Abstract
The Ichneumonoidea (Ichneumonidae and Braconidae) is an incredibly diverse superfamily of parasitoid wasps that includes species that produce virus-like entities in their reproductive tracts to promote successful parasitism of host insects. Research on these entities has traditionally focused upon two viral genera Bracovirus (in Braconidae) and Ichnovirus (in Ichneumonidae). These viruses are produced using genes known collectively as endogenous viral elements (EVEs) that represent historical, now heritable viral integration events in wasp genomes. Here, new genome sequence assemblies for 11 species and 6 publicly available genomes from the Ichneumonoidea were screened with the goal of identifying novel EVEs and characterizing the breadth of species in lineages with known EVEs. Exhaustive similarity searches combined with the identification of ancient core genes revealed sequences from both known and novel EVEs. One species harbored a novel, independently derived EVE related to a divergent large double-stranded DNA (dsDNA) virus that manipulates behavior in other hymenopteran species. Although bracovirus or ichnovirus EVEs were identified as expected in three species, the absence of ichnoviruses in several species suggests that they are independently derived and present in two younger, less widespread lineages than previously thought. Overall, this study presents a novel bioinformatic approach for EVE discovery in genomes and shows that three divergent virus families (nudiviruses, the ancestors of ichnoviruses, and Leptopilina boulardi Filamentous Virus-like viruses) are recurrently acquired as EVEs in parasitoid wasps. Virus acquisition in the parasitoid wasps is a common process that has occurred in many more than two lineages from a diverse range of arthropod-infecting dsDNA viruses.
Keywords: Ichneumonoidea, virus, endogenous virus element (EVE), polydnaviruses (PDVs), parasitoid wasp genomes
Significance
Parasitoid wasps are an extremely diverse group of animals that are known to harbor Endogenous Virus Elements (EVEs) that produce virions or virus-like particles of key importance in wasps’ parasitism success. However, the prevalence and diversity of independently acquired EVEs in parasitoid wasp lineages have remained largely uncharacterized on a widespread scale. This study represents an important first step and hints at the untapped diversity of EVEs in parasitoid wasps via the identification of several virus endogenization events from diverse groups of double-stranded DNA viruses.
Introduction
Although viruses have long been viewed as pathogenic organisms, there is now ample evidence that viruses can confer important benefits to their hosts and have played a major role in the evolution of life on earth (Rossignol et al. 1985; Schmitt and Breinig 2002; Malmstrom et al. 2005; Moran et al. 2005; Brown et al. 2006; Dunlap et al. 2006; Barton et al. 2007; Ryabov et al. 2009; Villarreal and Witzany 2010; Goic and Saleh 2012; Strand and Burke 2020). Further, in most sequenced eukaryotic genomes, there is evidence of viral gene “footprints” (Feschotte and Gilbert 2012), suggesting viral endogenization is common and could play an important role in genome and organismal evolution. Although viral endogenization refers to the integration and vertical transmission of virus-derived genetic material into the germline of a host organism, viral domestication is a more specialized example of endogenization. Virus domestication is a phenomenon in which endogenization immobilizes virus replication genes to be retained in wasp genomes over time, and often confers a new function that benefits the eukaryotic organism (Burke and Strand 2012; Pichon et al. 2015; Gauthier et al. 2018). Understanding how beneficial viral associations evolve is essential for a holistic view on the evolution of eukaryotic organisms (Roossinck 2011; Villarreal 2015).
Perhaps some of the most complex examples of viral domestication occur in parasitic wasps belonging to the exceptionally diverse superfamily Ichneumonoidea (Braconidae + Ichneumonidae) with more than 44,000 described species (Yu et al. 2012). As parasitoids, these wasps lay their eggs in or on other insect “hosts,” where their progeny feed and complete the immature stages of development, resulting in the death of the host. Conservative estimates suggest that one in ten animals is a parasitoid wasp (Askew 1971) and more recent estimates exceed these already astonishing numbers (Jones et al. 2009; Rodriguez et al. 2013; Forbes et al. 2018). Among parasitoid wasps, Ichneumonoidea are particularly diverse, comprising ∼28% of all Hymenoptera, ∼3% of all terrestrial, multicellular life (Chapman 2009; Yu et al. 2012), and parasitizing a broad range of insects and other arthropods. To facilitate host invasion, ichneumonoid wasps are known to employ viral associations in the form of endogenous viral elements (EVEs), in which elements of viral genomes become permanently integrated into the genomes of wasps (Bézier et al. 2009; Volkoff et al. 2010; Béliveau et al. 2015; Pichon et al. 2015; Burke, Simmonds, et al. 2018). Multiple gene products encoded by EVEs produce virions or virus-like particles (VLPs) in ovaries of these wasps, which are injected into hosts during parasitism. Based upon experimental studies in a number of representative species, it is thought that these viruses function in the promotion of successful parasitism (Salt 1965; Rotheram and Salt 1973; Edson et al. 1981; Beckage et al. 1994) and may contribute to the immense diversity of these parasitic wasps. Viruses or VLPs function in the delivery of virulence molecules such as DNA or proteins that suppress host immune defenses or alter host development and behavior in ways that facilitate survival, growth, and development of the larval parasitoid (Reineke et al. 2006; Lee et al. 2009; Strand 2012; Darboux et al. 2019). There are too few genetically characterized large double-stranded DNA (dsDNA) viruses to be able to identify the immediate descendants of viral ancestors of viruses or VLPs produced by wasps; however, phylogenetic analyses show that most are related to viruses that play a pathogenic role (Stasiak et al. 2005; Bézier et al. 2009; Pichon et al. 2015; Burke, Simmonds, et al. 2018).
Polydnaviruses (PDVs, belonging to the family Polydnaviridae, Strand and Drezen 2012) are EVEs documented to occur within select but diverse clades within Ichneumonoidea. In Braconidae, they are present in the microgastroid complex (sensuSharanowski et al. 2011) comprising at least 6 subfamilies with more than 3900 described species. In Ichneumonidae, PDVs have been discovered in two families: Campopleginae and Banchinae, with more than 2,200 and 1,750 described species, respectively (Yu et al. 2012). Most large dsDNA viruses have genes that can be divided into two categories: 1) replication genes, which encode essential replication machinery and are conserved among genomes and 2) virulence genes of diverse origins whose products interact with host defenses and are gained and lost much more rapidly (Yutin et al. 2009; Rohrmann 2011; Kawato et al. 2018). PDVs have two components that are dispersed within the genomes of wasps: replication genes and proviral segments (the regions of the genome that are packaged into virions and contain virulence genes). The replication machinery for these viruses is not packaged into virions, making PDVs replication-defective and thus reliant on the wasp for replication (Bézier et al. 2009; Volkoff et al. 2010; Bézier et al. 2013; Burke et al. 2014; Strand and Burke 2014). The inheritance of permanently integrated PDVs is predicted to produce genetically unique, but related, EVEs in each wasp species within the three major clades of PDV-carrying wasps (Whitfield and Asgari 2003).
Despite such strong functional similarity, evidence shows that known PDVs have at least two unique origins. The family Polydnaviridae is divided into two genera: Bracovirus and Ichnovirus (Strand and Drezen 2012). The morphology of the PDVs found in Ichneumonidae (ichnoviruses) and Braconidae (bracoviruses) are vastly different (Stoltz and Whitfield 1992), and genomic sequences of the replication machinery show affinity to different classes of viruses (insect beta-nudiviruses and related baculoviruses for bracoviruses; relatives of nucleocytoplasmic large DNA viruses [NCLDVs] for ichnoviruses) (Bézier et al. 2009; Volkoff et al. 2010; Béliveau et al. 2015). In the Braconidae, bracoviruses have been localized histologically in species across the monophyletic microgastroid complex (Whitfield 1997), suggesting a single PDV origin in this clade ∼100 Ma (Murphy et al. 2008).
In addition to these PDVs, several types of EVEs have been recently discovered within Ichneumonoidea. For example, in the ichneumonid Venturia canescens (Campopleginae), viral replication genes were co-opted from the insect alpha-nudiviruses (related but distinct from the PDV progenitors, the beta-nudiviruses) and used to produce VLPs in wasp ovaries (Pichon et al. 2015). The V. canescens genome lacks the genes required to make a capsid to house viral DNA in virions, preventing delivery of virulence genes but allowing delivery of wasp-derived virulence proteins into hosts within VLPs (Feddersen et al. 1986; Pichon et al. 2015). Through wasp genome sequencing, an independent acquisition of viral genes from the alpha-nudiviruses was recently identified in Fopius arisanus (Braconidae), the first recognized incidence of viral genome integration in the subfamily Opiinae (Burke, Simmonds, et al. 2018). Other reports of reproductive gland-associated viruses in the Ichneumonoidea have been published (>35), but are limited to the identification of virions in wasp tissues and do not include any genetic analyses (Lawrence 2005; Suzuki and Tanaka 2006). Two very recent studies have documented the presence of EVEs outside of the Ichneumonoidea in parasitoid species belonging to the Figitidae and Chalcididae (Di Giovanni et al. 2020; Zhang et al. 2020). Recently discovered EVEs have not been assigned to Polydnaviridae because thus far, no VLPs produced package DNAs and because the family is polyphyletic and likely to be revised in the future. These data suggest that viral co-option events may be more common in parasitoids than previously thought.
Thus far, genetic discovery of these EVEs has been sporadic. Comprehensive exploration of the number of integration events and the rules governing their acquisition and function requires improvements in genomic pipelines for viral identification. In this study, we seek to better understand the diversity of viral origins in ichneumonoid wasps through developing a comparative genomics approach for EVE discovery. Using new genome sequence data sets in combination with publicly available genome assemblies from species belonging to Ichneumonoidea, this research has three objectives: 1) to develop a method for the identification of endogenous virus elements derived from diverse large dsDNA viruses; 2) to examine the breadth of PDV incidence in lineages known to produce bracoviruses and ichnoviruses; and 3) to identify novel EVEs likely to produce virions or VLPs in species in which they have not been described previously. Screening of 17 genomes from parasitoids within the Ichneumonoidea (including 11 new genome assemblies) identified both familiar and novel EVEs in a number of species. These results challenge existing assumptions about the species distribution and origins of ichnoviruses and highlight the discovery of a new family of dsDNA viruses that are common EVE progenitors.
Results
Sequencing and Assembly Generated 11 New Draft Genome Sequences for Parasitoid Wasps
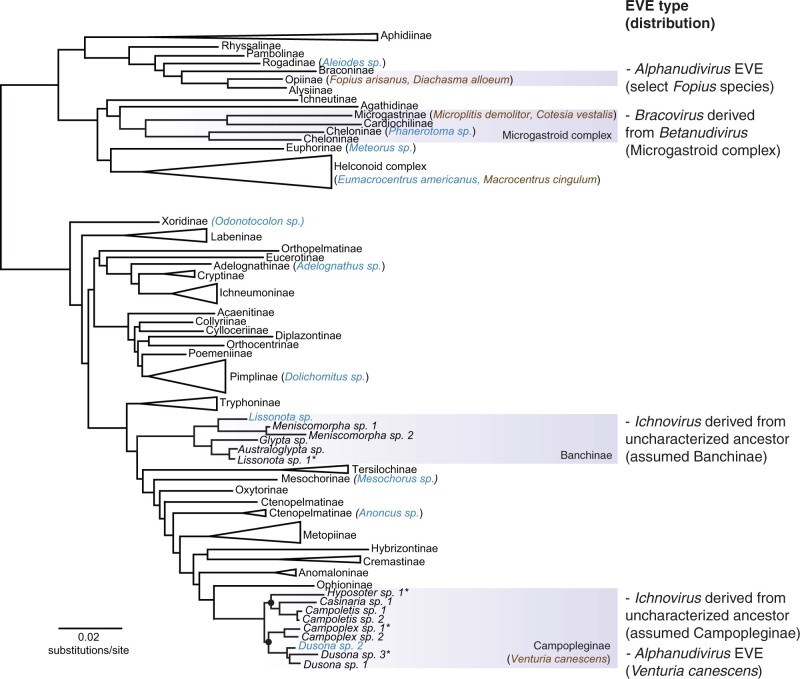
To accomplish the objective of identifying known and novel EVEs in parasitoid genomes, it was necessary to screen new genomic data for a range of wasp species that had both expected and unknown presence of EVEs (table 1 and fig. 1). In the literature, it is taken for granted that EVEs are present within entire clades of parasitoid species (e.g., the microgastroid lineage of Braconidae or the Campopleginae or Banchinae subfamilies of the Ichneumonidae), which sets up expectations for the identification of EVEs in particular species with newly generated genome assemblies (table 1, fig. 1). For newly sequenced taxa, it was expected that bracovirus genes would be found within Phanerotoma sp. (subfamily Cheloninae in the microgastroid lineage) and ichnovirus genes (e.g., IVSPER genes) within Lissonota sp. (Banchinae) and Dusona sp. 2 (Campopleginae, hereafter Dusona sp.). It was unknown whether the remaining eight species would have EVEs in their genomes.
Table 1.
