Abstract
In vertebrates, the RNA-binding protein (RBP) dead end 1 (DND1) is essential for primordial germ cell (PGC) survival and maintenance of cell identity. In multiple species, Dnd1 loss or mutation leads to severe PGC loss soon after specification or, in some species, germ cell transformation to somatic lineages. Our investigations into the role of DND1 in PGC specification and differentiation have been limited by the absence of an available antibody. To address this problem, we used CRISPR/Cas9 gene editing to establish a transgenic mouse line carrying a DND1GFP fusion allele. We present imaging analysis of DND1GFP expression showing that DND1GFP expression is heterogeneous among male germ cells (MGCs) and female germ cells (FGCs). DND1GFP was detected in MGCs throughout fetal life but lost from FGCs at meiotic entry. In postnatal and adult testes, DND1GFP expression correlated with classic markers for the premeiotic spermatogonial population. Utilizing the GFP tag for RNA immunoprecipitation (RIP) analysis in MGCs validated this transgenic as a tool for identifying in vivo transcript targets of DND1. The DND1GFP mouse line is a novel tool for isolation and analysis of embryonic and fetal germ cells, and the spermatogonial population of the postnatal and adult testis.
Keywords: RNA immunoprecipitation, male germ cell, dead end 1, RNA-binding protein, spermatogonia, meiosis
Characterization of this novel transgenic mouse showed that DND1GFP is expressed throughout the pre-meiotic germline, supports growing evidence in the field of germ cell heterogeneity, and demonstrated the tagged allele can be used for identification of DND1 targets.
Introduction
RNA-binding proteins (RBPs) play a critical role in post transcriptional regulation, particularly in the germ line [1, 2]. Regulatory roles include transcript trafficking, sequestration, and degradation [3]. Norman Hecht pioneered work on the role of RBPs during spermatogenesis [4] followed by many groups, who have contributed to our understanding of the requirement of RBPs for development of spermatogonial stem cells (SSCs) into sperm in the adult testis, including polypyrimidine tract-binding protein 2 (PTBP2) [5], translin (TSN) [6], Y box protein 2 aka MSY2 (YBX2) [7], Pumilio RNA-binding family member 1 (PUM1) [8], and nanos C2HC-type zinc finger 2 (NANOS2) [9]. In addition, there is a substantial body of work in the mouse supporting the vital role of RBPs in specification of primordial germ cells (PGCs) and regulation of cell identity transitions during differentiation into prospermatogonia including NANOS2 [10], nanos C2HC-type zinc finger 3 (NANOS3) [11], deleted in azoospermia-like (DAZL) [12], and dead end 1 (DND1) [13].
Dnd1 is a member of this group of germ cell–associated RBPs that are critical for germ cell survival and development in mouse [14–16] and other vertebrates, including zebrafish [17, 18], Atlantic salmon [19], rat [20], and frog [21, 22]. Dnd1 belongs to a family of RBPs that have two tandem RNA recognition motifs (RRMs) followed by a double-stranded recognition motif (DSRM or dsRBD) [23]. The structure of these domains determines the function of DND1 in germ cells. DND1 has been shown to prevent microRNA-mediated transcript degradation by protectively binding to transcript targets [24]. Conversely, in association with another RBP, NANOS2, DND1 has been shown to transport targets to P-bodies for degradation via the CCR4-NOT complex [10, 15, 25].
Loss of Dnd in zebrafish results in the transdifferentiation of PGCs to other lineages during their migration to the gonad [17], suggesting that Dnd protein plays a critical role, at least during this period, in maintaining PGC identity. Some of the most compelling evidence for the requirement of DND1 in murine prospermatogonial development comes from studies of the classic Ter mutation (Dnd1Ter) in mice [26–30]. The Ter mutation is a single nucleotide polymorphism (SNP) in exon 2 of Dnd1 that encodes a premature stop, expected to truncate DND1 [13]. When the Dnd1Ter mutation is present, in the context of the 129 strain of mice, males have a higher incidence of testicular teratomas that arise as early as E15.5 [31–33]. Work by multiple groups has shown that teratoma formation is correlated with a failure of germ cells to enter cell cycle arrest, silence expression of pluripotency genes, and activate the male differentiation pathway [28–30, 34]. Our recent work demonstrated that Ter mutant germ cells also fail to initiate expression of many chromatin modifiers, which may be required for the epigenetic changes associated with differentiation and stabilization of prospermatogonial fate [26, 34]. Studies of Dnd1Ter have provided valuable insight into stem cell dynamics during development and served as a model for understanding testicular germ cell tumors (TGCT), which account for about 95% of the testicular cancers that affect about 10 000 men and boys per year in the US [35–38].
A recent structural analysis of DND1 uncovered a mechanistic novelty governed by the proximity of the tandem RRMs (RRM1 and RRM2) (Supplementary Figure S1) [23]. The novel RRM structure results in a powerful 2-step grab and clamp mechanism for target transcript binding. The SNP that defines the Ter mutation occurs in the RRM2 sequence of Dnd1 exon 2. Loss of the clamp function of DND1 may partially explain the phenotype of the Dnd1Ter/Ter mutants. However, there are still many unanswered questions about the normal role of DND1 in the germ line. The absence of good antibodies against DND1 has impeded progress on defining the role of functional DND1 in germ cells.
To characterize the role of functional DND1 in murine germ cell development, we used CRISPR/Cas9 technology to produce a transgenic DND1GFP fusion allele. We used this mouse line to characterize the expression of DND1 throughout murine male and female germ line development and to perform RNA immunoprecipitation (RIP) experiments followed by RT-qPCR to validate in vivo some of the in vitro targets previously reported by us and others [25, 26, 29, 34, 39, 40]. Here, we show that DND1GFP expression begins embryonically during PGC specification and migration. DND1GFP is expressed in a heterogeneous pattern in the germ line until meiotic entry of fetal female germ cells (FGCs), and postnatal and adult male germ cells (MGCs). RIP–RT–qPCR experiments show that the DND1GFP fusion allele can be used to immunoprecipitate DND1 in vivo with its bound mRNA targets. Collectively, these findings indicate this new transgenic line will be a valuable tool for the community.
Materials and methods
Generation of transgenics
Donor plasmid construction
5′ and 3′ homology arms were PCR amplified from mouse genomic DNA (gDNA, donor plasmid primers listed in Table 1). Enhanced green fluorescent protein (EGFP) cDNA was PCR amplified from a pEGFP plasmid (Dr. Terry Lechler) (EGFP amplification primers listed in Table 1). 5′ homology arm, 3′ homology arm, and EGFP PCR fragments were subcloned into pL452 (Frederick Cancer Institute) at the EcoRI and BamHI sites by In-Fusion cloning (Takara Bio, catalog #638909).
Table 1.
Transgene primers.
| Primer name | Orientation | Sequence |
|---|---|---|
| Donor Plasmid 5′ Homology Arm | FOR | CCA AGC TAT CGA ATT CTG TGG CAT CTG CAC AGA AG |
| Donor Plasmid 5′ Homology Arm | REV | CTT GCT CAC CAT GGTAGC TCT TTC CAA AGT CTT ACA G |
| Donor Plasmid 3′ Homology Arm | FOR | GAC GAG CTG TAC AAG ATG CAG TCA AAA CGG GAG TG |
| Donor Plasmid 3′ Homology Arm | REV | CCG CTC TAG AAC TAG TCC ATT GCG GCC ACT TTC TG |
| EGFP cDNA from pEGFP | FOR | ACC ATG GTG AGC AAG GGC GAG |
| EGFP cDNA from pEGFP | REV | CTT GTA CAG CTC GTC CAT G |
| General eGFP insertion | FOR | GTT TCC AGT GTC CAT TC |
| General eGFP insertion | REV | CCT CCA GGG CTG CCT TG |
| Allele-specific insertion | FOR | CTG AGC AAA GAC CCC AAC GAG AAG |
| Allele-specific insertion | REV.1 | CCT GCC AAA CCG AGT GGT AGG TAC |
| Allele-specific insertion | REV.2 | CAC TCT TCT CTG CTC AGG GCT GTG |
| Southern Blot Probe Template | FOR | GAT CTC CTT TGA CCT GAG CCT TGC |
| Southern Blot Probe Template | REV | CCT CTA ATC TTT GCT CTA CAA TGG |
Pronuclear injection and embryo transfer
Standard microinjection methods were used [41]. Briefly, single-guide RNA (sgRNA) was synthesized by in vitro transcription (Supplementary Figure S1C). The microinjection reagent was generated by combining sgRNA (5 ng/μL), Cas9 mRNA (10 ng/μL) (TriLink, catalog #L-7606), and donor plasmid (5 ng/μL). Reagents were injected into the pronucleus of fertilized F2 mouse embryos from the B6SJLF1/J strain, which was purchased from Jax (stock# 100012). Injected embryos were transferred into the oviducts of surrogate females at the one-cell stage. Surrogates were mated with vasectomized males the night prior to embryo transfer. Surrogate mothers delivered and raised their offspring. Biopsies were collected from progeny before weaning to identify founders.
