Abstract
Aerosol transmission is one of the three major transmission routes of respiratory viruses. However, the dynamics and significance of the aerosol transmission route are not well understood, partially due to the lack of rapid and efficient tools for on-the-spot detection of airborne viruses. We report a hand-held device that integrates a 3D-printed sample preparation unit with a laminated paper-based RNA amplification unit. The sample preparation unit features an innovative reagent delivery scheme based on a ball-based valve capable of storing and delivering reagents through the rotation of the unit without manual pipetting, while the paper-based unit enables RNA enrichment and reverse transcription loop-mediated isothermal amplification (RT-LAMP). We have determined the detection limit of the integrated sample-preparation/amplification device (SPAD) at 1 TCID50 H1N1 influenza viruses in 140 µL aqueous sample. Further, we integrated SPAD with a previously reported viable virus aerosol sampler (VIVAS), a water-vapor-based condensational growth system capable of collecting aerosolized virus particles (Pan et al., 2016) [1]. Using the combined VIVAS-SPAD platform, we have demonstrated the collection/detection of lab-generated, airborne H1N1 influenza viruses in 65 min, suggesting that the platform has a potential for detecting and monitoring airborne virus transmission during outbreaks. The effective sampling and rapid detection of airborne viruses by the sample-to-answer platform will also help us better understand the dynamics and significance of aerosol transmission of infectious disease.
Keywords: Sample preparation, Influenza virus, Isothermal amplification, Paper microfluidics, Airborne, Valve
GRAPHICAL ABSTRACT
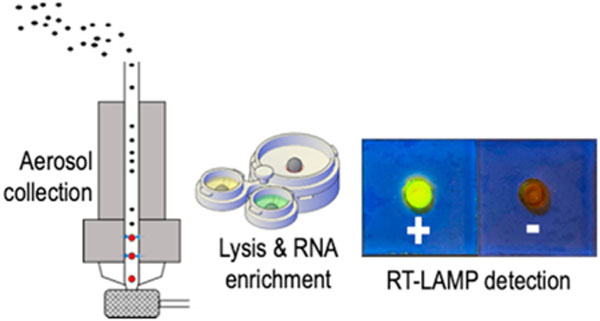
1. Introduction
Respiratory pathogens, such as influenza viruses, are transmitted through three primary routes: (a) inhalation of pathogen-containing aerosols, (b) droplet infection, and (c) contact transmission [2]. Understanding aerosol transmission dynamics is of great importance, as the risk of respiratory disease transmission through this route is exceptionally high in population-dense areas, such as hospitals, schools, airports and industrial animal farms [3–7]. However, the relative importance of the aerosol transmission route among the three is controversial, due to the limitations in the sampling and detection methods available for nanometer-sized viruses [6–11]. Moreover, traditional methods to detect the viruses collected from aerosols, for example by either viral culture or polymerase chain reactions (PCR), are time-consuming and labor-intensive. The requirement for a well-equipped laboratory as well as highly trained personnel makes it unrealistic in the field or in resource-limited settings.
Compared to the progress in the detection of pathogenic agents present in aqueous samples [12–14], detecting airborne pathogens collected using air samplers is still a challenge, especially due to the low number of pathogens in air combined with lack of automated platforms with efficient sampling and rapid detection [8,15–19]. In particular, due to the low pathogen content typically present in aerosols, a large volume of aerosols needs to be concentrated into a sub-milliliter-sized liquid to be detectable by biosensors or the like [8]. We reported a viable virus aerosol sampler (VIVAS) as an efficient collector for lab-generated, airborne virus aerosols [1]. In VIVAS, the diameter of virus particles in the aerosol being collected is enlarged from nanometer-size to micron-size through condensation of water on their surfaces, making it possible to collect aerosolized particles from 8 nm to 10 µm [20,21]. The micron-sized particles with a hydration sphere may contain an individual virion or multiple virions, depending on their particle size and the nebulization medium used [22]. Using VIVAS, we have demonstrated the successful collection of a variety of viable human respiratory viruses in a student infirmary during a late-onset influenza virus outbreak in 2016 as well as coronavirus disease 2019 (COVID-19) [6,7]. High collection efficiency and a proven ability to collect real-world virus aerosols makes the VIVAS an ideal candidate for becoming a part of airborne virus detection system without requiring labor-intensive and time-consuming viral culture.
An ideal airborne virus detection system in the field or in resource-limited settings requires an efficient aerosol collector as well as a rapid, sensitive detector for collected viruses. Moreover, these two components must be integrated in a way to operate with minimum manual intervention [8]. Rapid immunoassay-based point-of-care (POC) tests such as lateral flow assays (LFA) and electrochemical sensors have been integrated with aerosol sampling systems to achieve in-line virus detection [23–25]. However, these immunoassay-based POC assays have lower sensitivities and specificities than nucleic acid testing (NAT) that carries out amplification and genetic identification [26–29]. Saito et al. reported an air sampling system for detection of chemical and biological warfare agents that integrated biosensors and a microfluidic PCR device [30], but they used a solution of Bacillus subtilis as a simulant to demonstrate the feasibility without any sample preparation step. Sui and his colleagues developed a microfluidic device that incorporated a membrane filter to collect bacteria, followed by lysis and loop-mediated isothermal amplification (LAMP) [31]. However, the device did not employ any step to concentrate nucleic acids and remove inhibitors, which can decrease the sensitivity of NAT. Indeed, sample preparation for NAT has long been recognized as a great challenge at POC since it is labor-intensive [32,33]. The most common sample preparation method for nucleic acid extraction uses high concentration chaotropic salts to facilitate the binding of nucleic acid to silica as a solid phase for extraction, thus requiring subsequent washing steps to remove the chaotropic salts for downstream amplification [34]. Numerous approaches have been made to automate this sample preparation step, including the use of microfluidic devices and complex instruments with robotic liquid handling system [32,33]. However, these platforms often require the use of a syringe pump or specific sample collection and thus lacks the flexibility to integrate with VIVAS.
In this work, we employ a liquid handling scheme enabled by ball-based valves for the storage and sequential delivery of liquid reagents through simple rotational movement. The ball valve concept is inspired by the dispensing mechanism of a ballpoint pen, in which ink is transferred onto paper when the metal ball at the tip is pressed while writing [35]. We also incorporate a laminated paper-based analytical device for RNA extraction [35–37]. Compared to other materials, paper is low-cost, flexible, easy to dispose of and capable of driving liquid without an external pump [38–40]. The resulting virus RNA enriched on the paper device is then detected by reverse transcription loop-mediated isothermal amplification (RT-LAMP). RT-LAMP is an isothermal amplification technique that is rapid (<30 min, compared to 2 h of PCR), sensitive (due to amplification), and specific (due to genetic identification); its shorter incubation time and more simplified thermal management than PCR make it advantageous in POC platforms [35,41,42]. The integration of these components results in an instrument-free, hand-held device for achieving the sample preparation and nucleic acid detection of RNA viruses.
