Graphical abstract
Keywords: mRNA, Self-amplifying mRNA (saRNA), Innate immune stimulation, Type I IFNs, Immune evasion strategies, mRNA vaccination
Abstract
The recent approval of messenger RNA (mRNA)-based vaccines to combat the SARS-CoV-2 pandemic highlights the potential of both conventional mRNA and self-amplifying mRNA (saRNA) as a flexible immunotherapy platform to treat infectious diseases. Besides the antigen it encodes, mRNA itself has an immune-stimulating activity that can contribute to vaccine efficacy. This self-adjuvant effect, however, will interfere with mRNA translation and may influence the desired therapeutic outcome. To further exploit its potential as a versatile therapeutic platform, it will be crucial to control mRNA’s innate immune-stimulating properties. In this regard, we describe the mechanisms behind the innate immune recognition of mRNA and provide an extensive overview of strategies to control its innate immune-stimulating activity. These strategies range from modifications to the mRNA backbone itself, optimization of production and purification processes to the combination with innate immune inhibitors. Furthermore, we discuss the delicate balance of the self-adjuvant effect in mRNA vaccination strategies, which can be both beneficial and detrimental to the therapeutic outcome.
1. Introduction
The recent FDA approval of the first messenger RNA (mRNA) vaccine formulations, developed by Pfizer/BioNTech and Moderna, to control the worldwide COVID-19 pandemic, created a new wave of attention for mRNA-based therapeutics [1]. Currently, several other mRNA-based COVID-19 vaccine formulations are still under investigation, containing either conventional non-amplifying mRNA or self-amplifying mRNA (saRNA) [2], [3], [4], [5]. These successful outcomes are the result of a long and tedious journey that started in 1961 when conventional mRNA was first discovered, followed by successful pre-clinical experiments in 1990 [6], [7], [8]. In contrast, the larger saRNA gained a lot of attention, mainly for vaccination purposes as first reported in 1994 due to is self-replicating ability [2], [3]. The latter results in enhanced and prolonged protein expression which overcomes the often transient nature of conventional mRNA therapeutics [2], [3], [4]. Besides its use in vaccination against infectious diseases or cancer, mRNA is currently explored for many other non-immunotherapy applications, including protein replacement therapy, cell reprogramming and gene editing [7], [9], [10]. To date, a variety of mRNA formulations for different therapeutic indications have entered clinical trials, studying their safety and efficacy [2], [9], [11].
Several concerns, such as extra- and intracellular instability, intracellular delivery and inherent immunogenicity, delayed the translation of mRNA therapeutics to the clinic. Over the last few years, substantial progress was made in the design of the mRNA molecule itself and the development of appropriate delivery vehicles to overcome these concerns [9], [12]. First, stability issues are tackled by modifications of the mRNA backbone structure, such as 5′ capping, 5′ and 3′ untranslated regions (UTR) optimization and poly(A) tail addition [10], [13]. Second, the development of an optimal carrier has been a continuous research focus to improve both the stability and cellular uptake efficiency of mRNA. Both viral and non-viral carriers have been tested, where non-viral nanoparticles have emerged as a safer alternative to their viral counterparts, with lipid-nanoparticles being the most clinically advanced [10], [11], [14], [15]. An overview of the available carrier systems, their composition and the administration route is outside the scope of this review but is excellently summarized elsewhere [9], [11].
A third but very important factor influencing the effect of mRNA therapeutics is the inherent innate immune-stimulating activity of mRNA, which can either support or hamper the therapeutic outcome. Non-immunotherapy-based applications suffer from this intrinsic innate immune stimulation as it induces an overall cellular antiviral state in most cell types, promoting mRNA degradation and inhibiting the translation process. On the contrary, vaccination purposes might benefit from additional innate immune stimulation. However, this concept is rather complex and mainly depends on finding the optimal balance between the different factors involved [16], [17]. The innate immune activation by mRNA therapeutics may also result in side effects ranging from flu-like symptoms to risks of autoimmune diseases [18]. Hence, there is a high need to control and fine-tune inherent innate immunogenicity to align it with the anticipated therapeutic effect and to guarantee the safety of mRNA therapeutics. In this review, we will provide an extensive overview of mRNA-induced innate immunity and more specifically the strategies to control these responses. First, we will present the structural differences between conventional and self-amplifying mRNA, each with their advantages and limitations. Second, we will provide a concise overview of the cellular pathways leading to this inherent innate immune stimulating activity. Third, an in-depth discussion of the current strategies explored to control mRNA immunity will be provided, together with their influence on the safety and efficacy of conventional mRNA and saRNA-based therapeutics for different applications. Generally, these strategies can be divided in three main categories, including modifications to the backbone structure, optimization of the production and purification processes, and the use of innate immune inhibitors as a supplement therapy. Finally, given the more ambiguous situation for mRNA-based vaccines, we will provide a concise overview of the current knowledge and discussion regarding the influence of mRNA-induced immune responses on vaccine efficiency.
2. mRNA versus self-amplifying mRNA
2.1. Conventional mRNA
Messenger RNA (mRNA) is a fairly simple and relatively small single-stranded structure of around 1000 – 5000 nucleotides. It is typically made up of five elements that are critical for its expression: a cap structure (m7GpppN), a 5′ untranslated region (5′ UTR), an open reading frame (ORF) encoding the gene of interest (GOI) flanked by the start and stop codon, a 3′ untranslated region (3′ UTR) and a poly(A) tail at the 3′ end of about 100 – 250 adenosine residues. In living cells, mRNA is transcribed from DNA by RNA polymerase, spliced and subsequently transported from the nucleus to the cytosol. Then, the ribosomes are recruited, resulting in the direct but transient expression of the encoded protein, during the life span of the mRNA molecules (Fig. 1 , left) [19].
Fig. 1.
Schematic representation of the structure of conventional mRNA and the structure and intracellular amplification of self-amplifying mRNA (saRNA). (Left) Conventional mRNA consists of five critical elements: a 5′ cap structure (m7GpppN), a 5′ untranslated region (5′ UTR), the gene of interest (GOI), a 3′ untranslated region (3′ UTR) and a poly(A) tail. After conventional mRNA delivery into the cell cytosol, the encoded protein is produced directly. (Right) saRNA consists of a 5′ cap structure, a 5′ UTR, the sequences of viral non-structural proteins (nsP1-4), a subgenomic promoter (SGP), the GOI, a 3′ UTR and a poly(A) tail. (A) When saRNA arrives in the host cell cytosol, nsP1-4 is translated and forms the early replication complex (B) that generates a complementary negative-sense RNA strand from the original saRNA. (C) In a later phase, the early replication complex is cleaved into the individual nsPs. These form the cleaved replicase that uses the negative-sense RNA strand as a template to generate new copies of the original genomic RNA and also recognizes the SGP and triggers the production of an enormous amount of subgenomic RNAs, (D) which eventually leads to the production of the protein of interest.
In analogy to naturally produced mRNA, synthetic mRNA is prepared by in vitro transcription (IVT) of a linearized plasmid DNA (pDNA) or PCR template containing the gene of interest and a promoter region for a bacteriophage T7, SP6 or T3 RNA polymerase. To start the IVT reaction, also the four ribonucleotide triphosphates (NTPs), a ribonuclease (RNase) inhibitor (to inactivate contaminating RNase), a pH buffer and Mg2+ as co-factor for the RNA polymerase should be provided. The transcription is initiated by binding of RNA polymerase to its promoter sequence, after which it links ribonucleotides together as it moves along the DNA template to form the complementary mRNA strand. The RNA transcript is completed when the enzyme runs off at the end of the template (runoff transcription), ready for the next round. At the end, the DNA template is fragmented by treatment with a DNase and the produced mRNA is purified from the remaining reaction compounds, such as the RNA polymerase and the unincorporated NTPs [20].
The simplicity of the mRNA construct and its relatively small size compared to saRNA are key features promoting the use of conventional mRNA. Due to its short half-life and inherent instability, however, only low and transient protein production levels could be achieved. To date, remarkable improvements in mRNA translation efficiency and stability have been made through sequence optimization, the incorporation of chemically modified nucleosides, and several advanced purification strategies. Yet, protein expression remains proportional to the number of mRNA molecules successfully delivered to the cytosol of the cells and relatively high doses are often required to obtain significant therapeutic effects [3], [13], [21], [22], [23].
2.2. Self-amplifying mRNA
When compared to conventional, non-amplifying mRNA, saRNA is a more complex and considerably larger (9000 – 12,000 nucleotides) structure as besides the basic elements found in an mRNA molecule (a cap, 5′ UTR, an ORF with GOI, 3′ UTR and poly(A) tail), saRNA also contains the coding sequences of a viral replicase complex, a genomic and a subgenomic (SG) promoter [3], [20], [24]. Most saRNAs are based on the genome of alphaviruses such as the Sindbis virus (SINV), Semliki Forest virus (SFV), and Venezuelan equine encephalitis virus (VEEV). Alphaviruses are a group of small, enveloped positive-stranded RNA viruses producing considerable amounts of subgenomic RNA encoding for viral structural proteins during their natural replication cycle in the host cell cytosol [25]. saRNAs benefit from the self-replicating ability of these alphaviruses by retaining their non-structural proteins (nsP1-4), which include the replication machinery, while replacing the viral structural proteins by the GOI, which renders the mRNA incapable of producing infectious viral particles [3]. Although saRNA clearly differs structurally from conventional mRNA, it is produced in the same manner during an IVT reaction as described above [3], [20].
Unlike conventional mRNA, the GOI cannot be immediately translated from the originally delivered saRNA molecule upon entry in the host cell. As depicted in Fig. 1 (right), after cytosolic delivery, the host cell machinery is immediately engaged for the translation of nsP1-4, which together make up the replicase complex. This complex attaches to the plasma membrane where so-called spherules, i.e. membrane-associated structures, are formed in which replication takes place protected from cellular defense mechanisms. In a first phase, the translated nsP1-4 polyprotein is cleaved by nsP2 to generate the early replication complex (replicase), which consists of the nsP1-3 polyprotein and associated nsP4 (Fig. 1 , A), and transcribes the original saRNA into its complementary negative-sense RNA strand (Fig. 1 , B). In a later phase, the whole replication complex is cleaved into the individual nsPs that join together to form the cleaved replicase. Now, the negative-sense RNA strand serves as a template for the production of new copies of the original genomic RNA but also for the production of a high amount of shorter, SG RNAs from a 26S SG promoter (Fig. 1 , C). Subsequently, high levels of the protein of interest are translated from these SG RNAs (Fig. 1 , D) [3], [26], [27]. Overall, it is clear that the replicase complex is a multifunctional complex, in which each of the non-structural proteins has their own specific function. nsP1 is an enzyme required for 5′ capping of viral RNA and serves as an anchor to tether the replicase complex to the plasma membrane. nsP2 has helicase activity to unwind the RNA duplex during replication, but also has an important protease activity cleaving the polyprotein into individual nsPs. The function of nsP3 is not completely understood yet, but it is certainly an essential compound in the replicase complex. nsP4 is an RNA-dependent RNA polymerase and structures the replicase complex [28].
Due to its auto-replicative ability, a higher protein expression level can be maintained and also the duration of protein expression is significantly extended [3], [22], [23]. For example, Brito et al. evaluated the expression of a cationic nanoemulsion-formulated saRNA in non-human primates after intramuscular delivery using secreted alkaline phosphatase (SEAP) as reporter gene. Expression from saRNA reached its peak expression 3 days post-administration and remained measurable at least 14 days after injection [29]. Recently, Leyman et al. found that conventional mRNAs reached their maximum expression 1 day after intradermal electroporation in pigs, followed by a steady decrease in expression until day 6, whereas the expression of saRNA was similar to that of conventional mRNA at day 1, but peaked at day 6 and persisted until day 12 [30]. Additionally, in the vaccination context, Vogel et al. showed that saRNAs can be delivered at lower concentrations than unmodified conventional mRNA while still achieving an equivalent protection against influenza viruses [31]. However, in comparison to conventional mRNA, saRNA is a more complex and much larger molecule, thus complicating its delivery. Recent efforts to reduce its size resulted in the generation of trans-amplifying RNAs in which the saRNA is split into two transcripts, one encoding nsP1-4 and the other encoding the GOI as a “transreplicon” [32], [33]. Beissert et al. found that mRNA translation from such a trans-amplifying RNA vaccine was as efficient as its saRNA counterpart and an influenza-hemagglutinin encoding trans-amplifying mRNA successfully induced protective immune responses in mice upon intradermal injection [33]. Nevertheless, the mRNA coding for the replicase is still longer than 7 kb, which is considerably larger than the typical conventional mRNA and will therefore also require delivery vectors with a high loading capacity for efficient RNA delivery. Furthermore, saRNA not only encodes the protein of interest but also viral nsPs. These viral proteins might potentially be immunogenic, which may limit the repeated use of saRNA-based therapeutics. However, little information is available regarding the immunogenicity of the replicase complex itself and additional studies should be conducted to further explore this [3], [20], [21], [23].
3. Innate immunity stimulation
Our innate immune system has evolved to recognize non-self molecules, such as viral single-stranded (ss) and double-stranded (ds) RNA, initiating a series of signaling pathways to fight the pathogen. Both conventional mRNA and saRNA are potent activators of the innate immune system, leading to interferon (IFN) secretion which hampers RNA translation and promotes mRNA degradation [10], [16]. saRNA might be somewhat more immunogenic than conventional mRNA, as during self-replication in the host cell’s cytosol dsRNA amplification intermediates are formed, which can be recognized as foreign [5], [21], [22]. Moreover, to retain saRNA functionality, some commonly applied strategies to reduce the innate immune-stimulating activity of conventional mRNA, like nucleoside base modification and sequence alteration (see Section 4.1), are not tolerated. Below, we will provide a concise overview of the initiated pathways and factors involved in this innate immune-stimulating process.
Activation of the innate immune system can be initiated by stimulation of pattern recognition receptors (PRRs) through binding with pathogen-associated molecular patterns (PAMPs). The mRNA binding PRRs can be classified according to their cellular locations as the cytosolic PRRs and endosomal PRRs. In the cytosol, the PRRs include the nucleotide oligomerization domain like receptors (NLRs) and the RIG-I-like receptors (RLRs). In the endosomes, the PRRs are represented by the toll-like receptors (TLRs). Upon mRNA recognition, a complex series of interacting signaling pathways is initiated, eventually leading to the production of type I IFNs and pro-inflammatory cytokines as presented in Fig. 2 [10], [16], [24], [34], [35], [36].
Fig. 2.
Innate immune pathways stimulated upon mRNA delivery. Simplified representation of mRNA recognition by both cytosolic and endosomal pattern recognition receptors (PRRs) leading to production of type I interferons (IFN) and pro-inflammatory cytokines. Subsequent activation of the interferon-α/β receptor (IFNAR) results in Janus Kinase - signal transducer activator of transcription (JAK-STAT) signaling leading to interferon stimulated genes (ISGs) production. Finally, PKR and OAS activation can result in diminished mRNA translation and enhanced mRNA degradation, respectively. Site of action of the inhibitors, discussed in Section 4.3., is visualized. Abbreviations: dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; TLR, Toll-like receptor; MDA-5, melanoma differentiation-associated protein 5; RIG-I, retinoic-acid-inducible gene I; MAVS, mitochondrial adaptor molecule; TRIF, TIR-domain-containing adaptor inducing IFN-b; MyD88, myeloid differentiation primary response gene 88; TRAF, TNF receptor-associated factor; IKK, IkB kinase; TBK1, TANK-binding kinase 1; IRF, interferon-regulatory factor; NEMO, NF-κB essential modulator; NF-κB, nuclear factor-kB; TYK2, tyrosine kinase 2; ISGs, Interferon Stimulated Genes; PKR, dsRNA-dependent protein kinase; EIF2a, eukaryotic translation initiator factor 2; OAS, 20–50-oligoadenylate synthetase.
All TLRs which have been identified in humans, differ in their cellular location and ligand specificity. Endosomal mRNA-binding TLRs are TLR3, binding to dsRNA, and TLR7/8, which recognize uridine-rich ssRNA (Fig. 2). Stimulation of TLR3 and TLR 7/8 leads to the activation of the adaptor proteins, TIR-domain-containing adaptor-inducing IFN-β (TRIF) and myeloid differentiation primary response gene 88 (MYD88) respectively. Subsequently, signaling pathways involving several mediators, such as TRAF proteins and the IkB kinase (IKK) complex, are initiated, resulting in the activation and nuclear import of the transcription factors IRF3, IRF7 and NF-κB. Ultimately, this coordinates the production of type I IFNs, a subset of interferon stimulated genes (ISGs) and pro-inflammatory cytokines, such as IL-6 and IL-12 [10], [16], [24], [34], [36], [37], [38]. The RLRs, RIG-I and melanoma differentiation-associated protein 5 (MDA-5), are RNA helicases both composed of three domains: the caspase activation and recruitment domain (CARD), the DExD/H box RNA helicase domain and a regulatory domain [39], [40]. In general, it is considered that activation of RIG-I is mediated by short dsRNA containing a 5′ di- or triphosphate end, while MDA-5 is activated by recognition of long dsRNA (Fig. 2) [10], [16], [24], [35]. Stimulation of both receptors leads to conformational changes which induce interaction of the CARD domain with the mitochondrial antiviral-signaling (MAVS) protein. Upon complex formation, a cascade of signaling pathways is initiated, finally resulting in transcription factor activation and transport to the nucleus [10], [16], [24], [35], [36], [39]. In contrast to the aforementioned RLRs, the precise role of a third member of the RLR family, laboratory of genetics and physiology 2 (LGP2), is less clear. LGP2 does not possess a CARD domain and is suggested to be involved in the regulation of RIG-I- and MDA-5-signaling. It is generally considered that LGP2 stimulates MDA-5 signaling, as recently confirmed by Duic et al., while it is suggested to block RIG-I signaling [10], [16], [39], [40], [41]. Another recent study by Sanchez and colleagues showed the involvement of the IFN-inducible dsRNA-dependent protein kinase activator A (PACT), and more specifically the PACT-LGP2 association, in the regulatory role of LGP2 [39]. Nonetheless, a lot of controversy remains and further in-depth studies are needed to precisely reveal the underlying molecular mechanisms of these regulatory effects [10], [39], [40], [41]. A fourth cytosolic PRR suggested to be associated with mRNA-mediated immunity is the NOD2 receptor, belonging to the NLR family (Fig. 2). Sabbah et al. showed that ssRNA activation of the cytoplasmatic NOD2 receptor leads to the MAVS-interaction-mediated triggering of the transcription factor IRF3, eventually inducing IFN-β secretion [10], [16], [24], [35], [40], [42].