Taxa Used for Genome Analysis
| Assembly Accession | Family | Higher Group | Subfamily | Genus Species | Expected or Known Viral Association |
|---|---|---|---|---|---|
| Braconidae | Microgastroid | Cheloninae | Phanerotoma sp. | Bracovirus | |
| Braconidae | Euphoroid | Euphorinae | Meterous sp. | Unknown | |
| Braconidae | Helconoid | Helconinae | Eumacrocentrus americanus | Unknown | |
| Braconidae | Cyclostomes s.s. | Rogadinae | Aleiodes sp. | Unknown | |
| Ichneumonidae | Ichneumoniformes | Adelognathinae | Adelognathus sp. | Unknown | |
| Ichneumonidae | Ophioniformes | Banchinae | Lissonota sp. | Ichnovirus | |
| Ichneumonidae | Ophioniformes | Campopleginae | Dusona sp. | Ichnovirus | |
| Ichneumonidae | Ophioniformes | Ctenopelmatinae | Anoncus sp. | Unknown | |
| Ichneumonidae | Ophioniformes | Mesochorinae | Mesochorus sp. | Unknown | |
| Ichneumonidae | Pimpliformes | Pimplinae | Dolichomitus sp. | Unknown | |
| Ichneumonidae | Xoridiformes | Xoridinae | Odontocolon sp. | Unknown | |
| Previously published accessions | |||||
| GCA_000956155.1 | Braconidae | Microgastroid | Microgastrinae |
Cotesia vestalis Andong |
Bracovirus |
| N/Aa | Braconidae | Microgastroid | Microgastrinae |
Cotesia vestalis Hangzhou |
Bracovirus |
| GCA_001412515.3 | Braconidae | Alysioid | Opiinae | Diachasma alloeum | None discovered |
| GCA_000806365.1 | Braconidae | Alysioid | Opiinae | Fopius arisanus |
Endogenous nudivirus |
| GCA_002156465.1 | Braconidae | Macrocentroid | Macrocentrinae |
Macrocentrus cingulum |
None discovered |
| GCA_000572035.2 | Braconidae | Microgastroid | Microgastrinae |
Microplitis demolitor |
Bracovirus |
| N/Ab | Ichneumonidae | Ophioniformes | Campopleginae | Venturia canescens |
Endogenous nudivirus |
Note.—Higher Group placement is taken from Sharanowski et al. (2011) and Sharanowski et al. (2021). Taxa not identified to species due to difficulty in accurate identification are listed as sp.
Genome available at http://www.insect-genome.com/waspbase/download/genome_message.php?species=Cotesia%20vestalis, last accessed May 2021.
Genome available at: http://bipaa.genouest.org/sp/venturia_canescens, last accessed May 2021.
Fig. 1.
Maximum likelihood tree of Ichneumonoidea from Sharanowski et al. (2021). Subfamilies with multiple representatives (except the microgastroid complex, Campopleginae, and Banchinae) have been collapsed for viewing subfamily relationships, the presence and distribution of EVEs, and representative species analyzed in this study. Subfamilies with previously published, genetically characterized EVEs are shaded in boxes, and to the right, independent origins of EVEs, the type of virus ancestor they are derived from, and their known or assumed distribution is indicated. Two nodes within the Campopleginae are highlighted showing that the subfamily is divided into two major clades. Species analyzed in this study are placed next to the subfamily to which they belong and are colored blue (new genome assemblies) and brown (previously published genome assemblies). Taxa marked with an asterisk after the name have uncertain species identification. Support for the tree is robust and can be viewed in detail in Sharanowski et al. (2021).
Illumina read counts from the 11 newly added genomes ranged from 58.9 to 88.7 million per library (supplementary table 2, Supplementary Material online). Given that the sequence data were generated from short insert libraries only, the assemblies yielded many contigs (ranging from 13,843 to 423,888) with relatively low N50 values (1,110 to 31,843 nucleotides) compared with the current standard for complete insect genome sequences that employ scaffolding with large insert sequencing libraries or long-sequence read technologies (supplementary table 2, Supplementary Material online). The cumulative sizes of scaffolds (a proxy for genome size) were similar for new and previously published high-quality assemblies. Benchmark Universal Single-Copy Ortholog (BUSCO) analysis revealed variation in the level of completeness of each new genome assembly, ranging from detection of 98.7% complete BUSCOs in Meteorus to detection of 26.8% complete and 46.4% fragmented BUSCOs (total 73.2%) in Anoncus (supplementary table 3, Supplementary Material online). Although some genome assemblies were relatively fragmented, overall the amount of sequence data was likely enough to cover a majority of the genomes of these wasps.
Identification of Genes of Viral Origin Using Exhaustive Sequence Similarity Searches Is Fraught with False Positives
To identify genes responsible for making virions or VLPs in wasp genomes, a method to identify endogenous virus elements (EVEs) derived from diverse large dsDNA viruses was needed. Although actively replicating viruses and lateral transfers of single genes from viruses into eukaryotic hosts can generate novel phenotypes of key importance (Dunlap et al. 2006; Aswad and Katzourakis 2012; Lavialle et al. 2013; Parker and Brisson 2019; Coffman et al. 2020), the focus here is upon identification of sets of viral replication genes that could be responsible for the production of virions or VLPs that are permanent key components of wasp biology and parasitism success. Homology searches of all Open Reading Frames (ORFs) extracted from genome assembly scaffolds against a custom database produced viral hits for 37 to 75 ORFs for species with previously published genomes, and 10 to 53 ORFs for new species (supplementary figs. 1 and 2 and table 4, Supplementary Material online). Hits were to viruses belonging to several major groups: dsDNA viruses, ssDNA viruses, ssRNA viruses, dsRNA viruses, Ortervirales, and unclassified viruses. As all genetically characterized examples of virions or VLPs produced by parasitoid wasps thus far originate from large dsDNA viruses, hits to other types of viruses were not analyzed further in this study. Most hits were to the dsDNA viruses, particularly to the families Nudiviridae, Baculoviridae, Poxviridae, Ascoviridae, Hytrosaviridae, Iridoviridae, and the Caudovirales. However, manual inspection of the annotations associated with these hits revealed that many of these proteins are retroviral proteins, inhibitor of apoptosis proteins, chitosanases, and transposases (supplementary table 4, Supplementary Material online). These genes do not encode the necessary components for building virions or VLPs and are frequently transferred between viruses and eukaryotes. Therefore, many or most of these genes have obscure origins and are unlikely to play a role in producing virions or VLPs that contribute to wasp parasitism success.
Targeted Searches for Ancient Core Genes Can Accurately Identify EVEs Likely to Produce Virions or VLPs in Wasp Genomes
The preceding results indicated that a different approach was necessary to identify sets of virus-derived genes that are likely to be involved in producing virions or VLPs. dsDNA viruses that infect insects can be categorized into several virus families belonging to at least two major groups, the monophyletic NCLDVs and Nuclear Arthropod-specific Large DNA Viruses (NALDVs) (Iyer et al. 2001; Wang et al. 2012). Although viruses generally use a very diverse set of strategies for replication, there exist at least six genes encoding replication components that are common to NALDVs and NCLDVs (Wang et al. 2012). These genes are present in most extant members of major families of viruses in these groups and are referred to as ancient core genes (although they may not share a common origin, Iyer et al. 2006; Wang et al. 2012).
To produce the key components necessary for the completion of virus replication and construction of virions or VLPs, it was rationalized that wasp genomes would need to contain at least some of the ancient core genes. To identify the presence or absence of ancient core genes, each set of ORFs with viral hits generated by the exhaustive method above was searched with methods that can detect homologs with very low sequence similarity by focusing upon patterns of conserved sites among protein sequences. Although genes encoding ichnovirus replication machinery may be related to NCLDV core genes, the protein sequences from IVSPERs were too divergent to be incorporated into our search strategies. Thus, searches for ancient core genes are expected to identify EVEs derived from NCLDVs and NALDVs, but not the ancestors of ichnoviruses.
PSI-BLAST and HMM searches among putative viral protein sequences from each wasp species identified a total of 11 homologs of ancient core genes in new genome assemblies and 22 in previously published genome assemblies (table 2, supplementary table 5, Supplementary Material online). Previously published genome sequences known to contain EVEs served as positive controls for the identification of ancient core genes using the method described here that relies upon limited homology from deep divergence events. Previous work involving manual annotation of EVEs in the Microplitisdemolitor, V. canescens, and F. arisanus genomes indicated that all contain five out of six of the ancient core genes (a viral DNA polymerase gene is often lost in wasp species with EVEs, Burke 2019). Homology searches identified genes encoding helicase, lef-8, lef-9, and p33 proteins in the set of putative viral proteins for each positive control species. However, lef-5 was identified in F. arisanus only and could not be identified in the two other species even when the full set of ORFs from each species was searched. This may be due to limited sequence similarity and the short sequence length of lef-5 genes, reducing the success of PSI-BLAST and HMMR searches targeting very diverse taxa. Although Cotesiavestalis females are known to produce bracovirus in their ovaries, no publication has yet provided a detailed annotation of EVEs in the C. vestalis genome. ORFs with homology to viral helicase, lef-8, and lef-9 genes were identified, as well as a viral DNA polymerase (DNApol), warranting further exploration of genes of viral origin in this species (see below). Two previously published wasp genomes that are not known to contain EVEs (Diachasmaalloeum and Macrocentruscingulum) were negative for all ancient core genes. These results indicate that PSI-BLAST and HMMR can reliably detect the presence or absence of viral ancient core genes in wasp genomes.
Table 2.
Matches to Ancient Core Genes Identified in Sequenced Wasp Genomes
| Species | DNA Polymerase B | Helicase/Helicase III(D5R-Like Helicase) | Lef-8/Rbp2 | Lef-9/Rbp1 | Lef-5/TFIIS-LikeTranscription Factor | P33/Sulfhydryl Oxidase | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PSI | HMM | PSI | HMM | PSI | HMM | PSI | HMM | PSI | HMM | PSI | HMM | |
| Adelognathus sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aleiodes sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anoncus sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dolichomitus sp. | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| Dusona sp. | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Eumacrocentrus americanus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lissonota sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mesochorus sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Meterous sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Odontocolon sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Phanerotoma sp. | 0 | 0 | 0 | 0 | 2a | 2a | 1 | 1 | 0 | 0 | 1 | 1 |
| Cotesia vestalis | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 |
| Diachasma alloeum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fopius arisanus | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 2b | 1 | 1 | 1 | 1 |
| Macrocentrus cingulum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microplitis demolitor | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| Venturia canescens | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 2b | 0 | 0 | 1 | 1 |
a Likely one gene fragmented across two contigs.
ORFs with hits are located next to each other on scaffolds and are fragmented due to pseudogenization or intron presence.
In new wasp genomes, identification of at least one ancient core gene provided enough evidence to justify further exploration, whereas the absence of any hits led to the conclusion that specific wasp species are unlikely to produce virions or VLPs and exclude these species from further analysis. Proteins encoded by ancient core genes were identified in Dolichomitus (DNApol, helicase, lef-8, lef-9), Dusona (lef-8, lef-9), and Phanerotoma (lef-8, lef-9, p33).
Manual examination of protein alignments for each putative ancient core gene identified in wasp species (aligned with homologs from representative large DNA viruses and EVEs) confirmed protein identity and revealed that almost all hits were full length. Exceptions included one of the two DNApol genes from C. vestalis Andong and one of the two lef-8 genes from Dolichomitus. lef-8 sequences from Phanerotoma had two ORFs matching this protein (N-terminal and C-terminal fragments), which lay on two different contigs, likely indicating broken assembly. The matches from Dusona were ORFs that were truncated compared with full-length lef-8 and lef-9 sequences and came from short scaffolds with low coverage, so were excluded from further analysis as likely contaminant sequences.