Here we detail our design, fabrication and testing of our sample-preparation/amplification device (SPAD). The SPAD contains 3D-printed sample preparation components including a collector and a buffer unit, and a laminated paper-based RNA amplification device. The performance of SPAD was first evaluated using aqueous samples spiked with H1N1 influenza virus to detect down to 1 TCID50 (median tissue culture infective dose) per device. We then demonstrated the use of SPAD with VIVAS to detect lab-generated, airborne H1N1 influenza viruses in 50 min after 15 min of aerosol collection. Our results suggest that SPAD can be combined with VIVAS for detecting and monitoring airborne infectious disease in population-dense areas during outbreaks. The effective sampling and rapid detection of airborne viruses has a potential to help us better understand the role aerosol transmission plays during future airborne infectious disease outbreaks [43].
2. Materials and methods
2.1. Virus preparation
MDCK (CCL-34; Madin-Darby Canine Kidney Epithelial Cells) were obtained from the American Type Culture Collection (Manassas, VA, USA) and were propagated as monolayers at 37 °C and 5% CO2 in Advanced Dulbecco’s Modified Eagle’s Medium (aDMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 2 mM L-alanyl-L-glutamine (GlutaMAX, Invitrogen), antibiotics (PSN consisting of 50 µg/mL penicillin, 50 µg/mL streptomycin, and 100 µg/mL neomycin, from Invitrogen), and 10% (v/v) low IgG, heat-inactivated gamma-irradiated fetal bovine serum (HyClone, Logan, Utah). Prior to use, the cell line was treated for 3 weeks with plasmocin and verified free of mycoplasma DNA by PCR. To create virus stocks, T75 flasks of newly confluent MDCK cells (about 8.2 × 106 cells/flask) were used. The serum-containing cell growth medium was removed, and replaced with 5 mL of serum-free aDMEM supplemented as previously described plus L-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-treated mycoplasma-free and extraneous virus-free trypsin (Worthington Biochemical Company, Lakewood, NJ), and the cells infected at a multiplicity of infection of approx. 0.05 by adding 100 µL of influenza virus at a concentration of 4 × 106 TCID50/mL and incubated in 5% CO2 at 33 °C. The virus strain used for this work was Influenza A/Mexico/4108/2009 (pH1N1), a wild-type H1N1 pandemic 2009 strain. The TPCK trypsin was used at a final concentration of 2 µg/mL. After 4 h of incubation, 3 mL of serum-free aDMEM supplemented with TPCK trypsin was added. After cytopathic effects (CPE) were observed in over 50% of cells, the cells were scraped, and the scraped cells and spent media was collected and frozen at −80 °C. The resulting virus stock that was used for the work presented here had a titer of 6.4 × 106 TCID50/mL.
2.2. Virus aerosol generation and collection
The aforementioned H1N1 influenza virus strain was used for virus aerosol generation and testing. All the sampling experiments were performed in a US Department of Agriculture inspected-and-approved BSL2-enhanced laboratory following BSL3 work practices. To generate virus aerosols, 10 mL of 1 × 105 TCID50/mL H1N1 influenza virus suspension in phosphate-buffered saline (PBS) plus 0.5% (w/v) bovine serum albumin (BSA) fraction V was used with a 6-jet BioAerosol Nebulizing Generator (BANG, CH Technologies) [44–46]. HEPA-filtered room air was used to provide air flow for the BANG. The schematic diagram of the testing system is shown in the Supplementary Material (Fig. S1).
The airborne virus detection scheme is illustrated in Fig. 1. VIVAS, a laminar-flow, water-based condensational growth system, was used to enlarge the virus aerosols generated by the BANG as described previously [6,20,21,46]. The conditioner of the VIVAS was cooled to 6 °C and the initiator was heated to 45 °C with 100% relative humidity to enlarge virus aerosols by water condensation onto their surfaces. The enlarged particles were impinged directly into the 3D-printed SPAD collector as shown in Fig. 1a. The collector contains a funnel with a 40-mm-opening to fit the nozzles of the VIVAS. A piece of laminated paper device was taped to the bottom of the collector for RNA immobilization as described in the RNA enrichment step.
Fig. 1.
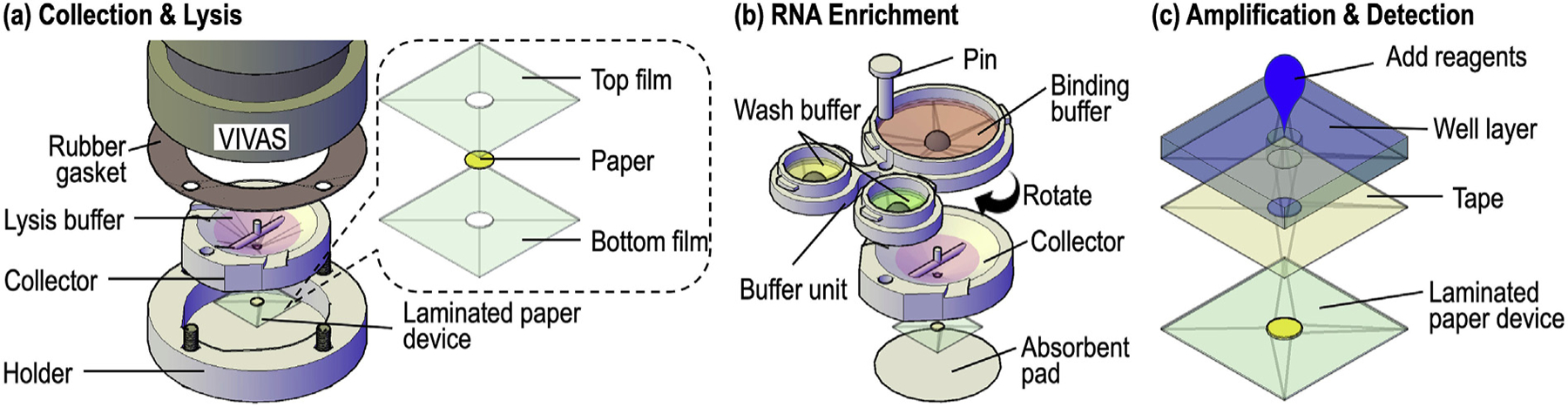
The airborne virus detection scheme includes three steps: collection & lysis, RNA enrichment, and amplification & detection. (a) Exploded view of the setup for airborne virus collection. The virus aerosols were enlarged by VIVAS and impinged directly into the lysis buffer housed in the SPAD collector (see Fig. 2 for the detail). A laminated paper device (with an exploded view in the inset on the right) was attached to the bottom of the collector for RNA enrichment. (b) After collection, the collector/paper device were separated from VIVAS, assembled with the buffer unit (top) and placed on top of an absorbent pad (bottom) for aerosol collection and virus detection. The buffer unit was then rotated to discharge the binding buffer into the collector (see Fig. 3 for the valving mechanism). The laminated paper device underneath the collector would enrich the virus RNA from the lysate while the waste was absorbed by the absorbent pad. Once the lysate filtration was completed, the buffer unit was rotated twice to discharge the two wash buffers sequentially. (c) The laminated paper device with enriched RNA was peeled from the collector and taped onto a well layer to form a RNA amplification device for RT-LAMP. After adding RT-LAMP amplification buffer and incubation, resultant amplicons could be detected colorimetrically. The 3D-printed sample preparation unit in (b) and the paper-based amplification unit in (c) form SPAD.
