Abstract
Skeletal muscle stem cells (MuSCs) are essential for efficacious muscle repair, making MuSCs promising therapeutic targets for tissue engineering and regenerative medicine. MuSCs are presented with a diverse and temporally defined set of cues from their microenvironment during regeneration that direct stem cell expansion, differentiation, and return to quiescence. Understanding the complex interplay among these biophysical and biochemical cues is necessary to develop therapies targeting or employing MuSCs. To probe the role of mechanical cues presented by the extracellular matrix, we leverage chemically defined hydrogel substrates with controllable stiffness and adhesive ligand composition to characterize the MuSC response to matrix cues presented during early and late phases of regeneration. We demonstrate that relatively soft hydrogels recapitulating healthy muscle stiffness promote MuSC activation and expansion, while relatively stiff hydrogels impair MuSC proliferation and arrest myogenic progression. These effects are seen on soft and stiff hydrogels presenting laminin-111 and exacerbated on hydrogels presenting RGD adhesive peptides. Soluble factors present in the MuSC niche during different phases of regeneration, prostaglandin E2 and oncostatin M, synergize with matrix-presented cues to enhance stem cell expansion on soft substrates and block myogenic progression on stiff substrates. To determine if temporally varied matrix stiffness reminiscent of the regenerating microenvironment alters MuSC fate, we developed a photoresponsive hydrogel system with accelerated reaction kinetics that can be rapidly softened on demand. MuSCs cultured on these materials revealed that the cellular response to a stiff microenvironment is fixed within the first three days of culture, as subsequent softening back to a healthy stiffness did not rescue MuSC proliferation or myogenic progression. These results highlight the importance of temporally controlled biophysical and biochemical cues in regulating MuSC fate that can be harnessed to improve regenerative medicine approaches to restore skeletal muscle tissue.
1. Introduction
Skeletal muscle stem cells (MuSCs), also known as satellite cells, inhabit a specialized niche between the plasma membrane of skeletal muscle fibers and the basal lamina surrounding the fibers.1,2 MuSCs normally exist in a quiescent state but activate rapidly upon injury to repair damaged muscle tissue.3 The central role of MuSCs in muscle regeneration has made them promising therapeutic targets for conditions spanning congenital muscular dystrophies, volumetric muscle loss, and aging-related muscle wasting (sarcopenia).4,5 Many proposed regenerative therapies require ex vivo manipulation of MuSCs, such as gene editing to correct mutations underlying muscular dystrophies or expansion and differentiation to generate engineered muscle tissues to replace lost tissue.6–8 To advance these therapeutic goals, an improved understanding of how microenvironmental cues regulate MuSC fate is required to ensure that functional stem cells are delivered to patients.
In the process of muscle regeneration, MuSCs experience a defined sequence of biochemical and biophysical cues to orchestrate stem cell activation, differentiation, and return to quiescence.2 Shortly after injury, soluble factors such as the pro-inflammatory metabolite prostaglandin E2 (PGE2) and hepatocyte growth factor (HGF) trigger activation and expansion of resident MuSCs that subsequently differentiate and fuse to repair damaged muscle fibers.9,10 To maintain the stem cell pool following regeneration, factors such as oncostatin M (OSM), Wnt7a, and collagen V play a critical role in promoting self-renewal and a return to quiescence.11–13
Biophysical cues provided by the extracellular matrix (ECM) also vary throughout the regeneration process. Injury results in a transient stiffening of the skeletal muscle tissue. For instance, one prior study reported that the elastic modulus of bulk mouse muscle tissue increased over the first three days following barium chloride injury and then subsequently returned to baseline.14 More recent studies investigating changes in local ECM mechanics within the MuSC niche suggest that microenvironmental stiffening occurs later during regeneration, beginning around 7–10 days post-injury.15,16 Concurrently, the cell-adhesive protein components of the ECM through which MuSCs sense matrix stiffness also change in abundance during regeneration. For example, early regeneration involves increased laminin-α1 production,17 while later phases of regeneration exhibit increased fibronectin deposition.18
We previously reported that MuSCs are acutely sensitive to microenvironmental stiffness, exhibiting optimal proliferation in vitro and engraftment in vivo after culture on substrates mimicking the stiffness of healthy skeletal muscle (E~12 kPa).19 Others have also reported that myoblasts, proliferative progenitor cells derived from MuSCs, are sensitive to the stiffness of cell culture substrates. Proliferation varies as a function of stiffness20 and optimal differentiation into myotubes is observed on substrates with stiffness similar to native tissue.21,22 We therefore hypothesized that mimicking the changes in mechanical properties of the ECM during muscle regeneration would alter MuSC fate in vitro.
While most engineered platforms present cells with a fixed stiffness, the stiffness of muscle tissue changes during regeneration.14 In a seminal study, Kloxin, et al. developed cell culture substrates with dynamically-tunable stiffness using hydrogels containing photocleavable ortho-nitrobenzyl (oNB) ester moieties that enabled controlled softening of the hydrogels by exposure to UV light.23 Similar oNB functionalities have been employed in other hydrogel systems to study how mesenchymal stromal cells respond to changes in matrix mechanics.24,25 One limitation of these studies is the use of relatively high energy 365 nm light to trigger hydrogel softening. While short-term exposure to low doses of 365 nm light has been shown to induce minimal cytotoxicity, longer exposure can cause DNA damage and cell death.26,27 Therefore, use of longer wavelength, lower energy light is desirable to minimize unintended perturbations. Fortunately, certain oNB esters, such as those used in these previous studies, can be cleaved with 405 nm light, albeit at a slower rate than with 365 nm exposure.23 The development of new oNB ester crosslinks that cleave more rapidly upon exposure to 405 nm light would thus enhance the bioselectivity of these approaches to tune material stiffness on demand and explore the reversibility of mechanically induced cell fate changes.
Here we leverage the control afforded by synthetic poly(ethylene glycol) (PEG) hydrogels crosslinked by the bioorthogonal strain-promoted azide-alkyne cycloaddition (SPAAC) reaction to present MuSCs with well-controlled biophysical cues mimicking the native regeneration process. PEG-based hydrogels crosslinked by step-growth polymerization are known to possess nearly ideal network connectivity,28–30 resulting in uniform presentation of mechanical cues to cells. Furthermore, the highly selective nature of the SPAAC reaction facilitates incorporation of cell-adhesive cues, including full-length ECM proteins and adhesive peptides, to mimic changes in ECM composition. We introduce a new oNB photocleavable crosslinker that is compatible with SPAAC crosslinking while cleaving ~5x faster in response to 405 nm light exposure, enabling rapid, cytocompatible softening of the hydrogels on demand to model the decrease in bulk tissue stiffness occurring during regeneration. Finally, we demonstrate that treatment of MuSCs on the engineered microenvironments with soluble factors mimicking the inflammatory (PGE2) and self-renewal (OSM) phases of regeneration synergizes with biophysical cues presented by the matrix to modulate MuSC fate. Our results identify optimal conditions for promoting proliferation of stem-like MuSCs while uncovering an unexpected temporal role of substrate stiffness in modulating MuSC activation.
2. Materials and Methods
2.1. Materials
Chemical precursors and solvents were purchased from Sigma-Aldrich, Fisher Scientific, or Acros Organics and used without further purification, unless otherwise noted. PEG precursors were purchased from JenKem Technology USA. Cell culture reagents were purchased from Thermo Fisher Scientific, unless otherwise noted.
2.2. Synthesis of hydrogel precursors
Detailed synthetic protocols are provided in the supporting information. The amine-reactive bicyclo[6.1.0]nonyne precursor BCN-NHS was prepared following known synthetic routes.31–33 The methyl substituted oNB crosslinker N3-Me-oNB-NHS (compound 8a) was synthesized following previously published protocols starting from acetovanillone.33,34 For synthesis of the tert-butyl substituted oNB crosslinker N3-tBu-oNB-NHS (compound 8b), 1-(4-hydroxy-3-methoxyphenyl)-2,2-dimethylpropan-1-one (compound 2) was prepared from o-dimethoxybenzene following a known synthetic route.35 Compound 2 was then used to synthesize N3-tBu-oNB-NHS by adapting the synthetic protocols used to synthesize N3-Me-oNB-NHS.33,34 Azide-functionalized multi-arm PEG macromers were prepared from 8-arm PEG-amine (MW~10 kDa or 20 kDa) by reacting with activated NHS ester azides under standard amide coupling conditions.31,36 To improve solubility and prevent polymer aggregation, 4-arm PEG-amines used to prepare BCN-functionalized macromers were first sulfated by reacting with Boc-L-cysteic acid followed by deprotection with trifluoroacetic acid.37 The 4-arm PEG-sulfo-amines were subsequently reacted with BCN-NHS under standard amide coupling conditions to afford 4-arm PEG-sulfo-BCN.31,36 PEG macromers were dialyzed against MilliQ-grade water and lyophilized prior to use. Azide-functionalized laminin-111 was prepared by adapting our previous protocols for laminin functionalization.19 Laminin was first dialyzed (MWCO~10 kDa) into sterile PBS and then reacted with a 50-fold molar excess of N3-PEG-NHS (MW~3.5 kDa) for 6 hours at room temperature. The reaction mixture was further dialyzed against sterile PBS to remove unreacted N3-PEG.
2.3. Rheological analysis
Multi-arm PEG precursors were separately dissolved in PBS to the desired polymer concentration. Azide- and BCN-containing macromers were mixed, immediately added to cylindrical silicone molds (8 mm diameter × 0.8 mm thickness), and incubated at 37°C for 30 minutes. The resulting hydrogels were transferred to PBS and allowed to equilibrate for at least 1 hour before mechanical testing. For dynamically softening hydrogels, following equilibration, the hydrogels were exposed to 405 nm light (~600 mW/cm2) for the desired time and then further allowed to equilibrate in PBS for an additional hour. Oscillatory rheology was performed on a ARG2 rheometer using a flat 8 mm geometry. Frequency sweeps from 0.1 to 10 Hz with 5% strain were collected for each sample, and the shear modulus (G’) was taken to be the average storage modulus over the linear range centered at 1 Hz, following our established protocol.19 Shear moduli were converted to Young’s moduli (E) by assuming that the PEG hydrogels are incompressible (i.e., Poisson’s ratio is ~0.5).
2.4. Reaction kinetics characterization
For analysis of solution phase reaction kinetics, multi-arm PEGs functionalized with methyl and tert-butyl oNB esters were separately dissolved in PBS to 20 mg/mL to achieve a similar concentration to the amount of oNB in the hydrogels. The solutions were exposed to 405 nm light (~600 mW/cm2), and aliquots were removed at predetermined time intervals. These aliquots were further diluted 20:1 in PBS to ensure a linear absorbance vs. concentration profile, and absorbance was measured using a NanoDrop 2000 microvolume spectrophotometer. Reaction progress was quantified using the normalized absorbance values at 275 nm.
