Abstract
Viruses hosted by uncultivated fungi have been poorly studied. We carried out studies to characterize a large dsRNA segment (~20 kbp) detected in the basidiomycetous, ectomycorrhizal fungus Hygrophorus penarioides. The dsRNA was gel-purified and its randomly amplified cDNA fragments were used for high throughput sequencing (HTS). Reads were de novo assembled and BLASTx analysis revealed sequence similarity to viruses of the family Endornaviridae. The 5' and 3' terminal sequences of the dsRNA segment were determined by performing RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE). The full-length cDNA sequence of the putative endornavirus comprises 16,785 nt and contains a single, long open reading frame which encodes for a polyprotein of 5522 aa with conserved domains for cysteine-rich region, helicase, glycosyltransferase, and RNA-dependent RNA polymerase. The virus was named Hygrophorus penarioides endornavirus 1 (HpEnV1). A BLASTp search performed using the polyprotein sequence revealed that the most closely related, fully sequenced endornavirus to HpEnV1 is Ceratobasidium endornavirus B.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-021-00500-8.
Keyword: Endornavirus, dsRNA; Ectomycorrhizal fungus; Hygrophorus penarioides
Introduction
Mycorrhizal mutualism is one of the fundamental symbiotic associations that significantly promoted the land colonization of plants since the Paleozoic era [1]. The vast majority of the present vascular plants form mycorrhizal associations with fungi [2]. Mycorrhizal symbiosis, which is primarily managed by a physical contact established between the hyphal network of a fungus and the root system of a plant, enables the exchange of various growth-promoting metabolic resources among partners. Typically, in this symbiosis, the mycorrhizal fungus renders inorganic decomposed material (e.g., phosphate and nitrogen) accessible to the host plant and further augments the resistance of its host to various stress factors. In return, the host plant caters for the organic nutrient requirements of the mycorrhizal fungus [1, 2]. By contributing to the soil nutrient cycling and formation of plant and microbial communities, mycorrhizal fungi continue to play a vital role in preserving the functioning and permanence of the land ecosystems [3].
The genus Hygrophorus belongs to the Hygrophoraceae family of the order Agaricales. The genus comprises nearly a hundred, basidiomycetous, ectomycorrhizal species, and its most characteristic features are often viscid pileus, waxy, often thick and distant lamellae, and hyaline, subglobose to ellipsoid spores [4]. Hygrophorus penarioides Jacobsson & E. Larss. is characterized by medium-sized white to cream-colored sporophore and having an ectomycorrhizal association with oak species. H. penarioides is easily distinguished from its closest relative H. penarius by its mycorrhizal partner preference and molecular phylogeny based on internal transcribed spacer (ITS) rDNA region [4, 5].
In contrast to their ecological importance, viruses inhabiting ectomycorrhizal fungi have been barely studied compared to the viruses of economically important fungi including the phytopathogenic and cultivated species. So far, mycoviruses from different evolutionary origins including viruses of the recognized families such as Partitiviridae, Totiviridae, Endornaviridae, and Narnaviridae as well as the proposed families such as Phlegiviridae, Fusagraviridae, Yadokariviridae, and Megatotiviridae have been reported from some ectomycorrhizal fungal species with basidiomycete and ascomycete origin [6–14].
The family Endornaviridae comprises viruses that are known to inhabit plants, fungi, and oomycetes [15]. Thus far, the family includes two recognized genera, Alphaendornavirus and Betaendornavirus, which members are differentiated according to their genome size, host range, and the presence of unique protein domains (www.ictv.global/report/endornaviridae). Members of the family Endornaviridae are composed of a single, unencapsidated positive-sense, single-stranded RNA molecule with a length of 9.7 to 17.6 kb. The only protein domain that is conserved among all endornaviruses is the RNA-dependent RNA polymerase (RdRp) domain. Besides RdRp domain, some but not all of the endornaviruses members encode other protein domains such as methyltransferase, helicase, glycosyltransferase, cysteine-rich region, phytoreo_S7 domain, and capsular polysaccharide synthase [15]. Endornaviruses are not known to form virion particles, and the RNA genome is thought to be associated with cytoplasmic membrane vesicles [16]. However, they usually establish persistent infections, and the presence of their horizontal transmission route in fungi has been reported [17].
According to the literature, the only virus reported thus far from the ectomycorrhizal basidiomycetous fungus Hygrophorus penarioides is an alphapartitivirus named Hygrophorus penarioides partitivirus 1 that was identified by high throughput sequencing approach [18]. Here, we report the first endornavirus inhabiting H. penarioides, which could contribute to our understanding of the evolution, diversity, and classification of endornaviruses.
