Abstract
Organic tea is more popular than conventional tea that originates from fertilized plants. Amino acids inorganic soils constitute a substantial pool nitrogen (N) available for plants. However, the amino-acid contents in soils of tea plantations and how tea plants take up these amino acids remain largely unknown. In this study, we show that the amino-acid content in the soil of an organic tea plantation is significantly higher than that of a conventional tea plantation. Glutamate, alanine, valine, and leucine were the most abundant amino acids in the soil of this tea plantation. When 15N-glutamate was fed to tea plants, it was efficiently absorbed and significantly increased the contents of other amino acids in the roots. We cloned seven CsLHT genes encoding amino-acid transporters and found that the expression of CsLHT1, CsLHT2, and CsLHT6 in the roots significantly increased upon glutamate feeding. Moreover, the expression of CsLHT1 or CsLHT6 in a yeast amino-acid uptake-defective mutant, 22∆10α, enabled growth on media with amino acids constituting the sole N source. Amino-acid uptake assays indicated that CsLHT1 and CsLHT6 are H+-dependent high- and low-affinity amino-acid transporters, respectively. We further demonstrated that CsLHT1 and CsLHT6 are highly expressed in the roots and are localized to the plasma membrane. Moreover, overexpression of CsLHT1 and CsLHT6 in Arabidopsis significantly improved the uptake of exogenously supplied 15N-glutamate and 15N-glutamine. Taken together, our findings are consistent with the involvement of CsLHT1 and CsLHT6 in amino-acid uptake from the soil, which is particularly important for tea plants grown inorganic tea plantations.
Subject terms: Plant molecular biology, Plant physiology
Introduction
Owing to its pleasant flavor and multiple health benefits, tea is the second most popular nonalcoholic beverage worldwide, second only to water. Tea quality largely depends on the contents of polyphenols, caffeine, and theanine in the new shoots used for tea processing. Previous studies have shown that the biosynthesis of these metabolites is related to N (N) conditions1–3, with high-quality tea being produced from plants grown under adequate N levels1,4,5. However, high N fertilization accelerates soil acidification6, which can lead to high accumulations of aluminum, fluorine, and heavy metals (e.g., lead and chromium) in tea leaves, posing potential risks to human health7. In recent decades, tea produced from organic plantations has increased in popularity8,9. For example, in China, during the past two decades, organic tea production has increased >45-fold9.
It has been shown that plants acquire N from the soil in the form of nitrate, ammonium, urea, and amino acids10, with amino acids representing a significant N pool in some soils11–13. Soil amino acids are derived mainly from exoenzymatic decomposition of proteins and peptides of decaying organisms14. Tea plants constitute perennial crops whose leaves are harvested and are periodically pruned 2~3 times per year to maintain vigorous vegetative growth. In this regard, such tea plantation pruning has the potential to produce ~8000 kg ha−1 of pruned litter annually15. This pruned litter contains high levels of amino acids and proteins that can be recycled following its decomposition in the soil. These pruned litter-derived amino acids may serve as an important N source to be taken up by tea plant root systems.
Plant cells, including those of the roots, take up nutrients through a combination of passive and active transport mechanisms. Channels and permeases can participate in passive uptake when soil nutrient concentrations are high, whereas proton-coupled transporters engage in secondary active transport under low-nutrient conditions16–18. Given that soil amino-acid levels are much lower than those within the cells of roots, plasma membrane-localized transporters are generally required for amino-acid uptake from soils18. At present, many plant amino-acid transporters have been identified19–23 and are grouped into two superfamilies: amino acid/auxin permeases (AAAPs) and amino acid-polyamine-choline transporters (APCs)16,24. The AAAP superfamily includes six families, amino-acid permeases (AAPs), lysine and histidine transporters (LHTs), proline transporters (ProTs), GATs (γ-aminobutyric acid transporters), auxin transporters (AUXs), and aromatic and neutral amino-acid transporters (ANTs), whereas the APC superfamily includes members of the cationic amino-acid transporter (CAT) and l-type amino-acid transporter (LAT) families. Amino-acid transporters involved in uptake from the soil belong mainly to the AAP, ProT, and LHT families25–28.
