Abstract
Paenibacillus species are Gram-positive bacteria that have been isolated from a diverse array of plant species and soils, with some species exhibiting plant growth-promoting (PGP) activities. Here we report two strains (S02 and S25) of a novel Paenibacillus sp. that were isolated from perennial ryegrass (Lolium perenne) seeds. Comparative genomics analyses showed this novel species was closely related to P. polymyxa. Genomic analyses revealed that strains S02 and S25 possess PGP genes associated with biological nitrogen fixation, phosphate solubilisation and assimilation, as well as auxin production and transportation. Moreover, secondary metabolite gene cluster analyses identified 13 clusters that are shared by both strains and three clusters unique to S25. In vitro assays demonstrated strong bioprotection activity against phytopathogens (Colletotrichum graminicola and Fusarium verticillioides), particularly for strain S02. A transcriptomics analysis evaluating nitrogen fixation activity showed both strains carry an expressed nif operon, but strain S02 was more active than strain S25 in nitrogen-free media. Another transcriptomics analysis evaluating the interaction of strains with F. verticillioides showed strain S02 had increased expression of core genes of secondary metabolite clusters (fusaricidin, paenilan, tridecaptin and polymyxin) when F. verticillioides was present and absent, compared to S25. Such bioactivities make strain S02 a promising candidate to be developed as a combined biofertiliser/bioprotectant.
Subject terms: Bacterial genomics, Bacterial transcription
Introduction
Microorganisms associated with plants (collectively the plant microbiome) are one of the key factors that determine plant health and productivity1. Plant growth-promoting (PGP) bacteria competitively colonise plant tissues and build beneficial interactions with plant hosts by acting as biofertilisers or bioprotectants2, such as commercial products based on Pseudomonas spp. and Rhizobium spp. that have been utilised globally of the past 30–40 years3. Compared with other bacteria, the endospore-forming Bacillus spp. and Paenibacillus spp. could survive longer in soils with varying conditions such as pH, temperature and salinity, representing promising candidates for agricultural usage as biologicals4.
Paenibacillus spp. are Gram-positive, facultative anaerobic bacteria that are commonly found in soil from diverse geographic environments4,5. As the type species of the genus Paenibacillus6, Paenibacillus polymyxa inhabits the rhizosphere or root tissues of a wide range of agricultural crops like wheat and barley7,8, agricultural pastures like perennial ryegrass9 and forest trees like pine and cedar10. P. polymyxa has been reported to promote the growth of many plants, with significant improvements in nutrient uptake and increases in total biomass and/or seeding height11–15. Genomic analyses of P. polymyxa strains have revealed that they are capable of acting as both biofertilisers and bioprotectants5,16–19. As biofertilisers, P. polymyxa possesses genes involved in biological nitrogen (N) fixation, phosphate solubilisation and assimilation, iron assimilation and auxin production. As bioprotectants, P. polymyxa carries gene clusters that synthesise bioactive compounds including polymyxin and fusaricidin, as well as novel clusters of unknown functions. Furthermore, P. polymyxa also plays an active role in the biotechnology sector due to its ability to produce cell wall degrading enzymes and exopolysaccharides20. Such diverse properties have made P. polymyxa a prominent commercially useful PGP bacterium for sustainable agriculture21,22.
The microbiome of perennial ryegrass (Lolium perenne L. cv. Alto) has been profiled recently by our laboratory, which suggested the presence of Paenibacillus spp. within the community23. This study aimed to confirm the presence of Paenibacillus spp. through genomic sequencing of the seed-associated bacterial community, and to isolate strains using selective media (antibiotics and nitrogen-free media). The biological nitrogen fixation ability of the community was assessed with nifH PCR. The genome of the isolated strains was assembled, which was in turn used to confirm the taxonomy and confirm the presence of PGP genes and secondary metabolite gene clusters. Further assays were conducted to determine the in vitro bioprotection activities against phytopathogens and to analyse the changes in transcriptome profiles associated with biological nitrogen fixation and early stage bacteria-pathogen interactions of the isolated strains.
Results
Seed-associated N-fixing bacterial strain detection and isolation
The presence of bacteria containing the nifH gene was confirmed from the seed of perennial ryegrass, as amplicons of the expected size (~ 400 bp) were produced by the nifH gene PCR. Amplicons were generated using DNA extracted from a suspension of ground perennial ryegrass seeds that was serially diluted (100–10–3), including from two of eight replicates of the 10–2 dilution (Supplementary Figure S1), while no amplicon was produced from all eight replicates of the 10–3 dilution. Amplicons were sequenced and identified as partial sequences of the nifH gene of P. polymyxa CR1 (Accession ID: CP006941.2, 1,087,670–1,088,026 bp; coverage = 97%, identity = 99%) using BLASTn search against the nt database.
Long read sequence data was also generated from the DNA of the seed suspension, and then classified by Kraken224. The reads had sequence homology to multiple bacterial species including Bacillus spp. (high read abundance), Pseudomonas spp., Massilia spp. and Paenibacillus spp. (low read abundance). Despite the low Paenibacillus spp. read abundance, a single 110 Kb read containing the entire P. polymyxa nif operon (nine genes) was identified (Supplementary Figure S2), which confirmed the presence of P. polymyxa in the two dilutions.
The confirmation of a P. polymyxa-like bacterial strain in the seed suspension provided guidance regarding its isolation and purification. P. polymyxa produces the antibiotic polymyxin, which is biocidal against Gram-negative bacteria25. Supplementing media with polymyxin B aided in the isolation of the low abundant Paenibacillus spp. strains from the dominant bacteria. Two bacterial strains (S02 and S25) were isolated using Burk’s N-free medium supplemented with polymyxin B. Both strains are rod-shaped and Gram-positive under microscopic examination and form heaped, small- to medium-sized colonies on agar plates. Strain S02 produces white and mucoid colonies, whilst strain S25 produces translucent colonies. Both strains were stored in 15% glycerol at − 80 ℃.
Genome sequencing, assembly and annotation
A total of 2,536,823,196 bp short reads and 13,203,686,400 bp long reads were generated for Paenibacillus sp. strains S02 and S25 (Supplementary Table S1). Complete circular genome sequences were produced for both strains. The genome size of Paenibacillus sp. S02 was 6,060,529 bp (5,310 CDSs), with a G + C content of 45.60%, while the genome size of Paenibacillus sp. S25 was 5,958,851 bp (5,177 CDSs), with a G + C content of 45.72% (Supplementary Table S2). There were no plasmids present in either strain.
Phylogeny and comparative genomics
The results of the nifH gene PCR and the mixed culture read analysis described in “Seed-associated N-fixing bacterial strain detection and isolation” section suggested that the isolated strains were closely related to the species P. polymyxa. The 16S ribosomal RNA genes showed both strains were phylogenetically related to P. polymyxa DSM36 (GenBank Accession: NR_117732.2) with a sequence homology of 99.45% and coverage of 100%. The close relationship between the two strains and P. polymyxa was further supported by genome-based identifications where both strains were classified by Kraken2 as P. polymyxa E681 (NCBI:txid 349520).
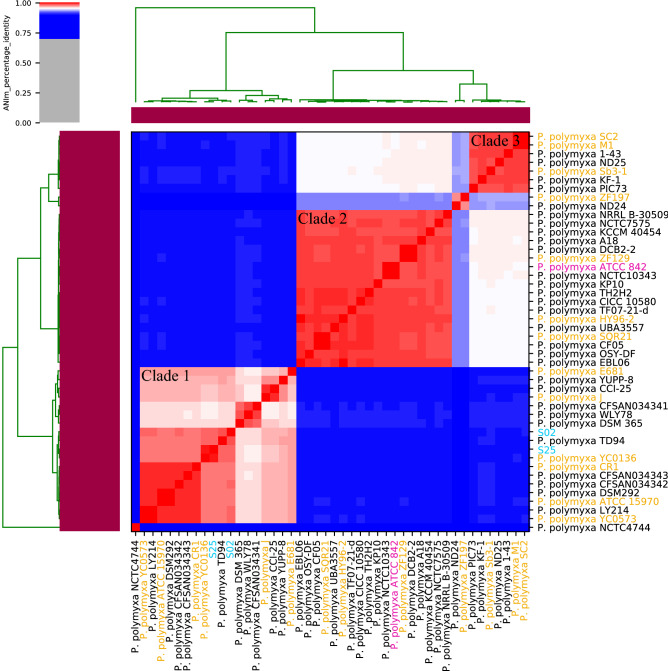
The average nucleotide identity (ANI) was compared between the genomes of the two isolated strains (S02 and S25) and 44 known P. polymyxa strains (Supplementary Table S3). The ANI dendrogram-heatmap revealed three major (Clades 1–3) and two minor clades (Fig. 1). The ANI between strains within the same clade was at least 95%. Among the three major clades, strains from Clade 2 and 3 had ANI < 95%, while Clade 1 was further separated from the other two clades (ANI < 91%). Paenibacillus sp. strains S02 and S25 were in Clade 1 and had an ANI of 97.78% to one another. Strain S02 was most similar to P. polymyxa TD94 (ANI = 98.11%) which was isolated from Scutellaria spp. rhizosphere26, while strain S25 was most similar to P. polymyxa YC0136 (ANI = 99.29%) which was isolated from tobacco rhizosphere19. Both strains were clustered with 16 known P. polymyxa strains isolated from various geographic regions including Asia, North America and Europe. The majority of the Clade 1 strains (14) were isolated from plant rhizosphere or soil, with the exceptions being P. polymyxa CCI-25 isolated from vermicompost18, and P. polymyxa J isolated from the phloem of a chilli plant. Clades 2 and 3 contained 18 and seven P. polymyxa strains respectively, and these strains were mainly isolated from plant rhizosphere or soil in Asia, Europe and North America. The type strain of P. polymyxa (ATCC 842) was placed in Clade 2. P. polymyxa ZF197 and ND24 formed a minor clade that had an ANI of 92.45–92.61% and 92.93–93.09% when compared to Clade 2 and 3, respectively. There was also another minor clade containing a single strain P. polymyxa NCTC4744 isolated from the UK, which had low ANI values (< 89%) when compared to the other strains.
