Significance
Decline of mitochondrial function may underlie the pathogenesis of many age-related diseases, such as Huntington’s disease (HD). Mitochondrial oxidative phosphorylation (OXPHOS) system is encoded partially by the mitochondrial genome (mtDNA). By investigating mtDNA in lymphoblast and blood samples of HD patients, we found that the expansion of OXPHOS-impairing mtDNA heteroplasmies (coexistence of mutated and wild-type mtDNA) is a molecular feature associated with the functional, motor, and cognitive aspects of HD progression, suggesting that improving mtDNA quality or restoring mitochondrial function could be a potential target for HD treatments.
Keywords: mitochondrial DNA, Huntington’s disease, sequencing
Abstract
Mitochondrial dysfunction is found in the brain and peripheral tissues of patients diagnosed with Huntington’s disease (HD), an irreversible neurodegenerative disease of which aging is a major risk factor. Mitochondrial function is encoded by not only nuclear DNA but also DNA within mitochondria (mtDNA). Expansion of mtDNA heteroplasmies (coexistence of mutated and wild-type mtDNA) can contribute to age-related decline of mitochondrial function but has not been systematically investigated in HD. Here, by using a sensitive mtDNA-targeted sequencing method, we studied mtDNA heteroplasmies in lymphoblasts and longitudinal blood samples of HD patients. We found a significant increase in the fraction of mtDNA heteroplasmies with predicted pathogenicity in lymphoblasts from 1,549 HD patients relative to lymphoblasts from 182 healthy individuals. The increased fraction of pathogenic mtDNA heteroplasmies in HD lymphoblasts also correlated with advancing HD stages and worsened disease severity measured by HD motor function, cognitive function, and functional capacity. Of note, elongated CAG repeats in HTT promoted age-dependent expansion of pathogenic mtDNA heteroplasmies in HD lymphoblasts. We then confirmed in longitudinal blood samples of 169 HD patients that expansion of pathogenic mtDNA heteroplasmies was correlated with decline in functional capacity and exacerbation of HD motor and cognitive functions during a median follow-up of 6 y. The results of our study indicate accelerated decline of mtDNA quality in HD, and highlight monitoring mtDNA heteroplasmies longitudinally as a way to investigate the progressive decline of mitochondrial function in aging and age-related diseases.
Huntington’s disease (HD) is a monogenic disorder caused by the expansion of cytosine–adenine–guanine trinucleotide (CAG) repeats in the HTT gene at chromosome 4p16.3 (1). Although HTT is expressed in various tissues, the brain, particularly the striatum, is vulnerable to mutant huntingtin (mHTT)-associated toxicity (2). The average age at onset of the characteristic motor symptoms of HD is between 40 and 50 y old, followed by a progressive decline of motor, cognitive, and psychiatric functions for an average of 20 y prior to death (3).
The biological processes that determine the onset and progression of HD are still elusive. Recent studies suggest that mitochondrial dysfunction may be involved in HD pathogenesis (4, 5). Mitochondria are subcellular organelles of eukaryotes, which play vital roles in maintaining energetic and metabolic homeostasis (6, 7). Evidence for mitochondrial dysfunction in HD was first reported in the postmortem brain of HD patients, which show low mitochondrial oxidative phosphorylation (OXPHOS) protein activity and energy deficits (8–10). Mitochondrial dysfunction was further found in peripheral tissues and cell lines of HD patients, such as blood, lymphoblasts, skeletal muscle, and skin fibroblasts (11–17).
Several molecular mechanisms have been proposed to connect mHTT to mitochondrial dysfunction. Studies in HD knockin mice indicate that toxic fragments derived from mHTT can suppress mitochondrial biogenesis and energy metabolism (18). mHTT has also been found to physically interact with mitochondria, reducing mitochondrial membrane potential (13, 19). Furthermore, mHTT may stimulate mitochondrial network fragmentation (20–22), and it has recently been found to impair mitophagy (23–28), an evolutionarily conserved quality control system in eukaryotes to selectively remove dysfunctional mitochondria (29). Perturbation of mitochondrial tubular networks, morphology, and mitophagy are pathological features common to various neurodegenerative diseases (30, 31).
Mitochondrial function is determined not only by the nuclear genome but also by the mitochondrial genome (mtDNA). Human mtDNA is a 16.6-kb circular DNA located within mitochondria. It encodes 13 evolutionarily conserved proteins in four of the five OXPHOS protein complexes (32). The accumulation of mtDNA mutations in somatic tissues has been suggested as a possible driver of age-related mitochondrial dysfunction (33). Transgenic mice with an increased level of mtDNA mutations manifest progeroid phenotypes and early neurodegeneration that resemble human aging (34, 35). Clonal expansion of preexisting mtDNA mutations in somatic tissues has also been shown to contribute to accelerated mitochondrial aging and OXPHOS defects in human diseases (36, 37).
Because there are multiple copies of mtDNA in a single cell, mutations can arise and coexist with wild-type mtDNA in a state called heteroplasmy, which has been linked to a variety of mitochondrial disorders in humans (32, 38). Our previous study on lymphoblasts from the 1,000 Genomes project indicates that about 90% of individuals in the general population carry at least one heteroplasmy in mtDNA, and purifying selection keeps most pathogenic heteroplasmies at a low fraction (39). Thus, when such a selective constraint on mitochondria is weakened under certain conditions (40), such as the presence of mHTT (20–28), these low-fraction pathogenic heteroplasmies may increase in their fractions in cells, culminating in dysfunctional mitochondria and related energy deficits (32).
In the current study, we hypothesized that HD progression is partially driven by the deterioration of mtDNA quality. Since HTT is universally expressed, and mitochondrial dysfunction has been repeatedly observed in peripheral tissues (11–16), we surmised that HD-associated mtDNA changes can be detected in peripheral tissues and cell lines of HD patients, such as blood-derived lymphoblasts, which are readily available in large patient cohorts and thus can provide increased power for identifying mtDNA changes in HD. To test this hypothesis, we employed a sensitive mtDNA targeted sequencing approach, STAMP (sequencing by targeted amplification of multiplex probes) (41), to assess mtDNA heteroplasmies in lymphoblast and longitudinal blood samples from HD patients and healthy control individuals in the European Huntington’s Disease Network’s REGISTRY study (hereafter referred to as REGISTRY) (42). We achieved ultradeep sequencing coverage on mtDNA in these samples and revealed an accelerated expansion of pathogenic mitochondrial DNA heteroplasmies in HD, illustrating a molecular feature underlying HD biology.
Results
High-Quality Mitochondrial Genome (mtDNA) Sequencing.
In total, we made mtDNA sequencing libraries with STAMP for 2,206 REGISTRY samples (1,830 lymphoblast samples and 376 blood samples; SI Appendix, Fig. S1). Among them, 2,107 (95.5%) with a median mtDNA sequencing coverage of consensus reads greater than 1,000-fold (X) were used for calling heteroplasmies (SI Appendix, Table S1). The average median coverage of consensus reads on mtDNA, after quality control for heteroplasmy analysis, was about 3,600× in lymphoblast samples and 6,100× in blood samples (SI Appendix, Table S2). According to the statistical power of STAMP in discriminating true low-fraction variants from sequencing errors in mtDNA (SI Appendix, Fig. S2), we called mtDNA heteroplasmies at variant allele fraction (VAF) ≥1% in lymphoblasts and at VAF ≥ 0.5% in blood samples, respectively.
We found that of the mtDNA heteroplasmies, which passed quality control filters in 17 lymphoblasts and 320 blood samples randomly selected as technical replicates, >95% were detectable at VAF ≥ 0.2% in the experiments performed on the same samples (SI Appendix, Table S3). mtDNA heteroplasmies detected also displayed high correlations (SI Appendix, Fig. S3), and nonsignificant differences, in their VAFs (SI Appendix, Table S3) between technical replicates.
Elevation of Pathogenic mtDNA Variant Dosages in Lymphoblasts of HD Patients.
We identified 9,729 heteroplasmies at 4,871 sites in mtDNA of 1,731 lymphoblasts that passed quality control for heteroplasmy analysis (Dataset S1). Of the 4,871 heteroplasmic sites, 2,790 (57%) were singletons, and another 1,779 (37%) were rare, detected in fewer than five samples. The average number of heteroplasmies was 5.6 in lymphoblasts of the current study. It was higher than an average of four heteroplasmies found in lymphoblasts from the 1000 Genomes project, which were detected with a lower average depth of coverage of 1,805× on mtDNA (39).