Once type I IFNs are expressed, they are transported to the extracellular environment where they can react with interferon-α/β receptors (IFNARs) in an auto- or paracrine fashion (Fig. 2) [10]. Type I IFNs are a family composed of many different members, of which IFN-α and IFN-β are the most characterized. They are recognized by a heterodimeric IFNAR consisting of the low-affinity IFNAR1 and high affinity IFNAR2 [34], [43], [44]. However, De Weerd et al. showed that specifically IFN-β can form a functional complex with IFNAR1, independent of IFNAR2, which can lead to unconventional signal transduction and selective gene expression [43], [45], [46]. Activation of the IFNAR1-IFNAR2 complex results in the initiation of the Janus kinase and signal transducer and activator of transcription (JAK-STAT) pathway. The Janus kinases JAK1 and TYK2 phosphorylate the STAT1 and STAT2 proteins, resulting in their dimerization and binding to IRF9 (Fig. 2). This three-component complex, also called IFN-stimulated gene factor 3 (ISGF3), migrates to the nucleus where it binds to promotor IFN-stimulated response elements (ISREs) leading to the transcription of ISGs. The latter encode a plethora of proteins involved in the innate immune pathways described above or in direct anti-viral responses [10], [34], [43], [44], [47], [48]. Also other STAT proteins can be phosphorylated and other complexes, such as STAT1 homodimers, can be formed that bind to IFN-γ-activated site (GAS) elements in the promotor region of ISGs instead. Researchers suggest that activation of different STAT molecules can lead to different responses. For instance, it has been suggested that STAT3 would be involved in hampering type I IFN responses, rather than stimulating them [34], [43], [44], [47], [48], [49]. Which STAT proteins are activated upon IFN-recognition by the IFNARs would be determined by different factors such as cell type and relative STAT protein levels [43], [47], [48]. Moreover, it is suggested that several other STAT-independent pathways are important in the IFN-signaling cascade and a combination is needed to induce IFN-I responses. Precise molecular mechanisms of both STAT (in)dependent signaling and complexity of these interactions still remain to be fully elucidated [34], [43], [44], [47], [48].
Upon expression of ISGs, several anti-viral responses can be initiated. Two important ISGs shown to be involved in this anti-viral responses upon dsRNA recognition, are dsRNA-dependent protein kinase (PKR) and 2′-5′-oligoadenylate synthetase (OAS) [10], [24]. PKR is a 68 kDa protein that can hamper mRNA translation through phosphorylation of the eukaryotic translation initiation factor 2 alpha subunit (eIF2α) (Fig. 2). Furthermore, several reports have shown that PKR can activate NF-κB, which is captured in the cytoplasm through binding of the IkBα protein. Dissociation of these two factors allows transportation of NF-κB to the nucleus where it is able to perform its role as a transcription factor. Precise mechanisms are still under investigation, but the NF-κB activating action of PKR Is suggested to be mediated by IKK complex interaction and to be independent of its kinase activity [10], [50], [51], [52], [53]. The OAS pathway is one of the first defined antiviral pathways following type I IFN stimulation. Upon dsRNA recognition, OAS produces 2′-5′-linked oligoadenylates which activate the RNase L enzyme, an endoribonuclease that leads to the degradation of ssRNA (Fig. 2) [10], [54], [55]. ADAR1 is another IFN-inducible dsRNA binding protein belonging to the adenosine deaminase acting on RNA (ADAR) family. ADAR1 can convert adenosine to inosine which induces mismatch base-pairing resulting in destabilization of dsRNA. Evidence suggests that ADAR1 is involved in suppression of RLR, PKR and OAS pathways, mediated by several processes which remain to be fully explained. This pro-viral activity is useful to inhibit autoimmunity caused by host dsRNA recognition but should be controlled during viral infection [10], [53], [56]. Of note, some ISGs can act as negative feedback regulators of the type I IFN immune response. For example, USP18 is known to prevent IFNAR activation as shown by Honke et al. where USP-18 secreting CD169+ macrophages displayed a lower type I IFN sensitivity upon viral infection [38], [57].
4. Strategies to control innate immune activation upon conventional and self-amplifying mRNA delivery
As described above, the innate immune-stimulating activity affects both the translation and integrity of conventional and self-amplifying mRNA. For non-immunotherapy applications, mRNA-induced immune stimulation is highly unwanted and should be avoided completely. In contrast, for vaccination applications, the situation is more ambiguous and mRNA-induced immune stimulation should rather be controlled than completely eliminated as discussed further in Section 5. Below, we will present an extensive overview of the strategies and methods that are currently being used to avoid or control the innate immune-stimulating activity of mRNA and saRNA, as presented in Fig. 3 .
Fig. 3.
Schematic overview of strategies to control innate immune activation upon mRNA and/or (+saRNA) delivery: A) Modifications to the mRNA backbone itself. Conventional mRNA contains a 5′ cap structure (m7GpppN), a 5′ untranslated region (5′ UTR), the gene of interest (GOI), a 3′ untranslated region (3′ UTR) and a poly(A) tail. This figure indicates which adaptations can be made to each of these regions to suppress innate immune recognition. B) Optimizing in vitro transcription (IVT) production and purification of mRNA and saRNA. During IVT, immunogenic byproducts, including dsRNAs, are formed next to the desired mRNA strand. To limit the dsRNA amount and reduce innate immune stimulation, their formation during IVT can be prevented or they can be removed after IVT. C) Innate immune inhibitors. A concise overview is given of innate immune inhibitors that have been used to reduce innate immune stimulation upon mRNA and/or saRNA delivery.
4.1. Modifications to the mRNA backbone itself
The first strategy to limit mRNA-induced immunity, comprises the modifications on the level of the pDNA template or the IVT mRNA or saRNA molecule itself, as summarized in Fig. 3 A.
4.1.1. 5′ cap
The 5′ cap structure (m7GpppN) is a regular characteristic of eukaryotic mRNAs, including viral RNAs such as those from alphaviruses [58]. It is typically composed of an N7-methylated guanosine linked to the first nucleotide of the RNA via a reverse 5′ to 5′ triphosphate bridge (cap 0) (m7G(5′)pppN1pN2p). In humans and other higher eukaryotes, the standard cap 0 structure is further modified to a cap 1 (m7G(5′)pppN1mpN2p) or cap 2 (m7G(5′)pppN1mpN2mp) structure by 2′-O-methylation of the first or both nucleotide riboses, respectively [59]. The 5′ cap structure is known to be required in various processes throughout the lifecycle of an mRNA, including pre-mRNA splicing, nuclear export, polyadenylation, initiation of translation by binding to the eukaryotic initiation factor (eIF) 4E and protection from 5′ to 3′ exonuclease cleavage. Moreover, a cap 1 or cap 2 structure may also prevent recognition of exogenous RNA by innate immune sensors, as it mimics the 5′ cap structure of eukaryotic mRNAs [58]. Recent studies found that a cap 1 structure is crucial to block RIG-I activation, whereas cap 0 and uncapped (5′ ppp) dsRNAs bind to RIG-I with similar affinity (Fig. 2) [59], [60]. Also, the cap 1 structure revealed to be important to evade recognition by MDA-5 [61] and the interferon-induced protein with tetratricopeptide repeats (IFIT) family of restriction factors [62]. Subsequently, as mRNA with other cap structures interact differently with PRRs, the cap structure can also influence protein expression from the IVT mRNA construct [63].
These cap structures can either be incorporated during the IVT production process of mRNA or saRNA (co-transcriptional capping) or after initial mRNA synthesis using recombinant vaccinia virus-derived capping enzymes (post-transcriptional capping) [13]. Up till now, the most reported cap analogues for co-transcriptional capping are the anti-reverse cap analogues (ARCAs) (3′-O-Me-m7G(5′)ppp(5′)G), which contain an additional methyl group on the 3′ position of the last nucleoside compared to cap 0 to prevent incorporation of the 5′ cap in the reverse orientation. However, the ARCA is not the method of choice for 5′ capping with regard to evading innate immune stimulation, as it generates an immunogenic cap 0 structure and results in a capping efficiency of only 70% due to competition with GTP nucleotides for incorporation into the mRNA. So, 30% of the IVT mRNA remains uncapped and contains a 5′ triphosphate at its 5′ end, which introduces additional immunogenicity and instability [13], [24], [64]. This fraction of uncapped mRNA can be reduced by phosphatase treatment to remove the triphosphates at the 5′ ends and avoid recognition by PRRs [65]. Quite recently, trimer analogues, such as Cleancap, have been introduced as an attractive alternative to ARCA for co-transcriptional capping with incorporation of a natural cap 1 or cap 2 structure and an increased capping efficiency of 90 – 99%, which is of interest to generate less immunogenic mRNA constructs [16], [66]. Interestingly, Trilink Biotechnologies also provides the CleanCap Reagent AU that has been specifically designed for saRNAs based on the genomes of positive-sense alphaviruses. Post-transcriptional capping is carried out using vaccinia virus-derived capping enzymes and can generate cap 0 and cap 1 structures. Although enzymatic capping can be highly efficient (100%) and can generate natural cap 1 structures, there is often a high variability in efficiency. Also, co-transcriptional capping is simpler, cheaper and easier to control compared with post-transcriptional enzymatic capping reactions [13], [24], [67].
4.1.2. Untranslated regions
The 5′ and 3′ UTRs in mRNA are important regulators of mRNA stability and translational efficiency [68], [69]. As the composition of the UTR sequences does not directly influence the immunogenicity of the conventional IVT mRNA or saRNA construct, we will just give a brief overview of UTRs that are often used [13]. A more in depth discussion can be found in the review of Kuhn et al. [70]. 3′ UTRs of α- and β-globin mRNAs are popular sequences, often incorporated in IVT mRNAs to increase its stability and translation [71], [72], [73]. The stabilizing effect of human β-globin 3′ UTRs sequences can even be further improved by incorporating two β-globin 3′ UTRs in a head-to-tail orientation [74]. Other similar efficacious UTR sequences include the 5′ UTR of human heat shock protein 70 [75], the UTRs of albumin [76] and viral UTRs from VEEV [77] and SINV [78]. Recently, Sahin’s group used a SELEX (systematic evolution of ligands by exponential enrichment) to identify new 3′ UTR sequences that augment protein expression from IVT mRNA. In total 64 double (d)UTR combinations were tested and the dUTRs carrying the mtRNR1 (mitochondrially encoded 12S rRNA) element in combination with AES (amino-enhancer of split) or human β-globin were found to be superior to the standardly used double β-globin in mRNA translation in human dendritic cells. Intravenous injection of liposome-formulated mRNA with an AES-mtRNR1 3′ UTR in mice was found to increase protein expression and the induction of antigen-specific T cells. For the reprogramming of human fibroblasts, mRNA containing the AES-mtRNR1 3′ UTR outperformed mRNA with other 3′ UTR variants [79]. Also for saRNA vaccines, the 5′ and 3′ UTRs can be optimized to improve translation, which is based on the evolution of naturally occurring alphaviruses [23].
4.1.3. Poly(A) tail
The 3′ end of the conventional IVT mRNA or saRNA is decorated by a poly(A) tail, which plays a key role in regulating mRNA stability. A long poly(A) tail is known to have a stabilizing function and naturally has a length of approximately 250 adenosine ribonucleotides in mammalian cells. In addition, the poly(A) tail is important for RNA translation by supporting the formation of a closed-loop state, which also impedes deadenylation and mRNA degradation, through association of poly(A)-binding proteins (PABPs) with both the poly(A) tail and the 5′ cap [80], [81]. A poly(A) tail is added to the IVT mRNA or saRNA either directly from the encoding DNA template (allowing precise control over the number of adenosines to incorporate) or post transcriptionally by using a recombinant poly(A) polymerase. However, the latter is not regularly done because of the high variability in poly(A) tail length generated between batches [9]. Most research focusses on estimating the impact of the length of the poly(A) tail on RNA translation and stability, but there is no general rule about its length on RNA translation. Some reported that longer poly(A) tails increase protein expression from mRNA in multiple cell types [74], [82], [83], whereas other suggested that longer poly(A) tails (≥100 adenosines) are not necessarily better [81], [84]. In contrast, the impact of poly(A) tail length on the immunogenicity of both non-amplifying and self-amplifying mRNA is less intensively studied. For example, one study of Koski et al. provided evidence that the immune-stimulatory activity of IVT mRNA could be lowered by enzymatic 3′-polyadenylation with a minimum of 150 adenosines, as increasing the length of the poly(A) tail generates mRNA with a reduced relative uridine content or shielded uridines in the sequence [85], [86].
4.1.4. Nucleoside base modifications
The use of modified nucleoside analogues is probably the most applied approach to prevent IVT mRNA from intracellular recognition by PRRs and consequently evade innate immunity. To date, 143 modified nucleosides have been characterized in endogenous RNAs and differences in nucleoside modifications can enable PRRs to detect foreign RNA of invading pathogens [87]. For example, RNA modifications are more abundant in mammalian rather than in bacterial mRNA and a subset of nucleoside modifications is unique to either bacterial or mammalian RNA [87], [88]. Therefore, it is not surprising that unmodified IVT mRNA has a stronger innate immune-stimulating activity and nucleoside base modifications have been introduced to silence recognition by innate immune sensors [88], [89].
Pioneering work in this area was done by Karikó et al. who demonstrated in primary dendritic cells (DCs) that upon mRNA transfection, TLR3, TLR7 and TLR8 activation could be reduced or completely eliminated with mRNA containing the following modifications either separately or in combination: 5-methylcytidine (m5C), N6-methyladenosine (m6A), 5-methyluridine (m5U), 2-thiouridine (s2U) or pseudouridine (Ψ) [88]. Especially replacement of uridine by Ψ seemed very promising, as it was shown to not only reduce the immunogenicity of mRNA, but to also enhance its translational capacity both in vitro and in vivo [90]. This increase in translational efficiency was later on shown to be aided by a reduced detection by PKR, a reduced PKR-mediated phosphorylation of eIF2α [91], [92], a reduced OAS-mediated activation of RNase L and an increase in resistance to RNase L-mediated degradation [93] (Section 3). Later on, many other studies were published in favor of modified mRNA, demonstrating its vast potential for therapeutic applications. For example, Kormann and colleagues found that replacement of 25% of uridine and cytidine with s2U and m5C synergistically diminished binding to PRRs, decreased innate immune activation and increased protein expression of erythropoietin (EPO)-encoding mRNA in vitro and in vivo in mice after intramuscular injection [94]. In a similar study by Karikó et al. on EPO-encoding mRNA it was shown that HPLC-purified (Section 4.2.3), codon-optimized (Section 4.1.5), Ψ-modified mRNA was clearly superior to its unmodified counterpart in inducing protein expression and limiting innate immune activation in mice, leading to increased erythropoiesis [95]. Mahiny et al. showed that m5C/s2U-modified mRNA encoding a zinc finger nuclease (ZFN) induced a more robust protein expression in mouse lungs than unmodified mRNA and corrected SP-B deficiency in mice [96]. Moreover, in 2015, Andries et al. reported on replacing uridine by N1-methylpseudouridine (m1Ψ) to reduce the intracellular innate immunity of mRNA. This modification seemed to increase translational capacity even more compared to Ψ-modified or m5C/Ψ-modified mRNA in vitro and in vivo [97]. Similarly, Devoldere et al. screened 16 different modifications in human retinal cells (in vitro) and found that m1Ψ-modified mRNA had the highest translational capacity of all modified mRNAs tested and also induced protein expression in bovine retinal explants and mouse retinas (in vivo) [98].
Although these findings suggest that mRNA modification is an inevitable prerequisite for the development of non-immunogenic, mRNA-based protein therapies, some other studies have been published that contradict this approach. For example, researchers of CureVac demonstrated that Ψ incorporation in sequence-optimized (Section 4.1.5), HPLC-purified (Section 4.2.3) mRNA reduced protein expression in HeLa cells and in mice after lipid nanoparticle (LNP)-mediated delivery. In contrast, when the mRNA sequence was not optimized, Ψ substitution did enhance protein production [76]. Kauffman et al. found that Ψ-modification of mRNA had no significant impact on protein expression and immunogenicity in mice when delivered systemically with liver-targeting C12-200 LNPs [99]. Hence, it is important to realize that mRNA modification does not necessarily increases protein expression and can even decrease it. This discrepancy may arise from variations in delivery system, route of administration, the level of innate immune sensing in the targeted cell types and mRNA properties like different UTRs, sequence optimization (Section 4.1.5) and HPLC purification (Section 4.2.3) [21], [24], [99]. Also, there is no control over the location where modified nucleosides are inserted in the mRNA strand. It has been shown that incorporation of modified nucleosides in translation-enhancing elements, such as UTRs and internal ribosomal entry sites (IRESs), impair their functionality [76].
In accordance with conventional IVT mRNA, also IVT saRNA is recognized as foreign by innate immune sensors. However, in contrast to conventional mRNA, the usage of modified nucleosides is not considered as an option to reduce its immune-stimulating activity. The effect of incorporating modified nucleosides into saRNA would already be lost after the first round of amplification. Moreover, it is expected to impair the self-replicating capacity of saRNA in the target cells [22].
4.1.5. Open reading frame
So far, we have discussed adaptations to the cap structure, UTRs, poly(A) tail and nucleotides of the IVT mRNA, but an important missing element in this overview is the ORF. This protein encoding region of conventional mRNA can be modified through codon optimization, in which rare codons are replaced by frequently used synonymous ones that have abundant cognate tRNA in the cytosol without altering the amino acid sequence of the encoded protein. Codon optimization is applied to increase mRNA stability and translation efficiency, but cannot be applied in general for mRNA-based therapeutics, as some proteins require slow translation for their proper folding, which is ensured by less frequent codons [13], [100], [101]. In addition, as codon optimization is not used as a tool to directly modulate the inherent immunogenicity of IVT mRNA, we will not discuss this topic further in detail, but some other reviews are available describing this thoroughly [102], [103], [104]. Furthermore, codon optimization also includes the replacement of GC-poor codons in the ORF by synonymous GC-rich codons, as this has been shown to increase steady-state mRNA levels in vitro [105] and protein expression in vivo, while reducing innate immunogenicity [76] as explained below.
