Significance
Communication between cells is essential for the development and homeostasis of tissues and prevents diseases, including cancers. The Notch and Jagged transmembrane proteins interact to regulate cell–cell communication in all multicellular animals. Defining their interactions is critical to understand Notch-associated disorders. While structural studies have focused on short regions of both proteins, it is unclear how their entire extracellular domains collectively engage to activate signaling. Here, we identify several unreported, interacting regions in the Notch1–Jagged1 full extracellular complex. We show that Notch1 and Jagged1 ectodomains are not fully extended and reveal that activation-determining regions, previously thought to be distal, engage directly. This interaction network redefines our knowledge on Notch activation and provides avenues for therapeutic advances.
Keywords: Notch1, Jagged1, cell signaling, cross-linking mass spectrometry, glycoprotein
Abstract
The Notch signaling system links cellular fate to that of its neighbors, driving proliferation, apoptosis, and cell differentiation in metazoans, whereas dysfunction leads to debilitating developmental disorders and cancers. Other than a five-by-five domain complex, it is unclear how the 40 extracellular domains of the Notch1 receptor collectively engage the 19 domains of its canonical ligand, Jagged1, to activate Notch1 signaling. Here, using cross-linking mass spectrometry (XL-MS), biophysical, and structural techniques on the full extracellular complex and targeted sites, we identify five distinct regions, two on Notch1 and three on Jagged1, that form an interaction network. The Notch1 membrane–proximal regulatory region individually binds to the established Notch1 epidermal growth factor (EGF) 8–EGF13 and Jagged1 C2–EGF3 activation sites as well as to two additional Jagged1 regions, EGF8–EGF11 and cysteine-rich domain. XL-MS and quantitative interaction experiments show that the three Notch1-binding sites on Jagged1 also engage intramolecularly. These interactions, together with Notch1 and Jagged1 ectodomain dimensions and flexibility, determined by small-angle X-ray scattering, support the formation of nonlinear architectures. Combined, the data suggest that critical Notch1 and Jagged1 regions are not distal but engage directly to control Notch1 signaling, thereby redefining the Notch1–Jagged1 activation mechanism and indicating routes for therapeutic applications.
Notch signaling plays a central role in developmental processes by determining cell fate decisions in tissues during development. In adults, these signals both determine differentiation and maintenance of neuronal and hematopoietic stem cells as well as regulate the immune system (1–4). Dysregulation often leads to debilitating diseases in humans, including congenital disorders and cancers (5–8). The mammalian Notch1 receptor is the prototypical member of the Notch protein family, which consists of four paralogs (Notch1 to Notch4) that all receive signals from the associated ligands Jagged1, Jagged2, Delta-like 1, and Delta-like 4 in trans (from adjacent cells) to initiate signaling or in cis (from the same cell) to inhibit signaling. The Notch1–Jagged1 receptor–ligand pair has been widely studied at functional, cellular, and molecular levels (4, 5). Both Notch1 and Jagged1 are type I transmembrane proteins with large modular extracellular segments that determine interaction specificity and control the activation of signaling. Notch1 has an extracellular segment of 209 kDa composed of 36 EGF repeats, followed by the negative regulatory region (NRR) at the membrane–proximal side, and differs from its paralogs in the number of EGF domains: from 36 for Notch2, 34 for Notch3, and 29 for Notch4. The Jagged1 ectodomain (139 kDa) is similar to that of Jagged2 and is composed of a C2 lipid-binding domain, a Delta/Serrate/Lag-2 (DSL) domain, 16 EGF repeats, and a cysteine-rich domain (CRD) at the membrane–proximal side.
The prevailing model for canonical Notch activation states that ligand binding at Notch1 EGF8–EGF12 and an endocytosis-induced pulling force (9–16), generated by the signal-sending cell on the Notch–ligand complex (17, 18), triggers a conformational change and proteolytic processing in the Notch NRR located 24 EGF domains downstream of the ligand-binding site (19–21). After Notch cleavage within the transmembrane domain (22, 23), the Notch intracellular domain translocates to the nucleus, where it regulates transcription (24). At the N-terminal side of Jagged1, the C2–EGF3 region is important for Notch1 binding (11, 25–28). A recent structural study demonstrated that the Notch1 EGF8–EGF12 region interacts in an antiparallel fashion through an extended interface with the Jagged1 C2–EGF3 region (11). Additional interactions add complexity to the mechanism of Notch activation and regulation. Notch–ligand, Notch–Notch, and ligand–ligand interactions in cis can both inhibit (29–31) or activate (32) signaling. In addition to the canonical, ligand-binding site on EGF8–EGF12 and the conformational change in the NRR, several other extracellular regions, such as EGF6, EGF25–EGF26, and EGF36, seem to play a role in Notch function (33–39). Also, the Jagged1 extracellular segment harbors additional functionality other than the C2–EGF3 region interacting to Notch. It has been suggested that the Jagged and Delta-like C2 domain binding to membranes has an important role in regulating ligand-dependent Notch signaling (26, 28). The CRD in Xenopus Serrate-1, a homolog of mammalian Jagged1, is required for Notch activation in primary neurogenesis (40). These studies indicate that several sites in the Notch and Jagged extracellular segments may contribute to Notch signaling and regulation.
Structural studies have revealed details of key interaction sites (11, 41) and indicate that flexibility is present to a certain extent in the Notch and Jagged ectodomains (28, 42). A low-resolution negative stain electron microscopy reconstruction of the Notch1 ectodomain resolved distinct globular dimer states, although this protein was purified in an unconventional manner (43). Backfolded models for the Notch ectodomain have also been suggested, based on genetic and interaction studies (36–38). Nonetheless, direct observations of ectodomain flexibility and nonextended architectures are limited. While Notch–Jagged interaction studies have focused predominantly on the well-established Notch1 EGF11–EGF12–Jagged1 C2–EGF3 regions, other sites may play a direct role in this intermolecular interaction. Structural and biophysical studies on the full extracellular portions of Notch and Jagged have, however, been limited because of the size, flexibility, and low-expression levels of the proteins, hampering the identification of several interacting regions.
In this study, we combine cross-linking mass spectrometry (XL-MS), quantitative interaction assays, and small-angle X-ray scattering (SAXS) on purified Notch1 and Jagged1 full ectodomains (Notch1fe–Jagged1fe), as well as shorter constructs, to probe the structure of the Notch1–Jagged1 complex and of the unliganded proteins (Fig. 1 A–D). This analysis reveals several, hitherto unreported, intra- and intermolecular interaction regions. We show that Jagged1 C2–EGF3, EGF8–EGF11, and CRD can all interact with Notch1 EGF33–NRR and that the Notch1 NRR is sufficient for the interaction with Jagged1 C2–EGF3. In addition, the Notch1 EGF8–EGF13 region directly interacts with Notch1 EGF33–NRR. XL-MS analysis suggested that four regions, C2–EGF1, EGF5–EGF6, EGF9–EGF12, and CRD, are in proximity within Jagged1, and we confirmed direct interactions for C2–EGF3 binding to EGF8–EGF11 and to CRD. These data, together with the SAXS analysis of the Notch1 and Jagged1 ectodomains, suggest that the proteins are not fully extended and indicate that regions in both proteins (i.e., Notch1 EGF8–EGF13, Notch1 EGF33–NRR, and Jagged1 C2–EGF3), previously shown to be important for Notch signaling, affect each other directly.
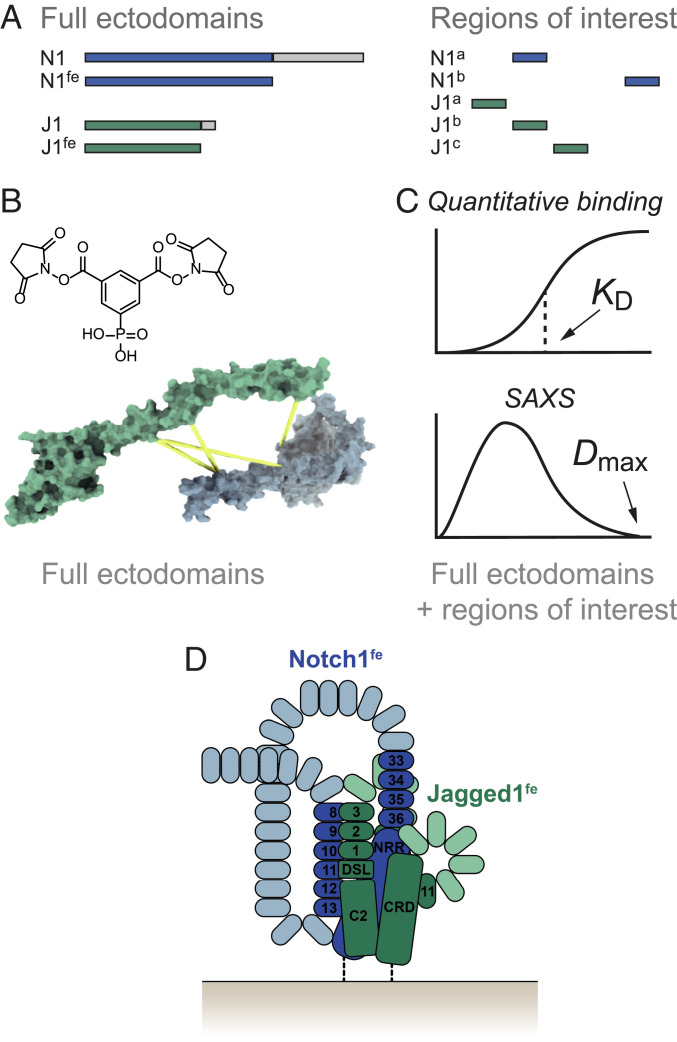
Fig. 1.
XL-MS and biophysical studies reveal an interaction network in the Notch1–Jagged1 complex. (A) Notch1fe, Jagged1fe, and targeted sites are expressed in HEK293 cells and purified by IMAC and SEC. (B) Identification of regions in proximity in the Notch1fe–Jagged1fe complex by XL-MS using PhoX (44). (C) The purified full ectodomain samples and shorter regions of interest are used in quantitative binding experiments to confirm direct interactions, and in SAXS studies. (D) The resulting data provide insights into the molecular architecture of the Notch1–Jagged1 complex, represented here as a schematic in a cis setting.
