Significance
Cell-surface glycans are dynamic indicators reporting cell physiological state and pathological processes including occurrence and progression of disease (e.g., cancer). For precise cancer diagnosis and treatment, however, it’s difficult to selectively visualize glycans of different cancer cells or cancer subtypes. Herein, we demonstrate the feasibility of cancer cell membrane–camouflaged azidosugar liposomes for cancer cell–selective metabolic glycoengineering. The biomimetic liposomes inherit the cell selectivity to homotypic cancer cells. Therefore, they cannot only realize selective glycan imaging of different cancer cells and breast cancer subtypes but can also sense lung metastasis of tumors. The strategy is also applicable to the use of tumor tissue–derived cell membranes, implying the clinical prospects for personalized medicine.
Keywords: membrane camouflage, cell selectivity, bioorthogonal chemistry, metabolic glycoengineering, imaging
Abstract
The dynamic change of cell-surface glycans is involved in diverse biological and pathological events such as oncogenesis and metastasis. Despite tremendous efforts, it remains a great challenge to selectively distinguish and label glycans of different cancer cells or cancer subtypes. Inspired by biomimetic cell membrane–coating technology, herein, we construct pH-responsive azidosugar liposomes camouflaged with natural cancer-cell membrane for tumor cell–selective glycan engineering. With cancer cell–membrane camouflage, the biomimetic liposomes can prevent protein corona formation and evade phagocytosis of macrophages, facilitating metabolic glycans labeling in vivo. More importantly, due to multiple membrane receptors, the biomimetic liposomes have prominent cell selectivity to homotypic cancer cells, showing higher glycan-labeling efficacy than a single-ligand targeting strategy. Further in vitro and in vivo experiments indicate that cancer cell membrane–camouflaged azidosugar liposomes not only realize cell-selective glycan imaging of different cancer cells and triple-negative breast cancer subtypes but also do well in labeling metastatic tumors. Meanwhile, the strategy is also applicable to the use of tumor tissue–derived cell membranes, which shows the prospect for individual diagnosis and treatment. This work may pave a way for efficient cancer cell–selective engineering and visualization of glycans in vivo.
Cell-surface glycans play pivotal roles in various physiological and pathological processes (1, 2). For example, they are involved in cell–cell communication, pathogen recognition, inflammation, and immune response (3, 4). In particular, the change of glycome in structure and expression may indicate cancer development and metastasis, such as aberrant MUC1 glycosylation used as a biomarker for diagnosis and prognosis of cancer (5, 6). Therefore, analysis of cellular glycans can augment our understanding of glycan-related biological processes and improve disease diagnosis and treatment. The metabolic glycan-labeling technique assisted by “bioorthogonal chemistry” has gained more prominence in probing glycans on live cells or in vivo (7, 8). The metabolic machinery can be harnessed to incorporate bioorthogonal group-contained sugars into cellular glycans (9, 10). In a second step, the installed functional group is covalently reacted with a complementary report probe. Although this chemical reporter technique has dominated glycosylation investigation thus far, selective imaging of glycans in certain cell types, especially cancer cells, is still far from being satisfactory (11). After an administration to mice, the azidosugars will be metabolized into many cell lines in various tissues, bringing about great interference on subsequent analysis (12).
To selectively achieve glycan imaging, two major methods have been exploited: the enzyme-activated sugar precursor and the liposome-assisted delivery (5, 9, 12–14). In the former case, peptide substrates have been used to cage unnatural azidosugar, which could be specifically activated by cancer-overexpressed enzymes. In the latter case, a ligand-targeted (e.g., folate or RGD), liposome-assisted strategy has been developed to deliver azidosugars and selectively visualize glycans in specific tissues, like tumors. Both of them pave ways for selectively probing cancer cell glycans. Though great efforts have been made, a dilemma is that the present methods may cause off-target effects on cancer cells or subtypes sharing the same biomarkers (13, 15–17). Unfortunately, the frequency of multiple primary tumors in a cancer population has increased to 17% over the past two decades, and cancer usually consists of many subtypes due to tumor heterogeneity (18, 19). Furthermore, given the systemic administration, synthetic carriers may encounter protein adsorption, unfavorable tissue distribution, and rapid elimination by the phagocyte system (20). Thus, it is imperative to develop a biocompatible azidosugars delivery system, which cannot only accomplish different cancer cell–selective glycan imaging but also distinguish cancer subtypes.
Cell membrane coating nanotechnology is becoming a prospective tool for targeting specific cells and diseased tissues (21, 22). Due to the abundant membrane receptors, immunocompatibility, and prolonged circulation time, cell membrane coating is superior to many synthetic surfaces (23). This top-down biomimetic approach makes use of cell membrane–derived vesicles to camouflage nanoparticles as “natural” and bestow them with inherent capabilities of source cells. When administrated into the body, cell membrane coating can reduce the nano–bio interaction caused by traditional synthetic surfaces. With the help of this emerging technique, researchers have made significant achievements, for instance, erythrocyte membrane–coated nanoparticles with prolonged blood circulation properties (24), platelet membrane–encapsulated nanomaterials displaying selective adhesion to impaired vasculatures (25), and leukocyte membrane–cloaked silica microparticles possessing endothelium-crossing features (26). Increasingly, cancer cell membrane–camouflaged nanomaterials become a burgeoning targeted strategy for cancer theranostics owing to the immune evasion and self-recognition ability (27–29).
In light of the merits of cancer cell membrane coating, herein, we fabricate cancer cell membrane–camouflaged azidosugar liposomes for selective glycan engineering of multiple tumors in vivo (Fig. 1). This strategy is facile, avoiding tedious chemical modification of metabolic sugar and synthesis of liposomes modified with ligand. In view of abundant membrane receptors, we hypothesize the cancer cell–biomimetic liposomes can 1) prevent protein corona formation, 2) exhibit immune evasion ability to prolong blood circulation and enhance azidosugars delivery efficacy in vivo, and 3) not only realize multiple cancer cell–selective glycan imaging but also distinguish different cancer subtypes in vivo.
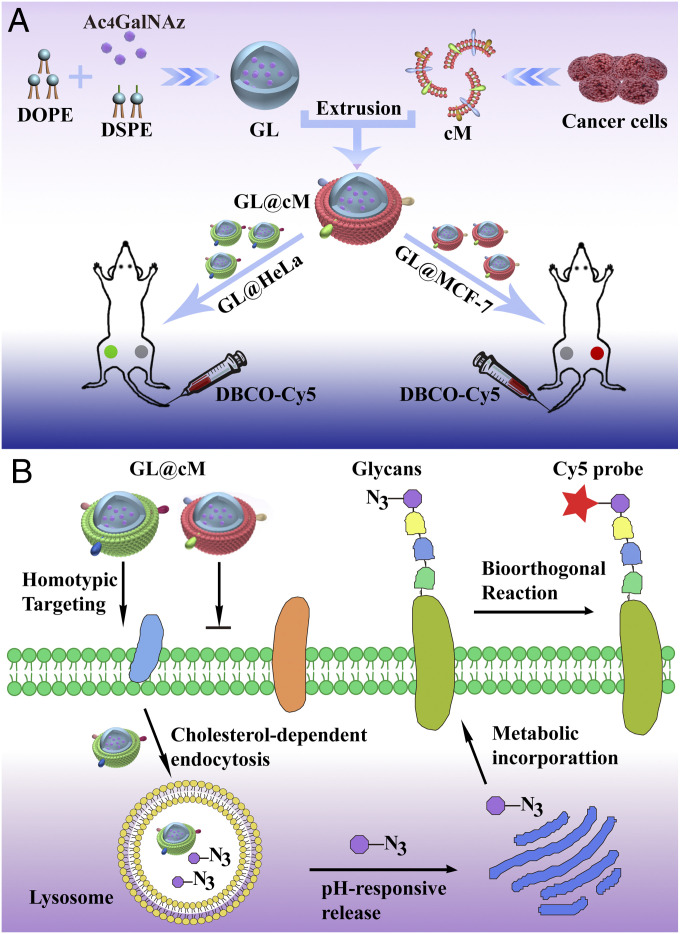
Fig. 1.
