Significance
Rice RNA polymerase V (Pol V) influences the accumulation of 24-nucleotide small interfering RNAs (24-nt siRNAs) in a locus-specific manner. Accumulation of 24-nt siRNAs is dependent on Pol V on loci with high CHH methylation levels, low CG and CHG methylation levels, and high active histone modifications. In contrast, a low methylation level in the CHH context, high methylation levels in CG and CHG contexts, and enrichment of repressive histone modifications predispose 24-nt siRNA accumulation to be independent of Pol V. DNA methylation is essential for 24-nt siRNA biosynthesis on Pol V–dependent loci but not on Pol V–independent loci. Our results suggest that monocot Pol V–transcribed scaffold RNAs are essential for reproductive development and 24-nt siRNA biosynthesis on euchromatin.
Keywords: RNA polymerase V, OsNRPE1, siRNA, rice
Abstract
RNA-directed DNA methylation (RdDM) functions in de novo methylation in CG, CHG, and CHH contexts. Here, we performed map-based cloning of OsNRPE1, which encodes the largest subunit of RNA polymerase V (Pol V), a key regulator of gene silencing and reproductive development in rice. We found that rice Pol V is required for CHH methylation on RdDM loci by transcribing long noncoding RNAs. Pol V influences the accumulation of 24-nucleotide small interfering RNAs (24-nt siRNAs) in a locus-specific manner. Biosynthesis of 24-nt siRNAs on loci with high CHH methylation levels and low CG and CHG methylation levels tends to depend on Pol V. In contrast, low methylation levels in the CHH context and high methylation levels in CG and CHG contexts predisposes 24-nt siRNA accumulation to be independent of Pol V. H3K9me1 and H3K9me2 tend to be enriched on Pol V–independent 24-nt siRNA loci, whereas various active histone modifications are enriched on Pol V–dependent 24-nt siRNA loci. DNA methylation is required for 24-nt siRNAs biosynthesis on Pol V–dependent loci but not on Pol V–independent loci. Our results reveal the function of rice Pol V for long noncoding RNA production, DNA methylation, 24-nt siRNA accumulation, and reproductive development.
DNA methylation plays an essential role in silencing transposons and repetitive elements. RNA-directed DNA methylation (RdDM) is the main mechanism for de novo DNA methylation in plants (1–9). In the canonical RdDM pathway, a transcription factor–like protein SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1, also known as DNA-BINDING TRANSCRIPTION FACTOR 1, DTF1) and four chromatin remodeling proteins, CLSY1 to CLSY4, facilitate RNA polymerase IV (Pol IV) targeting and transcription (10–14). Pol IV transcribes targets into 25- to 50-nt small RNAs (P4-RNAs) (15–19), which are immediately transcribed into double-stranded RNAs (dsRNAs) by RNA-dependent RNA polymerase 2 (RDR2) (19, 20). The dsRNAs are cleaved by DICER-LIKE 3 (DCL3) to generate 24-nucleotide small interfering RNAs (24-nt siRNAs) (19, 21), which are loaded into ARGONAUTE 4 (AGO4) protein to form the silencing complex (22). Pol V recruits AGO4 via the AGO hook motif in the C-terminal domain (CTD). The siRNAs in AGO4 are believed to base pair with the nascent RNAs transcribed by Pol V (23, 24). In this process, DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) is recruited to the targets and catalyzes DNA methylation (25).
Pol V transcription and targeting depend on the DDR complex, which contains DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), and RNA-DIRECTED DNA METHYLATION 1 (RDM1) (23, 24, 26). The recruitment of Pol V to targets requires the binding of SU(VAR)3-9 homologs SUVH2 and SUVH9 to methylated DNA (27, 28). Recent research has revealed that even on non-RdDM loci, Pol V transcription is dependent on the DDR complex, SUVH2, and SUVH9 (29). Pol V transcripts contain a common uracil (U) at position 10, which is complementary to the 5′ adenine (A) of the 24-nt siRNAs bound in the AGO4 protein (30), suggesting that AGO4 can slice Pol V transcripts. The slicing activity of AGO4 has been found to be essential for DNA methylation and siRNA production on a subset of RdDM targets (31, 32). These results indicate that the slicing of Pol V might be a key signal that initiates efficient de novo methylation.
In Arabidopsis, tomato, and Brassica rapa, the accumulation of nearly all 24-nt heterochromatic siRNAs is dependent on Pol IV, but accumulation of only a subset of siRNAs is dependent on Pol V (33–37). On the non-long terminal repeat (non-LTR) retrotransposons and helitrons, siRNA biosynthesis tends to depend on Pol V in Arabidopsis (34), suggesting that transposon identity might be a factor that determines whether siRNA production is dependent on Pol V. Based on the different roles of Pol IV and Pol V in siRNA production, researchers have hypothesized that Pol V could function as a component in a self-reinforcing loop in siRNA production and de novo DNA methylation (33, 34). According to this hypothesis, if the initial signal for recruitment of Pol IV-RDR2 is sufficiently strong, siRNA biosynthesis is independent of Pol V; if the initial signal is weak, however, the effect of Pol V on siRNA production will be pronounced (34). This hypothesis was supported by the finding that the accumulation of AGO4-dependent siRNAs on a subset of RdDM loci requires Pol V, DRM2, and SHH1 (32). The locus-specific role of Pol V in siRNA production has been found in several plant species, but the underlying mechanism is not known.
Here, we used a genetic screen to identify and clone OsNRPE1 as a key component in the silencing of transgene in rice. Mutation in OsNRPE1, which encodes the largest subunit of Pol V, greatly reduced seed setting, suggesting that Pol V as well as OsRDR2 and Pol IV play a vital role in the reproductive development of rice (38–40). Depletion of Pol V led to a substantial loss of CHH methylation on short transposons near genes. Moreover, siRNA accumulation was Pol V–dependent in a locus-specific manner. Analysis of DNA methylation contexts demonstrated that high CHH methylation combined with low CG and CHG methylation predisposes siRNA production to be dependent on Pol V, while siRNA production on loci with high CG and CHG methylation and with low CHH methylation tends to be independent of Pol V. In addition to observing the context bias for DNA methylation, we also observed the enrichment of H3K9me1 and H3K9me2 on loci that do not depend on Pol V for siRNA production and the enrichment of multiple active histone modifications on loci that depend on Pol V for siRNA production. The transcriptional levels are much higher for genes adjacent to loci that depend on Pol V for siRNA production than for those adjacent to Pol V–independent loci. Our findings demonstrate that the degree to which siRNA production depends on Pol V in rice is probably determined by DNA methylation contexts and histone modifications. Rice has developed multiple epigenetic mechanisms to fine-tune gene expression in order to ensure proper development.
Results
Map-Based Cloning of Five Elements Mountain 3.
