously reported promoter region deletion, c.-13413_2982-344del [24]; two were compound heterozygous for L750V and c.2810+2T>C [24]; one was compound heterozygous for L750V and c.2471+1G>A, p.L825*; and one was compound heterozygous for p.Tyr872* (Y872*) and c.2810+2T>C.
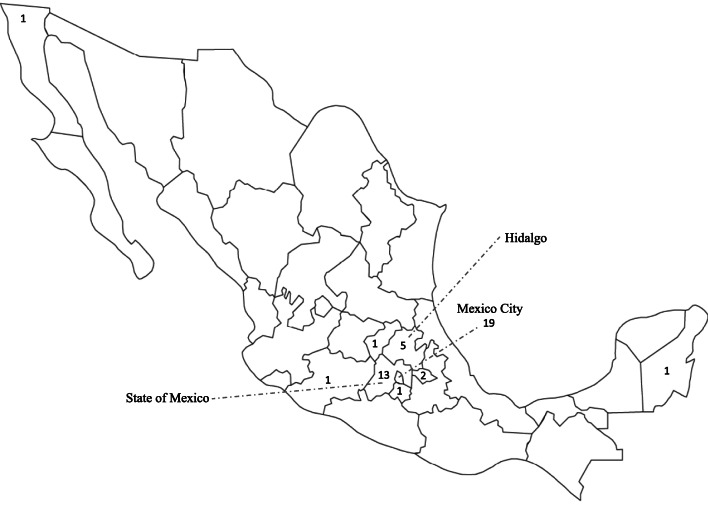
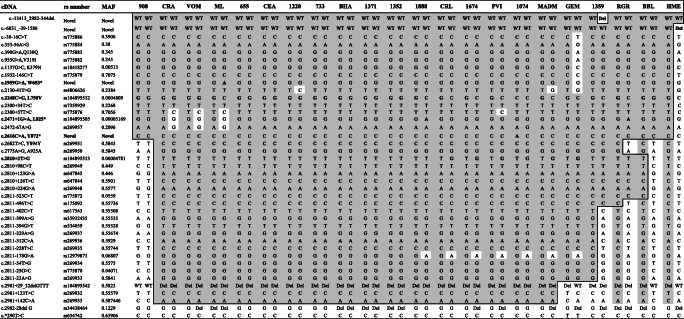
The states of origin of 44 unrelated patients analyzed on DNA from blood or molar tissues in this study or in Estrada et al. [16] with at least one copy of the L750V were available and are provided on the Mexican map in Fig. 1, which shows an important clustering of these patients in the state of Mexico City where they were recruited and also in some neighboring states. Haplotype analysis of all the SNPs and variants that are covered by our Sanger sequencing demonstrated the inheritance of the L750V mutation on a shared haplotype between patients from various Mexican states (Table 2), from rs775886 to rs269933 spanning 18,296 bp. We note that the shared haplotype is certainly larger; however, in Table 2, we included only the single nucleotide polymorphisms that are covered by our Sanger sequencing.
Fig. 1.
Geographical distribution of patients carrying L750V in Mexican states. The numbers refer to unrelated patients
Table 2.
