Abstract
Diabetes is an emerging health condition globally and is suggested to have a direct connection with the gut microbiota that determine our metabolic outcomes. Sensitivity to insulin and glucose metabolism is normal in healthy people as compared to those people who cannot maintain their glucose metabolism. One of the reasons of the differences is that healthy people have different microbiome that leads to achieve more short chain fatty acids and make up more branched amino acids, while the gut microbiota of the other group of people are more likely to produce compounds that affects glucose metabolism. Herein, this review will present the research related to the impact of gut microbes on diabetes carried out in the past decade. The review focus on the relation between gut microbiota and Type-1 Diabetes (T1D), Type-2 Diabetes (T2D), and how gut microbiota could be an alternative therapy for treatment of diabetes.
Keywords: Diabetes, Insulin Resistance, Microbiota, Metagenomics, Inflammation
1. Introduction
Human body is inhabited by numerous microbes that resides on and within the human body. These microorganisms constitute our microbiota, and their complete genome is referred to as microbiome. Microorganisms existing from 10 to 100 trillion in numbers, survive in the adult gut with approximately 1000 species (Sabit et al., 2021, Hajjo and Geva-Zatorsky, 2020, Fan and Pedersen, 2021). Most of the bulk of these microorganisms live in the bowel. The major constituents of gut flora are mainly bacteria along with few viruses and fungi (Erdogan and Rao, 2015).
The impact of gut microbiota in diabetes has been the talk of the researchers for the past few years. Several investigations have been done to study this relationship and the effect of gut microbiota on diabetes as it has become a health burden around the globe (Lynch and Pedersen, 2016, Tang et al., 2017). Therefore, it is very crucial to recognize and understand the mechanism and causes of diabetes to find a treatment. Also, it has been analyzed that the gut microbiota functions as a supplementary endocrine system (Pascale et al., 2018, Aron-Wisnewsky et al., 2020). With time it has been understood that human gut has variety of microorganisms that influences number of diseases including diabetes. Two main types of Diabetes Miletus (DM) include Type-1 Diabetes (T1D) and Type-2 Diabetes (T2D). In T1D, T cells attack β-cell of islets, which in turn results in inadequate insulin production. Islets of beta cell fails in T2D where the body becomes immune to the insulin it creates which in turn causes obesity as well. Although DM is caused due to the genetic factors, another factor is the microbiota of the human gut which plays a vital role in the progression of both T1D and T2D.
2. Consortium of gut microbiota
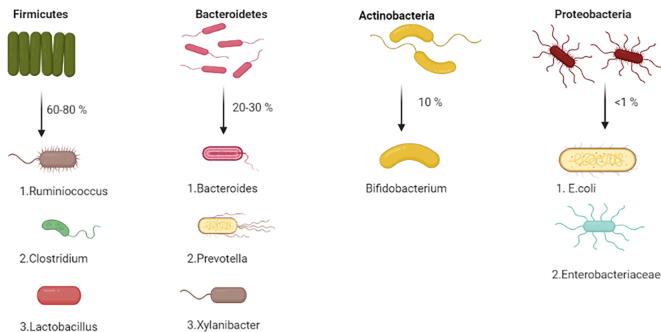
Human intestine is inhabitant to trillions of not only bacteria but viruses, fungi and archaea, that makeup a complex ecological community with which an individual lives in a symbiotic manner all his life (Hoffmann et al., 2013). The gastrointestinal system represents the biggest connection between the microbial environment existing inside the human and the exterior world environment. Gut microbiota has a pivotal role in regulating the systemic and intestinal immune and metabolic homeostasis (Needell and Zipris, 2016). The consortium of gut microbiota is connected to the host genetics and various other conditions like food habits, exposure to drugs, toxins or stresses. Composition of gut microbiota majorly include gram negative bacteria and one phylum of gram-positive organisms as well (Lagier et al., 2012). Four major phyla of bacteria that are residents of human gut are: (Fig. 1)
-
a)
Firmicutes (mostly gram-positive bacteria) account for 60–80%
-
b)
Bacteroidetes (Gram Negative bacteria) accounts for 20–30%
-
c)
Actinobacteria (Gram negative bacteria) accounts for 10%
-
d)
Proteobacteria (Gram negative bacteria) very less commonly found accounts for less than 1%
Fig. 1.
The four major phyla of human gut bacteria.
During early childhood, actinobacteria (Bifidobacterim genus) dominates. But with growing age, these bacteria gain variety of new strains due to the influence of diet and diseases (Mika et al., 2015). Composition of gut microbiota depends on various factors. Some of these factors include age, diet, geographical area, sex etc. It can be even modulated by use of antibiotics, probiotics and prebiotics.
An important concern in this area is to understand the methods of measuring microbiota. Analysis of gut microbiota was initially relied only on culture methods and identification was done by conventional methods of morphological, biochemical, and phenotypic tests. Culture of fecal microbiota is the most primitive method, but it has not proved to be reliable as the majority of the microbes are anaerobic and a strict environment needs to be created while processing the culture methods. With the advent of 16 s ribosomal RNA and whole genome approach, this work has become much more quantitative and elaborative (Le Chatelier et al., 2013). But this technique fails to classify microbes at species level (Rajendhran and Gunasekaran, 2011). Polymerase chain reaction is one of the widely used processes for estimation of microbes that cannot be cultured. Few commonly used procedures include:
2.1. Gnotobiotic
It is a process to study gut microbes in vivo using germ free animals. In this process, after caesarean section, animals are placed in an isolator where food, water and air is sterile and then they are exposed to the microorganism which is needed to be studied and is implanted to the GI tract of that animal. These animals are animals with defined microbiological status. Then comparisons can be done between the gut microbiota of these sterile animals and the conventional animals with normal microbiota (Singhvi et al., 2020, Gou et al., 2021).
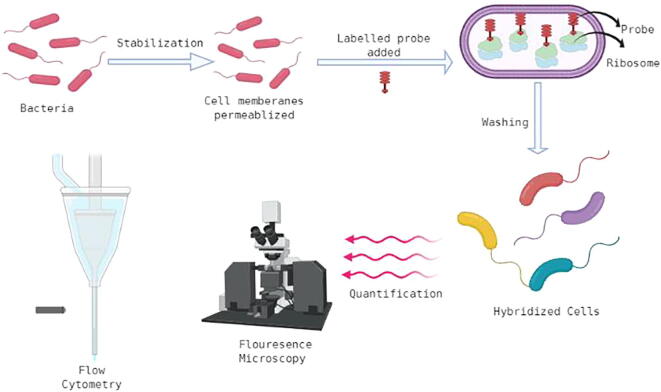
2.2. Fluorescence in situ hybridization (FISH)
FISH can mark a variety of bacterial communities in host tissue and reveal the organization of these microbes in their place. It is a quantitative procedure where rRNA probes marked with fluorescence are used. This imaging technique can further be collaborated with mass spectrometry imaging to understand the spatial location of the bacterium. Fluorescence microscopy can also be used for detection and visualization. An important quality of this visually pleasing technique is that it provides an intermediary clarity between DNA study and chromosomal analysis, while also keeping in place information at the single-cell level (Volpi and Bridger, 2018) (Fig. 2).
