Abstract
This unit describes several established genetic approaches for Corynebacterium diphtheriae, the causative agent of diphtheria that provided acknowledgedly key evidence for Koch’s postulates on the germ theory. First, it includes a detailed gene deletion method that generates non-polar, in-frame markerless deletion mutants, utilizing the levansucrase SacB as a counter-selectable marker. Second, it provides a thorough protocol for rescuing deletion mutants using E. coli-Corynebacterium shuttle vectors. Finally, a Tn5 transposon mutagenesis procedure is described. In principle, these protocols can be used for other Corynebacterium species including Corynebacterium glutamicum and Corynebacterium matruchotii.
Basic Protocol 1: Gene deletion in C. diphtheriae
Basic Protocol 2: Complementation of a mutant strain
Basic Protocol 3: Tn5 transposon mutagenesis of C. diphtheriae
Keywords: Corynebacterium, genetic manipulation, Tn5 transposon, mutagenesis, gene deletion
INTRODUCTION
More than 120 Corynebacterium species with available genome assemblies are listed in NCBI, at https://www.ncbi.nlm.nih.gov/genome. The most well-known among these high GC-content Gram-positive bacteria are Corynebacterium diphtheriae and Corynebacterium glutamicum, with the former widely recognized for its disease caused by the exotoxin diphtheria toxin and the latter for its industrial value of amino acid productions (Cozzi et al., 2011). Facile genetic tools for Corynebacterium species have been credited to the initial development of vectors for gene deletion and complementation in C. glutamicum (Ankri, Reyes, & Leblon, 1996; Eikmanns, Kleinertz, Liebl, & Sahm, 1991; Schafer et al., 1994). Later, Tn5 transposon mutagenesis was developed for C. diphtheriae, C. matruchotii, and C. glutamicum (D. M. Oram, Avdalovic, & Holmes, 2002; Suzuki et al., 2006; Wang, Hayes, Vestling, & Takayama, 2006).
Here, in Basic Protocol 1 we describe how to generate a non-polar, in-frame, markerless gene deletion mutant in C. diphtheriae. In Basic Protocol 2, we provide a detailed procedure of how to construct a complementing plasmid to rescue a deletion mutant. Finally, in Basic Protocol 3, we describe how to perform Tn5 transposon mutagenesis and identify Tn5 insertion.
CAUTION:
Corynebacterium diphtheriae is a Biosafety Level 2 (BSL-2) pathogen. All procedures must be performed following guidelines for handling pathogenic microbes.
STRATEGIC PLANNING
C. diphtheriae strain NCTC13129 and its isogenic strains are streaked from frozen −80°C stock vials on heart infusion agar (HIA) plates and incubated at 37°C for 16 to 24 hours prior to any experiments; these plate can be stored at 4°C for about a week. Heart infusion broth for growing corynebacteria in liquid cultures needs also to be prepared in advance. If needed, kanamycin or chloramphenicol is added to these media to the final concentration of 25 μg/ml or 2 μg/ml, respectively.
E. coli DH5α and S17–1 strains are grown on Luria-Bertani (LB) agar plates or in LB broth at 37°C for 12 hours. Depending whether these strains carry a vector with an antibiotic-resistant gene, kanamycin or chloramphenicol is added to these media to the final concentration of 50 μg/ml or 10 μg/ml, respectively. Agar and broth media are also prepared in advance, so are plasmid DNA from these vectors using commercially available kits. Competent cells (E. coli and C. diphtheriae) are prepared, using the rubidium chloride method for E. coli (Mülhardt, 2007) and a combination of tween-80 and glycine for C. diphtheriae (H. Ton-That & O. Schneewind, 2003), respectively, and kept at −80°C prior to usage.
BASIC PROTOCOL 1: GENE DELETION IN C. diphtheriae
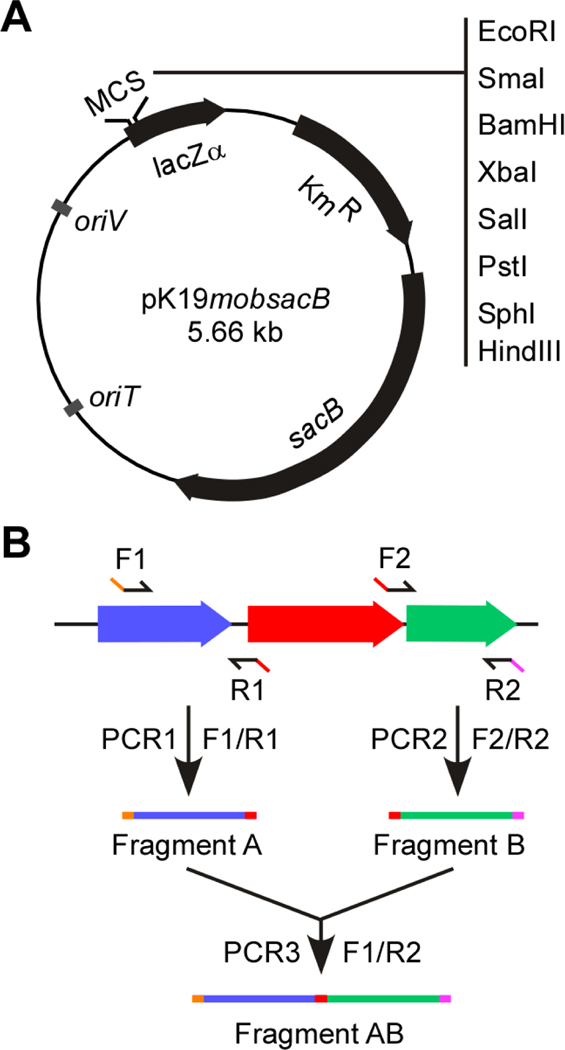
In 1994, Schäfer and colleagues described some small mobilizable vectors based on the E. coli plasmids pK18 and pK19 that are used to generate gene deletion mutants in C. glutamicum (Schafer et al., 1994). Since then, these vectors have widely been used in genetic manipulation in several Corynebacterium species including C. diphtheriae and C. matruchotii (Luong et al., 2018; H. Ton-That & O. Schneewind, 2003). One of the vectors utilized is pK19mobsacB, which contains many useful features including a multiple cloning site within a lacZα fragment, a kanamycin resistant cassette (KanR), mob (DNA region for mobilization by RP4), and sacB coding for levansucrase (Fig. 1A). When expressed in C. diphtheriae, levansucrase SacB metabolizes sucrose, leading to accumulation of toxic byproducts; thus, SacB has often been used as a counterselective marker (H. Ton-That & O. Schneewind, 2003).
Figure 1: Vector and procedure used for generation of a gene deletion cassette.
(A) Presented is pK19mobsacB, a non-replicating vector in C. diphtheriae used to generate gene deletion mutants in this organism. This vector contains a kanamycin resistant gene (KmR), sacB, mutiple cloning sites (MCS) within a lacZα fragment, and origins of replicon oriV and oriT; adapted from (H. Ton-That & O. Schneewind, 2003). (B) To delete a gene of interest (red), two sets of primers (F1/R1 and F2/R2) are designed for two PCR reactions PCR1 and PCR2 that generate 1-kb fragments A and B, respectively. Restriction enzyme sites are incorporated into primers F1 and R2 (orange and pink). Primers R1 and F2 contain a complementary sequence permitting annealing of fragments A and B to produce fragment AB during the third PCR reaction with primers F1 and R2.
