Significance
The TATA box-binding protein (TBP) is highly conserved throughout eukaryotes and plays a central role in the assembly of the transcription preinitiation complex (PIC) at the promoter of genes. We show here that TBP can interact with nucleosomes containing the Widom-601 sequence at two different positions. One position comprises a TATA box-like sequence near the end of nucleosomal DNA, and the other position is GC-rich and located near the nucleosome dyad. TBP–nucleosome complexes must be resolved before PIC assembly can occur but may persist during mitosis when chromatin is condensed, possibly explaining how TBP can act as a bookmarking factor that transfers transcriptional memory.
Keywords: gene transcription, RNA polymerase II, nucleosome, chromatin, structural biology
Abstract
The TATA box-binding protein (TBP) is highly conserved throughout eukaryotes and plays a central role in the assembly of the transcription preinitiation complex (PIC) at gene promoters. TBP binds and bends DNA, and directs adjacent binding of the transcription factors TFIIA and TFIIB for PIC assembly. Here, we show that yeast TBP can bind to a nucleosome containing the Widom-601 sequence and that TBP–nucleosome binding is stabilized by TFIIA. We determine three cryo-electron microscopy (cryo-EM) structures of TBP–nucleosome complexes, two of them containing also TFIIA. TBP can bind to superhelical location (SHL) –6, which contains a TATA-like sequence, but also to SHL +2, which is GC-rich. Whereas binding to SHL –6 can occur in the absence of TFIIA, binding to SHL +2 is only observed in the presence of TFIIA and goes along with detachment of upstream terminal DNA from the histone octamer. TBP–nucleosome complexes are sterically incompatible with PIC assembly, explaining why a promoter nucleosome generally impairs transcription and must be moved before initiation can occur.
Transcription by RNA polymerase II (Pol II) begins with the assembly of a preinitiation complex (PIC) on promoter DNA around the transcription start site (1–3). It is generally thought that one of the first steps in PIC assembly is the binding of the TATA box-binding protein (TBP) to promoter DNA. TBP is a subunit of the multiprotein complex transcription factor (TF) IID, which is important for promoter recognition (4, 5). TBP can also be delivered to the promoter via its binding to the transcriptional coactivator SAGA (5, 6). The very high conservation of TBP throughout eukaryotes befits the central role of TBP in transcription initiation.
The structure of TBP revealed a symmetric, saddle-shaped DNA-binding fold that was predicted to sit astride the DNA duplex (7) or to follow the DNA minor groove (8). Structures of TBP–DNA complexes showed that TBP bends DNA by 90°, widens the minor groove, and introduces kinks at either end of the bound DNA region (9, 10). The TBP–DNA interface is primarily hydrophobic and includes the concave surface of the TBP saddle and the edges of the DNA bases. TBP initially forms an unstable complex with unbent DNA that is slowly converted to a stable complex containing bent DNA (11). The TBP–DNA complex can initiate PIC assembly by binding TFIIA and TFIIB on its upstream side and downstream side, respectively (12–16). Structural studies of the PIC showed that TBP resides on the outside and TBP-induced DNA bending facilitates the arrangement of general transcription factors on the Pol II surface (17–26).
PIC assembly is generally thought to occur on DNA that is free of nucleosomes (27, 28). Indeed, it was long known that the presence of nucleosomes impairs PIC assembly and inhibits transcription initiation (29–31). Binding of TBP to DNA is severely impaired by incorporation of a TATA box sequence into a nucleosome, and this is affected by the orientation of the TATA sequence relative to the surface of the histone octamer (32). TBP and TFIIA can, however, bind a TATA box that is located within linker DNA at the edge of a nucleosome (33). Here, we show that TBP can bind a nucleosome core particle at two different positions and report structures that elucidate TBP–nucleosome interactions and their stabilization by TFIIA. We then discuss implications of these findings for transcription initiation and the possible role of TBP as a bookmarking factor that may enable transcriptional memory through mitosis.
Results
TBP Can Stably Bind a Nucleosome.
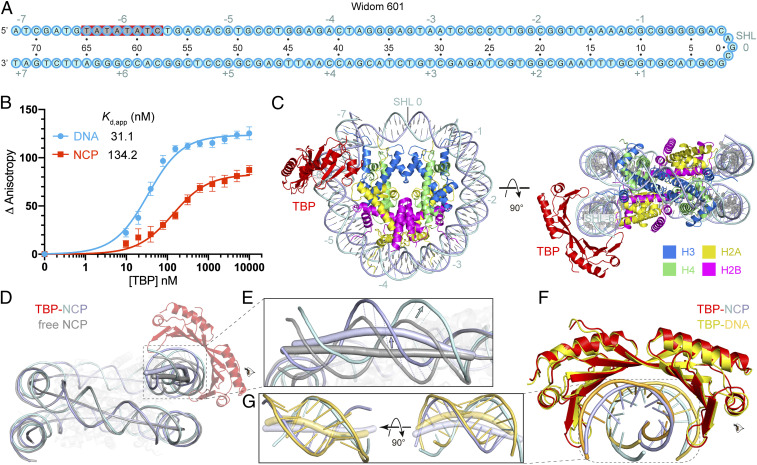
A stable nucleosome core particle (NCP) can be reconstituted with the use of a 145-bp DNA containing the Widom-601 sequence (34). The Widom-601 sequence comprises a TATA-like motif that is located at superhelical location (SHL) –6 within a reconstituted nucleosome (Fig. 1A). This TATA-like motif (TATATATC) deviates only in one nucleotide from the consensus TATA box sequence (TATAWAWR; W indicates A/T and R indicates A/G) (35). To our knowledge, it has not been explored whether this TATA sequence is recognized by TBP and how such recognition may be modulated by the presence of a histone octamer.
