Significance
Transfer RNA (tRNA) transfer appropriately activated amino acids to the end of the growing peptide chain, playing significant roles in primary metabolism. Importantly, tRNA have also been shown to function outside of translation. In this report, we present the characterization of a new class of aminoacyl-tRNA (aa-tRNA)–dependent enzyme, BlsK, from Streptomyces griseochromogenes. BlsK catalyzes an aa-tRNA–dependent leucyl transfer reaction in blasticidin S biosynthesis. BlsK contains a [3Fe-4S] cluster that is necessary for its function and unprecedented in previously identified tRNA-dependent transferases. Because BlsK does not show similarity to any known functional protein, it may represent a type of aa-tRNA–dependent enzyme. As such, this work has broad implications for both tRNA-dependent and iron–sulfur–containing enzymes.
Keywords: tRNA-dependent enzymes, natural product, iron–sulfur cluster, leucyl transfer reaction
Abstract
Blasticidin S is a peptidyl nucleoside antibiotic. Its biosynthesis involves a cryptic leucylation and two leucylated intermediates, LDBS and LBS, have been found in previous studies. Leucylation has been proposed to be a new self-resistance mechanism during blasticidin S biosynthesis, and the leucyl group was found to be important for the methylation of β-amino group of the arginine side chain. However, the responsible enzyme and its associated mechanism of the leucyl transfer process remain to be elucidated. Here, we report results investigating the leucyl transfer step forming the intermediate LDBS in blasticidin biosynthesis. A hypothetical protein, BlsK, has been characterized by genetic and in vitro biochemical experiments. This enzyme catalyzes the leucyl transfer from leucyl-transfer RNA (leucyl-tRNA) to the β-amino group on the arginine side chain of DBS. Furthermore, BlsK was found to contain an iron–sulfur cluster that is necessary for activity. These findings provide an example of an iron–sulfur protein that catalyzes an aminoacyl-tRNA (aa-tRNA)–dependent amide bond formation in a natural product biosynthetic pathway.
Amide bond formation in natural product biosynthesis can be catalyzed by nonribosomal peptide synthetases (NRPSs) and ATP-grasp ligases including the ATP-dependent activation of amino acid substrates. The carboxyl group is activated by phosphorylation and adenylation, respectively, in ATP-grasp ligase and NRPS catalyzed reactions (1). Recently, a new family of enzymes has been found to catalyze amide bond formation using aminoacyl-transfer RNA (aa-tRNA) as an activated cosubstrate. aa-tRNA plays a profound role in cells connecting the messenger RNA (mRNA) and protein synthesis at the ribosome in primary metabolism (2). Interestingly, recent studies have revealed that aa-tRNA can also be involved in natural product biosynthesis (3). aa-tRNA–dependent enzymes involved in natural product biosynthesis mainly form three groups: amide bond-forming ligases homologous with FemX peptidyltransferases from cell wall biosynthesis (4), synthase enzymes including the cyclodipeptide synthase family (5, 6), and dehydratase enzymes in RiPP (ribosomally synthesized and posttranslationally modified peptide) synthesis (7).
Blasticidin S (BS), a representative of the amino hexose pyrimidine nucleoside antibiotics, was first isolated from Streptomyces griseochromogenes (8). The structure of BS features a C2′, C3′-dehydrated pyrose ring that is connected with cytosine at C1′ and β-arginine at C4′ (9). BS shows a broad spectrum of biological activities and had been widely used as a fungicide to protect rice from blast diseases (10, 11). Presently, BS is commonly used as an efficient selection antibiotic for transformed mammalian cells that express appropriate resistance genes (12, 13).
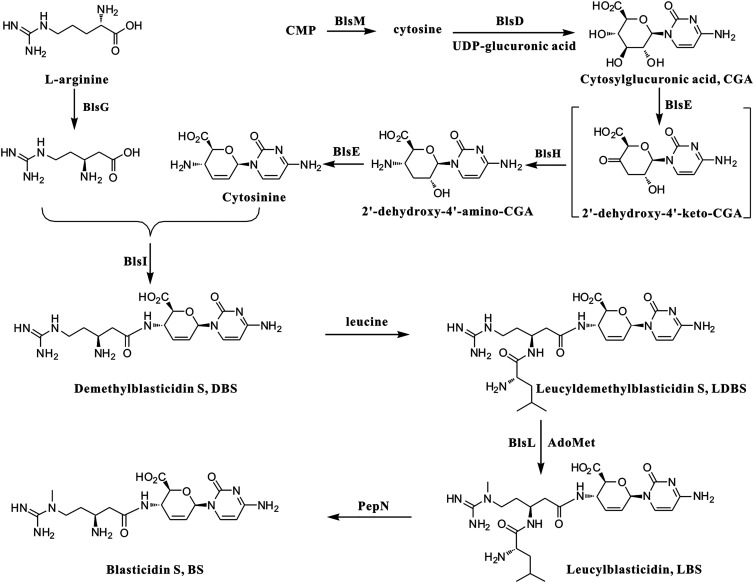
The structural features and commercial value of BS stimulated interest in its biosynthetic studies. Early feeding experiments and characterization of related metabolites determined that glucose, cytosine, L-arginine, and methionine are the basic precursors for BS biosynthesis (14). The biosynthesis gene cluster was reported in 2003 and enabled a proposal for the BS biosynthetic pathway (Fig. 1) (15). However, difficulty carrying out gene knockout experiments in the native producer, S. griseochromogenes, hindered the further characterization of each biosynthetic step. More recently, the BS biosynthetic cluster was engrafted into the chromosome of Streptomyces lividans to generate the genetically stable mutant strain WJ2, which is able to produce BS and thereby facilitating in vivo studies of the function of BS biosynthetic genes (13). Gene deletions in WJ2 determined the essential genes for BS biosynthesis including blsD-blsL that are transcribed in one direction and the divergently transcribed blsM. The first committed step of BS biosynthesis is the hydrolysis of cytidine monophosphate by BlsM to produce free cytosine, which is then condensed with UDP-glucuronic acid by BlsD to form cytosyl-glucuronic acid (CGA) (15, 16). The process from CGA to cytosinine, a putative intermediate for BS biosynthesis, remains to be determined. A radical SAM protein BlsE and an aminotransferase BlsH are possibly involved in this transformation (13, 15). The β-arginine moiety of BS is derived from L-arginine through a radical-mediated reaction catalyzed by BlsG, a 2,3-arginine aminomutase (15, 17). It remains unknown whether BlsI, a putative ligase, is involved in the formation of DBS (demethylblasticidin S) by coupling of the carboxyl group of β-arginine and the amido group at C4′ of cytosinine since mutant WJ2∆blsI only accumulates the very early intermediate CGA. It is worth noting that DBS cannot be directly methylated to BS by the N-methyltransferase BlsL, which was confirmed by in vitro biochemical characterization (18).
Fig. 1.
The proposed biosynthetic pathway of BS. CMP: Cytidine 5-monophosphate; AdoMet: S-Adenosylmethionine.
Addition of leucine to DBS at the β-amino group of the arginine side chain forms leucyldemethylblasticidin S (LDBS) which is then methylated by BlsL to form the penultimate product leucylblasticidin S (LBS) (18). Finally, PepN catalyzes the maturation of BS biosynthesis via the hydrolysis of the leucyl group of LBS (19). The cell toxicity of LDBS and LBS are much lower than DBS and BS. Therefore, it is proposed that the circuitous modifications of BS and DBS with a leucyl group function as the self-resistance mechanism during BS biosynthesis (16). Attempts to form LDBS from DBS in a S. griseochromogenes cell-free system were not successful. A plausible reason for the failure of LDBS formation was that even if LDBS could be formed, it would be readily hydrolyzed back to DBS by the conserved aminopeptidase PepN in the cell-free system (16, 19).