(A) Illustration of the construction of biomimetic Ac4GalNAz liposomes (GL@cM) for in vivo progenitor cell-selective glycan imaging. The left tumor is HeLa and the right is MCF-7. (B) Scheme of the path of GL@cM for homotypic cell-selective metabolic glycan labeling.
Results and Discussion
Synthesis and Characterization of Biomimetic GL@cM.
To verify our design, the cancer cell membrane–coated azidosugar liposomes were constructed with the following steps: 1) preparing azidosugar liposomes, 2) collecting cancer cell membranes, and 3) camouflaging azidosugar liposomes with cancer cell membranes (Fig. 1). Specially, the azidosugar liposomes were prepared with 1,2-Distearoyl-sn-Glycero-3-Phosphoethanolamine-poly(ethylene glycol)-amine (DSPE-PEG-NH2), 1,2-dioleoyl-sn-glycero-3-phoshoethanolamine (DOPE), and N-azidoacetylgalactosamine-tetraacylated (Ac4GalNAz) by a thin-film hydration method (30, 31). Ac4GalNAz was extensively applied in investigating mucin-type O-linked glycosylation, whose form variations were associated with most epithelial cancers (32). The final encapsulation efficiency of azidosugar was ∼36.4%, and the loading capacity was 20.8% as identified by high-performance liquid chromatography (HPLC) analysis (SI Appendix, Table S1). The nanometer-sized azidosugar liposomes (defined as GL) were exhibited by transmission electron microscopy (Fig. 2A), which was smaller than their hydrodynamic diameter (Fig. 2E). The Zeta potential was about +5.04 mV (SI Appendix, Fig. S4). The morphology and size of azidosugar liposomes scarcely changed within 24 h in phosphate-buffered saline (PBS) at pH 7.4 (SI Appendix, Figs. S1 and S2A). However, in intracellular acidic environments of lysosomes (pH 4.5 to 5) (23), the DOPE in liposomes could transfer from a lamellar phase to a fusogenic hexagonal phase, resulting in liposome disintegration and cargo release (33). The increased dynamic light scattering (DLS) size demonstrated a quick phase transformation in liposomes within 6 h when immersed in pH 5.0 PBS, which was consistent with previous reports (SI Appendix, Fig. S2B) (34). Then, the pH-sensitive functionality of liposomes was appraised by monitoring the cargo release profile in PBS at pH 7.4 and 5.0. Rhodamine B (denoted as RB) was used as simulated cargo whose loading ratio was 15.6% as determined by ultraviolet–visible spectroscopy. According to the profile, the RB release at pH 5.0 was more rapid than pH 7.4 over time. The cargo release percentage reached 72% at pH 5.0 after 12 h, while it was only 29% at pH 7.4 (Fig. 2D). These results verified that the liposomes were able to demonstrate pH-sensitive cargo release in mild acidic environments.
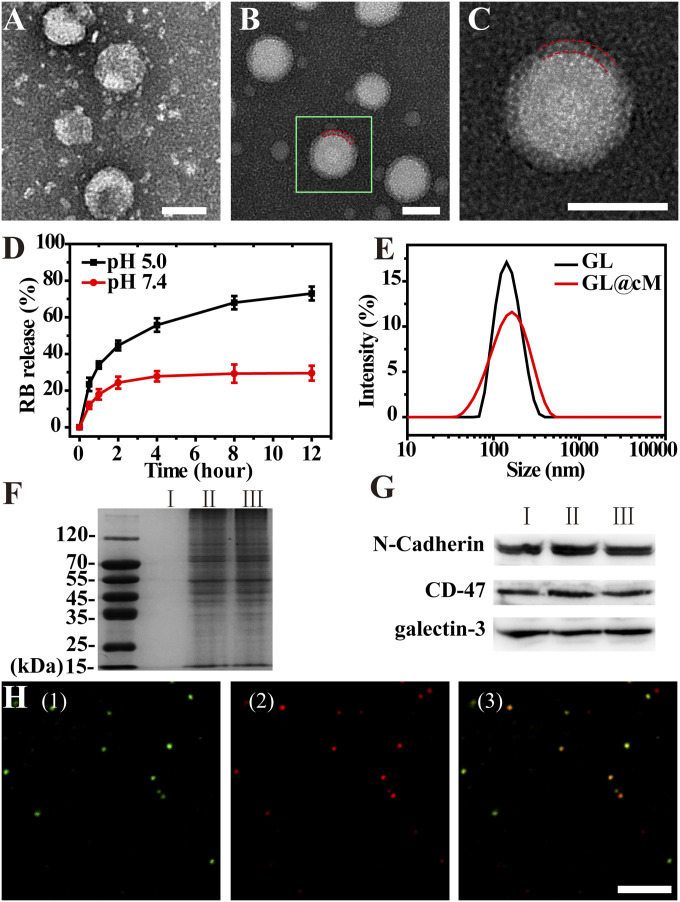
Fig. 2.
Characterization of GL and GL@cM. Transmission electron microscopy pictures of (A) GL and (B) GL@cM. (Scale bar: 100 nm.) (C) Magnification of one segment of B. The red dashed lines showed the membrane coating. (Scale bar: 100 nm.) (D) RB release from liposomes in PBS at different pH. (E) The DLS size distribution of GL and GL@cM. (F) Protein profiles in I: GL, II: cM, and III: GL@cM characterized by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. (G) Western blotting analysis of I: cancer cell lysate, II: cM, and III: GL@cM. (H) Confocal laser scanning microscopy pictures of GL@cM (1) membrane labeled with fluorescein isothiocyanate (FITC) (green), (2) liposomes encapsulated with RB (red), and (3) the merged image. (Scale bar: 2 μm.) The cM was derived from HeLa cells.
Next, the cancer cell membrane fragments (cM) were collected according to previous reports (SI Appendix, Fig. S3) (27). Self-assembly of the membrane fragments on the GL surface was driven by ultrasonication and repeated extrusion to get cancer cell membrane–camouflaged Ac4GalNAz liposomes (termed GL@cM) (mass fraction of Ac4GalNAz was 18.1% as identified by HPLC) (SI Appendix, Table S1). Various characterizations were conducted to validate the successful preparation of GL@cM. After membrane decoration, the size of GL increased from 142 to 164 nm (Fig. 2E). The Zeta potential of GL@cM was about −12.4 mV, nearly equal to membrane fragments (SI Appendix, Fig. S4). Compared to GL, the size of GL@cM remained almost constant in pH 7.4 PBS for 7 d (SI Appendix, Fig. S5). The improved stability might be ascribed to the enhanced electrostatic repulsion and stabilizing effect of hydrophilic surface glycans on cell membranes (30, 35). More than 90% GL@cM was successfully cloaked by the cM according to a colocalization in Fig. 2H. The partial colocalization of green membranes and red liposomes encapsulated with RB (RB-L) in cells suggested that RB-L@cM were more likely taken up via the endocytic pathway and then released as cargo due to the pH-sensitive capacity (SI Appendix, Fig. S6) (27). Subsequently, sodium dodecyl sulfate–polyacrylamide gel electrophoresis verified the reservation of membrane proteins in GL@cM, whereas no protein was detected in GL (Fig. 2F). Furthermore, the presence of specific homologous adhesion proteins like galectin-3 and N-cadherin on GL@cM was affirmed by Western blotting analysis (Fig. 2G), indicating the possibility of specific recognition between GL@cM and source cancer cells via the homologous binding mechanism (36, 37).
Reduced Protein Corona Formation and Antiphagocytosis of Cancer Cell Membrane–Cloaked Liposomes.