Fig. 2.
Schematic representation of fluorescence in situ hybridization.
2.3. Gradient gel electrophoresis
It includes two methods: denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE). DGGE and TGGE both uses chemical gradient or temperature gradient to denature the sample while moving through an acryl amide gel. Same sized gene sequences are separated by DGGE according to their varying capability to denature, that is evaluated by their base pair sequence, while TGGE is dependent on temperature which leads to alteration in structure resulting to separation of nucleic acids. (Das and Dash, 2019).
2.4. 16S rDNA sequencing
16S rDNA sequencing is useful for identification of rare bacteria that are difficult to identify using traditional methods, with genus identification in > 90% of cases and species identification in 65–83% of cases. 16S rDNA sequencing is considered a solution to improve our understanding of the epidemiology and pathogenic function of gut bacteria, which was previously impossible. In clinical microbiology laboratories, 16S rDNA sequencing will continue to play an important role in identifying unusual gut microbiota and bacteria with uncertain characteristics (Woo et al., 2007, Khaled et al., 2019).
DNA libraries of 16S rDNA sequences are generated by multiplying bacterial DNA from the given samples. Microbial diversity is analyzed by sequencing and comparison of sequences that are available in the libraries (Johnson et al., 2019).
3. Gut microbiota and Type-1 diabetes (T1D)
Type 1 diabetes is a chronic disease where autoreactive T- cells attack pancreatic cells of islets of β-cells that results in insufficient production of insulin. In this condition body is unable to process glucose due to lack of insulin. Glucose from the food cannot make its way to the cells and hence, high level of glucose keeps on circulating in our blood. These high glucose levels in blood can cause short term and long-term problems. T1D is the outcome of an interlinkage between varying degrees of genetic susceptibility and environmental factors. Recently gut microbiota is known to be one of the environmental factors and has been shown to have an effect on T1D and cause the development of this disease (Murri et al., 2013). There are evidence showing that gut microbiota can contribute to development of gut associated lymphoid tissues and also enhance immunity to pathogens (Kamada et al., 2013). Even, research has been done both on animal and human models of T1D and non-diabetic models and proved that human microbiome is corelated with the development of T1D (Nielsen et al., 2014, Vaarala, 2013). Type 1 diabetes mellitus (T1D) usually originates in the early years of life, a stage at which the intestinal microbiota is developing. The microbial composition, however, increases with age, productivity, and reaches the highest complicatedness in adults (Ringel-Kulka et al., 2013). Children advancing to T1D have a reduced abundance of bacteria that manufacture butyrate or lactate following the emergence of the first disease-predictive autoantibodies. Many studies are done on children who develop T1D and their gut microbiota has been compared with gut microbiota of the normal children (Murri et al., 2013). This was studied for the first time in Finland in 2011 where 16 s pyrosequencing was used for research analysis with the help of their stool samples. The modern 16 s pyrosequencing technology suggested an increased ratio of Bacteriodetes:Firmicutes as compared to the control. Also, shot-gun metagenomics analysis showed a high proportion of mucin degrading and butryte producing bacteria in controls as compared to the cases. But the ratio of other short chain fatty acid producing bacteria were higher in a case study (Brown et al., 2011). Shotgun metagenomics provide advanced method that is culture-independent to study the existing bacterial flora where the entire genomes of the sample is sequenced, resulting in identification up to the species level of bacteria (Morgan and Huttenhower, 2014).
In patients with T1D, there are several reports that equate the gut microbiota to that found in healthy individuals. One of the earliest of such study was done in 2013, where gut microbiota of 16 children with T1D was compared with 16 children without T1D. Reduction in the number of Actinobacteria and Firmicutes and the ratio of Firmicutes to Bacteroidetes was observed, with an improvement in the number of Bacteroidetes as compared to safe controls in children without T1D (Murri et al., 2013). The study also reported a raise in levels of Clostridium, Bacteroides and Veillonella and a decrease in Lactobacillus, Bifidobacterium, the Blautia coccoides–Eubacterium rectale group and Prevotella in the children with T1D.
The correlation of altered gut microbiota with β-cell autoimmunity has been seen in pediatric group who are at potential chance of developing T1D (Murri et al., 2013). Alteration of microbial occurrence in childhood would lead to abnormal microbial prevalence in adulthood that may have an immense effect in diversity and richness of particular species of bacteria. All these studies indicate that altered gut microbiota are strongly linked to β-cell autoimmunity and T1D in either composition or/and function, while geographical and ethnic diversity can affect the diversity of the gut microbiota of humans.
Apart from all this, it is investigated that segmented filamentous bacteria (SFB) are related with the development of Th17 (T- helper 17 cells) cells and may have influence in protecting female normal mice from developing diabetes by Th17 cells existing in the small intestine. Although, another study indicated that the impact of SFB on the prevalence of diabetes in mice was also correlated with other gut bacteria, as they were not shielded from the development of diabetes by single SFB colonization in these mice (Yurkovetskiy et al., 2013). Some of the bacterial species can develop acquired immunity and amend the health condition of the host. According to a study by Yang and his group, SFB initiates Th17 cells development in a mouse where antigen specific CD4 + T cells in the intestines developed into Th1 or Th17 cells. This report indicates specifically that associated antigens obtained from intestinal bacteria command the development of antigen-specific T cells. Such findings indicate that microbial peptides derived from commensal bacteria can trigger antigen-specific T cells. (Yang et al., 2014).
4. Gut microbiota and Type 2 diabetes (T2D)
Two interrelated problems may be the cause of Type 2 Diabetes (T2D):
-
(i)
Insulin resistance in muscle, fat and the liver cells as these cells don't respond to insulin in a normal way.
-
(ii)
Insufficient insulin produced by the pancreas.
To provide the best therapeutic strategy for clinically treating T2D patients, different factors such as BMI (body mass index), race and age are considered. Studies have shown that the function of gut microbiota in the control of T2D production is important. Normally, sugar moves into our cells but in case of T2D it gets accumulated in our bloodstream thereby increasing blood sugar and thus making the insulin-producing beta cells in the pancreas secrete more insulin. Consequently, such cells turn into disabled and stop making required insulin needed by body. An interference or change in the diversity of the microbiome is directly associated to T2D (Adeshirlarijaney and Gewirtz, 2020), explaining the influence of symbiotic microbiota in host homeostasis (David et al., 2014). Beli et al. (2018) reported that diabetic retinopathy can be prevented by intermittent fasting in mice by redesigning the microbiome towards bacteria that produce tauroursodeoxycholate and activating the Takeda G-protein-coupled receptor 5. It is reported that the gut microbiota has a close interaction with various systems like inflammatory, renal, cardiovascular, and endocrine systems. (Kanbay et al., 2018, Mafra and Fouque, 2015).
