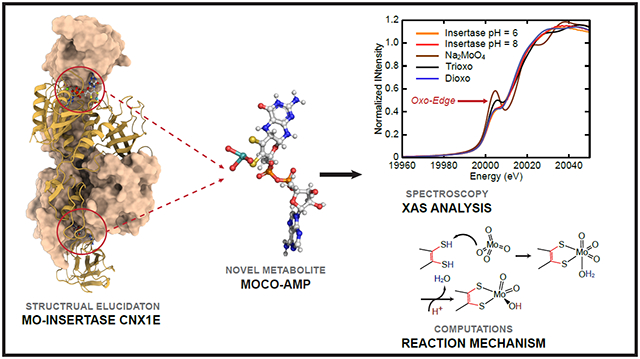
Abstract
The molybdenum cofactor (Moco) is found in the active site of numerous important enzymes that are critical to biological processes. The bidentate ligand that chelates molybdenum (Mo) in Moco is the pyranopterin dithiolene (molybdopterin, MPT); however, neither the mechanism of molybdate insertion into MPT nor the structure of Moco prior to its insertion into pyranopterin molybdenum enzymes is known. Here we report this final maturation step, where adenylated MPT (MPT-AMP) and molybdate are the substrates. X-ray crystallography of the Arabidopsis thaliana Mo-insertase variant Cnx1E S269D D274S identified adenylated Moco (Moco-AMP) as an unexpected intermediate in this reaction sequence. X-ray absorption spectroscopy revealed the first coordination sphere geometry of Moco trapped in the Cnx1E active site. We have used this structural information to deduce a mechanism for molybdate insertion into MPT-AMP. Given their high degree of structural and sequence similarity, we suggest that this mechanism is employed by all eukaryotic Mo-insertases.
Keywords: Molybdenum cofactor, Moco, Moco biosynthesis, Molybdenum insertion, X-ray crystallography, X-ray absorption spectroscopy, EXAFS
Graphical Abstract
Molybdenum cofactor (Moco) dependent enzymes (Mo-enzymes) are ubiquitous, and are commonly found in archaea, bacteria, and eukaryota1-3. These enzymes are involved in numerous redox reactions that contribute to the global sulfur, carbon and nitrogen cycles4, and they are vitally important to a variety of life processes3. This last point is underscored by the fact that specific Moco dependent enzymes are essential to human life. A lack of sulfite oxidase (SOX) activity in humans results in the accumulation of highly toxic sulfite, which subsequently leads to the onset of severe neurological disorders and eventual early childhood death5. Regarding their global importance, the Moco dependent nitrate reductases (NR) are essential enzymes in autotrophic organisms such as plants, and defects in the NR structural gene, or within the Moco biosynthesis pathway, cause severe growth phenotypes6-11. Moco biosynthesis is catalyzed by an evolutionarily old and highly conserved multi-step pathway12. Consequently, this pathway is found in the last universal common ancestor of all life forms (LUCA) where Moco-dependent enzymes likely functioned to assimilate carbon, catalyze oxygen independent hydroxylation reactions, and contribute to energy metabolism13.
Moco biosynthesis is subdivided into four major steps (Figure 1)12. In the first step of the pathway, GTP is converted to 3’,8-cH2GTP14,15, which is subsequently processed to yield cyclic pyranopterin monophosphate (cPMP16). Work with the model plant Arabidopsis thaliana (A. thaliana) showed that cPMP synthesis takes place in the mitochondria, and an ABC type exporter is required for cPMP export into the cytosol of the cell17. Given the high degree of functional conservation, this is assumed to be valid for all eukaryotes12. In the second step of the pathway, cPMP is converted into molybdopterin (MPT). This reaction is characterized by the sequential introduction of the MPT dithiolene sulfurs18, where after each reaction cycle the MPT-synthase (precisely its small subunits) have to be re-sulfurated19. Work on the first and second steps of the Moco biosynthesis pathway were primarily performed using E. coli enzymes, which serve as model enzymes here20. Elucidation of the third and fourth steps in the pathway primarily involved the A. thaliana molybdenum insertase (Mo-insertase) Cnx1 serving as a model enzyme21. Early work with fungal Mo-insertase mutants identified these to be functionally rescued when grown in the presence of millimolar concentrations of sodium molybdate22-24, rendering molybdate complexation by the MPT dithiolene to be independent from an enzyme catalyst under conditions where excess molybdate is available. In the third step, MPT is adenylated by the Mo-insertase G-domain to yield adenylated MPT (MPT-AMP). MPT-AMP is a substrate of the Mo-insertase E-domain, which catalyzes the insertion of molybdate into the MPT dithiolene25. Early25-27 and more recent28-30 work identified this reaction to depend on a critical subset of steps that must occur prior to the actual molybdate insertion reaction. In our more recent work, we identified Cnx1E to possess two mutually exclusive molybdate binding sites, which we assigned as functioning in molybdate binding and insertion, respectively29. These molybdate binding sites are in close proximity to what we previously proposed to be the dithiolene chelate component of bound MPT-AMP28,29. However, a critical question remains regarding the nature of the individual steps and the energetics that define the reaction coordinate for molybdate insertion into MPT-AMP. Recently, a structural analysis of the GephE ADP complex suggested that the active site residue Asp580 is required for functionality, while earlier genetic work identified the homologous aspartate residue in the fungal Mo-insertase CnxE (A. nidulans31) to be vitally important. Furthermore, the corresponding Cnx1 exchange variant (D274E) was consistently found to be catalytically inactive30.
Figure 1: The four steps of molybdenum cofactor biosynthesis.
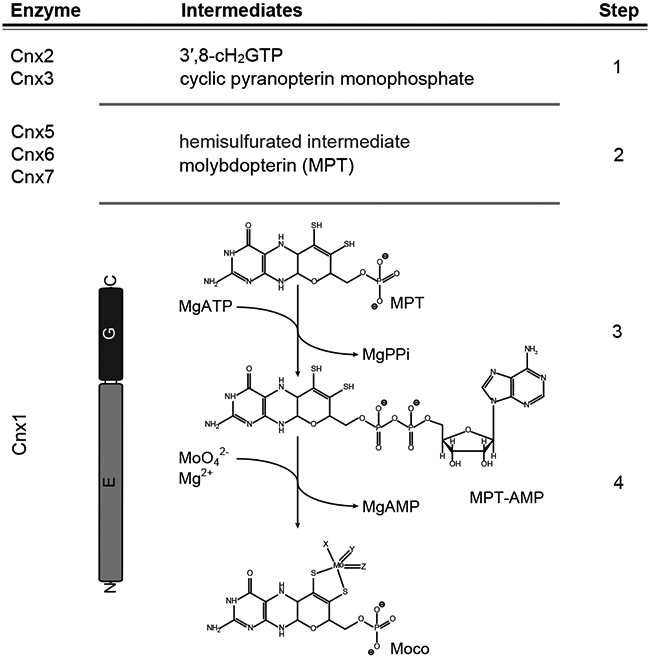
Schematic representation of the plant (Arabidopsis thaliana) molybdenum cofactor (Moco) biosynthesis pathway. The catalyzing enzymes and reaction intermediates are given. In the first step of the pathway, GTP is converted into 3′,8-cH2GTP14,15, which is subsequently turned over into cyclic pyranopterin monophosphate (cPMP16). cPMP is the substrate of the MPT-synthase complex (Cnx6 + Cnx712) catalyzing the formation of a hemisulfurated intermediate and finally molybdopterin (MPT)18. After each reaction cycle, the MPT synthase needs to be re-sulfurated19, which is catalyzed by Cnx5 in Arabidopsis12. MPT is a substrate of the molybdenum insertase (Mo-insertase) Cnx150. Here, the Cnx1-G domain catalyzes the formation of adenylated MPT (MPT-AMP26,27), which is used as the substrate for the E-domain catalyzed molybdate insertion25,26 and results in the formation of physiologically active Moco. Both the reaction mechanism describing molybdate insertion into the MPT dithiol and the first coordination sphere geometry of Moco following this last step in cofactor biosynthesis are unknown.