When nanoparticles are injected into blood, they adsorb proteins to form a coating called a protein “corona” (35). The acquired corona affects the biological identity of nanoparticles, causes elimination by phagocytes, and ultimately decides the biodistribution of nanoparticles (38). To investigate whether the biomimetic GL@cM nanoparticles could prevent proteins from forming a corona on the GL@cM suface, we incubated GL@cM and GL with fluorescent IgG, one abundant serum protein. Compared to bare GL, the GL@cM reduced IgG adsorption by 2.4-fold (Fig. 3A). Furthermore, they were incubated with 50% fetal bovine serum (FBS). The digital picture and DLS size distribution revealed that bare GL could not keep stable and would aggregate due to the protein adsorption after incubating with 50% FBS for 12 h (Fig. 3B and SI Appendix, Fig. S7). Whereas GL@cM showed barely noticeable changes, implying the significant role of the biomimetic membrane for reducing protein adsorption in serum.
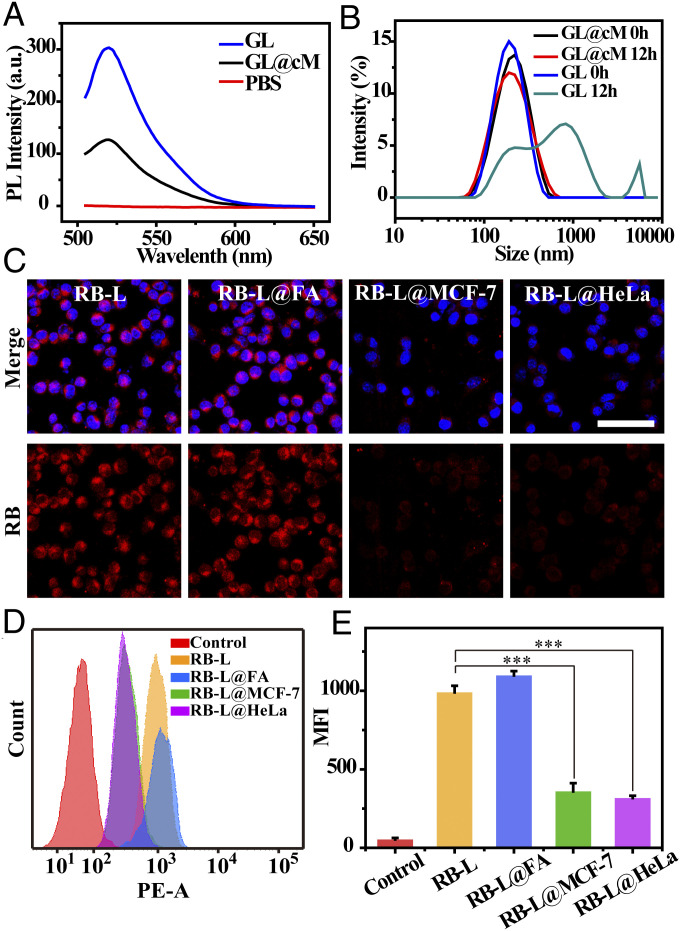
Fig. 3.
Biomimetic GL@cM reduced surface corona formation and phagocytosis. (A) Fluorescence spectrometric analysis of fluorescent IgG asorbed by GL, and GL@cM nanoparticles. (B) The DLS size changes of GL and GL@cM after 50% FBS incubation for 12 h. (C) Confocal laser scanning microscopy pictures of RAW264.7 cells incubated with RB-L, RB-L@FA, RB-L@MCF-7, and RB-L@HeLa for 4 h. The cell nucleus was stained with Hoechst (blue). Liposomes were encapsulated with Rhodamine B (red). (Scale bar: 50 μm.) (D and E) Flow cytometry assessment of RAW264.7 cells treated with RB-L, RB-L@FA, RB-L@MCF-7, and RB-L@HeLa for 4 h. Data are presented as mean ± SD (n = 3). Asterisks indicate significant differences (***P < 0.001).
An ideal delivery carrier should also possess gratifying immune compatibility, reducing the potential risks caused by macrophage activation through the phagocytosis (39). Accumulated evidence revealed that cancer cell membranes camouflaged nanomaterial-reserved integrin-associated protein CD47, which mediated a “do not eat me” signal to escape phagocytosis (40). The presence of CD47 on GL@cM was identified by Western blotting analysis (Fig. 2G). For testifying the immune evasion capability of GL@cM, fluorescent Rhodamine B was chosen as the azidosugar substitute encapsulated by liposome@cM (termed RB-L@cM). The bare RB-L, RB-L@FA, RB-L@MCF-7, and RB-L@HeLa loaded with equal amounts of Rhodamine B were cultured with RAW264.7 cells for 4 h before imaging. As revealed in Fig. 3C, a clear red fluorescence was monitored in RAW264.7 cells treated with RB-L. In contrast, just a dim red fluorescence was observed in RAW264.7 cells in RB-L@MCF-7 and RB-L@HeLa groups. However, RB-L@FA could not escape phagocytosis by macrophage according to the distinct red fluorescence in RAW264.7 cells. Furthermore, the quantitative flow cytometry analysis displayed the same results (Fig. 3 D and E). In detail, uptake of RB-L@MCF-7 and RB-L@HeLa were just 32.7 and 28.2% of the RB-L, indicating a desirable immune evasion capability of RB-L@cM. The above outcomes demonstrated that the liposomes camouflaged with cancer cell membranes possessed stealth ability to avoid systemic clearance of the phagocytes, which would facilitate metabolic glycan labeling in vivo.
Multiple Receptors Mediated Cancer Cell–Selective Glycan Imaging.
For verifying the selectivity of RB-L@cM to homotypic cancer cells, the cellular internalization of RB-L@cM was evaluated upon incubating with different cell lines. Thus, the RB-L@HeLa was incubated with HeLa, MCF-7, A549, 4T1, and CT26 cells. Intriguingly, as revealed by confocal imaging and flow cytometry analysis, the HeLa cells exhibited the prominent selectivity of RB-L@HeLa (SI Appendix, Figs. S8 and S9), an ∼8.8-fold mean fluorescence intensity (MFI) increase, much higher than other heterotypic cells (1.3- to 1.8-fold MFI increase). Gratifyingly, when RB-L was cloaked with MCF-7 cell membranes, more considerable internalization by the MCF-7 cell was monitored, the MFI of which increased 6.9-fold (SI Appendix, Figs. S10 and S11). These results manifested the progenitor cell selectivity of cancer cell membrane coating even if various cancer cells were present.
The encouraging homotypic-targeting ability of membrane-camouflaged liposomes stimulated us to evaluate their effects on cancer cell–selective glycan imaging. HeLa and MCF-7 cells were chosen for following experiments because of their similar levels of glycans labeling (SI Appendix, Fig. S12). Both HeLa and MCF-7 cells exhibited metabolic glycoengineering in a dose-dependent manner as labeled with the aza-dibenzocyclooctyne-Cy5 conjugate (DBCO-Cy5) via bioorthogonal chemistry.
According to previous reports, the folate-modified liposomes could effectively carry azidosugar to folate receptor (FR)-positive cancer cells, conducive to selective glycan labeling (9). Compared with the folate-modified liposomes (GL@FA), cancer cell membrane–camouflaged liposomes possessed abundant receptors related with homotypic targeting, such as N-cadherin, galectin-3, and so on (36, 40). Hence, we wondered if the delivery efficacy of membrane-coated liposomes was superior to folate-modified liposomes. As convinced by fluorescence micrographs and flow cytometry analysis, the delivery efficacy of GL@HeLa was 1.7-fold higher than GL@FA (Fig. 4 A–C). Western blot analysis also verified the generation of more cell-surface azido groups when treated with GL@HeLa (SI Appendix, Fig. S13). These results indicated that multiple receptor-mediated homotypic targeting was more powerful than single-ligand targeting. Furthermore, HeLa cells were incubated with various concentrations of GL@HeLa for 24 h (calculated based on azidosugar concentration). The fluorescence intensity increased in a concentration-dependent manner (SI Appendix, Figs. S14 and S15). Pronounced fluorescence intensity was attained at a concentration as low as 12.5 μM, reaffirming the remarkable azidosugars delivery efficacy mediated by multiple receptors (SI Appendix, Fig. S15).