To attach the SPAD collector to VIVAS, a holder was designed to fix the collector onto VIVAS’ aerosol outlet with four screws (Fig. 2b). A rubber gasket was used to form an air-tight seal to prevent aerosol leakage during sampling (Figs. 1a and 2c). We used fluorescent aerosols to demonstrate the collection result. Fig. 2d shows the enlarged and collected droplets in the collector deposited by the 32 nozzles of VIVAS. The droplets collected at the bottom of funnel formed a pool while some droplets collected near the top of funnel remained separate to reflect the arrangement of the corresponding VIVAS’ nozzles [20,21].
Fig. 2.
Illustration and photographs of those components for virus aerosol collection. (a) Illustration of the VIVAS process and the collected particles into SPRA. (b) Integration of the SPAD collector with VIVAS using a 3D-printed device holder. (c) Photograph of the collector, device holder, and rubber gasket assembled together using four screws. (d) Photograph of collected fluorescent droplets in the collector by VIVAS.
As described previously [1], an air flow at 6 L per minute (LPM) was introduced into VIVAS. A negative control sample using 10 mL PBS plus 0.5% (w/v) BSA in the BANG was first collected for 15 min. Three virus aerosol collections with virus solution in the BANG were performed for 15 min each. A lysis buffer (buffer AVL, QIAGEN) of 560 µL was preloaded to the SPAD collector. The lysis buffer protected viral RNA from RNase degradation and deactivated the collected virus to reduce the generation of biohazardous wastes. Between each virus aerosol collection, two washing steps were performed to prevent the result of the next collection being affected by a previous collection. The first wash was a 5-min collection of 0.01% sodium dodecyl sulfate (SDS, ThermoFisher Scientific) solution in the BANG. The SDS as a surfactant flushed away the virus aerosols left in VIVAS. The second wash was a 25-min collection of molecular-biology grade water to flush away the SDS residual.
2.3. RT-LAMP amplification
To test the primer set used for RT-LAMP amplification, RNA was extracted from 140 µL of H1N1 influenza virus samples using a QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA) following the manufacturer’s protocol. The extracted RNA was eluted with 60 µL of buffer AVE (QIAGEN) and stored at −80 °C before use.
Each 25 µL of RT-LAMP assay contained 2.5 µL of 10X isothermal amplification buffer, 1.4 mM dNTPs, 6 mM MgSO4, 2.5 µL of 10X primer mix, 8 U Bst 2.0 WarmStart® DNA polymerase, 7.5 U WarmStart® RTx reverse transcriptase, and 1 µL RNA sample. Except for the dNTPs from ThermoFisher Scientific (MA, USA), other reagents used in the RT-LAMP assays were obtained from New England Biolabs (Ipswich, MA, USA). The primer mix contained 1.6 µM F1P/B1P, 0.2 µM F3/B3, and 0.4 µM LF/LB. Their sequences are listed in the Supplementary Material (Table S1) [47]. These primers were purchased from Integrated DNA Technologies (Coralville, Iowa, USA). RT-LAMP was performed at 63 °C for 30 min in a Bio-Rad Mycycler® (Bio-Raid, CA, USA). Aliquots of the reaction products were electrophoresed in a 2% agarose gel, followed by imaging using a Gel Doc™ EZ system (Bio-Rad).
To verify that the 25-min incubation period used with the H1N1 influenza virus primers was appropriate, a real-time RT-LAMP assay was carried out by adding 0.5 µL of 10X concentrated SYBR green I nucleic acid gel stain in dimethyl sulfoxide (ThermoFisher Scientific) and 0.5 µL ROX reference dye (ThermoFisher Scientific) to the 25 µL RT-LAMP reaction buffer. The fluorescence signal from the RT-LAMP reactions was subsequently measured using a QuantStudio 3 real-time PCR system (ThermoFisher Scientific).
2.4. Paper-based device for RNA enrichment and amplification
For RNA enrichment using SPAD, we employed lamination technique [35–37] to prepare the laminated paper device that functioned as a filter for RNA isolation, in a way similar to the commercially available nucleic acid purification spin column [48]. The chaotropic-salt-based buffers from a QIAamp Viral RNA Mini Kit (QIAGEN) were used to improve RNA binding to the paper substrate. A well layer was attached to the laminated paper device as a sample well to form the paper-based RNA amplification device (Fig. 1). Instead of centrifugal force as used for the spin column, sample filtration was powered by capillary forces generated in porous paper and the absorbent pad beneath the device.
Three types of paper materials, FTA® classic card (ThermoFisher Scientific), Whatman™ 1 chromatography paper (ThermoFisher Scientific), and Whatman™ GF/F glass microfiber filter paper (ThermoFisher Scientific) were evaluated for fabricating the laminate paper-based RNA enrichment device. The FTA® card is a commercially available filter paper specifically developed to extract, bind, and preserve nucleic acids from blood, plant and animal tissue extracts and other sources according to the manufacturer [49]. The chromatography paper is an untreated, high quality cellulose fiber paper. The GF/F glass microfiber filter is a paper designed for nucleic acid purification. The device was made by sandwiching a piece of paper material between two thermoplastic films as shown in the inset of Fig. 1a. The paper piece of 3.5-mm-diameter was made using a steel puncher. Two layers of thermoplastic films with a 3-mm-diameter hole were shaped by cutting a section of 75-µm-thick polyester thermal bonding lamination film (Lamination Plus, Kaysville, UT, USA) using a Graphtec Craft Robo-S cutting plotter (Graphtec Corporation, Yokohama, Japan). The paper and the top and bottom films were then aligned and passed through a heated laminator (GBC® Catena 65 Roll Laminator, GBC, Lake Zurich, IL, USA), which was set at a rolling speed of “1” with the temperature at 220 °F.
Aliquots containing 10, 1, and 0.1 TCID50 H1N1 influenza virus lysate per µL were made with the virus stock solution and molecular-biology-grade water and lysis buffer AVL (QIAGEN) at the ratio of 1:4 and stored at −80 °C for testing paper-based RNA amplification. To compare the RNA capture efficiency of laminated paper-based RNA amplification device made of FTA® card, chromatography paper, or glass microfiber paper, a serial dilution of 70 µL H1N1 influenza virus lysate was used to compare the limit of detection (LOD) of each material. An ethanol (100%) solution of 56 µL was mixed into the diluted virus lysate before introducing to the device. A solution of AW1 (Qiagen) and AW2 (Qiagen) of 100 µL each was then filtered through the paper device sequentially to purify the RNA captured by the paper device. RT-LAMP amplification and gel electrophoresis were performed as described previously to detect the captured RNA.
2.5. RNA enrichment using SPAD
After the 15-min sampling period, the collector was separated from VIVAS by loosening screws (Fig. 2c). The collector was then assembled with the buffer unit by inserting a pin through one hole in both buffer unit and collector unit as shown in Fig. 1b. The SPAD was designed to perform sample preparation and RNA enrichment from the collected viruses, without using lab tools such as a pipette. The collector is for collecting the virus aerosols and lysing viruses while the buffer unit is for housing the binding buffer (molecular-biology grade ethanol) and the two wash buffers (AW1 & AW2, QIAGEN) for RNA purification and enrichment.