Fig. 1.
Structure of TBP–nucleosome complex shows TBP binding to SHL –6. (A) Widom-601 sequence. The TATA box-like sequence is highlighted in gray. Red background indicates observed TBP–DNA contacts. Cyan numbers show SHL positions on the nucleosome with the central base pair denoted as SHL 0. Black numbers indicate DNA nucleotide positions relative to SHL 0. (B) TBP binds to Widom-601 DNA (blue) or NCP (red), as monitored by the change in relative FA. Error bars reflect the SD from three experimental replicates. Apparent dissociation constant (Kd,app) values are provided (see SI Appendix, Table S1 for fitting parameters and error). (C) Structure of the TBP–NCP complex. Numbers denote SHL locations. (D) Superimposition of the TBP–NCP structure with Widom-601 NCP (gray; PDB ID code 3LZ0) (36) aligned on histones. The DNA duplex axes are shown as tubes. The axis of TBP-bound nucleosomal DNA is colored in light blue, and the axis of unbound nucleosomal DNA is colored in gray. (E) Enlarged view of the DNA region around SHL –6 that is distorted upon TBP binding. Dashed lines denote the apparent movement of the DNA backbone of the TATA box. The directions of movement are indicated with arrows. (F) Comparison with TBP–DNA structure (yellow; PDB ID code 1VTL) (9, 10) based on superposition of the TBP structures. (G) Two views of the TATA box trajectories in TBP–NCP and TBP–DNA structures. The axes of DNA are shown as tubes and bases as sticks.
To investigate whether TBP binds a nucleosome reconstituted with the Widom-601 sequence, we measured the binding affinity of full-length TBP from the yeast Saccharomyces cerevisiae to free Widom-601 DNA or reconstituted nucleosomes using fluorescence anisotropy (FA) assays and 5′ FAM-labeled DNA (Materials and Methods). TBP bound to both free DNA and nucleosomal DNA, with apparent dissociation constants of 31.1 ± 8.5 nM for free DNA and 134.2 ± 28.7 nM for the nucleosome (Fig. 1B). These results showed that TBP could bind the nucleosome, albeit with an approximately fourfold lower affinity compared to free Widom-601 DNA.
Structure of the TBP–Nucleosome Complex.
We next used cryo-electron microscopy (cryo-EM) analysis to investigate how TBP binds the nucleosome. We assembled a TBP–NCP complex by mixing TBP and NCP at a molar ratio of 2:1, cross-linked with glutaraldehyde, and prepared cryo-EM grids. Cryo-EM data were collected on a Titan Krios microscope (FEI) with a K3 detector (Gatan) (Materials and Methods). A three-dimensional (3D) reconstruction of the complex was obtained from 36,784 particles at a resolution of 3.4 Å (SI Appendix, Fig. S1). The reconstruction revealed a saddle-shape density for TBP located at SHL –6 of the nucleosome where the TATA-like motif is present (SI Appendix, Figs. S1 and S3 A and D).
The structure shows that TBP uses its concave surface to interact with the minor groove of the nucleosomal TATA box-like sequence (Fig. 1C), similar to what was first seen in TBP–DNA structures (9, 10). Comparison with the free NCP structure (36) shows that binding of TBP to the nucleosome pulls DNA off the surface of the histone octamer by up to ∼8 Å at base pairs –72 to –57 (Fig. 1 D and E and SI Appendix, Movie S1). The structure of TBP is highly similar to that observed in the TBP–DNA complex (Fig. 1F), whereas the structure of the DNA is clearly distinct (Fig. 1G). Compared to the DNA conformation observed in isolated TBP–DNA complexes (9, 10), DNA is less bent and less unwound, and the minor groove is less widened (Fig. 1G). In summary, the local TATA DNA structure in the TBP–NCP complex is intermediary between the fully distorted DNA observed in the TBP–DNA complex and the partially distorted DNA observed in an unbound NCP. Partial distortion of the DNA may explain the lower affinity of TBP to the NCP compared to free DNA (Fig. 1B).
Structure of a TBP–TFIIA–Nucleosome Complex.
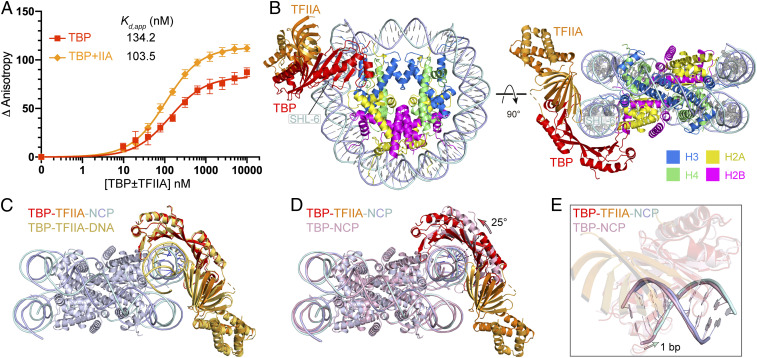
Because TFIIA is known to stabilize TBP binding to TATA DNA (37, 38), we tested whether TFIIA can enhance TBP binding to the nucleosome. Electrophoretic mobility shift assays showed that the addition of TFIIA increases the affinity of TBP to the nucleosome (SI Appendix, Fig. S2A). To corroborate these findings, we measured the binding affinity using FA assays and found that TFIIA enhanced TBP binding to the nucleosome ∼1.3-fold (Fig. 2A). These results are consistent with a biochemical study that showed binding of TBP and TFIIA to the edge of a nucleosome (33).