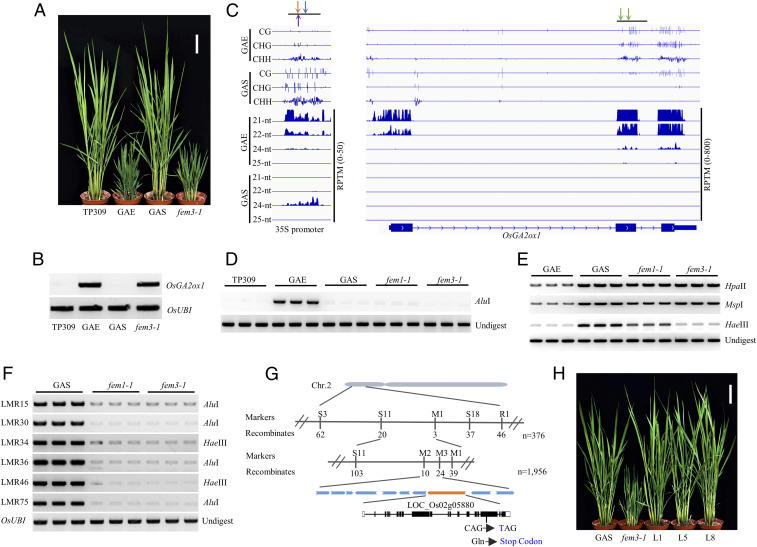
OsGA2ox1 encodes a gibberellin metabolism enzyme (41), and its ectopic expression in a japonica variety, TP309, resulted in transgenic plants (GAE) showing GA deficiency and dwarfism (38) (Fig. 1 A and B and SI Appendix, Fig. S1A). Spontaneous silencing of the transgene 35S::OsGA2ox1 in the transgenic plants resulted in plants (GAS) with normal plant height (38) (Fig. 1 A and B and SI Appendix, Fig. S1A). Small-RNA sequencing (sRNA-seq) revealed that OsGA2ox1 produced abundant 21- and 22-nt siRNAs and relatively few 24-nt siRNAs on the exons of OsGA2ox1 in GAE (Fig. 1C). Whole-genome bisulfite sequencing (WGBS) found that on 3′ two exons of OsGA2ox1, there was methylation on CG, CHG, and CHH in GAE (Fig. 1C). In contrast, there was methylation on CG but not on CHG or CHH in TP309 (SI Appendix, Fig. S1B), suggesting that siRNAs are involved in establishing non-CG methylation on OsGA2ox1 in GAE. Because the siRNAs on OsGA2ox1 exons were eliminated, non-CG methylation disappeared in GAS (Fig. 1C). Chop-PCR assay confirmed that CHH methylation on OsGA2ox1 is greater in GAE than in TP309 and is absent in GAS (Fig. 1D).
Fig. 1.
Identification of the fem3-1 mutant and map-based cloning of FEM3. (A) The morphologies of TP309, GAE, GAS, and fem3-1 plants. (Scale bar, 10 cm.) (B) Transcript levels of OsGA2ox1 as determined by semi–qRT-PCR in the indicated genotypes. OsUbiquitin (LOC_Os03g13170) served as the control. (C) Integrative genome browser image showing the methylation levels and siRNA abundance on the 35S promoter and OsGA2ox1 in GAE and GAS. The arrows indicate cutting sites (orange for MspI; purple for HpaII; blue for HaeIII; and green for AluI). (D and E) DNA methylation levels on OsGA2ox1 (D) and the 35S promoter (E) in GAE, GAS, fem1-1, and fem3-1 as determined by Chop-PCR assay. Undigested DNA served as the control. (F) DNA methylation levels on six RdDM loci in GAS, fem1-1, and fem3-1 as determined by Chop-PCR assay. The template of the PCR was digested by AluI or HaeIII. OsUbiquitin (LOC_Os03g13170) served as the control with undigested DNA as template. (G) Diagram indicating the map-based cloning of FEM3. The recombinant number and molecular markers (Dataset S1) are indicated Below and Above the bars, respectively. The changes in DNA and protein sequences in fem3-1 are indicated in blue. (H) Morphologies of the indicated genotypes. (Scale bar, 10 cm.)
In GAE, the 35S promoter also produced abundant 21- and 22-nt siRNAs and relatively few 24-nt siRNAs (Fig. 1C); there were low DNA methylation levels in CG, CHG, and CHH on the 35S promoter in GAE (Fig. 1C). In GAS, the 21- and 22-nt siRNAs disappeared but 24-nt siRNAs increased. The DNA methylation levels on CG, CHG, and CHH increased substantially in GAS (Fig. 1C). Chop-PCR assay confirmed that DNA methylation in three contexts was higher in GAS than in GAE (Fig. 1E), suggesting that transcriptional gene silencing of 35S::OsGA2ox1 probably occurs in GAS.
After using ethyl methyl sulfonate to treat GAS seeds, we isolated five elements mountain 3 (fem3) rice plants with restored overexpression of 35S::OsGA2ox1 (Fig. 1 A and B and SI Appendix, Fig. S1A). In the fem3 mutant, the CHH methylation level on the 35S promoter was lower than in GAS (Fig. 1E), while the levels of CG and CHG methylation in fem3 were similar to those in GAS, suggesting that FEM3 probably regulates gene silencing by controlling CHH methylation on the 35S promoter. On the gene body of OsGA2ox1, the low methylation levels in the CHH context in GAS were also evident in the fem3 mutant (Fig. 1D).
Beside the exogenous 35S promoter, six endogenous RdDM loci (38) were examined for the effect of FEM3 on CHH methylation. Chop-PCR assay showed that, like in fem1, CHH methylation in fem3 was substantially lower than in GAS (Fig. 1F), demonstrating that FEM3 controls CHH methylation on both transgene and endogenous genomic loci.
We performed map-based cloning of FEM3 using 978 F2 plants with a GA-deficient phenotype that were generated from a cross between fem3-1 heterozygous plants and Taichuang native 1 (TN1), an indica variety. The location of the candidate gene was narrowed to a region of ∼68 kb on the short arm of chromosome 2, which contains 10 putative genes (Fig. 1G). Sequencing detected a single-base substitution (C-T) on the seventeenth exon of LOC_Os02g05880, which resulted in a premature stop codon and a truncated protein that lacked five repeats and a DEFECTIVE CHLORLOPLASTS AND LEAVES (DeCL) domain (Fig. 1G and SI Appendix, Fig. S1C). To confirm that the point mutation in LOC_Os02g05880 was responsible for the fem3-1 mutant phenotypes, we transformed its genomic DNA fragment with its 2-kb promoter into fem3-1 plants for a complementary test. We obtained 18 independent transgenic lines including 15 positive and 3 negative lines. All positive line plants displayed wild-type (WT)-like plant height, and the three negative lines plants were similar to fem3-1. The three selected positive lines restored 35S::OsGA2ox1 silencing and plant height (Fig. 1H and SI Appendix, Fig. S1 D and E). In addition, the CHH methylation levels at the 35S promoter and at six endogenous loci were also restored in the complemented plants (SI Appendix, Fig. S1 F and G). These results demonstrated that mutation in LOC_Os02g05880 is responsible for overexpression of OsGA2ox1 in the fem3-1 mutant.
Pol V Regulates Important Agronomic Traits.
LOC_Os02g05880 encodes the largest subunit of RNA polymerase V, OsNRPE1 (SI Appendix, Fig. S2A), and the rice genome contains a homologous gene of OsNRPE1, OsNRPF1 (LOC_Os01g73430) (42, 43) (SI Appendix, Fig. S2A). Both OsNRPE1 and OsNRPF1 have A through H domains and a DeCL domain, which are the typical domains of the largest subunit of RNA polymerase (SI Appendix, Fig. S2B). The two proteins differ, however, in that the CTD of OsNRPE1 contains five imperfect repeats of 51 amino acids, which are absent in the CTD of OsNRPF1 (SI Appendix, Fig. S2B). Because the repeats are required for interaction with AGO4 and DNA methylation in Arabidopsis (44, 45), we hypothesized that OsNRPF1 might not be involved in RdDM. The spatiotemporal expression patterns of OsNRPE1 and OsNRPF1 were examined by qRT-PCR. Both OsNRPE1 and OsNRPF1 were predominately expressed in the inflorescence and anther (SI Appendix, Fig. S2C). However, the transcriptional levels were much higher for OsNRPE1 than for OsNRPF1 in various tissues (SI Appendix, Fig. S2C), suggesting that OsNRPE1 likely functions as the main and largest subunit of Pol V in rice.