The intermediacy of LBS and LDBS hints at the necessity of an extra ligase other than BlsI in BS biosynthesis (15). In this report, we disclose the mechanism of the cryptic leucylation involved in BS biosynthesis. By combining in vivo gene inactivation and in vitro biochemical assays, we demonstrate that BlsK catalyzes the tRNA-dependent transfer of a leucyl group to the DBS β-arginine moiety by directing leucyl-tRNALeu to the BS biosynthetic pathway. More interestingly, BlsK is determined to be an iron–sulfur enzyme that does not show similarity with any known iron–sulfur enzymes. BlsK contains a [3Fe-4S] cluster that is critical for its activity. An iron–sulfur protein was shown to be involved in the amide bond formation. These results pave the way to fully understand the self-resistance and biosynthesis of BS.
Results
BlsK Is Necessary for the BS Biosynthesis.
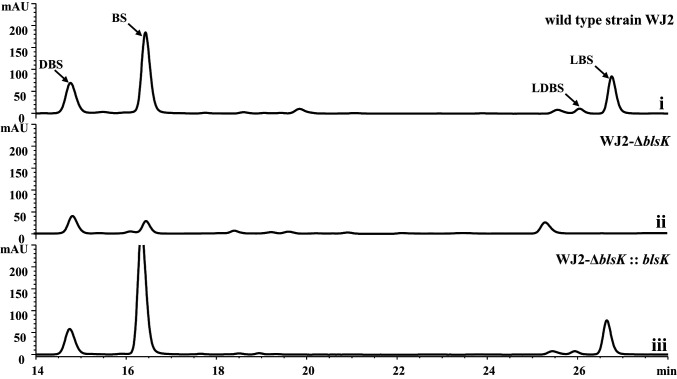
Two leucylated intermediates, LDBS and LBS, have been found to be involved in BS biosynthesis. To date, the mechanism of the peptide bond formation between leucine and the β-arginine amine is still unknown. The most common strategy used to form peptide bonds in secondary metabolism is through the NRPS system (20). According to the reported BS biosynthetic cluster, there is no NRPS-like enzyme involved in BS formation, indicating that a different mechanism may be involved (21). It was suggested that BlsK is the most likely candidate for the attachment of a leucyl group in BS biosynthesis (15). The recently published genome sequence of S. griseochromogenes ATCC 14511 indicates that the blsK sequence is 1,875 bp instead of the previously reported 1,740 bp (21). When queried in the National Center for Biotechnology Information (NCBI) database, BlsK shares no close homology with any proteins of known function, and structural prediction by HHpred also did not identify any homologous proteins with known structures (22). Cone et al. reported that BlsK shows modest sequence similarity with lysyl-tRNA synthetase and might be involved in leucyl transfer in BS biosynthesis (15). To explore the function of blsK in the BS biosynthetic gene cluster, blsK was deleted in S. lividans WJ2. High performance liquid chromatography (HPLC) analysis of the fermentation culture of the ΔblsK mutant strain showed that the production of BS was dramatically decreased indicating that the BS biosynthesis was significantly affected (Fig. 2). Furthermore, intermediates containing the leucyl group, LDBS and LBS, which accumulated in wild-type WJ2, could not be detected in the ΔblsK mutant. To exclude a possible polar effect, the mutant strain was complemented with the integrated shuttle plasmid pIB139 containing blsK under the control of the ermE promoter. The production of BS was restored to a similar level as the wild-type strain and the two leucyl intermediates, LDBS and LBS, were again observed (Fig. 2). Collectively, these data demonstrate that BlsK is involved in BS biosynthesis, and the disappearance of the leucylated intermediates suggests BlsK might function in the attachment of the leucyl group.
Fig. 2.
HPLC analysis of blsK knockout and its complementation experiments in strain WJ2: (i) wild-type strain WJ2 as control; (ii) blsK gene knockout strain WJ2-ΔblsK (WXK3); and (iii) blsK complementation strain WJ2-ΔblsK:: blsK (WXK4).
In Vitro Characterization of BlsK.
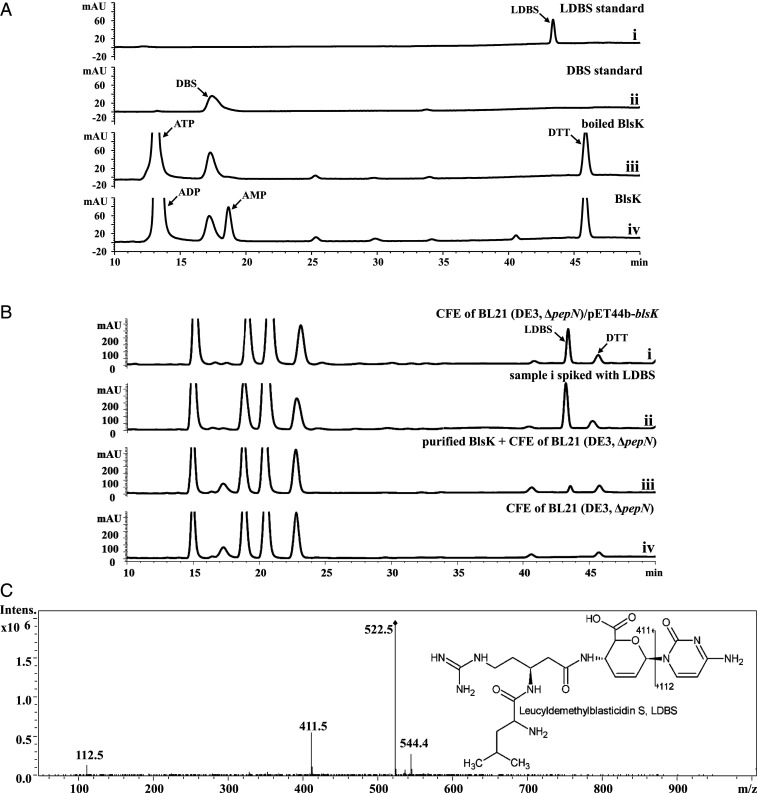
In order to directly investigate the exact role of BlsK in the biosynthesis of BS, recombinant BlsK was overexpressed as a C-terminal His-tagged fusion protein and purified to homogeneity. Due to the proposed function of BlsK as a leucyl transferase, we first assessed activity in vitro with DBS plus leucine and ATP as potential substrates. ATP was added, because the carboxyl group of leucine must be activated for amide bond formation and the activation is often ATP dependent (20). However, no formation of LDBS was observed after the assay mixture was incubated in 37 °C for half an hour (Fig. 3A). Regardless of how assay conditions were changed, such as longer incubation times or varied pH, no LDBS could be detected. To exclude the possibility that the activity was lost during the purification process, Escherichia coli BL21 (DE3, ΔpepN)/pET44b-blsK was cultured, induced with IPTG, and the cell-free extract (CFE) was directly incubated with DBS, leucine, and ATP. Given that E. coli PepN is able to excise the leucyl group from LDBS (19), E. coli BL21 (DE3, ΔpepN) was used to avoid the interference of PepN activity (SI Appendix, Fig. S1). Under these conditions, HPLC analysis revealed efficient production of LDBS, which was confirmed by spiking with an authentic LDBS standard and liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis (Fig. 3 B and C). When the CFE of E. coli BL21 (DE3, ΔpepN)/pET44b was used as control, no production of LDBS was detected, indicating the formation of LDBS was BlsK dependent (Fig. 3B). The above results clearly show that BlsK in the CFE has the ability to catalyze the leucyl transfer, indicating that either the purified BlsK lost activity after purification or it requires some cofactor(s) from the CFE of E. coli BL21 (DE3, ΔpepN) to carry out its function. To discriminate between these two possibilities, purified BlsK was incubated with DBS, leucine, ATP, and CFE of E. coli BL21 (DE3, ΔpepN). HPLC analysis indicated the generation of LDBS, although in relatively low efficiency (Fig. 3B). Together, these results clearly demonstrate that BlsK is active in vitro, and the CFE of E. coli BL21 (DE3, ΔpepN) facilitates the transfer of the leucyl group to DBS.
