Dear Editor,
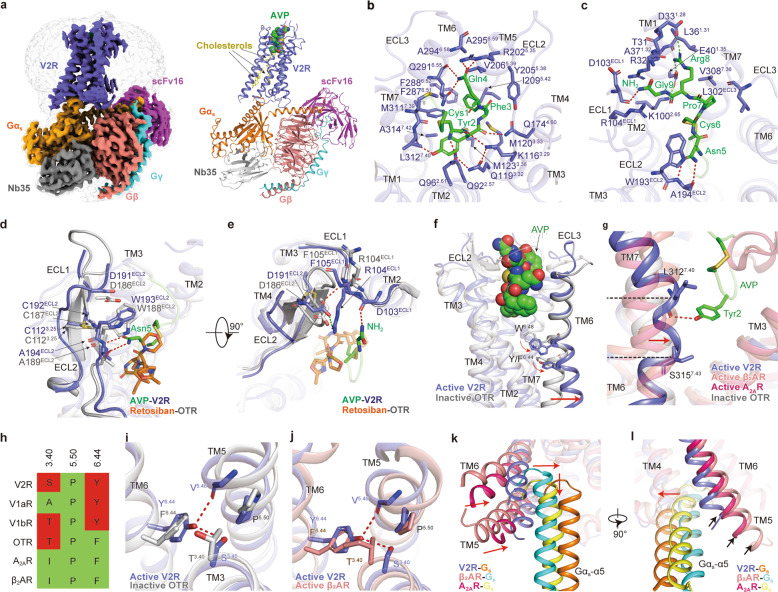
Vasopressin type 2 receptor (V2R) belongs to the vasopressin (VP)/oxytocin (OT) receptor subfamily of G protein-coupled receptors (GPCRs), which comprises at least four closely related receptor subtypes: V1aR, V1bR, V2R, and OTR.1 These receptors are activated by arginine vasopressin (AVP) and OT, two endogenous nine-amino acid neurohypophysial hormones, which are thought to mediate a biologically conserved role in social behavior and sexual reproduction.2 V2R is mainly expressed in the renal collecting duct principal cells and mediates the antidiuretic action of AVP by accelerating water reabsorption, thereby playing a vital role in controlling water homeostasis. Moreover, numerous gain-of-function and loss-of-function mutations of V2R have been identified and are closely associated with human diseases, including nephrogenic syndrome of inappropriate diuresis (NSIAD) and X-linked congenital nephrogenic diabetes insipidus (NDI).3 Thus, V2R has attracted intense interest as a drug target. However, due to a lack of structural information, how AVP recognizes and activates V2R remains elusive, which hampers the V2R-targeted drug design. Here, we determined a 2.6 Å resolution cryo-EM structure of the full-length, Gs-coupled human V2R bound to AVP (Fig. 1a; Supplementary information, Table S1). The Gs protein was engineered based on mini-Gs that was used in the crystal structure determination of the Gs-coupled adenosine A2A receptor (A2AR) to stabilize the V2R–Gs protein complex (Supplementary information, Data S1).4 The final structure of the AVP–V2R–Gs complex contains all residues of AVP (residues 1–9), the Gαs Ras-like domain, Gβγ subunits, Nb35, scFv16, and the V2R residues from T31 to L3398.57 (superscripts refer to Ballesteros–Weinstein numbering5). The majority of amino acid side chains, including AVP, transmembrane domain (TMD), all flexible intracellular loops (ICLs) and extracellular loops (ECLs) except for ICL3 and G185–G188 in ECL2, were well resolved in the model, refined against the EM density map (Fig. 1a; Supplementary information, Figs. S1–S3). The complex structure can provide detailed information on the binding interface between AVP and helix bundle of the receptor, as well as the receptor–Gs interface.
Fig. 1. Cryo-EM structure of the AVP–V2R–Gs complex.