Purification of genomic DNA to verify insertion
Progeny gDNA was purified from tail biopsies. PCR on genomic DNA was performed using LA Taq DNA polymerase (TaKaRa) with EGFP primers to detect EGFP insertion (Supplementary Figure S1A and B, general EGFP insertion primers listed in Table 1). Allele-specific PCR was performed on founder gDNAs to confirm insertion of EGFP specifically in the Dnd1 locus (Supplementary Figure S1D, allele-specific primers listed in Table 1). Southern analysis was performed on 10 μg of PstI-digested gDNA to verify copy number of the insertion. The 5′ probe template was synthesized by PCR amplification of mouse gDNA (Southern blot primers listed in Table 1). The map of the EGFP insertion into Dnd1 locus is included as a SnapGene file in the supplements to this paper.
Genotyping
PCR was performed in a 10-μL reaction mixture containing ready-to-use Taq polymerase master mix (MyTaq Red Mix, BIOLINE BIO-25043), water, primers, and gDNA isolated from mouse ear biopsies. The primers for routine detection of the EGFP insertion into the Dnd1 locus are listed in Table 2. The reaction resulted in a 963 bp amplicon for mutants carrying the EGFP insertion in the Dnd1 locus and a 244 bp amplicon for the wild-type Dnd1 allele. The primer pair distinguished between wild type, heterozygous, and homozygous mutants (Supplementary Figure S1E). The PCR program was as follows: initial denaturation for 3 min at 95 °C followed by 35 cycles of 30 s at 95 °C, 30 s at 60 °C, and 60 s at 72 °C and a final extension of 5 min at 72 °C. PCR products were run on 1.5% agarose gel for 30 min at 150 V for detection.
Table 2.
Genotyping primers.
| Genotyping | Orientation | Sequence |
|---|---|---|
| Dnd1-Gfp | FOR | GGA ATC TTC CAG AGG CAG AA |
| Dnd1-Gfp | REV | TTC ACC CTC TCA CAC CAT TG |
Colony maintenance and timed matings
All mice were housed in accordance with National Institutes of Health (NIH) guidelines, and experiments were conducted with the approval of the Duke University Medical Center (DUMC) Institutional Animal Care and Use Committee (IACUC). B6SJLF1/J strain founders (JAX stock# 100012) were bred with CD1 mice (Charles River Labs strain code 022). Heterozygous progeny were bred to produce homozygous offspring. Homozygous offspring were intercrossed to maintain a homozygous hybrid strain colony (DND1EGFP/EGFP, hereafter written simply as DND1GFP). Approximately every six generations, homozygous males were outbred with CD1 females. New heterozygous progeny were then inbred to restore a homozygous hybrid strain colony. For timed matings, homozygotes were intercrossed, and females were checked for plugs and staged as day E0.5 if positive.
Antibodies
Listed in Table 3.
Table 3.
Antibodies.
| Against | Raised in | Conjugate | [Conc] | Vendor | Catalog # |
|---|---|---|---|---|---|
| GFP | Chicken | - | 1:5000 | Abcam | ab13970 |
| Chicken | Donkey | AF488 | 1:1000 | Jackson ImmunoResearch | 703-545-155 |
| Goat | Donkey | AF647 | 1:1000 | Invitrogen Thermo Fisher Sci | A21447 |
| Rabbit | Donkey | AF647 | 1:1000 | Jackson ImmunoResearch | 711-605-152 |
| Rabbit | Donkey | Cy3 | 1:1000 | Jackson ImmunoResearch | 711-165-152 |
| Rat | Donkey | AF488 | 1:1000 | Invitrogen Thermo Fisher Sci | A21208 |
| Rat | Donkey | Cy3 | 1:1000 | Jackson ImmunoResearch | 712-165-153 |
| cKIT | Goat | - | 1:100 | R&D Systems | AF1356 |
| FOXL2 | Goat | - | 1:250 | Novus Biologicals | NB100-1277 |
| GATA4 | Goat | - | 1:500 | Santa Cruz | sc-1237 |
| GFRa1 | Goat | - | 1:100 | R&D Systems | AF560 |
| DCP2 | Rabbit | - | 1:300 | Millipore | ABE2901 |
| DDX4 | Rabbit | - | 1:500 | Abcam | ab13840 |
| Nanog (Mouse) | Rabbit | - | 1:200 | Repro Cell Inc. | RCAB0002P-F |
| SOX2 | Rabbit | - | 1:1000 | Chemicon/EMD Millipore | AB5603 |
| SOX9 | Rabbit | - | 1:2000 | Millipore | AB5535 |
| STRA8 | Rabbit | - | 1:1000 | Michael Giswold Lab, WSU | [1] |
| E-cadherin | Rat | - | 1:500 | Zymed Laboratories | 13-1900 |
| Endomucin | Rat | - | 1:250 | Santa Cruz | sc-65495 |
| TRA98 | Rat | - | 1:1000 | Abcam | ab82527 |
| GFP | Rabbit | - | 1:1000 | Invitrogen Thermo Fisher Sci | A-6455 |
| GAPDH | Goat | - | 1:2500 | R&D Systems | AF5718 |
| Chicken | Donkey | HRP | 1:10000 | Jackson ImmunoResearch | 703-035-155 |
| Goat | Chicken | HRP | 1:50000 | R&D Systems | HAF019 |
Whole mount immunofluorescence
Whole mount immunofluorescence was performed for stages E7.5–E9.5 as described in [42] and for stages E11.5–E18.5 as described in [43, 44]. For E7.5–E9.5, whole embryos were collected and fixed in 4% paraformaldehyde while rocking for 30 min at room temperature. For E11.5–E18.5, embryonic gonads were dissected from embryos and fixed in the same manner. All samples were washed in PBS for 30–60 min, rocked for 15 min at room temperature at each step of the dehydration gradient (25%, 50%, 75%, and 100% methanol in PBS), and then stored in 100% fresh methanol at −20 °C. Prior to immunofluorescent staining, all samples were incubated at room temperature while rocking for 15 min at each step of the rehydration gradient (75%, 50%, and 25% methanol in PBS) and washed twice in PBS for 15 min each. Samples were incubated rocking at room temperature in a permeabilizing solution for 1 h followed by incubation in blocking solution for 1 h and then transferred to 4 °C in blocking solution with primary antibodies overnight. The following day, samples were washed by rocking three times in wash solution at room temperature for 30 min each and then incubated at 4 °C in blocking solution with secondary antibodies and Hoechst overnight. On day 3, samples were washed rocking three times in wash solution at room temperature for 30 min each, mounted in polyvinyl alcohol (PVA aka Mowiol) mounting medium, and cover-slipped for imaging on an SP8 Leica DM6000CS upright confocal microscope with affiliated Leica software (LAS AF 3). All tile stitching was done with Leica software, and remaining image processing was done with FIJI (version 2.1.0/1.53c) and Adobe Photoshop (version 22.0.0). Solutions for gonads E14.5–E18.5 are as follows: permeabilizing (2% Triton X-100 in PBS); blocking (1% Triton X-100, 3% BSA, and 10% Horse Serum in PBS); washing (1% Triton X-100 in PBS). Solutions for embryos at E7.5–E9.5 and gonads E11.5–E13.5 are as follows: permeabilizing (0.1% Triton X-100 in PBS); blocking (0.1% Triton X-100 in 3% BSA, and 10% Horse Serum in PBS); washing (0.1% Triton X-100 in PBS).
Cryosection immunofluorescence
Tissue preparation for cryosectioning and immunofluorescent staining was performed as described previously with some modifications [44, 45]. Briefly, postnatal testes at P0, P3, P7, and P10 were dissected from neonates and fixed in 4% paraformaldehyde rocking at 4 °C overnight. Adult ovaries were dissected and fixed in 4% paraformaldehyde rocking at 4 °C overnight. Adult testes were dissected and fixed (unpunctured) in 4% paraformaldehyde rocking at 4 °C overnight. All samples were washed rocking in PBS for 1 h and taken through a 4-step sucrose gradient (5%, 10%, 20%, and 50% sucrose in PBS). Samples were embedded in OCT and stored at −80 °C. After at least 24 h of storage at −80 °C, samples were sectioned at 8 μm (testes) or 20 μm (ovaries) and transferred to slides. For staining, boundaries were drawn around samples using an ImmEdge pen (Vector Laboratories, catalog #H-4000) to contain solutions. Samples were incubated at room temperature in blocking solution (0.1% Triton X-100, 3% BSA, and 10% Horse Serum in PBS) for 1 h, incubated at 4 °C in blocking solution with primary antibodies overnight, washed (washing solution: 0.1% Triton X-100, 3% BSA, and 1% Horse Serum in PBS) at room temperature 3 times for 10 min each, incubated at room temperature in blocking solution with secondary antibody and Hoechst for 1 h, and washed at room temperature 3 times for 10 min each. Samples were mounted in PVA and cover-slipped for imaging on an SP8 Leica DM6000CS upright confocal microscope with affiliated Leica software (LAS AF 3). All tile stitching was done with Leica software, and remaining image processing was done with FIJI (version 2.1.0/1.53c) and Adobe Photoshop (version 22.0.0).
RT-qPCR primers
Listed in Table 4.
Table 4.
