Significance
Bacteria rely on RNA-binding and RNA-remodeling proteins to regulate gene expression posttranscriptionally. RNA-dependent nucleoside-triphosphatases of the DEAH/RHA family constitute important posttranscriptional gene regulatory proteins in bacteria, but their molecular mechanisms are presently poorly understood. Here, we show that the DEAH/RHA protein, HrpA, from Escherichia coli is an RNA helicase and that its helicase activity is required to modulate bacterial survival under diverse antibiotics treatments. HrpA crystal structures in different functional states, cross-linking/mass spectrometry, and structure-guided functional analyses indicate that alternative interdomain contacts facilitate large-scale domain movements that are required for RNA binding, translocation, and unwinding. Our findings portray HrpA as a molecular ratchet that translocates single-stranded RNA through a central orifice, thereby displacing a complementary strand.
Keywords: RNA-dependent NTPase/RNA helicase, co-/posttranscriptional gene regulation, antibiotics resistance, X-ray crystallography, structural biology
Abstract
Many bacteria harbor RNA-dependent nucleoside-triphosphatases of the DEAH/RHA family, whose molecular mechanisms and cellular functions are poorly understood. Here, we show that the Escherichia coli DEAH/RHA protein, HrpA, is an ATP-dependent 3 to 5′ RNA helicase and that the RNA helicase activity of HrpA influences bacterial survival under antibiotics treatment. Limited proteolysis, crystal structure analysis, and functional assays showed that HrpA contains an N-terminal DEAH/RHA helicase cassette preceded by a unique N-terminal domain and followed by a large C-terminal region that modulates the helicase activity. Structures of an expanded HrpA helicase cassette in the apo and RNA-bound states in combination with cross-linking/mass spectrometry revealed ratchet-like domain movements upon RNA engagement, much more pronounced than hitherto observed in related eukaryotic DEAH/RHA enzymes. Structure-based functional analyses delineated transient interdomain contact sites that support substrate loading and unwinding, suggesting that similar conformational changes support RNA translocation. Consistently, modeling studies showed that analogous dynamic intramolecular contacts are not possible in the related but helicase-inactive RNA-dependent nucleoside-triphosphatase, HrpB. Our results indicate that HrpA may be an interesting target to interfere with bacterial tolerance toward certain antibiotics and suggest possible interfering strategies.
To cope with fast-changing environmental conditions, bacteria rapidly adjust their cellular proteomes. As changes in protein levels often do not perfectly correlate with changes in messenger RNA (mRNA) levels, gene expression programs are not only regulated on the transcriptional but also on the posttranscriptional level (1). Noncoding RNAs and RNA-binding proteins (RBPs) are the main mediators of posttranscriptional gene regulation in bacteria (2). RNA-dependent nucleoside-triphosphatases (rdNTPases) are a major group of posttranscriptionally active RBPs that can impact, among others, RNA processing, RNA decay, ribosome biogenesis, and translation initiation by binding and remodeling RNAs and RNA–protein complexes (RNPs) (3, 4). Thereby, they can influence quorum sensing and biofilm formation (5), mediate adaptation to diverse stress conditions (6–10), modulate virulence and infectivity (11–13), alter susceptibility of bacteria to antimicrobials (14, 15), and buffer adverse effects of mutations (16).
In vitro, many rdNTPases can unwind RNA duplexes in an NTP-dependent (most often ATP-dependent) manner [i.e., act as RNA helicases (17, 18)]. However in vivo, some of these enzymes can also anneal RNAs (19, 20), stably clamp RNAs (21), initiate buildup of higher-order RNPs (22, 23), or displace RNA-bound proteins (24) or RNA-bound RNPs (25, 26) from RNAs. All of these activities depend on the presence of RecA-like NTP-binding/NTPase domains that form the rdNTPase cores. The RecA domains comprise parallel-stranded β-sheets surrounded by α-helices and harbor up to about a dozen conserved sequence motifs that are involved in NTP binding/hydrolysis, RNA/RNP binding/remodeling, and functionally connecting these activities (17, 18). Additional domains and regions can modulate the functions of the RecA cores in diverse ways (27).
Based on the presence and conservation of particular motifs, the presence of auxiliary domains and the biochemical activities, nucleic acid–dependent NTPases have been divided into six superfamilies (SFs) (17, 18). The vast majority of known rdNTPases belong to SF2. SF2 enzymes have been further subdivided into several families and subfamilies. rdNTPases that exhibit an Asp-Glu-Ala-Asp or Asp-Glu-variable-His sequence in their motifs belong to the DEAD-box and DEAH/RHA families of SF2, respectively (17, 18). The molecular mechanisms of DEAD-box proteins have been thoroughly studied in both prokaryotes and eukaryotes (3, 4, 17, 18, 28), whereas our present knowledge about DEAH/RHA protein mechanisms has been predominantly derived from studies of eukaryotic enzymes (29, 30).
Structural analyses of eukaryotic DEAH/RHA proteins, such as Chaetomium thermophilum Prp22 (31), Saccharomyces cerevisiae Prp43 (32–34), Drosophila melanogaster MLE (35), or Mus musculus Dhx37 (36) have revealed a core comprising two RecA-like domains with auxiliary winged helix (WH), helical bundle/ratchet (HB), and oligonucleotide/oligosaccharide-binding (OB) domains forming a ring-like arrangement with the core domains. In DEAH/RHA proteins that act as RNA helicases, this architecture is required for processive RNA unwinding (37, 38). RNA unwinding involves translocation of a loading strand of an RNA duplex across the RecA domains, with the helicase moving in 3′ to 5′ direction on the loading strand and with concomitant exclusion of the displaced strand (37, 38). In the process, the RecA-like domains predominantly contact the sugar-phosphate backbone of the loading strand and thus do not generally provide substrate specificity (18). Details of the translocation and unwinding mechanisms can differ in different DEAH/RHA enzymes (30) due to differences in particular motifs or due to specific insertions or terminal extensions in some of these proteins (33, 35, 39–41).
In bacteria, DEAH/RHA NTPases have been divided into three subfamilies: HrpA-like, HrpB-like, and Z5898-like enzymes (3, 4), referring to the corresponding Escherichia coli factors. The cellular functions and RNA-related activities of these proteins are still poorly understood. HrpA was shown to strongly influence the fimbrial attachment system in E. coli by regulating mRNA processing of its subunits (42). The hrpA deletion mutants in E. coli (14, 15), Burkholderia multivorans (43), and Pseudomonas aeruginosa (44) showed an enhanced sensitivity toward various antibiotics. Deletion of hrpA in the Lyme disease—causing pathogen, Borrelia burgdorferi, led to altered mRNA and protein levels and abrogated tick-mediated infectivity of mice (12, 13). In the citrus plant pathogen Xanthomonas citri, the hrpB deletion mutant showed reduced adhesion and biofilm formation on surfaces, increased cell dispersion on solid medium, and delayed development of disease symptoms (45). Recent crystal structures and structure-based functional analyses from our group (46) and others (47) have revealed that HrpB proteins are RNA-dependent NTPases but that they lack RNA helicase activity in isolation. The NTPase activity of HrpB depends on the length and structure of the bound RNA (48). Deletion of the z5898 gene in the pathogenicity island of the enterohemorrhagic E. coli O157:H7 resulted in reduced levels of flagellin and impaired bacterial motility (49). These findings suggest important roles for bacterial DEAH/RHA enzymes in various stress responses, environmental adaptation, surface interaction, and virulence.
Here, we characterized E. coli (ec) HrpA as a 3 to 5′ RNA helicase. An E. coli hrpA mutant was impaired in survival compared to a wild-type (WT) strain after exposure to certain antibiotics. Near-WT levels of survival could be restored in the hrpA mutant only by complementation with helicase-active ecHrpA variants, suggesting a key role of ecHrpA in environmental adaptation mechanisms based on its helicase activity. We show that HrpA helicase activity is embodied in an expanded N-terminal DEAH/RHA-like helicase cassette and further modulated by a large, HrpA-specific C-terminal region (CTR). We present crystal structures of the expanded ecHrpA helicase cassette in the apo and RNA-bound states, revealing large-scale domain movements during substrate binding. In line with targeted functional analyses, the structures suggest how HrpA might translocate RNA to achieve duplex unwinding via ratchet-like domain rearrangements.
Results
ecHrpA Modulates Antibiotics Tolerance of E. coli.
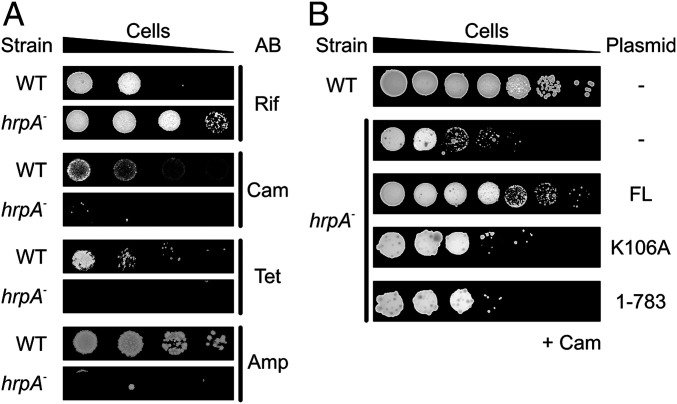
A previous high-throughput screen for mutant phenotypes associated with single-gene deletions revealed altered susceptibility of an hrpA-deficient E. coli strain (BW25113) to sublethal doses of certain antibiotics (50). The molecular mechanisms underlying this effect are presently unknown. To further characterize hrpA-dependent phenotypes under antibiotics exposure, we generated an E. coli K12 strain with a deletion of the chromosomal hrpA gene (MC4100 hrpA::kan). The survival of the hrpA mutant was analyzed in comparison to the isogenic MC4100 parent strain in the presence of selected antibiotics that interfere with transcription (rifampicin), protein synthesis (chloramphenicol and tetracyclin), or cell wall synthesis (ampicillin). In the absence of antibiotics, the hrpA mutant showed a similar survival as the WT strain, suggesting that ecHrpA is not essential for viability or that it is redundant. In the presence of rifampicin, hrpA mutant cells showed an enhanced survival compared to WT cells (Fig. 1A). However, the hrpA mutant was strongly impaired in survival in the presence of chloramphenicol, tetracyclin, or ampicillin (Fig. 1A). These results show that the hrpA gene bestows on E. coli increased tolerance toward certain antibiotics that target the translation machinery or cell wall synthesis but can also cause higher sensitivity toward other antibiotics, such as the RNA polymerase inhibitor rifampicin.
Fig. 1.