a Orthogonal view of the density map (left panel) and model (right panel) for the AVP–V2R–Gs complex. V2R, slate blue; AVP, green; Gs, orange; Gβ, salmon; Gγ, cyan; Nb35, gray; scFv16, magenta; cholesterols, yellow. b, c Interactions between AVP and residues in V2R-binding pocket. d, e A comparison of the conformational change of ECL1 and ECL2 in AVP-bound V2R structure and retosiban-bound OTR (PDB: 6TPK) inactive structure. f Conformational changes of TM6 and conserved W6.48 and Y/F6.44 of V2R compared to inactive OTR. Red arrows indicate the movement of TM6 and the side chains of W6.48 and Y/F6.44 in V2R relative to inactive OTR. g Conformational change of TM7 in AVP–V2R structure relative to inactive OTR, β2AR–Gs, and A2AR–Gs structures. Red arrow indicates the movement of TM7 in V2R. h Sequence comparison of conserved P5.50-I3.40-F6.44 motif among V2R, V1aR, V1bR, OTR, A2AR, and β2AR. Polar residues are colored in red and nonpolar residues in green. Conformational changes of AVP-bound V2R structure resulted from substitutions in the conserved P5.50-I3.40-F6.44 motif relative to inactive OTR (i) and active β2AR (j) structures, respectively. k, l A conformational comparison of receptor helical bundle and Gαs α5 helix among AVP–V2R–Gs, β2AR–Gs, and A2AR–Gs complexes. Red arrows indicate the movements of intracellular tips of TMs5, 6, and α5 helix of Gαs from V2R–Gs relative to other complexes. Black arrows label the cytoplasmic ends of TM5 in these structures. H-bonds and salt bridges are depicted as red and green dashed lines, respectively. The disulfide bonds are shown as yellow sticks. The structural alignment is based on receptors.
AVP occupies an orthosteric binding pocket in the TMD bundle composed of all TM helices and ECLs (Fig. 1b, c; Supplementary information, Table S2). The most notable conformational feature of AVP is the tocin ring formed by a disulfide bridge between the first and sixth cysteine residues, presenting a “spoon-like” conformation (Supplementary information, Fig. S4a, b). The cyclic spoon head inserts deeply into the TMD core, while the C-terminal spoon tail stretches toward the ECLs of the receptor. Cys1P of AVP and Q962.61, K1163.29, Q1193.32 of V2R form a stabilizing H-bond network, consistent with the decreased potency of the receptor mutants with these residues mutated (Fig. 1b; Supplementary information, Fig. S4c, d). The hydroxyl group of Tyr2P forms an H-bonds with the main chain oxygen of L3127.40, whereas its main chain CO group forms another H-bond with Q1744.60 (Fig. 1b; Supplementary information, Fig. S4c). Phe3P buries in a hydrophobic cleft constituted by M1203.33, M1233.36, Y2055.38, V2065.39, I2095.42, F2876.51, F2886.52, and Q2916.55 (Fig. 1b; Supplementary information, Fig. S4c). Other polar contacts are seen between Gln4P and R2025.35, Gln4P and the backbone and side chain oxygens of Q2916.55, as well as Asn5P and the main chain of A194ECL2 (Fig. 1b, c; Supplementary information, Fig. S4c). Arg8P forms salt bridges with D331.28 and E401.35, the latter forms an extra salt bridge with K1002.65 (Fig. 1c; Supplementary information, Fig. S4c). Additionally, the C-terminal amide of AVP points to ECL1, stacking against the R104ECL1 guanidinium group and H-bonding to the backbone CO group of D103ECL1 (Fig. 1c; Supplementary information, Fig. S4c). These interactions between AVP and residues in V2R binding pocket were confirmed through alanine mutagenesis studies using cAMP accumulation assays (Supplementary information, Fig. S4d–g and Table S4).
This structure also provides a template to study the selectivity of AVP and OT. OT has two amino acid substitutions (Phe3P to Ile3P and Arg8P to Leu8P) (Supplementary information, Fig. S5a), and exhibits a much lower selectivity against V2R as opposed to AVP.6 By docking OT into the V2R binding pocket, a weaker hydrophobic interaction network is created for Ile3P compared to its equivalent residue Phe3P in AVP (Supplementary information, Fig. S5b). Additionally, the hydrophobic leucine at position 8 no longer shows the polar interactions with the receptor as compared with Arg8P of AVP (Supplementary information, Fig. S5c). Thus, the peptide-binding pocket of V2R defines a relatively unfavorable binding environment for OT in contrast to AVP.
A “DCWA” sequence in ECL2 is highly conserved throughout VP/OT receptor subfamily but is not a feature of other class A GPCRs.7 Interestingly, structural comparison of the AVP–V2R–Gs complex with inactive OTR (PDB: 6TPK)8 reveals a notable conformational transition of ECL2 from β-hairpin to a flexible loop (Fig. 1d). In this conserved sequence, W193ECL2 and A194ECL2 directly contact with AVP. Asn5P may push and stack against W193ECL2 and overcome the probable steric clash between Asn5P and W188ECL2 of OTR, the cognate residue of V2R, which may confer this conformational transition (Fig. 1d). Although D191ECL2 and C192ECL2 do not directly interact with AVP, the former forms an H-bond to the F105ECL1 amide and a salt-bridge to R104ECL1 and the latter forms a conserved disulfide bond with C1123.25. These interactions, in turn, stabilize ECL1 and ECL2 conformations, thereby facilitating the interactions between ECL1 and the C-terminal amide of AVP (Fig. 1e; Supplementary information, Fig. S4f, g and Table S4).