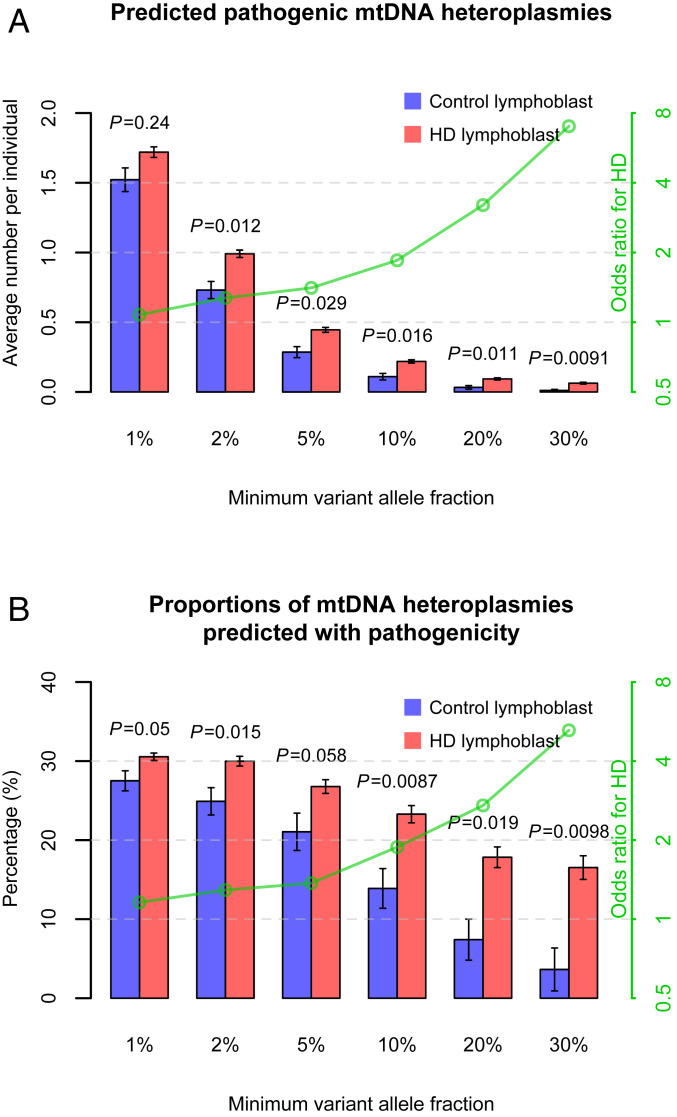
We compared mtDNA heteroplasmies in lymphoblasts between 1,549 HD patients and 182 control individuals. To assess whether there was an overrepresentation of pathogenic heteroplasmies in HD lymphoblasts relative to controls, we determined the pathogenicity of variants in protein-coding and RNA-coding regions of human mtDNA based on a variety of sources, including known disease associations, bioinformatic pathogenicity predictions, and variant frequency in the general population (Dataset S1). We found that HD patients possessed more predicted pathogenic heteroplasmies of medium and high fractions (VAF ≥ 2%, P = 0.012) in lymphoblasts compared to control individuals (Fig. 1A). The elevation of pathogenic heteroplasmy incidence in HD lymphoblasts became more pronounced when calculated with only high-fraction heteroplasmies, showing a rise in odds ratios for HD from 1.3, when computed at VAF ≥ 2%, to 7.0, when computed at VAF ≥ 30% (P = 0.0091; Fig. 1A), in logistic regression analyses. Similar odds ratios of predicted pathogenic heteroplasmies for HD were also observed among the subset of lymphoblasts from young and middle-aged individuals (age < 55 y; SI Appendix, Fig. S4 A and B), including 887 HD patients and 138 control individuals (average age: 43.8 y in patients vs. 44.4 y in controls).
Fig. 1.
mtDNA variant incidence in lymphoblasts of HD patients and control individuals. The results are shown by using bars in A for average numbers of predicted pathogenic heteroplasmies per individual, and in B for proportions of mtDNA heteroplasmies that were predicted with pathogenicity, in lymphoblasts of HD patients and control individuals. The values on the x axes refer to the minimum VAF of the heteroplasmies used in the analyses, from a low fraction at 1% to a high fraction at 30%. Error bars represent SEM. SEM in B was estimated using 1,000 bootstrap resamples of control lymphoblasts or HD lymphoblasts. The P values for mtDNA heteroplasmies from the logistic regression analyses of the disease status are shown above the bars in A. The P values from Fisher’s exact test are shown above the bars in B. The effects of mtDNA heteroplasmies, as odds ratios for HD, are illustrated with the green lines indicated by the values on the green y axes on a logarithmic scale.
Interestingly, the incidence of all mtDNA heteroplasmies, or those not predicted to be pathogenic, did not dramatically increase in HD lymphoblasts compared to control lymphoblasts, regardless of their VAFs (P ≥ 0.065; SI Appendix, Fig. S5 A and C). These results indicate that the overall mtDNA heteroplasmy load and fraction distribution were not affected by HD. In contrast, proportions of heteroplasmies that were predicted with pathogenicity differed significantly between HD lymphoblasts and control lymphoblasts (Fisher’s exact test, P = 0.05; Fig. 1B). Of note, only 3.6% of the heteroplasmies of VAF ≥ 30% detected in control lymphoblasts were pathogenic. This proportion was more than quadrupled in HD lymphoblasts (16.5%; Fisher’s exact test, P = 0.0098; Fig. 1B).
Pathogenic mtDNA Variant Dosages in HD Lymphoblasts Increase with Disease Stages.
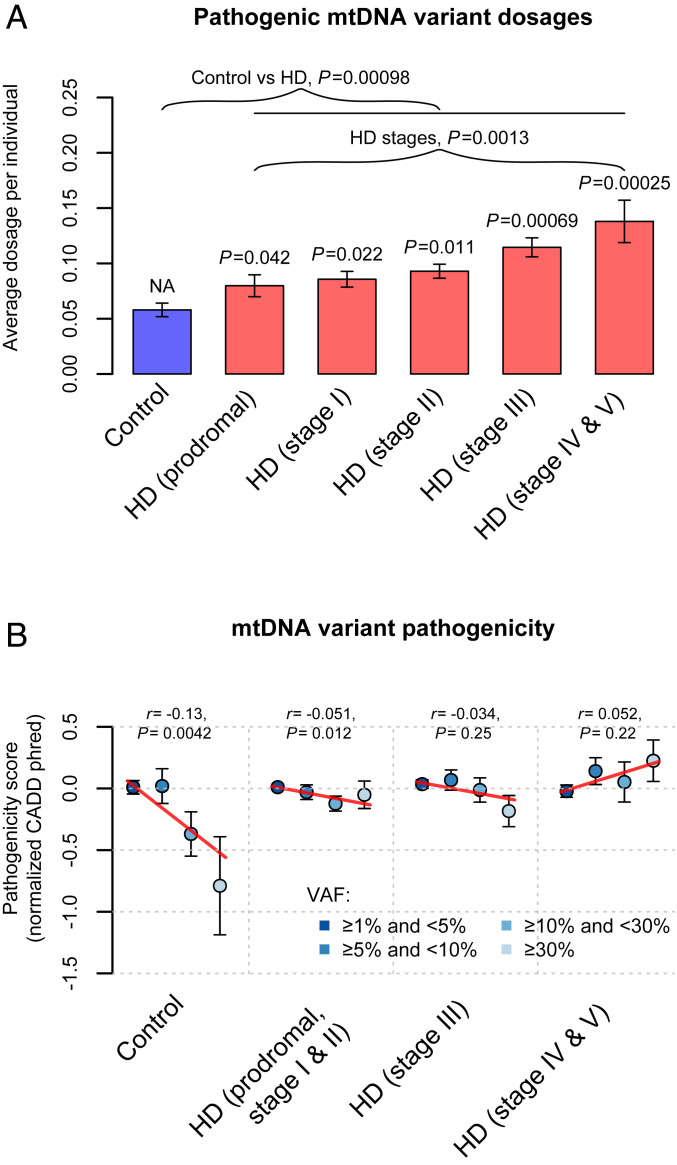
Since multiple mtDNA heteroplasmies can be present in one sample, for each sample, we computed the variant dosage of heteroplasmies by using the sum of the VAFs of all heteroplasmies identified. Again, HD lymphoblasts exhibited significantly increased dosages of predicted pathogenic heteroplasmies (P = 0.00098; Fig. 2A), but similar variant dosages of all heteroplasmies and heteroplasmies without pathogenicity predictions (P ≥ 0.47; SI Appendix, Fig. S5 B and D), compared to control lymphoblasts.
Fig. 2.
mtDNA variant dosages and pathogenicity in lymphoblasts of HD patients and control individuals. (A) Bar plots of the average variant dosages of predicted pathogenic heteroplasmies. The P values for mtDNA variant dosages from the logistic regression analyses of disease status are indicated above the bars representing the corresponding HD stages. In the linear regression analyses of disease stages, HD stages were treated as a continuous dependent variable with integer values from 1 to 5. NA, not applicable. (B) The average pathogenicity of nonsynonymous heteroplasmies stratified by their variant allele fractions (VAFs) in lymphoblasts. The dots representing different VAF categories were depicted by using colors indicated in the legend. Pearson’s r between the heteroplasmic VAF and the pathogenicity score, as well as the corresponding P value, are shown in each panel of B. The CADD scores are shown with the inverse normal transformed values, which increase with the chance of a heteroplasmy being pathogenic. The red lines in B represent the fitted regression lines for the VAF categories and the pathogenicity scores. NA, not applicable. Error bars in A and B represent SEM.