In this protocol, we describe a detailed procedure to a generate non-polar, in-frame, markerless mutant in C. diphtheriae by double-crossover homologous recombination using pK19mobsacB based on a previously published protocol (H. Ton-That & O. Schneewind, 2003). This vector harboring a deletion construct is delivered to C. diphtheriae from E. coli S17–1 by conjugation. Integration of this plasmid into the C. diphtheriae by a first crossover event is selected by kanamycin. The second crossover, leading to excision of the plasmid, is selected by sucrose. In principle, this protocol is applicable to generate gene deletion mutants in C. matrochutii as previously reported (Luong et al., 2018). RNA preparation and RT-PCR are described to evaluate the gene expression for the deleted gene and its adjacent genes.
Materials
- Custom primers for gene deletion (Fig. 1B) (see Primer Design below)
- Forward F1
- Reverse R1
- Forward F2
- Reverse R2
-
Chromosomal DNA of strain NCTC 13129 (see Basic Protocol 2)
Plasmid pK19mobsacB (Schafer et al., 1994) (see Fig. 1A); available at Nova lifetech Limited (Cat # 10633)
0.2 ml PCR tube
- PCR amplification reagents
- Phusion kit (NEB, Cat. # M0530S)
- 10 mM dNTP mix
Heat blocks at 42°C, 65°C and 55°C
Thermal cycler
- Equipment for gel electrophoresis
- Electrophoresis chambers
- Power suppliers
- Gel casting trays
Qiagen DNA Miniprep kit (Cat # 27106)
- Reagents for DNA isolation and purification
- 1% dissolved agarose gel in TAE buffer
- TAE Buffer (see Reagents and Solutions)
- 1kb plus DNA marker (user’s preference)
- Ethidium bromide (EtBr) (1mg/ml)
- 10X gel loading dye (see Reagents and Solutions)
- Gel DNA Recovery kit (ZymoResearch, Cat. # D4001 or D4001T)
- DNA Binding Buffer (ZymoResearch, Cat. # D4004–1-L)
HF restriction enzymes from NEB and 10X CutSmart buffer
NanoDrop spectrophotometer
- Ligation reagents:
- T4 DNA 10X Ligase Buffer (NEB)
- T4 DNA Ligase (NEB)
SOC medium (see Reagents and Solutions)
Competent cells of E. coli DH5α and S17–1
HF Restriction Enzymes from NEB and 10X CutSmart buffer
LB broth (Reagents and Solutions)
LB agar plates (Reagents and Solutions)
Kanamycin (FisherScientific, Cat # BP906–5) (50 mgl/ml)
Apex Tag Red 2X Master Mix (Genesee Scientific, Cat # 42–137)
5% BSA and 5% monosodium glutamate, sterile-filtered
Nalidixic acid (FisherScientific, Cat. # BP908–25), 35 mg/ml in 0.1 M NaOH
HIA plates (Reagents and Solutions)
Heart infusion (HI) broth (Reagents and Solutions)
C. diphtheriae strain NCTC 13129
37° and 30°C air incubators
Sterilized scrapers, spreaders and tooth picks
10% sucrose (sterile)
- RNA preparation reagents:
- NucleoSpin® RNA (Macherey-Nagel, Cat. # 740955.10 / .50 / .250)
- 14.3 M β-mercaptoethanol
- 2 ml Bead Tubes Type B (MP Biomedicals, Cat. # SKU 116911100)
- 70% Ethanol
BeadBug microtube homogenizer (Benchmark Scientific, Cat. # D1030)
-
cDNA preparation reagents:
Random primers (Invitrogen, Cat. #48190–011)
10 mM dNTP mix (NEB)
M-MLV Reverse Transcriptase (Invitrogen, Cat. # 28025013)
RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen, Cat. #10777–019)
-
qPCR reagents and supplies
-
Primer sets for target genes and 23S rRNA:
23S rRNA Forward: GCCGCTTTAATGGGCGAAC
23S rRNA Reverse: GGGACTAGTGATCCGGCACC
iTaq Universal SYBR Green Supermix (Bio-rad, Cat. #1725120)
96-well white qPCR plates and adhesive sealing films
-
CFX Connect Real-Time PCR Detection System (Bio-rad, Cat. # 1855201)
Construction of a gene deletion cassette
1. Design two primer sets (F1/R1) and F2/R2); see Materials for PCR amplification of ~1 kb-flanking regions upstream and downstream of a gene of interest, respectively (Fig. 1B). Since C. diphtheriae is a high GC organism, the melting temperature of each primer set should be similar and around 60°C. Generally, primers are designed to generate 1-kb fragments (A and B) for a high frequency of homologous recombination; nonetheless, at a minimum 300-kb fragments are needed.
-
2. Perform two PCR reactions, using chromosomal DNA of strain NCTC 13129, generating 2 fragments A and B (Fig. 1B).
-
PCR reaction (50 μl)
10 μl 5X Phusion HF buffer
1 μl dNTP (10 mM)
1 μl Forward primer F1 or F2 (10 pmol/μl)
1 μl Reverse primer R1 or R2 (10 pmol/μl)
1 μl Chromosomal DNA (70 ng; see Basic Protocol 2, Steps 1–17)
1 μl Phusion DNA polymerase
35 μl MilliQ water
-
Thermal cycler program:
Step 1: 98°C, 3 min
Step 2: [98°C, 20 sec; Tm (°C), 15 sec; 72°C, 20 sec] 35 cycles
Step 3: 72°C, 5 min
Step 4: 12°C, hold
-
3. Separate the PCR products by gel electrophoresis and purify the two fragments by gel extraction as per the manufacturer’s instructions (Gel DNA Recovery kit; ZymoResearch).
-
4. Perform crossover-PCR, using the two purified PCR products above as templates, linking fragments A and B to generate AB (Fig. 1B)
-
PCR reaction (50 μl):
10 μl 5X Phusion HF buffer
1 μl dNTP (10 mM)
1 μl Primer F1 (10 pmol/μl)
1 μl Primer R2 (10 pmol/μl)
50 ng Fragment A (template 1)
50 ng Fragment B (template 2)
1 μl Phusion DNA polymerase
MilliQ water added to 50 μl
-
Thermal cycler program
Step 1: 98°C, 3 min,
Step 2: [98°C, 15 sec; Tm (°C), 15 sec; 72°C, 1 min] 35 cycles]
Step 3: 72°C, 5 min,
Step 4: 12°C, hold
-
-
5. Purify the linked fragment after PCR amplification by gel extraction, digest the fragment by appropriate resstriction enzymes, and ligate into pK19mobsacB.
-
Digestion of vector or PCR fragment
2 to 3 μg Vector or purified PCR fragment
3 μl 10X CutSmart Buffer
1. μl Restriction enzyme 1
1 μl Restriction enzyme 2
Add water to a final volume of 30 μl. Incubate the reaction at 37°C for 2–3 h. Stop the enzyme reactions by incubating the tubes at 65°C for 20 min. Purify the linearized vector by gel extraction, and measure DNA concentrations by NanoDrop.