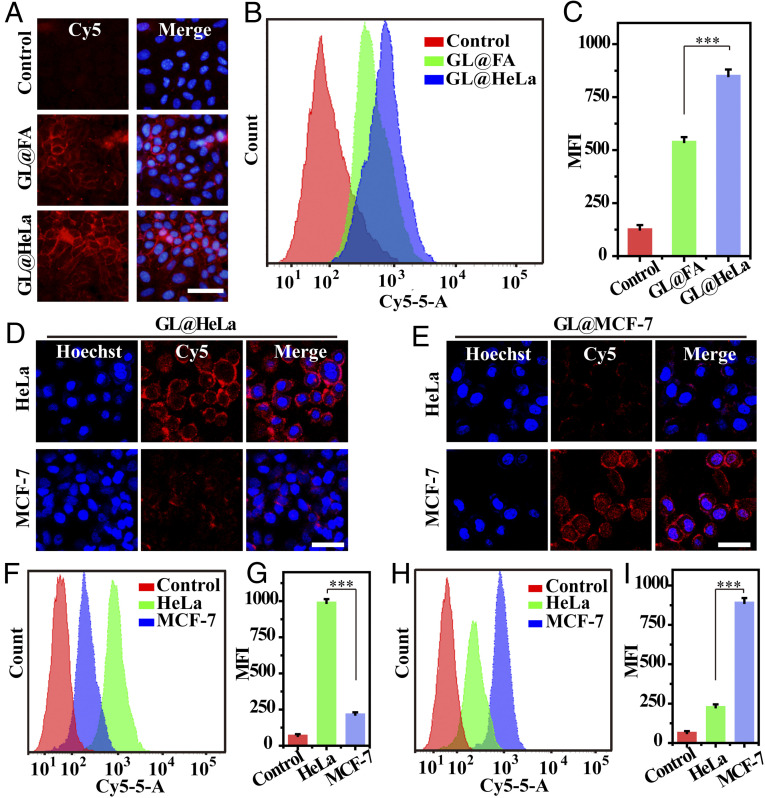
Fig. 4.
Cell-selective glycan imaging mediated by multiple receptors of GL@cM. (A) Fluorescence micrographs of HeLa incubated with 50 µM GL@FA or 50 µM GL@HeLa. (B and C) Quantitative flow cytometry assessment of A. (D and E) CLSM images of cells incubated with 50 µM GL@HeLa and GL@MCF-7, respectively. (F and G) Flow cytometry assessment of cells incubated with 50 µM GL@HeLa. (H and I) Flow cytometry assessment of cells incubated with 50 µM GL@MCF-7. The cell nucleus was stained with Hoechst (blue). Cells were labeled with DBCO-Cy5 for both imaging and flow cytometry assessment. (Scale bars: 50 μm.) Data are presented as mean ± SD (n = 3). Asterisks indicate significant differences (***P < 0.001).
Next, the biomimetic azidosugar liposomes were used for cell-selective glycan imaging in vitro. When treated with GL@HeLa, HeLa cells showed apparent fluorescent signals on the surface; however, much weaker fluorescence was observed on the MCF-7 cell surface (Fig. 4D). Similarly, when treated with GL@MCF-7, MCF-7 cells displayed much stronger fluorescence on the surface than HeLa cells (Fig. 4E). Moreover, the selective efficiency was quantified by flow cytometry. The metabolic labeling results of the GL@HeLa group showed that the fluorescent signal of HeLa cells was about 6.7-fold higher than MCF-7 cells (Fig. 4 F and G). In contrast, opposite targeting selectivity was observed in the GL@MCF-7 group, in which the fluorescence intensity of MCF-7 cells was 5.3-fold more than HeLa cells (Fig. 4 H and I). However, though GL@FA was able to target FR-positive cells rather than FR-negative A549 cells, they could not realize cell-selective glycan imaging between HeLa and MCF-7 because both of them were FR positive (SI Appendix, Figs. S16 and S17) (15, 41). These data demonstrated that the cancer cell membrane coating was indispensable for exclusive cell-selective glycan imaging.
Internalization Pathway and Release Mechanism.
For further clarifying the endocytosis mechanism of membrane-camouflaged liposomes, HeLa cells were incubated with RB-L@HeLa and different endocytosis pathway inhibitors, including chlorpromazine (CHL, inhibitor of clathrin-mediated endocytosis), amiloride (AMI, inhibitor of Na+/H+ pump–related macropinocytosis), and methyl-beta-cyclodextrin (MBD, inhibitor of cholesterol-dependent endocytosis) (29, 42). As validated by flow cytometry analysis, inhibitor MBD substantially impeded endocytosis of nanoparticles, suggesting the uptake of RB-L@HeLa was primarily mediated by cholesterol-dependent endocytosis pathways (SI Appendix, Fig. S18). The similar result was obtained when MCF-7 cells were treated with RB-L@MCF-7, implying the internalization of RB-L@MCF-7 was through the same pathways (SI Appendix, Fig. S19). What’s more, MBD was added into the cell medium when HeLa cells were incubated with GL@HeLa. The following DBCO-Cy5 labeling reaffirmed that the metabolic incorporation of Ac4GalNAz was notably prevented (SI Appendix, Fig. S20). Afterward, we observed the intracellular track of membrane-camouflaged liposomes to visualize their transport (SI Appendix, Fig. S21). At first, RB-L@HeLa quickly attached to HeLa cell surfaces within 1 h as a result of homotypic recognition of membrane receptors. Then, many intracellular puncta were observed and colocalized with lysosomes at 2 h, implying GL@HeLa got into the lysosomes. At 4 h, the distinct red fluorescence indicated the nanoparticles were well colocalized with lysosomes. After 8 h, however, the red fluorescence gradually diffused to the cytoplasm over time, implying the successful release of cargos in mild acidic lysosomes. The colocalization of RB-L@HeLa with endosomes showed consistent results (SI Appendix, Fig. S22). Therefore, we deduced that the GL@cM could target progenitor cells via homotypic recognition of membrane receptors, enter cells through cholesterol-dependent endocytosis pathways, and get into endosomes/lysosomes, where the azidosugars were released, and then intercept the salvage pathways of monosaccharides (Fig. 1B) (9).
Visualizing Multiple Tumor Glycans Efficiently and Selectively In Vivo.
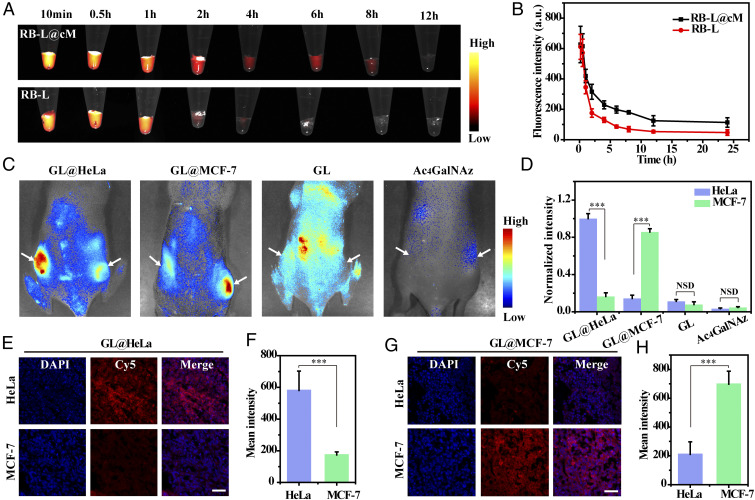
The prolonged circulation time of azidosugars was facilitated to accumulate in tumor tissues and realize efficient tumor labeling in vivo (28). Thus, we investigated the blood circulation spans of bare RB-L and RB-L@cM. After intravenous injection of RB-L and RB-L@cM into healthy mice, blood specimens were gathered at different time points for fluorescence imaging and fluorescence spectra analysis. The RB-L@cM displayed notably prolonged circulation time and high reservation in blood circulation compared to the RB-L, owing to the superior stealth property of cancer cell membrane camouflage (Fig. 5 A and B). In addition, ex vivo imaging exhibited that the displayed fluorescent signals in major organs (liver, spleen, and kidney) of the RB-L@cM group were weaker than the uncoated RB-L group, which might be attributed to the decreased cells uptake caused by membrane coating (SI Appendix, Fig. S23) (43).