In this work, we describe the identification of an unexpected Moco precursor viz. adenylated molybdenum cofactor (Moco-AMP), which provides detailed insight into the underlying mechanism of molybdate insertion into the MPT-AMP dithiolene. Remarkably, ATP does not appear to be required as an energy transfer molecule for the actual molybdate insertion process, but appears to be required for the positioning of MPT within the Cnx1E active site. Although the Cnx1E co-structure has been solved here to high resolution (PDB: 6Q32), the precise nature of the Moco-AMP molybdenum coordination environment could not be determined. To address this critical structural issue and provide much-needed insight into the insertion mechanism, we performed a detailed Mo K-edge X-ray absorption spectroscopic (XAS) study of the Cnx1E Moco-AMP complex. The Cnx1E Moco-AMP X-ray absorption near-edge structure (XANES) data have been analyzed in the context of XANES data from small molecule structural analogs of the Moco-AMP Mo site. This comparative analysis yielded the discovery of a dioxo-Mo structure for Cnx1E bound Moco-AMP. This structure is further revealed to be a Mo(VI) [(MPT)MoO2(OH)]1− site by extended X-ray absorption fine structure (EXAFS) experiments, and represents a unique structure observed for any Moco-dependent pyranopterin Mo enzyme32. Defining the nature of the Mo coordination geometry has finally allowed us to deduce the mechanism for molybdate insertion into MPT-AMP; a process that underlies Moco formation. We have thus characterized the Mo-insertase model enzyme Cnx1E from the plant A. thaliana and suggest our proposed mechanism to be valid for all eukaryotic Mo-insertases, since they are both structurally and functionally conserved.
Results and Discussion
X-ray crystallography.
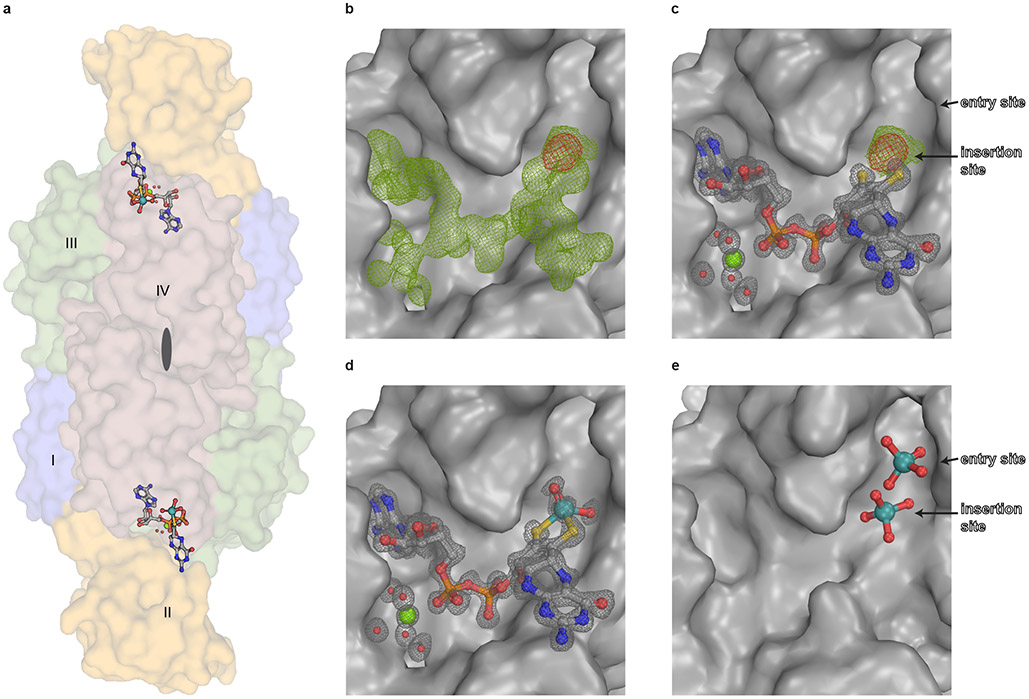
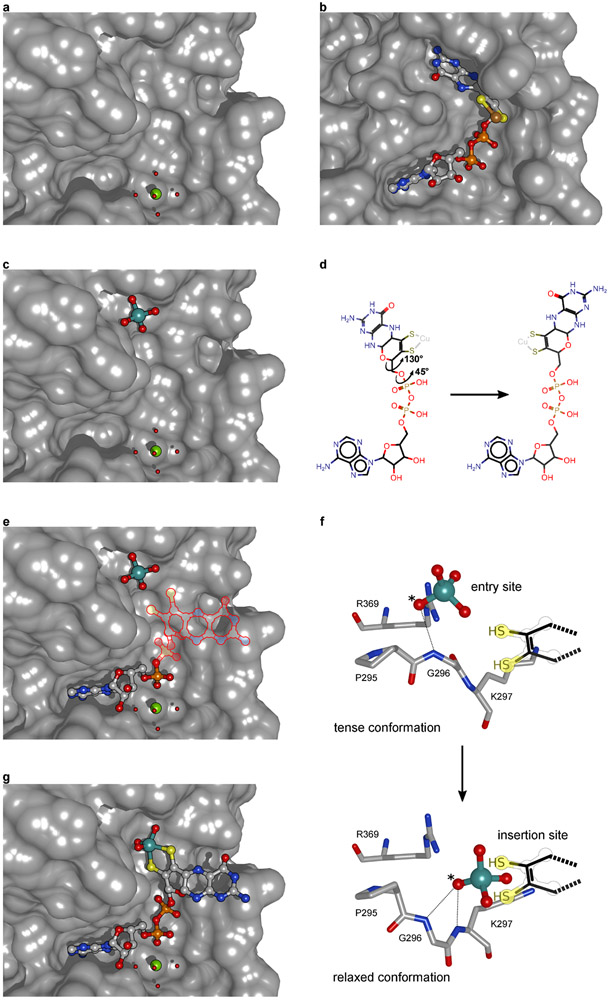
The Cnx1E active site loop30 was targeted by structure-guided mutagenesis experiments to yield the substrate accumulating Cnx1E variant S269D D274S (Supplementary Fig. 1 and 2), the structure of which has been determined by X-ray crystallography to 1.51 Å resolution (PDB: 6Q32; Rfree=0.176; Supplementary Table 1) and is reported here. Within the enzyme-substrate complex, MPT-AMP is bound in a shallow cavity that is located at the interface between Cnx1E sub-domains III (SIII) and II′ (SII′) (Fig. 2A, Supplementary Fig. 3 and 4). Our earlier work identified Cnx1E MPT-AMP binding to correlate with the capability of Cnx1E to form a dimeric enzyme28, a finding which is fully consistent with the identified position of MPT-AMP at the interface of the dimeric enzyme as is shown here (Fig. 2A). Previously, Cnx1E was characterized to bind Moco/MPT with a rather low affinity (KD) in the μM range33. We recently suggested that the low binding affinity for Moco/MPT may be due to the lack of AMP, which when covalently linked to MPT (MPT-AMP) could function to anchor MPT within the Cnx1E active site28,29. Adjacent to the bound MPT-AMP molecule, we observe additional well-defined electron density that is located within bond-length distance to the MPT-AMP dithiolene chelate (Fig. 2B, C). The nature of this electron density is consistent with either a dioxo [(MPT)MoO2(OH)]1− species or a trioxo [(MPT)MoO3]2− species with a distorted square-pyramidal coordination geometry. The presence of a molybdenum atom at the center of this additional electron density is confirmed by anomalous diffraction data (Fig. 2B, C). This finding was unexpected and led us to the conclusion that Cnx1E variant S269D D274S accumulates the novel Moco precursor adenylated Moco (Moco-AMP, Fig. 2D). Analysis of its coordination environment revealed that the Moco-AMP pterin moiety almost exclusively interacts with subdomain II′ (Supplementary Fig. 3 and 4), thus complementing the subdomain III AMP binding function that was suggested to be essential for positioning of the otherwise loosely bound pterin moiety within the E-domain active site28,29. Here, molybdate is initially bound in the vicinity of a strictly conserved set of amino acids29, awaiting its relocation to the catalytically productive site. In addition to molybdate, various other oxyanions can induce cooperative MPT-AMP binding to Cnx1E. However, only molybdate is accepted as a substrate for the insertion reaction25,29, thus rendering molybdate binding and insertion to be distinctly separate processes29.
Figure 2: Structure of Cnx1E variant S269D D274S with bound ligands.
