Significance
To transcribe DNA sequences into RNA, RNA polymerase (RNAP) first binds to promoter DNA. The promoter sequence dictates both binding strength and rate of remodeling the DNA duplex by RNAP to open 13 base pairs in the start site region. This open “bubble” allows complementary nucleotides to pair with template-strand bases and be incorporated into RNA. All sequence-specific, RNAP-promoter contacts must break for RNAP to escape and elongate the RNA, but how this occurs is not well understood. Here, we report rate constants for each initiation step at the λPR promoter at different temperatures. We analyze these data to obtain insights into which open complex initiates and when RNAP-promoter contacts are disrupted, allowing bubble collapse, duplex formation, and promoter escape.
Keywords: transcription, initiation, regulation, thermodynamics, kinetics
Abstract
Transcription initiation is highly regulated by promoter sequence, transcription factors, and ligands. All known transcription inhibitors, an important class of antibiotics, act in initiation. To understand regulation and inhibition, the biophysical mechanisms of formation and stabilization of the “open” promoter complex (OC), of synthesis of a short RNA–DNA hybrid upon nucleotide addition, and of escape of RNA polymerase (RNAP) from the promoter must be understood. We previously found that RNAP forms three different OC with λPR promoter DNA. The 37 °C RNAP-λPR OC (RPO) is very stable. At lower temperatures, RPO is less stable and in equilibrium with an intermediate OC (I3). Here, we report step-by-step rapid quench-flow kinetic data for initiation and growth of the RNA–DNA hybrid at 25 and 37 °C that yield rate constants for each step of productive nucleotide addition. Analyzed together, with previously published data at 19 °C, our results reveal that I3 and not RPO is the productive initiation complex at all temperatures. From the strong variations of rate constants and activation energies and entropies for individual steps of hybrid extension, we deduce that contacts of RNAP with the bubble strands are disrupted stepwise as the hybrid grows and translocates. Stepwise disruption of RNAP-strand contacts is accompanied by stepwise bubble collapse, base stacking, and duplex formation, as the hybrid extends to a 9-mer prior to disruption of upstream DNA–RNAP contacts and escape of RNAP from the promoter.
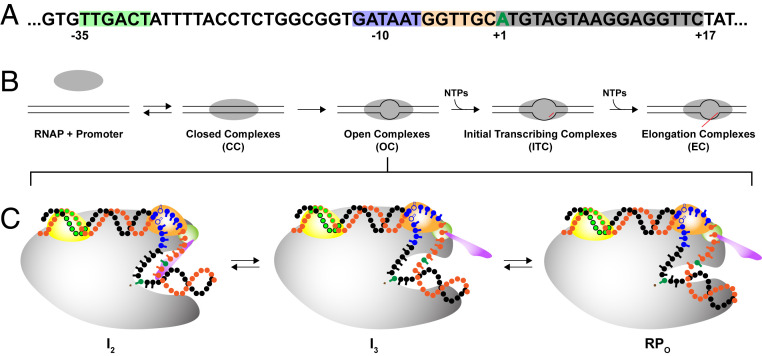
Transcription of DNA information into RNA is fundamental to all life. In prokaryotes, all transcription is performed by a single multisubunit RNA polymerase (RNAP). Gene expression is highly regulated, and much of this regulation occurs in the steps of transcription initiation. Promoter sequences, transcription factors, ligands, and conditions are key regulatory variables (1–3). All known transcription inhibitors, an important class of antibiotics, act in initiation (4). Fig. 1A shows the sequence and the key regions of λPR-promoter DNA. These include the -35 and -10 hexamers, the 6 bp discriminator region between the -10 region and the transcription start site (TSS, +1), and the initial transcribed region (ITR).
Fig. 1.
Transcription initiation at λPR promoter. (A) Nontemplate strand sequence of the λPR promoter studied here, in which the ITR is modified to eliminate the incorporation of CMP before position +17, so transcription halts at a 16-mer RNA when CTP is withheld. The −35 (green), −10 (blue), and discriminator (tan) elements and the start site (+1; green) and ITR (gray) are highlighted. (B) Summary of stages in productive initiation by RNAP holoenzyme (α2ββ’ωσ70). Key aspects of the mechanisms of two of these stages (OC and ITC) are determined in this study. (C) Schematic representation of OC species at the λPR promoter at 37 °C: intermediates I2 and I3 and stable 37 °C complex RPO. As illustrated, interactions of in-cleft and downstream elements of RNAP with the discriminator and ITR, weak or absent in I2, are stronger in I3 and much stronger in RPO at 37 °C (10). RNAP core (gray) is shown with relevant σ70 subunits 1.1 (purple), 1.2 (green), 2 (light orange), and 4 (yellow). Linking domains between subunits are not shown for clarity. The template DNA strand is shown in black and the nontemplate strand in orange, with the −35 (light green), −10 (blue), and start site (dark green) indicated.
Stages of productive initiation are summarized in Fig. 1B. Specific binding of RNAP to duplex (closed) promoter DNA forms an ensemble of closed complexes (CC), including an initial closed intermediate (designated RPC) and a series of more advanced closed intermediates (collectively called I1), in which the promoter DNA is remodeled. “Isomerization” of the ensemble of CC intermediates, including the opening of the DNA from the −10 region to the TSS, forms a series of open complexes (OC), including an initial unstable open intermediate (I2), which at λPR promoter converts to more stable species (intermediate I3, stable 37 °C complex RPO), all with the same open region but with different interactions involving the discriminator strands and the ITR (for representations of these OC, see Fig. 1C). Nucleotide triphosphates (NTPs) complementary to the template DNA sequence are bound and the corresponding monophosphates (NMP) incorporated into an RNA–DNA hybrid in a series of initial transcription complexes (ITCs). The hybrid translocates into the cleft with each step of RNA extension, stressing and disrupting RNAP-promoter contacts so that RNAP escapes from the promoter in the transition from initiation to elongation.
Initiation in all likelihood is regulated at all of these stages. To understand regulation, the detailed mechanisms of these stages must be understood. Recent structural (5–9), kinetic–mechanistic (10–19), and high-throughput sequencing studies (20, 21) have greatly advanced our understanding of initiation, but much remains to be learned for the rational discovery of drug targets and for the design of synthetic promoters optimized for specific applications in molecular biology and medical biotechnology (22).