Fig. 5.
In vivo selective fluorescence imaging of tumor-associated glycans using GL@cM. (A) Fluorescence images of blood samples from mice injected with RB-L and RB-L@cM. (B) In vivo pharmacokinetic curves of RB-L and RB-L@cM as quantified by fluorescence spectroscopy (n = 3). (C) In vivo fluorescence visualization of mice with tumors administrated with GL@HeLa (60 mg ⋅ kg−1), GL@MCF-7 (60 mg ⋅ kg−1), GL (60 mg ⋅ kg−1), and Ac4GalNAz (60 mg ⋅ kg−1) on 4 consecutive days. DBCO-Cy5 was intravenously injected into the mice on day 5. (D) Quantitative analysis of the normalized fluorescence intensity. Data are presented as mean ± s.d. (n = 3). (E and F) Fluorescence photographs and intensity analysis of tumor sections from mice injected with GL@HeLa. (G and H) Fluorescence photographs and intensity analysis of tumor sections from mice injected with GL@MCF-7. Cell nucleus was stained with DAPI. Fluorescence intensity of confocal microscopy images was analyzed by using Nikon Eclipse Analysis software. Data were presented as mean intensity (n = 10). (Scale bars: 50 µm.) Asterisks indicate significant differences (NSD: no significant difference, ***P < 0.001).
Next, we studied whether GL@cM could achieve cancer cell–selective glycan visualization in living mice. Thus, the mouse models were inoculated with HeLa tumor (left flank) and MCF-7 tumor (right flank) to imitate multiple primary tumors, which showed similar size (SI Appendix, Figs. S24A and S25). The tumor growth rates and body weight were not influenced by the administration of different nanomaterials (SI Appendix, Figs. S25 and S26). Particularly, the tumor-bearing mice were intravenously administrated with different cancer cell membrane–camouflaged azidosugar liposomes, once a day for 4 consecutive days. On day 5, DBCO-Cy5 was intravenously injected to image azidosugars on tumor cell surfaces by in vivo bioorthogonal click chemistry (SI Appendix, Fig. S24B). Interestingly, when injected with GL@HeLa, substantial fluorescence signals were exhibited in the HeLa tumor, the normalized intensity of which was 6.1-fold higher than the MCF-7 tumor (Fig. 5 C and D). Thus, it confirmed that GL@HeLa was able to actively recognize and “home” to the homotypic HeLa tumor while “bypassing” the coexisting heterotypic MCF-7 tumor to a large extent (27). Similarly, the selective glycan imaging of the MCF-7 tumor could be realized by substituting the HeLa cell membrane for the MCF-7 cell membrane to cloak the azidosugar liposomes. As illustrated in Fig. 5 C and D, the MCF-7 tumors exhibited 6.0-fold higher fluorescence intensity over the HeLa tumor, manifesting the role of homotypic cell membranes in tumor self-targeting. The tumor tissue sections and ex vivo analysis of tumors were consistent with above data, indicating the selectively expressed azido groups in homotypic tumors (Fig. 5 E–H and SI Appendix, Fig. S27). These results corroborated our design that the membrane-camouflaged azidosugar liposomes were capable of realizing multiple tumor-selective imaging of glycans by an initiative targeting behavior. In addition, bare GL (i.v.) and Ac4GalNAz (i.p.) were administrated into tumor-bearing mice as a comparative study. Unfortunately, minimal labeling effects were observed for both HeLa and MCF-7 tumors, which might result from the fast clearance of GL by phagocytes and the short circulation spans of small molecule Ac4GalNAz (Fig. 5 C and D). Cell toxicity and hematoxylin and eosin–stained sections of main organs and tumors suggested ignorable toxicity of membrane-camouflaged liposomes within an experimental dose (SI Appendix, Figs. S28 and S29). In brief, our results validated cancer cell membrane–camouflaged liposomes not only markedly enhanced the delivery of azidosugar to tumor tissues but also actualized tumor-selective glycan imaging in a homotypic-targeting manner. Both of the factors played pivotal roles in achieving effective and selective glycan imaging of tumor tissues.
Selectively Visualizing Glycans of Breast Cancer Subtypes.
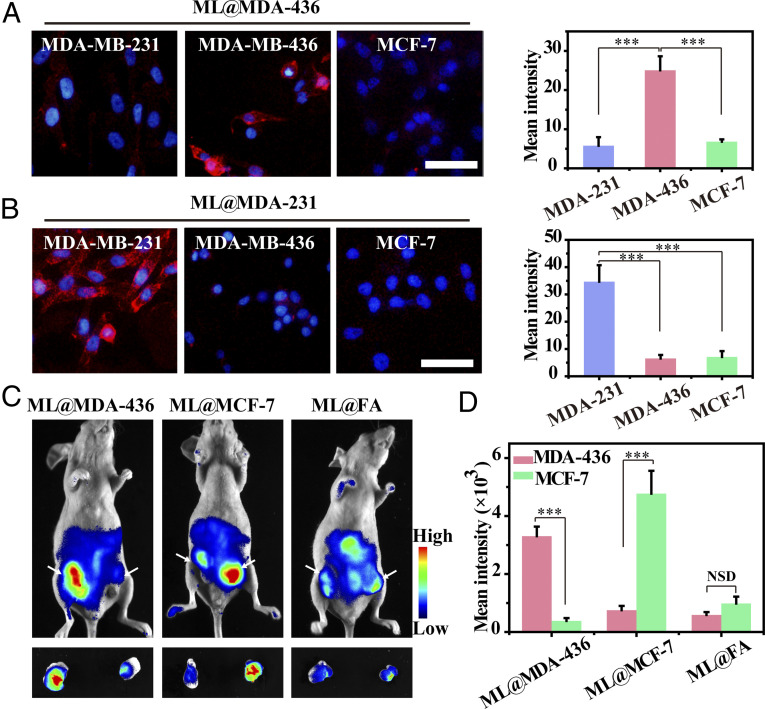
Breast cancer is one of the highly heterogeneous tumors, and each subtype has a different prognosis and treatment response (44). Triple-negative breast cancer (TNBC), one subtype of breast cancer, is considered to be with the worst prognosis due to high metastatic nature (45). Due to similar molecular markers shared by TNBC and other breast cancer subtypes as well as insufficiently expressed tumor targets of TNBC, it is a challenge to distinguish TNBC from other breast cancer subtypes (46, 47). Therefore, we investigated the possibility of cancer cell membrane coating technology for selectively labeling glycans of different breast cancer subtypes (29). Two different TNBC subtypes (BRCA mutant cell MDA-MB-436 and BRCA nonmutant cell MDA-MB-231) and Luminal A subtype (MCF-7 cell) were chosen to verify our design. Moreover, N-Azidoacetylmannosamine-tetraacylated (Ac4ManNAz) was used to prepare membrane-coated Ac4ManNAz liposomes (ML@cM) for probing cancer cell-surface sialoglycoconjugates, which validated the universality of the biomimetic liposome as a carrier for diverse azidosugars (SI Appendix, Table S1). As illustrated in Fig. 6A, MDA-MB-436 cells showed obvious fluorescence signals when treated with ML@MDA-436, 4.3-fold and 3.7-fold higher than MDA-MB-231 and MCF-7 cell group. The outcomes suggested that these biomimetic liposomes not only differentiated TNBC from other breast cancer subtypes but also distinguished different TNBC subtypes. When treated with ML@MDA-231 or ML@MCF-7, a similar result was obtained that the homotypic cells displayed over 4.9-fold or 7.8-fold higher fluorescence on the cell surface than that of heterotypic cells (Fig. 6B and SI Appendix, Fig. S30). However, ML@FA could not realize cell-selective glycan imaging between these breast cancer cells because all of them expressed folate receptors (SI Appendix, Fig. S31) (48).
