Abstract
Tuberculosis (TB) caused by Mycobacterium tuberculosis (MTB) is a chronic infectious disease. Interferon-gamma (IFN-γ) is an important cytokine imparting resistance to mycobacterial diseases. It is believed that IFN-γ and Interleukin-10 (IL-10) play divergent roles in the host immune system against MTB infection. IL-10 is an important inhibitory cytokine and helps balancing the inflammatory and immune responses. IL-10 is involved in down regulation of Th1 cytokines, MHC class II antigen and co-stimulatory molecular expression on macrophages, while IFN-γ results in macrophage activation allowing them to exert the microbicidal role. The objectives were to find out the association of IL-10 (−1082 A/G) and IFN-γ (+874 A/T) single nucleotide polymorphisms (SNPs) with extrapulmonary tuberculosis in ethnic Kashmiri population. A total of 100 extrapulmonary tuberculosis cases and 102 healthy controls were analyzed for IL-10 (−1082 A/G) and IFN- γ (+874 A/T) SNPs using Allele-Specific PCR. We found a significant association of IFN-γ + 874 ‘TT’ genotype with extrapulmonary tuberculosis (p = 0.006) and in case of IL-10 (−1082 A/G) we found a significant association with extrapulmonary tuberculosis under recessive model (GG vs GA + AA) (p = 0.03) in Kashmiri population. IL-10 (−1082 A/G) and IFN-γ (+874 A/T) have a significant association with extrapulmonary tuberculosis in ethnic Kashmiri population.
Keywords: Interferon-gamma (IFN-γ), Interleukin-10 (IL-10), Single nucleotide polymorphism (SNP), Mycobacterium tuberculosis (MTB), Extrapulmonary tuberculosis (EPTB)
1. Introduction
Tuberculosis is a challenging health problem throughout the world. This dreadful disease caused by Mycobacterium tuberculosis (MTB) affects one third of world population. According to World Health Organization nearly 10.4 million new cases and 1.4 million deaths were reported in 2019 (Chakaya et al., 2021). It is the leading cause of death from an infectious disease in adults (Furin et al., 2019, Ranaivomanana et al., 2018). Among the various infectious diseases, TB has a distinctive feature of being almost exclusicely transmitted through airTB primarily affects the lungs, although it can also affect other tissues or organs (Alvarez and McCabe, 1984, Maiolini et al., 2020). When the organs other than lungs (e.g., pleura, lymph nodes, abdomen, genitourinary tract etc) are involved it is known as Extrapulmonary tuberculosis (EPTB) (Lee, 2015). Various studies report a high proportion (20–53%) of cases presenting with EPTB in all the cases of TB (Kang et al., 2020). The mechanism of MTB following entry from pulmonary tuberculosis to EPTB is not well understood. Following entry of tuberculosis from lungs, invasion of the epithelial layer by alveolar macrophages and production of various cytokines such as IFN-γ, TNF-α etc occurs. After MTb infection, IFN-γ is essential in maintaining mononuclear cell inflammation. The innate cytokine and chemokine response and phagocyte activation is initiated with the help of TNF-α (Domingo-Gonzalez et al., 2017). Cytokines activate the inflammatory cells resulting in the formation of granuloma which contains pathogen (Gideon et al., 2015, Orme and Basaraba, 2014). It has been found that the biomarkers help to improve EPTB diagnosis and help in improving global tuberculosis control. Both EPTB and TB pathogenesis is believed to involve genetic and host immune factors (Caws et al., 2008).
A study showed different cytokine production in patients recovered from pulmonary tuberculosis when it was compared to EPTB (Fiske et al., 2012, Hasan et al., 2009). Cytokines type-1 interferon-γ and Interleukin-12 (IL-12) are important factors for restriction of infection with MTB. Other factors such as type 2 cytokines IL-10 are also essential part of effective control of inflammation in the host (Murray and Young, 1999). During the dormant TB stage, IL-10 an effective TH-2 regulatory cytokine plays a very important role and increased production of this cytokine enhances reactivation of disease in mice and decrease in cell-mediated immunity against the intracellular infection (Möller and Hoal, 2010, Turner et al., 2002).
Moreover, several single nucleotide polymorphisms located in IL-10 (1082 A/G) particularly in its promoter region have been linked with the aberrant expression of this cytokine that may distort the balance of Th1/Th2 with important impact in tuberculosis infection. The higher levels of 1L-10 may avoid collateral tissue damage mostly in the lungs (Gonzalez-Gay et al., 2005).
A SNP (+874 A/T) (rs2430561) found in the first intron of human Interferon gamma [IFN-γ] gene, which helps in the production of cytokines has an overall impact on the outcome of the tuberculosis infection (Moran et al., 2007). This polymorphic variation has shown different results with tuberculosis disease severity and susceptibility (Sousa-Vasconcelos et al., 2015). In some populations IFN-γ gene (+874 A/T) (rs2430561) shows significant association with tuberculosis but in other populations it doesn’t which might be due to dictinct ethnicities (Matos et al., 2007, Pacheco et al., 2008).