- Clean-up of the digested PCR fragment
- Add 2 volumes (60 μl) of DNA Binding Buffer (ZymoResearch) to the digested solution and mix well before loading the mixture onto a Zymo-Spin column in a collection tube. Using a microfuge, spin it at 10,000 rpm for 30 sec and discard the filtrate.
- Wash the column by adding 200 μl of DNA wash buffer to the column and spin at 12,000 rpm for 30 sec. Discard the flow-through. Repeat the wash once more.
- Place a new 1.5 ml collection tube under the column. Add 15 μl pre-warmed (~55°C) DNA elution buffer directly to the center of column. Wait for 30 to 60 sec before centrifuging for 1 min at 12000 rpm to elute DNA. Check the DNA concentration by NanoDrop.
-
Ligation reaction (molar ratio of insert and vector at least 5:1 or 10:1 if possible)
50~100 ng Vector DNA (5.6 kb)
90~180 ng Insert DNA (2 kb)
1 μl 1X ligation Buffer
1 μl T4 DNA ligase
Add water to a final volume of 20 μl. Incubate the reaction at 25°C for 2–3 h.
-
6. Thaw a tube of 50 μl E. coli DH5α competent cells on ice. Add 10 μl of the ligation mix (step 5C) to the competent cells; incubate the cells on ice for 20 min prior to heat-shock treatment at 42°C for 40 sec; and immediately incubate the cells on ice before adding 250 μl of SOC medium kept at room temperature.
7. Incubate the transformed cells at 37°C with gentle shaking for 1 h; harvest the cell pellet by centrifugation using a microfuge at 15,000 rpm for 3 min; discard 150 μl of the supernatant; resuspend in the remaining supernatant and spread the entire suspension onto an LB agar plate containing 50 μg/ml kanamycin (Kan) (selection for pK19mobsacB) or appropriate antibiotics if different vectors are used; and incubate the plate overnight at 37°C.
8. Pick 10 to 20 transformants using sterile pipette tips and transfer them to a LB agar plate containing the same antibiotic, i.e. Kan, by making a short streak. After a few hours of incubation at 37°C, the transferred colonies are ready for colony-PCR.
-
9. Perform colony-PCR to check the presence of the insert, using the forward and reverse primers A and D
-
Colony-PCR: In each PCR tube, add 8 μl of sterile water; suspend cells from a single colony (Step 8) using pipette tips; add 1 μl of each primer (F1 and R2; see Materials), followed by adding 10 μl of Apex Taq Red 2X master mix; mix well by vortex; and perform a quick spin in a microfuge prior to PCR-amplification.
Note: Melting temperature (Tm) is primer-dependent and calculated in the presence of 1.5 mM MgCl2, and the extension time is calculated using 30 sec per kb.
-
Thermal cycler program
Step 1: 95°C, 8 min
Step 2: [95°C, 20 sec; Tm, 15 sec; 72°C, extension time] 25 cycles
Step 3: 72°C, 3 min
Step 4: 12°C, hold
-
10. Check the PCR products for the presence of the insert with its correct size by agarose gel electrophoresis.
11. Choose 3 colonies verified from Step 10 for plasmid DNA miniprep using Qiagen plasmid preparation kits according to the manufacturer’s instructions, followed by plasmid digestion using the same restriction enzymes mentioned in Step 5a.
-
12. Plasmid DNA of verified clones is used for transformation with E. coli S17–1.
Transformation: Add 1 μl of plasmid DNA from Step 11 to 50 μl of E. coli S17–1 competent cells; perform the heat-shock procedure (see Step 6); and plate 50 μl of the cell suspension onto an LB agar plate containing Kan (50 μg/ml).
13. Pick 2 Kan-resistant colonies for storage at −80°C in 1 ml of 5% BSA (w/v) and 5% (w/v) monosodium glutamate (sterile filtered).
Bacterial conjugation and selection of gene deletion mutants
14. Inoculate 3 ml-cultures of C. diphtheriae in HIB (see Materials) and E. coli S17–1 harboring pK19mobsacB with a gene deletion construct as described above in LB supplemented with kanamycin (50 μg/ml) at 37°C overnight with shaking.
15. Harvest the cell pellets from 1.5 ml of each culture above by centrifugation at 6,000 x g for 3 min; resuspend each cell pellet in 0.5 ml of HIB; combine them together into one Eppendorf tube; and quickly vortex.
16. Pellet the cell mixture by centrifugation at 6,000 x g for 3 min and incubate undisturbed at 30°C for 1 h.
17. Discard 800 μl of the supernatant and thoroughly mix the cells in the remaining medium by pipetting.
18. Transfer the cell suspension onto a HI agar plate as a single spot and incubate the plate at 30°C to allow conjugation to occur overnight.
19. The next day, add 1 ml of HIB onto the plate, scrape off the cells and gently resuspend the cells by pipetting.
20. Spread 200 μl of the cell suspension onto a HI agar plate containing nalidixic acid (35 μg/ml) and Kan (25 μg/ml) (HIANal35Kan25), and incubate at 30°C for 2–3 days or until colonies are visible.
21. Pick 2 to 5 colonies using sterile toothpicks by streaking onto a new HI agar plate containing the same antibiotics as listed above and incubate the plate at 30°C for 2–3 days.
-
22. Pick 2 colonies for storage at −80°C in 1 ml of 5% BSA and 5% monosodium glutamate (see Step 13).
Note: Perform colony-PCR, using primers F1 and R2, to verify these co-integrates (plasmid integrated into the chromosome).
23. Inoculate 1 ml cultures from an integrate colony in HIB without any antibiotics at 30°C overnight. The next morning, spread 50 μl of the overnight cultures onto an HI agar plate containing 10% sucrose (HIA10S).
24. Pick at least 20 colonies using sterile toothpicks and patch each colony onto a HIA10S plate, following by patching with the same toothpicks onto a HIANal35Kan25 plate. Inoculate the plates at 30°C overnight.
25. Using primers F1 and R2, perform colony-PCR with at least 10 patched colonies from the HIA10S plate that are sensitive to nalidixic acid and kanamycin (growth on the sucrose plate and no growth on the HIANal35Kan25 plate) for the loss of the gene of interest, i.e. the presence of 2-kb amplicons.
26. Further confirm these mutant clones by RT-PCR (see below, steps 28–53) or western blotting if antibodies are available.
27. Pick 2 confirmed mutants for storage at −80°C in 1 ml of 5% BSA and 5% monosodium glutamate.
RNA isolation and RT-PCR
28. Inoculate 3-ml overnight cultures in HIB from a single colony of the mutant strain above (applicable to any corynebacterial strains).
29. Harvest the cells by centrifugation at 6,000 x g for 10 min.
30. Resuspend the cell pellets in 0.8 ml of RA1 buffer and 8 μl of 14.3 M β-mercaptoethanol. Note: 10–20 mM DTT or TCEP can be substituted for β-mercaptoethanol.
31. Transfer the above cell suspension to ice-cold Bead Tubes (Type B) and place on ice for at least 5 min.
32. Lyse the cells by mechanical disruption using a BeadBug Microtube Homogenizer for 30 sec, with speed setting of 4000, for 3–4 times. Keep the tubes on ice for 1 min after each time of disruption.