Results
XL-MS of the Notch1–Jagged1 Complex Reveals a Mosaic of Interaction Sites.
To determine which regions, beyond the canonical Notch1EGF8–12–Jagged1C2–EGF3 interaction site, are involved in receptor–ligand binding, we probed Notch1fe–Jagged1fe with XL-MS (Figs. 1 A–D and 2 A and B and SI Appendix, Table S1 and Datasets S1 and S2). Two variants of Jagged1 were used: a wild-type version (Jagged1fe,wt) and one with five-point mutations in the Jagged C2 region (Jagged1fe,HA) that provide higher-affinity binding to Notch1 EGF8–EGF12 when incorporated in a Jagged1 C2–EGF3 construct (11). In surface plasmon resonance (SPR) experiments, in which Notch1fe is coupled at the C terminus to the sensor surface to achieve a close-to-native topology (see Materials and Methods), Notch1fe–Jagged1fe,HA interact with a dissociation constant (KD) of 1 µM, and Jagged1fe,HA interacts with similar affinity to the EGF8–EGF13 portion of Notch1, while no interaction was measured between Jagged1fe,wt and Notch1 EGF8–EGF13 at 1 µM (Fig. 2 C and D and SI Appendix, Fig. S1 A and B).
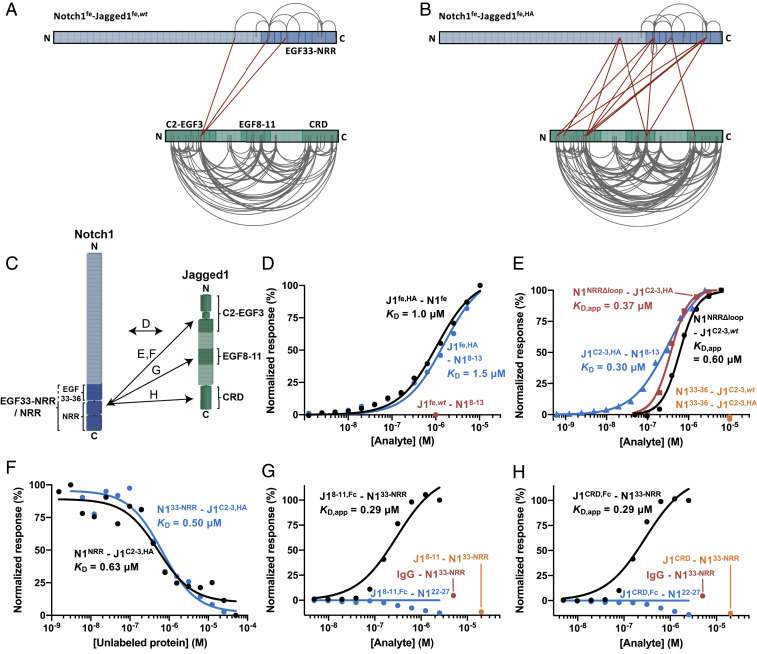
Fig. 2.
The Notch1 C-terminal region interacts with Jagged1C2–EGF3, Jagged1EGF8–11, and Jagged1CRD in the Notch1fe–Jagged1fe complex. (A and B) Overview of the detected distance constraints from the XL-MS experiments for wild-type (A) and high-affinity (B) versions of Jagged1fe. (C) Schematic representation of the interactions reported in D–H, based on the XL-MS data and quantitative binding experiments. (D) SPR equilibrium-binding plots of Jagged1fe,HA to Notch1fe (black) and to Notch1EGF8–13 (blue). Jagged1fe,wt does not interact with Notch1EGF8–13 at 1 µM (red). (E) SPR equilibrium-binding plots of Notch1NRRΔloop to wild-type (black) and to high-affinity (red) versions of Jagged1C2–EGF3 and of Jagged1C2–EGF3,HA to Notch1EGF8–13 (blue). Notch1EGF33–36 does not bind to Jagged1C2–EGF3 constructs (orange). A Hill coefficient of 2 is used to model the Notch1NRRΔloop–Jagged1C2–EGF3 interactions (see also Materials and Methods). (F) MST-binding curve of Notch1NRR (black) and Notch1EGF33–NRR (blue) to Jagged1C2–EGF3,HA. (G and H) SPR equilibrium-binding plots indicate the interaction of dimerized Jagged1EGF8–11,Fc (G) and dimerized Jagged1CRD,Fc (H) to Notch1EGF33–NRR (black), but not to Notch1EGF22–27, that acts as negative control (blue). Nondimerized versions do not interact at 20 µM (orange). The Fc domain does not interact with Notch1EGF33–NRR, as shown by the IgG control at 5 µM (red).
Purified Notch1 and Jagged1 full ectodomain proteins were incubated at a 1:1 molar ratio to induce complex formation (i.e., Notch1fe–Jagged1fe,wt and Notch1fe–Jagged1fe,HA) and cross-linked with the lysine-targeting PhoX cross-linking reagent (44). In subsequent steps, the samples were subjected to deglycosylation, enriched for cross-linked peptides by immobilized metal affinity chromatography (IMAC), and finally analyzed by liquid chromatography coupled to tandem mass spectrometry. From three independent replicates for each complex, we detected 166 unique distance restraints for Notch1fe–Jagged1fe,wt and 232 for Notch1fe–Jagged1fe,HA. As an additional step to reduce false positives and remove distance constraints arising from nonspecific aggregation, we solely retained restraints detected in at least two out of three replicates (45). This reduced the output to 113 and 164 restraints for Notch1fe–Jagged1fe,wt and Notch1fe–Jagged1fe,HA, respectively (Fig. 2 A and B). For both complex samples, few intralinks were detected for Notch1fe (9 for Notch1fe–Jagged1fe,wt and 12 for Notch1fe–Jagged1fe,HA;SI Appendix, Fig. S2 A–D). The number of intralinks for Jagged1fe was, however, significantly larger and increased by 38% for the mutant (100 for Notch1fe–Jagged1fe,wt and 138 for Notch1fe–Jagged1fe,HA). A similar trend was visible in the number of intermolecular connections between Notch1 and Jagged1, in which three interlinks were detected for Notch1fe–Jagged1fe,wt and 13 for Notch1fe–Jagged1fe,HA. This identification of intra- and interlinks suggests that the mutant protein, Jagged1fe,HA, assisted by the stronger interaction between the two molecules, adopts a less flexible conformation, compared to Jagged1fe,wt, and provides more efficient complex formation that is beneficial for our approach (46).
The interlinks reveal that in the Notch1fe–Jagged1fe complex, three Jagged1 regions (C2–EGF1, EGF10, and CRD) are in proximity to the Notch1 EGF29–NRR site, with most interlinks arising from the Jagged C2–EGF1 region. The XL-MS experiments do not reveal any cross-links or monolinks between Notch1 EGF8–EGF12 and Jagged1 C2–EGF3 (Fig. 2 A and B and SI Appendix, Fig. S2A), the well-established interaction site (11) for which we find a KD of 0.3 µM by SPR using the high-affinity variant of Jagged1 C2–EGF3 (Fig. 2E and SI Appendix, Fig. S1C). There are two possible explanations for the lack of links to Notch1 EGF8–EGF12. 1) The two lysine residues in Notch1 EGF8–EGF12, Lys395 and Lys428, are occluded in the Notch1fe–Jagged1fe complex or 2) the lysines are occluded from the cross-linking reaction by O-linked glycans such as O-fucose residues, which we show are present in our Notch1 sample (SI Appendix, Fig. S2E and Dataset S3). The shotgun mass spectrometric analysis of noncross-linked Notch1fe covers the segment containing the two lysine residues within the Notch1 EGF8–EGF12 region, indicating that the relevant peptides can be identified (SI Appendix, Fig. S2A). A large part of the Notch1fe EGF repeat region is decorated with O-linked glycosylation sites, with an average of 1.5 sites per EGF domain, based on sequence prediction (47), and we cannot fully exclude that the glycans prevent the cross-linking reaction. Notably, however, 25 cross-links are identified in the Notch1 EGF29–EGF36 region, predicted to contain slightly less O-linked glycosylation sites (i.e., 1.1 sites per EGF domain) (47). Combined, these observations suggest that Notch1 EGF8–EGF12 is hidden in the folded Notch1 full ectodomain. Although the XL-MS analysis has not revealed all the interacting regions on Notch1 in the Notch1fe–Jagged1fe complex, it does indicate that the Notch1 C-terminal region plays an important role in the interaction with Jagged1.
Notch1 NRR Directly Interacts with Jagged1 C2–EGF3.
To further investigate interacting regions, we generated shorter Notch1 and Jagged1 constructs (SI Appendix, Fig. S3) and probed them by SPR and microscale thermophoresis (MST). The Notch1EGF33–NRR site interacts directly with Jagged1C2–EGF3 in MST (Fig. 2F and SI Appendix, Fig. S1G) and in SPR (SI Appendix, Fig. S4 A–C), and this interaction is independent of the high-affinity mutations in the C2 domain of Jagged1 (Table 1 and SI Appendix, Fig. S4 A–D). Jagged1C2–EGF3 is required and sufficient for the interaction with Notch1EGF33–NRR (SI Appendix, Fig. S4 A–F). The Notch1EGF33–NRR–Jagged1C2–EGF3 binding site was further defined to Notch1NRR, that binds with a KD of 0.6 µM to Jagged1C2–EGF3,HA, measured in solution by MST (Fig. 2F and SI Appendix, Fig. S1F), while Notch1EGF33–36 by itself does not interact with either Jagged1C2–EGF3,wt or Jagged1C2–EGF3,HA (Fig. 2E). In the NRR, a large, unstructured loop (consisting of 38 residues) that contains the heterodimerization S1 cleavage site (19, 48) is not required for interaction (Fig. 2E and SI Appendix, Fig. S1D). In addition, the interaction is not affected by the high-affinity mutations in Jagged1C2–EGF3, as the KD values determined by SPR for Notch1NRRΔloop binding to Jagged1C2–EGF3,wt or to Jagged1C2–EGF3,HA are similar (Fig. 2E and SI Appendix, Fig. 1 D and E). Docking of the Notch1NRR–Jagged1C2–EGF3 complex, using the intermolecular cross-links as restraints, suggests that Jagged1 domains DSL–EGF1 engage Notch1 NRR (SI Appendix, Fig. S5) (11). However, given that the two cross-link sites on Notch1 NRR are both in the flexible heterodimerization loop, which we show is not involved in the interaction (Fig. 2E), there is ambiguity in the location of the Notch1NRR–Jagged1C2–EGF3 interaction site. Taken together, our interaction data on the smaller Notch1 and Jagged1 portions show that the Notch1 NRR is responsible for the interaction with the Jagged1 C2–EGF3 region.