Fig. 2.
Structure of TBP–TFIIA–nucleosome complex with TBP located at SHL –6 (TBP–TFIIA–NCPSHL-6). (A) Changes in relative FA show that TFIIA stabilizes TBP binding to the nucleosome. Error bars reflect the SD from three experimental replicates. Apparent dissociation constant (Kd,app) values are provided (see SI Appendix, Table S1 for fitting parameters and error). (B) Overall structure. (C) Comparison to TBP–TFIIA–DNA structure (PDB ID code 1YTF) (13, 14) after superposition of TBP proteins. (D) Comparison to TBP–NCP structure (Fig. 1C) after superposition of histones shows sliding of TBP along DNA that results in a 25° apparent rotation (arrow). (E) Comparison to TBP–NCP structure (Fig. 1C) after superposition of TBP proteins shows a shift of DNA by 1 bp (arrow).
To visualize how TFIIA stabilizes TBP binding to a nucleosome, we mixed TBP, TFIIA, and NCP at a molar ratio of 2.5:3:1, and subjected the cross-linked TBP–TFIIA–NCP complex to cryo-EM analysis (SI Appendix, Fig. S2 B–G). After particle classification, a 3D reconstruction of the TBP–TFIIA–NCP complex was obtained at 3.0-Å resolution. The reconstruction revealed unambiguous densities for TBP and TFIIA at SHL –6 of the nucleosome (SI Appendix, Figs. S2 and S3 B and E). TBP is again bound to SHL –6 and TFIIA is located adjacently (Fig. 2B). As in TBP–TFIIA–DNA structures (13, 14), the β-barrel domain of TFIIA binds to the outer β-strand of the N-terminal TBP region, forming a continuous β-sheet (Fig. 2C). TFIIA also contacts DNA ∼3 bp upstream of the TATA box. The structure is consistent with the observed stabilization of the TBP–NCP complex by TFIIA binding.
Comparison of the TBP–TFIIA–NCP structure with the TBP–NCP structure reveals that TFIIA binding leads to a sliding of TBP along the DNA minor groove in the upstream direction by 1 bp, altering the relative orientation of TBP and NCP by ∼25° (Fig. 2 D and E and SI Appendix, Movie S1). The direction of sliding is reversed compared to that observed with TBP–TFIIA–DNA crystal structures, in which TBP had slid by 2 bp downstream compared to the TBP–DNA complex (13, 14). Thus, TFIIA can use binding energy to induce sliding of TBP along both free and nucleosomal DNA. In the context of the nucleosome, TBP sliding is required for TFIIA to bind, as modeling shows that TFIIA would otherwise clash with the adjacent DNA gyre. The direction of sliding likely depends on both sequence context and steric restraints.
TBP Binds an Alternative Position on the Nucleosome.
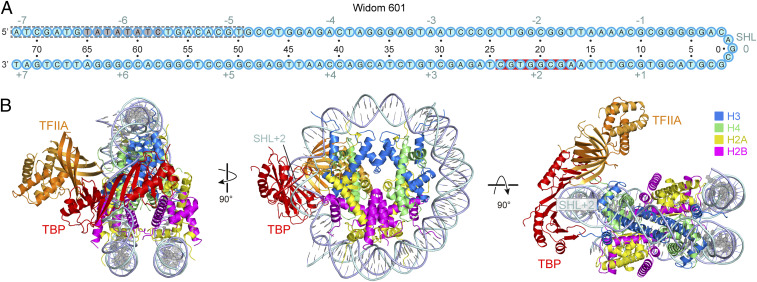
Further particle classification of the TBP–TFIIA–NCP cryo-EM data led to an additional structure that we refined to 2.9-Å resolution (SI Appendix, Fig. S2 B–G). This structure showed TBP binding to SHL +2 of the nucleosome and contained TFIIA bound over the nearby H3–H4 heterodimer on the nucleosome disk (Fig. 3 A and B). In this structure, 23 bp of terminal DNA (SHL –7 to –5) were detached from the histone octamer, apparently because TBP at SHL +2 clashes with the second DNA gyre at SHL –6, as shown by superposition of the free NCP structure (Fig. 3 A and B and SI Appendix, Fig. S4A). Modeling also showed that simultaneous binding of TBP to SHL +2 and SHL –6 would lead to detachment of DNA near SHL –6 due to steric constraints (SI Appendix, Fig. S4B). Hereafter, this alternative structure is referred to as TBP–TFIIA–NCPSHL+2, and our initial structure as TBP–TFIIA–NCPSHL-6.
Fig. 3.
Structure of TBP–TFIIA–nucleosome complex with TBP located at SHL +2 (TBP–TFIIA–NCPSHL+2). (A) Cartoon depiction of the Widom-601 sequence. The TATA box-like sequence is highlighted in gray. Red background indicates observed TBP–DNA contacts. The gray dashed lines indicate the region of detached terminal DNA upon binding of TBP–TFIIA at SHL +2 position. Cyan numbers indicate SHL positions on the nucleosome with the central base pair denoted as SHL 0. Black numbers indicate nucleotide positions with respect to the SHL 0. (B) Three views of the overall structure.
