Abstract
Biofilms that form on implanted medical devices cause recalcitrant infections. The early events enabling contaminating bacteria to evade immune clearance, before a mature biofilm is established, are poorly understood. Live imaging in vitro demonstrated that Staphylococcus aureus sparsely inoculated on an abiotic surface can go undiscovered by human neutrophils, grow, and form aggregates. Small (~50 μm2) aggregates of attached bacteria resisted killing by human neutrophils, resulting in neutrophil lysis and bacterial persistence. In vivo, neutrophil recruitment to a peritoneal implant was spatially heterogenous, with some bacterial aggregates remaining undiscovered by neutrophils after 24 hours. Intravital imaging in mouse skin revealed that attached S. aureus aggregates grew and remained undiscovered by neutrophils for up to three hours. These results suggest a model in which delayed recruitment of neutrophils to an abiotic implant presents a critical window in which bacteria establish a nascent biofilm and acquire tolerance to neutrophil killing.
Keywords: Staphylococcus aureus, biofilm, neutrophil, implant-associated infection, microscopy
Introduction
The bacterial or fungal biofilm state is often responsible for chronic, persistent infections on implanted surfaces and is an area of great concern to the medical community [1–4]. Biofilms that mature on implanted medical devices become recalcitrant to both antimicrobial therapies and host immune defenses [5–7]. Decades of research into novel biomaterial surfaces and antimicrobial coatings have yet to alleviate the issue. We suspect that an improved understanding of the innate immune response to an emergent biofilm is necessary to inform better strategies for preventing these serious infections.
Staphylococcus aureus readily forms biofilms on biotic and abiotic surfaces and has been a persistent threat to public health both in hospital-associated and more recently, community-associated infections [8, 9]. In addition to causing skin and soft tissue infections, sepsis, and necrotizing pneumonia, both methicillin-resistant and sensitive strains of S. aureus are common culprits in implant-associated infections, especially periprosthetic joint infections (PJI) and orthopedic implants [9–13]. Two-stage revision, one of the most used procedures to treat PJI, is expensive, prolonged (6 – 12 weeks), and only has a success rate between 65 to 90 percent [14, 15]. As is often the case in PJI, removal of an infected implant is frequently necessary to resolve biofilm infections [12, 14, 15]. In addition to causing serious harm to the patient, joint infections significantly increase economic costs, with studies finding total joint arthroplasty infection can triple or quadruple costs in Europe and the United States, respectively [16, 17].
In addition to forming biofilms, S. aureus expresses potent immunomodulatory virulence factors, making it an ideal organism to study host-pathogen interactions on a surface [18]. Mature S. aureus biofilms have been shown to utilize several mechanisms to modulate host immune responses such as inducing premature neutrophil extracellular trap (NET) formation [19], inhibiting complement [20], preventing phagocytosis [21, 22] and attenuating the anti-bacterial activity of macrophages [23]. These and other studies have identified numerous strategies used by mature S. aureus biofilms to interfere with the normal host immune defenses. Nevertheless, it is unclear how, and at what stage in biofilm formation tolerance to host immune defenses manifests.
Previously, we have demonstrated that clearance of S. aureus aggregates attached to an abiotic surface in vitro depended on high densities of human neutrophils [24]. In addition, the aggregate size appears to play a role in the efficiency of phagocytosis [25]. Therefore, it is reasonable to hypothesize that clearance of a contaminated surface in vivo would require fast recruitment and high densities of neutrophils. Neutrophil recruitment in vivo is thought to occur within a few hours (2 – 4 h) after infection or injury [26, 27] and this timeframe may serve as a critical window for bacteria to establish biofilm aggregates. To our knowledge, this time scale of recruitment has not been investigated in the context of an implanted surface. In this present study, we hypothesized that delayed neutrophil recruitment to a S. aureus-contaminated implant site allows for nascent biofilm formation and tolerance to host defenses in vivo.
For successful clearance of contaminating bacteria to occur, we hypothesize that the following steps must occur: (i) Neutrophils must be recruited to the contaminated surface, via host- or pathogen-derived signals; (ii) Neutrophils must adhere to and patrol the surface in search of bacteria; (iii) Contaminating bacteria must be discovered by recruited neutrophils; and (iv) Neutrophils must phagocytose and successfully kill the discovered bacteria. We contend that these events must occur in a timely manner, before S. aureus biofilm aggregates develop tolerance to host defenses.
In this work, we investigated how small (~5–100 μm2) S. aureus aggregates evade phagocytosis and killing by neutrophils by challenging adherent bacteria with human neutrophils in vitro during the early stages of biofilm formation. We additionally explored the dynamics of early neutrophil recruitment to a S. aureus contaminated surface using in vivo intravital imaging and neutrophil localization using a peritoneal implant model. Of note, we observed that human neutrophils readily clear lone cells or small groups of S. aureus. However, S. aureus rapidly becomes resilient through the formation of multicellular aggregates. In vivo, we observed that neutrophil recruitment is spatially and temporally heterogenous and can take several hours to occur, potentially providing sufficient time for contaminating microorganisms to mount mechanical and leukotoxic defenses prior to engagement by leukocytes.
Materials and Methods
Bacteria and neutrophil preparation
Staphylococcus aureus strain AH2547 (HG001 + pCM29, courtesy of Alex Horswill), a known biofilm-forming strain [36] and MW2-GFP (MW2 + pCM29) [37] with constitutive expression of a green fluorescent protein, was grown overnight in tryptic soy broth supplemented with 10 μg/ml chloramphenicol. Overnight cultures were centrifuged for 5 minutes at 4000 rpm, rinsed, resuspended in phosphate buffered saline (PBS), and serially diluted. Cells were attached to a 4-chambered glass bottom petri dish (Cellvis, CA, USA) to facilitate live-cell imaging. To attach cells, varied concentrations of cells were added to the surface in PBS at a volume of 10 μL. After 30 minutes of incubation at 37°C, unattached bacteria were gently rinsed from the surface with PBS. Each chamber of the petri dish was filled with 1 ml of 10% fresh human serum in Hank’s Balanced Salt Solution (HBSS) with Ca2+ and Mg2+ to simultaneously coat the surface with serum and opsonize bacteria and incubated at 37°C for 30 min. Human neutrophils were isolated from heparinized venous blood obtained from healthy donors following a standard IRB-approved protocol as described previously. All donors provided written consent to participate in the study. Neutrophils were isolated under endotoxin-free conditions (<25 pg ml−1) and purity (<1% PBMC contamination) and viability (<2% propidium iodide positivity) of neutrophil preparations were assessed by flow cytometry as previously described [38, 39]. Neutrophils were kept on ice until stained with LysoBrite™ Red (AAT Bioquest, CA, USA) according to the manufacturer’s instructions. Propidium iodide was added to the bulk medium just prior to neutrophil addition at a concentration of 5 μg/ml. To enumerate surviving bacteria, bacteria were removed from the surface using a sterile scraper and pipetting, vortexed in dilution tubes with PBS, and plated on tryptic soy agar in triplicate. Plates without detectable bacteria were counted as 0.5 colonies prior to log transformation.
Microscopy
A Leica SP5 inverted confocal scanning microscope was utilized for all in vitro imaging. GFP-tagged bacteria and neutrophils were excited with the 488 nm and 561 nm laser lines, respectively. A LiveCell (Pathology Devices, CA, USA) environmental chamber system was utilized to maintain 5% CO2, 20% O2, 50% humidity, and 37°C for sample incubation duri ng imaging. Image stacks with 1-μm z-slices were recorded sequentially at 1–2 min intervals over a 4-h time course using a 20X objective lens. At least two fields of view from each chamber of the dish were generally imaged per experiment. Each well was then imaged using a 7 × 7 stitched tile scan image with a 10x objective to quantify total amount of bacteria and count neutrophils remaining on the surface.
Image analysis
MetaMorph version 7.8.13 (Molecular Devices) image analysis software was used to measure the change in bacterial biomass by quantifying the threshold area of bacterial green fluorescence over the period of 4 h. Maximum growth rates were calculated by fitting an exponential curve to the data within log phase. Movies were prepared with Imaris version 8.0, 9.2, or 9.3 (Bitplane). To quantify neutrophil motion, the “Spots” module in Imaris was utilized to track 3D-objects over time, display their path, and analyze their motion. Neutrophils were automatically identified as “Spots” based on size and manually edited and confirmed when needed. Motion was tracked using the Brownian motion algorithm. To quantify the fraction of a field of view “patrolled” by a neutrophil over 4 hours, neutrophil tracks determined by the “Spots” module were set to be cylinders 10.87 μm in diameter. The image was then imported into MetaMorph, and the percentage of the field of view covered by a neutrophil track was determined by thresholding.
For the analysis of individual neutrophil-bacteria interactions, the “Spots” feature was used to identify neutrophils and propidium iodide staining events and the “Surfaces” feature was used to dynamically track bacterial growth. From the 4 experiments, two fields of view for each condition were analyzed. 8 interactions per field of view were analyzed for control wells (8 fields of view × 8 interactions, n = 64) while all interactions that could reasonably be distinguished in the head start wells were analyzed (8 fields of view, n = 70 interactions). Measurements of the bacterial aggregate were recorded at the start and end of each interaction with a neutrophil, and propidium iodide staining or aggregate breakup events were cataloged. The percentage of aggregates inducing PI staining was calculated by dividing the number of aggregates that caused PI staining of at least one phagocytosing neutrophil by the total number of aggregates discovered by a neutrophil in each field of view.
Maximum growth rates of individual aggregates were calculated by fitting an exponential curve to volume measurements calculated using “Surfaces” in Imaris. Aggregates that were not detected for at least 88% of the frames were discarded. To determine the change in bacterial biomass over time from in vivo experiments, Imaris surface output files were analyzed in MATLAB and the total GFP volume (μm3) was calculated for each frame. Percent change in biomass was calculated by comparing the last frame to the first frame for the final continuous field of view in each position. In vivo growth rates were calculated by fitting an exponential curve to the data collected in a continuous field of view that appeared to be out of lag phase and free of significant signal noise (such as due to animal movement).
Leukocyte counts from SEM images were produced by manually counting all visible cells that could be distinguished as distinct leukocytes and normalizing to the field of view area.
All images and videos from obtained from intravital microscopy were analyzed using Imaris version 9.5.1. GFP S. aureus clusters were quantified over time using the surface function and volume and neutrophil recruitment was tracked using the spot function. For quantification of neutrophil migration within the top 20 μm of collagen, spots were filtered for any spot entering the defined area of interest, top 20 μm of collagen, in the z-plane. Discovery frequency was quantified by applying a filter to the GFP bacteria surface “intensity mean” of tdTomato channel and thresholding was automatically applied by default settings.