(A) Side view of the Cnx1E physiological dimer in Conolly-surface representation and transparent. The Cnx1E subdomains are colored as follows: subdomain I = slate-blue, subdomain II = yellow-orange, subdomain III = forest green, subdomain IV = dark salmon. Subdomains are indicated (I-IV) for monomer A. (B-E) Electron density of Cnx1E bound ligands. (B) Composite-omit map at 1.5 σ (green) and anomalous map at 3.5 σ (red) of the electron density found in the active site of Cnx1E variant S269D D274S. (C) Feature-enhanced map at 1.5 σ (gray) calculated in the presence of MPT-AMP. Composite map (green) and anomalous map (red) indicate the presence a molybdenum atom near the dithiolene motif. (D) Feature-enhanced map at 1.5 σ calculated in the presence of modeled Moco-AMP. (E) Oxo-anion binding pocket of Cnx1E as seen in the wildtype structure (PDB: 6ETF)29. (A-E) Ligands are shown in ball and stick representation or spheres (magnesium water complex). The following color code was applied: light gray, carbon; red, oxygen; blue, nitrogen; orange, phosphorous; yellow, sulfur; green, magnesium; teal, molybdenum.
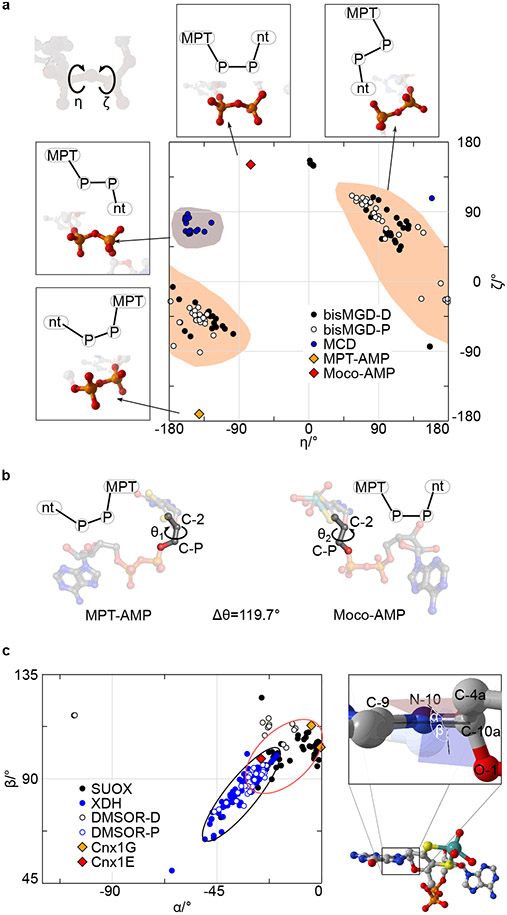
Analogous to Moco-AMP, other molybdenum dinucleotide cofactors, including molydopterin cytosine dinucleotide (MCD) and bis-molybdopterin guanosine dinucleotide (bis-MGD), are commonly found in the active sites of numerous prokaryotic Mo enzymes30. Both MCD and bis-MGD are synthesized in a reaction step that is subsequent to the release of mature Moco from the bacterial Cnx1E homolog by dedicated enzymes that only exist in prokaryotes30. Consequently, Moco-AMP constitutes the first example of a Moco dinucleotide found in eukaryotic organisms. The phosphoric acid anhydride (phosphoanhydride) that connects the two nucleotides can be characterized by its two dihedral angles, η and ζ, which differ among Moco-AMP, MCD, and bis-MGD but are similar between Moco-AMP and MPT-AMP bound to Cnx1G26 (Fig. 3A, Supplementary Table 2). As a consequence of these phosphoric acid anhydride η and ζ values, the nucleotide moieties in MPT-AMP and Moco-AMP are arranged in a “pseudo-cis” conformation relative to an imaginary axis that connects the two phosphorous atoms of the anhydride bond, while the MCD and bis-MGD are in a “pseudo-trans” conformation. However, the orientation of the AMP moiety with respect to Moco in Cnx1E is different from that between AMP and MPT in Cnx1G, which requires a rotation by approximately Δθ=120° around the bond formed between the molybdopterin atom C-2 and the C-P carbon atom connected to the phosphate group (Fig. 3B, Supplementary Fig. 12. This rotation must occur during the transition of MPT-AMP from Cnx1G to Cnx1E because the conformation present in Cnx1G cannot be accommodated by Cnx1E28. The nature of the Mo-dithiolene bonding in Moco-AMP closely resembles that found in Moco-containing enzymes that belong to the sulfite oxidase family34, and this is apparent by the fact that their chiral volumes are of the same sign. However, the conformation of the pyranopterin backbone, as characterized through the dihedral angles α (−26.0°) and β (98.6°), cannot be assigned clearly to one of the two conformations that are typical for the Mo-enzyme families35 (Fig. 3C). In contrast to MCD and bis-MGD, Moco-AMP is relevant only as a cofactor biosynthesis intermediate. Therefore, the identification of Moco-AMP is in complete agreement with the latest model of E-domain functionality28-30. Moco-AMP was found to be bound by both monomers of the Cnx1E dimer, and this is consistent with the current model of E-domain ancestry, which suggests that throughout evolution the Mo-insertase G-domain encoding gene was duplicated36 to form subdomain III, which is opposing subdomain II′ in the dimer (Fig. 2A). Subdomain III retained the ability to bind AMP but lost a helical active site element that is part of the G-domain pterin binding site (Supplementary Fig. 3). This Cnx1G element confines the range of orientations that the MPT-AMP pterin moiety can adopt. The evolutionary loss of this element within the Cnx1E subdomain III is expected to enable the pterin in Cnx1E bound MPT-AMP to adopt the characteristic E-domain conformation, which we suggest is essential for facilitating the catalyzed insertion reaction25,30.
Figure 3: Moco-AMP dihedrals.
(A) A plot of ζ (Pπ–OB–P5’–O5’) vs η (Oπ–Pπ–OB–P5’) for Moco-AMP and the bis-MGD, MCD, and MPT-AMP structures in the Protein Data Bank. The most common regions are highlighted for bis-MGD and MCD. The schematic drawings indicate the orientations of MPT and the second nucleotide relative to each other. (B) Reorientation of MPT-AMP during transition from Cnx1G to Cnx1E. (C) Phosphoric acid anhydride dihedrals found in molybdopterin dinucleotides. A plot of α (C9–N10–C10a–C4a) vs β (C9–N10–C10a–O1) for Moco-AMP and other Moco structures found in the Protein Data Bank. The black and red ellipses indicate the favored regions for pyranopterins in members of the xanthine dehydrogenase and the sulfite oxidase family, respectively.
X-ray absorption spectroscopy.
