Abstract
Liquid biopsies are becoming popular for managing a variety of diseases due to the minimally invasive nature of their acquisition, thus potentially providing better outcomes for patients. Circulating tumor cells (CTCs) are among the many different biomarkers secured from a liquid biopsy, and a number of efficient platforms for their isolation and enrichment from blood have been reported. However, many of these platforms require manual sample handling, which can generate difficulties when translating CTC assays into the clinic due to potential sample loss, contamination, and the need for highly specialized operators. We report a system modularity chip for the analysis of rare targets (SMART-Chip) composed of three task-specific modules that can fully automate processing of CTCs. The modules were used for affinity selection of the CTCs from peripheral blood with subsequent photorelease, simultaneous counting, and viability determinations of the CTCs and staining/imaging of the CTCs for immunophenotyping. The modules were interconnected to a fluidic motherboard populated with valves, interconnects, pneumatic control channels, and a fluidic network. The SMART-Chip components were made from thermoplastics via microreplication, which lowers the cost of production making it amenable to clinical implementation. The utility of the SMART-Chip was demonstrated by processing blood samples secured from colorectal cancer (CRC) and pancreatic ductal adenocarcinoma (PDAC) patients. We were able to affinity-select EpCAM expressing CTCs with high purity (0–3 white blood cells/mL of blood), enumerate the selected cells, determine their viability, and immunophenotype the cells. The assay could be completed in <4 h, while manual processing required >8 h.
Keywords: affinity enrichment, circulating tumor cells, immunophenotyping, impedance sensing, integrated modular system, microfluidics, liquid biopsy
Graphical Abstract
Liquid biopsies are generating significant interest in the medical community due to their minimally invasive nature of acquisition and the fact that they can enable decisions on managing a variety of diseases (i.e., precision medicine).1,2 Liquid biopsy markers include, but are not limited to, rare cells such as circulating tumor cells (CTCs), cell-free molecules for example, cell-free DNA (cfDNA) and microRNA (miRNA), and extracellular vesicles (EVs).
Analyzing liquid biopsy markers can be challenging due to the multiple processing steps involved in the assay and the low mass of the marker that is typically isolated from a clinical sample, especially for early stages of the disease. For typical liquid biopsy assays, the relevant markers must first be enriched from the sample because in most cases they are a vast minority in a mixed population. Following the enrichment, the liquid biopsy markers must be further analyzed to secure the necessary clinical information to guide patient management. For example, in the case of CTCs, the enriched cells must be stained with a panel of markers (i.e., CD45, pan-cytokeratins, and DAPI) for immunophenotypic identification. Typically, this is done by off-loading the enriched sample. Due to the low abundance of liquid biopsy markers (CTCs in patients’ blood can range from 1 to 1000 CTCs per mL),3,4 marker loss or contamination caused by manual sample handling during multiple processing steps may result in false negative/positive results and the need for well-trained operators that can hamper the transition of liquid biopsy assays into the clinic.
Several new microfluidic technologies have been reported for the enrichment of liquid biopsy markers. For example, enrichment of CTCs can utilize different techniques such as deterministic lateral displacement (DLD) or inertial focusing,5,6 size-based filtration,7,8 or affinity selection.9–11 Unfortunately, enrichment microfluidics is typically the only step that the chip performs. Thus, there is a need for process step integration so that all of the necessary assay steps can be performed without requiring operator intervention to obviate issues associated with sample loss and contamination as well as minimizing the need for highly trained operators.
Integrated microfluidic systems can be broadly categorized into centrifugal, monolithic, and modular systems. Centrifugal systems consist of a compact disk-like format with fluidic channels and chambers that use centrifugal forces to manipulate fluids.12–14 Centrifugal microsystems have been developed for CTC isolation.15–17 Potential challenges are related to the limited space restricting the number of operational steps that can be integrated into the system.18,19 In a monolithic configuration, multiple functional units are situated on a single wafer.20–22 Therefore, modifications or changes in the processing steps require redesigning the entire system.23 Monolithic systems have been reported for CTC analysis.24–26 For example, a CTC affinity isolation strategy was combined with size-based separation.27–29 A modular configuration consists of a set of task-specific modules integrated together using a fluidic motherboard.30–34 Modular systems have also been reported for CTC analysis.35–37 The ability to be reconfigured to match the application need, selection of materials specific to optimize the performance of each module, and the selection of a fabrication protocol appropriate for the module have made modular microfluidic systems attractive for clinical assays.33
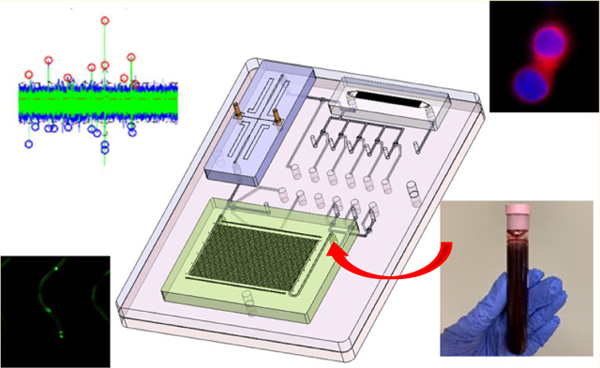
We report a system modularity chip for the analysis of rare targets (SMART-Chip), which contained three task-specific modules connected to a fluidic motherboard that can be used for the comprehensive analysis of liquid biopsy markers secured from clinical samples using CTCs as an example. Three task-specific modules consisted of a CTC enrichment module, impedance module, and imaging module. The SMART-Chip could perform the following steps: (i) affinity-select CTCs; (ii) release of the enriched biomarkers using a photocleavable (PC) linker;38 (iii) counting of the released cells using an impedance sensor that also determined cell viability; and (iv) immunophenotyping.39
The CTC enrichment module consisted of an array of high-aspect-ratio sinusoidally shaped microchannels arranged in a z-configuration40 and provided high cell recovery (>80%) with exquisite purity (>85%).41 The impedance module counted unlabeled single cells passing through a pair of electrodes to assess their viability and count them. The imaging module was used to physically entrap the enriched cells against pore structures, stain, and image the cells for phenotypic identification.39 The modules were connected to a fluidic motherboard, while valving structures present on the motherboard were programmed to direct the fluids precisely through the microchannels to automate sample processing.
We previously reported a microfluidic system for the enrichment of CTCs directly from a patient’s blood sample and their subsequent immunophenotyping.37 In that example, a series of microfluidic chips were connected via capillary tubing, which required plugging/unplugging different units during the assay that significantly complicated system operation. This was obviated in the present report by using membrane-based valves on a fluidic motherboard to control operation to minimize the need for operator intervention during the assay.
To establish the proof of concept of the SMART-Chip, healthy donor blood spiked with cancer cells (SKBR3) was analyzed by the system. Clinical validation of the system was performed by processing blood samples obtained from CRC and PDAC patients. The SMART-Chip enabled enrichment and post-enrichment analysis in a closed environment to minimize sample loss and contamination with no operator intervention required except for sample addition. Moreover, the SMART-Chip was made using thermoplastics via microreplication. Thus, the system can be mass-produced with high compliancy,42 making it appropriate for one-time use applications as required for in vitro diagnostics. An additional benefit of the modular system we report herein is that the individual modules can also be used as standalone units.