Figure 1.
Phylogeny of Paenibacillus species based on a heatmap with row and column dendrograms from the average nucleotide identity (ANI) of genomes of Paenibacillus sp. strains S02 and S25 and 44 P. polymyxa strains from NCBI. Clustering across the dendrograms were based on overall genomic sequence similarity, forming three major clades and two minor clades (intra-cluster ANI > 95%). Clade 1 contained Paenibacillus sp. strains S02 and S25 as well as 16 known P. polymyxa strains. Clade 2 and 3 contained 18 and seven known P. polymyxa strains, respectively. P. polymyxa ZF197 and ND24 formed a minor clade, and P. polymyxa NCTC4744 formed another minor clade. Blue label: Paenibacillus sp. strains isolated in this study. Yellow label: P. polymyxa strains with complete circular genome sequences. Purple label: The type strain of P. polymyxa.
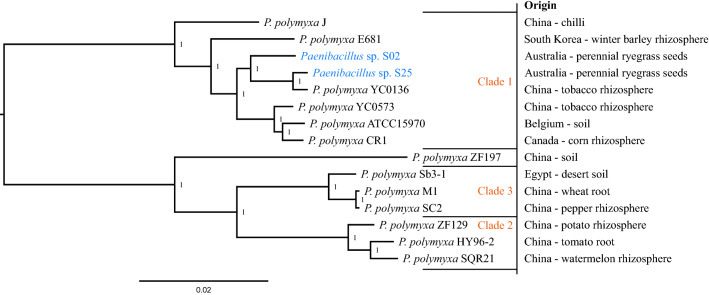
A pan genome Roary27 analysis was conducted comparing the sequence similarity of genes shared by Paenibacillus sp. strains S02 and S25, along with 13 other P. polymyxa strains with complete genomes available on NCBI. The analysis identified 2059 shared genes by all 15 strains. A maximum-likelihood phylogenetic tree was inferred based on the sequence homology of the shared genes (Fig. 2). The topology of the tree consisted of three major clades and was consistent with the ANI analysis (Fig. 1, Clade 1–3). All clades were separated with a strong local support value (100%). Clade 1 consisted of eight strains, including the two Paenibacillus sp. strains (S02 and S25), and were separated from Clades 2 and 3 at the root node. Clades 2 and 3 formed adjoining clades on the same primary root node, and each had three strains. Strain ZF197 also clustered with Clade 2 and 3 but formed its own branch. Clade 1 consisted of strains from across a broad geographic range, including Asia (China, South Korea), the Pacific (Australia), North America (Canada) and Europe (Belgium), whereas Clades 2 and 3 were largely from Asia (China), except strain Sb3-1 in Clade 3 that was isolated from Egypt. All strains were either associated with plants or soil.
Figure 2.
Phylogeny of Paenibacillus species based on a pan genome Roary analysis of strains S02 and S25 and 13 P. polymyxa strains with complete circular genome sequences. The maximum-likelihood tree was inferred based on 2059 genes conserved among 15 genomes. Values shown next to branches were the local support values calculated using 1,000 resamples with the Shimodaira-Hasegawa test. Paenibacillus sp. strains S02 and S25 clustered with other six P. polymyxa strains in Clade 1. Clades 2 and 3 separated from Clade 1 at the root node, and consisted of three P. polymyxa strains each.
Plant growth-promoting genes
The genomes of both Paenibacillus sp. strains (S02 and S25) were assessed for the presence of 30 plant growth-promoting (PGP) genes and were found to possess a comprehensive set of genes (Supplementary Table S4). A 10.54 Kb region containing a nif operon of nine genes (nifB/H/D/K/E/N/X, hesA/moeB and nifV) was identified, including all the genes necessary for encoding the Mo-nitrogenase (molybdenum-dependent nitrogenase) that catalyses biological nitrogen fixation28. In addition, 16 genes associated with phosphate solubilisation and assimilation were identified, including the glucose-1-dehydrogenase (gcd) gene for inorganic phosphate solubilisation29, the phn cluster of nine genes for organic phosphate (phosphonates) solubilisation2 and the phosphate-specific transport system of six genes for phosphate assimilation30. However, the gluconic acid dehydrogenase (gad) gene for inorganic phosphate solubilisation29 was not found in either of the two strains. Additionally, genes involved in indole-3-acetic acid (IAA) production and transportation were identified, including the ipdC gene that encodes a key enzyme in the IAA biosynthetic pathway31, as well as three auxin efflux carrier genes. Sequence comparison of the PGP genes between the two strains (S02 and S25) showed sequence similarity of 95.39–99.54%, while the two strains showed sequence similarity of 94.69–99.78% when compared to P. polymyxa strain CR1 (Supplementary Table S5).
Secondary metabolite genes
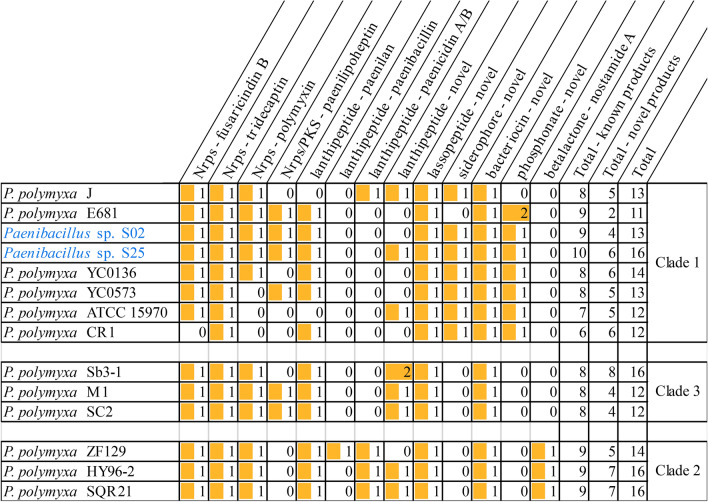
Genes associated with secondary metabolite production were identified using antiSMASH32. The analyses identified 16 clusters (designated C1–C16) consisting of 13 clusters that were shared by both strains and three clusters that only strain S25 possessed (Supplementary Table S6). All clusters contained all the genes (core/additional biosynthetic genes, regulatory genes, transport-related genes and other genes) required for complete function.
Secondary metabolite gene clusters that were shared by both strains consisted of four that were identical to known clusters, including three nonribosomal peptide synthetase (Nrps) clusters (C1, fusaricidin B; C10, tridecaptin; C15, polymyxin) and one lanthipeptide cluster (C7, paenilan). The products of all four clusters have been reported to have antimicrobial bioactivities25,33–35. A further four clusters had partial sequence homology to known clusters included a lassopeptide cluster (C5), a Nrps cluster (C6), a Nrps/transAT-polyketide synthase (PKS) cluster (C11) and a Nrps/Type III (T3) PKS/transAT-PKS cluster (C14), which had homology to paeninodin, marthiapeptide A, paenilipoheptin and aurantinin B/C/D, respectively. Among these four clusters, cluster C11 had the highest similarity (S02, 73%; S25, 76%) to a known cluster of P. polymyxa E681 that produces paenilipoheptin36. There were also five clusters that appear novel based on sequence homology, including a siderophore cluster (C2), a bacteriocin cluster (C3), a Nrps/transAT-PKS cluster (C4), a Nrps-like cluster (C9) and a phosphonate cluster (C16).
Paenibacillus sp. S25 had three unique secondary metabolite gene clusters that were missing in the genome of Paenibacillus sp. S02, including a lanthipeptide cluster (C8) and two novel Nrps clusters (C12, C13). While the two Nrps clusters appear novel based on sequence homology, the lanthipeptide cluster had a similarity of 71% to a known paenicidin B cluster, which was a novel lantibiotic peptide active against Gram-positive bacteria produced by Paenibacillus terrae35.