Table 1.
Summary of measured affinities
| Notch1NRRΔloop | Notch1NRR | Notch1EGF33–NRR | Jagged1fe,HA | Jagged1C2–EGF3,HA | Jagged1EGF8–11,Fc | Jagged1CRD,Fc | |
| Jagged1C2–EGF3,wt | 0.60 ± 0.03 | n.d. | 28 ± 2 | n.d. | n.d. | 0.34 ± 0.08 | 0.93 ± 0.13 |
| Jagged1C2–EGF3,HA | 0.37 ± 0.02 | 0.63 ± 0.18* | 0.50 ± 0.19* 15 ± 2 | n.d. | n.d. | 0.33 ± 0.07 | 0.57 ± 0.07 |
| Jagged1C2–EGF7,wt | n.d. | n.d. | 19 ± 3 | n.d. | n.d. | 0.40 ± 0.11 | 0.89 ± 0.12 |
| Jagged1C2–EGF13,wt | n.d. | n.d. | 8 ± 2 | n.d. | n.d. | 0.17 ± 0.03 | 0.26 ± 0.04 |
| Jagged1fe,wt | n.d. | n.d. | n.d. | n.d. | n.d. | 0.22 ± 0.06 | 0.68 ± 0.10 |
| Notch1fe | n.d. | n.d. | n.d. | 1.0 ± 0.1 | n.d. | n.d. | n.d. |
| Notch1EGF8–13 | n.d. | n.d. | 115 ± 8 | 1.5 ± 0.2 | 0.30 ± 0.03 | n.d. | n.d. |
| Notch1EGF33–NRR | n.d. | n.d. | n.d. | n.d. | n.d. | 0.29 ± 0.09 | 0.29 ± 0.10 |
All values are expressed in micromolars and derived from MST (*) or SPR experiments. n.d., not determined. Ligands and analytes are in the left column and top row, respectively.
Notch1 EGF33–NRR Contains Low-Affinity Sites for Jagged1 EGF8–EGF11 and Jagged1 CRD.
The XL-MS data of Notch1fe–Jagged1fe,HA indicates that two additional regions in Jagged1, EGF10 and CRD, are in proximity to the Notch1 EGF33–NRR site (Fig. 2B). SPR-binding experiments confirm the direct interactions to Notch1EGF33–NRR, albeit with much lower affinity than the Jagged1 C2–EGF3 region, with no binding of Jagged1EGF8–11 or Jagged1CRD to Notch1EGF33–NRR observed at a concentration of 20 µM (Fig. 2 G and H). To enhance a possible weak affinity, we employed a widely used strategy for cell- and surface-binding assays of artificially dimerizing proteins (49) that has previously been used to measure Notch interactions (25, 37). Fc-tagged versions of Jagged1EGF8–11 and Jagged1CRD, that are covalently dimerized by the Fc tag, interact both with an apparent dissociation constant (KD,app) of 0.29 µM to Notch1EGF33–NRR (Fig. 2 G and H and SI Appendix, Fig. S1 H and I).
Notch1fe Is Flexible and Has Intramolecular Interactions.
SAXS analysis coupled to size-exclusion chromatography (SEC-SAXS) shows that monomeric Notch1fe is a flexible molecule (Fig. 3 A and B), has a radius of gyration (Rg) of 105 ± 0.4 Å (Fig. 3C), and a maximum distance (Dmax) of 380 Å (Fig. 3D). This suggests that Notch1fe does not exist as an extended molecule, as it would have a Dmax of 1,027 Å for a fully elongated Notch1fe (see Materials and Methods), but instead has a nonlinear architecture. Backfolded models were previously suggested based on genetic (38) and interaction data (36, 37), in which the EGF domain connections were determined to confer flexibility to the Notch1 extracellular region (42). In addition, two parts in Notch1, EGF8–EGF13 and EGF33–NRR, interact with a KD of 115 µM (Fig. 3E and SI Appendix, Fig. S1J). While this is a relatively low affinity for an intermolecular interaction (i.e., as in a Notch1 dimer), it may be possible that these regions interact directly in an intramolecular fashion within the same Notch1 molecule. Overall, the nonlinear architecture suggests that EGF domains may become buried in the fully folded molecule, providing further support to the data obtained by XL-MS.
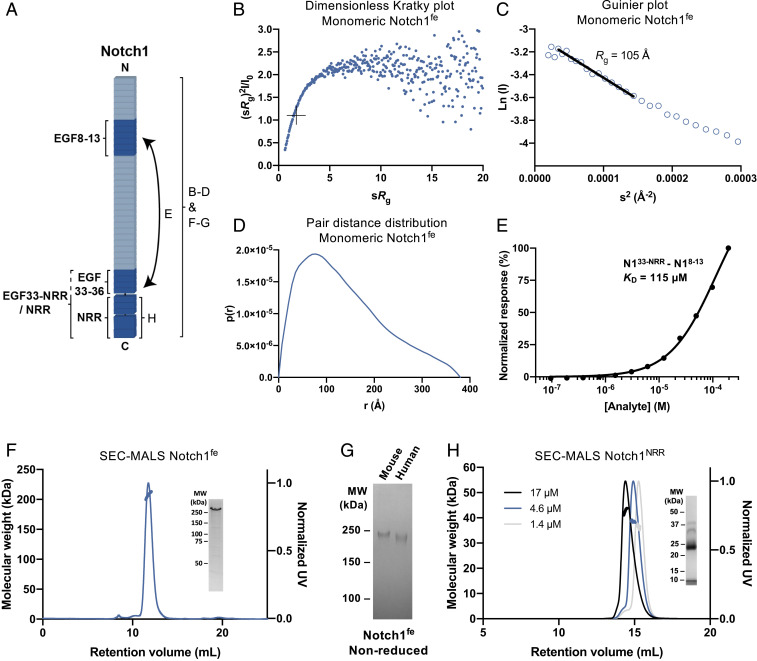
Fig. 3.
Notch1fe is flexible and the NRR dimerizes weakly. (A) Schematic representation of the interaction and biophysical experiments on regions reported in B–H. (B–D) Structural analysis of monomeric Notch1fe from SEC-SAXS, including the Dimensionless Kratky plot with crosshairs indicating the peak position for a globular protein (B), Guinier plot with a black line indicating the fit used to derive the Rg (C), and pair distance distribution function (D). (E) SPR equilibrium-binding plot of Notch1EGF33–NRR to Notch1EGF8–13. (F) SEC-MALS analysis of Notch1fe shows a monomeric and monodisperse sample (thick lines indicate the molecular weight, left axis). (Inset) Coomassie-stained SDS-PAGE of purified Notch1fe in reducing conditions. UV, ultraviolet. (G) Coomassie-stained SDS-PAGE of purified Notch1fe in nonreducing conditions. MW, molecular weight. (H) SEC-MALS analysis of Notch1NRR at three concentrations, determined at elution, shows a monomer–dimer equilibrium (thick lines indicate the molecular weight, left axis). (Inset) Coomassie-stained SDS-PAGE of purified Notch1NRR in reducing conditions, note that Notch1NRR is processed at the S1 cleavage site into two fragments of 8 and 27 kDa.
Notch1 Dimerizes through the NRR.
Human Notch1fe is a monomer at a concentration of 0.26 µM and has a molecular weight of 209 ± 2.4 kDa (Fig. 3F). This correlates well with the theoretical molecular weight of 200 to 220 kDa that is dependent on the glycosylation state (33, 50). Although mouse Notch1fe has an additional cysteine at EGF25, it does not form a covalent homodimer (Fig. 3G). Interestingly, our XL-MS data showed that Notch1fe can form dimers, which can be detected by XL-MS when the same residue in the protein sequence is linked by two different peptides induced by, for example, a missed cleavage. One self-link at lysine residue 1,314 in EGF34 arises from an intermolecular Notch1–Notch1 interaction (Fig. 2B). In addition, the Notch1 NRR itself (Notch1NRR) undergoes weak concentration-dependent dimerization during SEC coupled to multiangle light scattering (SEC-MALS) analysis at concentrations ranging from 1.4 to 17 µM (Fig. 3H). Dimerization of the NRR has previously been reported for Notch3 and was predicted for the Notch1 NRR, based on similarities in crystal packing comparing the NRR of Notch3 and Notch1 (51–53). The NRR-controlled dimerization of Notch3 may maintain the receptor in an autoinhibited state before ligand binding (53). We determined a crystal structure of the S1-cleaved mouse Notch1 NRR (SI Appendix, Fig. S6 A–C; Protein Data Bank [PDB]: 7ABV) that shows the same dimerization interface as its human ortholog (51, 52). N-linked glycans, that do not seem to interfere with dimerization, are visible in the electron density at position N1489, as also reported previously (54), and additionally at position N1587 (SI Appendix, Fig. S6A). Taken together, the XL-MS analysis on Notch1fe and dimerization of Notch1NRR indicate that Notch1 can dimerize through the membrane–proximal region.
Jagged1fe Has a Nonlinear Architecture and Oligomerizes.
