Significance
Ligand-dependent conformational switches are ubiquitous in biological macromolecules, from allosteric proteins to RNA riboswitches. Molecular design of artificial switches provides a general strategy to test relationships between macromolecular structure and function. The present study exploited recent structures of complexes between an ancestral signaling protein (insulin) and the ectodomain of its cellular receptor to insert a metabolite-regulated switch into the hormone. Whereas binding of ligands often stabilizes structure, this design envisioned metabolite-dependent “opening” of a closed, inactive insulin conformation. Assessment of hormone-directed receptor autophosphorylation and a downstream signaling cascade in liver-derived cells demonstrated that binding of metabolite (a monosaccharide) enabled hormonal signaling. These results suggest a mechanism-based strategy to design “smart” glucose-responsive analogs to more safely treat insulin-dependent diabetes mellitus.
Keywords: hormone–receptor recognition, protein engineering, receptor tyrosine kinase, diabetes mellitus, insulin pharmacology
Abstract
Insulin-signaling requires conformational change: whereas the free hormone and its receptor each adopt autoinhibited conformations, their binding leads to structural reorganization. To test the functional coupling between insulin’s “hinge opening” and receptor activation, we inserted an artificial ligand-dependent switch into the insulin molecule. Ligand-binding disrupts an internal tether designed to stabilize the hormone’s native closed and inactive conformation, thereby enabling productive receptor engagement. This scheme exploited a diol sensor (meta-fluoro-phenylboronic acid at GlyA1) and internal diol (3,4-dihydroxybenzoate at LysB28). The sensor recognizes monosaccharides (fructose > glucose). Studies of insulin-signaling in human hepatoma-derived cells (HepG2) demonstrated fructose-dependent receptor autophosphorylation leading to appropriate downstream signaling events, including a specific kinase cascade and metabolic gene regulation (gluconeogenesis and lipogenesis). Addition of glucose (an isomeric ligand with negligible sensor affinity) did not activate the hormone. Similarly, metabolite-regulated signaling was not observed in control studies of 1) an unmodified insulin analog or 2) an analog containing a diol sensor without internal tethering. Although secondary structure (as probed by circular dichroism) was unaffected by ligand-binding, heteronuclear NMR studies revealed subtle local and nonlocal monosaccharide-dependent changes in structure. Insertion of a synthetic switch into insulin has thus demonstrated coupling between hinge-opening and allosteric holoreceptor signaling. In addition to this foundational finding, our results provide proof of principle for design of a mechanism-based metabolite-responsive insulin. In particular, replacement of the present fructose sensor by an analogous glucose sensor may enable translational development of a “smart” insulin analog to mitigate hypoglycemic risk in diabetes therapy.
Insulin and insulin-like growth factors (IGFs) bind to a cognate set of specific cellular receptors (the insulin receptor [IR] and homologous Type 1 IGF receptor [IGF-1R]) to regulate vertebrate metabolism and development (1–3). IR and IGF-1R are heterodimeric disulfide-linked (α-β)2 dimers wherein extracellular α-subunits contain respective hormone-binding elements and transmembrane β-subunits contain intracellular tyrosine kinase (TK) domains (SI Appendix, Fig. S1) (4). Binding of the hormone to the ectodomain leads to transmission of a structural signal across the plasma membrane, leading to autophosphorylation of the intracellular TK domains (5) and in turn to activation of branching postreceptor signaling pathways (6, 7). Broadly conserved among metazoans (8), this system is of central importance in diverse diseases, including diabetes mellitus (DM), acromegaly, and cancer (9–12). Since its landmark discovery in Toronto 100 y ago (13, 14), insulin replacement therapy has been a therapeutic mainstay in DM (15).
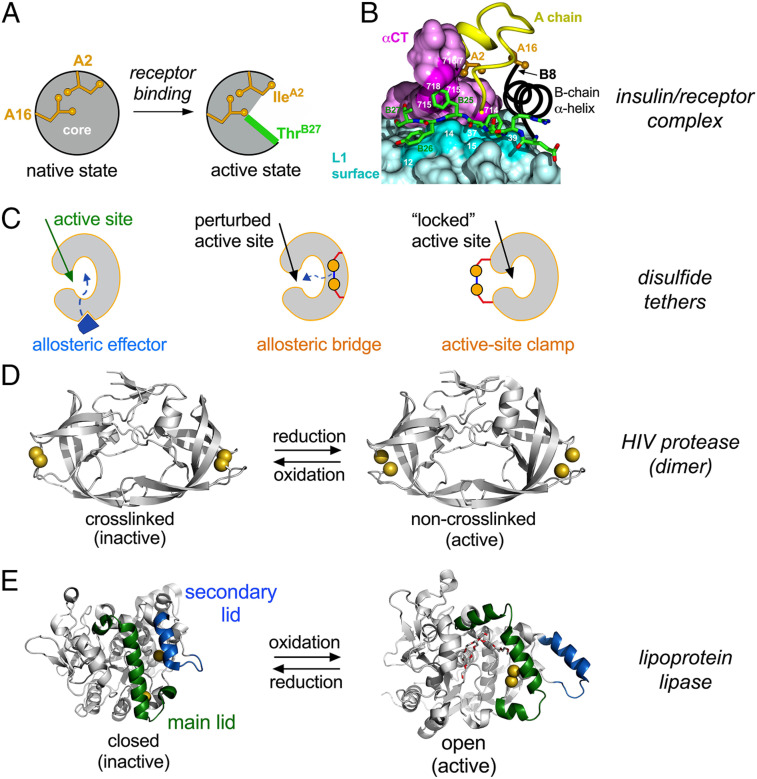
Recent advances in structural biology have established that, in isolation, insulin and the IR each adopt autoinhibited conformations (SI Appendix, Fig. S2) (2, 16). The free hormone folds as a small globular domain (SI Appendix, Fig. S3) (17) but in part “opens” on receptor binding (Fig. 1A) (16). In the open state, the C-terminal B-chain β-strand (residues B24 to B28) detaches from insulin’s α-helical core to insert between receptor elements L1 and αCT' in dimer-related IR α-subunits (Fig. 1B) (2, 5, 16, 18). The conserved “aromatic triplet” in this segment (PheB24, PheB25, and TyrB26) (17, 19) thus pivots between structural roles: contributing to folding and assembly of the closed state (20–22) and to receptor binding in the open state (5, 18). B-chain detachment also exposes conserved nonpolar surfaces—within the N-terminal A-chain α-helix (IleA2 and ValA3) and central B-chain α-helix (ValB12 and LeuB15)—to enable their engagement at the L1/αCT' interface (2, 16). Similar structural features have been observed in homologous model complexes between IGF-I and IGF-1R (3, 23).
Fig. 1.
Molecular switches in proteins. (A and B) Insulin conformation “opens” on receptor binding. (A) Schematic depiction of closed state (free hormone at Left) and active, open state (receptor-bound conformation at Right). Sidechains IleA2 and LeuA16 are shown in gold. The green segment in open state represents detached segment B20 to B27. (B) Cocrystal structure of insulin bound to a fragment of the IR ectodomain [the μIR complex (16)] showing displaced position of the B20 to B27 segment (with green carbon atoms) in groove between IR domains αCT (purple) and L1 (powder blue). Residue-specific contacts in αCT and L1 are shown in darker purple and darker powder blue, respectively (16). Panels are adapted from ref. 86 with permission. (C) Models depicting inhibition of an active site in varying degrees by (Left to Right) allosteric effector, allosteric bridge, or active-site clamp. (D) Tethering by engineered disulfide bridge inactivates HIV protease (28). The pairwise Cys-substituted noncrosslinked enzyme is active (Right) whereas the tethered protein exhibits impaired activity (Left). HIV protease dimer is shown as gray ribbon with sulfur atoms as gold spheres; coordinates were obtained from PDB entries 4DQG and 4DQC. (E) Disulfide tether activates lipoprotein lipase (29). Closed conformation (Left) was modeled using the crystal structure of wild-type Bacillus stearothermophilus lipase 1; PDB entry 1KU0) with pairwise substitutions F206C and A191C (PyMOL). Open conformation (Right) was modeled using crystal structure of bacterial Thermoalkalophilic lipase (PDB entry 2W22); Triton X-100 detergent molecules are shown by red and white sticks. In this structure, the two Cys substituents can bridge within an active conformation. In both conformations, the lipases are as cartoon gray ribbons whereas main and secondary “lids” are shown in green and blue, respectively. Sulfur atoms of interest shown as gold spheres.