In Arabidopsis, the LHT family members LHT1 and LHT6 have been shown to be critical for amino-acid uptake by plant roots25,29. AtLHT1 displays uptake activity for glutamine (Gln), Ala, Glu, and Asp but not for Arg or Lys. AtLHT6 is involved in the uptake of acidic amino acids (Gln, Ala, and probably phenylalanine) but does not seem to transport basic or other neutral amino acids25. Studies devoted to amino-acid transporters have generally been performed on annual plant species, with much less information available on the molecular mechanisms underlying amino-acid transport in perennial species30–33.
In this study, we measured the amino-acid composition in the soil at a normal (common) tea plantation and an organic tea plantation. Furthermore, we fed tea plants Glu, the most abundant amino acid detected in the soil and found that it was efficiently absorbed and utilized by these plants. We then cloned seven CsLHT genes and determined that CsLHT1 and CsLHT6 were able to transport a broad spectrum of amino acids. Arabidopsis lines overexpressing CsLHT1 and CsLHT6 were found to exhibit increased uptake of exogenously supplied amino acids. In addition, expression patterns and subcellular localization studies provided support for the hypothesis that CsLHT1 and CsLHT6 play important roles in amino-acid uptake into the roots of tea plants.
Materials and methods
Plant culture and amino-acid treatments
Tea plant (Camellia sinensis L.) seeds were grown in plastic pots filled with a mixture of soil (40%) and vermiculite (60%). All the pots were irrigated weekly with tap water. The tea plant growth conditions included 16 h of light and 8 h of darkness, 70% relative humidity, and daytime and nighttime temperatures of 25°C and 18°C, respectively. After germination and growth for 100 d, healthy plants with similar crown sizes and heights were selected for studies performed under hydroponic culture. The hydroponic culture method was as described previously by Yang et al.34. After 30 d in hydroponic culture, healthy plants with similar crown size and height were selected for amino-acid treatments. Nine seedlings were used for each treatment. For long-term feeding experiments, tea seedlings were transplanted into an N-deficient nutrient solution and allowed to grow for 2 d; afterward, a 1 mM Glu solution was added, and the seedlings were allowed to grow for 5 d. The addition of no Glu served as the control. The tea plant roots were then collected for amino-acid content analysis. For short-term feeding experiments, tea seedlings were transplanted into an N-deficient nutrient solution and allowed to grow for 2 d; afterward, a 2 mM 15N-Glu solution was added, and the seedlings were allowed to grow for 6 h or 24 h. The addition of no 15N-Glu served as a control. The tea plant roots were then collected for gene expression analysis and 15N content determination using a DeltaV isotope ratio mass spectrometer (IRMS; Thermo Fisher Scientific, USA).
Amino-acid feeding to Arabidopsis was performed as previously described by Lee et al.17. Arabidopsis seeds were rinsed in 70% (v/v) ethanol for 1 min followed by sterilization in a solution containing 5% (v/v) NaClO for 15 min. The seeds were then washed four times using sterile water and then vernalized in water for 3 d at 4°C. For amino-acid uptake analysis, the seeds were cultured on 1/2-strength MS solid media for 1 week. The seedlings were then transferred to N-free 1/2-strength MS media that included either 1 mM 15N-Glu or 1 mM 15N-Gln and allowed to grow for 6 h. These Arabidopsis seedlings were then collected to determine the 15N content using a DeltaV isotope ratio mass spectrometer. The Arabidopsis growth conditions included 16 h of light and 8 h of darkness, 70% relative humidity, and daytime and nighttime temperatures of 21°C and 18°C, respectively.
Soil sampling and amino-acid extraction and analysis
Amino-acid extraction was performed as previously described by Li et al.35. Soil samples were collected from both a control (normal) fertilized tea plantation (Shizipu), located in Xuancheng, and an organic tea plantation (Dayangdian), located in Hefei, Anhui Province, China. The normal (control) tea plantation was fertilized, annually, whereas at the organic tea plantation chemical fertilizer had not been applied during the past 3 consecutive years. Soil samples were collected as follows: the humus in the surface soil was removed, and then soil to a depth of 10 cm was then collected, using a soil sampler; roots and other litter were then removed and 10 g soil samples were taken for drying, with three independent replicates being performed. Subsequently, 10 ml of water was added to each 4 g aliquot of dried soil and then incubated at 70°C for 12 h to extract amino acids. The samples were then cooled to room temperature, followed by centrifugation at 6000 × g for 10 min, after which the supernatants were then filtered through a 0.22 μm membrane. The filtrates were subsequently analyzed via an amino acid analyzer (Hitachi, L-8900, Japan).