TBP Binding to GC-Rich Nucleosomal DNA.
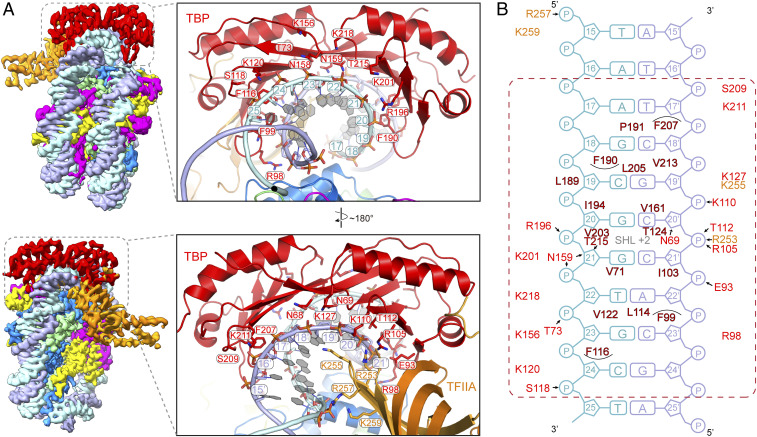
Binding of TBP to SHL +2 was surprising because the NCP is GC-rich in this region (Fig. 3A). The high resolution of nucleosomal DNA in our TBP–TFIIA–NCPSHL+2 structure (2.6 to 2.9 Å) revealed the nature of DNA bases and provided detailed insights into how TBP binds to GC-rich DNA (SI Appendix, Figs. S2G and S3 C and F). TBP binding at SHL +2 does not alter local DNA conformation except that it widens the minor groove slightly (SI Appendix, Fig. S4A). Compared with the TBP–TFIIA–DNA structure, the DNA covered by TBP is less bent and the base pairs are much less inclined (SI Appendix, Fig. S4 C and D).
TBP–DNA interactions are clearly distinct from those observed in the TBP–TFIIA–DNA structure (13, 14) (Fig. 4). First, the hydrophobic contacts and hydrogen bonds formed at both ends of the interface are shifted (Fig. 4B). Second, the two pairs of phenylalanine residues that insert between DNA base pairs in the TFIIA–TBP–DNA structure (13, 14) do not penetrate DNA, are instead located above the edges of the DNA base pairs, and contact only the ribose moieties and base edges. As a consequence, DNA does not show the kinks observed in free DNA (SI Appendix, Fig. S4D). Furthermore, TFIIA residues Arg257 and Lys259 contact the phosphate backbone 1 to 3 bp closer to the TBP-binding site when compared to the TBP–TFIIA–DNA structure (Fig. 4B).
Fig. 4.
Protein–DNA interface in the TBP–TFIIA–NCPSHL+2 structure shows how TBP binds to GC-rich DNA. (A) Two views onto the edge of the nucleosome are shown on the Top and Bottom that are related by a 180° rotation around a vertical axis. On the Left, the cryo-EM maps are shown. On the Right, protein residues involved in DNA interactions are shown as stick models. (B) Schematic representation of protein–DNA interactions. Putative hydrogen bonds are shown as arrows. Residues from TBP and TFIIA are colored in red and orange, respectively. Residues involved in hydrophobic interactions are in dark red. The dashed red rectangle indicates the 8 bp involved in TBP contacts (compare Fig. 3A).
To investigate why TBP can bind to SHL +2 in the absence of a TATA-like sequence, we subjected the free NCP structure obtained from the same cryo-EM dataset (SI Appendix, Fig. S2D) to a global analysis of the DNA base pair step and groove geometry parameters. We found that SHL +2 showed the largest minor groove widths and the highest roll angles among all SHLs (SI Appendix, Fig. S4E). Provided that TBP binding generally distorts DNA, the predistorted DNA conformation at SHL +2 can explain why TBP preferentially binds to this site. The parameters obtained for SHL –2 were similar, although the roll angles were lower, possibly explaining why TBP prefers SHL +2 over the highly similar SHL –2. In summary, our three TBP–nucleosome structures provided unexpected and detailed insights into how TBP can bind to a nucleosome at sites with different base composition, and how this interaction is modulated by adjacent binding of TFIIA.
Discussion
The surprising finding of our work is that TBP can form stable complexes with a nucleosome that can be structurally resolved. The obtained structures show that TBP can bind at SHL –6, recognizing a TATA sequence and allowing for adjacent binding by TFIIA. TBP can alternatively bind at SHL +2, which is GC-rich in our structure. Binding of TBP at SHL +2 is only observed when TFIIA binds adjacently and goes along with detachment of upstream nucleosomal DNA. The conformation of bound DNA in TBP–nucleosome structures is closer to A-form DNA due to the restrictions within the nucleosome, which is distinct in base pair inclination to that in all known structures of TBP or TFIID bound to TATA-containing or TATA-less promoter DNAs (9, 10, 19, 21–25). Our structural analyses suggest that TBP binding is facilitated by prebending of the DNA within the nucleosome, which enables TBP interaction even with a GC-rich region. TBP binding can increase local DNA bending on the nucleosome at SHL –6, but the full 90° bend cannot be achieved, suggesting why TBP affinity for nucleosome-bound DNA is lower than for unbound DNA.