Autoinhibited conformations of proteins and their functional opening represent solutions to an evolutionary tension between competing biophysical and biological constraints (24, 25). On the one hand, natural selection has favored protein sequences that exhibit efficient foldability with sufficient native-state stability (including kinetic barriers) to avoid toxic misfolding (26) and amyloidogenesis (27). On the other hand, biological function often exploits dynamic mechanisms, such as in enzymatic catalysis (28, 29) or (as here) the transmission of conformational change by transmembrane receptors (5, 18). Insulin exemplifies this tension (22, 30–33): its closed state confers foldability, mediates native self-assembly, and protects from aggregation-coupled misfolding (25) whereas its open state triggers allosteric receptor reorganization as the first step in transmembrane signaling (5). An evolutionary innovation—a protective hinge (16)—thus emerged as an evolutionary hallmark of the vertebrate insulin-IGF family (34–36).
The present study tests the functional relationship between insulin’s hinge and cellular signaling by the holoreceptor. Our approach was stimulated by exploitation of engineered disulfide bridges as probes of dynamic coupling in enzymatic catalysis (Fig. 1C) as illustrated in unrelated systems (HIV protease and lipoprotein lipase; Fig. 1 D and E) (28, 29). In those studies, pairwise Cys substitutions provided reversible redox-regulated tethers to insert a cleavable allosteric bridge or clamp. Although such an approach is inapplicable to insulin (as both the hormone and receptor contain native disulfide bridges required for function), might the essential idea be extended to an artificial switch? Because the logic of this approach is independent of molecular embodiment, we sought to exploit an exogenous ligand to regulate insulin’s protective hinge (16).
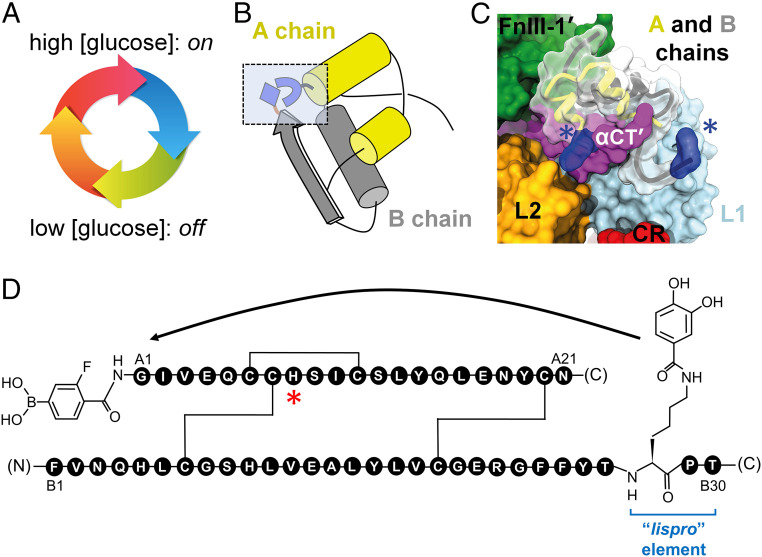
Our strategy derived from a long-standing (and to date unmet) goal in insulin pharmacology: design of a glucose-responsive insulin (GRI; Fig. 2A) to protect patients from hypoglycemia (37, 38). One potential design scheme envisioned a glucose-cleavable tether between the C terminus of the B chain and N terminus of the A chain (Fig. 2B), thereby providing an insulin analog that would be closed and inactive at low glucose concentration but openable and active at high glucose concentration (39). The resulting cycle of conformational states (Fig. 2A) would in principle be reversible (depending on metabolic state), and its implementation would reflect the signaling conformation of the insulin–IR complex (Fig. 2C) (16). Although to our knowledge such a GRI design has not been reduced to practice, we took advantage of an alternative monosaccharide sensor (recognition of fructose by phenylboronic acid [PBA]; Fig. 2D and SI Appendix, Fig. S4) (40, 41). Our results demonstrate selective fructose-dependent IR autophosphorylation in a human liver–derived cell line [HepG2; (42)], leading to activation of an appropriate downstream kinase cascade (43) and physiologic transcriptional regulation of metabolic target genes (44–46). These findings validate a functional coupling between insulin’s hinge-opening and transmembrane signaling and provide proof of principle for a reversible mechanism-based design of a “smart” insulin.
Fig. 2.
Design of FRI. (A) Glucose-regulated conformational cycle in which a monosaccharide acts as a competitive ligand to regulate a conformational switch between closed state (inactive in absence of ligand; Bottom) and open state (active in presence of ligand; Top). (B) Ribbon model of insulin (T-state monomer). The box indicates sites of chemical modification showing “switch” elements in closed conformation due to intramolecular DHBA/m-fPBA ester bonding. (C) Structural model of FRI binding to insulin receptor. Blue volumes and asterisks highlight the protein adducts. A and B chains are shown as yellow and gray ribbons, respectively. IR domains are shown as surfaces by color code: L1 (powder blue), L2 (gold), CR (red), FnIII-1' (green), and dimer-related αCT' (purple). (D) GlyA1 α-amino group is modified by fPBA, whereas the ε-amino group of LysB28 (in lispro B chain with “KP switch”; ProB28→Lys and LysB29→Pro) is linked to an aromatic 1,2-diol (DHBA). Weakened IR-binding affinity due to the A1-adduct is compensated by favorable substitution ThrA8→His (asterisked; 51).
Results
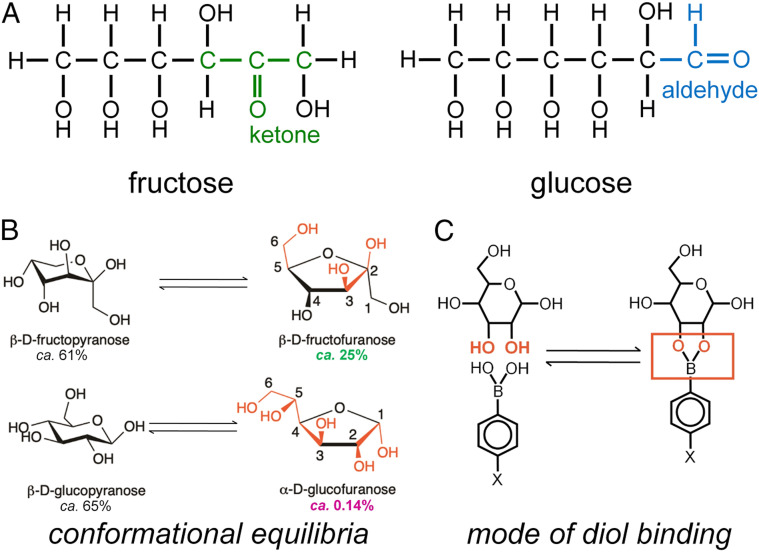
The switchable insulin analog (designated FRI; fructose-responsive insulin) contains meta-fluoro-PBA* (m-fPBA) as a diol sensor linked to the α-amino group of GlyA1 (at left in Fig. 2D) and an aromatic diol (3,4-dihydroxybenzoic acid; DHBA) attached to the ε-amino group of LysB28 of insulin lispro† (“KP” element; blue bracket in Fig. 2D). Although fructose and glucose each contain diols (Fig. 3A), the sensor preferentially binds to aligned 1,2-diol groups as found in β-D-fructofuranose and α-D-glucofuranose (Fig. 3B) (47, 48). Affinity of m-fPBA is higher for fructose than glucose due to salient differences in respective conformational equilibria (Fig. 3B and SI Appendix, Fig. S4); binding is covalent but reversible (Fig. 3C). To compensate for impairment of IR-binding affinity generally associated with N-linked adducts at GlyA1 (49, 50), ThrA8 was substituted by His (red asterisk in Fig. 2D), a favorable substitution found in avian insulins (51). Control analogs were provided by 1) insulin KP, 2) a KP derivative containing an A1-linked m-fPBA but not the B28 diol (diol-free control; DFC), and 3) a peptide bond between LysB28 and GlyA1 in a des-[B29, B30] template (52). The latter [a covalent “closed” state (53)] was inactive (SI Appendix, Fig. S5).
Fig. 3.
Monosaccharide structures and mode of binding to PBA. (A) Linear structures of fructose and glucose. (B) PBA binds most strongly to aligned 1,2-diol elements as in the β-D-furanose conformation of fructose (Top right; 25% occupancy) or α-D-furanose conformation of glucose (Bottom right; 0.14% occupancy). Respective conformational equilibria thus favor selective binding to fructose (47, 48). Key cis-hydroxyl groups are shown in red. Percent populations are as indicated. (C) Schematic binding mode of PBA to a 1,2-diol element in a monosaccharide (red). Panel B is adapted from ref. 87.
Western Blot Assays Demonstrated Fructose-Dependent Signaling.