Survival assays. (A) Survival of the indicated E. coli strains in the presence of the indicated antibiotics (AB). WT, MC4100 strain; hrpA−, MC4100 hrpA::kan strain; Rif, rifampicin; Cam, chloramphenicol; Tet, tetracycline; and Amp, ampicillin. (B) Survival assays monitoring restoration of chloramphenicol tolerance upon complementation of the E. coli hrpA mutant with the indicated ecHrpA variants. FL, full-length ecHrpA; K106A, full-length ecHrpA bearing a K106A exchange; 1-783, ecHrpA1-783. Survival assays were performed by spotting serial dilutions of the bacterial overnight cultures on LB agar plates in the absence or presence of the antibiotics to observe colonies.
ecHrpA Is an RNA Helicase with 3 to 5′ Directionality.
ecHrpA is a putative RNA helicase (42). However, we and others have recently investigated the related ecHrpB protein, whose orthologs had also been tentatively annotated as RNA helicases (45); while ecHrpB exhibited RNA-stimulated NTPase activity, it failed to unwind diverse model RNAs in vitro (46, 47). Moreover, some eukaryotic DEAH/RHA helicases require G-patch proteins for helicase activity (51). We therefore systematically tested nucleic acid binding, NTPase, and unwinding activities of isolated ecHrpA variants.
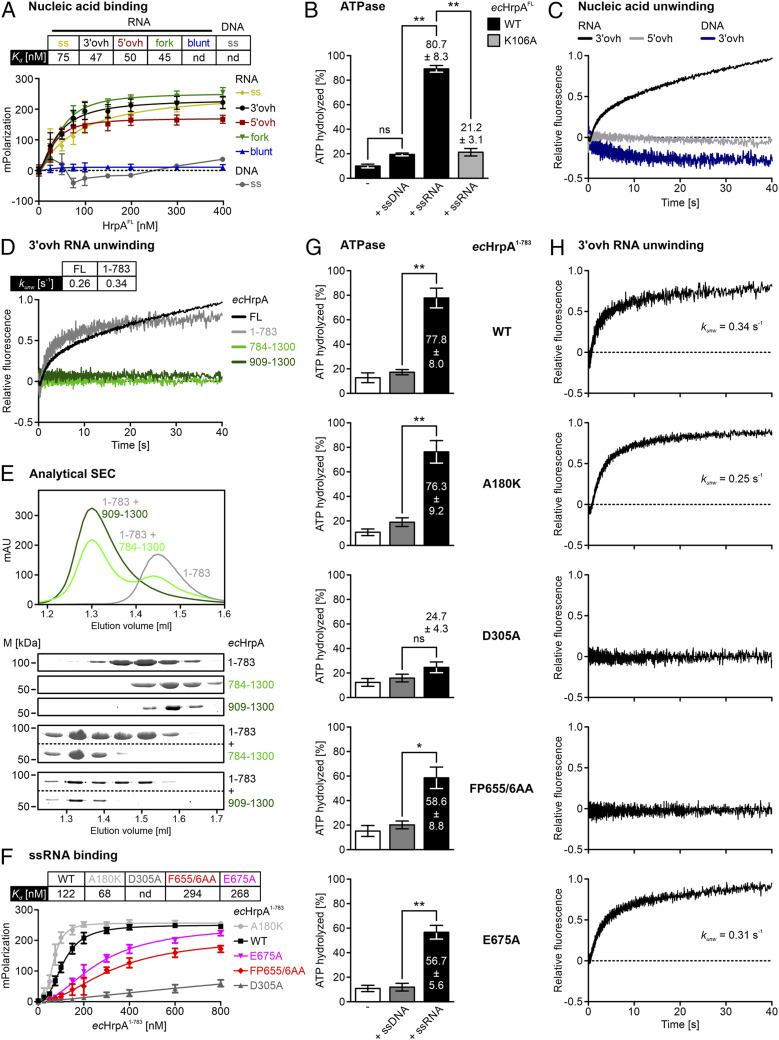
Fluorescence anisotropy measurements revealed that full-length (FL) ecHrpA bound a 15-nucleotide (nt) single-stranded (ss) RNA, double-stranded (ds) RNAs that contained 15-nt ss 3 or 5′ overhangs, as well as a forked RNA with neighboring 15-nt 3 and 5′ overhangs with high affinities (Fig. 2A and reference SI Appendix, Table S1 for oligonucleotides employed). In contrast, the protein did not associate with blunt-ended dsRNA or ssDNA (Fig. 2A). ecHrpAFL showed weak intrinsic ATPase activity that was stimulated approximately eightfold in the presence of ssRNA, while ssDNA had no effect (Fig. 2B). Conserved sequence motif I in the RecA1-like domains of DEAH/RHA proteins is essential for the NTPase activity, with an invariant lysine residue in motif I (K106 in ecHrpA) required for NTP binding (52); as expected, ecHrpAFL,K106A showed markedly reduced stimulated ATPase activity compared to WT ecHrpAFL (Fig. 2B). Using a stopped-flow/fluorescence–based assay (46, 53), we next monitored RNA unwinding by ecHrpAFL. The protein efficiently unwound a 12–base pair (bp) dsRNA bearing a 31-nt 3′ overhang, but we did not observe any strand separation when using 5′-overhang RNA or 3′-overhang DNA as substrates (Fig. 2C). These analyses show that ecHrpAFL is a 3 to 5′ directional RNA helicase in vitro.
Fig. 2.
Biochemical characterization of ecHrpA variants. (A) Fluorescence anisotropy measurements monitoring binding of ecHrpAFL to the indicated nucleic acids. ssRNA, yellow; 3′-overhang (3′ovh) RNA, black; 5′-overhang (5′ovh) RNA, red; forked RNA, green; blunt-ended dsRNA, blue; 3′ovh DNA, gray. Data points represent means ± SEM of three independent experiments using the same biochemical samples. Kd values listed are derived from curve fits; nd, not determined. (B) Quantification of thin-layer chromatography analyses monitoring intrinsic and nucleic acid–stimulated ATPase activities of ecHrpAFL or ecHrpAFL,K106A. Nucleic acids added are indicated below the bars. Quantified ATPase activities (% hydrolyzed) are indicated Above the boxes. Values represent means ± SEM of three independent experiments using the same biochemical samples. Significance indicators (unpaired Student’s t test): **P ≤ 0.01; ns, not significant. (C) Results from stopped-flow/fluorescence analyses monitoring ecHrpAFL-mediated unwinding of 3′ovh RNA (black), 5′ovh RNA (gray), or 3′ovh DNA (blue). (D) Unwinding of 3′ovh RNA by ecHrpAFL (black), ecHrpA1-783 (gray), ecHrpA784-1300 (dark green), or ecHrpA909-1300 (light green) monitored by stopped-flow/fluorescence assays. kunw, amplitude-weighted unwinding rate constants. (E) Absorbance traces (Top) and sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis (Bottom) of analytical SEC runs of ecHrpA1-783, ecHrpA784-1300, and ecHrpA909-1300, as well as of ecHrpA1-783/ecHrpA784-1300 and ecHrpA1-783/ecHrpA784-1300 mixtures. M, molecular mass markers [kDa]. (F) Fluorescence anisotropy measurements monitoring binding of ssRNA by the indicated ecHrpA1-783 variants (WT, D305A). Data points represent means ± SEM of three independent experiments using the same biochemical samples. Kd values listed are derived from curve fits; nd, not determined. (G) Quantified thin-layer chromatography analyses monitoring intrinsic and nucleic acid–stimulated ATPase activities of the ecHrpA1-783 variants indicated on the Right (WT, A180K, D305A, FP655/6AA, and E675A). Nucleic acids added are indicated Below the bars. Quantified ATPase activities (% hydrolyzed) are indicated in or Above the boxes. Values represent means ± SEM of three independent experiments using the same biochemical samples. Significance indicators (unpaired Student’s t test): *P ≤ 0.05; **P ≤ 0.01; ns, not significant. (H) Unwinding of 3′ovh RNA by the ecHrpA1-783 variants indicated on the Left (WT, A180K, D305A, FP655/6AA, and E675A) monitored by stopped-flow/fluorescence assays. kunw, amplitude-weighted unwinding rate constants.
The N-terminal ∼700 residues of ecHrpA exhibit homology to the helicase cassettes of eukaryotic DEAH/RHA proteins, suggesting that the N-terminal portion of ecHrpA embodies the helicase activity of the enzyme. Consistent with a DEAH/RHA-like helicase cassette in the N-terminal half of ecHrpAFL, digestion of the protein with trypsin gave rise to a stable fragment encompassing residues 1 to 783 (ecHrpA1-783), as revealed by mass spectrometric fingerprinting. Recombinant ecHrpA1-783 unwound 3′-overhang RNA, while recombinant C-terminal fragments, ecHrpA784-1300 and ecHrpA909-1300, did not exhibit any RNA unwinding activity (Fig. 2D).
The amplitude-weighted unwinding rate constant, kunw, of ecHrpA1-783, extracted from the biphasic unwinding trajectory (54), was increased by about 30% compared to ecHrpAFL (Fig. 2D and SI Appendix, Fig. S1 and Table S2), suggesting that the ecHrpA-specific CTR modulates the helicase activity of the N-terminal DEAH/RHA cassette. We therefore tested whether these two parts of the protein directly contact each other. Indeed, separately produced ecHrpA1-783 and ecHrpA784-1300 or ecHrpA909-1300 comigrated in analytical size-exclusion chromatography (SEC), and the combined fragments eluted earlier than either one alone (Fig. 2E), suggesting formation of stable complexes. Together, the above findings suggest that the first 783 residues of ecHrpA represent a stably folded unit that encompasses a DEAH/RHA protein–like helicase cassette, whose helicase activity is modulated by the large C-terminal extension due to direct, intramolecular interactions of the N- and C-terminal parts of the protein.
ecHrpA NTPase/Helicase Activity Is Required for Chloramphenicol Tolerance.
To test whether NTPase/helicase activities of ecHrpA play a role in mediating antibiotics tolerance, we introduced plasmids encoding ecHrpA variants into MC4100 hrpA::kan cells. The survival defect of the hrpA mutant in the presence of chloramphenicol was alleviated to a large extent when WT hrpA was expressed from a plasmid (Fig. 1B). In contrast, the chloramphenicol sensitivity of the hrpA mutant was mitigated to only a small extent by a plasmid-borne hrpAK106A allele that encoded an NTPase-deficient ecHrpAK106A variant (Fig. 1B). Likewise, ectopically produced ecHrpA1-783 did not restore chloramphenicol tolerance of the hrpA mutant to a comparable level as the ecHrpAWT (Fig. 1B), despite its higher amplitude-weighted unwinding rate constant compared to ecHrpAFL. Together, the above results suggest that the ecHrpA NTPase/helicase activity and the ecHrpA-specific CTR are required to enhance tolerance toward certain antibiotics.
ecHrpA1-783 Constitutes Conserved and Unique Domains.