The assembled device was first placed on top of a piece of cellulose absorbent pad (Kimtech Science) to provide capillary forces driving fluid flows. The binding buffer (560 µL ethanol) was discharged first to enhance RNA binding to the paper in the laminated paper device. The process of flowing the mixture through the paper took about 15 min, depending on the volume of sample collected. After all the lysate was filtered through the laminated paper device, the buffer unit was rotated to align and dispense the first wash buffer (250 µL AW1) to the collector, and then the buffer unit was rotated again, and the second wash buffer (250 µL AW2) was discharged and filtered through in the same manner to remove inhibitors from the captured RNA. The two wash steps took about 5 min each to complete.
A liquid dispensing scheme using a simple fluid control valve was developed to trigger the release of reagents from each reservoir of the buffer unit to the collector of SPAD (Fig. 3). The ball valve concept was inspired by the dispensing mechanism of a ballpoint pen, in which ink is transferred onto paper when the metal ball at the tip is pressed while writing [35]. A 5/16-inch-diameter opening was created at the bottom of the funnel-shaped reservoir to house a stainless-steel ball (McMaster-Carr). This opening was designed in a way that the ball could function as a plug to keep the liquid from flowing out. The opening size was chosen to accommodate the layer resolution of the 3D printer and allow the ball to have a 2-mm vertical displacement during valve operation for dispensing the stored liquid. To prevent possible ball movement and reagent spillage during transportation, a lid with a lure-lock thread and a piece of O-ring (McMaster-Carr) was used to “lock” the ball valve (Fig. 3a, Fig. S2a). The lid contained a peg underneath that pressed the ball against the opening while the thread was tightened. As illustrated by a short movie in the Supplementary Material (Video S1), the valve could achieve a leak-free seal when the lid was properly assembled. After transportation, the lid can be loosened up or removed to allow the valve operation. As shown in Fig. 3b, the ball valve could be triggered by rotating the buffer unit along the pin to align the reservoir with the collector when the ball was pushed up by a pillar in the center of the collector. The operation of the ball valve was demonstrated in Video S2.
Fig. 3.
Ball-based valve mechanism for liquid dispensing. (a) A ball is used to block the opening at the bottom of the reservoir. A peg protruding from the lid is used to lock the ball in place during storage and transportation. (b) After the lid is removed or loosened, the reservoir and the collector are assembled together through a pin. When the reservoir is rotated to align with the collector, the valve is actuated to discharge the solution housed in the reservoir as the ball is pushed up by the pillar in the middle of the collector. The cartoon on the right shows valve’s opening and fluid flowing down upon the ball being pushed up.
These sample preparation components were fabricated by a commercial 3D printer, Ultimaker 3 (Ultimaker, Geldermalsen, Netherlands), using polylactic acid (PLA) filament with polyvinyl alcohol (PVA) as support material. The print layer height was set to 0.06 mm and the infill density was set to 100%. A photo of these SPAD parts, with a U.S. quarter for the size comparison, was shown in Fig. S2b. It is conceivable that these parts can also be manufactured using injection molding or other methods. This sample preparation unit can be reusable if desired, though the detection unit must be disposable.
2.6. RT-LAMP amplification in SPAD
After RNA enrichment, the laminated paper-device was peeled from the collector and taped to a well layer to form a device to conduct RT-LAMP amplification (Fig. 1c). The well layer was a 3-mm-thick, 2 cm × 2 cm square, cut from a piece of clear polycarbonate sheet (McMaster-Carr, Elmhurst, IL) using a milling machine (Sherline Products, Vista, California). A 3-mm-diameter hole was made in both the polycarbonate sheet and the double-sided adhesive tape (3 M 9087 white bonding tape, R. S. Hughes, Sunnyvale, CA), the holes were aligned with the laminated paper device. After the well layer was attached, a piece of adhesive tape (Fellows®) was attached to the bottom of the device, and 25 µL RT-LAMP amplification buffer (as described above) was loaded to the well of the device, followed by sealing the device with a piece of adhesive tape on the top for evaporation control. The sealed device was incubated at (63 ± 0.5) °C for 25 min in an Isotemp 105 water bath (ThermoFisher Scientific) for DNA amplification. The amplicons were analyzed using either gel electrophoresis or SYBR green for instrument-free detection as discussed below. Note that we used a water bath for this work, but it is possible to use a battery-operated coffee mug to achieve RT-LAMP in the field as we demonstrated previously [35].
After RT-LAMP amplification, 1 µL SYBR green (10,000X concentrate SYBR green I nucleic acid gel stain, ThermoFisher Scientific) was added to the amplicons, and the results were readable by naked-eye. Alternatively, an ULAKO blue LED flashlight (Amazon, WA, USA) powered by 1 AA battery was used to excite the green fluorescence from the amplicon-SYBR complexes. The resulting color of the solution was imaged using a smart phone. A piece of brown-tainted translucent plastic film was taped in front of the phone camera lens to filter out the blue light from the flashlight.
Viral RNA enrichment and RT-LAMP amplification in SPAD was first evaluated with H1N1 influenza viruses spiked in water of the same volume range of those collected by the VIVAS at 6 LPM for 15 minutes. H1N1 influenza viruses spiked in 140 µL water were lysed using 560 µL lysis buffer AVL (Qiagen) before pipetting onto the collector of SPAD. The volume ratio of (sample volume):(lysis solution):(binding solution) was kept at 1:4:4 as instructed in the QIAamp Viral RNA Mini Kit.
3. Results and discussion
3.1. RT-LAMP reaction time
The QIAamp Viral RNA Mini Kit (QIAGEN) was used as a benchmark standard to purify RNA from aqueous samples spiked with 10, 100 and 1000 TCID50 H1N1 virus. The resultant purified virus RNA was employed to estimate the RT-LAMP reaction time using the QuantStudio-3 real-time amplification system (Fig. 4). All the wells containing virus RNA were observed with a signal that reached a plateau within 10–18 min, and no non-specific amplification was observed during the 30-min incubation period for the no-template control (NTC). This result suggested that 25-min incubation was sufficient to detect the virus RNA in our device. Note that LAMP amplification involves many complicated reaction steps, thus the absolute signal is not necessarily linear with the original RNA amount. Instead, the threshold time is used in the literature to correlate with the copy number of genetic materials [50]. The linear-regression calibration curve in Fig. 4b indicated that quantitative airborne virus detection is feasible.
Fig. 4.
