Significance
Progress in understanding and treating inflammatory diseases relies on accurate characterization of the molecular pathology. From application of nanoparticle tracking analysis, we expose a fundamental insight in oligomeric protein structure as polyvalent stimulants of autoreactive B cells. Quantification of hydrodynamic radii implicates these large proteins as part of vascular damage, which is an important source of morbidity and death in consequence of autoimmunity. Dysregulated alterations in protein concentration have been often sought as an explanation for diseases. Our work highlights now protein size as a key parameter in the inflammatory response both in and outside lymphoid organs. In this way, our findings may have wide implications for understanding disease associations based on polymerization and hydrodynamic radius changes of biological macromolecules.
Keywords: nanotechnology, autoimmunity, mannan-binding lectin, systemic lupus erythematosus
Abstract
Nanotechnology enables investigations of single biomacromolecules, but technical challenges have limited the application in liquid biopsies, for example, blood plasma. Nonetheless, tools to characterize single molecular species in such samples represent a significant unmet need with the increasing appreciation of the physiological importance of protein structural changes at nanometer scale. Mannose-binding lectin (MBL) is an oligomeric plasma protein and part of the innate immune system through its ability to activate complement. MBL also serves a role as a scavenger for cellular debris, especially DNA. This may link functions of MBL with several inflammatory diseases in which cell-free DNA now appears to play a role, but mechanistic insight has been lacking. By making nanoparticle tracking analysis possible in human plasma, we now show that superoligomeric structures of MBL form nanoparticles with DNA. These oligomers correlate with disease activity in systemic lupus erythematosus patients. With the direct quantification of the hydrodynamic radius, calculations following the principles of Taylor dispersion in the blood stream connect the size of these complexes to endothelial inflammation, which is among the most important morbidities in lupus. Mechanistic insight from an animal model of lupus supported that DNA-stabilized superoligomers stimulate the formation of germinal center B cells and drive loss of immunological tolerance. The formation involves an inverse relationship between the concentration of MBL superoligomers and antibodies to double-stranded DNA. Our approach implicates the structure of DNA–protein nanoparticulates in the pathobiology of autoimmune diseases.
Autoimmune diseases involve a chronic inflammatory response, which often leads to severe organ damage. Innate immunity, especially the complement system, serves a crucial role in instructing the adaptive immune response by facilitating transport of antigen into the lymph nodes and spleen (1–3). Engineered nanoparticles are targets for innate immunity (4, 5). So far it remains unclear whether physiologically formed nanoparticles also can manipulate the immune response or even be a source of autoinflammatory pathologies. Systemic lupus erythematosus (SLE) is a prototypic and potentially fatal multisystem autoimmune disease (6). B cells develop antibodies (Ab) to double-stranded (ds) DNA and spread a loss of host self-tolerance in expanding germinal centers (GCs) (7), which increases the SLE disease activity index (SLEDAI) (6). A missing link remains in our understanding of how dsDNA becomes a strong antigenic stimulus in the GC environment. Furthermore, the physiological significance of such stimulation has recently gained attention from the increasing number of other inflammatory diseases where cell-free DNA appear to play role (8).
Mannose (or mannan)-binding lectin (MBL) is part of innate immunity against infections along with other proteins of the complement system. It also contributes to aberrant chronic inflammation with damage to vascularized host tissue, especially the kidney (9). A recent study showed that MBL binds glycosylated proteins in a nanoparticulate vaccine formulation. This augments immunity by strengthening the formation of antigen-reactive GCs (10). MBL binds dsDNA, presumably as a scavenger of cellular debris (11). In animal models of SLE, MBL deficiency is protective (12), while clinical studies did not associate MBL with SLE (13, 14).
The 26-kDa MBL protein chain contains a tertiary structure with an N-terminal cysteine-rich segment, followed by a collagen-like region and a C-terminal Ca2+-dependent carbohydrate recognition domain (15, 16). Assembly of the collagen triple helix makes a structural trimer unit (MBL3) (17, 18), which further oligomerizes into polydisperse structures with 3 to 8× MBL3 units. In purified recombinant protein, atomic force microscopy (AFM) identified forms as large as 16 trimer units (16× MBL3) (19), and small-angle X-ray scattering spectroscopy (SAXS) characterized anisotropic oligomers with an ∼150-nm long axis (20). Especially the SAXS data suggested a type of oligomerization, which involves juxtaposition of a number of ∼6× MBL3 units, into resulting molecular ultrastructures that we here term superoligomers. Unfortunately, AFM and SAXS are unsuited for analyzing the MBL structural organization in complex samples such as liquid biopsies, including blood (21). Consequently, it is unclear if superoligomeric (sp)MBL exists in vivo and what its relevance is to human disease.
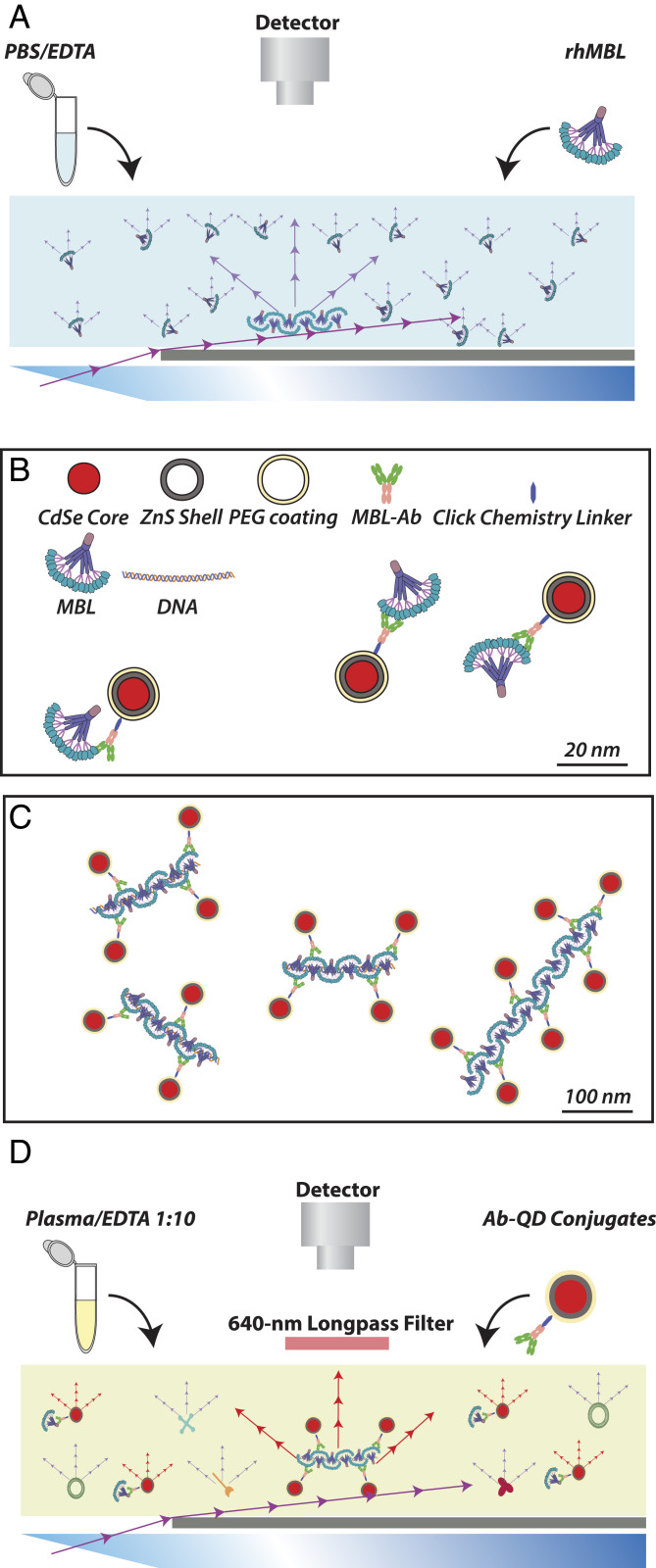
Nanoparticle tracking analysis (NTA) relates the Brownian motion of particles in liquid to their hydrodynamic radius. Motion is tracked by particle scattering of projected laser light (Fig. 1A) or by emission from long-lived fluorescent dyes (Fig. 1D), as recently demonstrated with biotinylated protein aggregates using streptavidin-coupled quantum dots (QDs) (22). Monoclonal Ab-conjugated QDs enable specific targeting of proteins in fluorescence microscopy (23). In principle, a similar application is possible in NTA to characterize changes in the structural organization of proteins, even in complex samples based on the emittance of the QDs (Fig. 1 B–D).