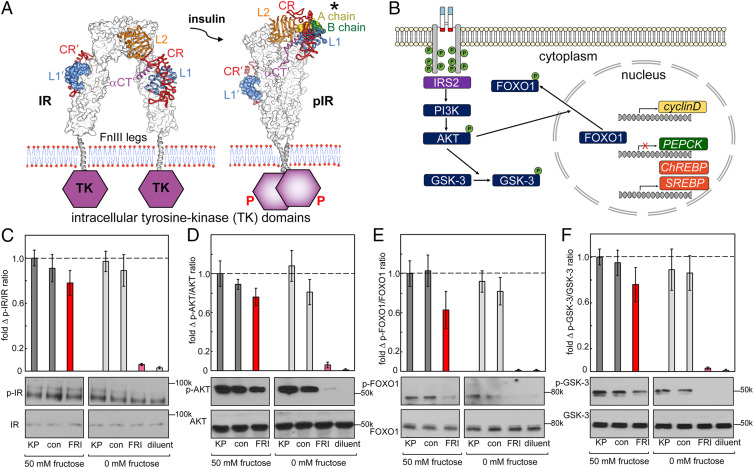
Structural studies suggest that insulin’s hinge-opening at a dimer-related αCT/L1 interface (2) is coupled to closure of IR ectodomain legs (16), leading to TK-mediated trans-phosphorylation and receptor activation (Fig. 4A) (18). Signal propagation was probed via a cytoplasmic kinase cascade and changes in metabolic gene expression (Fig. 4B) in HepG2 cells (42, 43). Control studies indicated that addition of 0 to 100 mM fructose or glucose did not trigger changes in signaling outputs (SI Appendix, Figs. S6 and S7). An overview of IR autophosphorylation (probed by anti-pTyr IR antibodies; Fig. 4C) and downstream phosphorylation of Ser-Thr protein kinase AKT (protein kinase B; ratio p-AKT/AKT), forkhead transcription factor 1 (p-FOXO1/FOXO1), and glycogen synthase kinase-3 (p-GSK-3/GSK-3) at a single hormone dose (50 nM) was provided by Western blot (WB; Fig. 4 D–F). In each case, WBs demonstrated fructose-dependent signaling by FRI and fructose-independent signaling by KP and DFC. The activity of FRI in the absence of fructose is low.
Fig. 4.
Fructose-dependent IR activation and downstream pathways. (A) IR ectodomain structures either unbound (Left) or as activated by insulin (Right), asterisk highlights the A chain (light green) and B chain (green) of insulin). On insulin binding, the ectodomain legs come together to enable TK transphosphorylation (red P’s). (B) Selected downstream IR signaling pathways as probed by WB. IR L1 and αCT domains are highlighted in blue and red; JM- and TK-domain phosphorylation sites are shown in schematic form by green circles. IR activation leads to AKT phosphorylation, which in turn phosphorylates and inhibits GSK-3; pAKT also phosphorylates FOXO in cytoplasm to enable transcriptional regulation of cell proliferation (cyclin D), gluconeogenesis (PEPCK), and fatty-acid synthesis (ChREBP and SREBP) in the nucleus. (C-F) WB assays probing the p-IR/IR (C), p-AKT/AKT (D), p-FOXO/FOXO (E), and p-GSK-3/GSK-3 (F) in the presence or absence of fructose (50 mM) in medium. Studies of FRI were conducted relative to lispro insulin (“KP”), a diol-free control analog (DFC; “con”) or buffer only (diluent; “dil”). The error bars in histograms indicate SEM. Representative gel images are displayed underneath corresponding columns in histograms; assays were performed in triplicate.
Plate Assays Demonstrated Ligand-Selective Signaling.
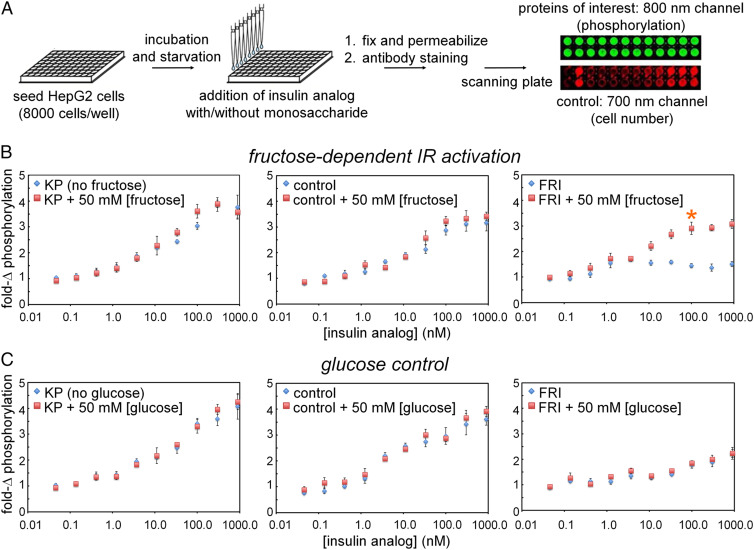
Quantitative dose-dependent and ligand-selective IR autophosphorylation were evaluated in a 96-well plate assay (Fig. 5A). FRI triggered a robust signal on addition of 50 mM fructose (asterisk at Right in Fig. 5B) whereas baseline activity in the absence of fructose was low. As expected, KP and DFC exhibited high signaling activity in the presence or absence of fructose (Left and Center in Fig. 5B, respectively). Ligand-dependent activation of FRI is specific to fructose as addition of 50 mM glucose did not influence its activity (nor the activities of KP and DFC; Fig. 5C). These data indicate that in 50 mM fructose FRI is almost as active as KP.
Fig. 5.
Dose–response studies of FRI activity. (A) Schematic outline of in-cell assays probing IR autophosphorylation on binding of insulin. Flowchart illustrates procedure to assess hormone-induced IR signaling via in-cell illumination assay. On binding of insulin analogs, autophosphorylation was evaluated via 800-nm emissions mediated by antibody-conjugated signals; 700-nm readout provided cell-number control. (B) Fructose dependence of pIR/IR ratio on hormone-binding. Whereas extent of IR autophosphorylation was independent of fructose on binding of insulin lispro (“KP”; Left) or a DFC analog (Middle), FRI (Right) was activated when incubated with 50 mM [fructose] or higher; baseline activity was low. Orange asterisk indicates significant effect on 100 nM [insulin analog] on FRI activity. The asterisk (*) indicates P < 0.05 with respect to the treatment difference (0 vs 50 mM fructose) at an FRI dose of 100 nM. (C) Control studies using glucose instead of fructose show that the FRI was not affected by presence or absence of glucose (Right). As expected, glucose did not modulate the high baseline activities of KP (Left) and the diol-free control (Middle).
PCR Assays Demonstrated Ligand-Selective Metabolic Gene Regulation.
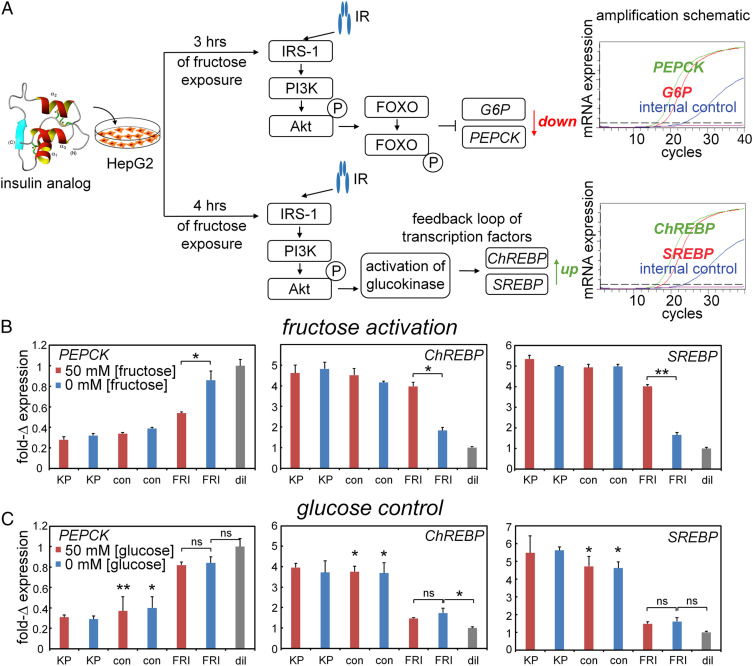
Insulin-signaling in hepatocytes extends to metabolic transcriptional regulation (Fig. 6 and 4B) (46, 54) as recapitulated in HepG2 cells (Fig. 6A). At hypoglycemic conditions, the cells exhibited stronger gluconeogenesis-related responses following insulin stimulation than at hyperglycemic conditions. In this protocol (Upper arm in Fig. 6A), FRI, when activated by fructose, regulated downstream expression of the gene encoding phosphoenolpyruvate carboxykinase (PEPCK; a marker for hormonal control of gluconeogenesis). Under normoglycemic conditions (Lower arm in Fig. 6A), FRI, when activated by fructose, regulated the genes encoding carbohydrate-response–element and sterol-response–element binding proteins (ChREBP and SREBP; markers for hormonal control of lipid biosynthesis). No fructose dependence was observed in control studies of KP and DFC (Left and Central panels of Fig. 6B); no effects were observed on addition of glucose instead of fructose (Fig. 6C). Control studies were undertaken in the absence of insulin analogs to assess potential confounding changes in metabolic gene expression on addition of 0 to 100 mM fructose (SI Appendix, Fig. S7A) or 0 to 100 mM glucose (SI Appendix, Fig. S7B). No significant effects were observed in either case, indicating that the present short-term fructose exposure (to activate FRI; schematic protocol in SI Appendix, Fig. S7C) is unassociated with the transcriptional signature of longer-term exposure (SI Appendix, Fig. S7D).