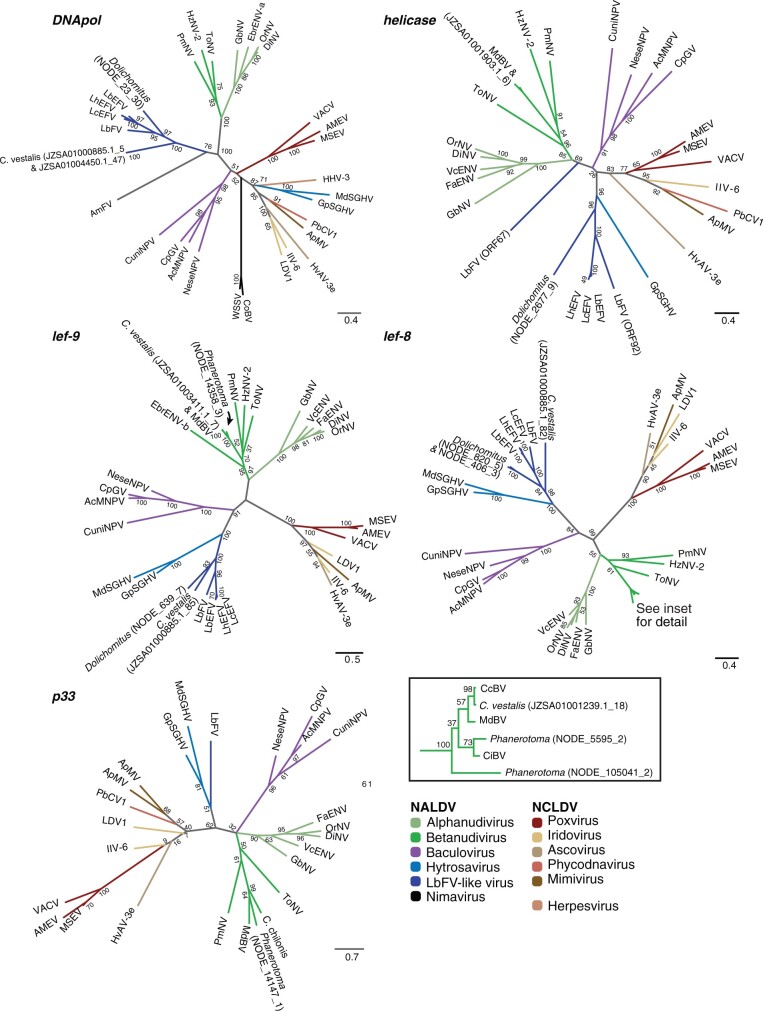
Phylogenetic reconstruction of ancient core gene trees revealed that genes of viral origin in each new wasp genome assembly came from a variety of sources (fig. 2 and supplementary table 6, Supplementary Material online). As expected, Phanerotoma lef-8, lef-9, and p33 sequences grouped with the bracoviruses. Dolichomitus DNApol, helicase, lef-8, and lef-9 sequences all grouped with protein sequences from Leptopilina boulardi Filamentous Virus (LbFV), a large dsDNA virus that is distantly related to hytrosaviruses and may represent a distinct virus family (Lepetit et al. 2017). The C. vestalis Andong genome contains at least two ORFs of viral origin for lef-8 and lef-9 each. Cotesiavestalis sequences fell into two groups: sequences clustered with bracoviruses (helicase, lef-8, and lef-9) or with LbFV (DNApol, lef-8, and lef-9). These data indicate that the sequenced C. vestalis Andong samples contain two separate sources of viral genes; bracoviruses and an LbFV-like entity.
Fig. 2.
Genes of viral origin in wasp genomes are related to genes in large dsDNA viruses. Maximum likelihood phylogenies constructed from protein sequences of individual genes are shown, with numbers on nodes indicating support from 100 bootstrap replicates. Colored branches indicate virus families or groups. The sequences used for alignment were obtained from Microplitis demoltior bracovirus (MdBV), Cotesia congregata bracovirus (CcBV), Chelonus insularis bracovirus (CiBV), a C. chilonis transcriptome, Eurytoma brunniventris endogenous nudivirus alpha (EbENV-a), Eurytoma brunniventris endogenous nudivirus beta (EbENV-b), Tipula oleracea nudivirus (ToNV), Heliothis zea nudivirus 2 (HzNV-2), Penaeus monodon nudivirus (PmNV), Venturia canescens endogenous nudivirus (VcENV), Drosophila innubila nudivirus (DiNV), Oryctes rhinoceros nudivirus (OrNV), Fopius arisanus endogenous nudivirus (FaENV), Gryllus bimaculatus nudivirus (GbNV), Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Cydia pomonella granulovirus (CpGV), Neodiprion sertifer nucleopolyhedrovirus (NeseMNPV), Culex nigripalpus nucleopolyhedrosis virus (CuniNPV), Glossina pallidipes salivary gland hytrosavirus (GpSGHV), and Musca domestica salivary gland hytrosavirus (MdSGHV), L. boulardi filamentous virus (LbFV), L. boulardi endogenous filamentous virus (LbEFV), Leptopilina heterotoma endogenous filamentous virus (LhEFV), Leptopilina clavipes endogenous filamentous virus (LcEFV), A. mellifera filamentous virus (AmFV), White spot syndrome virus (WSSV), Chionoecetes opilio bacilliform virus (CoBV), Amsacta moorei entomopoxvirus (AMEV), Melanoplus sanguinipes entomopoxvirus (MSEV), Vaccinia virus (VACV), Invertebrate iridescent virus 6 (IIV-6), LDV1, Trichoplusia ni ascovirus 3e (TnAV-3e), Paramecium bursaria chlorella virus 1 (PbCV1), Acanthamoeba polyphaga mimivirus (ApMV), Human herpesvirus 3 (HHV-3), supplementary table S5, Supplementary Material online. Sequences from the IVSPERs of the ichnovirus in Glypta fumiferanae and nimavirus p33 homologs were not included because they were too divergent compared with the other included sequences. Scale bars indicate substitutions per amino acid residue. For ORFs found in wasp genomes, the accession number or scaffold name is followed by an underscore and the ORF number.
Bracovirus Nudivirus-Like Replication Genes Were Detected in Species in the Microgastroid Complex
As described above, bracoviruses were expected in the previously unannotated C. vestalis genome assembly and the new Phanerotoma genome assembly. These findings provide two proof-of-concept data sets to validate the approach for the identification of nudivirus-like replication genes beyond just the ancient core genes. Although genes from proviral segments are undoubtedly present in the data sets, their annotation was not attempted due to the fragmented nature of the genome assemblies and the lack of conserved genes that could be used as a proviral segment diagnostic. The first strategy used for the identification of nudivirus-like replication genes involved alignment of ORFs from each genome assembly to the custom diamond database. The second strategy was designed to identify an expanded set of nudivirus-like EVEs in each genome assembly, and involved adding nudivirus-like replication genes from M. demolitor, Cotesiacongregata, and Chelonusinanitus to the custom diamond database and repeating the diamond search. The existing nudivirus-like replication gene models from bracoviruses are the products of careful manual annotation of sequence data. These manually curated sequences are more likely to generate database search hits compared with searches against the set of insect virus genomes available in public databases because they are less divergent than other nudiviruses; these genes are the products of a single integration event and share a common ancestor ∼100 Ma (Murphy et al. 2008).
Using the first search strategy, 18 ORFs similar to nudivirus and baculovirus genes were identified on 18 contigs in the Phanerotoma assembly ranging in size from 885 to 12,791 bp. The GC content of these contigs was not significantly different from BUSCO-containing contigs, but on average their coverage was slightly higher (supplementary fig. 2, Supplementary Material online, 28x compared with 9x, P < 0.05). The second strategy for identification of nudivirus-derived genes found 13 of the original 18 ORFs plus 37 additional genes (55 total, supplementary table 7, Supplementary Material online). A search of all ORFs in the C. vestalis Andong genome against the custom diamond database revealed 29 ORFs matching to nudivirus or baculovirus coding sequences located on 24 contigs. The GC content of contigs containing nudivirus-like genes was not significantly different than BUSCO-containing contigs (mean 29.7% compared with 29.2%, respectively, P = 0.30). It was not possible to create scaffold coverage and GC content plots for the C. vestalis population from Andong, South Korea, because the sequencing reads used to make the genome assembly were not publicly available. A broader search using the second strategy as described above identified 13 of the original 29 ORFs, plus an additional 78 hits (106 total, supplementary table 7, Supplementary Material online).
To assess the completeness of the set of bracovirus replication genes identified in each assembly, a tally of the genes conserved in bracoviruses and also nudiviruses that are presumed to be essential for virion production was conducted. The M. demolitor genome contains at least one copy of 19 of the 33 nudivirus core genes (18 of these are also core genes in baculoviruses, Burke 2019). All of the 19 genes conserved in nudiviruses and M. demolitor were present in the set of nudivirus-like genes identified in Phanerotoma, and only one gene, pif-4, was missing in the C. vestalis Andong assembly. These results indicate that the methods used were appropriate for the identification of the presence of EVEs that are likely to produce bracoviruses. However, the genome assemblies were sufficiently fragmented that there was not enough information to confidently assess the placement of genes or the presence of synteny between nudivirus-like genes in microgastroid species. The fragmented nature of the new genome assemblies and the coarse nature of the methods used made it possible that a complete list of EVE genes in the Phanerotoma and C. vestalis Andong genomes was not fully identified. A more detailed and comprehensive analysis will be appropriate if and when assemblies with larger scaffold sizes are available for these species.
Ichnovirus Structural Protein Encoding Regions (IVSPERs) Were Detectable but Not Always Present Where Expected
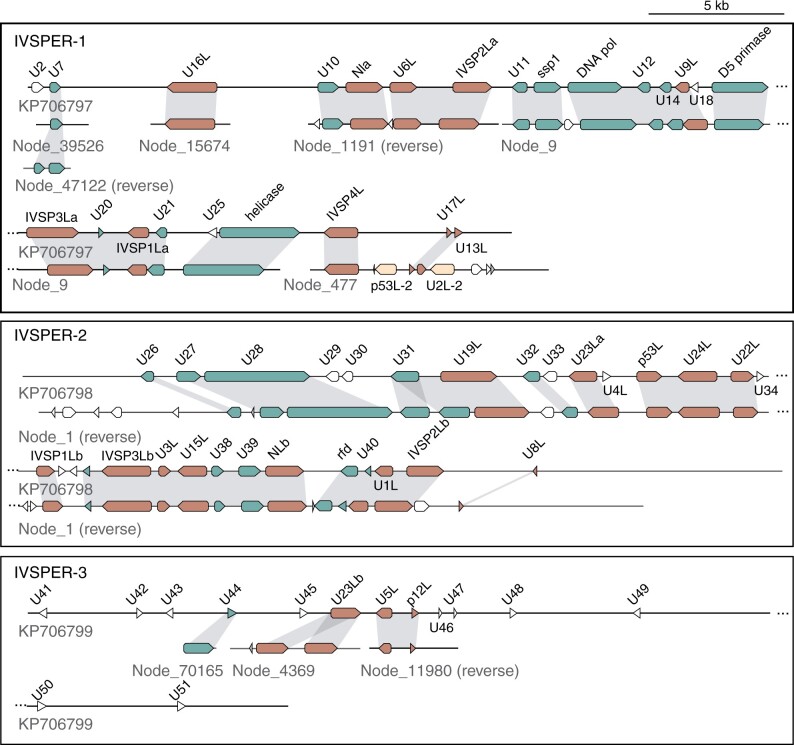
Given that no ancient core genes have been documented as consistently present in ichnovirus-producing wasp genomes to date, the strategy used for the identification of ichnovirus structural protein encoding region (IVSPER) genes in wasp genome assemblies relied upon homology to the IVSPERs from Hyposoterdidymator and Glyptafumiferanae. IVSPER genes were identified in the Lissonota genome assembly associated with ten scaffolds ranging in size from 1,227 to 44,485 bp (supplementary fig. 2, Supplementary Material online). The protein sequence percentage identity to G. fumiferanae IVSPER genes from diamond searches was 28–86%, with an average of 57%. Like the scaffolds containing EVEs in F. arisanus and M. demolitor, in Lissonota the IVSPERs were located on scaffolds that did not significantly differ in coverage from scaffolds that contain BUSCO genes (supplementary fig. 2, Supplementary Material online). This indicates that as in other IV-producing wasps, the IVSPER genes are integrated into the genome of Lissonota (Volkoff et al. 2010; Béliveau et al. 2015). Scaffolds containing IVSPERs had a slightly higher GC content (39.7%) compared with BUSCO scaffolds (37.8%, P < 0.05). Lissonota IVSPERs had high levels of synteny with IVSPERs previously identified in the banchine G. fumiferanae, indicating that the viral integration event occurred in a shared ancestor of these wasp species (fig. 3, NCBI accession JABMBS000000000). Twenty-four genes were shared between these two species and were also present in the IVSPERs in H. didymator, whereas another 25 were present in the two banchine species only. The four genes linking IVs to NCLDVs (ssp1, DNApol, D5 primase, and helicase) were present in Lissonota as well as G. fumiferanae, where they were discovered.
Fig. 3.
IVSPER genes identified in the Lissonota genome and synteny with Glypta fumiferanae. Glypta fumiferanae sequences are shown above with Lissonota sequences shown below. Homologous genes with synteny between the two species are indicated by gray shading. Some genes are present in banchine and campoplegine species (G. fumiferanae, Lissonota, and Hyposoter didymator; colored orange) whereas others are present only in the two banchine species presented here (blue). Genes colored beige are homologous to other IVSPER genes but are not detected in syntenous regions of the G. fumiferanae genome.
There were no significant hits to any ORFs from the remaining ten new genome assemblies or the previously published genome assemblies. The only exception was a match to a hypothetical protein (U44, AKD28091.1) in the V. canescens, M. cingulum, F. arisanus, and Lissonota ORFs. BLASTP of the G. fumiferanae U44 against the NCBI nr database with default parameters revealed that this protein has strong similarity to other hymenopteran proteins, making it unclear whether the ORFs identified are viral in origin, or merely wasp genes. The absence of hits to IVSPER genes in Dusona is notable because this species belongs to the Campopleginae, where all taxa were assumed to have ichnoviruses. The absence of IVSPERs and related genes in Mesochorus (Mesochorinae) and Anoncus (Ctenopelmatinae) is also notable as they share a common ancestor with the Campopleginae and Banchinae (Bennett et al. 2019; Sharanowski et al. 2021). The absence of IVSPERs in Dusona suggests that IVs might actually be limited to a subset of species within the campoplegine wasps.