Materials and methods
Sampling of C. fulgens ascocarp
A single basidiocarp of H. penarioides ANK Akata 7305 which grows under a sessile oak tree (Quercus petraea (Matt.) Liebl.) was sampled in the Belgrad Forest located in Istanbul province, North-Western Turkey during a field study performed on the 30th of October 2019. The collected basidiocarp specimen was surface-sterilized by sequential treatment with 2% sodium hypochlorite for 2 min and 70% ethanol for 10 s. After surface sterilization, the basidiocarp specimen was sluiced with sterile distilled water.
dsRNA preparation, cDNA synthesis, and PCR amplification
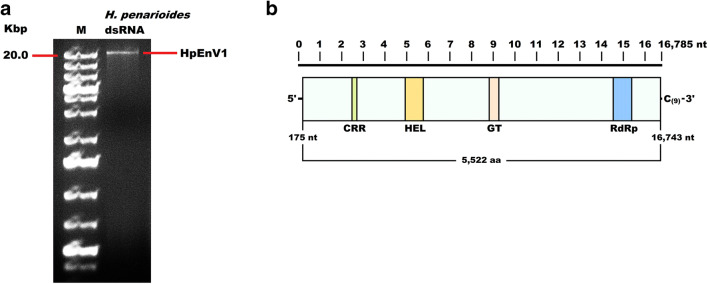
Double-stranded RNA was extracted from the basidiocarp using a cellulose-column-based Double-RNA Viral dsRNA Extraction Kit (iNtRON Biotechnology). The extracted dsRNA was sequentially treated with DNase I and S1 nuclease (Promega) for removing the remaining genomic DNA and host RNA. The nuclease-treated dsRNA sample was separated on a 1% agarose gel, and the dsRNA segment corresponding to the 20 Kbp fragment of the DNA marker (Fig. 1a) was excised and purified using a GeneJET PCR Purification Kit (Thermo Fisher). The purified dsRNA sample was reverse transcribed into cDNA using the primer-dN6 (5’-CCTGAATTCGGATCCTCCNNNNNN-3’) and Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the instruction of the manufacturer. The synthesized cDNA was used as a template for random PCR (rPCR) which was performed using 1 mM of rPCR primer (5'-CCTGAATTCGGATCCTCC-3') and DreamTaq DNA Polymerase (5 U/μL) (Thermo Fisher) as described previously [19]. rPCR amplicon was purified using a GeneJET PCR Purification Kit (Thermo Fisher) and 500 ng of the purified amplicon was delivered to the Novogene (Cambridge, UK) for library construction, and high-throughput sequencing of 150 bp long paired ends with a sequencing depth of ≥ 100X on a Novaseq 6000 platform (Illumina).
Fig. 1.
a Electrophoresis of the DNase I and S1 nuclease treated dsRNA extracted from H. penarioides ANK Akata 7305 in a 1% agarose gel. Lane M, DNA molecular weight marker. b Genome organization of Hygrophorus penarioides endornavirus 1 (HpEnV1). The rectangular box represents the large ORF, whereas colored boxes depict the Cysteine-rich region (CRR), viral helicase1 (Hel), Glycosyltransferase (GT), and RNA dependent RNA polymerase (RdRp). UTR regions are also shown
Bioinformatic analyses of the sequence data
For contig generation, HTS-derived raw reads were assembled de novo by using the CLC Genomic Workbench version 20.0.2 (Qiagen) software. Assembly parameters were selected as word size of 26, default bubble size of 50, automatic paired distance estimation, and a minimum contig length of 200 nt. Assembled contigs and their deduced amino acid sequences were analyzed by batch BLASTx and BLASTp, respectively, to identify virus-related contigs (e-value < 1). The NCBI conserved domain search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to find out virus-related domains including RNA-dependent RNA polymerase, helicase, and glycosyltransferase.
Deduced amino acid sequences of virus-related contigs were aligned using ClustalW in MEGA X software [20] and analyzed to find the best evolution model. Maximum likelihood (ML) phylogenetic tree was generated by using the Nearest-Neighbor-Interchange (NNI) algorithm for the ML heuristic method by performing 1000 bootstrap replicates. Pairwise identity analyses of HpEnV1 polyprotein and conserved domains with corresponding sequences of selected endornaviruses were conducted using the NCBI BLASTp and Clustal Omega online sequence alignment tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins, https://www.ebi.ac.uk/Tools/msa/clustalo/) with default parameters.