RESULTS AND DISCUSSION
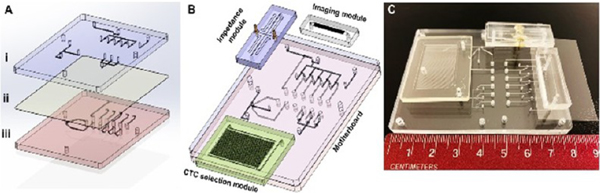
A picture of the SMART-Chip is shown in Figure 1. It contained three task-specific modules: affinity selection module for CTCs isolation using antibodies attached to the walls of the sinusoidal channels via a PC linker and their subsequent release using blue light, single-cell impedance sensor module, and imaging module. The imaging module was made from PDMS via soft lithography and covered with a thin glass plate. The other two modules were made from a thermoplastic. The CTC selection module was made from COC because COC offers high loading of antibodies following UV/O3 activation of the COC that results in high target cell recovery.40 The impedance module was made from PMMA but could have been made from any thermoplastic. Both modules were produced via hot embossing. The fluidic motherboard was made from PMMA by direct high-precision micromilling. In the following sections, a description of the design and performance of the fluidic motherboard and task-specific modules are provided.
Figure 1.
Design and assembly of the SMART-Chip. (A) Both sides of a PDMS membrane layer (ii) and PMMA layers that contained microfeatures (i – fluidic layer and iii – pneumatic control layer) were UV/O3-treated for 10 min (22 mW/cm2). The PDMS membrane was sandwiched between the two PMMA layers. The valve seats and displacement chambers were aligned, and pressure (165 psi) was applied to conformally seal the three layers that comprised the motherboard. (B) 3D image of three task-specific modules (CTC selection module, impedance module, and imaging module) interfaced to the fluidic motherboard. (C) Picture of the assembled SMART-Chip including the three task-specific modules.
Fluidic Motherboard.
The fluidic motherboard performed the following functions: (i) a structural element to which all task-specific modules were connected; (ii) a path for transferring fluidic information to the task-specific modules; (iii) ports for sample and reagent introduction; and (iv) valving for precise directional fluid flow through the integrated system during the assay.
The fluidic motherboard consisted of three layers (Figure 1A). The fluidic network layer (i) was made from PMMA and also contained the valve seats. The pneumatic layer (iii), which was also made from PMMA, contained control lines and displacement chambers for valve actuation. Finally, the middle layer (ii) was made from a PDMS elastomer and served as the membrane for the 11 valves situated on the motherboard. The three layers were assembled after modifying their surfaces with UV/O3.43
Modules were connected to the fluidic motherboard (Figure 1B,C) using interconnects,31 which consisted of conical receiving ports in the motherboard and modules (Figure S1A,B) and Tefzel tubing, which was inserted into mating ports on the modules and motherboard to create the fluidic connection (Figure S1C) with minimal unswept volume (measured value ~20 nL, which was <1% of the total interconnect volume; Figure S1D).44
The valves incorporated into the SMART-Chip were normally closed, and we could apply additional pneumatic closing pressure to keep the valve closed under high volume flow rates.43 Applying vacuum to the control layer changed the position of the PDMS membrane to open the valve (Figure S2A). We prepared a single valve unit (Figure S2B) into a PMMA substrate and evaluated its performance by measuring the outlet flow rate at different forward pressures (Figure S2C). The valve was operated under two different pneumatic closing pressures, 25 and 35 kPa. At 25 kPa pneumatic closing pressure, the valve was able to impede the fluid passage until the forward fluidic pressure reached 40 kPa (Figure S2D). At 35 kPa closing pressure, the valve was able to hold the forward fluidic pressure up to 60 kPa without leakage (Figure S2E).
CTC Selection Module.
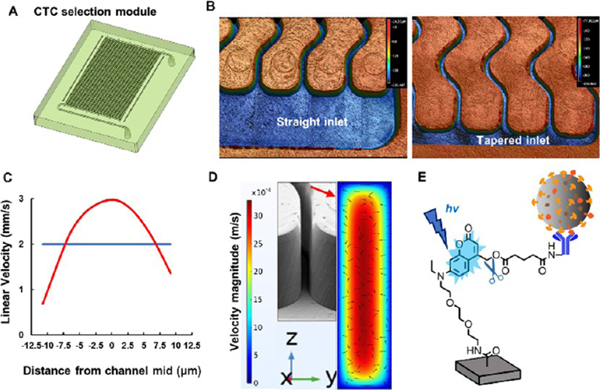
The CTC selection module has been reported previously by our group and consisted of an array of sinusoidal microchannels (150 μm depth; 25 μm width, 30 mm length; Figure 2A).41 The attractive nature of this design is that it can process whole blood directly yielding high recovery of CTCs with high purity.40 The selection module was replicated in COC because this material has excellent optical properties that enable efficient UV/O3 activation of the high-aspect-ratio microchannels for high loads of antibody onto the activated channels using the PC linker40 as well as high transmissivity in the blue region of the electromagnetic spectrum to allow for efficient CTC release.38
Figure 2.
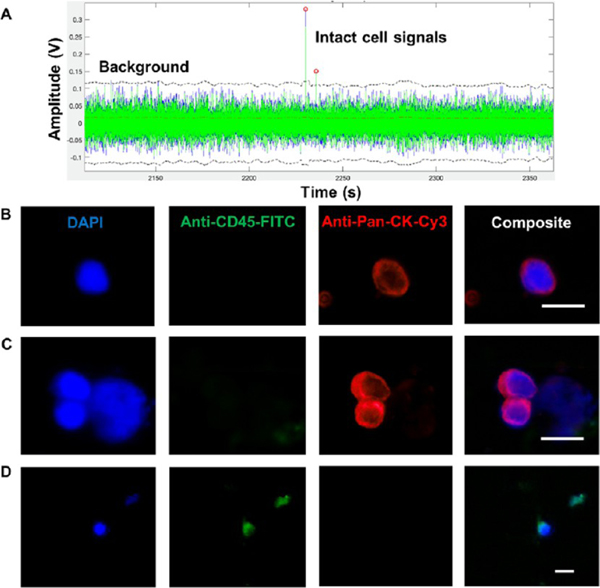
CTC selection module with high-aspect-ratio sinusoidal microchannels (aspect ratio = 6). (A) 3D design of the CTC selection module. The effective length of the selection channels was 3 cm. (B) 3D images of straight and tapered geometries for the inlet/outlet channels. The inlet/outlet channels were positioned orthogonal to the sinusoidal channel array (z-configuration). These images were acquired using a rapid scanning confocal microscope (Keyence). (C) COMSOL simulation of the average particle velocity (n = 10) in one sinusoidal channel over a distance of 1 cm. Particles were given an initial velocity of 2 mm/s (blue line), and as they traveled through the selection channel, their velocities changed with respect to the distance from the wall (red line). (D) COMSOL simulation with arrows indicating the direction and velocity magnitude of the fluid in the selection channel. As the fluid travels through the sinusoid apex, the fluid is directed toward both the inner and outer walls of the channel increasing the probability of antigen–antibody interaction. The arrows show the velocity profile in the yz direction. (E) Coumarin-based photocleavable linker that was used to immobilize antibodies to the cell selection module’s surface. After the affinity selection of the CTCs, the PC linker was exposed to blue light (400–450 nm) to cleave the linker and release the antibody–CTC complex from the module’s wall.