Novel Filamentous Virus-Like Genes Were Identified in the C. vestalis and Dolichomitus Genomes
The identification of several ancient core genes related to LbFV genes in the C. vestalis Andong and Dolichomitus genomes provided a strong hint that these species contain EVEs or are “contaminated” with sequences from an actively replicating viral infection. When possible, a variety of characteristics were assessed to determine whether LbFV-like genes found in C. vestalis and Dolichomitus were likely to be endogenous or exogenous with respect to wasp chromosomes, including gene content and architecture, sequence read coverage and GC content, the presence of heterozygous alleles, and prevalence among individual wasps (see Materials and Methods). The presence of virus-derived genes in the C. vestalis and Dolichomitus genome assemblies motivated expansion of the search for LbFV-like genes beyond the ancient core genes initially identified to look for signatures of wasp genome integration. In order to effectively identify LbFV-like genes in wasp genomes, better annotation of genes in LbFV was attempted. Previous studies identified homologs for DNApol, helicase, lef-8, lef-9, pif-0 (p74), pif-2, pif-5, ac81, and odv-e66 in LbFV (Lepetit et al. 2017; Kawato et al. 2018; Di Giovanni et al. 2020). Using HMMs built with homologs from other insect-infecting large dsDNA viruses, it was possible to identify the following additional genes in LbFV: lef-4 (ORF107), 38 K (ORF19), and pif-1 (ORF32) (supplementary table 8, Supplementary Material online).
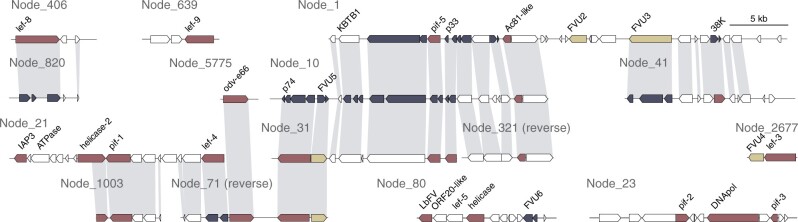
A total of 18 contigs in the Dolichomitus genome assembly (ranging in size from 312 bp to 39.5 kb) contained genes related to LbFV. These contigs were too short in length to determine whether any genes with eukaryotic architecture were flanking the virus-derived genes. The GC content of these contigs was significantly lower than BUSCO-containing contigs (mean 37.4% compared with 39.6%, P < 10−4), and on average their coverage was higher (28x compared with 7x, P < 10−9 on log10 transformed values, supplementary fig. 2, Supplementary Material online). After manual annotation of these contigs, diamond analysis revealed that 24 of the total 149 ORFs had similarity to LbFV genes, including DNApol, lef-3, lef-4, lef-5, lef-8, lef-9, pif-0, pif-1, pif-2, pif-3, pif-5, 38 K, helicase, helicase2, and ac81 (fig. 4, NCBI accession JAAXZA000000000). Although the cumulative size of the “viral” contigs was 199 kb, the nondegenerate size was approximately 114 kb because many contigs were repetitive, with up to three divergent copies of distinct genes present among the set (fig. 4). BLASTP local alignment identity for LbFV protein-coding sequences to translated Dolichomitus ORFs ranged from 22.9% to 42.7% (mean 31.2%, median 32%), whereas identity among paralogous LbFV-like Dolichomitus ORFs was 30.4–95.83% (average 62.8%, median 62.9%). Comparison of the Dolichomitus and C. vestalis genes (see below) with viral architecture flanking LbFV-like genes revealed homologs of six genes that are common to the two wasp genome assemblies but are not present in LbFV and were given the gene names Filamentous Virus Unknown 1 through 6 (fig. 4). The divergence of viral genes in Dolichomitus compared with LbFV often made it difficult to determine whether an ORF represents a full-length gene. However, homology between ORFs on different contigs in Dolichomitus revealed that some ORFs represent fragments of pseudogenized genes (fig. 4). Of all of the ORFs predicted on the 18 Dolichomitus contigs, 33 are fragments of a total of 14 pseudogenized genes (ranging from 1 to 6 per contig). There was no significant difference in coverage for contigs that do or do not contain pseudogenized genes (average 49x compared with 38x, respectively, P = 0.19). On four contigs, core genes that are considered essential for replication were inactivated: 38K on Node_1, p74 and pif-5 on Node_10, lef-4 on Node_71 and lef-8 on NODE_820. The presence of degenerated essential genes provides strong evidence that at least these four LbFV-like contigs are endogenous in the Dolichomitus genome. These contigs are not likely to be part of an archetypal virus genome actively replicating in Dolichomitus, because the products of these genes would be nonfunctional or absent and are essential for virion formation. The presence of several syntenous copies of viral genes could be the product of multiple integration or genome locus duplication events that have substantially diverged over time.
Fig. 4.
Regions of the Dolichomitus genome similar to the L. boulardi Filamentous Virus (LbFV). Homologous genes with synteny between contigs are indicated by gray shading. Genes with homology to LbFV are highlighted red, genes with homology to LbFV-like genes in Cotesia vestalis are shaded yellow, and other predicted genes are colored white. FVU = Filamentous Virus Unknown protein encoding gene. ORFs shown in deep purple represent pieces of pseudogenes that can be found intact on other contigs. Three short nodes with incomplete gene sequences were not included in this figure (NODE_51377 and NODE_267127 containing pif-0, and NODE_236835 containing lef-9).
Heterozygosity was also examined in contigs from the Dolichomitus genome assembly. The sequence data generated from Dolichomitus were generated from DNA isolated from a single individual diploid female. An exogenous haploid virus would be expected to be devoid of heterozygous sites, whereas heterozygous sites are expected in the genome of a single diploid female. Additional evidence for endogenization of the contigs containing viral genes would be the presence of heterozygous alleles in these contigs, indicative of their residing within wasp chromosomes. Sequences from a contaminating Wolbachia genome initially removed from the wasp genome sequence assembly were also scrutinized for heterozygous sites as a negative control. In BUSCO gene-containing contigs, 1.20 heterozygous alleles were detected per kilobase of sequence (present in 800 out of 1,254 contigs) compared with 4.51 in contigs (in 14 out of 18 contigs) containing filamentous virus-related genes (supplementary table 9, Supplementary Material online). Three of the four contigs containing filamentous virus-related genes that lacked heterozygous sites were the shortest of the contigs possessing these types of genes (those not shown in fig. 5), offering a possible explanation for the absence of heterozygous sites. There was no significant correlation between the number of heterozygous sites found and the corresponding sequence read coverage for a given contig (Pearson correlation, t16 = 1.9, P = 0.08), and many of the higher coverage contigs contained heterozygous sites. In contrast, close to zero heterozygous sites (a total of 5 sites found in 2 of 108 contigs, 0.00451 heterozygous alleles per kilobase) were detected in Wolbachia contigs.
Fig. 5.
Regions of the Cotesia vestalis Andong genome similar to the LbFV. Genes with homology to LbFV are highlighted red, pseudogenes as a deep purple. Genes with homology to LbFV-like genes in Dolichomitus are shaded yellow or purple if pseudogenized (FVU = Filamentous Virus Unknown protein encoding gene). Other predicted genes are shaded in gray. Genes with blue background shading have eukaryotic structure (introns and exons), and of these, genes with dark blue colored exons are “high confidence” given evidence from EST alignments.
For heterozygous single nucleotide polymorphisms (SNPs) located in the diploid wasp chromosomes, there should be just two haplotypes (combinations of nearby variable sites) among linked sites. Our analysis identified haplotype blocks only if they were inferred to be diploid. Although both BUSCO and filamentous virus gene-containing contigs had numerous diploid haplotype blocks of average length 99 and 283 bp, respectively, only one out of 108 Wolbachia contigs contained a single diploid haplotype block (two heterozygous sites separated by 42 bp, supplementary table 10, Supplementary Material online). Cumulatively, these results strongly indicate that all of the contigs containing filamentous virus-derived genes are part of the wasp genome and that Dolichomitus has an endogenous virus element. Given the conclusion that all of the LbFV-like contigs are endogenous in the Dolichomitus genome, it is noted that at least one copy of each LbFV-like gene with essential functions would be intact across the contigs cumulatively. Thus, it is theoretically possible that this Dolichomitus wasp produced VLPs by assembling protein products from intact LbFV-like genes that are dispersed throughout the Dolichomitus genome.
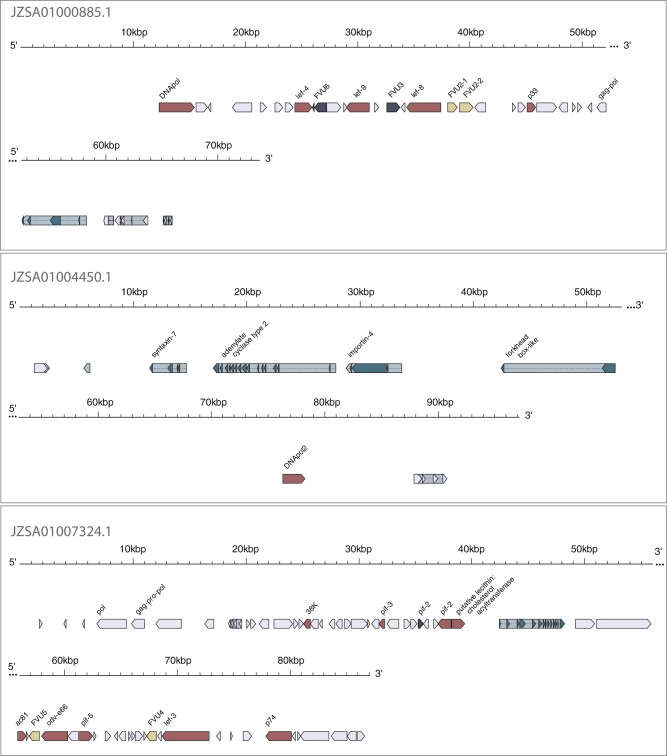
The genome of C. vestalis Andong was available on NCBI in an unannotated state because the assembly is somewhat fragmented. In addition to genes originating from bracoviruses, HMM and PSI-BLAST searches identified four additional ancient core genes in the C. vestalis Andong genome (two copies of DNApol; lef-8 and lef-9). The results from diamond and HMM searches revealed nine more LbFV-like genes in the C. vestalis Andong genome. Genes similar to LbFV genes were identified on three C. vestalis Andong genomic scaffolds ranging in size from 73.7 to 97.2 kb (fig. 5, accession doi:10.15482/USDA.ADC/1504545). DNApol, lef-4, lef-8, lef-9, and 25 other neighboring genes encoding hypothetical proteins with typical viral architecture (short, closely spaced, no introns) are located on the first scaffold (JZSA01000885.1) next to four eukaryotic genes encoding uncharacterized proteins: two with similarity to hymenopteran species and another with similarity to Helicoverpa armigera. The second scaffold (JZSA01004450.1) contains a partial (50% full length) copy of a viral DNApol gene situated among seven genes with eukaryotic architecture and with strong similarity to other hymenopteran genes, including syntaxin-7, adenylate cyclase type 2, importin-4, and forkhead box-like genes (fig. 5). The final scaffold (JZSA01007324.1) contains lef-3, 38K, p74, pif-2, pif-5, ac81, odv-e66, putative lecithin: cholesterol acyltransferase genes, and 52 other genes with viral architecture. Interspersed are two genes that have eukaryotic architecture with significant similarity to genes encoding uncharacterized proteins in other hymenopteran species (fig. 5). Alignment of sequence expression data provided evidence that 6 of the 13 eukaryotic genes are expressed and thus represent high-confidence annotations. The GC content of nudivirus-like and LbFV-like gene-containing contigs in this assembly was not significantly different from BUSCO-containing contigs (29.7%, 30.0%, and 29.2%, respectively, F2,995 = 0.4, P = 0.65, supplementary fig. 3, Supplementary Material online).
The second scaffold containing a partial copy of the viral DNApol gene may represent an integration event in which part of the viral DNApol gene was transferred to a different region of the wasp genome. This partial DNApol gene is identical in sequence to the full-length copy of the first scaffold, indicating that they diverged very recently, or that the partial gene is an assembly error. The location of viral genes on the first and third scaffolds could be indicative of two different scenarios. The first scenario is viral endogenization, in which LbFV-like genes are present in the wasp genome in two different locations; likely representing primary integration events constituting large portions of the LbFV-like ancestor’s genome. For both of these putative primary integration events, the ends of strings of genes with viral architecture are associated with remnants of retroviral genes such as pol, gag-pol, and gag-pro-pol, suggesting retroelements as a possible source for entry of viral genes into the wasp genome (Desjardins et al. 2008). A second scenario is infection with a filamentous virus, in which the LbFV-like scaffolds belong to an exogenous filamentous virus genome containing some horizontally transferred genes of eukaryotic origin.
