Abstract
The current COVID-19 pandemic, that has caused more than 100 million cases as well as more than two million deaths worldwide, demands the development of fast and accurate diagnostic methods despite the lack of available samples. This disease mainly affects the respiratory system of the patients and can lead to pneumonia and to severe cases of acute respiratory syndrome that result in the formation of several pathological structures in the lungs. These pathological structures can be explored taking advantage of chest X-ray imaging. As a recommendation for the health services, portable chest X-ray devices should be used instead of conventional fixed machinery, in order to prevent the spread of the pathogen. However, portable devices present several problems (specially those related with capture quality). Moreover, the subjectivity and the fatigue of the clinicians lead to a very difficult diagnostic process. To overcome that, computer-aided methodologies can be very useful even taking into account the lack of available samples that the COVID-19 affectation shows. In this work, we propose an improvement in the performance of COVID-19 screening, taking advantage of several cycle generative adversarial networks to generate useful and relevant synthetic images to solve the lack of COVID-19 samples, in the context of poor quality and low detail datasets obtained from portable devices. For validating this proposal for improved COVID-19 screening, several experiments were conducted. The results demonstrate that this data augmentation strategy improves the performance of a previous COVID-19 screening proposal, achieving an accuracy of 98.61% when distinguishing among NON-COVID-19 (i.e. normal control samples and samples with pathologies others than COVID-19) and genuine COVID-19 samples. It is remarkable that this methodology can be extrapolated to other pulmonary pathologies and even other medical imaging domains to overcome the data scarcity.
Keywords: X-ray portable device, COVID-19, Data augmentation, Screening, CycleGAN, Deep learning
Graphical abstract
1. Introduction
The novel coronavirus SARS-CoV-2, that causes the COVID-19 disease, was first detected in Wuhan (province of Hubei) at the end of 2019 (Huang et al., 2020). This pathogen is highly contagious, reason why it was rapidly spread worldwide causing more than 120 millions of infections and more than 2.65 million deaths (Coronavirus Resource Center, Johns Hopkins, 2020). This rapid spread of the coronavirus forced the World Health Organization (WHO) to declare COVID-19 as a global pandemic on March 11, 2020. This critical situation has pointed out the importance of developing fast and accurate diagnostic methods to reduce the significant load that the health care services are currently experiencing and to overcome the problem of the clinicians diagnostic subjectivity, scenario where computer-aided diagnostic tools should be taken into account.
Regarding the clinical diagnosis of pulmonary pathologies, the chest X-ray imaging modality has been widely used, as it is cheap, quick and easy to perform. In this sense, the chest X-ray modality has demonstrated its usefulness to diagnose common pulmonary diseases and issues like lung nodules, pneumonia, edema, fibrosis, cardiomegaly or pneumotorax, among others (Xu, Wu, & Bie, 2019). In the context of the COVID-19 pandemic, radiologists are strongly recommended to use chest X-ray images for a preliminary assessment of the lung damage rather than any other imaging modality.
Generally, given the relevance of the COVID-19 disease, we can find some contributions from the state of the art to solve several tasks. As reference, these works perform useful tasks in the application field as lung segmentation (Teixeira et al., 2021, Vidal et al., 2021), screening and classification (Abbas et al., 2020, Asif et al., 2020, Basu et al., 2020, Duong et al., 2020, Ismael and Şengür, 2021, Misra et al., 2020, de Moura et al., 2020, Ozturk et al., 2020, Sharma et al., 2020, Yeh et al., 2020). These works are characterized for using deep learning strategies to distinguish among COVID-19 and other scenarios (healthy or pathological), providing satisfactory results. Moreover, some of the proposed methodologies include GradCAM (Selvaraju et al., 2017) to visualize the most relevant regions to focus on the images under the point of view of the training models, adding explicability to the method and providing an useful explanation for the clinicians to understand the decisions of the algorithm. In particular, some other contributions use conventional machine learning algorithms as Support Vector Machines (SVMs) in specific parts of their workflows (Mahdy et al., 2020, Novitasari et al., 2020).
Considering the recent emergence of the COVID-19 disease and the critical situation of the healthcare services, it exists a lack of sufficiently large COVID-19 chest X-ray image datasets. Normally, to overcome the problem of data scarcity, the works perform conventional data augmentation transforms like random rotations, translations or pixel intensity changes. While this is an interesting strategy that has demonstrated a great potential in many domains, including medical imaging, the new transformed images could be insufficiently representative of the present notable variability that exists in the image domain. Moreover, we can find several proposals that have addressed the synthetic image generation to perform the oversampling of the original dataset, taking advantage of particular GAN architectures in other similar domains, as Frid-Adar et al., 2018, Han et al., 2018 and Shin et al. (2018).
On the other hand, to develop robust and accurate computer-aided methodologies, the availability of big amounts of data is generally a critical aspect, specially in a pandemic scenario. In this sense, some few works have addressed the oversampling of chest X-ray images datasets taking into account the synthetic image generation to increase the size of the original datasets. Particularly, as reference, a Cycle Generative Adversarial Network, often denoted as CycleGAN, was used to perform an oversampling over a chest X-ray images dataset for only the analysis of pneumonia (Malygina, Ericheva, & Drokin, 2019).
Directly related with the analysis of the COVID-19 using chest X-ray images, (Zebin, Rezvy, & Pang, 2021) proposed the use of a CycleGAN to perform an oversampling using the COVID-19 Image Data Collection dataset (Cohen et al., 2020). In a similar way, the work from Singh, Pandey, and Babu (2021) take into consideration a GAN architecture different from the CycleGAN to generate only COVID-19 synthetic images. Therefore, both works partially address the problem, without considering all the classes for oversampling, which may bias the final learning process. Furthermore, unlike with the fixed X-ray devices mostly used for making the previously mentioned COVID-19 chest X-ray datasets, the recommendation from the American College of Radiology (ACR) is to use portable chest X-ray devices in order to reduce the transmission of COVID-19 (Kooraki, Hosseiny, Myers, & Gholamrezanezhad, 2020). However, although these devices are very flexible, making the captures easy to perform, the obtained images have a poor quality, providing less details. In this sense, a poor quality set of images makes the diagnosis even more difficult to carry out. Similarly, automatic image analysis approaches are also more challenging to develop. Using chest X-ray images from portable devices as input, the work from De Moura et al. (2020) demonstrated that convolutional neural networks in an end-to-end training approach are capable of distinguishing among patients with healthy lungs, lungs with pathologies others than COVID-19 and COVID-19 itself, obtaining an acceptable performance using chest X-ray images. On the other hand, (Morís, de Moura, Novo, & Ortega, 2021) demonstrated the suitability of using CycleGAN architectures to perform an oversampling generating novel relevant synthetic images augmenting the size of three different classes: healthy, COVID-19 and other pulmonary diseases different from COVID-19. In the same way, this approach was conducted on a low quality chest X-ray images dataset that was obtained from portable devices used in a real clinical context.
In this paper, we propose a novel fully automatic methodology for the improvement of COVID-19 screening using different data augmentation approaches based on the CycleGAN architecture, a deep learning-based network for synthetic image generation. To this end, we combined 3 complementary CycleGAN architectures to simultaneously generate a new set of synthetic portable chest X-ray images from 3 different scenarios (normal, pathological and genuine COVID-19) without the necessity of paired data. Additionally, in order to make an exhaustive validation of the methodology, we used 4 different CycleGAN configurations (Unet with 7 downsampling blocks, denoted as Unet-128, Unet with 8 downsampling blocks, denoted as Unet-256, ResNet with 6 residual blocks, denoted as ResNet-6 and ResNet with 9 residual blocks, denoted as ResNet-9) that are tested with each scenario, thus evaluating 12 different approaches. Then, this new set of synthetic images is added to the original dataset to increase its dimensionality, taking advantage of the data augmentation to improve the training in the COVID-19 screening process. This data augmentation is very powerful, as one of the main targets of the CycleGAN is to achieve a realistic appearance for the synthetic X-ray images it generates. It is remarkable the great advantage that provides the use of unpaired data, as the image translations can be performed without pairing cases as well as without image alignments. In the same way, this idea provides a great versatility in this problem as well as in other potential problems. To validate our proposal, exhaustive experiments were conducted using a chest X-ray dataset obtained in real clinical practice from portable devices. In this line, this study demonstrates the potential of introducing new synthetic images in low-dimensional datasets to improve the training of the COVID-19 screening, despite the lower quality and level of detail of the images obtained from portable devices.