33. Centrifuge the tube at 13000 x g for 1 min and transfer to lysate to ice-cold 1.5 ml-tubes.
34. Filtrate the lysates by applying into a NucleoSpin® Filter (violet ring) and centrifuging at 11,000 x g for 1 min.
35. Add an equal volume of 70% ethanol to the filtrated lysates and mix by pipetting.
36. RNA binding – Load the samples to NucleoSpin® RNA Columns (light blue ring) and centrifuge at 11,000 x g for 30 sec.
37. Desalting – Add 350 μl MDB (Membrane Desalting Buffer) and centrifuge at 11,000 x g for 1 min.
38. DNA removal – First, mix 10 μl rDNase (provided from the kit) to 90 μl Reaction Buffer; then transfer 95 μl of the mixture onto the center of the silica membrane of the column; and incubate at room temperature for 15 min.
39. 1st wash – Add 200 μl Buffer RAW2 and centrifuge at 11,000 x g for 30 sec.
40. 2nd wash – Add 600 μl Buffer RA3 and centrifuge at 11,000 x g for 30 sec.
41. 3rd wash – Add 250 μl Buffer RA3 and centrifuge at 11,000 x g for 2 min.
42. RNA elution – Add 40–60 μl RNase-free H2O and centrifuge at 11,000 x g for 1 min to elute RNA.
43. Measure RNA concentrations using a NanoDrop spectrophotometer and store RNA samples at −80°C for future usage.
-
44. Add the following components to a nuclease-free PCR tube:
250 ng of random primers
500 ng of total RNA
1 μl of 10 mM dNTP Mix
Nuclease-free water to 12 μl
46. Incubate the tube at 65°C for 5 min and then keep it on ice for 2 min.
47. Briefly centrifuge and add 4.0 μl of 5X First-Strand Buffer, 2.0 μl μl of 0.1 M DTT, and 1 μl RNaseOUT Recombinant Ribonuclease Inhibitor (40 units/μl).
48. Gently mix the content and incubate at 37°C for 2 min.
49. Add 1 μl of M-MLV Reverse Transcriptase, mix gently, and incubate at 25°C for 10 min, then 37°C for 50 min, and finally at 70°C for 15 min.
50. Store the resulting cDNA samples at −80°C for future usage.
51. Dilute each stock cDNA with a ratio of 1:100 and 1:1,000,000 for genes of interest and 23S rRNA, respectively.
52. Add to 4 μl of the diluted cDNA samples 5 μl of iTAQ SYBR green supermix (2X) and 1 μl of primer mix (forward and reverse primers for target genes and 23S rRNA)
53. Seal the plates with optical transparent film and quick spin for 30 sec to remove any bubbles and sediment all the mixture to the bottom wells.
-
53. Run RT-PCR reaction with the following program, using a CFX Connect Real-Time PCR Detection System (Bio-Rad) and check the products by gel electrophoresis for the presence or absence of the amplicons.
Step 1: 95°C, 3 min, 1 cycle
Step 2: 95°C, 10 sec; 60°C, 10 sec; 39 cycles
Add the following steps for melting curve analysis if needed
Step 3: 95°C, 10 sec
Step 4: 65°C, 5 sec
Step 5: increase from 65°C to 95°C in increments of 0.5°C
BASIC PROTOCOL 2: COMPLEMENTATION OF A MUTANT STRAIN
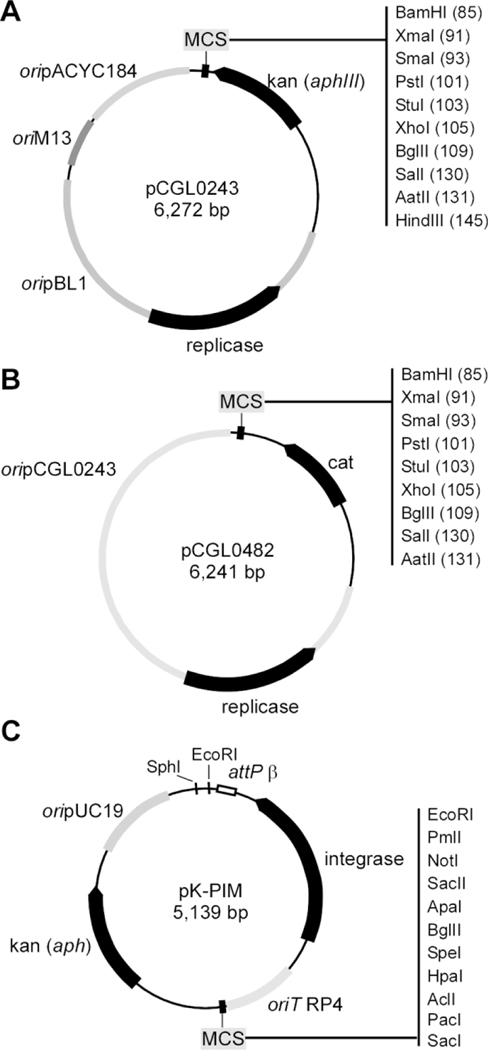
pCGL0243 and pCGL0482 (Ankri et al., 1996) are two E. coli/Corynebacterium shuttle vectors commonly used for complementation of mutant strains in C. diphtheriae, with the former harboring a kanamycin resistance gene and the latter a chloramphenicol resistance gene (Fig. 2A-2B). While pCGL0243 and pCGL0482 are multiple-copy plasmids, the bacteriophage-base plasmid pK-PIM (Fig. 2C) integrates into the corynebacterial chromosome via either attB1 or attB2 site, whichever is available, often as a single copy (M. Oram, Woolston, Jacobson, Holmes, & Oram, 2007). For complementation, all three vectors can be introduced into the cytoplasm of C. diphtheriae by electroporation; however only pK-PIM can be delivered via conjugation.
Figure 2: Vectors for complementation in C. diphtheriae.
(A) The E. coli/Corynebacterium shuttle vector pCGL0243 contains a kanamycin-resistant gene (aphIII), origins of replicon oripBL1, oriM13 and oripACY184, and multiple cloning sites (MCS). (B) As a derivative of pCLG0243, pCGL0482 harbors a chloramphenicol-resistant gene (cat) and some common features of the former plasmid. (C) The integration plasmid pK-KIM contains a kanamycin-resistant gene, MCS, integrase, and origins of replicon oriT RP4 and oripUC19.