IFN-γ and other cytokines can result in macrophage activation and activate plasma extravasations (Döffinger et al., 2004). Previous studies have shown that production of IFN-γ helps in prevention of infection by MTB. Several studies have reported that mice with defunct IFN-γ gene are more vulnerable to develop tuberculosis. The replacement of functional gene into lungs imparts tuberculosis resistance (Cooper et al., 1993, Harapan et al., 2013, Moreira et al., 2000). Recent studies have shown that both humans & macaque monkeys with higher IFN-γ levels [post two months MTB infection] were found to be more vulnerable to active tuberculosis predisposition (Lin et al., 2009, Möller et al., 2010).
Futher, IFN-γ (+874 A/T) has been shown to be associated with tuberculosis development in several other populations [including Sicilians, South Africa, Hong Kong and Chinese and Spanish] (Lio et al., 2002, López-Maderuelo et al., 2003, Winek et al., 2008). However, in Malawian, Houston, West African and South Indian populations, no such association has been found (Fitness et al., 2004, Tso et al., 2005).
One of the key factors responsible for above-mentioned discrepancies is ethnic difference. Hence, it is important to study the polymorphic variations in different ethnic populations to have a better understanding of molecular genetics of a disease. Since, in Kashmiri population there hasn’t been any such study till date, keeping in view the distinct ethnicity and geographical isolation of our population, we aimed to study the role of IFN-γ [(+874 A/T) and IL-10 (−1082 A/G)] SNPs in EPTB in Kashmiri population.
2. Materials and methods
2.1. Subjects
The current study included 100 newly diagnosed EPTB cases. The demographic characteristics of study subjects are given in Table 1. The samples were collected before starting anti-tuberculosis medicines. Mostly patients attending the general medicine, neurology, cardiology, urology, nephrology, at Sheri-I-Kashmir Institute of Medical Sciences, Srinagar, Kashmir, were included in our study. The research work was conducted in the department of Immunology and Molecular Medicine. All the cases were ethnic Kashmiri patients who were proven cases of EPTB.
Table 1.
Demographic characteristics of the study subjects.
| Characteristics | Cases (N = 100) | Controls (N = 102) | p-value |
|---|---|---|---|
| Age ≤40 >40 |
51 49 |
60 42 |
0.32 |
| Male Female |
40 60 |
69 33 |
0.3 |
| Rural Urban |
75 25 |
80 22 |
0.62 |
| Smoker Non-smoker |
25 75 |
36 66 |
0.12 |
| Lifestyle Active Sedentary |
79 21 |
85 15 |
0.35 |
The diagnosis was based on the PCR. The patients did not have any immune-mediated disorders and all patients were negative for HIV, Hepatitis B and Hepatitis C. However, one patient was HIV positive and later that patient expired having Co-MTB infection with the lower CD4 count. In our study, the healthy control group consisted of 102 healthy blood donors having no history of TB or any immune-related disorder, without any clinical symptoms of tuberculosis. In order to avoid possible effects of population stratification, cases and controls for the study were unrelated, matched for age and sex besides, all the healthy controls belonged to ethnic Kashmiri population.
The power of study was found to be >80% by Quanto software. The subject information sheet was taken from questionnaires, patient files and investigation reports. The Institutional Ethics Committee of Sheri-i-Kashmir Institute of Medical Sciences, Srinagar approved the study. Each patient gave a written informed consent. After taking proper consent, 3–5 ml of blood was collected [in EDTA vial] from each subject (from both cases and controls) and stored at −20 until further processesing. Gene jet genomic DNA purification kit was used for DNA extraction and quality of the purification of extracted DNA was determined by spectrophotometer. Further, each sample was run on ethidium bromide stained 1% agarose gel electrophoresis in order to check the integrity of DNA.
2.2. Molecular analysis
Polymerase chain reaction amplification of IL-10 (−1082 A/G) and IFN- γ (+874 A/T) was done using specific primers. A 25 μl final PCR mixture contained 50 ng of genomic DNA, 10× of Taq buffer, primers [0.4 mM of each], dNTPS [dATP, dCTP, dGTP, dTTP 50 mM of each], Mgcl2 [1.5 mM] and 1U of taq DNA polymerase (Fermentas). The PCR was performed using 10 cycles (95 °C for 1 min, 95 °C for 15 s, 62 °C for 50 s, and 72 °C for 40 s), 62 °C for 50 s, & 72 °C for 40 s), followed by 20 cycles (95 °C for 20 s, 56 °C for 50 s, & 72 °C for 50 s). A 2% agarose gel was prepared and PCR products were run until bands could be easily visualized under UV illuminator. The primers used for amplification of SNPs [IFN- γ (+874 A/T) (rs2430561) and IL-10 (−1082 G/A) (rs1800896)] are given in Table 2.
Table 2.