Fig. 1.
NTA for detection of MBL oligomers as purified protein and in bioliquids. (A) Principles of detection in NTA based on light scatter. Raw protein, including MBL, scatters laser light, which permits tracking of diffusion in a simple medium without other scattering constituents. (B and C) Design of a QD-based reporter system for detection of MBL. Commercially available 20-nm QDs with a Cd · Se core, Zn · S shell, and a pegylated surface with a chemistry linker enabling covalent coupling to glycan structure in IgG was conjugated to monoclonal Ab to either human MBL or murine MBL-C (or isotypic control Ab) of choice (B). The overall dimensions of the reporter permitted repeated binding to spMBL, here shown with a ligand (dsDNA)-driven lateral oligomerization (C). In both B and C, all items are drawn approximately to scale (scale bars indicated). (D) Detection in NTA based on QD emission. With the MBL Ab-QD reporter, the fluorescent emission at 655 nm permits detection among other constituents of a complex bioliquid such as diluted plasma.
Here, we used NTA to characterize the structural variation of MBL oligomers in plasma from SLE patients. The concentration of spMBL is vastly increased in patients, amplified by MBL’s interaction with dsDNA. The spMBL concentration correlates directly with disease activity as may be explained by the large hydrodynamic radii of the oligomers, which pushes spMBL into blood vessel walls, adding to the vascular damage seen in SLE. An even wider link to SLE morbidity derives from the influence of spMBL on the autoimmune pathology, mechanistically supported by an animal model of SLE where spMBL is connected to the formation of autoreactive GC B cells.
Results
Characterization of spMBL.
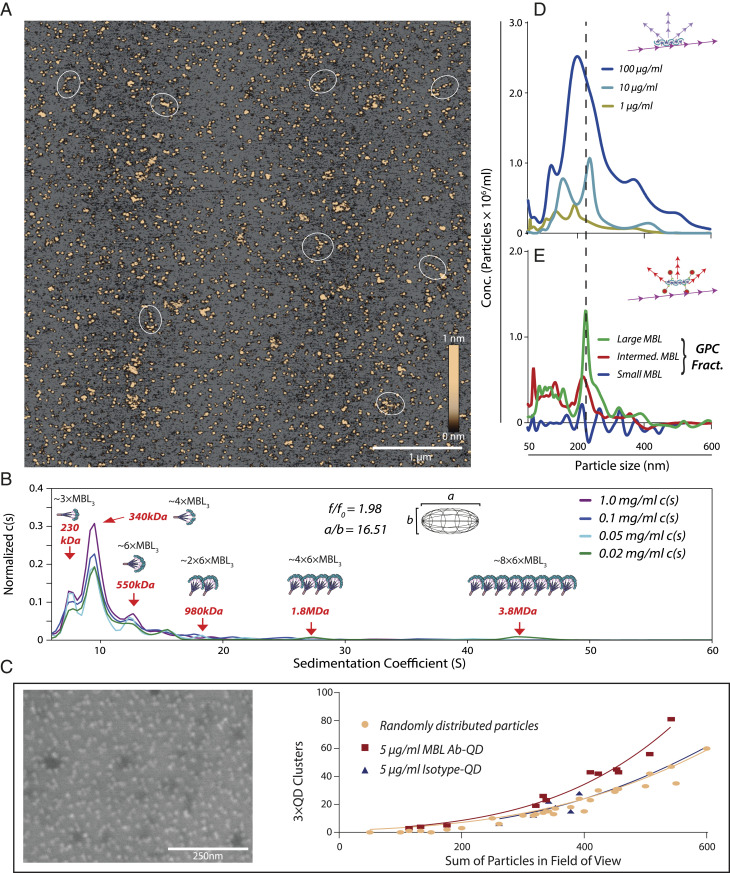
To characterize the geometric size of individual MBL oligomers in vitro, we performed AFM on clinical-grade recombinant human (rh)MBL applied to a modified mica surface. The mean height of all particles was 0.968 ± 0.011 nm (Fig. 2A), within the range of 1.46 ± 0.57 nm reported earlier (24). Among the particles, it was possible to identify a minor population of highly elongated spMBL with long axes of 100 to 200 nm (Fig. 2A) in agreement with dimensions previously reported by SAXS (20).
Fig. 2.
Characterization of spMBL. (A) AFM imaging of rhMBL. Oligomers with long axes of ∼100 to 200 nm are indicated with white circles. (B) SVAUC of rhMBL with the normalized abundance of oligomers, c(s), plotted a function of the sedimentation coefficient. (C) Specificity of MBL AB-QD. rhMBL was dispersed on gold surfaces and analyzed using SEM. Deposited proteins produce a contrast enhancement, showing several dark ∼40-nm deposits surrounded by a corona of MBL Ab-QDs. SEM experiments on titanium surfaces consisted of two separate surfaces analyzed in six different zones for a total of 12 images for each condition. The number of clusters of three or more QDs within a radius of 100 nm from center to center was plotted as a function of the total number of QDs (SI Appendix, Fig. S3 D–G). (D and E) NTA analysis of rhMBL. NTA of unfractionated MBL oligomers in scatter detection mode (D). Following separation of MBL by GPC (SI Appendix, Fig. S4) into small, intermediate, and large oligomers, the fractions were analyzed by NTA using MBL Ab-QD reporters in fluorescent detection mode with subtraction of an isotype-QD control signal (E). NTA data show the average from three experiments with five separate recordings.
Sedimentation velocity (SV) analytical ultracentrifugation (AUC) is a classic tool for establishing the size distribution of proteins in solution. We performed SVAUC on rhMBL and found the well-known oligomers of 3× MBL3 (∼230 kDa) and 4× MBL3 (∼340 kDa) as well as much larger oligomers at 1.8 MDa (∼24× MBL3) and 3.8 MDa (∼48× MBL3) (Fig. 2B and SI Appendix, Fig. S1). Species above 20 S accounted for ∼8% of the total signal. We observed an average frictional ratio close to 2 (1.98) consistent with a high degree of elongation in the observed structures of the MBL oligomers seen in the AFM data. The elongated oligomers remained stable in the presence of human serum supporting their physiological presence, and the slightly increased size distribution indicates an intact interaction with serum proteins (SI Appendix, Fig. S2 A–C). In the presence of serum, a sharp peak was formed in the sedimentation profile with a coefficient of ∼16 S (SI Appendix, Fig. S2C), which is consistent with ∼3 to 6× MBL3 oligomers in complex with MBL-associated serine proteases (MASP) as judged from structural investigations of these complexes (25).
In order to validate an Ab-dependent size-sensitive QD reporter system as shown in Fig. 1 B–D, we performed scanning electron microscopy (SEM) images of rhMBL using QDs coupled with monoclonal Ab (131-1) to MBL (MBL Ab-QDs). The MBL oligomers, visible as enhanced contrast on the gold surfaces, were surrounded by the MBL Ab-QDs (Fig. 2C). Importantly, in similar experiments on titanium surfaces, where the QDs (but not the protein) delivered optimal contrast differences (SI Appendix, Fig. S3 A–C), the specificity of the MBL Ab-QDs was validated from comparison with images of isotypic control Ab-coupled QDs or in silico–randomized distributions of QDs (Fig. 2C and SI Appendix, Fig. S3 D–G).