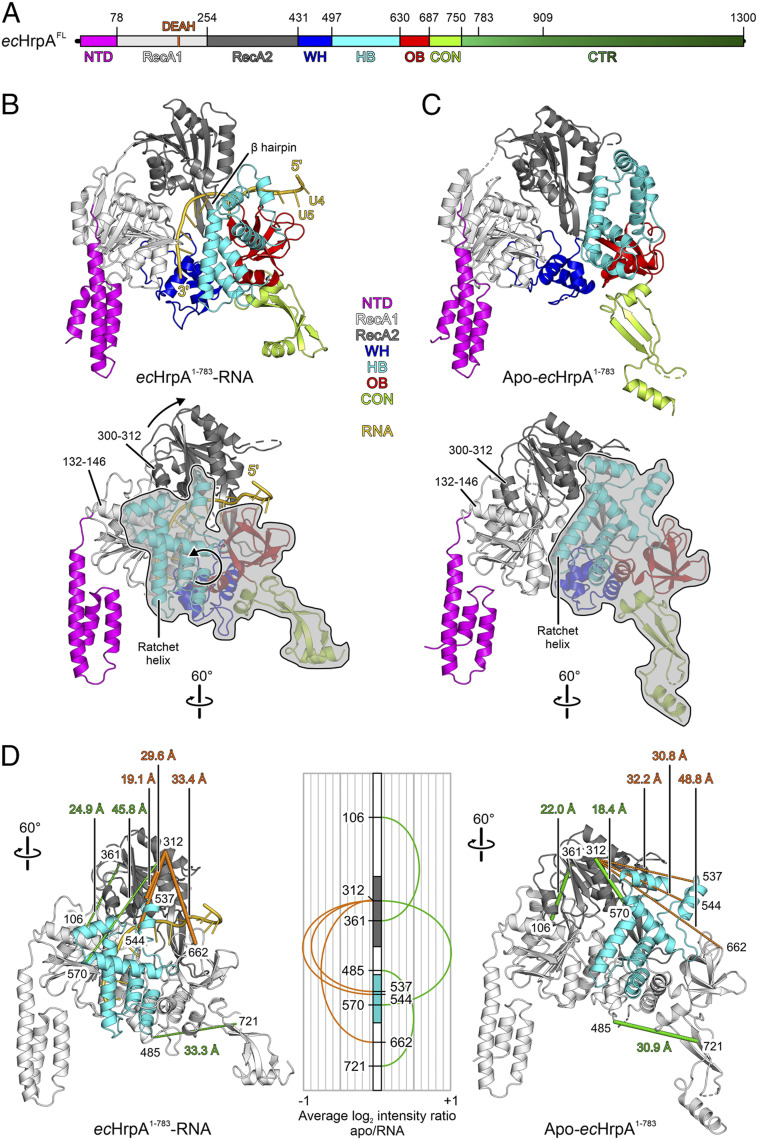
To compare the structure and molecular mechanisms of a bacterial DEAH/RHA RNA helicase with its better-investigated eukaryotic counterparts, we determined a crystal structure of ecHrpA1-783 in complex with a U15 ssRNA at 3.2-Å resolution (SI Appendix, Table S3) (55). ecHrpA1-783-U15 crystals contained four ecHrpA1-783-U15 complexes in an asymmetric unit, whose structures were virtually identical (pairwise rmsd of 0.45 to 0.57 Å for 699 to 722 pairs of common C-α atoms).
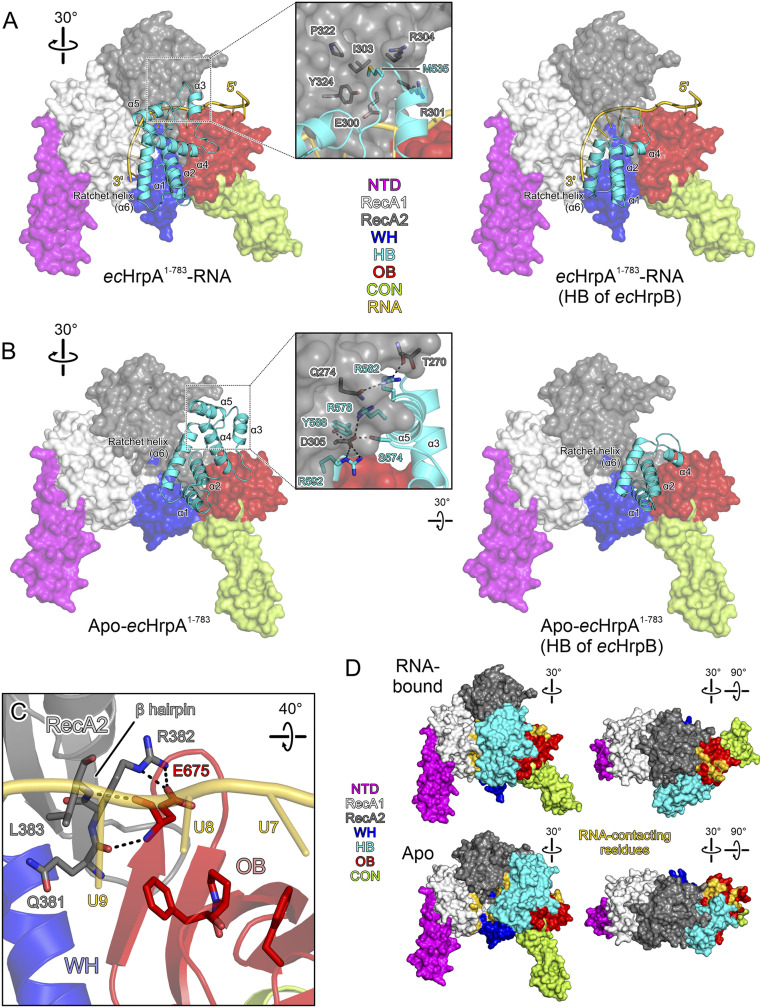
The most-complete model of RNA-bound ecHrpA1-783 comprises residues 2 to 755 with residues 263 to 272 lacking well-defined electron density. The structure of ecHrpA1-783 can be divided into seven domains (Fig. 3 A and B) [i.e., an N-terminal domain (NTD; residues 2 to 78), two RecA-like domains (RecA1, residues 79 5 to 254; RecA2, residues 255 to 431), a WH domain (residues 432 to 497), an HB domain (residues 498 to 630), an OB domain (residues 631 to 687), and a connector domain (CON, residues 688 to 755) that links up to the CTR not contained in the structure].
Fig. 3.
ecHrpAFL domain architecture and ecHrpA1-783 structures. (A) Domain architecture of ecHrpAFL. Numbers above the scheme represent domain borders or borders of recombinantly produced ecHrpA fragments. Color coding of domains/regions is maintained in the following figures. N-terminal domain (NTD), magenta; RecA1 domain, light gray; RecA2 domain, gray; helical bundle (HB) domain, cyan; winged-helix (WH) domain, blue; oligonucleotide/oligosaccharide-binding (OB) domain, red; connector domain (CON), lime green; and CTR, darker shades of green. (B) Cartoon plot of RNA-bound ecHrpA1-783 in two orientations. RNA, gold. Selected structural elements described in the text and the RNA directionality are pointed out. Arrows in the Lower panel represent movements of RecA2 and WH-HB-OB-CON modules compared to the apo-state conformation shown in C. Rotation symbols in this and all following figures represent views relative to the Top. (C) Cartoon plot of apo-ecHrpA1-783 in two orientations identical to (B) after superpositioning of the RecA1 domains. The outlines around the WH-HB-OB-CON modules in the Lower panels of (B) and (C) emphasize the large-scale rearrangement of this part of the protein relative to the NTD-RecA1 and RecA2 modules. (D) Mapping of cross-linked residues on domains that undergo conformational rearrangements on the structures of RNA-bound ecHrpA1-783 (Left) and apo-ecHrpA1-783 (Right). RecA2 domain, dark gray; HB domain, cyan; remainder of the protein, light gray. Orientations identical to (B, C), Bottom. Cross-links more abundant in RNA-bound ecHrpA1-783 are shown by thick orange dashed lines, and cross-links more abundant in apo-ecHrpA1-783 are shown by thick green dashed lines. Cross-links less abundant in RNA-bound ecHrpA1-783 are shown by thin green dashed lines, and cross-links less abundant in apo-ecHrpA1-783 are shown by thin orange dashed lines. Cross-linked residues are identified by numbers (all lysines), and Cα-Cα distances of cross-linked residues are indicated (as determined based on the crystal structures). Central panel, mapping of observed cross-links between residues on domains that undergo conformational rearrangements on a linear representation of ecHrpA1-783. Numbers, cross-linked residues. Cross-links more abundant in RNA-bound ecHrpA1-783 are represented by orange arches extending to the Left, cross-links more abundant in apo-ecHrpA1-783 are represented by green arches extending to the Right. The heights of the arches represent the average log2 intensity ratios of cross-links in apo-ecHrpA1-783 relative to RNA-bound ecHrpA1-783, as deduced from the analysis of mixed isotope-labeled/unlabeled samples.
The RecA1/2, HB, WH, and OB domains are arranged in a circle, with the WH and OB domains sandwiching a prominent β-hairpin (residues 368 to 387) that projects from the RecA2 domain; NTD and CON protrude as two “legs” on opposite sides of the central cassette (Fig. 3B). The NTD consists of a three-helix bundle that is appended to the N-terminal edge of the central RecA1 β-sheet (Fig. 3B), forming a distinctive feature among DEAH/RHA proteins with known structures. The folds of the RecA1/2, HB, WH, and OB domains resemble those of analogous domains in the NTPase/helicase cassettes of ecHrpB (46, 47) and of eukaryotic DEAH/RHA helicases (29). The ecHrpA1-783 CON comprises a three-stranded, antiparallel β-sheet capped by α-helices, resembling a similar element of ecHrpB (46, 47) and eukaryotic Dhx37 (36).
ecHrpA1-783 Fully Encircles an RNA Loading Strand.