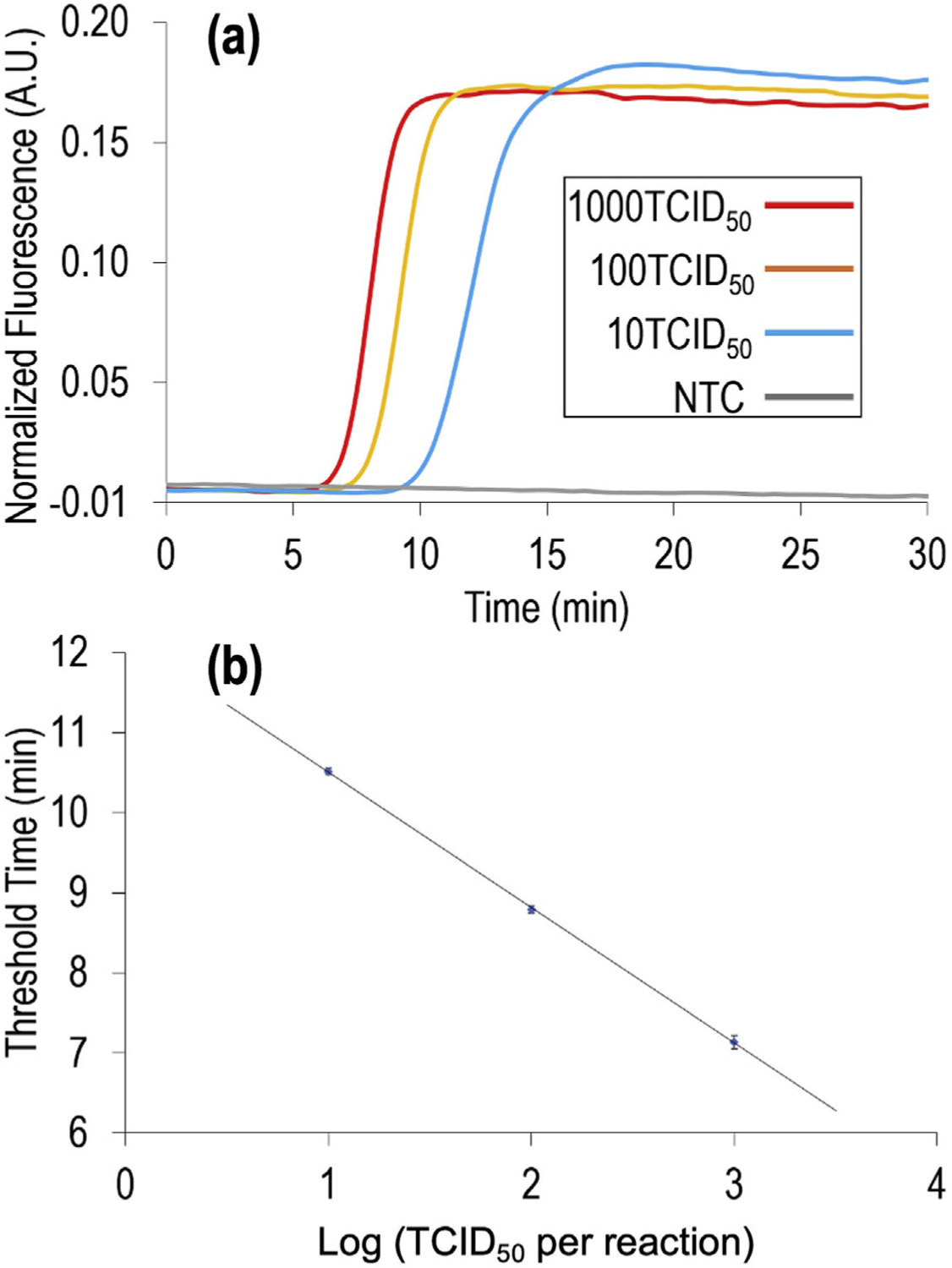
Real-time RT-LAMP amplification for H1N1 influenza virus RNA. (a) Normalized fluorescent signal of 10, 100, and 1000 TCID50 H1N1 virus genome equivalents as a function of RT-LAMP time. NTC, no-template control. (b) Calibration curve showing the threshold cycling time (Ct) as a function of TCID50 in each reaction (in log scale). The Ct values were provided by the instrument. The results were generated from 3 replicates of each concentration of H1N1 virus RNA samples. The error bars indicate one standard deviation.
3.2. Paper-based RNA enrichment and amplification
Paper-based devices made of FTA® card, glass microfiber and chromatography paper were tested with different concentrations of H1N1 virus lysate to determine the limit of the detection (LOD) of each device. The results (Table 1 & Figs. S3–S5) indicate that the devices made of chromatography paper have the lowest LOD, detecting spiked samples of 0.8 TCID50 influenza virus, while the device made of FTA® card and glass microfiber detected only 5 TCID50 and above. Note that TCID50 is a value obtained by using a series of dilution of a viral fluid to infect a number of cell culture in a well plate; after incubation, the percentage of infected wells is observed for each dilution, which is used to calculate the median tissue culture infective dose (i.e., TCID at 50%).
Table 1.
Paper material comparison for detection of H1N1 influenza virus.
| Virus concentration (TCID50) | 50 | 25 | 10 | 5 | 2.5 | 1 | 0.9 | 0.8 | 0.7 | NTC |
|---|---|---|---|---|---|---|---|---|---|---|
| FTA® card | Y | Y | Y | Y | N | N | N | |||
| Glass microfiber paper | Y | Y | Y | Y | N | N | N | |||
| Chromatography paper | Y | Y | Y | Y | Y | Y | Y | Y | N | N |
Note: Virus concentration in TCID50 per device; NTC, no-template control; Y (yes) or N (no) indicates whether virus RNA was detected or not.
Untreated cellulose has been used as a nucleic acid isolation material since the 1960s, though its use is far less common than silica [51–53]. This is possibly due to the complicated steps of assembling cellulose powder into a column often involved. Those manual steps make the cellulose powder less desirable compared to the ready-to-use silica column. The laminated paper-based RNA amplification device, on the other hand, is easy to make, of low cost, and does not require complicated manual steps during use. Our results suggest that the cellulose chromatography paper can function as an RNA extraction substrate. As a result, all the following experiments were performed using devices made from chromatography paper.
3.3. Detection of influenza virus in aqueous solutions
H1N1 influenza viruses spiked in 140 µL water were lysed using 560 µL lysis buffer AVL (Qiagen) and loaded to SPAD for RNA enrichment and RT-LAMP amplification. The RNA enrichment and amplification process took about 25 min each to complete, after which the result could be read using SYBR green dye and blue LED flashlight without lab instruments. As shown in Fig. 5, we successfully detected 1 TCID50 H1N1 influenza virus per 140 µL sample using the SPAD in 50 min. The SYBR green-DNA-complex absorbs blue light and emits green light, resulting in a light-yellow color when observed under the ambient light and a bright green fluorescence under blue LED. We chose SYBR green as it detects the amplicons directly [54], while other colorimetric RT-LAMP methods such as the hydroxynaphthol blue [55], the leuco crystal violet [56], and the phenol red [57] detect the by-products of amplification. Note that the test only gives a binary yes/no answer (i.e., presence or absence of viruses). As illustrated in Fig. 4, similar fluorescent signals were observed for different concentrations of viruses. However, they took different time to reach the signal plateau as illustrated in Fig. 3a. Also note that RT-LAMP produces a mixture of amplicons, thus it does not have one specific gel band as with PCR.
Fig. 5.