Using CRISPR/Cas9 technology, we created osnrpe1, osnrpf1, and osnrpe1/osnrpf1 mutants in the Nipponbare background. For osnrpe1 and osnrpf1 mutations, one-base insertion or deletion near the protospacer-adjacent motif caused a premature stop codon and a truncated protein (SI Appendix, Fig. S2D). Chop-PCR assay showed that the osnrpe1 and osnrpe1/osnrpf1 mutants exhibited hypomethylation on six of the tested RdDM loci but that DNA methylation levels in osnrpf1 single mutants were not reduced (SI Appendix, Fig. S2E), suggesting that OsNRPF1 is probably nonfunctional in mediating DNA methylation.
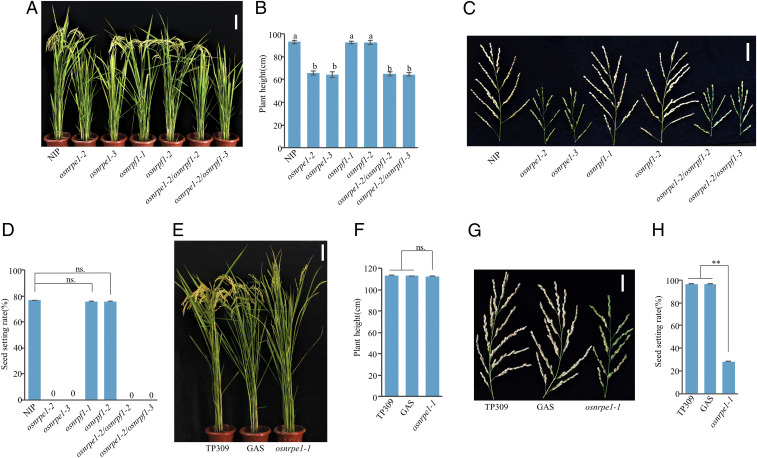
osnrpe1 and osnrpe1/osnrpf1 plants but not osnrpf1 plants were significantly shorter than WT plants (Fig. 2 A and B). In addition, neither osnrpe1 nor osnrpe1/osnrpf1 plants produced seeds, while seed setting was normal in osnrpf1 plants (Fig. 2 C and D). The encoded protein’s structure, expression pattern, role in DNA methylation, and effects on plant morphology demonstrate that OsNRPE1 but not OsNRPF1 encodes the largest subunit of Pol V in rice. We obtained osnrpe1-1 (fem3-1) without the transgene by backcrossing with TP309 WT plants and segregating out 35S::OsGA2ox1 in the F2 population. The stature of osnrpe1-1 plants was quite similar to that of control plants of TP309 and GAS (Fig. 2 E and F); in contrast, osnrpe1 plants were shorter than WT plants in the Nipponbare background (Fig. 2 A and B). Moreover, osnrpe1-1 produced some seeds even though its seed setting rate was 70% lower than that of the WT (Fig. 2 G and H). The difference in osnrpe1 performance in Nipponbare versus TP309 can probably be attributed to the weak allele for osnrpe1-1 in TP309 and to differences in the genetic background. Overall, the phenotypic analysis of osnrpe1 mutants in Nipponbare and TP309 indicated that OsNRPE1 but not OsNRPF1 functions as the largest subunit of Pol V and plays a vital role in reproductive development, which is consistent with a previous hypothesis that OsNRPF1 encodes the largest subunit of Pol VI (43).
Fig. 2.
OsNRPE1 regulates important agronomic traits. (A) The morphologies of Nipponbare, osnrpe1, osnrpf1, and osnrpe1/osnrpf1 plants. (Scale bar, 10 cm.) (B) Plant height of the indicated genotypes. Values are means ± SD; means with different letters are significantly different (P < 0.05) according to Fisher’s least significant difference (LSD). (C) Seed setting phenotype of the indicated genotypes. (Scale bar, 5 cm.) (D) Seed setting rate in various genotypes (n = 10). Values are means ± SD; ns. indicates that the compared means were not significantly different (P > 0.05). (E) Morphologies of TP309, GAS, and osnrpe1-1 without 35S::OsGA2ox1. (Scale bar, 10 cm.) (F) Plant height of TP309, GAS, and osnrpe1-1 (n = 10). Values are means ± SD; ns. indicates that the compared means were not significantly different (P > 0.05). (G) Seed setting phenotype in osnrpe1-1 and control plants. (Scale bar, 5 cm.) (H) Seed setting rate in osnrpe1-1 and control plants (n = 10). Values are means ± SD; ** indicates that the compared means are significantly different at P < 0.01 according to Student’s t test.
Pol V Regulates Genome-Wide CHH Methylation.
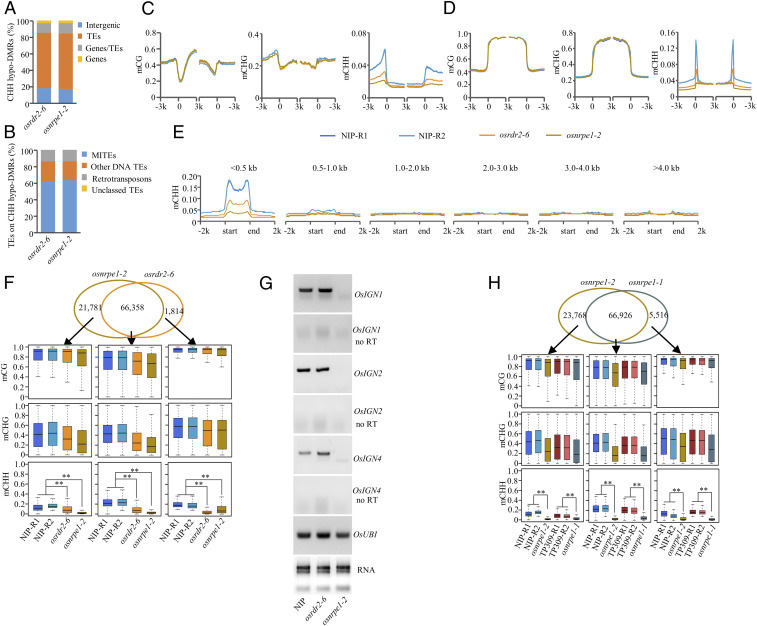
To determine the effect of OsNRPE1 on the methylome, we performed WGBS on 18-d-old seedlings of osnrpe1-2, osrdr2-6, and Nipponbare. The whole-genome methylation levels of CG and CHG were similar in osnrpe1-2 and the WT. However, the CHH methylation level was lower in osnrpe1-2 than in the WT (WT: 3.10%, osnrpe1-2: 1.60%) (SI Appendix, Fig. S3A and Dataset S2). Consistent with the latter finding, CHH hypomethylated differentially methylated regions (hypo-DMRs) were the dominant DMRs in terms of abundance and length in osnrpe1-2 (SI Appendix, Fig. S3 B and C). About 66.6% of the CHH hypo-DMRs of osnrpe1-2 were located on transposable elements (TEs) (Fig. 3A). Among the TEs that overlapped with CHH hypo-DMRs, MITEs were the main targets (Fig. 3B), which is quite similar to that in osrdr2 (38). The methylation levels of CG and CHG on both genes and TEs in osnrpe1-2 were similar to those in the WT (Fig. 3 C and D). The CHH methylation level, however, was substantially lower in osnrpe1-2 than in the WT, especially on the borders of genes and TEs (Fig. 3 C and D). We divided TEs into six subgroups based on length (<0.5 kb, 0.5 to 1.0 kb, 1.0 to 2.0 kb, 2.0 to 3.0 kb, 3.0 to 4.0 kb, and >4.0 kb) and found that CHH methylation was dependent on OsNRPE1 and was mainly located on short TEs (Fig. 3E). The dependency of OsNRPE1 on CHH methylation is reminiscent of FEM1, which encodes OsRDR2 (38).