Jagged1fe,HA has a weak propensity to dimerize. Up to a concentration of 1.6 µM, Jagged1fe,HA is a monomer with a molecular weight of 137 ± 0.2 kDa (Fig. 4 A and B) that correlates well with the theoretical molecular weight of 120 to 140 kDa, depending on the glycosylation state (55, 56). At higher concentrations, Jagged1fe forms oligomers (Fig. 4 C–E). In sedimentation velocity analytical ultracentrifugation (SV-AUC), at 5 µM, 19% of Jagged1fe,HA consists of oligomers, and this increases to 31% at 20 µM (Fig. 4C). Concentration-dependent dimerization is also supported by batch SAXS analysis. At 5 µM, the Rg of Jagged1fe,HA is 81.2 ± 0.8 Å, and this increases to 102 ± 0.4 Å at 42 µM (Table 2 and Fig. 4D), indicating that more Jagged1fe,HA dimers or larger oligomeric species are present at a higher concentration.
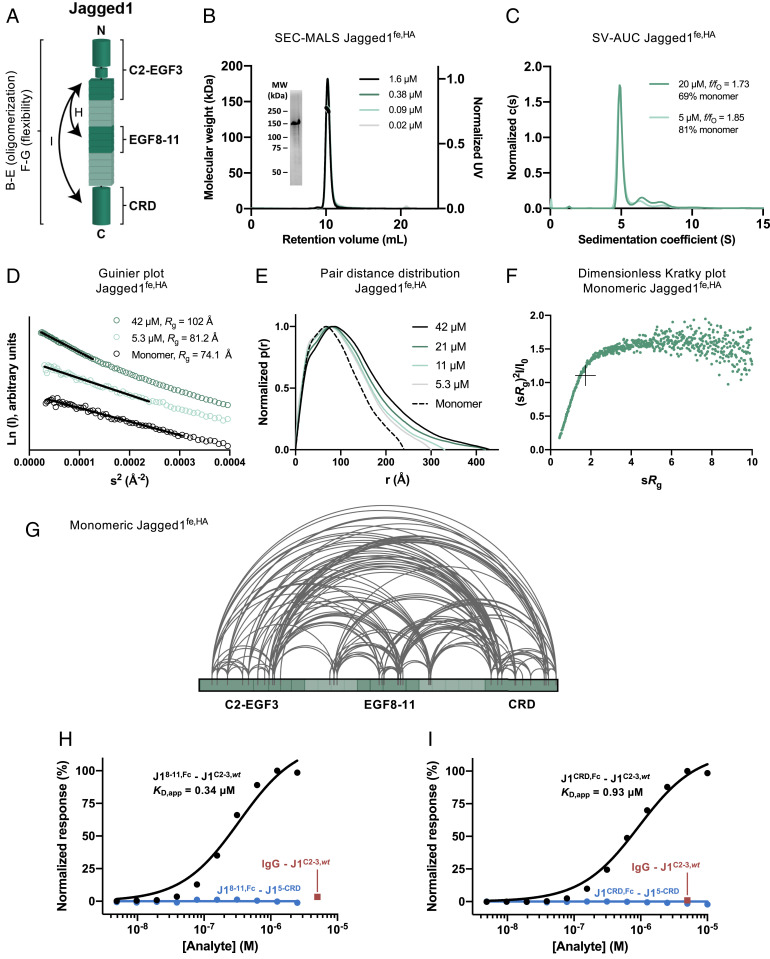
Fig. 4.
Jagged1fe,HA is not fully extended, is flexible and oligomerizes weakly. (A) Schematic representation of the interactions and biophysical experiments on regions reported in B–I. (B) SEC-MALS analysis of Jagged1fe,HA at four concentrations, determined at elution, shows overlapping monomeric and monodisperse peaks (thick lines indicate the molecular weight, left axis). (Inset) Coomassie-stained SDS-PAGE of the purified sample in reducing conditions. MW, molecular weight; UV, ultraviolet. (C) SV-AUC analysis shows that Jagged1fe,HA oligomerizes in a concentration-dependent manner. (D–F) SAXS analysis of Jagged1fe,HA in batch and from monomeric SEC-SAXS fractions, including the Guinier plot with black lines indicating the fits used to derive the Rg (D), pair distance distribution function (E), and dimensionless Kratky plot with crosshairs indicating the peak position for a globular protein (F). (G) Overview of the detected distance constraints from the XL-MS experiments for monomeric Jagged1fe,HA. (H and I) SPR equilibrium-binding plots indicate the interaction of Jagged1EGF8–11,Fc (H) and Jagged1CRD,Fc (I) to Jagged1C2–EGF3 (black), but not to Jagged1EGF5–CRD, that acts as negative control (blue). The Fc domain does not interact with Jagged1C2–EGF3,wt, as shown by the IgG control at 5 µM (red).
Table 2.
Structural parameters derived from SAXS experiments
| Concentration (µM) | Rg (Å) Guinier | sRg range used in Guinier for Rg | Rg (Å) P(r) | Dmax (Å) | I0 (cm−1) | |
| Notch1fe SEC-SAXS | n/a | 105 ± 0.2 | 0.62–1.26 | 113 | 380 | 0.047 ± 5.7 × 10−4 |
| Jagged1fe,HA SEC-SAXS | n/a | 74.1 ± 0.6 | 0.44–1.29 | 74.3 | 240 | 0.07 ± 4.4 × 10−4 |
| Jagged1fe,HA Batch | 42 | 102 ± 0.4 | 0.49–1.15 | 110 | 430 | 0.26 ± 5.8 × 10−4 |
| 21 | 96.4 ± 0.7 | 0.49–1.08 | 103 | 430 | 0.23 ± 8.7 × 10−4 | |
| 11 | 89.2 ± 1.0 | 0.49–1.10 | 90.2 | 330 | 0.19 ± 1.1 × 10−3 | |
| 5.3 | 81.2 ± 0.8 | 0.45–1.25 | 85.3 | 300 | 0.16 ± 1.2 × 10−3 | |
| Jagged1EGF8–11 Batch | 230 | 31.7 ± 0.1 | 0.62–1.12 | 32.7 | 120 | 0.044 ± 6.4 × 10−5 |
| 115 | 31.3 ± 0.1 | 0.69–1.22 | 32.7 | 115 | 0.045 ± 8.4 × 10−5 | |
| 58 | 31.5 ± 0.1 | 0.56–1.26 | 32.8 | 115 | 0.046 ± 1.0 × 10−4 | |
| 29 | 32.7 ± 0.4 | 0.64–1.16 | 32.5 | 110 | 0.047 ± 3.0 × 10−4 | |
| Jagged1CRD Batch | 167 | 24.1 ± 0.0 | 0.40–1.09 | 24.3 | 90 | 0.036 ± 2.6 × 10−5 |
| 83 | 23.3 ± 0.0 | 0.18–1.16 | 23.3 | 82 | 0.036 ± 3.0 × 10−5 | |
| 42 | 22.6 ± 0.1 | 0.21–1.29 | 22.8 | 78 | 0.036 ± 4.4 × 10−5 | |
| 21 | 22.7 ± 0.1 | 0.21–1.30 | 22.8 | 75 | 0.035 ± 7.5 × 10−5 |
SAXS batch data I0 have been normalized by the sample concentration to allow for the comparison between samples. Nonnormalized I0 values are available on SASBDB under the accession codes defined in Data Availability. n/a, not applicable.
We used SEC-SAXS to separate monomeric Jagged1fe,HA from oligomeric species. The region at the right side of the Jagged1fe,HA elution peak (i.e., at larger retention volume) was selected for further analysis, as this region most likely represents a monomeric fraction. Jagged1fe,HA has an Rg of 74.1 ± 0.6 Å (Fig. 4D) and a Dmax of 240 Å (Fig. 4E). The normalized Kratky plot indicates that structural flexibility is present in the Jagged1 ectodomain (Fig. 4F). SAXS analysis of smaller Jagged1 portions, Jagged1EGF8–11 and Jagged1CRD (SI Appendix, Fig. S7 A–H), show that both samples do not change their oligomeric state at different concentrations (Table 2 and SI Appendix, Fig. S7 B and F). While Jagged1EGF8–11 is flexible (SI Appendix, Fig. S7D), Jagged1CRD is compact and globular (SI Appendix, Fig. S7H). The measured Dmax of 240 Å indicates that monomeric Jagged1fe,HA is not extended, as a fully elongated Jagged1 ectodomain would have a maximum dimension of 585 Å (see Materials and Methods). In agreement with the SAXS data, the XL-MS analysis suggests that the extracellular region of Jagged1 is not fully extended (Fig. 2 A and B). The detected distance restraints arise from either intra- or intermolecular Jagged1fe interactions, as Jagged1fe may be dimerizing in this experiment. To isolate the intramolecular cross-links from the ambiguous intra- and intermolecular cross-links, we repeated the cross-linking experiment with Jagged1fe,HA and separated monomeric Jagged1fe,HA from cross-linked Jagged1fe,HA oligomers by SEC (SI Appendix, Fig. S2B) and analyzed the cross-links of both fractions by mass spectrometry (MS). The data indicate that four regions of the Jagged1 extracellular segment (C2–EGF2, EGF5–EGF6, EGF9–EGF12, and CRD) are in proximity within the same Jagged1fe,HA molecule, as most identified cross-links are present in the monomeric (as well as in the oligomeric) fraction (Fig. 4G and SI Appendix, Fig. S2C). Most of these intramolecular cross-links are also found in the Notch1fe–Jagged1fe,wt and Notch1fe–Jagged1fe,HA XL-MS datasets, indicating that these intramolecular cross-links are independent of Notch1fe binding.