A structural comparison of the AVP–V2R–Gs complex with the inactive OTR supports the contention that our structure is indeed in the active state. AVP triggers the rotameric change of “toggle switch” W2846.48, initiating the rotation and a notable outward displacement of TM6, the hallmark of GPCR activation (Fig. 1f; Supplementary information, Fig. S6a).9 Intriguingly, a distortion of TM7 helix was observed between L3127.40 and S3157.43. This distorted conformation appears to be stabilized by the H-bond between Tyr2P of AVP and the main chain CO group of L3127.40 in the receptor, drawing TM7 toward the peptide-binding pocket core (Fig. 1g). [Phe2]AVP, a synthesized AVP analog with a substitution of Tyr by Phe that abolishes this H-bond, showed decreased agonistic activity by 28-fold,10 implying that the polar interaction between Tyr2P and the receptor is critical to AVP-induced V2R activation. Specifically, F6.44 and I3.40, residues in the conserved “P-I-F” motif in class A GPCRs, are substituted by polar residues Y2806.44 and S1273.40 in V2R, respectively (Fig. 1h). AVP binding triggers the rotation of Y2806.44 relative to the inactive OTR (Fig. 1i), subsequently forming a featured H-bond network between Y2806.44/S1273.40 and the backbone CO group of V2135.46 in contrast to active β2AR (PDB: 3SN6),11 which probably stabilizes the active conformation of the receptor (Fig. 1j). The disease-associated mutations S1273.40F and Y2806.44C in V2R deactivate the receptor,12,13 while the replacement of F2846.44 in OTR by the V2R-equivalent tyrosine converts AVP from a partial to a full agonist,14 supporting the hypothesis that these unconventional “P-I-F” residues play critical roles in V2R activation. Furthermore, the cytoplasmic end of TM4 is one helical turn shorter than that of the inactive OTR,8 Gs-coupled β2AR11 and A2AR4 (PDB: 5G53), thereby releasing a longer ICL2. The ICL2 protrudes towards TM1 and adopts a C-shaped conformation (Supplementary information, Fig. S6b). This C-shaped loop is stabilized by H-bonds formed between H802.45 and the backbone CO group of H155ICL2, as well as W71ICL1 and backbone CO group of A154ICL2, and further by an intra-loop polar interaction network (Supplementary information, Fig. S6c–e), which may maintain ICL2 in a specific conformation and probably affect the ICL2–Gαs interaction pattern.
TM5 and TM6 with the outward movement of the cytoplasmic ends open a cytoplasmic cavity together with TM2, TM3, and helix 8 to accommodate the α5 helix of Gαs, constituting a major V2R–Gs interface (Supplementary information, Fig. S7a). One additional interface relates to ICL2 that interacts with α5 helix, αN-β1 junction, and the top of the β3-strand of Gαs, presumably stabilized by hydrophobic contacts (Supplementary information, Fig. S7b). In contrast to the Gs-coupled β2AR and A2AR structures, the cytoplasmic ends of TM5 and TM6 in V2R display a smaller amplitude of outward displacements (5.8 Å for TM5 and 8.7 Å for TM6 relative to β2AR,11 2.5 Å for TM5 and 7.2 Å for TM6 relative to A2AR,4 measured at Cα carbon of residues 5.67 and 6.30, respectively) (Fig. 1k, l). Also, a noticeable shift of Gs protein in the AVP–V2R–Gs complex may partly be caused by relatively inward positions of TM5 and TM6 that push the entire Gs heterotrimer shifts in the same direction. Besides, the α5 helix of Gαs in the AVP–V2R–Gs complex shifts half a helical turn away from the 7TM core (Fig. 1k). These structural differences altogether create a smaller cytoplasmic cavity to accommodate Gs protein. Indeed, the solvent-accessible surface area (SASA) of the V2R–Gαs interface (943.32 Å2) is smaller than that of β2AR–Gαs (1,030 Å2) and A2AR–Gαs interfaces (1,276 Å2) (Supplementary information, Fig. S7c). Interestingly, compared to Gs-coupled β2AR and A2AR, V2R exhibits a 1–2 helical turns shorter TM5 cytoplasmic end, which may be caused by a consecutive P-G-P sequence (P238, G239, and P240, residues known as α-helix breakers), resulting in fewer interactions between TM5 of V2R and Gαs subunit (Fig. 1l; Supplementary information, Fig. S8).