RT-qPCR primers.
| Gene name | FOR | REV | Ref. |
|---|---|---|---|
| Canx | GAC ATG ACT CCT CCT GTA AAC CCT | CGT CCA TAT CCT CAT CCC AAT CCT | [2] |
| Ccne1 | GCC CTC TGA CCA TTG TGT CC | GCA CCA CTG ATA ACC TGA GAC CT | [3] |
| Ddx4 | TAC TGT CAG ACG CTC AAC AGG A | ATT CAA CGT GTG CTT GCC CT | [3] |
| Dnd1 | GCC CTG GTA GAA GGT CAG TCA C | GCC CTG TTC CTA AAC ACT TGG TC | [3] |
| eGfp | CTT CTT CAA GTC CGC CAT GCC | GGT GTT CTG CTG GTA GTG GTC | [3] |
| Gapdh | GGT GAA GGT CGG TGT GAA CG | CTC GCT CCT GGA AGA TGG TG | [3] |
| Gata4 | AAA CGG AAG CCC AAG AAC CT | GCC CAT AGT GAG ATG ACA GCC | [4] |
| Kat7 | GAT GCC CGC TGT ATC ATA ACC | TCC TTA TAC CGC AGC TGT CTC TCT | [5] |
| Lefty2 | CGT GCA ATG TGC AGA AGC AGA A | AAT GAC ATG GGC AAA GCT GCC A | [6] |
| Oct4 | GGA GGA AGC CGA CAA CAA TGA | TCC ACC TCA CAC GGT TCT CAA | [3] |
| Ret | ATG GAG AGG CCA GAC AAC TG | GTG AGT CCG AAG GTG TGG AT | [7] |
| Sox9 | TCC AGC AAG AAC AAG CCA CAC | TCT CGT TCA GCA GCC TCC AG | [4] |
RNA immunoprecipitation, RNA extraction, and RT-qPCR (RIP–RT–qPCR)
RIP experiments were performed as previously described with adjustments to account for the very low input inherent in experiments with fetal testes [46, 47]. Briefly, timed matings between DND1GFP homozygotes were established. Fetal testes at either E14.5 or E18.5 were dissected away from the mesonephros and collected. Pooled testes (6–16 testes total for a given biological replicate) were washed in DEPC PBS and incubated at 37 °C in fresh clean Gibco TrypLE (Thermo Fisher Scientific, catalog #12563029) for 7 or 10 min depending on the stage (E14.5 or E18.5, respectively). TrypLE was inactivated with ice cold DEPC PBS and aspirated off testes. Testes were then resuspended in ice cold DEPC PBS and pipetted vigorously to dissociate cells. Cells were pipetted through a 0.32-μm cell strainer into a fresh Eppendorf tube and pelleted by 4 °C centrifugation at 1000 x g for 10 min. The cell pellet was resuspended in 20-μL complete Poly Lysis Buffer (PLB) [47], incubated on ice for 5 min, and stored at −80 °C for at least 24 h. PLB cell suspension was then thawed on ice and cellular debris was pelleted by 4 °C centrifugation at 21 000 x g for 10 min. After centrifugation, 3 μL of supernatant was collected for RNA extraction as input sample. Remaining 17 μL of supernatant was used for immunoprecipitation via incubation at 4 °C on a tube rotator in NT2 Buffer [47] with washed GFP-trap magnetic agarose beads (ChromoTek, catalog #gtma-20) for 2–4 h. After immunoprecipitation, beads were pelleted, washed in NT2 buffer, and pelleted again for RNA extraction as immunoprecipitation (IP) sample. RNA from input and IP samples was extracted using the RNeasy Micro Kit per the manufacturer’s protocol (Qiagen, catalog #74004). cDNA was synthesized from extracted RNA using the Verso cDNA synthesis kit (Thermo Fisher Scientific, catalog #AB4100A). cDNA from input and IP samples at E14.5 and E18.5 was used in RT-qPCR as described in [43]. Briefly, RT-qPCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, catalog #1725124). Each cDNA sample was run in technical triplicate on a QuantStudio 6 Flex System Real-Time PCR system (Applied Biosystems) for 45 cycles of: 95 °C for 15 s, 59 °C for 30 s, and 72 °C for 30 s. RT-qPCR primers are listed in Table 4.
RIP–RT–qPCR analysis
Threshold cycle (Ct) values were calculated using StepOne software (version 2.2; Applied Biosystems). ΔCt values were calculated relative to the housekeeping gene Gapdh [29]. Expression values were calculated as a function of 2-ΔCt. Expression values for IP samples from biological replicates (n = 3) were standardized to the mean normalized expression value for biological replicates of input samples to determine fold change (FC). Resulting FC in expression for IP vs input of each transcript target at each time point was used to determine enrichment.
Western blot
Protein extraction from input and RIP was done using components (10x Cell Lysis Buffer #9803, 3x Blue Loading Buffer #56036, 30x Reducing Agent #14265) of the Cell Signaling Technology Western Blotting Kit (#12957) as per the manufacturer’s protocol. Samples were run in a Mini-PROTEAN Tetra Cell (Bio-Rad) at 100 V for 15 min and 75 V for 85 min on a 4–20% Mini-PROTEAN TGX Precast 10-well, 50 μL gel (Bio-Rad #4561094). Transfer was done overnight at 4 °C and 40 V from the gel onto a 0.20-μm nitrocellulose membrane. Following transfer and blocking, membranes were incubated overnight with one of the following primary antibodies: rabbit anti-GFP or goat anti-GAPDH (Table 3). Incubation with the corresponding secondary antibody conjugated to peroxidase (anti-rabbit-HRP or anti-goat-HRP) (Table 3), and detection by chemiluminescence was carried out as described by the manufacturer (Clarity Max Western ECL Substrate, Bio-Rad #1705062).
Fluorescence-associated cell sorting and RT-qPCR
Stage E18.5 testes were collected as described in the RIP, RNA Extraction, and RT-qPCR (RIP–RT–qPCR) method section. However, after TrypLE treatment, testes were dissociated in 3% BSA in DEPC PBS. After dissociation, the cell suspension was passed through a 0.32-μm cell strainer. Collected cells were then sorted on presence and absence of GFP (GFP(+) and GFP(−)) on a B-C Astrios cell sorter. GFP(+) and GFP(−) cells were sorted directly into RLT buffer from RNeasy Micro Kit (Qiagen, catalog #74004). RNA extraction and RT-qPCR were performed as described in the RIP, RNA Extraction, and RT-qPCR (RIP–RT–qPCR) method section. RT-qPCR primers are listed in Table 4. Expression values were calculated as described in the RIP–RT–qPCR Analysis method section.
Statistical analysis
RIP–RT–qPCR and RT-qPCR data were plotted in GraphPad Prism (version 8.0) for statistical significance testing and visualization. Student’s t-test was used for all analyses in GraphPad Prism (version 8.0).
Quantification of germ cells
Fiji ROI manager was used to extract quantitative data about the populations of germ cell expressing markers [DND1, germ cell nuclear acidic protein (GCNA), DEAD box helicase 4 (DDX4)] from 1.5-μm sagittal optical sections at the approximate middle of stained E12.5 ovaries (n = 3) and testes (n = 3, see breakouts Figure 2I and J and Supplementary Figure S2A–D) [48]. Briefly, the Fiji multiselection tool was used to mark all DND1-expressing cells in the green-channel for the total DND1 count. The marked green-channel was then duplicated into the blue-channel containing GCNA stain where all marks from the green-channel were removed that did not co-localize with GCNA stain for the DND1–GCNA count. The resulting marked blue/green-channel was then duplicated into the red-channel containing DDX4 with removal of marks from the blue/green-channel that did not co-localize with DDX4 stain for the DND1–GCNA–DDX4 count. For the total GCNA count, the marks of the blue/green-channel were duplicated into a fresh blue-channel containing GCNA stain with marks added to annotate all previously unannotated GCNA-staining cells. The total GCNA count blue-channel was duplicated into a fresh red-channel containing DDX4 stain with marks from the blue-channel being removed that did not co-localize with DDX4 stain for the GCNA–DDX4 count. The marked green-channel was also duplicated into a fresh red-channel containing DDX4 stain where all marks from the green-channel were removed if they did no co-localize with DDX4 stain for the DND1–DDX4 count. For the total DDX4 count, the marks for the green/red-channel and blue/red-channel were copied and merged with overlapping duplicate marks removed, and all unmarked DDX4-expressing cells were then annotated. Quantification of cells expressing only 2 of the 3 markers (DND1, GCNA, DDX4) were confirmed by overlaying the marks for cells expressing the third marker onto the channel with marked cells co-expressing the first and second marker together, essentially this was a subtraction of marks for triple positive cells from all quantifications that were intended to capture only double positive cells. Quantification of cells expressing only 1 of the 3 markers (DND1, GCNA, DDX4) was done using basic arithmetic: the two counts for cells double positive for the first marker and one of the other two markers were subtracted from the total count for a marker (A_total—AB—AC = A_only). An overlay of the marks for the DND1–GCNA–DDX4 counts was used in all two-channel annotations as a double check that would help avoid accidental removal of marks for triple positive cells in double stain quantification and duplicate counting and prevent discrepancies between counts (work was also checked in Excel by viewing the coordinates of every annotation exported from the Fiji ROI manager). The total number of germ cells was calculated by summation of the total count for the first marker, the double positive count of the second and third markers, the single positive count for the second marker, and the single positive count of the third marker (A_total + BC + B_only + C_only = GC_total). This total number of germ cells was calculated thrice using a different marker as the first marker aka A_total (DND1, GCNA, DDX4) for each run. This robust verification of starting with any marker as the first marker resulted in the same final total germ cell count each of the three times. All of these steps were performed independently on optical sections of E12.5 ovaries (n = 3) and testes (n = 3) from DND1GFP mice stained with antibodies against GFP, GCNA, and DDX4.