We used SPR to verify that the regions identified by XL-MS interact directly. Constructs consisting of the Jagged1 regions C2–EGF3, EGF8–EGF11, and CRD reveal direct interactions between Jagged1C2–EGF3 and Jagged1EGF8–11 and between Jagged1C2–EGF3 and Jagged1CRD, supporting the XL-MS results. The interactions are weak, as covalent dimerization by Fc-fusion was required to measure binding. Fc-Jagged1EGF8–11 and Fc-Jagged1CRD bound to Jagged1C2–EGF3,wt, with a KD,app of 0.34 and 0.93 µM, respectively (Fig. 4 H and I and SI Appendix, Fig. S1 K and L). The C2–EGF3 region is required and sufficient for these interactions. Both Fc-Jagged1EGF8–11 and Fc-Jagged1CRD do not interact with Jagged1EGF5–CRD that is lacking the C2–EGF3 region (Fig. 4 H and I), and affinities are similar for larger constructs that include the C2–EGF3 region (i.e., Jagged1C2–EGF7, Jagged1C2–EGF13, and Jagged1fe) (Table 1). In addition, the Jagged1 high-affinity mutations (11) do not affect this interaction (Table 1). Taken together, the SPR and XL-MS data indicate that the EGF8–EGF11 and CRD regions interact intramolecularly with the C2–EGF3 region within the Jagged1 molecule.
Discussion
Two regions in Notch, EGF11–EGF12 and NRR, have been widely studied because of their critical role in Notch signaling (10, 15, 20, 21, 48, 57) and represent the minimal requirements for ligand-dependent Notch activation (19, 58). Transcellular ligand binding at the Notch1 EGF8–EGF12 site, positioned far away from the NRR in the primary sequence, and subsequent Notch1–ligand endocytosis generate a mechanical pulling force (9–14, 16) that could be transmitted via EGF13–EGF36 to the NRR, in which it triggers a conformational change to expose the S2 site for proteolytic cleavage (19–21). Ligand binding in cis can inhibit Notch activation (29–31), while it was recently shown that it could also stimulate Notch activation (32), although it is not clear if and how endocytosis plays a direct role in this setting. These studies raise the question of how the different regions within Notch1 and Jagged1 interact.
Here, we show that the Jagged1 C2–EGF3 segment is in close proximity to the Notch1 NRR in the Notch1fe–Jagged1fe complex, that Notch1EGF8–13 and Notch1NRR can interact directly with the C2–EGF3 region in Jagged1, and that Notch1EGF8–13 interacts with Notch1EGF33–NRR (Fig. 5A). We confirm that the Notch1 ectodomain has regions of flexibility (36, 37, 42), which suggests that the EGF8–EGF13 and the EGF33–NRR segments in Notch1 can interact intramolecularly. In addition to the importance of the canonical ligand-binding site, EGF8–EGF12, and the proteolytic activation site, NRR, in Notch, other regions have previously been proposed to play a role in Notch function (33, 34, 36–39). Intramolecular interactions have been determined between Notch EGF8–EGF12 and EGF22–EGF27 (36) and were suggested to occur for Notch EGF8–EGF12 and EGF25–EGF26 by demonstrating that antibodies targeting EGF25–EGF26 prevent Jagged1-mediated full length Notch activation (37). In a deletion study, Notch EGF25–EGF36 was shown to play a role in the interaction with Serrate (38). Specific regions on Notch, namely EGF24–EGF26 (39), O-linked fucosylation on EGF26 (34), and O-fucose extension with GlcNAc on EGF6 and EGF36 (33), play a role in Jagged/Serrate-mediated signaling. Some of these studies highlight the importance of the membrane–proximal region of the Notch ectodomain [e.g., EGF25–EGF36 (38) and EGF36 (33)]. These sites are next to or include the region we identify in Notch1–Jagged1 and Notch1–Notch1 interactions by XL-MS and quantitative binding assays (Figs. 2 A, B, and E and 3E and SI Appendix, Fig. S4B). The interaction of Jagged1 C2–EGF3 with the membrane–proximal Notch1 NRR fits well with the previously shown lipid-binding role of the Jagged1 C2 domain and the requirement of C2-lipid binding for optimal Notch activation (26, 28). In addition, the interactions of Jagged1CRD with Jagged1C2–EGF3 and with Notch1EGF33–NRR (Figs. 2 B and H and 4I) support the finding that the CRD is involved in signaling (40). Collectively, our work and that of others indicate that several sites in the Notch and Jagged extracellular segments contribute to Notch–Jagged interactions and signaling.
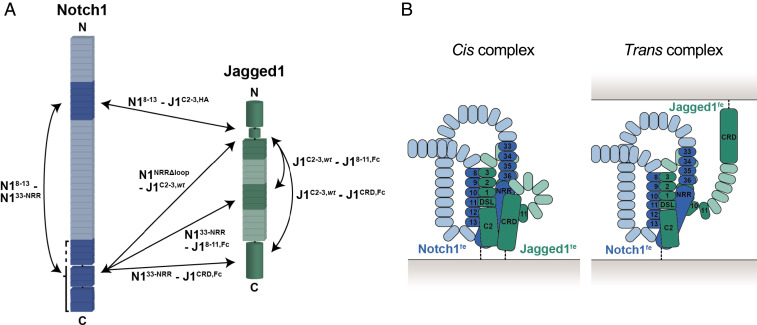
Fig. 5.
Summary of the reported direct interactions and possible architectures of the complex. (A) Inter- and intramolecular interactions based on the XL-MS and quantitative interaction experiments are indicated by double arrows. (B) Schematic architectures of the Notch1–Jagged1 full ectodomain complex based on the interaction data shown in A, represented in a cis or trans setting. Not all interactions might occur simultaneously, as reflected by the trans complex in which Jagged1 CRD is not contributing to interactions. The domains enabling backfolding have not been determined experimentally.
The various segments have different interaction strengths. The interaction of the Notch1 ectodomain and that of Jagged1 is weak but strengthened by a pulling force (11). The mutation of five residues in the Jagged1 C2 domain increases the affinity of the Jagged1 ectodomain for the Notch1 ectodomain to 1 µM (Fig. 2D), indicating that the Jagged1 C2 domain plays an important role in the interaction with Notch1. Surprisingly, the measured interaction between Notch1NRRΔloop and Jagged1C2–EGF3 also has a KD of about 1 µM and is not dependent on the high-affinity mutations (Fig. 2E). While this interaction may be influenced in the SPR experiment by an avidity effect, arising from dimerization of the NRR, the interaction measured between Notch1NRR and Jagged1C2–EGF3,HA in solution using MST also shows a KD of around 1 µM (Fig. 2F). The interaction of the larger Notch1EGF33–NRR with Jagged1C2–EGF3 shows a similar affinity with a KD of 0.5 µM measured by MST (Fig. 2F), whereas it is 30-fold weaker in the surface-based SPR method (Table 1 and SI Appendix, Fig. S4B), which indicates that the context of this interaction may be important. Taken together, these data show that the NRR in the Notch1 ectodomain is in direct contact to the Jagged1 C2–EGF3 region in the Notch1fe–Jagged1fe complex and suggest that ligand binding is directly coupled to Notch activation or regulation.
The setting at the cell surface or between two cells may dictate how Notch1 and Jagged1 interact. In our experiments, we cannot discriminate between cis and trans interactions, and it may be possible we see both types of interactions simultaneously (Fig. 5B). For example, the interaction of the membrane proximal regions (i.e., Notch1 EGF33–NRR and Jagged1 CRD) seems more likely in a cis setting with both molecules expressed on the same cell. At the same time, the receptor and the ligand may undergo homomeric interactions on the cell surface which influences Notch signaling further (43, 53, 59–62). Besides the C2–EGF3 region, we have identified additional Jagged1 segments, namely EGF8–EGF11 and CRD, that interact intermolecularly with Notch1 EGF33–NRR, as well as intramolecularly with Jagged1 C2–EGF3 (Fig. 5A), and these regions could have a role in the clustering of Jagged1 and the Notch1–Jagged1 complex on or between cells. The interactions that we identify as intramolecular (i.e., Notch1 EGF8–EGF13 with EGF33–NRR and Jagged1 C2–EGF3 with EGF8–EGF11 and CRD) may instead be used for intermolecular interactions when the proteins are expressed in a cell-surface setting. The role of the interactions in the function of Notch1 and Jagged1, whether they are intra- or intermolecular, occur in cis or in trans and, simultaneously or not, will need to be determined. In addition, it is currently not clear whether the Notch1 NRR—Jagged1 C2–EGF3 and Notch1 EGF8–EGF13—NRR interactions are common features for the Notch and DSL family members. Interestingly, despite differences in domain composition, these three regions are present in all members (i.e., all Notch paralogs contain the EGF8–EGF13 and NRR segments) and all DSL ligands have the C2–EGF3 region in common. Our data indicate that a mosaic of interaction sites is present, both on Notch1 and on Jagged1. Targeting these interactions may reveal their role in Notch signaling and could have potential for therapeutic applications to treat Notch-associated disorders.
Materials and Methods
Generation of Constructs and Mutagenesis.
Notch1 and Jagged1 constructs were generated by PCR using mouse Notch1 (Addgene 41,728), human Notch1 (kind gift of Warren Pear, University of Pennsylvania, Philadelphia, PA), and mouse Jagged1 (Image clone 6,834,418) as templates. All constructs are mouse version unless stated otherwise. Notch1fe (residue numbers 19 to 1,717) was subcloned in pUPE106.03 (U-Protein Express BV, cystatin secretion signal peptide, and N-terminal His6-tag), Notch1fe (19 to 1,728, human version), Notch1EGF8–13 (294 to 526), Notch1EGF22–27 (828 to 1,058), Notch1EGF33–36 (1,267 to 1,426), Notch1EGF33–NRR (1,267 to 1,717), Notch1NRR (1,446 to 1,717) with and without its unstructured loop (1,622 to 1,659), Jagged1fe (31 to 1,067), Jagged1C2–EGF3 (31 to 334), Jagged1C2–EGF7 (31 to 485), Jagged1C2–EGF13 (31 to 741), Jagged1EGF5–13 (374 to 741), Jagged1EGF5–CRD (374 to 1,067), Jagged1EGF8–11 (487 to 665), and Jagged1CRD (857 to 1,067) were subcloned in pUPE107.03 (U-Protein Express BV, cystatin secretion signal peptide, and C-terminal His6-tag). Jagged1 mutations (S32L, R68G, D72N, T87R, and Q182R) based on Luca et al. (11) were introduced using Q5 site-directed mutagenesis to generate Jagged1fe,HA (31 to 1,067) and Jagged1C2–EGF3,HA (31 to 334) constructs. In several figures, Notch1 and Jagged1 constructs are referred to as N1 and J1, respectively, and EGF repeats are referred to as their number (i.e., J1C2–3 for Jagged1C2–EGF3).