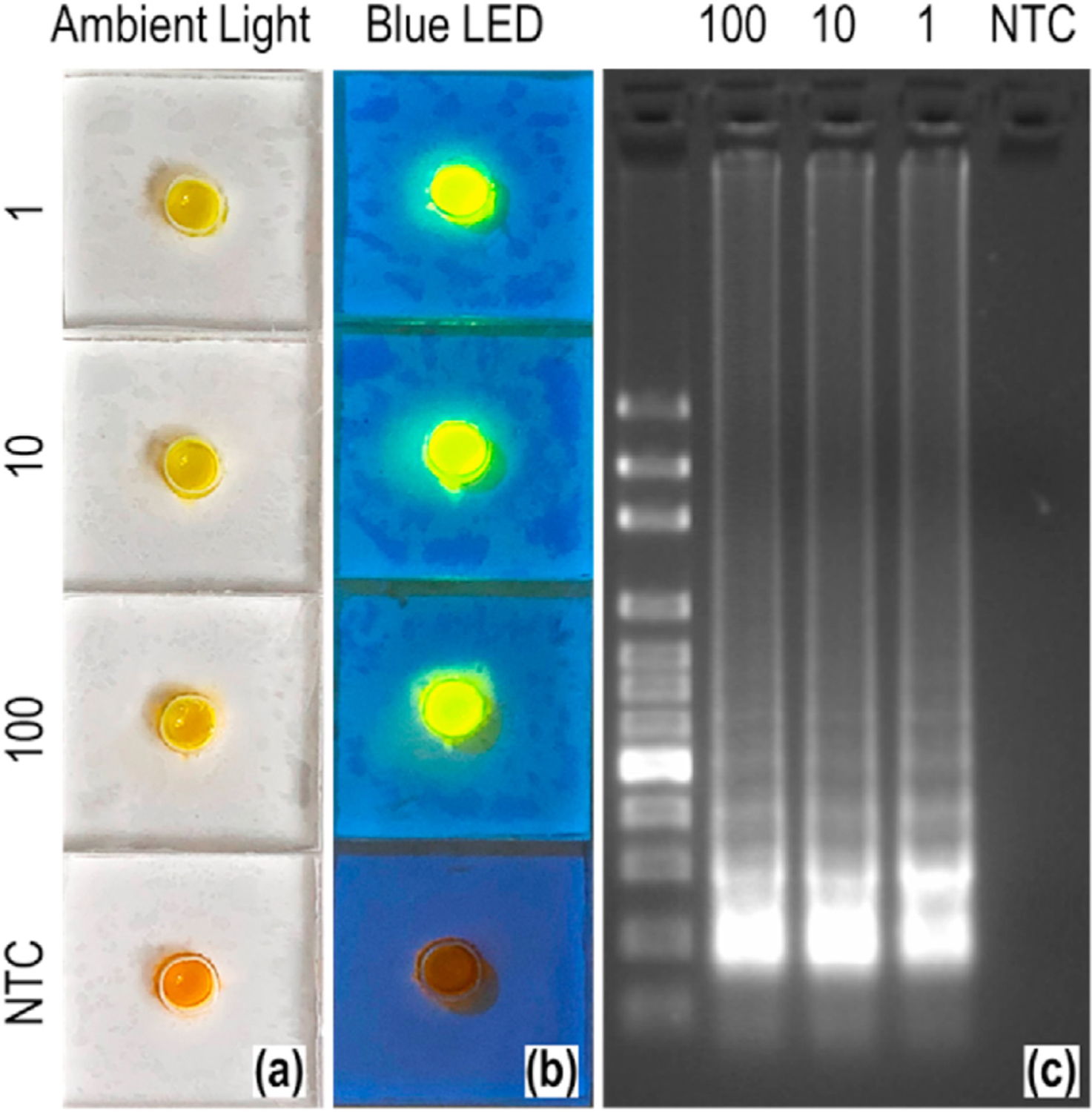
Detection of H1N1 influenza virus spiked in water. (a) Pictures of the detection units under ambient light, taken by a cell phone. (b) Photographs of the detection units illuminated by a blue LED flashlight. The amount of TCID50 H1N1 virus spiked in each device as well as the negative control (NTC) is indicated on the left side of each devices in (a) and (b). The emission observed outside the wells are optical effects resulting from light transmission into transparent plastics and the angles taking the pictures. (c) Gel electrophoresis image of the amplicons from each device. The sample for each lane is indicated at the top: 100 bp DNA ladder, 100, 10, 1 TCID50 and NTC. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Detection of airborne influenza viruses
The average collection volume of our test system for a 15-min sampling using 10 mL PBS/BSA media in BANG and 6 LPM air flow rate for VIVAS was determined to be 143 ± 25 µL by weighing the collector before and after collection. As a result, 560 µL lysis buffer was used in the SPAD collector for the H1N1 influenza virus detection experiment, according to sample-to-lysis solution ratio recommended by the sample preparation kit manufacturer (similar to the 140 µL of sample used in Section 3.3). A virus concentration of 1.0 × 105 TCID50/mL H1N1 influenza virus in PBS/BSA was used in the aerosol generator (BANG) and the resulting aerosol was sampled in triplicates by VIVAS. The virus solution consumed by BANG in the 15 min collection is 1367 ± 287 µL. According to the infectious H1N1 virus collection efficiency reported by our previous work [46], the amount of collected H1N1 influenza virus was about (1.09 ± 0.23) x 105 TCID50.
Our system successfully detected airborne H1N1 influenza viruses in lab-generated aerosols (Fig. 6). No non-specific amplification was observed in the negative control collection of PBS/BSA aerosols. Strong fluorescence signals were observed from all the triplicates of the H1N1 virus aerosol collections. These results also illustrate the reproducibility of valves, assays, and the overall SPAD system. Including the sampling time (15 min), the virus detection process in its entirety took around 65 min to complete, making our system a fast and effective method to study airborne virus transmission, screen the presence of a certain type of airborne virus in the environment, and help guide possible infection mitigation.
Fig. 6.
Detection of airborne H1N1 influenza virus through the combined use of SPAD and VIVAS. (a) Photographs of the detection units under ambient light, taken by a cell phone. (b) Photographs of the detection units illuminated by a blue LED flashlight. The negative control (NTC) and three aerosol samples (S1–S3) are labelled on the left side of each devices in (a) and (b). (c) Gel electrophoresis image of the amplicons from each device. The sample for each lane is indicated at the top: 100 bp DNA ladder, NTC, S1, S2, S3. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions
By combining the use of SPAD for sample preparation and RT-LAMP amplification with VIVAS for virus aerosols collection, we detected lab-generated, airborne H1N1 influenza viruses in ~1 h. This approach features a two-step concentration for sporadic airborne viruses: (a) the concentration of liters of virus aerosols into a ~140 µL aqueous sample, and (b) the enrichment of lysed virus RNA onto the laminated paper device. The two-step concentration grants our approach the sensitivity required to detect airborne viruses using a rapid and specific RT-LAMP method. Moreover, our laminated paper device could process a flexible amount of collected aerosol samples, allowing us to increase the sensitivity by increasing the collection time, i.e., the volume of aerosol collected by VIVAS, if necessary. In addition, we have demonstrated the superior efficacy of chromatograph paper to the commercial FTA® card and Whatman™ glass microfiber for RNA filtration via chaotropic agents, as well as high portability of our SPAD device for RNA virus detection.
Future directions of this work include the collection of real-world influenza virus aerosols in places such as infirmaries and classrooms [6]. Our approach can be further developed for detecting other airborne viruses. A part of the approach has been adapted for detecting airborne SARS-CoV-2 virus [43] and the overall approach is currently being modified for the same purpose. The limit of detection of our SPAD device at 1 TCID50 is lower than 3 TCID50, which is believed to be human infectious dose of the influenza A virus from aerosols [58]. The limit of detection is also lower than 35.4 TCID50, which is the average amount in one cubic meter of air collected in a healthcare center, a day-care center, and airplanes during a flu season [58]. In addition to detecting airborne viruses, SPAD can be adapted to detect viruses from non-airborne samples, including aqueous solutions such as blood, urine, and saliva.