In conclusion, we solved the cryo-EM structure of Gs-coupled V2R bound to its endogenous ligand AVP. In this complex, AVP presents a unique cyclic conformation formed by an intramolecular disulfide bond and engages the orthosteric binding pocket of V2R in a ligand-specific mode. AVP-induced distortion of TM7 and a unique polar network formed by equivalent residues in the “P-I-F” motif may differentiate activation of V2R from other class A GPCRs. A smaller amplitude of the outward displacement of TM5 and TM6 and the concomitant shift of Gs protein not only distinguish V2R from its counterparts but also represent a diversified Gs coupling mechanism. Additionally, our structure is valuable to the mechanistic understanding of V2R mutation-associated diseases (Supplementary information, Fig. S9), while systematic investigations are required to enrich our knowledge in this regard. Together, our findings provide a framework for understanding AVP recognition and V2R activation, thereby offering a structural template for drug design targeting V2R.
Accession codes
The corresponding coordinates and cryo-EM density map have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 7DW9, and in EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code EMD-30877.
Supplementary information
Acknowledgements
We thank all staff members of the Cryo-EM Centre, Southern University of Science and Technology for their assistance in data collection. This work was partially supported by the Ministry of Science and Technology (China) grant (2018YFA0507002 to H.E.X.); National Natural Science Foundation of China (31770796 to Y.J., 81872915 to M,-W.W., 31600606 to X.Z., and 81773792 to D.Y.); National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (2018ZX09711002-002-002 to Y.J., 2018ZX09735-001 to M.-W.W., and 2018ZX09711002-002-005 to D.Y.); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to H.E.X.); CAS Strategic Priority Research Program (XDB37030103 to H.E.X.); Start-up funding by Fudan University (Q.Z.); Wellcome Trust 209407/Z/17/Z; National Key R&D Program of China (2016YFA0501100 to X.Z.); Guangdong Provincial Key Laboratory of Brain Connectome and Behavior (2017B030301017 to X.Z.); CAS Key Laboratory of Brain Connectome and Manipulation (2019DP173024 to X.Z.).
Author contributions
F.Z. prepared samples for cryo-EM grid preparation; F.Z. and X.M. performed cryo-EM data collection and processing; F.Z., T.C., X.M., and X.Z. built and refined the structure model; C.Y. and D.Y. conducted functional studies and data analysis; Q.Z. and X.H. carried out docking analysis; W.Y. engineered the mini-Gs protein; F.Z. and Y.J. prepared the bulk of figures, performed the structural analysis, and drafted the manuscript; Y.J., H.E.X., and M.-W.W. supervised the studies, analyzed the data, and wrote the manuscript with inputs from all co-authors; P.W. supervised the EM studies.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Fulai Zhou, Chenyu Ye, and Xiaomin Ma
Contributor Information
Peiyi Wang, Email: wangpy@sustech.edu.cn.
H. Eric Xu, Email: eric.xu@simm.ac.cn.
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
Yi Jiang, Email: yijiang@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00480-2.
References
- 1.Barberis C, Mouillac B, Durroux T. J. Endocrinol. 1998;156:223–229. doi: 10.1677/joe.0.1560223. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson ZR, Young LJ. Science. 2008;322:900–904. doi: 10.1126/science.1158668. [DOI] [PubMed] [Google Scholar]
- 3.Makita N, Manaka K, Sato J, Iiri T. Vitam. Horm. 2020;113:79–99. doi: 10.1016/bs.vh.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter B, Nehme R, Warne T, Leslie AG, Tate CG. Nature. 2016;538:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballesteros JA, Weinstein H. Methods Neurosci. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 6.Tahara A, et al. Br. J. Pharmacol. 1998;125:1463–1470. doi: 10.1038/sj.bjp.0702220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conner M, et al. J. Biol. Chem. 2007;282:17405–17412. doi: 10.1074/jbc.M702151200. [DOI] [PubMed] [Google Scholar]
- 8.Waltenspuhl Y, Schoppe J, Ehrenmann J, Kummer L, Pluckthun A. Sci. Adv. 2020;6:eabb5419. doi: 10.1126/sciadv.abb5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q, et al. Elife. 2019;8:e50279. doi: 10.7554/eLife.50279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisniewski K, et al. J. Med. Chem. 2011;54:4388–4398. doi: 10.1021/jm200278m. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen SG, et al. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdélyi L, Szalai L, Sziráki A, Balla A, Hunyady L. Endocrine Abstracts. 2017;49:EP804. [Google Scholar]
- 13.Wenkert D, et al. Mol. Cell Endocrinol. 1996;124:43–50. doi: 10.1016/S0303-7207(96)03926-3. [DOI] [PubMed] [Google Scholar]
- 14.Chini B, et al. FEBS Lett. 1996;397:201–206. doi: 10.1016/S0014-5793(96)01135-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.