Next, we examined how the elevated variant dosages of predicted pathogenic heteroplasmies in HD lymphoblasts would relate to HD clinical stages. Among the 1,549 HD patients, 1,524 had information on the Unified Huntington’s Disease Rating Scale (UHDRS ’99) total functional capacity (TFC), total motor scores, and diagnostic confidence levels recorded in the REGISTRY clinical database within about 1 y of the sample collection. There were 156 patients in the prodromal stage of HD (43) (UHDRS diagnostic confidence level < 4). The remaining 1,368 patients were grouped into different disease stages based on their TFC scores (44). Among them, 766 were in the early stages, including 321 in stage I (TFC score ≥ 11) and 445 in stage II (7 ≤ TFC score < 11), 404 were in the middle stage (stage III: 4 ≤ TFC score < 7), and 198 were in the late stages (stage IV/V: TFC score ≤ 3). Of note, substantial increases in the variant dosages of predicted pathogenic mtDNA heteroplasmies had already been revealed in lymphoblasts of HD patients in the prodromal and early stages (P ≤ 0.042; Fig. 2A), which became more prominent among patients in the middle and late stages (P ≤ 0.00069). Accordingly, there was a significant association between pathogenic mtDNA variant dosages in lymphoblasts and advancing disease stages among the 1,524 HD patients (P = 0.0013; Fig. 2A) as well as among the 1,368 manifest HD patients (P = 0.0084).
The increase of pathogenic mtDNA variant dosages with disease stages could result from relaxation of purifying selection on mtDNA heteroplasmies in HD lymphoblasts. To support this, in control samples we found a negative correlation between the VAFs of nonsynonymous heteroplasmies, which can alter the amino acid sequences of OXPHOS protein complexes, and their Combined Annotation Dependent Depletion (CADD) pathogenicity scores (r = −0.13, P = 0.0042; Fig. 2B). These results substantiate our observations from the general population (39), indicating that purifying selection may prevent the expansion of pathogenic heteroplasmies in lymphoblast mtDNA. In contrast, the degree of purifying selection on mtDNA heteroplasmies diminished in HD patients in the prodromal, early, and middle stages, with mild negative correlations between the VAFs of nonsynonymous heteroplasmies and their pathogenicity (Fig. 2B). In late-stage HD patients, we even detected a slight but nonsignificant positive correlation of pathogenicity with the VAFs of nonsynonymous heteroplasmies (Fig. 2B), suggesting complete loss of purifying selection on mtDNA heteroplasmies in the late stages of the disease.
The observed increases in pathogenic variant dosages and the relaxation of purifying selection on mtDNA heteroplasmies in HD lymphoblasts also persisted among young and middle-aged individuals (age < 55 y; SI Appendix, Fig. S4 C and D). Taken together, these results indicate that decline of mtDNA quality could be a molecular signature reflecting clinical progression of HD.
Pathogenic mtDNA Variant Dosages in HD Lymphoblasts Are Linked to Clinical Phenotypes and Disease Burden.
Clinical progression of HD is characterized by deterioration of motor, cognitive, and psychiatric functions. We thus sought to investigate how predicted pathogenic mtDNA heteroplasmies in lymphoblasts of HD patients could reflect these functional declines and correspond to HTT-related genetic burden. In addition to UHDRS TFC and total motor scores (n = 1,524), we retrieved UHDRS symbol digit modalities test (SDMT) scores (n = 1,266) from the REGISTRY database to assess the severity of cognitive signs in HD patients (42, 43).
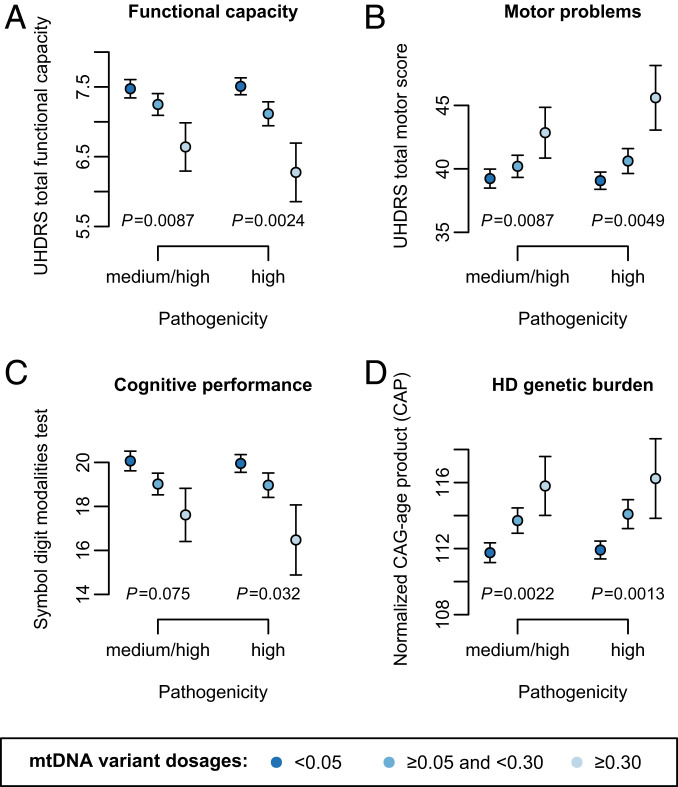
We found that pathogenic mtDNA variant dosages in lymphoblasts displayed significant associations with worsened disease severity, as measured on functional capacity (P = 0.0087; Fig. 3A) and motor scales (P = 0.0087; Fig. 3B), and a suggestive association with decreased cognitive performance in SDMT (P = 0.075; Fig. 3C). These associations were strengthened when we focused on mtDNA heteroplasmies predicted to have high pathogenicity (P = 0.0024–0.032; Fig. 3 A–C), which supports our conclusion that loss of selective constraints on mtDNA leads to elevated pathogenicity of mtDNA heteroplasmies during HD progression.
Fig. 3.
Associations of pathogenic mtDNA variant dosages with HD clinical phenotypes and genetic burden. The pathogenic mtDNA variant dosages were computed using either heteroplasmies with medium or high pathogenicity or heteroplasmies with only high pathogenicity. The significance levels of the associations of pathogenic mtDNA variant dosages with HD clinical phenotypes are shown in A for UHDRS TFC score, in B for total motor score, and in C for symbol digit modalities test score, all of which were assessed with adjustment for age, sex, and CAG repeat length. The significance levels of the associations with HD genetic burden are shown in D for normalized CAG-age product, which were assessed without adding CAG repeat length and age as covariates. The mean ± SEM of the phenotypes in the lymphoblasts with low (<0.05), medium-to-high (0.05–0.3), and high pathogenic mtDNA variant dosages (≥0.3) are illustrated in each panel.
Furthermore, we noted significant correlations between pathogenic mtDNA variant dosages and HD disease burden, which we computed as a normalized product between CAG repeat length and age (n = 1,524; P ≤ 0.0022; Fig. 3D). Inspired by such an observation, we subsequently assessed age-dependent changes in mtDNA heteroplasmies by using linear models comprising age, CAG repeat length, and their interaction as predictors for mtDNA heteroplasmies in HD lymphoblasts. As a result, we found that the interaction effect of elongated CAG repeat length and advancing age was positive, and significant in predicting both variant dosages (P ≤ 0.011) and incidence (P ≤ 0.014) of predicted pathogenic heteroplasmies in addition to the effect of age (P ≤ 0.0046; Table 1). Of note, expanded CAG repeat length also showed a substantial main impact on the increase of variant dosages (P = 0.035) and incidence (P = 0.04) of heteroplasmies predicted to have high pathogenicity (Table 1). In contrast, neither CAG repeat length nor its interaction with age was found to affect variant dosages or incidence of heteroplasmies that were not predicted to have medium or high pathogenicity (P ≥ 0.23), while the effects of age remained significant (P ≤ 7.4 × 10−5; Table 1).
Table 1.
Age- and CAG-dependent changes of mtDNA heteroplasmies in lymphoblasts of HD patients
| Variables | mtDNA variant pathogenicity | Age | CAG repeat length | Age × CAG repeat length | |||
| Beta (SE) | P | Beta (SE) | P | Beta (SE) | P | ||
| mtDNA variant dosages | M/H | 0.012 (0.003) | 0.00011 | 0.018 (0.012) | 0.15 | 0.0014 (0.0006) | 0.011 |
| H | 0.012 (0.003) | 5.8 × 10−5 | 0.025 (0.012) | 0.035 | 0.0015 (0.0005) | 0.0062 | |
| others | 0.013 (0.003) | 7.4 × 10−5 | 0.003 (0.013) | 0.82 | 0.0003 (0.0006) | 0.55 | |
| mtDNA variant incidence | M/H | 0.009 (0.003) | 0.0046 | 0.014 (0.012) | 0.26 | 0.0014 (0.0005) | 0.013 |
| H | 0.010 (0.003) | 0.00065 | 0.024 (0.012) | 0.040 | 0.0013 (0.0005) | 0.014 | |
| others | 0.015 (0.003) | 4.8 × 10−6 | 0.002 (0.013) | 0.87 | 0.0007 (0.0006) | 0.23 | |
The variant incidence and dosages of mtDNA heteroplasmies were inverse normal transformed (INV) and were further adjusted for sex and sequencing coverage. The values of age and CAG repeat length were centered at the population mean. The associations were assessed by using the model: INV dosage/incidence ∼ age + CAG_length + age × CAG_length. H, high pathogenicity; M/H, medium or high pathogenicity; others, not predicted with medium or high pathogenicity. Values of P < 0.05 are highlighted in bold type.