Primers used for Amplification of IFN-γ ((+874 A/T) & IL-10 (−1082 A/G).
| SNP | Primer sequence | Amplicon size |
|---|---|---|
|
IL-10 (−1082 A/G) |
(ANTISENSE) 5′-CAGCCCTTCCATTTTACTTTC-3′ *G (SENSE) 5′-TACTAAGGCTTCTTTGGGAG-3′ *A (SENSE) 5′-CTACTAAGGCTTCTTTGGGAA-3′ |
550 bp |
|
IFN-γ (+874 A/T) |
(ANTISENSE) 5′- TCAACAAAGCTGATACTCCA-3′ *A (SENSE) 5′-TTCTTACAACACAAAATCAAATCA-3′ *T (SENSE) 5′-TTCTTACAACACAAAATCAAATCT-3′ |
261 bp |
2.3. Procedure for PCR amplification Mycobacterium tuberculosis by MPB64
DNA amplification was carefully done, avoiding any contamination. The forward and reverse primers used are given below [Forward primer- MPB64-F-5′TCCGCTGCCAGTCGTCTTCC3′ and Reverse primer- MPB64-R- GTCCTCGCGAGTCTAGGCCA]. The amplicon size was 245 bp.
2.4. Statistical analysis
For statistical analysis, the genotype and allelic frequency distributions of polymorphisms in the control and EPTB patient groups were compared using the χ2 test. When the assumption of the χ2 test was violated (i.e., when one cell had an expected count of <1, or >20% of the cells had an expected count of <5), Fisher's exact test was used. Odds ratios (ORs) with 95% confidence intervals (CIs) were determined for the disease susceptibility of specific genotypes and alleles. Results were considered statistically significant when the probability of findings occurring by chance was <5% (P < 0.05). The statistical analysis was done with SPSS v 20 and online software via http://vassarstats.net.
3. Results
3.1. Participants
The study included 100 EPTB cases and 102 healthy controls. Both patients and control groups were from ethnic Kashmiri population (North India). There were 60 Women and 40 Men in the patient group (Female / Male ratio = 1.5). The mean average age in the patient group was ±45.9 years, gender or age-releated differences were insignificant between the groups (p > 0.05). Among 102 patients with EPTB, smokers consisted of 25 (25%) patients. The Hardy–Weinberg equilibrium (HWE) analysis showed both SNPs to be in HWE in the control group (p > 0.05).
3.2. Pattern of the different genotypes
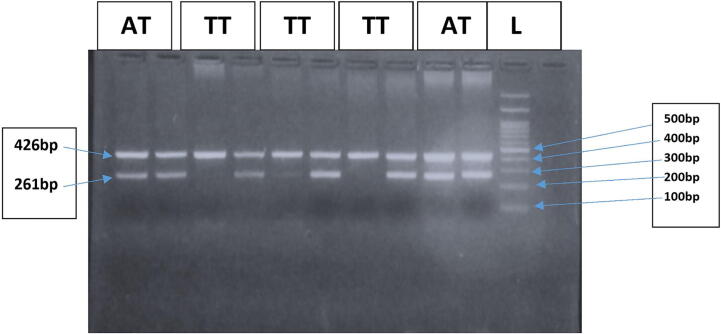
Genotypes of IFN-γ gene (+874 A/T) (rs2430561) and IL-10 (1082 A/G) (rs 1800896) were defined by Allele-Specific PCR method (Fig. 1, Fig. 2). The IFN-γ (+874 A/T) (rs2430561) genotypes were based on two different patterens of bands as 261 bp (Allele specific band) and 426 bp (internal control). For each sample, a two-tube reaction was setup (one tube containing primer specific for ‘A’ allele and second tube containing primer specific for ‘T’ allele, internal control was used as a confirmation that the PCR has functioned successfully. Presence or absence of bands at 261 bp in the two tubes was observed. If bands were present at 261 bp in both the tubes it represented “AT” genotype, if only first tube contained a band at 261 bp it represented “AA” genotype and if only second tube contained the band at 261 bp it depicted “TT” genotype (Fig. 1).
Fig. 1.
Representative gel picture showing different IFN-gamma (+874) genotypes. Lane 11 represents 100 bp DNA marker. Lanes 1/2 & 9/10 represent AT genotype. Lanes 3/4, 5/6 & 7/8 represent TT genotype. Band at 462 bp depicts internal control.
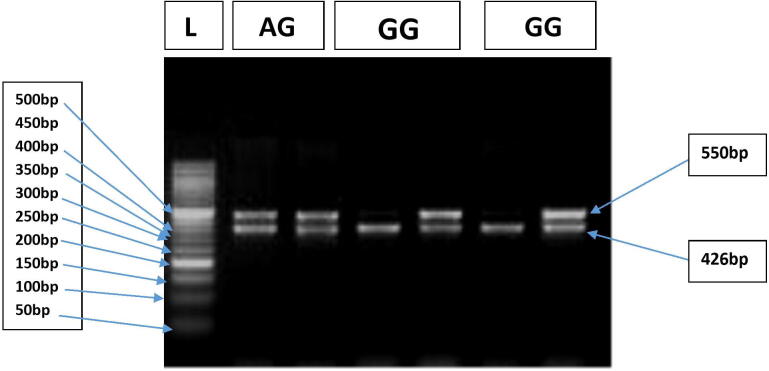
Fig. 2.