Figure 2.
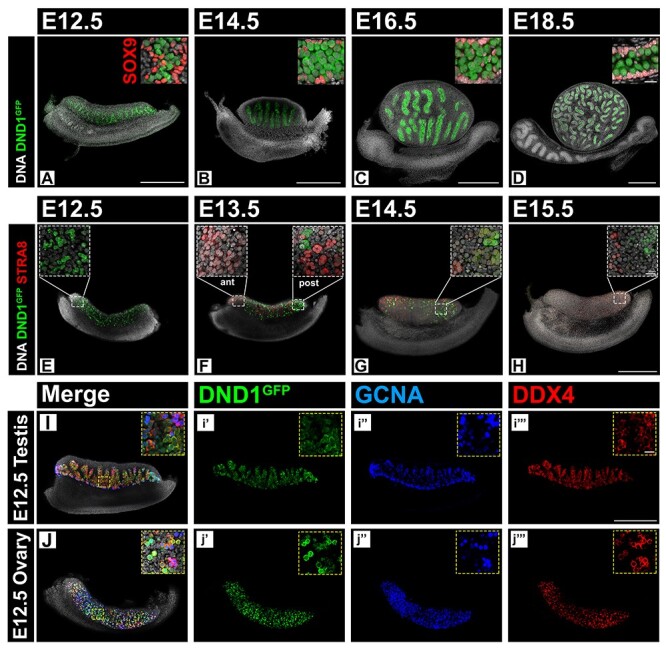
DND1GFP is expressed in male and female germ cells prior to FGC meiotic entry. (A–D) Whole mount time course in the fetal testis (E12.5, E14.5, E16.5, and E18.5) showing continued MGC-specific DND1GFP (green) expression. SOX9 (red) marks Sertoli cells bounding testis cords. A confocal section, enlarged in top right corner, shows negative correlation of DND1 and SOX9 expression (breakouts in Figure S3). (E–H) Whole mount time course in the fetal ovary (E12.5, E13.5, E14.5, and E15.5). DND1GFP (green) expression is negatively correlated with the initiation of meiosis marked by STRA8 (red). Anterior (ant), posterior (post) (breakouts in Figure S4). (I–J) Exemplary whole mount images of E12.5 testis and E12.5 ovary co-labeled with DND1GFP and pan-germ cell markers GCNA (blue) and DDX4 (red). Demonstration of heterogeneity of stain for all three markers in germ cells at higher magnification in insets. Scale bar all 20x images 500 μm; scale bar all 40x inset images 20 μm (scale D inset for A–D, scale H inset for E–H). Scale bar in H for E–H. Scale bar in i”’ for I and J.
Quantification of the null hypothesis, a uniform distribution, was modeled using Bernoulli distributions. Parameters were set as series of three binary questions (DND1+/−, GCNA+/−, DDX4+/−). Probability for success of each question was set as P = 0.5, with a stipulation that there must be at least one success (aka positive for at least one marker). This results in seven possible categories (DND1, GCNA, DDX4, DND1/GCNA, GCNA/DDX4, DND1/DDX4, DND1/GCNA/DDX4). Five thousand “cells” were used in each run of the simulation, with n = 100 runs performed. The resulting average of 100 runs was used as the cumulative tabulated count of the null hypothesis to compare to collected data. Script for model was written and run in Google Colaboratory using Python [49, 50]. Annotated script can be found at GitHub (https://gist.github.com/valext34/e40e0cc28a4631bfe8f87d212e045a2b).
Cumulative tabulated counts for germ cells in ovaries and testes, and the null hypothesis were loaded into Rstudio [51, 52]. Eulerr was used to generate Euler plots of each of the three cumulative data sets [53].
Results
CRISPR-mediated knock-In of an EGFP tag into the Dnd1 locus
To facilitate characterization of expression and function of DND1, we used a CRISPR/cas9 strategy to introduce an EGFP tag into the endogenous Dnd1 locus (Supplementary Figure S1A–C). Previous experiments showed that a Dnd1 allele carrying a FLAG tag at the amino terminus (N terminus) of the protein was functional in HEK293 cells [26]. Consequently, an sgRNA was designed to target the 5′ end of Dnd1 exon 1 in frame. Founders were identified by Southern blotting and verified using EGFP and allele-specific primers (Supplementary Figure S1D). The founder line was bred to homozygosity confirmed with genotyping (Supplementary Figure S1E). Homozygous mice were used for all timed matings in the experiments described here. There was no evidence of disruption to testis cord or seminiferous tubule development at any stage due to DND1GFP homozygosity.
DND1 is expressed in primordial germ cells shortly after specification, and during migration and population of the bipotential gonad
Consistent with previous results from in situ hybridization assays [13], DND1GFP was detected at E7.5 in a population of cells in the proximal epiblast at the base of the allantois, some of which co-expressed the PGC marker SOX2 [SRY (sex determining region Y)-box 2] (Figure 1A), and in migratory PGCs some of which co-stained with Nanog homeobox (NANOG) at E8.5 (Figure 1B). Migratory PGCs positive for DND1GFP were also detected at E9.5 (Figure 1C). Expression of DND1GFP was sustained in both sexes as PGCs entered the bipotential gonad [gonadal somatic cells are marked with GATA binding protein 4(GATA4)] (Figure 1D).
Figure 1.
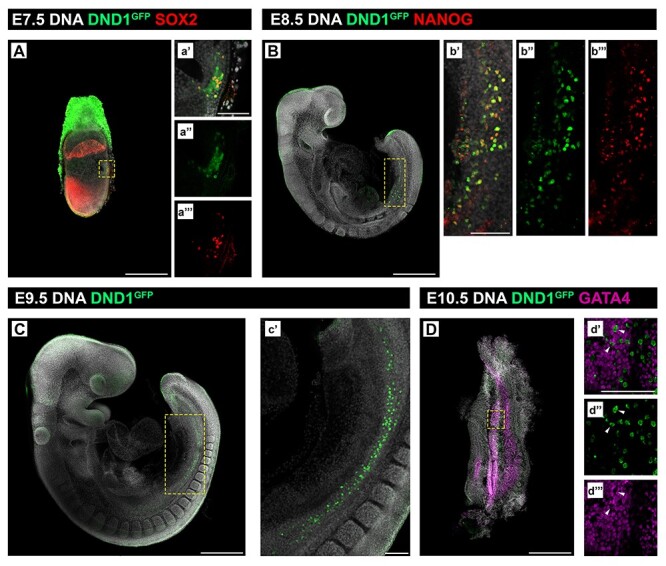
DND1GFP is expressed by PGCs soon after their specification in the E7.5 embryo. (A–D) Whole mount embryonic time course showing DND1GFP (green) positive PGCs (A) at E7.5 at the base of the allantois, co-stained with SOX2 (red); (B, C) at E8.5 and E9.5, migrating along the gut mesentery, co-stained with NANOG (red) in (B); (D) at E10.5 migrating into the bipotential gonad stained with GATA4 (magenta). Wedges mark negative correlation of DND1 and GATA4 in d’, d”, and d”’. (A–D) Scale bar is 500 μm. Boxed region is expanded in a’, a”, a’”, b’, b”, b’”, c’, d’, d”, and d”’; scale bar in a’, b’ c’, and d’ is 100 μm.
At E12.5, DND1GFP was expressed in MGCs and FGCs (Figure 2A, E, I, J). However, levels of DND1GFP varied among both MGCs and FGCs. DND1 was not co-detected in some cells labeled with either of two pan-germ cell markers, GCNA [54, 55] and DDX4 [56, 57] (Figure 2I and J, Supplementary Figure S2A–E), and less than a quarter of germ cells labeled with all three markers (Figure 2I and J insets, Supplementary Figure S2A–D insets and S2E). Quantification of germ cells from optical sections of E12.5 ovaries and testes labeling with just 1, only 2, or all 3 markers (DND1, GCNA, DDX4) showed a slight bias against expression of only 1 marker. At E12.5, DND1 was expressed in 69% of FGCs and 62% of MGCs as opposed to 58% predicted by the null hypothesis of uniform distribution. The most common overlap in the ovary was co-expression of DND1 and DDX4 (24%), whereas the most common overlap in the testis was co-expression of DDX4 and GCNA (29%). The only populations that were statistically different than predicted by the null hypothesis were DND1-only and GCNA-only populations in the testis (data not shown).
DND1 is stably expressed in MGCs throughout testis development but declines as FGCs enter meiosis
Samples were collected spanning the duration of fetal gonad development (E12.5, E14.5, E16.5, and E18.5) for whole mount imaging. At all fetal time points, MGCs within testis cords expressed DND1GFP (Figure 2A–D), while Sertoli cells (labeled with SOX9 (SRY (sex determining region Y)-box 9)) did not (Figure 2A–D insets). Expression analysis of GFP(+) and GFP(−) cells sorted from dissociated E18.5 DND1GFP testes, supported conclusions from imaging analysis in the fetal testis. Compared to GFP(−) cells, GFP(+) cells highly expressed Dnd1 and eGfp as well as established germ cell markers [Oct4 (aka Pou5f1, POU domain, class 5, transcription factor 1) and Ddx4] but not a somatic marker (Gata4) (Supplementary Figure S5).