Fig. 6.
Selectively visualizing glycans of breast cancer subtypes. Fluorescence imaging and quantitative examination of diverse subtypes of breast cancer cells incubated with 50 µM (A) ML@MDA-436 and (B) ML@MDA-231, respectively. Subsequently, cells were labeled with DBCO-Cy5. Cell nucleus was stained with Hoechst (blue). (Scale bars: 50 μm.) A quantitative examination of fluorescence images was analyzed by ImageJ software. Data were displayed as average intensity (n = 10). (C) In vivo fluorescence visualization of mice with tumors administrated with ML@MDA-436 (60 mg ⋅ kg−1), ML@MCF-7 (60 mg ⋅ kg−1), and ML@FA (60 mg ⋅ kg−1) on 4 consecutive days. DBCO-Cy5 was intravenously injected into mice on day 5. (D) Quantitative analysis of the intensity from whole-body fluorescence imaging. Data are presented as mean ± SD (n = 3). Asterisks indicate significant differences (NSD: no significant difference, ***P < 0.001).
Furthermore, the mice were inoculated with an orthotopic MDA-MB-436 tumor (left flank) and an MCF-7 tumor (right flank) simultaneously to develop multiple breast cancer subtype models. In vivo experiments revealed that when injected with ML@MDA-436 and ML@MCF-7, multitudinous fluorescence signals appeared in the homotypic tumor, whose fluorescence intensity was 8.6-fold and 6.4-fold higher than that of heterotypic tumors, respectively (Fig. 6 C and D). ML@FA was administrated into the tumor-bearing mice as a comparative group. Regrettably, slight labeling effects were observed in MDA-MB-436 and MCF-7 tumors, which might result from its quick clearance by phagocytes (Fig. 6 C and D). The targeting ability of ML@cM to MDA-MB-436 and MCF-7 was over 5.6-fold and 4.8-fold higher than ML@FA, respectively (Fig. 6D). These encouraging results suggested that the cancer cell membrane coating held tremendous promise for selectively labeling glycans of diverse breast cancer subtypes, conducive to molecular classification and precise theranostics of breast cancers.
Application in Probing Tumor-Spontaneous Metastasis.
Tumor metastasis, the malignant cell translocation from primary tumors to distant tissues, is responsible for 90% of mortality in cancer patients (49). Thus, we assessed the performance of cancer cell membrane–coated Ac4GalNAz liposomes for tracking the variation of tumor glycans and studying the correlation of tumor mucin glycosylation and spontaneous metastasis. To explore this, the 4T1 orthotopic mammary tumor-spontaneous metastasis model was developed (40). Then, GL@4T1 was used for tracking newly synthesized mucin-type glycans at different period of tumor development. After 6, 16, and 26 d of tumor growth, the mice were intravenously injected with GL@4T1 in equivalent doses for a consecutive 4 d and then injected with DBCO-Cy5 for imaging on day 10, 20, and 30, respectively. We found that the fluorescence signal of the primary tumors enhanced with time, suggesting a time-dependent increase of biosynthesis of mucin-type glycans in the process of tumor growth (SI Appendix, Fig. S32). As time went by, notable metastatic nodules were found in the lung on day 30, suggesting the metastasis of primary tumors (SI Appendix, Fig. S33 A and B). Meanwhile, dramatically enhanced fluorescence intensity was observed in the lung with metastatic nodules in the 30-d group, implying the GL@4T1 could trace and realize the glycan labeling of metastatic tumors (SI Appendix, Fig. S34 A and B). The outstanding performance of GL@4T1 for targeting primary tumors and lung metastasis was due to the homotypic-targeting potential of cell membrane coating and the significantly prolonged circulation time with the immune evasion effects. Finally, we sought to investigate the change of mucin-type glycoproteins in primary tumors during tumor growth and metastasis. Tumor tissues were isolated from mice on day 10, 20, and 30 for Western blotting analysis. The Western blotting analysis displayed an increased intensity over time and a significantly increased biosynthesis of mucin-type glycoproteins on day 30, which might be associated with aberrant overexpression and glycosylation of metastasis-associated mucins (SI Appendix, Fig. S35) (50). Combined with gel-based proteomic identification using tandem mass spectrometry, the glycoproteins as biomarkers could be used for evaluating metastasis and prognosis of tumors, even screening therapeutic targets (12, 51, 52). Thus, the cancer cell membrane–coated azidosugar liposomes were suitable for analysis of glycoproteins associated with primary tumor metastasis, which might advance our understanding of cancer metastasis and treatment.
Cancer Tissue–Derived Cell Membranes for Selective Glycan Labeling.
In consideration of interpatient heterogeneity of tumors and clinical safety, utilizing patient tumor tissue cell membrane–based nanocarriers has a future for personalized diagnosis and therapy (53, 54). Therefore, we tried to study the feasibility of using tumor tissue–derived cell membranes for homotypic targeting. Firstly, 4T1 and B16F10 cells were isolated from 4T1 and B16F10 tumor tissues and cultured for further membrane extraction (SI Appendix, Fig. S36). Then, the GL@4T1 and GL@B16 were prepared for metabolic glycan labeling in cells and in vivo (left is 4T1 tumor and right is B16F10 tumor) (SI Appendix, Fig. S37 A and B). In vitro and in vivo results all indicated GL@4T1 and GL@B16 realized specific glycan visualization of homotypic tumors rather than heterotypic tumors (SI Appendix, Figs. S38 and S39). The imaging efficiency and selectivity of homotypic tumors were superior to GL@FA and bare GL, implying the feasibility of tumor tissue–derived cell membranes for clinical application.
Furthermore, we wondered whether there were differences between autologous tumor-derived GL@4T1 (named as GL@4T1-au) and allogeneic tumor-derived GL@4T1 (named as GL@4T1-al) when used for metabolic glycan labeling. In vitro labeling experiments revealed that the cells treated with GL@4T1-al displayed slightly lower labeling performance, about 18% lower than cells treated with GL@4T1-au (SI Appendix, Fig. S40). To test the difference in vivo, 4T1 tumor-bearing mice were injected with GL@4T1-au and GL@4T1-al. As labeled with the DBCO-Cy5, all of the 4T1 tumor tissues exhibited apparent fluorescence of tumor glycans (SI Appendix, Fig. S41). Moreover, the tumor fluorescence intensity of GL@4T1-au–treated mice slightly increased as compared with other mice. This might be ascribed to the individual variations in tumor-specific receptor expression caused by interindividual, intertumor heterogeneity (55, 56).
Discussion
In summary, we have designed and constructed cancer cell membrane–camouflaged azidosugar liposomes to realize selective imaging of tumor glycans in vivo. Owing to cancer cell membrane coating, the biomimetic azidosugar liposomes revealed reduced protein corona formation and admirable antiphagocytosis by macrophages. This immune evasion ability extended the blood circulation time of biomimetic azidosugar liposomes. Moreover, homotypic cancer cell–selective glycan labeling was accomplished through multiple receptor-mediated targeting, the targeting efficacy of which was 1.7-fold higher than single-ligand targeting strategy in vitro. In vivo data validated that azidosugar liposome@cM achieved multiple tumor-selective glycan imaging in a homotypic-targeting manner, whose imaging efficiency was over 3.4-fold higher than azidosugar liposome@FA in athymic nude mice and immune competent mice.