Representative gel picture showing different IL-10 (−1082) genotypes. Lane 1 shows 50 by DNA marker. Lanes 2,3 represent AG genotype Lanes 4,5 and 6,7 represent GG genotype. Band at 462 bp depicts the internal control.
The IL-10 (−1082 A/G) (rs1800896) genotypes were defined by Allele specific PCR based on two different patterens of bands as 550 bp (Allele specific band) and 426 bp (internal control band). For each sample, a two-tube reaction was setup (one tube containing primer specific for A allele and second tube containing primer specific for G allele, internal control was used as a confirmation that the PCR has functioned successfully. Presence or absence of bands at 550 bp in the two tubes was observed. If bands were present at 550 bp, in both the tubes it represented “AG” genotype, if only first tube contained a band at 550 bp it represented “AA” genotype and if only second tube contained band at 550 bp it depicted “GG” genotype (Fig. 2.)
3.3. IFN-γ (+874 A/T) (rs2430561) polymorphic analysis
In our study, we evaluated the genotype distribution & allele frequencies of IL-10 (−1082 A/G) & IFN-γ (+874 A/T) (rs2430561) polymorphisms in healthy individuals and patients with EPTB.
For IFN-γ (+874 A/T), ‘AA’ genotype was found in 52% cases and 35.2% in controls. ‘AT’ genotype was found in 40% cases, 47% controls, and ‘TT’ genotype in 8% cases and 17.6% controls. There was a statistically significant difference in the distribution of genotypic and allelic frequencies of IFN-γ (+874 A/T) SNP in cases and controls [OR, 0.58; 95% CI (0.32–1.05); P = 0.014] and [OR, 0.55; 95% CI (0.37–0.84); P = 0.006] [Table 3]. Indicating that IFN- γ (+874 ‘TT’ genotype & ‘T’ allele might act as protective variants against EPTB in Kashmiri population and that IFN-γ (+874 A/T) polymorphism might play a crucial role in the pathogenisis of EPTB. Further, the use of dominant and co-dominant models revealed that ‘TT’ & combined ‘TA + TT’ genotype was associated with reduced risk of EPTB compared to ‘AA’ genotype (p < 0.05) is given in Table 6.
Table 3.
Genotypic distribution of IFN-γ & IL-10 gene polymorphism in cases & controls.
| SNP | Genotype |
Cases (n = 100) |
Controls (n = 102) |
Odds Ratio (OR) | p-value |
|---|---|---|---|---|---|
| IFN- γ (A/T) | AA | 52 (52%) | 36 (35.2%) | 1.0 (Reference) | – |
| AT | 40 (40%) | 48 (47.0%) | 0.58 (0.32–1.05) | 0.9 | |
| TT | 08 (8%) | 18 (17.6%) | 0.31 (0.12–0.78) | 0.014 | |
|
Allele A T |
200 144 56 |
204 120 84 |
1.0 (Reference) 0.55 (0.37–0.84) |
0.006 |
|
| IL-10 (A/G) | AA | 28 (28%) | 32 (31.3%) | 1 (Reference) | – |
| AG | 70 (70%) | 60 (58.8%) | 1.33 (0.72–2.5) | 0.43 | |
| GG | 2 (2%) | 10 (9.8%) | 0.23 (0.04–1.13) | 0.1 | |
|
Allele A G |
200 126 74 |
204 124 80 |
1 (Reference) 0.91 (0.6–1.3) |
0.68 |
|
Table 6.
Association between IL and 10 (−1082 A/G) SNP and IFN-γ (A/T) with extrapulmonary tuberculosis under different models.
| SNP | Model | Genotype |
Cases 100 |
Controls 102 |
OR | p-value |
|---|---|---|---|---|---|---|
| IL10 (A/G) | Co-Dominant | AA GA GG |
28 (28) 70 (70) 2 (2) |
32 (31.3) 60 (58.8) 10 (9.8) |
1.33 (0.72–2.5) 0.23 (0.04–1.13) |
Ref. 0.43 0.1 |
| Dominant | AA GA + GG |
28 (28) 72 (72) |
32 (31.3) 70 (68.63) |
01.17 (0.64–2.15) |
Ref. 0.45 |
|
| Recessive | GA + AA GG |
98 (98) 2 (2) |
92 (90.2) 10 (9.8) |
0.19 (0.04–0.88) |
Ref. 0.03 |
|
|
IFN-γ (A/T) |
Co-Dominant | AA TA TT |
52 (52) 40 (40) 08 (8) |
36 (35.2) 48 (47) 18 (17.6) |
0.58 (0.32–1.05) 0.31 (0.12–0.78) |
Ref. 0.09 0.014 |
| Dominant | AA TA + TT |
52 (52) 48 (48) |
36 (35.3) 66 (64.7) |
0.50 (0.29–0.88) |
Ref. 0.02 |
|
| Recessive | TA + AA TT |
92 (92) 8 (8) |
84 (82.35) 18 (17.65) |
0.40 (0.17–0.98) |
Ref. 0.05 |
|
The association of IFN-γ (+874 A/T) with clinical parameters showed no statistical significance at all (Table 4).
Table 4.