Summarized, these are the main contributions that our work represents:
-
•
Fully automatic proposal considering a dataset obtained in real clinical practice in the context of poor quality and low detail chest X-ray images obtained from portable devices.
-
•
An oversampling strategy without the requirement of a paired dataset that uses non-trivial transformations, providing a set of useful samples that represent the remarkable variability of the domain.
-
•
Robust and effective methodology for COVID-19 screening in a data scarcity context, improving the performance of the state-of-the-art.
-
•
Exhaustive experimentation using 4 different CycleGAN configurations, demonstrating the suitability and robustness of the proposed methodology through a comprehensive analysis.
-
•
To the best of our knowledge, this represents the only work to improve COVID-19 screening on portable chest X-ray images by using complementary CycleGAN architectures to perform the data augmentation.
The present document is organized as follows: Section 2 “Materials”, describes the resources that are necessary to reproduce our work. Section 3 “Methodology”, details an explanation of the considered strategy and the different proposed scenarios, the used algorithms and the specific parameters for each experiment. Section 4 “Evaluation”, explains the considered metrics for the experimental validation. Section 5 “Results”, shows the obtained results after the experimental validation of the work was performed. Section 6 “Discussion”, shows a detailed analysis of the previously obtained results. In this analysis, we present the most remarkable aspects of our proposed methodology. Finally, Section 7 “Conclusions”, which includes a summary of the main contributions and remarkable aspects of our work and its experimental validation.
2. Materials
2.1. CHUAC dataset
In this work, the used dataset was obtained from the real clinical practice of a hospital in A Coruña, Galicia (Spain). Specifically, 720 chest X-ray images were retrieved from the radiology service of the Complejo Hospitalario Universitario de A Coruña (CHUAC). All the images were obtained from 2 different portable chest X-ray devices: an Agfa dr100E GE, and an Optima Rx200. The acquisition procedure was performed with the patient in a supine position, recording a single anterior–posterior projection. In particular, the X-ray tube is connected to a flexible arm so it can be extended over the patient and, on the other side, an X-ray film holder or an image recording plate is placed under the patient to make the image capturing. This imaging was performed in medical wings that were particularly dedicated for treatment and monitoring of suspicious or diagnosed COVID-19 cases. All the images were obtained during the first peak of the pandemic in 2020. In particular, the dataset is composed of 240 COVID-19 cases, 240 images from patients without COVID-19 but diagnosed with other pulmonary diseases that can present characteristics similar to COVID-19 and 240 images from normal control patients that include healthy subjects but also may include other pathological conditions that are not related with the characteristics of COVID-19 disease. The resolution of the samples is variable, having images of 1523 × 1904, 1526 × 1753, 1526 × 1754, 1526 × 1910, 949 × 827, 950 × 827 and 950 × 833 pixels.
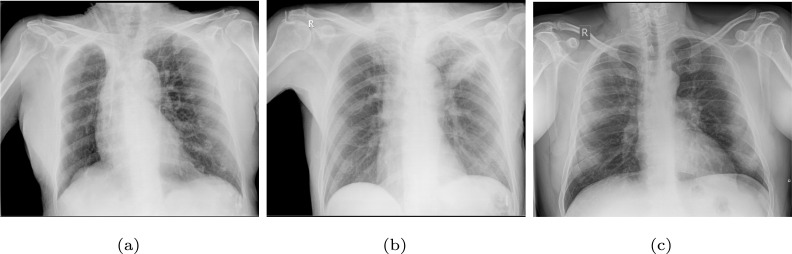
This retrospective study was approved by an ethics committee with code 2020-007. The whole dataset was anonymized before being received by the staff outside of the CHUAC radiology service, in order to protect the privacy of the patients. Images were carefully visually inspected by members of the CHUAC radiology service staff, in order to find symptoms of lungs affectation. In addition, all the suspected COVID-19 cases were checked with an external RT-PCR test, in order to support that first visual inspection. In the same way, the images were stored in private servers taking into account the appropriate security protocols. The access to the information was restricted to the project members. The particular used protocols have been reviewed by the hospital board as well as an agreement with the hospital management was constituted. As reference, Fig. 1 shows 3 representative examples of portable chest X-ray images for 3 different scenarios: normal patient, patient diagnosed with pulmonary pathologies (others than COVID-19) and patient diagnosed with COVID-19.
Fig. 1.
Representative examples of portable chest X-ray images. (a) Chest X-ray image of a normal patient. (b) Chest X-ray image of a patient diagnosed with pulmonary pathologies (others than COVID-19). (c) Chest X-ray image of a patient diagnosed with COVID-19.
3. Methodology
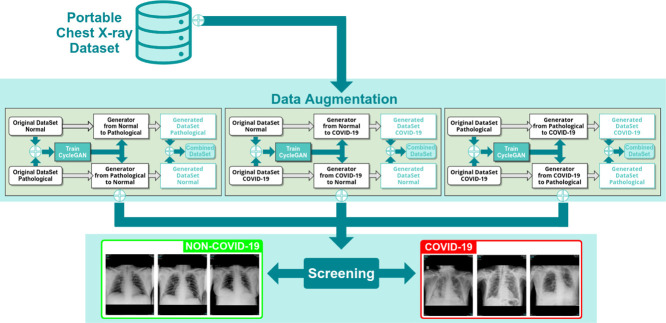
The proposed methodology is composed by a set of complementary modules for the simultaneous oversampling to produce novel portable chest X-ray images and the subsequent screening of COVID-19. Fig. 2 shows the main workflow of the proposed methodology, which is composed of 2 modules. In particular, the first module is responsible of the synthetic image generation, where 3 complementary CycleGAN architectures have been considered and adjusted. The second module performs the COVID-19 screening task, taking advantage of the augmented dataset obtained after the generated samples are added to the original small dataset in the training phase to improve the screening process. The first part of the methodology is described in detail in Section 3.1 and the second part of the methodology is described in Section 3.2.
Fig. 2.
Main workflow of the proposed methodology, which is comprised of two modules: one module for simultaneous data augmentation and another responsible for the screening tasks.
Finally, there is a common aspect that affects both parts of the methodology. It should be remarked that all the images of the original dataset are resized to a standard size of 256 × 256 × 3, which is the appropriate resolution to work with, regarding the particular network architectures used for this experimentation.
3.1. Computational approaches for data augmentation
In order to produce a new set of portable chest X-ray images, 3 different and complementary approaches are considered to perform this data augmentation task using an unsupervised strategy and without the necessity of paired data. These 3 computational approaches are the result of all the possible combinations between normal, pathological and COVID-19 samples.
3.1.1. Normal vs pathological approach
For this first scenario, the set of normal X-ray images is considered as well as the set of pathological X-ray images. Thus, 2 different translation tasks are performed. In the first case, the whole set of normal X-ray images is translated to a synthetic pathological representation of them. On the other hand, the whole set of pathological X-ray images is translated to a synthetic healthy representation of them.
3.1.2. Normal vs COVID-19 approach
In this second scenario, the normal X-ray images are converted to their hypothetical COVID-19 scenario and vice versa. In this way, the model should introduce pathological structures compatible with COVID-19 in the original normal X-ray image for the first translation. Hence, for the second translation, the model should remove the COVID-19 pathological structures to convert original images to their hypothetical normal X-ray representation.