T2D patients show an altered intestinal microbiota where Bacteroidetes:Firmicutes ratio is decreased and also some other bacteria (e.g., Bifidobacteria) are decreased but there is an increase in some endotoxins-producing gram-negative bacteria that modify the metabolic activity of host (Qin et al., 2012, Wu et al., 2010, Larsen et al., 2010). Furthermore, the accumulation of inflammatory molecules like flagellin and peptidoglycans that are derived from gut- bacteria in the intestine hasten the inflammation in T2D (Kootte et al., 2012).
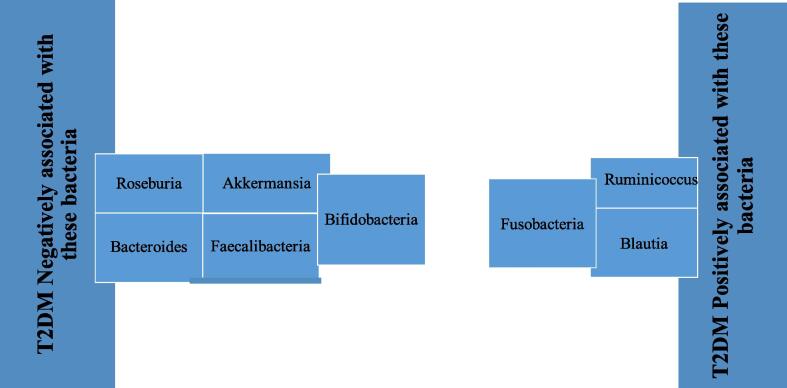
Fig. 3 shows the genera of bacteria that are both negatively and positively associated with T2D (Gao et al., 2018, Candela et al., 2016, Sedighi et al., 2017, Wu et al., 2010, Wu et al., 2017, Barengolts et al., 2018, Xu et al., 2015, Pedersen et al., 2016). Most of the research has reported a negative association of bifidobacteria with T2D except one report that indicates the opposite result (Sasaki et al., 2013). Almost all the animal studies show that Bifidobacterium (which is the most protective microbe against T2D) that habituate the human gut plays an important and protective role in T2D (Kikuchi et al., 2018, Aoki et al., 2017). Also, Bacteroides show negative association with the disease in some cases (Candela et al., 2016, Lippert et al., 2017, Yamaguchi et al., 2016) while some reports suggest tha they are positively associated with the disease (Murphy et al., 2017, Sun et al., 2018). But the reason for this positive association was the involvement of some treatment like metformin (Malik et al., 2018). Further investigations on species level of bacteroides; Bacteroides intestinalis, Bacteroides 20–3 and Bacteroides vulgatus showed a decreased level in T2D patients and Bacteroides stercoris were enhanced after sleeve gastrectomy surgery in T2D patients (Zhang et al., 2013, Karlsson et al., 2013). These findings clearly indicate that bacteriodes have a significant role in glucose metabolism.
Fig. 3.
Representative of bacteria positively and negatively associated with T2D.
Lactobacillus genus is a diversified genus containing the largest number of species in the human gut among the class of probiotic bacteria. It appears to have species-specific or even strain-specific effects on T2D, which might be the reason why genus level analysis lacks consistency amongst studies using lactobacillus (He et al., 2018). Various species of lactobacillus like L. acidophilus, L. gasseri, L. salivarius were increased and L. amylovorus was decreased in T2D patients (Graessler et al., 2013, Karlsson et al., 2013, Forslund et al, 2015). Several species of this genus also work as probiotics. L. plantarum, L. reuteri, L. casei, L. curvatus, L. gasseri , L. paracasei, L.rhamnosus, L. sakei proved to be useful as probiotics in T2D mice models (Martinic et al., 2018, Balakumar et al., 2018, Park et al., 2013, Lim et al., 2016).
According to one of the studies conducted with 345 T2D Chinese patients and healthy controls, a metagenome-wide association research (MGWAS) was conducted to examine the taxonomic changes in T2D-related intestinal microbiota and found that patients with T2D had mild intestinal microbial dysbiosis, a decrease in some butyrate-producing bacteria and a rise in various opportunistic pathogens (Qin et al., 2012).
5. Features of microbiota that may lead to the development of T2D
5.1. Endotoxins
A moderate increase in endotoxins (also called as lipopolysaccharides or LPS) like cytokines (IL-6, IL-1) which impairs insulin interaction with its receptor is produced by cell wall of gram-negative bacteria and are associated with T2D. The causative role of LPS after infusion of LPS into mice fed a regular diet induced insulin resistance in the liver, glucose tolerance, and increased adipose tissue weight. In macrophages, LPS binds to the CD14/TLR4 receptor and induces an increase in pro-inflammatory molecule production. But when LPS injections were induced in mice without CD14/TLR4, mice failed to develop these characteristics without development of T2D, thus indicating the role of CD14/TLR4 receptors for LPS (Pussinen et al., 2011). These conditions suggest that if there is any change in gram negative gut microbiota, it may lead to a change in intestinal permeability which is directly linked to resistance to insulin.
5.2. Brown adipose tissue (BAT)
BAT encourages immunity to insulin. Brown fat cells arise in white adipose tissue (WAT) by a mechanism called browning in cold environments or during exercise. (Wu et al., 2012). Such cells are called as beige fat cells. Upon antibiotic treatment in animal models, the beige fat formation in adipose tissue eradicates microbiota due to which there is increase in glucose tolerance and insulin sensitivity. These effects are reversed in mices where microbes treated with antibiotics are recolonized (Suárez-Zamorano, 2015).
5.3. Drugs and T2D
Metformin treatment in T2D patients modifies gut microbiota due to its gastrointestinal effects according to some studies (Forslund et al., 2015). Metformin treatment in T2D patients has helped in improving glycemic index and gut microbiota diversity (Wu et al., 2017). Metformin works by inhibiting mitochondrial function via respiratory chain complex I or glycerophosphate dehydrogenase, and/or amelioration of glucagon-induced cAMP. If administered intravenously, metformin does not control hyperglycemia. This indicates that the intestine is an important site of metformin action (Adeshirlarijaney and Gewirtz, 2020). Many researchers have suggested metformin as a therapy for patients with T2D. Not only this, but metformin has also been proposed as a treatment of gestational diabetes and T2D prevention in pre-diabetic individuals (Wu et al., 2017, Gui et al., 2013).
Acarbose, another drug which lowers postprandial blood glucose level has also been known to have an impact on microbiota composition. After alteration of gut microbiota in patients with T2D condition on treatment with acarbose, an increased concentration of B. longum and decreased concentration of lipopolysaccharides was observed (Su et al., 2015) while in another study butyricicoccus, Phascolarctobacterium, and Ruminococcus decreased but Lactobacillus, Faecalibacterium, and Dialister increased after acarbose intake in prediabetic patients (Zhang et al., 2017).