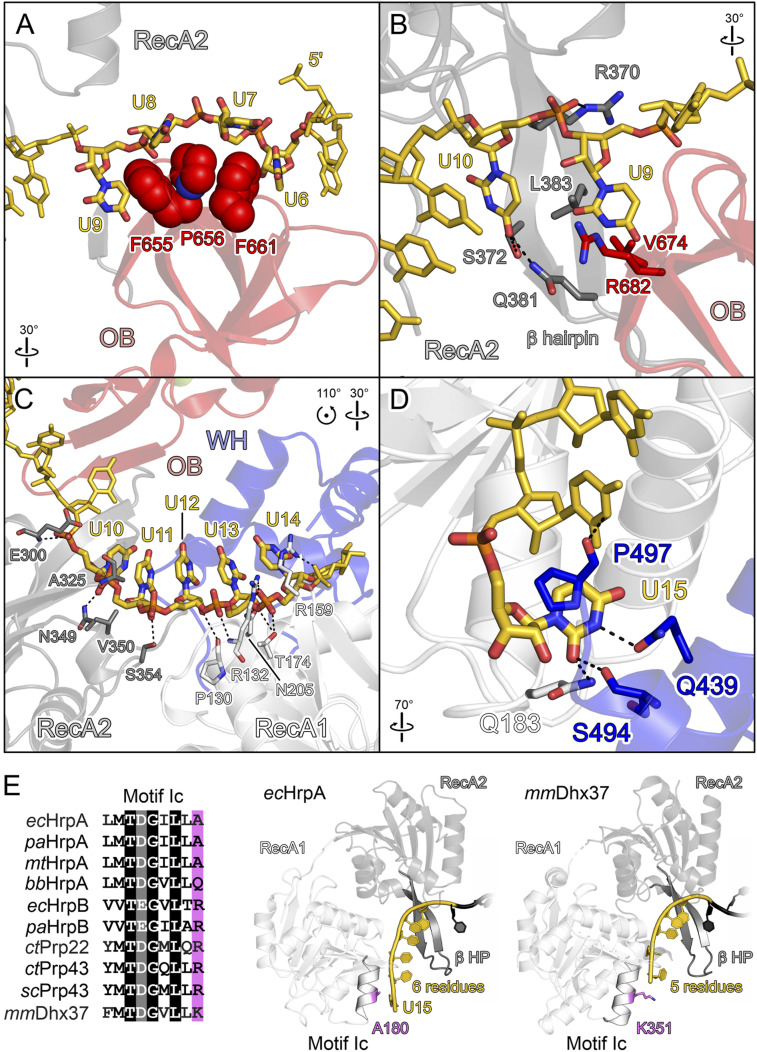
Of the 15 nucleotides of U15 RNA, 12 are well defined in the electron density; we arbitrarily assigned the most 3′ terminal residue defined in the density as nucleotide 15. The RNA is embedded in a central tunnel of the helicase cassette, running across the OB domain, between the RecA-like domains and the HB domain, and across the WH domain (Fig. 3B). Residues U4 and U5 at the 5′ end of the visible RNA region do not exhibit any direct contacts to the protein (Fig. 3B). Residues U6 to U9 lie along one flank of the OB domain at the entrance to the RNA-binding tunnel (Fig. 4A). The nucleobases of U6 to U9 wrap around F655, P656, and F661 (the “stacking triad”), with F661 and F655 redirecting the RNA by intervening between nucleotides U6/U7 and U8/U9, respectively (Fig. 4A). A similar stacking triad is also found in Prp22 (31) and Dhx37 (36) but is lacking [e.g., in Prp43 (33)] and thus does not represent a universally conserved feature of DEAH/RHA helicases. To test the importance of the OB stacking triad in ecHrpA, we produced an ecHrpA1-783 variant, in which both F655 and P656 were exchanged for alanines. The ssRNA affinity of ecHrpA1-783,FP655/6AA was reduced about 2.4-fold (Fig. 2F) and its RNA-dependent ATPase activity was diminished by about 25% (Fig. 2G), while RNA unwinding was completely abrogated (Fig. 2H). These observations indicate an important role of the OB stacking triad for helicase activity that does not merely root in reduced RNA affinity or RNA-stimulated ATPase activity.
Fig. 4.
RNA binding by ecHrpA1-783. (A–D) Close-up views of ecHrpA1-783 residues that contact RNA from the 5′ to the 3′ end. Protein regions are shown as semitransparent cartoons. Interacting protein and RNA residues are highlighted as spheres or sticks and color-coded by atom type. In this and the following figures: carbon, as the respective protein/RNA domain/region; nitrogen, blue; oxygen, red; phosphorus, orange. Dashed black lines, hydrogen bonds or salt bridges. (A) RNA residues U6 to U9 wrapping around the OB stacking triad (F655, P656, and F661). (B) Nucleotides U9 and U10 binding across the continuous β-sheet formed between the prominent RecA2 β-hairpin and the OB domain. (C) RNA residues U10 to U14 binding across the RecA1/2 RNA binding surfaces. (D) Nucleotide U15 bound at an ecHrpA-specific pocket formed from residues of the RecA1 and WH domains. (E) Multiple sequence alignment (Left) and RNA binding (Right) to motif Ic in bacterial and eukaryotic DEAH/RHA proteins; ec, E. coli; pa, P. aeruginosa; mt, Mycobacterium tuberculosis; bb, B. burgdorferi; ct, C. thermophilum; sc, S. cerevisiae; mm, M. musculus. In contrast to other DEAH/RHA proteins, the small alanine at the C terminus of motic Ic in the majority of HrpA orthologs allows binding of an additional RNA nucleobase (U15). Divergent C-terminal motif Ic residues, violet.
U9 is positioned between residues V674 and R682 of the OB domain, and U9 and U10 are splayed out across a continuous β-sheet formed by the OB domain and the prominent RecA2 β-hairpin (Fig. 4B). Base-stacked residues U10 to U14 interact with residues of the RecA1 and RecA2 domains, predominately via the RNA backbone (Fig. 4C). RecA2 residues N349 (motif V), V350 (motif V), and E300 (motif IV) interact with U10, while A325 (motif IVa-HL) contacts U11. U12 is bound to RecA2 residue S354 (motif V) and RecA1 residue P130 (motif Ia). U13 is bound to RecA1 via interactions with T174 (motif Ic), N205, and R132 (motif Ia). R159 (motif Ib-HT) and P497 of the WH domain contact the backbone and nucleobase of U14. Except for P497 and N205, these residues are part of the conserved helicase core motifs involved in RNA binding (SI Appendix, Fig. S2).
U15 shows a number of interactions to four residues from the RecA1 (Q183) and WH (Q439, S494, and P497) domains (Fig. 4D). These residues do not belong to any of the canonical helicase motifs and are not highly conserved among DEAH/RHA proteins. In the RNA complex structures of Prp22 (31) and Dhx37 (36), there is no RNA residue that adopts a U15-equivalent binding pose, suggesting that U15-binding elements represent a unique feature of ecHrpA. Sequence and structural comparisons showed that a small alanine at the C terminus of motif Ic of ecHrpA1-783 (A180) leaves enough space for the U15 base to approach RecA1 and WH domain residues, while Prp22 and Dhx37 exhibit arginine or lysine residues at the equivalent position (Fig. 4E). When we exchanged A180 in ecHrpA1-783 for a Dhx37-like lysine, the RNA affinity increased about 1.8-fold (Fig. 2F), the RNA-dependent ATPase activity was largely unaffected (Fig. 2G), and the amplitude-weighted unwinding rate constant was moderately reduced to the level of ecHrpAFL (Fig. 2H). These data suggest that a Dhx37-like lysine at position 180 may foster additional RNA backbone contacts, and that the alternative, less-stable accommodation of the U15 base in the case of ecHrpA1-783 weakly supports RNA duplex unwinding.
ecHrpA1-783 Undergoes Large Conformational Changes upon RNA Binding.
To visualize possible conformational changes in ecHrpA1-783 upon RNA binding, we determined a crystal structure of apo-ecHrpA1-783 at 2.7-Å resolution (SI Appendix, Table S3) (56). Apo-ecHrpA1-783 crystals contained one ecHrpA1-783 molecule in an asymmetric unit. The final apo-ecHrpA1-783 model contained residues 5 to 756 with gaps between residues 251 and 254, 264 and 270, 475 and 483, and 711 and 715, where the electron density was poorly defined.
The spatial domain arrangements differ drastically in RNA-bound and apo-ecHrpA1-783 (Fig. 3 B and C and Movie S1). Section-wise superpositioning revealed that these rearrangements involve relative movements of three segments—NTD/RecA1, RecA2, and WH/HB/OB/CON. NTD/RecA1, RecA2, and WH/HB/OB/CON modules remain largely unaltered (rmsds of 1.03, 1.36, and 1.14 Å for 236, 158, and 296 pairs of common C-α atoms, respectively), showing that they rearrange as approximate rigid bodies.
Upon RNA binding, RecA2 rotates away from RecA1 about a pivot point located in the RecA1/2 linker (G251); one edge of RecA2, made up by α-helix300-312, which abuts α-helix132-146 of RecA1 in the apo-state, is displaced from RecA1 by about 12 Å (Fig. 3 B and C, Lower). Thereby, the RecA1/RecA2 RNA–binding surface is splayed out and prepared to accommodate six nucleotides of an extended RNA strand (Fig. 4 C and D). Concomitantly, the WH/HB/OB/CON module rotates about a pivot point located at the beginning of the WH domain (L433) with the HB domain moving toward RecA1; in the process, the N-terminal end of a long HB α-helix (residues 588 to 607; the so-called “ratchet helix”) and one tip of the HB domain (α-3) are repositioned by ∼25 and ∼28 Å, respectively (Fig. 5 A and B).
Fig. 5.
Transient intramolecular contacts stabilize alternative conformations. (A and B, Left) Surface views of ecHrpA1-783 in the RNA-bound (A) and apo (B) states with the HB domain shown as a cartoon, illustrating large-scale domain movements. (Insets) Alternative interactions networks between the RecA2 and HB domains, stabilizing the RNA-bound (A) or apo (B) state. Interacting residues are shown as sticks and colored by atom type; sulfur, yellow. Dashed black lines, hydrogen bonds or salt bridges. (Right) Models in which the HB domain of ecHrpB (PDB ID: 6EUD) was transplanted onto ecHrpA1-783 in the RNA-bound (A) or apo (B) state, showing that the truncated ecHrpB HB domain cannot mediate ecHrpA-equivalent interactions with RecA2 in either conformation. Helices in the ecHrpB HB domain are labeled in analogy to the helicases in the ecHrpA1-783 HB domain. (C) Interaction network around E675 connecting the OB-domain β-sheet with the prominent β-hairpin of the RecA2 domain in the RNA-bound state of ecHrpA1-783. Interacting residues are shown as sticks and colored by atom type. Dashed black lines, hydrogen bonds or salt bridges. (D) Accessibility of RNA-binding residues (gold) in the RNA-bound (Top) and apo (Bottom) states of ecHrpA1-783.
Alternative Interdomain Contacts Facilitate the Conformational Changes.