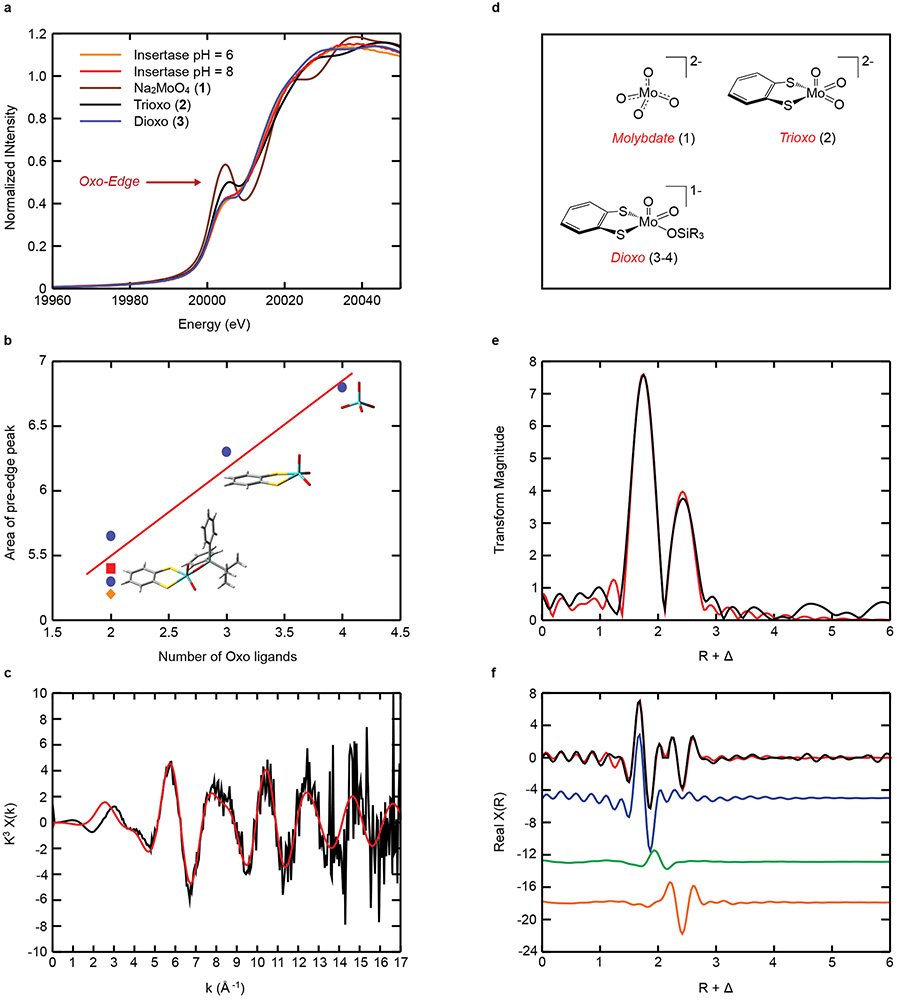
Mo K-edge XANES data have been collected in order to probe the nature of the Mo center of Moco-AMP in Cnx1E S269D D274S at pH 6 and pH 8 (Fig. 4A). These data display an intense pre-edge feature at ~20,006 eV built on the rising Mo K-edge at ~20,016 eV. The intensity of these “oxo edge” pre-edge features has previously been correlated with the number of oxo donor ligands that are coordinated to the Mo ion37,38, with a greater number of oxo ligands leading to a correspondingly greater pre-edge intensity. Thus, the XAS intensity in the pre-edge region is expected to be a sensitive probe of the number of Mo-oxo bonds in Cnx1E bound Moco-AMP, providing a competitive advantage over X-ray crystallography in discerning terminal oxo ligands from hydroxide and aqua ligands. In order to more accurately assess the oxidation state and the number of oxo ligands bound to Mo in Cnx1E bound Moco-AMP, we have compared the enzyme XANES data with sodium molybdate, Na2MoO4 (1), and three synthetic Mo(VI)-dithiolene analog complexes: [(bdt)MoO3](NEt4)2 (2)39, [(bdt)MoO2(OSiPh2tBu)](NEt4) (3)40, and [(bdt)MoO2(OSiPh3)](NEt4) (4)41 (bdt = benzene-1,2-dithiolate) (see Supplementary Fig. 6-9). The nearly identical rising edge inflection point energies observed for Cnx1E bound Moco-AMP proteins and molecules 1-4 clearly show that the Mo ion in Cnx1E Moco-AMP is in the Mo(VI) oxidation state. The strong relationship between the number of oxo ligands bound to Mo and the Mo K-edge “oxo edge” intensity is quantitatively represented in Fig. 4B, where the integrated area for each individual pre-edge peak is plotted as a function of the number of terminal oxo donors. The remarkable similarity between the pre-edge intensities and the overall XANES structure of Cnx1E bound Moco-AMP, 3, and 4 strongly support a Mo(VI) cis dioxo coordination geometry for the Moco-AMP Mo site, which we can evaluate at higher resolution using Mo K-edge EXAFS (Figure 4C-4E, and Supplementary Fig. 5). The EXAFS analysis required the inclusion of a single non-oxo light atom scatter in order to obtain a high-quality fit to the data. Inspection of the real part of the FT EXAFS in Figure 4E shows that this light atom scatterer contributes to the overall shape of this function in the R + Δ = 2.00 Å – 2.25 Å range, providing opposite phase components with respect to the Mo-oxo and Mo-S contributions. The 2.03 Å – 2.12 Å bond distance determined for the light atom scatterer is most consistent with a coordinated hydroxyl ligand (Mo-OH)42 since Mo-aqua bonds are anticipated to be markedly longer (ca. 2.35 Å)42,43. Although a weakly coordinated aqua ligand cannot be completely ruled out, our DFT calculations on [(MPT)MoO2(OH)]1− and (MPT)MoO2(OH2) computational models for Moco yield Mo-OH and Mo-OH2 bond lengths of 1.96 Å and 2.42 Å, respectively, supporting our argument for a coordinated hydroxide (Supplementary Fig. 10). The bond lengths (R) and the number of scatterers of a given atom type (N) determined from the Moco-AMP EXAFS analysis are compared with crystal structure values for trioxo model compound 239, dioxo model compound 340, and a dioxo computational model (listed in Supplementary Table 3). EXAFS parameters for various values of N at pH 6 and pH 8 are presented in Supplementary Tables 4-5. Although there is precedence for a [(MPT)MoO3]2− site in the C207S variant of human sulfite oxidase44 we could not obtain satisfactory fits to the EXAFS data for a trioxo structure. Additionally, the trioxo structure is inconsistent with our analysis of the “oxo-edge” intensity for Moco in Cnx1E bound Moco-AMP (Figure 4A-4B). In summary, the combined computational, XANES, and EXAFS data strongly support a [(MPT)MoO2(OH)]1− structure for Moco-AMP, which bears close resemblance to the 5-coordinate structures determined for model compounds 3 and 4 that were previously characterized by X-ray crystallography39,41.
Figure 4: Mo K-edge XAS spectra.
(A) Comparison of the XANES region of the Mo K-edge XAS data for Cnx1E variant S269D D274S at pH=6 and pH=8 with model compounds 1-3, which possess 4, 3, and 2 terminal oxo ligands, respectively. (B) Integrated area of the “oxo-edge” pre-edge peaks vs. the number of oxo ligands. The four blue dots (●) represent the model compounds 1-4. Pre-edge peak areas for the insertase proteins at pH6 (■) and pH8 (♦) are also shown. The Mo, S, O, C, and H atoms of the model compounds are represented in cyan, yellow, red, gray, and white, respectively. (C-E) Mo K-edge EXAFS data for Cnx1E variant S269D D274S at pH = 8. (C) EXAFS oscillations in k-space; (D) The small molecule analog complexes 1-4 used in this study. (E) Fourier transforms of the EXAFS data presented in panel C; (F) The real part of the Fourier transform. The data is presented as black lines and the best fits to the data are depicted in red. The blue, green and orange colored lines in Panel E are the individual contributions from the oxo, OH, and S scattering paths, respectively. The Fourier transformed data have been phase-corrected using Mo-oxo backscattering. The bond length resolution (ΔR) expected from EXAFS data is defined by ΔR ≥ π/2Δk44. Therefore, given the range of our k-space data (2.5 – 12 Å−1) we should ideally be able to resolve bond length differences that are greater than ~ 0.17 Å. This resolution allows one to distinguish between Mo-oxo, Mo-thiolate, and Mo-hydroxide/water ligands. Please see Figure S5 for the Cnx1E variant S269D D274S Mo K-edge EXAFS data at pH = 6.
Molybdate insertion into Moco.