Fig. 3.
Functional analysis of BlsK in vitro. (A) Assay with purified BlsK: (i) the LDBS standard; (ii) DBS standard; (iii) boiled BlsK as a negative control; and (iv) assay with 1 mM ATP, 10 mM leucine, and 250 μM DBS. (B) Assay of BlsK with CFE: (i) CFE of BL21 (DE3, ΔpepN)/pET44b-blsK with DBS; (ii) sample i spiked with LDBS standard; (iii) purified BlsK adding CFE of BL21 (DE3, ΔpepN) with DBS, and (iv) CFE of BL21 (DE3, ΔpepN) with DBS. In total, 1 mM ATP and 5 mM leucine were added in all groups. (C) MS/MS spectrum of the product of BlsK-catalyzed reaction (sample [B] 1): chemical structure of LDBS with the key fragmentation pattern shown. The reaction conditions were pH 8.0 and 37 °C for overnight.
BlsK Is a leucyl-tRNA–Dependent Transferase.
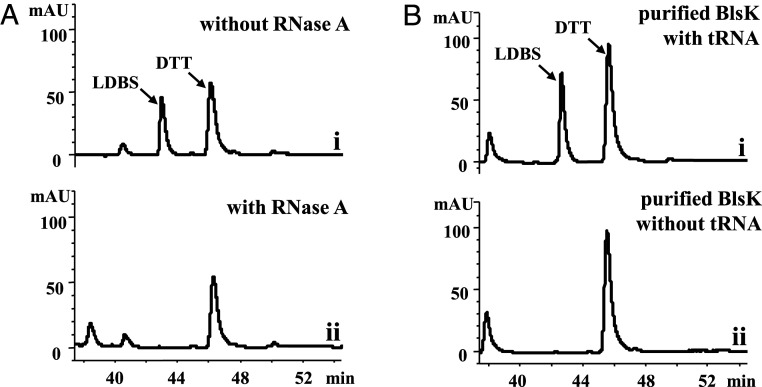
To gain further insight into the mechanism of BlsK and explore the exact reasons that purified BlsK cannot catalyze the leucyl transfer without CFE, we looked for the active component(s) in E. coli BL21 (DE3, ΔpepN) CFE necessary for BlsK to transfer the leucyl group. Based on some similarity of BlsK with lysyl-tRNA synthetase, we speculated that tRNA in the CFE might help BlsK carry out its function (15). To test this hypothesis, we first investigated whether tRNA is necessary for the activity of BlsK. RNase A was added to BL21 (DE3, ΔpepN) CFE to decompose endogenous tRNA. When the RNase A–treated CFE was incubated with purified BlsK, DBS, leucine, and ATP, no LDBS was produced, suggesting that BlsK catalyzes the leucyl transfer reaction in a tRNA-dependent manner (Fig. 4A). To further confirm the role of tRNA, E. coli total tRNA was added to the reaction mixtures (purified BlsK, DBS, leucine, and ATP). Under these conditions, LDBS was produced efficiently (Fig. 4B). Together these results show that tRNA is necessary for the BlsK-catalyzed leucyl transfer reaction. We propose that leucine is first activated as leucyl-tRNA and then transferred to DBS to form LDBS. However, no exogenous leucyl-tRNA synthetase (LeuRS) was added to the reaction mixture, indicating that BlsK may catalyze the leucyl-tRNA formation and then transfer the leucyl group from leucyl-tRNA to DBS consecutively. In addition, there is the possibility that the endogenous LeuRS of E. coli was copurified with BlsK and is responsible for the leucine activation when purified BlsK was used in the in vitro activity assay.
Fig. 4.
The catalytic activity of BlsK requires tRNA. (A) Effect of RNase A on BlsK activity: (i) positive control of BL21 (DE3, ΔpepN) CFE with BlsK and (ii) BL21 (DE3, ΔpepN) CFE treated by RNase A with BlsK. (B) Assay of purified BlsK with tRNAs: (i) adding 1 mg/mL tRNAs to the purified BlsK reaction system and (ii) the control without tRNAs. In total, 1 mM ATP, 5 mM leucine, and 20 mM MgCl2 were added in all reactions, pH 8.0, 37 °C overnight.
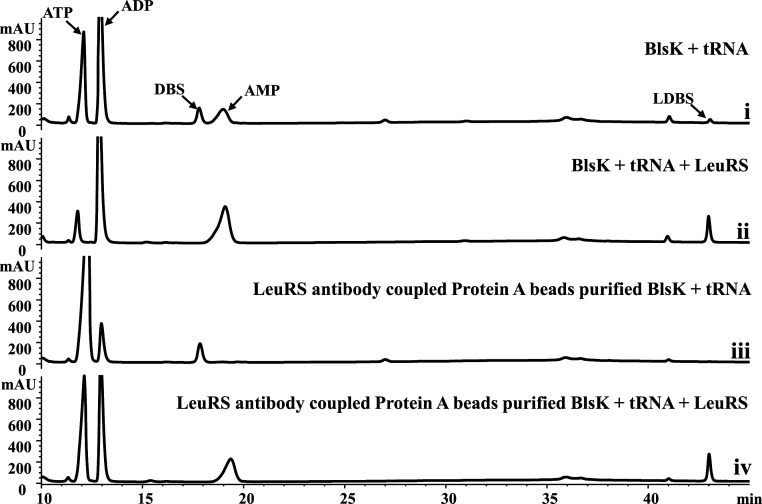
To investigate whether BlsK has leucyl-tRNA synthase activity, purified LeuRS was added to the previous reaction system (purified BlsK, DBS, leucine, E. coli total tRNAs, and ATP). The production of LDBS was increased by about 10-fold, indicating the important role of LeuRS in the leucyl transfer process (Fig. 5 and SI Appendix, Fig. S2). No LDBS was produced when BlsK was not added in the control experiment (SI Appendix, Fig. S2). Thus, we suspect that BlsK purified by affinity chromatography is likely to contain trace amounts of LeuRS, which is responsible for the activation of leucine in the original in vitro BlsK activity assay lacking exogenous addition of LeuRS. In order to confirm this speculation, an antibody against E. coli LeuRS was generated and coupled to protein A beads then used to remove any potential contaminated LeuRS from affinity and size-exclusion chromatography–purified BlsK. This further-purified BlsK was then assayed for LDBS production without the addition of LeuRS. Indeed, BlsK itself cannot transfer leucine to DBS in the presence DBS, leucine, E. coli total tRNAs, and ATP (Fig. 5). When LeuRS was added to the reaction mixture, the formation of LDBS was restored (Fig. 5). Hence, our results strongly suggest that BlsK catalyzes the transfer of leucine to DBS via leucyl-tRNA, whose formation is catalyzed by LeuRS in primary metabolic pathway. BlsK itself does not have leucyl-tRNA synthase activity.
Fig. 5.