Materials
Wizard Genomic DNA Purification Kit (Promega Cat. #A1120)
C. diphtheriae NCTC 13129 (ATCC)
HIB media (see Reagents and Solutions)
1.5 ml Eppendorf tubes
50 mM EDTA, pH 8.0
Lysozyme (Sigma Cat. # 17651); 10 mg/ml
Mutanolysin (Sigma Cat.# M9901–10KU); 5 units/μl
Nuclei Lysis Solution
80°C and 65°C water baths
RNAse A
Protein Precipitation Solution
Isopropanol
70% Ethanol
DNA Rehydration Solution
NanoDrop spectrophotometer
Sterile MilliQ H2O
2X HIBTW (Reagents and Solutions)
HTGS (Reagents and Solutions)
High-speed Centrifuge (Beckman Avanti J-E centrifuge with JA14.5 rotor)
15% cold glycerol
Custom primers (forward and reverse primers; 10 pmol/μl)
-
PCR amplification reagents
Phusion kit (NEB, Cat. # M0530S)
mM dNTP mix
pCGL0243 (KanR) or pCGL0482 (CamR) shuttle vector (100–150 ng/μl)
pKPIM (KanR) integration vector (350–450 ng/μl)
0.2 ml PCR tubes
-
Reagents for DNA isolation and purification
1% dissolved agarose gel in TAE buffer
TAE Buffer (see Reagents and Solutions)
1kb plus DNA marker (user’s preference)
Ethidium bromide (EtBr) (1mg/ml)
10X gel loading dye (see Reagents and Solutions)
Gel DNA recovery kit (ZymoResearch Cat. # D4001 or D4001T)
DNA Binding Buffer (ZymoResearch Cat. # D4004–1-L)
10 % glycine (filter-sterilized, kept at 4°C)
50% sucrose (filter-sterilized)
50 ml conical tubes
1.5 ml Eppendorf tubes
Dry ice/alcohol bath
HF Restriction enzymes and 10X CutSmart buffer (NEB)
-
Ligation reagents:
T4 DNA 10X Ligase Buffer (NEB)
T4 DNA Ligase (NEB)
SOC medium (see Reagents and Solutions)
E.coli DH5α competent cells
42°C heat block
Sterile spreader
Apex Taq Red 2X master mix (Genesee Scientific, Cat. # 42–137)
5% BSA and 5% monosodium glutamate, filter-sterilized
C. diphtheriae competent cells
0.2 cm electroporation cuvette (Fisherbrand, Cat. # FB102)
20% glucose (filter-sterilized)
HIB (Reagents and Solutions)
HIB containing 1% glucose (Add 1 ml of 20% glucose into 19 ml of sterile HIB)
E.coli S-17 competent cells
HIA plates containing 25 μg/ml of Kanamycin (HIA Kan25)
HIA plates containing 35 μg/ml of nalidixic acid, 25 μg/ml of kanamycin (HIA Nal35 Kan25)
LB plate containing 50 μg/ml of Kanamycin (LB Kan50) or 10 μg/ml of chloramphenicol
(LB Cam10)
Electroporation device (Bio-rad Gene Pulser Xcell)
C. diphtheriae chromosomal DNA isolation
Chromosomal DNA can be isolated using commercially available kits. The following procedure is used with the Wizard Genomic DNA Purification Kits with minor modifications.
1. Harvest corynebacterial cells (strain NCTC 13129) from 1.5 ml of overnight cultures in HIB by centrifugation at 10,000 rpm for 5 min.
2. Discard the supernatant and suspend the cell pellet in 480 μl of 50 mM EDTA, pH 8.0
3. Digest the cell wall by adding 60 μl of 10 mg/ml Lysozyme and 60 μl of Mutanolysin (300 units) to the cell suspension above gentle mixing; incubate at 37°C with gentle shaking for at least 2 h.
4. Centrifuge at 10,000 rpm for 5 min and remove the supernatant.
5. Add 600 μl of Nuclei Lysis Solution and gently resuspend the cell pellet by pipetting.
6. Incubate at 80°C in a water bath for 5 min to lyse the cell, then cool to room temperature.
7. Add 3 to 6 μl RNaseA (4mg/ml) to the cell lysate and mix by inverting the tube.
8. Incubate at 37°C for 1 hr
9. Add 200 μl of Protein Precipitation Solution to the lysate. Quickly vortex for 5 sec to make sure the solution is fully mixed.
10. Chill the tube on ice for 5 min
11. Obtain the supernatant by centrifugation at 15,000 rpm for 5 min and transfer the supernatant to another sterile Eppendorf tube.
12. Add 600 μl of room temperature isopropanol to the supernatant and gently mix the tube by inverting it a few times.
13. Spin down the DNA at 15,000 rpm for 5 min. (Use Sharpie to mark the location of white pellet on the tube)
14. Gently pour out the S/N and add 600 μl room temperature 70% ethanol to rinse the inside of the tube by gently inverting the tube a couple times before centrifuging the tube at 15,000 rpm for 5 min again.
15. Remove the alcohol and allow the pellet to air dry for 30 min.
16. Add 100 μl of DNA Rehydration Solution and incubate at 65°C for 1 hr to dissolve the DNA. Alternatively, the DNA can be rehydrated by incubation overnight at room temperature or 4°C.
17. Check the DNA concentration using NanoDrop spectrophotometer.
Preparation of C. diphtheriae competent cells
18. Inoculate 10 ml cultures of C. diphtheriae in 1X HIBTW (see Reagents and Solutions) at 37°C overnight.
19. The next morning, dilute the overnight cultures in 50 ml HTGS (see Reagents and Solutions) to have starting OD600 of 0.1; grow cells at 37°C with shaking until OD600 between 0.4 and 0.6 (roughly 3 to 4 h).
20. Chill the culture flasks on ice for at least 15 min before transferring the cultures to a sterile 50ml conical tube, which is also kept on ice.
21. Harvest the cells by centrifugation at 4°C at 6,000 x g for 15 min in Beckman Avanti J-E centrifuge (JA 14.5 rotor).
22. Decant the supernatants and completely suspend the cell pellets in 20 ml of sterile pre-chilled 15% glycerol (no clumping).
23. Wash the cells by centrifugation at 6,000 x g for 15 min and resuspension with 20 ml of sterile cold 15% glycerol, followed by centrifugation.
24. Discard the supernatants and resuspend the cell pellets in 2 ml of sterile pre-chilled 15% glycerol.
25. Make 100 μl aliquots of the competent cells in pre-chilled 1.5 mL Eppendorf tubes, snap-freeze the aliquots by dipping into a dry ice/alcohol bath, and store them at −80°C for future usage.
Construction of a complementing clone
26. Design forward and reverse primers (see Materials) for PCR-amplification of a gene of interest including its promoter region from C. diphtheriae chromosomal DNA, with appropriate restriction enzyme sites on each end of the primers.
-
27. Perform PCR and purify the PCR products by gel extraction according to the manufacturer’s kit instructions.
-
PCR reaction
10.0 μl 5X Phusion HF buffer
1.0 μl dNTP (10 mM)
1.0 μl forward primer (10 pmol/μl)
1.0 μl reverse primer (10 pmol/μl)
0.5 μl chromosomal DNA (70 ng) (see Steps 1–17)
1.0 μl Phusion polymerase
Add H2O to 50 μl
-
PCR program
Step 1: 98°C, 3 min
Step 2: [98°C, 20 sec; Tm, 15 sec; 72°C, extension time] 35 cycles
Step 3: 72°C, 3 min
Step 4: 12°C, hold
-
28. Digest the PCR products and an appropriate vector (pCGL0342, pCGL483 or pK-PIM) with the restriction enzymes mentioned in Step 26; see Step 5 (Basic Protocol 1) for digestion and purification.
29. Perform ligation reactions with the purified PCR products and vector in Step 28 (see Step 5, Basic Protocol 1, for ligation).