In apo-ecHrpA1-783, an electrostatic interaction network ensues between RecA2 residues (T270, Q274, and D305) and the HB α-5 (S574, R578, and R582) and ratchet (Y588 and R592) helices, which is disrupted in the RNA-bound state; instead, in the RNA-bound state, M535 of the HB α-2 to α-3 loop is positioned in a pocket on RecA2 formed by residues E300, R301, I303, R304, P322, and Y324 (Fig. 5 A and B, Insets). Thus, HB residues differentially contact the RecA2 domain in the apo and RNA-bound states, leading to a pronounced ratcheting movement of the WH/HB/OB/CON module relative to the RecA1/2 core (Movie S1).
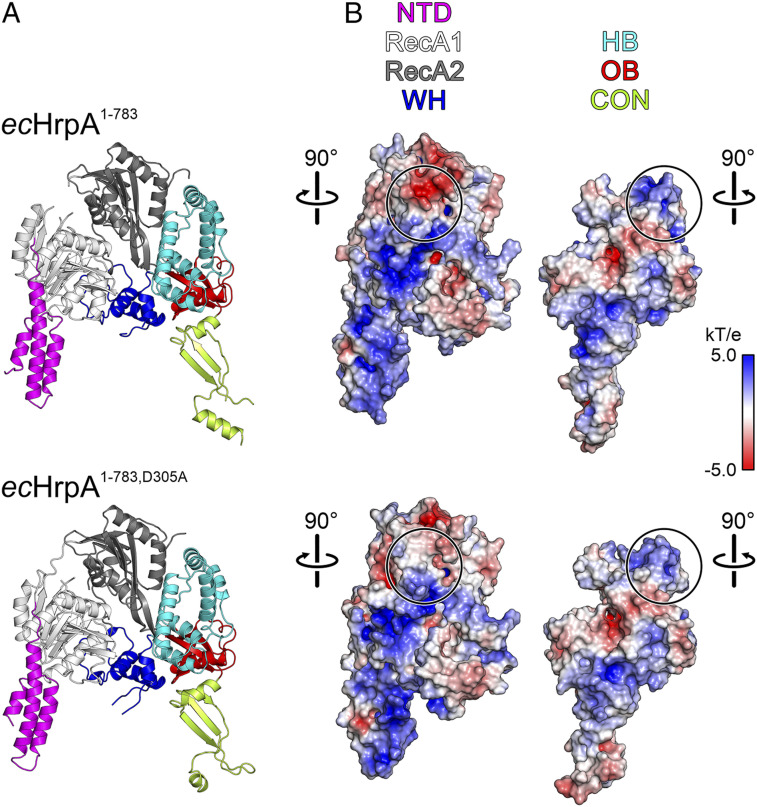
To test if the RecA2–HB interaction network observed in the apo state is important for ecHrpA activities, we introduced a D305A exchange, which is expected to disrupt this interaction network (Fig. 5B, Inset). Strikingly, RNA binding, RNA-stimulated ATPase, and helicase activities were completely abrogated in ecHrpA1-783,D305A (Fig. 2 F–H). In a crystal structure that we determined at 2.6-Å resolution (SI Appendix, Table S3) (57), apo-ecHrpA1-783,D305A adopted the same overall conformation as WT apo-ecHrpA1-783 (Fig. 6A) However, apart from disrupting a local RecA2–HB interaction network, the D305A exchange significantly reduces the negative electrostatic potential on the RecA2 surface that faces a positively charged surface on the HB domain in WT apo-ecHrpA1-783 (Fig. 6B).
Fig. 6.
(A) Cartoon representation of ecHrpA1-783 (Top) and ecHrpA1-783,D305A (Bottom), showing similar overall structures. (B) Book view on the electrostatic potentials covering the intramolecular contact surfaces between the NTD-RecA1/2-WH (Left) and HB-OB-CON (Right) portions of ecHrpA1-783 (Top) and ecHrpA1-783,D305A (Bottom). Black circles on the NTD-RecA1/2-WH parts mark the surface areas around D305 (Top) or A305 (Bottom), highlighting loss of local negative surface potential upon the D305A residue exchange. Black circles on the HB-OB-CON part mark surfaces interacting intramolecularly with the D305/A305 regions.
Repositioning of the WH/HB/OB/CON module upon RNA binding places the OB domain next to the RecA2 domain, allowing it to form a continuous β-sheet with the prominent RecA2 β-hairpin. The OB E675 residue now contacts Q381, L383 (via its backbone), and R382 (via its side chain) of RecA2 (Fig. 5C). The repositioned OB domain provides an extension to the RecA1/RecA2 RNA–binding surface so that its stacking triad is appropriately positioned to engage RNA near the 5′ end (Fig. 5C). When we introduced an E675A exchange in ecHrpA1-783, predicted to weaken RecA2–OB contacts that only ensue in the RNA-bound state (Fig. 5C), RNA binding and RNA-stimulated ATPase activities were reduced about 2.2- and 1.4-fold, respectively [i.e., to a similar extent as observed for ecHrpA1-783,FP655/6AA with a modified OB stacking triad (Fig. 2 F and G)]. These observations are consistent with a role of E675 in expanding the RecA-domain RNA binding platform; the effects are most likely moderate because the E675A exchange only affects side chain but not backbone contacts (Fig. 5C). However, the RNA unwinding activity of the E675A variant was essentially unaffected (Fig. 2H), in stark contrast to complete abrogation of helicase activity in ecHrpA1-783,FP655/6AA, underscoring an additional role of the OB stacking triad for ecHrpA helicase activity.
Conformational Flexibility of ecHrpA1-783 in Solution.
Conformations observed in crystal structures can be influenced by crystal packing. To test if ecHrpA1-783 undergoes conformational changes upon RNA binding in solution as suggested by our crystal structures, we conducted cross-linking/mass spectrometry (CLMS) analyses (Dataset S1). To this end, we used isotopically labeled and unlabeled versions of the primary amino group cross-linker, disuccinimidyl suberate (DSS; 30-Å theoretical cutoff distance). Apo-ecHrpA1-783 and ecHrpA1-783-U15 were independently cross-linked with labeled or unlabeled DSS. Samples of the two states treated with differently labeled DSS were then mixed in all constellations, digested by trypsin, and analyzed by liquid chromatography coupled to multidimensional mass spectrometry (LC-MS/MS) to directly compare relative abundancies of the same cross-linked peptides. Indeed, we found changes in cross-link abundancies in agreement with changes in residue distances upon RNA binding as observed in the crystal structures. The RNA-bound state was characterized by more-abundant cross-links of K312 (RecA2) to K537 (HB), K544 (HB), and K662 (OB) (Fig. 3D, Left; thick orange dashed lines; SI Appendix, Table S4). In contrast, cross-links of K106 (RecA1), K312 (RecA2), and K485 (WH) to K361 (RecA2), K570 (HB), and K721 (OB), respectively, were more abundant in the apo-state (Fig. 3D, Right; thick green dashed lines; SI Appendix, Table S4). Thus, upon RNA binding, K570 (HB) seems to move away from K312 (RecA2), while K662 (OB) approaches K312 more closely, fully consistent with the rotation of the WH/HB/OB/CON module suggested by the crystal structures.
Discussion
Physiological Functions of ecHrpA.
Eukaryotes express multiple DEAH/RHA proteins that have important and, in many cases, pleiotropic roles in posttranscriptional gene regulation (58). In contrast, these proteins are less widespread in bacteria, and most bacteria that encode them harbor only one or a few homologs (3, 4). Results presented here suggest that one DEAH/RHA family member in E. coli, HrpA, plays important roles in modulating antibiotics tolerance, and that these effects depend on the NTPase/helicase activity and on a large accessory CTR. These findings are in line with the identification of an HrpA homolog from P. aeruginosa as a modulator of the cellular response to the translation inhibitor azithromycin (44). Interestingly, the E. coli hrpA mutant had diverse effects on antibiotics tolerance depending on the substance tested. While the hrpA mutant showed an increased susceptibility toward compounds that target translation or cell wall synthesis, its survival was improved when challenged with the transcription inhibitor, rifampicin. These findings parallel observations noted by ref. 50.
The large CTR (>600 residues) of ecHrpA that we show is required to elicit chloramphenicol tolerance can directly contact the N-terminal helicase unit and modulate its helicase activity. Such modulation of helicase activity from a distance may be brought about, for example, by the CTR expanding the RNA-binding surface of the helicase region, occupying RNA-binding regions on the helicase region, altering access to the ATP binding sites, or offering binding sites that tune helicase-related conformational changes. Effects of accessory domains on RNA binding, nucleotide transactions, or conformational flexibility have been suggested to be at work in other RNA helicases/rdNTPases, such as Mtr4 (41), eIF4AIII (22, 23), and Brr2 (39), respectively. However, as the helicase-active ecHrpA1-783 fragment does not efficiently complement the hrpA mutant, we suggest that the CTR elicits its role in antibiotics tolerance by forming a landing pad for yet-to-be-identified ecHrpA cofactors or by mediating recruitment of ecHrpA to particular RNA substrates. Interestingly, ecHrpA was shown to interact with ribosomal proteins (59). Furthermore, Salmonella enterica HrpA has been found to associate with the small ribosomal subunit (60), and a recent GradSeq analysis in E. coli revealed an association of ecHrpA with 70S ribosomes or polysomes (61). It is conceivable that an interaction of HrpA with ribosomes, mediated, for example, via the CTR, directly alters the effect of translation inhibitors, while the effect of HrpA on other antibiotics may result from HrpA–ribosome/polysome interactions that elicit changes in the levels of certain proteins. Analyses of other DEAH/RHA enzymes that exhibit C-terminal extensions support these notions. For example, the related DEAH/RHA NTPase HrpB from P. aeruginosa can bind short RNAs via its NTPase region but requires its C-terminal extension to engage structured RNAs (48); furthermore, the CTR of Dhx37 was shown to act as a binding platform for protein cofactors that regulate the protein’s activity and thereby influence ribosome biogenesis (36).
Significance of the Unique Apo-ecHrpA1-783 Conformation.
We showed that ecHrpA1-783 can undergo large-scale domain rearrangements upon RNA binding. The only other DEAH/RHA protein for which structures of apo and RNA-bound/nucleotide-free states are available is Prp22 (31). Structural comparisons reveal much more limited domain rearrangements in Prp22 upon RNA binding; moreover, opposite to the situation in ecHrpA1-783, the HB domain of Prp22 turns away from RecA1 upon RNA binding (Movie S2). As both proteins exhibit very similar RNA-bound states, the differences are due to the unique apo conformation of ecHrpA1-783. Our CLMS data suggest that a conformation resembling the apo state observed in one of our crystal structures can be adopted in solution. Furthermore, a functional relevance of the apo state is strongly supported by the large defects in RNA binding, ATPase, and helicase activities elicited by the D305A mutation, as D305 mediates intimate RecA2–HB domain contacts in apo-ecHrpA1-783 but is not engaged in any interactions in the RNA-bound state.