Secondary metabolite gene cluster analyses were also conducted using the 13 P. polymyxa strains with complete circular genome sequences (Fig. 1, yellow labels), and their presence was compared to Paenibacillus sp. strains S02 and S25. The total number of secondary metabolite gene clusters of each strain varied between 11 to 16 (Table 1). Clade-specific clusters were identified, including one associated with a novel siderophore and another associated with a novel phosphonate being exclusive to Clade 1 strains, and a nostamide A (betalactone) cluster being exclusive to Clade 2 strains. Moreover, the paenilipoheptin cluster was only identified in some Clade 1 and 3 strains but no Clade 2 strain. More than half of the identified clusters were Nrps or PKS, including a tridecaptin cluster that was shared by all strains regardless of the phylogenetic clades. The fusaricidin B cluster was identified in all strains except P. polymyxa CR1, and the polymyxin cluster was identified in all strains except P. polymyxa strains YC0573, ATCC 15,970 and CR1. Nrps/PKS clusters producing novel products were also identified in all strains. Lanthipeptide clusters producing both novel and known products were also widely distributed in all strains. Specifically, the paenilan cluster was identified in all strains except P. polymyxa strains J and ATCC 15,970 (Clade 1), however the paenibacillin cluster and the paenicidin A/B cluster were less common. The paenibacillin cluster was only identified in P. polymyxa ZF129 (Clade 2), and the paenicidin A/B cluster was only identified in all Clade 2 strains and one Clade 1 strain (P. polymyxa J). Other widely distributed secondary metabolite gene clusters included one novel lassopeptide cluster and one novel bacteriocin cluster.
Table 1.
Secondary metabolite gene clusters identified in Paenibacillus sp. strains S02 and S25 and 13 P. polymyxa strains with complete circular genome sequences.
The type and product of each secondary metabolite gene cluster were shown in the first row. Numbers are the total count of each secondary metabolite gene cluster. The length of the orange bar represents the total count of each secondary metabolite gene cluster.
Novel: Similarity ≤ 70% when compared to the most similar known cluster in the antiSMASH database.
Nrps: Nonribosomal peptide synthetase.
PKS: Polyketide synthase.
Clade 1/2/3: Clades identified in phylogeny and comparative genomics study (“Phylogeny and comparative genomics” section).
Bioprotection assay (in vitro)
A dual culture in vitro assay was established to compare the biocidal activity of Paenibacillus sp. strains S02 and S25 against three fungal pathogens, Colletotrichum graminicola, Fusarium verticillioides and Microdochium nivale. Paenibacillus sp. S02 significantly (P < 0.05) reduced the average colony diameter of the fungal pathogens C. graminicola and F. verticillioides compared to the blank control and Paenibacillus sp. S25 (Supplementary Table S7, Supplementary Figure S3). It reduced the growth of C. graminicola and F. verticillioides by up to 74.9% and 56.9%, respectively. Paenibacillus sp. S25 significantly (P < 0.05) reduced the growth of F. verticillioides by 9.6% compared to the blank control, however no similar activity was observed for C. graminicola. Neither of the two strains could significantly reduce the average colony diameter of M. nivale.
Transcriptome sequencing and analysis—N-fixation activity assay
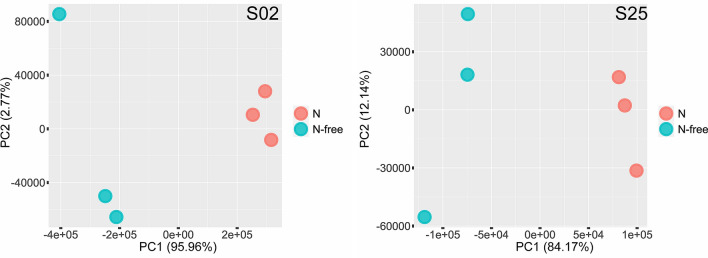
A transcriptome sequencing experiment was designed to confirm the expression of the nif operon of Paenibacillus sp. strains S02 and S25 when the nitrogen source (NH4Cl) was removed from the media. A 150 bp PE library prepared from cDNA from strains used in the N-fixation activity assay generated an average of 13.8 million clean reads per sample. Differential gene expression (DGE) analysis successfully identified genes that were differentially expressed under different conditions in the assay. A total of 5,059 and 4,745 genes passed the abundance filter for strain S02 and S25, respectively, and were used in the subsequent DGE analysis. Among those genes, 2,467 and 2,479 genes were differentially expressed when nitrogen was removed from the media for strain S02 and S25, respectively. Biological replicates of the nitrogen treatment (+ / −) formed distinctive clusters along the PC1 axis for both strains, suggesting the presence/absence of nitrogen impacted the transcriptome profiles of both strains (Fig. 3). Specifically, when nitrogen was present in the media, the nif operon comprising nine genes was expressed by both strains, and there was no significant difference in expression levels of any nif gene between the two strains. However, the expression levels of the nif operon varied when nitrogen was removed from the media (Table 2). Gene expression levels when nitrogen was removed were represented as fold-changes in relation to the expression levels when nitrogen was present. The expressions of all nine genes of the nif operon of Paenibacillus sp. S02 were upregulated by 8.62–22.50 folds. For Paenibacillus sp. S25, the nifB/H/D/K/E genes were differentially expressed with 1.76- to 3.90-fold increase. The remaining four genes of the nif operon also had fold-changes in expression level, however they failed to be considered as differentially expressed (q-value < 0.05 and absolute fold-change ≥ 1.5). Such results confirmed that both strains likely carry a biologically functional nif operon that enables biological nitrogen fixation.
Figure 3.
PCA plots of transcriptome profiles of Paenibacillus sp. strains S02 (left) and S25 (right) when grown in media with nitrogen (N) and without nitrogen (N-free) in the N-fixation activity assay. Percentage variance explained by each axis are given in brackets. Distinctive clusters indicate that the presence/absence of a nitrogen source (NH4Cl) in the media changed the transcriptome profiles of both strains.
Table 2.
Changes in expression levels of the nif operon of Paenibacillus sp. strains S02 and S25 when NH4Cl was removed from the media.
| nif genes | S02 | S25 |
|---|---|---|
| Fold-change | Fold-change | |
| nifB | 22.50* | 2.46* |
| nifH | 20.21* | 3.90* |
| nifD | 15.80* | 2.06* |
| nifK | 17.51* | 2.01* |
| nifE | 15.86* | 1.76* |
| nifN | 18.16* | 1.59 |
| nifX | 8.62* | 1.19 |
| hesA/moeB | 15.13* | 1.56 |
| nifV | 11.01* | − 1.46 |
*Genes that were differentially expressed (q-value < 0.05 and absolute fold-change ≥ 1.5) when nitrogen was removed from the media.
Transcriptome sequencing and analysis—Bacteria-pathogen interactions assay
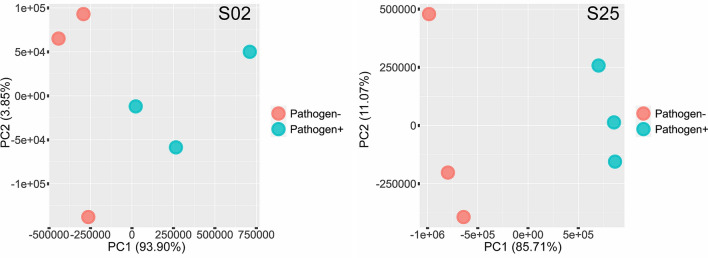
A transcriptome sequencing experiment was designed to explore the early stage interactions between Paenibacillus sp. strains S02 and S25 and the fungal pathogen F. verticillioides. A 150 bp PE library prepared from cDNA from strains used in the bacteria-pathogen interactions assay generated an average of 31.1 million clean reads per sample. DGE analysis successfully identified genes that were differentially expressed under different conditions in the assay. A total of 5,201 and 4,817 genes passed the abundance filter for strain S02 and S25, respectively, and were used in the subsequent DGE analysis. Among those genes, only 61 genes were differentially expressed by strain S02 when F. verticillioides was present. In contrast, 2,706 genes were differentially expressed by strain S25 when F. verticillioides was present. Moreover, distinctive clustering of the three biological replicates based on transcriptome profiles was identified for strain S25 along the PC1 axis, but the clustering was not as evident for strain S02, particularly when the pathogen was present (Fig. 4). The Clusters of Orthologous Groups (COGs) of proteins37 encoded by the genes mentioned above rendered an overview of functions of those genes. For Paenibacillus sp. S02, 29 genes were associated with cellular processes and signalling, and 14 genes were associated with metabolism. There were also 18 genes with unknown functions. For Paenibacillus sp. S25, 368 genes were associated with cellular processes and signalling, and 889 genes were associated with metabolism. In addition, there were 416 genes associated with information storage and processing, 43 genes associated with multiple function groups and 990 genes with unknown functions. Given the complexity of transcriptome profiles, this study focused on the expression levels of the core biosynthetic genes of secondary metabolite gene clusters identified in “Secondary metabolite genes” section to demonstrate the early stage bacteria-pathogen interactions.
Figure 4.
PCA plots of transcriptome profiles of Paenibacillus sp. strains S02 (left) and S25 (right) when grown in media with F. verticillioides (Pathogen+) and without F. verticillioides (Pathogen−) in the bacteria-pathogen interactions assay. Percentage variance explained by each axis are given in brackets. Distinctive clusters indicate that the presence/absence of F. verticillioides in the media changed the transcriptome profiles of strain S25 (along the PC1 axis).
When F. verticillioides was absent, Paenibacillus sp. S02 showed increased expressions of the core biosynthetic genes of secondary metabolite gene clusters compared to Paenibacillus sp. S25. Gene expression levels of those genes of strain S02 were represented as fold-changes in relation to the expression levels of the corresponding homolog of each gene of strain S25 (Table 3). Among the 44 core biosynthetic genes of secondary metabolite gene clusters shared by the two strains, the majority (41) were expressed in increased levels ranging from 1.92- to 486.58-fold increase, including the four clusters that produce known antimicrobial compounds (C1, C7, C10 and C15). Only one cluster had a decreased expression, with C9 exhibiting a 7.42-fold decrease in the expression level. Two core biosynthetic genes of cluster C4 were not differentially expressed when comparing the two strains.