On the other hand, control lymphoblasts did not display an age-dependent increase in the variant dosages and incidence of predicted pathogenic heteroplasmies (P ≥ 0.3; SI Appendix, Table S4), even though the impact of age on other, nonpathogenic heteroplasmies was found significant (P ≤ 0.02). These results imply that the expansion of heteroplasmies with damaging consequences is largely suppressed in lymphoblasts expressing normal HTT. Collectively, these results indicate that elongated CAG repeats in HTT may accelerate the age-dependent increase of pathogenic heteroplasmies in lymphoblasts, in accord with the biochemical evidence that mHTT impairs mitochondrial quality control and causes bioenergetic deficits (11, 30).
Expansion of Preexisting mtDNA Heteroplasmies in Blood of HD Patients.
To directly investigate changes in mtDNA heteroplasmies during HD progression, we performed STAMP on longitudinal blood samples from 188 HD patients collected from two visits 5 to 9 y apart (median = 6 y) in REGISTRY. We called mtDNA heteroplasmies at VAF ≥ 0.5% in these samples. We found that mtDNA from seven individuals showed an excess of heteroplasmies (n ≥ 14) at known polymorphic sites of mtDNA, which could be caused by low-level contamination with other DNA samples. As such, we focused on the remaining 181 HD patients for the following analysis, of whom 169 were not in the late stages of the disease at baseline.
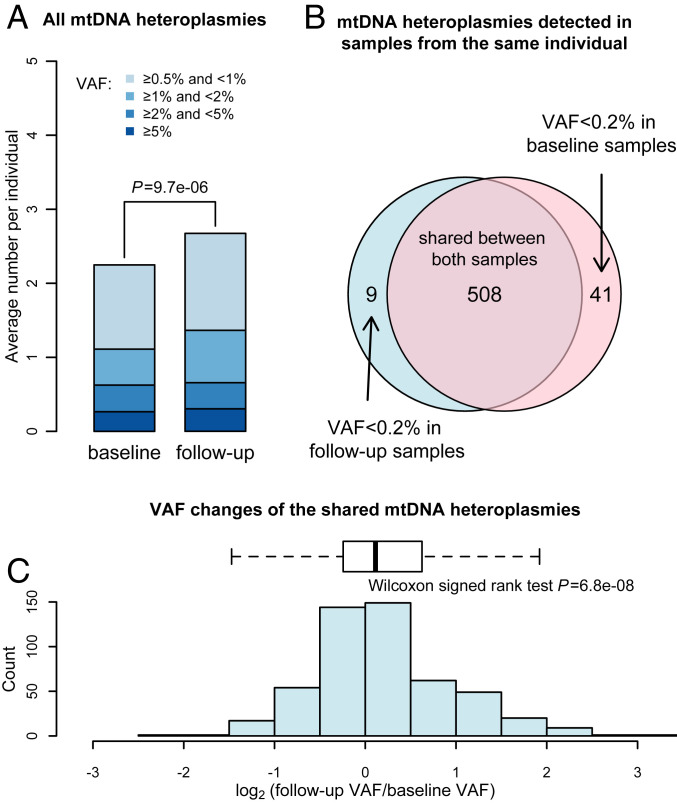
We observed a roughly 19% increase in the incidence of heteroplasmies, from an average of 2.25 at baseline to an average of 2.67 at follow-up (paired t test, P = 9.7 × 10−6; Fig. 4A). Among the 558 mtDNA heteroplasmies with VAF ≥ 0.5%, 508 (91%) were shared between the baseline and follow-up samples (Fig. 4B). Another 21 heteroplasmies (4%; Dataset S2) could be considered possibly shared, which were used in the sensitivity analysis of preexisting very low-fraction heteroplasmies (SI Appendix, Supplemental Discussion 2).
Fig. 4.
mtDNA variant incidence and fraction changes detected in longitudinal blood samples of HD patients. (A) Bar plots of the incidence of mtDNA heteroplasmies detected in the baseline and follow-up blood samples. The relative incidence of heteroplasmies in different variant allele fraction (VAF) categories was depicted using colors indicated in the legend. The P value from paired t test of the overall heteroplasmy incidence is shown. (B) Venn diagram of mtDNA heteroplasmies detected in samples from the same individuals. The light blue circle represents heteroplasmies identified in the baseline sample. The red circle represents heteroplasmies identified in the follow-up samples. The overlapping region shows the share of 508 mtDNA heteroplasmies with VAF ≥ 0.2% in both samples. (C) Histogram and box plots of the distribution of the VAF changes of the 508 shared mtDNA heteroplasmies during the follow-up.
Of the 449 heteroplasmic sites detected, 407 (91%) were unique to one of the 181 HD patients, suggesting that they were not recurrent heteroplasmies driven by a selective advantage in the hematopoietic system (45). There was also a high correlation in VAFs of heteroplasmies detected between the baseline and follow-up samples (r > 0.99; P < 2.2 × 10−16). Therefore, most heteroplasmies detected in the follow-up samples must have already existed in the hematopoietic system, specifically, in long-lived stem or progenitor cells, since the time of the baseline visit. Interestingly, our results reveal that the increase of detectable heteroplasmies in follow-up samples can largely be attributed to the expansion of preexisting mtDNA heteroplasmies in the hematopoietic system. We found a statistically significant increase of VAFs of these preexisting heteroplasmies when comparing their VAFs between the baseline and follow-up samples (Wilcoxon signed-rank test, P = 6.8 × 10−8; Fig. 4C). This increase might be linked to the advance of age or disease stage among these HD patients.
Expansion of Pathogenic mtDNA Heteroplasmies in Blood Parallels the Progression of HD Stages and Clinical Phenotypes.
To investigate the influence of HD progression on the expansion of preexisting mtDNA heteroplasmies, we divided the 169 HD patients, who were not in the late stages at baseline, into two groups, including 134 who experienced progression of disease stage at follow-up, and 35 showing a slow progression of the disease with a stable stage at follow-up. Duration of follow-up did not significantly differ between these two groups (t test, P = 0.17). We detected 359 preexisting heteroplasmies in 120 progressed-stage patients and 107 in 28 stable-stage patients. Of them, 56 and 16 heteroplasmies were predicted to be pathogenic in 38 progressed-stage patients and 13 stable-stage patients, respectively.
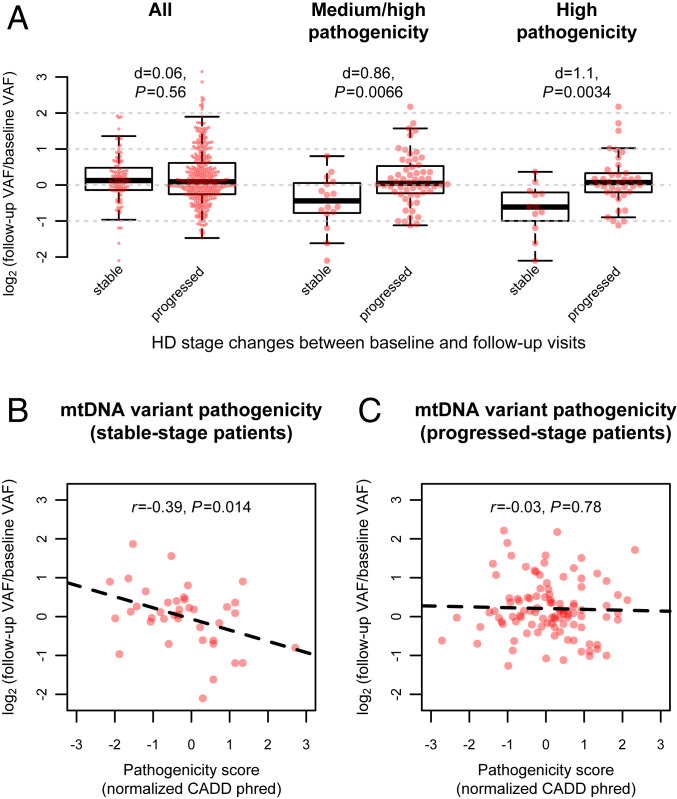
Among all the preexisting heteroplasmies, the degree of VAF changes at follow-up was comparable in progressed-stage patients and stable-stage patients (Cohen’s d = 0.06; t test, P = 0.56; Fig. 5A). In contrast, the VAF changes of predicted pathogenic heteroplasmies displayed a significant increase in progressed-stage patients compared to those in stable-stage patients (Cohen’s d ≥ 0.86; t test, P ≤ 0.0066; Fig. 5A). As reassuring evidence that the pathogenicity of these heteroplasmies, rather than other uncontrolled confounding factors, contributed to the observed difference, we found that the VAF changes of heteroplasmies without pathogenicity annotations did not correspond to the stage changes among the 51 patients (Cohen’s d = −0.05; t test, P = 0.78) carrying pathogenic heteroplasmies.
Fig. 5.