Shared haplotype between patients
MAF minor allele frequency in gnomAD database. In column cDNA, bold character indicates pathogenic variants
Since NLRP7 is highly rich in Alu repeats and so far, nine of its 80 reported mutations are mediated by Alu recombination (https://infevers.umai-montpellier.fr/web/), which can be easily missed when using only Sanger sequencing, we attempted to retrieve archived FFPE tissues from patients with no mutations to re-evaluate the diagnosis of their HMs and determine whether they are diploid biparental. Among the patients who were recruited in Mexico, we were able to retrieve four products of conception (POCs), two from each of patients 29 and 30. Morphological and genotypic evaluation of two POCs from patient 29 demonstrated that one is a triploid dispermic PHM and the other lacked morphological features of molar pregnancies and we revised its diagnosis to miscarriage (Table 1). Multiplex microsatellite genotyping of this miscarriage demonstrated its diploid biparental genome and SNP microarray confirmed the diagnosed and demonstrated the absence of aneuploidy [22]. Therefore, this patient did not have RHMs (Table 1). The two POCs from patient 30 fulfilled the morphological diagnosis of CHM and both were found diploid androgenetic monospermic by multiplex microsatellite genotyping. From a third patient, 31, no tissues could be retrieved, but one of her POCs had been karyotyped and found to be tetraploid 92,XXYY. Of the patients referred to the RI-MUHC, only one patient was negative for NLRP7 mutations and four of her molar conceptions were available for genotype analysis and were found diploid androgenetic monospermic. This patient was later analyzed by exome sequencing and found to have biallelic mutation in MEI1 [15]. Therefore, the data on the POCs of these four patients explain the absence of NLRP7 mutations in them since biallelic NLRP7 mutations are associated with RHMs that are diploid biparental (Table 1). In conclusion, of the five patients with no NLRP7 mutations, only four had RHMs, which brings the number of patients with RHMs recruited in Mexico to thirty and the total number of analyzed and reviewed patients in this study to forty-three.
Discussion
Recurrent molar pregnancy is a rare disease. However, in the current study along with that of Estrada et al. [16], we report on a total of 70 unrelated patients with RHMs of Mexican origin (30 recruited in Mexico, 13 referred to the RI-MUHC, 7 molar tissues, and 20 reported in Estrada et al.). To our knowledge, this is the largest series from a single country and suggests a higher frequency of RHMs in Mexico than in other countries. This finding is in line with a previous report describing a higher frequency of RHMs in Mexico as compared to western countries.
Here, we describe the results of mutation analysis on 30 new unrelated patients with RHMs recruited in Mexico, seven molar tissues from seven unrelated patients with RHMs, and review mutation analysis on 13 unrelated patients of Mexican origin referred to the RI-MUHC. Of the 43 analyzed patients, excluding the molar tissues, L750V was present in homozygous or heterozygous state in 37 (86%) of them (Table 3). These data make the L750V the most frequent NLRP7 mutation reported to date and are in agreement with its presence at a minor allele frequency (MAF) of 0.025 in control subjects from Mexico [16] and 0.00310 in Latino population reported in gnomAD v2.1.1 (135 out of 35,430) (gnomAD (broadinstitute.org)) and Varsome (3 out of 848) (Varsome The Human Genomics Community).
Table 3.
Distribution of mutations in 43 Mexican patients with RHMs
| Biallelic NLRP7 mutations | Number of patients |
|---|---|
| L750V homozygous | 24 (55.8%) |
| L750V, c.2810+2T>G | 8 (18.6%) |
| L750V, c.2471+1G>A | 2 (4.6%) |
| L750V, c.-13413_2982-344del | 1 (2%) |
| L750V, c.-6831_-39-1586del | 1 (2%) |
| L750V,W653* | 1 (2%) |
| Number of patients with ≥ 1 L750V | 37 (86%) |
| Y872X, c.2810+2T>G | 1 (2%) |
| R390Afs*26 homozygous | 1 (2%) |
| RHMs and no mutations in NLRP7 | 4 (9.3%) |
| Biallelic MEI1 mutations | 1 (2%) |
| Number of patients with RHMs | 43 |
In addition, this study revealed a second founder variant, c.2810+2T>G in the Mexican population that was present in eight unrelated patients (Table 3). This mutation is also reported in databases with a MAF in Latino population of 0.0002892 (10 out of 34,574) in gnomAD v.2.1.1 and 0.0004 in Varsome. Of note, that L750V and c.2810+2T>G both appear to be specific for Mexican/Latino population (Varsome) and have never been reported in patients with RHMs or healthy subjects from other populations. However, the c.2471+1G>A mutation has been reported in patients of Pakistani, Indian, and Chinese origin, and this study revealed its presence for the first time in two unrelated Mexican patients, which is not unexpected since the Mexican population consists of a mixture of Native American inhabitants (56.4%), European migrants (41.8%), and West Africans (1.8%) [25]. Ruiz-Linares et al. [26] estimated individual ancestry proportions in different countries from Latin America and found that in the Mexican population, Native American ancestry is highest in the center/south of the country where the highest number of patients with L750V was observed. This suggests that L750V may have been inherited from the Native American population that remains to be demonstrated in future studies.