LeuRS is necessary for the activation of leucine in the BlsK-catalyzed leucyl transfer reaction: (i) Ni-NTA–purified BlsK incubated with DBS and E.coli total tRNAs; (ii) Ni-NTA–purified BlsK incubated with DBS, E.coli total tRNAs, and LeuRS; (iii) LeuRS antibody–Protein A bead–purified BlsK incubated with DBS and E.coli total tRNAs; and (iv) LeuRS antibody–treated BlsK incubated with DBS, E.coli total tRNAs, and LeuRS. In total, 1 mM ATP, and 5 mM leucine were added in all reaction mixtures. Reaction conditions: 37 °C for 1 h.
To further prove that BlsK can directly use leucyl-tRNALeu as a substrate, we generated tRNALeu by in vitro transcription, which was then aminoacylated by LeuRS to obtain the leucyl-tRNALeu. The results showed that when leucyl-tRNALeu was added to reaction mixtures containing BlsK and DBS, LDBS was generated, and the amount of LDBS produced was proportional to the amount of added leucyl-tRNALeu (SI Appendix, Fig. S3). This clearly indicates BlsK is a leucyltransferase that steals the activated leucyl-tRNALeu formed by LeuRS and then transfers the leucyl group to DBS to generate LDBS. At the same time, we found that the amount of LDBS produced is comparable using either mature E. coli tRNAs or the tRNA obtained from in vitro transcription as cosubstrates (SI Appendix, Fig. S3), indicating that posttranscriptional modification does not affect recognition of tRNA by BlsK.
BlsK Contains a [3Fe-4S] Cluster.
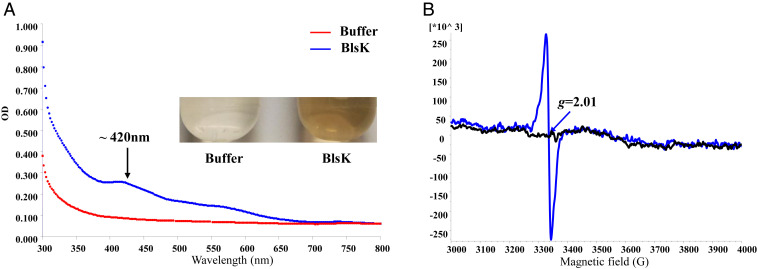
Purified BlsK is brownish in color, with a broad absorption band at around 420 nm in the ultraviolet-visible (UV-visible) spectrum (Fig. 6A). Addition of dithiothreitol (DTT) darkened the color of BlsK slightly and increased the UV-visible absorption, while addition of sodium dithionite bleached the brownish color of BlsK and abolished the absorption around 420 nm (SI Appendix, Fig. S4). Although the sequence of BlsK does not show similarity with any known iron–sulfur protein, its color and features within the UV-visible spectrum are characteristic of iron–sulfur proteins. To confirm the presence of iron–sulfur cluster and characterize the cluster, chemical composition analysis and electron paramagnetic resonance (EPR) were used. Considering most iron–sulfur clusters in iron–sulfur proteins are sensitive to oxygen and not stable under aerobic conditions, BlsK was purified and further reconstituted under anoxic conditions and chemical analysis was used to measure the contents of iron and sulfur in the protein. This revealed 3.5 ± 0.4 Fe and 4.1 ± 0.2 S atoms per molecule of BlsK, indicating the existence of iron–sulfur cluster. For a more accurate analysis of the iron and sulfur content, inductively coupled plasma mass spectrometry (ICP-MS) analysis was employed and demonstrated that one molecule of BlsK contained 3.2 ± 0.2 Fe and 4.3 ± 0.3 S atoms, suggesting a possible [3Fe-4S] cluster in BlsK. To confirm this hypothesis, EPR analysis was performed. The EPR results showed that BlsK had a signal of g = 2.01 (S = 1/2) (Fig. 6B), which is characteristic of [3Fe-4S]+ clusters. When the reducing agent sodium dithionite was added, the EPR signal disappeared, indicating that [3Fe-4S]+ was reduced to [3Fe-4S]0, which is EPR silent (Fig. 6). The activity of BlsK was not affected with the reduction of [3Fe-4S]+ by dithionite (SI Appendix, Fig. S5). Collectively, UV-visible and EPR spectra combined with iron and sulfur contents analysis are most consistent with the presence of [3Fe-4S] cluster in BlsK.
Fig. 6.
Spectroscopic characterization of recombinant BlsK. (A) Color and UV-visible spectrum analysis of BlsK protein. The blue curve represents BlsK protein and red curve is the buffer control. (B) EPR analysis of BlsK with (black) and without (blue) addition of 1 mM sodium dithionite. EPR parameters: microwave frequency, 9.39 GHz; microwave power, 2 mW; modulation amplitude, 3 Gauss; modulation frequency, 100 kHz; and temperature, 10 K. The protein concentration used was 10 mg /mL.
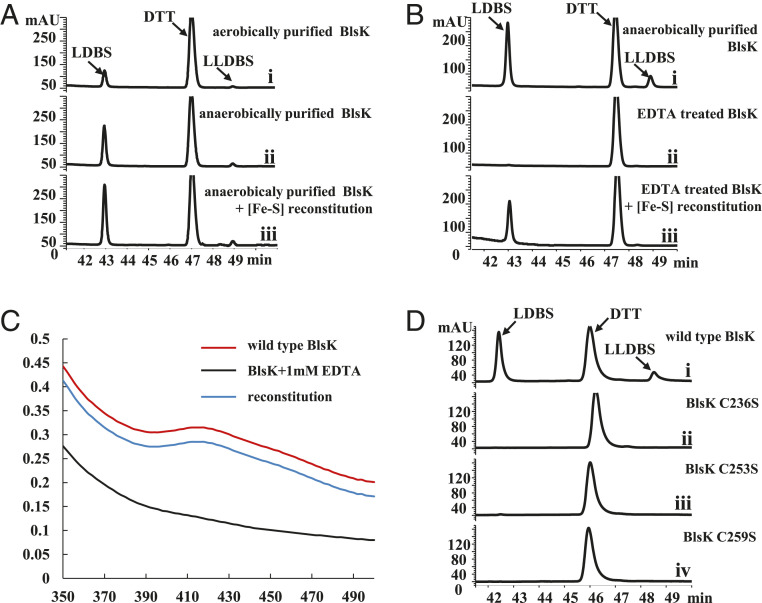
To further characterize the [3Fe-4S]+ cluster of BlsK, we identified the amino acid residues involved in the binding of the [3Fe-4S] cluster. As BlsK does not show similarity with known iron–sulfur proteins, MetalPredator was used to predict the metal-binding sites but gave no information about BlsK. The most common iron–sulfur cluster binding sites are cysteine residues (23). In order to determine which cysteine residues in BlsK may be involved in the coordination of the [3Fe-4S] cluster, we conducted site-directed mutagenesis of cysteine residues in BlsK. All nine cysteine residues in BlsK were mutated to serine residue respectively. Except for BlsK (C482S), all the other BlsK mutants were expressed as soluble proteins and purified similarly with wild-type BlsK. Among these soluble mutants, BlsK (C236S), BlsK (C253S), and BlsK (C259S) lost the brown color, and the peak around 420 nm in the UV-visible spectra disappeared, indicating these three mutants lost the [3Fe-4S]+ cluster (SI Appendix, Fig. S6A). Concomitant with the disappearance of the characteristic UV-visible absorption band, the EPR spectra of these three BlsK mutants lost peaks centered at g = 2.01 (SI Appendix, Fig. S6B). Instead, a signal for Fe3+ at g = 4.3 in the EPR spectra appeared, indicating the mutants could chelate the ferric ion and possibly form incomplete iron–sulfur clusters. Therefore, the data indicate the iron–sulfur cluster in BlsK might be coordinated by C236, C253, and C259.