Fig. 6.
Fructose-dependent transcriptional signaling. (A) Schematic outline of qPCR-based assays of insulin-regulated gene regulation: (Top) insulin-dependent repression of gluconeogenic gene PEPCK and (Bottom) insulin-dependent activation of lipid-biosynthetic genes ChREBP and SREBP. (B and C) Transcriptional responses specific to PEPCK (Left) versus ChREBP and SREBP (Middle and Right, respectively) on addition of 50 mM fructose (B) or 50 mM glucose (C). Decreased downstream accumulation of PEPCK mRNA and increased ChREBP and SREBP mRNA reflect the activation stimulated by testing analogs in cellular metabolism pathways. Insulin lispro (“KP”) and DFC (“con”) exhibited no monosaccharide-dependent changes in their high baseline activities. Asterisks (*) and (**) indicate P value < 0.05 and < 0.01. The “ns” indicates P value > 0.05 (SI Appendix, Tables S1 and S2). Control studies were undertaken in the absence of insulin analogs to exclude potential confounding effects of 0 to 100 mM monosaccharides on metabolic gene expression (SI Appendix, Fig. S7).
Ligand-Binding to FRI Affects Protein Structure.
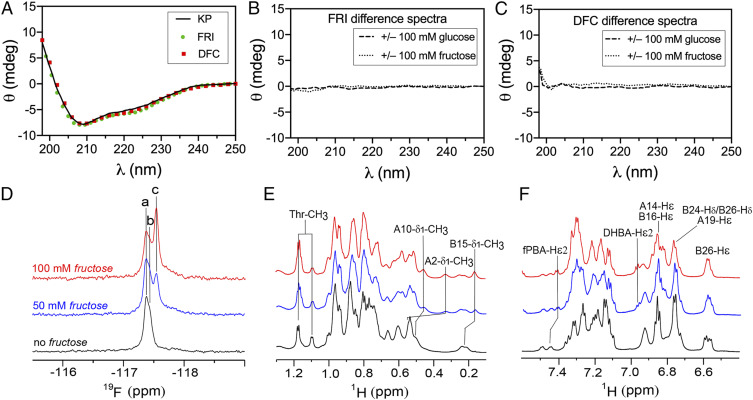
Far-UV circular dichroism (CD) spectra of FRI and DFC are indistinguishable from parent analog KP (Fig. 7A), indicating that secondary structure is not affected by the modifications at A1 and B28 (Fig. 2D). Difference CD spectra calculated on addition of 100 mM fructose or glucose were in each case featureless (Fig. 7 B and C and SI Appendix, Fig. S8). High-resolution NMR spectroscopy [as enabled by the monomeric KP template (55)] corroborate essential elements of the intended fructose-selective switch as follows (SI Appendix, Supplementary Discussion):
-
1)
19F-NMR spectra monitored fructose sensor. The fluorine atom in m-fPBA provided an NMR-active nucleus. Addition of 0 to 100 mM fructose led to an upfield change in 19F-NMR chemical shift in slow exchange on the NMR time scale (Fig. 7D and SI Appendix, Figs. S9A, S10 and Table S3). This upfield shift presumably reflects displacement of an aromatic diol by a nonaromatic ligand. No change in FRI 19F chemical shift was observed on addition of glucose (SI Appendix, Fig. S9B and Table S3). Although an analogous 19F resonance was observed in the NMR spectrum of DFC, its chemical shift did not change on addition of glucose or fructose (SI Appendix, Fig. S9 C and D and Table S3). Presumptive NMR resonance assignments are given in SI Appendix, Table S3. Interestingly, a broadened 19F signal was observed in ligand-free DFC, probably due to conformational exchange or self-association; this signal sharpened on addition of ligand (fructose or glucose). Dual 19F- and 1H NMR–monitored titration (Fig. 7 D–F and SI Appendix, Fig. S10 A and B and S11 A and B) and natural-abundance 1H-13C heteronuclear single quantum coherence (HSQC) spectra (Fig. 8) provided further evidence of a specific interaction between FRI and fructose.
-
2)
1H-13C 2D HSQC spectra monitored “closed” conformation of ligand-free FRI. One-dimensional (1D) 1H and 1H-13C HSQC spectra of DFC were similar to those of parent analog KP (SI Appendix, Fig. S12 A and B), excepting methyl resonances of IleA2 and ValA3 (adjacent to the GlyA1-attached m-fPBA). Patterns of 1H-13C chemical shifts of FRI and DFC were also similar (SI Appendix, Fig. S12 C and D). Those NMR features provided evidence that FRI and DFC retain a native-like structure. However, in FRI, the resonances of IleA2, ValA3, LeuB11, ValB12, and LeuB15 exhibited larger chemical shift differences (relative to KP) than in DFC (SI Appendix, Fig. S12D and Table S4). These findings suggest that FRI exhibits a local change in conformation and/or dynamics, presumably due to the intended DHBA/m-fPBA tether. We envision that constraining the C-terminal B-chain segment alters aromatic ring currents affecting the central B-chain α-helix (via TyrB26-LeuB11, TyrB26-ValB12, and PheB24-LeuB15 packing) and N-terminal A-chain helix (via native-like TyrB26-IleA2 and TyrB26-ValA3 packing).
-
3)
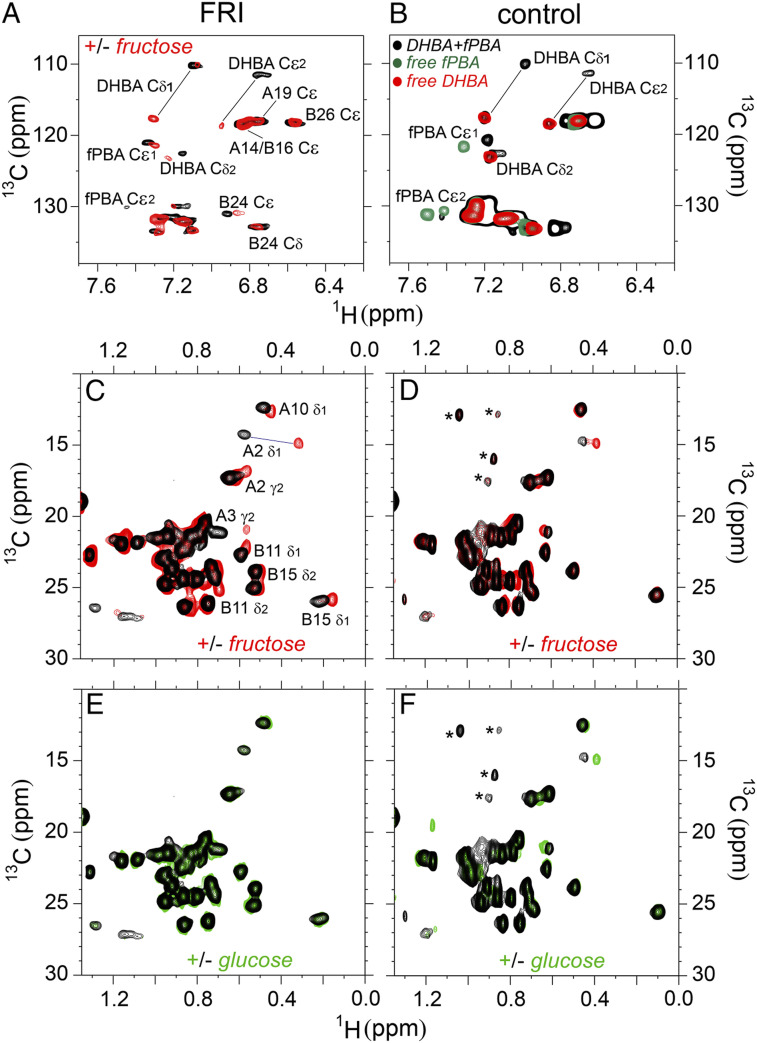
Aromatic 1H-13C two-dimensional (2D) HSQC spectra monitor hinge-opening. 1H-13C HSQC spectra provide probes of aromatic resonances in FRI’s DHBA/m-fPBA adducts in the absence of fructose (black cross-peaks in Fig. 8A) and in the presence of 100 mM fructose (red cross-peaks in Fig. 8A). Significant chemical shift changes in both 1H/13C dimensions were observed (SI Appendix, Table S5–S8). Resonance assignments were corroborated by model studies of m-fPBA– and DHBA-modified peptides (Fig. 8B). DHBA chemical shifts in fructose-free FRI are similar to those in the complex of model peptides, whereas such chemical shifts in fructose-bound FRI are similar to that of free DHBA-modified octapeptide (SI Appendix, “NMR Studies”). In addition, methyl resonances sensitive to addition of fructose exhibited a trend toward corresponding chemical shifts observed in spectra of insulin lispro and ligand-free DFC (SI Appendix, Fig. S13 and Table S4–S6). Together, these NMR features provide evidence that in FRI the LysB28-attached DHBA binds GlyA1-linked m-fPBA in absence of fructose, but this tether is displaceable by fructose.