Two genome assemblies are available for C. vestalis on NCBI. The two C. vestalis assemblies generated from wasp populations located in Andong, South Korea (ASM95615v1) and Hangzhou, China could reveal whether LbFV-like genes are present in both populations of this wasp species (ASM167554v1). Full-length versions of all of the genes of viral origin were present in both assemblies (except ac81, which was only present at 80% length in the Hangzhou assembly, located at the end of a scaffold) with nucleotide sequence identity ranging from 97% to 99%. Although the Andong assembly had three contigs that contained LbFV-like genes, regions of the genome containing LbFV-like genes were more fragmented in the Hangzhou assembly (found on 13 shorter contigs with average size of 7.6 kb). As mentioned above, raw reads were not available for the Andong assembly to generate read coverage information. Mapping sequencing reads to the Hangzhou assembly revealed the average coverage of LbFV-like gene-containing contigs (494x) was substantially and significantly higher than BUSCO-containing contigs (117x), whereas nudivirus gene-containing contigs (147x) and BUSCO-containing contigs had similar average coverage (F2,987 = 131.9, P < 2e−16 on log10 transformed values and Tukey’s HSD comparison of means, supplementary fig. 2, Supplementary Material online). The GC content of nudivirus-like gene-containing contigs in the Hangzhou assembly was not significantly different than BUSCO-containing contigs (mean 29.3% compared with 29.2%, respectively, Tukey’s HSD, supplementary fig. 2, Supplementary Material online). However, the mean GC content of LbFV-like gene-containing contigs in the Hangzhou assembly was slightly lower at 26.6% (comparison of GC content for all three scaffold types: F2,987 = 5.2, P < 0.01). None of the genes with eukaryotic architecture on the LbFV-like gene-containing contigs in the Andong assembly were detected in LbFV-like contigs in the Hangzhou assembly.
The presence of slightly divergent genes of viral origin in both assemblies suggests that both wasp populations have been infected with a virus of similar ancestry in the past. Given the lack of sequence depth coverage for the C. vestalis Andong genome, it is difficult to confidently conclude that the LbFV-like genes identified are endogenous in this population of C. vestalis. In contrast, the high coverage and differing GC content of LbFV-like gene-containing contigs in the C. vestalis Hangzhou assembly is strongly suggestive of an exogenous virus infection. More contiguous genome assemblies will be necessary to confirm whether LbFV-like genes are exogenous or endogenous in the two populations of C. vestalis.
EVEs in Parasitoid Wasp Species Are Distantly Related to Other Arthropod DNA Viruses
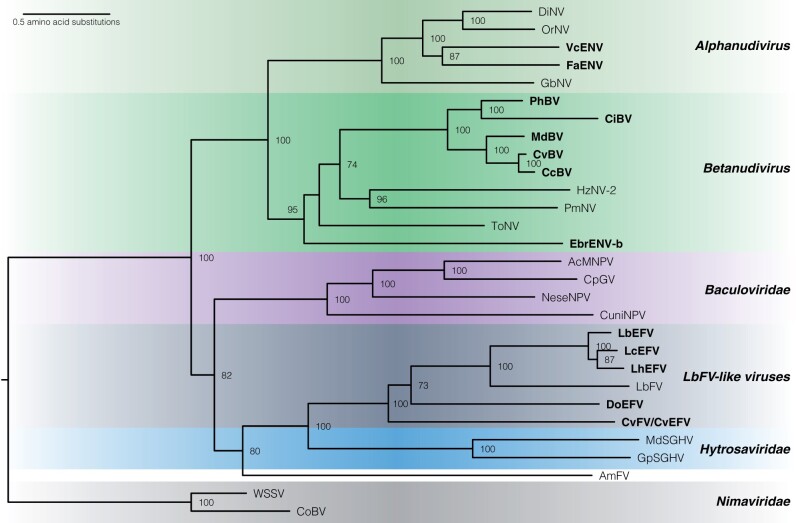
After annotation of the genes of viral origin in all of the new wasp genome assemblies, a list of genes emerged as conserved in many if not all viruses and EVEs derived from within the NALDVs. In addition to the ancient core genes (DNApol, helicase, lef-5, lef-8, lef-9, and p33), the recent description of nimavirus EVEs present in crustacean genomes identified pif-0, pif-1, pif-2, pif-3, and pif5 as core genes present in NALDVs (Kawato et al. 2018). Based upon new data from EVEs related to LbFV, ac81 can be added to a core gene set for viruses that infect insects (supplementary table 8, Supplementary Material online). Protein sequences from these 12 genes were used to construct a multigene phylogeny and examine the relationships between viruses and EVEs (fig. 6).
Fig. 6.
Phylogenetic analysis of arthropod-infecting large dsDNA viruses and parasitoid EVEs. Relationships were derived using a maximum likelihood analysis from 12 core genes with a total of 5003 characters from concatenated amino acid sequences. Bootstrap values over 50% are indicated near the relevant node. Scale bar indicates average number of amino acid substitutions per site. Names of EVEs are highlighted with bold type. Taxa are named as in figure 1, with the addition of Phanerotoma bracovirus (PhBV), Cotesia vestalis bracovirus (CvBV), DoEFV, and CvFV/CvEFV. EbrENV-a was omitted because only one sequence (DNA polymerase) was available.
As expected, bracoviruses from the Cheloninae (Phanerotoma and C. inanitus) were most closely related to each other, as were bracoviruses from both wasps belonging to Cotesia within the Microgastrinae (C. congregata and C. vestalis) (fig. 6). The EVEs related to LbFV are referred to as Dolichomitus Endogenous Filamentous Virus (DoEFV) and C. vestalis Filamentous Virus or Endogenous Filamentous Virus (CvFV/CvEFV). These taxa are as divergent from each other as they are from LbFV. Despite their names, the filamentous viruses from L. boulardi and Apismellifera are separated on the phylogeny by hytrosaviruses and do not appear to belong to the same virus family (fig. 6).
Discussion
This study investigated the presence and distribution of EVEs in one of the most astonishing radiations on Earth (parasitoid wasps from the Ichneumonoidea) and hints at the untapped number and diversity of viral acquisition events. As of early 2019 when this study began, only five genomes from the Ichneumonoidea were available in the National Center for Biotechnology Information database, but with advances in sequencing technologies, rapid growth of this number is expected. The availability of genome sequence data from 11 new species representing diverse lineages from within the Ichneumonoidea provided a unique opportunity to identify genes of viral origin. The identification of sets of genes involved in viral replication is suggestive of the production of virions or VLPs that have a significant impact upon parasitoid wasp biology. Further, characterizing the presence or absence of EVEs in relatives of wasp species known to produce virions or VLPs provides information about the origin of these features and their species distribution.
A new approach was needed in order to screen wasp genomes for genes of viral origin in a consistent and high-throughput manner. The approaches designed here were validated using six previously published genomes from the Ichneumonoidea with known presence or absence of EVEs. The first, simplest approach used to identify genes of viral origin was to screen all ORFs against a streamlined form of the NCBI nr database. However, this screen resulted in false positives due to the presence of many hits to viral genes that are likely to be frequently transferred between eukaryotic and viral genomes. The second approach therefore focused upon identification of ancient core genes, or genes that serve essential functions in the production of virions in dsDNA viruses. With this approach, it was possible to verify the presence of bracoviruses in the previously published genomes of M. demolitor and C. vestalis. The presence of genes derived from nudivirus acquisition events was also recovered with this method in F. arisanus and V. canescens. The ancient core gene approach also accurately showed that there are no virus core gene acquisitions in the D. alloeum or M. cingulum genomes. In the future, this approach could be extended to parasitoid wasp genomes as they become available, for example the recently published C.congregata, Lysiphlebus fabarum, and Aphidius ervi genomes (Dennis et al. 2020, Gauthier et al. 2021).
Having established a method that could pinpoint which species to focus upon further, the new genome assemblies were searched for the presence of ancient core genes. If ancient core genes were identified, their evolutionary history was reconstructed with phylogenetic trees with representatives of the diversity of large dsDNA viruses. Ancient core genes are not necessarily monophyletic, in fact, many seem to be acquired from eukaryotic replication systems independently in NALDVs and NCLDVs (Kool et al. 1994; Iyer et al. 2004, 2006, 2003). Thus, the weak support at basal nodes of the phylogenetic trees in figure 2 was expected, and rather than attempting to elucidate relationships between ancient divergence events in viral evolution, these trees functioned to categorize viral endogenization events into groups with viral ancestors.
As expected, a bracovirus was identified in the microgastrine braconid, Phanerotoma, and an ichnovirus in the banchine ichneumonid, Lissonota. The screen for ancient core genes then identified two major findings that were not expected a priori: 1) the presence of LbFV-related genes in C. vestalis (Braconidae: Microgastrinae) and Dolichomitus (Ichneumonidae: Pimplinae) genomes and 2) the absence of an ichnovirus in the campoplegine ichneumonid, Dusona. The preceding results will be discussed further below.
Both Nudiviruses and LbFV-Like Viruses Are Common Endogenous Virus Progenitors
Prior to this study, endogenous viruses discovered in parasitoid wasp species revealed that virus co-option from nudivirus ancestors has occurred independently on at least four occasions (Bézier et al. 2009; Pichon et al. 2015; Burke, Simmonds, et al. 2018; Zhang et al. 2020), all in koinobiont endoparasitic wasps. Out of all of the major groups of insect-infecting viruses, the predominance of nudivirus ancestors of EVEs suggested specific, as yet unknown conserved features of nudiviruses may predispose this group to integration and establishment in wasp genomes (Strand and Burke 2020, 2014). Recently, the first endogenization event from an ancestor other than nudiviruses and the ichnovirus ancestor was identified in L.boulardi (Figitidae), a koinobiont endoparasitoid wasp that produces VLPs or Mixed Strategy Endocytic Vesicles (MSEVs) (Rizki and Rizki 1990; Heavner et al. 2017; Di Giovanni et al. 2020). The ancestor of the EVE in L. boulardi (henceforth referred to as LbEFV) was related to a behavior-manipulating filamentous virus that also infects L. boulardi (LbFV), although the two are relatively divergent (Di Giovanni et al. 2020). This virus was only recently identified and although it clearly belongs to the NALDVs, it is so divergent from other virus families that it most likely represents the first discovered member of a new virus family (Lepetit et al. 2017).
This study identified further independently derived instances of endogenization of an LbFV-like entity into parasitoid wasp genomes, making LbFV-like viruses a second group of viruses that seem to be predisposed to endogenization. Both C. vestalis and Dolichomitus genome assemblies possess intact versions of many of the genes necessary to make virions or VLPs, justifying future investigation of the reproductive secretions of these wasp species. The LbFV-like virus in Dolichomitus also represents the first endogenized virus in an idiobiont ectoparasitoid, suggesting that this virus may function differently for parasitism success. Additionally, given that C. vestalis already produces a bracovirus, investigation of the role of LbFV-like genes in this species could address whether LbFV-like virions are also produced and how both types of virus might impact parasitism success and/or behavior. Our evidence is inconclusive regarding whether the LbFV-like genes in C. vestalis are endogenous or exogenous in two different wasp populations. More contiguous genome assemblies for this species will resolve this question. Previous studies have demonstrated distinct population structure, low gene flow, and reproductive isolation among both widespread populations of C. vestalis from around the world (Rincon et al. 2006) and from various populations from China (Wei et al. 2017), suggesting the possibility of recent speciation. It is noted that nonbracovirus filamentous particles have been observed in Cotesia species previously. Filamentous virions with similar morphology have been observed in C. congregata and C. marginiventris (as well as the ichneumonid Eriborus terebrans (formerly Diagema terebrans)) (Townes 1965; Krell 1987; Styer et al. 1987; Hamm et al. 1990). Although no genetic data are available for the filamentous viruses in other Cotesia species, these viruses and LbFV have similar morphogenesis (capsid production in cell nuclei, acquisition of an envelope in the cytoplasm) and may be derived from the same type of viral ancestor (de Buron and Beckage 1992; Varaldi et al. 2006).
“Ichnoviruses” Represent Two Independent, Relatively Recent Virus Acquisition Events
The genus “Ichnovirus” was coined to describe a type of virus that replicates in wasp ovaries and is injected into host insects during oviposition (Lefkowitz et al. 2018). This type of virus has a lenticular capsid surrounded by two unit membrane envelopes and a polydisperse genome including multiple double-stranded, circular DNAs of different sizes and coding capacities (Stoltz et al. 1984). At that time, it was not known that ichnoviruses were actually heritable endogenous entities, unlike archetypal viruses. It is more appropriate to group and name EVEs according to their independent evolutionary origins, which requires knowledge of the evolutionary history of wasp species that possess EVEs. The hyperdiversity of species within the Ichneumonidae has made their evolutionary relationships historically difficult to resolve. However, recent studies (Bennett et al. 2019; Klopfstein et al. 2019; Sharanowski et al. 2021) have confirmed results from a single-gene phylogeny (Quicke et al. 2009) to show that Campopleginae and Banchinae are not closely related to each other, despite both belonging to the informal lineage, Ophioniformes. Two previous studies have described the IVSPERs in a single member of each of these subfamilies, and from extensive overlap of the catalog of IVSPER genes, concluded that these EVEs came from similar, if not identical virus ancestors (Volkoff et al. 2010; Béliveau et al. 2015). Béliveau et al (2015) suggested two evolutionary scenarios giving rise to the presence of IVSPERs in these two divergent wasp subfamilies. First, that there was a single IV ancestor, in which an NCLDV integrated into the genome of an ancestor of banchine and campoplegine wasps. If no IVs are observed in wasp species belonging to the subfamilies that also descend from the common ancestor of banchine and campoplegine wasps, it implies that the capacity to produce IVs was lost in all of these species, although traces of IVSPER genes could remain in their genomes. The second scenario involves two separate integration events involving very similar viral ancestors (related to NCLDVs) that took place separately in the ancestors of banchine and campoplegine wasps.