Many examples exist of the regulation of initiation at the CC level (i.e., rates and extents of CC formation and isomerization) by promoter sequences, factors, ligands, and conditions (2–4). Regulation of initiation at the OC level is potentially also significant but not at all well understood. Correlations of OC stability or lifetime with TSS selection (23), promoter output (24), and RNA–DNA hybrid length for RNAP escape have been proposed (15). The three λPR OC in Fig. 1C differ in lifetime and stability by four orders of magnitude (lifetimes ranging from ∼1 s (I2) to ∼13 h (RPO) at 37 °C) (11, 15, 25). The question of which OC is/are capable of productive initiation in the presence of NTPs has not been addressed previously for any promoter. As shown schematically in Fig. 1C, the λPR OC differ in the strength of in-cleft interactions with the discriminator strands and of interactions of the downstream duplex (ITR) with downstream mobile elements (DMEs) of RNAP (11, 26–28). The difference in stability between RPO and I3 is known to decrease at temperatures below 37 °C, but otherwise, I3 is much less characterized than RPO and I2. Formation equilibrium constants and lifetimes of OC at the well-studied T7A1 and rrnB P1 promoters are ∼1 and 3 orders of magnitude less than for λPR. These OCs are proposed to be similar in their downstream interactions to λPR I3 and I2, respectively (11).
For the transition from initiation to elongation to occur, specific RNAP-promoter contacts (29) must be disrupted by translocation of the RNA–DNA hybrid into the upstream cleft (6, 16, 30–34). Disruption of these RNAP-promoter contacts results in the collapse of (and duplex formation by) the upstream portion of the initiation bubble (−1 to −11 for λPR), escape of RNAP from the promoter, and dissociation of the σ70 subunit (12, 35). Escape of RNAP from the λPR promoter occurs after synthesis of an initial 10-mer RNA (15, 35).
To investigate these processes in the escape of RNAP, we previously determined the step-by-step kinetics and mechanism of NMP incorporation into the growing RNA–DNA hybrid at the λPR promoter at 19 °C up to the escape point (16). This information, not available for any other promoter DNA or multisubunit RNAP, parallels that obtained for the single-subunit T7 bacteriophage RNAP (36). From an analysis of the repeated pattern of small and larger rate constants found for successive steps of RNA extension in initial transcription, we proposed that disruption of RNAP-promoter contacts occur in a stepwise manner prior to promoter escape (16).
Here, we report the 25 and 37 °C kinetics of initial transcription at the λPR promoter. Rapid quench mixing is used to determine overall rates of full-length (FL) RNA synthesis and rate constants for the individual steps of NMP incorporation into the RNA–DNA hybrid at two NTP conditions for comparison with 19 °C results. A change in the initial step of the mechanism at 37 °C, as compared to 19 °C, reveals that the stable 37 °C OC (RPO) cannot initiate and that an intermediate in RPO formation (I3) is the initiation complex. From analysis of the temperature dependence of these rate constants, we deduce when specific contacts of RNAP with single-stranded (ss) and duplex regions of promoter DNA are disrupted as the hybrid translocates into the cleft, resulting in the collapse of the initiation bubble, duplex formation, and escape of RNAP from the promoter.
Results
Unexpected Effects of Temperature on the Rate of Transcription Initiation at the λPR Promoter.
Time courses spanning more than two orders of magnitude (≤0.5 to ≥90 s) of transcription initiation by Escherichia coli RNAP at the λPR promoter at 37 and 25 °C were obtained by rapid mixing at two different sets of NTP concentrations (designated “low UTP” and “high UTP”) for comparison with previous results at 19 °C (16). The initial transcribed sequence (Fig. 1A), specifying a RNA that starts with pppApU, is modified from that of λPR to eliminate the need for CTP until position +17, causing RNA synthesis by productive complexes to pause after 16-mer production when CTP is omitted. The competitor heparin, added with the NTP mixture, ensures single-round productive initiation by preventing reinitiation by any dissociated RNAP.
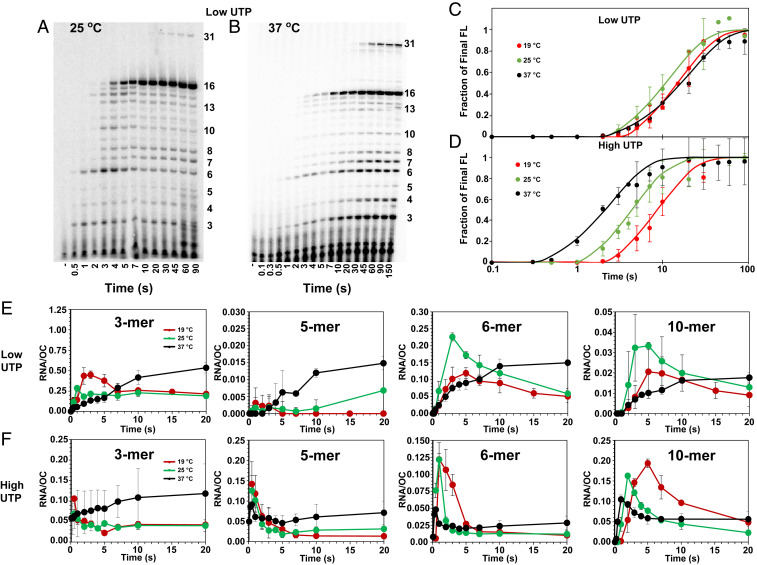
Fig. 2 A and B show representative polyacrylamide gel electrophoresis (PAGE) separations of RNAs present in samples quenched at a series of time points during initiation and the transition to elongation at the low-UTP condition (final concentrations 10 μM UTP, 200 μM ATP and GTP (no CTP), and 17.5 nM α-32P-UTP) at 25 and 37 °C. SI Appendix, Fig. S1 shows representative gels from initiation kinetics experiments at high UTP (final concentrations 200 μM UTP and ATP, 10 μM GTP (no CTP), and labeling with 17.5 nM α-32P-GTP). Amounts of each RNA length at each time are quantified by 32P-UTP or 32P-GTP incorporation. Efficient incorporation of the α-32P label into the transcript is achieved by use of a low concentration (10 μM) of the corresponding unlabeled NTP.
Fig. 2.