3.1.3. Pathological vs COVID-19 approach
Finally, for the third possible scenario, the set of the whole pathological X-ray images is considered alongside the whole set of COVID-19 X-ray images. In a similar way as the previous scenarios, 2 translations are performed. Thus, pathological X-ray images are translated to their hypothetical COVID-19 representation and vice versa. Hence, in the first translation, the model should remove the pathological structures caused by a particular pulmonary disease different from COVID-19, then adding structures compatible with the COVID-19 affectation. In the opposite way, the second translation should remove the COVID-19 pathological structures from the chest X-ray images, then adding pathological structures related with other pulmonary diseases different from COVID-19.
3.1.4. Network architectures and training details
For the automatic generation of synthetic portable chest X-ray images, we propose the use of a deep learning strategy, a CycleGAN (Zhu, Park, Isola, & Efros, 2017) architecture with 4 different configurations: ResNet with 6 residual blocks (denoted as ResNet-6), ResNet with 9 residual blocks (denoted as ResNet-9), Unet with 7 downsampling blocks (denoted as Unet-128) and Unet with 8 downsampling blocks (denoted as Unet-256). To do this, in this work, CycleGAN learns the correlation between the input and output images to generate a set of new chest X-ray images using an unpaired dataset. Furthermore, the cyclic design of this architecture allows for a reverse transformation, i.e. the model can convert a generated chest X-ray image into an original chest X-ray image. In this line, CycleGAN architectures were commonly used in medical image analysis for image-to-image generation tasks (Harms et al., 2019, Ma et al., 2020, Russ et al., 2019) due to its flexibility, robustness and promising results on similar problems.
As every GAN architecture, the CycleGAN is characterized for using two different types of models: the generators and the discriminators. The generator (also known as generative model) is the part of the architecture that performs the synthetic image generation. On the other hand, the discriminative model is the part of the architecture that learns to determine if the generated synthetic images have a realistic appearance. Thus, the generative model is trained in order to generate realistic synthetic samples that must be classified by the discriminative model as real samples.
Regarding the networks architectures, both generators use an encoder–decoder while the discriminators use an encoder. Particularly, for this work, 4 different encoder–decoder architectures are considered for both generative models: Unet with 7 downsampling blocks (denoted as Unet-128), Unet with 8 downsampling blocks (denoted as Unet-256), ResNet with 6 residual blocks (denoted as ResNet-6) and ResNet with 9 residual blocks (denoted as ResNet-9). On the other hand, the discriminative models have only one configuration as they always consider a 70 × 70 PatchGAN architecture.
Particularly, CycleGAN must learn two mapping functions, that work between two classes: and . Thus, input samples belong to one of these classes. We can denote as the set of images that belong to class A and denote as the set of images that belong to class B. Moreover, the data distribution is denoted as and . In this way, generators can be denoted as mapping functions and . On the other hand, discriminators are called as and , respectively, where distinguish images from the synthetic images and where distinguish images from the synthetic images .
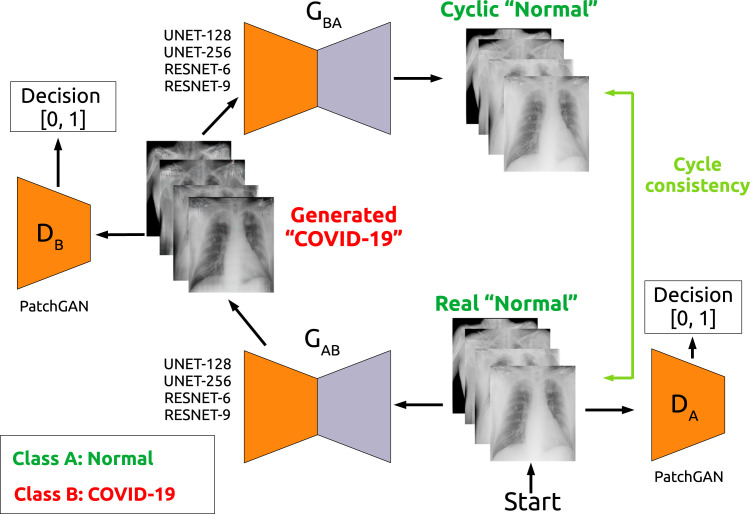
Fig. 3 shows an example of the CycleGAN workflow for the scenario Normal vs COVID-19 to translate Normal samples to COVID-19 samples, where we can see that this architecture is composed of 2 generative models as well as 2 discriminative models. In order to train the 4 models that are required by the CycleGAN architecture, first of all it is necessary to translate the image from class A to class B (task that is performed by the model). Then, this generated image (that should be classified as class B) is used as input for the model, in order to perform the reverse task, obtaining a generated synthetic image that should be classified as class A. In this sense, the final generated image (also known as cycle A) should be the same as the initial real input image from class A. To assure that, the CycleGAN architecture introduces the concept of cycle consistency, i.e. a loss that compares the cycle A image against the original input image from class A.
Fig. 3.
An illustration of the CycleGAN architecture that was adapted for the experiments of this work for the Normal vs COVID-19 scenario.
The CycleGAN objective is expressed considering two types of terms: the adversarial loss and the cycle consistency loss. The adversarial loss is applied in both mapping directions, using the least-square loss to compute the difference between the expected output and the predicted output. In this way, denoting the adversarial loss as , considering the as the generative model and as the discriminative model, we train to minimize the expression defined in Eq. (1).
| (1) |
On the other hand, taking as reference, we train the discriminative models to minimize the expression defined in Eq. (2).
| (2) |
Regarding the cycle consistency loss, denoted as , considering both pathways from class A to class B and vice versa, this loss is defined as in Eq. (3).
| (3) |
where two weight parameters and are defined. Each one refers to the given weight to the cycle consistency loss for class A and class B, respectively.
Additionally, the considered CycleGAN implementation also defines another loss which is called identity loss. Assuming that, as an example, the mapping function is able to translate images from class A to class B, then it is plausible to think that the function will also return a synthetic image from class B when receiving a real input image from class B. Therefore, it is also reasonable to think that both images should be the same. Taking these ideas into account, the expression of the identity loss can be seen in Eq. (4).
| (4) |
where defines the weight given to the identity loss in the final objective.
Finally, considering all the losses previously described, the full objective is expressed as can be seen in Eq. (5).
| (5) |
Then, the aim of the networks weights optimization is to minimize the expression defined in Eq. (6).
| (6) |
Regarding the training settings, the same parameters apply for all the considered CycleGAN configurations. Particularly, each model is trained from scratch during 250 epochs with a mini-batch size of 1, using the Adam algorithm (Kingma & Ba, 2014), considering a constant learning rate of and decay rates of and , setting the following values for the loss weights: , and . The whole set of 720 images was used for training.
3.2. Computational approaches for the screening tasks
In this second part of the proposed methodology, we designed computational approaches for the analysis of the degree of separability between the synthetic X-ray images and the suitability of the newly generated images for the COVID-19 screening process. In the case of the analysis of the synthetic X-ray images, 3 possible scenarios were considered: (I) the separability between normal and pathological; (II) the separability between normal and COVID-19; and (III) the separability between pathological and COVID-19 X-ray images.
Once this separability is analyzed, the novel set of generated X-ray images will be added to the original dataset, in order to perform a data augmentation process. Then, a comparison with a baseline approach will demonstrate the improved performance of this oversampling strategy in terms of the COVID-19 screening. For this purpose, we analyzed the degree of separability between NON-COVID-19 and COVID-19 categories. For this analysis, only the training and validation sets contain a combination between the original and the generated X-ray images. The test set only consists of genuine X-ray images from the original dataset.