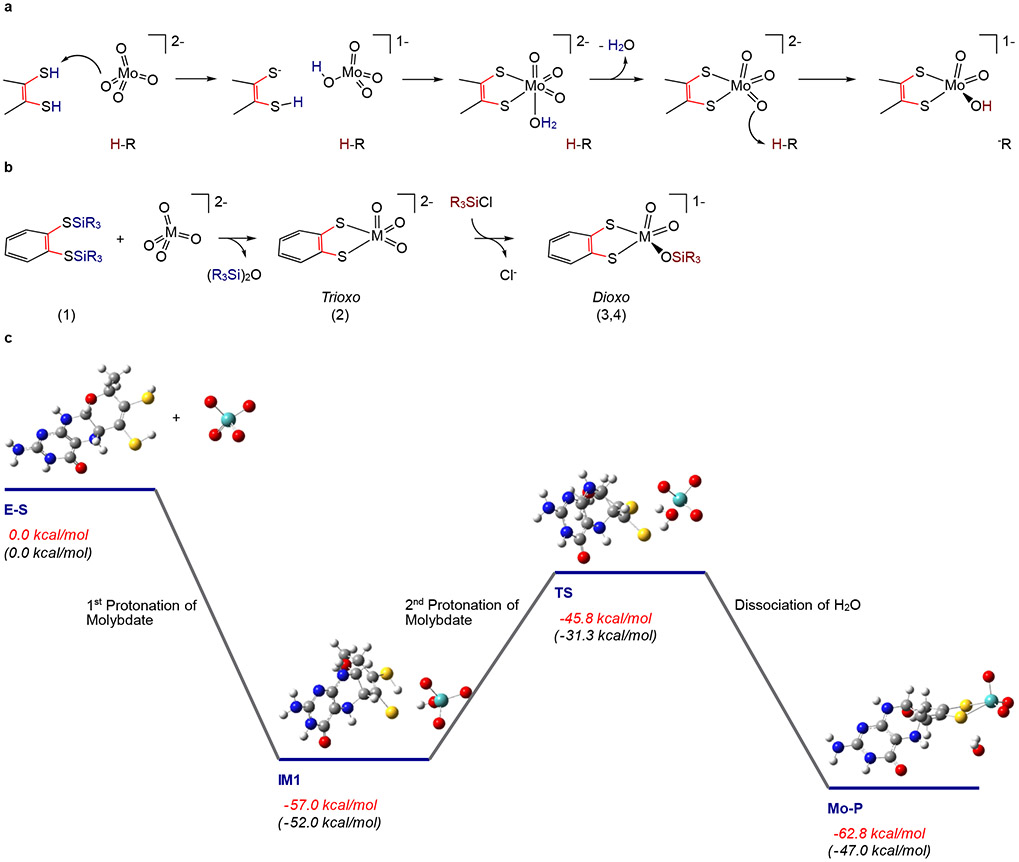
Identification of a [(MPT)MoO2(OH)]1− structure in Cnx1E S269D D274S allows for a mechanism to be posited regarding how molybdate is incorporated into Moco (Figure 5A). A key component of the mechanism is that molybdate (molybdic acid: pKa1 = 3.9; pKa2 =4.4 @ 30C)43,45 must be protonated prior to, or concomitantly with its insertion into the MPT dithiolene. The side chains of Arg (pKa = 12.5), Ser (pKa = 15.0), and Lys (pKa = 10.8), which are most proximal to a molybdate oxo in the binding site (3.0 – 4.7Å; Supplementary Fig. 3 and Fig. 4), could function as proton donors. However, the relatively high pKa values of these side chains would require that they are significantly lowered within the Cnx1E active site in order to protonate molybdate. The diacid form of the MPT dithiolene could also supply the necessary protons, with molybdate properly positioned in the binding pocket through a combination of hydrogen bonding and electrostatic interactions involving these amino acids. Although the pKas for the MPT dithiolene protons are not known, the corresponding dithiolene pKa values for 1,2-benzenedithiol (bdt) are known (pKa1 = 6.5; pKa2 =10.9), with the dithiolene pKa1 being markedly lower than the amino acids in the vicinity of the molybdate binding site. It is known from model chemistry that dithiolene protons (e.g. H2bdt) can labilize strong Mo-oxo bonds, resulting in the elimination of water and insertion of Mo into the dithiolene.40 As a result, we suggest that in Cnx1E the MPT dithiolene supplies the two protons that are required to labilize one oxo ligand from MoO42− and eliminate a water molecule. This proposal is based on (1) the need to deprotonate the dithiolene in order to coordinate Mo, (2) the immediate proximity of the two dithiolene S-H protons to a single terminal oxo ligand of molybdate, and (3) the relatively high acidity of the dithiolene protons compared to nearby amino acids. Thus, the double-protonation of a single molybdate oxygen would function to trigger the release of a water molecule from molybdate, resulting in Mo insertion and the formation of a trioxo [(MPT)MoO3]2− intermediate which, upon an additional protonation step, leads directly to the formation of [(MPT)MoO2(OH)]1−; a structure that is fully consistent with the results of our X-ray crystallographic and XAS data. Remarkably, this biosynthetic structure-based mechanistic proposal for molybdenum incorporation into MPT-AMP bound to Cnx1E is related to the route by which trioxo-M and dioxo-M inorganic model compounds (M = Mo, W; Supplementary Fig. 6-8) can be chemically synthesized (Figure 5B)39-41. For the small molecule analog synthesis, silyl groups (-SiR3) function as proton analogs and the silylation reactions used in the synthesis of 2 effectively mimic the double-protonation of molybdate by the diprotic acid form of the MPT dithiolene to yield [(MPT)MoO3]2−. However, an important difference between the small molecule synthetic route and the Cnx1E catalyzed insertion of molybdate is the markedly greater Si-O bond enthalpy in the model system compared to the H-O bond enthalpy (e.g. the formation of (R3Si)2O (model) vs. H2O (enzyme)). A lower thermodynamic driving force for the formation of a trioxo [(MPT)MoO3]2− species in Cnx1E likely contributes to the requirement for further protonation, leading to the more thermodynamically stable [(MPT)MoO2(OH)]1− structure that we observe experimentally.
Figure 5: Proposed reaction mechanism for Cnx1E catalyzed Mo insertion.
(A) The mechanism of molybdate insertion consistent with X-ray crystallographic and XANES/EXAFS studies on Cnx1E variant S269D D274S. (B) Synthetic scheme for the synthesis of trioxo-(Mo/W) and dioxo-W small molecule analogs of the Cnx1E Mo site showing a remarkable similarity to the proposed mechanism for molybdate insertion catalyzed by Cnx1E. Note that in the small molecule synthetic scheme [SiR3]+ is the functional equivalent of the proton (H+) in the enzyme mechanism. (C) Computed reaction coordinate for the formation of the molybdenum cofactor that is consistent with the mechanism described in A. (E-S) A doubly protonated molybdopterin (MPT) is the likely candidate for activating one of the molybdate Mo-oxo bonds to eliminate water. Although the first protonation is barrierless (IM 1), the second protonation step (IM1→TS→Mo-P) to eliminate water and bind Mo to MPT (yielding [(MPT)MoO3]2−) occurs with a moderate activation barrier. The XAS results show that the initial [(MPT)MoO3]2−product must be further protonated at one of the oxo ligands to yield the [(MPT)MoO2(OH)]1− structure identified in Cnx1E bound Moco-AMP. Formation of [(MPT)MoO2(OH)]1− is expected to prevent the potential back reaction of Mo-P to IM1. Energies in red are the computed total energies, while those in black are the computed Gibbs free energies.
The elegant chemical synthesis of trioxo and dioxo small molecule analogs of the Cnx1E Mo site provides a direct chemical precedent for our proposed insertion mechanism39,41, and we have evaluated this mechanism in Cnx1E by performing linear transit reaction coordinate computations at the DFT level of theory. Specifically, these computations have used our new XAS determined structural details to assess the energetics (total energy and Gibbs free energy) of the individual steps involved in the insertion mechanism (Figure 5C). The initial protonation of molybdate by an MPT dithiolene proton to yield [MoO3(OH)]1− is calculated to be barrierless. This leads to the formation of IM1, which is stabilized by 57.0 kcal/mol relative to E-S. The second protonation requires an activation barrier of 11.2 kcal/mol and leads to the formation of the initial [(MPT)MoO3]2− product (Mo-P) complex with elimination of water, which is only stabilized relative to IM1 by 5.8 kcal/mol. The Gibbs free energies computed along this coordinate display similar behavior with an activation barrier of 20.7 kcal/mol and Mo-P being slightly destabilized relative to IM1 by 5.0 kcal/mol. The fact that both IM1 and the trioxo Mo-product are nearly isoenergetic poses a potential problem with respect to the thermodynamic stability of the trioxo form of Moco. Therefore, we postulate that a final protonation step of the trioxo Mo-product, which results in the formation of [(MPT)MoO2(OH)]1−, must be critical for the overall catalytic efficiency of the insertion reaction. This final protonation event will drive the equilibrium to the [(MPT)MoO2(OH)]1− product side, and effectively prevent a back reaction that leads to Mo-P and the potential for dissociation of Mo from MPT (i.e. IM1). The Cnx1E amino acids Ser328, Arg369, and Ser400 are located at distances of 2.7, 2.9, and 2.8 Å, respectively, with respect to Mo-P (measured without regard for the hydrogen atoms). These distances are within a range compatible with their potential role of protonating [(MPT)MoO3]2−. However, the lower pKa value for Arg renders Arg369 the most likely candidate to protonate [(MPT)MoO3]2− and form [(MPT)MoO2(OH)]1− (Supplementary Fig. 11). We cannot eliminate the possibility that protonation occurs through Ser328 or Ser400 since their actual pKa values could be lower than expected in the protein. However, our assessment of the local protein environment does not provide any evidence that their pKa values are deviate from literature reference values. We note that the Cnx1E amino acids Arg369, Ser328, and Ser400 are all strictly conserved among various eukaryotic Mo-insertases29, providing strong support for their proposed function in the Cnx1E active site.
Moco-AMP as an intermediate of the Moco biosynthesis pathway.