Step-by-step kinetics of transcription initiation from λPR promoter at different NTP concentrations and temperatures. (Top Left) PAGE separations of individual 32P-labeled RNA bands (from 3-mer to 16-mer and 31-mer) observed as a function of time during productive and nonproductive initiation at 25 (A) and 37 °C (B). Lanes span the time range from 0.5 to 90 s or 0.1 to 150 s after adding NTPs and heparin to OC formed by premixing RNAP and λPR-promoter DNA (SI Appendix, Methods). Gels shown are for the low-UTP condition: 200 μM ATP and GTP, 10 μM UTP, and 17.5 nM α-32P-UTP. CTP is omitted, causing transcription to pause at a 16-mer RNA, before readthrough occurs to synthesize longer transcripts (15). Representative high-UTP (200 μM ATP and UTP, 10 μM GTP, and 17.5 nM α-32P-GTP) gels at 25 and 37 °C are shown in SI Appendix, Fig. S1. (Top Right) Time courses (log scale) of synthesis of FL (>10-mer) RNA at 19 [red (15)], 25 (green), and 37 °C (black) at low UTP (C) and high UTP (D). (Bottom) Comparisons of linear time courses for representative short RNAs (3-mer, 5-mer, 6-mer, and 10-mer) synthesized by productive and nonproductive complexes at low UTP (E) and high UTP (F) at 19 [red (15)], 25 (green), and 37 °C (black). At each time point, the average amount of each RNA present (per OC) is shown with the estimated uncertainty.
Because RNAP escapes from this λPR promoter in the conversion of 10-mer to 11-mer, all RNAs greater than 10-mer in length are defined as FL RNA (15). The transient accumulation of 12-mer and 13-mer may result from the reduction in rate constants for the subsequent steps caused by the coupling of translocation to disruption of σ70-core RNAP contacts. Readthrough by misincorporation results in extension of the 16-mer RNA to the position of the next C in the transcript at +32, near the fragment end. Transcription occurs slowly near a fragment end, so this second pause is effectively a stop point.
FL RNA Synthesis.
The kinetics of synthesis of FL RNA, determined by summing phosphorimager data for RNA species longer than 10-mer, are shown in Fig. 2 C and D. These compare time courses (log scale) of FL RNA synthesis for low-UTP and high-UTP conditions at 25 and 37 °C with previous results at 19 °C. Results are the averages of two to four independent experiments at each condition, including those in Fig. 2 A and B and SI Appendix, Fig. S1. In all cases, these kinetics are well described as a short lag phase, followed by a first order (single exponential) approach to a plateau value with rate constant kFL, as observed previously at 19 °C (16). Values of the lag time and kFL at high-UTP and low-UTP conditions at each temperature are given in SI Appendix, Table S3. At the plateau, 0.5 ± 0.15 FL RNA are synthesized per OC, demonstrating that ∼50% of the λPR OC population at all three temperatures are productive and capable of promoter escape, as previously observed at 19 and 37 °C (15, 16).
Arrhenius plots of ln kFL versus 1/T are shown in SI Appendix, Fig. S2. At the high-UTP–low-GTP condition, ln kFL decreases monotonically but not linearly with 1/T, indicating a positive Arrhenius activation energy that decreases with increasing temperature. At the low-UTP–high-GTP condition, all kFL values are much smaller, despite a similar number of U and G bases incorporated into the FL transcript (3 U and 4 G by 11-mer formation, Fig. 1A). In addition, kFL is larger at 25 °C than at 37 °C, corresponding to a negative activation energy in this temperature range (SI Appendix, Fig. S2) and indicating a change in the mechanism with increasing temperature. These observations can all be explained by an additional step early in the mechanism of FL RNA synthesis that is very significant at 37 °C but not at 19 °C and is favored at high-UTP concentration (see Discussion).
Transient Short RNA Intermediates in FL RNA Synthesis.
Fig. 2 E and F show the time evolution of amounts of four different short RNAs (3-mer, 5-mer, 6-mer, and 10-mer) formed transiently in FL RNA synthesis by productive complexes at low-UTP and high-UTP conditions at 37 and 25 °C. Previously published results at 19 °C are shown for comparison. The results plotted are averages obtained from the analysis of multiple gels, such as those in Fig. 2 A and B, and are normalized per OC.
At 25 °C, at both low UTP and high UTP, Fig. 2 E and F show that amounts of all four RNA species increase rapidly and then decrease in the first 10 s after NTP addition, consistent with previous observations at 19 °C and indicating significant transient intermediates on the pathway to productive synthesis (16). Each transient occurs earlier at 25 °C than at 19 °C at both NTP conditions, as expected since most reaction rates increase with increasing temperature. Each transient occurs earlier at high UTP than at low UTP at both temperatures, expected because UTP is a reactant in the first step of initial transcription (pppApU synthesis).
At 37 °C and low UTP, however, no significant transient population of any intermediate is observed in FL RNA synthesis. At 37 °C and high UTP, transient populations of some longer RNA intermediates (5-mer, 6-mer, 9-mer, and 10-mer) are detected, but no transients for shorter RNAs (3-mer and 4-mer) are observed, and transient peak amounts of longer RNAs are small by comparison to lower temperatures. These observations, and the slower synthesis of FL RNA at low UTP at 37 °C than at 25 °C (Fig. 2C), indicate that the population of λPR OC at 37 °C, unlike at lower temperatures, is unable to productively bind the initiating NTP and synthesize pppApU without undergoing a conformational change. This conformational change is sufficiently unfavorable or slow to serve as a bottleneck and desynchronize subsequent steps of short RNA synthesis, so fewer transients are observed.
The stability of the λPR OC is more than 30-fold greater at 37 °C than at 19 °C (27, 37). We previously proposed that the population distribution of λPR OC also changed with temperature, shifting from the very stable 37 °C RPO, with strong downstream interactions between RNAP DME and duplex DNA extending to +20, to a mixed population of RPO and the I3 intermediate OC (Fig. 1C) at lower temperature (27). If only I3 and not RPO can productively bind the two initiating NTP, then the differences in rates of FL RNA synthesis between 37 °C and lower temperatures are readily explained. In the Discussion, this proposal is incorporated into the initiation mechanism and used to analyze the kinetics of transient and FL RNA synthesis by productive complexes.
Discussion
Evidence for an OC Conformational Change Prior to NTP Binding at 37 °C but not at 19 °C.