Determination of amino-acid contents in tea plant roots
Amino acids were extracted from 50 mg aliquots of freeze-dried tea plant roots using 5 ml of double-distilled water by boiling at 100°C for 20 min. The samples were then cooled to room temperature before centrifugation at 6000 × g for 10 min, after which the supernatants were then filtered through a 0.22 μm membrane. The filtrates were analyzed as described above.
Total RNA extraction and real-time quantitative RT-PCR analysis
Real-time quantitative RT-PCR was performed, as previously described by Li et al.35. For tissue-specific expression analysis, total RNA was isolated from buds, stems, vascular bundles, first leaves, leaf veins of the first and third leaves, leaf veins of the third and fifth leaves, leaf veins of the fifth leaf, and roots using an RNAprep Pure Plant Plus Kit (polysaccharides & polyphenolics-rich) (TIANGEN, Beijing, China). The RNA concentration and integrity were evaluated via a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and confirmed via gel electrophoresis. Total RNA (1 μg) was reverse-transcribed with Oligo dT primer using a HiScript® II One Step RT-PCR Kit (Vazyme, China). qRT-PCR was performed on a Bio-Rad CFX96 in conjunction with SYBR Green I dye (Vazyme, China), while qPCR was performed on a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Primers were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The results were normalized to those of a housekeeping gene, CsGAPDH, and calculated by the comparative Ct method36. The primers used for real-time quantitative RT-PCR are listed in Supplementary Table S1.
CsLHT expression in yeast cells
To isolate CsLHT genes, PCR was performed using cDNA derived from the roots of the tea plant cultivar Shuchazao (Camellia sinensis cv. Shuchazao). Primers were designed based on the CsLHT nucleotide sequences deposited in the tea plant genome database. For yeast complementation and uptake studies, CsLHT genes were cloned into a yeast pYES2 expression vector (Invitrogen, Carlsbad, CA) and confirmed by sequencing. The primers used for cloning are listed in Supplementary Table S1.
The method of plant gene transformation into yeast cells was as previously described by Hirner et al.37 and Dong et al.38. For amino-acid uptake studies, the Saccharomyces cerevisiae mutant strain 22Δ10α (MATα gap1-1 put4-1 uga4-1 can1::HisG lyp1/alp1::HisG hip1::HisG dip5::HisG gnp1Δ agp1Δ ura3-1) and wild-type strain 23344c were employed. Recombinant plasmids were transferred into these 22Δ10α mutants using a yeast transformation kit (Zymo Research, USA). Synthetic dropout (SD/-Ura) media were used, with 2% glucose as the carbon source for selection and normal growth and with 2% galactose for inducing the expression of CsLHT genes. Yeast cells were grown on nitrogen (N)-free media supplemented with ammonium sulfate or the respective test amino acid as the N source.
Yeast amino-acid transport assays
Yeast amino-acid transport assays were performed, as previously described by Hirner et al.37 and Dong et al.38, with some modifications. For amino-acid uptake experiments, yeast cells were inoculated into YNB media consisting of 2 mM (NH4)2SO4 and grown to an OD600 of 1.0. An aliquot of these cells was then used to inoculate 50 mL of fresh media, after which the cells were allowed to grow overnight. The cells were then harvested at the desired optical density, washed three times with N-free growth media, and collected in the same media. The method for 15N isotope detection, using a DeltaV IRMS (Thermo Fisher Scientific, USA), was as previously described39.