Fig. 3.
OsNRPE1 controls genome-wide CHH methylation. (A) Genomic location of CHH hypo-DMRs in osrdr2-6 and osnrpe1-2. (B) TE categories associated with CHH hypo-DMRs in osrdr2-6 and osnrpe1-2. (C and D) DNA methylation levels of CG, CHG, and CHH on genes (C) and on TEs (D) in Nipponbare, osrdr2-6, and osnrpe1-2. The average methylation levels within each 100-bp interval are plotted. (E) CHH methylation levels on TEs of different lengths in Nipponbare, osrdr2-6, and osnrpe1-2. (F, Top) Venn diagram showing the overlap of CHH hypo-DMRs in osrdr2-6 and osnrpe1-2. (Bottom) DNA methylation levels of CG, CHG, and CHH in the indicated genotypes on osrdr2-6–specific, osnrpe1-2–specific, and overlapped CHH hypo-DMRs as indicated by box plots. ** indicates that compared plots are significantly different at P < 0.01 (Fisher’s LSD). (G) RT-PCR analysis of Pol V–dependent transcripts on loci OsIGN1, OsIGN2, and OsIGN4 in Nipponbare and in the osrdr2-6 and nrpe1-2 mutants. (H, Top) Venn diagram showing the overlap of CHH hypo-DMRs in osnrpe1-2 and osnrpe1-1. (Bottom) DNA methylation levels of CG, CHG, and CHH on osnrpe1-2–specific, osnrpe1-1–specific, and overlapped CHH hypo-DMRs as indicated by box plots. ** indicates that compared plots are significantly different at P < 0.01 (Fisher’s LSD).
A Venn diagram showed that 75.29% of the CHH hypo-DMRs in osnrpe1-2 overlapped with those in osrdr2-6 (Fig. 3F). On the osnrpe1-2–specific CHH hypo-DMRs, the CHH methylation level was significantly lower in osrdr2-6 than in the WT. On the osrdr2-6–specific CHH hypo-DMRs, the CHH methylation level was significantly lower in osnrpe1-2 than in the WT (Fig. 3F), indicating that OsNRPE1 and OsRDR2 function together in de novo methylation on RdDM loci in rice.
To determine whether OsNRPE1 is responsible for producing scaffold transcripts in RdDM, we selected six loci (SI Appendix, Fig. S4 A and B). On three loci OsIGN3, OsIGN5, and OsIGN6, the transcript levels in the osnrpe1 mutant were comparable to those in the WT (SI Appendix, Fig. S4C). On OsIGN1, OsIGN2, and OsIGN4, the transcripts were largely down-regulated in the osnrpe1-2 mutant relative to the WT and the osrdr2-6 mutant (Fig. 3G), suggesting that OsNRPE1-dependent transcripts probably function in RdDM.
In TP309, we used CRISPR/Cas9 technology to knockout two OsNRPD1 genes in order to create the pol iv mutants (SI Appendix, Fig. S5A). Like the osnrpe1-1 mutant, the pol iv mutants had significantly reduced fertility (SI Appendix, Fig. S5 B and C). We then conducted WGBS with 18-d-old seedlings of osnrpe1-1 (without 35S:OsGA2ox1), pol iv (osnrpd1a-1/osnrpd1b-1), and two types of WT (TP309-replicate 1 was WT, and TP309-replicate 2 was GAS). The whole-genome methylation level of CHH was lower in osnrpe1-1 than in the WTs (TP309-R1: 2.60%, TP309-R2: 2.40%, osnrpe1-1: 1.60%, pol iv: 1.70%; SI Appendix, Fig. S5D and Dataset S2). Consistent with the whole-genome methylation, CHH hypo-DMRs were the main type of DMRs for osnrpe1-1 and pol iv (SI Appendix, Fig. S5E). In addition, the total length and number of CHH hypo-DMRs were similar in osnrpe1-1 and pol iv (SI Appendix, Fig. S5 E and F). Like in Nipponbare, the majority of CHH hypo-DMRs in osnrpe1-1 and pol iv were located on TEs, and among them, MITEs were major targets (SI Appendix, Fig. S5 G and H). The substantially reduced CHH methylation levels in osnrpe1-1 and pol iv suggested that the high CHH methylation on gene borders and TEs was dependent on Pol IV and Pol V (SI Appendix, Fig. S5 I and J). The CHH methylation mainly occurred on short TEs, which is also dependent on Pol IV and Pol V (SI Appendix, Fig. S5K). More than 79.59% of the CHH hypo-DMRs of osnrpe1-1 were shared by pol iv (SI Appendix, Fig. S5L), suggesting that Pol IV and Pol V act together in RdDM in rice. In addition, about 92.39% of the CHH hypo-DMRs of osnrpe1-1 overlapped with those of osnrpe1-2 (Fig. 3H), suggesting that RdDM targets are largely conserved in different varieties of rice.
Pol V Controls 24-nt siRNA Accumulation in a Locus-Specific Manner.
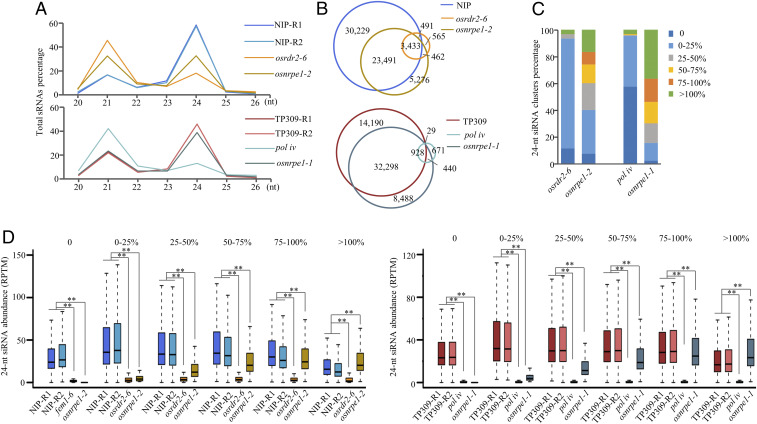
To understand the role of Pol V in the regulation of 24-nt siRNA accumulation on a genome scale, we performed sRNA-seq with 18-d-old seedlings of osnrpe1-1, osnrpe1-2, osrdr2-6, and pol iv mutants, and of the WTs of Nipponbare and TP309. More than 20 million raw reads were obtained for each of the genotypes (Dataset S2). After low quality reads and the structural RNA were removed, the clean sRNA reads were mapped to the rice genome (RGAP7.0). The abundance of 24-nt siRNAs was calculated using the reads per ten million mapped reads (RPTM) normalized by all unique mapped reads. In the Nipponbare background, the accumulation of 24-nt siRNAs was substantially reduced in osrdr2-6 and moderately reduced in osnrpe1-2 (Fig. 4A). In the TP309 background, the accumulation of 24-nt siRNAs was almost eliminated in pol iv but was only slightly decreased in osnrpe1-1 (Fig. 4A).
Fig. 4.