Changes of mtDNA variant fractions and pathogenicity in blood during HD progression. (A) Box plots of the variant allele fraction (VAF) changes of preexisting mtDNA heteroplasmies in blood samples of HD patients with and without a progression of disease stage during the follow-up. The P values from t test and Cohen’s d are shown for the difference between stable-stage patients and progressed-stage patients, which were computed using (from Left to Right in A) all heteroplasmies, heteroplasmies with medium or high pathogenicity, or heteroplasmies with only high pathogenicity. Each red dot in A indicates one heteroplasmy with its VAF change during the follow-up indicated by the value on the y axis. (B and C) The correlations between the VAF changes of preexisting nonsynonymous heteroplasmies and their CADD pathogenicity scores among (B) stable-stage patients and among (C) progressed-stage patients. The CADD scores are shown with the inverse normal transformed values, which increase with the chance of a heteroplasmy being pathogenic. The dashed lines represent the fitted regression lines.
Interestingly, in stable-stage patients, predicted pathogenic heteroplasmies exhibited a decrease in their VAFs at follow-up (Wilcoxon signed-rank test, P ≤ 0.029), suggesting effective purifying selection on mtDNA. We also noted a negative correlation between the degree of VAF changes and the pathogenicity (CADD phred scores) of nonsynonymous heteroplasmies (r = −0.39, P = 0.014; Fig. 5B) in stable-stage patients, indicating that heteroplasmies with a higher pathogenic potential were subject to stronger purifying selection. The evidence for purifying selection on mtDNA disappeared among progressed-stage patients, displaying a slight increase in the VAFs of pathogenic heteroplasmies at follow-up (Wilcoxon signed-rank test, P ≥ 0.23), and a weak and nonsignificant correlation between the VAF changes and pathogenicity of nonsynonymous heteroplasmies (r = −0.03; Fig. 5C). These results suggest that loss of purifying selection on preexisting heteroplasmies in mtDNA may contribute to the expansion of their fractions during HD progression.
Next, we assessed how the expansion of predicted pathogenic heteroplasmies in blood would relate to clinical phenotypic data recorded at baseline and follow-up visits of these patients. By using mixed-effects models, we found significant associations of the VAF changes of predicted pathogenic heteroplasmies with the follow-up clinical phenotypes, including TFC scores (P ≤ 0.0044), total motor scores (P ≤ 0.024), and SDMT scores (P ≤ 0.011), but not with the baseline clinical phenotypes (Table 2). Moreover, among the 51 patients carrying pathogenic heteroplasmies, no apparent associations were found between the follow-up clinical phenotypes and the VAF changes of other, nonpathogenic heteroplasmies (Table 2). These results point to an overall decline of mtDNA quality in the hematopoietic system during clinical progression of HD, which is independent of the effect of normal aging on heteroplasmy expansion.
Table 2.
Associations of mtDNA variant fraction changes in blood with HD clinical phenotypes
| Variables | mtDNA variant pathogenicity (N)* | TFC score | Total motor score | SDMT score | |||
| Beta (SE) | P | Beta (SE) | P | Beta (SE) | P | ||
| Model 1 (baseline phenotypes) | M/H (72) | 0.005 (0.037) | 0.90 | 0.010 (0.008) | 0.20 | 0.009 (0.011) | 0.41 |
| H (49) | 0.033 (0.047) | 0.49 | 0.018 (0.011) | 0.12 | 0.015 (0.013) | 0.25 | |
| Others (145) | −0.023 (0.026) | 0.38 | 0.002 (0.006) | 0.70 | −0.007 (0.009) | 0.42 | |
| Model 2 (follow-up phenotypes) | M/H (72) | −0.13 (0.037) | 0.0011 | 0.015 (0.006) | 0.021 | −0.050 (0.013) | 0.00046 |
| H (49) | −0.15 (0.050) | 0.0044 | 0.019 (0.008) | 0.024 | −0.042 (0.015) | 0.011 | |
| Others (145) | −0.014 (0.029) | 0.62 | 0.001 (0.005) | 0.83 | −0.002 (0.015) | 0.92 | |
The associations were assessed by using the following linear mixed-effects model: log2(VAF follow-up/VAF baseline) ∼ score + age + sex + CAG_length + followup_duration + (1|patient_id). In the analyses of the follow-up clinical phenotypes, the baseline phenotype and the baseline disease stage were considered as additional fixed-effect covariates in the model. H, high pathogenicity; M/H, medium or high pathogenicity; others, not predicted with medium or high pathogenicity in HD patients carrying pathogenic heteroplasmies. Values of P < 0.05 are highlighted in bold type.
The number of mtDNA heteroplasmies used for analysis; due to missing phenotypes in either the baseline sample or the follow-up sample, 1 pathogenic heteroplasmy and 2 nonpathogenic heteroplasmies were not included in the analyses of total motor scores, and 2 pathogenic heteroplasmies and 32 nonpathogenic heteroplasmies were not included in the analyses of SDMT scores.
Furthermore, we noted a significant positive effect of CAG repeat length on the VAF changes of 29 pathogenic heteroplasmies (linear mixed-effects model, P = 0.0034; SI Appendix, Table S5) identified among early-stage patients with moderate motor symptoms at baseline (TFC score ≥ 7 and total motor score < 25). However, this effect was weakened and became nonsignificant among all patients (SI Appendix, Table S5), implying that HTT may exert its impact on mtDNA early in HD pathogenesis. These results further suggest that elongated CAG repeats in HTT may promote the expansion of pathogenic heteroplasmies.
Discussion
In the current study, we applied ultradeep sequencing to assessing mitochondrial genome (mtDNA) heteroplasmies in lymphoblasts and longitudinal blood samples of HD patients. Our sequencing data provide more evidence to support the existence of purifying selection on mtDNA (39), which suppresses the expansion of pathogenic mtDNA heteroplasmies in cells and therefore could be an important mechanism that ensures cellular mitochondrial function during aging.
The increased pathogenic mtDNA variant dosages in HD lymphoblasts and their positive association with worsened disease severity indicate that mitochondrial quality control is impaired in HD. This notion of defective mtDNA quality control in HD was also supported by the data from longitudinal blood samples of HD patients whose clinical phenotypes progressed over time. They showed an expansion of preexisting pathogenic heteroplasmies, probably from early or midlife mtDNA mutations in the hematopoietic system. Moreover, we performed sensitivity analyses and found that our results were not affected by possibly fixed pathogenic heteroplasmies in lymphoblasts as well as preexisting very low-fraction pathogenic heteroplasmies in blood samples (detailed in SI Appendix, Supplemental Discussion 1 and 2, Fig. S11, and Table S11) .
Most predicted pathogenic heteroplasmies were singletons (62%) or doubletons (20%) among HD lymphoblasts (SI Appendix, Fig. S6A). The most frequent one was identified at m.3244 (n = 21), two nucleotides away from the fourth most prevalent one at m.3242 (n = 12; SI Appendix, Fig. S6B). The variant hotspot at m.3242–m.3244, located in the MT-TL1 [tRNALeu(UUR)] gene, has been shown to have increased incidence in the putamen from aged donors relative to young donors (46). At this site, m.3243 (A>G or A>T) occurred four times in HD samples but was absent in controls. m.3243A>G is the most prevalent and studied pathogenic mtDNA mutation, which causes mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) (47), and is a risk factor for human aging (48). Heteroplasmic fraction of m.3243A>G determines its phenotypic manifestation (49–51). In cybrid cell models, increasing heteroplasmic fraction of m.3243A>G alters mitochondrial metabolic and redox status, and subsequently induces global changes in transcriptome and epigenome of nuclear DNA (50, 51). At a high fraction of m.3243A>G, cells exhibit molecular phenotypes resembling those found in neurodegenerative diseases, including HD (50, 51). There were 13 other heteroplasmies detected in >0.4% (n ≥ 7) of HD lymphoblasts, which were scattered over eight genes from three OXPHOS complexes (SI Appendix, Fig. S6B). Of note, m.10197G>A in MT-ND3 (SI Appendix, Fig. S6B) is a confirmed pathogenic mutation for Leigh-like syndrome (52). High fraction of m.10197G>A has been found to reduce complex I activity in muscle and ATP production in skin fibroblasts (52).
We found that the locations of pathogenic heteroplasmies, in HD samples, spanned all 13 protein-coding genes in mtDNA. The average number and dosages of pathogenic heteroplasmies from each of the four OXPHOS complexes showed strong correlations with the total number of nonsynonymous sites in mtDNA genes (R2 ≥ 0.98; SI Appendix, Fig. S6 C and D). Comparable effects of variant dosages on HD stages were further identified in pathogenic heteroplasmies from all four OXPHOS complexes (P for heterogeneity ≥ 0.53; SI Appendix, Table S6). However, our analysis had limited power (11–22%) to reveal significant association for complexes III and V (SI Appendix, Table S6), as only 10% and 7% of nonsynonymous sites in mtDNA are used to encode these two complexes, respectively (SI Appendix, Fig. S6C). Overall, our data support contribution of multiple mtDNA genes and OXPHOS complexes to mitochondrial dysfunction in HD. These results may also explain heterogenous findings on dysfunctional OXPHOS complexes in tissues and cell lines from HD patients (8–10, 14, 53).