Whole mount confocal imaging of DND1GFP females, spanning the period just before meiotic initiation (E12.5) and during the wave of initiation [E13.5–E15.5; detected using the meiotic marker stimulated by retinoic acid gene 8 (STRA8)], showed that expression of DND1GFP declined as STRA8 expression was established. The anterior to posterior wave of STRA8 expression was less clear in these samples than previously reported, but STRA8 expression was negatively correlated with DND1GFP (Figure 2E–H). DND1GFP disappeared from the most anterior FGCs at E13.5, was still detected in patches of cells at the posterior pole at E14.5, and was scarcely detectable by E15.5 (Figure 2E–H insets). To determine whether DND1GFP expression persists in some cells or is regained in the ovary postnatally, we stained ovaries at P2 (data not shown) and P90 (Supplementary Figure S6) and found no evidence of DND1GFP persistence in female germ cells after birth.
DND1 expression is negatively correlated with MGC meiotic entry during both the postnatal first wave and adult cyclic spermatogenesis
To track the pattern of DND1 expression in the testis after birth, we stained sections of postnatal testes from P1 to P10, during the first wave of spermatogenesis (Figure 3). Co-staining with GCNA demonstrated that prospermatogonia were positive for both GCNA and DND1 from P1 to P10, with only basal germ cells double positive at P10 (Figure 3D, H, L). DND1GFP+, GCNA+ cells became positive for the undifferentiated spermatogonia marker GFRa1 (glial cell line–derived neurotrophic factor family receptor alpha 1) by P3 (Figure 3A and B) [58, 59]. By P10, DND1GFP+ and GFRa1+ populations resolved with cells retaining both markers (Figure 3D and Supplementary Figure S7D, d’). At P6, as a subset of prospermatogonia quickly differentiates, DND1GFP+ cells became positive for the differentiating spermatogonia marker cKIT (KIT proto-oncogene receptor tyrosine kinase) [60–63]. The cKIT+ cells retained expression of DND1GFP at P10 (Figure 3E–H). At P3 and P6, there were DND1GFP+ cells that co-labeled with the marker of meiotic initiation, STRA8 (Figure 3I–L), However, by P10, these populations were distinct: STRA8+ cells, still positive for GCNA (Figure 3L), migrated luminally, whereas DND1GFP+ cells remained in close association with the basal lamina (Figure 3L).
Figure 3.
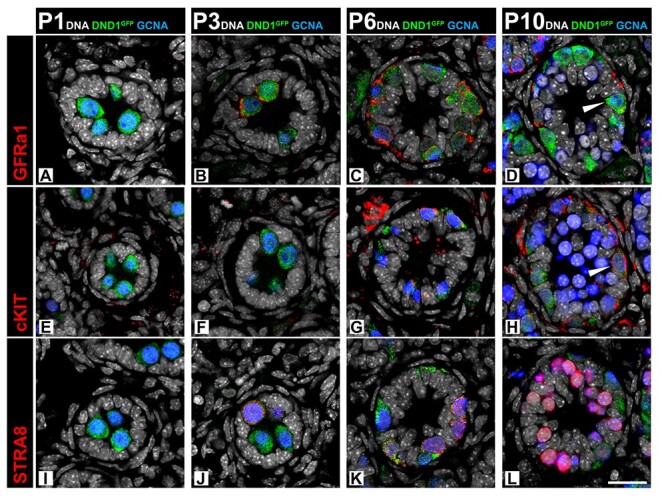
Expression of DND1GFP is negatively correlated with meiotic entry in MGCs during postnatal initiation of the first spermatogenic wave. Time course of DND1GFP expression in sections from the postnatal testis (P1, P3, P6, P10) during initiation of the first wave of spermatogenesis. (A–D) DND1GFP is expressed in undifferentiated prospermatogonia, which begin to co-express GFRa1 at P3 (red). By P10, cells express GFRa1 or DNDGFP, with cells also co-expressing both (wedge). (E–H) Differentiating spermatogonia express cKIT (red) by P6, and cells retain cKIT and DND1GFP at P10. (I–L) Expression of DNDGFP overlaps with the STRA8 positive population (red) at P3 and P6, but the populations are nearly distinct by P10. Breakouts in Figure S7. Scale bar in L is 20 μm and for all images.
Similar to expression of DND1GFP in the postnatal testis, expression in the adult testis was present in the spermatogonial population and negatively correlated with meiotic entry (Figure 4). DND1 was co-expressed with the undifferentiated Type A spermatogonial marker GFRa1 in a subset of cells (Figure 4A and B). DND1 was also expressed in differentiating spermatogonia marked with cKIT (Figure 4C and D). Unlike the postnatal first wave of spermatogenesis, meiotic entry in adult spermatogenesis, as marked by STRA8, shows a sharp delineation between cells expressing DND1 and those expressing STRA8 (Figure 4E and F). In summary, DND1 is present in the major three types of spermatogonia in the mouse (Type A, intermediate, and Type B) as evidenced by the presence of GFP in stage VII–VIII, II–IV, and V–VI tubules, respectively (Figure 4G).
Figure 4.
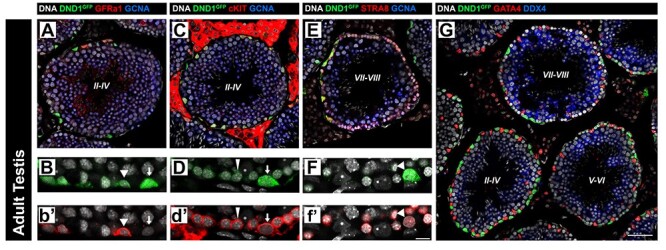
Expression of DND1GFP is negatively correlated with meiotic entry in spermatogonia during adult spermatogenesis. DND1GFP is expressed in undifferentiated (GFRa1, red, A-B, arrowheads) and differentiating (cKIT, red, C-D, arrows and wedges (intermediate and Type B, respectively)) spermatogonia in stages II–IV tubules (A-D, G), but declines during late differentiation in stages V–VI tubules (G) and is negatively correlated with meiotic entry (STRA8, red E-F) in stages VII–VIII tubules (E-G). GCNA or DDX4 (germ cells, blue), GATA4 (Sertoli and Leydig cells, red). Staging for each tubule is centered in tubule in Roman numerals. Breakouts in (Figure S8). Scale bar in G is 50 μm and for A, C, E, and G, B, D, and F. 60x representative segments of the basement membrane of seminiferous tubules of same stage as 40x images above (A, C, and E). The intensity of DND1GFP varies among cells. Scale bar in f’ is 20 μm and for B, D, and F.
The GFP tag on DND1 does not affect co-localization with P-body component DCP2
DND1 has been previously shown to co-localize with P-bodies as part of the CCR4-NOT deadenylation complex with NANOS2 [15]. This complex has been shown to recruit transcripts to P-bodies for degradation [10]. To confirm that the DND1GFP fusion tag did not interfere with the ability of DND1 to localize to P-bodies, we co-stained DND1GFP E15.5 testes with an antibody against decapping mRNA 2 (DCP2), the active mRNA decapping component of P-bodies [64–68]. E15.5 was chosen as previous work has demonstrated this to be the zenith of DCP2 presence in MGCs [10]. Similar to reports by others in the adult testis, at E15.5, DND1 signal was observed throughout the nucleus and cytoplasm of prospermatogonia [69]. However, concentrations of DND1GFP co-localized with DCP2 puncta in these cells (Figure 5A and B arrows), indicating that the GFP tag does not prevent DND1 from localizing to P-bodies.
Figure 5.
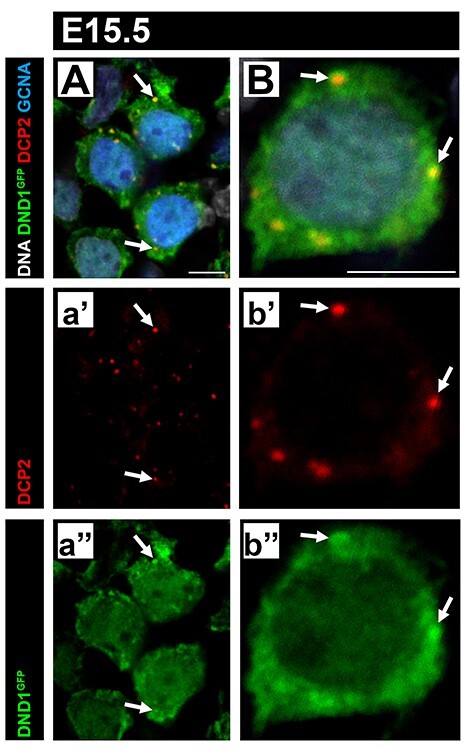
DND1GFP is expressed throughout the cytoplasm and nucleus with puncta that co-localize with the P-bodies. (A, B) Representative images from sections of E15.5 DND1GFP testes. Some regions of concentrated GFP (green) signal co-localize with DCP2 puncta (red, arrows), which mark P-bodies, GCNA (blue, germ cells). Both scale bars are 5 μm.