2.5. Muscle stem cell isolation
All animal protocols were approved by the Stanford University Administrative Panel on Laboratory Animal Care (APLAC) and experiments were performed in compliance with the institutional guidelines of Stanford University. Myogenin reporter mice were generated by breeding Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat mice harboring the floxP-STOP-floxP Sun1-GFP construct (JAX #021039)38 with Myogtm1.1(cre/ERT2)Lepr mice expressing Cre-ERT2 under control of the myogenin promoter (JAX #025668).39 Mice heterozygous for both alleles were used for all experiments. To mark myogenic cells that were already committing in vivo and exclude these cells from hydrogel experiments, mice were treated with tamoxifen by intraperitoneal injection of 100 μL of a 20 mg/mL solution daily for 3 days, followed by a recovery period of at least 3 weeks. Muscle stem cells were then isolated from the dissected hindlimb muscles of the reporter mice following our published procedures.19,40 Briefly, muscles were digested with collagenase and dispase, mechanically dissociated with a gentleMACS Octo Dissociator, and passed through a 45 μm cell strainer. The cell suspension was then incubated with PE-Cy7 conjugated antibodies against CD45, CD11b, CD31, and Sca1 (BD Biosciences; lineage markers, Lin) to label non-muscle cell types, in addition to fluorescently tagged antibodies to label MuSCs: integrin-α7-PE (Ablab) and CD34-Alexa647 (BD Biosciences). The cell suspension was sorted by FACS to deplete non-muscle lineage cells and enrich MuSCs (Lin−/integrin-α7+/CD34+/GFP−).
2.6. Muscle stem cell culture and analysis
PEG hydrogels for MuSC culture were cast directly in glass bottom 24-well plates (Cellvis). The bottoms of the wells were first functionalized with azides by incubating with a solution of 95:5:0.5 ethanol/acetic acid/(3-azidopropyl)trimethoxy silane31 for 10 minutes, washing extensively with ethanol, and drying in a biosafety cabinet. PEG precursors were dissolved in PBS and sterilized by filtration through a 0.22 μm filter. Working on ice to slow gelation, PEG-sulfo-BCN and either laminin-PEG-N3 (100 μg/mL final concentration) or cycloRGD-N3 (500 μM final concentration) were mixed together, followed by addition of azide-functionalized PEGs. The crosslinking PEG solution (200 μL/well) was pipetted into the well plate, and the plate was then centrifuged at 2000×g for 10–15 minutes at room temperature to uniformly coat the well surface. The plates were incubated for 30 minutes at 37°C to complete crosslinking and then equilibrated in cell culture medium at 37°C prior to cell seeding. Freshly isolated MuSCs were immediately seeded onto hydrogels at a density of 2000 cells/well. To facilitate live cell fluorescence microscopy, MuSCs were cultured in FluoroBrite DMEM supplemented with 15% fetal bovine serum (FBS), 1x L-glutamine, 1x sodium pyruvate, 1x nonessential amino acids, 1x penicillin/streptomycin, and 2.5 ng/mL FGF-2. The medium was supplemented daily with 4-hydroxytamoxifen (1 μM) to enable Cre-mediated recombination and expression of the Sun1-GFP reporter in committing MuSCs. Media was replaced on days 3 and 5. For soluble factor experiments, cells were treated with either DMSO vehicle or PGE2 (10 ng/mL) at the time of seeding and then either fresh media or media containing OSM (100 ng/mL) on days 3 and 5 of culture. For dynamic hydrogel softening experiments, stiff controls and oNB containing gels were exposed to 405 nm light (~300 mW/cm2) for two minutes. Live cell images were collected on days 3 and 7 on a Zeiss AxioObserver inverted microscope with a motorized scanning stage and environmental stage incubator, collecting a 7×7 tiled region near the center of the gels with 20x magnification. Images were stitched using a custom GPU-accelerated image processing pipeline, which allows for highly accurate and performant flatfield correction, high-dimensional image alignment/registration, deconvolution, stitching, chromatic aberration correction, and background subtraction.41
2.7. Immunofluorescence analysis of MuSC fate
After 7 days of culture on hydrogels, MuSCs were fixed with 4% paraformaldehyde in PBS. Cells were permeabilized with PBS plus 0.25% (v/v) Triton X-100 (PBST), blocked with 5% bovine serum albumin (BSA) and 5% goat serum (GS) in PBST, and incubated with primary antibodies against Pax7 (Developmental Studies Hybridoma Bank, 2 μg/mL) and MyoD (Santa Cruz Biotechnology, clone G-1, 1:200) diluted in 2.5% BSA and 2.5% GS in PBST. Samples were washed with PBST and then incubated with secondary antibodies (Cy3 goat anti-mouse IgG2b and AF647 goat anti-mouse IgG1, Jackson ImmunoResearch, 1:500) diluted in 2.5% BSA and 2.5% GS in PBST. Samples were washed with PBST and imaged on a Zeiss AxioObserver inverted microscope equipped with a 7-channel ZEISS Colibri 7 light source for multichannel fluorescence imaging. Tiled image regions were collected near the center of the gels and were stitched into composite images using our custom image processing pipeline described above.
2.8. Analysis of MuSC Viability
For live/dead viability assays, MuSCs derived from wild-type (GFP-) mice were isolated and cultured on hydrogels as described above. At days 3 and 7, MuSCs were treated with calcein-AM (1 μM) and ethidium homodimer (1 μM) diluted in FluoroBrite DMEM supplemented as above for 30 minutes prior to fluorescence imaging. To determine the fraction of apoptotic cells, MuSCs were fixed with 4% paraformaldehyde in PBS and immunostained as described above, using a primary antibody against cleaved caspase-3 (Cell Signaling Technology, clone 5A1E, 1:400) and an AF647 goat anti-rabbit secondary antibody (Jackson ImmunoResearch, 1:500).
2.9. Generation of fluorescent reporter fibroblast cell line
MRC-5 human fetal lung fibroblasts and HEK 293T cells were purchased from ATCC. For routine culture, both MRC-5s and HEK 293Ts were cultured in high-glucose DMEM, supplemented with 10% FBS and 1x penicillin/streptomycin and passaged by trypsinization. Lentiviral expression plasmids encoding H2B-GFP with puromycin resistance (plasmid #69550) and LifeAct-mRuby2 with blasticidin resistance (plasmid #84384)42 were purchased from Addgene. Lentivirus was packaged by transfecting a 10 cm2 dish of HEK 293Ts with one of the fluorescent reporter plasmids (3.83 μg) and the packaging plasmids Delta8.9 (2.87 μg) and PMD1G (1.91 μg) using Fugene6 transfection reagent (Promega). Viral supernatants were collected after 48 and 72 hours and filtered (0.4 μm) to remove cell debris. The unconcentrated supernatants were diluted 1:1 with fresh media to transduce MRC-5s. To produce the dual reporter cells, MRC-5s were first transduced with the LifeAct-mRuby2 reporter and selected with blasticidin (4 μg/mL). The cells were then subsequently transduced with the H2B-GFP reporter and selected with puromycin (Invivogen, 1 μg/mL).
2.10. Time lapse microscopy on softening hydrogels
For time lapse microscopy, hydrogels were fabricated within glass bottom 12-well plates (Cellvis). The wells were functionalized with azides as described above. Separately, 12 mm circular glass coverslips were passivated with Sigmacote, washed with ethanol, and dried. PEG precursors were separately dissolved in PBS and sterile filtered (0.22 μm). CycloRGD-N3 dissolved in PBS was added to the PEG-sulfo-BCN to achieve a final concentration in the gels of 500 μM, incubated at room temperature for 10 minutes, and then mixed with the azide-bearing PEG component. The crosslinking mixture (25 μL) was pipetted onto the azide-functionalized glass bottom. The passivated coverslip was placed on top of the precursor solution, and the gel was crosslinked at 37°C for 30 minutes. The well was flooded with PBS and incubated at 37°C for one hour to enable easy removal of the coverslip, affording a flat hydrogel surface for cell culture. After washing with PBS, the wells were seeded with the MRC-5 reporter cells at a density of 25,000 cells/well in FluoroBrite DMEM supplemented with 10% FBS, 1x L-glutamine, and 1x penicillin/streptomycin. Cells were allowed to adhere overnight. Time lapse microscopy was performed using a Zeiss AxioObserver inverted microscope with a motorized scanning stage and environmental stage incubator, collecting a 7×7 tiled region near the center of the gels with 20x magnification. Acquisition was paused for exposure to 405 nm light (~600 mW/cm2 for 1 minute) to enable hydrogel softening.
Tiled regions were stitched into composite images using our custom image processing pipeline described above. Using ImageJ software, the stitched images were cropped to be uniform in dimension, and masks of the objects in the individual channels were obtained. The GFP channel was used to detect the cell nuclei based on H2B-GFP expression, and the DsRed channel was used to detect the cell cytoplasm based on LifeAct-mRuby2 expression. Spatial variations in background fluorescence intensity were removed using a rolling ball filter, followed by processing with a median filter (despeckle) to remove pixel-level noise. The DsRed channel was further processed with a Gaussian Blur filter for smoothing. The resulting images were converted to binary masks by implementing Huang’s fuzzy thresholding method.43 Objects outside of the expected cell size range were discarded. Only those objects identified as GFP+ nuclei that were surrounded by cytoplasm were selected for further analysis. Binary images were created, consisting of cell cytoplasm containing nuclei, and were used to quantify the cell spread area.
2.11. Statistical analysis
At least 3 independent replicates were used for each experiment. Statistical analysis was performed using GraphPad Prism 8. Comparisons between two experimental groups with a single varying parameter were performed using two-tailed Student’s t-tests. Comparisons among more than two experimental groups with two varying parameters were performed using two-way analysis of variance (ANOVA) with Bonferroni post-hoc testing. Statistical assessment of synergy among hydrogel stiffness, adhesive ligand composition, and soluble factor treatment was performed using three-way ANOVA. For quantification of reaction kinetics, first-order exponential association or decay models were fit to the data, and the kinetic parameters were compared using an extra sum-of-squares F test.
3. Results
3.1. Soft hydrogels promote MuSC activation and expansion
The elastic modulus of healthy skeletal muscle is ~12 kPa.14,19,21 Following injury, the stiffness of the bulk muscle tissue has been reported to transiently increase during regeneration.14 To mimic these different tissue stiffness regimens in engineered microenvironments, hydrogels were prepared from multi-arm PEG macromers separately functionalized with azide or bicyclo[6.1.0]nonyne (BCN) groups that enable crosslinking by the bioorthogonal SPAAC reaction. The elastic moduli of these materials can be tuned over a broad range spanning physiologically relevant stiffness by varying the total polymer content of the hydrogels as well as the molar ratio of BCN-bearing macromers to azide-bearing macromers. By modulating these two variables, “soft” hydrogels with elastic moduli similar to native muscle tissue (E~12 kPa) and “stiff” hydrogels with elastic moduli similar to stiffer regenerating and fibrotic tissue (E~40 kPa) were prepared (Supporting Figure S1). To mimic the ECM composition during the initial phases of regeneration, purified laminin-111 was functionalized with linear PEG-azides and incorporated into hydrogels during crosslinking. Alternatively, to mimic the fibronectin-rich ECM characteristic of later phases of regeneration, azide-bearing cyclic RGD peptides were incorporated into the gels during crosslinking.