Our findings raise the question whether and when TBP–nucleosome complexes exist in cells. Comparison of our TBP–nucleosome structures with PIC structures shows major clashes (SI Appendix, Fig. S5), excluding that TBP–nucleosome complexes allow for PIC assembly. Indeed, PIC assembly in yeast cells relies on downstream movement of the +1 nucleosome, which liberates the TBP-binding sites and is achieved by ATP-dependent chromatin remodeling enzymes, mainly the essential SWI/SNF-type remodeler RSC (39). Depletion of RSC from cells leads to an upstream shift of the +1 nucleosome by ∼20 to 40 bp, moving the TATA box into the nucleosome (39, 40). RSC depletion strongly decreased TBP occupancy overall, but a portion of promoters retained TBP (39), raising the possibility that TBP may bind TATA sequences within a nucleosome before RSC acts. Modeling TBP onto RSC–nucleosome structures (41, 42) shows that TBP at SHL –6 clashes with the ATPase module of RSC, but TBP at SHL –7 can be accommodated between the ARP module and the DNA-interacting module (DIM) of RSC (SI Appendix, Fig. S6 A and B).
To examine the relative location of TBP and nucleosomes in vivo, we reanalyzed available genome-wide chromatin immunoprecipitation–exonuclease digestion (ChIP-exo) occupancy data for yeast TBP and TFIIA (Toa1 subunit), and genome-wide nucleosome positioning data (43, 44). In metagene plots, TBP and TFIIA occupancy showed peaks ∼80 to 90 bp upstream of the +1 nucleosome dyad, near the edge of the nucleosome (SI Appendix, Fig. S7). A subset of inducible genes with TATA sequences in their promoters, however, showed high occupancy for TBP and TFIIA within the nucleosome near its upstream edge (SI Appendix, Fig. S7 A and B), maybe reflecting TBP binding to a TATA box at SHL –6. In mammalian cells, TBP binding is related to SWI/SNF-induced nucleosome sliding and PIC assembly at the inducible IFN-β promoter during gene activation (45). However, genome-wide occupancy data were obtained from a cell population, and it is likely that most of the ChIP signal for TBP reflects PICs in a context when the nucleosome is depleted. Therefore, it remains to be studied further how TBP and chromatin remodelers cooperate to enable PIC assembly in vivo.
Our results also have implications for the TBP-binding regulatory factors Mot1 and NC2 (46, 47). Mot1 is an ATPase that can displace TBP from promoter DNA (48). Superposition of the Mot1–TBP structure (49) onto our TBP–NCP structures shows that the N-terminal domain of Mot1 can be accommodated without clashes, as required to displace TBP (SI Appendix, Fig. S8A). Superposition of the Mot1–TBP–DNA–NC2 structure (50) onto our TBP–TFIIA–NCP structures shows that Mot1 clashes with TFIIA, implying that TFIIA needs to be removed for Mot1 to bind (SI Appendix, Fig. S8B). NC2 is a negative regulator of transcription initiation that sequesters the TATA-bound TBP by blocking TFIIB binding to prevent PIC assembly (51). Superposition of the TBP–NC2–DNA structure onto our TBP–NCP structures shows that NC2 clashes with the H3–H4 dimer and thus NC2 cannot bind in the context of a TBP–nucleosome complex (SI Appendix, Fig. S8B).
The formation of TBP–nucleosome complexes may also be relevant for understanding the observation that TBP can act as a “bookmarking” factor for genes during mitosis (52–54). TBP can remain bound globally to active promoters during mitosis when chromatin is condensed and can facilitate transcription reactivation in daughter cells after mitosis (53). TBP also allows for dephosphorylation and inactivation of condensin near such bookmarked promoters by recruiting the PP2A phosphatase, thereby inhibiting chromatin compaction (54). During mitosis, a nucleosome may be incorporated at promoter regions, but this may not prevent TBP binding. Instead, formation of TBP–nucleosome complexes as observed here may be involved in transferring transcriptional memory through mitosis (reviewed in ref. 55). However, it is unclear how TBP would be stabilized on chromosomes. Recently, it was shown that the TBP-like factor TBPL2, which does not assemble into TFIID complex but stably associates with TFIIA, prefers to bind TATA-like motifs in oocyte-specific promoters to establish the maternal transcriptome in metazoan oocytes (56). In addition, TBPL2 is one of eight transcription factors that together convert pluripotent stem cells into oocyte-like cells (57). It is therefore possible that TBP-like factors may under some circumstances bind nucleosomes to initiate events that open chromatin locally.
Materials and Methods
Protein Expression and Purification.
Preparation of S. cerevisiae TBP and TFIIA was essentially as described (20). Briefly, full-length C-terminal histidine-tagged TBP was expressed in Escherichia coli BL21 (DE3) RIL strain and purified with affinity chromatography and anion-exchange chromatography. TBP was further purified by gel filtration and concentrated to 8.1 mg⋅mL−1. TFIIA subunits Toa1 and Toa2 were coexpressed with a C-terminal histidine tag on Toa2 in BL21 (DE3) RIL strain. TFIIA was purified in a similar way as TBP and concentrated to 7.1 mg⋅mL−1.
Nucleosome Reconstitution.