RNA immunoprecipitation experiments validate in vivo DND1 transcript
We next aimed to determine whether the DND1GFP fusion protein could be used in vivo for RIP assays to immunoprecipitate transcripts previously identified as targets of DND1 in vitro. We selected several potential targets previously identified in vitro [25, 26, 40]. These potential targets (1) contained DND1 DAUBAW-binding sites as defined by DO-RIP seq and/or (2) showed changes in expression in Dnd1Ter/Ter mutants compared to wild type [26], and all candidates also had predicted 3’UTR DND1-binding sites (Table 5). Calnexin (Canx), cyclin E1 (Ccne1), and lysine acetyltransferase 7 (Kat7) were previously shown to be bound in vitro and showed up-regulation (Canx, Ccne1) or down-regulation (Kat7) in Dnd1Ter/Ter mutants. Lefty-right determination factory 2 (Lefty2) and ret proto-oncogene (Ret) showed strong expression changes in Dnd1Ter/Ter mutants compared to wild type [26] and had numerous 3’UTR motifs present (Table 5). Lefty2 is part of the Tgfβ superfamily, a group previously shown to be at least partially regulated by DND1 [25]. Ret is a critical factor to fetal MGC survival [70]. Canx, aside from being a predicted target, is also a standard housekeeping gene for the gonad and was included as a positive control [71]. Sox9, which has predicted 3’UTR sites for DND1 but is not expressed in germ cells, was included as a negative control (Table 5).
Table 5.
Potential target info.
| Gene name | DO-RIP-seq “target”1 | 3’UTR2 | Dnd1Ter vs WT3 |
|---|---|---|---|
| Sox9 | n.d. | 11 | n/a |
| Canx | Yes | 16 | Up |
| Lefty2 | n.d. | 15 | Up |
| Ret | n.d. | 15 | Down |
| Ccne1 | Yes | 1 | Up |
| Kat7 | Yes | 13 | Down |
1Transcript bound by DND1-FLAG in transfected HEK cells [8].
2Motif presence in 3′UTR of mouse transcript sequence.
3Differential gene regulation (up or down) in Dnd1Ter MGCs versus Wild Type (WT) [8].
n.d.: Not determined due to lack of presence in HEK cells during DO-RIP-seq.
n/a: Not applicable due to lack of expression in germ cells.
We performed small-scale RIP–RT–qPCR on pooled testes from a single Dnd1GFP/GFP litter at each stage based on previously established methods [46, 47]. Ability to immunoprecipitate DND1GFP was confirmed by Western blotting (Supplementary Figure S9A). RIP was performed at E14.5, a stage when MGCs are entering G0 cell cycle arrest, for which we had MGC-specific transcriptome information, and E18.5, a stage late in MGC G0 arrest. At E14.5 and E18.5, RIP–RT–qPCR showed significant enrichment only of Lefty2. Although levels of Ret and Ccne1 enriched above background at both time points, neither was significant. Despite Canx and Kat7 enrichment in vitro, neither enriched at either stage in vivo, similar to the negative control, Sox9, (Figure 6, expression data Supplementary Figure S9B and C). Significance of transcript enrichment was determined by comparison to lack of enrichment of both Sox9 and Canx at E14.5 or E18.5.
Figure 6.
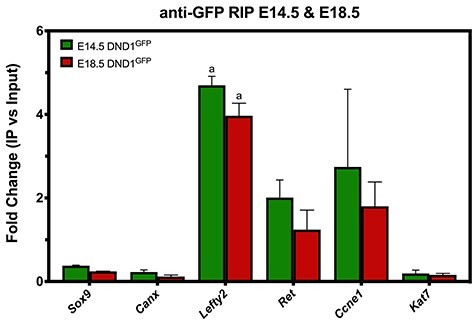
The DND1GFP allele can be used for RIP to validate predicted mRNA targets. RIP was performed using antibodies against GFP on testis extracts at E14.5 and E18.5, followed by RT-qPCR for predicted targets identified in previous RIP studies performed in vitro. Enrichment of target candidates shown here represents genes from functional categories: non-germ-cell nontarget control (Sox9), housekeeping gene with multiple DND1-binding sites on transcript (Canx), pluripotency (Lefty2), male fate commitment (Ret), cell cycle regulation (Ccne1), chromatin modification (Kat7). Enrichment was calculated as the fold change (FC) difference between a transcript’s mean normalized expression value (2-ΔCT) in input and in RIP, (ΔCT was calculated as difference between nontarget (Gapdh) and gene of interest), n = 3, and bars are mean ± SEM. Statistical significance (t-test) “a” significant enrichment to both Sox9 and Canx at E14.5 or E18.5.
Discussion
Due to the lack of an available antibody against DND1, detection of Dnd1 expression has relied on in situ hybridization or RT-qPCR. Utilizing our new DND1GFP reporter mouse, we showed that this RBP was expressed in newly specified PGCs at E7.5, based on the marker SOX2, and was present in the germ line during PGC migration to the urogenital ridge, based on the marker NANOG. Overlap between each of these markers and DND1-GFP was incomplete, which could reflect heterogeneity among germ cells during their specification and migration. Heterogeneity among germ cells was also detected soon after germ cells populated the gonad using three markers, DND1GFP with DDX4 and GCNA, the latter two of which are widely used as pan-germ cell markers. Once sex-specific gonad development begins, DND1GFP declined in XX germ cells, while expression was maintained in XY germ cells. DND1 expression was detected in a population of neonatal MGCs that take up residence at the periphery of testis cords and was retained in a population of spermatogonia in the adult testis. We demonstrated that Lefty2 was significantly enriched as an in vivo target of DND1 at both E14.5 ad E18.5, indicating that this tagged allele can be used in RIP to analyze the mRNA targets of DND1.
There is a large body of work on the effects on Dnd depletion. Early knockdown of xDnd in Xenopus embryos [21] resulted in migratory failure and ultimate loss of PGCs before they reached the gonad. In zebrafish, morpholinos against DND led to apoptotic blebbing as well as transdifferentiation of some germ cells to somatic lineages [17, 18, 72], strongly suggesting that DND1 safeguards the identity of germ cells and prevents them from assuming somatic fate. Targeted disruption of Dnd1 in mice resulted in embryonic lethality [14], while conditional knockout at E13.5 led to loss of germ cells [15]. These previous experiments demonstrated the importance of DND1 to the identity and survival of the germ line in both male and female embryos. Our new DND1GFP mouse line will be a valuable tool for isolating a subpopulation of germ cells and taking advantage of recently advanced techniques for molecular and epigenetic studies of very small populations of cells, tracing their biology from their earliest specification.
The reason for the heterogeneous overlap of DND1GFP signal with the canonical germ cell markers GCNA and DDX4 is unknown. However, it seems clear, at least at E12.5, that (1) none of these markers stain the entire germ cell population and (2) that there is significant heterogeneity among fetal germ cells in both the male and female gonad. Some previous studies reported evidence that fetal germ cells are heterogeneous [73, 74]. Care should be taken in relying on antibody staining since staining differences could result from differences in efficiency or background of the antibodies or from the plane of section. However, recent single-cell sequencing experiments have provided further support for the idea that distinct populations exist [74–76]. Differences might arise from pulsatile behavior of markers, from cells occupying different stages of the cell cycle, or cells occupying distinct microenvironments in the ovary or testis. Some investigators have suggested that there is a unique subpopulation of germ cells destined to become the definitive oogonia or spermatogonial stem cells. Recent studies reporting on some germ cell subpopulations in the fetal testis have demonstrated differences in cell fate and stemness [73, 74, 76].
Our results show a negative correlation between DND1 expression and meiotic entry. This relationship is striking in the female gonad where time course imaging analysis showed that DND1 signal was negatively correlated with STRA8 expression and declined in an anterior to posterior pattern synchronously with entry into meiosis. This negative relationship was also observed in the male germ line both postnatally during the first wave of spermatogenesis and during adult spermatogenesis. The difference in the timing of DND1 loss in germ cells between postnatal and adult testis is most likely due to the fundamental difference between direct entry of an SSC to meiosis in the first wave and the slower transition of an SSC through the subsets of differentiating spermatogonia prior to meiotic entry during adult spermatogenesis. Similar findings in adult differentiating spermatogonia were recently reported [69]. Whether these observations are causal or coincidental remains unknown. However, the negative correlation with meiotic entry is consistent with a role for DND1 in preserving the pluripotent identity of PGCs and blocking their differentiation. Future studies to elucidate the nature of this relationship should focus on perturbing or accelerating normal meiotic entry in all three instances (fetal FGC, postnatal MGC, and adult MGC) to define how DND1 levels are affected. Initial experiments could take advantage of established methods for altering meiotic entry [77, 78]. Alternatively, over-/under-expression of DND1 around meiotic entry through a transgenic system would also determine how DND1 levels relate to initiation of meiosis [79].