MuSC fate is regulated by a series of transcription factors that direct key steps in the myogenic progression, including stem cell activation and differentiation.1 Muscle stem cells express a characteristic transcription factor, Pax7, in their quiescent state prior to injury. These quiescent MuSCs do not express MyoD or myogenin, two other myogenic transcription factors that are involved in MuSC responses to injury. Following injury, MuSCs activate and begin expressing MyoD, which is required for myogenic differentiation. However, MyoD expression is not sufficient to commit MuSCs to differentiate, as MyoD+ progenitors are proliferative and can return to quiescence.1,44 MyoD is involved in regulating the expression of myogenin, which is expressed in progenitors that are committed to differentiation. Myogenin expression promotes terminal differentiation and fusion of myogenic progenitors into multi-nucleated myofibers.
To facilitate tracking of MuSC fate (Figure 1A), MuSCs were isolated from transgenic reporter mice harboring GFP fused to the nuclear envelope protein Sun1 downstream of a loxP-STOP-loxP cassette that express tamoxifen-inducible Cre recombinase driven by the myogenin promoter. Accordingly, in the presence of 4-hydroxytamoxifen, myogenin promoter activity results in excision of the STOP codon and expression of the GFP signal, marking cells that are committed to myogenic differentiation and expression of myogenin. To ensure only non-committed myogenic cells were isolated, mice were dosed with tamoxifen daily for 3 days. To identify committed MuSCs, the 3 days of tamoxifen treatment were followed by a 22-day rest period during which committed cells were labeled by Sun1-GFP expression. MuSCs were isolated from the hindlimb skeletal muscles of the reporter mice following our published protocols.19,40 Purified MuSCs were obtained by FACS by selecting for integrin-α7+/CD34+/GFP− cells. Freshly isolated MuSCs were plated on soft (E~12 kPa) or stiff (E~40 kPa) hydrogels that were functionalized with either laminin-111 or RGD peptides to promote cell adhesion and mechanosensing, mimicking the different ECM stiffness and composition that are observed during regeneration (Figure 1B). Following seven days in culture with daily 4-hydroxytamoxifen treatment, the samples were fixed and immunostained for the MuSC transcription factor Pax7 and the activated myogenic progenitor transcription factor MyoD to assess myogenic progression in conjunction with the myogenin GFP reporter.
Figure 1.
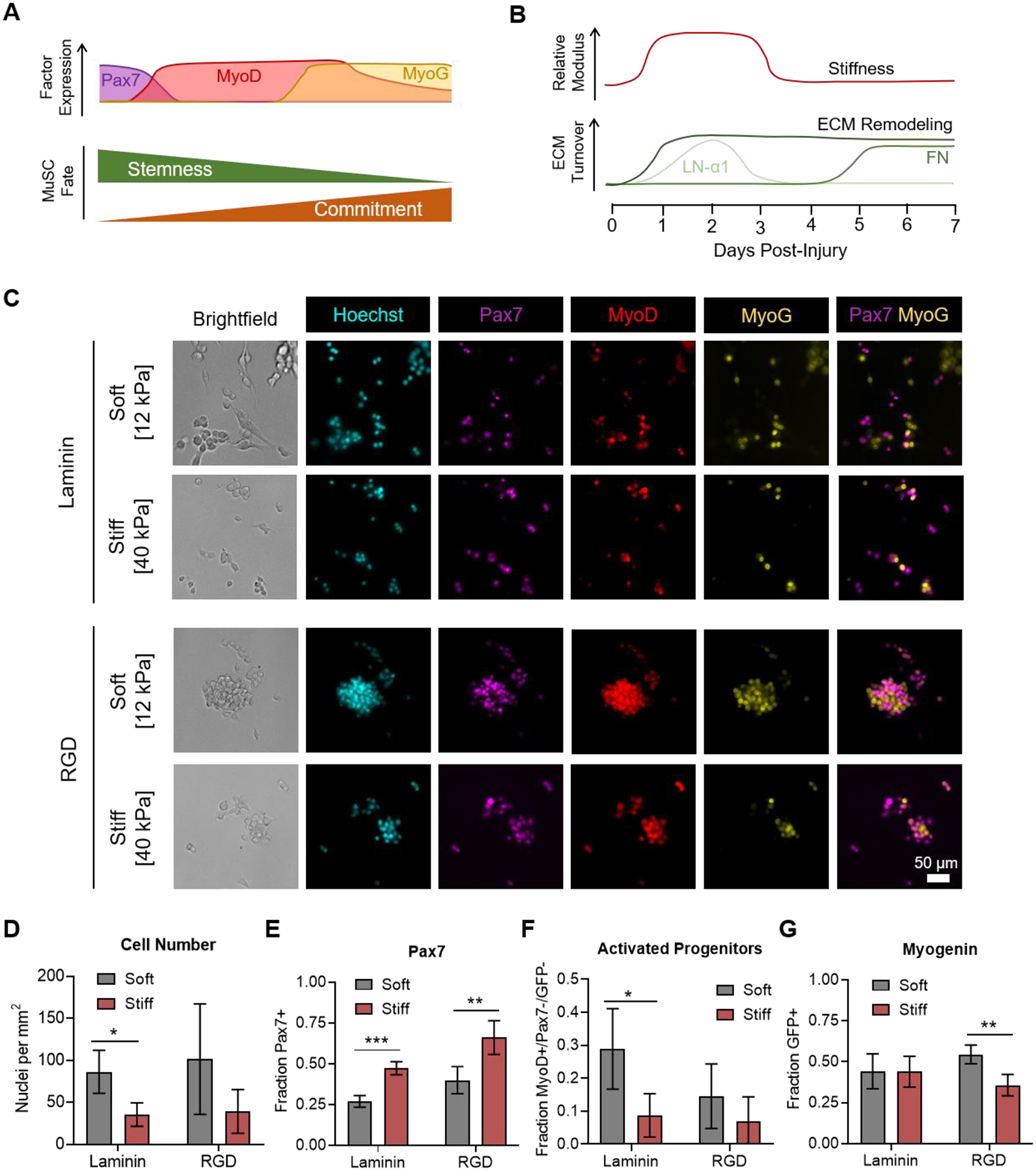
Culture on soft hydrogels promotes MuSC expansion and activation. (A) Schematic depicting the expression of myogenic transcription factors (Pax7, MyoD, and myogenin (MyoG)) as myogenesis progresses from quiescent stem cells to activated progenitors and commitment to differentiation. (B) Schematic depicting temporal changes in ECM properties (stiffness and protein composition) as a function of regeneration time. LN-α1 = laminin-α1; FN = fibronectin. (C) Immunofluorescence microscopy images characterizing the expression of Pax7 and MyoD by immunostaining and expression of the Sun1-GFP myogenin fluorescent reporter (MyoG) on hydrogels with controlled stiffness and adhesive ligand presentation. (D) Quantification of cell number, revealing increased MuSC expansion on soft gels. (E) Quantification of the fraction of cells expressing Pax7, revealing an arrested, more quiescent-like phenotype on stiffer gels. (F) Quantification of the fraction of cells that are activated progenitors, those positive for MyoD but negative for Pax7 and myogenin reporter expression, revealing improved activation on soft gels. (G) Quantification of the fraction of cells expressing the myogenin reporter, revealing a combined effect of stiffness and ligand presentation on commitment. In D-G, data are presented as mean ± s.d. n = 4. *p<0.05, **p<0.01, ***p<0.001, two-tailed Student’s t-test.
MuSCs cultured on soft hydrogels exhibited enhanced activation and proliferation when compared to culture on stiff hydrogels (Figure 1C–F). This finding is consistent with our previous studies in which we observed optimal MuSC expansion and engraftment when cultured on hydrogels with physiological stiffness (E~12 kPa).19 On laminin-presenting gels, MuSC numbers were significantly greater on soft hydrogels (Figure 1D). A similar trend was observed for the RGD-presenting hydrogels. The expression pattern of myogenic transcription factors suggests that the difference in proliferation was mediated by a larger pool of activated progenitors on the soft hydrogels. Notably, MuSCs on laminin appeared to preferentially adhere to the substrate rather than forming cell-cell contacts with other MuSCs. In contrast, MuSCs on RGD matrices exhibited increased cell aggregation and formed more cell-cell contacts. On stiff gels, proliferation was impeded but a greater fraction of these growth arrested MuSCs expressed Pax7 than on soft gels (Figure 1E), suggesting impaired activation on stiff microenvironments. On soft hydrogels, a larger fraction of MuSCs expressed MyoD without co-expression of Pax7 and myogenin, suggesting the presence of a highly proliferative progenitor pool (Figure 1F, Supporting Figure S2). Commitment to differentiation, as measured by myogenin reporter expression, was not sensitive to stiffness on laminin presenting gels, while a lower fraction of MuSCs on stiff RGD-presenting gels expressed the myogenin reporter when compared to soft RGD-presenting gels (Figure 1G).
To confirm that the decreased MuSC numbers on stiff substrates are due to impaired proliferation and not increased cell death, we assessed cell viability on days 3 and 7 of culture. Staining with calcein-AM to mark live cells and ethidium homodimer to mark dead cells revealed that MuSCs cultured on all substrates remained highly viable (>94%) throughout the culture duration, with no differences among substrates (Supporting Figure S3). Furthermore, immunostaining for cleaved caspase-3 revealed that the vast majority (>99%) of cells were not apoptotic, which was consistent across all substrates tested (Supporting Figure S4). Taken together, these results indicate that soft hydrogels promote MuSC activation and proliferation to expand the stem cell pool, while stiff hydrogels block activation and commitment, resulting in a population of less proliferative, Pax7-expressing cells.
3.2. Soluble niche factors synergize with matrix cues to promote expansion of MuSCs on soft gels
During muscle regeneration, MuSCs are presented with a temporally defined sequence of biochemical cues that can be roughly divided into an inflammatory phase and a self-renewal phase (Figure 2A).2 We recently identified two soluble factors that are key mediators of these two phases of regeneration. During the inflammatory phase, the lipid metabolite prostaglandin E2 (PGE2) promotes MuSC survival and expansion.9 Later during regeneration, the cytokine oncostatin M (OSM) promotes maintenance of the stem cell pool by acting as a potent quiescence inducer.11 As the presentation of these two factors is tightly controlled during regeneration, we hypothesized that the sequential treatment of MuSCs with PGE2 and OSM would enable improved expansion of MuSCs on engineered microenvironments (Figure 2B).
Figure 2.