Table 3.
Fold-changes in expression levels of the core biosynthetic genes of secondary metabolite gene clusters of strain S02 compared to strain S25 when F. verticillioides was absent.
| ID | Type | Most similar known cluster (similarity) | Paenibacillus sp. S02 | |
|---|---|---|---|---|
| Gene ID | Fold-change | |||
| C1 | Nrps | Fusaricidin B (100%) | KAI36_00078 | 486.58* |
| KAI36_00083 | 20.36* | |||
| C2 | Siderophore | – | KAI36_00955 | 11.71* |
| KAI36_00956 | 13.93* | |||
| KAI36_00959 | 4.64* | |||
| C3 | Bacteriocin | – | KAI36_01103 | 46.95* |
| C4 | Nrps transAT-PKS | – | KAI36_01166 | 18.88* |
| KAI36_01170 | 2.43* | |||
| KAI36_01172 | 1.93* | |||
| KAI36_01173 | 3.31* | |||
| KAI36_01175 | 2.35* | |||
| KAI36_01176 | 2.30* | |||
| KAI36_01178 | 2.27* | |||
| KAI36_01179 | 1.92* | |||
| KAI36_01180 | – 1.25 | |||
| KAI36_01181 | 1.27 | |||
| C5 | Lassopeptide | Paeninodin (40%) | KAI36_01236 | 134.38* |
| KAI36_01240 | 37.82* | |||
| C6 | Nrps | Marthiapeptide A (33%) | KAI36_01339 | 239.45* |
| KAI36_01340 | 159.93* | |||
| KAI36_01341 | 119.29* | |||
| C7 | Lanthipeptide | Paenilan (100%) | KAI36_01558 | 60.94* |
| KAI36_01560 | 271.12* | |||
| KAI36_01562 | 69.08* | |||
| C9 | Nrps-like | – | KAI36_01944 | – 7.42* |
| C10 | Nrps | Tridecaptin (100%) | KAI36_02333 | 7.74* |
| KAI36_02334 | 32.36* | |||
| C11 | Nrps transAT-PKS | Paenilipoheptin (S02, 73%; S25, 76%) | KAI36_02506 | 17.20* |
| KAI36_02507 | 29.10* | |||
| KAI36_02508 | 23.54* | |||
| KAI36_02509 | 22.65* | |||
| KAI36_02510 | 10.63* | |||
| C14 | Nrps T3PKS transAT-PKS | Aurantinin B/C/D (35%) | KAI36_03362 | 118.38* |
| KAI36_03363 | 46.83* | |||
| KAI36_03365 | 125.65* | |||
| KAI36_03366 | 98.69* | |||
| KAI36_03367 | 31.92* | |||
| KAI36_03368 | 77.19* | |||
| KAI36_03371 | 134.46* | |||
| KAI36_03372 | 113.90* | |||
| C15 | Nrps | Polymyxin (100%) | KAI36_04684 | 9.61* |
| KAI36_04687 | 8.61* | |||
| KAI36_04688 | 9.86* | |||
| C16 | Phosphonate | – | KAI36_05277 | 15.10* |
Clusters in bold: Known antimicrobial compounds.
*Genes that were differentially expressed (q-value < 0.05 and absolute fold-change ≥ 1.5) when comparing the two strains.
The presence of F. verticillioides had different effects in expressions of the core biosynthetic genes of secondary metabolite gene clusters on the two strains. Gene expression levels when F. verticillioides was present were represented as fold-changes in relation to the expression levels when F. verticillioides was absent (Table 4). For Paenibacillus sp. S02, only three of 44 core biosynthetic genes of secondary metabolites were differentially expressed, while for Paenibacillus sp. S25, 32 of 51 genes were differentially expressed. Despite clusters C1, C7, C10 and C15, which produce known antimicrobial compounds, being shared by both strains, the expression levels of core biosynthetic genes of secondary metabolite varied. For instance, the four genes of the polymyxin cluster (C15) were not differentially expressed by strain S02 but were differently expressed with 1.45- to 2.61-fold decrease by strain S25. Strain S25 had a gene of the fusaricidin B cluster (C1) that was differentially expressed with a 5.21-fold decrease, and two genes of the tridecaptin cluster (C10) that were differentially expressed (one with a 1.92-fold decrease and the other with a 1.79-fold increase), whereas these gene clusters were not affected in strain S02. The three core biosynthetic genes of the paenilan cluster (C7) were not differentially expressed in either strain. As for the four clusters that had the best matches in the antiSMASH gene cluster database (C5, C6, C11 and C14), although the core genes had differing levels of fold-change, the trend of changes (i.e. upregulated or downregulated) were consistent between strains. Similar consistencies were also observed from the core biosynthetic genes of three shared novel clusters (C4, C9 and C16). Furthermore, the core gene of the bacteriocin cluster (C3) was not differentially expressed in either strain. However, two genes of the siderophore cluster (C2) of strain S25 were differentially expressed with a 2.06- and 2.60-fold increase, respectively, unlike strain S02. Moreover, amongst the three novel clusters (C8, C12 and C13) that were unique to strain S25, two (C12 and C13) had differentially expressed core genes.
Table 4.
Fold-changes in expression levels of the core biosynthetic genes of secondary metabolite gene clusters identify in Paenibacillus sp. strains S02 and S25 when F. verticillioides was present.
| ID | Type | Most similar known cluster (similarity) | Paenibacillus sp. S02 | Paenibacillus sp. S25 | ||
|---|---|---|---|---|---|---|
| Gene ID | Fold-change | Gene ID | Fold-change | |||
| C1 | Nrps | Fusaricidin B (100%) | KAI36_00078 | − 1.28 | KAI37_00078 | − 1.50 |
| KAI36_00083 | − 1.36 | KAI37_00083 | − 5.21* | |||
| C2 | Siderophore | – | KAI36_00955 | − 1.23 | KAI37_00927 | 2.06* |
| KAI36_00956 | − 1.15 | KAI37_00928 | 2.60* | |||
| KAI36_00959 | − 1.12 | KAI37_00931 | 1.04 | |||
| C3 | Bacteriocin | – | KAI36_01103 | 1.10 | KAI37_01049 | − 1.19 |
| C4 |
Nrps transAT− PKS |
– | KAI36_01166 | − 1.25 | KAI37_01130 | − 1.17 |
| KAI36_01170 | − 1.89* | KAI37_01134 | − 1.08 | |||
| KAI36_01172 | − 1.54 | KAI37_01136 | − 1.43 | |||
| KAI36_01173 | − 1.65* | KAI37_01137 | − 1.49 | |||
| KAI36_01175 | − 1.64 | KAI37_01139 | − 1.52 | |||
| KAI36_01176 | − 1.48 | KAI37_01140 | − 1.56 | |||
| KAI36_01178 | − 1.41 | KAI37_01142 | − 1.51* | |||
| KAI36_01179 | − 1.40 | KAI37_01143 | − 1.55 | |||
| KAI36_01180 | − 1.43 | KAI37_01144 | − 2.34* | |||
| KAI36_01181 | − 1.33 | KAI37_01145 | − 1.90* | |||
| C5 | Lassopeptide | Paeninodin (40%) | KAI36_01236 | − 1.20 | KAI37_01200 | − 3.63* |
| KAI36_01240 | − 1.30 | KAI37_01204 | − 2.07 | |||
| C6 | Nrps | Marthiapeptide A (33%) | KAI36_01339 | 1.19 | KAI37_01293 | 4.34* |
| KAI36_01340 | 1.15 | KAI37_01294 | 2.43* | |||
| KAI36_01341 | 1.12 | KAI37_01295 | 2.02* | |||
| C7 | Lanthipeptide | Paenilan (100%) | KAI36_01558 | − 1.04 | KAI37_01518 | 1.32 |
| KAI36_01560 | − 1.00 | KAI37_01520 | 1.10 | |||
| KAI36_01562 | 1.02 | KAI37_01522 | 1.18 | |||
| C8 | Lanthipeptide | Paenicidin B (71%) | KAI37_01661 | − 1.73 | ||
| KAI37_01663 | − 1.19 | |||||
| C9 | Nrps− like | – | KAI36_01944 | 1.81* | KAI37_01854 | 2.07* |
| C10 | Nrps | Tridecaptin (100%) | KAI36_02333 | 1.02 | KAI37_02322 | − 1.92* |
| KAI36_02334 | 1.00 | KAI37_02323 | 1.79* | |||
| C11 |
Nrps transAT− PKS |
Paenilipoheptin (S02, 73%; S25, 76%) |
KAI36_02506 | 1.14 | KAI37_02476 | 15.43* |
| KAI36_02507 | 1.16 | KAI37_02477 | 23.42* | |||
| KAI36_02508 | 1.16 | KAI37_02478 | 11.77* | |||
| KAI36_02509 | 1.21 | KAI37_02479 | 7.92* | |||
| KAI36_02510 | 1.11 | KAI37_02480 | 3.23* | |||
| C12 | Nrps | – | KAI37_02516 | -1.91* | ||
| C13 |
Nrps betalactone |
– | KAI37_02623 | 1.50* | ||
| KAI37_02624 | 2.02* | |||||
| KAI37_02633 | 2.10* | |||||
| C14 |
Nrps T3PKS transAT-PKS |
Aurantinin B/C/D (35%) | KAI36_03362 | − 1.29 | KAI37_03372 | 1.71 |
| KAI36_03363 | − 1.35 | KAI37_03373 | − 1.78 | |||
| KAI36_03365 | − 1.32 | KAI37_03375 | − 1.66* | |||
| KAI36_03366 | − 1.34 | KAI37_03376 | − 1.82* | |||
| KAI36_03367 | − 1.26 | KAI37_03377 | − 2.00* | |||
| KAI36_03368 | − 1.05 | KAI37_03378 | − 1.78* | |||
| KAI36_03371 | − 1.05 | KAI37_03381 | N/A | |||
| KAI36_03372 | − 1.03 | KAI37_03382 | − 4.68* | |||
| C15 | Nrps | Polymyxin (100%) | KAI36_04684 | − 1.02 | KAI37_04566 | − 2.61* |
| KAI37_04567 | − 2.33* | |||||
| KAI36_04687 | 1.10 | KAI37_04570 | − 1.45* | |||
| KAI36_04688 | 1.12 | KAI37_04571 | − 1.92* | |||
| C16 | Phosphonate | – | KAI36_05277 | − 1.38 | KAI37_05154 | − 1.77* |
Clusters in bold: Known antimicrobial compounds.