An array of 50 sinusoidal microchannels were organized in the so-called z-configuration. In this arrangement, inlet and outlet channels were positioned orthogonal to the sinusoidal micro-channel array. We have demonstrated that uniform translational velocity in the sinusoidal channels is required for high CTC recovery because the cell velocity must be close to an optimal value based on the binding kinetics of the antigen–antibody pair as well as the delivery rate of CTCs to the surface-bound antibodies.41,45 Uniformity of the flow depends upon the proper design of the inlet/outlet channels (Figure 2B).46 COMSOL simulations showed that tapered inlet/outlet channels provided uniform cell translational velocity within the sinusoidal channels compared to nontapered inlet/outlet channels.47
COMSOL simulations showed that a cell traveling close to the channel wall had a reduced linear velocity as compared to distances further from the wall due to fully developed laminar flow (Figure 2C), which increased the probability of antigen–antibody interaction. The centrifugal force acting on a 16 μm diameter cell at a velocity of 3 mm/s traveling through a sinusoidal channel having a radius of curvature of 125 μm (at the apex of the curve section) was calculated to be 0.162 pN.47 This centrifugal force caused the cell to be pushed toward the wall, which increased its interaction probability with the surface-bound capture antibodies. Moreover, as the cell traveled through alternating curves, the force acting on the translating cell resulted in a net centrifugal force being directed toward the channel walls. However, the lift forces generated by the channel walls directed the cells away from the wall. The two opposing forces resulted in a net force toward the sinusoidal channel wall due to the higher centrifugal forces (Figure 2D).
CTCs were affinity-selected using the appropriate antibody, such as anti-EpCAM antibodies. UV/O3 treatment generated surface-confined carboxylic acid groups that can react with primary amine containing entities using EDC/NHS chemistry48,49 and a Coumarin-based heterobifunctional PC linker (Figure 2E). The anti-EpCAM-PC linker selects CTCs with high recovery (73%) and purity (>85%). The affinity-selected CTCs were released for downstream analysis rapidly (2 min) and efficiently (>90%) by exposing the selection module to blue light (λmax = 412 nm, 32 ± 4 mW/cm2). Visible blue light as opposed to UV-light was used to minimize any damage to the nucleic acids contained within a CTC.50,51
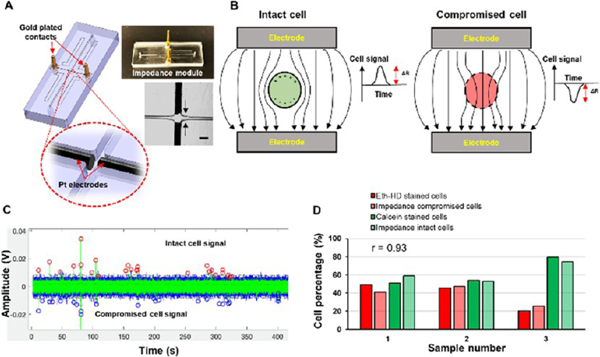
Impedance Sensor Module.
Impedance sensing can be used as a label-free strategy for the detection of single cells.41 The impedance sensor module consisted of a single fluidic channel and two Pt electrodes placed into microchannels positioned orthogonally to the fluidic channel (Figure 3A). At low waveform frequencies (<100 kHz), cells are treated as insulating particles due to high cell membrane resistance and a signal produced by the passage of an intact cell through the detector can be expressed as:52,53
where ΔR is the change in resistance between the electrode pair, ρc is the resistivity of the cell, ρm is the resistivity of the medium, dc is the cell diameter, and Ael is the area of the electrodes. When no cell was present between the electrodes, the signal was proportional to the resistance of the buffer. Every cell that has an intact cell membrane and passing between the electrodes displaces a finite volume of the buffer. The volume displaced by a cell, which has an intact cell membrane, has a higher resistance than the corresponding volume of the buffer alone producing a positive impedance signal in our case. When the cell membrane was compromised, a lower resistivity than the buffer results in. Thus, the impedance signal had a negative polarity (Figure 3B).
Figure 3.
Impedance module and single cell impedance sensing. (A) Impedance module with Pt electrodes and gold-plated electrical contacts. Pt electrodes were situated orthogonal to the fluidic channel with ~50 μm distance. The scale bar is equivalent to 100 μm. (B) Cells with an intact membrane have a higher resistance (Rcell) than the buffer (Rsol) and ΔR > 0, and the detector produced a positive signal. When the membrane was compromised, Rcell < Rsol, making ΔR < 0. This gave a negative polarity signal. (C) SKBR3 cells with an intact cell membrane gave positive polarity signals (red circles), while compromised cells yielded negative signals (blue circles). (D) Cells recovered after the impedance sensing were subjected to Eth-HD1 and Calcein-AM staining. The results were compared to the impedance signals for compromised and intact cells. Strong correlation (r = 0.93) was observed between the two methods. The impedance traces were collected for cells suspended in 1× TG buffer (pH 8.3). The measurements were recorded at a frequency of 40 kHz.
To verify that the polarity of the impedance signal was indicative of membrane integrity, freshly harvested SKBR3 cells were suspended in 1× TG buffer. Cells were flowed through the impedance module while recording the electrical signal (Figure 3C). The cells were recovered following their passage through the impedance module, and their viability was evaluated using cell viability fluorescent markers ethidium homodimer 1 (Eth-HD1) and Calcein-AM. The results obtained from the impedance sensing and staining methods were compared with the results shown in Figure 3D. The percentages of Eth-HD1-stained cells were similar to the percentages of impedance negative signals with a Pearson correlation of 0.93 (n = 3), and the percentages of Calcein-AM-stained cells were similar to the percentages of impedance positive signals with a Pearson correlation of 0.93 (n = 3). Thus, impedance sensing can be used as a label-free strategy to count cells and at the same time assess their viability.
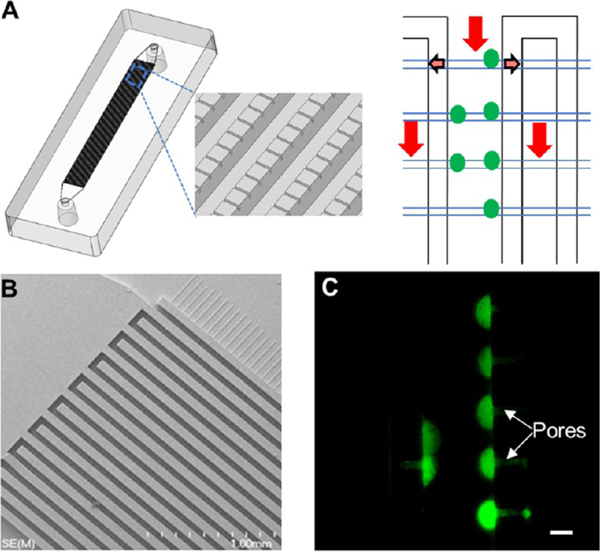
Imaging Module.