Peritoneal implant model
The peritoneal implant model was performed as previously described with modifications [28, 29, 40]. Overnight cultures of S. aureus strain HG001 were grown in TSB for 20 h with shaking at 150 rpm and resuspended in 5 ml TSB before dilution. Sterile silicone tubes (i.d. 4 mm, o.d. 6 mm, length 4 mm) were incubated in a diluted overnight culture of S. aureus in 0.9% saline (OD600 = 0.1) for 1 hour at 100 rpm. Female C57bl/6JRj mice were purchased from Janvier and allowed two weeks to acclimate prior to surgery. Mice were anaesthetized with Hypnorm-midazolam (Hypnorm [0.315 mg fentanyl citrate/ml and 10 mg fluanisone/ml] and midazolam [5 mg/ml] and sterile water [1:1:2]) via subcutaneous injection in the groin area. 1 ml sterile PBS was injected into the mouse peritoneum 4h prior to implantation. Implants were inserted into the peritoneal cavity via a 1 cm groin incision. The mice were given bupivacaine and Temgesic for postoperative pain and euthanized by cervical dislocation at the end of the experiment. Several implants were removed and fixed in 2% glutaraldehyde for analysis by scanning electron microscopy while the remaining implants were put in 2 ml 0.9% NaCl and kept on ice until further processing. To quantify colony forming units on implants pre-insertion, representative silicone tubes were sonicated in 0.9% saline and dilutions were plated on tryptic soy agar. To determine the number of neutrophils recruited to the peritoneum, a lavage was performed by injecting 5 ml of PBS into the peritoneal cavity and gently massaging the abdomen before withdrawing the peritoneal fluid (PF) [41]. PF was stored on ice until preparing the samples for determination of recovered neutrophils by flow cytometry. To enumerate the neutrophils in the PF a 100 μl sample was added to a TruCount Tube (340334, Becton Dickinson). Ten μl of Anti-Ly-6C-FITC, mouse (130-102-295, Miltenyi Biotec), Anti-Ly-6G-APC, mouse (130-102-342, Miltenyi Biotec), and CD45-PerCP, mouse (130-102-469, Miltenyi Biotec) were added and the samples were incubated on ice for 30 min in the dark before fixation by addition of 1 ml of 10% (v/v) FACS lysing solution (349202, Becton Dickinson). Stained and fixed samples were analyzed by flow cytometry using a FACSCanto II (Becton Dickinson). Light scatter and fluorescence parameters for >10 000 events were recorded after gating on forward light scatter and fluorescence for CD45 staining to exclude debris, cell aggregates and bacteria. The instrument was calibrated with CSTbeads (Becton Dickinson, Franklin Lakes, NJ, USA). All experiments were authorized by the National Animal Ethics Committee, Denmark, Permit number 2018-15-0201-01500.
Scanning electron microscopy
Specimens were fixed in 2% glutaraldehyde in 0.05 M sodium phosphate buffer, pH 7.4. Following 3 rinses in 0.15 M sodium phosphate buffer (pH 7.4) specimens were post fixed in 1% OsO4 in 0.12 M sodium cacodylate buffer (pH 7.4) for 2 h. Following a rinse in distilled water, the specimens were dehydrated in 100% ethanol according to standard procedures and critical point dried (Balzers CPD 030) with CO2. The specimens were subsequently mounted on stubs using double adhesive carbon tape (Ted Pella) as an adhesive and sputter coated with 6 nm gold (Leica ACE 200). Specimens were examined with a Quanta 3D SEM (FEI, Eindhoven, The Netherlands) operated at an accelerating voltage of 2 kV.
Intravital imaging in a murine skin flap model
Animal experiments were performed with male adult 7–8-wk-old mice and all experimental animal protocols were approved by the University of Calgary Animal Care Committee and followed guidelines established by the Canadian Council for Animal Care (protocol number AC19-0138). All mice were housed under specific pathogen-free conditions and received sterilized rodent chow and water ad libitum. CatchupIVM-red mice in which neutrophils are tagged with a tdTomato red fluorescent protein were anesthetized and kept at body temperature using a heating pad [31]. A jugular catheter was inserted to maintain anesthesia and deliver systemic treatments. The dorsal flank skin was exteriorized on the right flank as previously described [26]. The tissue was covered with a cover slip that had been seeded with GFP-tagged S. aureus at surface densities similar to those used in in vitro experiments (103–104 CFU/cm2, pregrown in RPMI medium, rinsed with HBSS). Superfusion buffer (HBSS without calcium, magnesium, or phenol red, ThermoFisher) was then perfused across the exteriorized skin tissue to keep the skin moist at a flow rate set to 0.17 ml/min.
Mice were imaged for up to 180 min after being anesthetized using a Leica SP8 upright multiphoton microscope. Laser excitation at 940 nm was used to excite tdTomato and GFP with external detectors (HyD-RLD2 BP 585/40 for tdTomato, HyD-RLD3 BP 525/50 for GFP) and second harmonic generation (external detector HyD-RLD4 BP 450/70) to visualize skin collagen. Laser power, detector settings and acquisition settings were maintained throughout each experiment. Blood vasculature was labeled intravascularly using Qtracker™ 655 Vascular Labels (ThermoFisher) with excitation/emission at 405–615/655 nm (external detector HyD-RLD1 BP 675/50).
A 3D tile scan (4×4 fields of view; each field of view 350 × 350 × 100 μm3) was first collected to get an overview of the GFP S. aureus aggregates on the coverslip. From there, 3 fields of view (350 × 350 × 80 μm3) were selected within the 3D tile scan to select the video positions. Images were collected every 45 s for up to 180 min.
Statistical treatment of data
Data were analyzed using GraphPad Prism version 8.4.2. Normality was assessed by the D’Agostino-Pearson or Shapiro-Wilk test. Normal data was tested with an unpaired student’s t test, one-way ANOVA with Tukey’s post hoc test, two-way ANOVA with Bonferroni’s or Tukey’s multiple comparisons test or Brown-Forsythe ANOVA with Dunnet’s T3 multiple comparisons test as appropriate. Otherwise, the Kruskal-Wallis test or Mann-Whitney test was applied. The test applied is described in each figure legend.
Data Availability
The raw and processed data required to reproduce these findings are available to download from https://datadryad.org/stash/share/WHrJoeXSaqg3jdFH9pOPwUlablkGwl2RVvFePLu3fc4.
Results
High surface densities of neutrophils are required for effective clearance of S. aureus
We utilized an in vitro confocal microscopy approach to observe interactions between adherent S. aureus and human neutrophils (polymorphonuclear leukocytes, PMNs). Green fluorescent protein (GFP) expressing S. aureus AH2547 was sparsely seeded on a glass surface, challenged with fluorescently labeled neutrophils, and continuously imaged for 4 h. In the absence of neutrophils, bacteria grew and formed dense aggregates on the surface with a collective specific growth rate of 0.80 ± 0.16 h−1. Interestingly, the growth rate of individual S. aureus aggregates was quite variable, ranging from −0.2 to 1.6 h−1 (Figure S1A). Clearance of bacteria was highly dependent on the density of neutrophils on the surface, with effective killing occurring under higher neutrophil conditions (Figures 1A and 1B). Low neutrophil conditions resulted in killing of some attached bacterial aggregates, but survival and prolific growth of the many undiscovered aggregates. Conversely, sufficiently high neutrophil concentrations allowed rapid discovery and elimination of adherent bacteria before aggregates were able to grow significantly (Figures 1A and 1B). Neutrophil surface densities greater than 10,000 neutrophils/cm2 often enabled effective clearance of bacteria, resulting in 1.5 – 2 log reductions in bacterial burden compared to control wells without neutrophils (Figure 1C). Clearance of the bacteria correlated well with the ability of neutrophils to “patrol” large surface areas (Figure 1D). Net bacterial growth as measured by GFP area within a given field of view (FOV) was halted when approximately 50% of the surface was patrolled and reductions in bacterial burden greater than one log were consistently observed at approximately 80% coverage of the surface by neutrophil tracks (Figure 1D). When densities of both neutrophils and bacteria were low, the outcome was stochastic and the result often depended on timely discovery of the bacteria, which we have previously described in a mathematical model [24]. Thus, we have observed that a high surface density of neutrophils is required to effectively discover and eliminate S. aureus sparsely adherent to an abiotic surface.
Figure 1:
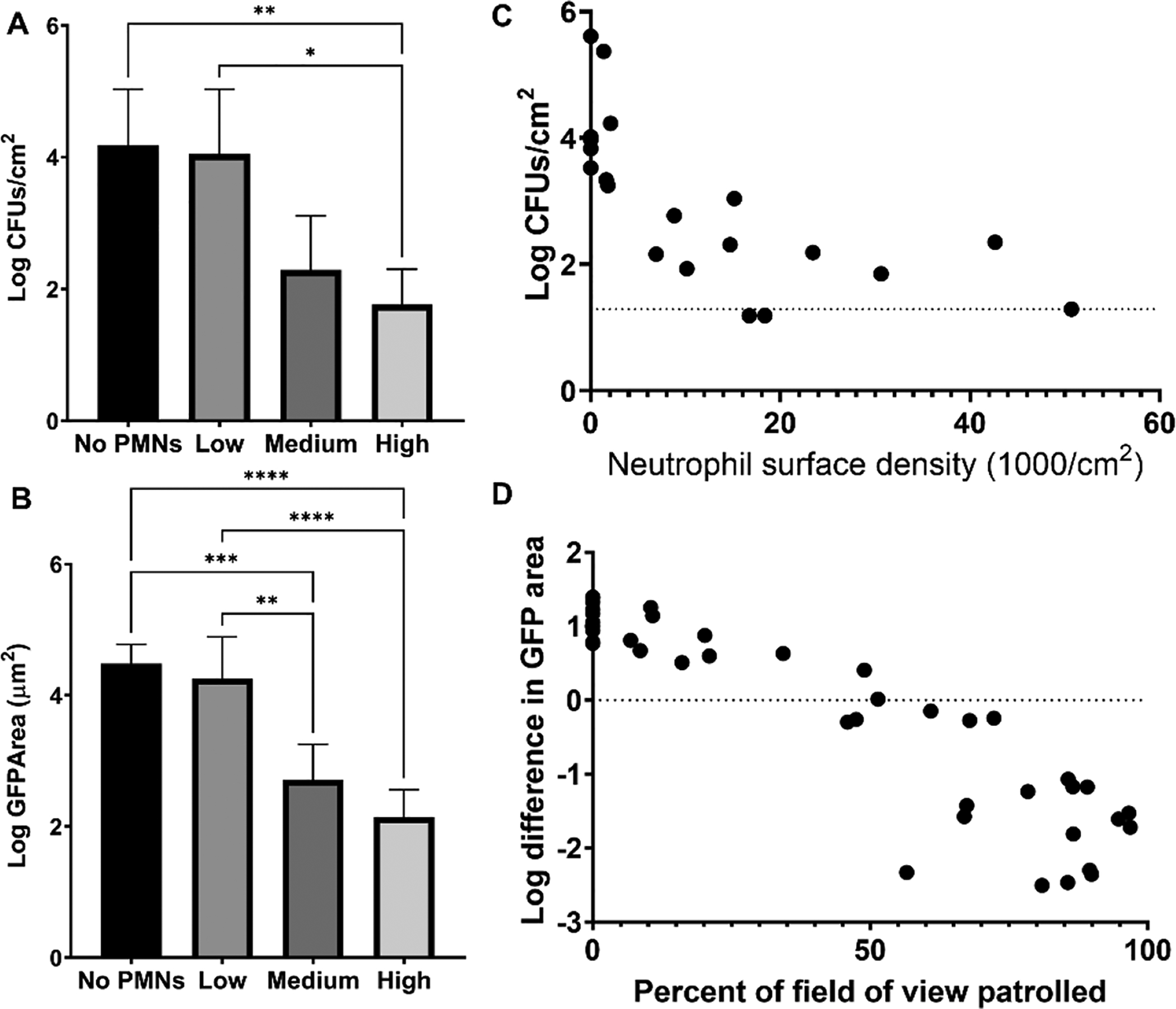
High neutrophil densities are required for adequate clearance of attached S. aureus in vitro. Approximately 2×103 CFUs/cm2 S. aureus GFP were initially added to the surface and challenged with PMNs for 4 h. (A) S. aureus CFUs recovered and (B) total remaining GFP signal from wells with no, low (1.7 ± 0.27 × 103 PMNs/cm2), medium (1.0 ± 0.31 × 104 PMNs/cm2), or high (2.8 ± 1.3 × 104 PMNs/cm2) numbers of PMNs. Data are mean ± SD (n=4–7. Kruskal-Wallis Test (panel A) and one-way ANOVA with Tukey’s post hoc test (panel B) **p<0.01, ***p<.001, **** p<0.0001). (C) CFUs recovered from challenge plotted against varied neutrophil surface densities. Dashed line represents limit of detection. n=5 independent experiments. (D) Log difference in GFP area per field of view as a function of the percentage of the field of view covered by neutrophil tracks. n=40 FOVs from 5 independent experiments.