The [3Fe-4S] Cluster Is Essential for the Function of BlsK.
It is well known that the most common function of iron–sulfur clusters in proteins is to participate in redox reactions (24). It is surprising to find an iron–sulfur cluster inside BlsK because its catalyzed reaction, amide bond formation, does not include electron transfer. Thus, it is worth further studying the role of the iron–sulfur cluster in BlsK. It is noteworthy that the iron–sulfur cluster in BlsK seems somewhat resistant to oxygen, as the aerobically purified BlsK has the characteristic UV-visible signal of [3Fe-4S]+ and is able to catalyze the transfer of leucyl group to DBS. However, the addition of DTT increased the activity of aerobically purified BlsK (SI Appendix, Fig. S7). When BlsK was purified and assayed anaerobically, its activity increased compared with the aerobically purified BlsK (Fig. 7A). When the anaerobically purified BlsK was further incubated with ferrous iron, sodium sulfide, and DTT, its activity was increased further (Fig. 7A). All these results suggest a possible relationship between the leucyl transfer activity of BlsK and the iron–sulfur cluster.
Fig. 7.
[3Fe-4S] is essential for the catalytic activity of BlsK. (A) HPLC analysis of BlsK-catalyzed reaction: (i) using BlsK purified under aerobic conditions; (ii) with BlsK purified under anaerobic conditions; and (iii) BlsK purified under anaerobic conditions with the [3Fe-4S] further reconstituted. (B) EDTA abolished the activity of BlsK: (i) without EDTA as control; (ii) BlsK treated with EDTA; and (iii) BlsK treated with EDTA following by the reconstitution of iron–sulfur cluster in vitro. (C) UV-visible analysis of BlsK treated with EDTA. (D) HPLC analysis of the activity of BlsK mutant proteins: (i) wild-type BlsK, and (ii–iv) represent C236S, C253S, and C259S mutant proteins, respectively. The concentration of BlsK in the reaction system was 10 μM. Enzyme assays in A and B were carried out under anaerobic conditions.
To further investigate whether the [3Fe-4S] cluster is essential for activity, EDTA was used to remove iron ions in BlsK. After incubation, BlsK lost its brownish color and became colorless, and the apo-BlsK lost the activity to transfer a leucyl group to DBS, demonstrating the importance of the [3Fe-4S] cluster to the activity of BlsK (Fig. 7B). Also, the UV-visible signal for [3Fe-4S]+ disappeared, indicating the loss of the [3Fe-4S]+ cluster (Fig. 7C). The apo-protein was then reconstituted with the addition of ferrous iron, sodium sulfide, and DTT in vitro. The reconstituted BlsK regained the brownish color and showed the typical absorption around 420 nm in the UV-visible spectrum seen with the untreated BlsK, indicating the successful reconstitution of the [3Fe-4S] cluster (Fig. 7C). In line with the expectations, the activity of the reconstituted BlsK was restored (Fig. 7B). In further support of the importance of the [3Fe-4S] cluster, activity assays showed that C236S, C253S, and C259S mutants, which do not contain functional [3Fe-4S] clusters, lost the capacity to catalyze LDBS formation (Fig. 7D). Circular dichroism (CD) spectra of these mutant proteins were very similar to that of wild-type BlsK, indicating that their secondary structures were similar (SI Appendix, Fig. S8). Thus, the loss of protein activity was due to the incompleteness of the [3Fe-4S] clusters. Viewed together, the above results clearly demonstrate that the [3Fe-4S] cluster is critical for the leucyl transfer activity of BlsK.
Surprisingly, as the activity of BlsK was increased with the in vitro [3Fe-4S] cluster reconstitution, an additional product peak appeared (Fig. 7A). When labeled leucine (α-carbon with 13C isotope) was used in the catalytic reaction, the high-resolution mass spectra demonstrated that the molecular weight of the product other than LDBS is 637.3329 ([M+H]+), which is 114 mass units greater than that of LDBS (containing one 13C) (the same molecular weight difference between LDBS (containing one 13C) and DBS), indicating attachment of another leucyl group (SI Appendix, Fig. S9). Also, the MS/MS result of the product is consistent with the proposed product, leucyl-LDBS (SI Appendix, Fig. S9). Moreover, we found that about 10% LDBS was produced using the anaerobic-purified and -reconstituted BlsK without the addition of tRNA compared with that in the presence of tRNA (SI Appendix, Fig. S10A). To exclude the possibility that a trace amount of tRNA purified with BlsK might interfere with the results, RNase A was added to the reaction system. However, LDBS could still be produced, similar with that in the control group without RNase A (SI Appendix, Fig. S10B). Also, the possibility that purified BlsK had leucyl group anchored on some residues was ruled out because incubation of only purified BlsK and DBS could not form LDBS (SI Appendix, Fig. S11). Leucyl-AMP could be generated by LeuRS when tRNA is not present. The results indicated that in addition to leucyl-tRNA, BlsK may utilize other leucyl-activated species, probably leucyl-AMP, as activated leucyl species although with a strong preference for leucyl-tRNALeu. A similar finding was observed in PacB in pacidamycin biosynthesis (25).
Discussion
An interesting feature in BS biosynthesis is the existence of a cryptic leucylation in the overall biosynthetic pathway. A leucyl group is first transferred to the β-amino group of the β-arginine side chain of the intermediate DBS and is then removed in the final biosynthetic step (15, 16). This cryptic enzymatic step has also been seen in several other natural product biosyntheses, such as chlorination during cyclopropyl amino acid biosynthesis and C-terminal methylation in thiostrepton biosynthesis (26, 27). We report here the identification of BlsK as a leucyl transferase in BS biosynthesis. The main purpose of the incorporation of the leucyl group during BS biosynthesis appears to be self-resistance, although it was found that the efficiency of BlsL, the N-methyl transferase in BS biosynthesis, is much higher with LDBS compared with DBS (16, 18). Thus, the function of BlsK is important to the BS biosynthesis. When blsK is deleted, BS production is significantly decreased. The small amount of BS that accumulates in the mutant strain may be derived from the direct methylation of DBS to BS. The addition of the leucyl group greatly increases the efficiency of N-methylation in BS biosynthesis catalyzed by BlsL (18). Thus, investigating the role and mechanism of leucylation is important to fully understand the biosynthesis of BS.
With the identification of LBS as a biosynthetic intermediate, BS biosynthesis requires two amino acid (arginine and leucine) activation enzymes. In the BS biosynthetic cluster, there are no NRPS-like genes, and blsI encodes an ATP-grasp enzyme, which could be used to activate one amino acid (16, 21). BlsI has been suggested to activate and ligate arginine to the cytosinine skeleton in the early step of BS biosynthesis (16). We could not identify another carboxyl activation enzyme in the BS cluster. Previous research suggested that BlsK could be a possible candidate enzyme (15). However, it does not show much similarity with other adenylating enzymes. Sequence and structural homology analysis indicate BlsK does not belong to any known protein family. In particular, HHpred-based analysis could only find some relevant domain templates for partial amino acid region of BlsK. Interestingly, through genetic and biochemical experiments, BlsK has been confirmed to be responsible for the leucyl transfer in an aa-tRNA–dependent manner. The finding of BlsK as a leucyl transferase adds an additional functional member to the aminoacyl transferases of the aa-tRNA–dependent protein family.