-
30. Perform transformation with the ligation mix (Step 29) and E. coli DH5α competent cells and select for a complementing plasmid (see Step 6–8, Basic Protocol 1).
Note: LB agar plates containing 50 μg/ml kanamycin are used for selecting pCGL0243 and pK-PIM; for pCGL0482, 10 μg/ml chloramphenicol is added to agar plates.
31. Perform colony-PCR to check the presence of the insert, using the forward and reverse primers mentioned in Step 26 (see Step 9–11, Basic Protocol 1).
32. Plasmid DNA of verified clones is isolated, using a Qiagen Plasmid Prep kit, and subjected to DNA sequencing to further confirm the insert.
33. Pick 2 positive clones of each construct for storage at −80°C in 1 ml of 5% BSA and 5% monosodium glutamate.
34. Thaw 100 μl competent cells on ice, add to the cells 300 to 500 ng plasmid DNA (Step 32), transfer the mixture to a 2-mm pre-chilled electroporation cuvette, and keep it on ice for 10 min (save the competent cell tube).
35. Perform electroporation with a Bio-Rad GenePulser Xcell with settings of 2.5 kV, 25 μF and 200 Ω.
36. Immediately transfer the electroporated cells to the saved tube containing 900 μl HIB supplemented with 1% glucose; incubate the tube at 37°C with gentle shaking for 1 h.
37. Spread 100 to 300 μl of the cell suspension onto an HI agar plate containing 25 μg/ml kanamycin and incubate the plate at 37°C for 48 h or until colonies appear.
38. Pick 2 colonies for storage at −80°C in 1 ml of 5% BSA and 5% monosodium glutamate. Note: If pK-PIM is used, it can be delivered into C. diphtheriae by conjugation. In that case plasmid DNA in Step 32 is used to transform E. coli S17–1 prior to performing bacterial conjugation (see Basic Protocol 1, Steps 14–20).
BASIC PROTOCOL 3: TN5 TRANSPOSON MUTAGENESIS OF C. diphtheriae
Tn5 transposon mutagenesis has been reported in many corynebacterial species including C. diphtheriae (D. M. Oram et al., 2002), C. matruchotii (Wang et al., 2006), and C. glutamicum (Suzuki et al., 2006). Tn5 transposon is commercially available as kits, which contain a stable Transposome complex, consisted of EZ-Tn5 transposase and EZ-Tn5<Kan-2> transposon. The transposome complex is delivered into the corynebacterial cytoplasm by electroporation and the Tn5 transformants are selected by kanamycin. Here, we describe a detailed protocol of Tn5 transposon mutagenesis for C. diphtheriae using an EZ-Tn5<Kan-2>Tnp Transposome kit obtained from Lucigen and a protocol of identifying Tn5 insertion based on thermal asymmetric interlaced PCR (TAIL-PCR)(Nakayama, Soma, Rahmutula, Ozawa, & Kanmatsuse, 2001; Singer T., 2003).
Materials
EZ-Tn5™ <Kan-2>Tnp Transposome (Lucigen, Cat. # TSM99K2)
GenePulserXcell (Bio-rad) for Electroporation
C. diphtheriae competent cells
0.2 cm electroporation cuvette (Fisherbrand, FB102)
HIA plate containing 25 μg/ml of kanamycin (HIA Kan25)
Sterile HIB containing 15% glycerol
15-ml conical tubes
HIA plates containing 25 μg/ml of kanamycin (HIAKan25)
15% glycerol
96-well plates
Apex Taq Red 2X master mix (Genesee Scientific, Cat. # 42–137)
Sterile water
-
Primers
Kan2-Tn5–1-F: TGCAGTTTCATTTGATGCTCGATGAG (919–944)
Kan2-Tn5–2-F: ACCTACAACAAAGCTCTCATCAACC (1127–1151)
AD-1-R: NGT CGA SWG ANA WGA A (N= A/G/C/T, S= G/C, W= A/T)
-
Reagents for DNA isolation and purification
1% dissolved agarose gel in TAE buffer
TAE Buffer (see Reagents and Solutions)
1kb plus DNA marker (user’s preference)
Ethidium bromide (EtBr) (1mg/ml)
10X gel loading dye (see Reagents and Solutions)
Gel DNA recovery kit (ZymoResearch Cat. # D4001 or D4001T)
Tn5 transposon mutagenesis
1. Thaw 100 μl of C. diphtheriae competent cells (see Steps 18–25, Basic Protocol 2) on ice.
2. Add 1 μl of EZ-Tn5<Kan-2>Tnp Transposome (Lucigen; 80 ng transposon DNA) to the competent cells and incubate the mixture on ice for 10 min before transferring the cell mixture to a pre-chilled 0.2-cm electroporation cuvette.
3. Perform electroporation (see Steps 35–36, Basic Protocol 2).
4. After electroporation, transfer the entire transformed cells to 9 ml HIB in a 15-ml conical tube. Spread 50-μl aliquots of the transformed cells onto HIAKan25 plates (~20 plates), and incubate them at 30°C until colonies are visible (1 to 2 days).
5. Pool all Kan-resistant colonies into a 15-ml conical tube containing ~10 ml HIBKan25 and 15% glycerol; make 1-ml aliquots and store them at −80°C for future usage. Alternatively, individual colonies can be stored in 98-well plates.
Identification of Tn5 insertion sites
-
6. Perform 1st PCR-amplication with individual Tn5 mutant colonies
-
PCR reaction
1 μl Kan2-Tn5–1-F primer (10 pmol/μl)
1 μl AD-1-R primer (10 pmol/μl)
8 μl H2O mixed with colony
10 μl 2X Apex Taq Red 2X mater mix
-
program
Step 1; 95°C, 8 min
Step 2: [95°C for 20 sec, annealing time for 25 sec, 72°C for 1min] 40 cycles
Note: Set Tm starting from 55°C and decreasing 0.5°C each cycle
-
-
7. Perform 2nd PCR-amplification with PCR products from Step 6
-
PCR reaction
4 μl PCR product from Step 6
4 μl Kan2-Tn5–1-F primer (10 pmol/μl)
4 μl AD-1-R primer (10 pmol/μl)
38 μl H2O
50 μl 2X Apex Taq Red 2X mater mix
Divide the reaction mix into 4 PCR tube with 25 μl each and place each tube inside a thermal cycler for each temperature setting.
-
Gradient PCR program
Step 1: 95°C, 3 min
Step 2: [95°C for 25 sec; Tm is set between 44° to 50°C for 20 sec; 72°C for 1 min] 30 cycles
Step 3: 72°C, 2 min
Step 4: 4°C, hold
-
-
8. Separate the PCR products by gel electrophoresis and extract the brightest bands for DNA sequencing.
Note: If no products are found, repeat the PCR program at Step 6 using the 4 PCR products from Step 6 as templates with primer Kan2-Tn5–1-F replaced by primer Kan2-Tn5–2-F.
REAGENTS AND SOLUTIONS
-
1. SOC medium (1 L)
20 g/L Bacto-tryptone
5 g/L Bacto-yeast extract
2.5 mM KCl
0.5 g/L NaCl (pH 7.0)
Add water to 1 L and adjust pH to 7.0 with 5N NaOH before autoclave.