3.2.1. Network architecture and training details
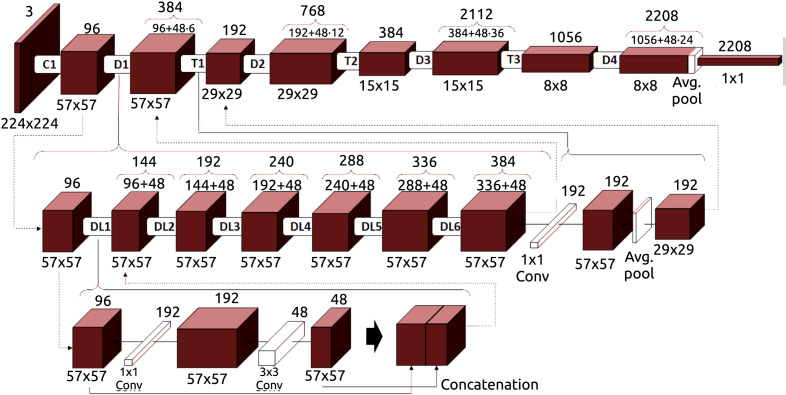
In this work, we used a Densely Connected Convolutional Network (DenseNet) (Huang, Liu, Van Der Maaten, & Weinberger, 2017) architecture to perform the screening tasks. For this purpose, an original structure of the DenseNet-161 architecture has been adapted. Fig. 4 shows the layout of this particular architecture, which is composed of 4 dense blocks, 3 transition layers that involve convolution and pooling, and a final classification layer. Each model is trained end-to-end with a cross-entropy loss function (Zhang & Sabuncu, 2018), computing the difference between the network output and the ground truth provided by the manual labeling. The cross-entropy loss expression can be seen in Eq. (7):
| (7) |
where denotes the ground truth and denotes the output of the network.
Fig. 4.
An illustration of the DenseNet-161 architecture that was adapted for the experiments of this work.
The same training details are applicable to all the computational approaches of the screening tasks. In particular, the dataset is randomly partitioned in 3 subsets, with 60% of the samples for training, 20% for validation and 20% for test. The trained model is initialized taking the weights from another model which was trained on the ImageNet (Deng et al., 2009) dataset. These weights were optimized during 200 epochs using the algorithm of Stochastic Gradient Descent (SGD) (Ketkar, 2017) with a mini-batch size of 4, a first-order momentum of 0.9 and a constant learning rate of . To evaluate the performance of each model, the whole process is repeated 5 times with a different random selection of the sample divisions, being calculated the mean cross-entropy loss and the mean accuracy to evaluate their global performance.
4. Evaluation
Regarding the evaluation of the proposed methodology, several classification performance metrics were taken into account, in order to provide the corresponding information under several points of view. In this sense, we considered the value of precision, recall, F1-score and accuracy. For this particular case, and assuming as reference the values of True Positives (), True Negatives (), False Positives () and False Negatives (), metrics can be computed as follows:
| (8) |
| (9) |
| (10) |
| (11) |
5. Results
In this section, we evaluated and compared several configurations to determine the best-performing network configuration. To do so, 4 different experiments were performed. The first 3 experiments are focused on the separability among the different classes of a set composed only of generated portable chest X-ray images. Thus, each experiment will evaluate the degree of separability of the normal class from the pathological class, the normal class from the COVID-19 class as well as the pathological class from the COVID-19 class, respectively. Moreover, for each considered scenario, the performance of the 4 network configurations for the CycleGAN architecture will be compared: Unet-128, Unet-256, Resnet-6 and Resnet-9.
Then, a fourth experiment will measure the degree of separability among NON-COVID-19 samples (i.e., normal patients and patients with pulmonary pathologies others than COVID-19) and COVID-19 samples taking advantage of the dataset obtained after the data augmentation was performed (i.e., after augmenting the original dataset with the novel set of generated images). Moreover, in this last experiment, the performance of the COVID-19 screening will be measured and compared using, respectively, the novel set of generated images from the Unet-128, the novel set of generated images from the Unet-256, the novel set of generated images from the ResNet-6 and the novel set of generated images from the ResNet-9. Now, we will proceed to detail the results obtained for each experiment.
Complementarily, an ablation study for the test stage was conducted taking as reference the best configuration obtained in the fourth experiment. In particular, this ablation analysis is composed of 5 additional experimental tests, in order to evaluate the performance improvement that the oversampling contributes with respect to the number of generated synthetic X-ray images that were used in the training stage and tested with only original images. For this purpose, we used 0% of generated images, 20% of generated images, 40% of generated images, 60% of generated images, 80% of generated images as well as 100% of generated images that was already tested in the fourth experiment.
1st experiment: analysis of the separability between normal and pathological generated samples.
In this first experiment, the capability of the CycleGAN models is proved to translate from normal to pathological and vice versa. In order to do this, we have taken into account the whole set of the novel 240 normal images as well as the whole set of the novel 240 pathological images from the generated dataset.
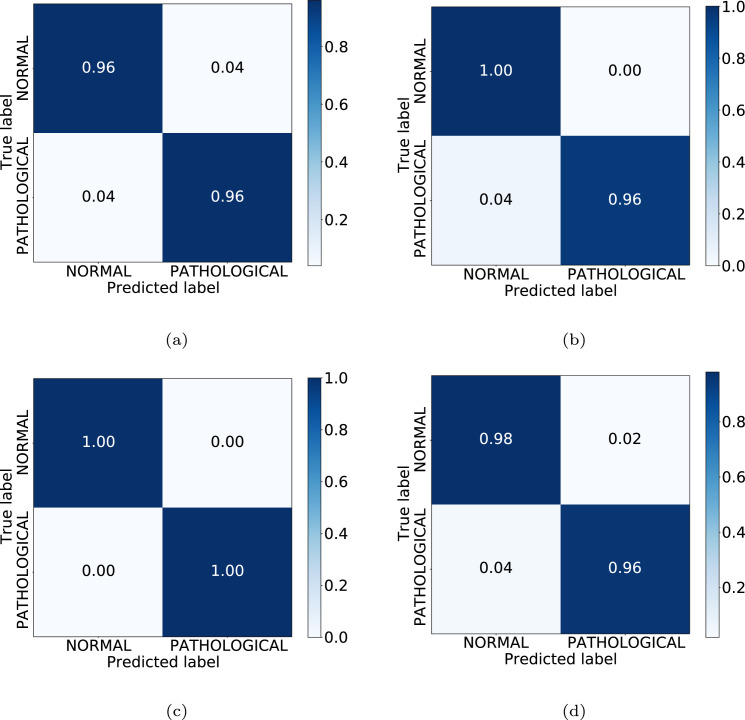
In Table 1, we can see the results obtained at the test stage. Finally, the accuracy values obtained for testing were 95.83%, 97.92%, 100.00% and 96.88% for Unet-128, Unet-256, ResNet-6 and ResNet-9, respectively. There, it can be observed that the portable chest X-ray images generated by the ResNet-6 model have the greatest grade of separability between classes, as can be seen with metrics of precision, recall, F1-score and accuracy values. Additionally, Fig. 5 shows the confusion matrices for the 4 considered CycleGAN architectures, representative of the performance in the testing, with correct classification ratios higher than 0.96 for all CycleGAN configurations and for both classes.
Table 1.
Test results for the separability among generated normal and pathological samples (1st scenario).
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| Unet-128 | Normal | 0.96 | 0.96 | 0.96 |
| Pathological | 0.96 | 0.96 | 0.96 | |
| Unet-256 | Normal | 0.96 | 1.00 | 0.98 |
| Pathological | 1.00 | 0.96 | 0.98 | |
| ResNet-6 | Normal | 1.00 | 1.00 | 1.00 |
| Pathological | 1.00 | 1.00 | 1.00 | |
| ResNet-9 | Normal | 0.96 | 0.98 | 0.97 |
| Pathological | 0.98 | 0.96 | 0.97 | |
Fig. 5.
Confusion matrices obtained in the 1st experiment for the 4 CycleGAN considered configurations. (a) Unet-128. (b) Unet-256. (c) ResNet-6. (d) ResNet-9.