Additional Conformational Changes Required for RNA Loading.
As it is unlikely that long RNAs thread end-on into the helicase, RNA substrates have to sneak laterally between the RecA2 and HB domains to gain access to the RNA binding surfaces on the RecA domains. However, both in the observed apo and RNA-bound states, the HB domain contacts the RecA2 domain, albeit via different surfaces; thus, additional conformational changes are required for initial RNA engagement. As the D305A mutation abrogates RNA binding while retaining the apo-state conformation, it is possible that a negatively charged residue at position 305 is required for ecHrpA1-783 to intermittently adopt an RNA-receptive conformation (e.g., it is conceivable that additional conformational changes are induced when RNA laterally approaches the apo-state RecA2–HB interface, and its negatively charged sugar-phosphate backbone leads to electrostatic repulsions with D305). This idea is supported by the loss of negative surface potential in the RNA binding–defective D305A variant (Fig. 6B).
Converting an RNA-Dependent NTPase to an RNA Helicase.
Curiously, the ecHrpA-related DEAH/RHA protein, ecHrpB, has been found to encompass RNA-stimulated NTPase but no helicase activity (46, 47). The results reported here suggest that differences in the HB and OB domains of ecHrpA and ecHrpB underlie this functional dichotomy. We showed that RNA contacts to the stacking triad in the ecHrpA1-783 OB domain (F655, P656, and F661) are essential for helicase activity, resembling Prp22 (31). ecHrpB, in contrast, exhibits residues at spatially equivalent positions (M519, L520, and D521) that do not support similar RNA interactions. Interestingly, ecHrpA1-783,FP655/6AA, in which this stacking triad is compromised as in ecHrpB, exhibits ecHrpB-like in vitro activities: it is active as an RNA-dependent ATPase but is unable to unwind RNA duplexes. Lack of a stacking triad in the OB domain of ecHrpB thus likely contributes to the lack of helicase activity in isolated ecHrpB.
In addition, the ecHrpA HB domain that exhibits altered interactions with RecA2 upon RNA binding comprises six α-helices (Fig. 5 A and B, Left). The ecHrpB HB domain is truncated, lacking equivalents of the third and fifth helix and bearing shortened equivalents of the fourth and sixth helix (Fig. 5 A and B, Right). Notably, the regions missing in the ecHrpB HB domain mediate alternative contacts to RecA2 in the RNA-bound and apo states of ecHrpA (Fig. 5 A and B, Left). These observations suggest that a truncated HB domain also contributes to the lack of helicase activity in isolated ecHrpB, as it does not support toggling of ecHrpB between conformations that resemble the ecHrpA-like apo and RNA-bound states.
The OB Domain as a Strand-Separation Device.
Details of the RNA-translocation and strand-separation mechanisms in DEAH/RHA helicases are not yet fully understood. Our ecHrpA1-783-RNA crystal structure shows that the F661, P656, and F655 stacking triad in the OB domain induces bends in the bound RNA strand between nucleotides U6/U7 and U8/U9 (Fig. 4A), such that a complementary strand cannot remain paired to the entire U6 to U9 region. NTPase-driven translocation of the bound strand would drive the stacking triad further into the RNA duplex region, sequestering the next 5′-located nucleotide from the displaced strand. Our observation that ecHrpA1-783,FP655/6AA with a disrupted stacking triad still shows RNA-stimulated ATPase activity, and thus RNA binding, but can no longer unwind RNA is consistent with this model.
The above model is also consistent with recent studies of eukaryotic DEAH/RHA proteins, which showed that the intimacy of interaction between the OB domain and the bound RNA differs between family members and is thought to be related to their differential dependencies on cofactors. Prp22 tightly attaches to the RNA via a stacking triad in the OB domain and is an active helicase in isolation (31). Prp43, in contrast, exhibits a truncated stacking triad that only loosely attaches to the 5′ end of the RNA (33) and requires G-patch cofactors Ntr1/TFIP11 or Pfa1/Sqs1 for helicase activity (51) that bind the OB domain (62). Our results, therefore, are consistent with the idea that the OB domain represents the primary strand-separating element of DEAH/RHA helicases that couples NTPase-powered RNA translocation to unwinding and, depending on the absence or presence of a stacking triad, requires support by a cofactor or not.
Implications for the Mechanisms of RNA Translocation and Unwinding.
Interestingly, apo-ecHrpB adopts a conformation resembling RNA-bound ecHrpA1-783 (SI Appendix, Fig. S3). As ecHrpB can bind RNA (46, 47), we suggest that the unique apo-ecHrpA–like conformation is not only adopted in the absence of RNA but that a similar conformation is visited intermittently during NTPase-driven RNA translocation and unwinding. In this scenario, D305 would be required as an intermediate latching point for the HB domain to toggle relative to the RecA domains. Compared to the RNA-bound state, the HB domain in the apo state is positioned more in the direction of the 5′ end of the bound RNA strand. Thus, converting from the RNA-bound to the apo conformation, the HB domain may slide in 3 to 5′ direction along the RNA, which is held on RecA1/2. NTPase-dependent conformational changes may then lead to weaker RNA binding to RecA1/2 and stronger RNA binding to the HB domain. After handover of RNA from RecA1/2 to the HB domain, ecHrpA may return to the conformation seen in the RNA-bound state, during which the HB domain could take RNA along. RecA1/2 would then be registered more toward the 5′ end of the bound RNA strand, and handing RNA back to RecA1/2 would complete the translocation cycle. In this model of action, the HB domain would exactly serve the function of a ratchet, as originally proposed for the Hel308 helicase (63).
HrpA as Target for Antibiotics.
The influence of HrpA on antibiotics tolerance not only in E. coli but also in other bacteria (14, 15, 43, 44, 50) suggests that HrpA may be an interesting target for compounds to interfere with the prevalence of antimicrobial resistance in bacterial pathogens. However, based on results presented here and elsewhere (50), inhibition of bacterial HrpA may also compromise the susceptibility toward certain antibiotics treatments, depending on the substances employed.
Based on our results, HrpA’s RNA-unwinding activity is essential to elicit these effects. While the RNA-binding stacking triad in the OB domain could, therefore, represent a target site for interfering substances, this and several other features of the helicase are recapitulated in eukaryotic DEAH/RHA enzymes, posing the danger of side effects by cross-reactive substances. A unique feature in HrpA is a conserved alanine at the C terminus of motif Ic (A180) that leads to the accommodation of an additional nucleobase (U15; Fig. 4E), but our mutational analysis suggests that this feature only weakly impacts HrpA helicase activity. The unique apo conformation of ecHrpA, in contrast, is unprecedented in previously studied eukaryotic DEAH/RHA enzymes and, based on our findings, is required for helicase function. Therefore, compounds that interfere with transient RecA2–HB interactions in the HrpA-specific apo conformation (Fig. 5B, Inset) may selectively inhibit HrpA.
Methods
Construction of an E. coli hrpA Mutant Strain.
E. coli K12 derivative, MC4100 (64), was used for construction of the hrpA mutant by using the one-step inactivation method (65). The hrpA gene was replaced by a PCR-amplified kanamycin resistance cassette via λ red recombinase, resulting in strain MC4100 hrpA::kan. Primers K1 and K2 (65) that bind within the kanamycin cassette and primers HrpA-up and HrpA-down that bind upstream and downstream of the cassette were used to verify the correct insertion. Primers employed are listed in (SI Appendix, Table S1).
Cloning.
A DNA fragment encoding ecHrpAFL (Gene ID: P43329) was PCR-amplified from E. coli DH5-α genomic DNA and inserted into the pETM-11 vector (EMBL Heidelberg) via sequence- and ligation-independent cloning (66), generating pETM-11-hrpA. DNA fragments encoding ecHrpA variants were PCR amplified from pETM-11-hrpA vector using inverse PCR (67).
A DNA fragment encoding ecHrpAFL was PCR amplified from pETM-11-hrpA and inserted into a pBAD LIC vector (8A) together with a 6His_BIO_6His_3xFLAG-tag using Gibson assembly (68). pBAD LIC (8A) vectors encoding ecHrpAFL,K106A or ecHrpA1-783 were generated from the ecHrpAFL-encoding pBAD LIC (8A) vector via inverse PCR.
Survival Assays on Agar Plates.
Several colonies of MC4100 or MC4100 hrpA::kan were used to inoculate 50 mL Luria–Bertani (LB) shaking cultures and incubated overnight at 37 °C. Overnight cultures were diluted to an optical density at 600 nm (OD600) of 2, and 5 µL diluted cultures, or further serial dilutions, were spotted on LB/agar plates lacking antibiotics or containing rifampicin (4.5 µg/mL), ampicillin (9 µg/mL), chloramphenicol (3.125 µg/mL), or tetracycline (0.9 µg/mL). Bacterial survival was monitored after incubation at 37 °C for 40 h to observe colonies.
For rescue experiments with complemented strains, MC4100 hrpA::kan mutant cells were complemented with pBAD LIC (8A) vectors containing different hrpA alleles. Survival assays were conducted as above, but liquid medium and LB agar plates additionally contained 2.5 µg/mL arabinose to induce protein production. To maintain pBAD LIC (8A) vectors, precultures contained 100 µg/mL ampicillin.
Recombinant Protein Production and Purification.
Proteins of interest (POIs) were produced with a TEV-cleavable N-terminal His6-tag by transforming E. coli Rosetta2 DE3 cells with pETM-11 expression vectors and cultivation in auto-inducing medium (69) at 37 °C for 4 h, followed by overnight incubation at 18 °C. Cells were harvested by centrifugation at an OD600 of ∼10.
Cell pellets were resuspended in 50 mM Tris HCl, pH 8.0, 500 mM NaCl, 2 mM β-mercaptoethanol, and supplemented with 0.02% (vol/vol) Nonidet P-40, 0.2 mg/mL lysozyme and 0.01 mg/mL DNase. The cell suspension was lysed by sonication using a Sonopuls HD 3100 μLtrasonic homogenizer (Bandelin). Cleared lysate was passed through 3 mL Ni2+-NTA agarose beads (Qiagen) in a gravity flow column to capture the His6-tagged POIs. After washing and elution with 400 mM imidazole, the N-terminal His6-tag was cleaved by incubation with TEV protease overnight. The samples were diluted to a final NaCl concentration of 100 mM and loaded on a HiPrep Heparin FF 16/10 column (GE Healthcare) equilibrated with 25 mM Tris HCl, pH 8.0, 50 mM NaCl, and 2 mM DTT. POIs were eluted with a linear gradient from 100 to 1,500 mM NaCl and were further purified by gel filtration on a 26/60 Superdex 200 or 26/60 Superdex 75 column (GE Healthcare) in 10 mM Tris HCl, pH 8.0, 200 mM NaCl, and 1 mM DTT. For production of selenomethionine (SeMet)-labeled ecHrpA1-783, transformed E. coli Rosetta2 DE3 cells were grown in minimal medium with methionine replaced by seleno-D,L-methionine, and the protein was purified in the same manner as the unlabeled POIs.