Add 5 ml of sterile 2 M MgCl2 and 20 ml of 1 M glucose to the above solution cooled to room temperature.
-
2. 10X gel loading dye (10 ml)
3.9 ml Glycerol
500 μl 10% SDS
200 μl 0.5 M EDTA
0.025 g Bromophenol blue
0.025 g Xylene cyanol
Add water to 10 ml
-
3. TAE buffer:
20 mM acetic acid
1 mM EDTA,
40 mM Tris-base, pH 8.5.
-
4. LB medium (1 L):
25 g LB broth, Miller (FisherScientific; Cat # BP1425)
Add water to 1 L and autoclave
-
5. LB Agar (500 ml):
12.5 g LB broth, Miller (FisherScientific; Cat # BP1425)
7.5 g Agar (Fisher, CAT BP1423–500)
Add water to 0.5 and autoclave.
-
6. HIB medium (1 L):
25 g Heart Infusion Broth (BD Biosciences; Cat # DF0038–17-7)
Add water to 1 L and autoclave
-
7. HIA (0.5 L):
12.5 g Heart Infusion Broth (BD Biosciences; Cat # DF0038–17-7)
7.5 g Agar (Fisher, Cat # BP1423–500)
Add water to 0.5 L and autoclave
-
8. 2X HIBTW (0.5 L)
25 g Heart Infusion Broth (BD Biosciences; Cat # DF0038–17-7)
2 ml Tween-80 (0.4%)
Add water to 0.5 L and autoclave
-
9. HTGS medium (50 ml)
25 ml sterile 2X HIBTW
10 ml 10% glycine (sterile-filtered)
15 ml 50% sucrose (sterile-filtered)
COMMENTARY
Background Information
Discovered by Klebs and Löffler as the causative agent of diphtheria (Murphy, 1996), the Gram-positive club-shaped bacillus C. diphtheriae secretes a potent binary toxin called diphtheria toxin, comprised of the active toxin A subunit and the B subunit that binds to the EGF-like growth factor (HB-EGF) on human epithelial cells (Rogers, Das, & Ton-That, 2011). This pathogen also produces covalently-linked pili assembled by the conserved sortase enzymes (Chang, Mandlik, Das, & Ton-That, 2011; Mandlik, Das, & Ton-That, 2008; Mandlik, Swierczynski, Das, & Ton-That, 2007; H. Ton-That & O Schneewind, 2003). These pili mediate bacterial adherence to pharyngeal epithelial cells (Mandlik et al., 2007) and are critical for bacterial virulence in experimental models of infection (Broadway et al., 2013; Reardon-Robinson et al., 2015). Additional adhesins have also been identified (Hirata Jr et al., 2008; Moreira, Mattos-Guaraldi, & Andrade, 2008; Ott et al., 2010), although their role in bacterial virulence in vivo are not known.
Thus, while it is clear that diphtheria toxin and pili are the two major virulence factors identified to date, a comprehensive view of corynebacterial adhesion/invasion, colonization and persistence is far from certain. It is noteworthy that C. diphtheriae has been an excellent model of gene regulation and iron acquisition with the prototypical regulator DtxR and of exotoxin with the archetypal diphtheria toxin (Tao, Schiering, Zeng, Ringe, & Murphy, 1994), as well as pilus assembly and oxidative protein folding in Gram-positive bacteria (Ramirez, Das, & Ton-That, 2020; Reardon-Robinson & Ton-That, 2016; Siegel, Liu, & Ton-That, 2016). Therefore, detailed protocols for genetic manipulations in C. diphtheriae described in this unit would be helpful to the scientific community.
Critical Parameters
Construction of complementing and deletion mutant strains
In cloning, the molar ratio of insert to vector needs to be at least 5 to 1, if not 10 to 1 to yield satisfactory ligation. In crossover PCR reactions, two purified templates should be added in equal molar amounts. For colony-PCR to verify deletion mutants, it is important to avoid contamination with E. coli or wild-type C. diphtheriae, which leads to false positives.
In designing primers for construction of gene deletion cassettes, attention is paid to avoid potential polar effects due to out-of-frame deletion. At the 5’ end of primers R1 and F2, the Tm of the overlapping sequence should be similar to that of primers F1 and R2. In addition, the high GC-content of the C. diphtheriae genome poses problems for primer design, such as high Tm, self-dimer formation, and secondary structures. This results in unspecific amplifications in initial and crossover PCR reactions. A single product will be ideal in each PCR step; otherwise, PCR buffer modification, e.g. addition of DMSO, MgCl2, optimization of annealing temperatures by gradient PCR, and primer redesign, are highly recommended. Furthermore, DNA purification by gel extraction can be used to obtain specific templates.
RNA preparation
When working with RNA, handle samples with care to minimize RNase contamination and RNA degradation. RNA yield may vary, depending on growth phase. To ensure all bacterial cells are lysed, it is important not to use more than 1 × 109 bacterial cells in the indicated volume. Furthermore, DNA contamination should be avoided if the extracted RNA is used for qPCR.
Anticipated Results and Troubleshooting
Basic Protocols 1 and 2
The concentration of gel-purified vectors, e.g. pCGL0243, is typically from 10 to 20 ng/μl or less, due to low efficiency of gel extraction for large DNA molecules. In order to reach the molar ratio of 5 to 1 (insert to vector), the concentration of purified inserts needs to be at least 30 ng/μl or higher, especially for the size of insert greater than 2 kb. We noticed that ligation reactions performed at 16°C overnight yield better results than at room temperature. The transformation procedure with pCGL0243 derivatives, i.e. with inserts, typically yields 3 to 4 × 104 CFU per μg DNA. This efficiency is reduced with larger inserts, so more competent cells and DNA are needed.
Colony-PCR is useful for initial screening of gene deletion mutants and transformants harboring recombinant vectors. However, conventional PCR with chromosomal DNA as template is needed for confirmation, as colony PCR may produce false-positive results. Additionally, digestion with restriction enzymes is recommended to verify inserts, and western blot and northern blot analyses are needed for confirming deletion mutants.
In conjugation, roughly 50 co-integrates can be obtained following 2–3 days of incubation. To improve efficiency, more cells can be used for plating after the conjugation step between E. coli and C. diphtheriae. Secondly, conjugation efficiency can also be improved by optimizing the cell number of the donor and recipient.
In allelic exchange, this procedure would generate an equal population of wild-type and mutant alleles, in theory. If a gene of interest encodes a factor important for a cellular process, wild-type alleles are obtained more often than the mutant ones in the screening step by colony-PCR. Therefore, a considerable number of colonies should be screened, e.g. 30–50. If more than 100 colonies are screened with all yielding wild-type alleles, it is most likely that this gene is essential. A conditional gene deletion may be employed.
In RNA isolation, the procedure typically yields 20–40 μg at a concentration of 0.5–1 μg/μl. RNA quality is determined via A260/A280 and A260/A230 measurements, and considered to be good with these values of ~2.0 and ~1.4, respectively. Extra cleaning with DNAase or re-purification is needed if contamination occurs.