In contrast to codon optimization, the following adaptation clearly affects the immune-stimulating activity of IVT conventional mRNA. The immunogenicity of IVT mRNA can be eliminated by minimizing or completely eliminating uridine content, as uridine-rich regions are known potent activators of several RNA sensors, including TLR7, TLR8 and RIG-I [106], [107], [108], [109]. In addition, uridine depletion is not necessarily restricted to the ORF of the mRNA but can be applied over the whole mRNA molecule. Minimizing or eliminating uridine content can be accomplished in several ways [86]. First of all, the IVT mRNA can be rendered completely non-immunogenic by replacing all uridines with Ψ, as was done for example in a study of Karikó and colleagues [95]. Secondly, uridine can also be replaced for 25% by s2U, resulting in mRNA with some residual immunogenicity as shown by Kormann et al. [94]. Thirdly, as also mentioned previously, the length of the poly(A) tail can be increased to diminish the relative uridine content or to shield the uridines present in the mRNA sequence [85]. Lastly, the uridine content in the IVT mRNA can be decreased by enrichment of the GC content. For example, Thess and colleagues designed an unmodified EPO-encoding mRNA by selecting GC-rich codons for each amino acid. This rendered the mRNA non-immunogenic and increased protein expression levels even more than its Ψ-modified counterpart. Also, LNP-mediated, intravenous delivery of their EPO-encoding mRNA construct in pigs and non-human primates resulted in a significant increase in serum EPO levels and reticulocyte counts as well as an elevation of the hematocrit, without detectable immunogenicity [76].
Distinct from conventional mRNA, the ORF of saRNA encodes four, viral nsPs essential for its self-replicating capacity. These nsPs are important controlling elements with regard to the saRNA biology and the host cell response [110]. Therefore, it might be interesting to introduce mutations in the nsPs to adapt both parameters. For example in the past, several groups reported on amino acid substitution mutations in the nsP2 protein to decrease the cytopathic effect of alphavirus-based saRNAs [111], [112], [113], [114]. Recently, Li et al. used an in vitro evolution approach in IFN-competent cells to identify mutations within the VEE nsPs that promote in situ expression of subgenomic RNA. Five mutations in nsP2 and nsP3 were found to have a significant impact on saRNA persistence and gene expression levels. Additionally, they checked whether these mutations change the innate IFN response to saRNA, which might declare their findings. Nevertheless, the best performing saRNAs in terms of protein expression were not those with the lowest interferon response [110]. Hence, mutations to the nsPs proteins can be considered to lower the innate immune-stimulating activity of saRNA but this is not straightforward and more research is still required.
4.1.6. mRNA secondary structures
Lastly, in several parts of the mRNA or saRNA, including the UTRs and the ORF, the mRNA secondary structure can have a significant impact on mRNA translation and immune-stimulating activity [16], [115]. These secondary structures in the single-stranded mRNA molecules can be easily formed through complementary self-interactions [116]. First of all, highly stable secondary structures in the 5′ UTR and the first 10 codons of the ORF should be avoided as they are known to reduce translation initiation efficiency and therefore overall protein production [117], [118], [119]. In contrast, Mauger et al. reported that an increased secondary structure, which was varied by the incorporation of modified nucleosides, in the ORF downstream of the first 30 nucleotides and in the 3′ UTR correlated with increased protein expression. They demonstrated that introducing modified uridines, such as the use of m1Ψ, stabilizes mRNA secondary structures, by forming more stable base pairs than uridine [119]. These structural changes might not only enhance the functional half-life of mRNA, but was recently also found to be a key determinant for mitigating hydrolytic degradation of mRNA [120]. However, highly stable and long secondary structures in the whole construct are unwanted and should be avoided as they can be recognized as dsRNAs by PRRs [16], [115].
4.2. Optimizing in vitro transcription (IVT) production and purification of mRNA and saRNA
4.2.1. General IVT production and purification of mRNA and saRNA
Both conventional mRNA and saRNA are synthesized by IVT of a linearized pDNA or PCR template, as described in Section 2.1. During IVT, also some byproducts are formed including short ssRNAs produced by abortive initiation events and dsRNAs, which have been identified as major triggers for the innate immune pathway by binding to TLR3, MDA-5 and RIG-I (Fig. 2) [5], [24], [121], [122]. Hence, mRNA or saRNA purification is a vital step to remove immunogenic byproducts and ensure a safe and fully functional end product (Fig. 3 B). Simple purification methods, such as LiCl or alcohol-based precipitation and silica membrane columns, are often used on small laboratory scale and efficiently remove free NTPs, proteins, salts and to a lesser extent short RNAs [20], [123]. However, when highly pure mRNA should be obtained for GMP production, chromatographic approaches are typically used as a first choice [20], high performance liquid chromatography (HPLC) [124], [125], anion-exchange chromatography [126], [127], size-exclusion chromatography [128], [129], and affinity chromatography with immobilized oligo(dT) [130]. Of interest, these chromatographic methods might be applicable for saRNA purification as well, but this is anything but straightforward due to its larger and more complex structure. At the moment, no commercially viable scalable purification process has been disclosed so far for saRNA purification [3], [5].
4.2.2. Prevention of immunogenic dsRNA byproduct formation during IVT
Recent studies revealed two main mechanisms through which dsRNA byproducts are formed. Both are based on aberrant activity of the RNA polymerase besides its normal function as DNA-dependent RNA polymerase, transcribing mRNA from a pDNA template. First of all, T7 RNA polymerase has an obscure RNA-dependent RNA polymerase activity [131], [132], [133], [134]. The synthesized RNA (runoff transcript) can serve as a template for T7 RNA polymerase leading to 3′ extension of the runoff transcript if there is sufficient 3′ complementarity to fold back on itself in cis, which results in the formation of intramolecular RNA duplexes [134]. In addition, during initial transcription, RNA polymerase produces short abortive RNA byproducts from 5 to 11 nucleotides in length [122], [135]. When these short fragments anneal to complementary sequences in the runoff transcript (in trans), this can prime complementary RNA synthesis from the primary transcripts resulting in the generation of dsRNA contaminants [136]. Secondly, the T7 RNA polymerase has promoter-independent activity and could wrongly initiate transcription from a promoter-less DNA end, resulting in the formation of an antisense RNA molecule that is fully complementary to the intended sense transcript. Hybridization of these two RNA molecules leads to the formation of long dsRNA contaminants in the IVT reaction mixture [137].
In a recent study, Mu et al. showed that for a few specific templates the formation of dsRNA species can be reduced during IVT by lowering Mg2+ concentrations or by using modified nucleosides [137]. However, since Mg2+ is a crucial element during IVT, reducing its concentration also affects the total mRNA yield, which is undesirable in the production of large quantities of mRNA. Another strategy to reduce the amount of dsRNA byproducts is the use of a thermostable T7 RNA polymerase in the IVT reaction. Wu et al. demonstrated that performing the IVT reaction of a couple of mRNAs at 50 °C with thermostable T7 RNA polymerases reduced the 3′ extension of the runoff product, and subsequently the formation of dsRNA byproducts without loss of the functional runoff transcript. Furthermore, the presence of a template-encoded poly(A) tail showed to reduce antisense dsRNA byproducts, but not the amount of 3′ extended dsRNA byproducts [138]. In another study of Gholamalipour et al., the addition of a DNA oligonucleotide complementary to the 3′ end of the expected runoff RNA has shown to restrain the production of 3′ extended dsRNA byproducts during an IVT reaction of mRNA [139]. However, this DNA oligonucleotide needs to be removed again after IVT, requiring another enzymatic step, which is unfavorable as it complicates the production process. It should be noted that to date, the concept of diminishing dsRNA formation by adaptation of several IVT reaction components has not been described specifically for saRNA and the optimal reaction conditions may differ from those for conventional mRNA due to the structural differences between both mRNA types.
4.2.3. Removal of dsRNA byproducts from IVT mRNA and saRNA
An alternative approach to reduce the number of dsRNA contaminants is to eliminate dsRNA byproducts after completion of the IVT reaction. The vast majority of known purification techniques for mRNA and saRNA as mentioned in Section 4.2.1 are ineffective in the removal of dsRNAs. In this context, a major breakthrough was achieved when Karikó and colleagues reported that dsRNA impurities could be eliminated from IVT mRNA by ion pair reversed-phase HPLC based on the longer retention time of dsRNAs. They demonstrated that reversed-phase HPLC purification eliminated the residual immune stimulating activity of Ψ- or m5C/Ψ-modified mRNA, rendering a completely non-immunogenic mRNA. In addition, reversed-phase HPLC purification of both modified and unmodified mRNA improved protein production significantly in primary cells [121]. In a recent study, Nelson et al. highlighted the importance to combine different strategies to eliminate innate immune recognition of IVT mRNA. They modified the mRNA by replacing uridine by m1Ψ and removed dsRNAs by reversed-phase HPLC purification and by alteration of some IVT components including the use of a custom NTP ratio instead of equimolar levels of each NTP. The combination of both resulted in the creation of an IVT mRNA nearly indistinguishable of injected PBS with regard to the innate immune activation in two mouse strains, whereas both approaches on their own were less effective at reducing immunogenicity [140]. Despite the clear potential of ion pair reversed-phase HPLC to remove dsRNA byproducts and reduce the immunogenicity of IVT mRNA, the need for expensive and specialized instrumentation, the long purification time, the use of toxic acetonitrile as eluent and the low recovery rate of only 50% impedes the cost effectiveness of the mRNA production and makes it less suitable for scaling up the process [121], [136].
Of interest, Baiersdörfer et al. reported on a cellulose-based chromatographic method that removes at least 90% of the dsRNA contaminants, based on the selective binding of dsRNA to cellulose in an ethanol-containing buffer. This new purification method is similarly effective to remove dsRNA contaminants as reversed-phase HPLC, but on the contrary is fast, cost effective, scalable and has a relatively high recovery rate between 65% and 85%. Upon cellulose-based chromatography, m1Ψ-containing IVT mRNA no longer induced IFN-α secretion in vivo when injected intravenously as lipoplex. In addition, elimination of dsRNA byproducts by cellulose-based purification showed to increase the in vivo translatability of IVT mRNA to a similar extent as reversed-phase HPLC purification, suggesting that these methods are equally effective and interchangeable for the removal of dsRNA contaminants from conventional IVT mRNA [136].
A third and last strategy to eliminate dsRNA byproducts after completion of the IVT reaction, is treatment of mRNA with RNase III, which specifically recognizes and cleaves dsRNAs of approximately 22 nucleotides in length. A recent in vivo study showed an improvement in cytotoxic killing of leukemia cells by chimeric antigen receptor (CAR)-expressing T cells generated with IVT mRNA, which was digested with RNase III [141]. The main disadvantage of this technique is the risk on erroneous cleavage by RNase III of double-stranded secondary structures formed by ssRNA. Moreover, an additional purification step is required to remove RNase III from the end product [136].
In contrast to conventional mRNA, saRNA cannot be purified from dsRNAs by ion pair reversed-phase HPLC, as shear forces would cause the longer saRNA to break. On the contrary, cellulose-based chromatography of saRNA has already been reported to efficiently remove dsRNA byproducts from saRNA in a recent study by Zhong and colleagues. They demonstrated that cellulose purified saRNA elicited a significantly lower type I IFN response in IFN-β reporter mice after intradermal electroporation in comparison to saRNA purified on silica-based columns, which was however not reflected by an increase in protein translation. Nevertheless, a clear improvement in efficiency of a saRNA vaccine against Zika virus was noticed, characterized by a triplication in antibody titers, a doubling of the cellular immune responses and an increase in seroconversion rate from 62,5% to 100% [142]. Furthermore, no studies have been published yet describing RNase III treatment for saRNA to remove dsRNA contaminants, but most likely this technique would also work for saRNA.
4.3. Innate immune inhibitors
As mentioned above, several backbone modifications to reduce innate immune stimulation, including nucleoside base modification and sequence alteration, cannot be applied for saRNA, as this would interfere with its functionality. Also for conventional mRNA, nucleoside modifications could possibly alter mRNA secondary structures, translation and protein folding. Therefore, the use of innate immune inhibitors is gaining attention to modulate the immune-stimulating activity of both conventional and self-amplifying mRNA (Fig. 3 C). Over the last few years, many inhibitors have been screened for both conventional and self-amplifying mRNA but, although they often possess a clear mode of action, not every inhibitor tested showed potential as an appropriate adjuvant [10], [21], [24], [143], [144]. In this section, we will provide a comprehensive overview of the most important results obtained up to date.
4.3.1. Viral immune evasion mechanisms
Viruses have developed a plethora of intelligent mechanisms to avoid recognition and killing by the host’s immune system, such as evading PRR detection, inhibiting IFN mediated effects and hampering cytokine function [145]. Inspired by these viral escape mechanisms, several viral proteins have been evaluated as potential innate immune inhibitors to improve mRNA translation.
4.3.1.1. Vaccinia virus proteins: B18R, E3 and K3
Vaccinia viruses (VVs) belong to the Poxviridae family, which are a family of large and double-stranded DNA viruses, primarily known for their use in smallpox vaccine formulations. The genome can be divided in a fixed central region and the lesser conserved terminal regions. The latter are responsible for encoding proteins involved in virulence intensity and immune response regulation such as B18R, E3 and K3 which have been evaluated for their role in IFN interference [146], [147].
B18R is a 60–65 kDa glycoprotein encoded by the Western Reserve strain of the VVs and acts as a decoy receptor to neutralize type I IFNs (Fig. 2). As such, the activation of IFNAR and subsequent pathway signaling is inhibited, resulting in reduced mRNA degradation and enhanced translation. This VV protein can be present in its soluble form or bound to the cell surface and is reactive towards human, cow, rabbit, rat and with lesser affinity mouse type I IFNs [146], [148], [149], [150], [151]. Several reprogramming studies have used this inhibitor as an adjuvant to non-amplifying and self-amplifying mRNA delivery, where it was either administered as the soluble protein in the cell culture medium or expressed in situ by co-transfecting B18R mRNA with mRNA encoding for the protein of interest (POI). Results indicated a positive effect on cell viability and high protein expression, which is useful for the repetitive transfections often required during cell reprogramming [152], [153], [154], [155], [156]. Furthermore, B18R was also useful as adjuvant therapy to modified mRNA delivery during these reprogramming studies [152], [153], [156]. Although modified mRNA is known to efficiently reduce innate immune responses, it has been opted that B18R is used to neutralize remaining type I IFNs [155], [157], [158]. Besides its use for reprogramming purposes, B18R has also been explored in several non-reprogramming studies, where it has demonstrated to decrease type I IFN production and increase mRNA translation [159], [160], [161].
It should be noted that the beneficial effect of B18R on cell viability could not be replicated by both Gomez et al. and Minnaert et al. [159], [160]. However, this could be explained by the fact that these studies did not involve multiple mRNA transfections in contrast to the cell reprogramming studies. As Michel et al. showed, the increase in cell viability caused by B18R was only generated starting from the 5th transfection [162]. Furthermore, conflicting results regarding the effect of B18R on protein expression and immune response suppression have been reported [149]. The research group of Byrne and Adjaye was not able to increase reprogramming transcription factor expression or to reduce immune activation in fibroblast cells in the presence of B18R [143], [163], [164]. Also Poleganov et al. observed no enhanced protein expression or reduced IFN-β production upon addition of B18R mRNA or B18R protein [155]. Again, Michel et al. only observed an increase in protein expression upon 2 consecutive mRNA transfections co-delivered with B18R mRNA but not with the B18R protein [162]. Taken together, it appears that the effect of B18R on both cell viability and protein expression is dependent on the transfection frequency and duration, the cell type, and the POI.
Besides B18R, two other VV proteins, E3 and K3, have been tested to block innate immune response activation during mRNA transfections, although their precise mode of action remains elusive. The 25 kDa protein E3 is suggested to act primarily by dsRNA binding, thereby preventing activation of anti-viral state pathways, and direct PKR inhibition which hampers homodimerization (Fig. 2) [146], [165], [166], [167]. Moreover, it was recently suggested by Mehta et al. that E3 would prevent apoptosis upon viral infection by stimulation of apoptosis inhibitor F1 expression. The latter is allegedly mediated by PKR inhibition as described before [168]. K3, a 10.5 kDa protein with 28% homology to eIF2α, is suggested to act as a competitive substrate for PKR (Fig. 2) [146], [165], [166], [167], [169].
Poleganov et al. delivered non-modified mRNA encoding for the immune evasion proteins B18R, E3 and K3 together with non-modified mRNA encoding for different reprogramming factors, to Human Foreskin Fibroblast (HFF) cells. The authors observed a three-fold increase in protein expression mediated by E3 and K3 mRNA delivery, which further increased to 4.6-fold upon the additional delivery of B18R mRNA, despite the fact that B18R mRNA alone remained without effect. Consequently, the authors measured an 80% decrease in IFN-β secretion due to E3 and K3, which even further reduced upon addition of B18R mRNA or the B18R protein. Additionally, the addition of the immune evasion proteins also prevented cell death upon multiple mRNA transfections [155]. Beissert et al. performed a similar follow-up study where mRNA encoding the same three immune evasion proteins was delivered, as an adjuvant therapy for saRNA transfection in HFF cells. They observed that E3 inhibited PKR very efficiently, but K3 inhibition of eIF2α phosphorylation was less pronounced. B18R efficiently neutralized IFNs but only showed, similar to K3, a moderate effect on protein expression. However, when all three VV proteins were combined, both B18R and K3 did provide an additive effect to E3, which was confirmed in vivo [165]. Finally, Liu and colleagues delivered modified E3, K3 and B18R encoding mRNA as a pretreatment before transfection of HFF and HepG2 cells with unmodified mRNA. All three VV proteins increased protein expression, with E3 being the most potent inhibitor, without inducing cell toxicity. Furthermore, a reduced IFN-β production was seen in both cell types upon E3 mRNA delivery, albeit for B18R and K3 this effect seemed cell-type dependent [167].
Taken together, the different VV proteins described above all show potential as innate immune inhibitors to enhance treatment efficiency both in terms of cell viability or protein expression. Future studies to elucidate the molecular mechanisms involved and to determine in vivo effects more in detail will provide insight about their future clinical potential.