Xenopus laevis histones and Widom-601 DNA template were prepared as described (58) with minor modifications as follows. 5′/6-FAM labeled DNA was generated by PCR using purified 145-bp Widom-601 DNA as a template and two primers (forward: 5′-FAM-ATCAGAATCCCGGTGCCG; reverse: ATCGATGTATATATCTGACAC). PCR products were ethanol-precipitated and purified by anion-exchange chromatography as described (59). Purified DNA was resuspended in TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0), and stored at 4 °C in the dark. Nucleosomes were reconstituted using purified histone octamers and DNA mixed at a 1:1 molar ratio, followed by salt gradient dialysis as described (58).
Electrophoretic Mobility Shift Assay.
Nucleosome, TBP, and TFIIA were incubated in electrophoretic mobility shift assay buffer (20 mM Hepes–Na 7.5, 75 mM KCl, 1 mM TCEP, 10% glycerol, 0.2 mg/mL BSA) for 30 min on ice and analyzed by native 6% 0.5× TBE PAGE. Each reaction contained 1 pmol of nucleosome and increasing amounts of protein (0, 0.5, 1, 2, 4, 8, and 16 pmol). Electrophoresis was carried out at 120 V for 2.5 h. Gels were stained with SyberGold (Invitrogen) and imaged with Typhoon 9500 FLA Imager (GE Healthcare Life Sciences).
Fluorescence Anisotropy Assay.
The 5′/6-FAM labeled nucleosome was diluted to 30 nM with reconstitution buffer. TBP and TFIIA were serially diluted in twofold steps in dilution buffer (30 mM Hepes–Na, pH 7.5, 225 mM KCl, 10% glycerol, 1 mM TCEP). Nucleic acids (5 µL, 5 nM final concentration) and TBP/TFIIA (10 µL, 30 nM to 30 µM final concentration) were mixed on ice and incubated for 10 min. The assay was brought to a final volume of 30 µL with 2× assay buffer (20 mM Hepes–Na, pH 7.5, 4 mM MgCl2, 1.5% [vol/vol] glycerol, 0.02 mg/mL BSA, 0.75 mM TCEP) and incubated for 20 min at RT in the dark (final conditions: 20 mM Hepes–NA, pH 7.5, 75 mM KCl, 2 mM MgCl2, 4% [vol/vol] glycerol, 1 mM TCEP, 10 μg/mL BSA). Twenty microliters of each solution were transferred to a Greiner 384 flat-bottom black small volume plate.
Fluorescence anisotropy was measured at room temperature with an Infinite M1000Pro reader (Tecan) with an excitation wavelength of 470 nm (±5 nm), an emission wavelength of 518 nm (±20 nm) and a gain of 80. All experiments were carried out in triplicate and analyzed with GraphPad Prism, version 6. Binding curves were fit with a single-site quadratic binding equation as follows:
where Bmax is the maximum specific binding, L is the concentration of DNA or nucleosome, x is the concentration of TBP or TBP/TFIIA, and Kd,app is the apparent dissociation constant. Error bars are representative of the SD from the mean of three experimental replicates.
Cryo-EM Grid Preparation and Data Collection.
For the TBP–NCP complex, TBP and nucleosome were mixed at a molar ratio of 2:1. For the TBP–TFIIA–NCP complex, TFIIA, TBP, and nucleosome were mixed at a molar ratio of 3:2.5:1. Samples were incubated for 1 h on ice in dialysis buffer (20 mM Hepes–Na, pH 7.5, 75 mM KCl, 1 mM TCEP). The mixture was cross-linked with 0.05% (vol/vol) glutaraldehyde on ice for 10 min and quenched with 10 mM Tris⋅HCl, pH 7.5, 8 mM aspartate, and 2 mM lysine for 10 min. The sample was dialyzed against dialysis buffer for 6 h at 4 °C.
Four microliters of dialyzed samples were applied to glow-discharged UltrAuFoil 2/2 grids (Quantifoil). Samples were incubated for 10 s, blotted for 4 s, and vitrified by plunging into liquid ethane via a Vitrobot Mark IV (FEI) at 4 °C and 100% humidity. Cryo-EM data were acquired on a G2 Titan Krios (FEI) transmission electron microscope operated at 300 keV, equipped with a K3 summit direct detector and a GIF quantum energy filter (Gatan). Automated data acquisition was carried out using SerialEM software at a nominal magnification of 81,000×, resulting in a physical pixel size corresponding to 1.05 Å. Movies of 40 frames were collected in counting mode over 2.4 s at a defocus range from 1.0 to 2.0 μm. The dose rate was 17 e− per Å2 per s, resulting in 1.02 e− per Å2 per frame. A total of 5,409 and 3,772 movies were collected for the TBP–NCP and TBP–TFIIA–NCP complexes, respectively.
Image Processing and Model Building.