N/A: Genes that didn’t pass the abundance filter described in “Transcriptome analysis” section.
*: Genes that were differentially expressed (q-value < 0.05 and absolute fold-change ≥ 1.5) when F. verticillioides was present.
Fungal transcripts of F. verticillioides were also assessed as a part of the bacteria-pathogen interactions assay. Transcript quantification using the transcriptome sequences of F. verticillioides 7600 as the reference reflected the differences in bioprotection activities between the two strains (Supplementary Table S8). The percentage of mapped reads in the treated samples for Paenibacillus sp. S02, which showed stronger bioprotection activities in the in vitro assay (“Bioprotection assay (in vitro)” section), was much lower than that for Paenibacillus sp. S25. Furthermore, amongst the treated samples for Paenibacillus sp. S02, the percentage of mapped reads of the replicate 1 (S02_treated_1) was even comparable to the untreated samples.
Discussion
Isolation and identification of novel Paenibacillus sp. strains
Two N-fixing bacterial strains were isolated in this study using a combined approach that took advantage of both microbiological techniques (N-free medium) and genomic resources (sequencing). Various N-free media have been widely used to isolate N-fixing microorganisms from natural environments on the basis that nitrogen is required by all organisms to survive28,38. However, being able to grow in N-free media does not necessarily guarantee the ability to fix atmospheric nitrogen by a microorganism. For instance, Streptomyces thermoautotrophicus UBT1 was initially reported as a N-fixing bacterium39, however further analysis revealed that the solidifying agent used to prepare the N-free medium contained 0.095% nitrogen content40. Hence, instead of being a N-fixing bacterium, this strain was a N-scavenger that could utilise nitrogen present in ultra-low concentrations. Furthermore, even if the selective medium is N-free, those scavengers could still grow in combination with other N-fixing microorganisms present, hence hindering the isolation process, which was observed in this study. The N-free medium and the nifH gene PCR used in this study confirmed the presence of N-fixing bacteria in the culture. Moreover, the sequencing results provided possible identities of the N-fixing bacteria, which greatly assisted the isolations of the two strains (S02 and S25). The long-read sequencing data of dilutions confirmed the presence of Paenibacillus spp. and also revealed the possible identity of other bacterial species in dilutions, leading to the addition of polymyxin B, which removed all N-scavengers in the culture.
Taxonomic identification of bacterial species based on the 16S ribosomal RNA and whole genome sequence homology suggested that the two strains used in this study (S02 and S25) were closely related to P. polymyxa. Furthermore, the ANI analysis showed that the two strains were not genetically identical to any of the known P. polymyxa strains, and hence represent two novel strains of Paenibacillus spp. There are currently (27/07/2020) genome sequences of 56 P. polymyxa strains publicly available on NCBI, none of which was originally isolated from Australia. Therefore, this is the first study that reported Australian strains of this species. P. polymyxa has been extensively studied, and some strains have been developed as commercially available biopesticides or biofertilisers5. The two strains (S20 and S05) reported by this study will be of great benefit to the local application of this bacterial species as they represent indigenous Paenibacillus sp. strains.
Interestingly, the phylogeny and comparative genomics analyses of this study suggested possible future taxonomic subdivision of the species P. polymyxa. The dendrograms based on ANI of P. polymyxa genomes and the maximum-likelihood tree based on the sequence homology of conserved genes of P. polymyxa genomes shared the same topology consisting of three major clades. Moreover, the comparative genomics analysis showed that the ANI values between strains from different clades were lower than 95%, which is the proposed prokaryotic species boundary41,42. While both Clade 1 and Clade 2 contained 18 P. polymyxa strains, the type strain of P. polymyxa, which is ATCC 842, was in Clade 2. There was no apparent patterns of geographic locations or environment origins of strains associated with each clade (Supplementary Table S3). Such results demonstrated that current P. polymyxa strains might need to be reclassified into three species based on ANI, including the “original” P. polymyxa species (Clade 2). The two novel Paenibacillus sp. strains (S02 and S25) are representatives of Clade 1 and represent a new Paenibacillus species. Similarly, the nine strains identified in Clade 3 also represent a new Paenibacillus species. Strains from the three clades were genetically closely related, and shared some PGP genes (e.g. IAA production)16 and secondary metabolite gene clusters (e.g. fusaricidin and tridecaptin, Table 1). However, the differences between strains from the three clades were demonstrated by the absence of some PGP genes from strains of a clade (e.g. the nif operon, Clade 3)16 and the presence of unique secondary metabolite gene clusters in strains of a clade (e.g. betalactone, Clade 2). Such phylogenomic reclassifications of P. polymyxa have been proposed in a previous study43. One of the reasons that caused this taxonomic ambiguity was the molecular markers used for taxonomic assignment. Prokaryotic taxonomy identification has been relying on the 16S ribosomal RNA over the last four decades, however the current standard is now shifting to genome-scale data44. For example, P. polymyxa E681 (Clade 1), which was isolated in 1990s and was the most studied P. polymyxa strain, was identified as P. polymyxa solely based on the 16S ribosomal RNA gene sequences43. However, the ANI analysis using its genome published in Kim, et al.8 only showed a similarity of 89.98% compared to the type strain P. polymyxa ATCC 842, whose genome was published in 201145. Molecular phylogeny based on the 16S ribosomal RNA gene sequences led to the discovery of the novel genus Paenibacillus46, which now comprised more than 200 species and this number is still growing4. With more and more Paenibacillus spp. genomes becoming available, molecular taxonomy based on genome-scale data including ANI will clarify the complexity of Paenibacillus species.
PGP genes and secondary metabolite gene clusters in the two Paenibacillus sp. strains
P. polymyxa has long been described as a PGP bacterium. The mode of action of plant growth promotion utilised by P. polymyxa has been proposed by Jeong, et al.43, including (1) direct promotion by producing phytohormones and enhancing nutrient uptake by plants, and (2) indirect promotion by providing bioprotection against phytopathogens. Method 1 is implemented via the expression of PGP genes. In this study, Paenibacillus sp. strains S02 and S25 were found to possess multiple PGP genes associated with biological nitrogen fixation, inorganic and organic phosphate solubilisation, phosphate assimilation and IAA production. Comparative genomics studies conducted by Eastman, et al.16 and Xie, et al.5 showed that these genes are highly conserved in P. polymyxa strains. For example, the nif operon carried by P. polymyxa strains contained nine genes and had a high sequence homology (> 80%)4. Interestingly, it has been reported that the ancestral Paenibacillus species could not fix atmospheric nitrogen, and the nif operon was acquired by N-fixing strains from possibly Frankia spp. via horizontal gene transfer26. Hence. these PGP genes were highly likely acquired by Paenibacillus spp. during the evolution to adapt to their plant-associated lifestyle17.
In this study, the two Paenibacillus sp. strains (S02 and S25) exhibited strong inhibitory activities against several fungal phytopathogens in in vitro assays. Such bioprotection (method 2) activities of P. polymyxa have been extensively studied. It has been discovered that P. polymyxa strains colonise plant roots and form biofilms, hence preventing the colonisation of phytopathogens22. Moreover, P. polymyxa strains also produce a wide variety of secondary metabolites associated with bioprotection. The two strains used in this study were found to carry 13 and 16 secondary metabolite gene clusters synthesising both antimicrobial ribosomal peptides, e.g. paenilan and paenicidin (lanthipeptide), and nonribosomal peptides including fusaricidin B, polymyxin and tridecaptin. Comparative analyses conducted in this study revealed that these clusters are conserved in P. polymyxa strains regardless of the phylogenetic clades. Furthermore, the compounds synthesised by more than half of the secondary metabolite gene clusters were novel, and they were commonly detected in many P. polymyxa strains5,16,17. These novel clusters represented a repository where more potential bioactive antimicrobial compounds could be discovered. Future research is required to identify and characterise these novel compounds.