During the past decade, a number of aa-tRNA–dependent enzymes have been found to be involved in natural products biosynthesis and can be mainly clustered into three groups. The first identified aa-tRNA–dependent enzyme in natural product biosynthesis is VlmA, which catalyzes ester bond formation using seryl-tRNASer as a direct substrate in valanimycin biosynthesis. Interestingly, the valanimycin cluster contains a seryl-tRNASer synthetase VlmL, which can provide seryl-tRNASer for valanimycin biosynthesis (28). Later, several aa-tRNA–dependent transferases were discovered, including PacB in pacidamycin biosynthesis (25), FzmI in fosfazinomycin biosynthesis (29), and ORF11 in BD-12 biosynthesis (30). All of these enzymes show homology with leucyl/phenylalanyl-tRNA protein transferases (L/F transferases) involved in the N-terminal modification for protein degradation in bacteria and the family of Fem transferases required for cell wall biosynthesis (31). Although this family of proteins show low sequence similarity, all the currently known members adopt the same GCN5-related N-acetyltransferase fold (3). Recently, a family of peptide–amino acyl tRNA ligases (PEARL) has been found to add amino acids to the C terminus of small peptides (32). It is noteworthy that PEARL catalyze the transfer reaction in an ATP- and tRNA-dependent way, which is different from other aa-tRNA–dependent transferases. According to its catalytic activity, BlsK belongs to the aminoacyl transferase group. However, BlsK does not show any similarity with this family of enzymes, indicating that it may represent a different type of aa-tRNA–dependent aminoacyl transferase. As with the majority of other biosynthetic pathways that contain an aa-tRNA–dependent aminoacyl transferase, the BS biosynthetic cluster does not include aa-tRNA synthetase-encoding genes. Thus, BlsK might hijack the leucyl-tRNALeu generated by LeuRS in the cell that is primarily used for protein synthesis with an unknown mechanism.
Another unique feature of BlsK is that the enzyme contains an iron–sulfur cluster that is necessary for aminoacyl transferase activity. The dependency of BlsK activity on a [3Fe-4S] cluster is consistent with the relatively low activity of purified BlsK with the CFE compared with the CFE from E. coli BL21 (DE3, ΔpepN)/pET44b-blsK. In the purification process, the [3Fe-4S] cluster in BlsK was gradually degraded. However, the exact role of the iron–sulfur cluster in the aa-tRNA–dependent leucyl transfer process remains unknown. In many proteins, [3Fe-4S] clusters are involved in electron transfer reactions (33). BlsK catalyzes amide formation, which does not involve electron transfer. It is possible that the iron–sulfur cluster of BlsK plays an important role in stabilization of the structure of BlsK. When BlsK was over-produced in iron-deficient condition, it formed inclusion bodies, indicating the structural importance of the iron–sulfur cluster in BlsK (SI Appendix, Fig. S12). Another possible function of the iron–sulfur cluster of BlsK is substrate recognition. It has been reported that iron–sulfur clusters in several proteins are responsible for the recognition of aa-tRNA (34). Thus, the iron–sulfur cluster of BlsK might be involved in its binding with leucyl-tRNALeu and beneficial to divert leucyl-tRNALeu from the protein biosynthesis machinery to BS biosynthesis.
The BlsK sequence is not homologous to any known iron–sulfur–containing proteins. Iron–sulfur clusters in most proteins are usually chelated by Cys residues. The BlsK sequence contains one CXXC motif, which is commonly involved in the coordination of iron–sulfur clusters in many iron–sulfur–containing proteins. However, mutational analysis demonstrated that the CXXC motif in BlsK is not important for the iron–sulfur cluster and mutation of three other Cys residues (C236S, C253S, and C259S) implicated their involvement in chelating the iron–sulfur cluster. This binding motif is not the same but similar with previous reported CXXA/GXXC(X)nCP motif in several [3Fe-4S] proteins (35). The characterization of BlsK will expand our knowledge about iron–sulfur family proteins.
In conclusion, using genetic and biochemical methods, we demonstrated that BlsK catalyzes a tRNA-dependent leucyl group transfer during BS biosynthesis. BlsK contains an iron–sulfur cluster that is necessary for activity and utilizes aa-tRNA–loaded leucine as a donor. To the best of our knowledge, BlsK is an aa-tRNA–dependent aminoacyl transferase containing an iron–sulfur cluster, which has not been found before. Therefore, it may present a different type of peptide bond-forming enzyme and expand the existing family of aa-tRNA–dependent aminoacyl transferases.
Methods
General Materials and Procedures.
All chemicals and reagents were purchased from commercial sources without further purification. Primers are listed in SI Appendix, Table S1. Strains and plasmids are listed in SI Appendix, Table S2. E. coli strains including DH10B, BW25113/pIJ790 (for λ RED recombination), and methylation-deficient host ET12567/pUZ8002 (for conjugal transfer) were cultivated at 37 °C in lysogeny broth (LB) or LB with agar medium. Soybean cake flour medium agar was used for sporulation and conjugation of Streptomyces strains at 30 °C. The fermentation of Streptomyces strains was performed in TSBY medium (3% tryptone soy broth medium, 0.5% yeast extract, and 10.3% sucrose, wt/vol) at 30 °C.
Inactivation of blsK and Its Complementation.
The gene inactivation of blsK was carried out on plasmid pJTU1780 containing a 35-kb fragment of the BS biosynthetic cluster (13) by the method of PCR-targeting according to Bertolt Gust (36). Namely, the aadA-oriT cassette containing 39-bp sequences complementary to the upstream/downstream blsK gene was PCR amplified using pIJ778 as a template. After purification, the cassette was introduced into BW25113/pIJ790/pJTU1780 to replace the blsK gene. The resulting blsK deletion construct was PCR verified and introduced into S. lividans WJ2 using E. coli ET12567/pUB307–mediated triparental conjugation. The double-crossover blsK deletion mutant S. lividans WXK3 (SpecRThioS) was PCR verified. To construct the complementary plasmid, the blsK gene was PCR amplified from pJTU1780 and placed under the ermE* promoter in an integrative plasmid pIB139. The resulting pIB139-ermE*-blsK was introduced into S. lividans WXK3 to obtain the complementation strain (S. lividans WXK4).
Fermentation and HPLC/LC-MS Analysis.
Fermentation of S. lividans–derived strains (WJ2, WXK3, and WXK4) and metabolite analysis were performed as previously described (18). In brief, the strains were fermented at 30 °C for 5 d, and the fermentation broth was collected by centrifugation at 5,000 rpm for 10 min. The pH of the collected supernatant was adjusted to 5.0 with oxalic acid and kept at room temperature for 30 min. After centrifugation at 10,000 rpm for 30 min, the supernatant was collected and further purified using Supelclean LC-SCX solid-phase extraction columns (500 /3 mL) (Supelco) as previously described (13). Samples were analyzed with an Agilent 1,200 HPLC system using a C18 column (4.6 mm × 250 mm, Aegla Technologies) under the following conditions: buffer A (water with 0.1% trifluoroacetic acid) and buffer B (methanol). Buffer B was increased from 5 to 40% over 40 min at a flow rate of 0.3 mL/min. The UV detection wavelength was 275 nm.
Overexpression and Purification of Proteins.