5.4. Changes in incretin secretion and Diabetes
According to a research, an increase in Bifidobacterium spp. is related to increased secretion of glucagon like peptide-1 (GLP-1) by the bowel that has favorable effects in reducing insulin resistance (Velasquez-Manoff, 2015).
5.5. Secondary bile acids and T2D
Secondary bile acids have been reported to have an insulin sensitizing role. Secondary bile acids are formed when some of the primary bile acids skip the mechanism of reabsorption through enterohepatic circulation. This is done majorly by the action of Firmicutes. In T2D subjects, there is a lower number of secondary bile acids as compared to healthy subjects which indicates that it might be related to impaired carbohydrate metabolism (Palau-Rodriguez et al., 2015).
5.6. Vitamins and diabetes
Few vitamins like choline and niacin can be broken down by some bacteria; Firmicutes, Actinobacteria, Proteobacteria and F.prausnitzii and due to oxidative stress their end products have been linked with diabetes developement (Palau-Rodriguez et al., 2015).
Microbiota has its own mechanism of affecting metabolism of T2D patients. They have an effect on inflammation, affects gut permeability, glucose metabolism, insulin sensitivity and overall homeostasis in human body. Levels of inflammatory proteins, cytokines and chemokines are elevated in T2D patients (Dagdeviren et al., 2017). Overexpression of cytokine in the muscle protects against age-related insulin resistance, so induction of IL-10 by Roseburia intestinalis, Bacteroides fragilis, Akkermansia muciniphila, Lactobacillus plantarum can contribute to improved glucose metabolism. IL-22 restores insulin sensitivity and alleviate diabetes and R.intestinalis can increase this anti-inflammatory cytokine production (Zhu et al., 2018).
Because of increased gut permeability in T2D patients, gut microbial products translocate in blood that leads to metabolic endotoxemia. Different gut microbiota have been reported to reduce gut permeability. One of such example of a probiotic bacteria is Akkermansia muciniphila, which decreases gut permeability using extracellular vesicles that improve intestinal tight junctions via AMPK activation in epithelium (Chelakkot et al., 2018).
Glucose metabolism is also influenced by gut microbiota. Bifidobacterium lactis affects glycogen synthesis. This potential probiotic also enhanced the translocation of glucose transporter-4 and insulin stimulated glucose uptake (Kim et al., 2014).
6. Gut microbiota as an alternate therapy for diabetes
Dysbiosis or dysbacteriosis is a microbial imbalance on or inside our body. A change in microbial dysbiosis can be achieved by supplementation of prebiotics and this can improve bifidobacterium abundance and as discussed before, bifidobacteria is positively associated with improved glucose tolerance (Fig. 4). Various weight loss surgeries like bariatric surgery alters the composition and diversity of the gut microbiota in humans, rats and mice significantly (Ryan et al., 2014, Tremaroli et al., 2015, Yang et al., 2016). Various researchers have suggested the strategies for the prevention of dysbiosis of the gut microbiota through supplementation of beneficial live bacteria (Hill et al., 2014). In another study conducted by Buchwald et al, it was noted that most of the patients after bariatric surgery had improved T2D manifestations. Probiotics when administered may provide health benefits to the host. One such example is in inflammatory bowel disease where probiotics provide those bacteria whose reduced abundance is linked with the disease such as F. prausnitzii, which is diminished in this disorder (Kesika et al., 2019). Probiotics related with diabetes that are mostly studied are from Bifidobacterium and Lactobacillus phyla. Among lactobacillus, L. rhamnosus, L. acidophilus, L. gasseri, and L. casei have anti-diabetic effects (Honda et al., 2012, Chen et al., 2014). Studies also indicate that administration of Bifidobacterium animalis, B. breve, and B. longum can improve glucose intolerance (Stenman et al., 2014). Few years back, Akkermansia muciniphila has also been studied to treat T2D, and this increases the intolerance of glucose in diabetic mice (De la Cuesta-Zuluaga et al., 2017).
Fig. 4.
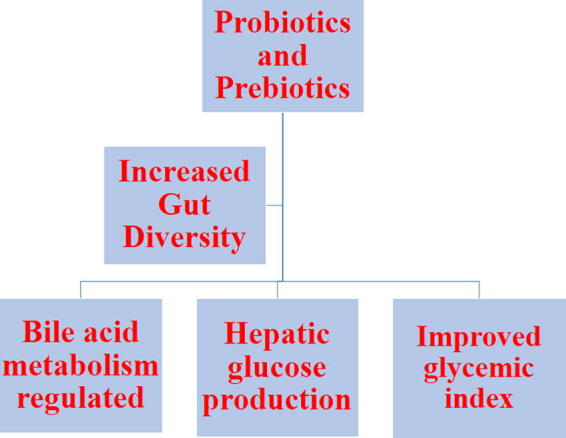
The potential of Microbiota as therapeutic alternative.
lipid profile, blood pressure, and other cardiovascular risks are related with T2D patients on probiotic supplementation (Kasińska and Drzewoski, 2015, Nikbakht et al., 2018). Probiotic supplements are known to have a control on the glycemic and inflammatory status of gestational diabetes (GDM) patients (Dallanora et al., 2018). By decreasing insulin resistance, probiotics regulate the glycemic condition and may decrease the development of inflammatory markers in people with T2D (Mazloom et al., 2013).
Fecal Microbiota Transplant (FMT) is one the advanced method that enhances sensitivity to insulin by influencing GM composition, hence alleviating diabetes. However, it demonstrates that FMT's beneficial effects are transient and highly dependent on the host's individual response (Wang et al., 2020). Ganesan K et al explained the beneficial results on gut microbiota pattern while transplantation of F. prausnitzii by restoring the structure and function of intestinal barrier. It may be used as a possible anti-inflammatory and anti-diabetic treatment (Ganesan et al., 2018).
A flavonoid, Quercitin, has been recently studied for its prebiotic capacity after testing on rats with induced diabetic peripheral neuropathy that showed that it affects intestinal dysbiosis by modulating the GM linked to the development of reactive oxygen species and the phenotypes of diabetic peripheral neuropathy (Xie et al., 2020).
Nowadays, new therapies like neuropathy and retinopathy that are associated with diabetes have gained importance. Such kinds of therapies may improve and help to diagnose the symptoms of patients.
7. Effect of diet on gut microbiota
Major components of human diet includes proteins, carbohydrates and fats and this diet influences the composition of gut microbiota in them. The degradation of proteins and carbohydrates results in end products like short chain fatty acids, acetate and butyrate in the intestinal gut tract. These metabolites have a major physiological impact on the health of the host. It is at the distal end of the colon where protein are degraded by proteolytic bacteria due to the conditions favorable for this bacteria. Ammonia, amino acids and amines are the end products of protein degradation. Malignant growths are related to high concentrations of ammonia (Hamer et al., 2012). Also, prebiotics are non-digestible food component that benefits the host by promoting microbiota development. Prebiotics are important for changing the composition of gut microbial communities and conferring several health benefits to the host. Few examples of prebiotics are insulin, fructooligosaccharides that function as essential stimulants for bacterial development (Schnorr et al., 2014). Use of prebiotics can improve the host’s health by altering the gut microbial composition.