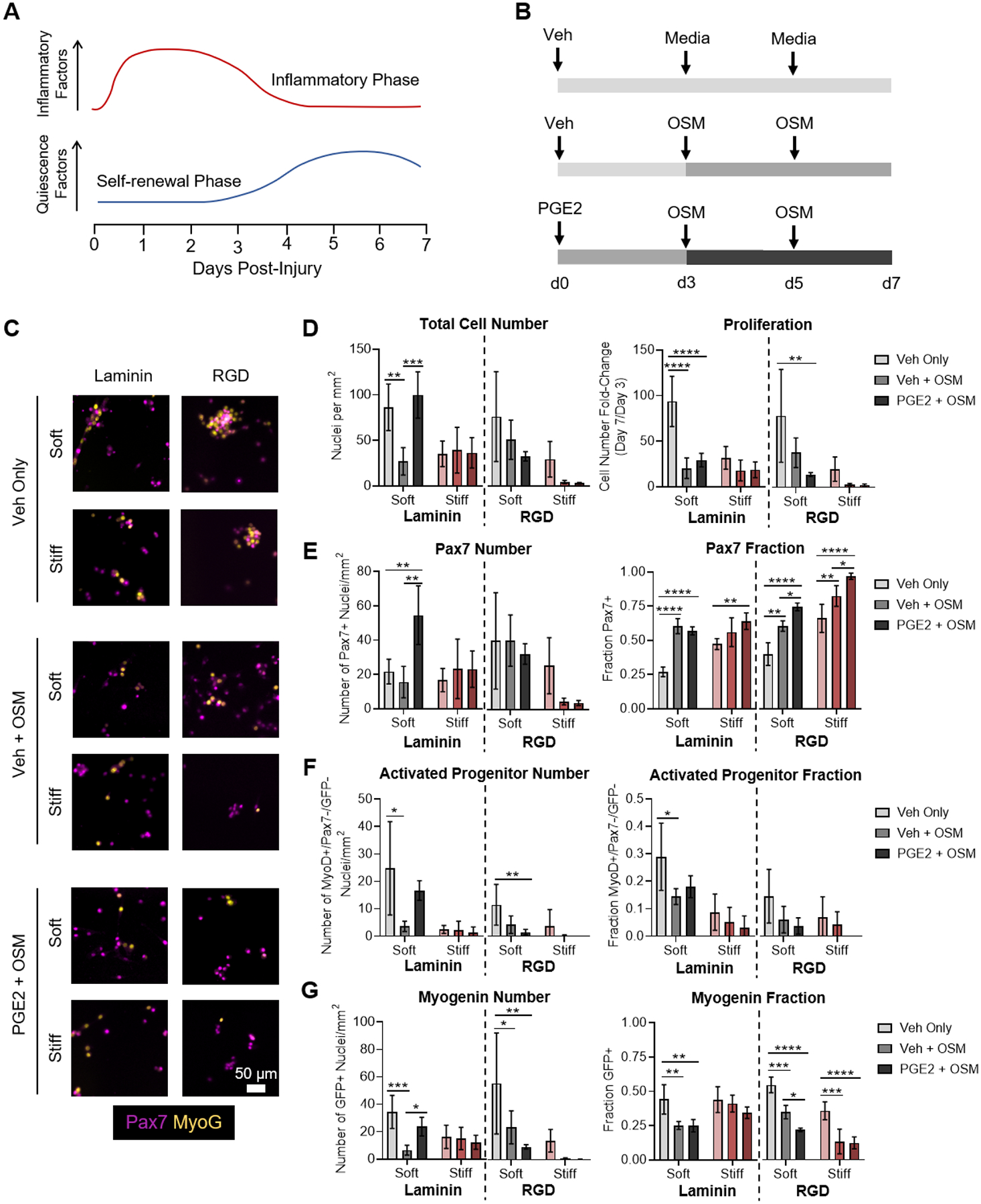
Soluble niche factors and ligand presentation synergize to enhance MuSC activation and expansion on soft hydrogels. (A) Schematic depicting temporal changes in expression of inflammatory factors and quiescence factors during regeneration. (B) Schematic depicting experimental setup treating cultured MuSCs with soluble niche factors that recapitulate the progression of regeneration. (C) Immunofluorescence microscopy images characterizing the expression of Pax7 by immunostaining and expression of the Sun1-GFP myogenin fluorescent reporter (MyoG). (D) Quantification of total cell number and fold-change in cell number between days 3 and 7. (E) Quantification of the number of Pax7+ cells and the fraction of total cells expressing Pax7, revealing optimal expansion of Pax7+ cells on soft, laminin-presenting gels with sequential PGE2 and OSM treatment. (F) Quantification of the number of activated progenitors (MyoD+/Pax7−/GFP−) and the fraction of total cells that are activated progenitors, revealing improved activation on soft gels that is suppressed by OSM treatment. (G) Quantification of the total number of myogenin reporter positive cells and the fraction of total cells expressing the myogenin reporter, revealing suppressed commitment to differentiation following OSM treatment. In D-G, data are presented as mean ± s.d. n = 3–4. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-way ANOVA with Bonferroni post-hoc test.
As soft, laminin-presenting hydrogels best enabled MuSC expansion, we first assessed whether PGE2 and OSM treatment would synergize with these permissive matrix cues to regulate MuSC fate. Treatment with only OSM beginning at day 3 of culture, to simulate the onset of the self-renewal phase, suppressed MuSC proliferation (Figure 2C,D, Supporting Figure S5), while increasing the fraction of cells that were positive for Pax7 (Figure 2E) in agreement with its function in vivo.11 OSM treatment simultaneously decreased the total number and fraction of cells that were MyoD+/Pax7−/MyoG− progenitors (Figure 2F) and committed myogenin reporter positive cells (Figure 2G). Decreased proliferation with increased Pax7 expression and decreased commitment toward differentiation is consistent with the expectation that OSM induces a more quiescent-like phenotype.
As we previously demonstrated that PGE2 treatment augments MuSC expansion,9 we hypothesized that sequential treatment with PGE2 followed by treatment with OSM would both increase proliferation and maintain a larger pool of stem-like, Pax7-expressing cells. On soft, laminin-presenting gels, MuSCs treated with a single dose of PGE2 at the time of seeding, followed by treatment with OSM starting at day 3, resulted in increased cell proliferation by day 3 (Supporting Figure S6) and an overall higher cell number by day 7 (Figure 2D). As with OSM treatment alone, a significantly larger fraction of the resulting cells was Pax7+ compared to the control treatment. Taken together with the increase in cell number afforded by early PGE2 treatment, this resulted in a large increase in the number of Pax7+ cells when compared with either the vehicle control or OSM treatment alone (Figure 2E). Furthermore, the fraction of myogenin reporter positive cells was also decreased relative to the control (Figure 2G), indicative of less commitment to differentiation. Thus, sequential treatment with PGE2 followed by OSM on soft, laminin-presenting cells resulted in enhanced expansion of stem-like cells.
To determine how matrix-presented cues such as stiffness and adhesive ligand presentation impact the MuSC response to PGE2 and OSM, the soluble factor treatments were also carried out on stiff laminin-presenting substrates and on both soft and stiff RGD-presenting substrates. The proliferation and myogenic progression of MuSCs cultured on stiff, laminin-presenting gels were largely insensitive to the presence of OSM or PGE2 (Figure 2C–G), using previously optimized concentrations.9,11 Similar to untreated cells, those treated with OSM or sequentially treated with PGE2 and OSM failed to activate, instead remaining more Pax7+. However, on RGD-presenting gels, OSM treatment and sequential PGE2 and OSM treatment altered MuSC proliferation and commitment (Figure 2C–G, Supporting Figure S7). On both soft and stiff RGD-presenting gels, OSM treatment resulted in decreased proliferation, similar to soft, laminin-presenting gels. Unexpectedly, sequential treatment with PGE2 followed by OSM further suppressed proliferation. Thus, on the stiff, RGD-presenting gels, proliferation was severely repressed, with cells expanding only ~3-fold between days 3 and 7 (Figure 2D). Both treatment with OSM and sequential treatment with PGE2 and OSM reinforced the arrested myogenic progression observed on untreated stiff, RGD-presenting gels, resulting in a very small number of cells after 7 days in culture that were nearly all Pax7+, but did not activate, proliferate, or commit (Figure 2E–G). Three-way ANOVA revealed statistically significant interaction terms for stiffness, cell-adhesive ligand composition, and soluble factor treatment (Supporting Figure S8), indicating that treatment with PGE2 and OSM synergizes with matrix properties to modulate MuSC proliferation and commitment. Neither treatment with OSM nor sequential treatment with PGE2 and OSM altered cell viability (Supporting Figure S9) or the fraction of apoptotic cells (Supporting Figure S10) across all conditions tested, indicating that the observed changes in cell number can be attributed predominantly to changes in proliferation. Taken together, these results suggest that the stiffness and composition of the substrate modulates MuSC responses to the soluble niche factors OSM and PGE2.
3.3. Development of dynamically softening substrates with improved reaction kinetics
We sought to test whether temporally changing stiffness reminiscent of in vivo regeneration could modulate MuSC fate. As bulk muscle stiffness was previously shown to increase shortly after injury, followed by a return to baseline after about 3 days,14 an ideal material to test the effects of dynamically changing stiffness would start out relatively stiff and then soften on demand. Because photocleavable oNB esters have previously been successfully used for this purpose,23–25 we decided to leverage this technology to probe the MuSC response to a softening microenvironment.
As oNB esters are sensitive to near-UV light, we sought to use the lowest energy wavelength of light that was practical, while also reducing the overall time of exposure, to limit potential cytotoxic effects that could confound our results. Longer wavelength 405 nm light can still trigger photolysis of oNB esters but requires longer exposures than 365 nm light.23 To increase the rate of photolysis in response to 405 nm light, we developed a novel bifunctional oNB ester crosslinker. As the alignment of the ester linkage with the nitro group on the aromatic ring is known to be required for photolysis to proceed, we were inspired by prior work demonstrating that substitution at the benzyl position of the oNB ester could improve reaction kinetics.35 We reasoned that substituting the commonly used methyl group at the benzyl position with a bulky tert-butyl group would sterically hinder rotation about the benzyl bond, enforcing alignment of the ester and nitro groups, increasing the reaction rate. By adapting known synthetic routes,33–35 bifunctional azide-bearing oNB ester crosslinkers with both methyl and tert-butyl substituents at the benzyl position were prepared (Figure 3).
Figure 3.