Sequencing of the 5’ and 3’ terminals using RLM-RACE
To determine the sequences of 5’- and 3’- terminals, the 3’ ends of the purified dsRNA were ligated to the oligonucleotide RLO (5’p-CATGGTGGCGACCGGTAG-NH2 3’) at 37 °C for 6 h followed by overnight treatment at 12 °C with T4 RNA ligase 1 (New England Biolabs). Oligo-ligated dsRNA was purified using a GeneJET PCR Purification Kit (Thermo Scientific) and reverse transcribed into cDNA using the primer RTP (5’-CTACCGGTCGCCACCATG-3’) and a Transcriptor First Strand cDNA Synthesis Kit (Roche) as described above. The 5’- and 3’-terminal sequences were PCR amplified with the sequence-specific reverse and forward primers, HpEV1-5 (5’-TTGTTGGAAAGTGGGTGGGT-3’) and HpEV1-3 (5’-CAATGAGAGCAGCCAACACC-3’) each of which was coupled with the RTP primer. The amplicons were cloned into pGEM-T Easy Vector (Promega) and sequenced with dideoxy chain termination method using the universal M13 primers at the laboratory of Eurofins Genomics (Germany).
Results and discussion
A total of 28,092 reads were used to assemble 169 contigs which were subjected to batch BLASTx searches against the protein database of NCBI GenBank. Among the analyzed contigs, the 16,829 nt long contig_1, assembled from 10,625 reads with a sequencing depth (coverage) of 189, exhibited homology to the sequences of various endornaviruses. Thus, the identified putative endornavirus was named Hygrophorus penarioides endornavirus 1 (HpEnV1). The 5’ and 3’ terminal sequences of HpEnV1 were determined by using RLM-RACE approach. The complete genome of HpEnV1 is 16,875 nt and contains a single, long open reading frame (ORF) encoding for a polyprotein of 5522 aa with a predicted molecular weight of 608.44 kDa. The genome sequence of HpEnV1 was deposited into NCBI GenBank under the accession number MW308217. The predicted 5’ UTR of HpEnV1 was 174 nt long. Although one in-frame AUG codon was found at nucleotides 16–18, online in silico analyses revealed that this codon is possibly not employed due to its inconvenience in efficient translation initiation. Compared with the consensus Kozak sequence of a model ectomycorrhizal fungus Laccaria bicolor N U C/G A H A A U G G/A C [21], the second AUG codon (G C C A G A A U G A G) of HpEnV1 is in the most convenient state for translation initiation and is estimated as the start codon for the translation of the long ORF. The 3′ UTR of HpEnV1 was 42 nt long and ended with a 9 nt long cytosine stretch which is typically found in many endornaviruses. A search for the presence of conserved domains in the polyprotein of HpEnV1 revealed four conserved domains, a cysteine-rich region (CRR) (1054–1110 aa), helicase (viral_helicase1, pfam01443) (1587–1851 aa), glycosyltransferase (YjiC, COG1819) (3194–3344 aa), and RNA-dependent RNA polymerase (RdRp_2, pfam00978) (5123–5397 aa) which are commonly found in endornaviruses (Fig. 1b). Pairwise identity analyses were performed using the HpEnV1 polyprotein and conserved domains with the corresponding sequences of selected endornaviruses (Table 1). Comparison with 14 complete genome sequences of members of the genus Alphaendornavirus revealed that the deduced HpEnV1 polyprotein exhibits a significant amino acid sequence homology to alphaendornaviruses, with Ceratobasidium endornavirus B being the most significantly (E value: 2e-89) similar virus with 33.79% similarity and 50% coverage rates. In regard to the conserved domains, RdRp, helicase, and glycosyltransferase domains of HpEnV1 polyprotein exhibit the highest sequence identities to the RdRp domain of Capsicum frutescens endornavirus 1, helicase domain of Helianthus annuus alphaendornavirus, and glycosyltransferase domain of Rhizoctonia solani endornavirus 1 with 46.44, 32.58, and 39.19 identity rates, respectively (Table 1). Furthermore, a phylogenetic tree constructed using the RdRp domains of endornaviruses placed HpEnV1 in an established clade with the alphaendornaviruses (Fig. 2). On the other hand, the cluster formed by HpEnV1 and Vicia faba alphaendornavirus should be interpreted cautiously as the branch support for this clade was low (bootstrap rate < 50%). Altogether, these results suggest that HpEnV1 is a new member of the genus Alphaendornavirus.
Table 1.