Large-Scale Expression and Purification.
Constructs were transiently expressed in N-acetylglucoaminyltransferase I–deficient, Epstein–Barr virus nuclear antigen 1–expressing human embryonic kidney 293 (HEK293) cells growing in suspension (U-Protein Express BV), allowing for homogeneous N-glycosylations of the oligomannose type. With an open search approach (see LC-MS and Data Analysis), we identified a core fucose modification (O-fucose) on four residues (T116, T194, T617, and T1362) (SI Appendix, Fig. S2E and Dataset S3). The medium was harvested 6 d after transfection, cells were spun down by 10 min of centrifugation at 1,000 × g, and cellular debris was spun down for 15 min at 4,000 × g. For human Notch1fe used in the SEC-MALS experiment, the supernatant was concentrated fivefold and diafiltrated against 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) pH 8.0, 500 mM NaCl, and 2 mM CaCl2 (IMAC A) using a Quixstand benchtop system (GE Healthcare) with a 10-kDa molecular weight cutoff (MWCO) membrane. Cellular debris were spun down for 10 min at 9,500 × g, and the concentrate was filtered with a glass fiber prefilter (Minisart, Sartorius). Protein was purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and eluted with a mixture of 60% IMAC A and 40% of 25 mM Hepes pH 8.0, 500 mM NaCl, 500 mM imidazole, and 2 mM CaCl2 (IMAC B). For all other constructs and experiments, cells were spun down by 10 min of centrifugation at 1,000 × g, cellular debris was spun down for 15 min at 4,000 × g, and protein was directly purified by Ni Sepharose excel (GE Healthcare) affinity chromatography. The protein was eluted with a mixture of 60% of IMAC C (same as IMAC A, except pH 7.4) and 40% of IMAC D (same as IMAC B, except pH 7.4) or with 100% of IMAC D. SEC was performed on either a Superose6 10/300 increase (GE Healthcare) or a Superdex200 10/300 increase (GE Healthcare) equilibrated in SEC buffer (20 mM Hepes pH 7.4, 150 mM NaCl, and 2 mM CaCl2). Protein purity was evaluated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. Protein was concentrated and then stored at −80 °C.
Protein Cross-linking with PhoX.
XL-MS was performed according to a previously optimized protocol (63). The optimal cross-linker concentration was established with SDS-PAGE. Cross-linking reactions were performed in triplicates with equimolar inputs of each protein for Notch1fe–Jagged1fe,wt and for Notch1fe–Jagged1fe,HA. A total of 42 µL protein solution, composed of the preincubated Notch1fe–Jagged1fe,wt or Notch1fe–Jagged1fe,HA complex at 5 µM in 20 mM Hepes pH 7.4, 150 mM NaCl, and 2 mM CaCl2, were mixed with 5 µL of crosslinker solution composed of 10 mM PhoX in pure DMSO. Final concentrations of Ca2+ and PhoX during the XL-MS experiment were therefore 1.8 and 1.1 mM, respectively. The sample mixtures were filtered through MWCO 10-kDa filters (Vivaspin) into 10 mM Tris pH 7.5 in a 3:1 ratio (volume:volume) to a final volume of 25 µL. Prior to protein digestion, samples were deglycosylated overnight with Deglycosylation Mix II (NEBB), which predominantly targets N-linked glycans. After deglycosylation, urea was added to a final concentration of 8 M followed by the addition of Tris(2-carboxyethyl)phosphine and chloroacetamide to a final concentration of 10 and 40 mM, respectively. Samples were incubated at 37 °C for 1 h, and then, proteolytic digestion was performed with LysC (Wako) for 4 h and trypsin (Promega) overnight. Resulting peptide mixtures were desalted with Oasis HLB plates (Waters), dried, and stored at −80 °C until further use.
Automated Fe(III)-IMAC–Based Enrichment.
Cross-linked peptides were enriched with Fe(III)-NTA 5 μL in an automated fashion using the AssayMAP Bravo Platform (Agilent Technologies). Fe(III)-NTA cartridges were primed with 250 μL 0.1% TFA in ACN and equilibrated with 250 μL loading buffer (80% ACN/0.1% TFA). Samples were dissolved in 200 μL loading buffer and loaded onto the cartridge. The columns were washed with 250 μL loading buffer, and the cross-linked peptides were eluted with 25 μL 10% ammonia directly into 25 μL 10% formic acid. Samples were dried down and stored in 4 °C until subjected to liquid chromatography MS (LC-MS). For LC-MS analysis, the samples were resuspended in 10% formic acid.
LC-MS and Data Analysis.
All MS data were acquired using an ultra-high-performance liquid chromatography 1290 system (Agilent Technologies) coupled online to an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). Peptides were trapped (Dr. Maisch Reprosil C18, 3 μm, and 2 cm × 100 μm) prior to separation on an analytical column (Agilent Poroshell EC-C18, 2.7 μm, and 50 cm × 75 μm). Trapping was performed by flushing in buffer A (0.1% volume/volume formic acid in water) for 10 min. Reversed phase separation was performed across a gradient of 10 to 40% buffer B (0.1% volume/volume formic acid in 80% volume/volume ACN) over 90 min at a flowrate of ∼300 nL/min. The instrument was operated in data-dependent fragmentation (MS2) mode with MS1 spectra recorded in the range 350 to 1,400 Th and acquired in the Orbitrap at a resolution of 60,000 with an AGC of 4 × 105 and a maximum injection time of 50 ms. For MS2, the cycle time was set to 3 s with a charge state inclusion set to 3 to 8 for the enriched fraction and 2 to 8 for the flow through. Dynamic exclusion was set to 12 s at 1.4 Th mass deviation. Stepped HCD was performed with the Ion Trap at NCE = 35 (±10%) and acquired in the Orbitrap at a resolution of 30,000 with AGC set at 1 × 105 maximum injection time to 120 ms.
To quality control whether the O-linked glycans were successfully incorporated during protein expression, we performed an open search against the full sequence of Notch1. Identifications were filtered on whether they conform to the correct precursor mass offset for fucose, identify the peptide with high confidence, contain diagnostic ion(s) indicative for fucose, and match the expected sequence motif. This analysis does not exclude that other sites are modified by O-linked glycans, as the data were not acquired in a mode geared toward glycan identification. The cross-linked peptides were analyzed with Thermo Proteome Discoverer (2.3.0.522) with incorporated XlinkX/PD nodes (63). The analysis was run with standard parameters in NonCleavable mode at 1% false discovery rate at the level of the cross-link spectrum match (CSM) and cross-link tables against a manually created database with the target proteins and 200 random decoy entries. As fixed modification carbamidomethyl (C) was set and as variable modification oxidation (M), acetyl (protein N-term), and Asn>Asp (N) (H−1, N−1, and O). As cross-linking reagent PhoX (C8, H3, O5, and P) was set, only cross-links detected in two out of three replicates were used for further analysis. The normal and monolinked peptides were analyzed with MaxQuant (1.6.17.0) (64). The analysis was run with standard settings applied using the same database to search the spectra. As fixed modification carbamidomethyl (C) was set, and as variable modification oxidation (M), acetyl (protein N-term), PhoX Tris (K) (C12, H14, N, O8, and P), PhoX H2O (K) (C8, H5, O6, and P), and Asn>Asp (N) (H−1, N−1, and O). Further downstream analysis and visual representation of the results was performed with the R scripting and statistical environment (65) using Circos (66) for data visualization.
Integrative Modeling and Docking of Notch1 NRR and Jagged1 C2–EGF3.
To the crystal structure of Notch1 NRR described here (PDB: 7ABV), the missing flexible loop modeled with trRosetta (67) was added (i.e., residues 1,622 to 1,659). A structure of mouse Jagged1 C2–EGF3 was generated by homology modeling in ITASSER (68), based on the rat high-affinity Jagged1 variant template (PDB: 5UK5) (11). Next, Notch1 NRR with the added loop and Jagged1 C2–EGF3 were docked together with three XL-MS–based restraints from these regions and defined as 5 to 25 Å distance restraints in the HADDOCK version 2.4 webserver (69) (SI Appendix, Fig. S5). The loop was defined as fully flexible, and the resulting outputs of the complex were examined in terms of scores with the emphasis on the biological relevance and restraints energy violations. University of California San Francisco ChimeraX (70) was used for visualization.
Surface Plasmon Resonance.
SPR ligand constructs subcloned in frame in pUPE107.62 (cystatin secretion signal peptide and C-terminal biotin acceptor peptide tag, followed by a C-terminal His6-tag) were biotinylated in HEK293 cells by cotransfection with Escherichia coli BirA biotin ligase, with a suboptimal secretion signal (in a pUPE5.02 vector) using a DNA ratio of 9:1 (sample:BirA, mass/mass). Additional sterile biotin (100 µL of 1 mg/mL Hepes-buffered biotin per 4 mL HEK293 culture) was supplemented to the medium. Protein was purified from the medium by Ni Sepharose excel (GE Healthcare) affinity chromatography. Purity was evaluated by SDS-PAGE and Coomassie staining. C-terminally biotinylated proteins were spotted on a P-STREP SensEye (Ssens) chip with a Continuous Flow Microspotter (Wasatch Microfluidics) using an 8 × 6 format. SEC buffer with 0.005% Tween 20 was used as a spotting buffer, and the coupling was quenched using 1 mM biotin in SEC buffer. Proteins were therefore C-terminally coupled to the chip to ensure a native topology. SPR experiments were performed on an MX96 Surface Plasmon Resonance imager (IBIS Technologies). Analytes in SEC buffer were flowed over the sensor chip, and SEC buffer with 0.005% Tween 20 was used as a running buffer. Temperature was kept constant at 25 °C. The data were analyzed using SprintX (IBIS Technologies) and Prism (Graphpad) and modeled with a 1:1 Langmuir-binding model to calculate the KD and the maximum analyte binding (Bmax). Since the NRR dimerizes, and bound with positive cooperativity to Jagged1C2–EGF3 when it was used as an analyte, we fitted SPR equilibrium-binding plots using a Hill equation with a Hill coefficient of 2. For the experiments in which full regeneration could not be achieved, the subsequent analyte injections were not zeroed in order to keep the Bmax constant (SI Appendix, Fig. S1 D, E, I, and L).