The imaging module39 was composed of two interleaving feed channels interconnected using an array of smaller channels positioned orthogonal to the feed channels (Figure 4A). Once the glass cover slip was bonded to the PDMS substrate, the small cross channels and the glass cover slip produced containment pores that could “entrap” cells larger than the pore size. PDMS and glass were selected to fabricate this module due to the low autofluorescence of glass and the simple O2/plasma treatment that can be used to bond the glass cover slip to the PDMS substrate. CTCs released from the selection module traveled through the impedance sensing module and were directed into the imaging module. CTCs, which are larger than the pore sizes, were physically entrapped near the pores and could be stained (DAPI, CD45, and pancytokeratins) and immunophenotyped for identification. This unique architecture enabled simple and rapid immunophenotyping of CTCs because entrapped cells were located within a common imaging plane and found at specific locations (i.e., near pore structures).
Figure 4.
Imaging module. (A) 3D image of a single bed imaging module populated with cell retaining pores. A cell suspension coming from the inlet (red arrow) of the imaging module entered into the outlet channels through the pores (faded arrows) while entrapping the cells at the inlet channels. (B) SEM image of lithographically patterned 2-level SU-8 mold for casting of PDMS to fabricate the imaging modules. (C) Isolation of live SKBR3 cells stained with Calcein-AM with a flow rate of 20 μL/min using the imaging module with 4 μm × 3.5 μm pore structures. Cells were retained near the pore structures. The image was taken using a 40× microscope objective. The scale bar is 15 μm.
Because the number of CTCs that can be affinity-selected from an epithelial cancer is limited (1–1000 CTCs per mL), single bed imaging modules employed 2400 or 7200 containment pores were used in this study (Figure S3A). Figure 4B shows an SEM of the fluidic network without a cover plate.
To establish the optimum design of the imaging module for CTC entrapment, we tested pore structures with 4, 6, and 8 μm widths and a fixed pore height of 3.5 μm at a 20 μL/min flow rate (Figure S3B,C). Live SKBR3 cells could be entrapped by the pores of 4 μm width (Figure 4C) with an efficiency of 98.4 ± 2.8% (n = 3). Six and 8 μm pore structures retained the live SKBR3 cells with an efficiency of 91.9 ± 13.3% (n = 3) and 82.6 ± 13.3 % (n = 3), respectively. Due to the higher efficiency of CTC entrapment, we used a pore size of 4 μm in this study.
Operation of the SMART-Chip.
Next, we evaluated the system’s ability to process whole blood spiked with CTCs. There are four major control steps associated with the assay including whole blood processing (Figure S4A), post selection wash (Figure S4B), impedance cell counting (Figure S4C), and immunophenotyping (Figure S4D). Each step was successfully carried out by the correct open/close state of the 11 valves as shown in Figure S4A–D. SKBR3 cells that have a high expression of EpCAM (Figure S5A) were used as a model for the CTCs to initially evaluate the operational capabilities of the SMART-Chip. Cells were spiked into healthy donor blood, and the SMART-Chip was used to isolate, count, assess viability, and immunophenotype the cells.
SKBR3 cell recovery (73%) was reported previously in the sinusoidal CTC selection modules that used the PC linker to immobilize anti-EpCAM antibodies (lower recovery of SKBR3 cells than of MCF-7 is due to differences in the EpCAM expression levels between these two cell lines).38 Affinity-enriched SKBR3 cells (Figure S5B) were then photoreleased by exposing the cell selection module to blue light (400–450 nm) for 2 min. Photoreleased cells were then directed through the impedance module and finally to the imaging module while recording the impedance signal (Figure S5C). Cells were physically entrapped in the imaging module, while the buffer solution passed to waste. Cells were then stained with anti-CD45 antibodies following fixing, permeabilizing, and staining with pan-cytokeratin. We counted 110 impedance signals in total, and all of them had a positive polarity (100% viable cells). We observed 112 SKBR3 cells in the imaging module (DAPI (+), FITC-CD45 (−), and Cy3-pan-CK (+), Figure S5D). We did not observe any white blood cells (CD45 (+) and pan-CK (−)) within the imaging module. In addition, we did not observe any DAPI (+) cells in the fluidic paths of the motherboard or at the interconnects, which implied that the SKBR3 cells were not trapped in or adhered to the fluidic network of the motherboard.
CTCs from Clinical Samples.
We isolated and enumerated CTCs from blood samples of patients diagnosed with metastatic colorectal cancer (mCRC) and pancreatic ductal adenocarcinoma (PDAC) using the SMART-Chip (see Table S1). Patients’ diseases were confirmed via computed tomography (CT) a week before CTC analysis and prior to surgery and chemotherapy.
Following affinity enrichment, CTCs were photoreleased for cell counting and immunophenotypic identification. In CRC patient 1, we were able to identify 7 impedance signals in total (part of the impedance signal trace is shown in Figure 5A) and 9 CTCs after immunophenotyping; this number falls within the range of CTCs we have enriched from mCRC patients using anti-EpCAM mAbs.54 All of the impedance signals were positive in polarity, which is in line with the fact that this patient had not received chemotherapy that can compromise cancer cell viability.54 Since first diagnosis, the patient’s serum was tested for carcinoembryonic antigen (CEA), a tumor marker predicting the treatment response and survival;55 CEA levels in this patient’s serum were normal (0.7–1.0 ng/mL). While an elevated serum CEA (>3 ng/mL) was found in 47% of CRC patients,55 for this patient, the CEA test was not informative. In blood collected from CRC patient 2, we detected 4 CTCs and all were determined to be nonviable based on the polarity of the signal from the impedance sensor. The CTC test was performed 6 months following hiperthermic intraperitoneal chemotherapy (HIPEC) administered to the patient following cytoreductive surgery to eliminate residual cancer cells from the abdominal cavity. For this patient before the cytoreductive surgery and HIPEC, we detected 5 times more CTCs (19 cells) than measured here with 85% of CTCs being viable. Three weeks following surgery and HIPEC, there was a significant switch in CTC viability as 85% of detected CTCs were identified as nonviable.
Figure 5.
Impedance counting and identification of CTCs by immunophenotyping. The CTCs were enriched from a blood sample secured from a metastatic colorectal cancer patient. Photoreleased CTCs were directed through the impedance and imaging modules. (A) A section of the impedance signal trace collected for EpCAM-enriched CTCs. Impedance signals ascribed to CTCs when the signal-to-noise ratio exceeded 7. All CTC-associated signals (7) were positive in polarity. (B–D) CTCs contained within the imaging module were stained with a panel of markers: DAPI (40 μg/mL), anti-CD45-FITC (2.5 μg/mL), and anti-pan-CK-Cy3 (0.01 mg/mL). CTCs were identified as DAPI (+), CD45 (−), and pan-CK (+), while white blood cells were identified as DAPI (+), CD45 (+), and pan-CK (−). (B) Single CTC; (C) cluster of CTCs; (D) a white blood cell. The images were taken using 20× and 40× objectives. Scale bars in the images are 15 μm.
In the metastatic PDAC patient, we detected 6 CTCs in the imaging module and 7 impedance signals. Four impedance signals were positive and three were negative in polarity, suggesting a compromised viability of ~40% of CTCs. For this patient, CTC analysis was performed at baseline (i.e., before chemotherapy). Typically, EpCAM (+) CTCs in PDAC patients range between 3 and 105/mL.54
We processed 2 mL of blood in these experiments, and 0–3 WBCs were observed following immunophenotyping. The high purity observed in this study is not surprising because we process blood at 40 dynes per cm2 shear rates that removes most white blood cells nonspecifically bound to the wall of the CTC selection module, and also, after photocleavage, those cells would most likely remain bound to the channel walls. The shear force applied in the post-selection wash (13 dynes per cm2) does not affect the interaction of CTC antigens with their cognizant antibodies.41,47 We observed individual CTCs (Figure 5B) and microclusters of CTCs (Figure 5C). CTC microclusters are recognized as important prognostic markers in cancer because their molecular characterization offers insights into the mechanisms of treatment resistance and increased metastatic potential.56,57 We also observed only a few white blood cells in the imaging module (Figure 5D). A healthy blood sample was processed as a control for these experiments, and no CTCs were identified (data not shown).