Our data support the second scenario involving two independent integration events in campoplegine and banchine wasp species. First, based upon the phylogeny generated by Sharanowski et al. (2021), two of the wasp species with new genome assemblies, Anoncus (Ctenopelmatinae) and Mesochorus (Mesochorinae), are “intervening taxa,” that is they do not belong to either of the IV-producing subfamilies but descend from the common ancestor of all of these subfamilies (all within Ophioniformes). Both of these genomes lack ancient viral core genes, common dsDNA virus replication genes, and IVSPER-like genes, intact or otherwise. Second, the unexpected absence of IVSPERs in Dusona suggests that ichnoviruses are not present in all campoplegine species, and may be limited to a clade containing Hyposoter, Tranosema, Campoletis, and Casinaria but excluding Dusona and possibly Campoplex (Pichon et al. 2015; Sharanowski et al. 2021 Volkoff et al. 2010). Determining the distribution of EVEs among species is important because it will help to inform whether other viral acquisition events are truly replacement events (such as the nudivirus in the campoplegine V. canescens, which is hypothesized to have replaced an ichnovirus) or represent acquisition into a wasp ancestor that ancestrally lacked an EVE. Third, the architecture of the IVSPERs within wasp genomes also provides evidence of recent acquisition. The IVSPERs in H. didymator, G. fumiferanae, and now Lissonota (all Banchines) are each located in three compact clusters within wasp genomes, respectively. This level of clustering is similar to the architecture observed for the recently derived endogenous nudivirus in F. arisanus (nine major clusters), and lies in contrast to the extensive spread of nudivirus-like genes in the M. demolitor genome representative of the bracovirus-producing microgastrine wasps (Burke et al. 2014; Burke, Simmonds, et al. 2018; Burke, Walden, et al. 2018). As the spread of virus-derived wasp genes throughout wasp genomes is thought to be a relatively neutral process that occurs over evolutionary time (Burke et al. 2014), the limited spread of the IVSPERs in wasp genomes indicates that their integration occurred relatively recently. Finally, there are morphological differences between the campoplegine and banchine ichnoviruses that also suggest their independent origins (Béliveau et al. 2015). Although a close relative of the “ichnoviruses” has yet to be discovered in a nonendogenous form, it is possible that the ichnovirus ancestor is also a common progenitor of endogenous associations in parasitoid wasps.
Concluding Remarks
This study has shown that sequencing parasitoid wasp genomes can reveal novel instances of EVE acquisitions in the Ichneumonoidea. The ancient core gene approach used here attempted to identify sets of endogenous virus-derived genes that encode the most conserved, key components necessary for virus replication and have the potential for producing virions or VLPs. Importantly, without histological or functional data, it is not yet possible to conclude that any of the cases of viral endogenization described in this manuscript are virus domestication events.
Identification of sets of ancient core genes provided the potential for the detection of endogenized virus genes derived from a large swath of diversity within the large dsDNA viruses likely to infect insects (NCLDVs + NALDVs). A similar approach could be developed for other virus groups (e.g., vertebrate-infecting viruses), but is most likely to be successful for groups with larger genomes and greater numbers of ancient core genes (as opposed to many RNA viruses that may only have a single ancient core gene, the RNA polymerase).
The generation of new genomic data from just 11 species and reanalysis of six publicly available genomes has identified at least one independently derived, novel EVE. Additionally, the new genomes generated sequence data for two more EVEs that were expected in two species based upon their clade membership. Following the proposed use of a standardized nomenclature toward binomial species names for viruses (ICTV) (Siddell et al. 2020), revision of the genera Bracovirus and Ichnovirus and the family Polydnaviridae should be considered to reflect their endogenous nature and their evolutionary history. Additionally, coining a name for the LbFV-like viruses would be useful given their prevalence in parasitoid wasps. Overall, the data presented in this study hint that the diversity of viral endogenization events extends much further and is more common than just two rare, ancient events previously referred to as bracoviruses and ichnoviruses. Rather, the acquisition of EVEs from different groups of viral pathogens creates an ample source of variation for the recurrent evolution of diverse parasitism-based virulence strategies in parasitoid wasps.
Materials and Methods
Wasp Species Sampling
Sharanowski et al. (2021) recently generated genome sequence data sets from 11 species for the purpose of resolving the phylogenetic relationships among species in the Ichneumonoidea. The sampling included seven species from Ichneumonidae and four species from Braconidae, all derived from different subfamilies to maximally capture diversity (table 1, supplementary table 1, Supplementary Material online). All DNA samples used for sequencing were derived from single adult females, except Odontocolon sp., which was sequenced from a single adult male. Additionally, seven publicly available genome assemblies from the Ichneumonoidea (five braconid and one ichneumonid species) with known presence or absence of viral associations served as controls for this study (table 1, Burke, Walden, et al. 2018; Shi et al. 2019; Tvedte et al. 2019; Geib et al. 2017; Leobold et al. 2018; Yin et al. 2018). Two genome assemblies were available for C.vestalis, representing populations from Andong, South Korea, and Hangzhou, China. The Andong assembly available in NCBI was generally better (fewer contigs, higher N50 values) and thus was preferentially used for genome analyses; however, the two assemblies currently have become comparable after a recent update of the Hangzhou assembly (Shi et al. 2019).
Genome Assembly
Illumina sequence reads generated from genomic DNAs from 11 wasp species (PE100) were screened with trimmomatic v.0.36, using the parameters “LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.” Any read pairs with overlapping 3ʹ ends were merged with pear v.0.9.8 using default settings. Paired reads, merged reads, and unpaired single reads were used as input for de novo assembly of contigs using SPAdes v.3.12.0 with default parameters, which combines assemblies generated with 21, 33, 55, 77, and 99 bp kmers (Bankevich et al. 2012).
Database Construction and Rapid Homology Searches for Curation of Assemblies and Identification of EVEs
A custom database was made to identify contigs that contain genes of viral origin and exclude contaminant contigs (Medd et al. 2018). The database contained all protein sequences from the NCBI refseq database (downloaded February 2019) from Hymenoptera, Lepidoptera, Diptera, Coleoptera, bacteria, archaea, nematodes, and fungi, and viral protein sequences from the NCBI nr database (downloaded February 2019). TaxonKit v.0.3.0 was used to generate lists of all NCBI taxonomy ID numbers for included groups of organisms (Shen and Ren 2021). The software csvtk v.0.15.0 (https://bioinf.shenwei.me/csvtk/) was used to obtain accession numbers from proteins derived from species with included taxonomy IDs using the NCBI database prot.accession2taxid.gz (downloaded February 2019). Protein sequences from included species were then extracted from the refseq and nr databases and made into a database with diamond v.1.0 (Buchfink et al. 2015). Sequences from hymenopteran genomes with known EVEs were excluded (6 species total) so that any genes with viral origin would have hits to viral proteins and be assigned to the correct taxonomic lineage (virus rather than insect). Protein sequences derived from genome assemblies generated in this study were not included in the database. Genes from complete polydnavirus genomes (proviral genome segments) were also excluded from the database because they do not contain viral replication genes and do contain many genes of eukaryotic origin. The final database contained 117,461,186 protein sequences.
ORFs were identified from genomic contigs (generated as described below) or scaffolds using emboss v.6.6.0 getorf with minimum size of 150 bp yielding between 247,340 and 691,147 ORFs per genome assembly (Rice et al. 2000). ORFs were searched against the database described above with diamond with an e-value threshold of 0.01, retaining a single top hit (parameters: –outfmt 6 qseqid pident length mismatch gapopen qstart qend sstart send evalue bitscore staxids salltitles –max-target-seqs 1 –max-hsps 1 –evalue 0.01). NCBI taxonomy IDs were included in diamond output reports. The results from these rapid homology searches were used to identify sequence contaminants and to exhaustively screen ORFs for viral hits as outlined in the following sections.
Removal of Sequence Contaminants and Assessment of Assemblies
Contigs smaller than 200 bp in size were removed from final assemblies. Blobtools v1.1 (Laetsch and Blaxter 2017) was used to retain only those contigs that were assigned to Arthropoda and Viruses or that had no hits. Assembly statistics were evaluated using quast v.5.0.2 (Gurevich et al. 2013). The completeness of each set of genome sequence contigs was analyzed by identifying the number of arthropod BUSCOs (Simão et al. 2015). BUSCO v.3.0.2 was run on the assembled contigs or genome scaffolds for previously published genomes (“-m geno”) to identify orthologs in the Insecta ortholog database version 9, using Nasonia models for gene prediction.
To determine the read coverage of assembled contigs, all reads used for sequence assembly were mapped to the contigs using bowtie2 v.2.3.4.1, and coverage calculated with samtools bedcov v.1.9 (Li et al. 2009; Langmead and Salzberg 2013). A similar process was used to determine read coverage of genome scaffolds in previously published genomes, except that sequence reads were downloaded from the NCBI Sequence Read Archive when available and processed using trimmomatic and pear as described above.
Identification of Genes of Viral Origin in Wasp Genomes
Identification of virus-derived genes in wasp genomes was done using several approaches. All 11 new and previously published genome assemblies were searched without any a priori expectation for the presence of virus-derived genes by: 1) exhaustively screening all ORFs from genome assemblies for viral hits against the curated database described above; and 2) searching all ORFs for matches to ancient core genes found in large dsDNA virus groups that are known to infect insects (see below). These methods were expected to nonexhaustively discover the presence of virus-derived genes to narrow down the list of species for which more detailed identification and annotation should be performed.
Exhaustive Screening of ORFs for Viral Hits
The first approach used was modified from Medd et al. (2018) and used exhaustive similarity searches of all possible ORFs (of a minimum size) against a curated database with diamond to identify sequences with similarity to viral proteins as described in the “database construction and rapid homology searches” section above. A virus taxonomy list of all possible NCBI taxonomy ID numbers describing viruses was generated using TaxonKit v.0.3.0 (Shen and Ren 2021). Any hits with NCBI taxonomy IDs that matched the virus taxonomy list were retrieved from the diamond output. The local alignment identity (percentage identity of the aligned portion of diamond hits) and e-value were recorded as evidence for homology between ORFs and protein sequences in the diamond database. Despite the fragmented state of the newly added genome assemblies, it was reasoned that the majority of genes of viral origin and architecture would be intact (not be broken into pieces across contigs) because N50 values were equal to or greater than the expected sizes of these genes (less than 1,000 base pairs on average).
Targeted Searches for Ancient Core Genes in dsDNA Viruses
In the second approach, the set of ORFs with viral hits from each wasp species was then queried for the presence of ancient core genes. In order to do this, Hidden Markov Models were constructed for six ancient core genes (DNApol, helicase, lef-5, lef-8, lef-9, and p33; Wang et al. 2012) to find deeply divergent matches. Protein sequences were collated for 24 representative viral species: AcMNPV, CpGV, NeseNPV, CuniNPV, ToNV, OrNV, HzNV-2, PmNV, DiNV, GbNV, GpSGHV, MdSGHV, LbFV, WSSV, CoBV, AMEV, MSEV, VACV, IIV-6, LDV1, HvAV-3e, PbCV1, ApMV, and HHV-3. Sequences were aligned using MUSCLE v3.8.31 (for use as PSSMs in PSI-BLAST) and made into HMMs using hmmbuild within HMMER v3.1b1 (Edgar 2004; Eddy 2011). HMMs were searched against ORFs with viral hits from each species using hmmsearch with default parameters. PSI-BLAST (v. 2.10.0) searches used ancient core gene PSSMs as queries, the set of ORFs with viral gene hits as a database (identified from exhaustive diamond searches described above), and the following parameters “-evalue 0.005 -outfmt “6 std qlen slen qcovs ppos.”” The dispersed nature of genes in all parasitoid EVEs discovered to date makes it likely that at least some virus-derived genes would be recovered, even though some genomes were poorly assembled.
Detailed Annotation of EVEs Using Homology to Other EVE Sequences
If sets of virus-derived genes were confidently identified in a wasp genome assembly using the above methods, more detailed annotation was employed for these species. Post hoc searches of wasp genome assembly ORFs against the previously excluded replication genes from bracovirus, ichnovirus, and other parasitoid EVEs were employed to further identify virus-derived genes that may not have been found in earlier searches due to sequence divergence from viral ancestors. To effectively identify virus-derived genes in wasp genomes expected to contain bracoviruses or ichnoviruses, several publicly available reference data sets were employed. The genes encoding the nudivirus-like structural components of bracovirus virions have been completely cataloged in M.demolitor and partially in C.congregata (both subfamily Microgastrinae ∼53 myo) and C.inanitus (subfamily Cheloninae, ∼85 myo) (Murphy et al. 2008; Bézier et al. 2009; Burke et al. 2014). The genes contributing to the production of ichnoviruses known as IVSPERs have been identified previously in wasps from the subfamily Campopleginae (H.didymator) and the subfamily Banchinae (G.fumiferanae) (Volkoff et al. 2010; Béliveau et al. 2015). Protein sequences from nudivirus-like replication genes from M. demolitor, C. congregata, and C. inanitus, and separately, protein sequences from IVSPERs in the ichnovirus-producing wasp species H.didymator and G.fumiferanae were added to the custom diamond database described above. All ORFs from each species were searched against the modified database (as above), and ORFs with top hits to sequences from bracovirus-producing or ichnovirus-producing wasp species were extracted from the output. Finally, when genes related to similar virus pathogens were found in multiple wasp genomes, these genes and surrounding ORFs were searched among the multiple wasp genomes to further identify and annotate virus-related genes.