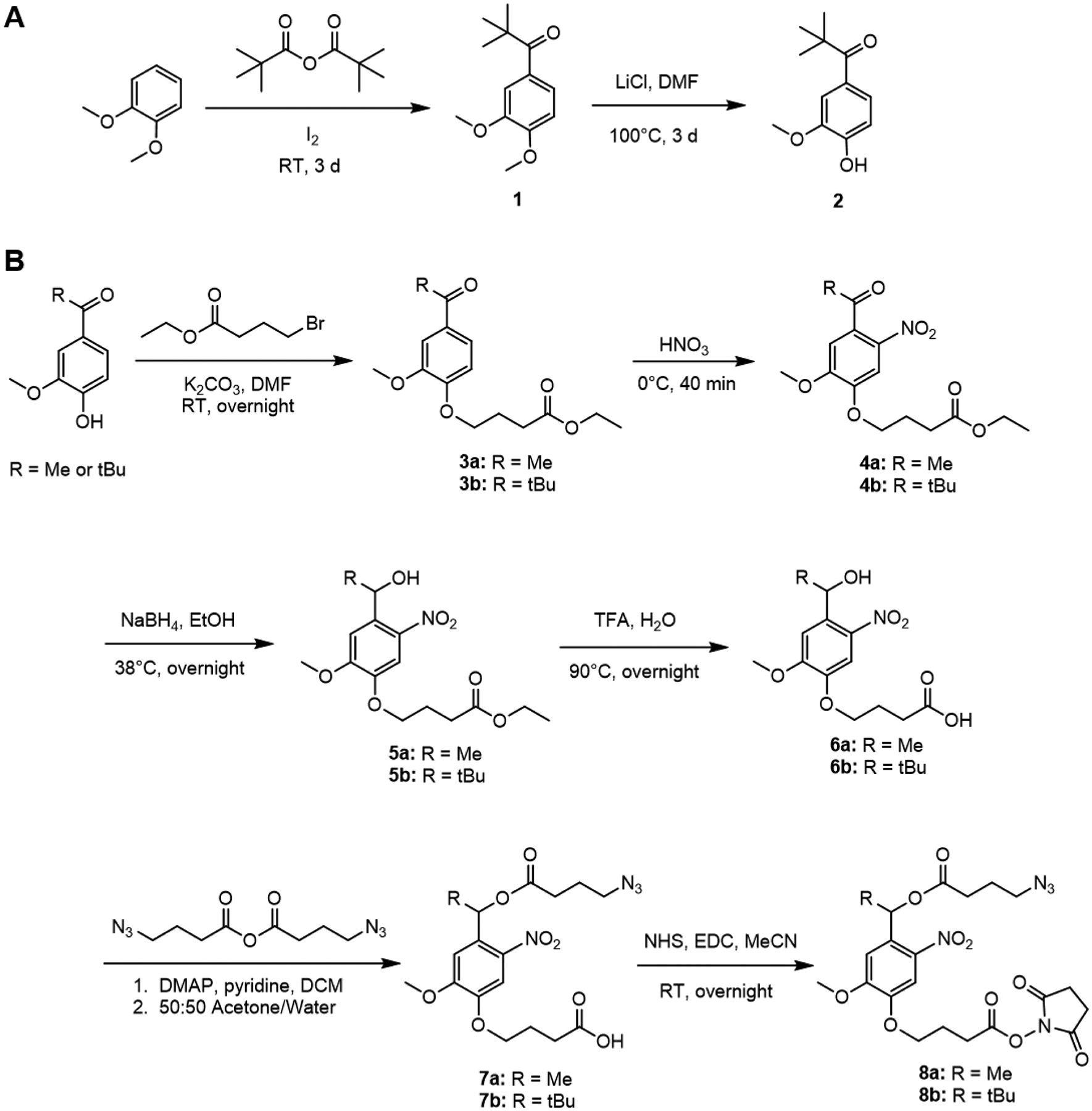
Synthesis of oNB crosslinkers. (A) Synthetic route to prepare tert-butyl substituted precursor. (B) Synthetic route to prepare methyl and tert-butyl substituted bifunctional oNB crosslinkers bearing amine-reactive NHS esters and BCN-reactive azides.
Eight-arm PEG macromers were subsequently synthesized wherein a subset of the azide groups used in crosslinking were connected to the PEGs via oNB linkages, allowing breakage of a fraction of the crosslinks upon light exposure to soften the gels (Figure 4A,B). The photolysis kinetics of the two oNB linkers were first assessed in solution phase using UV-visible light spectroscopy (Figure 4C,D). Upon illumination with 405 nm light, the cleavage reaction for the tert-butyl substituted oNB materials rapidly reached completion, with a reaction half-time on the order of 30 s (Figure 4E,F). In contrast, methyl substituted materials exhibited a reaction half-time on the order of 150 s (Figure 4F). Fitting a first-order association curve to the spectroscopy data revealed a ~5-fold increase in the reaction rate for the tert-butyl substituted material (Figure 4G). To confirm that the enhanced solution phase kinetics applied to crosslinks that are part of a hydrogel network, gels prepared from tert-butyl or methyl substituted oNB materials were subjected to rheological testing after exposure to fixed doses of 405 nm light. The decrease in elastic modulus for the tert-butyl samples saturated after ~60 s, whereas saturation for the methyl samples occurred around 300 s (Figure 4H). Fitting a first-order exponential decay curve to the rheology data confirmed a ~4-fold increase in softening rate for the tert-butyl substituted materials (Figure 4I,J). The initial and softened stiffness of the material was tuned by controlling the total polymer content of the hydrogels within a physiologically relevant range (Supporting Figure S11).
Figure 4.
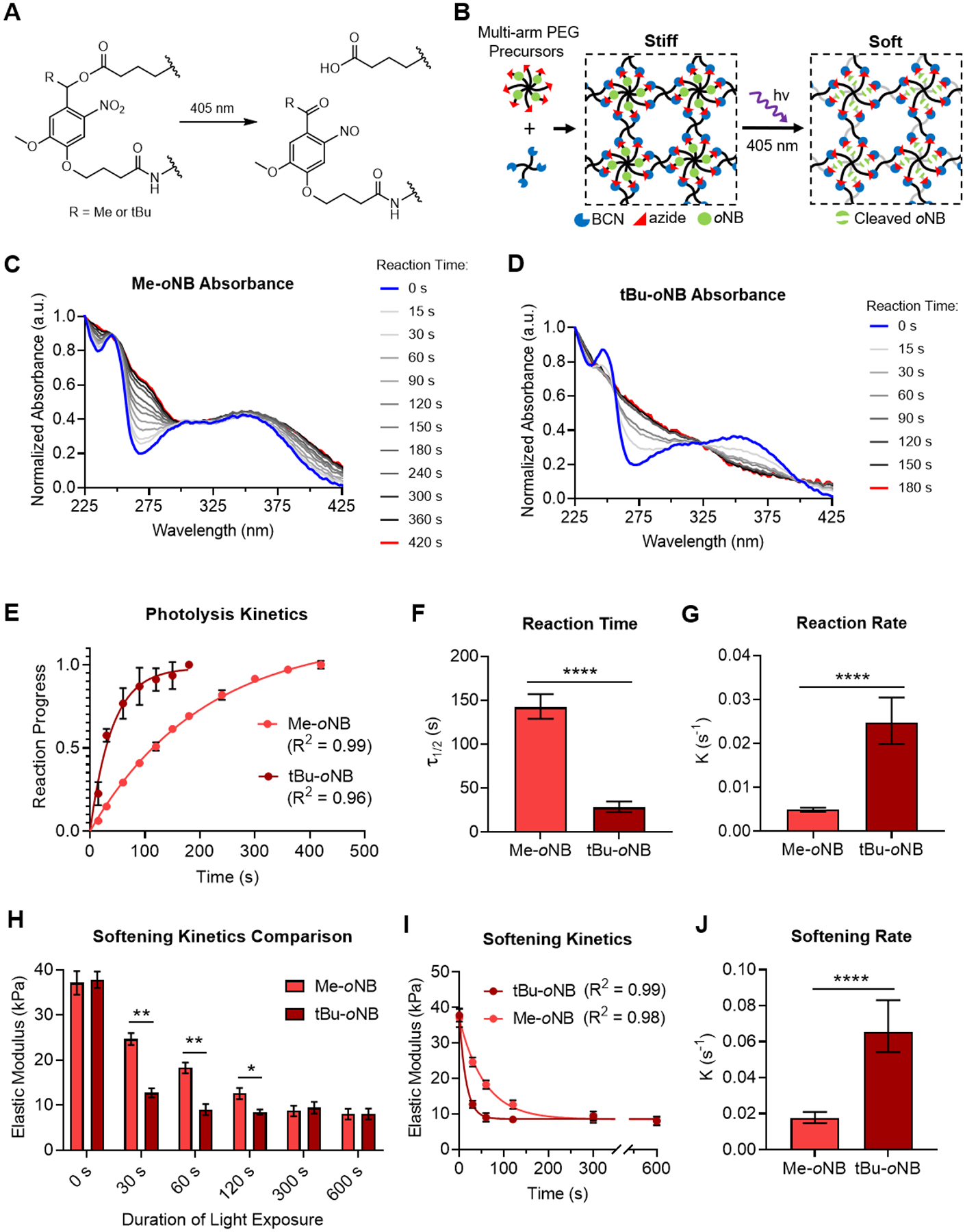
Substitution with tert-butyl moiety increases the softening rate of photoresponsive hydrogels. (A) Schematic of oNB ester hydrolysis reaction triggered by exposure to 405 nm light. (B) Idealized schematic of dynamically softening hydrogels comprised of oNB ester functionalized PEG macromers. Exposure to 405 nm light results in cleavage of the oNB esters, decreasing crosslink density and softening the network. Polymer chains that contribute to network elasticity are shown in black, while those that no longer contribute after photolysis are in gray. UV-visible light absorbance spectra for (C) methyl substituted and (D) tert-butyl substituted oNB ester photolysis after fixed durations of light exposure. (E) Solution phase photolysis kinetics were characterized by fitting a first-order exponential association curve to the extent of reaction progress obtained from the normalized absorbance at 275 nm over time. (F) Reaction half-time (τ1/2) and (G) reaction rate (k) determined from the curve fitting analysis in E. (H) Elastic moduli of hydrogels prepared from PEGs containing methyl and tert-butyl substituted oNB ester crosslinks after controlled durations of exposure to 405 nm light. (I) Softening kinetics were characterized by fitting a first-order exponential decay curve to the rheology data. (G) Softening rate (k) determined from the curve fitting analysis in I. In E, H, and I, data are presented as mean ± s.d. n = 3. In H, *p<0.05, **p<0.01, two-tailed Student’s t-test with Holm-Sidak multiple comparisons correction. In F, G, and J, data are presented as best-fit values with 95% confidence intervals. n = 3. ****p<0.0001, extra sum-of-squares F test.
3.4. Cells respond to dynamic softening of cell culture substrates
Using our materials capable of on-demand softening with improved reaction kinetics, we tested if cells could sense the dynamic changes in elastic modulus on these new materials. As fibroblasts have well-characterized responses to culture on substrates of different stiffness, we used human fetal lung fibroblasts for these confirmatory studies. Fibroblasts spread extensively and develop well-defined actin stress fibers on relatively stiff gels, while remaining poorly spread on soft gels. To track real-time changes in fibroblast spreading upon hydrogel softening, we generated a stable cell line that was lentivirally transduced to express a H2B-GFP fusion protein to label the nuclei green and the actin-binding LifeAct peptide fused to mRuby2 to label the actin cytoskeleton red. The fibroblasts were seeded onto RGD-presenting PEG gels, allowed to adhere overnight, and imaged by time lapse fluorescence microscopy (Figure 5).
Figure 5.