Movie stacks were motion-corrected, CTF corrected, and dose-weighted using Warp (60). Particles were auto-picked by Warp, yielding 448,679 and 1,756,032 particle images for TBP–NCP and TBP–TFIIA–NCP, respectively. Image processing was performed with RELION 3.0.5 (61). Particles were extracted using a box size of 2562 or 2002 pixels, and normalized. Reference-free 2D classification was performed to remove poorly aligned particles. An ab initio model generated from cryoSPARC (62) was used for subsequent 3D classification. All classes containing nucleosome density were combined and used for a global 3D refinement. A reconstructed map at 3.2-Å resolution was obtained from 196,518 particles for TBP–NCP. To obtain an improved density map for TBP, particles were subjected to focused 3D classification without image alignment. All classes containing TBP or TBP–TFIIA density were subjected to CTF refinement, Bayesian polishing, and 3D refinement. Postprocessing of refined models was performed using automated B-factor determination in RELION and reported resolutions were based on the gold-standard Fourier shell correlation 0.143 criterion (–67.51 Å2, –54.20 Å2, and –49.14 Å2 B-factor for TBP–NCP, TBP–TFIIA–NCPSHL–6, and TBP–TFIIA–NCP SHL+2, respectively). For the TBP–TFIIA–NCP complex, a reconstructed map at 3.0-Å resolution was obtained from 85,777 particles with TBP–TFIIA–NCPSHL–6 and another map at 2.9-Å resolution from 130,350 particles with TBP–TFIIA–NCPSHL+2. The density of TBP–TFIIA was further improved by applying signal subtraction and focused refinement. Local resolution estimates were obtained using the built-in local resolution estimation tool of RELION and previously estimated B-factors.
The structural models were built into the density of the final reconstructions with the best local resolutions for TBP or the TBP–TFIIA complex. A nucleosome structure with 145-bp Widom 601 DNA (PDB ID code 3LZ0) (36) and the crystal structure of the TBP–DNA (PDB ID code 1YTB) (10) or TBP–TFIIA–DNA (PDB ID code 1NH2) (16) were placed into the density maps by rigid-body fitting in Chimera. The structures were manually adjusted in COOT. The models were subjected to alternating real-space refinement and manual adjustment using PHENIX (63, 64) and COOT (65), resulting in very good stereochemistry (see SI Appendix, Table S2) as assessed by Molprobity (66).
Bioinformatics Analysis.
Published TBP (Spt15) and TFIIA (Toa1) ChIP-exo data, and yeast (S. cerevisiae) genome features, including four groups of protein-coding genes (RP, STM, TFO, UNB) and their +1 nucleosome dyad annotations were taken from ref. 44. TBP annotated peak file was downloaded from https://github.com/CEGRcode/2021-Rossi_Nature/blob/master/04_ChExMix_Peaks/Spt15_CX.bed, and TFIIA annotated peak file from https://github.com/CEGRcode/2021-Rossi_Nature/blob/master/04_ChExMix_Peaks/Toa1_CX.bed. The corresponding TBP and TFIIA filtered .bam files were downloaded from http://www.yeastepigenome.org with sample numbers 8,599 and 14,838, respectively. To identify genes that are targeted by TBP or TFIIA, we overlapped the annotated peaks (1,872 peaks for TBP, 1,587 peaks for TFIIA) with gene promoter regions (TSS +/− 200 bp). This resulted in 103 RP, 284 STM, 483 TFO, 291 UNB genes targeted by TBP and 103 RP, 283 STM, 342 TFO, 203 UNB genes targeted by TFIIA. For TBP and TFIIA coverage calculation, only Read_1 reads were used and the reads were extended to 8-bp length from the 5′ end (the exonuclease stop site). The strand-specific pile-up coverages were then calculated and combined. All genes were aligned at the corresponding +1 nucleosome dyads to generate the metagene plot. For heatmap, genes were sorted by the distance between annotated peaks summit and +1 nucleosome dyads in a decrease manner. All processing was carried out using the R/Bioconductor environment.
Supplementary Material
Acknowledgments
We thank C. Dienemann and U. Steuerwald for maintaining the EM facility, F. Grabbe and R. Yu for help with TBP and TFIIA protein purification, E. Oberbeckmann and M. Lidschreiber for advice on bioinformatic analysis, and S. Schilbach and F. Wagner for discussion. L.X. was supported by the International Max Planck Reseach School for Genome Science. P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP2191, and EXC 2067/1-390729940) and the European Research Council Advanced Investigator Grant CHROMATRANS (Grant Agreement 882357).
Footnotes
Competing interest statement: P.C. declares that he has been a coauthor with K.-P.H. on a recent publication. Both laboratories contributed samples to a third group that was leading the published study. The P.C. and K.-P.H. laboratories did not directly collaborate and there is no conflict of interest.
See Profile, e2111728118, in vol. 118, issue 31.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108859118/-/DCSupplemental.
Data Availability
Three cryo-EM structures have been deposited in the Electron Microscopy Data Bank (accession codes EMD-12897, 12898, 12899, and 12900) and Protein Data Bank (accession codes 7OH9, 7OHA, 7OHB, and 7OHC). All other study data are included in the article and/or supporting information. Previously published data were used for this work (yeastepigenome 8599 and 14838) (44).