Neutrophil recruitment to a peritoneal implant is heterogeneous
To observe spatial patterns of immune cell recruitment to a contaminated biomaterial in vivo, we utilized a previously described murine peritoneal implant model [28, 29]. A silicone tube was homogeneously coated with a low density of S. aureus and inserted into the mouse peritoneum via groin incision. Mice were euthanized 24 h post implantation and the implants were removed for analysis. We estimated the initial bacterial burden on the implant surface to be approximately 4.1 ± 1.7 × 105 CFUs/implant via plate count and 1.2 ± 0.51 × 106 cells/cm2 based on counts from scanning electron microscopy (SEM) micrographs (n = 9 FOVs from 2 implants) (Figure 2A). After 24 h in the peritoneum, the mean log reduction of viable bacterial counts recovered from implants (referenced to the pre-insertion count) was 1.98 ± 0.73 (n = 11). Flow cytometry of immune cells collected by peritoneal lavage showed that neutrophils were highly prevalent in the mouse peritoneum at 24 hpi, constituting 53 ± 6.8 % of host cells (n = 8 mice), whereas the background levels of neutrophils in animals that did not receive an implant was 0.7 ± 0.8 % (n = 8). The concentration of neutrophils in peritoneal lavage fluid was 4.3 ± 3.3 × 103 cells/ml in animals that did not receive an implant and rose to 4.1 ± 0.9 × 105 cells/ml in animals harboring inoculated implants for 24 h. Interestingly, although the distribution of S. aureus appeared uniform across the implant surface prior to insertion, recruitment of immune cells to the implant was spatially heterogeneous. By SEM, we observed the formation of distinct patches of host material on the implant surface (Figure 2B). Examination of these patches at higher magnification revealed aggregations of leukocytes (Figure 2C). We estimated the local surface density of these leukocyte patches to be 6.2 ± 5.6 × 105 leukocytes/cm2 based on counts of observable cells in areas where leukocytes were present (n = 10 FOVs from 2 implants) (Figure 2F). Several FOVs were devoid of leukocytes (Figure 2F). Magnification of areas lacking these leukocyte patches revealed that S. aureus aggregates remained on the surface with little to no interaction with recruited immune cells (Figure 2D). Additionally, while the SEM images of implants pre-insertion revealed mostly single or paired groupings of S. aureus cells (Figure 2A), the areas without recruited immune cells on explanted surfaces contained clusters of S. aureus with larger numbers of cells, suggesting growth (Figure 2E). These observations support the hypothesis that some bacteria on a contaminated surface may escape detection due to heterogeneous recruitment of immune cells, providing these cells an opportunity to develop into more resilient aggregates.
Figure 2:
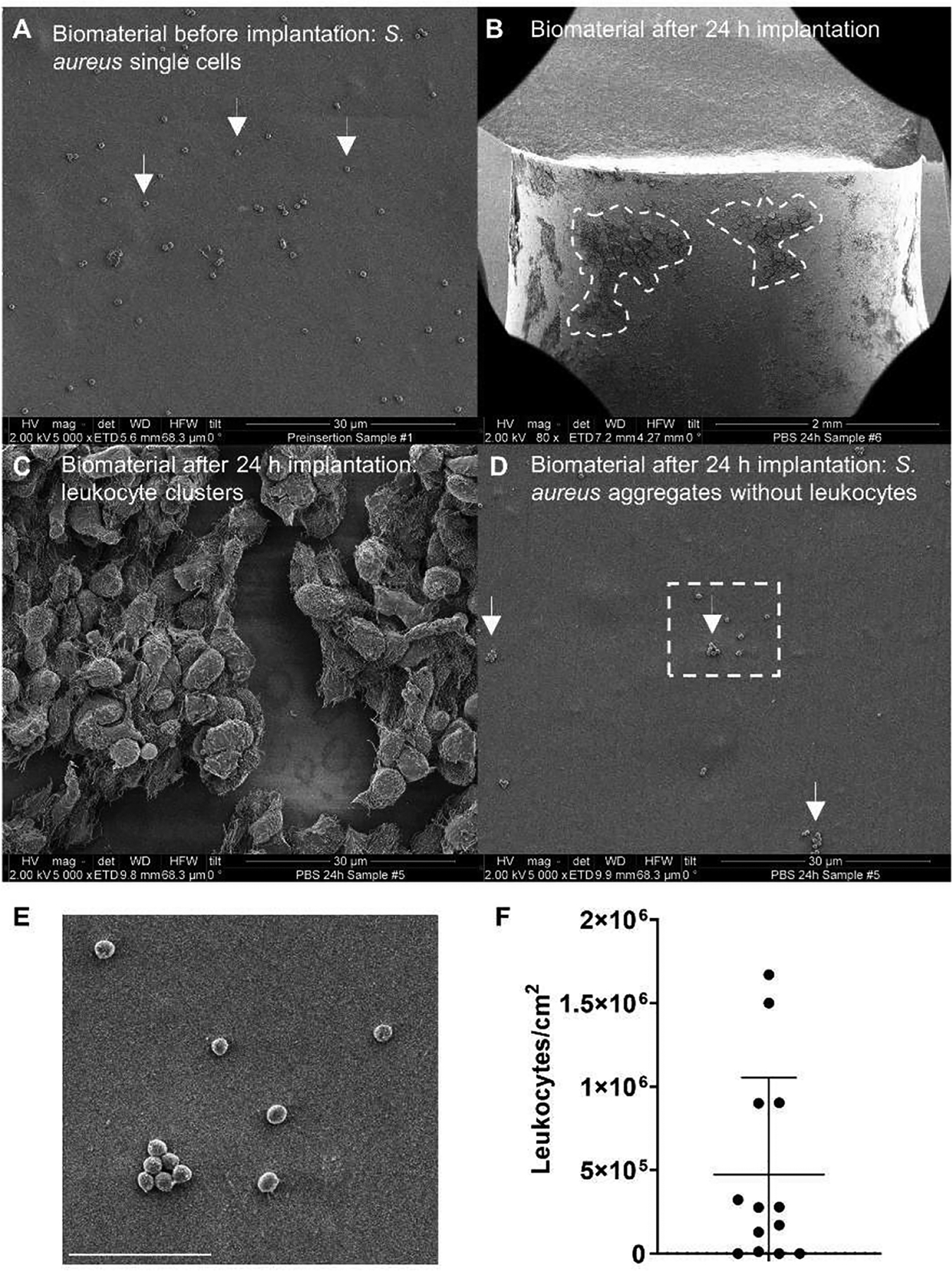
Immune cell recruitment to an inoculated peritoneal implant is heterogeneous. (A) SEM micrographs showing adherent S. aureus cells on the surface of a silicone tube prior to implantation into the mouse peritoneum. (B-D) Representative SEM micrographs from silicone tubes removed from the mouse peritoneal cavity after 24 h. (B) At low (80x) magnification, distinct areas of significant immune cell recruitment can be observed as dark patches on the implant surface (highlighted by dashed lines). (C) Higher magnification (5000x) reveals the dark patches outlined in (B) to be leukocyte clusters. (D) Higher magnification (5000x) reveals sections of the implant devoid of leukocytes. (E) Inset of the region denoted by the white box in (D). Scale bar = 5 μm. (F) Estimated leukocyte numbers on the implant surface from SEM images, normalized to FOV area (n=13 FOVs).
S. aureus aggregates resist clearance by neutrophils in vitro
We have observed previously that S. aureus aggregates discovered by neutrophils later in an in vitro experiment tended to be more tolerant to neutrophil clearance [24]. Therefore, we sought to further investigate and quantify the effect of aggregate size and timing of discovery on S. aureus survival of neutrophil challenge. S. aureus was sparsely seeded on glass and then allowed to form aggregates for 4 h prior to the addition of neutrophils. In control wells without neutrophils, preformed S. aureus aggregates grew into larger aggregates on the surface after 4 h of imaging (Fig. 3Ai–ii). Clusters of S. aureus that were given a 4 h head start prior to the addition of neutrophils resisted clearance, resulting in large aggregates remaining on the surface at the end of the experiment (Figure 3Aiii–iv). However, CFU-matched single-cell S. aureus were readily cleared by the neutrophils within 4 h (Figure 3Avii–viii). We quantified the log CFU of S. aureus/cm2 in the presence and absence of neutrophils for both head start (4 h) and no head start (single-cell S. aureus) and found that in the presence of neutrophils, there was a significant reduction of S. aureus recovered from the surface post-imaging (Figure 3B). Furthermore, neutrophils were significantly more effective at clearing single-cell S. aureus compared to preformed aggregates (Figure 3C). This suggests that surface attachment alone is not an inherent barrier to neutrophil clearance, but within a short period of time S. aureus aggregates can develop tolerance to neutrophils.
Figure 3:
S. aureus aggregates become resilient to neutrophil clearance after four hours of growth in vitro. S. aureus (green) clusters were seeded at ~102 CFUs/cm2 and grown for 4 h (HS) or seeded at ~103 CFUs/cm2 and grown for 0.5 h (No HS) prior to the start of imaging, with or without neutrophil (red) addition (~2×104 PMNs/cm2). (A) Representative microscopy images at t = 0 h and t = 4 h. (B) Log CFUs/cm2 recovered from wells post imaging. (C) Log difference in CFUs/cm2 (black points) and GFP area (white points). Data are mean ± SD (n=4 independent experiments. Paired student’s t-test *p<0.05, **p<0.01.).
We observed that some aggregates failed to decrease in size after co-localization with a neutrophil and that some neutrophils disappeared during this interaction. These observations led us to investigate whether larger S. aureus aggregates were lysing neutrophils. We added propidium iodide (PI), a membrane impermeable dye, to our assay to investigate whether neutrophil membrane integrity was compromised upon interaction with large S. aureus aggregates. Neutrophils interacting with larger aggregates often stained extensively with PI, suggesting these aggregates were able to damage neutrophil membrane integrity (Video S1). Neutrophils became PI positive at a much higher frequency when S. aureus aggregates were given a head start prior to the addition of neutrophils (Figures 4A–C). Under the head start condition, 86 ± 19% of aggregates in a given FOV induced PI staining of at least one neutrophil while we did not observe any single cells of S. aureus causing PI staining of neutrophils in this assay (Figure 4C, Video S1). We analyzed the outcome for each bacterial aggregate present in a field of view and found that aggregates greater than roughly 50 μm2 caused frequent neutrophil lysis and aggregates greater than 75 μm2 caused neutrophil lysis in all of the interactions observed in these experiments (Figure 4D). Larger aggregates tended to persist, as aggregates with initial area greater than 75 μm2 at the time of interaction with a neutrophil often failed to decrease or even grew in size from the time of discovery (Figure 4E). We also observed on occasion that large aggregates could be broken up or fractured into smaller pieces by multiple neutrophils and that these smaller pieces were more readily cleared (Video S2). These events did not seem to depend solely on the size of the aggregate (Figure S2A). Rather, aggregates appeared to fracture when a high number of neutrophils attempted to phagocytose a portion of the aggregate over the course of the experiment (Figure S2B). Non-fractured aggregates tended to grow slightly over the 4 h observation period following discovery by the first neutrophil, however when fracture occurred, there was a slight decrease in the total volume of all fractured pieces (Figure S2C). This suggests a better outcome when neutrophils are successfully able to fragment a portion of the bacterial cluster compared to when the clusters remain intact.