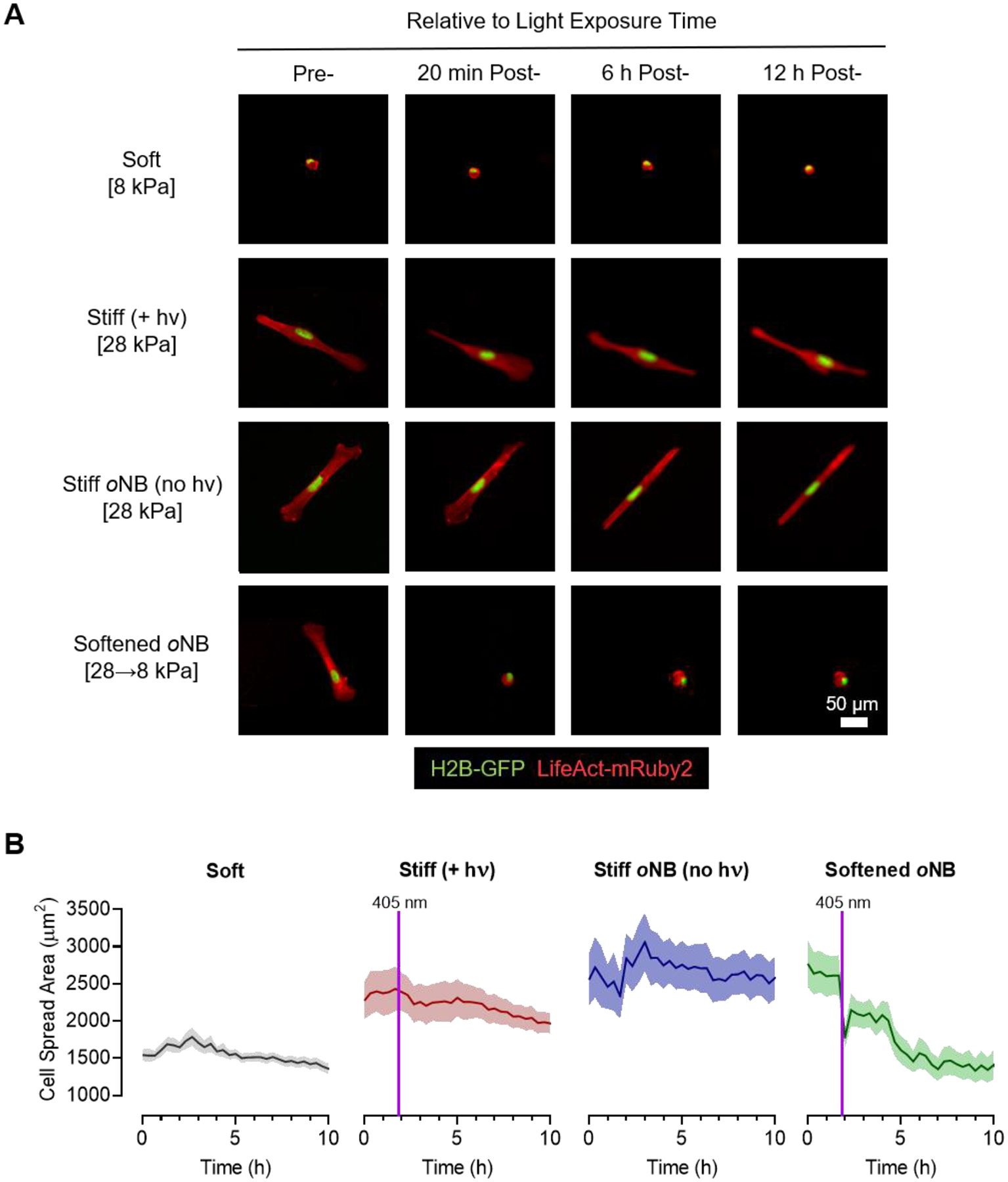
Optimized dynamically softening hydrogels enable real-time tracking of cellular responses to changing matrix stiffness. (A) Representative time lapse fluorescence microscopy images of H2B-GFP and LifeAct-mRuby2 expressing MRC-5 fibroblasts on soft control gels, stiff control gels without oNB crosslinks that were exposed to 405 nm light, stiff control gels with oNB crosslinks that were not exposed to light, and initially stiff oNB-containing gels that were softened by exposure to 405 nm light. (B) Quantification of cell spread area from time lapse microscopy reveals a rapid decrease in cell spreading following exposure to 405 nm light on the oNB-containing materials. In B, data are presented as mean with s.e.m. error envelopes. Greater than 100 individual cells were analyzed for each condition.
Fibroblasts seeded on relatively soft (E~8 kPa) control gels adhered well but maintained a rounded morphology throughout the time lapse experiment. Stiff (E~28 kPa) control gels that did not contain photocleavable oNB crosslinks but were exposed to 405 nm light to simulate the softening stimulus retained a highly spread and elongated morphology following light exposure, indicating that cell spreading was not altered by light exposure alone. Similarly, fibroblasts seeded on a second stiff (E~28 kPa) control, oNB-containing materials that were not exposed to 405 nm light, maintained a highly spread morphology throughout the experiment. In contrast, fibroblasts seeded on oNB materials that were initially stiff (E~28 kPa) started off well-spread but rapidly rounded up following softening to ~8 kPa (Figure 5A). Quantifying changes in spreading behavior by tracking cell spread area over time confirmed the poor spreading observed on soft gels, the consistently well-spread morphology observed on stiff controls, and the rapid loss of spreading following softening on the dynamically tunable materials (Figure 5B). These experiments confirmed that cells are able to sense real-time changes in substrate stiffness on our tert-butyl oNB materials.
3.5. Transient exposure to stiff microenvironments blocks MuSC proliferation and commitment
Having established a hydrogel cell culture platform that enables rapid on demand softening in response to 405 nm light, we sought to determine how mimicking the transition from a stiff to soft microenvironment during muscle regeneration impacts MuSC fate in vitro. As above, MuSCs were isolated from Sun1-GFP myogenin reporter mice and cultured on hydrogels of controlled stiffness presenting RGD adhesive peptides. Three control materials consisted of soft (E~12 kPa) hydrogels, stiff (E~40 kPa) hydrogels without oNB linkages that were exposed to 405 nm of light after 3 days in culture, and stiff (E~40 kPa) hydrogels containing oNB linkages but not exposed to light (Figure 6A). MuSCs were additionally cultured on oNB-containing hydrogels that were initially stiff (E~40 kPa) and then subsequently softened after 3 days of culture to ~12 kPa. To determine if sequential delivery of soluble niche cues was able to synergize with the dynamic change in stiffness, additional experimental groups included treatment with OSM at day 3 and sequential treatment with PGE2 upon initial seeding followed by treatment with OSM at day 3 (Figure 6B).
Figure 6.
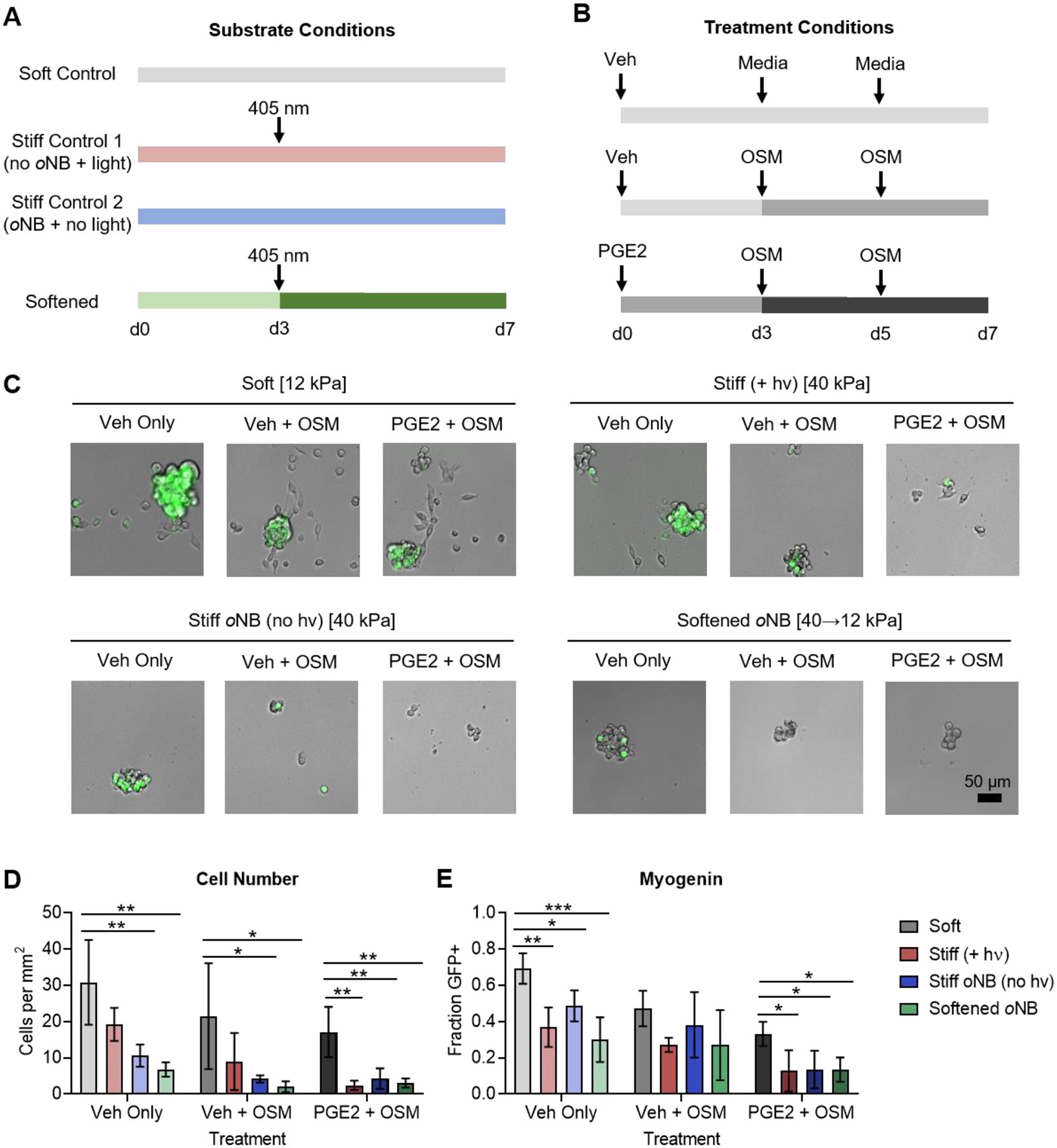
Dynamic softening reveals that the MuSC response to stiffness is fixed within the first three days of culture. (A) Schematic depicting the substrate conditions used for MuSC culture. (B) Schematic depicting the soluble factor treatments for MuSCs cultured on the four substrate conditions. (C) Brightfield and fluorescence microscopy of MuSCs cultured on the four substrate conditions described in A treated with soluble niche factors as described in B. The green signal corresponds to the Sun1-GFP myogenin reporter. (D) Quantification of total cell number reveals decreased proliferation in cells cultured on stiff controls and dynamically softened gels compared to soft controls. Treatment with OSM or with PGE2 followed by OSM further suppressed proliferation. (E) Quantification of the fraction of cells positive for myogenin reporter expression reveals decreased commitment for cells cultured on stiff controls and dynamically softened gels compared to soft controls. Treatment with OSM or with PGE2 followed by OSM further suppressed commitment. In D and E, data are presented as mean ± s.d. n = 3–4. *p<0.05, **p<0.01, ***p<0.001, two-way ANOVA with Bonferroni post-hoc test.