As earlier studies25,26 suggested that MPT-AMP hydrolysis is a prerequisite for molybdate insertion into the MPT dithiolene, we expected a hydrolysis-inactive Cnx1E variant (i.e. Cnx1E S269D D274S) to accumulate both of its substrates: MPT-AMP and molybdate. To our surprise these substrates were not observed, but rather Moco-AMP was found to be bound in the active site. This observation proves that molybdate insertion into the MPT dithiolene is independent of MPT-AMP hydrolysis, and therefore does not require the energy stored within MPT-AMPs phosphor-anhydride bond. This finding is in line with fact that premature MPT-AMP hydrolysis would negate AMP’s anchoring function and adversely affect the efficiency of Cnx1E. However, although Moco-AMP is stable in Cnx1E S269D D274S, we cannot rule out that MPT-AMP hydrolysis in wildtype Cnx1E occurs immediately upon molybdate insertion, rendering Moco-AMP as transient intermediate that quickly proceeds to form Moco. In this respect, Moco-AMP would resemble 3’,8-cH2GTP, the short-lived precursor of cPMP that was structurally characterized through co-crystallization with accumulating active site variants15.
Conclusions
The A. thaliana Mo-insertase Cnx1E functions as molecular scaffold, and is required to align MPT-AMP and molybdate in a catalytically productive geometry. Within MPT-AMP, the AMP moiety functions as an anchor to enable the enzyme to properly position MPT in the active site. While simultaneously maintaining a low Moco/MPT binding affinity46, this strategy would also allow for efficient release of the Moco product. Upon AMP directed positioning of MPT within the Cnx1E active site, Mo insertion is initiated by protonation of the molybdate ion and results in Mo binding to the MPT dithiolene leading to Moco formation. This mechanistic sequence displays a remarkable similarity to the methods by which di-oxo and tri-oxo small molecule analogs of Moco are chemically synthesized39,41, indicating that the underlying principles of molybdate insertion into dithiolene ligands are universal. Subsequent to Mo insertion into MPT-AMP, hydrolysis is required to liberate Moco from the anchoring AMP molecule (Extended Data Fig. 1). Interestingly, when molybdate is replaced by its tungstate congener neither the insertion into the MPT dithiolene chelate nor the hydrolysis reaction occur27,29. Hence we conclude that Moco formation affects a minor re-arrangement of the phosphoric acid anhydride bond, and this renders Moco-AMP compatible with Mg2+ driven hydrolysis25,30. Upon hydrolysis of the phosphoric acid anhydride bond, both AMP and Moco are released from Cnx1E25. When Moco is biosynthesized under fully-defined in vitro conditions, it is physiologically active25 and is accepted by various user enzymes. This leads to the conclusion that the Mo first coordination sphere geometry present in Moco-AMP is also fully compatible with all cellular downstream processes that Moco is subjected to, including Moco transfer, storage and insertion into apo-enzymes47-49. Given the high degree of structural and functional conservation amongst eukaryotic Mo-insertases21,28, we expect that the reaction mechanism described in this work is valid for all eukaryotic Mo-insertases.
METHODS
Cloning of cnx1e Variant s269d d274s –
cnx1e variant s269d d274s was generated by overlap extension PCR and following a protocol published earlier1. Mutagenesis primers used were Cnx1E_S269D_D274S_FusB_for (5′-ggtggtgttgatatgggagacaggtcattcgtcaagc-3′) and Cnx1E_S269D_D274S_FusA_rev (5′-gcttgacgaatgacctgtctcccatatcaacaccacc-3′).
Expression and Purification of Cnx1E –
Expression of Cnx1E wildtype and variant S269D D274S was carried out as described1. For biochemical characterization Cnx1E was purified under aerobic conditions, following established protocols2. For crystallization experiments cell lysis and subsequent purification steps were performed under anaerobic conditions. To enhance the Moco-AMP occupancy, preceding crystallization, for variant S269D D274S in vitro MPT-AMP loading was carried out essentially as described earlier1.
Moco metabolite detection and quantification –
Moco metabolite detection and quantification were carried out as described previously3.
Inductively coupled Plasma Mass Spectrometry (ICP-MS) –
The molybdenum content of wild type Cnx1E and Cnx1E variants was quantified using an Agilent 7700 Series ICP-MS machine (Agilent Technologies) as described earlier2.
Crystallization, data collection and structure determination of Cnx1E variant S269D D274S –
Prior to crystallization, protein preparations were anaerobically buffer exchanged to 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 2% (v/v) Glycerol. Cnx1E S269D D274S was crystallized by sitting-drop vapor diffusion method at 294 K in an anaerobic chamber containing a 95% Ar / 5% H2 atmosphere. Crystals were obtained with a reservoir solution containing 0.1 M MES pH 6.5, 24 %(v/v) PEG400. To all crystallization solutions 500 μM sodium dithionite was added. Crystals were harvested and cryo-protection was accomplished by transferring crystals into reservoir solution containing 10 % (v/v) MPD. Diffraction data were collected at 12,400 eV (0.99987 Å) and 7,100 eV (1.74626 Å) at the Stanford Synchrotron Radiation Lightsource (SSRL) and at the Advanced Light Source (ALS, Berkley). Obtained data sets were indexed and integrated using iMosflm4 and XDS5 and scaled with StarAniso6 taking into account the anisotropic diffraction. Phase information was derived and transplanted from the structure of MPT-AMP-free Cnx1E wt (PDB 5G2R1). Structural refinement and rebuilding were carried out using Buster7 and COOT8. Electron density maps were calculated using FFT (CCP49). Images of protein structures were generated with PyMol10 and UCSF Chimera11. The files containing the structure factors and the structural models were deposited within the Protein Data Bank with accession number 6Q32 (Cnx1E S269D D274S). The complete data collection and refinement statistics are summarized in Table S1.
X-ray Absorption Spectroscopy –
X-ray absorption spectroscopy of Mo K-edge (20,000 eV) for Cnx1E S269D D274S samples were collected at beamline 7-3 of the Stanford Synchrotron Radiation Lightsource (SSRL). Beamline 7-3 is equipped with a Si(220) double-crystal monochromator and a rhodium-coated vertical collimating mirror upstream of the monochromator. X-ray absorption data for the protein samples were collected under the Mo Kα fluorescence mode, while the reference absorption spectra of a Mo metal foil were collected simultaneously under the transmittance mode. The intensity of the incident X-ray was monitored with an argon-filled ion chamber and detuned ~20% away from its maximum to remove the adverse 2nd order harmonic. The incident light passed a Sollar slit and a Zr-3 metal filter onto the sample, and thereafter the florescence emitted from the sample was collected with a 30-element germanium array detector. The protein samples were freshly prepared and filled into the Delrin cells (~150 μL), which were subsequently sealed with thin sulfur-free Kapton tape, and then kept frozen. The temperature for data acquisition was maintained around 10K with an Oxford Instrument liquid helium flow cryostat. The energy of incident light was calibrated to the first reflection point (20,000 eV) of the Mo foil reference spectrum. The energy range for data collection starts from 19,692 eV to k = 18 Å above the Mo K-edge. Each spectrum in the manuscript is an average of six data sweeps. The X-ray absorption data were processed with the XAS analysis software DEMETER (version 0.9.25).12 Energy calibrations, averaging, and normalizations were performed using Athena to produce the X-ray absorption near edge spectra (XANES). The sample energy was calibrated with respect to the second inflection point (20,010 eV) of the molybdenum foil spectrum, and the energy threshold (E0) was set to the second inflection point of the sample (20,016 eV). The extended X-ray absorption fine structure (EXAFS) analysis was carried out in Artemis, in which IFEFF6 was imbedded to calculate the theoretical phase and amplitude functions, and to model the scattering paths. All data sets were fit in k space using k3 weighting, and the k- and R-ranges for fitting models to the data were k = 2.5 - 12 and R = 1 - 3, respectively. S0 was defined as 0.97 and the phase correction was the Mo-oxo scattering path. All other parameters were allowed to float. The resulting fit quality was judged by the R factor. No data smoothing or further data manipulation was performed.
Computational Studies Addressing Mechanism –
All model structures were geometry optimized using Gaussian 09, (revision C.01)13 at the density functional theory (DFT) level utilizing the B3LYP hybrid exchange-correlation functional. A 6-31g* basis set was used for all light atoms, and the LANL2DZ basis set with a LANL2 effective core potential was used for Mo. Solvation effects were taken into account using the polarizable continuum model (PCM) in the SCRF method with a modified dielectric value of 4 to mimic the protein environment. Transition state searches using the QST3 method and a single negative frequency was observed for the TS structure.