Supplementary Material
HIGHLIGHTS.
A sample preparation device for the storage and sequential delivery of reagents.
A paper-based analytical device for virus RNA enrichment and amplification.
Integration of the devices with an aerosol collector for virus detection.
Detection of influenza viruses, with potential to study their airborne transmission.
Acknowledgment
This work is supported in part by US National Science Foundation (IDBR-1353423 and CBET-2030844), US National Institutes of Health (R01AI158868), Florida Department of Health (7ZK22 and 7JK07), and the University of Florida, USA.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2021.338542.
References
- [1].Pan M, Eiguren-Fernandez A, Hsieh H, Afshar-Mohajer N, Hering SV, Lednicky J., et al. , Efficient collection of viable virus aerosol through laminar-flow, water-based condensational particle growth, J. Appl. Microbiol 120 (2016) 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jones RM, Brosseau LM, Aerosol transmission of infectious disease, J. Occup. Environ. Med 57 (2015) 501–508. [DOI] [PubMed] [Google Scholar]
- [3].Shakoor S, Mir F, Zaidi AK, Zafar A, Hospital preparedness in community measles outbreaks-challenges and recommendations for low-resource settings, Emerg. Health Threats J 8 (2015) 24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tran K, Cimon K, Severn M, Pessoa-Silva CL, Conly J, Aerosol generating procedures and risk of transmission of acute respiratory infections to healthcare workers: a systematic review, PloS One 7 (2012), e35797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bertran K, Balzli C, Kwon YK, Tumpey TM, Clark A, Swayne DE, Airborne transmission of highly pathogenic influenza virus during processing of infected poultry, Emerg. Infect. Dis 23 (2017) 1806–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pan M, Bonny TS, Loeb J, Jiang X, Lednicky JA, Eiguren-Fernandez A., et al. , Collection of viable aerosolized influenza virus and other respiratory viruses in a student health care center through water-based condensation growth, mSphere 2 (2017) e00251–17, 10.1128/mSphere.00251-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lednicky JA, Shankar SN, Elbadry MA, Gibson JC, Alam MM, Stephenson CJ, et al. , Collection of SARS-CoV-2 virus from the air of a clinic within a university student health care center and analyses of the viral genomic sequence, Aerosol and Air Quality Research 20 (2020) 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fronczek CF, Yoon JY, Biosensors for monitoring airborne pathogens, J. Lab. Autom 20 (2015) 390–410. [DOI] [PubMed] [Google Scholar]
- [9].Fabian P, McDevitt JJ, Houseman EA, Milton DK, Airborne influenza virus detection with four aerosol samplers using molecular and infectivity assays: considerations for a new infectious virus aerosol sampler, Indoor Air 19 (2009) 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Prather KA, Wang CC, Schooley RT, Reducing transmission of SARS-CoV-2, Science 368 (2020) 1422–1424. [DOI] [PubMed] [Google Scholar]
- [11].Pan M, Lednicky JA, Wu CY, Collection, particle sizing and detection of airborne viruses, J. Appl. Microbiol 127 (2019) 1596–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chang YF, Wang WH, Hong YW, Yuan RY, Chen KH, Huang YW, et al. , Simple strategy for rapid and sensitive detection of avian influenza A H7N9 virus based on intensity-modulated SPR biosensor and new generated antibody, Anal. Chem 90 (2018) 1861–1869. [DOI] [PubMed] [Google Scholar]
- [13].Huang S, Abe K, Bennett S, Liang T, Ladd PD, Yokobe L., et al. , Disposable autonomous device for swab-to-result diagnosis of influenza, Anal. Chem 89 (2017) 5776–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang H, Henry C, Anderson CS, Nogales A, DeDiego ML, Bucukovski J., et al. , Crowd on a chip: label-free human monoclonal antibody arrays for serotyping influenza, Anal. Chem 90 (2018) 9583–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ladhani L, Pardon G, Meeuws H, van Wesenbeeck L, Schmidt K, Stuyver L., et al. , Sampling and detection of airborne influenza virus towards point-of-care applications, PloS One 12 (2017), e0174314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Takenaka K, Togashi S, Miyake R, Sakaguchi T, Hide M, Airborne virus detection by a sensing system using a disposable integrated impaction device, J. Breath Res 10 (2016), 036009. [DOI] [PubMed] [Google Scholar]
- [17].Kovar D, Farka Z, Skladal P, Detection of aerosolized biological agents using the piezoelectric immunosensor, Anal. Chem 86 (2014) 8680–8686. [DOI] [PubMed] [Google Scholar]
- [18].Xiong C, Zhou X, Wang J, Zhang N, Peng WP, Chang HC, et al. , Ambient aerodynamic desorption/ionization method for microparticle mass measurement, Anal. Chem 85 (2013) 4370–4375. [DOI] [PubMed] [Google Scholar]
- [19].Therkorn J, Thomas N, Scheinbeim J, Mainelis G, Field performance of a novel passive bioaerosol sampler using polarized ferroelectric polymer films, Aerosol. Sci. Technol 51 (2017) 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hering SV, Stolzenburg MR, A method for particle size amplification by water condensation in a laminar, thermally diffusive flow, Aerosol Sci. Technol 39 (2005) 428–436. [Google Scholar]
- [21].Hering SV, Stolzenburg MR, Quant FR, Oberreit DR, Keady PB, A laminar-flow, water-based condensation particle counter (WCPC), Aerosol Sci. Technol 39 (2005) 659–672. [Google Scholar]
- [22].Pan MH, Carol L, Lednicky JA, Eiguren-Fernandez A, Hering S, Fan ZH, et al. , Determination of the distribution of infectious viruses in aerosol particles using water-based condensational growth technology and a bacteriophage MS2 model, Aerosol Sci. Technol 53 (2019) 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sung G, Ahn C, Kulkarni A, Shin WG, Kim T, Highly efficient in-line wet cyclone air sampler for airborne virus detection, J. Mech. Sci. Technol 31 (2017) 4363–4369. [Google Scholar]
- [24].Bhardwaj J, Kim MW, Jang J, Rapid airborne influenza virus quantification using an antibody-based electrochemical paper sensor and electrostatic particle concentrator, Environ. Sci. Technol 54 (2020) 10700–10712. [DOI] [PubMed] [Google Scholar]
- [25].Kim HU, Min J, Park G, Shin D, Sung G, Kim T., et al. , Electrochemical detection of airborne influenza virus using air sampling system, Aerosol Air Qual Res 18 (2018) 2721–2727. [Google Scholar]
- [26].Posthuma-Trumpie GA, Korf J, van Amerongen A, Lateral flow (immuno) assay: its strengths, weaknesses, opportunities and threats, A literature survey, Analytical and bioanalytical chemistry 393 (2009) 569–582. [DOI] [PubMed] [Google Scholar]