References
- 1.Roeder R. G., 50+ years of eukaryotic transcription: An expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 26, 783–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramer P., Organization and regulation of gene transcription. Nature 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hahn S., Young E. T., Transcriptional regulation in Saccharomyces cerevisiae: Transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel A. B., Greber B. J., Nogales E., Recent insights into the structure of TFIID, its assembly, and its binding to core promoter. Curr. Opin. Struct. Biol. 61, 17–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timmers H. T. M., SAGA and TFIID: Friends of TBP drifting apart. Biochim. Biophys. Acta. Gene Regul. Mech. 1864, 194604 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Soffers J. H. M., Workman J. L., The SAGA chromatin-modifying complex: The sum of its parts is greater than the whole. Genes Dev. 34, 1287–1303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolov D. B., et al., Crystal structure of TFIID TATA-box binding protein. Nature 360, 40–46 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Chasman D. I., Flaherty K. M., Sharp P. A., Kornberg R. D., Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 8174–8178 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J. L., Nikolov D. B., Burley S. K., Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365, 520–527 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Kim Y., Geiger J. H., Hahn S., Sigler P. B., Crystal structure of a yeast TBP/TATA-box complex. Nature 365, 512–520 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Zhao X., Herr W., A regulated two-step mechanism of TBP binding to DNA: A solvent-exposed surface of TBP inhibits TATA box recognition. Cell 108, 615–627 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Nikolov D. B., et al., Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377, 119–128 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Geiger J. H., Hahn S., Lee S., Sigler P. B., Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272, 830–836 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Tan S., Hunziker Y., Sargent D. F., Richmond T. J., Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381, 127–151 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Kosa P. F., Ghosh G., DeDecker B. S., Sigler P. B., The 2.1-Å crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc. Natl. Acad. Sci. U.S.A. 94, 6042–6047 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bleichenbacher M., Tan S., Richmond T. J., Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 332, 783–793 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Kostrewa D., et al., RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009). [DOI] [PubMed] [Google Scholar]
- 18.He Y., Fang J., Taatjes D. J., Nogales E., Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y., et al., Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plaschka C., et al., Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Schilbach S., et al., Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aibara S., Schilbach S., Cramer P., Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 594, 124–128 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Rengachari S., Schilbach S., Aibara S., Dienemann C., Cramer P., Structure of the human Mediator-RNA polymerase II pre-initiation complex. Nature 594, 129–133 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Abdella R., et al., Structure of the human Mediator-bound transcription preinitiation complex. Science 372, 52–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., et al., Structural insights into preinitiation complex assembly on core promoters. Science 372, eaba8490 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Robinson P. J., et al., Structure of a complete Mediator-RNA polymerase II pre-initiation complex. Cell 166, 1411–1422.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaudreau L., Schmid A., Blaschke D., Ptashne M., Hörz W., RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell 89, 55–62 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Sainsbury S., Bernecky C., Cramer P., Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Knezetic J. A., Luse D. S., The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell 45, 95–104 (1986). [DOI] [PubMed] [Google Scholar]
- 30.Lorch Y., LaPointe J. W., Kornberg R. D., Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210 (1987). [DOI] [PubMed] [Google Scholar]
- 31.Workman J. L., Roeder R. G., Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51, 613–622 (1987). [DOI] [PubMed] [Google Scholar]
- 32.Imbalzano A. N., Kwon H., Green M. R., Kingston R. E., Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370, 481–485 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Godde J. S., Nakatani Y., Wolffe A. P., The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 23, 4557–4564 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Basehoar A. D., Zanton S. J., Pugh B. F., Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699–709 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Vasudevan D., Chua E. Y. D., Davey C. A., Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Buratowski S., Hahn S., Guarente L., Sharp P. A., Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Imbalzano A. N., Zaret K. S., Kingston R. E., Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J. Biol. Chem. 269, 8280–8286 (1994). [PubMed] [Google Scholar]
- 39.Kubik S., et al., Sequence-directed action of RSC remodeler and general regulatory factors modulates +1 nucleosome position to facilitate transcription. Mol. Cell 71, 89–102.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Klein-Brill A., Joseph-Strauss D., Appleboim A., Friedman N., Dynamics of chromatin and transcription during transient depletion of the RSC chromatin remodeling complex. Cell Rep. 26, 279–292.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye Y., et al., Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner F. R., et al., Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 579, 448–451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee H. S., Pugh B. F., Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi M. J., et al., A high-resolution protein architecture of the budding yeast genome. Nature 592, 309–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomvardas S., Thanos D., Nucleosome sliding via TBP DNA binding in vivo. Cell 106, 685–696 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Gadbois E. L., Chao D. M., Reese J. C., Green M. R., Young R. A., Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc. Natl. Acad. Sci. U.S.A. 94, 3145–3150 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Werven F. J., et al., Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22, 2359–2369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auble D. T., et al., Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8, 1920–1934 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Wollmann P., et al., Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature 475, 403–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butryn A., et al., Structural basis for recognition and remodeling of the TBP:DNA:NC2 complex by Mot1. eLife 4, e07432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamada K., et al., Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106, 71–81 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Chen D., Hinkley C. S., Henry R. W., Huang S., TBP dynamics in living human cells: Constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13, 276–284 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teves S. S., et al., A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7, e35621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing H., Vanderford N. L., Sarge K. D., The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 10, 1318–1323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palozola K. C., Lerner J., Zaret K. S., A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell Biol. 20, 55–64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu C., et al., TBPL2/TFIIA complex establishes the maternal transcriptome through oocyte-specific promoter usage. Nat. Commun. 11, 6439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamazaki N., et al., Reconstitution of the oocyte transcriptional network with transcription factors. Nature 589, 264–269 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Dyer P. N., et al., Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Wang H., Farnung L., Dienemann C., Cramer P., Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding. Nat. Struct. Mol. Biol. 27, 8–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine P. V., et al., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Three cryo-EM structures have been deposited in the Electron Microscopy Data Bank (accession codes EMD-12897, 12898, 12899, and 12900) and Protein Data Bank (accession codes 7OH9, 7OHA, 7OHB, and 7OHC). All other study data are included in the article and/or supporting information. Previously published data were used for this work (yeastepigenome 8599 and 14838) (44).