The accumulation of 24-nt siRNA partially depends on OsNRPE1 in rice. (A) Size distribution of sRNAs in Nipponbare, osrdr2-6, and osnrpe1-2 (Upper), and in TP309, pol iv, and osnrpe1-1 (Lower). (B) Venn diagram indicating 24-nt siRNA cluster numbers and their overlap in Nipponbare, osrdr2-6, and osnrpe1-2 (Upper) or in TP309, pol iv, and osnrpe1-1 (Lower). (C) Ratios of 24-nt siRNA clusters in different groups based on the abundance in the mutant relative to that in the WT. (D) Abundance of 24-nt siRNAs in the indicated genotypes in six subsets of 24-nt siRNA clusters as indicated by box plots. ** indicates that compared plots are significantly different at P < 0.01 (Fisher’s LSD).
We merged the 24-nt siRNAs within 100-base pair (100-bp) windows, and the loci with an RPTM value >12 were defined as 24-nt siRNA clusters. A total of 57,644 24-nt siRNA clusters were defined in the Nipponbare WT, but only 4,951 siRNA clusters were defined in the osrdr2-6 mutant (Fig. 4B). The osnrpe1-2 mutant, however, retained a majority of the siRNA clusters of the WT (Fig. 4B). We defined 47,445 24-nt siRNA clusters in the TP309 WT and only 2,068 in the pol iv mutant (Fig. 4B). Again, osnrpe1-1 retained most of the siRNA clusters of the WT (Fig. 4B). These results suggested that 24-nt siRNA production is fully dependent on OsRDR2 and Pol IV but only partially dependent on Pol V in rice. Interestingly, osnrpe1-1 generated 8,928 new 24-nt siRNA clusters in the TP309 background, and osnrpe1-2 generated 5,738 new 24-nt siRNA clusters in the Nipponbare background (Fig. 4B). We then combined the 24-nt siRNA clusters in the WT and mutants in the same genetic background. We obtained 63,947 24-nt siRNA clusters in Nipponbare and 57,044 in TP309. The combined 24-nt siRNA clusters in the same genetic background were divided into six subsets based on the ratio of siRNA abundance in mutants compared to that in the WT (0, 0 to 25%, 25 to 50%, 50 to 75%, 75 to 100%, and >100%; SI Appendix, Fig. S6). In the Nipponbare background, more than 93% of the 24-nt siRNA clusters belonged to 0 and 0 to 25% subsets in osrdr2-6; however, only 40% belonged to the same subsets in osnrpe1-2 (Fig. 4C). In pol iv, about 96% of 24-nt siRNA clusters had siRNA abundances that were <25% of that in the WT background of TP309. However, only about 16% of 24-nt siRNA clusters in osnrpe1-1 had siRNA abundances that were <25% compared to the TP309 WT (Fig. 4C).
As illustrated by the box plots of the six subsets of 24-nt siRNA clusters in the osnrpe1 mutant, almost all of the 24-nt siRNAs were eliminated in osrdr2-6 and pol iv (Fig. 4D), suggesting that 24-nt siRNA biogenesis is fully dependent on OsRDR2 and Pol IV. The accumulation of siRNAs in osnrpe1 was eliminated, partially lost, fully retained, or even increased in a subset-specific manner in both backgrounds (Fig. 4D). Together, the results indicate that the accumulation of 24-nt siRNAs is fully dependent on OsRDR2 and Pol IV but partially dependent on Pol V.
DNA Methylation Is Required for 24-nt siRNA Accumulation on Pol V–Dependent Loci.
Total cytosine methylation levels and CG, CHG, and CHH methylation levels were significantly reduced in all subsets of 24-nt siRNA clusters in osnrpe1-1 and osnrpe1-2 except CG methylation in the 75 to 100% and >100% subsets in osnrpe1-2 (SI Appendix, Fig. S7 A and B). For total C, CG, CHG, and CHH contexts, the reduction of methylation levels in osnrpe1 relative to the WT gradually declined from OsNRPE1 fully dependent 24-nt siRNA clusters to OsNRPE1 fully independent 24-nt siRNA clusters in both Nipponbare and TP309 backgrounds (Fig. 5A). In addition, the methylation reduction pattern in osrdr2 and pol iv was similar to that in osnrpe1 (SI Appendix, Fig. S7 C and D), suggesting that RdDM is more important for maintaining DNA methylation in subsets of 24-nt siRNA clusters that depend on Pol V than in subsets of 24-nt siRNA clusters that do not depend on Pol V.
Fig. 5.
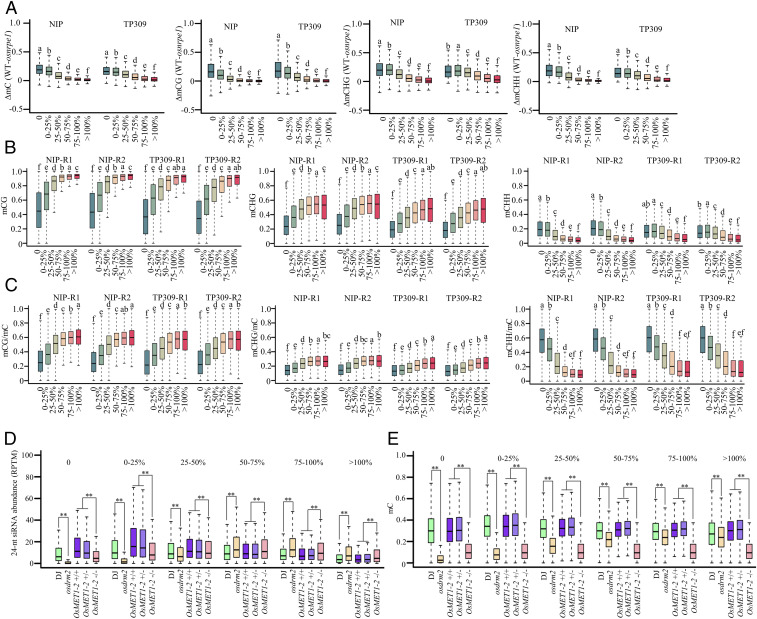
DNA methylation context affects the dependency of 24-nt siRNA accumulation on Pol V. (A) Reduction in methylation levels for the osnrpe1 mutant relative to the WT in total cytosine, CG, CHG, and CHH in six subsets of 24-nt siRNA clusters in backgrounds of Nipponbare and TP309. (B) DNA methylation levels of CG, CHG, and CHH in six subsets of 24-nt siRNA clusters in WTs of Nipponbare and TP309. (C) Ratios of mCG/mC, mCHG/mC, and mCHH/mC in total methylated cytosine in six subsets of 24-nt siRNA clusters in backgrounds of Nipponbare and TP309. (D) Abundance of 24-nt siRNAs in the indicated genotypes in six subsets of 24-nt siRNA clusters of osnrpe1-2 as indicated by box plots. (E) Total cytosine methylation levels in the indicated genotypes in six subsets of 24-nt siRNA clusters of osnrpe1-2 as indicated by box plots. Within each of the two or four groups of six box plots in each panel, plots with different letters are significantly different at P < 0.05 according to Fisher’s LSD. ** indicates that compared plots are significantly different at P < 0.01 (Fisher’s LSD).