Two patients [3 and 20], the first with a homozygous L750V and the second with L750V and c.2810+2T>G, had each a live birth from a spontaneous conception that led to healthy children. These observations are in agreement with previous ones documenting the occurrence of a total of 13 live births [8, 19, 23, 24, 27, 28], observed mostly in patients with mutations that have mild functional consequences on the protein such as missense, splice, or sometimes protein-truncating mutations at the end of the protein [27]. Among the 13 reported live births, 12 children were reported to be healthy and only one was reported with various morphological abnormalities [28]. Despite these relatively encouraging outcomes, spontaneous live births from such patients are extremely rare and account for approximately 1.5% of all their conceptions [29]. Because the primary defect in patients with biallelic NLRP7 mutations is in their oocytes, ovum donation has been proposed to these patients as their best reproductive option. To date, eight such patients, including three reported in this study, patients 733, 1224, and 1878, and another in a patient that we previously reported in Estrada et al. [16], have achieved successful pregnancies from donated ova and conceived ten healthy live births [27, 30, 31].
Based on the above data and the replicated strong founder effect for L750V, if Sanger sequencing were to be used for mutation analysis, we propose to begin the analysis by sequencing exon 6 of NLRP7. If the patient is negative for the common mutation, completing the gene sequencing is then recommended. Genetic counseling of patients with biallelic NLRP7 mutations must consider the age of the patients, the risk of neoplastic degeneration, which occurred in 29% of the 31 patients recruited in Mexico, the scarcity of spontaneous live births in these patients, and the benefit of oocyte donation. Spontaneous live births have been observed in 13 patients; however, we still do not know if these children are at a higher risk for imprinting disorders. It is therefore important to keep in mind that the earliest known defect in patients with biallelic NLRP7 mutations is the impaired establishment of maternal methylation marks in their oocytes. In addition, biallelic mutations in another member of the subcortical maternal complex, PADI6, which have been shown to cause female infertility, early embryonic arrest during preimplantation development [32], and miscarriages and HM [23, 33] were recently documented in patients with Beckwith-Wiedemann and Silver-Russell syndromes [34, 35]. Therefore, a close follow-up of the pregnancies of patients with biallelic NLRP7 mutations is highly recommended and may help monitoring for imprinting disorders which may lead to a broad spectrum of clinical manifestations.
Supplementary information
(XLSX 13 kb)
Author contribution
All authors contributed to the study conception and design. Material preparation was performed by Maryam Rezaei, Irma Monroy, Mechtouf Nawel, Javier Pérez, Elsa Moreno, Yolotzin Valdespino, Carolina Galaz, and Guadalupe Razo. Data collection was performed by Monica Aguinaga, Carolina Galaz, Daniela Medina D, Raúl Piña, and Rima Slim. Analyses were performed by Maryam Rezaei, Irma Monroy, Mechtouf Nawel, Javier Pérez, and Guadalupe Razo. The first draft of the manuscript was written by Mónica Aguinaga and all authors commented on previous versions of the manuscript. Rima Slim revised the work critically for important intellectual content and approved the version to be published. All authors read and approved the final manuscript.
Funding
This work was supported by the Canadian Institute of Health Research MOP130364 and by the Instituto Nacional de Perinatologia, Mexico City.
Data Availability
All data and materials are available upon request.
Code availability
Not applicable.