Our data suggest that elongated CAG repeats in HTT may affect mitochondrial quality control. In line with our finding, it has been demonstrated in numerous cellular and animal models of HD that mHTT can impair mitophagy, through disruption of the autophagic receptor p62-mediated cargo recognition (23, 26), limiting the transport and fusion of autophagosome to lysosome (27), and recruitment of valosin-containing protein to mitochondria (28). Moreover, mHTT has been implicated in the biological pathways that regulate mitochondrial morphology and tubular networks, since it may bind to the mitochondrial fission protein, DRP1, and alter the levels of fusion proteins to favor mitochondrial fission over fusion (20–22). Changes in mitochondrial fission and fusion dynamics and loss of effectiveness of the mitophagic apparatus in identifying and removing dysfunctional mitochondria may collectively relax selective constraints on mtDNA, precipitating the expansion of pathogenic mtDNA heteroplasmies in cells (40).
Moreover, our results imply that HTT-related genetic burden may not completely account for the impairment of mitochondrial quality control in HD. Other modifiers of HD progression may also play a role in this process. Some previous studies (54–56) suggested that age at onset of HD was associated with genetic variants in PPARGC1A, which encodes PGC-1alpha, a key regulator of mitochondrial energy metabolism (57). PGC-1alpha was found to be suppressed in the presence of mHTT (18). Knocking out PCG-1alpha in animal models not only reduced mitochondrial content but also attenuated lysosomal capacity and mitophagy flux (58). Recent genome-wide association studies (GWASs) (59, 60) indicated that genetic variants in RRM2B may modulate age at onset of HD. Deleterious mutations in RRM2B cause mitochondrial disorders showing abnormal mtDNA maintenance (61). Moreover, genetic variants in mitochondrial fission genes showed an enrichment of associations with the timing of HD onset in a pathway-based analysis in GWASs of over 4,000 HD patients (59). This result was, however, not replicated in GWASs performed on additional two HD patient cohorts (60). Taken together, these results point to a possible interaction between nuclear DNA-encoded mitochondrial genes and mtDNA in the pathogenesis and progression of HD.
In addition to HTT’s role in mitochondrial quality control, HTT has been investigated in the context of other mitochondrial characteristics, such as mitochondrial biogenesis and oxidative damage (18, 62). In lymphoblast and blood samples, we measured mtDNA content in relation to the amount of nuclear DNA as a proxy for mitochondrial biogenesis by using both STAMP and a quantitative PCR-based method. We found that mtDNA content did not correlate with the variant dosages of predicted pathogenic heteroplasmies (SI Appendix, Table S7) in lymphoblasts, or with the expansion of preexisting pathogenic heteroplasmies in blood samples of HD patients (SI Appendix, Table S8; further discussed in SI Appendix, Supplemental Discussion 3, Figs. S12 and S13). Therefore, mHTT’s impact on mtDNA quality control may be independent of its impact on mitochondrial biogenesis.
The decline of mtDNA quality in HD lymphoblasts and blood samples may not be a consequence of oxidative damage to mitochondria in HD (62). We found similar patterns of base changes mtDNA heteroplasmies in lymphoblasts and blood samples of HD patients, compared to lymphoblasts of control individuals, showing high transition to transversion ratios of >13 (SI Appendix, Fig. S7). The minimal proportions of transversion base changes in mtDNA of HD samples are suggestive of replication errors or base deamination in mtDNA rather than damage associated with oxidative stress (37, 63). Indeed, oxidative stress could result from reactive oxygen species produced by defective electron transport complexes in OXPHOS system if encoded by mtDNA harboring pathogenic heteroplasmies (40).
These lines of evidence stress the importance of mitochondrial quality control during clinical progression of HD. Intriguingly, effective purifying selection on mtDNA heteroplasmies was detected among patients with a slow disease progression in the analysis of blood samples. It suggests that the decline of mtDNA quality is not an irreversible process in HD. Modulating mitochondrial network dynamics and mitophagy pathway genes, such as DRP1, HDAC6, PINK1, and GAPDH, has been shown to effectively correct mHTT-associated toxicity in cell and animal models (24, 28, 64, 65). Therefore, future studies, which aim to investigate genes and compounds that could restore or improve mitochondrial quality control in the context of HD, are needed for the identification of new therapeutics.
The interpretation of the role of blood mtDNA heteroplasmies in the pathogenesis of HD is limited in the current study. However, peripheral blood and related cell lines have been repeatedly used as a surrogate for studying HD’s impact (11–13). Peripheral blood of HD patients also reveals transcriptomic changes resembling those of striatum and prefrontal cortex (66). Interestingly, energy metabolism is one of the significantly down-regulated pathways in HD that are shared between brain (67) and blood (11), and correlates with HD severity (66). Therefore, the pattern of mtDNA heteroplasmies in blood cells may be indicative of systemic, mtDNA quality and mitochondrial function in HD and during human aging. Furthermore, the difference in incidence and fractions of mtDNA heteroplasmies between lymphoblasts and blood samples in this study agrees with the observations from previous cell studies, which showed higher numbers of mtDNA heteroplasmies in cell lines than those in the parental tissues (37, 68, 69). Recently, the propagation of mtDNA heteroplasmies has been demonstrated in hematopoietic cells using various single-cell sequencing technologies (70). These technological advances will provide an unprecedented opportunity to directly study the changes of mtDNA at a single-cell level and their impact on cellular phenotypes in HD and other age-related diseases.
In sum, our large-scale deep-sequencing study illustrates mtDNA changes in the hematopoietic system during HD progression, echoing a theme of defective mitochondrial quality control in HD. Our data on mtDNA heteroplasmies reveal that decline in mtDNA quality results from multiple genes and occurs early in HD pathogenesis. These changes may be overlooked in biochemical assays that target a specific mitochondrial OXPHOS complex. Hence, harnessing mtDNA variation, measured by using a sensitive method such as STAMP, in peripheral tissues could provide an accessible biomarker for progression of HD and potentially other age-related disorders.
Materials and Methods
Ethics Statement.
REGISTRY is a multicenter, prospective observational study of HD in Europe (42) (http://www.euro-hd.net/html/registry). It was approved by the local ethics committees of REGISTRY sites (42) and was conducted in accordance with the Declaration of Helsinki. Participants of REGISTRY gave informed written consent according to the International Conference on Harmonization–Good Clinical Practice guidelines. For participants who lacked the capacity to consent, study sites adhered to country-specific guidelines to obtain consent. For data protection and confidentiality, all participants were assigned a unique pseudonym that does not contain any identifying information.
Study Participants.
Genomic DNA was extracted from established lymphoblast cell lines and peripheral blood samples of participants in REGISTRY (42). Based on the availability of lymphoblast DNA, we selected 1,630 HD patients with CAG repeat length in HTT > 36, and 200 age- and sex-matched individuals with normal HTT as healthy controls, for the study of mitochondrial genome (mtDNA) heteroplasmies in relation to HD and associated phenotypes. We selected 188 HD patients with DNA from two independent blood samples, collected at least 5 y apart, for the study of mtDNA heteroplasmy changes during disease progression. The study design and aims are illustrated in SI Appendix, Fig. S1. The demographics, HD-related phenotypes, and mtDNA sequencing characteristics are summarized in SI Appendix, Tables S1 and S2. The related power calculation for sample sizes is provided in SI Appendix, Fig. S8.
HTT CAG repeat length was determined as per the REGISTRY study protocol (42). Data on HD-related clinical phenotypes were collected using the UHDRS ’99 (43) for functional (TFC scores), motor (total motor scores), and psychiatric and cognitive signs (symbol digit modalities test scores) at annual or biennial visits of participants in REGISTRY. The clinical phenotypes closest to and within about 1 y of the time of lymphoblast or blood collection were considered for the analyses in the current study. A diagnostic confidence score of motor abnormalities on the UHDRS of less than 4 (not unequivocal) was used to define the prodromal stage (43). Disease stages after motor onset were derived from the UHDRS TFC scores (44). The disease burden score (3) was calculated as a normalized product of CAG repeat length and chronological age for each HD patient by using the following equation: (CAG repeat length – 30) × age × 100/627.
Mitochondrial Genome Sequencing.
We used STAMP (41) to sequence mtDNA of REGISTRY samples. The experimental and computational workflow of STAMP are illustrated in SI Appendix, Figs. S9 and S10, respectively, and are described in detail in SI Appendix, Supplemental Methods 1–4. In brief, the entire 16.6-kb human mtDNA was first captured with 46 pairs of probes designed with specific sequences complementary to the heavy or light strand sequences of mtDNA and five pairs of probes targeting single-copy nuclear DNA regions (SI Appendix, Fig. S9). The captured DNA fragments were then amplified using p5i5 and p7i7 indexing primers with individual barcode. The obtained STAMP libraries were sequenced with 2 × 250 paired-end reads on HiSeq 2500. Phi-X DNA library was spiked in at 5% to increase the overall complexity of the libraries. For samples with insufficient sequencing depth, such as blood samples with low mtDNA content, we performed PCR on an additional 1.5 to 3 μL of the capture products, sequenced the resulting libraries, and pooled the reads from the same sample for analysis. Detailed information on primer sequences and captured mtDNA fragments of STAMP is provided in SI Appendix, Table S9.