MST.
Jagged1C2–EGF3,HA in SEC buffer was labeled with NT-547 dye (NanoTemper Technologies) according to the manufacturer’s instructions. Unlabeled Notch1EGF33–NRR and Notch1NRR in SEC buffer were serially diluted from 50 µM to 3.0 (Notch1EGF33–NRR) or 1.5 nM (Notch1NRR) and incubated with 50 nM labeled Jagged1C2–EGF3,HA in the presence of 0.025% Tween 20 for 15 min at room temperature. Samples were transferred to Standard Treated Capillaries (NanoTemper Technologies) and run at 50% excitation power on a Monolith NT.115 (NanoTemper Technologies) at a constant temperature of 25 °C. KD was determined according to the law of mass action using the program MO Affinity Analysis (NanoTemper Technologies) and results were plotted using Prism (Graphpad).
Small-Angle X-Ray Scattering.
Notch1fe SEC-SAXS experiments were carried out at the European Synchrotron Radiation Facility (ESRF) beamline BM29. A total of 500 µL 8.1-µM human Notch1fe were loaded on a Superose6 10/300 increase column (GE Healthcare) equilibrated in SEC buffer, via a high-performance liquid chromatography (HPLC) system (Shimadzu). A stable background signal was confirmed before measurement. Measurements were performed at room temperature at a flow rate of 0.5 mL/min. SAXS data were collected at a wavelength of 0.99 Å using a sample-to-detector (Pilatus 1M, Dectris) distance of 2.85 m. The scattering of pure water was used to calibrate the intensity to absolute units. A total of 2,000 frames of 2 s each were collected, and data reduction was performed automatically using the EDNA pipeline (71). Frames with a stable Rg (± 10%) and buffer frames were selected for further analysis using Chromixs (72). Data were analyzed in Primus (73) and Scatter (74), and results were plotted in Prism (Graphpad). The maximum dimension of 1,027 Å for a theoretical elongated Notch1 ectodomain was calculated as follows: an average of 27 Å for the 36 EGF repeats (11) and 55 Å for the NRR (51).
Jagged1fe,HA SEC-SAXS experiments were carried out at the Diamond Light Source (DLS) beamline B21 operating at an energy of 12.4 keV and using a sample-to-detector (Eiger 4M, Dectris) distance of 4.01 m. A total of 45 µL of 42-µM Jagged1fe,HA were loaded on a Superose6 3.2/300 increase (GE Healthcare), equilibrated in SEC buffer via an HPLC system (Agilent). A stable background signal was confirmed before measurement. Measurements were performed at room temperature at a flow rate of 0.075 mL/min. The scattering of pure water was used to calibrate the intensity to absolute units. A total of 620 frames of 3 s each were collected, and data reduction was performed automatically using the DAWN pipeline (75). Frames with a stable Rg and buffer frames were selected for further analysis using Chromixs (72). Data were analyzed in Primus (73) and Scatter (74), and results were plotted in Prism (Graphpad).
Jagged1EGF8–11, Jagged1CRD, and Jagged1fe batch SAXS experiments were carried out at the DLS beamline B21 operating at an energy of 12.4 keV and using a sample-to-detector (Eiger 4M, Dectris) distance of 4.01 m. The scattering of pure water was used to calibrate the intensity to absolute units. Data reduction was performed automatically using the DAWN pipeline (75). Frames were averaged after being manually inspected for radiation damage, the scattering of the SEC buffer was subtracted, and intensities were normalized by the concentration. Data were analyzed in Primus (73) and Scatter (74), and results were plotted in Prism (Graphpad). The maximum dimension of 585 Å for a theoretical elongated Jagged1 ectodomain was calculated as follows: 160 Å for the C2–EGF3 region as measured from its crystal structures (11, 26), an average of 27 Å for each of the remaining 13 EGF domains (11), and 75 Å as determined for the C-terminal CRD by SAXS (SI Appendix, Fig. S7G).
Multiangle Light Scattering.
SEC-MALS was performed using a Superose6 10/300 increase (GE Healthcare) column for Notch1fe (human version) or a Superdex 10/300 increase (GE Healthcare) column for Jagged1fe,HA and Notch1NRR, equilibrated in SEC buffer. For molecular weight characterization, light scattering was measured with a miniDAWN TREOS MALS detector (Wyatt Technology) connected to an RID-10A differential refractive index monitor (Shimadzu) for quantitation of the protein concentration. Chromatograms were collected, analyzed, and processed on the ASTRA software suite (Wyatt Technology). A specific refractive index increment (dn/dc) of 0.1800 was calculated for Notch1fe based on six N-glycosylation sites of the oligomannose type and 55 O-glycosylation sites (two sugar moieties per site), 0.1814 for Jagged1fe,HA based on nine N-glycosylation sites and 16 O-glycosylation sites (four O-glucosylation sites extended with two xylose moieties each and 12 O-fucosylation sites), and 0.1828 for Notch1NRR based on two N-glycosylation sites.
Crystallization and Data Collection.
The Notch1 NRR was crystallized by sitting-drop vapor diffusion at 18 °C by mixing 200 nL protein solution containing a mixture of Notch1NRR and Jagged1C2–EGF3,HA at 8.5 mg/mL in SEC buffer and 100 nL reservoir solution, composed of 2.0 M sodium chloride and 0.1 M sodium acetate pH 4.6. The protein solution was deglycosylated beforehand using EndoHf 1:100 (volume/volume) overnight at room temperature in SEC buffer. The crystal was harvested and flash cooled in liquid nitrogen in the presence of reservoir solution supplemented with 25% glycerol. The dataset was collected at 100 K at the DLS beamline I03 (λ = 1.06998 Å).
Structure Solution and Refinement.
The data were processed by the autoPROC pipeline (76) consisting of XDS (77), POINTLESS (78), AIMLESS (79), CCP4 (80) and STARANISO (81). The structure was solved by molecular replacement by searching for one copy of PDB: 3ETO (51). After molecular replacement, the model was improved by manual model building in Coot (82) and refinement with REFMAC (83). Validation was performed using MolProbity (84).
Analytical Ultracentrifugation.
SV-AUC experiments were carried out in a Beckman Coulter Proteomelab XL-I analytical ultracentrifuge with An-60 Ti rotor (Beckman) at 40,000 rpm. Jagged1fe,HA at 5 µM and at 20 µM were measured in SEC buffer at 20 °C. Either 12- (5 µM sample) or 3-mm (20 µM sample) centerpieces with quartz windows were used. Absorbance was determined at 280 nm using SEC buffer as a reference. A total of 800 scans per cell were collected and analyzed in continuous c(s) mode in SEDFIT (85). Buffer density and viscosity were determined with SEDNTERP as 1.0061 g/mL and 0.010314 Pa/s, respectively.
Supplementary Material
Acknowledgments
We thank the staff of the DLS beamline B21, the staff of the ESRF beamline BM29, Dimphna H. Meijer, and Mercedes Ramírez-Escudero for assistance with SAXS data collection and the staff of the DLS beamline I03 for assistance with X-ray diffraction data collection. We thank Nadia Leloup for help with Notch1 SEC-MALS experiments and Dominique Thies-Weesie for help with SV-AUC experiments. This project has received funding from the following: the European Research Council under the European Union’s Horizon 2020 research and innovation program with Grant Agreement 677500 (to B.J.C.J.); the research program TA with Project 741.018.201 (to R.A.S.), which is partly financed by the Dutch Research Council; and the European Union Horizon 2020 program INFRAIA project Epic-XS Project 823839 (to R.A.S.). This work benefited from access to the Netherlands Cancer Institute, an Instruct-European Research Infrastructure Consortium (ERIC) centre, with financial support provided by Instruct-ERIC (proposal identifier [PID] 10025). This work has been supported by iNEXT (PID 6764), funded by the Horizon 2020 program of the European Union.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102502118/-/DCSupplemental.
Data Availability
The MS raw data, SAXS, and crystal structure data have been deposited in ProteomeXchange Consortium via the PRIDE partner repository, Small-Angle Scattering Biological Data Bank (SASBDB), and PDB. The MS data are made availabe at the PRIDE with identifier PXD023072. All SAXS data are made available at the SASBDB with the accession codes SASDJG8 (Monomeric Notch1fe), SASDJ38 (Monomeric Jagged1fe,HA), SASDJ48 (5.3 µM Jagged1fe,HA), SASDJ58 (11 µM Jagged1fe,HA), SASDJ68 (21 µM Jagged1fe,HA), SASDJ78 (42 µM Jagged1fe,HA), SASDJ88 (29 µM Jagged1EGF8–11), SASDJ98 (58 µM Jagged1EGF8–11), SASDJA8 (115 µM Jagged1EGF8–11), SASDJB8 (230 µM Jagged1EGF8–11), SASDJC8 (21 µM Jagged1CRD), SASDJD8 (42 µM Jagged1CRD), SASDJE8 (83 µM Jagged1CRD), and SASDJF8 (167 µM Jagged1CRD). Coordinates and structure factors for S1-cleaved mouse Notch1 NRR have been deposited to the PDB with accession code 7ABV.