Extended Data
Extended Data Fig. 1. The proposed mechanism of molybdate insertion into the MPT dithiolene moiety.
(A) Ground state of Cnx1E with a Mg2+-water complex bound. Mg2+ constitutes the cofactor for the Cnx1E catalyzed reaction25,30. (B) Conformation of MPT-AMP bound in the active site of Cnx1G26. (C) First state of the catalysis with the substrate molybdate bound to the entry site. The presence of molybdate in the entry site induces an unfavorable (tense) backbone conformation in Cnx1E29. (D) The conformation of MPT-AMP found in Cnx1G is not compatible with the Cnx1E active site28. Hence, the MPT-AMP pterin moiety must re-orient upon transfer from Cnx1G to Cnx1E, mostly involving two bond rotations. (E) Second state of the catalytic cycle with molybdate bound to the entry site and active site bound MPT-AMP. The orientation of the pterin moiety was derived from active site bound Moco-AMP identified in this work. (F) The transfer of molybdate from the entry to the insertion site is assisted by the relaxation of the tense backbone conformation29. According to our model of molybdate insertion, one molybdate oxygen ligand is two-fold protonated by the MPT dithiolene function and released as water. Simultaneously, the sulfur to molybdenum bonds are formed: This mechanism begins with a proton transfer from the diacid form of the MPT dithiolene side chain to a molybdate oxygen. A second, less activated protonation on the same oxygen atom would then yield a [(MPT)MoO3(OH2)]2− transition state. This [(MPT)MoO3(OH2)]2−species is expected to possess a highly labile aqua ligand, which would subsequently dissociate from Mo along the reaction coordinate to yield a more stable trioxo [(MPT)MoO3]2− intermediate. A final protonation of basic [(MPT)MoO3]2− by a suitable Cnx1E active site residue (most likely residue Arg369) yields our XAS derived [(MPT)MoO2(OH)]1− structure for Cnx1E Moco-AMP (for details see Figure 5 and supplementary Figure 11). The protonated oxygen atom is marked by an asterisk. (G) The Cnx1E active site with bound Moco-AMP.
Supplementary Material
ACKNOWLEDGEMENTS
Correspondence and requests for materials should be addressed to M.L.K and T.K. We thank Adelina Calean (TU Braunschweig) for excellent support with ICP-MS analysis. M.L.K. gratefully acknowledges support of this research by the National Institutes of Health (Grant No. GM-057378 to MLK). M.L.K. and J.Y. thank the UNM Center for Advanced Research Computing, supported in part by the National Science Foundation, for providing computing resources used in this work. Work at Caltech was supported by the National Institutes of Health (NIH) grant GM045162. We acknowledge the Gordon and Betty Moore Foundation and the Beckman Institute at Caltech for their support of the Molecular Observatory at Caltech and the staff at Beamline 12-2 and 7-3. M.L.K. and J.Y. acknowledge the Stanford Synchrotron Radiation Lightsource, which is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. R.R.M. and T.K. gratefully acknowledge the support of this research by the Deutsche Forschungsgemeinschaft (GRK 2223/1).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
SUPPLEMENTARY
Includes details of experimental data, procedural details, synthesis and characterization data, NMR spectra, the X-ray absorption spectroscopy (XAS) experiment, XAS data analysis, and electronic structure computations. Supplementary figures and tables are freely accessible on nature.com.
DATA AVAILABILITY STATEMENT
The protein structure data that support the findings of this study are publicly available from the Worldwide Protein Data Bank (https://www.rcsb.org/) with accession number 6Q32. We deposited the protein structure along with the structure factor data file that allows for the re-computing and re-evaluation of the structure. Figures 2, 3, S3, S4, and S12 are associated with these raw data. Manuscript datasets are available as Supplementary Data files, which are freely accessible on nature.com.
REFERENCES FOR MAIN TEXT
- 1.Hille R, Schulzke C & Kirk ML in RSC Metallobiology Series (eds Garner CD, Sun H, Wedd A, & Ciurli SL) (The Royal Society of Chemistry, Cambridge, UK, 2017). [Google Scholar]
- 2.Kirk ML & Stein B in Comprehensive Inorganic Chemistry II (Second Edition) (eds Reedijk Editors-in-Chief: Jan & Poeppelmeier Kenneth) 263–293 (Elsevier, 2013). [Google Scholar]
- 3.Hille R, Hall J & Basu P The Mononuclear Molybdenum Enzymes. Chemical Reviews 114, 3963–4038, doi: 10.1021/cr400443z (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stiefel EL The biogeochemistry of molybdenum and tungsten. Molybdenum and Tungsten: Their Roles in Biological Processes 39, 1–29 (2002). [PubMed] [Google Scholar]
- 5.Schwahn BC et al. Efficacy and safety of cyclic pyranopterin monophosphate substitution in severe molybdenum cofactor deficiency type A: a prospective cohort study. Lancet 386, 1955–1963, doi: 10.1016/s0140-6736(15)00124-5 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Mendel RR & Schwarz G Molybdenum cofactor biosynthesis in plants and humans. Coord. Chem. Rev 255, 1145–1158, doi: 10.1016/j.ccr.2011.01.054 (2011). [DOI] [Google Scholar]
- 7.Unkles SE et al. The Aspergillus nidulans cnxABC locus is a single gene encoding two catalytic domains required for synthesis of precursor Z, an intermediate in molybdenum cofactor biosynthesis. J. Biol. Chem 272, 28381–28390, doi: 10.1074/jbc.272.45.28381 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Appleyard M et al. The Aspergillus nidulans cnxF gene and its involvement in molybdopterin biosynthesis - Molecular characterization and analysis of in vivo generated mutans. J. Biol. Chem 273, 14869–14876, doi: 10.1074/jbc.273.24.14869 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Unkles SE, Heck IS, Appleyard M & Kinghorn JR Eukaryotic molybdopterin synthase - Biochemical and molecular studies of Aspergillus nidulans cnxG and cnxH mutants. J. Biol. Chem 274, 19286–19293, doi: 10.1074/jbc.274.27.19286 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Millar L et al. Deletion of the CnxE gene encoding the gephyrin-like protein involved in the final stages of molybdenum cofactor biosynthesis in Aspergillus nidulans. Molecular Genetics and Genomics 266, 445–453 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Probst C et al. Genetic characterization of the Neurospora crassa molybdenum cofactor biosynthesis. Fungal Genetics and Biology 66, 69–78, doi: 10.1016/j.fgb.2014.02.004 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Mendel RR The Molybdenum Cofactor. J. Biol. Chem 288, 13165–13172, doi: 10.1074/jbc.R113.455311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss MC et al. The physiology and habitat of the last universal common ancestor. Nature Microbiology 1, doi: 10.1038/nmicrobiol.2016.116 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Hover BM, Loksztejn A, Ribeiro AA & Yokoyama K Identification of a Cyclic Nucleotide as a Cryptic Intermediate in Molybdenum Cofactor Biosynthesis. J. Am. Chem. Soc 135, 7019–7032, doi: 10.1021/ja401781t (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hover BM, Tonthat NK, Schumacher MA & Yokoyama K Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proc. Natl. Acad. Sci. U. S. A 112, 6347–6352, doi: 10.1073/pnas.1500697112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wuebbens MM & Rajagopalan KV Structural Characterization of a Molybdopterin Precursor. J. Biol. Chem 268, 13493–13498 (1993). [PubMed] [Google Scholar]
- 17.Teschner J et al. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell 22, 468–480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wuebbens MM & Rajagopalan KV Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J. Biol. Chem 278, 14523–14532, doi: 10.1074/jbc.M300453200 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Pitterle DM, Johnson JL & Rajagopalan KV Invitro Synthesis of Molybdopterin From Precursor-Z Using Purified Converting Factor : Role of Protein-Bound Sulfur in Formation of the Dithiolene. J. Biol. Chem 268, 13506–13509 (1993). [PubMed] [Google Scholar]
- 20.Leimkuhler S, Wuebbens MM & Rajagopalan KV The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord. Chem. Rev 255, 1129–1144, doi: 10.1016/j.ccr.2010.12.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruse T Eukaryotic Molybdenum Insertases. Encyclopedia of Inorganic and Bioinorganic Chemistry, 1–6, doi:doi: 10.1002/9781119951438.eibc2736 (2020). [DOI] [Google Scholar]
- 22.Arst HN, Macdonald DW & Cove DJ Molybdate Metabolism in Aspergillus nidulans. Mutations Affecting Nitrate Reductase and/or Xanthine Dehydrogenase. Molecular and General Genetics 108, 129–145, doi: 10.1007/bf02430519 (1970). [DOI] [PubMed] [Google Scholar]