Association between IFN-γ (+874 A/T) SNP and various clinical parameters of extrapulmonary tuberculosis patients.
| PARAMETER | AA | TA + TT | p-value |
|---|---|---|---|
|
AGE <40 (51) >40 (49) |
27 (52.94%) 30 (61.22%) |
24 (47.06%) 19 (38.78%) |
0.42 |
|
GENDER Females (60%) Males (40%) |
30 (50%) 27 (67.5%) |
30 (50%) 13 (32.5%) |
0.1 |
|
FAMILY HISTORY Positive (16%) Negative (84%) |
10 (62.5%) 48 (57.14%) |
6 (37.5%) 30 (42.86%) |
1 |
|
SMOKING DETAILS YES 25 (25%) NO 75 (75%) |
18 (72%) 38 (50.67%) |
7 (28%) 37 (49.33) |
0.1 |
|
DWELLING AREAS RURAL (75%) URBAN (25%) |
48 (64%) 09 (36%) |
27 (36%) 16 (64%) |
0.02 |
3.4. IL-10 (−1082 A/G) (rs1800896) polymorphic analysis
For IL-10 (−1082 A/G) (rs1800896) SNP, ‘AA’ genotype was found in 28% cases and 31.3% in controls. ‘AG’ genotype was found in 70% cases and 58.8% controls while as ‘GG’ genotype was found in 2% cases and 9.8% controls. We found no significant differences in genotypic and allelic frequencies of IL-10 (−1082 A/G) (rs1800896) in cases and controls [OR, 1.33; 95% CI (0.72–2.5); P = 0.43] and [OR, 0.91; 95% CI (0.6–1.3); P = 0.68] [Table 3]. Furthermore, IL-10 (−1082 A/G) did not reveal any significant association with any of the clinical parameters studied (Table 5). However, the use of recessive model revealed that ‘GG’ genotype was associated with lower risk of EPTB compared to combined ‘GA + AA’ genotype (p < 0.05) is given in Table 6.
Table 5.
Association between IL and 10 (−1082 A/G) SNP and various clinical parameters of extrapulmonary tuberculosis patients.
|
IL-10 PARAMETER |
AA | GA + GG | p-value |
|---|---|---|---|
|
AGE ≤40 (51%) >40 (49%) |
11 (21.57%) 10 (20.4%) |
40 (78.43%) 39 (79.60%) |
1 |
|
GENDER Females (60%) Males (40%) |
10 (16.67%) 11 (27.5%) |
50 (83.33%) 29 (72.5%) |
0.21 |
|
FAMILY HISTORY Positive (16%) Negative (84%) |
2 (12.5%) 19 (22.62%) |
14 (90%) 65 (77.38%) |
0.5 |
|
SMOKING DETAILS YES (25%) NO (75%) |
5 (20%) 16 (21.33%) |
19 (80%) 59 (78.67%) |
1 |
|
DWELLING AREAS RURAL (75%) URBAN (25%) |
16 (21.33%) 5 (20%) |
59 (78.67%) 20 (80%) |
1 |
4. Discussion
To our knowledge, this is a first comparative study of cytokine markers in EPTB patients in Kashmiri population (North India). The case-control studies involving selected loci across ethnicities are helpful in recognizing alleles, which are associated with disease severity and suceptibility. The importance of genetics in tuberculosis pathogensis has now become evident with ethnicity playing a pivotal role. The single nucleotide polymorphism (SNP) association studies are used in identification of genes causing diseases in humans and variation in drug responses between different individuals. Major medical benefits could be derived from such research areas. Establishing an association between various polymorphisms and certain diseases could result in the identification of molecular markers, which could act as specific signatures of certain diseases and help in the early diagnosis. Furthermore, elucidation of drug response in comparison to individual genetic make-up can be used to develop genome based medicines, that are more beneficial, have minimum side effects and higher efficacy for each individual (Shastry, 2007).
Therefore, the present study aimed to find out the association of polymorphic variations in selected candidate genes in relation to EPTB susceptibility in ethnic Kashmiri population (North India). The association of Cytokine gene polymorphisms with tuberculosis has shown variable results in different populations. IL-10 (−1082 A/G) (rs 1800896) and IFN-γ (+874 A/T) (rs2430561) SNPs are the most common polymorphisms studied in EPTB. There are reports that the ‘A’ allele of IFN-γ (+874 A/T) SNP is more common in patients than T allele in many populations [Italian, South African, Hong-kong, Chinese and Spanish] (Rossouw et al., 2003, Selvaraj et al., 2008, Tso et al., 2005).
Our study shows that the ‘TT’ genotype and ‘T’ allele is over-represented in healthy controls than in patients with EPTB. Moreover, a study in Colombia has shown that the IFN-gamma +874 T allele is significantly associated with localized pleural disease. Studies in different ethnic groups have shown variable results regarding the association of tuberculosis with cytokine gene SNPs (López-Maderuelo et al., 2003).