Based on the relationship between OsNRPE1-affected DNA methylation and OsNRPE1-affected siRNA clusters, we hypothesized that the DNA methylation features may be involved in the effect of OsNRPE1 on 24-nt siRNA accumulation. We analyzed the methylation levels in the three DNA sequence contexts of the six subsets of 24-nt siRNA clusters of osnrpe1 mutants in WT plants and found that the CG and CHG methylation levels gradually increased from OsNRPE1 fully dependent subsets of 24-nt siRNA clusters to OsNRPE1 fully independent subsets in both Nipponbare and TP309 backgrounds (Fig. 5B). The pattern in the CHH context was opposite to that in the CG and CHG contexts; that is, the methylation level in the CHH context progressively decreased from 24-nt siRNA clusters that were fully dependent on Pol V to 24-nt siRNA clusters that were fully independent of Pol V (Fig. 5B). For total cytosine methylation levels in the Nipponbare background, in contrast, the methylation levels did not display the trend displayed by CG, CHG, and CHH (SI Appendix, Fig. S7E). The methylation level of total cytosine in the 0% subset of 24-nt siRNA clusters, however, was significantly lower than in other subsets of 24-nt siRNA clusters in the Nipponbare background (SI Appendix, Fig. S7E). In the TP309 background, the methylation levels of all cytosine in the subsets of 0 and 0 to 25% of 24-nt siRNA clusters were significantly lower than those in other subsets of 24-nt siRNA clusters (SI Appendix, Fig. S7E). Together, these results demonstrate that 24-nt siRNA accumulation on loci with higher CHH methylation levels and lower CG and CHG methylation levels tends to be more dependent on Pol V than 24-nt siRNA accumulation on loci with higher CG and CHG methylation levels and lower CHH methylation levels.
In addition, the ratio of mCG/mC and mCHG/mC gradually increased from 24-nt siRNA clusters that were fully dependent on Pol V to 24-nt siRNA clusters that were fully independent of Pol V (Fig. 5C). The ratio of mCHH/mC, however, gradually decreased from 24-nt siRNA clusters that were fully dependent on Pol V to 24-nt siRNA clusters that were fully independent of Pol V (Fig. 5C). For all cytosines, the contents of CG and CHG increased, but the content of CHH decreased from 24-nt siRNA clusters that were fully dependent on Pol V to 24-nt siRNA clusters that were fully independent of Pol V (SI Appendix, Fig. S7F). The trend of cytosine content throughout different subsets of 24-nt siRNA clusters was similar to that of methylated cytosine content (Fig. 5C and SI Appendix, Fig. S7F). Overall, the GC content was lower on Pol V–dependent 24-nt siRNA clusters than on Pol V–independent 24-nt siRNA clusters in both backgrounds (SI Appendix, Fig. S7G). Furthermore, 24-nt siRNA clusters that were fully dependent on Pol V were shorter than clusters that were fully independent of Pol V (SI Appendix, Fig. S7H), indicating that the features of DNA sequence might regulate the dependency of 24-nt siRNA biogenesis on Pol V.
To evaluate whether DNA methylation determines the dependency of 24-nt siRNA accumulation on Pol V, we first analyzed the siRNAs and methylome of the osdrm2 mutant (46, 47). On Pol V fully dependent loci, siRNA accumulation was almost totally eliminated in the osdrm2 mutant. On Pol V fully independent loci, by contrast, siRNA abundance was greater in the osdrm2 mutant than in the WT (Fig. 5D). The dependency of siRNA accumulation on OsDRM2 was similar to that on Pol V. Total cytosine methylation levels and CG, CHG, and CHH methylation levels were significantly reduced in all subsets of 24-nt siRNA clusters in the osdrm2 mutant (Fig. 5E and SI Appendix, Fig. S8A). For total C, CG, CHG, and CHH contexts, the reduction of methylation levels in osdrm2 relative to the WT gradually declined from Pol V fully dependent 24-nt siRNA clusters to Pol V fully independent 24-nt siRNA clusters (SI Appendix, Fig. S8B), which is also similar to that in osnrpe1, osrdr2, and pol iv mutants.
We next analyzed siRNAs and the methylome in the osmet1-2 mutant in which the majority of CG methylation is lost (48). Total cytosine methylation level and CG, CHG, and CHH methylation levels were significantly reduced in all subsets of 24-nt siRNA clusters in the osmet1-2 mutant (Fig. 5E and SI Appendix, Fig. S8 A and B). In subsets of 0, 0 to 25%, and 25 to 50%, the siRNA abundance in osmet1-2 mutant was significantly lower than in the WT or the heterozygote. In subsets of 50 to 75%, 75 to 100%, and >100%, by contrast, siRNA abundance was significantly higher in the osmet1-2 mutant than in the WT or the heterozygote (Fig. 5D). Overall, on Pol V–dependent loci, DNA methylation is required for siRNA accumulation.
Histone Modifications Are Associated with 24-nt siRNAs Accumulation on Pol V–Independent Loci.
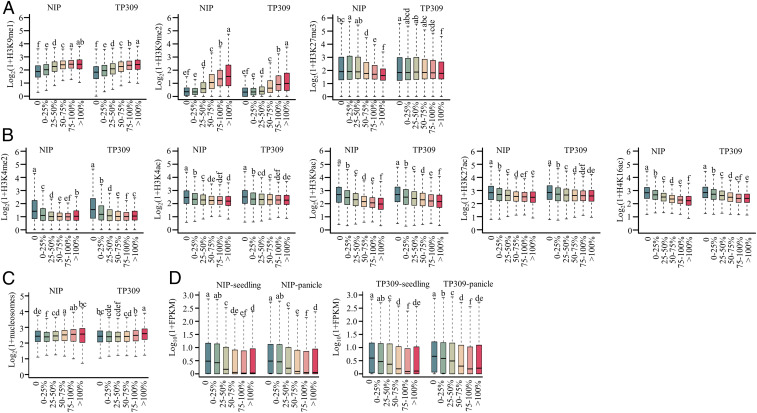
To determine whether other epigenetic marks regulate 24-nt siRNA production on Pol V–independent loci, we analyzed various histone modifications in the six subsets of 24-nt siRNA clusters. The levels of two repressive epigenetic marks, H3K9me1 and H3K9me2, gradually increased from 24-nt siRNA clusters that were fully dependent on Pol V to clusters that were fully independent of Pol V (Fig. 6A). The levels of another repressive epigenetic mark, H3K27me3, gradually decreased from 24-nt siRNA clusters that were fully dependent on Pol V to clusters that were fully independent of Pol V in the Nipponbare but not in the TP309 background (Fig. 6A), suggesting that Pol V–independent clusters tend to locate on stable but not conditional heterochromatin. In contrast, the active epigenetic marks H3K4me2, H3K4ac, H3K9ac, H3K27ac, and H4K16ac gradually decreased from 24-nt siRNA clusters that were fully dependent on Pol V to clusters that were fully independent of Pol V (Fig. 6B). The nucleosome occupancy slightly increased from 24-nt siRNA clusters that were fully dependent on Pol V to clusters that were fully independent of Pol V (Fig. 6C). Consequently, the messenger RNA (mRNA) levels of genes adjacent to 24-nt siRNA clusters were gradually decreased from clusters that were fully dependent on Pol V to clusters that were fully independent of Pol V in both seedlings and panicles (Fig. 6D). These results indicate that repressive histone modifications of H3K9me1 and H3K9me2 likely recruit Pol IV-RDR2 to produce 24-nt siRNAs on Pol V–independent loci.
Fig. 6.
Histone modifications, nucleosome occupancy, and transcriptional levels in six subsets of 24-nt siRNA clusters. (A) Relative enrichment of repressive histone modifications (H3K9me1, H3K9me2, and H3K27me3) in six subsets of 24-nt siRNA clusters in backgrounds of Nipponbare and TP309. (B) Relative enrichment of active histone modifications (H3K4me2, H3K4ac, H3K9ac, H3K27ac, and H4K16ac) in six subsets of 24-nt siRNA clusters in backgrounds of Nipponbare and TP309. (C) Nucleosome occupancy in six subsets of 24-nt siRNA clusters in backgrounds of Nipponbare and TP309. (D) Transcriptional levels of genes associated with 24-nt siRNA clusters within six subsets in backgrounds of Nipponbare and TP309. Within each of the two groups of six box plots in each panel, plots with different letters are significantly different at P < 0.05 according to Fisher’s LSD.