Furthermore, some P. polymyxa strains have been reported to produce volatile compounds that promote plant growth by utilising both methods described above. For example, the volatile compounds emitted by P. polymyxa E681 increased the total leaf surface area of Arabidopsis seedlings and induced plant resistance to Pseudomonas syringae47. Thirteen volatile compounds produced by P. polymyxa WR-2 not only inhibited the growth of F. oxysporum, but also prevented the germination of F. oxysporum spores48. Hence, further studies are needed to identify and characterise bacterial volatile compounds produced by the novel Paenibacillus sp. strains S02 and S25.
Transcriptome sequencing confirmed the activities of the nif operon and provided insights into the early stage bacteria-pathogen interactions
Transcriptome sequencing revealed that the presence/absence of nitrogen greatly affected the transcriptome profiles of the two Paenibacillus sp. strains (S02 and S25). The expression levels of all nine genes of the nif operon were regulated, and mostly increased, for both strains when nitrogen was removed from the medium. However, the level of changes of those genes caused by removing nitrogen varied between the two strains. There were much higher increases in expression levels of the nif genes in strain S02 when compared with strain S25. Given that the two strains shared similar expression levels of the nif operon before the removal of nitrogen, it could be concluded that strain S02 is more active in biological nitrogen fixation. Such increased bioactivity could be explained by the difference in growth kinetics. It has been found that strain S02 grows faster than strain S25 in both Burk’s medium and Nutrient Broth (based on OD600 readings after 24 h). It could be postulated that Paenibacillus sp. S02 requires more nitrogen to meet the demand of cell multiplication, which leads to increased bioactivities of the nif operon when nitrogen is removed from the growth medium. To validate this increased activity, further experiments using the acetylene reduction assay or similar methods49 are required to quantify the N-fixation rate of both strains.
The two Paenibacillus sp. strains (S02 and S25) were bioactive against F. verticillioides (growth reduction) in the in vitro assays, with strain S02 being significantly more bioactive than S25. Genomic analyses demonstrated that the two strains were capable of producing at least two antifungal secondary metabolite compounds, i.e. fusaricidin50 and bacterial siderophore51, as well as a wide range of novel secondary metabolite compounds. We proposed a hypothesis that these compounds were associated with the bioprotection activities against F. verticillioides exhibited by both strains. To support results from the bioassay and genomic analysis, a transcriptome sequencing experiment was designed to explore the early stage interactions between the two strains and the pathogen. The stronger bioprotection activities of strain S02 were reflected by the percentages of mapped F. verticillioides transcripts in the treated samples. One of the biological replicates of strain S02 even produced a mapping rate that was comparable to the untreated samples. Moreover, the treated samples of strain S02 were plated on Potato Dextrose Agar plates, and no visual evidence of fungal growth was observed for that biological replicate (Supplementary Figure S4). Such results suggested that, for this specific biological replicate, the growth of F. verticillioides was highly likely completely inhibited. Furthermore, the results of DGE analysis showed that the core biosynthetic genes of secondary metabolite gene clusters were actively expressed by strain S02 comparing to strain S25 before the introduction of F. verticillioides, including clusters producing two known antifungal compounds (C1: fusaricidin B; C2: bacterial siderophore) as well as clusters producing novel compounds (e.g. C6 and C14), which may have also contributed to the bioactivity. Hence, we proposed a hypothesis that the stronger bioprotection activities provided by strain S02 was related to the higher concentrations of antifungal bioactive compounds that were produced even before the introduction of F. verticillioides. In addition, changes in transcriptome profiles suggested that strain S02 was more resilient to the introduction of F. verticillioides when compared with strain S25. Such highly active and stable transcriptome profiles would make strain S02 a promising candidate to be developed as a bioprotection agent. Future experiments, including the in vitro and in planta assays and field assays, are needed to further validate the potential of this strain. The bioactive compounds produced by the two strains should also be identified, purified and characterised.
It is notable that 32 of 51 core biosynthetic genes of secondary metabolite clusters of strain S25 were differentially expressed when F. verticillioides was present, with 17 being downregulated including those from clusters producing known antifungal compounds. Given this strain was bioactive against F. verticillioides in the in vitro assays, a possible explanation was the incubation time. Whilst the two strains and F. verticillioides were co-incubated for five days in the in vitro bioassays, they were only co-incubated for six hours before RNA was extracted in the transcriptome sequencing experiment. The short incubation time may not be enough for strain S25 to produce significant amounts of antifungal compounds. It has been reported that the expressions of secondary metabolites of Bacillus spp. were enhanced when fungal pathogens (including some Fusarium spp.) were present52, however such information is still missing for P. polymyxa and F. verticillioides. It is possible that the decreased expressions of those genes observed in this study was related to the bacteria-pathogen interactions. Future studies should incorporate a longer incubation time or a series of time points, to further elucidate the changes in transcriptome profiles of the two strains after the introduction of F. verticillioides.
The two Paenibacillus sp. strains reported by this study were genetically highly similar (ANI = 97.78%) despite the apparent differences in bioactivities and transcriptome profiles. Interestingly, our laboratory has previously isolated and characterised three novel Xanthomonas sp. strains from the perennial ryegrass microbiome which were also genetically similar but phenotypically different53. Hence, we proposed a hypothesis that plant hosts recruit and take advantages of multiple genetically similar strains of the same species with a diverse range of bioactivities. Future studies, especially in planta assays, are required to further characterise those strains to gain a deeper understanding of the perennial ryegrass microbiome.
Materials and methods
Seed-associated N-fixing bacterial strain detection and isolation
A PCR assay was designed to detect the presence of seed-associated N-fixing bacteria by amplifying the nifH gene of nitrogenase. Perennial ryegrass seeds (L. perenne, cv. Alto, with standard endophytes) were sourced from Barenbrug Agriseeds, New Zealand with licences that comply with local and national regulations. Approximately 1,000 seeds were washed using sterile water and then ground and soaked in 30 mL of Burk’s N-free medium (MgSO4, 0.2 g/L; K2HPO4, 0.8 g/L; KH2PO4, 0.2 g/L; CaSO4, 0.13 g/L; FeCl3, 0.00145 g/L; Na2MoO4, 0.000253 g/L; sucrose, 20 g/L). The suspension was incubated for two days at 26 ℃ and 200 rpm, and then serial diluted using sterile Burk’s N-free medium (1:10, 100 µL in 900 µL, eight replicates per dilution). Genomic DNA was extracted from the 10–2 and 10–3 dilutions using a Wizard Genomic DNA Purification Kit (A1120, Promega, Madison, WI, USA), and assessed for quality on a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). PCR conditions were as per Gaby and Buckley54. In brief, OneTaq@ Hot Start 2 × Master Mix (M0484, Promega, Madison, WI, USA) was used with a universal nifH gene PCR primer pair (IGK3: 5’- GCIWTHTAYGGIAARGGIGGIATHGGIAA-3’; DVV: 5’-ATIGCRAAICCICCRCAIACIACRTC-3’; final concentration = 0.4 µM)54 and 50 ng of extracted DNA. For the no template control, nuclease-free water was used. For the positive control, the genomic DNA of Rhizobium leguminosarum bv. trifolii WSM132555 was used. PCR products (~ 400 bp) were visualised on an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA), sequenced by Macrogen and then analysed using BLAST56.
Genomic DNA of dilutions that produced PCR amplicons were sequenced using long read sequencing technology. A library was prepared using the Oxford Nanopore Technologies (ONT) ligase-based library preparation kit (SQK-LSK109, ONT, Oxford, UK) and sequenced on a MinION Mk1B platform (MIN-101B) with R10 flowcells (FLO-MIN110). Genomic sequence data (raw read signals) were basecalled using ONT’s Guppy software (Version 3.4.3, HAC basecalling model), and assessed for quality using NanoPlot57. Basecalled data was filtered to remove adapter sequences using Porechop (Version 0.2.3, https://github.com/rrwick/Porechop), while reads shorter than 300 bp and the worst 5% of reads (based on quality) were discarded using Filtlong (Version 0.2.0, https://github.com/rrwick/Filtlong). Sequencing reads were taxonomically classified by Kraken224 using a custom database containing all completed bacterial reference genomes in NCBI (20/03/2020) and were analysed using BLAST56. In addition, 50 µL of those dilutions were inoculated into vials containing 5 mL of Burk’s N-free medium supplemented with 1.6 g/L agar and incubated for up to five days at 26 ℃. Cultures were checked daily for a band of microbial growth below the surface of medium, which indicated the presence of N-fixing bacteria38. Microbes were streaked onto Burk’s N-free medium supplemented with 15 g/L agar and Burk’s N-free medium supplemented with 15 g/L agar and 100 IU/mL polymyxin B (P4932-1MU, Sigma-Aldrich, St. Louis, MO, USA) and incubated for up to five days at 26 ℃ to isolate pure colonies.