2nd experiment: analysis of the separability between normal and COVID-19 generated samples.
The second experiment proved the capability of the CycleGAN models to perform a translation from normal to COVID-19 and vice versa. To do so, the whole set of novel generated 240 normal images alongside the set of novel generated 240 COVID-19 images was taken into account.
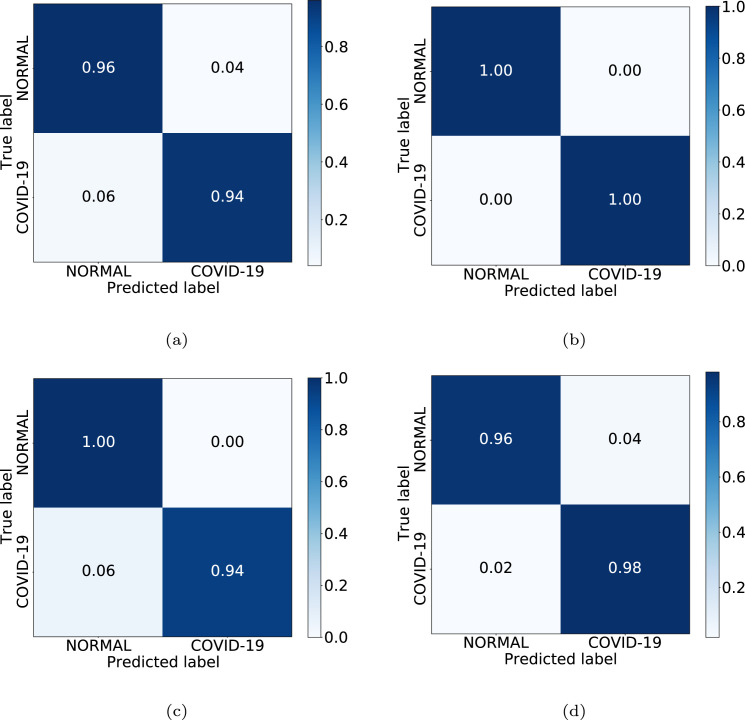
In Table 2, we can see the results obtained at the test stage. For this scenario, the accuracy values for test, in the same order as the previous cases, were as follows: 94.79%, 100.00%, 96.88% and 96.88%. In this experiment, as we can see, the set of synthetic images with the best grade of separability corresponds to those generated by the Unet-256 model. Additionally, Fig. 6 shows the confusion matrices for the 4 considered CycleGAN architectures, representative of the performance in testing, always obtaining a correct classification ratio higher than 0.94 for all scenarios and for both classes.
Table 2.
Test results for the separability among generated normal and COVID-19 samples (2nd scenario).
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| Unet-128 | Normal | 0.94 | 0.96 | 0.95 |
| COVID-19 | 0.96 | 0.94 | 0.95 | |
| Unet-256 | Normal | 1.00 | 1.00 | 1.00 |
| COVID-19 | 1.00 | 1.00 | 1.00 | |
| ResNet-6 | Normal | 0.93 | 1.00 | 0.97 |
| COVID-19 | 1.00 | 0.94 | 0.97 | |
| ResNet-9 | Normal | 0.98 | 0.96 | 0.97 |
| COVID-19 | 0.96 | 0.98 | 0.97 | |
Fig. 6.
Confusion matrices obtained in the 2nd experiment for the 4 CycleGAN considered configurations. (a) Unet-128. (b) Unet-256. (c) ResNet-6. (d) ResNet-9.
3rd experiment: analysis of the separability between pathological and COVID-19 generated samples.
For the third possible scenario, the CycleGAN models were tested in order to evaluate their performance in translation tasks from pathological to COVID-19 and vice versa. Particularly, the whole set of novel generated 240 pathological samples was taken into account alongside the whole set of novel generated 240 COVID-19 samples.
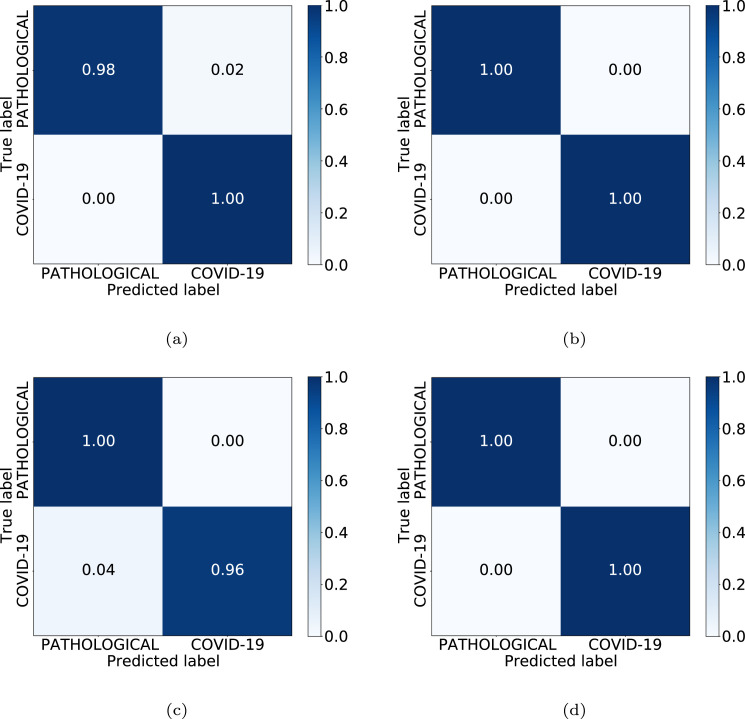
In Table 3, we can see the results obtained at the test stage. The accuracy values achieved for the Unet-128, Unet-256, ResNet-6 and ResNet-9 in the test set were, respectively, 98.96%, 100.00%, 97.92% and 100.00%. In this case, as we can see, the best performance is achieved when considering the synthetic images generated by the Unet-256 as well as when considering the set of images generated by the ResNet-9. In both cases, the precision, recall, F1-score and accuracy metrics achieve a value of 1.0, representing the maximum degree of separability between the classes. Additionally, Fig. 7 shows the confusion matrices for the 4 considered CycleGAN architectures, representative of the performance in the testing. In this case, we always obtained correct classification ratios higher than 0.96.
Table 3.
Test results for the separability among generated pathological and COVID-19 samples (3rd scenario).
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| Unet-128 | Pathological | 1.00 | 0.98 | 0.99 |
| COVID-19 | 0.98 | 1.00 | 0.99 | |
| Unet-256 | Pathological | 1.00 | 1.00 | 1.00 |
| COVID-19 | 1.00 | 1.00 | 1.00 | |
| ResNet-6 | Pathological | 0.96 | 1.00 | 0.98 |
| COVID-19 | 1.00 | 0.96 | 0.98 | |
| ResNet-9 | Pathological | 1.00 | 1.00 | 1.00 |
| COVID-19 | 1.00 | 1.00 | 1.00 | |
Fig. 7.
Confusion matrices obtained in the 3rd experiment for the 4 CycleGAN considered configurations. (a) Unet-128. (b) Unet-256. (c) ResNet-6. (d) ResNet-9.
4th experiment: analysis of the COVID-19 screening considering the new set of generated images.
In this experiment, we analyzed the degree of separability between NON-COVID-19 and COVID-19 categories, to measure the improvement of oversampling in the screening process. As said, for this analysis, only the training and validation sets contain a combination between the original and the generated chest X-ray images. The test set only includes chest X-ray images from the original dataset.