Declarations
Ethics approval
The study was approved by the Instituto Nacional de Perinatologia (INPer) Review Board, study number: 212250-3220-11108-01-14 and the McGill Institutional Review Board (IRB# A01-M07-03A). This study was performed in line with the principles of the Declaration of Helsinki.
Consent to participate
All patients provided written consent to participate in our study.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Savage PM, Sita-Lumsden A, Dickson S, Iyer R, Everard J, Coleman R, Fisher RA, Short D, Casalboni S, Catalano K, Seckl MJ. The relationship of maternal age to molar pregnancy incidence, risks for chemotherapy and subsequent pregnancy outcome. J Obstet Gynaecol. 2013;33(4):406–411. doi: 10.3109/01443615.2013.771159. [DOI] [PubMed] [Google Scholar]
- 2.Hui P, Buza N, Murphy KM, Ronnett BM. Hydatidiform moles: genetic basis and precision diagnosis. Annu Rev Pathol. 2017;12:449–485. doi: 10.1146/annurev-pathol-052016-100237. [DOI] [PubMed] [Google Scholar]
- 3.Lurain JR, Sand PK, Carson SA, Brewer JI. Pregnancy outcome subsequent to consecutive hydatidiform moles. Am J Obstet Gynecol. 1982;142(8):1060–1061. doi: 10.1016/0002-9378(82)90798-0. [DOI] [PubMed] [Google Scholar]
- 4.Sebire NJ, Fisher RA, Foskett M, Rees H, Seckl MJ, Newlands ES. Risk of recurrent hydatidiform mole and subsequent pregnancy outcome following complete or partial hydatidiform molar pregnancy. BJOG. 2003;110(1):22–26. doi: 10.1046/j.1471-0528.2003.02388.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim JH, Park DC, Bae SN, Namkoong SE, Kim SJ. Subsequent reproductive experience after treatment for gestational trophoblastic disease. Gynecol Oncol. 1998;71(1):108–112. doi: 10.1006/gyno.1998.5167. [DOI] [PubMed] [Google Scholar]
- 6.Yapar EG, Ayhan A, Ergeneli MH. Pregnancy outcome after hydatidiform mole, initial and recurrent. J Reprod Med. 1994;39(4):297–299. [PubMed] [Google Scholar]
- 7.Eagles N, Sebire NJ, Short D, Savage PM, Seckl MJ, Fisher RA. Risk of recurrent molar pregnancies following complete and partial hydatidiform moles. Hum Reprod. 2015;30(9):2055–2063. doi: 10.1093/humrep/dev169. [DOI] [PubMed] [Google Scholar]
- 8.Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, Hanash S, Rouleau GA, Slim R. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38(3):300–302. doi: 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen NMP, Khawajkie Y, Mechtouf N, Rezaei M, Breguet M, Kurvinen E, Jagadeesh S, Solmaz AE, Aguinaga M, Hemida R, Harma MI, Rittore C, Rahimi K, Arseneau J, Hovanes K, Clisham R, Lenzi T, Scurry B, Addor MC, Bagga R, Nendaz GG, Finci V, Poke G, Grimes L, Gregersen N, York K, Bolze PA, Patel C, Mozdarani H, Puechberty J, Scotchie J, Fardaei M, Harma M, Gardner RJMK, Sahoo T, Dudding-Byth T, Srinivasan R, Sauthier P, Slim R. The genetics of recurrent hydatidiform moles: new insights and lessons from a comprehensive analysis of 113 patients. Mod Pathol. 2018;31(7):1116–1130. doi: 10.1038/s41379-018-0031-9. [DOI] [PubMed] [Google Scholar]