The blsK gene was amplified from plasmid pJTU1780 and cloned into pET44b (Novagen) digested by NdeI and EcoRI. The correct recombinant vector pET44b-blsK was verified by sequencing and was transformed into E. coli BL21 (DE3) for protein expression. The overnight BL21 (DE3)/pET44b-blsK culture was inoculated (1% vol/vol) into fresh LB medium containing 100 µg/mL ampicillin and cultured at 37 °C until the optical density at 600 nm (OD600) reached 0.4 to 0.6. Cells were then cooled to 16 °C and induced with IPTG (final concentration: 0.4 mM) for 16 h. Cells were harvested by centrifugation and stored at −80 °C until further use. The C-6×His protein BlsK was purified either in the open space or in an anaerobic glove box (Coy Laboratory Products Inc.) using Ni-NTA agarose followed by mono Q and Superdex-200 26/60 size-exclusion chromatography when needed. The procedures for the construction of the E. coli LeuRS expression vector and the subsequent protein purification were the same as those above for BlsK. A modified Immunoaffinity Chromatography method was applied to remove the small amount of contaminated LeuRS in purified BlsK. The LeuRS antibody was first bound to Protein A agarose beads, and then the purified BlsK was mixed with the beads, followed by centrifugation to remove the Protein A agarose beads/antibody/LeuRS complex. The supernatant was further purified by Ni-NTA column to obtain BlsK without LeuRS. Site-directed mutagenesis of BlsK was done by overlap extension PCR using the pET44b-blsK as template (primers are listed in SI Appendix, Table S1). The PCR product was treated with DpnI to remove the methylated template DNA. The mixture was then transformed into DH10B, and the site-mutated plasmids were verified by sequencing and transformed into BL21 for overexpression. The protein concentration was determined by the bicinchoninic acid (BCA) Protein Assay Kit (Sangon Biotech Co., Ltd.).
In Vitro Reconstitution and Characterization of the Iron–Sulfur Cluster in BlsK.
The reconstitution of the [3Fe-4S] cluster was performed under anoxic conditions according to the following procedure: purified BlsK (200 μM) was incubated with 5 mM DTT on ice for 30 min, then 1.2 mM (sixfold protein concentration) (NH4)2Fe(SO4)2 was added and incubated for 1 h followed by adding 1.2 mM Na2S and incubation on ice for another 1 h. The excess reagent was removed by a PD-10 desalting column (GE Healthcare) using Tris⋅HCl buffer (50 mM, pH 8.0) containing 100 mM NaCl, 10 mM DTT, and 10% glycerol. The resulting protein fraction was collected and used for in vitro assay or stored at −80 °C.
Ferrozine assays were used to quantify the iron content of BlsK. Reagent A (50 μL; 0.6 M HCl, 2.25% KMnO4) was added to 100 μL protein solution and digested at 60 °C for 2 h. After cooling to room temperature, 10 μL reagent B (6.5 mM ferrozine, 13.1 mM neocuproine, 2 M ascorbic acid, and 5 M ammonium acetate) was added to the mixture and incubated for 30 min. The absorbance of 562 nm was measured using a Biotek Synergy 2 plate reader (Biotek). The standard curve of Fe content was measured with a standard solution (6 μg/mL ferrous ethylenediamine sulfate).
For the content of free S, 75 μL of freshly prepared zinc acetate (1%, wt/vol) was added to 25 μL of protein solution, and 3.75 μL of NaOH (12%, wt/vol) was added immediately after mixing. The solution was gently mixed and placed at room temperature for 15 min, then 18.75 μL N,N-dimethyl-p-phenylene-diamine monohydrochloride (dissolved in 5 M HCl, 0.1%, wt/vol,) and 37.5 μL FeCl3 (23 mM, dissolved in 1.2 mM HCl) were added, mixed thoroughly until the solution was colorless, and centrifuged at 12,000 rpm for 15 min. The absorption at 670 nm of the supernatant was measured with a Biotek Synergy 2 plate reader. The standard curve of S content was measured with S standard solution reagents (Na2S 0.2 mg/mL, dissolved in 0.1 M NaOH). In order to reconfirm the content of Fe and S, ICP-MS (iCAP Q, Thermo Fisher Scientific) was used. UV-visible scanning of BlsK was performed on the Biotek Synergy 2 plate reader (Biotek) between 300 to 700 nm. X-band EPR spectra of BlsK were recorded with Bruker EMX plus 10 /12 (Bruker Corp.), equipped with an Oxford ESR910 Liquid Helium cryostat spectrometer (Oxford Instruments plc) at 10K. The CD spectra of BlsK and its mutant proteins were measured using a CD spectrophotometer (J-815, Jasco International, Co. Ltd.). The assay buffer (50 mM Tris⋅HCl buffer pH 8.0, 100 mM NaCl, 10 mM DTT, and 10% glycerol) was used for the EPR, CD, and UV-visible scanning experiments.
Enzymatic Assay of BlsK and HPLC/LC-MS Analysis.
For in vitro assay of BlsK, the reactions were conducted in 100 μL Tris⋅HCl buffer pH 8.0 (50 mM pH, 8.5, 100 mM NaCl) containing 10 μM BlsK, 2 μM LeuRS, 1 mM ATP, 10 mM Leu, 250 μM DBS, 1 mg/mL tRNAS, 20 mM MgCl2, 10 mM KCl, and 10 mM DTT at 37 °C for various incubation times. For the preparation of CFE used in the activity assay, the E. coli strain BL21 (DE3, ΔpepN)/pET44b-blsK was used. After induction by IPTG for 16 h, the appropriate amount of cells was collected and resuspended in cell lysate buffer (50 mM Tris HCl pH 8.0, 0.3 M NaCl, 10% glycerol), sonicated, and centrifuged at 4 °C at 12,000 rpm for 30 min. The supernatant was used as the CFE. CFE of the E. coli strain BL21 (DE3, ΔpepN) was similarly prepared (without IPTG induction). The assays were stopped by adding an equal volume of chloroform to denature the protein, and the aqueous fractions were collected after centrifugation. The reaction products were analyzed by HPLC and LC-MS using the TC-C18 (2) column (4.6 mm × 250 mm, Agilent). The following HPLC conditions were used: buffer A (water with 20 mM ammonium acetate, pH 5.5) and buffer B (methanol). Buffer B was increased from 10 to 18% within 0 to 22 min and further increased to 90% from 22 to 50 min and then held for 5 min at a flow rate of 0.3 mL/min. The absorbance was monitored at 275 nm. The molecular weight of the compound corresponding to the peak position was analyzed and confirmed by LC-MS using an Agilent 1,100 LC/MSD (mass selective detector) with positive mode electrospray ionization.
In Vitro Transcription of tRNA and Preparation of Leucyl-tRNALeu
The DNA sequence of tRNALeu (CUC) (from S. griseochromogenes strain ATCC 14511) containing the T7 promoter and terminator was synthesized and cloned into pUC57 between the EcoRI and BamHI sites by GenScript Biological Technology Co., Ltd. The template for in vitro transcription was prepared by PCR with the primers of Leu-F and Leu-R. For in vitro transcription, the HiScribe T7 High Yield RNA Synthesis Kit (NEB) was used. After transcription, DNase I (20 u/mL) was added to digest the DNA template, then an equal volume of precooled phenol (pH 7.9) was added and vortexed for 2 min to denature proteins. The supernatant was collected by centrifugation at 12,000 rpm for 10 min, and 2.5 times the volume of precooled ethanol was added. The mixture was placed at −40 °C overnight and centrifuged at 12,000 rpm for 30 min to precipitate tRNA. The precipitated tRNA was washed twice with 70% ethanol and dried in a fume hood. In order to fold the tRNA correctly, 5 mM MgCl2 was added to dissolve the precipitate and then denatured in a water bath at 85 °C for 10 min. In the end, the tRNA was cooled to room temperature and stored at −40 °C for later use.