-
4)
Methyl 1H-13C 2D HSQC spectra monitor protein core. Aliphatic 1H-13C spectra (56) reflect tertiary structure as probed by upfield-shifted methyl resonances. Changes in cross-peak chemical shifts were observed in FRI on overlay of spectra acquired in the absence of an added monosaccharide (black cross-peaks in Fig. 8C) or on addition of 100 mM fructose (red cross-peaks; SI Appendix, Fig. S14). Fructose-binding accentuated upfield 1H secondary shifts (horizontal axis in Fig. 8C and SI Appendix, Table S5) with smaller changes in 13C chemical shifts (vertical axis, SI Appendix, Table S6). These changes presumably reflect altered aromatic ring currents within insulin’s core (57). Control studies of DFC suggested that such chemical shift changes require the interchain DHBA/m-fPBA tether (Fig. 8D and SI Appendix, Tables S9–S11); in these spectra, changes were restricted to IleA2 immediately adjoining the sensor. Addition of 50 mM glucose caused essentially no changes in 1H-13C fingerprints of FRI or DFC in accordance with the fructose selectivity of m-fPBA (black and green cross-peaks in Fig. 8 E and F and SI Appendix, Tables S5–S6 and S9–S11).
Fig. 7.
Spectroscopic studies of FRI structure. (A) Far-UV CD spectra of insulin lispro (KP; black lines), FRI (green circles) and DFC (red squares) in 50 mM KCl and 10 mM phosphate (pH 7.4) at 25 °C without added glucose or fructose. (B and C) Difference spectra of FRI (B) and the DFC (C) +/− 100 mM glucose (dashed line) or +/− 100 mM fructose (dotted line). (D) 19F-NMR titration of FRI in the absence of a monosaccharide (black) and on addition of 50 mM (blue) and 100 mM (red) fructose. Peaks a and b indicated fructose-free 19F signals of m-fPBA; peak c is specific to the fructose complex. (E) 1H-NMR titration of FRI in methyl region: line broadening and changes in chemical shifts were observed on fructose-binding. Well-resolved methyl resonance assignments are as labeled (Top). Color code is as in panel (D). (F) 1H-NMR titration of FRI in aromatic region; selected resonance assignments are provided (Top). Spectra were acquired at a 1H frequency of 700 MHz (19F frequency of 658 MHz) at pD 7.8 (direct meter reading) and 25 °C. The protein concentration was ca 0.4 mM.
Fig. 8.
1H-13C HSQC spectra reveal fructose-dependent closed-open conformational transition and protein structure change in FRI on fructose-binding. (A) Aromatic region of 1H-13C HSQC spectra of FRI in absence of fructose are shown in black and on addition of 100 mM fructose in red. (B) Model study of DHBA-fPBA interaction. Aromatic 1H-13C HSQC spectrum of free DHBA-modified octapeptide is shown in red, free m-fPBA–modified octapeptide in green and their complex in black. (C) Methyl region of 1H-13C HSQC correlation spectra of FRI and (D) its DFC in absence of a saccharide are shown in black and in the presence of 100 mM fructose in red. (E) Methyl-aliphatic region of 1H-13C HSQC spectra of FRI and (F) DFC acquired in absence of a saccharide in black and in the presence of 50 mM glucose in green. Data were acquired at a 1H frequency of 600 MHz for FRI or 700 MHz for DFC at pD 7.4 (direct meter reading) and 25 °C. Asterisks in panel (D) and (F) indicate contaminants (also see SI Appendix, Fig. S15).
Discussion
Engineering of a ligand-regulated switch within a protein requires 1) a ligand-binding element and 2) a mechanism-coupling ligand-binding to a functional step. The present application to insulin exploited the hinge-opening mechanism through which the native hormone interacts with its receptor (Fig. 1 A and B) (16). Coupling between IR-binding and ligand sensing was provided by an internal interchain tether displaced by the ligand (fructose) (40, 47, 48). Our results provide evidence that hinge-opening is required for hormone-triggered receptor autophosphorylation and downstream signaling.
Site-1 and Site-2 Insulin Complexes.
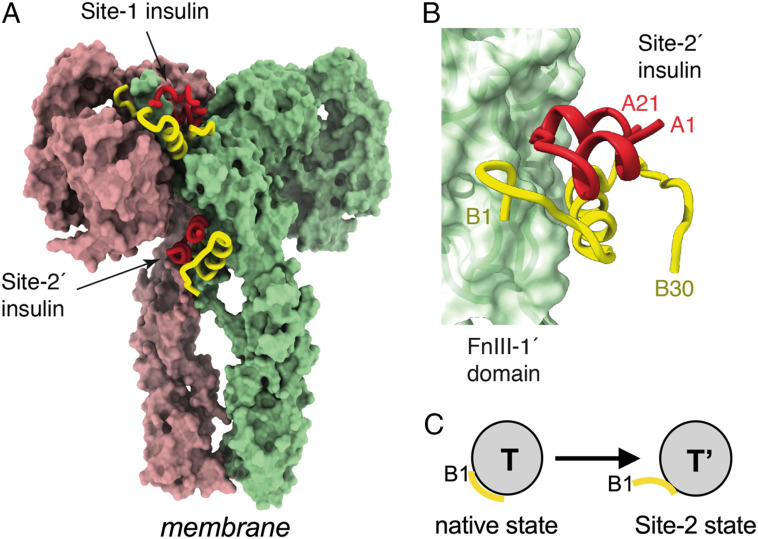
Recent cryogenic electron microscopy (cryo-EM)–derived structures of ligand-saturated hormone-ectodomain and hormone-holoreceptor complexes revealed, in contrast to prior structures, two additional insulin molecules bound to domains FnIII-1 and FnIII-1′, respectively (58) (Fig. 9A and SI Appendix, Table S12). These insulins—designated insulin 2 and 2′ in relation to an alternative “Site-2” binding surface proposed by De Meyts (59) and Schäffer (60)—exhibit closed T-like conformations (Fig. 9 B and C). Long the subject of speculation (5), insulin's Site-2–binding surface was shown by Gutmann et al. (58) to include respective central regions of the A and B chains (residues A10 to A17 and HisB10, GluB13, and LeuB17), unexpectedly extended by a displaced N-terminal B-chain segment (B1 to B4; Fig. 9C).‡ Although the function of Site 2 is not well understood, kinetic studies have suggested that it may represent an initial encounter complex (1, 61). The low activity of the fructose-free FRI and inactivity of B28-A1 single-chain insulin (SI Appendix, Fig. S5) suggest that Site-2 binding cannot in itself trigger receptor autophosphorylation.
Fig. 9.
Structure of the insulin-saturated IR ectodomain. (A) IR ectodomain showing the disposition of two of the four insulins bound under insulin-saturated conditions (the remaining two insulins are obscured from view but are pseudosymmetry related to those shown). Receptor monomers are shown in light green and pink, respectively, whereas insulin A and B chains are shown in red and yellow, respectively. (B) Interaction of insulin with receptor Site 2', located on the surface of domain FnIII-1'. Colors are as in panel (A). Coordinates were obtained from PDB entry 6SOF (58). (C) Schematic depiction on subtle conformational change in T state on Site-2 binding to ectodomain stalk. Yellow segment indicates residues B1 to B4.
Engineered Tethers in Proteins.