Amino-acid uptake assays were initiated by adding yeast cells (4 mL aliquots) into tubes containing 200 µM 15N-labeled Glu. The yeast cells were collected at 30°C after 0, 2, 5, 10, 20, and 30 min of incubation. Kinetic analyses of amino-acid uptake were initiated by adding 4 mL mixtures of yeast cells into tubes containing 10, 20, 50, 100, 200, or 500 µM 15N-labeled Glu for CsLHT1-expressing cells and 0.5, 1, 2, 5, 10, or 20 mM 15N-labeled Glu for CsLHT6-expressing cells. These yeast cells were collected at 30°C after 10 min of incubation. Amino-acid uptake under different pH levels was initiated by adding 4 mL mixtures of yeast cells into tubes containing 200 µM 15N-Glu in media at a pH of 4, 5, 6, 7, or 8. Inhibitor experiments were performed under the same conditions, except that the incubation medium also included 0.1 mM carbonyl cyanide-m-chlorophenyl hydrazone (CCCP) or 0.1 mM diethylstilbestrol (DES). However, for competition experiments, the medium also included 2 mM unlabeled competitive amino acids. For these assays, yeast cells were collected at 30°C after 10 min of incubation.
Subcellular localization of CsLHT1 and CsLHT6
Subcellular localization of amino-acid transporters was performed, as described previously by Santiago et al.40 and Li et al.41, with some modifications. To investigate subcellular localization, the coding sequences of CsLHT1 and CsLHT6 were amplified and cloned into a PK7WGFS2.0-GFP vector for transient expression in Nicotiana benthamiana leaves according to a previous report35. Three days after infiltration, GFP fluorescence was examined via an Olympus FV1000 confocal laser scanning microscope (Olympus Corporation, Beijing, China) using excitation and emission wavelengths of 484 nm and 507 nm, respectively.
Overexpression of CsLHT1 and CsLHT6 in Arabidopsis
The coding DNA sequences of CsLHT1 and CsLHT6 were cloned into a pB2WG7 vector downstream of the Cauliflower mosaic virus 35 S promoter and then introduced into Agrobacterium tumefaciens (GV3101). Gene transformation into Arabidopsis was performed, as previously described by Zhang et al.42. The floral-dip method was used to generate CsLHT1- and CsLHT6-overexpressing Arabidopsis lines, as well as control plants carrying a pB2WG7,0 empty vector.
Results
Amino-acid composition in tea plantation soils
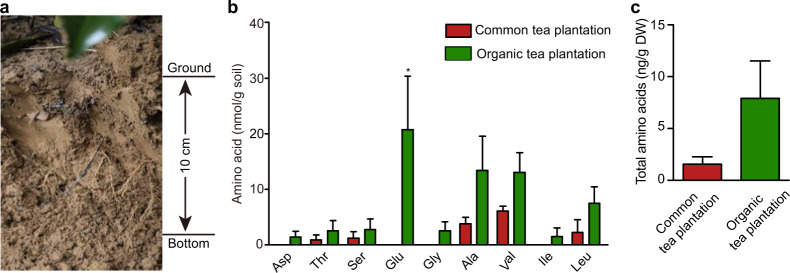
Amino acids have been detected in many soil types11–13. However, there is little information on the amino-acid contents in tea plantation soils. In this study, soil samples were collected from control (common) tea plantation and an organic tea plantation to assess their amino-acid contents (Fig. 1a). Amino acids, including glutamate (Glu), alanine (Ala), valine (Val), leucine (Leu), threonine (Thr), serine (Ser), aspartate (Asp), glycine (Gly), and isoleucine (Ile), were detected in these soils (Fig. 1b). The levels of Glu, Ala, Val, and Leu were higher than those of the other amino acids. The total amino-acid content in the soils was 1–8 μg/g dry soil, with the amino-acid contents in the organic tea plantation soil being higher than those in the conventional tea plantation soil (Fig. 1b, c). These results confirmed the presence of amino acids in the soils of tea plantations, which is consistent with findings in other ecosystems, and implied a more important role of soil amino acids inorganic tea plantations.