To investigate if spMBL were amenable to characterization by NTA by light scattering (Fig. 1A), we applied it directly on unfractionated rhMBL (Fig. 2D). The size distribution of the detectable particles showed a peak near 200 nm (Fig. 2D), in good agreement with the spMBL species presenting a long axis of 200 to 300 nm as also shown above. A similar result came from analysis with MBL Ab-QD reporters (Fig. 1C) applied to fractions from gel-permeation chromatography (GPC), sorting the MBL oligomers into small, intermediate, and large species (SI Appendix, Fig. S4). Only the fraction with the largest oligomers produced a strong signal, again peaking at a size of 220 nm (Fig. 2E). The small 20-nm increment in size compared to raw MBL (Fig. 2D) is attributable to the use MBL Ab-QD reporters. The experiment also excluded that MBL Ab-QD reporters are any quantitative source of particle size increments or cross-linking of MBL. Taken together, this showed that NTA with MBL Ab-QD was capable of reporting spMBL size, with the hydrodynamic radius in acceptable agreement with previous results from SAXS (20) as well as further orthogonal techniques presented here including the geometric size directly measured by AFM. To investigate if spMBL associated with MASP, we mixed the MBL with catalytically inactive MASP-1. As also found for SVAUC, a sharp peak was formed at 40 nm corresponding to a complex of 3 to 6× MBL3 with two ∼5-nm overhangs from the inclusion of MASP-1 dimer (25) (SI Appendix, Fig. S2D). For the larger spMBL oligomers in the range of 100 to 200 nm, a small shift in size was detectable as a shoulder on the size distribution profile upon mixture with MASP-1 (SI Appendix, Fig. S2E), corresponding to an increase in the median size of ∼7 nm (SI Appendix, Fig. S2F).
The spMBL Concentration Is Increased in SLE Patients and Correlates with SLEDAI.
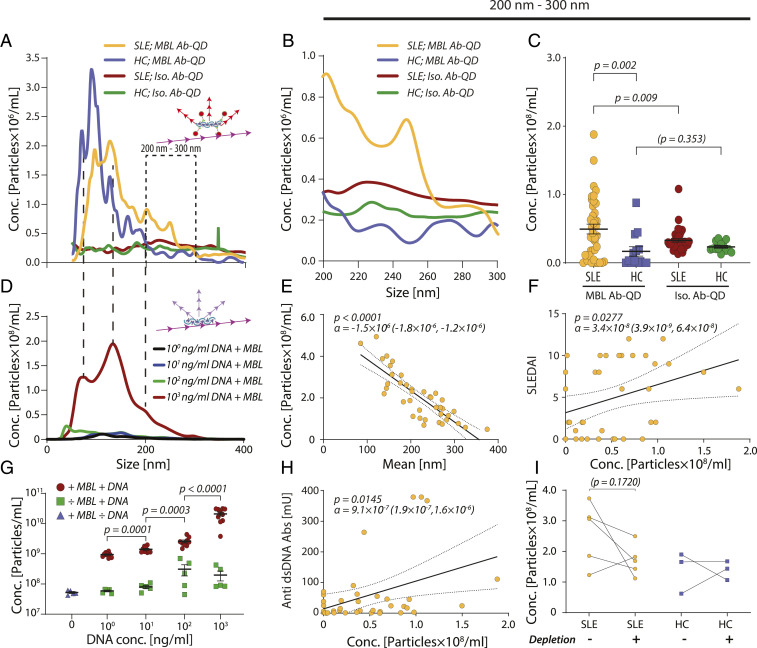
The detection of spMBL by NTA with Ab-QD reporters raised the possibility of discovering previously unnoticed changes in MBL from characterization of plasma samples. A recent study of a cohort of 372 patients and 170 healthy controls (HCs) found no difference in the total (mass) concentration of MBL between the two groups (14). We selected a representative sample of this cohort with 39 patients and 14 controls with similar age and mean MBL plasma total concentration (SI Appendix, Fig. S5 A and B) and a distribution with low and high SLEDAI scores for the patients (SI Appendix, Table S1). The samples were analyzed with MBL Ab-QD or the isotypic control IgG1 also used in the SEM analysis (Fig. 2C). Particles in the size interval of 50 to 400 nm (Fig. 3A) were quantified by NTA with the lower limit set to exclude unbound reporters and small MBL oligomers (SI Appendix, Fig. S6 A–C). The presence of spMBL in plasma was confirmed by the significant difference between MBL Ab-QD and isotypic control reporters (Fig. 3 A–C). The concentration of spMBL was in the femtomolar range (0.5 × 108 particles/mL ∼ 83 fM), compared to the nanomolar range of the most abundant forms of MBL (2 µg/mL with an average composition of 4× MBL3 ∼ 8.2 nM). The specific MBL signal for HCs was lost for sizes larger than 200 nm where MBL Abs and isotypic control reporters produced similar signals (Fig. 3 A–C). By contrast, the concentration of MBL oligomers in the interval 200 to 300 nm was significantly higher for SLE patients, both in comparisons with HCs and between MBL Ab-QD and QDs with isotypic control (Fig. 3 B and C).
Fig. 3.
Characterization of spMBL in SLE and HC plasma. (A–C) NTA of MBL Ab-QDs or isotypic control-QD size distributions in plasma from patients (n = 39) or HCs (n = 14) (A). Particle concentrations in the size interval of 200 to 300 nm (B). Comparisons between the spMBL concentration in SLE and HC plasma in a two-tailed Welch’s t test for the size regimen 200 to 300 nm. The solid lines indicate the mean value, and error bars indicate SEM (C). (D and G) Formation of spMBL from addition of dsDNA to rhMBL. Features were compared with SLE and HC profiles by hatched lines (D). Linear regression (solid line and CI with dotted lines) between spMBL concentration and particle mean size in SLE patients and p stating the likelihood of α = 0 (E). Linear regression between SLEDAI and particle concentration in the 200- to 300-nm range (F). Detected particles as a function of dsDNA concentrations compared in a Mann–Whitney U test (n = 10) (G). (H) Linear regression between concentrations of anti-dsDNA Abs and spMBL concentration in SLE plasma for the 200- to 300-nm range. (I) Influence of vesicle depletion on spMBL concentration. Six SLE and three HC plasmas were used and compared using a paired t test with pairing indicated with lines.
The mean size of the MBL Ab-QD particles correlated negatively with the concentration of detected particles (Fig. 3E). The lack of correlation between the total MBL and spMBL concentrations exclude spontaneous oligomerization as a major source of spMBL formation (SI Appendix, Fig. S5D). An alternative explanation is that spMBL forms from smaller units through stabilization of the superoligomers limited by additional constituents. Depletion of lipids from the plasma only moderately changed the concentration of spMBL in SLE plasma, not supporting lipids as a critical part of complexes (Fig. 3I). Chosen from the known ability of MBL to bind dsDNA and the important role of dsDNA in SLE, we then applied increasing concentrations of dsDNA to a fixed total concentration of MBL and measured the formation of spMBL in vitro (Fig. 3 D and G). A substantial increase in the concentration of superoligomers occurred when dsDNA was added, both illustrating dsDNA ligand-induced superoligomerization of MBL and dsDNA as a limiting factor in this process. The size distribution for the dsDNA–MBL particles (Fig. 3D) had several curve features in common with spMBL from HC and SLE plasma (Fig. 3A), supporting that particles purely consisting of MBL and dsDNA in vitro take comparable sizes to spMBL found in plasma. dsDNA on its own did not produce comparable particles (Fig. 3G). Measurement of the zeta potential of the dsDNA–MBL particles compared to pure MBL particles showed an increase in negative charge of the particles (SI Appendix, Fig. S7A) consistent with application of the anionic DNA, which shields positively charged side chains in MBL upon binding (11). The dynamic light scattering instrument (DLS) applied for the measurement gave a size distribution in agreement with the NTA measurements also showing an increase in MBL particle size upon application of DNA (SI Appendix, Fig. S7B).
We then investigated if the concentration of spMBL particles with a size between 200 to 300 nm is a predictor of disease activity (SLEDAI score). We found that the spMBL concentration correlated positively with SLEDAI (Fig. 3F), while the total concentration of MBL showed no correlation (SI Appendix, Fig. S5C). This shows a link between SLE disease severity and the concentration of spMBL. The plasma concentration of anti-dsDNA Abs is determined as part of SLEDAI (SI Appendix, Table S1) (6). The concentrations of spMBL and anti-dsDNA Abs correlated positively (Fig. 3H). Since both anti-dsDNA Abs and spMBL are dsDNA binding, it prompted the question what relationship exists between them and disease severity. A significant linear correlation was established between the SLEDAI score and the ratio between the spMBL and anti-dsDNA Ab concentrations (SI Appendix, Fig. S5F). The negative correlation suggested that an excess of spMBL increased disease severity and, vice versa, an excess of anti-dsDNA Abs lowered the severity. This is a nontrivial analytical result since both spMBL and dsDNA correlated positively with SLEDAI in the patients (Fig. 3F and SI Appendix, Fig. S5E).