Basic Protocol 3
One transposition reaction with roughly 80 ng of Tn5 Transposome typically yields 45,000 to 50,000 colonies. This efficiency can be improved by optimizing conditions to make better competent cells and transposome.
Time Considerations
For Basic Protocols 1 and 2, generation of a deletion mutant can take 2 to 3 weeks; construction of a gene deletion cassette is generally a rate-limiting step. The cloning procedure would take a week. Normally, RNA purification can be finish within a few hours. For Tn5 transposition in Basic Protocol 3, the entire procedure would take less than 3 days. To individually store Tn5 clones in 96-well plates, extra time and lab personnel are needed.
ACKNOWLEDGEMENTS
We would like to thank Emily Peluso and Matthew Scheible for critical review and discussion of this manuscript. Work related to this manuscript was supported by the National Institute of Dental & Craniofacial Research of the National Institutes of Health under the award numbers DE025015 and DE017382 (to H.T.-T).
LITERATURE CITED
- Ankri S, Reyes O, & Leblon G. (1996). Electrotransformation of highly DNA-restrictive corynebacteria with synthetic DNA. Plasmid, 35(1), 62–66. [DOI] [PubMed] [Google Scholar]
- Broadway MM, Rogers EA, Chang C, Huang IH, Dwivedi P, Yildirim S, . . . Ton-That H. (2013). Pilus gene pool variation and the virulence of Corynebacterium diphtheriae clinical isolates during infection of a nematode. J Bacteriol, 195(16), 3774–3783. doi: 10.1128/JB.00500-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Mandlik A, Das A, & Ton-That H. (2011). Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol Microbiol, 79(5), 1236–1247. doi: 10.1111/j.1365-2958.2010.07515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi R, Malito E, Nuccitelli A, D’Onofrio M, Martinelli M, Ferlenghi I, . . . Rinaudo CD (2011). Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. FASEB J, 25(6), 1874–1886. doi:fj.10–174797 [pii] 10.1096/fj.10-174797 [DOI] [PubMed] [Google Scholar]
- Eikmanns BJ, Kleinertz E, Liebl W, & Sahm H. (1991). A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene, 102(1), 93–98. [DOI] [PubMed] [Google Scholar]
- Hirata R Jr, Pereira GA, Filardy AA, Gomes DL, Damasco PV, Rosa AC, . . . Mattos-Guaraldi AL (2008). Potential pathogenic role of aggregative-adhering Corynebacterium diphtheriae of different clonal groups in endocarditis. Braz J Med Biol Res, 41(11), 986–991. doi:S0100-879X2008001100007 [pii] [DOI] [PubMed] [Google Scholar]
- Luong TT, Tirgar R, Reardon-Robinson ME, Joachimiak A, Osipiuk J, & Ton-That H. (2018). Structural Basis of a Thiol-Disulfide Oxidoreductase in the Hedgehog-Forming Actinobacterium Corynebacterium matruchotii. J Bacteriol, 200(9). doi: 10.1128/JB.00783-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Das A, & Ton-That H. (2008). The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci U S A, 105(37), 14147–14152. doi: 10.1073/pnas.0806350105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, & Ton-That H. (2007). Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol, 64(1), 111–124. doi: 10.1111/j.1365-2958.2007.05630.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LO, Mattos-Guaraldi AL, & Andrade AF (2008). Novel lipoarabinomannan-like lipoglycan (CdiLAM) contributes to the adherence of Corynebacterium diphtheriae to epithelial cells. Arch Microbiol, 190(5), 521–530. doi: 10.1007/s00203-008-0398-y [DOI] [PubMed] [Google Scholar]
- Mülhardt C. (2007). Molecular Biology and Genomics: Elsevier Science & Technology. [Google Scholar]
- Murphy JR (1996). Corynebacterium diphtheriae. In Baron S(Ed.), Medical Microbiology. 4th edition (Vol. Chapter 32). Galveston (TX): University of Texas Medical Branch at Galveston. [PubMed] [Google Scholar]
- Nakayama T, Soma M, Rahmutula D, Ozawa Y, & Kanmatsuse K. (2001). Isolation of the 5’-flanking region of genes by thermal asymmetric interlaced polymerase chain reaction. Med Sci Monit, 7(3), 345–349. [PubMed] [Google Scholar]
- Oram DM, Avdalovic A, & Holmes RK (2002). Construction and characterization of transposon insertion mutations in Corynebacterium diphtheriae that affect expression of the diphtheria toxin repressor (DtxR). J Bacteriol, 184(20), 5723–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M, Woolston JE, Jacobson AD, Holmes RK, & Oram DM (2007). Bacteriophage-based vectors for site-specific insertion of DNA in the chromosome of Corynebacteria. Gene, 391(1–2), 53–62. doi:S0378–1119(06)00749–9 [pii] 10.1016/j.gene.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott L, Holler M, Gerlach RG, Hensel M, Rheinlaender J, Schaffer TE, & Burkovski A. (2010). Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells. BMC Microbiol, 10, 2. doi:1471–2180-10–2 [pii] 10.1186/1471-2180-10-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez NA, Das A, & Ton-That H. (2020). New Paradigms of Pilus Assembly Mechanisms in Gram-Positive Actinobacteria. Trends Microbiol, (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon-Robinson ME, Osipiuk J, Jooya N, Chang C, Joachimiak A, Das A, & Ton-That H. (2015). A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol, 98(6), 1037–1050. doi: 10.1111/mmi.13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon-Robinson ME, & Ton-That H. (2016). Disulfide-Bond-Forming Pathways in Gram-Positive Bacteria. J Bacteriol, 198(5), 746–754. doi: 10.1128/JB.00769-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Das A, & Ton-That H. (2011). Adhesion by pathogenic corynebacteria. Adv Exp Med Biol, 715, 91–103. doi: 10.1007/978-94-007-0940-9_6 [DOI] [PubMed] [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, & Puhler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene, 145(1), 69–73. [DOI] [PubMed] [Google Scholar]
- Siegel SD, Liu J, & Ton-That H. (2016). Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol, 34, 31–37. doi: 10.1016/j.mib.2016.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T. BE (2003). High-Throughput TAIL-PCR as a Tool to Identify DNA Flanking Insertions (Grotewold E Ed. Vol. 236): Humana Press. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Okai N, Nonaka H, Tsuge Y, Inui M, & Yukawa H. (2006). High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl Environ Microbiol, 72(5), 3750–3755. doi:72/5/3750 [pii] 10.1128/AEM.72.5.3750-3755.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Schiering N, Zeng HY, Ringe D, & Murphy JR (1994). Iron, DtxR, and the regulation of diphtheria toxin expression. Mol Microbiol, 14(2), 191–197. [DOI] [PubMed] [Google Scholar]
- Ton-That H, & Schneewind O. (2003). Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol, 50(4), 1429–1438. doi: 10.1016/j.tim.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Wang C, Hayes B, Vestling MM, & Takayama K. (2006). Transposome mutagenesis of an integral membrane transporter in Corynebacterium matruchotii. Biochem Biophys Res Commun, 340(3), 953–960. doi:S0006–291X(05)02847–0 [pii] 10.1016/j.bbrc.2005.12.097 [DOI] [PubMed] [Google Scholar]