4.3.1.2. Influenza A virus proteins: NS1 and PB1-F2
The influenza A virus (IAV) belongs to the Orthomyxoviridae virus family and is a single-stranded RNA virus, consisting of eight RNA segments. Similar to VVs, several mechanisms have been developed by the IAVs to evade the innate immune response, including the expression of proteins, which might inhibit triggers and signaling pathways of the innate immunity. Below, we will discuss two noteworthy non-structural IAV proteins and their influence on innate immunity [170], [171].
The non-structural protein 1 (NS1) is encoded by the eighth mRNA segment of the influenza A virus and consists of both an RNA binding domain and an effector domain. Innate immune signaling is prevented through several mechanisms depending on the virus strain of origin including inhibition of IRF3, direct PKR inhibition and OAS inhibition by dsRNA binding (Fig. 2). Moreover, upon viral infection, NS1 proteins encoded by some virus strains can also inhibit the cleavage and polyadenylation factor 30 (CPSF30) function, which interferes with pre-mRNA processing in the nucleus. Hence, the transport to the cytoplasm is inhibited, which therefore hampers the expression of many genes, such as ISGs. However, several additional immune response suppression mechanisms mediated by NS1 still require further study [167], [170], [172], [173], [174]. Phua et al. co-delivered unmodified mRNA encoding for six different NS1 proteins, derived from different virus strains and subtypes, together with unmodified luciferase mRNA in fibroblasts and observed an increase in luciferase protein expression by all NS1 proteins. Although IFN reduction was similar for all NS1 proteins, NS1-TX91 and NS1-VN showed a higher protein expression in comparison to the others, which was attributed to their CPSF30 inhibiting activity. Furthermore, the authors could increase the protein expression of modified mRNA upon addition of modified NS1-TX91 mRNA. In vivo, however, the effects were not as outspoken as compared to in vitro settings [175]. In a later study, the same research group delivered eGFP encoding unmodified mRNA together with NS1-HK encoding unmodified mRNA to several cell types and noticed enhanced eGFP expression in BJ fibroblasts, HepG2, RAW 264.7 and pMEF cells, accompanied by a reduced IFN production. Furthermore, cell viability increased upon NS1 mRNA delivery, both after single and multiple mRNA transfections. However, when testing several NS1 proteins, originating from different virus strains, this was only observed for the NS1 proteins that did not inhibit CPSF30. Of note, these effects appeared to be cell type- and dose-dependent, so further in-depth research is required [172]. Recently, Liu et al. pretreated HepG2 and BJ Fibroblasts with modified mRNA encoding NS1 and the three VV proteins E3, K3 and B18R before transfection with unmodified luciferase mRNA. The authors showed a superior behavior for NS1 and no synergistic effect of the VV immune evasion proteins could be noticed, explained by a possible overlap in function between E3 and NS1 [167]. Although more studies are needed to further elucidate the immune evasion mechanisms of NS1 and to determine efficacy and safety in vivo, the abovementioned results show great potential for this protein to enhance mRNA translation.
A second potentially interesting non-structural IAV protein is PB1-F2, which has been linked to several effects such as type I IFN response suppression. However, most of the effects seem to be virus strain dependent. Although Le Goffic et al. described an increase in type I IFN activity mediated by PB1-F2, several more recent studies have contradicted this observation [174], [176], [177], [178]. A type I IFN antagonist activity could be observed for both the PB1-F2 wild type (WT) protein and the mutated PB1-F2 N66S protein, containing a serine instead of an arginine residue at position 66 [174], [177], [178]. The molecular mechanisms underlying this type I IFN inhibitory effect are still not fully elucidated but would be situated in the RIG-I pathway, where association between RIG-I and MAVS is suggested to be hampered, which would mainly interfere with the IRF3 function (Fig. 2) [174], [177], [178]. Although more in-depth mechanistic studies are needed, it might be worthwhile investigating the potential of PB1-F2, both in its WT and mutated form, to enhance IVT mRNA translation and protein expression.
4.3.1.3. MERS-CoV ORF4a and PIV-5V self-amplifying mRNA inhibitors
The studies described above demonstrate that applying two different mRNAs, i.e. encoding the protein of interest on the one hand and the immune inhibitor on the other, has shown good potential to reduce IVT mRNA-induced innate immune responses. However, the fact that cells have to receive both mRNA formulations remains a difficulty. Recently, Blakney et al. provided an interesting alternative to overcome this hurdle. They established saRNA constructs containing coding regions for both the POI and an innate immune inhibitor. An in vitro screen of several different inhibitors encoded by the saRNA showed the superiority of the MERS-CoV ORF4a and PIV-5 V protein in different human cell lines. The MERS-CoV ORF4a protein binds to dsRNA, which hampers PACT activation of the RIG-I and MDA-5 receptors, while the PIV-5 V protein binds to MDA-5 directly. As such, these proteins could possibly interfere with IFN production. The mechanism of increased protein expression was studied in MRC5 cells and they indeed found a decrease of NF-κB and IRF3 in the early stages upon transfection. The in vitro results were however not confirmed in vivo, as neither of the proteins led to a substantial and significant increase in protein expression in both BALB/c and C57BL/6 mice. In contrast, both proteins were able to increase the percentage of transfected cells in human skin explants [4]. Nevertheless, this innate immune evasion strategy remains interesting to explore. However, future studies should include more in-depth research on the delivery system dependency, anti-vector immunity, possible combination of inhibitors and effects in humans [4].
4.3.2. Non-viral immune evasion mechanisms
Besides the use of viral immune evasions proteins, many other non-viral molecules have been tested for their potential to reduce immune responses and increase protein expression, of which we will provide a selection below.
4.3.2.1. RNA Interference-mediated immune suppression
Small interfering RNAs (siRNAs) are short dsRNA sequences that can be designed to silence any gene in a sequence specific manner [10]. The research group of Angel et al. combined mRNA for cell reprogramming with an siRNA cocktail targeting different immune-related molecules and found that knockdown of IFN-β, PKR and STAT2 completely recovered cell viability upon multiple mRNA transfections [179]. These studies highlight the complexity of the immune responses and the fact that inhibiting multiple factors might be needed to evade immune responses efficiently [24]. Furthermore, Lee et al. showed the use of short hairpin RNAs (shRNAs) to inhibit the function of TLR3, TRIF and MYD88. Although the authors performed the experiments to elucidate the role of these receptor and transcription factors in pluripotency induction, this mechanism could also be explored to suppress immune-component activity beneficial for treatment efficiency [180]. At last, the use of micro RNAs (miRNAs), small non-coding RNA sequences, could also be explored. Zhang et al. demonstrated the potential of miR-29c to protect the deubiquitinating enzyme A20. The latter is involved in the suppression of NF-κB and IRF signaling pathways, thereby inhibiting the innate immune response during IAV infections. Results showed that miR-29c was capable to protect A20 mRNA against RISC-degradation by acting as an RNA decoy. Moreover, A20 protein expression was enhanced leading to reduced pro-inflammatory cytokine expression [171]. Although this study does not involve IVT mRNA transfection, the function of miRNAs in reducing synthetic mRNA-related innate immune responses might be worthwhile investigating. Yet, it is important to consider that siRNA, shRNA and miRNA molecules might also activate the immune system, which possibly counteracts their effects. However, chemical modification of these compounds can possibly overcome this problem [181], [182], [183].
4.3.2.2. Small molecule inhibitors
Several small molecules have been identified that can inhibit key components of the innate immune pathways, thereby possibly reducing IFN production. However, Liu et al. tested 15 different small molecular compounds and found that none of them enhanced mRNA transfection efficiency in human fibroblasts [144]. Also Drews et al. tested several small molecules to suppress innate immune responses in HFF cells, but neither chloroquine, Pepinh-TRIF, Pepinh-MYD88, Trichostatin A or the VV protein B18R induced a considerable reduction of the innate immune response as measured by expression of innate immune associated genes [143]. On the other hand, Awe et al. showed that BAY11, a IKK complex inhibitor, was more potent than the TBK1 and IKKε inhibitor BX795 to increase protein expression and modified mRNA stability, mediated by reduction of NF-κB expression [163].
Also the integrated stress response inhibitor (ISRIB), a molecule capable of enhancing translation by improving eIF2B complex formation, demonstrated to enhance protein expression in the early transfection stages (0–6 h) upon luciferase mRNA delivery [184], [185]. The authors did notice, however, that the corticosteroid dexamethasone was more capable of increasing translation efficiency in the intermediate to late stages upon transfection. When both components were delivered together a two-phase increase in protein expression was observed. When dexamethasone was used for further in vivo studies, it improved luciferase expression after intravenous delivery of dexamethasone-palmitate loaded mRNA lipoplexes, possibly attributed to its NF-κB inhibiting activity [185]. A very recent study of Zhong et al. tested another corticosteroid, clobetasol propionate, which was able to reduce type I IFN response and significantly enhanced expression upon intradermal electroporation of a self-amplifying mRNA vaccine against the Zika virus [142].
Taken together, the use of viral and non-viral innate immune inhibitors is an interesting strategy to consider during the development of mRNA-based therapeutics for different applications. The inhibitors can be used as an adjuvant to self-amplifying mRNA, modified and unmodified conventional mRNA. Moreover, they can provide an added value when multiple mRNA transfections are required, for instance with cellular reprogramming. Finally, inhibitors can be delivered as soluble proteins or as mRNA constructs, possibly co-encapsulated with the therapeutic mRNA in a nanoparticle carrier. However, which inhibitor should be selected and how it should be used is cell type and therapy dependent.
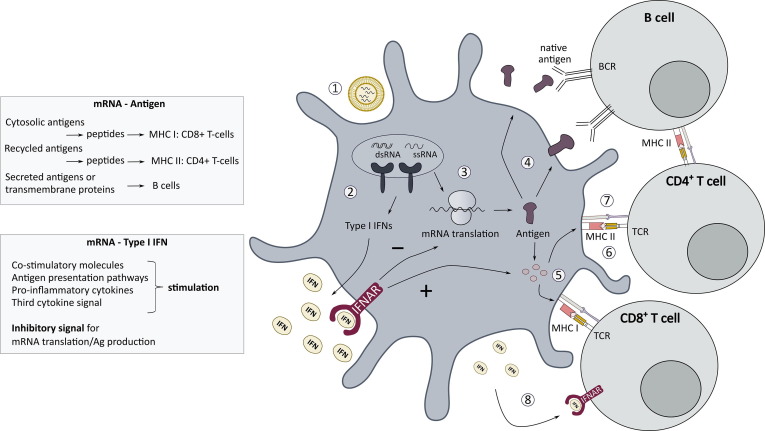
5. mRNA vaccines
Vaccines can prevent against a variety of infectious diseases, by priming the immune system which is then able to recognize and destroy the pathogen upon future encounter. Most of the current vaccines are based on attenuated or inactivated pathogens, or recombinant produced antigens. Over the recent years, many researchers have recognized the advantages of conventional and self-amplifying mRNA-based vaccines for both infectious diseases and cancer treatment [3], [5], [21], [23], [35], [186], [187]. First, the mRNA sequence can be easily and rapidly adapted to express the antigen of choice, which is very convenient for personalized treatment with cancer vaccines or to provide quick responses against emerging infectious diseases [16], [17], [21], [23], [35], [36]. Second, depending on the mRNA vaccine design and immunogenicity profile, mRNA vaccines have shown great versatility to treat different disease indications, ranging from infectious diseases, cancer treatment, and even autoimmune diseases [188], [189], [190]. At last, mRNA does not require penetration into the nucleus to perform its action, making it useful for transfection of slow or non-dividing cells, such as dendritic cells (DCs) [35]. Despite this plethora of advantages, some attention points remain. As mRNA is a labile molecule, in vitro and in vivo stability should be improved by for example optimizing the mRNA backbone structure, the delivery route and especially by formulating the mRNA into nanoparticles. Several different administration routes and delivery carriers have been explored to improve the in vivo stability and to promote efficient intracellular delivery [17], [21], [30], [191], [192], [193], [194]. Another key challenge in mRNA vaccine development is to find the optimal balance between the mRNA-mediated innate immune stimulation and antigen expression, which should be fine-tuned depending on the specific therapeutic application. This interesting topic has been also discussed in several other reviews [16], [17], [34], [192], [195].
As outlined in Section 3, type I IFNs and many other proteins, encoded by ISGs are produced upon the cellular entrance of synthetic mRNA by activation of PRR signaling pathways [10], [16], [34], [37], [38]. On the one hand, type I IFN signaling induced by synthetic mRNA plays a key role in the immune activation of both innate and adaptive immune cells, mimicking the immune events upon a natural infection with viruses. On the other hand, the type I IFN reaction can inhibit the translation of mRNA and could therefore also work detrimental for the vaccine efficiency. Moreover, the inflammatory effects of type I IFNs can be a culprit for the safety of mRNA vaccines, as they play - in all likelihood - an important role in the reactogenicity of mRNA vaccines [17], [18], [21], [34], [35], [36], [196]. Therefore, the key towards a successful mRNA vaccine might be a well-balanced innate immune response, while avoiding excessive innate immune triggering in order to sustain the protein expression needed to reach the antigenic threshold for T cell and B cell activation. However, it is not completely understood how to hit this sweet spot. This challenge becomes somewhat more complicated, since the carrier system, such as LNPs, might not only contribute to the mRNA delivery efficiency, but can also possess intrinsic innate immune effects that can influence and maybe even overrule the innate immune activity of mRNA vaccines [11], [197], [198]. Moreover, the mRNA vaccine design may have to be tailored to the specific therapeutic application and the associated desired adaptive immune response. Indeed, differential kinetics and amount of mRNA encoded antigen expression and type I IFNs might be required for the generation of antibody producing B cells, helper CD4+ T cells or cytotoxic CD8+ T cells [199], [200], [201], [202]. A more in-depth overview about the role of this self-adjuvant effect on the efficacy of mRNA vaccines and the use of other immune adjuvants, will be provided below. After all, as mRNA vaccination has truly gained more attention since the recent COVID-19 pandemic, this subject is more than ever important to consider.
5.1. Type I IFN-mediated effects: Beneficial or detrimental for mRNA vaccines?
A first requirement for sufficient mRNA vaccine efficiency is that the antigen expression needs to reach the antigenic threshold for T cell and B cell activation [202], [203], [204]. After the mRNA translation process, preferentially taken place in antigen presenting cells (APCs), produced antigens can, depending on their signaling properties, accumulate in the cytosolic compartment, be secreted into the extracellular matrix environment, or be exposed at the cellular surface as a membrane protein antigen. The latter two transport mechanisms make the antigenic proteins accessible for B cell recognition [205], [206]. Secreted antigens can also be recycled into APCs, thereby entering the major histocompatibility complex (MHC) II pathway to be presented to CD4+ T cells [207], [208]. Produced antigens in the cytosol can be directly processed into antigenic peptides, followed by their presentation in MHC-I complexes to CD8+ T cells [17], [34], [209]. Besides this antigen signal, other stimulatory signals are needed in order to activate adaptive immune cells [210]. This includes their interactions with co-stimulatory molecules (e.g. CD40, CD80 and CD86) and pro-inflammatory cytokines, as illustrated in Fig. 4 . The type I IFN reaction, induced by mRNA vaccines, may trigger several stimulatory effects on both innate and adaptive immune cells that can contribute to the vaccine efficacy. This self-adjuvant effect of mRNA vaccines promotes the activation of APCs, as type I IFN signaling leads to the upregulation of co-stimulatory molecules, stimulates antigen presentation pathways, and activates the production of pro-inflammatory cytokines and chemokines. Moreover, type I IFNs can directly act as a third cytokine signal for T cell activation. However, at the same time, type I IFNs can interfere with the mRNA translation process, lowering the antigen availability, and therefore in some cases might negatively impact the vaccine outcome [17], [34], [35], [201], [211], [212], [213], [214], [215].
Fig. 4.
From mRNA delivery to immunity activation. Upon mRNA delivery to APCs (1), mRNA sensing by PRRs leads to the production of type I IFNs, ISGs and pro-inflammatory cytokines (2). A crucial step is that mRNA is efficiently translated resulting in the production and accumulation of antigen proteins in the cytosol (3). Subsequently, antigen proteins can be secreted in the extracellular environment or presented as a transmembrane protein to B cells, depending on their signaling properties (4). Moreover, antigen processing to peptides occurs by several pathways (5), which are presented via the MHC-I and MHC-II molecules to CD8+ and CD4+ T cells, respectively (6). Interaction of T cells with co-stimulatory molecules on the APC surface serves as a second necessary signal for T cell activation (7). All of these activation signals are boosted by type I IFNs. Moreover, type I IFNs, present in the extracellular environment, can interact with their receptor on the T cell surface, thereby acting as a direct third activation signal (8). Abbreviations: BCR, B cell receptor; TCR, T cell receptor.
In the context of the development of mRNA vaccines for COVID-19, the focus was laid on the generation of neutralizing antibody responses against the SARS-COV-2 Spike glycoprotein [205]. At the end of last year, two COVID-19 mRNA vaccines, BNT162b2 and mRNA-1273, were granted authorization for emergency use. These COVID-19 mRNA vaccines are composed of HPLC-purified, m1Ψ-containing mRNA formulated in LNPs to allow their delivery upon intramuscular administration [205]. By mitigating the type I IFN activity, e.g. using modified uridines and extensive purifications methods, more durable protein expression and thus prolonged antigen availability can be achieved [205]. In general, a prolonged mRNA expression and continuous presentation of antigens via MHC-II molecules is expected to promote the induction of helper CD4+ T cells and to elicit Germinal Center (GC) reactions. In these GCs, formed in secondary lymphoid tissues such as in the spleen and lymph nodes, B cells undergo cell divisions, affinity maturation and isotype switching. Eventually, this will determine the quality and magnitude of the antibody response [202]. Lederer et al. demonstrated that intramuscular immunization with a SARS-CoV-2 (m1Ψ) mRNA-LNP vaccine in mice induced robust spike-specific GC B cells and follicular CD4+ T cell responses [216]. Moreover, a recent study report provided first clinical evidence that vaccination with the SARS-CoV-2 mRNA vaccine, BNT162b2, in humans induces a robust and persistent GC B cell response in the draining axillary lymph nodes of all participants after boosting [217].