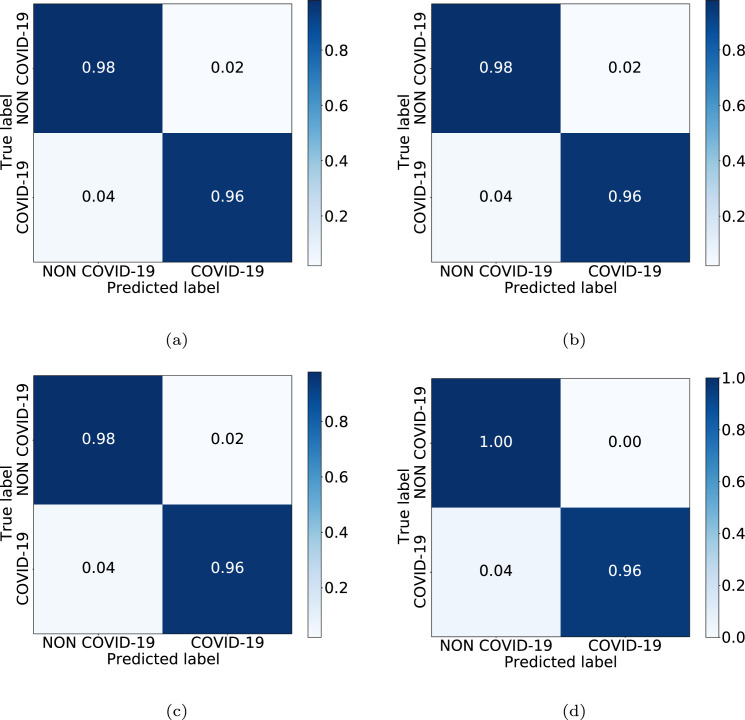
Table 4 presents a comparative analysis of the performance at the test stage using precision, recall, and F1-score measures. In this case, as we can see, the best performance is achieved by using the original images combined with the images generated by ResNet-9 to train the model. Particularly, the values of precision, recall and F1-score were, respectively, 0.98, 1.00, 0.99 for NON-COVID-19 class and 0.99, 0.99, 0.99 for COVID-19 class. Complementarily, the accuracy obtained by this configuration for the test set was 98.61%. Additionally, Fig. 8 shows the confusion matrices for the 4 considered CycleGAN architectures, representative of the performance in the testing. Here, models obtained values of correct classification ratio higher than 0.96 for all cases.
Table 4.
Results of the COVID-19 screening analysis using the original images combined with the new generated images to train the model.
| Model | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| Unet-128 | NON-COVID-19 | 0.98 | 0.98 | 0.98 |
| COVID-19 | 0.96 | 0.96 | 0.96 | |
| Unet-256 | NON-COVID-19 | 0.98 | 0.98 | 0.98 |
| COVID-19 | 0.96 | 0.96 | 0.96 | |
| ResNet-6 | NON-COVID-19 | 0.98 | 0.98 | 0.98 |
| COVID-19 | 0.96 | 0.96 | 0.96 | |
| ResNet-9 | NON-COVID-19 | 0.98 | 1.00 | 0.99 |
| COVID-19 | 1.00 | 0.96 | 0.98 | |
Fig. 8.
Confusion matrices obtained in the 4th experiment for the 4 CycleGAN considered configurations. (a) Unet-128. (b) Unet-256. (c) ResNet-6. (d) ResNet-9.
Ablation study for the test stage: analysis of the performance improvement with respect to the number of generated images used for the oversampling.
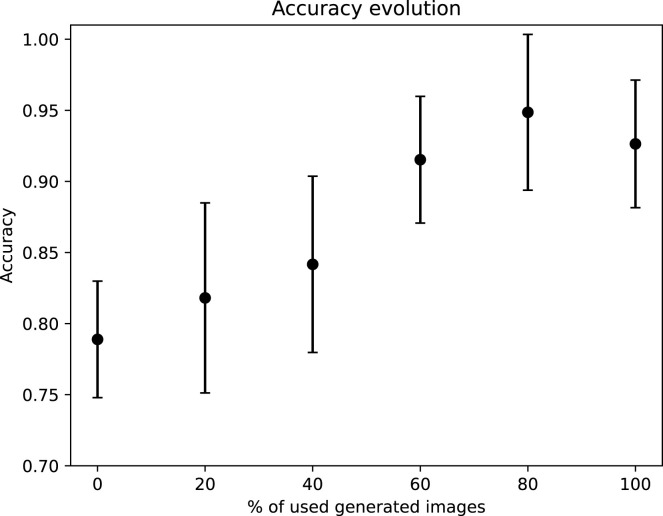
This study is composed of 5 additional experimental tests, in order to evaluate the performance improvement that the oversampling contributes with respect to the number of generated images that were used. For this purpose, we have taken the ResNet-9 as reference (the model that achieved the best performance in the fourth experiment). The first test was conducted without using generated images, the second test was conducted using the 20% of the whole amount of generated images (288 images in total), the third test was conducted using the 40% of the generated images (576 images), the fourth test was performed with the 60% of the generated images (864 images), the fifth test was done using the 80% of the whole amount of generated images (1152 images) and all these experimental tests are compared against the case with the whole amount of generated images (1440 images). To evaluate the performance of each model, the whole process is repeated 5 times with a different random selection of the sample divisions, being calculated the mean accuracy to evaluate their global performance in test. Fig. 9 depicts the results of these experimental tests showed in terms of the mean and the standard deviation accuracy values in the test stage. There, it can be clearly seen that the performance improves notably as the amount of generated images increases. Additionally, it is remarkable that the accuracy improvement starts to converge moreless at 60%, obtaining the best results at 80%. Furthermore, the standard deviation keeps stable during the whole study. This demonstrates that, even with a small amount of generated images, the performance improvement is clearly noted, being more notable as this amount increases until the accuracy converges to a high value.
Fig. 9.
Analysis of the performance improvement in the test stage with respect to the number of generated synthetic images used for the oversampling in the training stage. The obtained results are represented by the mean and the standard deviation of the accuracy values.
6. Discussion
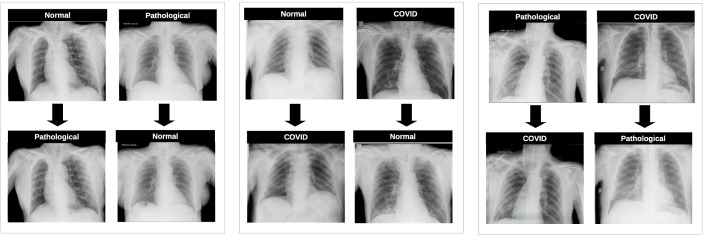
Regarding the results the obtained for the first three experiments, which represent the validation of the separability between the generated images, we can observe that the classification model is able to accurately distinguish normal from pathological, normal from COVID-19 as well as pathological from COVID-19 cases, independently of the CycleGAN configuration used to generate the synthetic images. In all the cases, the metrics of accuracy are over 90% but, in this particular set of evaluations, it is unclear which is the CycleGAN architecture that achieves the best global performance, as the highest degree of separability for the first experiment (healthy vs pathological scenario) was achieved by the ResNet-6, the highest grade of separability for the second experiment (healthy vs COVID-19 scenario) was achieved by the Unet-256 and the highest grade of separability for the third experiment (pathological vs COVID-19 scenario) was achieved simultaneously by the Unet-256 alongside the ResNet-9. Fig. 10 depicts some representative examples of the X-ray images generated for all the possible scenarios showing a qualitative assessment of the remarkable and well-synthesized differences in lung regions for all the cases that were analyzed in this work. The method generated 480 images for each scenario, then having a total of synthetic images, with 480 images for each class.
Fig. 10.
Examples of generated images for all the possible scenarios. 1st block, Normal vs Pathological scenario. 2nd block, Normal vs COVID-19 scenario. 3rd block, Pathological vs COVID-19 scenario.
In terms of the COVID-19 screening, we can see that the separability between the NON-COVID-19 class (i.e., normal and pathological samples) and the COVID-19 class achieves a high performance in terms of precision, recall and F1-score metrics as well as accuracy for the whole set of proposed CycleGAN architectures. It should be noted that these experiments were conducted by training with a mixed set of original and generated images while the test set only contains original images, in order to make a fair comparison with the baseline. In terms of effectiveness, it can be concluded that all the CycleGAN architectures achieved similar values, though the ResNet-9 achieves a slightly higher performance. Moreover, using this CycleGAN configuration, we performed an ablation study for the test stage, demonstrating that the proposed data augmentation strategy improves the screening performance by increasing the number of samples.