Figure 4:
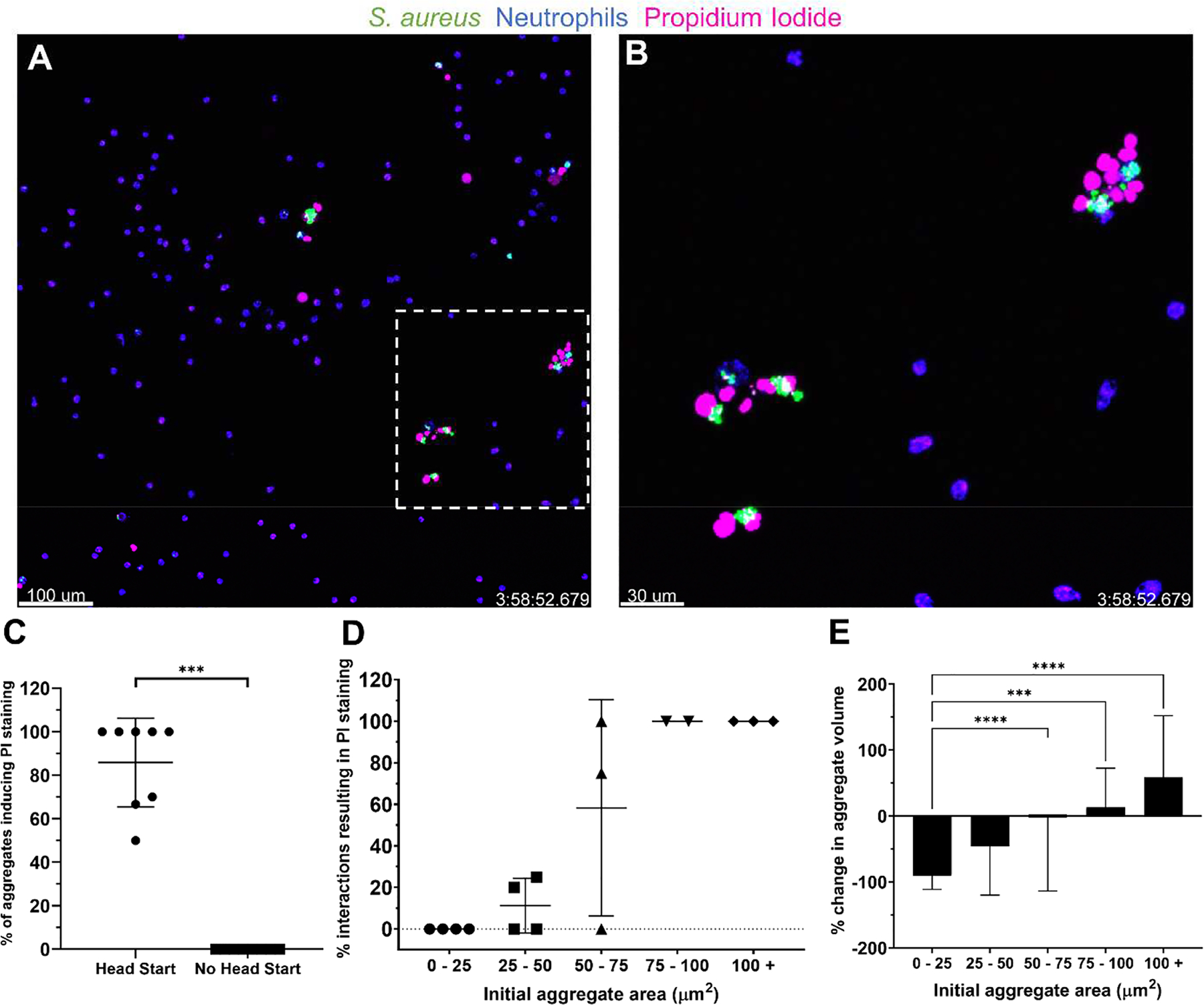
Larger S. aureus aggregates cause neutrophil lysis and resist clearance in vitro. (A) S. aureus aggregates (green) were given 4 h to grow prior to the addition of neutrophils (blue). Representative images at t = 4 h show propidium iodide (PI) staining (pink) of neutrophils interacting with larger, persistent clusters of S. aureus. (B) Enlarged image of the inset in (A). (C) Percentage of S. aureus aggregates causing PI staining of at least one neutrophil. Data are mean ± SD (n=8 FOVs per group from 4 independent experiments. Mann-Whitney test ***p<.001). (D) Rates of PI staining of neutrophils as a function of the bacterial aggregate area at the beginning of the interaction. Each point represents the calculated average of all interactions within the size boundaries from a given experiment. The number of observed interactions per point ranged from n = 1–19 collected from 16 FOVs from 4 independent experiments. (E) Change in volume of an aggregate from the first frame in which a neutrophil phagocytoses it to the last frame that it is visible. Data are mean ± SD (Bins contain 6–58 measurements from 16 FOV from 4 independent experiments. Kruskal-Wallis test ***p<0.001, ****p<0.0001).
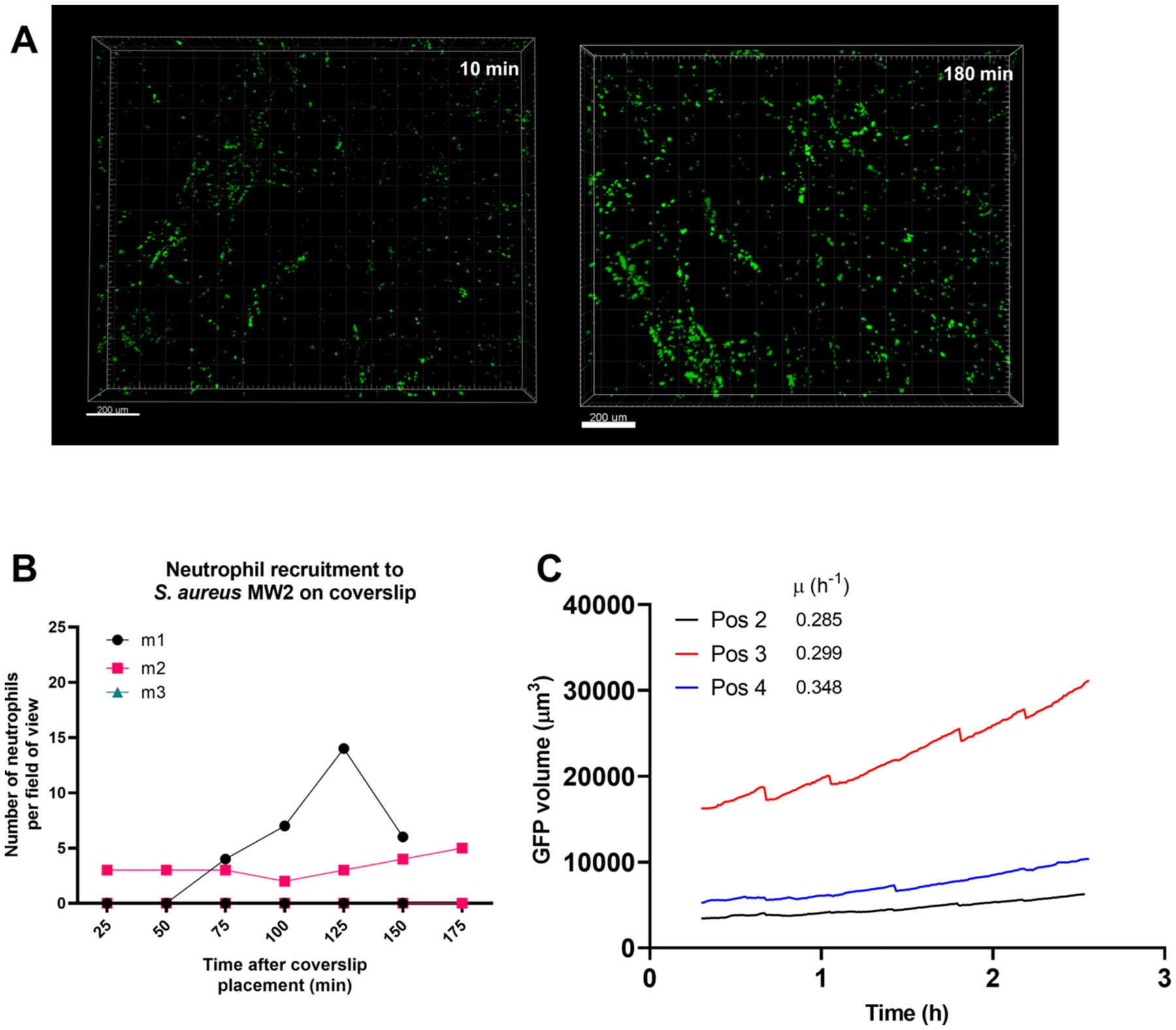
Minimal neutrophil recruitment to a S. aureus-contaminated coverslip in vivo
We adapted a murine skin intravital microscopy model [26, 30] to visualize in vivo neutrophil behavior on a coverslip inoculated with bacteria in real time using resonant-scanning multiphoton intravital microscopy. This design paralleled the in vitro experiments described above. Approximately 104 CFUs of S. aureus AH2547 GFP were seeded onto a glass coverslip and allowed to grow for 4 h to form aggregates (head start) and the contaminated coverslip was interfaced with the dermis. With the multiphoton system, dermal collagen was visualized via second harmonic generation (SHG), neutrophils were fluorescently labeled with tdTomato in the CatchupIVM-red reporter mouse [31] and S. aureus was visualized with GFP. Previously, it has been shown that neutrophils are robustly recruited to a localized S. aureus infection in the skin within 2 h [26], so we asked whether neutrophils were recruited in a similar timeline to S. aureus aggregates on a coverslip. Over the course of 180 min, there was minimal neutrophil recruitment to the S. aureus coverslip surface (Figures 5A–C, Video S3). In many FOVs, we observed zero or one neutrophil and the one neutrophil was often deeper in the collagen away from the S. aureus aggregates (Figure 5A). In the rare occurrence where we saw neutrophil recruitment (Figure 5B), we quantified how many neutrophils migrated towards the S. aureus aggregates within the top 20 μm of collagen (Figures 5D–E). We recorded three FOVs per mouse (from n = 4 independent experiments) and observed around 10 neutrophils migrate into the top 20 μm of collagen in only two of the twelve FOVs (Figure 5D), suggesting very few neutrophils are recruited to the S. aureus-contaminated coverslip in vivo during these early time points. Of those neutrophils that were able to migrate towards the coverslip surface, we further quantified how many associated with S. aureus aggregates (Figure 5F) and found again that few neutrophils were interacting with S. aureus (Figures 5F and 5G, Video S4). To confirm these results, we repeated the experiment using a clinically relevant methicillin-resistant S. aureus strain MW2 and again observed minimal neutrophil recruitment (Figure S3). Using single-cell S. aureus attached to the coverslip for 30 min (no head start), we observed no neutrophil recruitment to the coverslip over 180 min (Figure 5H). In many of the videos we occasionally observed one or two neutrophils, but this was transient as the neutrophils seemed to be passing by in deeper skin capillaries (data not shown). Importantly, there was no neutrophil recruitment to a sterile coverslip (Figure 5I). At the end of the 180 min of imaging, a fluorescent vascular dye was injected to visualize dermal blood vessels and capillaries within the dermal collagen (Figure 5J). This larger stitched image highlights that there was no neutrophil recruitment to a sterile coverslip, though we were able to detect one or two neutrophils within capillaries (Figure 5Ki–ii) and within larger dermal vessels (Figure 5Kiii).