In contrast, the use of “immunogenic” unmodified mRNA has been the preferred choice in the design of mRNA vaccines for cancer vaccination, where the main focus is to elicit cytotoxic CD8+ T cell responses to eliminate tumor cells [205]. In light of this, several conventional mRNA vaccination studies for cancer immunotherapy highlighted the role of type I IFNs to elicit cytotoxic T cells and therefore their influence on vaccine efficiency. Additionally, it is important to remark that type I IFNs have also shown to possess direct antitumor effects, exerted via two mechanisms: inhibition of tumor cell growth and angiogenesis [205]. Kranz et al. illustrated that intravenous injection of RNA-lipoplexes induced IFN-α secretion leading to strong CD8+ T cell responses. When an IFNAR blocking antibody was administered, the T cell effector response dropped and antitumor immunity was decreased in contrast to mice not treated with IFNAR antibodies [218]. Furthermore, Broos and colleagues observed a decrease in mRNA expression upon intravenous administration of mRNA-RNAiMAX complexes, due to type I IFN signaling. However, the tumor regression and cytotoxic T cell response was considerably lower in IFNAR deficient mice, corroborating the role of type I IFN secretion. Of note, it is important to mention that both intramuscular and subcutaneous injection did not lead to the desired CD8+ T cell expansion and activity [219]. The latter suggests an important role for the administration route on the final outcome. Additionally, the carrier system needs to be optimized according to the route of administration, as different anatomical and physiological barriers for successful mRNA delivery will be encountered. For instance, as also shown by Hassett et al., mRNA lipoplexes composed with the cationic lipid DOTAP fail to obtain protein expression when these particles are intramuscularly injected [220]. However, these particles are effective to induce CD8+ T cells upon intravenous administration, by successfully targeting and transfecting APCs in the spleen [196], [218].
The impact of type I IFN signaling on the vaccine outcome was shown to be different for saRNA vaccines. The antigen expression kinetics from saRNA differs from conventional mRNA, as it is preceded by the translation of the replicase complex and as it is more intense and long lasting [3], [20]. Also, due to structural and possible productional differences between mRNA and saRNA, their innate immune-stimulating activity might not be equally strong. Hence, the intensity and timing of the elicited innate immune response and the final outcome of type I IFNs on vaccine efficiency might differ as well. Pepini and colleagues revealed that intramuscular injection of a saRNA-based vaccine induced an early and robust type I IFN and ISG response in mice, which negatively impacted initial antigen expression. In IFNAR knockout mice, reporter antigen expression was significantly higher and sustained upon intramuscular LNP-mediated saRNA delivery. Moreover, a saRNA vaccine encoding respiratory syncytial virus (RSV) F protein was shown to be more potent in IFNAR knockout compared to wild-type mice, as illustrated by an increase in CD8+ T cell responses and in total IgG anti-RSV F antibody titer [213]. Additionally, Zhong et al. reported on the negative effect of type I IFNs on saRNA vaccine efficiency. They showed that intradermal electroporation of their saRNA vaccine encoding the pre-membrane and envelope (prM-E) glycoproteins of Zika virus (ZIKV) elicited higher ZIKV E specific CD8+ T cell responses, antibody titers and seroconversion rates in IFNAR1 knockout mice in comparison to wild-type mice [221].
It should be noted that the different experimental conditions used in the many, sometimes contradictory, studies available make it difficult to compare results with one another. It has however been suggested that the amount of type I IFN secretion as well as the timing of type I IFN signaling are crucial for the final outcome [16], [34], [208], [212], [219], [222], [223]. Depending on whether the type I IFNs act before or after T cell priming, the effects can be detrimental or beneficial respectively, as discussed by De Beuckelaer et al. [34]. T-cell activation followed by or simultaneous with type I IFN signaling stimulates clonal proliferation and differentiation. In contrast, when the opposite order of events occurs, proliferation might be inhibited and T cell apoptosis can take place. An explanation for this complex mechanism was found in the STAT signaling pathway. When T cells are activated by antigen recognition, the transcription factor STAT4 is activated. Subsequent IFNAR stimulation can re-activate STAT4, thereby inducing proliferation and blocking apoptosis. Although, if IFNAR stimulation occurs before T cell priming, the STAT1 protein can be upregulated resulting in apoptosis and inhibition of proliferation [34], [215], [223]. In fact, both the administration route and carrier can determine the order of signaling and the effect of type I IFNs on CD8+ T cell responses, making it important aspects of the vaccine formulation [16], [34], [224].
5.2. Adjuvant therapies
All things considered, both antigen expression and innate immune stimulation are crucial for the vaccine efficiency [16], [17], [219]. The self-adjuvant effect of mRNA therapeutics is one of the aspects that contributed to the increasing interest in conventional and self-amplifying mRNA-based vaccines. However, type I IFN production can come at a cost of both non-amplifying and self-amplifying mRNA translation, which can work detrimental for the vaccine efficiency. Currently, researchers continuously aim to develop optimal mRNA vaccine formulations where the antigen is adequately expressed and innate immune responses are contributing to the final outcome without hampering the safety. The question however remains, if only the application of the mRNA modifications is enough to reach this goal. Besides adaption of the mRNA structure as extensively described in this review and the optimization of delivery vehicles, many researchers have focused on the use of other immune-stimulatory compounds. Especially, in the field of cancer immunotherapy, this could be of interest to increase CD8+ T cell response [205]. We provide some examples below but for an extensive list of the explored options, we refer to [16], [17], [21], [34].
As mentioned above, the use of modified uridines can improve the intracellular mRNA stability and translation efficiency by reduction of PRR signaling, but can also come at a cost of T cell priming and functioning [19], [34], [225]. Therefore, Verbeke et al. explored the use of the TLR4-agonist monophosphoryl lipid A (MPLA), as an adjuvant to nucleoside-modified mRNA-loaded liposomes. Results indicated that type I IFN signaling was extremely reduced using nucleoside-modified mRNA, while CD8+ T cell responses were maintained if MPLA was incorporated in the modified mRNA liposomes [17], [196]. A later study by the same researchers, revealed the potential of intravenously injected mRNA Galsomes, liposomes containing nucleoside-modified mRNA encoding the antigen of interest and α-galactosylceramide, a glycolipid to promote anti-tumor immunity. The authors observed a more potent CD8+ T cell response compared to liposomes containing immunogenic unmodified mRNA or modified mRNA alone, resulting in substantial tumor regression [226]. Furthermore, CureVac has developed the RNActive® vaccine formulation, which consists of naked sequence-engineered mRNA supplemented with mRNA-protamine complexes to induce immune responses upon TLR7 activation. In order to benefit from both considerable antigen expression and immune stimulation, an appropriate ratio between free mRNA and mRNA-protamine complexes should be maintained [17], [21], [34], [227]. Another vaccine adjuvant formulation, TriMix, contains a mixture of three different mRNA molecules encoding CDL40, CD70 and constitutive active TLR4 (caTRL4) that boost DC activation [17], [34], [35]. TriMix and tumor antigen mRNA co-electroporated in DCs progressed as a treatment strategy for melanoma [228]. Recently, a phase II clinical trial was completed where patients received TriMixDC-MEL together with the checkpoint inhibitor ipilimumab, to block negative feedback signals in the generated immune response, long term follow-up results indicate an overall survival of 28% after 5 years and a clear correlation between the clinical outcome and the tumor-antigen-specific CD8+ T cell responses was observed [229], [230]. Furthermore, this adjuvant therapy co-delivered with HIV immunogen sequence (HTI)-encoding mRNA was explored as treatment option for HIV patients. Promising results were gathered upon intranodal administration during preclinical studies and the formulation entered phase I and phase II clinical trials. However, the latter was terminated due to insufficient immunity induction compared to placebo [231], [232], [233]. At last, a very recent and interesting study by Tse et al. evaluated a mutated version of the stimulator of interferon genes (STINGV155M) as an adjuvant for mRNA-based cancer immunotherapy. Mice were vaccinated by intramuscular injection of LNPs containing mRNA encoding the antigen of choice together with mRNA encoding the adjuvant. They observed a strong CD8+ T cell response and increased vaccine efficiency in terms of tumor growth and survival. It is however remarkable that the adjuvant effect is linked to the production of type I IFNs, which has been considered detrimental for antigen expression when generated as part of the self-adjuvant effect of the antigen-encoding-mRNA. However, as discussed above, the authors acknowledge the timing and amount of type I IFN production as critical factors for the fate of CD8+ T cell responses. To address this issue, several mass ratios of antigen/adjuvant were tested for their potential to induce CD8+ T cell activation [223]. Future studies, focused especially on the influence of adjuvant therapies on the mRNA expression kinetics, would be useful.
Taken together, mRNA vaccine development is all about achieving optimal contribution of the different elements involved. Researchers agree that several aspects such as administration route, mRNA design and formulation, cell type, and translation from animals to humans will define which pathways and effects are triggered [17], [34], [192], [196]. Different options to optimize vaccine efficiency have been explored. The question however remains if optimal vaccine efficiency can be obtained solely by fine-tuning of the mRNA molecule or if there is room for improvement with other immune adjuvant therapies. Moreover, an interesting but often neglected aspect to take into account is the adjuvant effect of the delivery vehicle itself. However, with the very recent approval of two mRNA vaccine formulations to control the COVID-19 pandemic, we believe a new era of mRNA vaccine development has started. The ongoing research by many scientists over the world will surely increase our knowledge which can possibly improve the progress of mRNA therapeutics in general.
6. Conclusions
During the last decade, important investments have been made in the research and development of mRNA- and saRNA-based therapeutics. Up till now, most preclinical and clinical reports on mRNA therapeutics focus on its potential for vaccination purposes against infectious diseases and cancer, but the IVT mRNA platform is assumed to be versatile and promising for other applications as well, including protein replacement therapy, gene editing, and cell fate determination and reprogramming. Although the applications are very diverse, some common hurdles should be taken into account in order to fully exploit their potential as therapeutics. In this review, we focus on one of these hurdles and discuss in depth the immune-stimulatory properties of IVT mRNA and saRNA.
It is important to realize that the therapeutic outcome of mRNA- and saRNA-based therapies can either benefit or suffer from RNA-induced immune stimulation depending on the treatment application. For non-immunotherapy applications, we can unambiguously state that mRNA-induced immune stimulation should be avoided. Moreover, the induced innate immune response is highly unfavorable for frequent mRNA administration, which might be required for protein replacement therapies and mRNA-based cell reprogramming protocols. In contrast, for vaccination approaches, the situation is more complex and not fully elucidated yet. Here, it will be crucial but challenging to find a balance between the negative impact of the induced type I IFNs on antigen expression and RNA amplification on the one hand, and the potential of type I IFNs to instigate B and T cell responses on the other making the mRNA to become its own adjuvant. Moreover, conflicting reports regarding the influence of type I IFNs on CD8+ T cell responses further increase complexity.
Consequently, a variety of strategies have been explored to control the innate immune response elicited by mRNA- and saRNA-based therapeutics and more in-depth research is still ongoing. The vast majority of studies on this topic investigate mRNA-based therapeutics, whereas only a few solely focus on reducing the innate immunogenicity of saRNA. To us this is not surprising because up till now, except for some saRNA-mediated cell reprogramming studies, the use of saRNA was restricted to vaccination approaches in which the self-adjuvant properties of saRNA were considered as an important benefit for vaccine potency. However, nowadays, it is generally realized that controlling the innate immune response to saRNA is of vital importance to obtain effective and safe saRNA-based vaccines in the end, and therefore researchers started looking into strategies to modulate the innate immune response to saRNA. Furthermore, if saRNA-based therapeutics would be explored in the future for other non-immunotherapy approaches, it will be even more crucial to reduce its immune-stimulating activity.
Currently, the methods that have been applied to limit the immune-stimulatory activity of IVT mRNA and saRNA can be divided into three main categories. Firstly, every element of the mRNA or saRNA backbone itself can be altered, which is a very convenient strategy to optimize its stability and to evade innate immune stimulation directly or indirectly. In this regard, the most frequently applied and successful approach is the incorporation of modified nucleosides. However, caution should be taken because conflicting studies report that incorporation of modified nucleosides did not impact or even negatively impacted protein expression. Once again, this highlights the complexity and shows that variations in delivery system, administration route, target cell and mRNA properties may also affect the evoked innate immune response. Secondly, there is no doubt that proper production and purification of the mRNA or saRNA are essential requirements to obtain a safe and effective end product, free of immunogenic byproducts such as short abortive RNA transcripts and dsRNAs. Thirdly, both conventional and self-replicating mRNA delivery can be combined with innate immune inhibitors to quell its immune-stimulating activity. This approach is an attractive alternative to other rather time consuming and expensive approaches described above. In addition, to retain saRNA functionality, neither nucleoside modification nor sequence alteration is tolerated, which increases the interest in the use of innate immune inhibitors. Both for mRNA and saRNA, promising results were obtained already in multiple studies with a variety of inhibitors, highlighting their potential and the need for further investigation. However, it is important to realize that the choice of inhibitor is also depending on the target cell type and the treatment application, which complicates the identification of innate immune inhibitors that might be applied universally for mRNA- and saRNA-based therapeutics.
It is clear that the mRNA- or saRNA-mediated innate immune response is a complex interplay of many factors, like the mRNA properties, target cell, the delivery vehicle and the administration route, which define the fate of the mRNA. The immune-stimulating activity of the carrier itself is often overlooked and future research on this topic would be of interest. Since many different players are involved in the innate immunity signaling pathway itself, it is not surprising that reducing the innate immune-stimulating activity of mRNA and saRNA is not straightforward and none of the methods described in this review can be regarded as the golden standard to achieve this goal. Moreover, a combination of different strategies will be typically required to modulate the innate immunogenicity of mRNA and saRNA according to the desired application. In conclusion, although numerous efforts have been made already with regard to innate immune sensing of mRNA and saRNA, there are still knowledge gaps requiring further research and head-to-head comparisons between mRNA platforms. We believe that progress in this field will be made soon, as the recent marketing of the mRNA-based vaccines developed by Pfizer/BioNTech and Moderna is an ultimate boost for the research and development of numerous other mRNA- and saRNA-based therapeutics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
An-Katrien Minnaert and Helena Vanluchene are doctoral fellows from the Research Foundation-Flanders, Belgium (FWO Vlaanderen) with grant numbers 1S28420N (FWO-SB) and 11A6320N (FWO) respectively. Rein Verbeke is supported by a UGent BOF postdoc grant (BOF20/PDO/041). The authors would like to acknowledge funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 810685 (DelNam project), the Research Foundation-Flanders, Belgium (FWO-Vlaanderen; grant No. G040319N and grant No. G016221N), the Research Foundation “Kom op Tegen Kanker” (grant No. FAF-C/2018/1213), Ghent University Concerted Research Action (grant No. BOF21/GOA/033) and the European Joint Programme on Rare Diseases (FWO-ERA-Net grant No. G0H7520N).