CONCLUSIONS
In this study, we reported a modular microfluidic system (SMART-Chip) that could carry out the entire processing steps required for analyzing liquid biopsy samples and used CTCs as an example. The processing steps carried out by the SMART-Chip included affinity selection, photorelease, enumeration, viability assessment, and immunophenotyping. The three task-specific modules integrated into the system included: (i) CTC selection module, (ii) impedance module, and (iii) imaging module that were connected to a fluidic motherboard using interconnects that minimized unswept volumes. The fluidic motherboard contained channels that provided fluidic paths between each module and a valving system that was pneumatically operated so that precise control of the fluid path through the system could be achieved for each processing step. Particularly attractive with the modular nature of this integrated system is that alternative modules can be “plugged” into the fluidic motherboard to process other liquid biopsy markers, such as cfDNA58 or EVs.59
A proof of concept was demonstrated by processing a healthy donor blood sample spiked with SKBR3 cells. The SMART-Chip was able to perform the entire assay yielding a highly pure isolate (0 WBCs per 1 mL of blood) with no observable loss of rare cells due to unswept volumes or nonspecific adsorption artifacts. This was affected by using the proper material for each module and motherboard, proper design of the fluidic interconnects, and engineering surfaces to make them accommodating for the biological sample, which was whole blood.
We also tested the clinical utility of the SMART-Chip by processing blood samples obtained from CRC and PDAC patients. We also found high purity in the rare cell isolates (0–3 WBCs per 2 mL of blood). Blood samples were processed at a throughput of 1.5 mL/h, but the throughput can be improved by using a 320 channel version of the CTC selection device (10 mL/h) without sacrificing the analytical figures-of-merit.37
The SMART-Chip and its modular format provided several advantages: (1) The system was made from plastics using microreplication so that all components could be produced in a high production mode and at low cost to make it appropriate for in vitro diagnostics that requires disposable devices. Even the membrane valves were all plastic. (2) There is full process automation of the assay with no requirement of transferring the sample from one device to another. For example, our manual method for CTC processing required off-loading the photoreleased cells into a microfuge tube, cytospinning the released cells onto a microscope slide, placing the microscope slide with deposited cells into an autostainer, and finally situating the slide on a microscope for fluorescence imaging. Thus, the SMART-Chip reduces sample loss and/or contamination, which is especially important for the analysis of rare targets. (3) The modular architecture of the SMART-Chip allows for reconfiguration by placing different fluidic modules onto the motherboard without requiring the need to re-engineer the entire system. As an example, the SMART-Chip can be reconfigured to isolate EVs from a liquid biopsy sample59 and count them using a nanocoulter counter.60,61 (4) The modular approach enables the ability to select the substrate material and fabrication modality to suit the task requirement. As an example, we used different materials for the modules of the SMART-Chip and each utilized a different fabrication method. (5) The SMART-Chip is flexible in its operational mode. For example, following selection and enumeration, the biomarkers can be shuttled to another module for molecular processing. (6) It has the ability to reduce the CTC assay time. In this case, manual processing of CTCs in our hands required ~8 h,37 and using the SMART-Chip the processing time was 3.5 h. (7) The individual modules can be used as standalone units as well without requiring any structural modifications.
We should note that in our past work using a plastic-based modular microfluidic system,31 the challenge was the polycarbonate membrane layer used, which resulted in poor process yield rates (yield rate < 20% for the polycarbonate membrane compared to 100% using the PMMA/PDMS/PMMA valves), the need for mechanical solenoids to actuate the polycarbonate valves due to polycarbonate’s higher Young’s modulus than PDMS’s, and the limited number of times the polycarbonate valves could be actuated (<10 compared to 77 herein).
We are now directing our efforts toward employing injection molding as the microreplication method for both the modules and motherboard due to the high-scale production capacity with reduced unit cost it offers compared to hot embossing, PDMS casting, and direct milling as used here.62,63 In addition, we are developing plastic modules that can provide molecular information from the enriched targets to also provide information on nucleic acids packaged into the rare cells.
METHODS
Reagents and materials used, experimental procedures for cell culture and antigen expression analysis, fabrication of the modules and fluidic motherboard, surface attachment of monoclonal antibodies using PC linkers, microfabrication of the motherboard and membrane valve and characterization, evaluating the relationship between impedance signal polarity and cell viability, and determination of cell retention efficiency in the imaging module are in the Supporting Information. Information on patient samples is in the Supporting Information as well.
SMART-Chip Assembly.
The motherboard was fabricated in three layers and consisted of a fluidic layer, pneumatic control layer (both of these layers were made of PMMA), and a membrane layer consisting of a PDMS elastomer (Figure 1A). The fluidic layer contained the fluidic network and valve seats (11 total valves in the system) as well as conical connecting ports for the three modules. The pneumatic control layer also contained displacement chambers. Conical ports (top diameter = 1.706 mm, bottom diameter = 1.444 mm, depth = 2.2 mm) were milled from the back of the PMMA fluidic layer (d = 0.045″, see Figure S1). These ports were used to connect individual modules to the motherboard. After direct milling of the required structures in the PMMA layers, the substrates were cleaned with 10% micro 90, IPA, and nanopure water and finally dried with N2. The PMMA fluidic layer and PDMS membrane were UV/O3-activated (22 mW/cm2) for 10 min, and the activated surface of the PDMS membrane was placed across the PMMA fluidic layer. This PMMA/PDMS assembly was then UV/O3-modified and aligned to the UV/O3-activated PMMA pneumatic control layer. The PMMA/PDMS/PMMA assembly was then subjected to pressure (165 psi) using a PHI Precision Press (City of Industry, CA) to facilitate compression bonding between the three layers (Figure 1B).
The CTC selection module, impedance module, and imaging module were connected to the motherboard (see Figure 1C) using semirigid Tefzel tubing.31 PEEK tubing was then connected to the motherboard as fluidic and pneumatic inputs, respectively. PEEK tubing (OD 1/16″) from the motherboard was connected to a solenoid control system (SCS). The SCS contained 11 solenoids (3/2way, Humphrey, M1533724VDC) connected to a manifold (Humphrey, 150 M12), and the manifold was connected to a pressure and vacuum source. Individual solenoids were connected to controller hardware (Instrumental Design Lab, KU) using 2-pin Molex C-GRID connectors. The controller hardware was interfaced to controlling software using a virtual serial port connected to a computer via a USB cable. Software was developed via Visual studio.
System Validation Using SKBR3 Cells as CTC Surrogates.