Genome sequencing
DNA was extracted from bacterial pellets (overnight cultures) using a Wizard Genomic DNA Purification Kit (A1120, Promega, Madison, WI, USA), and assessed for quality (average molecular weight ≥ 30 Kb) on an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Genomic sequencing libraries (short reads) were prepared from the DNA using a PerkinElmer NEXTFLEX Rapid XP DNA-Seq Kit (Cat# NOVA-5149-03) and sequenced on an Illumina NovaSeq 6000 platform. Genomic sequence data (raw reads) were assessed for quality and filtered to remove any adapter and index sequence, and low-quality bases using fastp58 with the following parameters: -w 8 -3 -5 -detect_adapter_for_pe. In addition, genomic sequencing libraries (long reads) were prepared from the DNA as per “Seed-associated N-fixing bacterial strain detection and isolation” section.
Genome assembly and classification
The whole genomes of bacterial strains were assembled with filtered long and short reads using Unicycler59. Long reads were used for primary assembly and to resolve repeat regions in the genome, whereas short reads were used to correct small base-level errors. Assembly graphs were visualised using Bandage60. Assembled genomes were taxonomically classified by Kraken224 as per “Seed-associated N-fixing bacterial strain detection and isolation” section.
Genome annotation and characterisation
The assembled genomes of bacterial strains were annotated using Prokka61 with a custom Paenibacillus protein database (based on Kraken2 classification) to predict genes and corresponding functions. Identification of secondary metabolite gene clusters from annotated genomes was conducted using antiSMASH32 with the following options: --clusterblast --asf --knownclusterblast --subclusterblast --smcogs --full-hmmer. The presence of PGP genes in the annotated genomes was conducted using BLAST56 (blastn and tblastn, e value > 1e−10). PGP genes previously reported in P. polymyxa strains5,16 were targeted (30 genes), including biological nitrogen fixation (nine genes), phosphate solubilisation and assimilation (17 genes) and indole-3-acetic acid production and auxin transportation (four genes). The PGP gene identification compared the sequence homology of genes from strains S02 and S25 with a closely related P. polymyxa strain CR1.
Phylogeny and comparative genomics
A comparative genomic analysis was performed by calculating the average nucleotide identity (ANI) of the genomes of isolated strains to 44 P. polymyxa genomes that were publicly available on NCBI (Supplementary Table S3) using a python package pyani (Version 0.2.8, https://widdowquinn.github.io/pyani/). Moreover, a pan-genome analysis was conducted using Roary27 to compare the isolated strains to 13 P. polymyxa strains with complete circular genome sequences (Fig. 1, yellow labels) and to identify shared genes. A maximum-likelihood phylogenetic tree was inferred using FastTree62 with Jukes-Cantor Joins distances, the Generalized Time-Reversible substitution model and the CAT approximation model. Local branch support values were calculated using 1000 resamples with the Shimodaira-Hasegawa test.
Bioprotection assay (in vitro)
An assay was conducted to assess the in vitro bioprotection activity of isolated strains against fungal pathogens of Poaceae species. Three fungal pathogens of Poaceae species (Supplementary Table S9) were obtained from the National Collection of Fungi (VPRI, Bundoora, Victoria, Australia). The setup of the in vitro assay was described in detail in Li, et al.53. Briefly, bacterial strains, which were drop-inoculated onto four equidistant points on a Nutrient Agar plate, and pathogens, which were placed at the centre of the plate as a plug containing actively growing hyphae, were co-incubated at 28 ℃ in the dark for five days. The diameter of the fungal colony was measured twice, and the average of the two readings was used for statistical analysis. Three plates were prepared for each treatment as biological replicates. Sterile medium was used as the blank control. Statistical analysis (One-way ANOVA and Tukey Test) was conducted using OriginPro 2020 (Version SR1 9.7.0.188) to detect the presence of any significant difference (P < 0.05) between treatments.
Transcriptome sequencing
Transcriptome sequencing experiments were designed to confirm the expression of the nif operon and to explore the early stage of bacteria-pathogen interactions. For the N-fixation activity assay, bacterial strains were cultured in Burk’s N-free medium overnight (OD600 = 1.0). Cultures were diluted using Burk’s N-free medium to OD600 = 0.7 and further cultured for six hours to produce actively growing cells for extracting high quality RNA. Burk’s N-free medium supplemented with 10 g/L NH4Cl was used for culturing the bacterial strains as the control. For the bacteria-pathogen interactions assay, bacterial strains and the phytopathogen VPRI42586a Fusarium verticillioides were cultured in Nutrient Broth overnight (OD600 = 1.0). Bacterial cultures were diluted using Nutrient Broth to OD600 = 0.7. 20 mL of such culture was mixed with 200 µL of the pathogen culture and was further incubated for six hours. For the control, the pathogen culture was replaced by sterile Nutrient Broth. Three biological replicates were prepared for each treatment.
Total RNA was extracted form cell pellets using a TRIzol Plus RNA Purification Kit (12183555, Thermo Fisher Scientific). On-column treatments were conducted using a PureLink DNase Kit (12185010, Thermo Fisher Scientific) to ensure the complete removal of genomic DNA that would affect the downstream analyses, and ribosomal RNA was depleted using a NEBNext rRNA Depletion Kit (E7860L, NEB, Ipswich, MA, USA). Directional RNA-seq libraries were prepared using a NEBNext Ultra II Directional RNA Library Prep Kit (E7765) and sequenced on an Illumina NovaSeq 6000 platform. RNA-seq data (raw reads) were assessed for quality and filtered as per described in “Genome sequencing” section.
Transcriptome analysis
Salmon63 was used to quantify transcripts using the clean RNA-seq reads with the following parameters: -l A --validateMappings --numBootstraps 1000 --seqBias. The references used for transcript quantification were the gene sequences generated by Prokka (“Genome annotation and characterisation” section) or the gene sequences of F. verticillioides 7600 downloaded from NCBI GenBank (Accession ID: SAMN02953630). A total of 1,000 rounds of bootstraps were performed during transcript quantification to minimise the impact of technical variations. Differential gene expression (DGE) analysis was conducted using a R package sleuth64. Likelihood ratio tests were conducted to detect the presence of any significant difference (q-value < 0.05) in transcript abundances between treatments, and Wald tests were conducted to determine an approximation of the fold-change in transcript abundances between treatments. Transcripts that were of ultra-low abundance (defined by having less than 20 mapped reads or were only present in less than three samples) were removed prior DGE analysis. The differentially expressed genes were defined to be significant at q-value < 0.05 and absolute fold-change ≥ 1.5.
Supplementary Information
Acknowledgements
Tongda Li received the La Trobe University Full-Fee Research Scholarship, the La Trobe University Postgraduate Research Scholarship and the DairyBio Scholarship. The authors wish to thank Dr. Jacqueline Edwards for access to the Victorian Plant Pathogen Herbarium and Desmond Auer for editing the manuscript.
Author contributions
T.S. conceptualised the study. T.L., T.S., and R.M. designed the experiment. T.L. and J.K. contributed to the laboratory work. T.L. prepared the manuscript. R.M., T.S., and G.S. reviewed and edited the manuscript. T.S. and R.M. supervised the study. G.S. contributed to the funding acquisition. All authors have read and agreed to the submitted version of the manuscript.
Data availability
Annotated genome sequences of strains S02 and S25 were deposited in the NCBI GenBank with the accession number PRJNA720481. The results of transcript quantification (raw reads count) were provided as supplementary datasets (Supplementary Table S10 and S11).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94820-2.
References
- 1.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 3.Berg G. Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009;84:11–18. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- 4.Grady EN, MacDonald J, Liu L, Richman A, Yuan Z-C. Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell. Fact. 2016;15:203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie J, et al. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 2016;6:21329. doi: 10.1038/srep21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trüper, H. The type species of the genus Paenibacillus Ash et al. 1994 is Paenibacillus polymyxa. Opinion 77. Judicial Commission of the International Committee on Systematics of Prokaryotes. Int. J. Syst. Evol. Microbiol.55, 17 (2005). [DOI] [PubMed]
- 7.Cherchali, A., Boukhelata, N., Kaci, Y., Abrous-Belbachir, O. & Djebbar, R. Isolation and identification of a phosphate-solubilizing Paenibacillus polymyxa strain GOL 0202 from durum wheat (Triticum durum Desf.) rhizosphere and its effect on some seedlings morphophysiological parameters. Biocatal. Agric. Biotechnol.19, 101087, 10.1016/j.bcab.2019.101087 (2019).
- 8.Kim JF, et al. Genome sequence of the polymyxin-producing plant-probiotic rhizobacterium Paenibacillus polymyxa E681. J. Bacteriol. 2010;192:6103. doi: 10.1128/JB.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holl, F. B., Chanway, C. P., Turkington, R. & Radley, R. A. Response of crested wheatgrass (Agropyron cristatum L.), perennial ryegrass (Lolium perenne) and white clover (Trifolium repens L.) to inoculation with Bacillus polymyxa. Soil Biol. Biochem.20, 19–24, 10.1016/0038-0717(88)90121-6 (1988).
- 10.Bal A, Anand R, Berge O, Chanway CP. Isolation and identification of diazotrophic bacteria from internal tissues of Pinus contorta and Thuja plicata. Can. J. For. Res. 2012;42:807–813. doi: 10.1139/x2012-023. [DOI] [Google Scholar]
- 11.Zhou C, et al. Paenibacillus polymyxa BFKC01 enhances plant iron absorption via improved root systems and activated iron acquisition mechanisms. Plant Physiol. Biochem. 2016;105:162–173. doi: 10.1016/j.plaphy.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Puri, A., Padda, K. P. & Chanway, C. P. Evidence of nitrogen fixation and growth promotion in canola (Brassica napus L.) by an endophytic diazotroph Paenibacillus polymyxa P2b-2R. Biol. Fertility Soils52, 119–125, 10.1007/s00374-015-1051-y (2015).