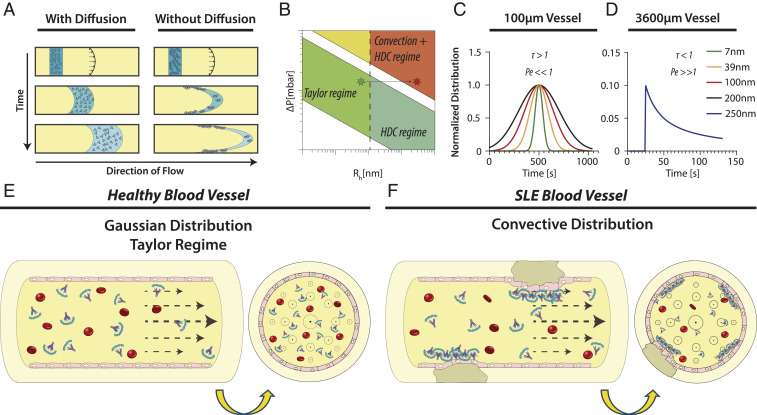
The surprisingly strong link between spMBL concentration and SLEDAI score raises the possibility that the hydrodynamic properties of spMBL play a direct role in SLE morbidities. The dispersion under flow of nanoparticulate colloidals are critically dependent on whether transport is dominated by diffusion or convection (26) (Fig. 4 A and B). We performed Taylor dispersion analysis (TDA) (27) using physiologically relevant blood vessel diameters and pressures and the previously experimentally determined properties of nanoparticulates (26). From these results, we noted that spMBL tend, in consequence of their large hydrodynamic radii, to have a convective dominated distribution (Fig. 4 C and D), especially in larger vessels, which are complement-mediated disease targets in SLE (28, 29) (Fig. 4 E and F).
Fig. 4.
Capillary distribution of spMBL. (A) The effect of diffusion on the distribution of particles in a capillary under constant pressure. (B) Different regimes in TDA experiments as a function of particle size and mobilizing pressure. Stars indicate the shift from Gaussian Taylor regime to convective driven distribution with correction for hydrodynamic chromatography (HDC) (21). (C) Taylorgrams calculated for nanoparticles at the same mobilization pressure with a Gaussian-driven distribution (Pe << 1 and τ > 1) (26, 27, 50). Capillary dimensions were set at 100-μm internal diameter ×50 cm total capillary length. This series of experiments corresponds to the horizontal arrow in B. (D) Taylorgram calculated for 250-nm nanoparticles in a large blood vessel with primarily convective distribution (Pe >> 1 and τ < 1). Capillary dimensions were set at 3,600 μm × 50 cm corresponding to the endpoint (red star) in the convective/HDC regime in B. (E) In healthy blood, MBL oligomers are relatively small and the Gaussian Taylor regimen dominates. (F) In SLE blood, the concentration of spMBL is increased, and a convective distribution is more prominent. As a consequence, the deposition of spMBL into the endothelial wall contributes to the formation of atherosclerotic plaques.
Formation of GC B Cells Is Associated with the spMBL Concentration.
To more specifically follow the relationship between spMBL and the autoimmune SLE response, we employed a murine lupus model, the 564Igi mouse. 564Igi mice carry a knockin of prerecombined VDJ and VJ segments in their immunoglobulin heavy chain and kappa light chain locus, respectively, which together encode antigen-specificity for ribonuclear protein complexes (30). On a C57BL/6J background, the mice develop a severe SLE-like phenotype with generation of anti-dsDNA Abs and disease symptoms.
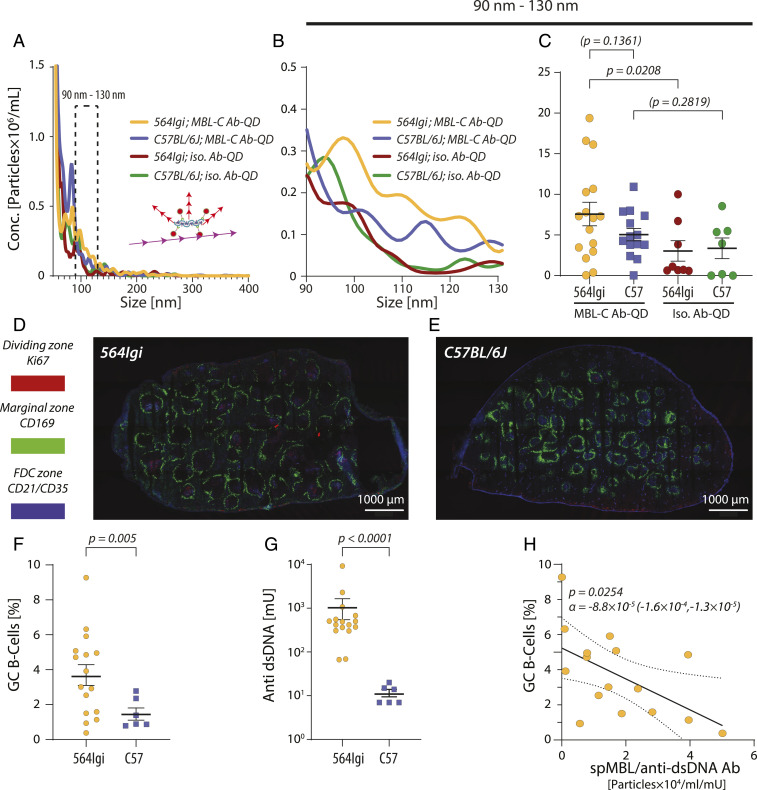
Plasma samples and spleen cells were collected from 16 564Igi and 14 C57BL/6J background control animals. Mice express two types of MBL, MBL-A and MBL-C, where MBL-C is closer both phylogenetically and structurally to human MBL (31). By targeting QD-labeled monoclonal Abs to murine MBL-C (MBL-C Ab-QD), we established a reporter system for analyzing spMBL-C with NTA (Fig. 5 and SI Appendix, Fig. S8 A–C). The particle concentrations seen were markedly lower in murine than in human plasma (Fig. 5A). By following the same strategy as for the human samples (Fig. 5 A–C), we identified a size interval of 90 to 130 nm, where the signal from MBL-C Ab-QD exceeded the signal from the isotypic Ab-QD control (Fig. 5B). Similar to the human samples, we saw a significant difference between MBL-C Ab-QD and isotypic control Ab-QD in the 564Igi mice. By contrast, no such difference was observed for the C57BL/6J animals (Fig. 5C). The mean concentration of spMBL was 1.5-fold higher in the diseased 564Igi mice compared with healthy C57BL/6J mice, similar in magnitude to the 2.7-fold difference between SLE patients and HCs (Fig. 3C).
Fig. 5.
spMBL-C in 564Igi mice with SLE-like disease and correlation with GC B cell formation. (A–C) MBL-C or isotypic Ab-QDs were mixed with plasma from 564Igi (n = 16) or C57BL/6J (n = 14) mice and characterized by NTA (A). Statistically significant differences were tested for the size interval 90 to 130 nm (B) in a two-tailed t test with Welch’s correction. Solid lines indicate mean value and error bars SEM (C). (D and E) Confocal microscopy of GCs in the spleens of 564Igi and C57BL/6J mice. The marginal zone (CD169+ macrophages), dark zone (dividing centroblasts positive for Ki67), and follicular dendritic cell (CD21/CD35+) zones were stained. Images were made by 4 × 7 tile scans acquired with a 10× objective at 512 × 512 (D) or 1,024 × 1,024 (E) pixel resolution using 10% overlap for each tile scan. (F and G) Quantification of GC B cells (F) and anti-DNA Abs (G) in 564Igi (n = 16) and C57BL/6J (n = 6) mice. Statistical significance was tested in a two-tailed Welch’s t test (F) or in a Mann–Whitney U test (G). (H) Correlation between GC B cell percentage and the ratio of spMBL and anti-dsDNA Abs concentrations. A linear correlation was calculated according to the expression α · [spMBL]/[Anti-dsDNA Abs] + β for 16 564Igi mice. The linear regression is indicated with a solid line, the CI is shown with dotted lines, and the P value states the likelihood of α = 0.