In the absence of additional soluble factors, MuSCs cultured on dynamically softened materials presenting RGD peptides behaved strikingly similar to MuSCs cultured on the stiff control materials (Figure 6C). Decreased total cell numbers were observed on both persistently stiff control materials, as well as materials that were initially stiff and subsequently softened at day 3 (Figure 6D), suggesting a diminished proliferation capacity. Furthermore, MuSCs that had spent any time exposed to a stiff microenvironment exhibited less commitment to myogenic differentiation, as evidenced by a lower fraction of cells expressing the myogenin reporter (Figure 6E). As above, the quiescence promoting sequential treatment of PGE2 and OSM further decreased cell numbers and the fraction of cells positive for the myogenin reporter on both the persistently stiff and transiently stiff hydrogels (Figure 6D,E). Taken together, these results suggest that even transient 3-day exposure to a stiff microenvironment is sufficient to impair MuSC proliferation and block activation and commitment to differentiation.
4. Discussion
Identifying optimal MuSC expansion conditions ex vivo remains a major challenge for musculoskeletal tissue engineering and regenerative medicine. Once MuSCs become activated and commit, they are rendered incapable of extensive muscle repair. Although such in vitro cultured myoblasts can differentiate and fuse to form tissue engineered constructs, they do not retain the ability to function as stem cells when transplanted in vivo. While freshly isolated MuSCs can engraft into the satellite cell niche and respond to injury by expanding and fusing to form myofibers, transplanted myoblasts lose the ability to engraft and repair.40,45 Engineered cell culture platforms are therefore needed to maintain MuSCs in a stem cell state ex vivo. Given the critical role that MuSCs play in maintaining muscle homeostasis throughout life, any regenerative approach employing MuSCs or MuSC-derived cells will need to provide patients with functional stem cells. This is true for gene editing to restore functional protein expression in muscular dystrophies or for engineering tissue replacements for large scale muscle loss.6,8
We previously demonstrated that the high stiffness of rigid plastic dishes typically used in cell culture impairs the ability of MuSCs to expand and subsequently engraft in vivo,19 implicating dysfunctional mechanosignaling as a key barrier to proper MuSC function. Consistent with our earlier work, here we report that soft, laminin-presenting hydrogel materials that recapitulate healthy muscle stiffness (E~12 kPa) best enable MuSC activation and proliferation, while maintaining a substantial pool of Pax7+ cells (Figure 1). We further demonstrate that soluble factors from the regenerating muscle niche augment MuSC expansion. Sequential treatment with PGE2 followed by OSM results in robust expansion of Pax7+ cells on soft, laminin-presenting hydrogels, with OSM, a quiescence inducer, apparently dominating over PGE2, a stimulant of proliferation (Figure 2).
In contrast, culture on physiologically stiff (E~40 kPa) hydrogels results in decreased MuSC proliferation and impairs myogenic progression (Figure 1). MuSCs cultured on stiff gels remain largely arrested in a dysfunctional non-proliferative, non-activated state. This response contrasts with previous studies demonstrating increased myoblast proliferation as stiffness increases.20 We hypothesize that the initial culture of myogenic progenitors on rigid plastic to generate myoblasts selects for a subset of cells that are capable of sustained proliferation on stiff substrates. However, in agreement with the data presented in this report, our previous studies using freshly isolated MuSCs revealed decreased proliferation on stiff substrates that corresponded to diminished in vivo engraftment.19 The process of engraftment likely requires transient activation and expansion to obtain an experimentally detectable number of stem cells, while retaining the ability to return to quiescence and respond to subsequent injury. The increased activation of MuSCs with simultaneous maintenance of Pax7 expression on soft, laminin-presenting materials suggests that these cells would indeed engraft better than the cells cultured on stiff gels with an arrested myogenic phenotype. While this hypothesis is consistent with our prior data, additional in vivo engraftment experiments will be required to confirm differences in MuSC function.
Gaining a better understanding of MuSC behavior in stiff microenvironments with varied matrix composition is also critically important for developing therapeutic strategies targeting MuSCs in the context of disease and aging, in which fibrosis of the muscle results in tissue stiffening and aberrant ECM deposition.46–51 We have previously implicated mechanosensing in MuSC dysfunction in aged mice. Culture on relatively soft substrates was necessary to render interventions blocking overactivated p38 MAP kinase signaling in aged cells effective at restoring engraftment capacity.52 As MuSCs lose regenerative capacity with age,53 it is plausible that increased microenvironmental stiffness results in arrested myogenesis as we observe here in vitro. Our observation that even transient exposure to stiff substrates impairs MuSC expansion suggests a form of “mechanical memory”24,54 that may impact the ability of aged MuSCs to respond to therapeutic interventions in a fibrotic microenvironment. Future studies are warranted to both uncover the prevalence of dysfunctional mechanosignaling pathways in aged MuSCs and molecular mechanisms that may confer a “mechanical memory.”
5. Conclusion
The use of well-defined engineered microenvironments enabled identification of synergistic interactions among substrate stiffness, adhesive ligand presentation, and soluble niche factors that regulate MuSC fate. Culturing freshly isolated MuSCs on soft hydrogels that mimic healthy muscle stiffness and present laminin enabled robust MuSC expansion. Stem cell expansion was further augmented by sequential treatment with PGE2 and OSM, resulting in a large pool of Pax7+ cells after 7 days of culture. In contrast, culture on stiff hydrogels, particularly those presenting RGD adhesive peptides, impaired MuSC proliferation and arrested myogenic progression. On stiff, RGD-presenting gels, sequential treatment with PGE2 and OSM reinforced the arrested phenotype, resulting in a diminished number of cells of which most failed to activate in culture. To determine whether temporally changing substrate stiffness altered MuSC proliferation and commitment, we developed a novel oNB-based hydrogel with improved softening kinetics. Culturing MuSCs on these dynamic hydrogels revealed that culture for 3 days on a stiff substrate sufficed to impair MuSC proliferation and block commitment. These results emphasize the importance of biophysical matrix cues in regulating MuSC fate for tissue engineering and regenerative medicine applications.
Supplementary Material
Acknowledgements
The authors would like to thank Prof. Sarah Heilshorn (Stanford Materials Science & Engineering) for use of chemical synthesis and mechanical testing equipment. C.M.M. was supported by the Open Philanthropy Project through a Life Sciences Research Foundation (LSRF) Postdoctoral Fellowship and by the Stanford ChEM-H Postdoctoral Training Program in Quantitative Mechanobiology. Y.X.W. was supported by the Canadian Institutes of Health Research and a K99 Pathway to Independence Award (K99NS120278) from the US National Institutes of Health (NIH). This study was supported by the Baxter Foundation, the Li Ka Shing Foundation, California Institute for Regenerative Medicine (CIRM) grant DISC2-10604 (to H.M.B.), and NIH grants R01AG020961 and R01AG069858 (to H.M.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
H.M.B. is a named inventor on patent applications held by Stanford University regarding PGE2 and muscle regeneration licensed to Myoforte Therapeutics. C.A.H., Y.X.W., and H.M.B. are named inventors on a patent application held by Stanford University for processing of multiplex microscopy images. H.M.B. is a cofounder of Myoforte Therapeutics, and receives consulting fees and has equity and stock options from Myoforte Therapeutics. H.M.B. is a cofounder of Rejuvenation Technologies, Inc., and has equity and stock options in the company. C.M.M. and I.A.F. declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
All processed data supporting the findings of this study are available within the paper and its supplementary information files. Raw data are available upon request. Correspondence and requests for materials should be addressed to H.M.B.
References:
- (1).Yin H; Price F; Rudnicki MA Satellite Cells and the Muscle Stem Cell Niche. Physiol Rev 2013, 93 (1), 23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Fuchs E; Blau HM Tissue Stem Cells: Architects of Their Niches. Cell Stem Cell 2020,27 (4), 532–556. 10.1016/j.stem.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cheung TH; Rando TA Molecular Regulation of Stem Cell Quiescence. Nat. Rev. Mol. Cell Biol 2013, 14 (6), 329–340. 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Blau HM; Daley GQ Stem Cells in the Treatment of Disease. N. Engl. J. Med 2019, 380 (18), 1748–1760. 10.1056/NEJMra1716145. [DOI] [PubMed] [Google Scholar]