Healthy blood samples were provided by the Biospecimen Repository Core Facility (BRCF) at KU Cancer Center under an IRB approved protocol (HSC #5929). SKBR3 cells were stained with a nuclear stain (DAPI, 40 μg/mL) and incubated for 15 min at room temperature. Modules were connected to the motherboard, and the SMART-Chip was washed with 0.5% BSA/PBS (0.1 mL, 20 μL/min) prior to blood infusion. Stained cells were spiked into healthy blood and hydrodynamically introduced (1 mL, 25 μL/min) into the CTC selection module that was surface-decorated with anti-EpCAM monoclonal antibodies via a photocleavable (PC) linker.38 Following blood infusion, the module was rinsed with 1 mL of 0.5% BSA/PBS (50 μL/min) to remove any nonspecifically bound material. Next, 1× Tris Glycine (TG) buffer (pH 8.3, 0.1 mL) was infused into the SMART-Chip at 20 μL/min. Blood and the wash buffers were directed to waste using the appropriate open/close status of the PDMS membrane valves. The CTC selection module was exposed to blue light (34 ± 4 mW/cm2, 400–450 nm) for 2 min to release the affinity selected SKBR3 cells and swept into subsequent modules.
The released cells were swept through the impedance module generating an electrical signal based on cell membrane integrity. Impedance signatures were counted when the signal-to-noise ratio exceeded 7, and cell viability was determined by the polarity of the impedance signal. The data was acquired using in-house built electronics.41
After the impedance module, the enriched cells were directed into the imaging module. Entrapped cells were stained using different fluorescently labeled markers as reported previously.37 The stained cells were imaged using a Keyence BZ-X710 microscope (Keyence Corporation, Itasca, IL, USA) equipped with BZ-X filters. Images were collected with 20× and 40× objectives using exposure times of 50 ms for DAPI (nuclear stain), 500 ms for FITC (CD45), and 1500 ms for Cy3 (cytokeratins). Images were analyzed using a BZ-X analyzer (Keyence Corporation).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support from the University of Kansas (KU) Cancer Center’s Biospecimen Repository Core for obtaining human specimens and the Instrumental Design Lab at KU for the valve control hardware and interface software. We are appreciative to patients and healthy donors for their blood donations. We would like to thank the synthetic biology core facility at KU for synthesizing the PC linker. We also thank the Physics machine shop at KU for milling fluidic motherboards. The authors are grateful for support from the KU Cancer Center’s Support Grant.
Funding
Funds came from R33-CA235597, P41-EB020594, P20-GM130423, and P30-CA168524.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.0c02728.
Reagents and materials used, device fabrication, experimental details, and supplementary Figures S1–S5 and Table S1 (PDF)
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acssensors.0c02728
Contributor Information
Thilanga N. Pahattuge, Department of Chemistry, University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States
Ian M. Freed, Department of Chemistry, University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States
Mateusz L. Hupert, BioFluidica, Inc., Lawrence, Kansas 66045, United States
Swarnagowri Vaidyanathan, Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States; Department of BioEngineering, University of Kansas, Lawrence, Kansas 66045, United States.
Katie Childers, Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States; Department of BioEngineering, University of Kansas, Lawrence, Kansas 66045, United States.
Malgorzata A. Witek, Department of Chemistry, University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States
Kumuditha Weerakoon-Ratnayake, Department of Chemistry, University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States.
Daniel Park, Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States; Department of Mechanical & Industrial Engineering, Louisiana State University, Baton Rouge, Louisiana 70803, United States.
Anup Kasi, Department of Medical Oncology, University of Kansas Medical Center, Kansas City, Kansas 66160, United States.
Mazin F Al-Kasspooles, Department of Medical Oncology, University of Kansas Medical Center, Kansas City, Kansas 66160, United States.
Michael C. Murphy, Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States; Department of Mechanical & Industrial Engineering, Louisiana State University, Baton Rouge, Louisiana 70803, United States.
Steven A. Soper, Department of Chemistry, University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multi-scale Systems for Precision Medicine, University of Kansas, Lawrence, Kansas 66045, United States; BioFluidica, Inc., Lawrence, Kansas 66045, United States; Department of BioEngineering, University of Kansas, Lawrence, Kansas 66045, United States; Department of Mechanical Engineering, University of Kansas, Lawrence, Kansas 66045, United States; Department of Cancer Biology and KU Cancer Center, University of Kansas Medical Center, Kansas City, Kansas 66160, United States.
REFERENCES
- (1).Macias M; Alegre E; Diaz-Lagares A; Patino A; Perez-Gracia JL; Sanmamed M; Lopez-Lopez R; Varo N; Gonzalez A. Liquid Biopsy: From Basic Research to Clinical Practice. In Advances in Clinical Chemistry, Vol 83; Makowski GS, Ed. 2018; Vol. 83, pp. 73–119. [DOI] [PubMed] [Google Scholar]
- (2).Jeffrey SS; Toner M. Liquid biopsy: a perspective for probing blood for cancer. Lab Chip 2019, 19, 548–549. [DOI] [PubMed] [Google Scholar]
- (3).Wang S; Thomas A; Lee E; Yang S; Cheng X; Liu Y. Highly efficient and selective isolation of rare tumor cells using a microfluidic chip with wavy-herringbone micro-patterned surfaces. Analyst 2016, 141, 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yap TA; Lorente D; Omlin A; Olmos D; De Bono JS Circulating tumor cells: a multifunctional biomarker. Clin. Cancer Res 2014, 20, 2553. [DOI] [PubMed] [Google Scholar]
- (5).Xiang N; Wang J; Li Q; Han Y; Huang D; Ni Z. Precise size-based cell separation via the coupling of inertial microfluidics and deterministic lateral displacement. Anal. Chem 2019, 91, 10328–10334. [DOI] [PubMed] [Google Scholar]
- (6).Fachin F; Spuhler P; Martel-Foley JM; Edd JF; Barber TA; Walsh J; Karabacak M; Pai V; Yu M; Smith K; Hwang H; Yang J; Shah S; Yarmush R; Sequist LV; Stott SL; Maheswaran S; Haber DA; Kapur R; Toner M. Monolithic chip for high-throughput blood cell depletion to sort rare circulating tumor cells. Sci. Rep 2017, 7, 10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Au SH; Edd J; Stoddard AE; Wong KHK; Fachin F; Maheswaran S; Haber DA; Stott SL; Kapur R; Toner M. Microfluidic isolation of circulating tumor cell clusters by size and asymmetry. Sci. Rep 2017, 7, 2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yang C; Zhang N; Wang S; Shi D; Zhang C; Liu K; Xiong B. Wedge-shaped microfluidic chip for circulating tumor cells isolation and its clinical significance in gastric cancer. J. Transl. Med 2018, 16, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kang Y-T; Kim YJ; Bu J; Chen S; Cho Y-H; Lee HM; Ryu CJ; Lim Y; Han S-W Epithelial and mesenchymal circulating tumor cell isolation and discrimination using dual-immunopatterned device with newly-developed anti-63B6 and anti-EpCAM. Sens Actuators B Chem 2018, 260, 320–330. [Google Scholar]