- 13.Puri A, Padda KP, Chanway CP. Seedling growth promotion and nitrogen fixation by a bacterial endophyte Paenibacillus polymyxa P2b–2R and its GFP derivative in corn in a long-term trial. Symbiosis. 2016;69:123–129. doi: 10.1007/s13199-016-0385-z. [DOI] [Google Scholar]
- 14.Anand R, Grayston S, Chanway C. N2-fixation and seedling growth promotion of lodgepole pine by endophytic Paenibacillus polymyxa. Microb. Ecol. 2013;66:369–374. doi: 10.1007/s00248-013-0196-1. [DOI] [PubMed] [Google Scholar]
- 15.Xu S, Kim B-S. Evaluation of Paenibacillus polymyxa strain SC09-21 for biocontrol of Phytophthora blight and growth stimulation in pepper plants. Trop. Plant. Pathol. 2016;41:162–168. doi: 10.1007/s40858-016-0077-5. [DOI] [Google Scholar]
- 16.Eastman AW, Heinrichs DE, Yuan Z-C. Comparative and genetic analysis of the four sequenced Paenibacillus polymyxa genomes reveals a diverse metabolism and conservation of genes relevant to plant-growth promotion and competitiveness. BMC Genomics. 2014;15:851. doi: 10.1186/1471-2164-15-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, et al. Comparative genome analysis and mining of secondary metabolites of Paenibacillus polymyxa. Genes Genet. Syst. 2020;95:141–150. doi: 10.1266/ggs.19-00053. [DOI] [PubMed] [Google Scholar]
- 18.Aleti G, et al. The draft genome sequence of Paenibacillus polymyxa strain CCI-25 encompasses high potential for secondary metabolite production. Genome Announc. 2016;4:e00366–e1316. doi: 10.1128/genomeA.00366-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, et al. Complete genome sequence of Paenibacillus polymyxa YC0136, a plant growth–promoting rhizobacterium isolated from tobacco rhizosphere. Genome Announc. 2017;5:e01635–e11616. doi: 10.1128/genomeA.01635-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lal S, Tabacchioni S. Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview. Indian J. Microbiol. 2009;49:2–10. doi: 10.1007/s12088-009-0008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padda, K. P., Puri, A. & Chanway, C. P. in Agriculturally important microbes for sustainable agriculture: Volume 2: Applications in crop production and protection (eds Vijay Singh Meena, Pankaj Kumar Mishra, Jaideep Kumar Bisht, & Arunava Pattanayak) 165–191 (Springer Singapore, 2017).
- 22.Timmusk S, Grantcharova N, Wagner EGH. Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 2005;71:7292–7300. doi: 10.1128/aem.71.11.7292-7300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannenbaum I, et al. Profiling the Lolium perenne microbiome: from seed to seed. Phytobiomes J. 2020;4:281–289. doi: 10.1094/pbiomes-03-20-0026-r. [DOI] [Google Scholar]
- 24.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SK, et al. Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J. Bacteriol. 2009;191:3350–3358. doi: 10.1128/jb.01728-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie, J.-B. et al. Comparative genomic analysis of N2-fixing and non-N2-fixing Paenibacillus spp.: Organization, evolution and expression of the nitrogen fixation genes. PLOS Genet.10, e1004231, 10.1371/journal.pgen.1004231 (2014). [DOI] [PMC free article] [PubMed]
- 27.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franche C, Lindström K, Elmerich C. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil. 2009;321:35–59. doi: 10.1007/s11104-008-9833-8. [DOI] [Google Scholar]
- 29.de Werra P, Pechy-Tarr M, Keel C, Maurhofer M. Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 2009;75:4162–4174. doi: 10.1128/AEM.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Z-C, Zaheer R, Finan TM. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 2006;188:1089–1102. doi: 10.1128/jb.188.3.1089-1102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 32.updates to the secondary metabolite genome mining pipeline Blin, K. et al. antiSMASH 5.0. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Jensen SE. Nonribosomal biosynthesis of fusaricidins by Paenibacillus polymyxa PKB1 involves direct activation of a D-amino acid. Chem. Biol. 2008;15:118–127. doi: 10.1016/j.chembiol.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Park JE, Kim HR, Park SY, Choi SK, Park SH. Identification of the biosynthesis gene cluster for the novel lantibiotic paenilan from Paenibacillus polymyxa E681 and characterization of its product. J. Appl. Microbiol. 2017;123:1133–1147. doi: 10.1111/jam.13580. [DOI] [PubMed] [Google Scholar]
- 35.Lohans CT, et al. Biochemical, structural, and genetic characterization of tridecaptin A1, an antagonist of Campylobacter jejuni. ChemBioChem. 2014;15:243–249. doi: 10.1002/cbic.201300595. [DOI] [PubMed] [Google Scholar]
- 36.Vater J, et al. Genome mining of the lipopeptide biosynthesis of Paenibacillus polymyxa E681 in combination with mass spectrometry: Discovery of the lipoheptapeptide paenilipoheptin. ChemBioChem. 2018;19:744–753. doi: 10.1002/cbic.201700615. [DOI] [PubMed] [Google Scholar]
- 37.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VLD. The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil. 2014;384:413–431. doi: 10.1007/s11104-014-2186-6. [DOI] [Google Scholar]
- 39.Ribbe M, Gadkari D, Meyer O. N2 fixation by Streptomyces thermoautotrophicus involves a molybdenum-dinitrogenase and a manganese-superoxide oxidoreductase that couple N2 reduction to the oxidation of superoxide produced from O2 by a molybdenum-CO dehydrogenase. J. Biol. Chem. 1997;272:26627–26633. doi: 10.1074/jbc.272.42.26627. [DOI] [PubMed] [Google Scholar]
- 40.MacKellar D, et al. Streptomyces thermoautotrophicus does not fix nitrogen. Sci. Rep. 2016;6:20086. doi: 10.1038/srep20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chun J, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 43.Jeong H, Choi S-K, Ryu C-M, Park S-H. Chronicle of a soil bacterium: Paenibacillus polymyxa E681 as a tiny guardian of plant and human health. Front. Microbiol. 2019;10:467. doi: 10.3389/fmicb.2019.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klenk HP, Goker M. En route to a genome-based classification of Archaea and Bacteria? Syst. Appl. Microbiol. 2010;33:175–182. doi: 10.1016/j.syapm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Jeong H, et al. Draft genome sequence of the Paenibacillus polymyxa type strain (ATCC 842), a plant growth-promoting bacterium. J. Bacteriol. 2011;193:5026. doi: 10.1128/JB.05447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ash C, Priest FG, Collins MD. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Van Leeuwenhoek. 1993;64:253–260. doi: 10.1007/BF00873085. [DOI] [PubMed] [Google Scholar]
- 47.Lee B, et al. Induced resistance by a long-chain bacterial volatile: Elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE. 2012;7:e48744. doi: 10.1371/journal.pone.0048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raza, W., Yuan, J., Ling, N., Huang, Q. & Shen, Q. Production of volatile organic compounds by an antagonistic strain Paenibacillus polymyxa WR-2 in the presence of root exudates and organic fertilizer and their antifungal activity against Fusarium oxysporum f. sp. niveum. Biol. Control80, 89–95, 10.1016/j.biocontrol.2014.09.004 (2015).
- 49.Das S, De TK. Microbial assay of N2 fixation rate, a simple alternate for acetylene reduction assay. MethodsX. 2018;5:909–914. doi: 10.1016/j.mex.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SH, et al. An antibiotic fusaricidin: a cyclic depsipeptide from Paenibacillus polymyxa E681 induces systemic resistance against Phytophthora blight of red-pepper. Phytoparasitica. 2012;41:49–58. doi: 10.1007/s12600-012-0263-z. [DOI] [Google Scholar]
- 51.Compant S, Duffy B, Nowak J, Clément C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrić S, Meyer T, Ongena M. Bacillus responses to plant-associated fungal and bacterial communities. Front. Microbiol. 2020;11:1350. doi: 10.3389/fmicb.2020.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li T, et al. Novel Xanthomonas species from the perennial ryegrass seed microbiome – Assessing the bioprotection activity of non-pathogenic relatives of pathogens. Front. Microbiol. 2020;11:1991. doi: 10.3389/fmicb.2020.01991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaby, J. C. & Buckley, D. H. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLOS One7, e42149 (2012). [DOI] [PMC free article] [PubMed]
- 55.Reeve, W. et al. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual Mediterranean clovers. Stand. Genomic Sci.2, 347–356, 10.4056/sigs.852027 (2010). [DOI] [PMC free article] [PubMed]
- 56.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Coster, W., D'Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: visualizing and processing long read sequencing data. Bioinformatics, 2666–2669. 10.1093/bioinformatics/bty149 (2018). [DOI] [PMC free article] [PubMed]
- 58.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comp. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 62.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Annotated genome sequences of strains S02 and S25 were deposited in the NCBI GenBank with the accession number PRJNA720481. The results of transcript quantification (raw reads count) were provided as supplementary datasets (Supplementary Table S10 and S11).