Fig. 1. Amino-acid contents in soils of a conventional (control) tea plantation and an organic tea plantation.
a Schematic diagram showing the upper region of the soil from which samples were collected in the present study. Soil samples up to 10 cm beneath the surface were collected. b Contents of various amino acids in soils collected from a conventional (control) and an organic tea plantation. c Total amino-acid contents in the soils of the two tea plantations. The data shown are the means ± SDs (n = three replicates)
Glu feeding increases amino-acid levels in tea plant roots
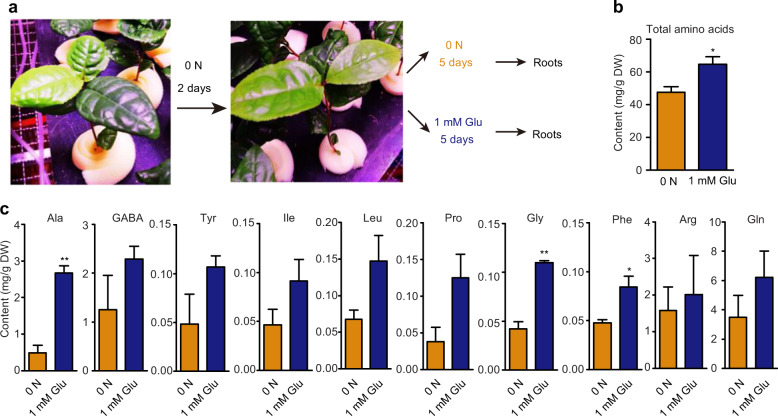
Plant roots can take up various amino acids, such as Asp, Glu, and Gln, over a wide range of concentrations from 0.01 μM to 5 mM37,43. Exogenous Glu can support seedling growth and increase the contents of Glu, Gln, Asp, Ala, Ser, Asn, and γ-aminobutyric acid (GABA) in rice roots44,45. As shown in Fig. 1b, Glu was the most abundant amino acid in the soil of the organic tea plantations investigated in our study (Fig. 1b). We speculated that Glu is an important amino acid that can be directly utilized by tea roots. Therefore, we fed tea plants with Glu, as the sole N source, to test whether tea plants could use amino acids from the media. In this experiment, hydroponically cultured tea plant seedlings were first subjected to an N deficiency (0 N) for 2 d and then provided 1 mM Glu for 5 d, with 0 N for 5 d serving as the control (Fig. 2a). The roots of these seedlings were collected for amino-acid analyses, and we found that 1 mM Glu feeding increased the total amino-acid contents in these roots, especially Ala, GABA, tyrosine (Tyr), Ile, Leu, proline (Pro), Gly, phenylalanine (Phe), and Gln (Fig. 2b, c). These findings support the hypothesis that tea plants can acquire Glu from media as an N source and metabolize it to increase the synthesis of other amino acids.
Fig. 2. Amino-acid contents in the roots of tea seedlings fed exogenous Glu or subjected to N stress.
a Schematic diagram of amino-acid feeding of hydroponically grown tea seedlings. Tea seedlings were first cultured under nitrogen deficiency (0 N) for 2 d, and then fed with 1 mM Glu for 5 d, with seedings kept under 0 N as the control. b Total amino-acid contents in the roots of tea seedlings under 0 N or fed 1 mM Glu. c Levels of various amino acids in the roots of tea seedlings under 0 N or fed 1 mM Glu. The data shown are the means ± SDs (n = 3). The asterisks represent statistical significance determined by Student’s t test (*p < 0.05; **p < 0.01)
Absorption of 15N-labeled Glu by tea roots and identification of CsLHT genes
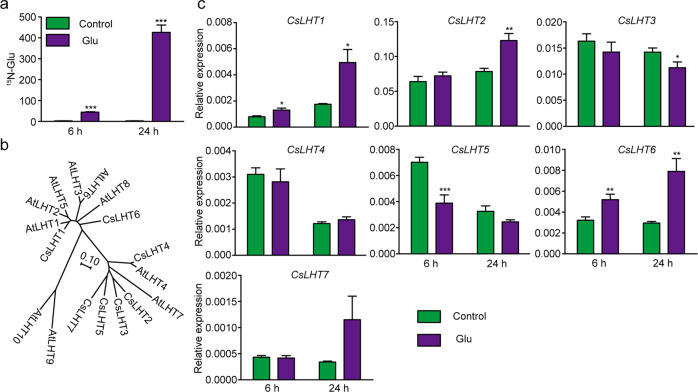
To test whether tea plant roots can acquire amino acids from the soil, we fed hydroponically cultured tea plant seedlings 15N-labeled Glu by adding 15N-Glu to the nutrient solution. After 6 h or 24 h, the 15N levels in the roots increased significantly compared with those in the control roots (Fig. 3a). These results indicated that tea seedlings can directly take up exogenously applied Glu.