References
- 1.Radtke F., MacDonald H. R., Tacchini-Cottier F., Regulation of innate and adaptive immunity by Notch. Nat. Rev. Immunol. 13, 427–437 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Louvi A., Artavanis-Tsakonas S., Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Ables J. L., Breunig J. J., Eisch A. J., Rakic P., Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12, 269–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray S. J., Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Siebel C., Lendahl U., Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235–1294 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Weng A. P., et al., Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Aster J. C., Pear W. S., Blacklow S. C., The varied roles of Notch in cancer. Annu. Rev. Pathol. 12, 245–275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mašek J., Andersson E. R., The developmental biology of genetic Notch disorders. Development 144, 1743–1763 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury F., et al., Defining single molecular forces required for Notch activation using Nano Yoyo. Nano Lett. 16, 3892–3897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovendahl K. N., Blacklow S. C., Gordon W. R., “The molecular mechanism of Notch activation” in Molecular Mechanisms of Notch Signaling, Borggrefe T., Giaimo B. D., Eds. (Springer International Publishing, 2018), pp. 47–58. [DOI] [PubMed] [Google Scholar]
- 11.Luca V. C., et al., Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 355, 1320–1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meloty-Kapella L., Shergill B., Kuon J., Botvinick E., Weinmaster G., Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell 22, 1299–1312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo D., et al., A mechanogenetic toolkit for interrogating cell signaling in space and time. Cell 165, 1507–1518 (2016). Correction in: Cell 169, 1357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., Ha T., Defining single molecular forces required to activate integrin and Notch signaling. Science 340, 991–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebay I., et al., Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell 67, 687–699 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Gordon W. R., et al., Mechanical allostery: Evidence for a force requirement in the proteolytic activation of Notch. Dev. Cell 33, 729–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols J. T., et al., DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176, 445–458 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parks A. L., Klueg K. M., Stout J. R., Muskavitch M. A., Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127, 1373–1385 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Gordon W. R., et al., Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14, 295–300 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Brou C., et al., A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Mumm J. S., et al., A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Kopan R., Ilagan M. X. G., The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 137, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G., et al., Structural basis of Notch recognition by human γ-secretase. Nature 565, 192–197 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Bray S. J., Gomez-Lamarca M., Notch after cleavage. Curr. Opin. Cell Biol. 51, 103–109 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Shimizu K., et al., Mouse Jagged1 physically interacts with Notch2 and other Notch receptors. Assessment by quantitative methods. J. Biol. Chem. 274, 32961–32969 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Chillakuri C. R., et al., Structural analysis uncovers lipid-binding properties of Notch ligands. Cell Rep. 5, 861–867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordle J., et al., A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 15, 849–857 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suckling R. J., et al., Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands. EMBO J. 36, 2204–2215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Álamo D., Rouault H., Schweisguth F., Mechanism and significance of cis-inhibition in Notch signalling. Curr. Biol. 21, R40–R47 (2011). [DOI] [PubMed] [Google Scholar]
- 30.D’Souza B., Miyamoto A., Weinmaster G., The many facets of Notch ligands. Oncogene 27, 5148–5167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprinzak D., et al., Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nandagopal N., Santat L. A., Elowitz M. B., Cis-activation in the Notch signaling pathway. eLife 8, e37880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakuda S., Haltiwanger R. S., Deciphering the fringe-mediated Notch code: Identification of activating and inhibiting sites allowing discrimination between ligands. Dev. Cell 40, 193–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampal R., Arboleda-Velasquez J. F., Nita-Lazar A., Kosik K. S., Haltiwanger R. S., Highly conserved O-fucose sites have distinct effects on Notch1 function. J. Biol. Chem. 280, 32133–32140 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holdener B. C., Haltiwanger R. S., Protein O-fucosylation: Structure and function. Curr. Opin. Struct. Biol. 56, 78–86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei Z., Baker N. E., Competition between Delta and the Abruptex domain of Notch. BMC Dev. Biol. 8, 4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A., Rangarajan A., Dighe R. R., Antibodies against the extracellular domain of human Notch1 receptor reveal the critical role of epidermal-growth-factor-like repeats 25-26 in ligand binding and receptor activation. Biochem. J. 449, 519–530 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Xu A., Lei L., Irvine K. D., Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J. Biol. Chem. 280, 30158–30165 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Lawrence N., Klein T., Brennan K., Martinez Arias A., Structural requirements for Notch signalling with delta and serrate during the development and patterning of the wing disc of Drosophila. Development 127, 3185–3195 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Kiyota T., Kinoshita T., Cysteine-rich region of X-Serrate-1 is required for activation of Notch signaling in Xenopus primary neurogenesis. Int. J. Dev. Biol. 46, 1057–1060 (2002). [PubMed] [Google Scholar]
- 41.Luca V. C., et al., Structural basis for Notch1 engagement of Delta-like 4. Science 347, 847–853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weisshuhn P. C., et al., Non-linear and flexible regions of the human Notch1 extracellular domain revealed by high-resolution structural studies. Structure 24, 555–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly D. F., et al., Molecular structure and dimeric organization of the Notch extracellular domain as revealed by electron microscopy. PLOS One 5, e10532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Steigenberger B., Pieters R. J., Heck A. J. R., Scheltema R. A., Pho X., PhoX: An IMAC-enrichable cross-linking reagent. ACS Cent. Sci. 5, 1514–1522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klykov O., van der Zwaan C., Heck A. J. R., Meijer A. B., Scheltema R. A., Missing regions within the molecular architecture of human fibrin clots structurally resolved by XL-MS and integrative structural modeling. Proc. Natl. Acad. Sci. U.S.A. 117, 1976–1987 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fürsch J., Kammer K.-M., Kreft S. G., Beck M., Stengel F., Proteome-wide structural probing of low-abundant protein interactions by cross-linking mass spectrometry. Anal. Chem. 92, 4016–4022 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi H., Haltiwanger R. S., Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 453, 235–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logeat F., et al., The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. U.S.A. 95, 8108–8112 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czajkowsky D. M., Hu J., Shao Z., Pleass R. J., Fc-fusion proteins: New developments and future perspectives. EMBO Mol. Med. 4, 1015–1028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor P., et al., Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc. Natl. Acad. Sci. U.S.A. 111, 7290–7295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon W. R., et al., Structure of the Notch1-negative regulatory region: Implications for normal activation and pathogenic signaling in T-ALL. Blood 113, 4381–4390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon W. R., et al., Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One 4, e6613 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X., et al., Insights into autoregulation of Notch3 from structural and functional studies of its negative regulatory region. Structure 23, 1227–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y., et al., Therapeutic antibody targeting of individual Notch receptors. Nature 464, 1052–1057 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Pandey A., Niknejad N., Jafar-Nejad H., Multifaceted regulation of Notch signaling by glycosylation. Glycobiology 31, 8–28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thakurdas S. M., et al., Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi). Hepatology 63, 550–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Handford P. A., Korona B., Suckling R., Redfield C., Lea S. M., “Structural insights into Notch receptor-ligand interactions” in Molecular Mechanisms of Notch Signaling, Borggrefe T., Giaimo B. D., Eds. (Springer International Publishing, 2018), pp. 33–46. [DOI] [PubMed] [Google Scholar]
- 58.Cordle J., et al., Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J. Biol. Chem. 283, 11785–11793 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Vooijs M., Schroeter E. H., Pan Y., Blandford M., Kopan R., Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J. Biol. Chem. 279, 50864–50873 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Hicks C., et al., A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J. Neurosci. Res. 68, 655–667 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Shimizu K., et al., Integrity of intracellular domain of Notch ligand is indispensable for cleavage required for release of the Notch2 intracellular domain. EMBO J. 21, 294–302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakamoto K., Chao W. S., Katsube K.-i, Yamaguchi A., Distinct roles of EGF repeats for the Notch signaling system. Exp. Cell Res. 302, 281–291 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Klykov O., et al., Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 13, 2964–2990 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Ihaka R., Gentleman R., R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996). [Google Scholar]
- 66.Krzywinski M., et al., Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J., et al., Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U.S.A. 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J., et al., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Zundert G. C. P., et al., The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Pettersen E. F., et al., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Incardona M.-F., et al., EDNA: A framework for plugin-based applications applied to X-ray experiment online data analysis. J. Synchrotron Radiat. 16, 872–879 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Panjkovich A., Svergun D. I., CHROMIXS: Automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics 34, 1944–1946 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003). [Google Scholar]
- 74.Förster S., Apostol L., Bras W., Scatter: Software for the analysis of nano- and mesoscale small-angle scattering. J. Appl. Cryst. 43, 639–646 (2010). [Google Scholar]
- 75.Filik J., et al., Processing two-dimensional X-ray diffraction and small-angle scattering data in DAWN 2. J. Appl. Cryst. 50, 959–966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vonrhein C., et al., Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winn M. D., et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tickle I. J., et al., Staraniso (Version 3.337 7, Glob. Phasing Ltd., Cambridge, UK, 2018).
- 82.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murshudov G. N., et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw data, SAXS, and crystal structure data have been deposited in ProteomeXchange Consortium via the PRIDE partner repository, Small-Angle Scattering Biological Data Bank (SASBDB), and PDB. The MS data are made availabe at the PRIDE with identifier PXD023072. All SAXS data are made available at the SASBDB with the accession codes SASDJG8 (Monomeric Notch1fe), SASDJ38 (Monomeric Jagged1fe,HA), SASDJ48 (5.3 µM Jagged1fe,HA), SASDJ58 (11 µM Jagged1fe,HA), SASDJ68 (21 µM Jagged1fe,HA), SASDJ78 (42 µM Jagged1fe,HA), SASDJ88 (29 µM Jagged1EGF8–11), SASDJ98 (58 µM Jagged1EGF8–11), SASDJA8 (115 µM Jagged1EGF8–11), SASDJB8 (230 µM Jagged1EGF8–11), SASDJC8 (21 µM Jagged1CRD), SASDJD8 (42 µM Jagged1CRD), SASDJE8 (83 µM Jagged1CRD), and SASDJF8 (167 µM Jagged1CRD). Coordinates and structure factors for S1-cleaved mouse Notch1 NRR have been deposited to the PDB with accession code 7ABV.