Amino acid identity (%) comparison of the polyprotein and conserved domains of Hygrophorus penarioides endornavirus 1 with corresponding sequences of selected endornaviruses
| Virus name | GenBank accession no. | Genome length (nt) | Sequence Identity (%) | |||
|---|---|---|---|---|---|---|
| PP | RdRp | Hel | GT | |||
| Ceratobasidium endornavirus B | NC_031463 | 23,635 | 33.79 | 45.97 | 27.74 | 25.24 |
| Vicia faba alphaendornavirus | NC_007648 | 17,635 | 21.47 | 42.22 | 25.31 | ND |
| Ceratobasidium endornavirus 1 | MN738550 | 21,587 | 30.27 | 40.56 | ND | 31.58 |
| Rhizoctonia solani endornavirus 1 | NC_040609 | 19,936 | 30.96 | 43.72 | ND | 39.19 |
| Rhizoctonia cerealis alphaendornavirus 1 | NC_022619 | 17,486 | 38.12 | 44.62 | 31.32 | ND |
| Capsicum frutescens endornavirus 1 | MN175322 | 14,846 | 37.99 | 46.44 | 26.41 | 19.19 |
| Cluster bean endornavirus 1 | NC_040825 | 12,895 | 36.45 | 43.07 | 27.82 | ND |
| Triticeae associated alphaendornavirus | MW091544 | 14,878 | 37.01 | 43.23 | 26.12 | 20.77 |
| Hot pepper alphaendornavirus | NC_027920 | 14,729 | 37.70 | 45.32 | 28.14 | 21.00 |
| Hordeum vulgare alphaendornavirus | MN107383 | 14,243 | 37.05 | 43.82 | 27.04 | ND |
| Helicobasidium mompa alphaendornavirus 1 | NC_013447 | 16,614 | 31.99 | 43.27 | 27.08 | 25.71 |
| Fagopyrum esculentum endornavirus 1 | LC500285 | 15,677 | 36.36 | 45.23 | 24.53 | 23.23 |
| Helianthus annuus alphaendornavirus | NC_040799 | 14,663 | 36.02 | 40.75 | 32.58 | 25.95 |
| Brown algae endornavirus 1 | LC521321 | 13,603 | 34.99 | 42.01 | 29.46 | 24.53 |
PP polyprotein; RdRp viral RNA-dependent RNA polymerase; Hel viral helicase1; GT glycosyltransferase; ND not detected
Highest identity rates are indicated with bold font
Fig. 2.
Phylogenetic tree of the RdRp domain of HpEnV1 and selected endornaviruses from plants and fungi. The alignment was generated using ClustalW algorithm and the best substitution model and a maximum likelihood phylogenetic tree were calculated using MEGA X. The best-fitted substitution model was calculated as LG + G. Values at the nodes indicate bootstrap values obtained for 1000 replicates. Branches with bootstrap values lower than 50 are not shown. RdRp sequence of the grapevine leafroll-associated virus (GenBank accession: AEW24402) was used as the outgroup sequence
Conclusion
Data presented in this study support the endornavirus nature of the large dsRNA molecule isolated from the ectomycorrhizal fungus Hygrophorus penarioides ANK Akata 7305. The organization of the genome and its close phylogenetic relatedness to other endornavirus members support this virus isolate, HpEnV1, as being a new member of the genus Alphaendornavirus within the family Endornaviridae. HpEnV1 is the first endornavirus identified in the basidiomycetous, ectomycorrhizal fungus H. penarioides.
Supplementary Information
(DOCX 23 kb)
Funding
This study was funded by the Ankara University, Coordinatorship of Scientific Research Projects with the project number 19B0430002.