The 564Igi mice developed strong cell proliferation in the GC as shown by confocal immunofluorescence microscopy of spleen sections. In addition, the follicles appeared larger with more predominant dividing zones (Fig. 5D) compared to C57BL/6J mice (Fig. 5E). To quantify the development of GCs, we measured the population of B220+CD95hiCD38low cells among the splenocytes by flow cytometry (SI Appendix, Fig. S9 A–C). The percentage of GC B cells in splenocytes was higher in 564Igi mice compared to C57BL/6J mice (Fig. 5F), which, together with an elevated concentration of Ab to dsDNA (Fig. 5G), validated the SLE-like phenotype. Inspired again by the shared ability of the anti-dsDNA Abs and MBL to bind dsDNA and the findings from the human cohort (SI Appendix, Fig. S5F), we considered the connection between the SLE phenotypic traits and the MBL-C superoligomers. A significant linear correlation was found between the percentage of GC B cells score and the ratio between the spMBL-C and anti-dsDNA Ab concentrations (Fig. 5H). Similar to the human disease, the negative correlation suggested that an excess of spMBL-C increased GC B cell formation while an excess of anti-dsDNA Abs decreased GC B cell proliferation.
Discussion
Engineered nanoparticles are potent stimulants of the immune response (4, 5) for reasons directly connected to their size and bottom-up synthesis resembling microbial threats, which activate especially innate immunity (32). By contrast, we know little about the influence of physiologically formed particles in human immunology because of several technical limitations, including the relatively weak forces governing their assembly (22) and heterogeneous surface interactions with several immunoactive proteins in bioliquids (33, 34). We open a perspective by showing that a reporter system of QDs and Abs enables direct detection of spMBL particles in plasma with several implications for understanding the autoimmune disease SLE.
Both in humans and experimental animal models, deficiencies in the complement system are usually linked with SLE or SLE-like symptoms (6). One exception is MBL, where knockout animals are instead protected from SLE (12). Our work offers at least two explanations for this phenotype, which strongly implicates MBL in the human disease. First, the large hydrodynamic radii make spMBL prone to accumulate on the surface of blood vessel walls. The pathophysiologic relevance is supported by vasculitis and renal damage as among the most important descriptors of disease severity (28) with a clear involvement of complement activation (29). We investigated the ability of MBL to bind MASP-1, one out of three proteases known to associate with MBL (15). Detection of formed complexes was easier for the smaller MBL oligomers. For the spMBL, a small increase in size was barely detectable but consistent with recent structural modeling of the interaction between MBL and the MASPs (25). Inclusion of MASP-1 would be expected to add a maximum of a 2 × 5 nm overhang from the MBL oligomer. In turn, a similar extension would probably be the case for the long axis of the spMBL. Precise theoretical or experimental estimation is difficult, however, due to the complex geometry of both the smaller MBL oligomers as well as spMBL. The ability to bind MASP-1 is important, especially in view of recent evidence of MASP activation through MBL/MASP intercomplex activation from juxtaposition of the complexes (25). The spMBL complexes, especially as revealed here by AFM, seem to enable such juxtaposition of MASP-carrying MBL. In the case of MASP-1, which is prone to autoactivate (35), one implication could be cleavage and activation of its several substrates in both the complement and coagulation system.
Second, our experiments showed that formation of spMBL is stabilized by dsDNA in vitro, with similar-sized spMBL particles seen to increase in plasma from SLE patients. Zeta potential measurements suggested that these particles carry a higher negative charge than naked MBL, in agreement, of course, with the expectation from complexing MBL with the polyanionic DNA. The higher charge would likely increase the colloidal stability of these particle, which may support their impact outside the vasculature, for instance in the spleen. Indeed, as shown in the 564Igi mouse model of SLE, a high concentration of spMBL relative to the dsDNA Abs concentration favors GC B cell formation. In humans, the same ratio also correlated negatively with the SLEDAI score. Anti-dsDNA Abs may limit the spMBL formation due to competition for dsDNA, in turn limiting the antigenic stimulus on dsDNA-reactive B cells. Anti-dsDNA Abs can be detected years prior to SLE manifestations, indicating that they are apathogenic or at least only weakly pathogenic even when a correlate with the disease is found (36). If the balance between spMBL and dsDNA Abs tilts toward more spMBL, as our measurements suggest to happen in severe SLE, the resulting GC activation may amplify by expansion of additional GC B cells with broader autoreactivity through epitope spreading, significantly worsening disease through loss of self-tolerance (7). Indeed, a scenario where spMBL and dsDNA-reactive Abs are competing, rather than mutually excluding, pro- and anti-inflammatory stimuli is reminiscent of other autoimmune diseases (37). Taken together, our findings make intriguing connections between formation of immunoactive nanoparticulates and human disease.
Materials and Methods
rhMBL and MASP-1.
Clinical-grade rhMBL was produced for NatImmune (Copenhagen, Denmark) using mammalian cell culture as described previously (38, 39). In brief, rhMBL was captured from the cell debris–deprived cell culture broth using a Glucosamine Sepharose 4FF column (GE Healthcare Life Science) and subsequently eluted with saline Tris buffer containing EDTA. The retained product was concentrated on a Source30Q column and desalted on a Sephacryl S-400 HR Column (GE Healthcare). The process also included a DNA removal step (using Benzonase) and several microfiltration and nanofiltration steps for bioburden and adventitious virus elimination. The rhMBL contained DNA from host cells for the expression (HEK293) below an assay sensitivity of 78.13 pg/mL. Inactive rhMASP-1 was made as described earlier with R504Q and S646A mutations in the catalytic domain and the natural proteolytic activation site replaced with an enterokinase site (35, 40).
AFM.
Inside a nitrogen-purged desiccator, freshly cleaved 10-mm mica discs (Ted Pella) on support were placed in a Petri dish immediately beside a 1.5-mL polypropylene tube containing 30 μL (3-Aminopropyl)triethoxysilane (APTES; Sigma-Aldrich). The desiccator was sealed off, and the surfaces were incubated for 30 min. After incubation, APTES was removed and the mica surfaces rinsed with ultrapure water (Milli-Q) by gentle aspiration, followed by drying with nitrogen. A total of 80 µl rhMBL with a concentration of 1 μg/μL in phosphate-buffered saline (PBS) was pipetted onto the APTES surface and incubated for 30 min in the Petri dish. The surface was then rinsed with ultrapure water by careful aspiration, followed by drying with nitrogen. The surfaces were imaged in a Multimode 8-HR atomic force microscope (Bruker) with a new, high-resolution Scanasyst-Air probe with nominal tip radius of 2 nm and spring constant at 0.4 N/m (Bruker) using tapping mode in air and a scan rate of 1 Hz. Images were made in areas of 5 × 5 μm (1,024 × 1,024 pixels) with three images taken at unique locations on the same surface. Scans were evaluated in NanoScope Analysis (Bruker) with images fitted to a third-order flattening curve to remove tilt and bow artifacts and further cleaned by removing spikes and streaks, both with a cutoff at 3 nm. Particle mean height was calculated from 378 particles taken from four images on different surfaces prior to the usage of the 2-nm tip but still using the same surface preparation protocol as for the images depicted in the article. To identify the particles, a mask with an artifact cutoff at 0.5 nm was used based on the signal-to-noise ratio obtained from mica surfaces without MBL.
QD Coupling of Ab.