On the other hand, the comparison of our method against the contribution of De Moura et al. (2020) can be seen in Table 5. It is important to remark that we use this previous work as reference as, to the best of our knowledge, represents the only COVID-19 screening proposal in the context of low quality images provided by portable X-ray devices. In this comparison, the CycleGAN architecture with the higher accuracy was taken into account, for this case, the ResNet-9. This table shows that the previous proposal of COVID-19 screening is being clearly outperformed by our new proposal, achieving values greater than 99% for precision, recall, F1-score and global accuracy, even obtaining a 1.0 or nearly values in some metrics.
Table 5.
Performance comparison between our proposal and the baseline.
| Proposal | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| De Moura et al. (2020) | NON-COVID-19 | 0.91 | 0.95 | 0.93 |
| COVID-19 | 0.88 | 0.80 | 0.84 | |
| Ours | NON-COVID-19 | 0.98 | 1.00 | 0.99 |
| COVID-19 | 0.99 | 0.99 | 0.99 | |
It is remarkable that these successful results can be obtained despite the lack of dimensionality of the used dataset and the poor quality of its images. Similarly, these results can be achieved despite the fact that the lung involvement of COVID-19 is very similar to the involvement of other lung diseases.
Another important aspect is that the human graders need a great resolution to properly visualize the fine details of the images, as chest X-ray is a challenging image modality to perform a diagnosis. This is also applicable to automatic methods. Therefore, it could be interesting to conduct experiments with different resolutions and understand the impact of this element on the synthetic image generation and classification performance. In this sense, a higher image resolution should represent a better level of detail of the generated images, thus improving the overall performance of the proposed methodology.
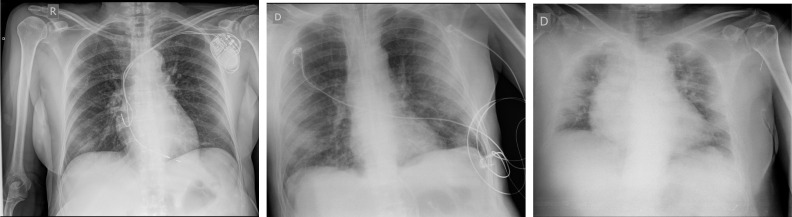
In addition, from a clinical practice point of view, there are some other aspects that can lead the automatic method to commit mistakes, as the artifacts that are introduced by the capture device (aspect that is more noticeable when using portable devices) and other artifacts related with the implants that patients can have as well as foreign objects as wires, or the tubes used for the mechanical ventilation. Other difficulties that the proposed methodology must overcome are related with the patient position, thing that can vary significantly among the samples. Despite all these related issues, the methodology is able to overcome them with acceptable results. Fig. 11 shows several examples of portable chest X-ray images with artifacts as well as low quality captures, which are representative of the complex scenarios that the models need to deal with.
Fig. 11.
Examples of portable chest X-ray images with artifacts and bad quality captures, representative of some complex scenarios the models must deal with.
7. Conclusions
COVID-19 is a disease that has already caused a large number of deaths worldwide. In order to mitigate this highly infectious pulmonary disease, one of the most important challenges is the development of fast and reliable diagnostic systems, despite the lack of huge datasets given the recent appearance of the disease and the exceptional and saturated situation of the healthcare services. In this sense, we address a relevant problem of data scarcity, which is widely well-known in many biomedical imaging domains, being even more crucial and important in the dramatic case of the COVID-19 disease.
In this work, we proposed an application of a CycleGAN oversampling to compensate the lack of COVID-19 portable chest X-ray samples. The considered oversampling paradigm is based on the idea of translating portable chest X-ray images from a normal scenario to a pathological one or vice versa in a fully automatic way, generating a set of useful and representative synthetic X-ray images. This set of images is then added to the original small dataset in order to increase its dimensionality in the training process. To evaluate the performance improvement that this novel set of X-ray images represents in the context of the COVID-19 screening, 5 complementary experiments were proposed, that can be divided in 3 phases.
Thus, the first 3 experiments, that correspond to the first phase, validated the separability among the generated images for all the possible scenarios of this work. In particular, these images were obtained from 4 different CycleGAN configurations: Unet-128, Unet-256, ResNet-6 and ResNet-9. On the other hand, the fourth experiment, that corresponds to the second phase, was carried out to validate the improvement of the COVID-19 screening performance, after the application of oversampling. The fifth and last experiment was conducted as an ablation study to analyze the impact of the data augmentation in the COVID-19 screening. The results of the first part of the methodology demonstrate that it exists a genuine separability among the generated images from the 3 considered classes: normal, pathological and COVID-19. Regarding the second part of the methodology, the results demonstrate a capability to obtain values near to 1.0 in several metrics as precision, recall and F1-score when distinguishing between NON-COVID-19 (i.e., normal and pathological samples) and COVID-19 portable chest X-ray images. Moreover, for the third part of the experimental validation, it is demonstrated that generally the performance of the model improves as the amount of used generated images increases, achieving a convergence as this value gets closer to the whole amount of generated X-ray images.
It should be remarked that this satisfactory performance can be achieved dealing with chest X-ray images from portable devices, that are widely used by the clinicians to diagnose the disease due to its versatility but that, in the opposite side, provide lower quality and less detailed images than those provided by fixed devices. In this sense, the study herein proposed demonstrates the potential of introducing novel synthetic images in low dimensional datasets to improve the COVID-19 detection in chest X-ray images, easing the application of this automatic screening method in a realistic clinical context.
As possible future works, several experiments could be conducted in order to evaluate the impact that higher chest X-ray image resolutions make on the performance. Moreover, we could also perform additional experiments proving several network architectures for image generation different from the CycleGAN. The CycleGAN architecture performs both translations simultaneously, from class A to class B and vice versa. Thus, it must exist a rigid relationship between both mapping functions. Contrarily, other network architectures for image generation are more flexible in this aspect. In this way, it could be interesting to understand the impact of this idea on the performance of the image generation models. Similarly, this could also be experimented in the case of the classification architectures. In this work, we use a DenseNet architecture, while it could be interesting to understand the impact of other network architectures on the classification performance. Finally, it should be noted that this model could be exploited in a transfer learning pipeline, using the models trained for image translation as pre-trained models for domain related applications. In the same way, the paradigm herein presented could be extrapolated to the study of other pulmonary diseases, image modalities and even other different medical domains to overcome situations of data scarcity, which is a very common and well-known issue in these particular domains and faced in many contributions.
CRediT authorship contribution statement
Daniel Iglesias Morís: Conceptualization, Methodology, Software, Writing – original draft, Writing – review & editing, Visualization. José Joaquim de Moura Ramos: Conceptualization, Methodology, Software, Validation, Writing – review & editing, Supervision. Jorge Novo Buján: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing – review & editing, Supervision, Project administration. Marcos Ortega Hortas: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding
This research was funded by Instituto de Salud Carlos III, Government of Spain, DTS18/00136 research project; Ministerio de Ciencia e Innovación y Universidades, Government of Spain, RTI2018-095894-B-I00 research project; Ministerio de Ciencia e Innovación, Government of Spain through the research project with reference PID2019-108435RB-I00; Consellería de Cultura, Educación e Universidade, Xunta de Galicia, Spain through the predoctoral and postdoctoral grant contracts ref. ED481A 2021/196 and ED481B 2021/059, respectively; and Grupos de Referencia Competitiva, grant ref. ED431C 2020/24; Axencia Galega de Innovación (GAIN), Xunta de Galicia, Spain, grant ref. IN845D 2020/38; CITIC, Centro de Investigación de Galicia, Spain ref. ED431G 2019/01, receives financial support from Consellería de Educación, Universidade e Formación Profesional, Xunta de Galicia, Spain , through the ERDF (80%) and Secretaría Xeral de Universidades (20%). Funding for open access charge: Universidade da Coruña/CISUG.