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brundrett MC, Tedersoo L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018;220(4):1108–1115. doi: 10.1111/nph.14976. [DOI] [PubMed] [Google Scholar]
- 2.Smith SE, Read DJ. Mycorrhizal symbiosis. 3. London: Academic Press; 2008. [Google Scholar]
- 3.Kariman K, Barker SJ, Tibbett M. Structural plasticity in root-fungal symbioses: diverse interactions lead to improved plant fitness. PeerJ. 2018;6:e6030. doi: 10.7717/peerj.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zajta E, Varga T, Kovács G, Dima B. New insights on Hygrophorus penarioides and H. penarius (Agaricales, Hygrophoraceae) from Hungary. Phytotaxa. 2019;392(2):127–139. doi: 10.11646/phytotaxa.392.2.2. [DOI] [Google Scholar]
- 5.LE Jacobsson S. Hygrophorus penarioides, a new species identified using morphology and ITS sequence data. Mycotaxon. 2007;99:337–343. [Google Scholar]
- 6.Petrzik K, Sarkisova T, Stary J, Koloniuk I, Hrabakova L, Kubesova O. Molecular characterization of a new monopartite dsRNA mycovirus from mycorrhizal Thelephora terrestris (Ehrh.) and its detection in soil oribatid mites (Acari: Oribatida) Virology. 2016;489:12–19. doi: 10.1016/j.virol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Sahin E, Akata I. Complete genome sequence of a novel mitovirus from the ectomycorrhizal fungus Geopora sumneriana. Arch Virol. 2019;164(11):2853–2857. doi: 10.1007/s00705-019-04367-x. [DOI] [PubMed] [Google Scholar]
- 8.Sahin E, Akata I, Keskin E. Novel and divergent bipartite mycoviruses associated with the ectomycorrhizal fungus Sarcosphaera coronaria. Virus Res. 2020;286:198071. doi: 10.1016/j.virusres.2020.198071. [DOI] [PubMed] [Google Scholar]
- 9.Sahin E, Keskin E, Akata I. Novel and diverse mycoviruses co-inhabiting the hypogeous ectomycorrhizal fungus Picoa juniperi. Virology. 2020;552:10–19. doi: 10.1016/j.virol.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Stielow B, Klenk HP, Menzel W. Complete genome sequence of the first endornavirus from the ascocarp of the ectomycorrhizal fungus Tuber aestivum Vittad. Arch Virol. 2011;156(2):343–345. doi: 10.1007/s00705-010-0875-x. [DOI] [PubMed] [Google Scholar]
- 11.Stielow B, Klenk HP, Winter S, Menzel W. A novel Tuber aestivum (Vittad.) mitovirus. Arch Virol. 2011;156(6):1107–1110. doi: 10.1007/s00705-011-0998-8. [DOI] [PubMed] [Google Scholar]
- 12.Stielow B, Menzel W. Complete nucleotide sequence of TaV1, a novel totivirus isolated from a black truffle ascocarp (Tuber aestivum Vittad.) Arch Virol. 2010;155(12):2075–2078. doi: 10.1007/s00705-010-0824-8. [DOI] [PubMed] [Google Scholar]
- 13.Stielow JB, Bratek Z, Klenk HP, Winter S, Menzel W. A novel mitovirus from the hypogeous ectomycorrhizal fungus Tuber excavatum. Arch Virol. 2012;157(4):787–790. doi: 10.1007/s00705-012-1228-8. [DOI] [PubMed] [Google Scholar]
- 14.Sutela S, Vainio EJ. Virus population structure in the ectomycorrhizal fungi Lactarius rufus and L. tabidus at two forest sites in Southern Finland. Virus Res. 2020;285:197993. doi: 10.1016/j.virusres.2020.197993. [DOI] [PubMed] [Google Scholar]
- 15.Valverde RA, Khalifa ME, Okada R, Fukuhara T, Sabanadzovic S, Ictv Report C. ICTV Virus Taxonomy Profile: Endornaviridae. J Gen Virol. 2019;100(8):1204–1205. doi: 10.1099/jgv.0.001277. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre A, Scalla R, Pfeiffer P. The double-stranded RNA associated with the '447' cytoplasmic male sterility in Vicia faba is packaged together with its replicase in cytoplasmic membranous vesicles. Plant Mol Biol. 1990;14(4):477–490. doi: 10.1007/BF00027494. [DOI] [PubMed] [Google Scholar]
- 17.Zheng L, Shu C, Zhang M, Yang M, Zhou E (2019) Molecular characterization of a novel endornavirus conferring hypovirulence in rice sheath blight fungus Rhizoctonia solani AG-1 IA strain GD-2. Viruses 11(2). 10.3390/v11020178 [DOI] [PMC free article] [PubMed]
- 18.Sahin E, Akata I. Full-length genome characterization of a novel alphapartitivirus detected in the ectomycorrhizal fungus Hygrophorus penarioides. Virus Genes. 2021;57:94–99. doi: 10.1007/s11262-020-01814-9. [DOI] [PubMed] [Google Scholar]
- 19.Darissa O, Willingmann P, Adam G. Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods. 2010;169(2):397–403. doi: 10.1016/j.jviromet.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemppainen M, Chowdhury J, Lundberg-Felten J, Pardo A. Fluorescent protein expression in the ectomycorrhizal fungus Laccaria bicolor: a plasmid toolkit for easy use of fluorescent markers in basidiomycetes. Curr Genet. 2020;66(4):791–811. doi: 10.1007/s00294-020-01060-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 23 kb)