To study the distribution of 24-nt siRNA clusters in the genome, we inspected chromosome 1 and chromosome 4 on which the representative heterochromatic regions are concentrated in the typical regions (46). For simplicity, we combined three Pol V–dependent 24-nt siRNA clusters (0, 0 to 25%, and 25 to 50%) into a <50% subgroup and three Pol V–independent 24-nt siRNA clusters (50 to 75%, 75 to 100%, and >100%) into a >50% subgroup. On heterochromatic regions, cluster numbers were similar for the two groups. On the euchromatic regions, in contrast, the number of 24-nt siRNA clusters was greater in the <50% subgroup than in the >50% subgroup (SI Appendix, Fig. S9). Together, these results suggested that heterochromatin and euchromatin environments are correlated with the dependence of 24-nt siRNA biosynthesis on Pol V.
Discussion
RdDM Is Essential for Reproductive Development in Rice.
The RdDM mechanism has been well studied in Arabidopsis, but the mutants of most RdDM genes have no obvious developmental defects in Arabidopsis (1–9). In other plant species, like maize, tomato, B. rapa, and Capsella rubella, RdDM mutants display pleiotropic developmental phenotypes, particularly for traits related to reproduction (7–9, 36, 37, 49–54). In rice, mutations in OsDCL3, OsDRM2, or OsNRPD1 change important agricultural traits, such as plant stature, flag leaf angle, tiller number, and fertility (39, 40, 46, 55, 56). Recent research demonstrated that maternal Pol IV–dependent siRNAs control seed development by regulating the expression of imprinting genes in Arabidopsis (57).
Here, we provided evidence that the mutation of OsNRPE1 (FEM3) fully blocked seed setting in the Nipponbare background and greatly reduced seed setting in the TP309 background (Fig. 2 C, D, G, and H). The role of OsNRPE1 (FEM3) in reproductive development is consistent with our previous work with OsRDR2 (FEM1), the mutation of which disrupted both male and female reproductive development and finally resulted in sterility (38). In the current study, the pol iv mutant in the TP309 background that was created by mutating two OsNRPD1 genes also had greatly reduced seed setting (SI Appendix, Fig. S5 A–C). The predominant expression of OsRDR2 (FEM1), OsNRPE1 (FEM3), and OsNRPD1 in reproductive tissues and the reproductive defects in osrdr2 (fem1), osnrpe1 (fem3), and pol iv mutants demonstrate the important role of RdDM in reproductive development in rice.
Most species of Poaceae have two homologous NRPE1 genes (42, 43). In the current study with rice, we investigated the homologous gene of OsNRPE1 (FEM3), OsNRPF1, which lacks the heptad repeats at the C terminus that are required for interaction with AGO4 and for mediated DNA methylation in Arabidopsis (44, 45). We found that the mRNA level is much lower for OsNRPF1 than OsNRPE1 (FEM3) and that the mutation in OsNRPF1 does not result in any visible developmental defects. In addition, our Chop-PCR assay indicated that OsNRPF1 is not involved in DNA methylation. Thus, only OsNRPE1 encodes the largest subunit of Pol V in rice, and the function of OsNRPF1 remains unknown (43).
Gene Silencing for 35S::OsGA2ox1.
The 35S promoter and OsGA2ox1 produced abundant 21- and 22-nt siRNAs but few 24-nt siRNAs in GAE. Consequently, DNA methylation on the 35S promoter and OsGA2ox1 was established, suggesting that noncanonical RdDM or moderate posttranscriptional gene silencing (PTGS) likely occurs in GAE. In GAS, by contrast, 21- and 22-nt siRNAs were absent on both the 35S promoter and OsGA2ox1. On the 35S promoter, 24-nt siRNAs were more abundant in GAS than in GAE. These results demonstrated that noncanonical RdDM or moderate PTGS for 35S::OsGA2ox1 in GAE changed into TGS in GAS, which is similar to the phase transition of epigenetic silencing for other transgenes or TEs (58). The 24-nt siRNAs increased on the 35S promoter but disappeared on OsGA2ox1 in GAS. From GAE to GAS, the change in the abundance of 24-nt siRNAs and non-CG methylation was opposite on the 35S promoter versus OsGA2ox1. The underlying mechanism for the difference warrants additional research.
24-nt siRNAs and DNA Methylation Form a Self-Reinforcing Loop on Euchromatic RdDM Loci.
During the initiation and establishment of RdDM, all cytosines including CG, CHG, and CHH are methylated by DRM2. Plants have specific mechanisms to ensure the maintenance of CG and CHG methylation. After DNA replication, however, DNA in the CHH context must be de novo methylated in the newly synthesized strand. The maintenance of CHH methylation is carried out by RdDM and CMT2 in Arabidopsis (59). The enrichment of H3K9me1 and H3K9me2 and the nucleosome is lower on Pol V–dependent loci than on Pol V–independent loci (SI Appendix, Fig. S10). In contrast, the enrichment of active histone marks is higher on loci with 24-nt siRNAs that are fully dependent on Pol V than on loci with 24-nt siRNAs that are fully independent of Pol V. We propose that Pol V–dependent and –independent 24-nt siRNA clusters belong to the euchromatic and heterochromatic RdDM loci, respectively. On euchromatic loci, DNA methylation in different contexts is interdependent, and DNA methylation in all contexts but not H3K9me2 recruits Pol IV-RDR2 and Pol V to catalyze DNA methylation. siRNA production and the establishment and maintenance of DNA methylation form a positive feedback loop. On heterochromatic loci, histone modifications like H3K9me2 but not DNA methylation are the main signals that recruit Pol IV-RDR2 to produce 24-nt siRNAs.
Possible Mechanisms Underlying Pol V Repression of siRNA Accumulation.
In addition, we found that nrpe1 mutants contained 5,738 and 8,928 more 24-nt siRNA clusters than the WT in the background of Nipponbare and TP309, respectively (Fig. 4B). Mutation in OsDRM2 and OsMET1-2 also increased 24-nt siRNA abundance on Pol V–independent loci. On those loci, the depletion of Pol V or OsDRM2 might increase chromatin access to Pol IV-OsRDR2 or Pol II-OsRDR6, which function in generating siRNAs. Another possibility is that a reduction in DNA methylation in RdDM mutants or osmet1-2 likely enhances the recruitment of Pol IV-OsRDR2 or Pol II-OsRDR6, thereby increasing 24-nt siRNA accumulation. Future study of siRNA profiles in mutants associated with Pol V (i.e., OsAGO4, OsIDN2, OsKTF1, OsDRD1, OsDMS3, and OsRDM1) is needed to clarify the repressive role of Pol V on siRNA production. Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis of Pol IV, OsRDR2, Pol II, OsRDR6, nucleosomes, and histone modifications like H3K9me2 in the mutants with increased siRNA production is needed to determine the mechanism by which Pol V, OsDRM2, and OsMET1 repress siRNA production. Determining the effect of Pol V, OsDRM2, and OsMET1 on the chromatin superstructure by Hi-C analysis will also increase our understanding of how Pol V represses 24-nt siRNA accumulation.
Materials and Methods
SI Appendix, SI Materials and Methods contains descriptions of performed experiments, including materials and growth conditions, genetic screen of five elements mountain mutants, map-based cloning of FEM3, phylogenetic analysis, RNA isolation and analysis of transcriptional levels, DNA methylation analyses of individual loci, and vector construction and plant transformation. It also includes descriptions of data analysis, including whole-genome bisulfite data analysis, sRNA-seq analysis, analysis of ChIP-seq, and gene expression analysis.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31671340), the Natural Science Foundation of Jiangsu Province (BK20170027), and the Jiangsu Collaborative Innovation Center for Modern Crop Production to D.-L.Y. The high-throughput sequencing data were analyzed on the high-performance computing platform of the Bioinformatics Center, Nanjing Agricultural University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100709118/-/DCSupplemental.