- (10).Zeinali M; Murlidhar V; Fouladdel S; Shao S; Zhao L; Cameron H; Bankhead A III; Shi J; Cuneo KC; Sahai V; Azizi E; Wicha MS; Hafner M; Simeone DM; Nagrath S. Profiling heterogeneous circulating tumor cells (CTC) populations in pancreatic cancer using a serial microfluidic CTC carpet chip. Adv. Biosyst 2018, 2, 1800228. [Google Scholar]
- (11).LeValley PJ; Tibbitt MW; Noren B; Kharkar P; Kloxin AM; Anseth KS; Toner M; Oakey J. Immunofunctional photo-degradable poly (ethylene glycol) hydrogel surfaces for the capture and release of rare cells. Colloids Surf., B 2019, 174, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Brassard D; Geissler M; Descarreaux M; Tremblay D; Daoud J; Clime L; Mounier M; Charlebois D; Veres T. Extraction of nucleic acids from blood: unveiling the potential of active pneumatic pumping in centrifugal microfluidics for integration and automation of sample preparation processes. Lab Chip 2019, 19, 1941–1952. [DOI] [PubMed] [Google Scholar]
- (13).Sunkara V; Kim C-J; Park J; Woo H-K; Kim D; Ha HK; Kim M-H; Son Y; Kim J-R; Cho Y-K Fully automated, label-free isolation of extracellular vesicles from whole blood for cancer diagnosis and monitoring. Theranostics 2019, 9, 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kim C-J; Ki DY; Park J; Sunkara V; Kim T-H; Min Y; Cho Y-K Fully automated platelet isolation on a centrifugal microfluidic device for molecular diagnostics. Lab Chip 2020, 20, 949–957. [DOI] [PubMed] [Google Scholar]
- (15).Park J-M; Kim MS; Moon H-S; Yoo CE; Park D; Kim YJ; Han K-Y; Lee J-Y; Oh JH; Kim SS; Park W-Y; Lee W-Y; Huh N. Fully automated circulating tumor cell isolation platform with large-volume capacity based on lab-on-a-disc. Anal. Chem 2014, 86, 3735–3742. [DOI] [PubMed] [Google Scholar]
- (16).Lee A; Park J; Lim M; Sunkara V; Kim SY; Kim GH; Kim M-H; Cho Y-K All-in-one centrifugal microfluidic device for size-selective circulating tumor cell isolation with high purity. Anal. Chem 2014, 86, 11349–11356. [DOI] [PubMed] [Google Scholar]
- (17).Aguirre GR; Efremov V; Kitsara M; Ducrée J. Integrated micromixer for incubation and separation of cancer cells on a centrifugal platform using inertial and dean forces. Microfluid. Nanofluid 2015, 18, 513–526. [Google Scholar]
- (18).Aeinehvand MM; Ibrahim F; harun SW; al-Faqheri W; Thio THG; Kazemzadeh A; Madou M. Latex micro-balloon pumping in centrifugal microfluidic platforms. Lab Chip 2014, 14, 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Aeinehvand MM; Weber L; Jiménez M; Palermo A; Bauer M; Loeffler FF; Ibrahim F; Breitling F; Korvink J; Madou M; Mager D; Martínez-Chapa SO Elastic reversible valves on centrifugal microfluidic platforms. Lab Chip 2019, 19, 1090–1100. [DOI] [PubMed] [Google Scholar]
- (20).Rodriguez-Moncayo R; Jimenez-Valdes RJ; Gonzalez-Suarez AM; Garcia-Cordero JL Integrated Microfluidic Device for Functional Secretory Immunophenotyping of Immune Cells. ACS sens. 2020, 5, 353–361. [DOI] [PubMed] [Google Scholar]
- (21).Lai X; Lu B; Zhang P; Zhang X; Pu Z; Yu H; Li D. Sticker Microfluidics: A Method for Fabrication of Customized Monolithic Microfluidics. ACS Biomater. Sci. Eng 2019, 5, 6801–6810. [DOI] [PubMed] [Google Scholar]
- (22).Huang S-P; Chuang Y-J; Lee W-B; Tsai Y-C; Lin C-N; Hsu K-F; Lee G-B An integrated microfluidic system for rapid, automatic and high-throughput staining of clinical tissue samples for diagnosis of ovarian cancer. Lab Chip 2020, 20, 1103–1109. [DOI] [PubMed] [Google Scholar]
- (23).Giménez-Gómez P; Fernández-Sánchez C; Baldi A. Microfluidic Modules with Integrated Solid-State Sensors for Reconfigurable Miniaturized Analysis Systems. ACS Omega 2019, 4, 6192–6198. [Google Scholar]
- (24).Karabacak NM; Spuhler PS; Fachin F; Lim EJ; Pai V; Ozkumur E; Martel JM; Kojic N; Smith K; Chen P.-i.; Yang J; Hwang H; Morgan B; Trautwein J; Barber TA; Stott SL; Maheswaran S; Kapur R; Haber DA; Toner M. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc 2014, 9, 694–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liu Z; Shu W; Chen R; Feng H; Hu Z; Wang Y; Tian G; Liu A; Chen Y. Integrated Microfluidic Chip for Rapid Capture and Drug Screening of Cancer Cells from Peripheral Blood. Integr. Cancer Biol. Res 2018, 2. [Google Scholar]
- (26).Dharmasiri U; Njoroge SK; Witek MA; Adebiyi MG; Kamande JW; Hupert ML; Barany F; Soper SA High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal. Chem 2011, 83, 2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chen K; Dopico P; Varillas J; Zhang J; George TJ; Fan ZH Integration of Lateral Filter Arrays with Immunoaffinity for Circulating-Tumor-Cell Isolation. Angew. Chem., Int. Ed 2019, 58, 7606–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Song Y; Shi Y; Huang M; Wang W; Wang Y; Cheng J; Lei Z; Zhu Z; Yang C. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem., Int. Ed 2019, 58, 2236–2240. [DOI] [PubMed] [Google Scholar]
- (29).Chen K; Amontree J; Varillas J; Zhang J; George TJ; Fan ZH Incorporation of lateral microfiltration with immunoaffinity for enhancing the capture efficiency of rare cells. Sci. Rep 2020, 10, 14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhuang B; Han J; Xiang G; Gan W; Wang S; Wang D; Wang L; Sun J; Li C-X; Liu P. A fully integrated and automated microsystem for rapid pharmacogenetic typing of multiple warfarin-related single-nucleotide polymorphisms. Lab Chip 2016, 16, 86–95. [DOI] [PubMed] [Google Scholar]
- (31).Wang H; Chen H-W; Hupert ML; Chen P-C; Datta P; Pittman TL; Goettert J; Murphy MC; Williams D; Barany F. Fully Integrated Thermoplastic Genosensor for the Highly Sensitive Detection and Identification of Multi-Drug-Resistant Tuberculosis. Angew. Chem., Int. Ed 2012, 51, 4349–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Millet LJ; Lucheon JD; Standaert RF; Retterer ST; Doktycz MJ Modular microfluidics for point-of-care protein purifications. Lab Chip 2015, 15, 1799–1811. [DOI] [PubMed] [Google Scholar]
- (33).Xie X; Maharjan S; Liu S; Zhang YS; Livermore C. A Modular, Reconfigurable Microfabricated Assembly Platform for Microfluidic Transport and Multitype Cell Culture and Drug Testing . Micromachines 2020, 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chen Y-S; Ma Y-D; Chen C; Shiesh S-C; Lee G-B An integrated microfluidic system for on-chip enrichment and quantification of circulating extracellular vesicles from whole blood. Lab Chip 2019, 19, 3305–3315. [DOI] [PubMed] [Google Scholar]