References
- 1.Meo S.A., Bukhari I.A., Akram J., Meo A.S., Klonoff D.C. COVID-19 vaccines: comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and moderna vaccines. Eur. Rev. Med. Pharmacol. Sci. 2021;25:1663–1669. doi: 10.26355/eurrev_202102_24877. [DOI] [PubMed] [Google Scholar]
- 2.Gómez-Aguado I., Rodríguez-Castejón J., Vicente-Pascual M., Rodríguez-Gascón A., Solinís M.Á., del Pozo-Rodríguez A. Nanomedicines to deliver mRNA: State of the art and future perspectives. Nanomaterials. 2020;10(2):364. doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brito L.A., Kommareddy S., Maione D., Uematsu Y., Giovani C., Berlanda Scorza F., Otten G.R., Yu D., Mandl C.W., Mason P.W., Dormitzer P.R., Ulmer J.B., Geall A.J. Self-Amplifying mRNA Vaccines. Elsevier Ltd. 2015 doi: 10.1016/bs.adgen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Blakney A.K., McKay P.F., Bouton C.R., Hu K., Samnuan K., Shattock R.J. Innate inhibiting proteins enhance expression and immunogenicity of self-amplifying RNA. Mol. Ther. 2021;29(3):1174–1185. doi: 10.1016/j.ymthe.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A.K. Blakney, S. Ip, A.J. Geall, An update on self-amplifying mRNA vaccine development, Vaccines Spec. Issue “The Past, Present Futur. MRNA Vaccines.” 9 (2021) 1–26. https://doi.org/https://doi.org/10.3390/vaccines9020097. [DOI] [PMC free article] [PubMed]
- 6.Brenner S., Jacob F., Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190(4776):576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 7.Hajj K.A., Whitehead K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:1–17. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 8.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science (80-) 1990;247(4949):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 9.Weng Y., Li C., Yang T., Hu B.o., Zhang M., Guo S., Xiao H., Liang X.-J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 10.Devoldere J., Dewitte H., De Smedt S.C., Remaut K. Evading innate immunity in nonviral mRNA delivery: Don’t shoot the messenger. Drug Discov. Today. 2016;21(1):11–25. doi: 10.1016/j.drudis.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Alfagih I.M., Aldosari B., AlQuadeib B., Almurshedi A., Alfagih M.M. Nanoparticles as adjuvants and nanodelivery systems for mRNA-based vaccines. Pharmaceutics. 2020;13:45. doi: 10.3390/pharmaceutics13010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27(4):710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahin U., Karikó K., Türeci Ö. MRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 14.Dewitte H., Verbeke R., Breckpot K., De Smedt S.C., Lentacker I. Nanoparticle design to induce tumor immunity and challenge the suppressive tumor microenvironment. Nano Today. 2014;9(6):743–758. doi: 10.1016/j.nantod.2014.10.001. [DOI] [Google Scholar]
- 15.Loomis K.H., Kirschman J.L., Bhosle S., Bellamkonda R.V., Santangelo P.J. Strategies for modulating innate immune activation and protein production of in vitro transcribed mRNAs. J. Mater. Chem. B. 2016;4(9):1619–1632. doi: 10.1039/C5TB01753J. [DOI] [PubMed] [Google Scholar]
- 16.Linares-Fernández S., Lacroix C., Exposito J.-Y., Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2020;26(3):311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- 18.Heine A., Juranek S., Brossart P. Clinical and immunological effects of mRNA vaccines in malignant diseases. Mol. Cancer. 2021;20:1–20. doi: 10.1186/s12943-021-01339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deering R.P., Kommareddy S., Ulmer J.B., Brito L.A., Geall A.J. Nucleic acid vaccines: Prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014;11(6):885–899. doi: 10.1517/17425247.2014.901308. [DOI] [PubMed] [Google Scholar]
- 20.Geall A.J., Mandl C.W., Ulmer J.B. RNA: The new revolution in nucleic acid vaccines. Semin. Immunol. 2013;25(2):152–159. doi: 10.1016/j.smim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27(4):757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom K., van den Berg F., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28(3-4):117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong Z., Mc Cafferty S., Combes F., Huysmans H., De Temmerman J., Gitsels A., Vanrompay D., Portela Catani J., Sanders N.N. mRNA therapeutics deliver a hopeful message. Nano Today. 2018;23:16–39. doi: 10.1016/j.nantod.2018.10.005. [DOI] [Google Scholar]
- 25.Rupp J.C., Sokoloski K.J., Gebhart N.N., Hardy R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015;96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietilä M.K., Hellström K., Ahola T. Alphavirus polymerase and RNA replication. Virus Res. 2017;234:44–57. doi: 10.1016/j.virusres.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Iavarone C., O’hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines. 2017;16(9):871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 28.Fros J., Pijlman G. Alphavirus infection: Host cell shut-off and inhibition of antiviral responses. Viruses. 2016;8(6):166. doi: 10.3390/v8060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., Dey A.K., Lilja A., Valiante N.M., Mason P.W., Mandl C.W., Barnett S.W., Dormitzer P.R., Ulmer J.B., Singh M., O'Hagan D.T., Geall A.J. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22(12):2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leyman B., Huysmans H., Mc Cafferty S., Combes F., Cox E., Devriendt B., Sanders N.N. Comparison of the expression kinetics and immunostimulatory activity of replicating mRNA, nonreplicating mRNA, and pDNA after intradermal electroporation in pigs. Mol. Pharm. 2018;15(2):377–384. doi: 10.1021/acs.molpharmaceut.7b00722. [DOI] [PubMed] [Google Scholar]
- 31.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., Reece S.T., Sahin U., Tregoning J.S. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26(2):446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blakney A.K., McKay P.F., Shattock R.J. Structural components for amplification of positive and negative strand VEEV splitzicons. Front. Mol. Biosci. 2018;5:1–10. doi: 10.3389/fmolb.2018.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beissert T., Perkovic M., Vogel A., Erbar S., Walzer K.C., Hempel T., Brill S., Haefner E., Becker R., Türeci Ö., Sahin U. A Trans-amplifying RNA vaccine strategy for induction of potent protective immunity. Mol. Ther. 2020;28(1):119–128. doi: 10.1016/j.ymthe.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Beuckelaer A., Grooten J., De Koker S. Type I interferons modulate CD8+ T cell immunity to mRNA vaccines. Trends Mol. Med. 2017;23(3):216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Pollard C., De Koker S., Saelens X., Vanham G., Grooten J. Challenges and advances towards the rational design of mRNA vaccines. Trends Mol. Med. 2013;19(12):705–713. doi: 10.1016/j.molmed.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Sergeeva O.V., Koteliansky V.E., Zatsepin T.S. mRNA based therapeutics – advances and perspectives. Biochem. 2016;81:709–722. doi: 10.1134/S0006297916070075. [DOI] [PubMed] [Google Scholar]
- 37.DeWitte-Orr S.J., Mossman K.L. dsRNA and the innate antiviral immune response. Future Virol. 2010;5(3):325–341. doi: 10.2217/fvl.10.18. [DOI] [Google Scholar]
- 38.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014;32(1):513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez David R.Y., Combredet C., Najburg V., Millot G.A., Beauclair G., Schwikowski B., Léger T., Camadro J.-M., Jacob Y., Bellalou J., Jouvenet N., Tangy F., Komarova A.V. LGP2 binds to PACT to regulate RIG-I– and MDA5-mediated antiviral responses. Sci. Signal. 2019;12(601):eaar3993. doi: 10.1126/scisignal.aar3993. [DOI] [PubMed] [Google Scholar]
- 40.Xiaojiao Fan and Tengchuan Jin, Structures of RIG-I-Like Receptors and Insights into Viral RNA Sensing, in: Struct. Immunol., 2019: pp. 157–188. https://doi.org/10.1007/978-981-13-9367-9_6. [DOI] [PubMed]
- 41.Duic I., Tadakuma H., Harada Y., Yamaue R., Deguchi K., Suzuki Y., Yoshimura S.H., Kato H., Takeyasu K., Fujita T. Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes. Nucleic Acids Res. 2020;48:11664–11674. doi: 10.1093/nar/gkaa935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabbah A., Chang T.H., Harnack R., Frohlich V., Tominaga K., Dube P.H., Xiang Y., Bose S. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009;10(10):1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581.Regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 45.Kaur S., Platanias L.C. IFN-β-specific signaling via a unique IFNAR1 interaction. Nat. Immunol. 2013;14(9):884–885. doi: 10.1038/ni.2686. [DOI] [PubMed] [Google Scholar]
- 46.de Weerd N.A., Vivian J.P., Nguyen T.K., Mangan N.E., Gould J.A., Braniff S.-J., Zaker-Tabrizi L., Fung K.Y., Forster S.C., Beddoe T., Reid H.H., Rossjohn J., Hertzog P.J. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 2013;14(9):901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- 47.van Boxel-Dezaire A.H.H., Rani M.R.S., Stark G.R. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25(3):361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber G. The molecular basis for differential type i interferon signaling. J. Biol. Chem. 2017;292(18):7285–7294. doi: 10.1074/jbc.R116.774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai M.H., Pai L.M., Lee C.K. Fine-tuning of type I interferon response by STAT3. Front. Immunol. 2019;10:1448. doi: 10.3389/fimmu.2019.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonnet M.C., Weil R., Dam E., Hovanessian A.G., Meurs E.F. PKR Stimulates NF-κB Irrespective of Its Kinase Function by Interacting with the IκB Kinase Complex. Mol. Cell. Biol. 2000;20(13):4532–4542. doi: 10.1128/MCB.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gil J., Alcamí J., Esteban M. Activation of NF-κB by the dsRNA-dependent protein kinase, PKR involves the IκB kinase complex. Oncogene. 2000;19(11):1369–1378. doi: 10.1038/sj.onc.1203448. [DOI] [PubMed] [Google Scholar]
- 52.Ishii T., Kwon H., Hiscott J., Mosialos G., Koromilas A.E. Activation of the IκBα kinase (IKK) complex by double-stranded RNA-binding defective and catalytic inactive mutants of the interferon-inducible protein kinase PKR. Oncogene. 2001;20(15):1900–1912. doi: 10.1038/sj.onc.1204267. [DOI] [PubMed] [Google Scholar]
- 53.Lamers M.M., van den Hoogen B.G., Haagmans B.L. ADAR1: “Editor-in-Chief” of Cytoplasmic Innate Immunity. Front. Immunol. 2019;10:1763. doi: 10.3389/fimmu.2019.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lohöfener J., Steinke N., Kay-Fedorov P., Baruch P., Nikulin A., Tishchenko S., Manstein D., Fedorov R. The activation mechanism of 2′-5′-oligoadenylate synthetase gives new insights into OAS/cGAS triggers of innate immunity. Structure. 2015;23(5):851–862. doi: 10.1016/j.str.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Drappier M., Michiels T. Inhibition of the OAS/RNase L pathway by viruses. Curr. Opin. Virol. 2015;15:19–26. doi: 10.1016/j.coviro.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Charles E., ADARs, Viruses and Innate Immunity, Assess. Eval. High. Educ. 37 (2012) 1–28. https://doi.org/10.1007/82_2011_148.
- 57.Honke Nadine, Shaabani Namir, Cadeddu Giuseppe, Sorg Ursula R, Zhang Dong-Er, Trilling Mirko, Klingel Karin, Sauter Martina, Kandolf Reinhard, Gailus Nicole, van Rooijen Nico, Burkart Christoph, Baldus Stephan E, Grusdat Melanie, Löhning Max, Hengel Hartmut, Pfeffer Klaus, Tanaka Masato, Häussinger Dieter, Recher Mike, Lang Philipp A, Lang Karl S. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat. Immunol. 2012;13(1):51–57. doi: 10.1038/ni.2169. [DOI] [PubMed] [Google Scholar]
- 58.Ramanathan Anand, Robb G. Brett, Chan Siu-Hong. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016;44(16):7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devarkar Swapnil C., Wang Chen, Miller Matthew T., Ramanathan Anand, Jiang Fuguo, Khan Abdul G., Patel Smita S., Marcotrigiano Joseph. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U. S. A. 2016;113(3):596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuberth-Wagner Christine, Ludwig Janos, Bruder Ann Kristin, Herzner Anna-Maria, Zillinger Thomas, Goldeck Marion, Schmidt Tobias, Schmid-Burgk Jonathan L., Kerber Romy, Wolter Steven, Stümpel Jan-Philip, Roth Andreas, Bartok Eva, Drosten Christian, Coch Christoph, Hornung Veit, Barchet Winfried, Kümmerer Beate M., Hartmann Gunther, Schlee Martin. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1–2’O-methylated self RNA. Immunity. 2015;43(1):41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Züst Roland, Cervantes-Barragan Luisa, Habjan Matthias, Maier Reinhard, Neuman Benjamin W, Ziebuhr John, Szretter Kristy J, Baker Susan C, Barchet Winfried, Diamond Michael S, Siddell Stuart G, Ludewig Burkhard, Thiel Volker. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr, Shi P.Y., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCaffrey A.P., Epitranscriptome R.N.A. Role of the 5’ Cap: Producing superior mRNA therapeutics through optimized capping. Genet. Eng. Biotechnol. News. 2019;39:59–61. doi: 10.1089/gen.39.05.17. [DOI] [Google Scholar]
- 64.Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl(3′-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 65.Pardi N., Muramatsu H., Weissman D., Karikó K. In vitro transcription of long RNA containing modified nucleosides. Methods Mol. Biol. 2013:29–42. doi: 10.1007/978-1-62703-260-5. [DOI] [PubMed] [Google Scholar]
- 66.Vaidyanathan S., Azizian K.T., Haque A.K.M.A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. - Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muttach F., Muthmann N., Rentmeister A. Synthetic mRNA capping. Beilstein J. Org. Chem. 2017;13:2819–2832. doi: 10.3762/bjoc.13.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrett Lucy W., Fletcher Sue, Wilton Steve D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012;69(21):3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatterjee S., Pal J.K. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/bc20080104. [DOI] [PubMed] [Google Scholar]
- 70.Kuhn A.N., Beißert T., Simon P., Vallazza B., Buck J., Davies B.P., Tureci O., Sahin U. mRNA as a versatile tool for exogenous protein expression. Curr. Gene Ther. 2012;12:347–361. doi: 10.2174/156652312802762536. [DOI] [PubMed] [Google Scholar]
- 71.Ross J., Sullivan T.D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. doi: 10.1182/blood.v66.5.1149.1149. [DOI] [PubMed] [Google Scholar]
- 72.Waggoner Shelly A., Liebhaber Stephen A. Regulation of α-Globin mRNA Stability. Exp. Biol. Med. 2003;228(4):387–395. doi: 10.1177/153537020322800409. [DOI] [PubMed] [Google Scholar]
- 73.Karikó K., Kuo A., Barnathan E.S. Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene Ther. 1999;6(6):1092–1100. doi: 10.1038/sj.gt.3300930. [DOI] [PubMed] [Google Scholar]
- 74.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci Ö., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 75.Vivinus S., Baulande S., Van Zanten M., Campbell F., Topley P., Ellis J.H., Dessen P., Coste H. An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur. J. Biochem. 2001;268:1908–1917. doi: 10.1046/j.1432-1327.2001.02064.x. [DOI] [PubMed] [Google Scholar]
- 76.Thess Andreas, Grund Stefanie, Mui Barbara L, Hope Michael J, Baumhof Patrick, Fotin-Mleczek Mariola, Schlake Thomas. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23(9):1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hayashi Toshinori, Lamba Deepak A., Slowik Amber, Reh Thomas A., Bermingham-McDonogh Olivia. A method for stabilizing RNA for transfection that allows control of expression duration. Dev. Dyn. 2010;239(7):2034–2040. doi: 10.1002/dvdy.22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garneau Nicole L., Sokoloski Kevin J., Opyrchal Mateusz, Neff C. Preston, Wilusz Carol J., Wilusz Jeffrey. The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells. J. Virol. 2008;82(2):880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orlandini von Niessen Alexandra G., Poleganov Marco A., Rechner Corina, Plaschke Arianne, Kranz Lena M., Fesser Stephanie, Diken Mustafa, Löwer Martin, Vallazza Britta, Beissert Tim, Bukur Valesca, Kuhn Andreas N., Türeci Özlem, Sahin Ugur. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol. Ther. 2019;27(4):824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eckmann Christian R., Rammelt Christiane, Wahle Elmar. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011;2(3):348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 81.Lima Sarah Azoubel, Chipman Laura B, Nicholson Angela L, Chen Ying-Hsin, Yee Brian A, Yeo Gene W, Coller Jeff, Pasquinelli Amy E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017;24(12):1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mockey Michael, Gonçalves Cristine, Dupuy Franck P., Lemoine François M., Pichon Chantal, Midoux Patrick. mRNA transfection of dendritic cells: Synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 2006;340(4):1062–1068. doi: 10.1016/j.bbrc.2005.12.105. [DOI] [PubMed] [Google Scholar]
- 83.Grier Alexandra E, Burleigh Stephen, Sahni Jaya, Clough Courtnee A, Cardot Victoire, Choe Dongwook C, Krutein Michelle C, Rawlings David J, Jensen Michael C, Scharenberg Andrew M, Jacoby Kyle. pEVL: A linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol. Ther. - Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jalkanen A.L., Coleman S.J., Wilusz J. Determinants and implications of mRNA poly(A) tail size - Does this protein make my tail look big? Semin. Cell Dev. Biol. 2014;34:24–32. doi: 10.1016/j.semcdb.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koski Gary K., Karikó Katalin, Xu Shuwen, Weissman Drew, Cohen Peter A., Czerniecki Brian J. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J. Immunol. 2004;172(7):3989–3993. doi: 10.4049/jimmunol.172.7.3989. [DOI] [PubMed] [Google Scholar]
- 86.Weissman Drew, Karikó Katalin. mRNA: Fulfilling the promise of gene therapy. Mol. Ther. 2015;23(9):1416–1417. doi: 10.1038/mt.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCown Phillip J., Ruszkowska Agnieszka, Kunkler Charlotte N., Breger Kurtis, Hulewicz Jacob P., Wang Matthew C., Springer Noah A., Brown Jessica A. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA. 2020;11(5) doi: 10.1002/wrna.v11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karikó Katalin, Buckstein Michael, Ni Houping, Weissman Drew. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 89.Eigenbrod Tatjana, Dalpke Alexander H. Bacterial RNA: An underestimated stimulus for innate immune responses. J. Immunol. 2015;195(2):411–418. doi: 10.4049/jimmunol.1500530. [DOI] [PubMed] [Google Scholar]
- 90.Karikó Katalin, Muramatsu Hiromi, Welsh Frank A, Ludwig János, Kato Hiroki, Akira Shizuo, Weissman Drew. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.B.R. Anderson, H. Muramatsu, B.K. Jha, R.H. Silverman, D. Weissman, K. Karikó, Nucleoside modifications in RNA limit activation of 2′-5′- oligoadenylate synthetase and increase resistance to cleavage by RNase L, Nucleic Acids Res. 39 (2011) 9329–9338. https://doi.org/10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed]