- (5).Judson RN; Rossi FMV Towards Stem Cell Therapies for Skeletal Muscle Repair. Npj Regen. Med 2020, 5 (1), 1–6. 10.1038/s41536-020-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Pini V; Morgan JE; Muntoni F; O’Neill HC Genome Editing and Muscle Stem Cells as a Therapeutic Tool for Muscular Dystrophies. Curr. Stem Cell Rep 2017, 3 (2), 137–148. 10.1007/s40778-017-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Quarta M; Cromie M; Chacon R; Blonigan J; Garcia V; Akimenko I; Hamer M; Paine P; Stok M; Shrager JB; Rando TA Bioengineered Constructs Combined with Exercise Enhance Stem Cell-Mediated Treatment of Volumetric Muscle Loss. Nat. Commun 2017, 8 (1), 15613. 10.1038/ncomms15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Langridge B; Griffin M; Butler PE Regenerative Medicine for Skeletal Muscle Loss: A Review of Current Tissue Engineering Approaches. J. Mater. Sci. Mater. Med 2021, 32 (1), 15. 10.1007/s10856-020-06476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ho ATV; Palla AR; Blake MR; Yucel ND; Wang YX; Magnusson KEG; Holbrook CA; Kraft PE; Delp SL; Blau HM Prostaglandin E2 Is Essential for Efficacious Skeletal Muscle Stem-Cell Function, Augmenting Regeneration and Strength. Proc Natl Acad Sci U A 2017, 114 (26), 6675–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rodgers JT; Schroeder MD; Ma C; Rando TA HGFA Is an Injury-Regulated Systemic Factor That Induces the Transition of Stem Cells into GAlert. Cell Rep. 2017, 19 (3), 479–486. 10.1016/j.celrep.2017.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sampath SC; Sampath SC; Ho ATV; Corbel SY; Millstone JD; Lamb J; Walker J; Kinzel B; Schmedt C; Blau HM Induction of Muscle Stem Cell Quiescence by the Secreted Niche Factor Oncostatin M. Nat. Commun 2018, 9 (1), 1531. 10.1038/s41467-018-03876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Le Grand F; Jones AE; Seale V; Scimè A; Rudnicki MA Wnt7a Activates the Planar Cell Polarity Pathway to Drive the Symmetric Expansion of Satellite Stem Cells. Cell Stem Cell 2009, 4 (6), 535–547. 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Baghdadi MB; Castel D; Machado L; Fukada S; Birk DE; Relaix F; Tajbakhsh S; Mourikis P Reciprocal Signalling by Notch–Collagen V–CALCR Retains Muscle Stem Cells in Their Niche. Nature 2018, 557 (7707), 714–718. 10.1038/s41586-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Safaee H; Bakooshli MA; Davoudi S; Cheng RY; Martowirogo AJ; Li EW; Simmons CA; Gilbert PM Tethered Jagged-1 Synergizes with Culture Substrate Stiffness to Modulate Notch-Induced Myogenic Progenitor Differentiation. Cell. Mol. Bioeng 2017, 10 (5), 501–513. 10.1007/s12195-017-0506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Moyle LA; Cheng RY; Liu H; Davoudi S; Ferreira SA; Nissar AA; Sun Y; Gentleman E; Simmons CA; Gilbert PM Three-Dimensional Niche Stiffness Synergizes with Wnt7a to Modulate the Extent of Satellite Cell Symmetric Self-Renewal Divisions. Mol. Biol. Cell 2020, 31 (16), 1703–1713. 10.1091/mbc.E20-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Silver JS; Günay KA; Cutler AA; Vogler TO; Brown TE; Pawlikowski BT; Bednarski OJ; Bannister KL; Rogowski CJ; Mckay AG; DelRio FW; Olwin BB; Anseth KS Injury-Mediated Stiffening Persistently Activates Muscle Stem Cells through YAP and TAZ Mechanotransduction. Sci. Adv 2021, 7 (11), eabe4501. 10.1126/sciadv.abe4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rayagiri SS; Ranaldi D; Raven A; Mohamad Azhar NIF; Lefebvre O; Zammit PS; Borycki A-G Basal Lamina Remodeling at the Skeletal Muscle Stem Cell Niche Mediates Stem Cell Self-Renewal. Nat. Commun 2018, 9 (1), 1075. 10.1038/s41467-018-03425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bentzinger CF; Wang YX; von Maltzahn J; Soleimani VD; Yin H; Rudnicki MA Fibronectin Regulates Wnt7a Signaling and Satellite Cell Expansion. Cell Stem Cell 2013, 12 (1), 75–87. 10.1016/j.stem.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Gilbert PM; Havenstrite KL; Magnusson KEG; Sacco A; Leonardi NA; Kraft P; Nguyen NK; Thrun S; Lutolf MP; Blau HM Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science 2010, 329 (5995), 1078–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Boontheekul T; Hill EE; Kong H-J; Mooney DJ Regulating Myoblast Phenotype Through Controlled Gel Stiffness and Degradation. Tissue Eng. 2007, 13 (7), 1431–1442. 10.1089/ten.2006.0356. [DOI] [PubMed] [Google Scholar]
- (21).Engler AJ; Griffin MA; Sen S; Bönnemann CG; Sweeney HL; Discher DE Myotubes Differentiate Optimally on Substrates with Tissue-like Stiffness : Pathological Implications for Soft or Stiff Microenvironments. J. Cell Biol 2004, 166 (6), 877–887. 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jiwlawat N; Lynch EM; Napiwocki BN; Stempien A; Ashton RS; Kamp TJ; Crone WC; Suzuki M Micropatterned Substrates with Physiological Stiffness Promote Cell Maturation and Pompe Disease Phenotype in Human Induced Pluripotent Stem Cell-Derived Skeletal Myocytes. Biotechnol. Bioeng 2019, 116 (9), 2377–2392. 10.1002/bit.27075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kloxin AM; Kasko AM; Salinas CN; Anseth KS Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science 2009, 324 (5923), 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yang C; Tibbitt MW; Basta L; Anseth KS Mechanical Memory and Dosing Influence Stem Cell Fate. Nat Mater 2014, 13, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rosales AM; Vega SL; DelRio FW; Burdick JA; Anseth KS Hydrogels with Reversible Mechanics to Probe Dynamic Cell Microenvironments. Angew Chem Int Ed 2017, 56 (40), 12132–12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ruskowitz ER; DeForest CA Proteome-Wide Analysis of Cellular Response to Ultraviolet Light for Biomaterial Synthesis and Modification. ACS Biomater. Sci. Eng 2019, 5 (5), 2111–2116. 10.1021/acsbiomaterials.9b00177. [DOI] [PubMed] [Google Scholar]
- (27).Herum KM; Choppe J; Kumar A; Engler AJ; McCulloch AD Mechanical Regulation of Cardiac Fibroblast Profibrotic Phenotypes. Mol Biol Cell 2017, 28, 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Malkoch M; Vestberg R; Gupta N; Mespouille L; Dubois P; Mason AF; Hedrick JL; Liao Q; Frank CW; Kingsbury K; Hawker CJ Synthesis of Well-Defined Hydrogel Networks Using Click Chemistry. Chem Commun 2006, 14, 2774–2776. [DOI] [PubMed] [Google Scholar]
- (29).DeForest CA; Polizzotti BD; Anseth KS Sequential Click Reactions for Synthesizing and Patterning Three-Dimensional Cell Microenvironments. Nat Mater 2009, 8, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Matsunaga T; Sakai T; Akagi Y; Chung U; Shibayama M SANS and SLS Studies on Tetra-Arm PEG Gels in As-Prepared and Swollen States. Macromolecules 2009, 42 (16), 6245–6252. [Google Scholar]
- (31).DeForest CA; Tirrell DA A Photoreversible Protein-Patterning Approach for Guiding Stem Cell Fate in Three-Dimensional Gels. Nat Mater 2015, 14, 523–531. [DOI] [PubMed] [Google Scholar]
- (32).Dommerholt J; Schmidt S; Temming R; Hendriks LJA; Rutjes FPJT; van Hest JCM; Lefeber DJ; Friedl P; van Delft FL Readily Accessible Bicyclononynes for Bioorthogonal Labeling and Three-Dimensional Imaging of Living Cells. Angew Chem Int Ed 2010, 49, 9422–9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Badeau BA; Comerford MP; Arakawa CK; Shadish JA; DeForest CA Engineered Modular Biomaterial Logic Gates for Environmentally Triggered Therapeutic Delivery. Nat Chem 2018, 10, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).DeForest CA; Anseth KS Photoreversible Patterning of Biomolecules within Click-Based Hydrogels. Angew Chem Int Ed 2012, 51 (8), 1816–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yamamoto S; Ikegami H; Yamaguchi K; Nakanishi J A Dynamic Biomaterial Based on a 2-Nitrobenzyl Derivative with a Tert-Butyl Substituent at the Benzyl Position: Rapid Response and Minimized Phototoxicity. ChemPhotoChem 2018, 2 (9), 786–790. [Google Scholar]
- (36).Madl CM; Katz LM; Heilshorn SC Tuning Bulk Hydrogel Degradation by Simultaneous Control of Proteolytic Cleavage Kinetics and Hydrogel Network Architecture. ACS Macro Lett 2018, 7 (11), 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Madl CM; Heilshorn SC Rapid Diels-Alder Crosslinking of Cell Encapsulating Hydrogels. Chem Mater 2019, 31 (19), 8035–8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mo A; Mukamel EA; Davis FP; Luo C; Henry GL; Picard S; Urich MA; Nery JR; Sejnowski TJ; Lister R; Eddy SR; Ecker JR; Nathans J Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 2015, 86 (6), 1369–1384. 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Southard S; Low S; Li L; Rozo M; Harvey T; Fan C-M; Lepper C A Series of Cre-ER(T2) Drivers for Manipulation of the Skeletal Muscle Lineage. Genes. N. Y. N 2000 2014, 52 (8), 759–770. 10.1002/dvg.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sacco A; Doyonnas R; Kraft P; Vitorovic S; Blau HM Self-Renewal and Expansion of Single Transplanted Muscle Stem Cells. Nature 2008, 456, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Palla AR; Ravichandran M; Wang YX; Alexandrova L; Yang AV; Kraft P; Holbrook CA; Schürch CM; Ho ATV; Blau HM Inhibition of Prostaglandin-Degrading Enzyme 15-PGDH Rejuvenates Aged Muscle Mass and Strength. Science 2020. 10.1126/science.abc8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Parker SS; Moutal A; Cai S; Chandrasekaran S; Roman MR; Koshy AA; Khanna R; Zinsmaier KE; Mouneimne G High Fidelity Cryopreservation and Recovery of Primary Rodent Cortical Neurons. eNeuro 2018, 5 (5), ENEURO.0135–18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Huang L-K; Wang M-JJ Image Thresholding by Minimizing the Measures of Fuzziness. Pattern Recognit. 1995, 28 (1), 41–51. 10.1016/0031-3203(94)E0043-K. [DOI] [Google Scholar]
- (44).Zammit PS; Golding JP; Nagata Y; Hudon V; Partridge TA; Beauchamp JR Muscle Satellite Cells Adopt Divergent Fates. J. Cell Biol 2004, 166 (3), 347–357. 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Montarras D; Morgan J; Collins C; Relaix F; Zaffran S; Cumano A; Partridge T; Buckingham M Direct Isolation of Satellite Cells for Skeletal Muscle Regeneration. Science 2005, 309 (5743), 2064–2067. 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- (46).Lacraz G; Rouleau A-J; Couture V; Söllrald T; Drouin G; Veillette N; Grandbois M; Grenier G Increased Stiffness in Aged Skeletal Muscle Impairs Muscle Progenitor Cell Proliferative Activity. PLoS One 2015, 10 (8), e0136217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Rosant C; Nagel MD; Perot C Aging Affects Passive Stiffness and Spindle Function of the Rat Soleus Muscle. Exp Gerontol 2007, 42, 301–308. [DOI] [PubMed] [Google Scholar]
- (48).Gao Y; Kostrominova TY; Faulkner JA; Wineman AS Age-Related Changes in the Mechanical Properties of the Epimysium in Skeletal Muscles of Rats. J Biomech 2008, 41, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kovanen V; Suominen H; Risteli J; Risteli L Type IV Collagen and Laminin in Slow and Fast Skeletal Muscle in Rats–Effects of Age and Life-Time Endurance Training. Coll Relat Res 1988, 8, 145–153. [DOI] [PubMed] [Google Scholar]
- (50).Hindle AG; Horning M; Mellish J-AE; Lawler JM Diving into Old Age: Muscular Senescence in a Large-Bodied, Long-Lived Mammal, the Weddell Seal (Leptonychotes Weddellii). J Exp Biol 2009, 212, 790–796. [DOI] [PubMed] [Google Scholar]
- (51).Paliwal P; Pishesha N; Wijaya D; Conboy IM Age Dependent Increase in the Levels of Osteopontin Inhibits Skeletal Muscle Regeneration. Aging 2012, 4, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Cosgrove BD; Gilbert PM; Porpiglia E; Mourkioti F; Lee SP; Corbel SY; Llewellyn ME; Delp SL; Blau HM Rejuvenation of the Muscle Stem Cell Population Restores Strength to Injured Aged Muscles. Nat Med 2014, 20 (3), 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Blau HM; Cosgrove BD; Ho ATV The Central Role of Muscle Stem Cells in Regenerative Failure with Aging. Nat Med 2015, 21 (8), 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Killaars AR; Grim JC; Walker CJ; Hushka EA; Brown TE; Anseth KS Extended Exposure to Stiff Microenvironments Leads to Persistent Chromatin Remodeling in Human Mesenchymal Stem Cells. Adv Sci 2019, 6 (3), 1801483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