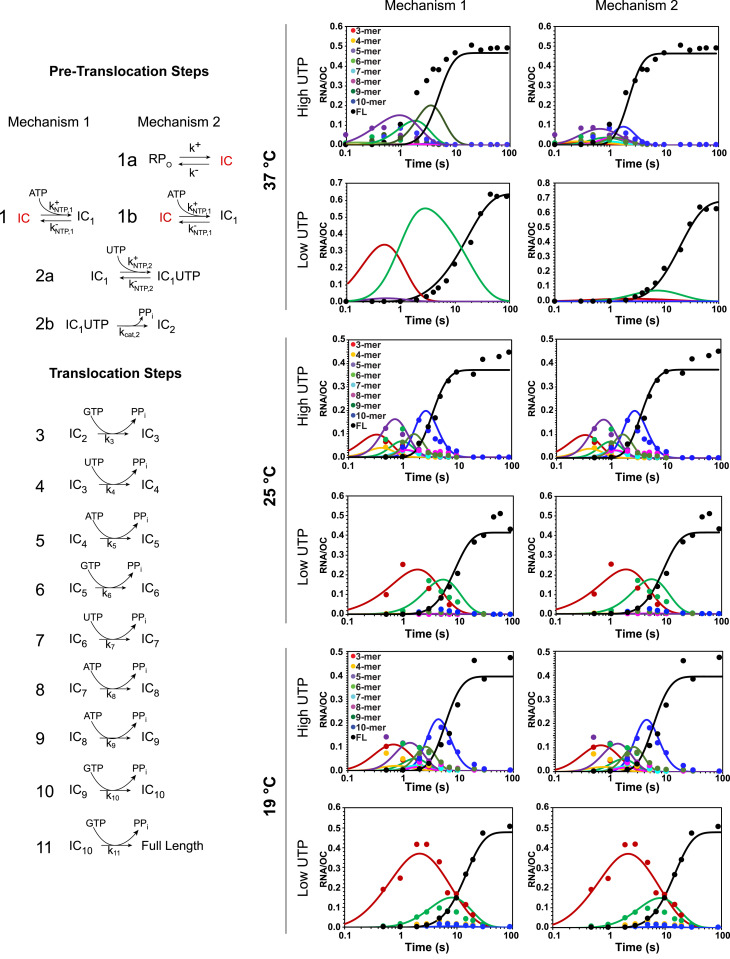
The mechanism previously used to interpret the kinetics of productive initiation at the λPR promoter at 19 °C, including the appearance and disappearance of short RNA intermediates and the synthesis of FL RNA (16), is shown as Mechanism 1 in Fig. 3. This mechanism begins with ordered, reversible binding of the substrates (ATP [+1] and UTP [+2]) to the binary RNAP-promoter OC, followed by irreversible catalysis to synthesize the dinucleotide pppApU. No evidence was obtained for a conformational change in the OC prior to NTP binding at 19 °C (16).
Fig. 3.
Fits of initiation kinetic data to step-by-step mechanisms. (Left) Two mechanisms differing in initial (pretranslocation) steps are shown. Mechanism 1, previously shown to describe λPR initiation data at 19 °C (15), is a minimal initiation mechanism for situations in which the stable OC is also the IC. Mechanism 2, proposed here to describe λPR initiation data at 37 °C in which the stable OC is RPO, includes an initial conformational change that converts RPO to the IC that binds NTP and productively initiates as in Mechanism 1. (Right) Comparison of fits (solid curves) to Mechanisms 1 and 2 of all λPR productive initiation data at 37, 25, and 19 °C for the two NTP conditions investigated (log time scale). Colors: 3-mer, red; 4-mer, yellow; 5-mer, purple; 6-mer, light green; 7-mer, light blue; 8-mer, pink; 9-mer, dark green; 10-mer, dark blue; and FL, black. The analyses of short RNA time courses (e.g., Fig. 2 E and F) to obtain the transients in FL RNA synthesis plotted here (points) are shown in SI Appendix, Figs. S3–S6. Rate constants ki, determined from these fits to Mechanism 2 at each temperature, are given in SI Appendix, Table S4.
Each subsequent step of initiation begins with reversible translocation. We previously deduced that most translocation steps in initiation are very unfavorable thermodynamically and rapidly reversible (16). Each translocation step in initiation requires the disruption of one downstream DNA–DNA base pair. In addition, translocation steps cause steric (6) and scrunching (31) stress as the RNA–DNA hybrid moves into the cleft, leading to disruption of RNAP-promoter contacts in many of these steps. Because translocation equilibrium constants for initiation steps are small, each step of the RNA–DNA hybrid extension up to the predicted point of escape is accurately quantified using a composite second-order rate constant ki (the analog of an enzymatic kcat/Km; see ref. 16), as shown in Mechanism 1 (Fig. 3). Much higher NTP concentrations would be necessary to approach a maximum velocity condition and thereby separate contributions of the overall equilibrium constant for the reversible translocation and NTP binding steps (analog of 1/Km) from the rate constant of the irreversible catalytic step (kcat). Fig. 3 shows the good fit of Mechanism 1 to the short RNA transients and FL RNA synthesis kinetics at 19 °C at both low- and high-UTP conditions.
Also shown in Fig. 3 are fits of productive initiation kinetic data to Mechanism 1 at 25 and 37 °C at low- and high-UTP conditions. At 25 °C, rate constants ki obtained from these fits accurately reproduce the short RNA transients and FL RNA synthesis kinetics. However, it is clear from Fig. 3 that Mechanism 1 is not sufficient to describe the kinetics of productive initiation at 37 °C. To obtain an accurate fit to the 37 °C kinetic data requires the addition of an unfavorable reversible step at the beginning of the mechanism, prior to initial ATP binding (Fig. 3, Mechanism 2, step 1a). This step represents a conformational change, which we propose is the conversion of the very stable 37 °C OC (RPO) to another OC conformation that we designate the initiating complex (IC). This step appears to be rapidly reversible and is characterized by the equilibrium constant K1a (for RPO → IC). All other steps of Mechanism 2 are the same as Mechanism 1. Fig. 3 also shows that use of Mechanism 2 to fit 19 and 25 °C datasets does not affect the quality of these fits and yields estimates of K1a at these temperatures.