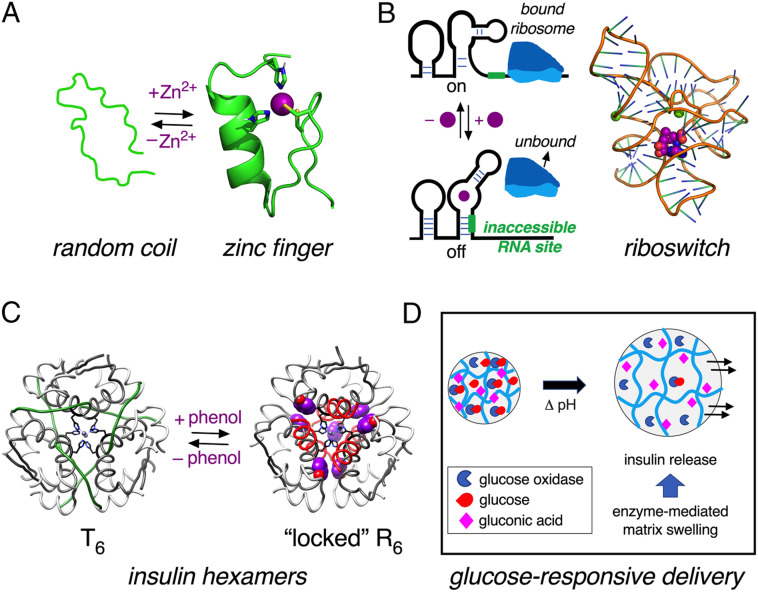
By analogy to engineered disulfide bridges as reducible probes of protein function (Fig. 1) (28, 29), we imagined a ligand-cleavable tether between insulin’s A and B chains as a redox-independent switch (SI Appendix, Supplementary Discussion). This design, making ligand-dependent hinge-opening possible, stands in contrast to classical ligand-binding motifs in proteins associated with stabilization of structure. Zn fingers and other Zn-binding motifs, for example, generally exhibit metal ion–dependent peptide folding (Fig. 10A) (62). Analogous metal ion–coupled folding of RNA underlies the function of riboswitches (Fig. 10B) (63), control motifs in untranslated messenger RNA (mRNA) regions (64). Insulin self-assembly itself (17) is stabilized by Zn2+ coordination (21), whereas the structure of each protomer within the T6 (2-Zn) hexamer (17) is similar to that of the native monomer (55). Binding of phenolic ligands to this hexamer triggers an allosteric transition, leading to the more-stable R6 state (Fig. 10C) (65). Containing an extended α-helix, the latter is preferred for pharmaceutical formulations as its greater stability (57) extends shelf life (66, 67). The present fructose-cleavage interchain tether in FRI provides a contrasting example of ligand-driven loss of structure or stability.
Fig. 10.
Switches in proteins can modulate degree of organization. (A) Schematic representation of “zinc finger” motif of a human enhancer binding protein (PDB entry 3ZNF) (62). Upon binding to Zn2+ ions, the random coil structure becomes more ordered, as shown by zinc finger structure on the Right. Side-chains of His and Cys residues are shown as sticks. (B) Left, schematic representation of a riboswitch that permits translation in the absence of ligand (Top) but inhibits when bound to ligand (Bottom) (64). Right, representative example of a riboswitch is provided by the crystal structure of Thermoanearobacter tengcongensis (Tte) metF SAM-I aptamer. Magnesium ions are represented as green spheres; ligand S-adenosyl-L-methionine (SAM) is shown as a CPK model in purple (88). (C) Phenol-induced reorganization of the insulin hexamer. Insulin hexamer adopts classical T6 conformation in presence of Zn2+ ions (Left) wherein residues B1 to B8 are extended (17). In the presence of phenol as an allosteric ligand, residues B1 to B8 adopt an α-helical conformation in R6 zinc hexamer (65). A- and B-chain ribbons are shown in light- and dark gray, respectively. The residues B1 to B8 shown in green for T6 (Left) and in red for R6 (Right). The phenolic ligands are shown as CPK models (purple); axial zinc ions are shown in blue. Coordinates were respectively obtained from PDB entries 1MSO (T6) and 1ZNJ (R6) (89, 90). (D) Schematic depiction of a glucose-responsive material. When given glucose as substrate, matrix-bound enzyme glucose oxidase converts glucose to gluconic acid, which in turn lowers pH to cause swelling of hydrogel, releasing encapsulated insulin (68, 91).
Translational Implications and Concluding Remarks.
Ligand-induced destabilization of structure has a long history of investigation in relation to glucose-responsive polymers, such as hydrogels designed to swell and release insulin on an increase in local glucose concentration (68, 69). A well-characterized embodiment is provided by polymer matrices embedded with glucose oxidase and insulin (box in Fig. 10D and SI Appendix, Supplementary Discussion). When the ambient glucose concentration is high, its enzymatic conversion to gluconic acid (in presence of oxygen) causes a reduction in pH, in turn swelling the matrix and enabling insulin release (68, 69). This “smart” materials approach to engineering a glucose-responsive subcutaneous depot addresses a long-sought but unmet medical need: how to reduce the risk of hypoglycemia (37, 38) in patients receiving insulin replacement therapy (15). Concerns related to hypoglycemia and its sequelae can limit glycemic targets in Type 1 and long-standing Type 2 DM.
The present monosaccharide-dependent disruption of an interchain tether in FRI extends to the nanoscale the goals of mesoscale glucose-responsive materials engineering. Its molecular design provides proof of principle for a minimal “smart” insulin nanotechnology in the absence of a polymer matrix and with mechanism unrelated to prior proposed unimolecular GRIs [SI Appendix, Supplementary Discussion (39, 70, 71)]. Whereas the fructose-free tethered state would resemble chemically cross-linked or single-chain insulin analogs (53, 72, 73)—long known to exhibit low activities—the fructose-bound open state is competent to bind IR via Site-1–associated detachment of the B24 to B30 segment from the α-helical core of the hormone (Fig. 1B) (2, 5, 16, 18). It would be of future interest to determine three-dimensional structures of the free analog, its fructose-bound state and in turn an IR ectodomain complex as an explicit visualization of ligand-regulated hinge-opening.
We anticipate that replacement of a PBA-based fructose sensor by a bona fide glucose sensor would provide a Site-1–based GRI of potential clinical utility. This scheme would provide a reversible conformational constraint regulating hormonal activity through changing metabolic conditions. Whereas the selectivity of PBA for fructose is in accordance with the conformational properties of monosaccharides (Fig. 3C and SI Appendix, Fig. S4), other types of monosaccharide-recognition elements have been described that recognize the distinctive arrangement of hydroxyl groups well populated among glucose isomers (71). It would be remarkable should recent advances in the structural biology of insulin-signaling enable such translational opportunities to be realized.
Materials and Methods
Further information regarding experimental procedures are given in SI Appendix.
Materials.
Wild-type insulin was purchased from Biodel. Insulin lispro was obtained from pharmaceutical vials of Humalog (Eli Lilly and Co.). Endoproteinase Lys-C was expressed as described (74). Diol-modified octapeptides and control octapeptide GFFYTKPT (residues B23 to B30 of insulin lispro) were synthesized using standard 9-fluoromethylmethoxycarbonyl (Fmoc)-protected amino acids with preloaded Fmoc-Thr(tBu)-Wang resin. Attachment of the diol adduct was confirmed using reverse-phase high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry. NHS-fPBA was synthesized as described (75). HisA8 miniproinsulin was expressed in Pichia pastoris and purified as described (74, 76). FRI and DFC were prepared by trypsin-catalyzed semisynthesis (SI Appendix, Scheme S1) and purified by HPLC (SI Appendix, Fig. S16–23); predicted molecular masses were confirmed by mass spectrometry (SI Appendix, Table S13).
Cell Culture.
HepG2 cells were cultured in Dulbecco's Modified Eagle Medium, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin as recommended by the American Type Culture Collection. A protocol employing 24-h serum starvation was adapted from Rege et al. (22); after starvation, cells were treated in parallel with insulin analogs in serum-free medium.
Real-Time qPCR Assays.
Following serum starvation, HepG2 cells were treated with medium containing an insulin analog (50 nM) for 8 h. In studies related to possible glucose responsiveness and lipid metabolism, cells were treated with analogs for 3 or 4 h in media containing either low or normal glucose concentrations. Gene-specific mRNA abundances were measured in triplicate by qPCR; samples were prepared as described by the vendor (Bio-Rad). Control studies were performed to assess transcriptional responses of HepG2 cells to short-term exposure (3 to 4 h; Fig. 6A) to 0 to 100 mM monosaccharides in insulin-free conditions (SI Appendix, Fig. S7).§
In-Cell pIR Immunoblotting.
The assay probed insulin-dependent IR activation via fluorescent readouts (22). HepG2 cells were seeded (∼8,000 cells/well) into a 96-well black plate with clear bottom and cultured (Fisher). Fixed cells were exposed to the primary antibody (10 µL anti-pTyr 4G10 into 20 mL blocking buffer) overnight at 4 °C. The secondary antibody (anti-mouse-IgG-800-CW antibody [Sigma] in 25 mL blocking buffer) was added after a wash. Fluorescence signals were detected on a LI-COR Infrared Imaging system (Odyssey).
Spectroscopy.
CD spectra were obtained using a Jasco spectropolarimeter. NMR spectra (1H and 19F) were acquired using Bruker 700 MHz spectrometer equipped with 1H, 19F, 13C, and 15N quadruple resonance cryoprobe or Bruker 600 MHz spectrometer at pD 7.4 (direct meter reading) at 25 °C; effective pH favors a tetrahedral boronate conformation suitable for diol binding. NMR data were processed with Bruker Topspin 4.0.5 and analyzed with NMRFAM-Sparky software (77). 1H-13C HSQC spectra were acquired at natural abundance with pulse sequence optimized for direct J-coupling constants of 145 Hz as a compromise between aliphatic and aromatic resonances.