Besides, cancer cell membrane–coated liposomes also played a role in selectively labeling glycans of different breast cancer subtypes, even TNBC subtypes, which shared similar molecular markers and expressed tumor targets insufficiently. Tumor-spontaneous metastasis models showed that GL@4T1 did well in labeling not only primary tumors but also metastatic tumors, providing a powerful solution to evaluate progression and search biomarkers for metastasis. Finally, the strategy for fabricating cancer cell membrane–cloaked azidosugar liposomes was expandable to the usage of cancer tissue–derived cell membranes. In vitro and in vivo study revealed that tumor tissue–derived GL@cM also successfully accomplished multiple tumor-selective glycan imaging in a homotypic-targeting manner, and the autologous tumor-derived GL@4T1 had better performance. These data indicate that the cell membrane coating strategy possesses potential for personalized diagnosis and treatment. Recently, it has been reported that per-acetylated azidosugars might induce nonspecific S-glyco modification during metabolic glycan labeling, likely causing false-positive results (57–59). In consideration of the poor membrane permeability of nonacetylated unnatural sugars, biomimetic liposomes can be used to realize efficient and accurate delivery of unprotected or partially protected azidosugars for metabolic glycan labeling.
While cell membrane coating technology shows encouraging performance, the future clinical applications of cancer cell membrane–cloaked azidosugar liposomes have a long way to go. In achieving the clinical translation of them, further systematic evaluation of potential short-term and long-term toxicity is imperative, even though our pilot toxicity investigation can guarantee the safety concerns of our biomimetic nanocarriers to some extent (60). In addition, cell membrane coating technology should be optimized and regularized to avoid immune responses from the blood complement system owing to incomplete or uneven coverage of nanomaterial (61). Scalability and consistency are also necessary demands for clinical translational application (61). At present, the study of biomimetic membrane-coated nanomaterials is at the stage of laboratory researches or early phase clinical trials. Further investigations are needed to seek low-cost and large-scale manufacturing of biomimetic membrane-coated nanomaterials for commercial supply because cell membrane components are rare and valuable. The standard protocols and infrastructures of creating biomimetic membrane-coated nanomaterials should be well established, including cell culture methods (e.g., adherent and suspension) (62), membrane isolation methods (e.g., specific isolation kits and high-speed centrifugation), and membrane coating technology (e.g., physical extrusion and sonication-based spontaneous formation) (21).
To realize personalized medicine, the access to real patients’ tumor membrane samples is another challenge. In clinical practice, two main ways are widely used to obtain tumor tissue samples: surgical resection and puncture biopsy (63). At present, tumors are still mainly removed by excisional biopsy for therapy and diagnosis, by which large massive tumor tissues can be obtained. Puncture biopsy has gradually displaced excisional biopsy as an authoritative diagnostic method for breast tumors and so on, which is safe and convenient to acquire superficial or deep tumor tissues (64). Of course, we have to admit there are some limitations of biopsies and surgical resections. For example, they are uncomfortable and increase the cost of patient care. Besides, even though the complications are low and controllable, biopsies are not without clinical safety concerns (65). Thus, we propose a less invasive alternative. Extracellular vesicles (EVs) secreted by tumors, such as exosomes and microvesicles, are prospective candidates for tumor targeting. In addition to cell culture medium, human tissues and fluids can be used as EV sources, indicating a more convenient accessibility (66). Delightedly, autologous dendritic cells, malignant cells, and ascites have been chosen as sources to produce autologous EV-based nanomaterials for successfully inducing immune response and targeting malignant cells (62). In light of these achievements, it is reasonable and practical to devise EV-based nanocarriers for cancer cell–selective engineering and visualization of glycans, avoiding complicated surgical operations. So, we envision that this biomimetic strategy, as a powerful tool for labeling tumor glycans, has tremendous potential for precisely probing tumor onsets and development.
Materials and Methods
Preparations of Ac4GalNAz Liposomes and Ac4ManNAz Liposomes.
Ac4GalNAz liposomes were created by a thin-film hydration method (30, 31). Briefly, Ac4GalNAz, DOPE, and DSPE-PEG-NH2 (2 mg, 2 mg, and 2 mg) were dissolved in methanol and dried into a thin film in a flask. Subsequently, the film was immersed in ultrapure water and sonicated at 4 °C for 4 min to form Ac4GalNAz liposomes. And then, they were extruded with a miniextruder using a polycarbonate membrane with a pore size of 200 nm for 10 cycles. Similarly, Ac4ManNAz liposomes (ML) were prepared in the same way.
In order to prepare azidosugar liposomes modified with folate (GL@FA or ML@FA), Ac4GalNAz or Ac4ManNAz, DOPE, and DSPE-PEG-folate (2 mg, 2 mg, and 2 mg) were dissolved in methanol and dried into a thin film in a round flask. Then, the film was hydrated, sonicated, and extruded with a miniextruder like before.
Preparation of cM.
The different cells were collected and suspended in lysing buffer containing membrane protein extraction reagent and incubated in an ice bath for 10 to 15 min. Then, the cells in lysing buffer were freeze thawed repeatedly. After centrifugation at 700 g for 10 min at 4 °C, the supernatant was further centrifuged at 14,000 g for 30 min to gather the membrane fragments. The membrane fragments were lyophilized overnight and stored at −80 °C.
Preparation of GL@cM and ML@cM.
The cM from 5 × 107 HeLa or other cells was mixed with 5 mg GL or ML and extruded through a polycarbonate membrane with pore sizes of 400 and 200 nm to prepare GL@cM and ML@cM.
Supplementary Material
Acknowledgments
Financial support was provided by the National Key Research and Development Program of China (2019YFA0709202), NSF of China (91856205, 21820102009, and 21871249), and the Key Program of Frontier of Sciences (Chinese Academy of Sciences QYZDJ-SSW-SLH052).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022769118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Laughlin S. T., Baskin J. M., Amacher S. L., Bertozzi C. R., In vivo imaging of membrane-associated glycans in developing zebrafish. Science 320, 664–667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Ding L., Ju H., In situ cellular glycan analysis. Acc. Chem. Res. 51, 890–899 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Lin L., et al., A bioorthogonal Raman reporter strategy for SERS detection of glycans on live cells. Angew. Chem. Int. Ed. Engl. 52, 7266–7271 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Wolfert M. A., Boons G.-J., Adaptive immune activation: Glycosylation does matter. Nat. Chem. Biol. 9, 776–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang P. V., Dube D. H., Sletten E. M., Bertozzi C. R., A strategy for the selective imaging of glycans using caged metabolic precursors. J. Am. Chem. Soc. 132, 9516–9518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui J., et al., Localized chemical remodeling for live cell imaging of protein-specific glycoform. Angew. Chem. Int. Ed. Engl. 56, 8139–8143 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Spiciarich D. R., et al., Bioorthogonal labeling of human prostate cancer tissue slice cultures for glycoproteomics. Angew. Chem. Int. Ed. Engl. 56, 8992–8997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grammel M., Hang H. C., Chemical reporters for biological discovery. Nat. Chem. Biol. 9, 475–484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y., et al., mechanistic investigation and multiplexing of liposome-assisted metabolic glycan labeling. J. Am. Chem. Soc. 140, 3592–3602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debets M. F., et al., Metabolic precision labeling enables selective probing of O-linked N-acetylgalactosamine glycosylation. Proc. Natl. Acad. Sci. U.S.A. 117, 25293–25301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie R., Hong S., Feng L., Rong J., Chen X., Cell-selective metabolic glycan labeling based on ligand-targeted liposomes. J. Am. Chem. Soc. 134, 9914–9917 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Xie R., et al., Targeted imaging and proteomic analysis of tumor-associated glycans in living animals. Angew. Chem. Int. Ed. Engl. 53, 14082–14086 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Shim M. K., et al., Cathepsin B-specific metabolic precursor for in vivo tumor-specific fluorescence imaging. Angew. Chem. Int. Ed. Engl. 55, 14698–14703 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Wang H., et al., Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat. Chem. Biol. 13, 415–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahrami B., et al., Folate-conjugated nanoparticles as a potent therapeutic approach in targeted cancer therapy. Tumour Biol. 36, 5727–5742 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Schottelius M., Laufer B., Kessler H., Wester H.