Fig. 3. Uptake of 15N-Glu by tea seedling roots and the response of CsLHT genes in the roots to Glu feeding.
a 15N-Glu content in the roots of tea seedlings fed exogenously with 2 mM 15N-Glu for 6 h or 24 h or in the roots of seedlings under 0 N (control). b Phylogenetic relationships among CsLHT and AtLHT genes. The genes and their sequence IDs are as follows: AtLHT1 (AT5G40780), AtLHT2 (AT1G24400), AtLHT3 (AT1G61270), AtLHT4 (AT1G47670), AtLHT5 (AT1G67640), AtLHT6 (AT3G01760), AtLHT7 (AT4G35180), AtLHT8 (AT1G71680), AtLHT9 (AT1G25530), AtLHT10 (AT1G48640), CsLHT1 (TEA026462), CsLHT2 (TEA021847), CsLHT3 (TEA033469), CsLHT4 (TEA029168), CsLHT5 (TEA016092), CsLHT6 (TEA003706), and CsLHT7 (TEA021821). Multiple sequence alignment of full-length proteins was performed, and the phylogenetic tree was constructed using MEGA 6 software. The scale bar represents a 10% amino-acid substitution rate. c Relative expression of CsLHT genes in the roots of tea seedlings fed exogenously with 2 mM 15N-Glu for 6 h or 24 h or in the roots of seedlings under 0 N (control). CsGAPDH was used as an internal control. The data represent the means ± SDs (n = 3). The asterisks represent statistical significance determined by Student’s t test (*p < 0.05; **p < 0.01; ***p < 0.001)
As AtLHT1 and AtLHT6 can mediate amino-acid uptake from the soil25,38, we hypothesized that CsLHTs are involved in amino-acid uptake from the soil in the tea plant. To test this hypothesis, we first identified LHT family members in the tea plant. To this end, we searched for “lysine histidine transporter” in the gene annotations of transcriptome data for tea plant roots38 and identified 13 putative CsLHTs. We next downloaded AtLHT sequence information from The Arabidopsis Information Resource website (www.arabidopsis.org) and used it to identify CsLHT homologs via an online BLAST search39 (http://tpia.teaplant.org/). In this way, we identified two additional CsLHTs. We successfully cloned seven of these 15 CsLHT genes, and they were named CsLHT1-CsLHT7 based on their homologies to AtLHTs (Fig. 3b).
Expression of CsLHT1, CsLHT2, and CsLHT6 in the roots was induced by Glu feeding
To investigate whether the expression of CsLHTs in the roots responds to amino acids in the soil, 100-day-old tea seedlings were transferred to N-deficient media supplemented with 2 mM Glu for 6 h or 24 h. Our results revealed that root expression of CsLHT1 and CsLHT6 was induced at both time points upon Glu feeding (Fig. 3c). Root expression of CsLHT2 was induced at 24 h, whereas CsLHT3, CsLHT4, CsLHT5, and CsLHT7 did not respond to or were repressed by Glu feeding. Hence, these CsLHT genes respond differentially to amino acids within the root media, which may reflect various roles for these genes in tea root physiology. Based on these findings, we proposed that CsLHT1, CsLHT2, and CsLHT6 likely function in the root uptake of amino acids from the soil.