8. Conclusion
According to various studies, intestinal dysbiosis is associated with the development of diabetes. Gut microbiota can offer new ways for intervention of the delay or arresting the process of diabetes. A broader global approach of analyzing gut microbiota may help us to understand the related pathogenic mechanisms of specific regulatory species. This may help to build tools for the prediction of the disease and develop strategies to modify or regulate the gut microbiota for therapeutic purposes. A very important point of interest is the energy metabolism which is the driving force in the pathogenesis of metabolic diseases and may promote T2D. Another approach is prebiotic supplementation which is very significant for the improvement of gut microbiota and body stability. For the long-term survival and proper functioning of the body, understanding the mechanism of association between diabetes and gut microbiota is mandatory.
Declaration of Competing Interest
The author declares that she has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The author thanks Imam Abdulrahman bin Faisal University for their support.
Footnotes
Peer review under responsibility of King Saud University.
References
- Adeshirlarijaney A., Gewirtz A.T. Considering gut microbiota in treatment of type 2 diabetes mellitus. Gut Microbes. 2020;11:253–264. doi: 10.1080/19490976.2020.1717719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, R., Kamikadi, k., Suda, W., Taki, H., Mikami, Y., Suganuma, N., Hattori, M., Koga, Y., 2017. A proliferative probiotic bifidobacterium strain in the gut ameliorates progression of metabolic disorders via microbiota modulation and acetate elevation. Sci Rep. 7, 43522. [DOI] [PMC free article] [PubMed]
- Aron-Wisnewsky J., Vigliotti C., Witjes J., Le P., Holleboom A.G., Verheij J., Nieuwdorp M., Clément K. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020;17(5):279–297. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- Balakumar M., Parbhu D., Sathishkumar C., Parbu P., Rokana N., Kumar R., Raghavan S., Soudaraijan A., Grover S., Batish V.K., Mohan V., Balsubramanyam M. Improvement in glucose tolerance and insulin sensitivity by probiotic strains of Indian gut origin in high-fat diet-fed C57BL/6J mice. Eur. J. Nutr. 2018;57(1):279–295. doi: 10.1007/s00394-016-1317-7. [DOI] [PubMed] [Google Scholar]
- Barengolts E., Green S.J., Eisenberg Y., Akbar A., Reddivari B., Layden T., Dugas L., Chlipala G. Gut microbiota varies by opioid use, circulating leptin and oxytocin in African American men with diabetes and high burden of chronic disease. PLoS ONE. 2018;13(3) doi: 10.1371/journal.pone.0194171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli E., Yan Y., Moldovan L., Vieira C.P., Gao R., Duan Y., Prasad R., Bhatwadekar A., White F.A., Townsend S.D., Chan L., Ryan C.N., Morton D., Moldovan E.G., Chu F., Oudit G.Y., Derendorf H., Adorini L., Wang X.X., Molina C.E., Mirmira R.G., Boulton M.E., Yoder M.C., Li Q., Levi M., Busik J.V., Grant M.B. Restructuring of the gut microbiome by intermittent fasting prevents retinopathy and prolongs survival in db/db mice. Diabetes. 2018;69:1867–1879. doi: 10.2337/db18-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.T., Davis-Richardson A.G., Giongo A., Gano K.A., Crabb D.B., Mukherjee N., Casella G., Drew J.V., Ilonen J., Knip M., Hyöty H., Veijola R., Simell T., Simell O., Neu J., Wasserfall C.H., Schatz D., Atkinson M.A., Triplett E.W. Gut microbiome metagenomics analysis suggests a functional model for development of autoimmunity for type 1 diabetes. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M., Biagi E., Soverini M., Consolandi C., Quercia S., Severgnini M., Peano C., Turroni S., Rampelli S., Pozzilli P., Pianesi M., Fallucca F., Brigidi P. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br. J. Nutr. 2016;116(1):80–93. doi: 10.1017/S0007114516001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Zhang Q., Dang H., Liu X., Tian F., Zhao J., Chen Y., Zhang H., Chen W. Antidiabetic effect of Lactobacillus casei CCFM0412 on mice with type 2 diabetes induced by a high-fat diet and streptozotocin. Nutrition. 2014;30(9):1061–1068. doi: 10.1016/j.nut.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Dallanora S., Medeiros de Souza Y., Deon R.G., Tracey C.A., Freitas-Vilela A.A., Roesch L.W., Mendes R.H. Do probiotics effectively ameliorate glycemic control during gestational diabetes? A systematic review. Arch. Gynecol. Obstet. 2018;298(3):477–485. doi: 10.1007/s00404-018-4809-2. [DOI] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., Biddinger S.B., Dutton R.J., Turnbough P.J. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cuesta-Zuluaga J., Mueller N.T., Corrales-Agudelo V., Velásquez-Mejía E.P., Carmona J.A., Abad J.M., Escobar J.S. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care. J. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Bridger J.M. FISH glossary: an overview of the fluorescence in situ hybridization technique. Biotechniques. 2018;45:4. doi: 10.2144/000112811. [DOI] [PubMed] [Google Scholar]
- Erdogan A., Rao S.S. Small intestinal fungal overgrowth. Curr. Gastroenterol. Rep. 2015;17(4):1–7. doi: 10.1007/s11894-015-0436-2. [DOI] [PubMed] [Google Scholar]
- Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nature Rev. Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- Forslund K., Hildebrand F., Nielsen T., Falony G., Chatelier E.L., Sunagawa S., Prifti E., Vieira-Silva S., Gudmundsdottir V., Pedersen H.K., Arumugam M., Kristiansen K., Voigt A.W., Vestergaard H., Hercog R., Costea P.I., Kultima J.R., Li J., Jørgensen T., Levenez F., Dore J., Consortium M.T., Nielsen H.B., Brunak S.B., Raes J., Hansen T., Wang J., Ehrlich S.D., Bork P., Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Zhu C., Li H., Yin M., Pan C., Huang L., Kong C., Wang X., Zhang Y., Qu S., Qin H. Dysbiosis signatures of gut microbiota along the sequence from healthy, young patients to those with overweight and obesity. Obesity (Silver Spring). 2018;26(2):351–361. doi: 10.1002/oby.22088. [DOI] [PubMed] [Google Scholar]