Figure 5:
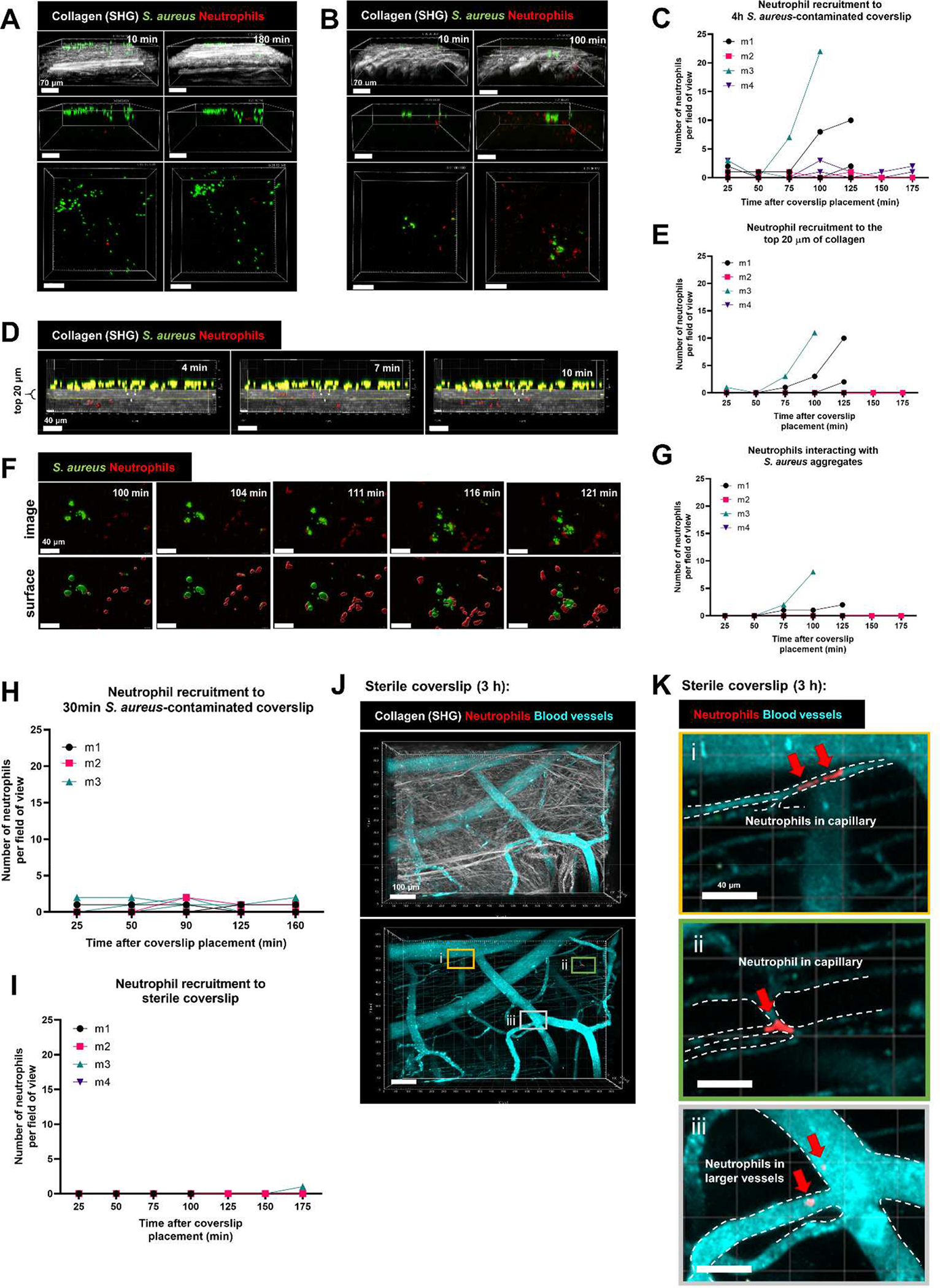
Minimal neutrophil recruitment to a S. aureus-contaminated coverslip in vivo. S. aureus aggregates were given 4 h to grow prior to placing the coverslip onto the dermal skin flap for intravital imaging. Representative 3D images of (A) minimal neutrophil recruitment and (B) moderate neutrophil recruitment at 10 min and 180 min after coverslip placement onto skin flap. Top: side view showing collagen (SHG) signal, middle: side view without collagen, bottom: top view without collagen. Scale bars = 70 μm. (C) Quantification of neutrophil recruitment to the 4 h S. aureus-contaminated coverslip. (D) Representative timelapse images of neutrophils (red spots) trafficking through the collagen into the top 20 μm. Scale bars = 40 μm. (E) Quantification of the number of neutrophils in the top 20 μm of collagen. (F) Representative timelapse images of neutrophils interacting with S. aureus aggregates at the coverslip surface over the course of 20 min. Scale bars = 40 μm. (G) Quantification of the number of neutrophils interacting with S. aureus. Quantification of (H) neutrophil recruitment to a single-cell 30-minute attached S. aureus-contaminated coverslip and (I) sterile coverslip. (J) Representative 3D stitched image of a sterile coverslip after 180 min of imaging, showing dermal collagen (white), blood vessels (cyan) and neutrophils (red). Bottom: identical 3D image without collagen. Scale bars = 100 μm. Three boxes (i, ii, iii) are selected to show one or two neutrophils in capillaries (K i, ii) and two neutrophils in larger dermal blood vessels (K iii). Scale bars = 40 μm. For C,E,G,H,I, n=3–4 mice from 3–4 independent experiments listed as m1, m2, m3, m4. Data show 3 fields of view per mouse.
Altogether, these data show that very few neutrophils are recruited to a S. aureus contaminated coverslip interfaced with the dermis over the course of 180 min in vivo. On the rare occasion where neutrophils migrated to the coverslip surface, they can interact with S. aureus aggregates, but the earliest this occurred was about 1–2 h after placing the coverslip onto the skin. In most FOVs, neutrophil recruitment was not observed over the imaging window of 180 min.
S. aureus aggregates grow on the coverslip in vivo
To understand the dynamics of bacterial growth in vivo, we measured the increase in surface volume of S. aureus aggregates over the course of imaging. It was clear that the bacterial aggregates became larger from the time the coverslip was added to the skin (Figure 6A) to the end of imaging at 180 min (Figure 6B). S. aureus aggregates were closely associated with the collagen surface (Figure 6C) suggesting they are in physical contact with the host dermal tissue. In vitro, S. aureus required supplementation of HBSS with serum to grow, demonstrating that host factors were required to support bacterial growth and that the superfusion buffer (HBSS) alone would not explain the observed growth (Fig. S1B and S1C). We measured the growth rate of S. aureus over the three hours of imaging and found the average growth rate to be 0.46 ± 0.45 h−1 (median 0.25 h−1) for strain AH2547, however significant heterogeneity was observed between FOVs (Figures 6D and 6E). Similar results were obtained with strain MW2 (Figures 6E, S3C). These data demonstrate that once the coverslip was placed in close contact with host tissue, bacteria started to grow and construct aggregate before neutrophils arrived.
Figure 6:
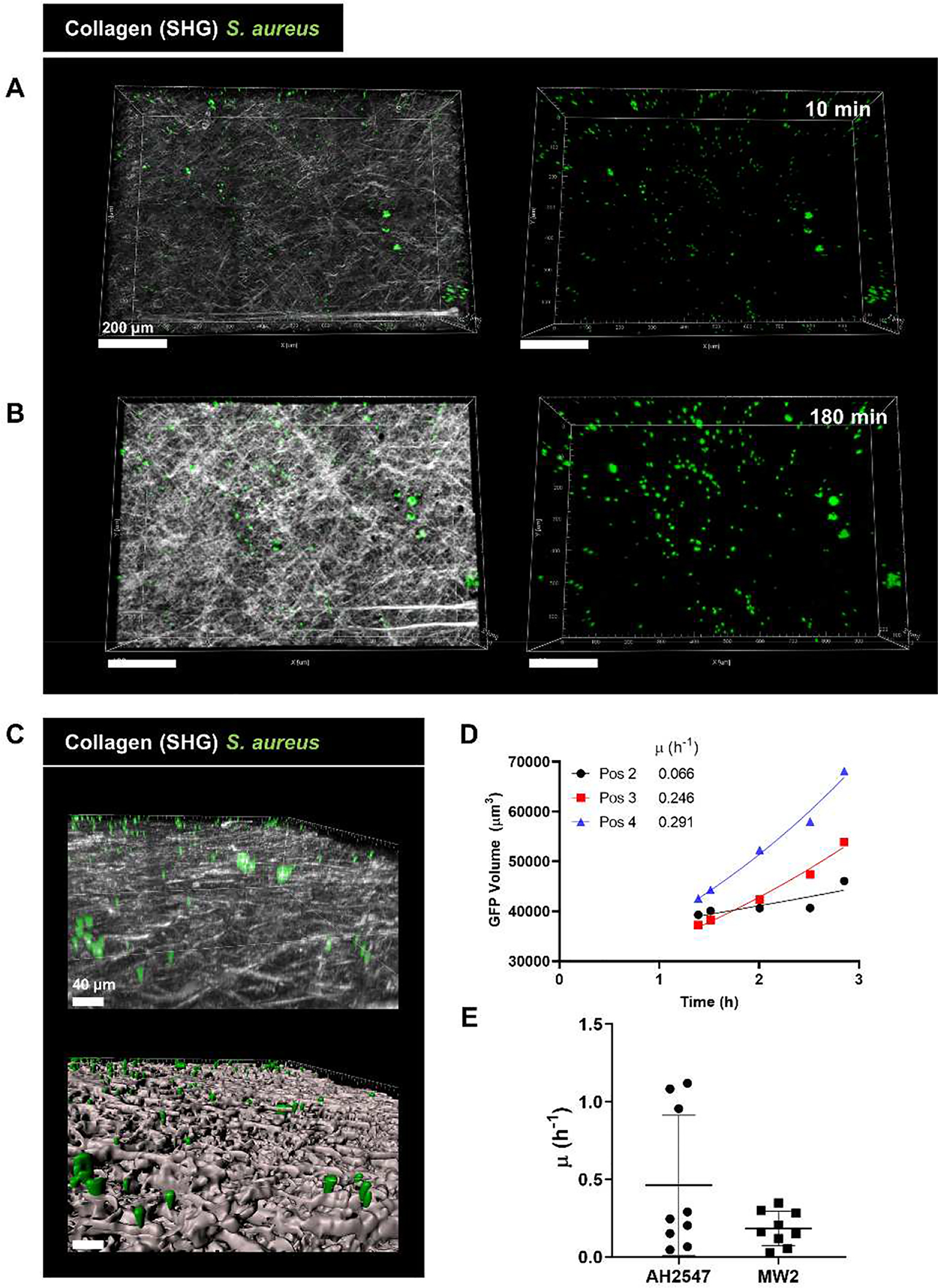
S. aureus aggregates grow on the coverslip in vivo. (A) Representative intravital image of a 3D region recorded at (A) start of imaging (10 min after coverslip addition) and (B) end of imaging (180 min after coverslip addition). Left: collagen (white) and S. aureus (green), right: S. aureus signal only. Scale bars = 200 μm. (C) 3D reconstruction of the coverslip surface coverslip showing GFP S. aureus in close contact with collagen. Top: 3D image, bottom: surface reconstruction with Imaris. Scale bars = 40 μm (D) Representative quantification of S. aureus growth in vivo over 180 min of imaging with an exponential curve fit to the data. Reported value μ represents the specific growth rate satisfying the equation X = X0eμt. Data shown includes representative points outside of lag phase from one mouse with three fields of view. (E) Growth rate values measured in vivo. Points represent value measured from a single FOV. Error bars indicate mean ± SD. (n=3 mice/condition with 3 FOV per mouse).