Nucleic Acid Binding Assays.
Fluorescence (5-FAM)–labeled ssRNA or ssDNA (5 nM) were titrated with increasing concentrations of HrpA (FL or truncations) in 40 mM Tris HCl, pH 7.5, 150 mM NaCl, 0.5 mM MgCl2, 1.5 mM DTT, and 46 mU/μL RNasin, and fluorescence anisotropy was quantified in 384-well plates via using a Victor plate reader (PerkinElmer). To extract binding constants, changes in anisotropy were plotted against protein concentration, and the data were fit to a single exponential Hill function (fraction bound = A[protein]n / ([protein]n + Kdn); A, fitted maximum of nucleic acid bound; n, Hill coefficient; Kd, dissociation constant) (70) using Prism (GraphPad Software, Inc.).
ATPase Assays.
HrpA variants were diluted to 2.5 μM in 40 mM Tris HCl, pH 7.5, 50 mM NaCl, 1.5 mM DTT, 0.1 mg/mL acetylated BSA, and 46 mU/μL RNasin without or with 2.5 μM nucleic acid (ssDNA, ssRNA, or dsRNA). The reactions were initiated by the addition of 1 mM ATP/MgCl2, supplemented with 0.0125 mCi/mL [α-32P]-ATP, and samples were incubated for 30 min at 37 °C. The reactions were stopped by the addition of an equivalent volume of 100 mM ethylenediaminetetraacetic acid, pH 8.0, and separated by thin-layer chromatography in 20% (vol/vol) ethanol, 6% (vol/vol) acetic acid, and 0.5 M LiCl. Chromatograms were scanned on a Storm 860 phosphorimager (GMI) and quantified by densitometry.
Helicase Activity Assays.
RNA/DNA unwinding was monitored by fluorescence stopped-flow measurements on an SX-20MV spectrometer (Applied Photophysics). RNA substrates contained the same 12–base pair duplex region and 31-nt single-stranded 3′- or 5′-overhangs as the DNA substrates (SI Appendix, Table S1). The short strand was labeled with Alexa Fluor 488, and the long strand, bearing the overhang, was labeled with the quencher Atto 540 Q, such that fluorophore and quencher resided close to each other on the blunt ends of the duplex regions after strand annealing (SI Appendix, Table S1). The fluorescence of Alexa Fluor 488 was excited at 465 nm, and the fluorescence emission was monitored after passing a 495-nm cutoff filter (KV 495, Schott). An increase in fluorescence was only observed when the Alexa Fluor 488–labeled strand was separated from the complementary strand carrying the quencher Atto 540 Q. For all experiments, 1 mM protein was mixed with 50 nM RNA in 40 mM Tris HCl, pH 7.5, 50 mM NaCl, and 0.5 mM MgCl2. The mixture was loaded into the syringe and incubated for 5 min at 25 °C. Protein–RNA mixture (60 μL) was then rapidly mixed with 60 µL 2 mM ATP/MgCl2 in the same buffer, and the fluorescence change was monitored over time.
Data from fluorescence measurement were baseline corrected by subtracting the starting fluorescence immediately after addition of ATP and normalized to the baseline-corrected maximum fluorescence. Fluorescence traces for ecHrpAFL/3′-overhang DNA, ecHrpAFL/5′-overhang RNA, ecHrpA784-1300/3′-overhang RNA, and ecHrpA909-1300/3′-overhang DNA, which did not show any unwinding, were normalized to the baseline-corrected maximum fluorescence of ecHrpAFL/3′-overhang RNA. Fluorescence traces for ecHrpA1-783,D305A and ecHrpA1-783,FP655/6AA, which did not show unwinding activity with 3′-overhang RNA, were normalized to the baseline-corrected maximum fluorescence of ecHrpA1-783/3′-overhang RNA. Three to five traces were averaged and plotted using Prism software (GraphPad). The data were fitted to a double exponential equation (fraction unwound = Afast ∙ (1 − exp(−kfastt)) + Aslow ∙ (1 − exp(−kslowt)); A, total unwinding amplitude; k, unwinding rate constants [s−1]; t, time [s]), as described previously (53, 54). The first second of data acquisition was excluded from curve fitting to account for the initial mixing periods. Amplitude-weighted unwinding rate constants were calculated as kunw=∑(Aiki2) / ∑(kiAi). Reference SI Appendix, Fig. S1 and Table S2 for curve fitting and extracted helicase parameters, respectively.
Crystallographic Procedures.
Crystals of ecHrpA1-783 (16.6 mg/mL), ecHrpA1-783,D305A (16.6 mg/mL), ecHrpA1-783,SeMet (16.9 mg/mL), or ecHrpA1-783 + U15 ssRNA (10 mg/mL, 1.5 molar excess of RNA) were grown via the sitting-drop vapor diffusion technique at 18 °C, with drops containing 1 µL of protein/protein–RNA solution and 1 µL of reservoir solution. Crystals of ecHrpA1-783 were obtained with 0.1 M Hepes-NaOH, pH 7.3, 0.2 M NaCl, and 9% [vol/vol] PEG400 as the reservoir solution. Crystals of ecHrpA1-783,D305A were obtained with 0.05 M Hepes-NaOH, pH 7.0, 0.1 M KCl, 0.01 M MgCl2, and 7% [wt/vol] PEG3350 as the reservoir solution. Crystals of HrpA1-783,SeMet were obtained with 0.1 M Hepes-NaOH, pH 7.3, 0.2 M NaCl, and 9% [wt/vol] PEG2000 as the reservoir. Crystals of ecHrpA1-783-U15 were grown with 0.1 M MES-NaOH, pH 6.0, 0.2 M CaCl2, and 8% [wt/vol] PEG6000 as the reservoir solution. Prior to flash cooling, crystals were transferred into cryoprotecting solution containing the respective mother liquor and additional 25% Ethylenglycol.
Synchrotron diffraction data were collected at 100 K on MX beamlines 14.1 and 14.2 of the BESSY II storage ring (71) and were processed with X-ray detector software (XDS) (72) via the XDSAPP GUI (73). Experimental phases for HrpA1-783,SeMet were determined via the single anomalous diffraction strategy using Phenix (74). The structure of HrpA1-783-U15 was solved by molecular replacement using structure coordinates for human Prp22 (PDB ID: 6I3P) (31) as a search model. Both structures were refined using alternating cycles of automated refinement with Phenix (75) and manual model building with Coot (76, 77). The ecHrpA1-783,D305A diffraction data were pseudomerohedrally twinned and refined with the twin law h,-k,-h-l. Translation–libration–screw refinement was employed for all structures. Structure figures were prepared using PyMOL (Schrödinger Inc.; https://pymol.org/2), and electrostatic surface potentials were calculated with APBS (78).
CLMS.
DSS efficiently reacts with primary amino groups, such as the ε-amino groups of lysine side chains. Apo-ecHrpA1-783 and RNA-bound ecHrpA1-783 were each treated with DSS that was either isotopically labeled with four deuterium atoms or did not contain an isotopic label by adding a 50-fold molar excess of the cross-linker and incubating the reaction mixtures for 30 min at room temperature. The reactions were quenched by the addition of 100 mM Tris HCl, pH 8.1, for 15 min. Apo-ecHrpA1-783 treated with labeled DSS and ecHrpA1-783-U15 treated with nonlabeled DSS, and vice versa, were mixed in equal amounts. The samples were prepared with the single-pot solid-phase–enhanced sample preparation (SP3) protocol (79) and digested overnight with trypsin in a 1:20 (wt/wt) ratio. SP3 beads were removed from the sample with a magnet, and the supernatant was dried in a vacuum centrifuge. The dried peptides were resuspended in 5% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid and subjected to LC-MS analysis on an UltiMate 3000 RSLCnano System and an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific). Peptides were separated on a C18 PepMap100 µ-Precolumn (0.3 × 5 mm, 5 µm; Thermo Fisher Scientific) connected to an in-house–packed C18 analytical column (75 µm × 300 mm, 1.9 µm, Reprosil-Pur 120C18-AQ; Maisch GmbH) using a 164-min gradient ranging from 10 to 45% buffer B (80% acetonitrile and 0.08% formic acid, vol/vol). Precursor scans were performed with a scan range from m/z 350 to 1,600, a resolution of 120,000 full width at half maximum (FWHM), 3 × 106 automatic gain control (AGC) target, and 25-ms maximum injection time. Each precursor scan was followed by 20 fragment ion scans of the most abundant precursors, fragmented with a normalized collision energy of 30, and acquired with a resolution of 30,000 FWHM, 1 × 10−5 AGC target and 120-ms maximum injection time. Doubly charged precursors were only considered for fragmentation in the m/z range from 500 to 1,600, while all precursors with a charge 3+ to 10+ were considered. Dynamic exclusion was set to 10 s.
The acquired data were submitted to a database search with plink version 2.3.9 (80) against the sequence of ecHrpA and common contaminants (in total 294 sequences) with the following parameters: DSS (138.068 Da) and heavy DSS (142.093 Da) as cross-linker, peptides between 6 and 60 amino acids and 600 to 6,000 Da, up to three missed cleavages, cysteine carbamidomethlylation as fixed and methionine oxidation as variable modification, 20-ppm peptide and fragment tolerance, 10-ppm filter tolerance, 5% FDR cutoff on spectral level, and separate calculation of FDR for intra- and interlinks. For quantification, “Labeling-Leiker etc.” was set to “None” for the light and “BS3_Labeling” for the heavy label. Quantification ratios per identified spectrum were summarized by calculating the mean and the median of all ratios supporting a particular cross-linked residue. Identifications were further filtered for at least four fragment ions for each of the peptides in a cross-link pair, a minimum score of 1 and more than three spectrum identifications. C-α distances were determined based on the apo-ecHrpA1-783 and ecHrpA1-783-U15 crystal structures using PyMOL. CLMS data are presented in Dataset S1.