- Gou W., Ling C.W., He Y., Jiang Z., Fu Y., Xu F., Miao Z., Sun T.Y., Lin J.S., Zhu H.L., Zhou H., Chen Y.M., Zheng J.S. Interpretable Machine Learning Framework Reveals Robust Gut Microbiome Features Associated with Type 2 Diabetes. Diabetes Care. 2021 doi: 10.2337/dc20-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J., Qin Y., Zhong H., Zhang J., Licinio J., Wong M.L., Xu A., Chavakis T., Bornstein A.B., Ehrhart-Bornstein M., Lamounier-Zepter V., Lohmann T., Wolf T., Bornstein S.R. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2013;13(6):514–522. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- Gui J., Liu Q., Feng L. Metformin vs insulin in the management of gestational diabetes: a meta-analysis. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0064585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjo H., Geva-Zatorsky N. Gut microbiota–host interactions now also brain-immune axis. Curr. Opin. Neurobiol. 2020;62:53–59. doi: 10.1016/j.conb.2019.10.009. [DOI] [PubMed] [Google Scholar]
- He, Y., Wu, W., Zheng, H., Li, P., McDonald, D., Sheng, H., Chen, M., Chen, Z., Ji, G.Y., Xi, Zheng, Z.D.X., Mujagond, P., Chen, X.J., Rong, Z.H., Chen, P., Lyu, L.Y., Wang, X., Wu, C.B., Yu, Xu, Y.J., Yin, J., Raes, J., Knight, R.., Ma, W.J., Zhou, H.W., 2018. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 24(10), 1532–5. [DOI] [PubMed]
- Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., Calder P.C., Sanders M.E. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Dollive S., Grunberg S., Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D., Lewis J.D., Bushman F.D. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Moto M., Uchida N., He F., Hashizume N. Anti-diabetic effects of lactic acid bacteria in normal and type 2 diabetic mice. J Clin Biochem Nutr. 2012;51(2):96–101. doi: 10.3164/jcbn.11-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson., J.S., Spakowicz, D.J., Hong,, Petersen, L.M., Johnson, J.S., Demkowicz, P., Chen, L., Leopold, S.R., Hanson, B.M., Agresta, H.O., Gerstein, M., Sodergren, E., Weinstock, G.M., 2019. Evaluation of 16s rRNA gene sequencing for species and strain level microbiome analysis, Nature Communications. [DOI] [PMC free article] [PubMed]
- Kamada N., Seo S.U., Chen G.Y., Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kanbay M., Onal E.M., Afsar B., Dagel T., Yerlikaya A., Covic A., Vaziri N.D. The crosstalk of gut microbiota and chronic kidney disease: role of inflammation, proteinuria, hypertension, and diabetes mellitus. Int. Urol. Nephrol. 2018;50:1453–1466. doi: 10.1007/s11255-018-1873-2. [DOI] [PubMed] [Google Scholar]
- Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Kasińska M.A., Drzewoski J. Effectiveness of probiotics in type 2 diabetes: a meta-analysis. Polish Archives of Internal Medicine. 2015;125(11):803–813. doi: 10.20452/pamw.3156. [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Ben Othman M., Sakamoto K. Sterilized bifidobacteria suppressed fat accumulation and blood glucose level. Biochem. Biophys. Res. Commun. 2018;501(4):1041–1047. doi: 10.1016/j.bbrc.2018.05.105. [DOI] [PubMed] [Google Scholar]
- Kootte R.S., Vrieze A., Holleman F., Dallinga-Thie G.M., Zoetendal E.G., de Vos W.M., Groen A.K., Hoekstra J.B.L., Stroes E.S., Nieuwdorp M. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes. Metab. 2012;14:112–120. doi: 10.1111/j.1463-1326.2011.01483.x. [DOI] [PubMed] [Google Scholar]
- Lagier J.C., Million M., Hugon P., Armougom F., Raoult D. Human gut microbiota: repertoire and variations. Front. Cell. Infect. Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Vogensen F.K., Berg F.W.J., Nielsen D.S., Andreasen A.S., Pedersen B.K., Abu Al-Soud W., Sørensen S.J., Hansen L.H., Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier, E., Nielson, T., Quin, J., Prifti, E., Hildebrand, F., Falony, G., Almeida, M., Arumugam, M., Batto, J.M., Kennedy, S., Leonard, P., Li, J., Burgdorf, K., Grarup, N., Jørgensen, T., Brandslund, I., Nielsen, H.B., Juncker, A.J., Bertalan, M., Levenez, F., Pons, N., Rasmussen, S., Sunagawa, S., Tap, J., Tims, S., Zoetendal, E.G., Brunak, S., Clément, K., Doré, J., Kleerebezem, M., Kristiansen, K., Renault, P., Sicheritz-Ponten, T., De Vos, W.M., Zucker, J.D., Raes, J., Hansen, T., Consortium, MHIT., Bork, P., Wang, J., Ehrlich, S.D., Pedersen, O., 2013. Richness of human gut microbiome corellates with metabolic markers. Nature. 500, 541-6. [DOI] [PubMed]
- Lim, S.M., Jeong, J.J., Woo, K.H., Han, M.J., Kim, D.H., 2016. Lactobacillus sakei OK67 ameliorates high-fat diet-induced blood glucose intolerance and obesity in mice by inhibiting gut microbiota lipopolysaccharide production and inducing colon tight junction protein expression. Nutr Res 2016;36(4):337–48 [DOI] [PubMed]
- Lippert K., Kedenko L., Antonielli L., Kedenko I., Gemeier C., Leitner M., Kautzky-Willer A., Paulweber B., Hackl E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef Microbes. 2017;8(4):545–556. doi: 10.3920/BM2016.0184. [DOI] [PubMed] [Google Scholar]
- Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- Mafra D., Fouque D. Gut microbiota and inflammation in chronic kidney disease patients. Clin Kidney J. 2015;8:332–334. doi: 10.1093/ckj/sfv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik F., Mehdi S.F., Ali H., Patel P., Basharat A., Kumar A., Ashok F., Stein J., Brima W., Malhotra P., Roth J. Is metformin poised for a second career as an antimicrobial? Diabetes Metab Res Rev. 2018;34(4) doi: 10.1002/dmrr.2975. [DOI] [PubMed] [Google Scholar]
- Martinic A., Barouei J., Bendiks Z., Mishchuk D., Heeney D.D., Martin R., Marco M.L., Slupsky C.M. Supplementation of lactobacillus plantarum improves markers of metabolic dysfunction induced by a high fat diet. J. Proteome Res. 2018;17(8):2790–2802. doi: 10.1021/acs.jproteome.8b00282. [DOI] [PubMed] [Google Scholar]
- Mazloom Z., Yousefinejad A., Dabbaghmanesh M.H. Effect of probiotics on lipid profile, glycemic control, insulin action, oxidative stress, and inflammatory markers in patients with type 2 diabetes: a clinical trial. Iranian J. Med. Sci. 2013;38(1):38–43. [PMC free article] [PubMed] [Google Scholar]
- Mika A., Van Treuren W., González A., Herrera J.J., Knight R., Fleshner M. Exercise is more effective at altering gut microbial composition and producing stable changes in lean mass in juvenile versus adult male F344 rats. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0125889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murri M., Leiva I., Gomez-Zumaquero J.M., Tinahones F.J., Cardona F., Soriguer F., Queipo-Ortuño M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan X.C., Huttenhower C. Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology. 2014;146:1437–1448. doi: 10.1053/j.gastro.2014.01.049. [DOI] [PubMed] [Google Scholar]