- (35).Lee TY; Hyun K-A; Kim S-I; Jung H-I An integrated microfluidic chip for one-step isolation of circulating tumor cells. Sens Actuators B Chem 2017, 238, 1144–1150. [Google Scholar]
- (36).Antfolk M; Kim SH; Koizumi S; Fujii T; Laurell T. Label-free single-cell separation and imaging of cancer cells using an integrated microfluidic system. Sci. Rep 2017, 7, 46507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kamande JW; Hupert ML; Witek MA; Wang H; Torphy RJ; Dharmasiri U; Njoroge SK; Jackson JM; Aufforth RD; Snavely A; Yeh JJ; Soper SA Modular microsystem for the isolation, enumeration, and phenotyping of circulating tumor cells in patients with pancreatic cancer. Anal. Chem 2013, 85, 9092–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Pahattuge TN; Jackson JM; Digamber R; Wijerathne H; Brown V; Witek MA; Perera C; Givens RS; Peterson BR; Soper SA Visible photorelease of liquid biopsy markers following microfluidic affinity-enrichment. Chem. Commun 2020, 56, 4098–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).M. Weerakoon-Ratnayake K; Vaidyanathan S; Larkey N; Dathathreya K; Hu M; Jose J; Mog S; August K; K. Godwin A; L. Hupert M; A. Witek M; A. Soper S. Microfluidic Device for On-Chip Immunophenotyping and Cytogenetic Analysis of Rare Biological Cells. Cell 2020, 9, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hupert ML; Jackson JM; Wang H; Witek MA; Kamande J; Milowsky MI; Whang YE; Soper SA Arrays of high-aspect ratio microchannels for high-throughput isolation of circulating tumor cells (CTCs). Microsyst. Technol 2014, 20, 1815–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Adams AA; Okagbare PI; Feng J; Hupert ML; Patterson D; Göttert J; McCarley RL; Nikitopoulos D; Murphy MC; Soper SA Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc 2008, 130, 8633–8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Tsao C-W Polymer Microfluidics: Simple, Low-Cost Fabrication Process Bridging Academic Lab Research to Commercialized Production. Micromachines (Basel) 2016, 7, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zhang W; Lin S; Wang C; Hu J; Li C; Zhuang Z; Zhou Y; Mathies RA; Yang CJ PMMA/PDMS valves and pumps for disposable microfluidics. Lab Chip 2009, 9, 3088–3094. [DOI] [PubMed] [Google Scholar]
- (44).Chen Y-W; Wang H; Hupert M; Witek M; Dharmasiri U; Pingle MR; Barany F; Soper SA Modular microfluidic system fabricated in thermoplastics for the strain-specific detection of bacterial pathogens. Lab Chip 2012, 12, 3348–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chang K-C; Hammer DA The forward rate of binding of surface-tethered reactants: effect of relative motion between two surfaces. Biophys. J 1999, 76, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Jackson JM; Hupert ML; Soper SA Discrete geometry optimization for reducing flow non-uniformity, asymmetry, and parasitic minor loss pressure drops in Z-type configurations of fuel cells. J. Power Sources 2014, 269, 274–283. [Google Scholar]
- (47).Jackson JM; Witek MA; Soper SA, Sinusoidal microchannels with high aspect ratios for CTC selection and analysis. Circulating Tumor Cells: Isolation and Analysis; Wiley: 2016, 85–126, DOI: 10.1002/9781119244554.ch4. [DOI] [Google Scholar]
- (48).McCarley RL; Vaidya B; Wei S; Smith AF; Patel AB; Feng J; Murphy MC; Soper SA Resist-free patterning of surface architectures in polymer-based microanalytical devices. J. Am. Chem. Soc 2005, 127, 842–843. [DOI] [PubMed] [Google Scholar]
- (49).Wei S; Vaidya B; Patel AB; Soper SA; McCarley RL Photochemically patterned poly(methyl methacrylate) surfaces used in the fabrication of microanalytical devices. J. Phys. Chem. B 2005, 109, 16988–16996. [DOI] [PubMed] [Google Scholar]
- (50).Rünger TM; Kappes UP Mechanisms of mutation formation with long-wave ultraviolet light (UVA). Photodermatol. Photoimmunol. Photomed 2008, 24, 2–10. [DOI] [PubMed] [Google Scholar]
- (51).Sage E; Girard P-M; Francesconi S. Unravelling UVA-induced mutagenesis. Photochem. Photobiol. Sci 2012, 11, 74–80. [DOI] [PubMed] [Google Scholar]
- (52).Sun T; Morgan H. Single-cell microfluidic impedance cytometry: a review. Microfluid. Nanofluid 2010, 8, 423–443. [Google Scholar]
- (53).Gawad S; Schild L; Renaud P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip 2001, 1, 76–82. [DOI] [PubMed] [Google Scholar]
- (54).Witek MA; Aufforth RD; Wang H; Kamande JW; Jackson JM; Pullagurla SR; Hupert ML; Usary J; Wysham WZ; Hilliard D; Montgomery S; Bae-Jump V; Carey LA; Gehrig PA; Milowsky MI; Perou CM; Soper JT; Whang YE; Yeh JJ; Martin G; Soper SA Discrete microfluidics for the isolation of circulating tumor cell subpopulations targeting fibroblast activation protein alpha and epithelial cell adhesion molecule. npj Precis. Oncol. 2017, 1, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Cho WK; Choi DH; Park HC; Park W; Yu JI; Park YS; Park JO; Lim HY; Kang WK; Kim HC; Cho YB; Yun SH; Lee WY Elevated CEA is associated with worse survival in recurrent rectal cancer. Oncotarget 2017, 8, 105936–105941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Amintas S; Bedel A; Moreau-Gaudry F; Boutin J; Buscail L; Merlio J-P; Vendrely V; Dabernat S; Buscail E. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int. J. Mol. Sci 2020, 21, 2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Hong Y; Fang F; Zhang Q. Circulating tumor cell clusters: What we know and what we expect (Review). Int. J. Oncol 2016, 49, 2206–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Campos CDM; Gamage SST; Jackson JM; Witek MA; Park DS; Murphy MC; Godwin AK; Soper SA Microfluidic-based solid phase extraction of cell free DNA. Lab Chip 2018, 18, 3459–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Wijerathne H; Witek MA; Jackson JM; Brown V; Hupert ML; Herrera K; Kramer C; Davidow AE; Li Y; Baird AE; Murphy MC; Soper SA Affinity enrichment of extracellular vesicles from plasma reveals mRNA changes associated with acute ischemic stroke. Commun. Biol 2020, 3, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Anderson W; Lane R; Korbie D; Trau M. Observations of Tunable Resistive Pulse Sensing for Exosome Analysis: Improving System Sensitivity and Stability. Langmuir 2015, 31, 6577–6587. [DOI] [PubMed] [Google Scholar]
- (61).Maas SL; Broekman ML; de Vrij J., Tunable resistive pulse sensing for the characterization of extracellular vesicles. In Exosomes and Microvesicles; Springer: 2017; pp. 21–33, DOI: 10.1007/978-1-4939-6728-5_2. [DOI] [PubMed] [Google Scholar]
- (62).Piotter V; Hanemann T; Ruprecht R; Hausselt J. Injection molding and related techniques for fabrication of microstructures. Microsyst. Technol 1997, 3, 129–133. [Google Scholar]
- (63).Heckele M; Schomburg WK Review on micro molding of thermoplastic polymers. J. Micromech. Microeng 2004, 14, R1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.