Our RIP–RT–qPCR results indicate that DND1GFP will be a valuable tool for understanding the role(s) of DND1 as an RBP. Here, we tested candidate target genes that harbored DND1-binding sites, were altered in Dnd1Ter mutants, and/or were identified as targets in previous RIP studies in cell lines [25, 26, 29, 40]. Our RIP experiments were conducted in vivo at two stages of development, at E14.5 (as MGCs enter mitotic arrest) and at E18.5 (near the end of fetal development). Although not all in vitro targets were confirmed, a gene in the Tgfβ superfamily (Lefty2) significantly enriched as a target of DND1 in vivo at E14.5 and E18.5. Members of the Tgfβ superfamily have been previously documented as DND1 targets by Yamaji et al. (2017) and in our work [25, 26]. DND1 targeting of Lefty2 is especially exciting, as DND1-mediated degradation of Lefty2 could be part of Lefty2 down-regulation that was recently shown as a requisite step in MGC differentiation by Nguyen and Laird [74]. Lefty2 is likely targeted for degradation by DND1 as cells enter mitotic arrest and down-regulate pluripotency. Conversely, Ret expression is on a strong upward trajectory at E14.5, and we predict that if Ret is a target, it would be protected by DND1 as MGCs initiate differentiation.
Since Canx was previously reported as a target of DND1 [26] and contains multiple DND1-binding sites, it was initially chosen as a housekeeping gene that could act as a positive control. Unexpectedly, we found that Canx failed to enrich at both E14.5 and E18.5, similar to Sox9, which is not expressed in germ cells. Kat7 expression was disrupted in Dnd1Ter/Ter mutants, and several binding sites for DND1 were detected in the transcript. However, despite mapping as a target of DND1 in vitro, Kat7 also failed to enrich in these experiments. It is possible that Kat7 is not directly targeted by DND1 in MGCs. However, an alternative explanation could be that the period for DND1 targeting of Kat7 was not encompassed by the two time points in MGC development (E14.5 and E18.5) reported on here.
Another very interesting question is how DND1 selects targets for protection versus targets for degradation. Saga and colleagues have shown that DND1 interacts with NANOS2 to bring targets to the CCR4-NOT complex for degradation [15, 25, 69]. Whether there are other protein partners of DND1 responsible for protective activity, or whether degradative and protective activities segregate along a developmental timeline, is unknown.
In summary, we used CRISPR technology to produce a novel transgenic mouse line carrying a GFP fusion tag on endogenous DND1. Imaging work in these mice showed that DND1 is expressed in the male germ line throughout fetal life but lost from the female germ line at the time of meiotic entry. The correlation between down-regulation of DND1 expression and meiotic initiation was also observed in the male germ line after birth in both the postnatal and adult testis. In general, the ability of the RIP–RT–qPCR method used here to successfully capture target information from such a small number of cells suggests that the GFP-tagged allele can be used for the unbiased in vivo identification of DND1 target RNAs and interacting proteins at multiple stages to gain further insight into the role DND1 plays during germ cell development.
Supplementary Material
Acknowledgements
We thank the Duke Light Microscopy Core Facility, Duke Cancer Institute Flow Cytometry Shared Resource, Rodent Genetic Engineering Services Team (formerly the Duke Transgenic Core Facility, a part of the “Rodent Cancer Models Shared Resource” supported in part by the DCI: CCSG NCI P30 CA014236, PI Michael Kastan, MD, PhD). We thank Dr. Terry Lechler (Duke University Medical Center) for the EGFP plasmid and Dr. Michael Griswold (Washington State University) for the STRA8 antibody.
Footnotes
† Grant Support: This work was supported by a Pilot grant from DCI to BC, an R37 from NICHD to BC, and an F32 from NIGMS to VAR.
Contributor Information
Victor A Ruthig, Department of Cell Biology, Duke University Medical Center, Durham, NC, USA.
Tetsuhiro Yokonishi, Department of Cell Biology, Duke University Medical Center, Durham, NC, USA.
Matthew B Friedersdorf, Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC, USA.
Sofia Batchvarova, Department of Cell Biology, Duke University Medical Center, Durham, NC, USA.
Josiah Hardy, Department of Cell Biology, Duke University Medical Center, Durham, NC, USA.
Jason A Garness, Department of Cell Biology, Duke University Medical Center, Durham, NC, USA.
Jack D Keene, Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC, USA.
Blanche Capel, Department of Cell Biology, Duke University Medical Center, Durham, NC, USA.
References
- 1. Qi H. RNA-binding proteins in mouse male germline stem cells: a mammalian perspective. Cell Regen (Lond) 2016; 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nguyen-Chi M, Morello D. RNA-binding proteins, RNA granules, and gametes: is unity strength? Reproduction 2011; 142:803–817. [DOI] [PubMed] [Google Scholar]
- 3. Sutherland JM, Siddall NA, Hime GR, McLaughlin EA. RNA binding proteins in spermatogenesis: an in depth focus on the Musashi family. Asian J Androl 2015; 17:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morales CR, Wu XQ, Hecht NB. The DNA/RNA-binding protein, TB-RBP, moves from the nucleus to the cytoplasm and through intercellular bridges in male germ cells. Dev Biol 1998; 201:113–123. [DOI] [PubMed] [Google Scholar]
- 5. Xu M, Hecht NB. Polypyrimidine tract-binding protein 2 binds to selective, intronic messenger RNA and microRNA targets in the mouse testis. Biol Reprod 2011; 84:435–439. [DOI] [PubMed] [Google Scholar]
- 6. Cho YS, Iguchi N, Yang J, Handel MA, Hecht NB. Meiotic messenger RNA and noncoding RNA targets of the RNA-binding protein Translin (TSN) in mouse testis. Biol Reprod 2005; 73:840–847. [DOI] [PubMed] [Google Scholar]
- 7. Yang J, Medvedev S, Reddi PP, Schultz RM, Hecht NB. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc Natl Acad Sci U S A 2005; 102:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr Biol 2012; 22:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Z, Shirakawa T, Ohbo K, Sada A, Wu Q, Hasegawa K, Saba R, Saga Y. RNA binding protein Nanos2 organizes post-transcriptional buffering system to retain primitive state of mouse spermatogonial stem cells. Dev Cell 2015; 34:96–107. [DOI] [PubMed] [Google Scholar]
- 10. Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci U S A 2010; 107:3594–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamaji M, Tanaka T, Shigeta M, Chuma S, Saga Y, Saitou M. Functional reconstruction of NANOS3 expression in the germ cell lineage by a novel transgenic reporter reveals distinct subcellular localizations of NANOS3. Reproduction 2010; 139:381–393. [DOI] [PubMed] [Google Scholar]
- 12. Gill ME, Hu YC, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci U S A 2011; 108:7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 2005; 435:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zechel JL, Doerner SK, Lager A, Tesar PJ, Heaney JD, Nadeau JH. Contrasting effects of Deadend1 (Dnd1) gain and loss of function mutations on allelic inheritance, testicular cancer, and intestinal polyposis. BMC Genet 2013; 14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suzuki A, Niimi Y, Shinmyozu K, Zhou Z, Kiso M, Saga Y. Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep 2016; 17:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q, Li Y, Yang S, Huang S, Yan M, Ding Y, Tang W, Lou X, Yin Q, Sun Z, Lu L, Shi H et al. CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat Cell Biol 2018; 20:1315–1325. [DOI] [PubMed] [Google Scholar]
- 17. Gross-Thebing T, Yigit S, Pfeiffer J, Reichman-Fried M, Bandemer J, Ruckert C, Rathmer C, Goudarzi M, Stehling M, Tarbashevich K, Seggewiss J, Raz E et al. The vertebrate protein dead end maintains primordial germ cell fate by inhibiting somatic differentiation. Dev Cell 2017; 43:704–15 e5. [DOI] [PubMed] [Google Scholar]
- 18. Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E. Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol 2003; 13:1429–1434. [DOI] [PubMed] [Google Scholar]
- 19. Wargelius A, Leininger S, Skaftnesmo KO, Kleppe L, Andersson E, Taranger GL, Schulz RW, Edvardsen RB. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci Rep 2016; 6:21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Northrup E, Zschemisch NH, Eisenblätter R, Glage S, Wedekind D, Cuppen E, Dorsch M, Hedrich HJ. The ter mutation in the rat Dnd1 gene initiates gonadal teratomas and infertility in both genders. PLoS One 2012; 7:e38001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T. Xenopus dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol 2006; 291:1–11. [DOI] [PubMed] [Google Scholar]
- 22. Aguero T, Jin Z, Chorghade S, Kalsotra A, King ML, Yang J. Maternal dead-end 1 promotes translation of nanos1 by binding the eIF3 complex. Development 2017; 144:3755–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duszczyk MM, Wischnewski H, Kazeeva T, Loughlin FE, von Schroetter C, Pradère U et al. The solution structure of dead end bound to AU-rich RNA reveals an unprecedented mode of tandem RRM-RNA recognition required for mRNA repression. bioRxiv 2019; 572156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kedde M, Strasser MJ, Boldajipour B, Vrielink JAFO, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, Lund AH, Perrakis A et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007; 131:1273–1286. [DOI] [PubMed] [Google Scholar]