- Murphy R., Tsai P., Jüllig M., Liu A., Plank L., Booth M. Differential changes in gut microbiota after gastric bypass and sleeve gastrectomy bariatric surgery vary according to diabetes remission. Obes. Surg. 2017;27(4):917–925. doi: 10.1007/s11695-016-2399-2. [DOI] [PubMed] [Google Scholar]
- Needell J.C., Zipris D. The role of the intestinal microbiome in type 1 diabetes pathogenesis. Curr Diab Rep. 2016;16:89. doi: 10.1007/s11892-016-0781-z. [DOI] [PubMed] [Google Scholar]
- Suárez-Zamorano Nicolas. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015;21:1497–1501. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen D.S., Krych L., Buschard K., Hansen C.H.F., Hansen A.X. Beyond genetics. Influence of dietary factors and gut microbiota on type 1 diabetes. FEBS Lett. 2014;588(22):4234–4243. doi: 10.1016/j.febslet.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Nikbakht E., Khalesi S., Singh I., Williams L.T., West N.P., Colson N. Effect of probiotics and synbiotics on blood glucose: a systematic review and meta-analysis of controlled trials. Eur. J. Nutr. 2018;57(1):95–106. doi: 10.1007/s00394-016-1300-3. [DOI] [PubMed] [Google Scholar]
- Palau-Rodriguez, M., Tulipani, S., Isabel Queipo-Ortũno, M., Urpi Sarda, M., Tinahones, F.J., Andres-Lacueva, C., 2015. Metabolomics Insights into the intricate gut microbial---host interaction in the development of obesity and type 2 diabetes. Front Microbiol. 6, 1151. [DOI] [PMC free article] [PubMed]
- Park D.Y., Ahn Y.T., Park S.H., Huh C.S., Yoo S.R., Yu R., Sung M.K., McGregor R.A., Choi M.S. Supplementation of lactobacillus curvatus HY7601 and lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0059470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A., Marchesi N., Marelli C., Coppola A., Luzi L., Govoni S., Giustina A., Gazzaruso C. Microbiota and metabolic diseases. Endocrine. 2018;61:357–371. doi: 10.1007/s12020-018-1605-5. [DOI] [PubMed] [Google Scholar]
- Pedersen C., Gallagher E., Horton F., Ellis R.J., Ijaz U.Z., Wu H., Jaiyeola E., Diribe O., Duparc T., Cani P.D., Gibson G.R., Hinton P., Wright J., Ragione R.L., Robertson M.L. Host-microbiome interactions in human type 2 diabetes following prebiotic fibre (galacto-oligosaccharide) intake. Br. J. Nutr. 2016;116:1869–1877. doi: 10.1017/S0007114516004086. [DOI] [PubMed] [Google Scholar]
- Kesika P., Sivamaruthi S., Chaiyasut C. FDo Probiotics Improve the Health Status of Individuals with Diabetes Mellitus? A Review on Outcomes of Clinical Trials. Biomed. Res. Int. 2019 doi: 10.1155/2019/1531567. 1531567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pussinen P.J., Havulinna A.K., Lehto M., Sundvall J., Salomaa V. Endotoxemia Is Associated With an Increased Risk of Incident Diabetes. Diabetes Care. 2011;34(2):392–397. doi: 10.2337/dc10-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D., Peng Y., Zhang D., Jie Z., Wu W., Qin Y., Xue Y., Li J., Han L., Lu D., Wu P., Dai Y., Sun X., Li Z., Tang A., Zhong S., Li X., Chen W., Xu R., Wang M., Feng Q., Gong M., Yu J., Zhang Y., Zhang M., Hansen T., Sanchez G., Raes J., Falony G., Okuda S., Almeida M., LeChatelier E., Renault P., Pons N., Batto J.M., Zhang Z., Chen H., Yang R., Zheng W., Li S., Yang H., Wang J., Ehrlich S.D., Nielsen R., Pedersen O., Kristiansen K., Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Rajendhran J., Gunasekaran P. Microbial phylogeny and diversity: Small subunit ribosomal RNA sequence analysis and beyond. Microbiol. Res. 2011;166:99–110. doi: 10.1016/j.micres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Das, S., Dash, H.R, 2019. Microbial Diversity in the Genomic Era Book, 679-699
- Ringel-Kulka, T., Cheng. J., Ringel, Y., Salojärvi, J., Carroll, I., Palva, A., de Vos, W.M., Satokari, R. Intestinal microbiota in healthy U.S. young children and adults--a high A High Throughput Microarray Analysis. 2013. PLoS ONE. 8(5), e64315 [DOI] [PMC free article] [PubMed]
- Ryan K.K., Tremaroli V., Clemmensen C., Kovatcheva-Datchary P., Myronovych A., Karns R., Wilson-Pérez H.E., Sandoval D.A., Kohli R., Bäckhed F., Seeley R.J. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabit H., Tombuloglu H., Rehman S., Almandil N.B., Cevik E., Abdel-Ghany S., Rashwan S., Abasiyanik M.F., Waye M.M. Gut microbiota metabolites in autistic children: An epigenetic perspective. Heliyon. 2021;7(1) doi: 10.1016/j.heliyon.2021.e06105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Ogasawara N., Funaki Y., Mizuno M., Iida A., Goto C., Koikeda S., Kasugai K., Joh T. Transglucosidase improves the gut microbiota profile of type 2 diabetes mellitus patients: a randomized double-blind, placebo-controlled study. BMC Gastroenterol. 2013;13:81. doi: 10.1186/1471-230X-13-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighi M., Razavi S., Navab-Moghadam F., Khamseh M.E., Alaei-Shahmiri F., Mehrtash A., Amirmozafari N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017;111:362–369. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- Singhvi N., Gupta V., Gaur M., Sharma V., Puri A., Singh Y., Dubey G.P., Lal R. Interplay of Human Gut Microbiome in Health and Wellness. Indian J Microbiol. 2020;60(1):26–36. doi: 10.1007/s12088-019-00825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman L.K., Waget A., Garret C., Klopp P., Burcelin R., Lahtinen S. Potential probiotic Bifidobacterium animalis spp. lactis 420 prevents weight gain and glucose intolerance in diet-induced obese mice. Benef Microbes. 2014;5(4):437–445. doi: 10.3920/BM2014.0014. [DOI] [PubMed] [Google Scholar]
- Su B., Liu H., Li J., Sunli Y., Liu B., Liu D., Zhang P., Meng X. Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J. Diabetes. 2015;7(5):729–739. doi: 10.1111/1753-0407.12232. [DOI] [PubMed] [Google Scholar]
- Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., Liu J., Deng Y., Xia J., Chen B., Zhang S., Yun S., Lian G., Zhang X., Zhang H., Bisson W.H., Shi J., Gao X., Ge P., Liu C., Krausz K.W., Nichols R.G., Cai J., Rimal B., Patterson A.D., Wang X., Gonzalez F.J., Jiang C. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018;24(12):1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.H., Kitai T., Hazen S.L. Gut microbiota in cardiovascular health and disease. Circ. Res. 2017;120:1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V., Karlsson F., Werling M., Ståhlman M., Kovatcheva-Datchary P., Olbers T., Fändriks L., le Roux C.W., Nielsen J., Bäckhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala O. Human intestinal microbiota and type 1 diabetes. Curr Diab Rep. 2013;13(5):601–607. doi: 10.1007/s11892-013-0409-5. [DOI] [PubMed] [Google Scholar]
- Velasquez-Manoff M. Gut microbiome: the peacekeepers. Nature. 2015;518:S3–S11. doi: 10.1038/518S3a. [DOI] [PubMed] [Google Scholar]
- Wu H., Esteve E., Tremaroli V., Khan M.T., Caesar R., Mannerås-Holm L., Ståhlman M., Olsson L.M., Serino M., Planas-Fèlix M., Xifra G., Mercader J.M., Torrents D., Burcelin R., Ricart R., Perkins R., Fernàndez-Real J.M., Bäckhed F. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- Wu X., Ma C., Han L., Nawaz M., Gao F., Zhang X., Yu P., Zhao C., Li L., Zhou A., Wang J., Moore J.E., Millar B.C., Xu J. Molecular characterization of the faecal microbiota in patients with type II diabetes. Curr. Microbiol. 2010;61(1):69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- Wu J., Wu J., Boström P., Sparks L.M., Ye L., Choi J.H., Giang A.H., Khandekar M., Virtanen K.A., Nuutila P., Schaart G., Huang K., Tu H., Lichtenbelt W.D.M., Hoeks J., Enerbäck S., Schrauwen P., Spiegelman B.M. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xu J., Lian F., Zhao L., Zhao Y., Chen X., Zhang X., Guo Y., Zhang C., Zhou Q., Xue Z., Pang X., Zhao L., Tong X. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015;9(3):552–562. doi: 10.1038/ismej.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Adachi K., Sugiyama T., Shimozato A., Ebi M., Ogasawara N., Funaki Y., Goto C., Sasaki M., Kasugai K. Association of intestinal microbiota with metabolic markers and dietary habits in patients with type 2 diabetes. Digestion. 2016;94(2):66–72. doi: 10.1159/000447690. [DOI] [PubMed] [Google Scholar]
- Yang P.J., Yang W.S., Nien H.C., Yang P.J., Yang W.S., Nien H.C., Chen C.N., Lee C.M. Duodenojejunal bypass leads to altered gut microbiota and strengthened epithelial barriers in rats. Obes. Surg. 2016;26:1576–1583. doi: 10.1007/s11695-015-1968-0. [DOI] [PubMed] [Google Scholar]
- Yang Y., Torchinsky M.B., Gobert M., Xiong H., Xu M., Linehan F., Alonzo J.L., Ng C., Chen A., Lin X., Sczesnak A., Liao J.J., Torres V.J., Jenkins M.K., Lafaille J.J., Littman D.R. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510(7503):152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., Chervonsky A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shen D., Fang Z., Jie Z., Qiu X., Zhang C., Chen Y., Ji L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Fang Z., Zhang C., Xia H., Jie Z., Han X., Chen Y., Ji L. Effects of acarbose on the gut microbiota of prediabetic patients: a randomized, double-blind, controlled crossover trial. Diabetes Ther. 2017;8(2):293–307. doi: 10.1007/s13300-017-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P.C., Lau, S.K., Ada, W.C., Lin, Curreem, S.O.T., Fung, A.M.Y., Yuen, K.w., 2007. Surgical site abscess caused by Lactobacillus fermentum identified by 16S ribosomal RNA gene sequencing. Diagn Microbiol Infect Dis. 58, 251-254 [DOI] [PubMed]
- Khaled R.A., Rehman S., Alnimr A., Diab A., Hawwari A., Tokajian S. Molecular typing of MRSA isolates by spa and PFGE, Journal of King Saud University –. Science. 2019;31(4):999–1004. [Google Scholar]
- Wang H., Lu Y., Yan Y., Tian S., Zheng D., Leng D., Wang C., Jiao J., Wang Z., Bai Y. Promising Treatment for Type 2 Diabetes: Fecal Microbiota Transplantation Reverses Insulin Resistance and Impaired Islets. Front. Cell. Infect. Microbiol. 2020;9:455. doi: 10.3389/fcimb.2019.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan K., Chung S.K., Vanamala J., Xu B. Causal Relationship between Diet-Induced Gut Microbiota Changes and Diabetes: A Novel Strategy to Transplant Faecalibacterium prausnitzii in Preventing Diabetes. Int. J. Mol. Sci. 2018;19:3720. doi: 10.3390/ijms19123720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Song W., Liang X., Zhang Q., Shi Y., Liu W., Shi X. Protective Effect of Quercetin on Streptozotocin-Induced Diabetic Peripheral Neuropathy Rats through Modulating Gut Microbiota and Reactive Oxygen Species Level. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110147. [DOI] [PubMed] [Google Scholar]
- Hamer H.M., De Preter V., Windey K., Verbeke K. Functional analysis of colonic bacterial metabolism: relevant to health? Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G1–G9. doi: 10.1152/ajpgi.00048.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdeviren S., Dagdeviren S., Jung D.Y., Friedline R.H., Noh H.L., Kim J.H., Patel P.R., Tsitsilianos N., Inashima K., Tran D.A., Hu X., Loubato M.M., Craige S.M., Kwon J.W., Lee K.W., Kim J.K. IL-10 prevents aging-associated inflammation and insulin resistance in skeletal muscle. FASEB J. 2017;31(2):701–710. doi: 10.1096/fj.201600832R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr S.L., Candela M., Rampelli S., Centanni M., Consolandi C., Basaglia G., Turroni S., Biagi E., Peano C., Severgnini M., Fiori J., Gotti R., Bellis G.D., Luiselli D., Brigidi P., Mabulla A., Marlowe F., Henry A.G., Crittenden A.N. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Song K., Shen Z., Quan Y., Tan B., Luo W., Wu S., Tang K., Yang Z., Wang X. Roseburia intestinalis inhibits interleukin17 excretion and promotes regulatory T cells differentiation in colitis. Mol. Med. Rep. 2018;17(6):7567–7574. doi: 10.3892/mmr.2018.8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelakkot C., Choi Y., Kim D.K., Park H.T., Ghim J., Kwon Y., Jeon J., Kim M.S., Jee Y.K., Gho Y.S., Park H.S., Kim Y.K., Ryu S.H. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018;50(2) doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Huh C.S., Choi I.D., Jeong J.W., Ku H., Ra J.H., Kim T.Y., Kim G.B., Sim J.H., Ahn Y.T. The anti-diabetic activity of bifidobacterium lactis HY8101 in vitro and in vivo. J. Appl. Microbiol. 2014;117(3):834–845. doi: 10.1111/jam.12573. [DOI] [PubMed] [Google Scholar]