These fits predict that K1a is extremely temperature dependent, greatly favoring RPO at 37 °C (K1a ≈ 0.01; 99% RPO) but favoring IC at 19 °C (K1a ≈ 5.3; more than 80% IC). A near-equimolar ratio of RPO and IC is predicted at 25 °C (K1a ≈ 0.70; ∼40% IC; ∼60% RPO). Good fits of 19 and 25 °C kinetic data to Mechanism 1 are obtained because a significant fraction of the OC population is initially in the IC conformation at these temperatures. The strong decrease in K1a with increasing temperature indicates that the enthalpy change for the conversion of RPO to IC is large in magnitude and negative; van ‘t Hoff analysis yields = −60 ± 20 kcal/mol. The standard free energy change for this conversion ranges from ∼2.7 kcal at 37 °C to ∼−1 kcal at 19 °C, and the corresponding entropy change is −200 ± 60 eu. Conversion of RPO to IC, therefore, shows near-complete enthalpy–entropy compensation, like many other protein processes.
Evidence that Only the I3 Intermediate OC and not RPO or I2 Initiates Transcription from λPR Promoter upon NTP Addition.
Evidence exists for two open intermediates (I2 and I3) on the pathway to formation of the RPO complex at the λPR promoter (Fig. 1C). The thermodynamic, kinetic, and footprinting information available for these intermediates support the proposal that IC is I3. I2 is unstable with respect to I3 and/or RPO at higher temperatures and unstable with respect to CCs at lower temperatures (11, 17, 26, 27). Conversion of I2 to I3 involves the folding of 100 to 150 amino acid residues of RNAP DME (27) and is thought to strengthen contacts between the proximal downstream duplex, the β lobe, and the β’ clamp (28). Conversion of I3 to RPO is thought to involve primarily an interaction of the downstream jaw and associated DME with the distal downstream duplex (+10 to +20), which serves to tighten the entire RNAP-promoter interface in the OC. The OC formed by the jaw deletion variant of RNAP and by downstream truncation variants of the promoter are thought to be models of I3; equilibrium constants for forming these variant OC are one to two orders of magnitude smaller than the binding of wild-type (WT) RNAP to FL promoter DNA at 37 °C. Hydroxyl radical footprinting of the OC with the jaw deletion RNAP variant reveals that the entire RNAP-promoter interface in the OC is less protected and hence “looser” and more hydrated than in the WT RNAP OC (28).
From this body of previous research, the stable OC population was proposed to be an equilibrium mixture of I3 and RPO, with RPO highly favored at 37 °C and I3 increasing in significance at lower temperature (10, 28), but the details of this were not known. Here, we find that the stable OC population is an equilibrium mixture of RPO and the IC initiation complex, with RPO favored at 37 °C and IC favored below 25 °C, indicating that IC is I3. In support of this, extrapolation of K1a (Mechanism 2) to lower temperature, assuming a temperature-independent enthalpy change for RPO → IC ( ≈ −60 kcal/mol), predicts that the IC:RPO population distribution for λPR at 10 °C, before NTP addition, is ∼99% IC and only 1% RPO. At 10 °C, Gries et al. (38) determined MnO4− footprints of both strands of the open region in the stable λPR OC, now identified as the IC initiation complex. In addition, salt upshifts were used to rapidly destabilize the 10 °C λPR OC and obtain a burst of I2, the least stable open intermediate, for MnO4− footprinting. Hence, the 10 °C λPR OC population, identified in the current research as 99% IC, is more stable and hence more advanced than I2 at 10 °C and therefore must be I3.
Kinetic–Mechanistic Evidence for Sequential Disruption of RNAP-Strand Contacts and Bubble Collapse in the Steps of Initiation.
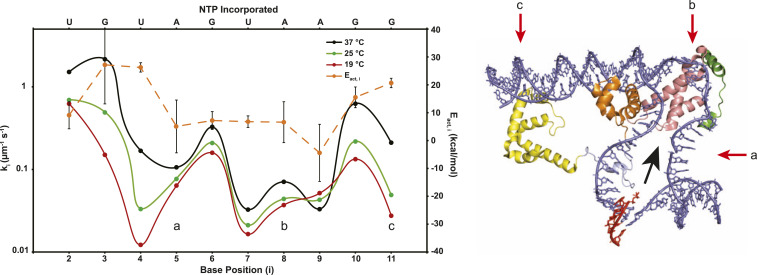
Fig. 4 (also SI Appendix, Table S4) compares rate constants ki for each step of incorporation of NMP into the RNA–DNA hybrid at 25 and 37 °C with 19 °C values. All are calculated using Mechanism 2 (Fig. 3); the 19 °C values are not significantly different from those obtained previously using Mechanism 1 (16). Steps of initiation that involve translocation show the same pattern of large and small ki values at all temperatures investigated. Three distinct regions of small ki are observed in Fig. 4. These correspond to 1) synthesis of 4-mer and 5-mer; 2) synthesis of 7-mer, 8-mer, and 9-mer; and 3) synthesis of 11-mer and are separated by single steps, with larger ki for synthesis of 3-mer, 6-mer, and 10-mer. In initiation, in which translocation is unfavorable because of steric and scrunching stress, ki is interpretable (to a good approximation) as the product of the equilibrium constants for translocation and NTP binding and the catalytic rate constant kcat (16).
Fig. 4.
(Left) Comparison of rate constants and activation energies for individual steps of initiation. Composite rate constants ki (left axis; micromolar−1 ⋅ seconds−1) for each step (i) of NMP incorporation and hybrid extension before the escape of RNAP are plotted on a log scale for the λPR promoter at 19 [red (15)], 25 (green), and 37 °C (black). At each temperature, differences in ki values are interpreted as differences in the equilibrium constant Ktr,I for translocation that occurs before NTP binding and catalysis in steps 3 to 11. Three regions of small ki (and therefore small Ktr, i) values, observed at all temperatures, are labeled a, b, and c for comparison with the framework ITC structure (Right). Arrhenius activation energies (Eact, i, orange, and right axis) of initial transcription steps, determined from the temperature dependences of the individual ki (SI Appendix, Fig. S7), are also shown. (Right) Structural representation of an ITC. This ITC framework structure (adapted from 4YLP) employed a promoter heteroduplex in the region of the initiation bubble and a 5-mer RNA hybridized to the template strand (RNA in red and promoter DNA in blue). Relevant regions of σ70 include σ1.2 (green), σ2 (pink), σ3 (orange), σ3.2 (light blue), and σ4 (yellow). Letters a, b, and c indicate the discriminator and −10 regions of the bubble strands and the −35 duplex, respectively. The expected direction of translocation of the hybrid into the RNAP cleft in initiation is indicated by the black arrow. Translocation is proposed to disrupt RNAP-promoter contacts with regions a, b, and c in the steps of initiation with small rate constants (indicated by the corresponding letters on the ki plot [Left]), resulting in stepwise bubble collapse.