- 23.Joshi MS, Johnson JL & Rajagopalan KV Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J. Bacteriol 178, 4310–4312, doi: 10.1128/jb.178.14.4310-4312.1996 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz G et al. The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 12, 2455–2471, doi: 10.1105/tpc.12.12.2455 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llamas A, Otte T, Multhaup G, Mendel RR & Schwarz G The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin - A novel route toward metal cofactor assembly. J. Biol. Chem 281, 18343–18350, doi: 10.1074/jbc.M601415200 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Kuper J, Llamas A, Hecht HJ, Mendel RR & Schwarz G Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430, 803–806, doi: 10.1038/nature02681 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Llamas A, Mendel RR & Schwarz N Synthesis of adenylated molybdopterin - An essential step for molybdenum insertion. J. Biol. Chem 279, 55241–55246, doi: 10.1074/jbc.M409862200 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Krausze J et al. Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor. Biochem. J 474, 163–178, doi: 10.1042/BCJ20160846 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Krausze J et al. The functional principle of eukaryotic molybdenum insertases. Biochem. J 475, 1739–1753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hercher TW et al. Insights into the Cnx1E catalyzed MPT-AMP hydrolysis. Biosci. Rep 40, doi: 10.1042/bsr20191806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heck IS et al. Mutational analysis of the gephyrin-related molybdenum cofactor biosyndietic gene cnxE from the lower eukaryote Aspergillus nidulans. Genetics 161, 623–632 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirk ML in Molybdenum and Tungsten Enzymes: Spectroscopic and Theoretical Investigations RSC Metallobiology Series No. 7 (eds Hille Russ, Schulzke Carola, & Kirk Martin L.) 13–67 (The Royal Society of Chemistry, Cambridge, UK, 2016). [Google Scholar]
- 33.Schwarz G, Boxer DH & Mendel RR in Chemistry and Biology of Pteridines and Folates (ed Pfleiderer W & Rokos H) 697–702 (Blackwell Science, 1997). [Google Scholar]
- 34.Schwarz G & Mendel RR Molybdenum Cofactor Biosynthesis and Molybdoenzymes Annual Review of Plant Biology 57, 623–647, doi: 10.1146/annurev.arplant.57.032905.105437 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Rothery RA, Stein B, Solomonson M, Kirk ML & Weiner JH Pyranopterin conformation defines the function of molybdenum and tungsten enzymes. Proc. Natl. Acad. Sci. U. S. A 109, 14773–14778, doi: 10.1073/pnas.1200671109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang S, Nichols J, Rajagopalan KV & Schindelin H The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin. Structure 9, 299–310, doi: 10.1016/s0969-2126(01)00588-3 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Kutzler FW et al. Single-crystal polarized X-ray absorption spectroscopy. Observation and theory for (MoO2S2)2-. J. Am. Chem. Soc 103, 6083–6088 (1981). [Google Scholar]
- 38.George GN et al. Structure of the Active Site of Sulfite Oxidase. X-Ray Absorption Spectroscopy of the Molybdenum (IV), Molybdenum (V), and Molybdenum (VI) Oxidation States. Biochemistry 28, 5075–5080, doi: 10.1021/bi00438a026 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Partyka DV & Holm RH Oxygen/sulfur substitution reactions of tetraoxometalates effected by electrophilic carbon and silicon reagents. Inorg. Chem 43, 8609–8616 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Wang JJ & Holm RH Silylation, sulfidation, and benzene-1,2-dithiolate complexation reactions of oxo- and oxosulfidomolybdates(VI) and -Tungstates(VI). Inorg. Chem 46, 11156–11164 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Lim BS, Willer MW, Miao MM & Holm RH Monodithiolene molybdenum(V,VI) complexes: A structural analogue of the oxidized active site of the sulfite oxidase enzyme family. J. Am. Chem. Soc 123, 8343–8349 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Harris HH, George GN & Rajagopalan KV High-Resolution EXAFS of the Active Site of Human Sulfite Oxidase: Comparison with Density Functional Theory and X-ray Crystallographic Results. Inorg. Chem 45, 493–495 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Liu XD, Cheng J, Sprik M & Lu XC Solution Structures and Acidity Constants of Molybdic Acid. Journal of Physical Chemistry Letters 4, 2926–2930, doi: 10.1021/jz401444m (2013). [DOI] [Google Scholar]
- 44.George GN, Garrett RM, Prince RC & Rajagopalan KV The Molybdenum Site of Sulfite Oxidase: A Comparison of Wild-Type and the Cysteine 207 to Serine Mutant using X-Ray Absorption Spectroscopy. J. Am. Chem. Soc 118, 8588–8592 (1996). [Google Scholar]
- 45.Minubayeva Z & Seward TM Molybdic acid ionisation under hydrothermal conditions to 300 degrees C. Geochim. Cosmochim. Acta 74, 4365–4374, doi: 10.1016/j.gca.2010.04.054 (2010). [DOI] [Google Scholar]
- 46.Schwarz G, Boxer DH & Mendel RR Molybdenum cofactor biosynthesis - The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem 272, 26811–26814, doi: 10.1074/jbc.272.43.26811 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Fischer K et al. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem 281, 30186–30194, doi: 10.1074/jbc.M603919200 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Ringel P et al. Biochemical Characterization of Molybdenum Cofactor-free Nitrate Reductase from Neurospora crassa. J. Biol. Chem 288, 14657–14671, doi: 10.1074/jbc.M113.457960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruse T et al. Identification and Biochemical Characterization of Molybdenum Cofactor-binding Proteins from Arabidopsis thaliana. J. Biol. Chem 285, 6623–6635, doi: 10.1074/jbc.M109.060640 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stallmeyer B, Nerlich A, Schiemann J, Brinkmann H & Mendel RR Molybdenum cofactor biosynthesis: The Arabidopsis thaliana cDNA cnx1 encodes a multifunctional 2-domain protein homologous to a mammalian neuroprotein, the insect protein Cinnamon and three Escherichia coli proteins. Plant Journal 8, 751–762, doi: 10.1046/j.1365-313X.1995.08050751.x (1995). [DOI] [PubMed] [Google Scholar]
REFERENCES FOR METHODS SECTION
- 1.Krausze J et al. Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor. Biochem. J 474, 163–178, doi: 10.1042/BCJ20160846 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Krausze J et al. The functional principle of eukaryotic molybdenum insertases. Biochem. J 475, 1739–1753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hercher TW et al. Insights into the Cnx1E catalyzed MPT-AMP hydrolysis. Biosci. Rep 40, doi: 10.1042/bsr20191806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leslie AGW & Powell HR in Evolving Methods for Macromolecular Crystallography. (eds Read Randy J. & Sussman Joel L.) 41–51 (Springer Netherlands; ). [Google Scholar]
- 5.Kabsch W. XDS. Acta Crystallographica Section D-Biological Crystallography 66, 125–132, doi: 10.1107/s0907444909047337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.STARANISO (Global Phasing Ltd., Cambridge, United Kingdom, 2017). [Google Scholar]
- 7.BUSTER v. 2.10.3 (Global Phasing Ltd., Cambridge, United Kingdom, 2017). [Google Scholar]
- 8.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography 66, 486–501, doi: 10.1107/s0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D-Structural Biology 67, 235–242, doi: 10.1107/s0907444910045749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The PyMOL Molecular Graphics System v. 1.5.0.4 (Schrödinger, LLC.). [Google Scholar]
- 11.Pettersen EF et al. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Ravel B & Newville M ATHENA, ARTEMIS, HEPHAESTUS: Data analysis for X-ray absorption spectroscopy using IFEFFIT. Journal of Synchrotron Radiation 12, 537–541, doi: 10.1107/S0909049505012719 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Gaussian 09 (Gaussian, Inc., Wallingford CT, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein structure data that support the findings of this study are publicly available from the Worldwide Protein Data Bank (https://www.rcsb.org/) with accession number 6Q32. We deposited the protein structure along with the structure factor data file that allows for the re-computing and re-evaluation of the structure. Figures 2, 3, S3, S4, and S12 are associated with these raw data. Manuscript datasets are available as Supplementary Data files, which are freely accessible on nature.com.