Data Availability
The following were uploaded to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) in our previous study (GEO: GSE130168): the bisulfite-seq datasets of Nipponbare and fem1-6 (osrdr2-6); the sRNA-seq datasets of Nipponbare, fem1-6, and TP309; and the RNA-seq datasets of Nipponbare and TP309. The following were uploaded to the NCBI GEO in the current study (GEO: GSE158711): the bisulfite-seq datasets of TP309, GAS, pol iv, fem3-1 (osnrpe1-1), and fem3-2 (osnrpe1-2) and the sRNA-seq datasets of pol iv, fem3-1 (osnrpe1-1), and fem3-2 (osnrpe1-2). The following data were downloaded from NCBI: the bisulfite-seq and sRNA-seq datasets of DJ and osdrm2 (GEO: GSE81436) (46) and of Nipponbare and osmet1-2 (SRP043447, SRP043449) (48). All other study data are included in the article and/or supporting information.
References
- 1.Zhang H., Zhu J. K., RNA-directed DNA methylation. Curr. Opin. Plant Biol. 14, 142–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzke M. A., Mosher R. A., RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Wendte J. M., Pikaard C. S., The RNAs of RNA-directed DNA methylation. Biochim. Biophys. Acta. Gene Regul. Mech. 1860, 140–148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matzke M. A., Kanno T., Matzke A. J., RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Erdmann R. M., Picard C. L., RNA-directed DNA methylation. PLoS Genet. 16, e1009034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Zhu J. K., New discoveries generate new questions about RNA-directed DNA methylation in Arabidopsis. Natl. Sci. Rev. 4, 10–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rymen B., Ferrafiat L., Blevins T., Non-coding RNA polymerases that silence transposable elements and reprogram gene expression in plants. Transcription 11, 172–191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow H. T., Chakraborty T., Mosher R. A., RNA-directed DNA Methylation and sexual reproduction: Expanding beyond the seed. Curr. Opin. Plant Biol. 54, 11–17 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Wambui Mbichi R., Wang Q. F., Wan T., RNA directed DNA methylation and seed plant genome evolution. Plant Cell Rep. 39, 983–996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith L. M., et al., An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19, 1507–1521 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., et al., DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. U.S.A. 110, 8290–8295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law J. A., et al., Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D. L., et al., Four putative SWI2/SNF2 chromatin remodelers have dual roles in regulating DNA methylation in Arabidopsis. Cell Discov. 4, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M., Palanca A. M. S., Law J. A., Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 50, 865–873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blevins T., et al., Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4, e09591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai J., et al., A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163, 445–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye R., et al., A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell 61, 222–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D. L., et al., Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 26, 66–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh J., Mishra V., Wang F., Huang H. Y., Pikaard C. S., Reaction mechanisms of Pol IV, RDR2, and DCL3 drive RNA channeling in the siRNA-directed DNA methylation pathway. Mol. Cell 75, 576–589.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haag J. R., et al., In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell 48, 811–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z., et al., Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi S., et al., Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wierzbicki A. T., Haag J. R., Pikaard C. S., Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wierzbicki A. T., Ream T. S., Haag J. R., Pikaard C. S., RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41, 630–634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong X., et al., Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong X., et al., DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 19, 870–875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson L. M., et al., SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z. W., et al., The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10, e1003948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuzuki M., et al., Broad noncoding transcription suggests genome surveillance by RNA polymerase V. Proc. Natl. Acad. Sci. U.S.A. 117, 30799–30804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W., et al., RNA-directed DNA methylation involves co-transcriptional small-RNA-guided slicing of polymerase V transcripts in Arabidopsis. Nat. Plants 4, 181–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y., et al., Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443, 1008–1012 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Wang F., Axtell M. J., AGO4 is specifically required for heterochromatic siRNA accumulation at Pol V-dependent loci in Arabidopsis thaliana. Plant J. 90, 37–47 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Pontier D., et al., Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosher R. A., Schwach F., Studholme D., Baulcombe D. C., PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3145–3150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee T. F., et al., RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7, 781–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouil Q., Baulcombe D. C., DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12, e1006526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grover J. W., et al., Maternal components of RNA-directed DNA methylation are required for seed development in Brassica rapa. Plant J. 94, 575–582 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Wang L., et al., Global reinforcement of DNA methylation through enhancement of RNA-directed DNA methylation ensures sexual reproduction in rice. bioRxiv [Preprint] (2020). 10.1101/2020.07.02.185371 (Accessed 3 July 2020). [DOI]
- 39.Debladis E., et al., Construction and characterization of a knock-down RNA interference line of OsNRPD1 in rice (Oryza sativa ssp japonica cv Nipponbare). Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L., et al., Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol. Plant 13, 851–863 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Lo S. F., et al., A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haag J. R., et al., Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep. 9, 378–390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trujillo J. T., Seetharam A. S., Hufford M. B., Beilstein M. A., Mosher R. A., Evidence for a unique DNA-dependent RNA polymerase in cereal crops. Mol. Biol. Evol. 35, 2454–2462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Shami M., et al., Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21, 2539–2544 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendte J. M., et al., Functional dissection of the Pol V largest subunit CTD in RNA-directed DNA methylation. Cell Rep. 19, 2796–2808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan F., et al., Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 171, 2041–2054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan F., et al., DDM1 represses noncoding RNA expression and RNA-directed DNA methylation in heterochromatin. Plant Physiol. 177, 1187–1197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu L., et al., Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. U.S.A. 111, 10642–10647 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkinson S. E., Gross S. M., Hollick J. B., Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 308, 462–473 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Wang Z., et al., Polymerase IV plays a crucial role in pollen development in Capsella. Plant Cell 32, 950–966 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erhard K. F. Jr, et al., RNA polymerase IV functions in paramutation in Zea mays. Science 323, 1201–1205 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Dorweiler J. E., et al., mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12, 2101–2118 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sidorenko L., et al., A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 5, e1000725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stonaker J. L., Lim J. P., Erhard K. F. Jr, Hollick J. B., Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 5, e1000706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei L., et al., Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 3877–3882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moritoh S., et al., Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 71, 85–98 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Kirkbride R. C., et al., Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 2761–2766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuerda-Gil D., Slotkin R. K., Non-canonical RNA-directed DNA methylation. Nat. Plants 2, 16163 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Zemach A., et al., The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following were uploaded to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) in our previous study (GEO: GSE130168): the bisulfite-seq datasets of Nipponbare and fem1-6 (osrdr2-6); the sRNA-seq datasets of Nipponbare, fem1-6, and TP309; and the RNA-seq datasets of Nipponbare and TP309. The following were uploaded to the NCBI GEO in the current study (GEO: GSE158711): the bisulfite-seq datasets of TP309, GAS, pol iv, fem3-1 (osnrpe1-1), and fem3-2 (osnrpe1-2) and the sRNA-seq datasets of pol iv, fem3-1 (osnrpe1-1), and fem3-2 (osnrpe1-2). The following data were downloaded from NCBI: the bisulfite-seq and sRNA-seq datasets of DJ and osdrm2 (GEO: GSE81436) (46) and of Nipponbare and osmet1-2 (SRP043447, SRP043449) (48). All other study data are included in the article and/or supporting information.