Recruited neutrophils discover some aggregates in vivo
Next, we asked whether neutrophils pre-recruited to the skin with the chemokine macrophage inflammatory protein 2 (MIP-2) would affect bacterial aggregate discovery and clearance. Mice were treated with MIP-2 4 h before the start of imaging (Figure 7A). In contrast to experiments without MIP-2 treatment (Figure 5A and 5B), many neutrophils were present in the dermis at the outset (Figure 7B and 7C). Over the course of imaging, we did not see a significant increase in the number neutrophils indicating that no additional neutrophils were recruited into the dermis during this time (Figure 7B, S4A and S4B). Within an individual animal, neutrophil recruitment to the coverslip was also heterogeneous where some fields of view had more neutrophils than other FOVs from the same mouse (Figures S4C and S4D, Video S5). Although we did see neutrophils enter the field of view during the 180 min window (Figure S4C and S4D), this was not due to an increase in total neutrophil recruitment (Figure 7B, S4A and S4B) but a change in neutrophil localization (Figure 7C). At 10 min many of the neutrophils were 50–100 μm away from the S. aureus aggregates but by 180 min, the neutrophils migrated towards the coverslip and were much closer to the aggregates (Figure 7C). Indeed, we observed a significant change in neutrophil localization as neutrophils migrated 50–100 μm towards the bacterial aggregates in the z-plane, which suggests that the existing neutrophils pre-recruited by MIP-2 addition were able to migrate towards S. aureus (Figures 7C and 7D). After a 30 min single-cell attachment, many neutrophils chemotaxed towards the S. aureus single cells and migrated very close to the coverslip interface with a mean distance of 26 μm away from the bacterial aggregates (Figure 7D). We next compared this to a head start experiment where we allowed S. aureus aggregates to grow for 4 h to the coverslip. Although neutrophils did migrate towards these larger aggregates, there was strikingly a significant decrease in neutrophil localization and these neutrophils were much further away from the aggregates, with a mean distance of 42 μm away (Figure 7D). The frequency of aggregate discovery by neutrophils was calculated using Imaris by applying a filter to the aggregate surface (Figure 7E, Video S6). When bacteria were only attached for 30 min, there was a significant increase in aggregate discovery between 10 min and 180 min (Figures 7F and 7G). At the 180 min time point, neutrophils more frequently discovered bacteria in the 30 minute attachment condition (45–75% discovery) compared to a 4 h biofilm (25–35% discovery) (Figures 7F, 7G, and S4E). Mice that were not treated with MIP-2 had negligible bacteria discovery (under 10% discovery) at the 180 min time point (Figures 7F, 7G and S4E). To identify any relationship between bacterial load and number of neutrophils recruited, we analyzed the trends of total bacterial surface volume at the 180 min time point against total number of neutrophils (Figure S4F). Interestingly, without MIP-2 pre-treatment we did not see a positive correlation between the number of neutrophils and the bacterial surface volume, suggesting that at these bacterial burdens and time points, increased bacterial biomass does not directly lead to increased neutrophil recruitment (Figure S4F).
Figure 7:
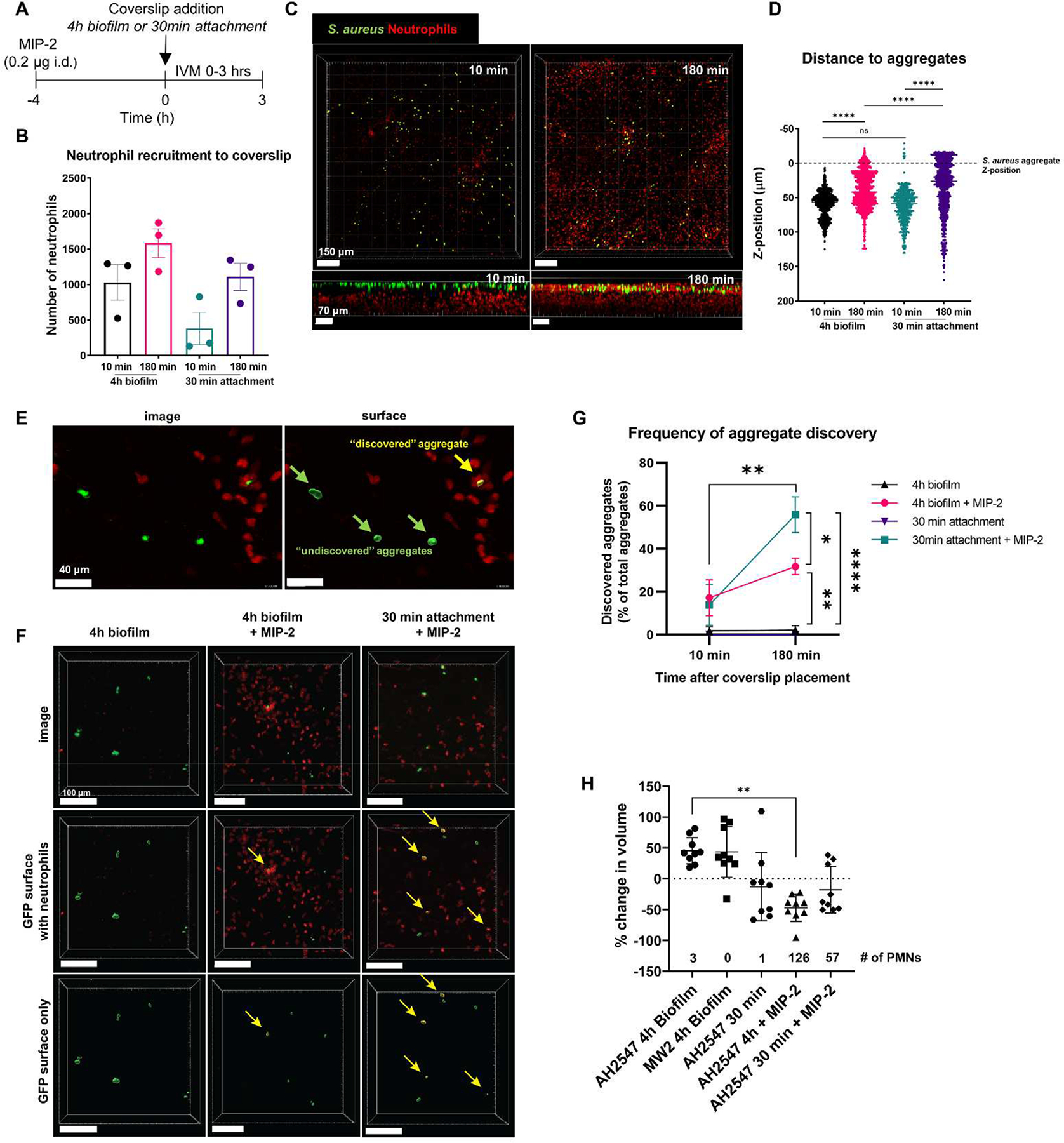
Pre-recruitment of neutrophils to the skin results in S. aureus aggregate discovery and increased clearance. (A) Experimental timeline for MIP-2 treatment and intravital microscopy. (B) Quantification of neutrophil recruitment at 10 min and 180 min after coverslip placement. Error bars indicate the mean ± SEM (n=3/condition, one-way ANOVA with Tukey’s post-hoc test). (C) Representative 3D stitched image showing neutrophil migration through the z-plane towards S. aureus on the coverslip at 10 min and 180 min after coverslip placement. Top panels show XY plane viewed from the top (scale bars = 150 μm), bottom panels show XZ plane viewed from the side (scale bars = 70 μm). (D) Quantification of neutrophil distance to S. aureus aggregates. Z-position was normalized to the averaged Z-position of S. aureus aggregates at each time point. Dashed line indicates the average z-position of S. aureus aggregates. Each dot represents one neutrophil. Data are mean ± SEM (n=3 mice/condition, one-way ANOVA p<0.0001, Tukey’s post hoc test **** p<0.0001). (E) Representative image showing one bacterial aggregate discovered by neutrophils and three other bacterial aggregates that remain undiscovered. Left: intravital image, right: surface reconstruction with a filter applied to label the discovered bacteria aggregate (yellow) and undiscovered aggregates (green). Scale bars = 40 μm. (F) Representative intravital images of aggregate discovery. Left: 4 h biofilm with no MIP-2 treatment. Middle: 4 h biofilm with MIP-2 treatment. Right: 30 min attachment with MIP-2 treatment. Yellow arrows point to discovered aggregates. Scale bars = 100 μm. (G) Quantification of aggregate discovery across the three conditions: 4 h biofilm, 4 h biofilm + MIP-2, 30 min attachment + MIP-2. Data show discovered aggregates as a percent of total aggregates at select time points 10 min and 180 min after coverslip placement. Data are mean ± SEM (n=3 mice/condition, Two-way RM ANOVA p<0.01, Tukey’s post hoc test *p<0.05, **p<0.01, **** p<0.0001 compared between groups at the indicated time point, Bonferroni’s multiple comparisons test **p<0.01 compared over time within the same group). (H) Percent change in GFP volume between the last frame and first frame for the final continuous FOV for each position. Numbers above the x-axis indicate the average number of neutrophils in the FOV at the end of imaging. Data mean ± SD (Each data point indicates one FOV, n=3 mice/condition, Kruskal-Wallis Test *p<0.05, ***p<0.001).
Overall, pre-recruitment of neutrophils to the skin with MIP-2 led to a marked reduction in S. aureus aggregate volume over the course of the experiment when S. aureus was given a head start to form aggregates (Figure 7H). S. aureus only given 30 min to form aggregates saw a slight but not statistically significant reduction in volume after MIP-2 treatment (Figure 7H). This demonstrates that neutrophils must be present in high densities in order to have an effective response to remove the contaminating bacteria. Due to the limited imaging window of 180 min, we never observed full clearance of bacteria from the coverslip even with pre-recruitment of neutrophils, and some bacterial aggregates were left undiscovered for the entire 180 min of imaging (Figure 7G).
Discussion
S. aureus is a major pathogen of implant-associated infections and serious complications can arise if the device is not surgically removed. Presently, it is unclear how small amounts of bacteria evade the host immune system and establish an infection. These early events in the interaction between host and pathogen are currently under-studied and we therefore investigated initial interactions between neutrophils and nascent Staphylococcus aureus biofilms in vitro and in vivo.
We hypothesized that successful clearance of contaminating bacteria from an implant surface requires that neutrophils are recruited to the surface, adhere to and patrol the surface, discover contaminating bacteria, and successfully phagocytose and kill the bacteria. We confirmed that neutrophils are able to kill surface attached S. aureus in vitro if a sufficient number of neutrophils are present on the surface and if S. aureus did not have time to form nascent biofilm aggregates. Deficiencies in neutrophil recruitment could permit biofilm formation in two ways: 1) Delayed recruitment could lead to the formation of neutrophil-tolerant biofilm aggregates prior to the appearance of a robust innate immune response, 2) An insufficient number of neutrophils “patrolling” an implanted surface could result in some of the attached S. aureus remaining undiscovered, again allowing aggregate formation. While we may expect in vivo to see directed recruitment of neutrophils to contaminating organisms due to chemotactic gradients produced by local cells that may initially discover contaminating microorganisms (macrophages, epithelial cells, etc.), we clearly observed in vivo that neutrophil recruitment can be highly heterogeneous. Thus, a high recruitment of neutrophils to an implanted surface is likely still important to enable full surface coverage and ensure that any contaminating microorganisms are destroyed promptly. Alternatively, during infection bacterial aggregates often exist in the peri-implant space and may not require direct neutrophil attachment to the surface for discovery. Robust recruitment to the peri-implant space to effectively discover these aggregates likely is still essential for clearance in this scenario.