CsLHT1 and CsLHT6 have the capacity to transport amino acids
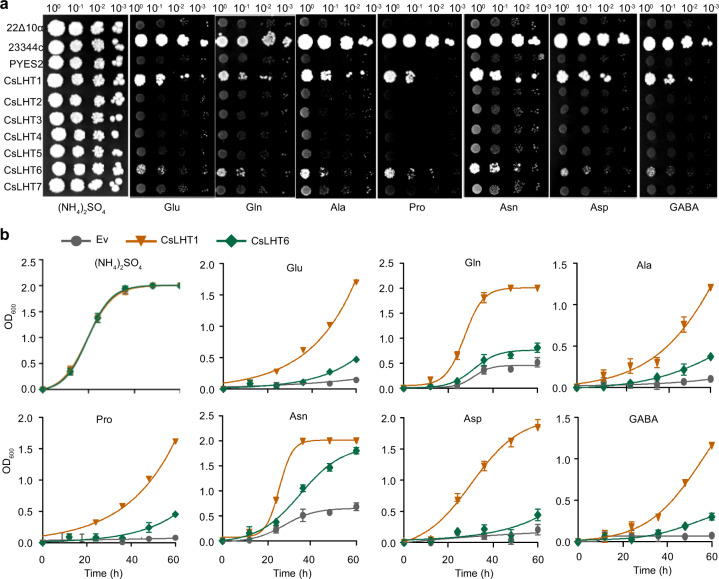
To identify the CsLHT(s) responsible for amino-acid uptake, we next tested whether these CsLHTs could transport amino acids. The yeast mutant strain 22Δ10α lacks 10 amino-acid transporter genes and cannot grow in media with amino acids (except arginine) constituting the sole N source46. Thus, we transferred the CsLHT genes individually into 22Δ10α and tested whether each transformant could grow in media with Glu, Gln, Ala, Pro, Asn, Asp or GABA as the sole N source. Interestingly, the expression of only CsLHT1 or CsLHT6 in this mutant enabled growth, indicating that CsLHT1 and CsLHT6 can transport these amino acids into yeast cells (Fig. 4a). Furthermore, according to these assays, yeast cells expressing CsLHT1 exhibited enhanced growth relative to that of cells expressing CsLHT6 (Fig. 4a). Similar findings were obtained from liquid culture assays (Fig. 4b). These results support the notion that CsLHT1 has a higher affinity for amino acids than does CsLHT6.
Fig. 4. CsLHT1 and CsLHT6 enable the growth of yeast strain 22Δ10α on media with amino acids constituting the sole N source.
Amino-acid transport-defective yeast strain 22Δ10α (genotype MATα gap1-1 put4-1 uga4-1 can1::HisG lyp1-alp1::HisG hip1::HisG dip5::HisG gnp1Δ agp1Δ ura3-1) was employed to analyze the growth of yeast cells expressing CsLHT genes and supplemented with the indicated amino acids (at 2 mM) as the sole N source. 23344 C was used as the wild-type yeast strain. Ammonium sulfate, at 2 mM, served as the control N source. pYES2 empty vectors were used. The CsLHT genes were cloned into pYES2 and then transferred into 22Δ10α. a Yeast growth on YNB solid media. b Yeast growth on YNB liquid media
CsLHT1 and CsLHT6 have different affinities for amino acids
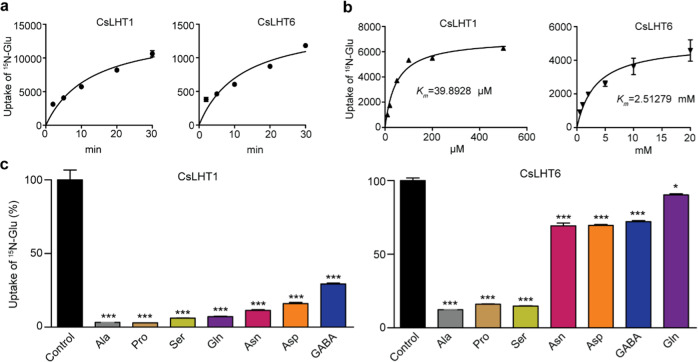
To assess the CsLHT1 and CsLHT6 affinities for amino acids, we next analyzed their transport kinetics for 15N-Glu. Here, the yeast mutant 22Δ10α, harboring CsLHT1 or CsLHT6, was fed 200 μM 15N-Glu, and the 15N taken up into these cells was measured after a 0, 2, 5, 10, 20, or 30 min incubation period (Fig. 5a). As expected, these transport assays demonstrated that 15N-Glu was absorbed and accumulated in the cells expressing CsLHT1 or CsLHT6. Importantly, 15N-Glu accumulation increased in the cells in a time-dependent manner. Moreover, 15N-Glu accumulation in cells expressing CsLHT1 was much higher than that in cells expressing CsLHT6. These results provided further support for the notion that CsLHT1 and CsLHT6 can transport amino acids, albeit with different affinities.