- 25. Yamaji M, Jishage M, Meyer C, Suryawanshi H, der E, Yamaji M, Garzia A, Morozov P, Manickavel S, McFarland HL, Roeder RG, Hafner M et al. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature 2017; 543:568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruthig VA, Friedersdorf MB, Garness JA, Munger SC, Bunce C, Keene JD, Capel B. The RNA-binding protein DND1 acts sequentially as a negative regulator of pluripotency and a positive regulator of epigenetic modifiers required for germ cell reprogramming. Development 2019; 146:dev175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stevens LC. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J Natl Cancer Inst 1973; 50:235–242. [DOI] [PubMed] [Google Scholar]
- 28. Cook MS, Coveney D, Batchvarov I, Nadeau JH, Capel B. BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1(Ter/Ter) mice. Dev Biol 2009; 328:377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cook MS, Munger SC, Nadeau JH, Capel B. Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development 2011; 138:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heaney JD, Anderson EL, Michelson MV, Zechel JL, Conrad PA, Page DC, Nadeau JH. Germ cell pluripotency, premature differentiation and susceptibility to testicular teratomas in mice. Development 2012; 139:1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stevens LC, Hummel KP. A description of spontaneous congenital testicular teratomas in strain 129 mice. J Natl Cancer Inst 1957; 18:719–747. [PubMed] [Google Scholar]
- 32. Dawson EP, Lanza DG, Webster NJ, Benton SM, Suetake I, Heaney JD. Delayed male germ cell sex-specification permits transition into embryonal carcinoma cells with features of primed pluripotency. Development 2018; 145:dev156612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stevens LC. Testicular teratomas in fetal mice. J Natl Cancer Inst 1962; 28:247–267. [PubMed] [Google Scholar]
- 34. Gu W, Mochizuki K, Otsuka K, Hamada R, Takehara A, Matsui Y. Dnd1-mediated epigenetic control of teratoma formation in mouse. Biol Open 2018; 7:bio032318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rajpert-De Meyts E, Jorgensen N, Brondum-Nielsen K, Muller J, Skakkebaek NE. Developmental arrest of germ cells in the pathogenesis of germ cell neoplasia. APMIS 1998; 106:198–204 discussion-6. [DOI] [PubMed] [Google Scholar]
- 36. Ulbright TM. Germ cell tumors of the gonads: a selective review emphasizing problems in differential diagnosis, newly appreciated, and controversial issues. Mod Pathol 2005; 18:S61–S79. [DOI] [PubMed] [Google Scholar]
- 37. von Hochstetter AR, Hedinger CE. The differential diagnosis of testicular germ cell tumors in theory and practice. A critical analysis of two major systems of classifiction and review of 389 cases. Virchows Archive A. Pathol Anat Histol 1982; 396:247–277. [DOI] [PubMed] [Google Scholar]
- 38. ACS . In: Society AC (ed.), American Cancer Society, Key statistics about testicular cancer. American Cancer Society; 2015. [Google Scholar]
- 39. Goudarzi M, Banisch TU, Mobin MB, Maghelli N, Tarbashevich K, Strate I, van den Berg J, Blaser H, Bandemer S, Paluch E, Bakkers J, Tolić-Nørrelykke IM et al. Identification and regulation of a molecular module for bleb-based cell motility. Dev Cell 2012; 23:210–218. [DOI] [PubMed] [Google Scholar]
- 40. Zhu R, Iacovino M, Mahen E, Kyba M, Matin A. Transcripts that associate with the RNA binding protein, DEAD-END (DND1), in embryonic stem (ES) cells. BMC Mol Biol 2011; 12:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Behringer R. Manipulating the mouse embryo: a laboratory manual. In: Cold Spring Harbor (ed.), 4th ed. New York: Cold Spring Harbor Laboratory Press; 2014. [Google Scholar]
- 42. DeFalco T, Bhattacharya I, Williams AV, Sams DM, Capel B. Yolk-sac-derived macrophages regulate fetal testis vascularization and morphogenesis. Proc Natl Acad Sci U S A 2014; 111:E2384–E2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin YT, Barske L, DeFalco T, Capel B. Numb regulates somatic cell lineage commitment during early gonadogenesis in mice. Development 2017; 144:1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Defalco T, Saraswathula A, Briot A, Iruela-Arispe ML, Capel B. Testosterone levels influence mouse fetal Leydig cell progenitors through notch signaling. Biol Reprod 2013; 88:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maatouk DM, Kellam LD, Mann MR, Lei H, Li E, Bartolomei MS, Resnick JL. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 2006; 133:3411–3418. [DOI] [PubMed] [Google Scholar]
- 46. Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 2006; 1:302–307. [DOI] [PubMed] [Google Scholar]
- 47. Nicholson CO, Friedersdorf MB, Bisogno LS, Keene JD. DO-RIP-seq to quantify RNA binding sites transcriptome-wide. Methods 2017; 118-119:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bisong E. Google Colaboratory. In: Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners. Berkeley, CA: Apress; 2019: 59–64. [Google Scholar]
- 50. Python Software Foundation . Python 3 Reference Manual. Scotts Valley, CA: CreateSpace; 2009. [Google Scholar]
- 51. RStudio Team . RStudio: Integrated Development for R. Boston, MA: RStudio, PBC; 2020. [Google Scholar]
- 52. R Development Core Team . R: A Lanuage and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 53. Larsson J. Eulerr: Area-proportional Euler and Venn diagrams with ellipses. 2020; R package version 6.1.0, https://cran.r-project.org/package=eulerr.
- 54. Tanaka H, Pereira LA, Nozaki M, Tsuchida J, Sawada K, Mori H, Nishimune Y. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int J Androl 1997; 20:361–366. [DOI] [PubMed] [Google Scholar]
- 55. Enders GC, May JJ 2nd. Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol 1994; 163:331–340. [DOI] [PubMed] [Google Scholar]
- 56. Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci U S A 1994; 91:12258–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse vasa-homologue protein during germ cell development. Mech Dev 2000; 93:139–149. [DOI] [PubMed] [Google Scholar]
- 58. Grasso M, Fuso A, Dovere L, de Rooij DG, Stefanini M, Boitani C, Vicini E. Distribution of GFRA1-expressing spermatogonia in adult mouse testis. Reproduction 2012; 143:325–332. [DOI] [PubMed] [Google Scholar]
- 59. Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 2009; 325:1394–1398. [DOI] [PubMed] [Google Scholar]
- 60. Ohbo K, Yoshida S, Ohmura M, Ohneda O, Ogawa T, Tsuchiya H, Kuwana T, Kehler J, Abe K, Schöler HR, Suda T. Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev Biol 2003; 258:209–225. [DOI] [PubMed] [Google Scholar]
- 61. Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc Natl Acad Sci U S A 2000; 97:8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c-kit in mouse undifferentiated and differentiating type a spermatogonia. Endocrinology 1999; 140:5894–5900. [DOI] [PubMed] [Google Scholar]
- 63. Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T, Nishikawa S. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 1991; 113:689–699. [DOI] [PubMed] [Google Scholar]
- 64. Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell 2007; 25:635–646. [DOI] [PubMed] [Google Scholar]
- 65. Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 2007; 8:9–22. [DOI] [PubMed] [Google Scholar]
- 66. Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell 2008; 32:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A 2002; 99:12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 2002; 21:6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Niimi Y, Imai A, Nishimura H, Yui K, Kikuchi A, Koike H, Saga Y, Suzuki A. Essential role of mouse dead end1 in the maintenance of spermatogonia. Dev Biol 2019; 445:103–112. [DOI] [PubMed] [Google Scholar]
- 70. Miles DC, van den Bergen JA, Wakeling SI, Anderson RB, Sinclair AH, Western PS. The proto-oncogene ret is required for male foetal germ cell survival. Dev Biol 2012; 365:101–109. [DOI] [PubMed] [Google Scholar]
- 71. van den Bergen JA, Miles DC, Sinclair AH, Western PS. Normalizing gene expression levels in mouse fetal germ cells. Biol Reprod 2009; 81:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wong TT, Zohar Y. Production of reproductively sterile fish by a non-transgenic gene silencing technology. Sci Rep 2015; 5:15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pui HP, Saga Y. Gonocytes-to-spermatogonia transition initiates prior to birth in murine testes and it requires FGF signaling. Mech Dev 2017; 144:125–139. [DOI] [PubMed] [Google Scholar]
- 74. Nguyen DH, Soygur B, Peng SP, Malki S, Hu G, Laird DJ. Apoptosis in the fetal testis eliminates developmentally defective germ cell clones. Nat Cell Biol 2020; 22:1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mayère C, Neirijnck Y, Sararols P, Stévant I, Kühne F, Chassot AA, Chaboissier M-C, Dermitzakis ET, Nef S. Single-cell transcriptomic reveals temporal dynamics of critical regulators of germ cell fate during mouse sex determination. bioRxiv 2019; 747279. [DOI] [PubMed] [Google Scholar]
- 76. Law NC, Oatley MJ, Oatley JM. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat Commun 2019; 10:2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hogarth CA, Evanoff R, Mitchell D, Kent T, Small C, Amory JK, Griswold MD. Turning a spermatogenic wave into a tsunami: synchronizing murine spermatogenesis using WIN 18,446. Biol Reprod 2013; 88:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bowles J, Feng CW, Spiller C, Davidson TL, Jackson A, Koopman P. FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev Cell 2010; 19:440–449. [DOI] [PubMed] [Google Scholar]
- 79. Nunez L, Mokkapati S, Yu C, Deng JM, Behringer RR, Matin A. Generation of a novel mouse strain with conditional, cell-type specific expression of DND1. Genesis 2019; 57:e23335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