Metrics Used to Determine Whether Virus-Derived Sequences Are Endogenous
Virus-derived sequences identified in wasp genome assemblies could be indicative of EVEs or “contamination” by sequences derived from an actively replicating viral infection. The latter would have a self-contained viral genome, that is contigs could be assembled into a single, circular, or linear dsDNA chromosome representing the viral genome of a nonintegrated infectious virus. Features of genome architecture and sequencing metrics were used to build support for the hypothesis that virus-derived sequences are integrated into a given wasp genome. First, it was determined whether virus-derived genes were interspersed among nonviral genes in the wasp genome. Virus-derived genes with “viral architecture” were defined as genes that had homology to genes in viral genomes, are short and most often intronless. Nonviral genes were defined as having homology to hymenopteran or other animal genes and had “eukaryotic architecture”: comprising longer genes most often containing introns. If novel virus-derived genes were identified in a wasp genome assembly, scaffolds upon which viral genes were identified were annotated with MAKER v. 3.01.02-beta for eukaryotic genes (Holt and Yandell 2011) and Prokka v. 1.12 for virus-derived genes (Seemann 2014) to see if there were clear signatures of flanking genes with eukaryotic architecture. If available, sequence expression data were aligned to gene annotations with GMAP v.2019-05-12 (Wu and Watanabe 2005) to determine whether they were expressed and represent high-confidence annotations. Second, unless extremely recently derived, EVEs are expected to be present in all individuals and populations of a given species of wasp. If available, different versions of wasp genome assemblies generated from geographically distinct populations of wasps were queried for homologous viral genes with TBLASTN v. 2.2.31 (using virus-derived gene protein-coding sequences as queries, the alternative population genome as a database, and default parameters including no e-value cutoff, Altschul et al. 1997). Third, characteristics of contiguous blocks of virus-derived genes were used to assess whether they were likely to be derived from EVEs or an actively replicating virus infection. Most core replication genes are present as a single copy in insect viral pathogens with dsDNA genomes (Yutin et al. 2009) whereas degraded, pseudogenized forms of viral genes can be found in genomes with EVEs. To assess whether viral genes were degraded, the sizes and completeness (length) of ORFs with virus hits were compared using BLASTP v. 2.2.31 alignments (default parameters and an e-value cutoff of 0.01) to protein-coding sequences within and among parasitoid genomes and in virus genomes. Fourth, sequence read coverage and GC content of contigs were used as further lines of evidence for differentiation of endogenous versus replicating viral sequences. As EVEs are integrated into wasp genomes, the sequence read coverage of contigs containing EVEs is expected to be similar to other contigs. However, high coverage could be observed from local chromosomal amplification of EVEs, which is known to occur in several parasitoid wasps (Louis et al. 2013; Burke et al. 2015; Di Giovanni et al. 2020). Coverage that is higher or lower than wasp genome contigs could also be an indication of the presence of viral DNA contaminating the insect DNA sequenced. EVEs that represent ancient integration events (such as bracoviruses, Burke and Strand 2012) are expected to have a GC content similar to the remainder of the wasp genome, whereas differing GC content between contigs with virus-derived genes and others could be indicative of recently acquired EVEs (whose ancestors may have had differing GC content) or actively replicating viral contaminants. Finally, as most new genome assemblies were generated from a single diploid female wasp, the presence (or absence) of heterozygous alleles expected to be present in diploid organisms can be used to differentiate between exogenous and endogenous sequences in parasitoid wasp genomes. An exogenous haploid virus is expected to be devoid of heterozygous sites; in contrast, heterozygous sites are expected in the genome of a single diploid female.
To identify heterozygous sites, sequence reads were mapped to genome assembly contigs following the read coverage calculation methods above, except that the reference contigs used were from genome assemblies prior to removal of sequence contaminants and unmerged quality-filtered reads were used. bcftools (v.1.10.2) mpileup was used to generate raw variant calls using a maximum read coverage setting of 50 (default parameters with -d 50, -C 50). Variant alleles were identified with bcftools call using the multiallelic algorithm, and filtered with bcftools filter to give heterozygous alleles with a minimum mapping quality of 20 (-i'%QUAL > 20 && AC/AN = 0.5'). This method identifies heterozygous sites with balanced allele frequencies and excludes sites that are unlikely to be heterozygous with unbalanced allele frequencies. In the Dolichomitus genome assembly, contigs from a contaminating Wolbachia genome were used to compare the number of heterozygous alleles in these contigs to contigs containing viral genes or BUSCO genes. Wolbachia contigs were identified using contigs assigned to “Proteobacteria” and “Bacteria-undef” by blobtools and filtered to retain hits that contained the word “Wolbachia.” Wolbachia contigs were further filtered to remove high and low coverage outliers with coverage greater or less than 1.5 times the interquartile range beyond median coverage. The number of heterozygous alleles detected per contig was tallied and contigs were categorized by type. To further test whether heterozygous sites likely belonged to the wasp, as opposed to multiple sequence variants of exogenous virus, heterozygous SNPs were “phased” into haplotype blocks using sequence read data. Haplotypes were identified using the list of heterozygous alleles and paired-end sequence reads mapped to the genome assembly with HapCUT2 (Edge et al. 2016). HapCUT2 assembles haplotypes assuming a diploid organism, that is, blocks are reported when only two haplotypes (“diploid haplotype blocks”) are detected for any given stretch of sequence. Therefore, under the scenario of viral endogenization, the presence and length of diploid haplotype blocks are expected to be similar for scaffolds containing virus-derived and BUSCO genes, and near absent for Wolbachia scaffolds. In contrast, under the scenario of diverse or evolving populations of haploid viral genomes (with a variable number of haplotypes for a given sequence), the number of diploid haplotype blocks is expected to be near zero and much shorter than blocks within scaffolds containing BUSCO genes. All of the above metrics were used when available and in conjunction with each other to provide an overall picture of the likelihood that virus-derived genes represent EVEs.
Phylogenetic Analyses to Determine the Closest Known Relatives of EVEs
To make single-gene trees, amino acid sequences from ORFs with viral hits and from endogenous viruses in wasp genomes were added to sequences from representative taxa used to make PSSMs and HMMs as above. BLASTP v. 2.2.31 of ORFs with viral hits against the NCBI nr database was used to identify any additional endogenous viral genes from parasitoid wasps to include in alignments (default parameters including no e-value cutoff, Altschul et al. 1997). Protein sequences from the EVEs in G.fumiferanae and p33 sequences from nimaviruses were excluded because they were extremely divergent and decreased alignment quality. Sequences were aligned using the MAFFT einsi algorithm (Katoh and Standley 2013), and maximum likelihood phylogenies were derived using the RAxML with the PROTGAMMALG model using default parameters and 100 bootstrap replicates (Stamatakis 2014).
A multigene phylogeny of NALDV representatives was generated by aligning protein sequences with MUSCLE (Edgar 2004), concatenated with FASconCAT-G (Kück and Longo 2014), and trimmed with default parameters and a gap threshold of 0.6 by TrimAl (Capella-Gutierrez et al. 2009). A maximum likelihood phylogeny was constructed for the concatenated alignment using RAxML with default parameters and 1,000 rapid bootstrap replicates in the CIPRES portal (Miller et al. 2010).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Michael Strand for feedback upon the manuscript. This work was supported by the US National Science Foundation (DEB-1542290 to G.R.B. and DEB-1916788 to G.R.B. and B.J.S.), the USDA National Institute of Food and Agriculture Hatch project (1013423 to G.R.B.), and the Natural Sciences and Engineering Research Council (NSERC) Discovery Program (B.J.S.).
Data Availability
The data underlying this article are available in the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov (accessions in supplementary table 1, Supplementary Material online), and the Ag Data Commons at https://data.nal.usda.gov (doi:10.15482/USDA.ADC/1504545), and can be accessed with accession numbers given throughout the manuscript, figures, tables, and supplementary information. Intermediate files from assemblies are available by request from the corresponding author.
Literature Cited
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew RR. 1971. Parasitic insects. London: Heinemann Educational Books. [Google Scholar]
- Aswad A, Katzourakis A.. 2012. Paleovirology and virally derived immunity. Trends Ecol Evol. 27:627–636. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ES, et al. 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447:326–329. [DOI] [PubMed] [Google Scholar]
- Beckage NE, Tan FF, Schleifer KW, Lane RD, Cherubin LL.. 1994. Characterization and biological effects of Cotesia congregata polydnavirus on host larvae of the tobacco hornworm, Manduca sexta. Arch Insect Biochem Physiol. 26(2–3):165–195. [Google Scholar]
- Béliveau C, et al. 2015. Genomic and proteomic analyses indicate that Banchine and Campoplegine polydnaviruses have similar, if not identical, viral ancestors. J Virol. 89(17):8909–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AMR, Cardinal S, Gauld ID, Wahl DB.. 2019. Phylogeny of the subfamilies of Ichneumonidae (Hymenoptera). J Hymenopt Res. 71:1–156. [Google Scholar]
- Bézier A, et al. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930. [DOI] [PubMed] [Google Scholar]
- Bézier A, et al. 2013. Functional endogenous viral elements in the genome of the parasitoid wasp Cotesia congregata: insights into the evolutionary dynamics of bracoviruses. Philos Trans R Soc Lond B Biol Sci. 368:20130047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, Le Chat L, De Paepe M, Taddei F.. 2006. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr Biol. 16:2048–2052. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. [DOI] [PubMed] [Google Scholar]
- Burke GR. 2019. Common themes in three independently derived endogenous nudivirus elements in parasitoid wasps. Curr Opin Insect Sci. 32:28–35. [DOI] [PubMed] [Google Scholar]
- Burke GR, Simmonds TJ, Sharanowski BJ, Geib SM.. 2018. Rapid viral symbiogenesis via changes in parasitoid wasp genome architecture. Mol Biol Evol. 35(10):2463–2474. [DOI] [PubMed] [Google Scholar]
- Burke GR, Simmonds TJ, Thomas SA, Strand MR.. 2015. Microplitis demolitor Bracovirus proviral loci and clustered replication genes exhibit distinct DNA amplification patterns during replication. J Virol. 89:9511–9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Strand MR.. 2012. Polydnaviruses of parasitic wasps: domestication of viruses to act as gene delivery vectors. Insects 3:91–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Walden KKO, Whitfield JB, Robertson HM, Strand MR.. 2018. Whole genome sequence of the parasitoid wasp Microplitis demolitor that harbors an endogenous virus mutualist. G3 (Bethesda) 8:2875–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Walden KKO, Whitfield JB, Robertson HM, Strand MR.. 2014. Widespread genome reorganization of an obligate virus mutualist. PLoS Genet. 10:e1004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AD. 2009. Numbers of living species in Australia and the world. Australian Government Department of the Environment and Energy. Available from: https://www.environment.gov.au/science/abrs/publications/other/numbers-living-species/contents. Accessed May 2021.