To transition from initiation to elongation, the specific contacts between RNAP and promoter DNA that were essential to form and stabilize the OC must be broken. Previously, we interpreted the pattern of 19 °C ki values in terms of the serial disruption of these promoter contacts. In this interpretation, differences in ki values arise from differences in the equilibrium constant for translocation at each nucleotide addition step, Ktr,i. The very similar patterns of small and large ki values observed in Fig. 4 at 37, 25, and 19 °C indicate that, at all three temperatures, interactions of RNAP with the discriminator strands (indicated as region a in Fig. 4) are broken in translocation preceding synthesis of 4-mer and 5-mer. Strong interactions of RNAP with the strands of the −10 region (region b in Fig. 4) are broken in translocation preceding synthesis of 7-mer, 8-mer, and 9-mer, and interactions of RNAP with the −35 and upstream regions (region c in Fig. 4) are disrupted in translocation prior to synthesis of an 11-mer.
Very different effects of temperature on ki values for different translocation steps of initiation are observed in Fig. 4. For early (3-mer, 4-mer) and late (10-mer, 11-mer) steps, ki values increase strongly with increasing temperature. For mid-initiation steps (5-mer, 6-mer, 7-mer, 8-mer), ki values increase more modestly with increasing temperature. Notably, rate constant k9 for 9-mer decreases with increasing temperature. Analysis of these temperature dependences yields Arrhenius activation energies Eact,i (Fig. 4 and SI Appendix, Table S5), which vary by 30 kcal from +26 to −4 kcal.
Eact,2 for incorporation of UTP into pppApU, a step that does not involve translocation stress, is ∼9 kcal, similar to that reported previously for steps of elongation by E. coli RNAP [10 to 13 kcal (39)]. Eact,3 and Eact,4 for 3-mer and 4-mer are significantly larger (∼26 kcal), while Eact values for the next five steps are quite small (∼6 kcal for synthesis of 5-mer, 6-mer, 7-mer, and 8-mer RNA and ∼−4 kcal for 9-mer). Contacts of RNAP with the strands of the upstream bubble (positions -1 to -11 relative to the +1 TSS) are proposed to be disrupted in many of these steps (16). An interpretation of ki and Eact,i values in terms of changes in base stacking in these steps is given in the section Identifying Contributions of Downstream DNA Melting and of Base Stacking and Duplex Formation of the Upstream Bubble to the Kinetics of Initiation Steps. Eact,10 and Eact,11 are larger than the previous Eact values, though not as large as Eact,3 and Eact,4.
From rate constants ki and activation energies Eact,i, the quasithermodynamic quantities , , and , for each step, can be determined if the hypothetical maximum second-order rate constant kmax for this process (at = 0) and its temperature dependence are known. These , , and are for conversion of the reactants in the i-th step (NTP, pretranslocated initiation complex) to the subsequent catalytic transition state. We assume that all steps have the same kmax and approximate it by the value for an orientation-corrected, diffusion-limited rate constant (kmax ≈ 103 μM-1 ⋅ s−1), neglecting the small temperature dependence of a diffusion-limited kmax in calculating values (40). Although the uncertainty in the appropriate value of kmax is probably one order of magnitude, this is of no consequence for the following analysis (see also SI Appendix, Tables S5 and S6) as long as kmax has the same value for each step.
For kmax ≈ 103 μM−1 ⋅ s−1, for incorporation of the initiating UTP into pppApU is ∼4.3 kcal, and values of for subsequent steps of initiation (3 ≤ i ≤ 11) are in the range ∼5.1 to ∼6.6 kcal (SI Appendix, Table S5). Values of and vary over much wider ranges (∼30 kcal range in = Eact,i; ∼100 eu range in ) (SI Appendix, Table S5). For incorporation of the initiating UTP into pppApU (2-mer synthesis), and are modest (9 kcal and 16 eu). The values of , , and include contributions from the thermodynamics of UTP binding, including stacking of UTP on ATP, and from the intrinsic activation quantities for the catalytic step, but do not include any contributions from translocation.
Values of and for subsequent steps involving translocation are very different from those of 2-mer synthesis. Values of and for 3-mer and 4-mer are much larger than for 2-mer, while values of and for 5-mer, 6-mer, 7-mer, and 8-mer are much smaller (SI Appendix, Table S5), and values of and for 9-mer are modestly negative. These steps involving translocation include those identified previously as ones in which contacts of the discriminator and −10 strands with RNAP are disrupted, freeing the initiation bubble strands. and for 10-mer and 11-mer are more comparable to 3-mer and 4-mer. Contacts that are disrupted in these steps are proposed to be with the duplex (−35 and upstream) and not with ssDNA. Hence, the unusual activation thermodynamics are confined to the steps that break RNAP contacts with the bubble strands.
Identifying Contributions of Downstream DNA Melting and of Base Stacking and Duplex Formation of the Upstream Bubble to the Kinetics of Initiation Steps.
Disruption of base-stacking interactions in the melting of double-helical DNA to two separated strands is a major determinant of the melting enthalpy (41). Bases in ss nucleic acids are highly stacked in solution at 0 °C and unstack noncooperatively with increasing temperature. The enthalpy change for base unstacking in ssDNA is ∼5 kcal ⋅ mol−1, and the enthalpy of duplex melting is predicted to vary from ∼5 kcal ⋅ (mol base pair)−1 under conditions in which the bases in the separated strands are fully stacked to ∼15 kcal ⋅ (mol base pair)−1 under conditions in which the bases in the separated strands are fully unstacked (41).