References
- Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2020;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif S., Wenhui Y., Jin H., Tao Y., Jinhai S. Classification of COVID-19 from chest X-ray images using deep convolutional neural networks. medRxiv. 2020 doi: 10.1101/2020.05.01.20088211. [DOI] [Google Scholar]
- Basu S., Mitra S., Saha N. 2020 IEEE symposium series on computational intelligence. IEEE; 2020. Deep learning for screening COVID-19 using chest X-ray images; pp. 2521–2527. [Google Scholar]
- Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. COVID-19 image data collection: Prospective predictions are the future. arXiv:2006.11988. [Google Scholar]
- Coronavirus Resource Center, Johns Hopkins J.P. 2020. COVID-19 dashboard by the center for systems science and engineering (CSSE) at John Hopkins university. Available from: https://coronavirus.jhu.edu/map.html. (Accessed 15 March 2021) [Google Scholar]
- De Moura J., García L.R., Vidal P.F.L., Cruz M., López L.A., Lopez E.C., et al. Deep convolutional approaches for the analysis of COVID-19 using chest X-ray images from portable devices. IEEE Access. 2020;8:195594–195607. doi: 10.1109/ACCESS.2020.3033762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE conference on computer vision and pattern recognition. IEEE; 2009. ImageNet: A large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- Duong L.T., Nguyen P.T., Iovino L., Flammini M. Deep learning for automated recognition of COVID-19 from chest X-ray images. medRxiv. 2020 doi: 10.1101/2020.08.13.20173997. [DOI] [Google Scholar]
- Frid-Adar M., Diamant I., Klang E., Amitai M., Goldberger J., Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321:321–331. doi: 10.1016/j.neucom.2018.09.013. https://www.sciencedirect.com/science/article/pii/S0925231218310749. [DOI] [Google Scholar]
- Han C., Hayashi H., Rundo L., Araki R., Shimoda W., Muramatsu S., et al. 2018 IEEE 15th international symposium on biomedical imaging. IEEE; 2018. GAN-based synthetic brain MR image generation; pp. 734–738. [Google Scholar]
- Harms J., Lei Y., Wang T., Zhang R., Zhou J., Tang X., et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Medical Physics. 2019;46(9):3998–4009. doi: 10.1002/mp.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G., Liu, Z., Van Der Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 4700–4708).
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. https://www.sciencedirect.com/science/article/pii/S0957417420308198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketkar N. Deep learning with Python: A hands-on introduction. Apress; Berkeley, CA: 2017. Stochastic gradient descent; pp. 113–132. [DOI] [Google Scholar]
- Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- Kooraki S., Hosseiny M., Myers L., Gholamrezanezhad A. Coronavirus (COVID-19) outbreak: What the department of radiology should know. Journal of the American College of Radiology. 2020;17(4):447–451. doi: 10.1016/j.jacr.2020.02.008. https://www.sciencedirect.com/science/article/pii/S1546144020301502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Liu Y., Cheng J., Zheng Y., Ghahremani M., Chen H., et al. International conference on medical image computing and computer-assisted intervention. Springer; 2020. Cycle structure and illumination constrained GAN for medical image enhancement; pp. 667–677. [Google Scholar]
- Mahdy L.N., Ezzat K.A., Elmousalami H.H., Ella H.A., Hassanien A.E. Automatic X-ray COVID-19 lung image classification system based on multi-level thresholding and support vector machine. medRxiv. 2020 doi: 10.1101/2020.03.30.20047787. [DOI] [Google Scholar]
- Malygina T., Ericheva E., Drokin I. International conference on analysis of images, social networks and texts. Springer; 2019. Data augmentation with GAN: Improving chest X-ray pathologies prediction on class-imbalanced cases; pp. 321–334. [Google Scholar]
- Misra S., Jeon S., Lee S., Managuli R., Jang I.-S., Kim C. Multi-channel transfer learning of chest X-ray images for screening of COVID-19. Electronics. 2020;9(9) doi: 10.3390/electronics9091388. [DOI] [Google Scholar]
- Morís D.I., de Moura J., Novo J., Ortega M. ICASSP 2021-2021 IEEE international conference on acoustics, speech and signal processing. IEEE; 2021. Cycle generative adversarial network approaches to produce novel portable chest X-rays images for COVID-19 diagnosis; pp. 1060–1064. [Google Scholar]
- de Moura J., Novo J., Ortega M. Fully automatic deep convolutional approaches for the analysis of COVID-19 using chest X-ray images. medRxiv. 2020 doi: 10.1016/j.asoc.2021.108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitasari D., Hendradi R., Caraka R., Rachmawati Y., Fanani N., Syarifudin M., et al. Detection of COVID-19 chest X-ray using support vector machine and convolutional neural network. Communications in Mathematical Biology and Neuroscience. 2020;2020:1–19. doi: 10.28919/cmbn/4765. [DOI] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. https://www.sciencedirect.com/science/article/pii/S0010482520301621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ T., Goerttler S., Schnurr A.-K., Bauer D.F., Hatamikia S., Schad L.R., et al. Synthesis of CT images from digital body phantoms using CycleGAN. International Journal of Computer Assisted Radiology and Surgery. 2019;14(10):1741–1750. doi: 10.1007/s11548-019-02042-9. [DOI] [PubMed] [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. 2017 IEEE international conference on computer vision. 2017. Grad-CAM: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [DOI] [Google Scholar]
- Sharma A., Rani S., Gupta D. Artificial intelligence-based classification of chest X-ray images into COVID-19 and other infectious diseases. International Journal of Biomedical Imaging. 2020;2020:1–10. doi: 10.1155/2020/8889023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.-C., Tenenholtz N.A., Rogers J.K., Schwarz C.G., Senjem M.L., Gunter J.L., et al. International workshop on simulation and synthesis in medical imaging. Springer; 2018. Medical image synthesis for data augmentation and anonymization using generative adversarial networks; pp. 1–11. [Google Scholar]
- Singh R.K., Pandey R., Babu R. Covidscreen: Explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Computing and Applications. 2021 doi: 10.1007/s00521-020-05636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira L.O., Pereira R.M., Bertolini D., Oliveira L.S., Nanni L., Cavalcanti G.D.C., et al. 2021. Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images. arXiv:2009.09780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal P.L., Moura J., Novo J., Ortega M. Multi-stage transfer learning for lung segmentation using portable X-ray devices for patients with COVID-19. Expert Systems with Applications. 2021 doi: 10.1016/j.eswa.2021.114677. https://www.sciencedirect.com/science/article/pii/S0957417421001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wu H., Bie R. CXNet-m1: Anomaly detection on chest X-rays with image-based deep learning. IEEE Access. 2019;7:4466–4477. doi: 10.1109/ACCESS.2018.2885997. [DOI] [Google Scholar]
- Yeh C., Cheng H.-T., Wei A., Liu K.-C., Ko M.-C., Kuo P.-C., et al. 2020. A cascaded learning strategy for robust COVID-19 pneumonia chest X-ray screening. ArXiv, abs/2004.12786. [Google Scholar]
- Zebin T., Rezvy S., Pang W. COVID-19 detection and disease progression visualization: Deep learning on chest X-rays for classification and coarse localization. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2021;51 doi: 10.1007/s10489-020-01867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Sabuncu M.R. 2018. Generalized cross entropy loss for training deep neural networks with noisy labels. ArXiv preprint arXiv:1805.07836. [Google Scholar]
- Zhu J.-Y., Park T., Isola P., Efros A. 2017 IEEE international conference on computer vision. 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks; pp. 2242–2251. [DOI] [Google Scholar]