We observed that S. aureus aggregates begin to demonstrate tolerance and the ability to lyse neutrophils at a relatively small aggregate size of approximately 50 μm2. Accordingly, it is plausible that S. aureus requires only a small amount of time, perhaps on the order of hours, prior to the arrival of host innate immune defenses in order to be able to mount a defense and tolerate neutrophil challenge. S. aureus aggregates appear to resist clearance by neutrophils both by the physical protection afforded by forming an aggregate and elaboration of virulence factors. Aggregates broken up by several neutrophils appeared to be slightly more susceptible to neutrophil killing, highlighting a benefit for the microorganisms of developing a structurally durable aggregate. After four hours of aggregate formation, S. aureus caused significant cytotoxic damage to interacting neutrophils in vitro, likely due to the production of pore-forming toxins such as γ-hemolysin or Panton-Valentine leukocidin, resulting in persistence of the bacteria [18, 19]. These behaviors are strikingly similar to examples of classical biofilm behavior that facilitates immune evasion [19, 22, 32]. However, here we have shown that tolerance to neutrophil clearance is occurring at a very early stage in biofilm development while previous work has primarily investigated immune cell interactions with mature in vitro biofilms. While these aggregates are quite small compared to a typical in vitro biofilm, aggregates of similar sizes have been reported in several types of human biofilm infections, such as implant associated infections [33]. These results suggest that contaminating S. aureus may be able to quickly establish a biofilm aggregate that can become relatively protected from early host immune defenses.
To investigate the dynamics of neutrophil recruitment in vivo, we utilized a peritoneal implant model and a dermal intravital imaging model to characterize and quantify leukocyte recruitment to contaminated surfaces. At 24 h after peritoneal implantation, we observed highly heterogenous recruitment of immune cells to a S. aureus contaminated silicone implant. While immune cells were strongly recruited to certain areas of the implant, SEM of seemingly bare areas only a few millimeters away revealed persistence of S. aureus cells. This observation suggests that discovery of bacteria on an implant surface could take longer than 24 h in some models, allowing a window for bacteria to grow and form more resilient aggregates. This mirrors our measurements in vitro (Figure 1D). We hypothesize that these leukocyte patches observed could develop in part due to the expression of chemotactic factors following neutrophil discovery and phagocytosis of bacteria or abiotic debris. This phenomenon has been described in other contexts as neutrophil swarming and is driven by LTB4, a molecule known to be produced by neutrophils [34]. LTB4-dependent neutrophil swarming was originally described during sterile injury, but recently neutrophil swarming was also observed during a S. aureus infection in the lymph node, indicating that neutrophils are capable of swarming in the presence of bacterial infection [35]. Indeed, we observed both in vitro and in vivo that on occasion once neutrophils encountered a S. aureus aggregate a neutrophil swarm formed (data not shown). The role of LTB4 in the control and clearance of S. aureus aggregates from a contaminated biomaterial is a mechanism we would like to pursue in future studies.
This study also used intravital microscopy to track the progression of S. aureus aggregate growth and neutrophil recruitment to the coverslip in vivo. First, we have demonstrated that once a contaminated coverslip is placed onto host tissue, bacteria are able to grow well before neutrophils can be recruited from the bloodstream. Robust neutrophil recruitment was not observed over 180 min of imaging, suggesting that a longer imaging window might be necessary to capture the onset of neutrophil recruitment in most cases. Although a previous S. aureus gel bead model resulted in neutrophil recruitment within 2 h [26], this disparity may have been due to a difference in dosage of bacteria (106 CFU/bead vs. 104 CFU/coverslip). It seems reasonable to assume that even the bacterial density utilized in our study would exceed the number of contaminating organisms present on an implant surface following surgery, suggesting delayed recruitment of neutrophils is plausible in human patients. Furthermore, differences in models (S. aureus bead vs. contaminated coverslip) could give rise to differences in neutrophil recruitment in vivo. We have also calculated the growth rate of S. aureus attached to a foreign object in vivo, which adds to the limited knowledge of in vivo bacterial growth rates reported. Since bacteria were able to grow significantly over the first 180 min, this indicates that early events are critical for bacteria to establish a protective niche.
We chose to use the chemokine MIP-2 as it is a potent chemoattractant to recruit neutrophils to sites of inflammation [27]. By pre-recruiting neutrophils, we were able to see aggregate discovery by neutrophils and a marked decrease in bacterial aggregate volume at the end of imaging, suggesting that neutrophils have the ability to clear contaminated surfaces in vivo if they reach the infected site quickly. However, future modification of the model will be necessary to follow up on this result, as we were unable to image for longer than three hours. It was also interesting that more neutrophils discovered aggregates that were only grown for 30 min compared to 4 h to form a biofilm, which highlights the importance of early bacterial discovery on contaminated implants before bacteria have the opportunity to form a biofilm.
Conclusion
In this work, we investigated the interactions between nascent S. aureus biofilms and human neutrophils in vitro and the recruitment dynamics of neutrophils in vivo. We observed a brief window of time where S. aureus aggregates are readily phagocytosed and killed by neutrophils if discovered, but these aggregates become more resilient once they exceed approximately 50 μm2. Leukocyte recruitment to a peritoneal implant was heterogeneous after 24 h and we observed many undiscovered bacteria remaining on the surface, suggesting that discovery of contaminating microbes can take up at least 24 h. We also observed in vivo with intravital microscopy that minimal numbers of neutrophils were recruited to a contaminated coverslip placed onto a dermal skin flap within 3 h. Even though pre-recruitment of neutrophils with MIP-2 resulted in a decrease in bacterial aggregate volume, some aggregates remained undiscovered by neutrophils, suggesting robust recruitment is required for sterilization.
Future studies should further investigate the time scales and distribution of neutrophil recruitment times to a contaminated surface in vivo and the potential roles of other immune cell types during the early stages of infection. Identification of mechanisms used by S. aureus to evade neutrophils through toxin production and aggregation could help to better inform pathogenesis and suggest therapeutic targets. Finally, our results raise the intriguing proposition of artificially “boosting” neutrophil recruitment near an implanted surface as a prophylactic measure and we aim to explore the viability of this treatment in a variety of clinically relevant models.
Supplementary Material
Figure S1: In vitro growth kinetics of S. aureus strain AH2547. (A) Individual S. aureus aggregate growth in 10% NHS is heterogeneous in vitro. Growth rate μ was calculated by fitting an exponential curve to volume measurements for continuously present aggregates in each field of view. (N = 302 individual aggregates measured from 8 FOVs from 4 independent experiments). (B) Log difference in GFP area for a given field of view from t = 0 to t = 6 h in Hank’s Balanced Salt Solution (HBSS) or HBSS with 10% normal human serum (NHS). Error bars indicate mean ± SD (N = 9 FOV per condition from 2 independent experiments. Unpaired t-test with Welch’s correction ****p<.0001). (C) FOV-averaged GFP area over time for S. aureus in HBSS or HBSS with 10% NHS. Error bars indicate mean ± SD (N = 9 FOVs per condition from 2 independent experiments).
Figure S2: S. aureus aggregates resist clearance by neutrophils through aggregation. (A) Initial aggregate area for cases where neutrophils fractured an aggregate into smaller pieces or when it was unable to break up the aggregate. Error bars indicate mean ± SD (N = 17 no fracture cases, n = 13 fracture cases, collected from 4 independent experiments. Mann-Whitney test ns = Not Significant). (B) Number of neutrophils interacting with a given aggregate over the 4 h observation period in cases where the aggregate did or did not fracture. Error bars indicate mean ± SD (N = 17 no fracture cases, n = 13 fracture cases, collected from 4 independent experiments. Mann-Whitney test ***p<.001). (C) Percent change in volume of an aggregate from the time of initial interaction with a neutrophil to the final frame where the aggregate is present. Percent change was calculated for aggregates that do not break up, the original aggregate when fracture occurs, the individual pieces broken off of a fractured aggregate, and the sum of the original fractured aggregate and its derivative pieces. Error bars indicate mean ± SD (N = 17 no fracture cases, n = 13 fracture cases from 4 independent experiments. Brown-Forsythe ANOVA with Dunnet’s T3 Multiple Comparisons test *p<.05, **p<.01).
Figure S3. Minimal neutrophil recruitment to S. aureus MW2 on a coverslip despite bacterial growth. (A) Representative intravital image of a 3D region taken at 10 min after coverslip addition to the mouse skin flap (left) and 180 min after coverslip addition (right). Scale bars = 200 μm. (B) Quantification of neutrophil recruitment to the 4 h S. aureus MW2-contaminated coverslip (n = 3 mice from 3 independent experiments, data show 3 fields of view per mouse). (C) Quantification of S. aureus MW2 growth in vivo over 180 min of imaging. Reported value μ represents the specific growth rate satisfying the equation X = X0eμt using data outside of lag phase (starting at 0.75 h). Data is representative from one mouse showing three fields of view.
Figure S4. Dynamics of neutrophil recruitment to the S. aureus-contaminated coverslip after MIP-2 treatment. Quantification of total neutrophils from 3D regions with (A) 4 h biofilm on coverslip and (B) 30 min attached S. aureus on coverslip showing changes in individual mice (n=3 per group). (C-D) Heterogeneity of neutrophil recruitment to the S. aureus coverslip with (C) 4 h biofilm or (D) 30 min attachment. Each line shows neutrophil recruitment from one field of view (3 fields of view per mouse, n=3 per group). (E) Frequency distribution of discovered S. aureus aggregates at 180 min post coverslip addition (n=3–4 per group). (F) Association between bacterial density and neutrophil recruitment at 180 min post coverslip addition (n=3–4 per group).
Video S1: Neutrophils (blue) that attempt to phagocytose large S. aureus (green) aggregates often stain with propidium iodide (red) indicating membrane damage (4 h).
Video S2: Neutrophils (red) can occasionally break up and disaggregate clusters of S. aureus (green), resulting in the destruction of the bacteria (4 h).
Video S3: Minimal neutrophil (red) recruitment to a S. aureus-contaminated coverslip (green) in vivo.
Video S4: Neutrophils (red) interacting with S. aureus-contaminated coverslip (green) in vivo.
Video S5: Pre-recruitment of neutrophils with MIP-2 to the skin still results in heterogenous neutrophil recruitment to the S. aureus coverslip.