Steps synthesizing 3-mer and 4-mer, the first two steps of initiation that begin with translocation and melting of one base pair at the downstream end of the initiation bubble, exhibit activation energies and enthalpies that are ∼17 kcal larger than for the step synthesizing 2-mer, which at the λPR promoter requires neither translocation nor base pair melting. Melting of one base pair can account for much of the 17 kcal differences between or versus , depending on the extent to which the bases after melting are unstacked.
Since subsequent steps of initiation also involve translocation and melting of one downstream DNA base pair, how can the very small (and in one case negative)values for the five subsequent steps of initiation (5-mer to 9-mer synthesis) be explained? SI Appendix, Table S5 shows that values of for 5-mer to 8-mer synthesis are ∼20 kcal less positive, and for 9-mer synthesis is ∼30 kcal less positive than for 3-mer and 4-mer synthesis.
Previously, to explain the pattern of small and large rate constants of these steps, we proposed that contacts of RNAP with the discriminator and −10 regions of the bubble strands are disrupted in a stepwise manner, beginning at the downstream end of the template and/or nontemplate discriminator strands, as the hybrid lengthens to a 9-mer. Here, to explain the unusual values of for 5-mer to 9-mer synthesis, we propose that the substantial stacking of bases in these strands accompanies the release of contacts with RNAP. A comparison of permanganate reactivities of thymines in the open region of the stable OC I3 and the unstable I2 revealed that most thymines in I3 are significantly more reactive and hence more exposed than in I2 (38). To explain this, we proposed that bases in the open region of I3 are much more unstacked than in I2 because of tighter binding of the strand backbones in the open region of I3 (38). Precedent for base unstacking in formation of a protein–ssDNA complex, in which the DNA backbone is strongly bound, is provided by the very stable SSB–ssDNA complex (42). Hence, base stacking is expected to accompany the disruption of contacts between RNAP and the open strands of the initiation bubble in steps of initiation involving translocation. The 20 to 30 kcal reductions in for 5-mer to 9-mer synthesis, compared to 3-mer and 4-mer synthesis, are consistent with the stacking of four bases in each step of synthesis of 5-mer, 6-mer, 7-mer, and 8-mer and stacking of six bases in synthesis of 9-mer (SI Appendix, Table S6).
We expected to observe a large negative contribution to from duplex formation by the bubble strands in one or more late steps of initiation. Formation of an 11 base pair duplex is predicted to contribute at least −50 kcal ⋅ mol−1 to if those strands are already stacked and as much as −150 kcal ⋅ mol−1 if they are initially unstacked. From the analysis in SI Appendix, Table S5, the only step that could include a negative enthalpy term of this magnitude is 9-mer formation, in which we identify a −30kcal contribution that could be from duplex formation from fully stacked strands. The smaller magnitude of this contribution (−30 versus −50 kcal ⋅ mol−1) could result from the need to break enthalpically, favorable interactions of the −7 and −11 bases on the nontemplate strand with aromatic residues in the base-binding pockets of sigma region 2 (29).
Alternatively, base pair formation may occur incrementally as the hybrid extends from a 5-mer to 9-mer. Base pair formation (including base stacking) in these steps would also explain their small activation enthalpies. Base pair formation would explain the finding (in the preceding paragraph; see also SI Appendix, Table S6) that base stacking increases in increments of four or six bases (i.e., two or three base pairs) in these steps. Base pair formation can also explain the offset between the first step with a small rate constant (4-mer synthesis, accompanied by disruption of some RNAP–discriminator strand interactions) and the first step with a small activation energy (5-mer synthesis, accompanied by formation of four base-stacking interactions or two base pairs). This offset would not be expected for ss stacking interactions but is explained if RNAP interactions with the downstream end of one discriminator strand are disrupted in 4-mer synthesis, while interactions with this region of the other strand are disrupted in 5-mer synthesis, allowing base pairing. A structural interpretation of activation enthalpies (SI Appendix, Table S5), in terms of step-by-step base pairing as RNAP-strand contacts are disrupted in initiation, is given in SI Appendix, Table S6. Because structural studies of initiation complexes to date have used heteroduplexes with an open initiation bubble, they provide no information about the extent of pairing of the bases that accompanies the synthesis of 5-mer or longer RNA in a productive initiation complex.
Conclusions
We find that the λPR initiation complex is the intermediate OC I3 (Fig. 1) and not the very stable RPO. Initiation at 37 °C, in which the λPR OC is predominantly RPO, requires an initial conformational change to form I3 before productive binding of both initiating NTPs. We also find that the disruption of RNAP contacts with the strands of the initiation bubble and bubble collapse occur stepwise in initiation, prior to disruption of RNAP contacts with the upstream duplex and escape of RNAP from the promoter. At a minimum, stepwise bubble collapse involves stepwise stacking of ss bases and may also involve stepwise base pairing. The activation thermodynamics provide no evidence that duplex formation by the bubble strands is delayed until the RNAP escape point (11-mer RNA formation), when interactions with the −35 region upstream duplex are proposed to be disrupted. Instead, the activation thermodynamics are consistent with duplex formation in one or more earlier steps, in which contacts of the bubble strands with the discriminator and −10 region are disrupted. Analogous quantitative studies of initiation kinetics and mechanism at other promoters and for λPR with different discriminator lengths and ITR are needed to characterize the relationship between promoter identity, initiation kinetics and mechanism, and the regulatory possibilities therein.
Materials and Methods
Details about reagents (buffers, enzymes, and DNA), initiation kinetic assays (single round in synthesis of FL RNA), and analysis of amounts of transient short RNAs from productive complexes and of stalled and released short RNA from nonproductive complexes are described in SI Appendix, with references to previous publications. Briefly, the λPR OC is preformed by incubation at the experimental temperature (25 or 37 °C) for 1 h. Preformed OC and initiation solution containing NTPs and heparin are mixed 1:1 at time 0 using a Kintek Corp Rapid Quench Flow, and quenched with 8 M urea and 15 mM EDTA at the times indicated. RNA products are visualized and quantified using PAGE, followed by phosphorimager analysis of incorporated α-32P-NTPs, as described previously (15). Fits were obtained using Kintek Explorer.