QD Ab coupling was done using SiteClick Qdot 655 Antibody Labeling Kit (Molecular Probes, S10453) according to manufactures instructions. In brief, Ab [either “Hyb 131-1” mouse IgG1 to human MBL (41), “14D12” rat IgG2a to mouse MBL-C (42), “X093101-2” mouse IgG1 isotype control (Agilient), or “RTK2758” rat IgG2a isotype control (Abcam)] was concentrated in Ab preparation buffer to a concentration of 2 mg/mL or above. Next, carbohydrates on the Ab were modified by the incubation with β-galactosidase for 4 h at 37 °C. Azide modification was achieved through incubation with uridine diphosphate glucose-GalT enzyme overnight at 30 °C. Ab with modified carbohydrates was purified and concentrated through a series of centrifugation steps using a molecular-weight cutoff membrane concentrator, and the buffer was simultaneously changed to Tris pH 7.0. Finally, 5′-dibenzocyclooctyne-modified QD nanocrystals were coupled overnight at 25 °C and until further use stored at 4 °C.
SEM.
We precleaned 8 × 8 mm pieces of precut 4-inch silicon wafers in oxygen plasma (in an MK II Advanced Vacuum, with settings at 50W and 0.1 mTorr for 120 s) and then coated via physical vapor deposition (electron-beam stimulated thermal evaporation in a CryoFox-GLAD Polyteknik A/S DK) covered with a layer of Ti (20 nm at 0.1 nm/s base pressure <1 × 10−7 Torr) or Au (20 nm at 0.1 nm/s with a 2 nm Ti adhesion layer base pressure <1 × 10−7 Torr) and then transferred to 48-well plates within 1 h (150687; Nunclon Delta Surface, ThermoFisher Scientific) containing 600 µl PBS with 1 mM EDTA (PBS/EDTA). A total of 400 µl PBS/EDTA was removed, and 100 µl GPC-fractionated large-sized MBL was added to the wells to a final concentration of 5 µg/mL. A 1:200 dilution of MBL-specific or isotype Ab-QD conjugate was added, and samples were incubated for 30 min at room temperature. Samples were washed by transfer of wafers into 600 µl PBS/EDTA twice and then into 600 µl distilled H2O twice before drying with pressurized nitrogen. Images were recorded in a Magellan 400 ultra-high resolution SEM (FEI), optimized for contrast using a 3 keV (for experiments with Au films) or 5 keV (for experiments with Ti films) beam with a nominal current of 50 pA (spot size ∼1 nm). The particle positions were tracked with ImageJ (NIH) “Analyze Particles function,” corrected by manually marking particles and then analyzed for nearest neighbor distance. Clusters with a maximum interparticle distance of 100 nm were considered as a single cluster. The clustering for experimental images was compared with random two-dimensional distributions of dots generated in silico with 0 to 2,000 dots.
AUC.
SV experiments with absorbance and interference detection were performed in a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter) by following standard protocols (43, 44). SV experiments with a titration of rhMBL concentrations in PBS 1 mM EDTA and fluorescein isothiocyanate–labeled anti-MBL (Hyb 131-1) in 10% human serum were carried out in an Optima XL-A analytical ultracentrifuge equipped with a fluorescence detection system with excitation at 488 nm using a 10-mW solid-state laser as the light source (AVIV Biomedical). Experiments with fluorescence detection system SV used FITC-conjugated Hyb 131-1 Ab to MBL and were set up according to the standard procedures (45). The experimental temperature was 20 °C. The samples were loaded into SV cell assemblies with 12- or 3-mm charcoal-filled Epon double-sector centerpieces and sapphire windows. The sample cells were loaded into a rotor for temperature equilibration for 2 to 3 h, followed by acceleration to full speed at 50,000 rpm. Continuous radial scans were initiated immediately using the selected detection systems. The accuracy of the determined s-values was ∼1% or better (43, 46).
NTA.
Samples for NTA were analyzed using a NanoSight NS300 system (Malvern Panalytical). The system was configured with a 405-nm laser, a high-sensitivity scientific complementary metal–oxide–semiconductor Orca Flash 2.8/Hamamatsu C11440 camera (Malvern Panalytical), a syringe pump, and for fluorescence measurements, a 650-nm long-pass filter was used. The sample chamber was washed twice with 1 mL PBS with 1 mM EDTA (PBS/EDTA) before each measurement. All samples were thoroughly mixed before measurement and were then injected into the sample chamber using 1-mL syringes. The measurement script comprised temperature control at 23 °C, followed by a 20 s flush at a flowrate mark 1000. Next, sample advancement was stabilized by a 120 s advancement at flowrate mark 10. Recordings were captured continuously during a steady flow at flowrate mark 10 with five 60-s recordings separated by 5-s lag time between each sample. The videos were collected and analyzed using NanoSight software (version 3.3 and 3.4 with a concentration upgrade; Malvern). Automatic settings were used for the minimal expected particle size, minimum track length, and blur setting. Camera sensitivity and detection threshold was adjusted according to sample composition and kept constant for all samples to be directly compared. For fluorescence mode, the camera level was set to maximum (mark 16), and the detection threshold was set close to minimum (mark 3). Pure protein was analyzed in PBS/EDTA with and without a 1:200 dilution of MBL-specific (Hyb 131-1) or isotype monoclonal Ab-QD reporters. Human plasma samples were analyzed at a 1:10 dilution in PBS/EDTA with a 1:200 dilution of MBL-specific or isotype Ab-QD reporters. Plasma samples from mice were analyzed in a 1:10 dilution in PBS/EDTA with a 1:20,000 dilution of MBL-C–specific (14D12) or isotype Ab-QD reporters. A 50-nm cutoff was established for all samples to exclude unbound QD conjugates as well as QD conjugates bound to smaller forms of MBL.
GPC.
Chromatography was carried out in the Äkta Pure system (GE Healthcare). A sample of rhMBL was centrifuged at 10,000 × g for 30 min, followed by application of 1 mL at a concentration of 1 mg/mL onto a 10 × 300 mm Superose6 Increase column (GE Healthcare). For size comparison, the column was calibrated with thyroglobulin (RH = 8.5 nm), ferritin (6.1 nm), IgG (5.8 nm), and human serum albumin (3.5 nm). The GPC was run in PBS/EDTA at a flow rate of 0.5 mL/min. The protein concentration of each fraction was assessed with 280 nm absorbance using a Nanodrop instrument (ThermoFisher).
SLE Patients and HCs.
SLE patients followed at the out-patient clinic at the Department of Rheumatology, Aarhus University Hospital, Denmark, were included for the study as previously described (14). Briefly, inclusion criteria were 1) age ≥ 18 and 2) fulfillment of the 1997 revised American College of Rheumatology classification criteria for SLE (47). Exclusion criteria were 1) incapacitation, 2) inability to understand Danish, 3) clinical and biochemical signs of infection, or 4) ongoing treatment for cancer or infection. The cohort is a cross-sectional cohort and represents patients in full remission as well as patients with active disease. The study was approved by the regional Danish Data Protection Agency (no. 1-16-02-339-13) and the Central Denmark Region Committee on Health Research Ethics (no. 1-10-72-214-13). All patients were given oral and written information about the project prior to inclusion and thereafter gave written consent. Patients were included according to the Helsinki Declaration. Blood samples were collected in EDTA plasma tubes (3675258; 10 mL; Alere Inc. or serum tubes [367896; 10 mL; Alere Inc.]), centrifuged for 10 min at 2,000 × g, and plasma was immediately collected, aliquoted, and frozen at −80 °C. Clinical data were recorded at the same time. For the present study, a subset of patient samples from the cohort was chosen at random, however, ensuring an even distribution between patients with an active disease (SLEDAI = 6 or above; n = 20) or patients in remission (SLEDAI = 2 or below; n = 19).
Lipid Vesicle Depletion.
Lipid vesicle depletion was performed on plasma samples using ExoQuick-TC (EXOTC10A-1; System Biosciences) according to manufactures instructions. In brief, 300 µl patient plasma was centrifuged for 15 min at 3,000 × g. Before overnight incubation, 60 µl Exoquick solution was added and mixed thoroughly. Samples were then centrifuged for 30 min at 1,500 × g and analyzed using MBL Ab-QD conjugates by NTA as described in section NTA.
Generation of Linearized DNA.