We developed a Python pipeline (the stamp toolkit) to process reads generated from STAMP (SI Appendix, Fig. S10) (41). In brief, paired-end reads were first demultiplexed into files for individual samples. Paired-end reads were then sorted into clusters of capture products according to the probe arm sequences identified. The resulting reads were aligned by using Burrows-Wheeler aligner (bwa mem, version 0.7.17) (71) to the complete human genome, including both nuclear DNA and mtDNA sequences (genome assembly GRCh38; downloaded from ftp://ftp.1000genomes.ebi.ac.uk), and in a second round to a modified mtDNA sequence, which had the final 120 bp copied to the start to accommodate alignment of D-loop-region reads in STAMP. Reads mapped to target regions were locally realigned by using freebayes (version 1.1.0) (72), and their base qualities were recalibrated by using samtools (version 1.6) (73). For paired-end reads with the same molecular barcode, the base information at corresponding sites of the alignments was merged using a Bayesian approach to generate a consensus read representing the captured DNA fragment (SI Appendix, Supplemental Method 1). The same method was also used to merge base information within the overlapping region of the paired-end reads. Reads from nuclear mitochondrial DNA segments were filtered out using both alignment and consensus read sequence methods (SI Appendix, Fig. S14 and Table S16) as described in SI Appendix, Supplemental Method 2. Finally, information on consensus reads was output into a pileup file by using samtools and was used to call mtDNA variants. The relative coverage of consensus reads from all mtDNA probe pairs was shown in SI Appendix, Fig. S15. mtDNA content was estimated by further using reads from five additional probe pairs targeting single-copy nuclear DNA regions (41). mtDNA content quantification and related quality control procedures (SI Appendix, Table S17), and comparisons with mtDNA content assessed by using a quantitative PCR-based method (SI Appendix, Table S12 and Fig. S16), are detailed in SI Appendix, Supplemental Method 4.
mtDNA Variant Identification.
mtDNA heteroplasmic variants (with VAF < 0.99) were determined by using the consensus reads that were aligned to mtDNA (mapping quality [MAPQ] ≥ 20; base alignment quality [BAQ] ≥ 30), and did not have an excess of nucleotide mismatches (>5 in the coding region and >8 in the D-loop region) compared to the individual’s major mtDNA sequence (41). Variants were further filtered based on a list of criteria to reduce false-positive calls, including 1) ≥100-fold (X) depth of coverage with ≥70% of the bases having BAQ ≥ 30; 2) not in low-complexity regions or low-quality sites of mtDNA; 3) ≥5 minor alleles detected; 4) a log likelihood quality score ≥5 computed with base quality scores (39); 5) comparable VAFs (Fisher’s exact test, P ≥ 10−4, and fold change, <5) when estimated using consensus reads constructed with and without duplicate paired-end reads; for variants with VAF < 1%, VAF was at least 0.2% among consensus reads constructed with duplicate paired-end reads; 6) the detected number of minor alleles significantly larger than the expected number of errors, which was estimated at a rate of 0.02% in STAMP (SI Appendix, Fig. S2A, exact Poisson test, P < 0.01/16569). To compare VAFs of a heteroplasmy between a pair of samples, we further required that, in both samples, this heteroplasmy was detected at a site that had at least 500 consensus reads with BAQ ≥ 30. The computational tools, parameters, and quality filters used for read alignment and variant detection are listed in SI Appendix, Table S10. The quality control procedures are described in detail in SI Appendix, Supplemental Method 3. mtDNA variant dosages were defined in the current study in order to represent the overall degree of heteroplasmy loads and fractions in mtDNA, analogous to the tissue-level mutation burden estimated for human nuclear DNA (74). It was computed in sample i as , where refers to the VAF of variant j detected in sample i. Only variants with VAF over the minimum detection threshold were used for the computation of dosages.
Haplogroup information of mtDNA in lymphoblast and blood samples was assigned by using haplogrep2 (version 2.1.1) (75). The distribution of mtDNA macrohaplogroups among lymphoblasts and blood samples of REGISTRY are shown in SI Appendix, Tables S13 and S15. The results regarding mtDNA haplogroups and HD are discussed in SI Appendix, Supplemental Discussion 4 (SI Appendix, Table S14 ).
Bioinformatic Analysis of mtDNA Variant Function.
The functional impact of mtDNA variants was determined by using the ANNOVAR pipeline (76). The population frequency of mtDNA polymorphisms in the general population was obtained from HmtDB (77). Pathogenicity of mtDNA variants (SI Appendix, Fig. S10) was evaluated by using CADD (78) and PON-mt-tRNA (79) for mtDNA-encoded OXPHOS genes and tRNAs, respectively. A list of disease-associated variants in mtDNA was collected from the MITOMAP website (80) and the ClinVar database (81) (last access, January 2020). Pathogenic variants were determined as 1) being reported with disease associations (“reported” or “confirmed”) in MITOMAP or (“pathogenic”) in ClinVar, or 2) having a CADD pathogenicity score > 20 (phred score, v1.3) in OXPHOS genes, or 3) a PON-mt-tRNA prediction as “likely pathogenic” or “pathogenic” in tRNAs. mtDNA variants without confirmed disease associations listed in MITOMAP and ClinVar were further required to not overlap with known mtDNA polymorphisms in apparently healthy populations of HmtDB (population frequency, <0.01%).
Moreover, based on the initial selection of variants annotated with medium or high pathogenicity, we prioritized variants with high pathogenicity by using more stringent criteria on the pathogenicity scores in 2) and 3). For OXPHOS genes, we required high-pathogenicity variants to have a CADD score >23 and to be verified by PolyPhen-2 (82) as “probably pathogenic” or by MutPred (83) to have a score >0.7, For tRNAs, high-pathogenicity variants predicted as “pathogenic” by PON-mt-tRNA had to be confirmed by MitoTIP (84) to have a score >16.25. These functional prediction scores have been used to study pathogenicity of mtDNA heteroplasmies and polymorphisms in the general population, and yield comparable annotations with each other (39, 79, 84, 85).
Statistical Analysis.
Associations of mtDNA variant incidence and dosages in lymphoblasts with disease status in all lymphoblasts were assessed by using logistic regression with adjustment for covariates including age, sex, and mtDNA sequence coverage. Associations of mtDNA variant incidence and dosages in HD lymphoblasts with disease stages and related continuous phenotype data were analyzed by using linear regression adjusted for age, sex, mtDNA sequence coverage, and CAG repeat length as an additional covariate. Age-dependent changes of mtDNA heteroplasmies in HD lymphoblasts were assessed by using linear model: age + cag_length + age × cag_length (detailed in SI Appendix, Supplemental Method 5).
In the analysis of longitudinal blood samples, we estimated the degree of expansion of a preexisting heteroplasmy by using a log2 fold change in the VAF of the same variant detected between the follow-up sample and the baseline sample. Its associations with HD-related phenotypes at baseline were analyzed by using a linear mixed-effects model adjusted for fixed effects of age, sex, duration of follow-up, and CAG repeat length, and a random effect of the patient identifier. Its associations with HD-related phenotypes at follow-up were tested similarly with further adjustment for the baseline phenotype and the baseline disease stage. The mixed-effects modeling and the significance level of the fixed effects were computed by using the lme4 (version 1.1) and lmerTest (version 3.0) R packages. Other statistical methods used are indicated next to the related results in the main text and SI Appendix. All statistical analyses were carried out with R (version 3.5.0). Two-tailed P values are reported.
Supplementary Material
Acknowledgments
We thank all participants of the European Huntington’s Disease Network’s REGISTRY study without whose dedication and commitment this work would not have been possible. We thank the CHDI Foundation and the European Huntington’s Disease Network for their support in this research project. We thank Andrew Cheung for assisting in mtDNA sequencing experiments. We also thank Drs. Martin Picard, Paul Soloway, Haiyuan Yu, Kimberly O’Brien, and Yiping Wang for critical reading and comments on the manuscript. This work was supported by CHDI.
Footnotes
Competing interest statement: P.J.S., Y.W., X.G., K.Y., and Z.G. are affiliated with Cornell University.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014610118/-/DCSupplemental.
Data Availability
Information on mtDNA heteroplasmies detected in lymphoblast samples and blood samples of the current study is provided in Datasets S1 and S2. The paired-end reads and mtDNA consensus reads of these samples have been deposited with the European Genome–Phenome Archive (EGA accession number EGAS00001004092). The pipeline used for analyzing STAMP sequencing data are available at https://github.com/mtstamp/stamp. All other study data are included in the article and/or supporting information.