- 10.Parry DA, Logan CV, Hayward BE, Shires M, Landolsi H, Diggle C, Carr I, Rittore C, Touitou I, Philibert L, Fisher RA, Fallahian M, Huntriss JD, Picton HM, Malik S, Taylor GR, Johnson CA, Bonthron DT, Sheridan EG. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89(3):451–458. doi: 10.1016/j.ajhg.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebire NJ, Savage PM, Seckl MJ, Fisher RA. Histopathological features of biparental complete hydatidiform moles in women with NLRP7 mutations. Placenta. 2013;34(1):50–56. doi: 10.1016/j.placenta.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15(3):416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akoury E, Zhang L, Ao A, Slim R. NLRP7 and KHDC3L, the two maternal-effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Hum Reprod. 2015;30(1):159–169. doi: 10.1093/humrep/deu291. [DOI] [PubMed] [Google Scholar]
- 14.Demond H, Anvar Z, Jahromi BN, Sparago A, Verma A, Davari M, Calzari L, Russo S, Jahromi MA, Monk D, Andrews S, Riccio A, Kelsey G. A KHDC3L mutation resulting in recurrent hydatidiform mole causes genome-wide DNA methylation loss in oocytes and persistent imprinting defects post-fertilisation. Genome Med. 2019;11(1):84. doi: 10.1186/s13073-019-0694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen NMP, Ge ZJ, Reddy R, Fahiminiya S, Sauthier P, Bagga R, Sahin FI, Mahadevan S, Osmond M, Breguet M, Rahimi K, Lapensee L, Hovanes K, Srinivasan R, van den Veyver IB, Sahoo T, Ao A, Majewski J, Taketo T, Slim R. Causative mutations and mechanism of androgenetic hydatidiform moles. Am J Hum Genet. 2018;103(5):740–751. doi: 10.1016/j.ajhg.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrada H, Buentello B, Zenteno JC, Fiszman R, Aguinaga M. The p.L750V mutation in the NLRP7 gene is frequent in Mexican patients with recurrent molar pregnancies and is not associated with recurrent pregnancy loss. Prenat Diagn. 2013;33(3):205–208. doi: 10.1002/pd.4036. [DOI] [PubMed] [Google Scholar]
- 17.Kou YC, Shao L, Peng HH, Rosetta R, del Gaudio D, Wagner AF, al-Hussaini TK, van den Veyver IB. A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod. 2008;14(1):33–40. doi: 10.1093/molehr/gam079. [DOI] [PubMed] [Google Scholar]
- 18.Slim R, Bagga R, Chebaro W, Srinivasan R, Agarwal N. A strong founder effect for two NLRP7 mutations in the Indian population: an intriguing observation. Clin Genet. 2009;76(3):292–295. doi: 10.1111/j.1399-0004.2009.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahadevan S, Wen S, Balasa A, Fruhman G, Mateus J, Wagner A, al-Hussaini T, van den Veyver IB. No evidence for mutations in NLRP7 and KHDC3L in women with androgenetic hydatidiform moles. Prenat Diagn. 2013;33(13):1242–1247. doi: 10.1002/pd.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian J, Deveault C, Bagga R, Xie X, Slim R. Women heterozygous for NALP7/NLRP7 mutations are at risk for reproductive wastage: report of two novel mutations. Hum Mutat. 2007;28(7):741. doi: 10.1002/humu.9498. [DOI] [PubMed] [Google Scholar]
- 21.Reddy R, Akoury E, Phuong Nguyen NM, Abdul-Rahman OA, Dery C, Gupta N, Daley WP, Ao A, Landolsi H, Ann Fisher R, Touitou I, Slim R. Report of four new patients with protein-truncating mutations in C6orf221/KHDC3L and colocalization with NLRP7. Eur J Hum Genet. 2013;21(9):957–964. doi: 10.1038/ejhg.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khawajkie Y, Mechtouf N, Nguyen NMP, Rahimi K, Breguet M, Arseneau J, Ronnett BM, Hoffner L, Lazure F, Arnaud M, Peers F, Tan L, Rafea BA, Aguinaga M, Horowitz NS, Ao A, Tan SL, Brown R, Buckett W, Surti U, Hovanes K, Sahoo T, Sauthier P, Slim R. Comprehensive analysis of 204 sporadic hydatidiform moles: revisiting risk factors and their correlations with the molar genotypes. Mod Pathol. 2020;33(5):880–892. doi: 10.1038/s41379-019-0432-4. [DOI] [PubMed] [Google Scholar]