Increased IFN-γ expression is associated with T allele; it has been shown that transcriptional factor NF-κβ has a binding preference for DNA containing IFN-γ 874 T allele, which increases the expression of this gene. Two studies have found that ‘T’ allele was associated with either milder form of pulmonary tuberculosis or with localized pleural tuberculosis (Palomino et al., 2007), but not with advanced pulmonary tuberculosis. Our study also confirms the significance of IFN-γ 874 A/T SNP in EPTB, here we show that ‘TT’ genotype and T allele of this SNP is associated with increased protection against active EPTB as compared to ‘AA’ genotype & ‘A’ allele.
IL-10 is an effective regulatory cytokine that has an important function during the dormant tuberculosis stage and higher production of IL-10 has shown suppression in [cell mediated] immunity against intracellular infection & also results in increased reactivation of disease in mice (Ansari et al., 2009). Moreover, many studies report that IL-10 polymorphisms in the promoter region [including IL-10 (−1082 A/G) polymorphism] increase the risk of the tuberculosis predisposition.
In our study, IL-10 (−1082 A/G) SNP did not show any association with EPTB (p > 0.05). Our results are in agreement with Dolores et al. where they didn’t find any association with pulmonary tuberculosis and IL-10 (−1082 A/G) SNP in Spanish population (López-Maderuelo et al., 2003). However, some studies show the association of IL-10 (−1082 A/G) SNP with tuberculosis (Liang et al., 2011). Since, it is known that the results of polymorphic studies vary considerably from population to population and ethnicity is an important determining factor. Furthermore, these discrepancies in results might be due to the heterogeneous frequency of SNPs in different populations (Ansari et al., 2009).
We would like to stress the importance of similar studies with functional analysis be carried in different populations in order to confirm our findings.
5. Conclusion
We conclude that the IFN-γ 874 ‘TT’ genotype & ‘T’ Allele have a strong association with the EPTB and might play a protective role against EPTB in Kashmiri population, whereas, IL-10 (−1082 A/G) ‘GG’ genotype is associated with reduced risk of EPTB compared to combined ‘GA + AA’ genotype.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
BAW, FS and DA drafted the experimental design. BAW and SS performed the experiments. AK, AY, MM, DA, FA, AD and RS helped in data collection, data analysis and initial draft of manuscript text. All authors read the manuscript before communication.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to thank the Deanship of Scientific Research at Taif University for their support through Researchers Supporting Project number (TURSP - 2020/222), Taif University, Taif, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Rajni Singh, Email: rsingh3@amity.edu.
Dil Afroze, Email: afroze@yahoo.com.
References
- Alvarez S., McCabe W.R. Extrapulmonary tuberculosis revisited: a review of experience at Boston City and other hospitals. Medicine (Baltimore) 1984;63:25–55. [PubMed] [Google Scholar]
- Ansari A., Talat N., Jamil B., Hasan Z., Razzaki T., Dawood G., Hussain R. Cytokine Gene Polymorphisms across Tuberculosis Clinical Spectrum in Pakistani Patients. PLOS One. 2009;4 doi: 10.1371/journal.pone.0004778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caws M., Thwaites G., Dunstan S., Hawn T.R., Lan N.T.N., Thuong N.T.T., Stepniewska K., Huyen M.N.T., Bang N.D., Loc T.H., Gagneux S., van Soolingen D., Kremer K., van der Sande M., Small P., Anh P.T.H., Chinh N.T., Quy H.T., Duyen N.T.H., Tho D.Q., Hieu N.T., Torok E., Hien T.T., Dung N.H., Nhu N.T.Q., Duy P.M., van Vinh Chau N., Farrar J. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakaya, J., Khan, M., Ntoumi, F., Aklillu, E., Fatima, R., Mwaba, P., Kapata, N., Mfinanga, S., Hasnain, S.E., Katoto, P.D.M.C., Bulabula, A.N.H., Sam-Agudu, N.A., Nachega, J.B., Tiberi, S., McHugh, T.D., Abubakar, I., Zumla, A., 2021. Global Tuberculosis Report 2020 – Reflections on the Global TB burden, treatment and prevention efforts. Int. J. Infect. Dis. 10.1016/j.ijid.2021.02.107 [DOI] [PMC free article] [PubMed]
- Cooper A.M., Dalton D.K., Stewart T.A., Griffin J.P., Russell D.G., Orme I.M. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döffinger R., Helbert M.R., Barcenas-Morales G., Yang K., Dupuis S., Ceron-Gutierrez L., Espitia-Pinzon C., Barnes N., Bothamley G., Casanova J.-L., Longhurst H.J., Kumararatne D.S. Autoantibodies to interferon-gamma in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2004;38:e10–14. doi: 10.1086/380453. [DOI] [PubMed] [Google Scholar]