-J., Ligands for mapping alphavbeta3-integrin expression in vivo. Acc. Chem. Res. 42, 969–980 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Dai B., Hu Y., Duan J., Yang X.-D., Aptamer-guided DNA tetrahedron as a novel targeted drug delivery system for MUC1-expressing breast cancer cells in vitro. Oncotarget 7, 38257–38269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogt A., et al., Multiple primary tumours: Challenges and approaches, a review. ESMO Open 2, e000172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y., et al., Detecting heterogeneity in and between breast cancer cell lines. Cancer Converg. 4, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knop K., Hoogenboom R., Fischer D., Schubert U. S., Poly(ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. Engl. 49, 6288–6308 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Fang R. H., Kroll A. V., Gao W., Zhang L., Cell membrane coating nanotechnology. Adv. Mater. 30, e1706759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alyami M. Z., et al., Cell-type-specific CRISPR/Cas9 delivery by biomimetic metal organic frameworks. J. Am. Chem. Soc. 142, 1715–1720 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Liu G., et al., Engineering biomimetic platesomes for pH-responsive drug delivery and enhanced antitumor activity. Adv. Mater. 31, e1900795 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., et al., Erythrocyte membrane cloaked metal-organic framework nanoparticle as biomimetic nanoreactor for starvation-activated colon cancer therapy. ACS Nano 12, 10201–10211 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Hu C. M., et al., Nanoparticle biointerfacing by platelet membrane cloaking. Nature 526, 118–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parodi A., et al., Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 8, 61–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J. Y., et al., Preferential cancer cell self-recognition and tumor self-targeting by coating nanoparticles with homotypic cancer cell membranes. Nano Lett. 16, 5895–5901 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Rao L., et al., Cancer cell membrane-coated upconversion nanoprobes for highly specific tumor imaging. Adv. Mater. 28, 3460–3466 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Cheng G., et al., Self-assembly of extracellular vesicle-like metal-organic framework nanoparticles for protection and intracellular delivery of biofunctional proteins. J. Am. Chem. Soc. 140, 7282–7291 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Cao H., et al., Liposomes coated with isolated macrophage membrane can target lung metastasis of breast cancer. ACS Nano 10, 7738–7748 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Wang H., et al., Targeted ultrasound-assisted cancer-selective chemical labeling and subsequent cancer imaging using click chemistry. Angew. Chem. Int. Ed. Engl. 55, 5452–5456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dube D. H., Prescher J. A., Quang C. N., Bertozzi C. R., Probing mucin-type O-linked glycosylation in living animals. Proc. Natl. Acad. Sci. U.S.A. 103, 4819–4824 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mura S., Nicolas J., Couvreur P., Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 12, 991–1003 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Van Bambeke F., et al., Biophysical studies and intracellular destabilization of pH-sensitive liposomes. Lipids 35, 213–223 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Cai R., et al., Corona of thorns: The surface chemistry-mediated protein corona perturbs the recognition and immune response of macrophages. ACS Appl. Mater. Interfaces 12, 1997–2008 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Chen Z., et al., Cancer cell membrane-biomimetic nanoparticles for homologous-targeting dual-modal imaging and photothermal therapy. ACS Nano 10, 10049–10057 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Iurisci I.et al. and Consorzio Interuniversitario Nazionale Per la Bio-Oncologia , Synthetic inhibitors of galectin-1 and -3 selectively modulate homotypic cell aggregation and tumor cell apoptosis. Anticancer Res. 29, 403–410 (2009). [PubMed] [Google Scholar]
- 38.Monopoli M. P., Åberg C., Salvati A., Dawson K. A., Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 7, 779–786 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Murray P. J., et al., Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun H., et al., Cancer-cell-biomimetic nanoparticles for targeted therapy of homotypic tumors. Adv. Mater. 28, 9581–9588 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Kumar M., et al., Cellular interaction of folic acid conjugated superparamagnetic iron oxide nanoparticles and its use as contrast agent for targeted magnetic imaging of tumor cells. Int. J. Nanomedicine 7, 3503–3516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W., et al., A C5 N2 nanoparticle based direct nucleus delivery platform for synergistic cancer therapy. Angew. Chem. Int. Ed. Engl. 58, 6290–6294 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Cao Z., Cheng S., Wang X., Pang Y., Liu J., Camouflaging bacteria by wrapping with cell membranes. Nat. Commun. 10, 3452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sørlie T., et al., Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 98, 10869–10874 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui L., et al., Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat. Biotechnol. 39, 357–367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subik K., et al., The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer (Auckl.) 4, 35–41 (2010). [PMC free article] [PubMed] [Google Scholar]
- 47.Thakur V., Kutty R. V., Recent advances in nanotheranostics for triple negative breast cancer treatment. J. Exp. Clin. Cancer Res. 38, 430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kazi J., et al., Folate decorated epigallocatechin-3-gallate (EGCG) loaded PLGA nanoparticles; in-vitro and in-vivo targeting efficacy against MDA-MB-231 tumor xenograft. Int. J. Pharm. 585, 119449 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Chaffer C. L., Weinberg R. A., A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Bhatia R., et al., Cancer-associated mucins: Role in immune modulation and metastasis. Cancer Metastasis Rev. 38, 223–236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheta R., et al., A metabolic labeling approach for glycoproteomic analysis reveals altered glycoprotein expression upon GALNT3 knockdown in ovarian cancer cells. J. Proteomics 145, 91–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie R., et al., In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc. Natl. Acad. Sci. U.S.A. 113, 5173–5178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bedard P. L., Hansen A. R., Ratain M. J., Siu L. L., Tumour heterogeneity in the clinic. Nature 501, 355–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng Z., Pu K., Improving cancer immunotherapy by cell membrane-camouflaged nanoparticles. Adv. Funct. Mater. 30, 2004397 (2020). [Google Scholar]
- 55.Feng L., Dong Z., Tao D., Zhang Y., Liu Z., The acidic tumor microenvironment: A target for smart cancer nano-theranostics. Natl. Sci. Rev. 5, 269–286 (2018). [Google Scholar]
- 56.Song W., et al., A cooperative polymeric platform for tumor-targeted drug delivery. Chem. Sci. (Camb.) 7, 728–736 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin W., et al., Artificial cysteine S-glycosylation induced by per-O-acetylated unnatural monosaccharides during metabolic glycan labeling. Angew. Chem. Int. Ed. Engl. 57, 1817–1820 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Hao Y., et al., Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nat. Commun. 10, 4065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin K., Zhang H., Zhao Z., Chen X., Protein S-glyco-modification through an elimination-addition mechanism. J. Am. Chem. Soc. 142, 9382–9388 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Rao L., et al., Hybrid cellular membrane nanovesicles amplify macrophage immune responses against cancer recurrence and metastasis. Nat. Commun. 11, 4909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sevencan C., et al., Cell membrane nanotherapeutics: From synthesis to applications emerging tools for personalized cancer therapy. Adv. Therap. 3, 1900201 (2020). [Google Scholar]
- 62.Elsharkasy O. M., et al., Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 159, 332–343 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Esteva-Socias M., et al., In search of an evidence-based strategy for quality assessment of human tissue samples: Report of the tissue Biospecimen Research Working Group of the Spanish Biobank Network. J. Transl. Med. 17, 370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakano S., Imawari Y., Mibu A., Otsuka M., Oinuma T., Differentiating vacuum-assisted breast biopsy from core needle biopsy: Is it necessary? Br. J. Radiol. 91, 20180250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bins S., et al., Implementation of a multicenter biobanking collaboration for next-generation sequencing-based biomarker discovery based on fresh frozen pretreatment tumor tissue biopsies. Oncologist 22, 33–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Escudé Martinez de Castilla P., et al., Extracellular vesicles as a drug delivery system: A systematic review of preclinical studies. Adv. Drug Deliv. Rev. 175, 113801 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.