Video S6: Frequency of aggregate discovery by neutrophils (red). Discovered aggregates are labeled yellow, undiscovered aggregates remain green.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 284:1318–1322. [DOI] [PubMed] [Google Scholar]
- [2].Gbejuade HO, Lovering AM, Webb JC. 2015. The role of microbial biofilms in prosthetic joint infections. Acta Orthop 86:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jacqueline C, Caillon J. 2014. Impact of bacterial biofilm on the treatment of prosthetic joint infections. J Antimicrob Chemother 69:i37–i40. [DOI] [PubMed] [Google Scholar]
- [4].Parsek MR, Singh PK. 2003. Bacterial Biofilms: An Emerging Link to Disease Pathogenesis. Annu Rev Microbiol 57:677–701. [DOI] [PubMed] [Google Scholar]
- [5].Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. 2003. Compromised Host Defense on Pseudomonas aeruginosa Biofilms: Characterization of Neutrophil and Biofilm Interactions. J Immunol 171:4329–4339. [DOI] [PubMed] [Google Scholar]
- [6].Stewart PS. 2015. Antimicrobial Tolerance in Biofilms. Microbiol Spectr 3: 10.1128/microbiolspec.MB-0010–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stewart PS, William Costerton J. 2001. Antibiotic resistance of bacteria in biofilms. The Lancet 358:135–138. [DOI] [PubMed] [Google Scholar]
- [8].Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S. 2013. National Burden of Invasive Methicillin-Resistant Staphylococcus aureus Infections, United States, 2011. JAMA Intern Med 173:1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kobayashi SD, DeLeo FR. 2009. An update on community-associated MRSA virulence. Curr Opin Pharmacol 9:545–551. [DOI] [PubMed] [Google Scholar]
- [10].Arciola CR, Campoccia D, Montanaro L. 2018. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol 16:397–409. [DOI] [PubMed] [Google Scholar]
- [11].Kourbatova EV, Halvosa JS, King MD, Ray SM, White N, Blumberg HM. 2005. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA 300 clone as a cause of health care-associated infections among patients with prosthetic joint infections. Am J Infect Control 33:385–391. [DOI] [PubMed] [Google Scholar]
- [12].Pulido L, Ghanem E, Joshi A, Purtill JJ, Parvizi J. 2008. Periprosthetic Joint Infection: The Incidence, Timing, and Predisposing Factors. Clin Orthop 466:1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R. 2007. Sonication of Removed Hip and Knee Prostheses for Diagnosis of Infection. N Engl J Med 357:654–663. [DOI] [PubMed] [Google Scholar]
- [14].Kurtz SM, Lau E, Watson H, Schmier JK, Parvizi J. 2012. Economic Burden of Periprosthetic Joint Infection in the United States. J Arthroplasty 27:61–65.e1. [DOI] [PubMed] [Google Scholar]
- [15].Senthi S, Munro JT, Pitto RP. 2011. Infection in total hip replacement: meta-analysis. Int Orthop 35:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kapadia BH, McElroy MJ, Issa K, Johnson AJ, Bozic KJ, Mont MA. 2014. The Economic Impact of Periprosthetic Infections Following Total Knee Arthroplasty at a Specialized Tertiary-Care Center. J Arthroplasty 29:929–932. [DOI] [PubMed] [Google Scholar]
- [17].Puhto T, Puhto A-P, Vielma M, Syrjälä H. 2019. Infection triples the cost of a primary joint arthroplasty. Infect Dis 51:348–355. [DOI] [PubMed] [Google Scholar]
- [18].Guerra FE, Borgogna TR, Patel DM, Sward EW, Voyich JM. 2017. Epic Immune Battles of History: Neutrophils vs. Staphylococcus aureus. Front Cell Infect Microbiol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bhattacharya M, Berends ETM, Chan R, Schwab E, Roy S, Sen CK, Torres VJ, Wozniak DJ. 2018. Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proc Natl Acad Sci 201721949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sultan AR, Swierstra JW, Toom NAL, Snijders SV, Maňásková SH, Verbon A, Wamel WJB van. 2018. Production of Staphylococcal Complement Inhibitor (SCIN) and Other Immune Modulators during the Early Stages of Staphylococcus aureus Biofilm Formation in a Mammalian Cell Culture Medium. Infect Immun IAI. 00352–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guggenberger C, Wolz C, Morrissey JA, Heesemann J. 2012. Two Distinct Coagulase-Dependent Barriers Protect Staphylococcus aureus from Neutrophils in a Three Dimensional in vitro Infection Model. PLOS Pathog 8:e1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Günther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, Wagner C, Hänsch GM. 2009. Host defence against Staphylococcus aureus biofilms infection: Phagocytosis of biofilms by polymorphonuclear neutrophils (PMN). Mol Immunol 46:1805–1813. [DOI] [PubMed] [Google Scholar]
- [23].Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus Biofilms Prevent Macrophage Phagocytosis and Attenuate Inflammation In Vivo. J Immunol 186:6585–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ghimire N, Pettygrove BA, Pallister KB, Stangeland J, Stanhope S, Klapper I, Voyich JM, Stewart PS. 2019. Direct Microscopic Observation of Human Neutrophil-Staphylococcus aureus Interaction In Vitro Suggests a Potential Mechanism for Initiation of Biofilm Infection on an Implanted Medical Device. Infect Immun 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alhede M, Lorenz M, Fritz BG, Jensen PØ, Ring HC, Bay L, Bjarnsholt T. 2020. Bacterial aggregate size determines phagocytosis efficiency of polymorphonuclear leukocytes. Med Microbiol Immunol 209:669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Harding MG, Zhang K, Conly J, Kubes P. 2014. Neutrophil Crawling in Capillaries; A Novel Immune Response to Staphylococcus aureus. PLOS Pathog 10:e1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P. 2010. Intravascular Danger Signals Guide Neutrophils to Sites of Sterile Inflammation. Science 330:362–366. [DOI] [PubMed] [Google Scholar]
- [28].Bjarnsholt T, van Gennip M, Jakobsen TH, Christensen LD, Jensen PØ, Givskov M. 2010. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. 2. Nat Protoc 5:282–293. [DOI] [PubMed] [Google Scholar]
- [29].van Gennip M, Christensen LD, Alhede M, Qvortrup K, Jensen PØ, Høiby N, Givskov M, Bjarnsholt T. 2012. Interactions between Polymorphonuclear Leukocytes and Pseudomonas aeruginosa Biofilms on Silicone Implants In Vivo. Infect Immun 80:2601–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yipp BG, Petri B, Salina D, Jenne CN, Scott BNV, Zbytnuik LD, Pittman K, Asaduzzaman M, Wu K, Meijndert HC, Malawista SE, de Boisfleury Chevance A, Zhang K, Conly J, Kubes P. 2012. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. 9. Nat Med 18:1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hasenberg A, Hasenberg M, Männ L, Neumann F, Borkenstein L, Stecher M, Kraus A, Engel DR, Klingberg A, Seddigh P, Abdullah Z, Klebow S, Engelmann S, Reinhold A, Brandau S, Seeling M, Waisman A, Schraven B, Göthert JR, Nimmerjahn F, Gunzer M. 2015. Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat Methods 12:445–452. [DOI] [PubMed] [Google Scholar]
- [32].Jensen PØ, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L, Moser C, Williams P, Pressler T, Givskov M, Høiby N. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology, 153:1329–1338. [DOI] [PubMed] [Google Scholar]
- [33].Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, Høiby N. 2013. The in vivo biofilm. Trends Microbiol 21:466–474. [DOI] [PubMed] [Google Scholar]
- [34].Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, Germain RN. 2013. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. 7454. Nature 498:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bogoslowski A, Butcher EC, Kubes P. 2018. Neutrophils recruited through high endothelial venules of the lymph nodes via PNAd intercept disseminating Staphylococcus aureus. Proc Natl Acad Sci 115:2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pabst B, Pitts B, Lauchnor E, Stewart PS. 2016. Gel-Entrapped Staphylococcus aureus Bacteria as Models of Biofilm Infection Exhibit Growth in Dense Aggregates, Oxygen Limitation, Antibiotic Tolerance, and Heterogeneous Gene Expression. Antimicrob Agents Chemother 60:6294–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Surewaard BGJ, Haas CJC de, Vervoort F, Rigby KM, DeLeo FR, Otto M, Strijp JAG van, Nijland R. 2013. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol 15:1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Saïd-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. 2005. Insights into Mechanisms Used by Staphylococcus aureus to Avoid Destruction by Human Neutrophils. J Immunol 175:3907–3919. [DOI] [PubMed] [Google Scholar]
- [39].Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR, Whitney AR, Otto M, DeLeo FR. 2009. The SaeR/S Gene Regulatory System Is Essential for Innate Immune Evasion by Staphylococcus aureus. J Infect Dis 199:1698–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Christensen LD, Moser C, Jensen PØ, Rasmussen TB, Christophersen L, Kjelleberg S, Kumar N, Høiby N, Givskov M, Bjarnsholt T. 2007. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153:2312–2320. [DOI] [PubMed] [Google Scholar]
- [41].Cavanagh JP, Granslo HN, Fredheim EA, Christophersen L, Jensen PØ, Thomsen K, Van Gennip M, Klingenberg C, Flaegstad T, Moser C. 2013. Efficacy of a synthetic antimicrobial peptidomimetic versus vancomycin in a Staphylococcus epidermidis device-related murine peritonitis model. J Antimicrob Chemother 68:2106–2110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: In vitro growth kinetics of S. aureus strain AH2547. (A) Individual S. aureus aggregate growth in 10% NHS is heterogeneous in vitro. Growth rate μ was calculated by fitting an exponential curve to volume measurements for continuously present aggregates in each field of view. (N = 302 individual aggregates measured from 8 FOVs from 4 independent experiments). (B) Log difference in GFP area for a given field of view from t = 0 to t = 6 h in Hank’s Balanced Salt Solution (HBSS) or HBSS with 10% normal human serum (NHS). Error bars indicate mean ± SD (N = 9 FOV per condition from 2 independent experiments. Unpaired t-test with Welch’s correction ****p<.0001). (C) FOV-averaged GFP area over time for S. aureus in HBSS or HBSS with 10% NHS. Error bars indicate mean ± SD (N = 9 FOVs per condition from 2 independent experiments).
Figure S2: S. aureus aggregates resist clearance by neutrophils through aggregation. (A) Initial aggregate area for cases where neutrophils fractured an aggregate into smaller pieces or when it was unable to break up the aggregate. Error bars indicate mean ± SD (N = 17 no fracture cases, n = 13 fracture cases, collected from 4 independent experiments. Mann-Whitney test ns = Not Significant). (B) Number of neutrophils interacting with a given aggregate over the 4 h observation period in cases where the aggregate did or did not fracture. Error bars indicate mean ± SD (N = 17 no fracture cases, n = 13 fracture cases, collected from 4 independent experiments. Mann-Whitney test ***p<.001). (C) Percent change in volume of an aggregate from the time of initial interaction with a neutrophil to the final frame where the aggregate is present. Percent change was calculated for aggregates that do not break up, the original aggregate when fracture occurs, the individual pieces broken off of a fractured aggregate, and the sum of the original fractured aggregate and its derivative pieces. Error bars indicate mean ± SD (N = 17 no fracture cases, n = 13 fracture cases from 4 independent experiments. Brown-Forsythe ANOVA with Dunnet’s T3 Multiple Comparisons test *p<.05, **p<.01).
Figure S3. Minimal neutrophil recruitment to S. aureus MW2 on a coverslip despite bacterial growth. (A) Representative intravital image of a 3D region taken at 10 min after coverslip addition to the mouse skin flap (left) and 180 min after coverslip addition (right). Scale bars = 200 μm. (B) Quantification of neutrophil recruitment to the 4 h S. aureus MW2-contaminated coverslip (n = 3 mice from 3 independent experiments, data show 3 fields of view per mouse). (C) Quantification of S. aureus MW2 growth in vivo over 180 min of imaging. Reported value μ represents the specific growth rate satisfying the equation X = X0eμt using data outside of lag phase (starting at 0.75 h). Data is representative from one mouse showing three fields of view.
Figure S4. Dynamics of neutrophil recruitment to the S. aureus-contaminated coverslip after MIP-2 treatment. Quantification of total neutrophils from 3D regions with (A) 4 h biofilm on coverslip and (B) 30 min attached S. aureus on coverslip showing changes in individual mice (n=3 per group). (C-D) Heterogeneity of neutrophil recruitment to the S. aureus coverslip with (C) 4 h biofilm or (D) 30 min attachment. Each line shows neutrophil recruitment from one field of view (3 fields of view per mouse, n=3 per group). (E) Frequency distribution of discovered S. aureus aggregates at 180 min post coverslip addition (n=3–4 per group). (F) Association between bacterial density and neutrophil recruitment at 180 min post coverslip addition (n=3–4 per group).
Video S1: Neutrophils (blue) that attempt to phagocytose large S. aureus (green) aggregates often stain with propidium iodide (red) indicating membrane damage (4 h).
Video S2: Neutrophils (red) can occasionally break up and disaggregate clusters of S. aureus (green), resulting in the destruction of the bacteria (4 h).
Video S3: Minimal neutrophil (red) recruitment to a S. aureus-contaminated coverslip (green) in vivo.
Video S4: Neutrophils (red) interacting with S. aureus-contaminated coverslip (green) in vivo.
Video S5: Pre-recruitment of neutrophils with MIP-2 to the skin still results in heterogenous neutrophil recruitment to the S. aureus coverslip.
Video S6: Frequency of aggregate discovery by neutrophils (red). Discovered aggregates are labeled yellow, undiscovered aggregates remain green.
Data Availability Statement
The raw and processed data required to reproduce these findings are available to download from https://datadryad.org/stash/share/WHrJoeXSaqg3jdFH9pOPwUlablkGwl2RVvFePLu3fc4.