The linearized DNA was generated by restriction digestion of the pOET1‐OAS3 plasmid (48). The set of plasmid backbone‐derived fragments was generated by digestion with NcoI (4,570 base pairs). After restriction digestion, the fragment was gel isolated. The size specificity of the restriction fragments was verified by automated gel electrophoresis on the Fragment Analyzer (AATI) using the High Sensitivity Next Generation Sequencing Fragment Analysis kit (DNF‐474, AATI) according to the manufacturer's instructions (49).
Zeta Potential and DLS Measurements.
The zeta potential of rhMBL and rhMBL in complex with linearized dsDNA was measured at 25 °C using a Zetasizer Nano ZS (Malvern Instruments). Samples for zeta potential measurements were measured in PBS/1 mM EDTA pH 7.4 diluted 1:10 in milli-Q water to reduce ionic strength for stable measurements. Each zeta potential measurement consisted of 11 to 13 technical replicates. For DLS determination of the particle size distribution, samples were diluted in PBS/1 mM EDTA pH 7.4. The temperature was calibrated for 30 s before measurement. For each measurement, the technical replicates consisted of 42 to 46 replicates.
564Igi Homozygous Mice and C57BL/6J Controls.
C57BL/6JRj mice were purchased from Janvier Labs. The 564Igi line (30) was kindly provided by Michael C. Carroll, Boston Children’s Hospital, Boston, MA, and maintained in-house. All mice were housed under specific pathogen-free conditions, using a 12-h light/dark cycle, and were fed standard chow and water ad libitum. All animal usage adhered to the European Community guidelines and was approved by the Danish Animal Experiments Inspectorate (protocol no. 2017-15-0201-01319).
Anti-dsDNA Ab Measurements.
Salmon sperm dsDNA (ThermoFisher) for coating was diluted to 500 µg/mL in PBS, filtered through a 0.45-μm filter (blocked with tris-buffered saline [TBS]/TW), and then further diluted to 100 µg/mL in PBS. Wells were coated by incubation overnight at 4 °C. Wells were then blocked with 200 µl TBS/1% (weight/[vol]volume) bovine serum albumin (BSA) for 1 h at room temperature (RT) following three times wash in TBS/Tween (TBS/TW). Controls were diluted 1/100 in TBS/TW/5mM EDTA/0.1% BSA (TBS/TW/EDTA/BSA). Samples were diluted 1/50 and 1/250 in TBS/TW/EDTA/BSA. Standard was diluted 1/300, and a further seven times, in threefold dilutions with TBS/TW/EDTA/BSA. Samples, controls, and standards were added in TBS/TW/EDTA/BSA and then incubated for 1 h at 37 °C. Plates were washed three times with TBS/Tween and incubated for 1 h at 37 °C with freshly prepared biotinylated Goat anti-mouse IgG (1010-08; SouthernBiotech) 1 µg/mL in TBS/Tween. Plates were then washed three times with TBS/Tween and incubated for 1 h at RT with Eu-labeled streptavidin freshly diluted 1/1,000 in TBS/Tween/5 mM EDTA. Plates were then washed three times with TBS/Tween and then added 200 µL enhancement solution. Plates were read by time-resolved fluorometry using a dissociation-enhanced lanthanide fluorescence immunoassay reader (Perkin-Elmer).
Germinal Center B Cell Quantification.
Mice were euthanized in anesthesia, and the spleen was removed and transferred to fluorescence-activated cell sorting (FACS) buffer (PBS with 2% [vol/vol] fetal bovine serum, 1 mM EDTA). Tissue was crushed with a pestle in 1.5-mL tubes and filtered through a 70-µm cell strainer. Cells were centrifuged at 200 × g for 5 min at 4°. Pellet was resuspended in 500 µL 1 × Red Blood Cell Lysis Buffer (155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA, pH∼7.3). After 3 to 5 min, 1 mL FACS buffer was added. Cells were collected at 200 × g for 5 min at 4°. Cells were resuspended in 750 µl FACS buffer. A 100-µl cell suspension was transferred to sample wells with spleen samples pooled to create a cell mixture for unstained, single-stain, and all-stain controls. All Ab dilutions were diluted 1/500 with a panel consisting of B220-Pacific Blue (558108; BD Biosciences), CD4-PerCP (561090; BD Biosciences), CD8-PerCP-Cy5.5 (100734; Biolegend), CD38-PE-Cy7 (102718; Biolegend), and CD95-PE (554258; BD Biosciences). The Live/Dead marker (65-0865-18; eFluor780, ThermoFisher) was diluted 1/2,000. A volume of 100-µl staining solution was added directly into each well and incubated for 30 min on ice in the dark. Plates were centrifuged as above, followed by resuspension of the cells in 200 µL FACS buffer. Washing step was repeated in 200 µl FACS buffer. All analyses were made on a BD LSRFortessa Flow Cytometer (BD Biosciences).
Immunostaining and Confocal Microscopy.
Spleens were harvested from 564Igi and C57BL/6J mice, embedded in optimal cutting temperature medium (Tissue-Tek, Sakura Finetec) and frozen. Sections measuring 15 to 20 µm in thickness were cut on a Cryostar NX70 cryostat (ThermoFisher) and mounted on SuperFrost+ glass slides. Sections were washed (PBS), fixed (4% Para-Formaldehyde), blocked (Tris-buffered saline), and permeabilized (PBS, 2% Fetal Bovine Serum, 0.1% Triton-X-100). Staining buffer (PBS, 2% Fetal Bovine Serum) containing anti–Ki-67-eFluor660 (50-5698-82; ThermoFisher Scientific), anti–CD21/35-Pacific Blue (123414; Biolegend), and anti–CD169-PE (142404; Biolegend) all diluted 1/500 was added to the sections and incubated overnight. Three washes with wash buffer (PBS, 0.05% Tween20) were performed, before mounting coverslips with Slowfade mounting medium (S36967; ThermoFisher). Imaging was performed on a Zeiss LSM800 confocal microscope (Carl Zeiss Microscopy GmbH).
TDA.
TDA describes the mass distribution of a plug of sample material when subjected to a pressure driven flow (27). The central parameters describing the sample dispersion include the following: radius of flow channel (blood vein in the current work), convection (flow rate determined by pressure), and diffusivity of the sample material. The diffusivity is inversely related to the hydrodynamic radius according to the Stokes–Einstein equation; large particles are therefore dispersed differently than small particles. The relative importance of convection over diffusivity can be rationalized from the Peclet number (Pe):
where a is the radius, u0 is the linear flow rate, and D is the diffusivity. τ is a dimensionless parameter related to the time required for the species to diffuse across the blood vein:
where t is the observation time.
For very large oligomers where no diffusion takes place (Pe >> 1) over the time frame of observation, the sample distribution simply adopts an exponentially declining distribution as has been previously described (26, 50). Provided diffusion dominates Pe <<1 and τ > 1, the sample distribution will adopt a symmetrical shape (26, 27).
Statistical Analysis and Reproducibility.
All statistical analyses were performed using Prism software (GraphPad Software). A t test with Welch’s correction for unequal variance was used for data set following a normal distribution according to QQ plot. For data sets not meeting these criteria, a Mann–Whitney U test was applied. A P value < 0.05 was considered to be statistically significant. All information on number of donors tested, number of experiments, and applied statistical tests are stated in figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Bent W. Deleuran and Dr. Per B. Fischer for advice and Bettina W. Grumsen, Annette G. Hansen, and Lisbeth Jensen for excellent technical assistance. Dr. Anne Færch Nielsen kindly commented on a draft of the manuscript. Flow cytometry was performed at the FACS Core Facility, Aarhus University, Denmark. Imaging was performed at the Imaging Core Facility, Aarhus University, Denmark. K.J.-M. and T.V.-J. were generously supported by grants from Aarhus University Research Foundation “NOVA” program (AUFF-E-2015FLS-9-6) and the Novo Nordisk Foundation (0058516). S.T., D.S.S., and S.E.D. acknowledge funding from the CellPAT center supported by the Danish National Research Foundation (DNRF135). L.H.K. acknowledges support from the Carlsberg Foundation. S.L. was supported by a PhD stipend from the Boehringer Ingelheim Fonds. A.T. was supported by grants from Lundbeck Fonden (R264-2017-3344) and the Danish Rheumatism Association (R166-A5554). This work was supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, NIH.