- 23.Rezaei M, Jagadeesh/Beena S, Bereke E, Aguinaga M, Qian J, Hadipour Z, et al. Novel pathogenic variants in PADI6, NLRP5, and NLRP7 in patients with hydatidiform moles and reproductive failure. Clin Genet. 2021. 10.1111/cge.13941. [DOI] [PubMed]
- 24.Reddy R, Nguyen NM, Sarrabay G, Rezaei M, Rivas MC, Kavasoglu A, et al. The genomic architecture of NLRP7 is Alu rich and predisposes to disease-associated large deletions. Eur J Hum Genet. 2016;24(10):1516. doi: 10.1038/ejhg.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, Balam-Ortiz E, del Bosque-Plata L, Velazquez-Fernandez D, Lara C, Goya R, Hernandez-Lemus E, Davila C, Barrientos E, March S, Jimenez-Sanchez G. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A. 2009;106(21):8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Linares A, Adhikari K, Acuna-Alonzo V, Quinto-Sanchez M, Jaramillo C, Arias W, et al. Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10(9):e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akoury E, Gupta N, Bagga R, Brown S, Dery C, Kabra M, et al. Live births in women with recurrent hydatidiform mole and two NLRP7 mutations. Reprod BioMed Online. 2015;31(1):120–124. doi: 10.1016/j.rbmo.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Sunde L, Vejerslev LO, Jensen MP, Pedersen S, Hertz JM, Bolund L. Genetic analysis of repeated, biparental, diploid, hydatidiform moles. Cancer Genet Cytogenet. 1993;66(1):16–22. doi: 10.1016/0165-4608(93)90142-9. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen NM, Slim R. Genetics and Epigenetics of recurrent hydatidiform moles: basic science and genetic counselling. Curr Obstet Gynecol Rep. 2014;3:55–64. doi: 10.1007/s13669-013-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher RA, Lavery SA, Carby A, Abu-Hayyeh S, Swingler R, Sebire NJ, Seckl MJ. What a difference an egg makes. Lancet. 2011;378(9807):1974. doi: 10.1016/S0140-6736(11)61751-0. [DOI] [PubMed] [Google Scholar]
- 31.Fallahi J, Razban V, Momtahan M, Akbarzadeh-Jahromi M, Namavar-Jahromi B, Anvar Z, Fardaei M. A novel mutation in NLRP7 related to recurrent hydatidiform mole and reproductive failure. Int J Fertil Steril. 2019;13(2):135–138. doi: 10.22074/ijfs.2019.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Shi Y, Fu J, Yu M, Feng R, Sang Q, Liang B, Chen B, Qu R, Li B, Yan Z, Mao X, Kuang Y, Jin L, He L, Sun X, Wang L. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99(3):744–752. doi: 10.1016/j.ajhg.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian J, Nguyen NMP, Rezaei M, Huang B, Tao Y, Zhang X, Cheng Q, Yang HJ, Asangla A, Majewski J, Slim R. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet. 2018;26(7):1007–1013. doi: 10.1038/s41431-018-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cubellis MV, Pignata L, Verma A, Sparago A, Del Prete R, Monticelli M, et al. Loss-of-function maternal-effect mutations of PADI6 are associated with familial and sporadic Beckwith-Wiedemann syndrome with multi-locus imprinting disturbance. Clin Epigenetics. 2020;12(1):139. doi: 10.1186/s13148-020-00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eggermann T, Kadgien G, Begemann M, Elbracht M. Biallelic PADI6 variants cause multilocus imprinting disturbances and miscarriages in the same family. Eur J Hum Genet. 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 13 kb)
Data Availability Statement
All data and materials are available upon request.
Not applicable.