Supplementary Material
Acknowledgments
We thank M. Otten, G. Pennypacker T. Gray, and A. Hagarman (Thermalin Inc.) for assistance with preparation of biosynthetic insulin analogs; A. Rundell (Thermalin Inc.) for initial biological characterization of FRI analogs; J. Harwood (Director, Purdue NMR Facility) for assistance with preliminary 19F-NMR spectroscopy; D. G. Anderson (Massachusetts Institute of Technology [MIT]), S. Dutta (Juvenile Diabetes Research Foundation [JDRF]), M. F. Ismail-Beigi (Case Western Reserve University [CWRU]), R. Langer (MIT), B. Smith (LaTrobe University), M. Strano (MIT), S. Sullivan (Helmsley Charitable Trust), and J. Whittaker (CWRU) for helpful discussion. M.A.W. acknowledges adjunct/courtesy appointments in the Department of Chemistry at Indiana University (Bloomington) and the Weldon School of Biomedical Engineering at Purdue University and thanks respective colleagues for stimulating discussions and encouragement. Y.-S.C. was supported by a postdoctoral fellowship from the JDRF and a pilot grant from the Diabetes Research Connection. N.B.P. was supported in part by the American Diabetes Association (Grants 7-13-IN-31 and 1-08-RA-149). This work was supported in part by grants from the JDRF, the Leona M. and Harry B. Helmsley Charitable Trust, and the NIH (R01 DK040949 and R01 DK127761). Walter and Eliza Hall Institute of Medical Research (WEHI) received Victorian State Government Operational Infrastructure Support and funding from the Australian National Health and Medical Research Council (NHMRC) Independent Research Institutes Infrastructure Support Scheme.
Footnotes
Competing interest statement: M.A.W. has equity in Thermalin Inc. (Cleveland, OH) at which he serves as Chief Innovation Officer; he has also been a consultant to Merck Research Laboratories and DEKA Research and Development Corp. N.B.P. is a consultant to Thermalin Inc. M.C.L.'s laboratory has a Research Agreement with Eli Lilly and Company to conduct research not connected to this publication.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
*The sequence (Fig. 2) contains a m-f-PBA at the α-amino group of GlyA1; the halogen shifts the pKa of PBA (75).
†Positioning of the Lys at B28 rather than its native position (B29) was motivated by the clinical utility of insulin lispro as a rapid-acting insulin analog (78). The “Lys-Pro switch” (ProB28→Lys and LysB29→Pro) destabilizes the dimer interface in the zinc hexamer to promote rapid absorption (79).
‡Although at or near the cryo-EM–defined Site-2 interface, classical synthetic studies established that the N-terminal residues of the B chain contribute little to insulin’s activity in vitro or in vivo (80–83).
§Prior studies of HepG2 cells established that their long-term exposure to high concentrations of fructose in culture (>2 days) can enhance expression of genes directing lipid biosynthesis (84, 85). Because such changes could in principle confound the present qPCR-based assays of ChREBP and SREBP mRNAs to monitor fructose-dependent activation of FRI, our protocol employed only short-term exposure (3 to 4 h; Fig. 6A). Control studies demonstrated no confounding effects under these conditions (SI Appendix, Fig. S7).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103518118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.De Meyts P., Whittaker J., Structural biology of insulin and IGF1 receptors: Implications for drug design. Nat. Rev. Drug Discov. 1, 769–783 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Menting J. G., et al., How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y., et al., How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun. 9, 821–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith B. J., et al., Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc. Natl. Acad. Sci. U.S.A. 107, 6771–6776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weis F., et al., The signalling conformation of the insulin receptor ectodomain. Nat. Commun. 9, 4420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi C. M., Emanuelli B., Kahn C. R., Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 (2006). [DOI] [PubMed] [Google Scholar]
- 7.De Meyts P., The insulin receptor and its signal transduction network. Endotext [Internet] (2016).
- 8.Skyler J., Atlas of diabetes (Springer Science & Business Media; ) 2012). [Google Scholar]
- 9.White M. F., Insulin signaling in health and disease. Science 302, 1710–1711 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Melmed S., Medical progress: Acromegaly. N. Engl. J. Med. 355, 2558–2573 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Belfiore A., Frasca F., Pandini G., Sciacca L., Vigneri R., Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 30, 586–623 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Du Z., Lovly C. M., Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 17, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banting F. G., Best C. H., The internal secretion of the pancreas. J. Lab. Clin. Med. 7, 465–480 (1922). [Google Scholar]
- 14.Bliss M., The Discovery of Insulin: Twenty-fifth Anniversary Edition (University of Chicago Press, Chicago, 2007), pp. 310. [Google Scholar]
- 15.Zaykov A. N., Mayer J. P., DiMarchi R. D., Pursuit of a perfect insulin. Nat. Rev. Drug Discov. 15, 425–439 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Menting J. G., et al., Protective hinge in insulin opens to enable its receptor engagement. Proc. Natl. Acad. Sci. U.S.A. 111, E3395–E3404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker E. N., et al., The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319, 369–456 (1988). [DOI] [PubMed] [Google Scholar]
- 18.Scapin G., et al., Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss M. A., Lawrence M. C., A thing of beauty: Structure and function of insulin’s “aromatic triplet”. Diabetes Obes. Metab. 20 (suppl. 2), 51–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blundell T., Dodson G., Hodgkin D., Mercola D., Insulin: The structure in the crystal and its reflection in chemistry and biology. Adv. Protein Chem. 26, 279–402 (1972). [Google Scholar]
- 21.Dodson G., Steiner D., The role of assembly in insulin’s biosynthesis. Curr. Opin. Struct. Biol. 8, 189–194 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Rege N. K., et al., “Register-shift” insulin analogs uncover constraints of proteotoxicity in protein evolution. J. Biol. Chem. 295, 3080–3098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menting J. G., et al., Structural congruency of ligand binding to the insulin and insulin/type 1 insulin-like growth factor hybrid receptors. Structure 23, 1271–1282 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Liu M., et al., Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes Obes. Metab. 20 (suppl. 2), 28–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobson C. M., Protein misfolding, evolution and disease. Trends Biochem. Sci. 24, 329–332 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Ke P. C., et al., Half a century of amyloids: Past, present and future. Chem. Soc. Rev. 49, 5473–5509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittal S., Cai Y., Nalam M. N., Bolon D. N., Schiffer C. A., Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease. J. Am. Chem. Soc. 134, 4163–4168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godoy C. A., et al., Disulfide engineered lipase to enhance the catalytic activity: A structure-based approach on btl2. Int. J. Mol. Sci. 20, 5245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss M. A., Diabetes mellitus due to the toxic misfolding of proinsulin variants. FEBS Lett. 587, 1942–1950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J., et al., Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes. Mol. Aspects Med. 42, 105–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arunagiri A., et al., Proinsulin misfolding is an early event in the progression to type 2 diabetes. eLife 8, e44532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rege N. K., et al., Evolution of insulin at the edge of foldability and its medical implications. Proc. Natl. Acad. Sci. U.S.A. 117, 29618–29628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner D. F., Chan S. J., An overview of insulin evolution. Horm. Metab. Res. 20, 443–444 (1988). [DOI] [PubMed] [Google Scholar]
- 35.Chan S. J., Cao Q.-P., Steiner D. F., Evolution of the insulin superfamily: Cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc. Natl. Acad. Sci. U.S.A. 87, 9319–9323 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Chan S., Steiner D. F., Insulin through the ages: Phylogeny of a growth promoting and metabolic regulatory hormone. Am. Zool. 40, 213–222 (2000). [Google Scholar]
- 37.Cryer P. E., Davis S. N., Shamoon H., Hypoglycemia in diabetes. Diabetes Care 26, 1902–1912 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Ismail-Beigi F., et al., Individualizing glycemic targets in type 2 diabetes mellitus: Implications of recent clinical trials. Ann. Intern. Med. 154, 554–559 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Bakh N. A., et al., Glucose-responsive insulin by molecular and physical design. Nat. Chem. 9, 937–943 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Springsteen G., Wang B., A detailed examination of boronic acid–diol complexation. Tetrahedron 58, 5291–5300 (2002). [Google Scholar]
- 41.Yan J., Springsteen G., Deeter S., Wang B., The relationship among pKa, pH, and binding constants in the interactions between boronic acids and diols—It is not as simple as it appears. Tetrahedron 60, 11205–11209 (2004). [Google Scholar]