- 94.Kormann Michael S D, Hasenpusch Günther, Aneja Manish K, Nica Gabriela, Flemmer Andreas W, Herber-Jonat Susanne, Huppmann Marceline, Mays Lauren E, Illenyi Marta, Schams Andrea, Griese Matthias, Bittmann Iris, Handgretinger Rupert, Hartl Dominik, Rosenecker Joseph, Rudolph Carsten. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29(2):154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 95.Karikó Katalin, Muramatsu Hiromi, Keller Jason M, Weissman Drew. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20(5):948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mahiny Azita J, Dewerth Alexander, Mays Lauren E, Alkhaled Mohammed, Mothes Benedikt, Malaeksefat Emad, Loretz Brigitta, Rottenberger Jennifer, Brosch Darina M, Reautschnig Philipp, Surapolchai Pacharapan, Zeyer Franziska, Schams Andrea, Carevic Melanie, Bakele Martina, Griese Matthias, Schwab Matthias, Nürnberg Bernd, Beer-Hammer Sandra, Handgretinger Rupert, Hartl Dominik, Lehr Claus-Michael, Kormann Michael S D. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat. Biotechnol. 2015;33(6):584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 97.Andries Oliwia, Mc Cafferty Séan, De Smedt Stefaan C., Weiss Ron, Sanders Niek N., Kitada Tasuku. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 98.Devoldere J., Peynshaert K., Dewitte H., Vanhove C., De Groef L., Moons L., Özcan S.Y., Dalkara D., De Smedt S.C., Remaut K. Non-viral delivery of chemically modified mRNA to the retina: Subretinal versus intravitreal administration. J. Control. Release. 2019;307:315–330. doi: 10.1016/j.jconrel.2019.06.042. [DOI] [PubMed] [Google Scholar]
- 99.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gustafsson Claes, Govindarajan Sridhar, Minshull Jeremy. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 101.Cannarozzi Gina, Schraudolph Nicol N., Faty Mahamadou, von Rohr Peter, Friberg Markus T., Roth Alexander C., Gonnet Pedro, Gonnet Gaston, Barral Yves. A role for codon order in translation dynamics. Cell. 2010;141(2):355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 102.Mauro Vincent P., Chappell Stephen A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014;20(11):604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mauro Vincent P. Codon optimization in the production of recombinant biotherapeutics: potential risks and considerations. BioDrugs. 2018;32(1):69–81. doi: 10.1007/s40259-018-0261-x. [DOI] [PubMed] [Google Scholar]
- 104.Hanson Gavin, Coller Jeff. Translation and protein quality control: codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19(1):20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kudla Grzegorz, Lipinski Leszek, Caffin Fanny, Helwak Aleksandra, Zylicz Maciej, Hurst Laurence D. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.S.S. Diebold, C. Massacrier, S. Akira, C. Paturel, Y. Morel, C. Reis e Sousa, Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides, Eur. J. Immunol. 36 (2006) 3256–3267. https://doi.org/10.1002/eji.200636617. [DOI] [PubMed]
- 107.V. Hornung, W. Barchet, M. Schlee, G. Hartmann, RNA recognition via TLR7 and TLR8, in: Handb. Exp. Pharmacol., 2008: pp. 71–86. https://doi.org/10.1007/978-3-540-72167-3-4. [DOI] [PubMed]
- 108.Saito Takeshi, Owen David M., Jiang Fuguo, Marcotrigiano Joseph, Gale Michael. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.F. Heil, H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, S. Bauer, Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8, Science (80-.). 303 (2004) 1526–1529. https://doi.org/10.1126/science.1093620. [DOI] [PubMed]
- 110.Li Y., Teague B., Zhang Y., Su Z., Porter E., Dobosh B., Wagner T., Irvine D.J., Weiss R. In vitro evolution of enhanced RNA replicons for immunotherapy. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-43422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frolov Ilya, Agapov Eugene, Hoffman Thomas A., Prágai Béla M., Lippa Mara, Schlesinger Sondra, Rice Charles M. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 1999;73(5):3854–3865. doi: 10.1128/JVI.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.S.A. Dryga, O.A. Dryga, S. Schlesinger, Identification of mutations in a Sindbis virus able to establish persistent infection in BHK cells. The importance of a mutation in the nsP2 gene, Virology. 228 (1997) 74–83. https://doi.org/10.1006/viro.1996.8364. [DOI] [PubMed]
- 113.Perri Silvia, Driver David A., Gardner Jason P., Sherrill Scott, Belli Barbara A., Dubensky Thomas W., Polo John M. Replicon vectors derived from sindbis virus and semliki forest virus that establish persistent replication in host cells. J. Virol. 2000;74(20):9802–9807. doi: 10.1128/JVI.74.20.9802-9807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Petrakova Olga, Volkova Eugenia, Gorchakov Rodion, Paessler Slobodan, Kinney Richard M., Frolov Ilya. Noncytopathic replication of venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J. Virol. 2005;79(12):7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shabalina S.A., Ogurtsov A.Y., Spiridonov N.A. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;34:2428–2437. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U. S. A. 1986;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.A.G. Hinnebusch, I.P. Ivanov, N. Sonenberg, Translational control by 5′-untranslated regions of eukaryotic mRNAs, Science (80-.). 352 (2016) 1413–1416. https://doi.org/10.1126/science.aad9868. [DOI] [PMC free article] [PubMed]
- 119.D.M. Mauger, B. Joseph Cabral, V. Presnyak, S.V. Su, D.W. Reid, B. Goodman, K. Link, N. Khatwani, J. Reynders, M.J. Moore, I.J. McFadyen, mRNA structure regulates protein expression through changes in functional half-life, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 24075–24083. https://doi.org/10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed]
- 120.Leppek K., Byeon G.W., Kladwang W., Wayment-steele H.K. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Under Rev. 2021 doi: 10.1038/s41467-022-28776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.K. Karikó, H. Muramatsu, J. Ludwig, D. Weissman, Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA, Nucleic Acids Res. 39 (2011) e142. https://doi.org/10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed]
- 122.Milligan John F., Groebe Duncan R., Witherell Gary W., Uhlenbeck Olke C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Edelmann Franziska Theresia, Niedner Annika, Niessing Dierk. Production of pure and functional RNA for in vitro reconstitution experiments. Methods. 2014;65(3):333–341. doi: 10.1016/j.ymeth.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 124.S. Pascolo, Messenger RNA-based vaccines, (2004) 1285–1294. [DOI] [PubMed]
- 125.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.-G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14(15):1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 126.L.E. Easton, Y. Shibata, P.J. Lukavsky, Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography, RNA. 16 (2010) 647–653. https://doi.org/10.1261/rna.1862210. [DOI] [PMC free article] [PubMed]
- 127.J. Koubek, K.F. Lin, Y.R. Chen, R.P. Cheng, J.J.T. Huang, Strong anion-exchange fast performance liquid chromatography as a versatile tool for preparation and purification of RNA produced by in vitro transcription, RNA. 19 (2013) 1449–1459. https://doi.org/10.1261/rna.038117.113. [DOI] [PMC free article] [PubMed]
- 128.Kim I., Mckenna S.A., Puglisi E.V., Puglisi J.D. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2007;13:289–294. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McKenna Sean A, Kim Insil, Puglisi Elisabetta Viani, Lindhout Darrin A, Aitken Colin Echeverría, Marshall R Andrew, Puglisi Joseph D. Purification and characterization of transcribed rnas using gel filtration chromatography. Nat. Protoc. 2007;2(12):3270–3277. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- 130.Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc. Natl. Acad. Sci. U. S. A. 1972;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Konarska Maria M., Sharp Phillip A. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989;57(3):423–431. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- 132.C.K. Biebricher, R. Luce, Template-free generation of RNA species that replicate with bacteriophage T7 RNA polymerase, EMBO J. 15 (1996) 3458–3465. https://doi.org/10.1002/j.1460-2075.1996.tb00712.x. [PMC free article] [PubMed]
- 133.Triana-Alonso Francisco J., Dabrowski Marylena, Wadzack Jörg, Nierhaus Knud H. Self-coded 3’ extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 1995;270(11):6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 134.Gholamalipour Y., Karunanayake Mudiyanselage A., Martin C.T. 3’ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character––RNA-Seq analyses. Nucleic Acids Res. 2018;46:9253–9263. doi: 10.1093/nar/gky796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martin Craig T., Muller Daniel K., Coleman Joseph E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988;27(11):3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 136.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. - Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mu X., Greenwald E., Ahmad S., Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu Monica Z., Asahara Haruichi, Tzertzinis George, Roy Bijoyita. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA. 2020;26(3):345–360. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Y. Gholamalipour, W.C. Johnson, C.T. Martin, Efficient inhibition of RNA self-primed extension by addition of competing 3’-capture DNA-improved RNA synthesis by T7 RNA polymerase, Nucleic Acids Res. 47 (2019) e118. https://doi.org/10.1093/nar/gkz700. [DOI] [PMC free article] [PubMed]
- 140.Nelson Jennifer, Sorensen Elizabeth W., Mintri Shrutika, Rabideau Amy E., Zheng Wei, Besin Gilles, Khatwani Nikhil, Su Stephen V., Miracco Edward J., Issa William J., Hoge Stephen, Stanton Matthew G., Joyal John L. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6(26):eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Foster Jessica B., Choudhari Namrata, Perazzelli Jessica, Storm Julie, Hofmann Ted J., Jain Payal, Storm Phillip B., Pardi Norbert, Weissman Drew, Waanders Angela J., Grupp Stephan A., Karikó Katalin, Resnick Adam C., Barrett David M. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response. Hum. Gene Ther. 2019;30(2):168–178. doi: 10.1089/hum.2018.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Z. Zhong, S. Mc Cafferty, L. Opsomer, H. Wang, H. Huysmans, S. De Temmerman, Joyca Lienenklaus, F. Catani, João Paulo Portela Combes, N.N. Sanders, Corticosteroids and cellulose purification improve respectively the in vivo translation and vaccination efficacy of self-amplifying mRNAs, Mol. Ther. (2021). [DOI] [PMC free article] [PubMed]
- 143.Drews Katharina, Tavernier Geertrui, Demeester Joseph, Lehrach Hans, De Smedt Stefaan C., Rejman Joanna, Adjaye James. The cytotoxic and immunogenic hurdles associated with non-viral mRNA-mediated reprogramming of human fibroblasts. Biomaterials. 2012;33(16):4059–4068. doi: 10.1016/j.biomaterials.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 144.Liu Yang, Krishnan Manoj N., Phua Kyle K.L. Suppression of mRNA nanoparticle transfection in human fibroblasts by selected interferon inhibiting small molecule compounds. Biomolecules. 2017;7(4):56. doi: 10.3390/biom7030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Simmons R.A., Willberg C.B., Paul K. Immune evasion by viruses. ELS. 2013:1–10. doi: 10.1002/9780470015902.a0024790. [DOI] [Google Scholar]
- 146.Perdiguero Beatriz, Esteban Mariano. The interferon system and vaccinia virus evasion mechanisms. J. Interf. Cytokine Res. 2009;29(9):581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 147.Smith G.L., Benfield C.T.O., Maluquer de Motes C., Mazzon M., Ember S.W.J., Ferguson B.J., Sumner R.P. Vaccinia virus immune evasion: Mechanisms, virulence and immunogenicity. J. Gen. Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 148.Liptáková Hana, Kontseková Eva, Alcamí Antonio, Smith Geoffrey L., Kontsek Peter. Analysis of an interaction between the soluble vaccinia virus-coded type I interferon (IFN)-receptor and human IFN-α1 and IFN-α2. Virology. 1997;232(1):86–90. doi: 10.1006/viro.1997.8527. [DOI] [PubMed] [Google Scholar]
- 149.Kim Yuriy G., Baltabekova Aliya Zh., Zhiyenbay Erzhan E., Aksambayeva Altynai S., Shagyrova Zhadyra S., Khannanov Rinat, Ramanculov Erlan M., Shustov Alexandr V., Schreiber Gideon. Recombinant Vaccinia virus-coded interferon inhibitor B18R: Expression, refolding and a use in a mammalian expression system with a RNA-vector. PLoS ONE. 2017;12(12):e0189308. doi: 10.1371/journal.pone.0189308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alcamí Antonio, Symons Julian A., Smith Geoffrey L. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 2000;74(23):11230–11239. doi: 10.1128/JVI.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Symons J.A., Alcamí A., Smith G.L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 152.Warren Luigi, Manos Philip D., Ahfeldt Tim, Loh Yuin-Han, Li Hu, Lau Frank, Ebina Wataru, Mandal Pankaj K., Smith Zachary D., Meissner Alexander, Daley George Q., Brack Andrew S., Collins James J., Cowan Chad, Schlaeger Thorsten M., Rossi Derrick J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Warren L., Ni Y., Wang J., Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci. Rep. 2012;2:1–7. doi: 10.1038/srep00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yoshioka Naohisa, Gros Edwige, Li Hai-Ri, Kumar Shantanu, Deacon Dekker C., Maron Cornelia, Muotri Alysson R., Chi Neil C., Fu Xiang-Dong, Yu Benjamin D., Dowdy Steven F. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13(2):246–254. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Poleganov Marco Alexander, Eminli Sarah, Beissert Tim, Herz Stephanie, Moon Jung-Il, Goldmann Johanna, Beyer Arianne, Heck Rosario, Burkhart Isabell, Barea Roldan Diana, Türeci Özlem, Yi Kevin, Hamilton Brad, Sahin Ugur. Efficient reprogramming of human fibroblasts and blood-derived endothelial progenitor cells using nonmodified RNA for Reprogramming and immune evasion. Hum. Gene Ther. 2015;26(11):751–766. doi: 10.1089/hum.2015.045. [DOI] [PubMed] [Google Scholar]
- 156.Mandal Pankaj K., Rossi Derrick J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 2013;8(3):568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- 157.Durruthy-Durruthy Jens, Briggs Sharon F., Awe Jason, Ramathal Cyril Y., Karumbayaram Saravanan, Lee Patrick C., Heidmann Julia D., Clark Amander, Karakikes Ioannis, Loh Kyle M., Wu Joseph C., Hoffman Andrew R., Byrne James, Reijo Pera Renee A., Sebastiano Vittorio, Zambidis Elias T. Rapid and efficient conversion of integration-free human induced pluripotent stem cells to GMP-grade culture conditions. PLoS ONE. 2014;9(4):e94231. doi: 10.1371/journal.pone.0094231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Quabius Elgar Susanne, Krupp Guido. Synthetic mRNAs for manipulating cellular phenotypes: An overview. N. Biotechnol. 2015;32(1):229–235. doi: 10.1016/j.nbt.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 159.Gomez Alejandro M., Ouellet Michel, Tremblay Michel J. HIV-1–triggered release of type I IFN by plasmacytoid dendritic cells induces BAFF production in monocytes. J. Immunol. 2015;194(5):2300–2308. doi: 10.4049/jimmunol.1402147. [DOI] [PubMed] [Google Scholar]
- 160.Minnaert A., Devoldere J., Peynshaert K., Vercruysse L., De Smedt S.C., Remaut K. Vaccinia virus protein B18R : Influence on mRNA immunogenicity and translation upon non-viral delivery in different ocular cell types. Pharmaceutics. 2021;13:1–18. doi: 10.3390/pharmaceutics13010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zangi Lior, Lui Kathy O, von Gise Alexander, Ma Qing, Ebina Wataru, Ptaszek Leon M, Später Daniela, Xu Huansheng, Tabebordbar Mohammadsharif, Gorbatov Rostic, Sena Brena, Nahrendorf Matthias, Briscoe David M, Li Ronald A, Wagers Amy J, Rossi Derrick J, Pu William T, Chien Kenneth R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31(10):898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Michel T., Golombek S., Steinle H., Hann L., Velic A., MacEk B., Krajewski S., Schlensak C., Wendel H.P., Avci-Adali M. Efficient reduction of synthetic mRNA induced immune activation by simultaneous delivery of B18R encoding mRNA. J. Biol. Eng. 2019;13:1–11. doi: 10.1186/s13036-019-0172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Awe J.P., Crespo A.V., Li Y., Kiledjian M., Byrne J.A. BAY11 enhances OCT4 synthetic mRNA expression in adult human skin cells. Stem Cell Res. Ther. 2013;4:1–12. doi: 10.1186/scrt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mah Nancy, Wang Ying, Liao Mei-Chih, Prigione Alessandro, Jozefczuk Justyna, Lichtner Björn, Wolfrum Katharina, Haltmeier Manuela, Flöttmann Max, Schaefer Martin, Hahn Alexander, Mrowka Ralf, Klipp Edda, Andrade-Navarro Miguel A., Adjaye James, Hoheisel Jörg D. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS ONE. 2011;6(8):e24351. doi: 10.1371/journal.pone.0024351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beissert Tim, Koste Lars, Perkovic Mario, Walzer Kerstin C., Erbar Stephanie, Selmi Abderraouf, Diken Mustafa, Kreiter Sebastian, Türeci Özlem, Sahin Ugur. Improvement of in vivo expression of genes delivered by self-amplifying RNA using vaccinia virus immune evasion proteins. Hum. Gene Ther. 2017;28(12):1138–1146. doi: 10.1089/hum.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Davies M V, Chang H W, Jacobs B L, Kaufman R J. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 1993;67(3):1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu Y., Chin J.M., Choo E.L., Phua K.K.L. Messenger RNA translation enhancement by immune evasion proteins: a comparative study between EKB (vaccinia virus) and NS1 (influenza A virus) Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-019-48559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mehta Ninad, Enwere Emeka K., Santos Theodore Dos, Saffran Holly A., Hazes Bart, Evans David, Barry Michele, Smiley James R., Sandri-Goldin Rozanne M. Expression of the vaccinia virus antiapoptotic F1 protein is blocked by protein kinase R in the absence of the viral E3 protein. J. Virol. 2018;92(19) doi: 10.1128/JVI.01167-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Davies M.V., Elroy-Stein O., Jagus R., Moss B., Kaufman R.J. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 1992;66(4):1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gao Shijuan, Song Liping, Li Jiandong, Zhang Zhenzhu, Peng Haiyan, Jiang Wei, Wang Qingtao, Kang Tiebang, Chen Shuai, Huang Wenlin. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell. Microbiol. 2012;14(12):1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 171.Zhang Xiaoyang, Dong Chunyan, Sun Xiaoning, Li Zhongyi, Zhang Maolin, Guan Zhenhong, Duan Ming. Induction of the cellular miR-29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochem. Biophys. Res. Commun. 2014;450(1):755–761. doi: 10.1016/j.bbrc.2014.06.059. [DOI] [PubMed] [Google Scholar]