- Domingo‐Gonzalez, R., Prince, O., Cooper, A., Khader, S.A., 2017. Cytokines and Chemokines in Mycobacterium tuberculosis Infection, in: Tuberculosis and the Tubercle Bacillus. John Wiley & Sons, Ltd, pp. 33–72. 10.1128/9781555819569.ch2 [DOI] [PMC free article] [PubMed]
- Fiske C.T., de Almeida A.S., Shintani A.K., Kalams S.A., Sterling T.R. Abnormal immune responses in persons with previous extrapulmonary tuberculosis in an in vitro model that simulates in vivo infection with Mycobacterium tuberculosis. Clin. Vaccine Immunol. CVI. 2012;19:1142–1149. doi: 10.1128/CVI.00221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitness J., Floyd S., Warndorff D.K., Sichali L., Malema S., Crampin A.C., Fine P.E.M., Hill A.V.S. Large-scale candidate gene study of tuberculosis susceptibility in the Karonga district of northern Malawi. Am. J. Trop. Med. Hyg. 2004;71:341–349. [PubMed] [Google Scholar]
- Furin J., Cox H., Pai M. Tuberculosis. The Lancet. 2019;393:1642–1656. doi: 10.1016/S0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- Gideon H.P., Phuah J., Myers A.J., Bryson B.D., Rodgers M.A., Coleman M.T., Maiello P., Rutledge T., Marino S., Fortune S.M., Kirschner D.E., Lin P.L., Flynn J.L. Variability in Tuberculosis Granuloma T Cell Responses Exists, but a Balance of Pro- and Anti-inflammatory Cytokines Is Associated with Sterilization. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gay M.A., Oliver J., Orozco G., Garcia-Porrua C., Lopez-Nevot M.A., Martin J. Lack of association of a functional single nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with susceptibility to biopsy-proven giant cell arteritis. J. Rheumatol. 2005;32:1510–1512. [PubMed] [Google Scholar]
- Harapan H., Fajar J.K., Wahyuniati N., Anand J.R., Nambaru L., Jamil K.F. Non-HLA gene polymorphisms and their implications on dengue virus infection. Egypt. J. Med. Hum. Genet. 2013;14:1–11. doi: 10.1016/j.ejmhg.2012.08.003. [DOI] [Google Scholar]
- Hasan Z., Jamil B., Ashraf M., Islam M., Yusuf M.S., Khan J.A., Hussain R. ESAT6-induced IFNgamma and CXCL9 can differentiate severity of tuberculosis. PloS One. 2009;4 doi: 10.1371/journal.pone.0005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, W., Yu, J., Du, J., Yang, S., Chen, H., Liu, J., Ma, J., Li, M., Qin, J., Shu, W., Zong, P., Zhang, Yi, Dong, Y., Yang, Z., Mei, Z., Deng, Q., Wang, P., Han, W., Wu, M., Chen, L., Zhao, X., Tan, L., Li, F., Zheng, C., Liu, H., Li, X., A, E., Du, Y., Liu, F., Cui, W., Wang, Q., Chen, X., Han, J., Xie, Q., Feng, Y., Liu, W., Tang, P., Zhang, Jianyong, Zheng, J., Chen, D., Yao, X., Ren, T., Li, Yan, Li, Yuanyuan, Wu, L., Song, Q., Yang, M., Zhang, Jian, Liu, Y., Guo, S., Yan, K., Shen, X., Lei, D., Zhang, Yanli, Yan, X., Li, L., Tang, S., 2020. The epidemiology of extrapulmonary tuberculosis in China: A large-scale multi-center observational study. PLOS One 15, e0237753. 10.1371/journal.pone.0237753 [DOI] [PMC free article] [PubMed]
- Lee J.Y. Diagnosis and Treatment of Extrapulmonary Tuberculosis. Tuberc. Respir. Dis. 2015;78:47. doi: 10.4046/trd.2015.78.2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Zhao Y.-L., Yue J., Liu J.-F., Han M., Wang H., Xiao H. Interleukin-10 gene promoter polymorphisms and their protein production in pleural fluid in patients with tuberculosis. FEMS Immunol. Med. Microbiol. 2011;62:84–90. doi: 10.1111/j.1574-695X.2011.00791.x. [DOI] [PubMed] [Google Scholar]
- Lin P.L., Rodgers M., Smith L., Bigbee M., Myers A., Bigbee C., Chiosea I., Capuano S.V., Fuhrman C., Klein E., Flynn J.L. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect. Immun. 2009;77:4631–4642. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio D., Marino V., Serauto A., Gioia V., Scola L., Crivello A., Forte G.I., Colonna-Romano G., Candore G., Caruso C. Genotype frequencies of the +874T→A single nucleotide polymorphism in the first intron of the interferon-γ gene in a sample of Sicilian patients affected by tuberculosis. Eur. J. Immunogenet. 2002;29:371–374. doi: 10.1046/j.1365-2370.2002.00327.x. [DOI] [PubMed] [Google Scholar]
- López-Maderuelo D., Arnalich F., Serantes R., González A., Codoceo R., Madero R., Vázquez J.J., Montiel C. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 2003;167:970–975. doi: 10.1164/rccm.200205-438BC. [DOI] [PubMed] [Google Scholar]