For the preparation of leucyl-tRNALeu, LeuRS was used, and the reaction buffer was 1 μM LeuRS, 200 μM leucine, 4 mM ATP, 20 mM MgCl2, 10 mM KCl, 1 mM DTT, 50 μM in vitro–transcribed tRNALeu, 100 mM Tris⋅HCl buffer (pH 7.2). After reacting at 37 °C for 1 h, a precooled equal volume of acidic phenol (pH 4.5) was added to the reaction mixture and vortexed thoroughly for 2 min. The supernatant was collected after centrifugation at 12,000 rpm for 10 min and a precooled equal volume of phenol: chloroform: isopropanol (25:24:1, pH 4.5) solution was added followed by vortexing for 2 min. The supernatant was collected after centrifugation at 12,000 rpm for 10 min and 1/20 volume of 3 M sodium acetate (pH 5.2), and 2.5 times the volume of precooled ethanol were added to precipitate leucyl-tRNALeu overnight at −40 °C. The mixture was centrifuged at 12,000 rpm for 30 min, and the precipitated leucyl-tRNALeu was washed twice with 70% ethanol. Finally, the leucyl-tRNALeu was dried in a fume hood and then dissolved in Hepes buffer (pH 6.8). The sample was stored at −40 °C for later use.
Supplementary Material
Acknowledgments
We would like to express our gratitude to Prof. Youli Xiao and his colleagues from the Institute of Plant Physiology and Ecology, Shanghai Chinese Academy of Sciences (CAS) for their help in the anaerobic purification of proteins. We also thank Prof. Xiaolong Zhou of Shanghai Institute of Biochemistry and Cell Biology for his help in the in vitro preparation of tRNA. The EPR analysis was performed on the Steady High Magnetic Field Facilities, High Magnetic Field Laboratory, CAS. This work was supported by grants from the National Key Research and Development Program of China (Grant 2018YFA0901900) and the National Natural Science Foundation of China (Grants 31871250, 31670034, and 31870026).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. K.M.-F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102318118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Goswami A., Van Lanen S. G., Enzymatic strategies and biocatalysts for amide bond formation: Tricks of the trade outside of the ribosome. Mol. Biosyst. 11, 338–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schimmel P., The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19, 45–58 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Moutiez M., Belin P., Gondry M., Aminoacyl-tRNA-utilizing enzymes in natural product biosynthesis. Chem. Rev. 117, 5578–5618 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Fonvielle M., et al., The structure of FemX(Wv) in complex with a peptidyl-RNA conjugate: Mechanism of aminoacyl transfer from Ala-tRNA(Ala) to peptidoglycan precursors. Angew. Chem. Int. Ed. Engl. 52, 7278–7281 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Moutiez M., et al., Specificity determinants for the two tRNA substrates of the cyclodipeptide synthase AlbC from Streptomyces noursei. Nucleic Acids Res. 42, 7247–7258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao T., et al., Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery. Nat. Commun. 9, 4091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortega M. A., et al., Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi S., Hirayama K., Ueda K., Sakai H., Yonehara H., Blasticidin S, a new antibiotic. J. Antibiot. (Tokyo) 11, 1–5 (1958). [PubMed] [Google Scholar]
- 9.Ōtake N., Takeuchi S., Endō T., Yonehara H., Chemical studies on blasticidin S. Agric. Biol. Chem. 30, 126–141 (1966). [Google Scholar]
- 10.Yamaguchi H., Yamamoto C., Tanaka N., Inhibition of protein synthesis by blasticidin S. I. Studies with cell-free systems from bacterial and mammalian cells. J. Biochem. 57, 667–677 (1965). [PubMed] [Google Scholar]
- 11.Yamaguchi H., Tanaka N., Inhibition of protein synthesis by blasticidin S. II. Studies on the site of action in E. coli polypeptide synthesizing systems. J. Biochem. 60, 632–642 (1966). [DOI] [PubMed] [Google Scholar]
- 12.Goyard S., Beverley S. M., Blasticidin resistance: A new independent marker for stable transfection of Leishmania. Mol. Biochem. Parasitol. 108, 249–252 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Li L., Wu J., Deng Z., Zabriskie T. M., He X., Streptomyces lividans blasticidin S deaminase and its application in engineering a blasticidin S-producing strain for ease of genetic manipulation. Appl. Environ. Microbiol. 79, 2349–2357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seto H., Yamaguchi I., Ōtake N., Yonehara H., Studies on the biosynthesis of blasticidin S: Part I. Precursors of blasticidin S biosynthesis part II. Leucylblasticidin S, a metabolic intermediate of blasticidin S biosynthesis. Agric. Biol. Chem. 32, 1292–1305 (1968). [Google Scholar]
- 15.Cone M. C., Yin X., Grochowski L. L., Parker M. R., Zabriskie T. M., The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: Sequence analysis, organization, and initial characterization. ChemBioChem 4, 821–828 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q., Cone M. C., Gould S. J., Mark Zabriskie T., Reevaluation of the final steps in the biosynthesis of blasticidin S by Streptomyces griseochromogenes and identification of a novel self-resistance mechanism. Tetrahedron 56, 693–701 (2000). [Google Scholar]
- 17.Prabhakaran P., Woo N. T., Yorgey P. S., Gould S. J., Biosynthesis of blasticidin S from L-. alpha.-arginine. Stereochemistry in the arginine-2, 3-aminomutase reaction. J. Am. Chem. Soc. 110, 5785–5791 (1988). [Google Scholar]
- 18.Wang X., Du A., Yu G., Deng Z., He X., Guanidine N-methylation by BlsL is dependent on acylation of beta-amine arginine in the biosynthesis of blasticidin S. Front. Microbiol. 8, 1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G., et al., The standalone aminopeptidase PepN catalyzes the maturation of blasticidin S from leucylblasticidin S. Sci. Rep. 5, 17641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitzer J., Steiner K., Amides in nature and biocatalysis. J. Biotechnol. 235, 32–46 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wu L., Chen G., Feng G., Complete genome sequence of Streptomyces griseochromogenes ATCC 14511T, a producer of nucleoside compounds and diverse secondary metabolites. J. Biotechnol. 249, 16–19 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Soding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi W., Cowan J. A., Structural, mechanistic and coordination chemistry of relevance to the biosynthesis of iron-sulfur and related iron cofactors. Coord. Chem. Rev. 255, 688–699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beinert H., Holm R. H., Münck E., Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 277, 653–659 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Zhang W., Ntai I., Kelleher N. L., Walsh C. T., tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc. Natl. Acad. Sci. U.S.A. 108, 12249–12253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaillancourt F. H., Yeh E., Vosburg D. A., O’Connor S. E., Walsh C. T., Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–1194 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Liao R., Liu W., Thiostrepton maturation involving a deesterification-amidation way to process the C-terminally methylated peptide backbone. J. Am. Chem. Soc. 133, 2852–2855 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Garg R. P., Qian X. L., Alemany L. B., Moran S., Parry R. J., Investigations of valanimycin biosynthesis: Elucidation of the role of seryl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 105, 6543–6547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Z., Wang K. A., van der Donk W. A., New Insights into the Biosynthesis of Fosfazinomycin. Chem. Sci. (Camb.) 7, 5219–5223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maruyama C., et al., tRNA-dependent aminoacylation of an amino sugar intermediate in the biosynthesis of a streptothricin-related antibiotic. Appl. Environ. Microbiol. 82, 3640–3648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung A. W., Leung C. C., Fahlman R. P., The determination of tRNALeu recognition nucleotides for Escherichia coli L/F transferase. RNA 20, 1210–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., van der Donk W. A., Nonribosomal peptide extension by a peptide amino-acyl tRNA ligase. J. Am. Chem. Soc. 141, 19625–19633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beinert H., Iron-sulfur proteins: Ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5, 2–15 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., et al., A [3Fe-4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 113, 12703–12708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontecave M., Iron-sulfur clusters: Ever-expanding roles. Nat. Chem. Biol. 2, 171–174 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Gust B., Kieser T., Chater K., PCR targeting system in Streptomyces coelicolor A3 (2). John Innes Centre 3, 1–39 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.