Supplementary Material
Acknowledgments
D.M.P. was supported by NIH Biotechnology Traineeship NIH Grant 5 T32 GM008349 and K.L.H. by NIH National Research Service Award postdoctoral fellowship NIH Grant GM 122303. We gratefully acknowledge the very helpful comments of the editor and reviewers and the support for this research from the abovementioned fellowships, University of Wisconsin–Madison, and NIH Grant GM R35-118100 (M.T.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021941118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Saecker R. M., Record M. T. Jr, Dehaseth P. L., Mechanism of bacterial transcription initiation: RNA polymerase – Promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 412, 754–771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haugen S. P., Ross W., Manrique M., Gourse R. L., Fine structure of the promoter-sigma region 1.2 interaction. Proc. Natl. Acad. Sci. U.S.A. 105, 3292–3297 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorman C. J., Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 41, 542–547 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Chen J., Boyaci H., Campbell E. A., Diverse and unified mechanisms of transcription initiation in bacteria. Nat. Rev. Microbiol. 19, 95–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., et al., Stepwise promoter melting by bacterial RNA polymerase. Mol. Cell 78, 275–288.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuo Y., Steitz T. A., Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell 58, 534–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L., Molodtsov V., Lin W., Ebright R. H., Zhang Y., RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription. Proc. Natl. Acad. Sci. U.S.A. 117, 5801–5809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin Y., et al., Structural basis of ribosomal RNA transcription regulation. Nat. Commun. 12, 528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glyde R., et al., Structures of bacterial RNA polymerase complexes reveal the mechanism of DNA loading and transcription initiation. Mol. Cell 70, 1111–1120.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruff E. F., et al., E. coli polymerase determinants of open complex lifetime and structure. J. Mol. Biol. 427, 2435–2450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruff E. F., Record M. T. Jr, Artsimovitch I., Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko J., Heyduk T., Kinetics of promoter escape by bacterial RNA polymerase: Effects of promoter contacts and transcription bubble collapse. Biochem. J. 463, 135–144 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Duchi D., et al., RNA polymerase pausing during initial transcription. Mol. Cell 63, 939–950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubin E. A., et al., Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. eLife 6, e22520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson K. L., et al., Mechanism of transcription initiation and promoter escape by E. coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 114, E3032–E3040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson K. L., et al., RNA polymerase: Step-by-step kinetics and mechanism of transcription initiation. Biochemistry 58, 2339–2352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreenivasan R., et al., Fluorescence-detected conformational changes in duplex DNA in open complex formation by E. coli RNA polymerase: Upstream wrapping and downstream bending precede clamp opening and insertion of the downstream duplex. Biochemistry 59, 1565–1581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen D., Manzano A. R., Rammohan J., Stallings C. L., Galburt E. A., CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase. Nucleic Acids Res. 47, 6685–6698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerner E., et al., Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 113, E6562–E6571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyduk E., Heyduk T., DNA template sequence control of bacterial RNA polymerase escape from the promoter. Nucleic Acids Res. 46, 4469–4486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkelman J. T., et al., XACT-seq comprehensively defines the promoter-position and promoter-sequence determinants for initial-transcription pausing. Mol. Cell 79, 797–811.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han L., et al., Development of a novel strategy for robust synthetic bacterial promoters based on a stepwise evolution targeting the spacer region of the core promoter in Bacillus subtilis. Microb. Cell Fact. 18, 96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L., et al., The mechanism of variability in transcription start site selection. eLife 6, e32038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyduk E., Heyduk T., Next generation sequencing-based parallel analysis of melting kinetics of 4096 variants of a bacterial promoter. Biochemistry 53, 282–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontur W. S., Saecker R. M., Davis C. A., Capp M. W., Record M. T. Jr, Solute probes of conformational changes in open complex (RPo) formation by Escherichia coli RNA polymerase at the lambdaPR promoter: Evidence for unmasking of the active site in the isomerization step and for large-scale coupled folding in the subsequent conversion to RPo. Biochemistry 45, 2161–2177 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kontur W. S., Capp M. W., Gries T. J., Saecker R. M., Record M. T. J. Jr, Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry 49, 4361–4373 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kontur W. S., Saecker R. M., Capp M. W., Record M. T. J. Jr, Late steps in the formation of E. coli RNA polymerase-λ P R promoter open complexes: Characterization of conformational changes by rapid [perturbant] upshift experiments. J. Mol. Biol. 376, 1034–1047 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drennan A., et al., Key roles of the downstream mobile jaw of Escherichia coli RNA polymerase in transcription initiation. Biochemistry 51, 9447–9459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., et al., Structural basis of transcription initiation. Science 338, 1076–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revyakin A., Ebright R. H., Strick T. R., Promoter unwinding and promoter clearance by RNA polymerase: Detection by single-molecule DNA nanomanipulation. Proc. Natl. Acad. Sci. U.S.A. 101, 4776–4780 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapanidis A. N., et al., Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Revyakin A., Liu C., Ebright R. H., Strick T. R., Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkelman J. T., et al., Crosslink mapping at amino acid-base resolution reveals the path of scrunched DNA in initial transcribing complexes. Mol. Cell 59, 768–780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feklistov A., Darst S. A., Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147, 1257–1269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stackhouse T. M., Telesnitsky A. P., Meares C. F., Release of the sigma subunit from Escherichia coli RNA polymerase transcription complexes is dependent on the promoter sequence. Biochemistry 28, 7781–7788 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Koh H. R., et al., Correlating transcription initiation and conformational changes by a single-subunit RNA polymerase with near base-pair resolution. Mol. Cell 70, 695–706.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saecker R. M., et al., Kinetic studies and structural models of the association of E. coli sigma(70) RNA polymerase with the lambdaP(R) promoter: Large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 319, 649–671 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Gries T. J., Kontur W. S., Capp M. W., Saecker R. M., Record M. T. Jr, One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Proc. Natl. Acad. Sci. U.S.A. 107, 10418–10423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mejia Y. X., Mao H., Forde N. R., Bustamante C., Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J. Mol. Biol. 382, 628–637 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg O. G., von Hippel P. H., Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 14, 131–160 (1985). [DOI] [PubMed] [Google Scholar]
- 41.Holbrook J. A., Capp M. W., Saecker R. M., Record M. T. Jr, Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: Interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry 38, 8409–8422 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Kozlov A. G., Lohman T. M., Adenine base unstacking dominates the observed enthalpy and heat capacity changes for the Escherichia coli SSB tetramer binding to single-stranded oligoadenylates. Biochemistry 38, 7388–7397 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.