- 172.Liu Yi, Chia Zhen Hua, Liew Johannes Nathaniel Min Hui, Or Shi Min, Phua Kyle K.L. Modulation of mRNA translation and cell viability by influenza A virus derived nonstructural protein 1. Nucleic Acid Ther. 2018;28(3):200–208. doi: 10.1089/nat.2017.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Krug R.M. Functions of the influenza A virus NS1 protein in antiviral defense. Curr. Opin. Virol. 2015;12:1–6. doi: 10.1016/j.coviro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dudek S.E., Wixler L., Nordhoff C., Nordmann A., Anhlan D., Wixler V., Ludwig S. The influenza virus PB1-F2 protein has interferon-antagonistic activity. Biol. Chem. 2011;392:1135–1144. doi: 10.1515/bc-2011-174. [DOI] [PubMed] [Google Scholar]
- 175.Phua K.K.L., Liu Y., Sim S.H. Non-linear enhancement of mRNA delivery efficiencies by influenza A derived NS1 protein engendering host gene inhibition property. Biomaterials. 2017;133:29–36. doi: 10.1016/j.biomaterials.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 176.Le Goffic Ronan, Bouguyon Edwige, Chevalier Christophe, Vidic Jasmina, Da Costa Bruno, Leymarie Olivier, Bourdieu Christiane, Decamps Laure, Dhorne-Pollet Sophie, Delmas Bernard. Influenza A virus protein PB1-F2 exacerbates IFN-β expression of human respiratory epithelial cells. J. Immunol. 2010;185(8):4812–4823. doi: 10.4049/jimmunol.0903952. [DOI] [PubMed] [Google Scholar]
- 177.G.M. Conenello, J.R. Tisoncik, E. Rosenzweig, Z.T. Varga, P. Palese, M.G. Katze, A Single N66S Mutation in the PB1-F2 Protein of Influenza A Virus Increases Virulence by Inhibiting the Early Interferon Response In Vivo, J. Virol. 85 (2011) 652–662. https://doi.org/10.1128/jvi.01987-10. [DOI] [PMC free article] [PubMed]
- 178.Varga Zsuzsanna T., Ramos Irene, Hai Rong, Schmolke Mirco, García-Sastre Adolfo, Fernandez-Sesma Ana, Palese Peter, Pekosz Andrew. The influenza virus protein PB1-F2 inhibits the induction of type i interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7(6):e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Angel Matthew, Yanik Mehmet Fatih, Tripp Ralph. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS ONE. 2010;5(7):e11756. doi: 10.1371/journal.pone.0011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee Jieun, Sayed Nazish, Hunter Arwen, Au Kin Fai, Wong Wing H., Mocarski Edward S., Pera Renee Reijo, Yakubov Eduard, Cooke John P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151(3):547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.M. Olejniczak, P. Galka-Marciniak, K. Polak, A. Fligier, W.J. Krzyzosiak, RNAimmuno: A database of the nonspecific immunological effects of RNA interference and microRNA reagents, Rna. 18 (2012) 930–935. https://doi.org/10.1261/rna.025627.110. [DOI] [PMC free article] [PubMed]
- 182.Meng Z., Lu M. RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front. Immunol. 2017;8:1–7. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mansoori Behzad, Mohammadi Ali, Shir Jang Solmaz, Baradaran Behzad. Mechanisms of immune system activation in mammalians by small interfering RNA (siRNA) Artif. Cells, Nanomedicine Biotechnol. 2016;44(7):1589–1596. doi: 10.3109/21691401.2015.1102738. [DOI] [PubMed] [Google Scholar]
- 184.H.H. Rabouw, M.A. Langereis, A.A. Anand, L.J. Visser, R.J. De Groot, P. Walter, F.J.M. Van Kuppeveld, Small molecule ISRIB suppresses the integrated stress response within a defined window of activation, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 2097–2102. https://doi.org/10.1073/pnas.1815767116. [DOI] [PMC free article] [PubMed]
- 185.Ohto T., Konishi M., Tanaka H., Onomoto K., Yoneyama M., Nakai Y., Tange K., Yoshioka H., Akita H. Inhibition of the inflammatory pathway enhances both the in vitro and in vivo transfection activity of exogenous in vitro-transcribed mRNAs delivered by lipid nanoparticles. Biol. Pharm. Bull. 2019;42:299–302. doi: 10.1248/bpb.b18-00783. [DOI] [PubMed] [Google Scholar]
- 186.Weide B., Carralot J.P., Reese A., Scheel B., Eigentler T.K., Hoerr I., Rammensee H.G., Garbe C., Pascolowz S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 187.Lundstrom K. Self-amplifying rna viruses as rna vaccines. Int. J. Mol. Sci. 2020;21:1–29. doi: 10.3390/ijms21145130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.C. Krienke, L. Kolb, E. Diken, M. Streuber, S. Kirchhoff, T. Bukur, Ö. Akilli-Öztürk, L.M. Kranz, H. Berger, J. Petschenka, M. Diken, S. Kreiter, N. Yogev, A. Waisman, K. Karikó, Ö. Türeci, U. Sahin, A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis, Science (80-.). 371 (2021) 145–153. https://doi.org/10.1126/science.aay3638. [DOI] [PubMed]
- 189.Sahin Ugur, Oehm Petra, Derhovanessian Evelyna, Jabulowsky Robert A., Vormehr Mathias, Gold Maike, Maurus Daniel, Schwarck-Kokarakis Doreen, Kuhn Andreas N., Omokoko Tana, Kranz Lena M., Diken Mustafa, Kreiter Sebastian, Haas Heinrich, Attig Sebastian, Rae Richard, Cuk Katarina, Kemmer-Brück Alexandra, Breitkreuz Andrea, Tolliver Claudia, Caspar Janina, Quinkhardt Juliane, Hebich Lisa, Stein Malte, Hohberger Alexander, Vogler Isabel, Liebig Inga, Renken Stephanie, Sikorski Julian, Leierer Melanie, Müller Verena, Mitzel-Rink Heidrun, Miederer Matthias, Huber Christoph, Grabbe Stephan, Utikal Jochen, Pinter Andreas, Kaufmann Roland, Hassel Jessica C., Loquai Carmen, Türeci Özlem. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585(7823):107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 190.Polack Fernando P., Thomas Stephen J., Kitchin Nicholas, Absalon Judith, Gurtman Alejandra, Lockhart Stephen, Perez John L., Pérez Marc Gonzalo, Moreira Edson D., Zerbini Cristiano, Bailey Ruth, Swanson Kena A., Roychoudhury Satrajit, Koury Kenneth, Li Ping, Kalina Warren V., Cooper David, Frenck Robert W., Hammitt Laura L., Türeci Özlem, Nell Haylene, Schaefer Axel, Ünal Serhat, Tresnan Dina B., Mather Susan, Dormitzer Philip R., Şahin Uğur, Jansen Kathrin U., Gruber William C. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Benteyn Daphné, Heirman Carlo, Bonehill Aude, Thielemans Kris, Breckpot Karine. mRNA-based dendritic cell vaccines. Expert Rev. Vaccines. 2015;14(2):161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 192.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 193.Dörrie J., Schaft N., Schuler G., Schuler-Thurner B. Therapeutic cancer vaccination with ex vivo rna-transfected dendritic cells—an update. Pharmaceutics. 2020;12:1–33. doi: 10.3390/pharmaceutics12020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.De Beuckelaer Ans, Pollard Charlotte, Van Lint Sandra, Roose Kenny, Van Hoecke Lien, Naessens Thomas, Udhayakumar Vimal Kumar, Smet Muriel, Sanders Niek, Lienenklaus Stefan, Saelens Xavier, Weiss Siegfried, Vanham Guido, Grooten Johan, De Koker Stefaan. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit Cytolytic T cell responses. Mol. Ther. 2016;24(11):2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cagigi A., Loré K. Immune responses induced by mrna vaccination in mice, monkeys and humans. Vaccines. 2021;9:1–14. doi: 10.3390/vaccines9010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Verbeke R., Lentacker I., Wayteck L., Breckpot K., Van Bockstal M., Descamps B., Vanhove C., De Smedt S.C., Dewitte H. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: Restoring the immunogenicity of immunosilent mRNA. J. Control. Release. 2017;266:287–300. doi: 10.1016/j.jconrel.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 197.K.K.L. Phua, S.K. Nair, K.W. Leong, Messenger RNA (mRNA) nanoparticle tumour vaccination, Nanoscale. 6 (2014) 7715–7729. https://doi.org/10.1039/c4nr01346h. [DOI] [PMC free article] [PubMed]
- 198.Zhang H., You X., Wang X., Cui L., Wang Z., Xu F., Li M., Yang Z., Liu J., Huang P., Kang Y., Wu J., Xia X. Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc. Natl. Acad. Sci. 2021;118:1–12. doi: 10.1073/pnas.2005191118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rabenstein Hannah, Behrendt Anne C., Ellwart Joachim W., Naumann Ronald, Horsch Marion, Beckers Johannes, Obst Reinhard. Differential kinetics of antigen dependency of CD4 + and CD8 + T cells. J. Immunol. 2014;192(8):3507–3517. doi: 10.4049/jimmunol.1302725. [DOI] [PubMed] [Google Scholar]
- 200.Braun D., Caramalho I., Demengeot J. IFn-α/β enhances BCR-dependent B cell responses. Int. Immunol. 2002;14:411–419. doi: 10.1093/intimm/14.4.411. [DOI] [PubMed] [Google Scholar]
- 201.Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C., Salazar A.M., Colonna M., Steinman R.M. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tam Hok Hei, Melo Mariane B., Kang Myungsun, Pelet Jeisa M., Ruda Vera M., Foley Maria H., Hu Joyce K., Kumari Sudha, Crampton Jordan, Baldeon Alexis D., Sanders Rogier W., Moore John P., Crotty Shane, Langer Robert, Anderson Daniel G., Chakraborty Arup K., Irvine Darrell J. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. 2016;113(43):E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.B.B. Au-Yeung, J. Zikherman, J.L. Mueller, J.F. Ashouri, M. Matloubian, D.A. Cheng, Y. Chen, K.M. Shokat, A. Weiss, A sharp T-cell antigen receptor signaling threshold for T-cell proliferation, Proc. Natl. Acad. Sci. 111 (2014). https://doi.org/10.1073/pnas.1413726111. [DOI] [PMC free article] [PubMed]
- 204.Henrickson Sarah E., Perro Mario, Loughhead Scott M., Senman Balimkiz, Stutte Susanne, Quigley Michael, Alexe Gabriela, Iannacone Matteo, Flynn Michael P., Omid Shaida, Jesneck Jonathan L., Imam Sabrina, Mempel Thorsten R., Mazo Irina B., Haining W. Nicholas, von Andrian Ulrich H. Antigen availability determines CD8+ T cell-dendritic cell interaction kinetics and memory fate decisions. Immunity. 2013;39(3):496–507. doi: 10.1016/j.immuni.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Verbeke Rein, Lentacker Ine, De Smedt Stefaan C., Dewitte Heleen. The dawn of mRNA vaccines: The COVID-19 case Rein. J. Control. Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Carrasco Yolanda R, Batista Facundo D. B-cell activation by membrane-bound antigens is facilitated by the interaction of VLA-4 with VCAM-1. EMBO J. 2006;25(4):889–899. doi: 10.1038/sj.emboj.7600944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Roche Paul A., Furuta Kazuyuki. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15(4):203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kuka M., De Giovanni M., Iannacone M. The role of type I interferons in CD4+ T cell differentiation. Immunol. Lett. 2019;215:19–23. doi: 10.1016/j.imlet.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Starbeck-Miller G.R., Harty J.T. The Role of Il-12 and type I Interferon in governing the magnitude of CD8 T cell responses, in. Crossroads Between Innate Adapt. Immun. 2015:;V:31–41. doi: 10.1007/978-3-319-15774-0_3. [DOI] [PubMed] [Google Scholar]
- 210.R.J. Noelle, M. Roy, D.M. Shepherd, I. Stamenkovic, J.A. Ledbetter, A. Aruffo, A 39-kDa protein on activated helper T cells binds CD40 and transduces the signal for cognate activation of B cells, Proc. Natl. Acad. Sci. 89 (1992) 6550–6554. https://doi.org/10.1073/pnas.89.14.6550. [DOI] [PMC free article] [PubMed]
- 211.Le Bon Agnes, Schiavoni Giovanna, D'Agostino Giuseppina, Gresser Ion, Belardelli Filippo, Tough David F. Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14(4):461–470. doi: 10.1016/S1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 212.Crouse Josh, Kalinke Ulrich, Oxenius Annette. Regulation of antiviral T cell responses by type i interferons. Nat. Rev. Immunol. 2015;15(4):231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 213.Pepini Timothy, Pulichino Anne-Marie, Carsillo Thomas, Carlson Alicia L., Sari-Sarraf Farid, Ramsauer Katrin, Debasitis Jason C., Maruggi Giulietta, Otten Gillis R., Geall Andrew J., Yu Dong, Ulmer Jeffrey B., Iavarone Carlo. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J. Immunol. 2017;198(10):4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McNab Finlay, Mayer-Barber Katrin, Sher Alan, Wack Andreas, O'Garra Anne. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15(2):87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Curtsinger Julie M., Valenzuela Javier O., Agarwal Pujya, Lins Debra, Mescher Matthew F. Cutting edge: type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J. Immunol. 2005;174(8):4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 216.Lederer Katlyn, Castaño Diana, Gómez Atria Daniela, Oguin Thomas H., Wang Sidney, Manzoni Tomaz B., Muramatsu Hiromi, Hogan Michael J., Amanat Fatima, Cherubin Patrick, Lundgreen Kendall A., Tam Ying K., Fan Steven H.Y., Eisenlohr Laurence C., Maillard Ivan, Weissman Drew, Bates Paul, Krammer Florian, Sempowski Gregory D., Pardi Norbert, Locci Michela. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53(6):1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Turner J.S., Halloran J.A.O., Kalaidina E., Kim W., Schmitz A.J., Lei T., Thapa M., Chen R.E., Case J.B., Amanat F., Rauseo A.M., Haile A., Xie X., Klebert M.K., Suessen T., Middleton W.D., Shi P.-Y., Krammer F., Teefey S.A., Diamond M.S., Presti R.M., Ellebedy A.H. SARS-CoV-2 mRNA vaccines induce robust plasmablast and germinal centre responses in humans. Under Rev. 2021 doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kranz Lena M., Diken Mustafa, Haas Heinrich, Kreiter Sebastian, Loquai Carmen, Reuter Kerstin C., Meng Martin, Fritz Daniel, Vascotto Fulvia, Hefesha Hossam, Grunwitz Christian, Vormehr Mathias, Hüsemann Yves, Selmi Abderraouf, Kuhn Andreas N., Buck Janina, Derhovanessian Evelyna, Rae Richard, Attig Sebastian, Diekmann Jan, Jabulowsky Robert A., Heesch Sandra, Hassel Jessica, Langguth Peter, Grabbe Stephan, Huber Christoph, Türeci Özlem, Sahin Ugur. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 219.Broos Katrijn, Van der Jeught Kevin, Puttemans Janik, Goyvaerts Cleo, Heirman Carlo, Dewitte Heleen, Verbeke Rein, Lentacker Ine, Thielemans Kris, Breckpot Karine. Particle-mediated intravenous delivery of antigen mRNA results in strong antigen-specific T-cell responses despite the induction of type I interferon. Mol. Ther. - Nucleic Acids. 2016;5:e326. doi: 10.1038/mtna.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. - Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhong Z., Portela Catani J.P., McCafferty S., Couck L., Van Den Broeck W., Gorlé N., Vandenbroucke R.E., Devriendt B., Ulbert S., Cnops L., Michels J., Ariën K.K., Sanders N.N. Immunogenicity and protection efficacy of a naked self-replicating mrna-based zika virus vaccine. Vaccines. 2019;7:1–17. doi: 10.3390/vaccines7030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Roth C., Cantaert T., Colas C., Prot M., Casadémont I., Levillayer L., Thalmensi J., Langlade-Demoyen P., Gerke C., Bahl K., Ciaramella G., Simon-Loriere E., Sakuntabhai A. A modified mRNA vaccine targeting immunodominant NS epitopes protects against dengue virus infection in HLA class I transgenic mice. Front. Immunol. 2019;10:1–14. doi: 10.3389/fimmu.2019.01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tse S.W., McKinney K., Walker W., Nguyen M., Iacovelli J., Small C., Hopson K., Zaks T., Huang E. mRNA-encoded, constitutively active STINGV155M is a potent genetic adjuvant of antigen-specific CD8+ T cell response. Mol. Ther. 2021;29:1–12. doi: 10.1016/j.ymthe.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Frederickson R., Herzog R.W. RNA-based vaccines and innate immune activation : Not too hot and not too cold. Mol. Ther. 2021;29:1–2. doi: 10.1016/j.ymthe.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Oberli Matthias A., Reichmuth Andreas M., Dorkin J. Robert, Mitchell Michael J., Fenton Owen S., Jaklenec Ana, Anderson Daniel G., Langer Robert, Blankschtein Daniel. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17(3):1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Verbeke R., Lentacker I., Breckpot K., Janssens J., Van Calenbergh S., De Smedt S.C., Dewitte H. Broadening the message: A nanovaccine co-loaded with messenger RNA and α-GalCer induces antitumor immunity through conventional and natural killer T cells. ACS Nano. 2019;13:1655–1669. doi: 10.1021/acsnano.8b07660. [DOI] [PubMed] [Google Scholar]
- 227.Kallen Karl-Josef, Heidenreich Regina, Schnee Margit, Petsch Benjamin, Schlake Thomas, Thess Andreas, Baumhof Patrick, Scheel Birgit, Koch Sven D, Fotin-Mleczek Mariola. A novel, disruptive vaccination technology Self-adjuvanted RNActive® vaccines. Hum. Vaccin. Immunother. 2013;9(10):2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wilgenhof S., Van Nuffel A.M.T., Benteyn D., Corthals J., Aerts C., Heirman C., Van Riet I., Bonehill A., Thielemans K., Neyns B. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann. Oncol. 2013;24(10):2686–2693. doi: 10.1093/annonc/mdt245. [DOI] [PubMed] [Google Scholar]
- 229.De Keersmaecker Brenda, Claerhout Sofie, Carrasco Javier, Bar Isabelle, Corthals Jurgen, Wilgenhof Sofie, Neyns Bart, Thielemans Kris. TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: Link between T-cell activation and clinical responses in advanced melanoma. J. Immunother. Cancer. 2020;8(1):e000329. doi: 10.1136/jitc-2019-000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wilgenhof Sofie, Corthals Jurgen, Heirman Carlo, van Baren Nicolas, Lucas Sophie, Kvistborg Pia, Thielemans Kris, Neyns Bart. Phase II study of autologous monocyte-derived mRNA electroporated dendritic cells (TriMixDC-MEL) plus ipilimumab in patientswith pretreated advanced melanoma. J. Clin. Oncol. 2016;34(12):1330–1338. doi: 10.1200/JCO.2015.63.4121. [DOI] [PubMed] [Google Scholar]
- 231.Guardo A.C., Joe P.T., Miralles L., Bargalló M.E., Mothe B., Krasniqi A., Heirman C., García F., Thielemans K., Brander C., Aerts J.L., Plana M. Preclinical evaluation of an mRNA HIV vaccine combining rationally selected antigenic sequences and adjuvant signals (HTI-TriMix) Aids. 2017;31:321–332. doi: 10.1097/QAD.0000000000001276. [DOI] [PubMed] [Google Scholar]
- 232.Leal L., Guardo A.C., Morón-López S., Salgado M., Mothe B., Heirman C., Pannus P., Vanham G., van den Ham H.J., Gruters R., Andeweg A., Van Meirvenne S., Pich J., Arnaiz J.A., Gatell J.M., Brander C., Thielemans K., Martínez-Picado J., Plana M., García F. Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS. 2018;32:2533–2545. doi: 10.1097/QAD.0000000000002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.de Jong W., Leal L., Buyze J., Pannus P., Guardo A., Salgado M., Mothe B., Molto J., Moron-Lopez S., Gálvez C., Florence E., Vanham G., van Gorp E., Brander C., Allard S., Thielemans K., Martinez-Picado J., Plana M., García F., Gruters R.A. Therapeutic vaccine in chronically Hiv-1-infected patients: A randomized, double-blind, placebocontrolled phase IIa trial with HTI-Trimix. Vaccines. 2019;7:1–13. doi: 10.3390/vaccines7040209. [DOI] [PMC free article] [PubMed] [Google Scholar]