- 42.Knowles B. B., Howe C. C., Aden D. P., Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209, 497–499 (1980). [DOI] [PubMed] [Google Scholar]
- 43.Hopkins B. D., Goncalves M. D., Cantley L. C., Insulin-PI3K signalling: An evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 16, 276–283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilkis S. J., Granner D. K., Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu. Rev. Physiol. 54, 885–909 (1992). [DOI] [PubMed] [Google Scholar]
- 45.Puigserver P., et al., Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Viscarra J., Kim S. J., Sul H. S., Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 16, 678–689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Angyal S. J., The composition and conformation of sugars in solution. Angew. Chem. Int. Ed. Engl. 8, 157–166 (1969). [Google Scholar]
- 48.Angyal S. J., “The composition of reducing sugars in solution: Current aspects” in Advances in Carbohydrate Chemistry and Biochemistry, Horton D., Ed. (Elsevier, 1991), 49, pp. 19–35. [Google Scholar]
- 49.Zahn H., Brandenburg D., Gattner H.-G., Molecular basis of insulin action: Contributions of chemical modifications and synthetic approaches. Diabetes 21(2, suppl.)468–475 (1972). [DOI] [PubMed] [Google Scholar]
- 50.Freychet P., Brandenburg D., Wollmer A., Receptor-binding assay of chemically modified insulins. Comparison with in vitro and in vivo bioassays. Diabetologia 10, 1–5 (1974). [DOI] [PubMed] [Google Scholar]
- 51.Yang Y., et al., An Achilles’ heel in an amyloidogenic protein and its repair: Insulin fibrillation and therapeutic design. J. Biol. Chem. 285, 10806–10821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaykov A. N., Mayer J. P., Gelfanov V. M., DiMarchi R. D., Chemical synthesis of insulin analogs through a novel precursor. ACS Chem. Biol. 9, 683–691 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Derewenda U., et al., X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue. J. Mol. Biol. 220, 425–433 (1991). [DOI] [PubMed] [Google Scholar]
- 54.De Meyts P., “The insulin receptor and its signal transduction network” in Endotext, K. R. Feingold et al., Eds. (MDText.com, Inc., South Dartmouth, MA, 2000). https://www.endotext.org/?s=The+insulin+receptor+and+its+signal+transduction+network. Accessed 14 July 2021. [PubMed]
- 55.Hua Q.-X., Jia W., Weiss M. A., Conformational dynamics of insulin. Front. Endocrinol. (Lausanne) 2, 48 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bodenhausen G., Ruben D. J., Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 (1980). [Google Scholar]
- 57.Jacoby E., Hua Q. X., Stern A. S., Frank B. H., Weiss M. A., Structure and dynamics of a protein assembly, 1H-NMR studies of the 36 kDa R6 insulin hexamer. J. Mol. Biol. 258, 136–157 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Gutmann T., et al., Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J. Cell Biol. 219, e201907210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Meyts P., The structural basis of insulin and insulin-like growth factor-I receptor binding and negative co-operativity, and its relevance to mitogenic versus metabolic signalling. Diabetologia 37 (suppl. 2), S135–S148 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Schäffer L., A model for insulin binding to the insulin receptor. Eur. J. Biochem. 221, 1127–1132 (1994). [DOI] [PubMed] [Google Scholar]
- 61.de Meyts P., Roth J., Neville D. M. Jr., Gavin J. R. 3rd, Lesniak M. A., Insulin interactions with its receptors: Experimental evidence for negative cooperativity. Biochem. Biophys. Res. Commun. 55, 154–161 (1973). [DOI] [PubMed] [Google Scholar]
- 62.Omichinski J. G., Clore G. M., Appella E., Sakaguchi K., Gronenborn A. M., High-resolution three-dimensional structure of a single zinc finger from a human enhancer binding protein in solution. Biochemistry 29, 9324–9334 (1990). [DOI] [PubMed] [Google Scholar]
- 63.Serganov A., Nudler E., A decade of riboswitches. Cell 152, 17–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Breaker R. R., Riboswitches and translation control. Cold Spring Harb. Perspect. Biol. 10, a032797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Derewenda U., et al., Phenol stabilizes more helix in a new symmetrical zinc insulin hexamer. Nature 338, 594–596 (1989). [DOI] [PubMed] [Google Scholar]
- 66.Brange J., Langkjœr L., “Insulin structure and stability” in Stability and Characterization of Protein and Peptide Drugs (1993), pp. 315–350. [DOI] [PubMed] [Google Scholar]
- 67.Rahuel-Clermont S., French C. A., Kaarsholm N. C., Dunn M. F., Chou C. I., Mechanisms of stabilization of the insulin hexamer through allosteric ligand interactions. Biochemistry 36, 5837–5845 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Miyata T., Uragami T., Nakamae K., Biomolecule-sensitive hydrogels. Adv. Drug Deliv. Rev. 54, 79–98 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Li X., et al., pH-sensitive peptide hydrogel for glucose-responsive insulin delivery. Acta Biomater. 51, 294–303 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Kaarsholm N. C., et al., Engineering glucose responsiveness into insulin. Diabetes 67, 299–308 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Jarosinski M. A., Dhayalan B., Rege N., Chatterjee D., Weiss M. A., ‘Smart’ insulin-delivery technologies and intrinsic glucose-responsive insulin analogues. Diabetologia 64, 1016–1029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hua Q. X., et al., Mini-proinsulin and mini-IGF-I: Homologous protein sequences encoding non-homologous structures. J. Mol. Biol. 277, 103–118 (1998). [DOI] [PubMed] [Google Scholar]
- 73.Hua Q.-X., et al., Design of an active ultrastable single-chain insulin analog: Synthesis, structure, and therapeutic implications. J. Biol. Chem. 283, 14703–14716 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glidden M. D., et al., An ultra-stable single-chain insulin analog resists thermal inactivation and exhibits biological signaling duration equivalent to the native protein. J. Biol. Chem. 293, 47–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chou D. H.-C., et al., Glucose-responsive insulin activity by covalent modification with aliphatic phenylboronic acid conjugates. Proc. Natl. Acad. Sci. U.S.A. 112, 2401–2406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kjeldsen T., Pettersson A. F., Hach M., Secretory expression and characterization of insulin in Pichia pastoris. Biotechnol. Appl. Biochem. 29, 79–86 (1999). [PubMed] [Google Scholar]
- 77.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DiMarchi R. D., et al., “Synthesis of a fast-acting insulin based on structural homology with insulin-like growth factor I” in Peptides: Proceedings of the Twelfth American Peptide Symposium, Smith J. A., Rivier J. E., Eds. (ESCOM Science Publishers B. V., Leiden, The Netherlands, 1992), pp. 26–28. [Google Scholar]
- 79.Campbell R. K., Campbell L. K., White J. R., Insulin lispro: Its role in the treatment of diabetes mellitus. Ann. Pharmacother. 30, 1263–1271 (1996). [DOI] [PubMed] [Google Scholar]
- 80.Brandenburg D., Des-PheB1-Insulin, ein kristallines Analogon des Rinderinsulins. Hoppe Seylers Z. Physiol. Chem. 350, 741–750 (1969). [DOI] [PubMed] [Google Scholar]
- 81.Saunders D. J., Offord R., Semisynthetic analogues of insulin. The use of N-substituted derivatives of methionine as acid-stable protecting groups. Biochem. J. 165, 479–486 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwartz G., Katsoyannis P. G., Synthesis of des(tetrapeptide B(1-4)) and des(pentapeptide B(1-5)) human insulins. Two biologically active analogues. Biochemistry 17, 4550–4556 (1978). [DOI] [PubMed] [Google Scholar]
- 83.Liu M., et al., Deciphering the hidden informational content of protein sequences: Foldability of proinsulin hinges on a flexible arm that is dispensable in the mature hormone. J. Biol. Chem. 285, 30989–31001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirahatake K. M., Meissen J. K., Fiehn O., Adams S. H., Comparative effects of fructose and glucose on lipogenic gene expression and intermediary metabolism in HepG2 liver cells. PLoS One 6, e26583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meissen J. K., Hirahatake K. M., Adams S. H., Fiehn O., Temporal metabolomic responses of cultured HepG2 liver cells to high fructose and high glucose exposures. Metabolomics 11, 707–721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pandyarajan V., et al., Aromatic anchor at an invariant hormone-receptor interface: Function of insulin residue B24 with application to protein design. J. Biol. Chem. 289, 34709–34727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peters J. A., Interactions between boric acid derivatives and saccharides in aqueous media: Structures and stabilities of resulting esters. Coord. Chem. Rev. 268, 1–22 (2014). [Google Scholar]
- 88.Trausch J. J., et al., Structural basis for diversity in the SAM clan of riboswitches. Proc. Natl. Acad. Sci. U.S.A. 111, 6624–6629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dodson G. G., Dodson E. J., Turkenburg J. P., Bing X., Molecular recognition in insulin assembly. Biochem. Soc. Trans. 21, 609–614 (1993). [DOI] [PubMed] [Google Scholar]
- 90.Smith G. D., Pangborn W. A., Blessing R. H., The structure of T6 human insulin at 1.0 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 59, 474–482 (2003). [DOI] [PubMed] [Google Scholar]
- 91.Raza F., et al., A review on recent advances in stabilizing peptides/proteins upon fabrication in hydrogels from biodegradable polymers. Pharmaceutics 10, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.