- Coffman KA, Harrell TC, Burke GR.. 2020. A mutualistic poxvirus exhibits convergent evolution with other heritable viruses in parasitoid wasps. J Virol. 94(8):e02059–19. doi:10.1128/JVI.02059-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darboux I, Cusson M, Volkoff A-N.. 2019. The dual life of ichnoviruses. Curr Opin Insect Sci. 32:47–53. [DOI] [PubMed] [Google Scholar]
- de Buron I, Beckage NE.. 1992. Characterization of a polydnavirus (PDV) and virus-like filamentous particle (VLFP) in the braconid wasp Cotesia congregata (Hymenoptera: Braconidae). J Invertebr Pathol. 59(3):315–327. [Google Scholar]
- Dennis AB, et al. 2020. Functional insights from the GC-poor genomes of two aphid parasitoids, Aphidius ervi and Lysiphlebus fabarum. BMC Genomics 21:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, et al. 2008. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni D, et al. 2020. A behavior-manipulating virus relative as a source of adaptive genes for Drosophila parasitoids. Mol Biol Evol. 37(10):2791–2807. doi:10.1093/molbev/msaa030. [DOI] [PubMed] [Google Scholar]
- Dunlap KA, et al. 2006. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci U S A. 103:14390–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge P, Bafna V, Bansal V.. 2016. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 27(5):801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson KM, Vinson SB, Stoltz DB, Summers MD.. 1981. Virus in a parasitoid wasp: suppression of the cellular immune response in the parasitoid’s host. Science 211:582–583. [DOI] [PubMed] [Google Scholar]
- Feddersen I, Sander K, Schmidt O.. 1986. Virus-like particles with host protein-like antigenic determinants protect an insect parasitoid from encapsulation. Experientia 42(11–12):1278–1281. [Google Scholar]
- Feschotte C, Gilbert C.. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 13:283–296. [DOI] [PubMed] [Google Scholar]
- Forbes AA, Bagley RK, Beer MA, Hippee AC, Widmayer HA.. 2018. Quantifying the unquantifiable: why Hymenoptera, not Coleoptera, is the most speciose animal order. BMC Ecol. 18(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Drezen J-M, Herniou EA.. 2018. The recurrent domestication of viruses: major evolutionary transitions in parasitic wasps. Parasitology 145:713–723. [DOI] [PubMed] [Google Scholar]
- Gauthier J, et al. 2021. Chromosomal scale assembly of a parasitic wasp genome reveals symbiotic virus colonization. Commun Biol. 4:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib SM, Liang GH, Murphy TD, Sim SB.. 2017. Whole genome sequencing of the braconid parasitoid wasp Fopius arisanus, an important biocontrol agent of pest tephritid fruit flies. G3 (Bethesda) 7:2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goic B, Saleh M-C.. 2012. Living with the enemy: viral persistent infections from a friendly viewpoint. Curr Opin Microbiol. 15:531–537. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm JJ, Styer EL, Lewis WJ.. 1990. Comparative virogenesis of filamentous virus and polydnavirus in the female reproductive tract of Cotesia marginiventris (Hymenoptera: Braconidae). J Invertebr Pathol. 55(3):357–374. [Google Scholar]
- Heavner ME, et al. 2017. Novel organelles with elements of bacterial and eukaryotic secretion systems weaponize parasites of Drosophila. Curr Biol. 27:2869–2877.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV.. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 75:11720–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L.. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156–184. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L.. 2003. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L.. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 146:11–31. [DOI] [PubMed] [Google Scholar]
- Jones OR, Purvis A, Baumgart E, Quicke DLJ.. 2009. Using taxonomic revision data to estimate the geographic and taxonomic distribution of undescribed species richness in the Braconidae (Hymenoptera: Ichneumonoidea). Insect Conserv Diver. 2(3):204–212. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawato S, et al. 2018. Crustacean genome exploration reveals the evolutionary origin of White Spot Syndrome Virus. J Virol. 93(3):e01144-18. doi:10.1128/jvi.01144-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfstein S, et al. 2019. Hybrid capture data unravel a rapid radiation of pimpliform parasitoid wasps (Hymenoptera: Ichneumonidae: Pimpliformes). Syst Entomol. 44(2):361–383. [Google Scholar]
- Kool M, Ahrens CH, Goldbach RW, Rohrmann GF, Vlak JM.. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci U S A. 91:11212–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell PJ. 1987. Replication of long virus-like particles in the reproductive tract of the ichneumonid wasp Diadegma terebrans. J Gen Virol. 68(5):1477–1483. [Google Scholar]
- Kück P, Longo GC.. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11:81. doi:10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML.. 2017. BlobTools: interrogation of genome assemblies. F1000Res. 6:1287. [Google Scholar]
- Langmead B, Salzberg SL.. 2013. Bowtie2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavialle C, et al. 2013. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 368:20120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PO. 2005. Non-poly-DNA viruses, their parasitic wasps, and hosts. J Insect Physiol. 51:99–101. [DOI] [PubMed] [Google Scholar]
- Lee MJ, et al. 2009. Chapter 5 Virulence factors and strategies of Leptopilina spp.: selective responses in Drosophila hosts. Adv Parasitol. 70:123–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz EJ, et al. 2018. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 46:D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leobold M, et al. 2018. The domestication of a large DNA virus by the wasp Venturia canescens involves targeted genome reduction through pseudogenization. Genome Biol Evol. 10:1745–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit D, Gillet B, Hughes S, Kraaijeveld K, Varaldi J.. 2017. Genome sequencing of the behaviour manipulating virus LbFV reveals a possible new virus family. Genome Biol Evol. 8(12):3718–3739. doi:10.1093/gbe/evw277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis F, et al. 2013. The bracovirus genome of the parasitoid wasp Cotesia congregata is amplified within 13 replication units, including sequences not packaged in the particles. J Virol. 87:9649–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom CM, McCullough AJ, Johnson HA, Newton LA, Borer ET.. 2005. Invasive annual grasses indirectly increase virus incidence in California native perennial bunchgrasses. Oecologia 145:153–164. [DOI] [PubMed] [Google Scholar]
- Medd NC, et al. 2018. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol. 4(1):vey009. doi:10.1093/ve/vey009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE). p. 1–8. doi:10.1109/gce.2010.5676129.
- Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H.. 2005. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci U S A. 102:16919–16926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy N, Banks JC, Whitfield JB, Austin AD.. 2008. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol Phylogenet Evol. 47:378–395. [DOI] [PubMed] [Google Scholar]
- Parker BJ, Brisson JA.. 2019. A laterally transferred viral gene modifies aphid wing plasticity. Curr Biol. 29:2098–2103.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon A, et al. 2015. Recurrent DNA virus domestication leading to different parasite virulence strategies. Sci Adv. 1:e1501150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quicke DLJ, Laurenne NM, Fitton MG, Broad GR.. 2009. A thousand and one wasps: a 28S rDNA and morphological phylogeny of the Ichneumonidae (Insecta: Hymenoptera) with an investigation into alignment parameter space and elision. J Nat Hist. 43(23–24):1305–1421. [Google Scholar]
- Reineke A, Asgari S, Schmidt O.. 2006. Evolutionary origin of Venturia canescens virus-like particles. Arch Insect Biochem Physiol. 61:123–133. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Rincon C, Bordat D, Löhr B, Dupas S.. 2006. Reproductive isolation and differentiation between five populations of Cotesia plutellae (Hymenoptera: Braconidae), parasitoid of Plutella xylostella (Lepidoptera: Plutellidae). Biol Cont. 36(2):171–182. [Google Scholar]
- Rizki RM, Rizki TM.. 1990. Parasitoid virus-like particles destroy Drosophila cellular immunity. Proc Natl Acad Sci U S A. 87:8388–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, et al. 2013. Extrapolations from field studies and known faunas converge on dramatically increased estimates of global microgastrine parasitoid wasp species richness (Hymenoptera: Braconidae). Insect Conserv Divers. 6(4):530–536. [Google Scholar]
- Rohrmann GF. 2011. Baculovirus molecular biology. Bethesda (MD: ): National Library of Medicine. National Center for Biotechnology Information. p. 188. [Google Scholar]
- Roossinck MJ. 2011. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol. 9:99–108. [DOI] [PubMed] [Google Scholar]
- Rossignol PA, et al. 1985. Enhanced mosquito blood-finding success on parasitemic hosts: evidence for vector-parasite mutualism. Proc Natl Acad Sci U S A. 82:7725–7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotheram S, Salt G.. 1973. The surface of the egg of a parasitic insect. I. The surface of the egg and first-instar larva of Nemeritis. Proc Royal Soc B Biol Sci. 183:179–194. [Google Scholar]
- Ryabov EV, Keane G, Naish N, Evered C, Winstanley D.. 2009. Densovirus induces winged morphs in asexual clones of the rosy apple aphid, Dysaphis plantaginea. Proc Natl Acad Sci U S A. 106:8465–8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt G. 1965. Experimental studies in insect parasitism XIII. The haemocytic reaction of a caterpillar to eggs of its habitual parasite. Proc Royal Soc B Biol Sci. 162:303–318. [DOI] [PubMed] [Google Scholar]
- Schmitt MJ, Breinig F.. 2002. The viral killer system in yeast: from molecular biology to application. FEMS Microbiol Rev. 26:257–276. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- Sharanowski BJ, Dowling APG, Sharkey MJ.. 2011. Molecular phylogenetics of Braconidae (Hymenoptera: Ichneumonoidea), based on multiple nuclear genes, and implications for classification. Syst Entomol. 36(3):549–572. [Google Scholar]
- Sharanowski BJ, et al. 2021. Phylogenomics of Ichneumonoidea (Hymenoptera) and implications for evolution of mode of parasitism and viral endogenization. Mol Phylogenet Evol. 156:107023. [DOI] [PubMed] [Google Scholar]
- Shen W, Ren H.. 2021. TaxonKit: a practical and efficient NCBI taxonomy toolkit. J Genet Genomics. doi:10.1016/j.jgg.2021.03.006. [DOI] [PubMed] [Google Scholar]
- Shi M, et al. 2019. The genomes of two parasitic wasps that parasitize the diamondback moth. BMC Genomics 20:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell SG, et al. 2020. Binomial nomenclature for virus species: a consultation. Arch Virol. 165(2):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak K, Renault S, Federici BA, Bigot Y.. 2005. Characteristics of pathogenic and mutualistic relationships of ascoviruses in field populations of parasitoid wasps. J Insect Physiol. 51:103–115. [DOI] [PubMed] [Google Scholar]
- Stoltz DB, Krell P, Summers MD, Vinson B.. 1984. Polydnaviridae—a proposed family of insect viruses with segmented, double-stranded, circular DNA genomes. Intervirology 21:1–4. [DOI] [PubMed] [Google Scholar]
- Stoltz DB, Whitfield JB.. 1992. Viruses and virus-like entities in the parasitic Hymenoptera. J Hymenopt Res. 1:125–139. [Google Scholar]
- Strand MR. 2012. Polydnavirus gene products that interact with the host immune system. In: Beckage NE, Drezen J-M, editors. Parasitoid Viruses. New York: Academic Press. p. 149–161. doi:10.1016/b978-0-12-384858-1.00012-6. [Google Scholar]
- Strand MR, Burke GR.. 2014. Polydnaviruses: nature’s genetic engineers. Annu Rev Virol. 1:333–354. [DOI] [PubMed] [Google Scholar]
- Strand MR, Burke GR.. 2020. Polydnaviruses: evolution and Function. Curr Issues Mol Biol. 34:163–182. [DOI] [PubMed] [Google Scholar]
- Strand MR, Drezen JM.. 2012. Family Polydnaviridae. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Amsterdam: Elsevier. p. 237–248. [Google Scholar]
- Styer EL, Hamm JJ, Nordlund DA.. 1987. A new virus associated with the parasitoid Cotesia marginiventris (Hymenoptera: Braconidae): replication in noctuid host larvae. J Invert Pathol. 50(3):302–309. [Google Scholar]
- Suzuki M, Tanaka T.. 2006. Virus-like particles in venom of Meteorus pulchricornis induce host hemocyte apoptosis. J Insect Physiol. 52(6):602–613. [DOI] [PubMed] [Google Scholar]
- Townes H. 1965. Nomenclatural notes on European Ichneumonidae (Hymenoptera). Pol Pismo Entomol. 35:409–417. [Google Scholar]
- Tvedte ES, et al. 2019. Genome of the parasitoid wasp Diachasma alloeum, an emerging model for ecological speciation and transitions to asexual reproduction. Genome Biol Evol. 11:2767–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaldi J, et al. 2006. Artifical transfer and morphological description of virus particles associated with superparasitism behaviour in a parasitoid wasp. J Insect Physiol. 52:1202–1212. [DOI] [PubMed] [Google Scholar]
- Villarreal LP. 2015. Force for ancient and recent life: viral and stem-loop RNA consortia promote life. Ann N Y Acad Sci. 1341:25–34. [DOI] [PubMed] [Google Scholar]
- Villarreal LP, Witzany G.. 2010. Viruses are essential agents within the roots and stem of the tree of life. J Theor Biol. 262:698–710. [DOI] [PubMed] [Google Scholar]
- Volkoff A-N, et al. 2010. Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLoS Pathog. 6:e1000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bininda-Emonds ORP, Jehle JA.. 2012. Nudivirus genomics and phylogeny. In: Garcia Romanowski ML V, editors. Viral genomes—molecular structure, diversity, gene expression mechanisms and host-virus interactions. Rijeka: IntechOpen. p. 33–52. doi:10.5772/27793. [Google Scholar]
- Wei S-J, et al. 2017. Different genetic structures revealed resident populations of a specialist parasitoid wasp in contrast to its migratory host. Ecol Evol. 7:5400–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Asgari S.. 2003. Virus or not? Phylogenetics of polydnaviruses and their wasp carriers. J Insect Physiol. 49:397–405. [DOI] [PubMed] [Google Scholar]
- Whitfield JB. 1997. Molecular and Morphological Data Suggest a Single Origin of the Polydnaviruses among Braconid Wasps. Naturwissenschaften. 84(11):502–507. [Google Scholar]
- Wu TD, Watanabe CK.. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- Yin C, et al. 2018. The genomic features of parasitism, polyembryony and immune evasion in the endoparasitic wasp Macrocentrus cingulum. BMC Genomics 19(1):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DYE, Van Achterberg C, Horstmann K, Yu D, van Achterberg C.. 2012. World Ichneumonoidea 2011. Taxonomy, biology, morphology and distribution. Available from: https://www.scienceopen.com/document?vid=55b95bf4-66fd-4cea-8b9b-27b4dbefc056. Accessed May 2021. [Google Scholar]
- Yutin N, Wolf YI, Raoult D, Koonin EV.. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 6:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Han G-Z.. 2020. Chalcid wasp paleoviruses bridge the evolutionary gap between bracoviruses and nudiviruses. Virology 542:34–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov (accessions in supplementary table 1, Supplementary Material online), and the Ag Data Commons at https://data.nal.usda.gov (doi:10.15482/USDA.ADC/1504545), and can be accessed with accession numbers given throughout the manuscript, figures, tables, and supplementary information. Intermediate files from assemblies are available by request from the corresponding author.