- Maiolini M., Gause S., Taylor J., Steakin T., Shipp G., Lamichhane P., Deshmukh B., Shinde V., Bishayee A., Deshmukh R.R. The War against Tuberculosis: A Review of Natural Compounds and Their Derivatives. Molecules. 2020;25 doi: 10.3390/molecules25133011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos, G.I., Covas, C. de J.F., Bittar, R. de C., Gomes-Silva, A., Marques, F., Maniero, V.C., Amato, V.S., Oliveira-Neto, M.P., Mattos, M. da S., Pirmez, C., Sampaio, E.P., Moraes, M.O., Da-Cruz, A.M., 2007. IFNG +874T/A polymorphism is not associated with American tegumentary leishmaniasis susceptibility but can influence Leishmania induced IFN-gamma production. BMC Infect. Dis. 7, 33. 10.1186/1471-2334-7-33 [DOI] [PMC free article] [PubMed]
- Möller M., Hoal E.G. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberc. Edinb. Scotl. 2010;90:71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Möller M., Nebel A., van Helden P.D., Schreiber S., Hoal E.G. Analysis of eight genes modulating interferon gamma and human genetic susceptibility to tuberculosis: a case-control association study. BMC Infect. Dis. 2010;10:154. doi: 10.1186/1471-2334-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A., Ma X., Reich R.A., Graviss E.A. No association between the +874T/A single nucleotide polymorphism in the IFN-gamma gene and susceptibility to TB. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Tuberc. Lung Dis. 2007;11:113–115. [PubMed] [Google Scholar]
- Moreira A.L., Tsenova L., Murray P.J., Freeman S., Bergtold A., Chiriboga L., Kaplan G. Aerosol infection of mice with recombinant BCG secreting murine IFN-gamma partially reconstitutes local protective immunity. Microb. Pathog. 2000;29:175–185. doi: 10.1006/mpat.2000.0382. [DOI] [PubMed] [Google Scholar]
- Murray P.J., Young R.A. Increased antimycobacterial immunity in interleukin-10-deficient mice. Infect. Immun. 1999;67:3087–3095. doi: 10.1128/iai.67.6.3087-3095.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme I.M., Basaraba R.J. The formation of the granuloma in tuberculosis infection. Semin. Immunol. 2014;26:601–609. doi: 10.1016/j.smim.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Pacheco A.G., Cardoso C.C., Moraes M.O. IFNG +874T/A, IL10 -1082G/A and TNF -308G/A polymorphisms in association with tuberculosis susceptibility: a meta-analysis study. Hum. Genet. 2008;123:477–484. doi: 10.1007/s00439-008-0497-5. [DOI] [PubMed] [Google Scholar]
- Palomino, J.C., Leao, S.C., Ritacco, V., 2007. Tuberculosis 2007; from basic science to patient care.
- Ranaivomanana P., Raberahona M., Rabarioelina S., Borella Y., Machado A., Randria M.J.D.D., Rakotoarivelo R.A., Rasolofo V., Rakotosamimanana N. Cytokine Biomarkers Associated with Human Extra-Pulmonary Tuberculosis Clinical Strains and Symptoms. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw M., Nel H.J., Cooke G.S., van Helden P.D., Hoal E.G. Association between tuberculosis and a polymorphic NFκB binding site in the interferon γ gene. The Lancet. 2003;361:1871–1872. doi: 10.1016/S0140-6736(03)13491-5. [DOI] [PubMed] [Google Scholar]
- Selvaraj P., Alagarasu K., Harishankar M., Vidyarani M., Nisha Rajeswari D., Narayanan P.R. Cytokine gene polymorphisms and cytokine levels in pulmonary tuberculosis. Cytokine. 2008;43:26–33. doi: 10.1016/j.cyto.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Shastry B.S. SNPs in disease gene mapping, medicinal drug development and evolution. J. Hum. Genet. 2007;52:871–880. doi: 10.1007/s10038-007-0200-z. [DOI] [PubMed] [Google Scholar]
- Sousa-Vasconcelos, P. da S., Seguins, W. da S., Luz, E. de S., Pinho, R.T. de, 2015. Pattern of cytokine and chemokine production by THP-1 derived macrophages in response to live or heat-killed Mycobacterium bovis bacillus Calmette-Guérin Moreau strain. Mem. Inst. Oswaldo Cruz 110, 809–813. 10.1590/0074-02760140420 [DOI] [PMC free article] [PubMed]
- Tso H.W., Ip W.K., Chong W.P., Tam C.M., Chiang A.K.S., Lau Y.L. Association of interferon gamma and interleukin 10 genes with tuberculosis in Hong Kong Chinese. Genes Immun. 2005;6:358–363. doi: 10.1038/sj.gene.6364189. [DOI] [PubMed] [Google Scholar]
- Turner J., Gonzalez-Juarrero M., Ellis D.L., Basaraba R.J., Kipnis A., Orme I.M., Cooper A.M. In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J. Immunol. Baltim. Md. 2002;1950(169):6343–6351. doi: 10.4049/jimmunol.169.11.6343. [DOI] [PubMed] [Google Scholar]
- Winek J., Rowinska-Zakrzewska E., Demkow U., Szopinski J., Szolkowska M., Filewska M., Jagodzinski J., Roszkowski-Sliz K. Interferon gamma production in the course of Mycobacterium tuberculosis infection. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2008;59(Suppl 6):751–759. [PubMed] [Google Scholar]