Footnotes
Competing interest statement: K.J.-M., A.T., K.S.-P., H.J.M., and T.V.-J. are inventors on a submitted patent application (PCT/EP2020/082837) related to the discoveries of the present paper and owned by Aarhus University.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106647118/-/DCSupplemental.
Data Availability
This study used data from a subset of the SLE cohort published previously (14). The selected data are shown in SI Appendix, Fig. S5A and B. All other study data are also included in the article and/or supporting information.
References
- 1.Gonzalez S. F., et al., Trafficking of B cell antigen in lymph nodes. Annu. Rev. Immunol. 29, 215–233 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Vorup-Jensen T., Jensen R. K., Structural immunology of complement receptors 3 and 4. Front. Immunol. 9, 2716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajic G., Yatime L., Sim R. B., Vorup-Jensen T., Andersen G. R., Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3. Proc. Natl. Acad. Sci. U.S.A. 110, 16426–16431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jalilian B., et al., Properties and prospects of adjuvants in influenza vaccination - Messy precipitates or blessed opportunities? Mol. Cell. Ther. 1, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moghimi S. M., Hunter A. C., Andresen T. L., Factors controlling nanoparticle pharmacokinetics: An integrated analysis and perspective. Annu. Rev. Pharmacol. Toxicol. 52, 481–503 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Kaul A., et al., Systemic lupus erythematosus. Nat. Rev. Dis. Primers 2, 16039 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Degn S. E., et al., Clonal evolution of autoreactive germinal centers. Cell 170, 913–926.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paludan S. R., Reinert L. S., Hornung V., DNA-stimulated cell death: Implications for host defence, inflammatory diseases and cancer. Nat. Rev. Immunol. 19, 141–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flyvbjerg A., The role of the complement system in diabetic nephropathy. Nat. Rev. Nephrol. 13, 311–318 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Tokatlian T., et al., Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 363, 649–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palaniyar N., et al., Nucleic acid is a novel ligand for innate, immune pattern recognition collectins surfactant proteins A and D and mannose-binding lectin. J. Biol. Chem. 279, 32728–32736 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Stuart L. M., Takahashi K., Shi L., Savill J., Ezekowitz R. A., Mannose-binding lectin-deficient mice display defective apoptotic cell clearance but no autoimmune phenotype. J. Immunol. 174, 3220–3226 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Troldborg A., et al., Levels in plasma of the serine proteases and associated proteins of the lectin pathway are altered in patients with systemic lupus erythematosus. J. Rheumatol. 42, 948–951 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Troldborg A., et al., The lectin pathway of complement activation in patients with systemic lupus erythematosus. J. Rheumatol. 45, 1136–1144 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Holmskov U., Thiel S., Jensenius J. C., Collections and ficolins: Humoral lectins of the innate immune defense. Annu. Rev. Immunol. 21, 547–578 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Weis W. I., Drickamer K., Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 65, 441–473 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Sheriff S., Chang C. Y., Ezekowitz R. A., Human mannose-binding protein carbohydrate recognition domain trimerizes through a triple alpha-helical coiled-coil. Nat. Struct. Biol. 1, 789–794 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Weis W. I., Drickamer K., Trimeric structure of a C-type mannose-binding protein. Structure 2, 1227–1240 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Jensenius H., et al., Mannan-binding lectin: Structure, oligomerization, and flexibility studied by atomic force microscopy. J. Mol. Biol. 391, 246–259 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Gjelstrup L. C., et al., The role of nanometer-scaled ligand patterns in polyvalent binding by large mannan-binding lectin oligomers. J. Immunol. 188, 1292–1306 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Vorup-Jensen T., Boesen T., Protein ultrastructure and the nanoscience of complement activation. Adv. Drug Deliv. Rev. 63, 1008–1019 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Chaturvedi S. K., et al., Measuring aggregates, self-association, and weak interactions in concentrated therapeutic antibody solutions. MAbs 12, 1810488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giepmans B. N., Adams S. R., Ellisman M. H., Tsien R. Y., The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Dong M., et al., Conformational changes in mannan-binding lectin bound to ligand surfaces. J. Immunol. 178, 3016–3022 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Degn S. E., et al., Complement activation by ligand-driven juxtaposition of discrete pattern recognition complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 13445–13450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamieh J., et al., Limits in size of Taylor dispersion analysis: Representation of the different hydrodynamic regimes and application to the size-characterization of cubosomes. Anal. Chem. 89, 13487–13493 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Taylor G., Dispersion of soluble matter in solvent flowing slowly through a tube Proc. R. Soc. Lond. A 219, 186–203 (1953). [Google Scholar]
- 28.Bombardier C., Gladman D. D., Urowitz M. B., Caron D., Chang C. H., Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 35, 630–640 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Trouw L. A., Pickering M. C., Blom A. M., The complement system as a potential therapeutic target in rheumatic disease. Nat. Rev. Rheumatol. 13, 538–547 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Berland R., et al., Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity 25, 429–440 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Hansen S., Thiel S., Willis A., Holmskov U., Jensenius J. C., Purification and characterization of two mannan-binding lectins from mouse serum. J. Immunol. 164, 2610–2618 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Vorup-Jensen T., On the roles of polyvalent binding in immune recognition: Perspectives in the nanoscience of immunology and the immune response to nanomedicines. Adv. Drug Deliv. Rev. 64, 1759–1781 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Vu V. P., et al., Immunoglobulin deposition on biomolecule corona determines complement opsonization efficiency of preclinical and clinical nanoparticles. Nat. Nanotechnol. 14, 260–268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F., et al., Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol. 12, 387–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Megyeri M., et al., Quantitative characterization of the activation steps of mannan-binding lectin (MBL)-associated serine proteases (MASPs) points to the central role of MASP-1 in the initiation of the complement lectin pathway. J. Biol. Chem. 288, 8922–8934 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giles B. M., Boackle S. A., Linking complement and anti-dsDNA antibodies in the pathogenesis of systemic lupus erythematosus. Immunol. Res. 55, 10–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldmann M., Brennan F. M., Maini R. N., Rheumatoid arthritis. Cell 85, 307–310 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Petersen K. A., et al., Phase I safety, tolerability, and pharmacokinetic study of recombinant human mannan-binding lectin. J. Clin. Immunol. 26, 465–475 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Vorup-Jensen T., et al., Recombinant expression of human mannan-binding lectin. Int. Immunopharmacol. 1, 677–687 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Pihl R., et al., ITIH4 acts as a protease inhibitor by a novel inhibitory mechanism. Sci. Adv. 7, eaba7381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drogari-Apiranthitou M., et al., The effect of mannan-binding lectin on opsonophagocytosis of Neisseria meningitidis. Immunopharmacology 38, 93–99 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Juul-Madsen K., et al., Size-selective phagocytic clearance of Fibrillar α-Synuclein through conformational activation of complement receptor 4. J. Immunol. 204, 1345–1361 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Ghirlando R., et al., Improving the thermal, radial, and temporal accuracy of the analytical ultracentrifuge through external references. Anal. Biochem. 440, 81–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desai A., Krynitsky J., Pohida T. J., Zhao H., Schuck P., 3D-printing for analytical ultracentrifugation. PLoS One 11, e0155201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaturvedi S. K., Ma J., Zhao H., Schuck P., Use of fluorescence-detected sedimentation velocity to study high-affinity protein interactions. Nat. Protoc. 12, 1777–1791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H., et al., A multilaboratory comparison of calibration accuracy and the performance of external references in analytical ultracentrifugation. PLoS One 10, e0126420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hochberg M. C., Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40, 1725 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Ibsen M. S., et al., The 2′-5′-oligoadenylate synthetase 3 enzyme potently synthesizes the 2′-5′-oligoadenylates required for RNase L activation. J. Virol. 88, 14222–14231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luecke S., et al., cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 18, 1707–1715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Latunde-Dada S., Bott R., Hampton K., Leszczyszyn O. I., Application of the exact dispersion solution to the analysis of solutes beyond the limits of Taylor dispersion. Anal. Chem. 87, 8021–8025 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used data from a subset of the SLE cohort published previously (14). The selected data are shown in SI Appendix, Fig. S5A and B. All other study data are also included in the article and/or supporting information.