- [27].Nicholson KG, Abrams KR, Batham S, Medina MJ, Warren FC, Barer M., et al. , Randomised controlled trial and health economic evaluation of the impact of diagnostic testing for influenza, respiratory syncytial virus and Streptococcus pneumoniae infection on the management of acute admissions in the elderly and high-risk 18- to 64-year-olds, Health Technol. Assess 18 (2014) 1–274, vii-viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vecino-Ortiz AI, Goldenberg SD, Douthwaite ST, Cheng CY, Glover RE, Mak C., et al. , Impact of a multiplex PCR point-of-care test for influenza A/B and respiratory syncytial virus on an acute pediatric hospital ward, Diagn. Microbiol. Infect. Dis 91 (2018) 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tromberg BJ, Schwetz TA, Perez-Stable EJ, Hodes RJ, Woychik RP, Bright RA, et al. , Rapid scaling up of covid-19 diagnostic testing in the United States - the NIH RADx initiative, N. Engl. J. Med 383 (2020) 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Saito M, Uchida N, Furutani S, Murahashi M, Espulgar W, Nagatani N., et al. , Field-deployable rapid multiple biosensing system for detection of chemical and biological warfare agents, Microsyst Nanoeng 4 (2018). [Google Scholar]
- [31].Liu Q, Zhang XL, Yao YH, Jing WW, Liu SX, Sui GD, A novel microfluidic module for rapid detection of airborne and waterborne pathogens, Sensor. Actuator. B Chem 258 (2018) 1138–1145. [Google Scholar]
- [32].Dineva MA, MahiLum-Tapay L, Lee H, Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings, Analyst 132 (2007) 1193–1199. [DOI] [PubMed] [Google Scholar]
- [33].Yin J, Suo Y, Zou Z, Sun J, Zhang S, Wang B., et al. , Integrated microfluidic systems with sample preparation and nucleic acid amplification, Lab Chip 19 (2019) 2769–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J, Rapid and simple method for purification of nucleic acids, J. Clin. Microbiol 28 (1990) 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jiang X, Loeb JC, Manzanas C, Lednicky JA, Fan ZH, Valve-enabled sample preparation and RNA amplification in a coffee mug for zika virus detection, Angew Chem. Int. Ed. Engl 57 (2018) 17211–17214. [DOI] [PubMed] [Google Scholar]
- [36].Cassano CL, Fan ZH, Laminated paper-based analytical devices (LPAD): fabrication, characterization, and assays, Microfluid. Nanofluidics 15 (2013) 173–181. [Google Scholar]
- [37].Liu W, Cassano CL, Xu X, Fan ZH, Laminated paper-based analytical devices (LPAD) with origami-enabled chemiluminescence immunoassay for cotinine detection in mouse serum, Anal. Chem 85 (2013) 10270–10276. [DOI] [PubMed] [Google Scholar]
- [38].Martinez AW, Phillips ST, Whitesides GM, Carrilho E, Diagnostics for the developing world: microfluidic paper-based analytical devices, Anal. Chem 82 (2010) 3–10. [DOI] [PubMed] [Google Scholar]
- [39].Noviana E, Carrao DB, Pratiwi R, Henry CS, Emerging applications of paper-based analytical devices for drug analysis: a review, Anal. Chim. Acta 1116 (2020) 70–90. [DOI] [PubMed] [Google Scholar]
- [40].Lam T, Devadhasan JP, Howse R, Kim J, A chemically patterned microfluidic paper-based analytical device (C-microPAD) for point-of-care diagnostics, Sci. Rep 7 (2017) 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Njiru ZK, Loop-mediated isothermal amplification technology: towards point of care diagnostics, PLoS Neglected Trop. Dis 6 (2012), e1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ganguli A, Mostafa A, Berger J, Aydin MY, Sun F, Ramirez SAS, et al. , Rapid isothermal amplification and portable detection system for SARS-CoV-2, Proc. Natl. Acad. Sci. U. S. A 117 (2020) 22727–22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lednicky JA, Lauzardo M, Fan ZH, Jutla A, Tilly TB, Gangwar M., et al. , Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients, Int. J. Infect. Dis 100 (2020) 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lednicky JA, Hamilton SB, Tuttle RS, Sosna WA, Daniels DE, Swayne DE, Ferrets develop fatal influenza after inhaling small particle aerosols of highly pathogenic avian influenza virus A/Vietnam/1203/2004 (H5N1), Virol. J 7 (2010) 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tuttle RS, Sosna WA, Daniels DE, Hamilton SB, Lednicky JA, Design, assembly, and validation of a nose-only inhalation exposure system for studies of aerosolized viable influenza H5N1 virus in ferrets, Virol. J 7 (2010) 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lednicky J, Pan MH, Loeb J, Hsieh H, Eiguren-Fernandez A, Hering S., et al. , Highly efficient collection of infectious pandemic influenza H1N1 virus (2009) through laminar-flow water based condensation, Aerosol Sci. Technol 50 (2016) IeIv. [Google Scholar]
- [47].Nakauchi M, Yoshikawa T, Nakai H, Sugata K, Yoshikawa A, Asano Y., et al. , Evaluation of reverse transcription loop-mediated isothermal amplification assays for rapid diagnosis of pandemic influenza A/H1N1 2009 virus, J. Med. Virol 83 (2011) 10–15. [DOI] [PubMed] [Google Scholar]
- [48].Purdy KJ, Embley TM, Takii S, Nedwell DB, Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method, Appl. Environ. Microbiol 62 (1996) 3905–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gustavsson I, Lindell M, Wilander E, Strand A, Gyllensten U, Use of FTA card for dry collection, transportation and storage of cervical cell specimen to detect high-risk HPV, J. Clin. Virol 46 (2009) 112–116. [DOI] [PubMed] [Google Scholar]
- [50].Nguyen DV, Nguyen VH, Seo TS, Quantification of colorimetric loop-mediated isothermal amplification process, Biochip J 13 (2019) 158–164. [Google Scholar]
- [51].Su X, Comeau AM, Cellulose as a matrix for nucleic acid purification, Anal. Biochem 267 (1999) 415–418. [DOI] [PubMed] [Google Scholar]
- [52].Barber R, The chromatographic separation of ribonucleic acids, Biochim. Biophys. Acta 114 (1966) 422–424. [DOI] [PubMed] [Google Scholar]
- [53].Moeller JR, Moehn NR, Waller DM, Givnish TJ, Paramagnetic cellulose DNA isolation improves DNA yield and quality among diverse plant taxa, Appl. Plant Sci 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Njiru ZK, Mikosza AS, Armstrong T, Enyaru JC, Ndung’u JM, Thompson AR, Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense, PLoS Neglected Trop. Dis 2 (2008) e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ma XJ, Shu YL, Nie K, Qin M, Wang DY, Gao RB, et al. , Visual detection of pandemic influenza A H1N1 Virus 2009 by reverse-transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye, J. Virol. Methods 167 (2010) 214–217. [DOI] [PubMed] [Google Scholar]
- [56].Priye A, Bird SW, Light YK, Ball CS, Negrete OA, Meagher RJ, A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses, Sci. Rep 7 (2017) 44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tanner NA, Zhang Y, Evans TC Jr., Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes, Biotechniques 58 (2015) 59–68. [DOI] [PubMed] [Google Scholar]
- [58].Nikitin N, Petrova E, Trifonova E, Karpova O, Influenza virus aerosols in the air and their infectiousness, Adv Virol 2014 (2014) 859090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