Supplementary Material
Acknowledgments
We are grateful to Nicole Holton, Laboratory of Structural Biochemistry, and Freie Universität Berlin for cloning the pBAD-vector; to Claudia Alings, Laboratory of Structural Biochemistry, and Freie Universität Berlin for help with crystallization; and to Christoph Weise, Laboratory of Protein Biochemistry, and Freie Universität Berlin for support in mass spectrometric fingerprinting. We acknowledge access to beamlines of the BESSY II storage ring via the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück-Centrum, the Leibniz-Institut für Molekulare Pharmakologie, and Charité–Universitätsmedizin Berlin. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Grant No. RTG 2473-1 to M.C.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100370118/-/DCSupplemental.
Data Availability
Structure factors and atomic coordinates have been deposited with the RSCB Protein Data Bank (https://www.rcsb.org) under accession codes 6ZWX (apo-ecHrpA1-783), 7AKP (apo-ecHrpA1-783,D305A), and 6ZWW (ecHrpA1-783-U15). All other data are contained in the main text or in the supporting information.
References
- 1.Perez-Rueda E., Martinez-Nuñez M. A., The repertoire of DNA-binding transcription factors in prokaryotes: Functional and evolutionary lessons. Sci. Prog. 95, 315–329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Assche E., Van Puyvelde S., Vanderleyden J., Steenackers H. P., RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 6, 141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khemici V., Linder P., RNA helicases in bacteria. Curr. Opin. Microbiol. 30, 58–66 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Redder P., Hausmann S., Khemici V., Yasrebi H., Linder P., Bacterial versatility requires DEAD-box RNA helicases. FEMS Microbiol. Rev. 39, 392–412 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Oun S., et al., The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol. 10, 157–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehnik-Habrink M., et al., DEAD-box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J. Bacteriol. 195, 534–544 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markkula A., Lindström M., Johansson P., Björkroth J., Korkeala H., Roles of four putative DEAD-box RNA helicase genes in growth of Listeria monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stress conditions. Appl. Environ. Microbiol. 78, 6875–6882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palonen E., et al., Requirement for RNA helicase CsdA for growth of Yersinia pseudotuberculosis IP32953 at low temperatures. Appl. Environ. Microbiol. 78, 1298–1301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandiani F., et al., Role of the five RNA helicases in the adaptive response of Bacillus cereus ATCC 14579 cells to temperature, pH, and oxidative stresses. Appl. Environ. Microbiol. 77, 5604–5609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng Y. T., Chiou N. T., Gogiraju R., Lin-Chao S., The protein interaction of RNA helicase B (RhlB) and polynucleotide phosphorylase (PNPase) contributes to the homeostatic control of cysteine in Escherichia coli. J. Biol. Chem. 290, 29953–29963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bäreclev C., Vaitkevicius K., Netterling S., Johansson J., DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression. RNA Biol. 11, 1457–1466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salman-Dilgimen A., Hardy P. O., Dresser A. R., Chaconas G., HrpA, a DEAH-box RNA helicase, is involved in global gene regulation in the Lyme disease spirochete. PLoS One 6, e22168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salman-Dilgimen A., Hardy P. O., Radolf J. D., Caimano M. J., Chaconas G., HrpA, an RNA helicase involved in RNA processing, is required for mouse infectivity and tick transmission of the Lyme disease spirochete. PLoS Pathog. 9, e1003841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu A., et al., Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: Generating an antibiotic bar code. Antimicrob. Agents Chemother. 54, 1393–1403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamae C., et al., Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190, 5981–5988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudan M., Schneider D., Warnecke T., Krisko A., RNA chaperones buffer deleterious mutations in E. coli. eLife 4, e04745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairman-Williams M. E., Guenther U. P., Jankowsky E., SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 20, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jankowsky E., RNA helicases at work: Binding and rearranging. Trends Biochem. Sci. 36, 19–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halls C., et al., Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 365, 835–855 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Q., Jankowsky E., ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44, 13591–13601 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Ballut L., et al., The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12, 861–869 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Andersen C. B., et al., Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Bono F., Ebert J., Lorentzen E., Conti E., The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Jankowsky E., Gross C. H., Shuman S., Pyle A. M., Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291, 121–125 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Bowers H. A., et al., Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA 12, 903–912 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theuser M., Höbartner C., Wahl M. C., Santos K. F., Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 113, 7798–7803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudolph M. G., Klostermeier D., When core competence is not enough: Functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol. Chem. 396, 849–865 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Cordin O., Banroques J., Tanner N. K., Linder P., The DEAD-box protein family of RNA helicases. Gene 367, 17–37 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Chen M. C., Ferré-D’Amaré A. R., Structural basis of DEAH/RHA helicase activity. Crystals (Basel) 7, 253 (2017). [Google Scholar]
- 30.Ozgur S., et al., The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 282, 850–863 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Hamann F., Enders M., Ficner R., Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 47, 4349–4362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y., Staley J. P., Andersen G. R., Nielsen K. H., Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA. RNA 23, 1110–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tauchert M. J., Fourmann J. B., Lührmann R., Ficner R., Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. eLife 6, e21510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walbott H., et al., Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 29, 2194–2204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prabu J. R., et al., Structure of the RNA helicase MLE reveals the molecular mechanisms for uridine specificity and RNA-ATP coupling. Mol. Cell 60, 487–499 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Boneberg F. M., et al., Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis. RNA 25, 685–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ledoux S., Guthrie C., Retinitis pigmentosa mutations in bad response to refrigeration 2 (Brr2) impair ATPase and helicase activity. J. Biol. Chem. 291, 11954–11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patrick E. M., Srinivasan S., Jankowsky E., Comstock M. J., The RNA helicase Mtr4p is a duplex-sensing translocase. Nat. Chem. Biol. 13, 99–104 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Absmeier E., et al., The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev. 29, 2576–2587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson R. N., et al., The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 29, 2205–2216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weir J. R., Bonneau F., Hentschel J., Conti E., Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc. Natl. Acad. Sci. U.S.A. 107, 12139–12144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo J. T., Choe J., Moseley S. L., HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 52, 1813–1826 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Nagata Y., et al., Identification of Burkholderia multivorans ATCC 17616 genetic determinants for fitness in soil by using signature-tagged mutagenesis. Microbiology (Reading) 160, 883–891 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Tan H., et al., PA3297 counteracts antimicrobial effects of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 7, 317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granato L. M., et al., The ATP-dependent RNA helicase HrpB plays an important role in motility and biofilm formation in Xanthomonas citri subsp. citri. BMC Microbiol. 16, 55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietrzyk-Brzezinska A. J., et al., Crystal structure of the Escherichia coli DExH-box NTPase HrpB. Structure 26, 1462–1473.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Xin B. G., Chen W. F., Rety S., Dai Y. X., Xi X. G., Crystal structure of Escherichia coli DEAH/RHA helicase HrpB. Biochem. Biophys. Res. Commun. 504, 334–339 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Hausmann S., et al., Auxiliary domains of the HrpB bacterial DExH-box helicase shape its RNA preferences. RNA Biol. 17, 637–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y., et al., An O island 172 encoded RNA helicase regulates the motility of Escherichia coli O157:H7. PLoS One 8, e64211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price M. N., et al., Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Robert-Paganin J., Réty S., Leulliot N., Regulation of DEAH/RHA helicases by G-patch proteins. BioMed Res. Int. 2015, 931857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Granneman S., Bernstein K. A., Bleichert F., Baserga S. J., Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell. Biol. 26, 1183–1194 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roske J. J., Liu S., Loll B., Neu U., Wahl M. C., A skipping rope translocation mechanism in a widespread family of DNA repair helicases. Nucleic Acids Res. 49, 504–518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Özeş A. R., Feoktistova K., Avanzino B. C., Baldwin E. P., Fraser C. S., Real-time fluorescence assays to monitor duplex unwinding and ATPase activities of helicases. Nat. Protoc. 9, 1645–1661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grass L. M., Wollenhaupt J., Barthel T., Loll B., Wahl M. C., Crystal structure of E. coli RNA helicase HrpA in complex with RNA. RSCB Protein Data Bank (PDB). https://www.rcsb.org/structure/6ZWW. Deposited 29 July 2020.
- 56.Grass L. M., Wollenhaupt J., Barthel T., Loll B., Wahl M. C., Crystal structure of E. coli RNA helicase HrpA. RSCB Protein Data Bank (PDB). https://www.rcsb.org/structure/6ZWX. Deposited 29 July 2020.
- 57.Grass L. M., Wollenhaupt J., Barthel T., Loll B., Wahl M. C., Crystal structure of E. coli RNA helicase HrpA-D305A. RSCB Protein Data Bank (PDB). https://www.rcsb.org/structure/7AKP. Deposited 1 October 2020.
- 58.Bourgeois C. F., Mortreux F., Auboeuf D., The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17, 426–438 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Butland G., et al., Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Smirnov A., et al., Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 113, 11591–11596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hör J., et al., Grad-seq shines light on unrecognized RNA and protein complexes in the model bacterium Escherichia coli. Nucleic Acids Res. 48, 9301–9319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studer M. K., Ivanović L., Weber M. E., Marti S., Jonas S., Structural basis for DEAH-helicase activation by G-patch proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 7159–7170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Büttner K., Nehring S., Hopfner K. P., Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 14, 647–652 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Casadaban M. J., Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104, 541–555 (1976). [DOI] [PubMed] [Google Scholar]
- 65.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M. Z., Elledge S. J., SLIC: A method for sequence- and ligation-independent cloning. Methods Mol. Biol. 852, 51–59 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Zhang B. Z., et al., An easy-to-use site-directed mutagenesis method with a designed restriction site for convenient and reliable mutant screening. J. Zhejiang Univ. Sci. B 10, 479–482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibson D. G., et al., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Ryder S. P., Recht M. I., Williamson J. R., Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 488, 99–115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller U., et al., The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 130, 141 (2015). [Google Scholar]
- 72.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sparta K. M., Krug M., Heinemann U., Mueller U., Weiss M. S., XDSAPP2. 0. J. Appl. Cryst. 49, 1085–1092 (2016). [Google Scholar]
- 74.Zwart P. H., et al., “Automated structure solution with the PHENIX suite” inStructural Proteomics, Kobe B., Guss M., Huber T., Eds. (Springer, 2008), pp. 419–435. [DOI] [PubMed] [Google Scholar]
- 75.Afonine P. V., et al., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hughes C. S., et al., Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Chen Z. L., et al., A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factors and atomic coordinates have been deposited with the RSCB Protein Data Bank (https://www.rcsb.org) under accession codes 6ZWX (apo-ecHrpA1-783), 7AKP (apo-ecHrpA1-783,D305A), and 6ZWW (ecHrpA1-783-U15). All other data are contained in the main text or in the supporting information.