References
- 1.Walker F. O., Huntington’s disease. Lancet 369, 218–228 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Roze E., et al., Huntington’s disease and striatal signaling. Front. Neuroanat. 5, 55 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross C. A., et al., Huntington disease: Natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 10, 204–216 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Victor M. B., et al., Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci. 21, 341–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmo C., Naia L., Lopes C., Rego A. C., Mitochondrial dysfunction in Huntington’s disease. Adv. Exp. Med. Biol. 1049, 59–83 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Friedman J. R., Nunnari J., Mitochondrial form and function. Nature 505, 335–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace D. C., A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.W. A. Brennan, Jr, Bird E. D., Aprille J. R., Regional mitochondrial respiratory activity in Huntington’s disease brain. J. Neurochem. 44, 1948–1950 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Gu M., et al., Mitochondrial defect in Huntington’s disease caudate nucleus. Ann. Neurol. 39, 385–389 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Browne S. E., et al., Oxidative damage and metabolic dysfunction in Huntington’s disease: Selective vulnerability of the basal ganglia. Ann. Neurol. 41, 646–653 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Seong I. S., et al., HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 14, 2871–2880 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Sawa A., et al., Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat. Med. 5, 1194–1198 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Panov A. V., et al., Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 5, 731–736 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Arenas J., et al., Complex I defect in muscle from patients with Huntington’s disease. Ann. Neurol. 43, 397–400 (1998).9506560 [Google Scholar]
- 15.Lodi R., et al., Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy. Ann. Neurol. 48, 72–76 (2000). [PubMed] [Google Scholar]
- 16.van der Burg J. M., Björkqvist M., Brundin P., Beyond the brain: Widespread pathology in Huntington’s disease. Lancet Neurol. 8, 765–774 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Gardiner S. L., et al., Bioenergetics in fibroblasts of patients with Huntington disease are associated with age at onset. Neurol. Genet. 4, e275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui L., et al., Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127, 59–69 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Wang J.-Q., et al., Dysregulation of mitochondrial calcium signaling and superoxide flashes cause mitochondrial genomic DNA damage in Huntington disease. J. Biol. Chem. 288, 3070–3084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J., et al., Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 19, 3919–3935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirendeb U. P., et al., Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 21, 406–420 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W., et al., Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 17, 377–382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Vicente M., et al., Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 13, 567–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil B., et al., PINK1-induced mitophagy promotes neuroprotection in Huntington’s disease. Cell Death Dis. 6, e1617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang S., Disatnik M.-H., Mochly-Rosen D., Impaired GAPDH-induced mitophagy contributes to the pathology of Huntington’s disease. EMBO Mol. Med. 7, 1307–1326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochaba J., et al., Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U.S.A. 111, 16889–16894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erie C., Sacino M., Houle L., Lu M. L., Wei J., Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur. J. Neurosci. 42, 1941–1951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., et al., VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat. Commun. 7, 12646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickles S., Vigié P., Youle R. J., Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guedes-Dias P., et al., Mitochondrial dynamics and quality control in Huntington’s disease. Neurobiol. Dis. 90, 51–57 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Burté F., Carelli V., Chinnery P. F., Yu-Wai-Man P., Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 11, 11–24 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Wallace D. C., Chalkia D., Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 5, a021220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun N., Youle R. J., Finkel T., The mitochondrial basis of aging. Mol. Cell 61, 654–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trifunovic A., et al., Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Edgar D., et al., Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 10, 131–138 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Payne B. A., et al., Mitochondrial aging is accelerated by anti-retroviral therapy through the clonal expansion of mtDNA mutations. Nat. Genet. 43, 806–810 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greaves L. C., et al., Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 10, e1004620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart J. B., Chinnery P. F., The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat. Rev. Genet. 16, 530–542 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Ye K., Lu J., Ma F., Keinan A., Gu Z., Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. U.S.A. 111, 10654–10659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn A., Zuryn S., The cellular mitochondrial genome landscape in disease. Trends Cell Biol. 29, 227–240 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Guo X., Wang Y., Zhang R., Gu Z., STAMP: A multiplex sequencing method for simultaneous evaluation of mitochondrial DNA heteroplasmies and content. NAR Genom. Bioinform. 2, lqaa065 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orth M.et al.; Investigators of the European Huntington’s Disease Network , Observing Huntington’s disease: The European Huntington’s Disease Network’s REGISTRY. PLoS Curr. 2, RRN1184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huntington Study Group , Unified Huntington’s disease rating scale: Reliability and consistency. Mov. Disord. 11, 136–142 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Shoulson I., Huntington disease: Functional capacities in patients treated with neuroleptic and antidepressant drugs. Neurology 31, 1333–1335 (1981). [DOI] [PubMed] [Google Scholar]
- 45.Samuels D. C., et al., Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 9, e1003929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams S. L., Mash D. C., Züchner S., Moraes C. T., Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet. 9, e1003990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goto Y., Nonaka I., Horai S., A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Tranah G. J., et al., Mitochondrial DNA m.3243A>G heteroplasmy affects multiple aging phenotypes and risk of mortality. Sci. Rep. 8, 11887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grady J. P., et al., mtDNA heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol. Med. 10, e8262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Picard M., et al., Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl. Acad. Sci. U.S.A. 111, E4033–E4042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kopinski P. K., et al., Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proc. Natl. Acad. Sci. U.S.A. 116, 16028–16035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tolomeo D., et al., Clinical and neuroimaging features of the m.10197G>A mtDNA mutation: New case reports and expansion of the phenotype variability. J. Neurol. Sci. 399, 69–75 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Polyzos A. A., McMurray C. T., The chicken or the egg: Mitochondrial dysfunction as a cause or consequence of toxicity in Huntington’s disease. Mech. Ageing Dev. 161 (Pt A), 181–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Che H. V. B., et al., Localization of sequence variations in PGC-1α influence their modifying effect in Huntington disease. Mol. Neurodegener. 6, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soyal S. M.et al.; European Huntington Disease Network , A greatly extended PPARGC1A genomic locus encodes several new brain-specific isoforms and influences Huntington disease age of onset. Hum. Mol. Genet. 21, 3461–3473 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Weydt P., Soyal S. M., Landwehrmeyer G. B., Patsch W.; European Huntington Disease Network , A single nucleotide polymorphism in the coding region of PGC-1α is a male-specific modifier of Huntington disease age-at-onset in a large European cohort. BMC Neurol. 14, 1–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang H., Ward W. F., PGC-1alpha: A key regulator of energy metabolism. Adv. Physiol. Educ. 30, 145–151 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Vainshtein A., Desjardins E. M., Armani A., Sandri M., Hood D. A., PGC-1α modulates denervation-induced mitophagy in skeletal muscle. Skelet. Muscle 5, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium , Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 162, 516–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium , CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell 178, 887–900.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorman G. S., Pitceathly R. D. S., Turnbull D. M., Taylor R. W., “RRM2B-related mitochondrial disease” in Mitochondrial Disorders Caused by Nuclear Genes (Springer, New York, 2013), pp. 171–182. [Google Scholar]
- 62.Johri A., Beal M. F., Antioxidants in Huntington’s disease. Biochim. Biophys. Acta 1822, 664–674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kennedy S. R., Salk J. J., Schmitt M. W., Loeb L. A., Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9, e1003794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo X., et al., Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J. Clin. Invest. 123, 5371–5388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guedes-Dias P., et al., HDAC6 inhibition induces mitochondrial fusion, autophagic flux and reduces diffuse mutant huntingtin in striatal neurons. Biochim. Biophys. Acta 1852, 2484–2493 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Hensman Moss D. J., et al., Huntington’s disease blood and brain show a common gene expression pattern and share an immune signature with Alzheimer’s disease. Sci. Rep. 7, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mochel F., et al., Early alterations of brain cellular energy homeostasis in Huntington disease models. J. Biol. Chem. 287, 1361–1370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang E., et al., Age-related accumulation of somatic mitochondrial DNA mutations in adult-derived human iPSCs. Cell Stem Cell 18, 625–636 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Perales-Clemente E., et al., Natural underlying mtDNA heteroplasmy as a potential source of intra-person hiPSC variability. EMBO J. 35, 1979–1990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ludwig L. S., et al., Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrison E., Marth G., Haplotype-based variant detection from short-read sequencing arXiv [Preprint] (2012). https://arxiv.org/abs/1207.3907 (Accessed 7 June 2019).
- 73.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martincorena I., et al., Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weissensteiner H., et al., HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44 (W1), W58–W63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rubino F., et al., HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res. 40, D1150–D1159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kircher M., et al., A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niroula A., Vihinen M., PON-mt-tRNA: A multifactorial probability-based method for classification of mitochondrial tRNA variations. Nucleic Acids Res. 44, 2020–2027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lott M. T., et al., mtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 44, 1–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Landrum M. J., et al., ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adzhubei I., Jordan D. M., Sunyaev S. R., Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 76, 7–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li B., et al., Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sonney S., et al., Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PLoS Comput. Biol. 13, e1005867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang R., Wang Y., Ye K., Picard M., Gu Z., Independent impacts of aging on mitochondrial DNA quantity and quality in humans. BMC Genomics 18, 890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Information on mtDNA heteroplasmies detected in lymphoblast samples and blood samples of the current study is provided in Datasets S1 and S2. The paired-end reads and mtDNA consensus reads of these samples have been deposited with the European Genome–Phenome Archive (EGA accession number EGAS00001004092). The pipeline used for analyzing STAMP sequencing data are available at https://github.com/mtstamp/stamp. All other study data are included in the article and/or supporting information.