Fig. 5. Kinetic and substrate specificity analyses of amino-acid transport mediated by CsLHT1 and CsLHT6.
a Time course of CsLHT1- and CsLHT6-mediated uptake of 15N-Glu into 22Δ10α mutant cells. b Determination of Km values of CsLHT1/6 for 15N-Glu in yeast. The data represent the means ± SDs (n = 3). c Substrate specificity of CsLHT1 and CsLHT6. 15N-Glu uptake was measured in the presence of a 10-fold excess of the indicated competitors. 15N-Glu uptake without competitive amino acids was set to 100%. The data represent the means ± SDs (n = 3). The asterisks represent statistical significance determined by Student’s t test (*p < 0.05; ***p < 0.001)
A 15N-Glu concentration series (10, 20, 50, 100, 200, and 500 µM) for CsLHT1 and one (0.5, 1, 2, 5, 10, and 20 mM) for CsLHT6 was also employed in our transport kinetics assays; 15N levels were measured in the cells after a 10 min incubation. Concentration-dependent accumulation of 15N-Glu in the cells was observed (Fig. 5b). The kinetic parameters calculated based on these 15N-Glu assays indicated Km values for CsLHT1 and CsLHT6 of 39.9 μM and 2.5 mM, respectively. These data indicated that CsLHT1 acts as a high-affinity-amino-acid transporter, whereas CsLHT6 is a low-affinity transporter.
To analyze CsLHT1 and CsLHT6 substrate specificities, competition experiments were next performed in which 15N-Glu uptake was measured in the presence of a 10-fold excess of unlabeled competitive amino acids. These assays showed that unlabeled Ala, Pro, Ser, Gln, Asn, Asp, and GABA significantly repressed 15N-Glu uptake by CsLHT1. In contrast, only Ala, Pro, and Ser significantly repressed 15N-Glu uptake by CsLHT6 (Fig. 5c). These findings indicated that, as a high-affinity-amino-acid transporter, CsLHT1 has broader substrate specificity than does the low-affinity-amino-acid transporter CsLHT6.
CsLHT1 and CsLHT6 are H+-coupled amino-acid transporters
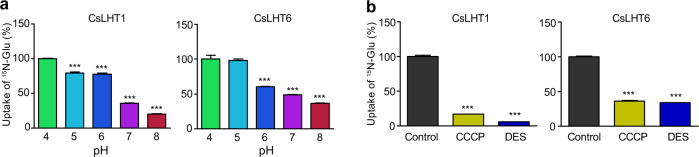
To explore the mechanism through which CsLHT1 and CsLHT6 transport amino acids, we next performed 15N-Glu uptake assays at pH levels of 4.0, 5.0, 6.0, 7.0, and 8.0. These assays revealed that CsLHT1 and CsLHT6 15N-Glu transport activity was high at low pH levels but was significantly lower at neutral and alkaline pH levels (Fig. 6a). Moreover, the addition of the H+-ATPase inhibitor DES and the protonophore CCCP nearly abolished 15N-Glu uptake by CsLHT1 and CsLHT6 (Fig. 6b). These findings support the hypothesis that CsLHT1 and CsLHT6 function as amino acid-proton cotransport systems.
Fig. 6. pH dependence of 15N-Glu uptake into 22Δ10α yeast mutants expressing CsLHT1 or CsLHT6.
a 15N-Glu uptake by CsLHT1 and CsLHT6 under various pH conditions. The uptake at a pH of 4 was set as 100%. The data represent the means ± SDs (n = 3). b 15N-Glu uptake by CsLHT1 and CsLHT6 in the presence of an H+-ATPase inhibitor, diethylstilbestrol (DES), and a protonophore, carbonyl cyanide-m-chlorophenyl hydrazone (CCCP). Yeast cells were incubated for 10 min in YNB media + 15N-Glu, with or without DES or CCCP. 15N-Glu uptake without DES or CCCP was used as a control. The data represent the means ± SDs (n = 3). The asterisks represent statistical significance determined by Student’s t test (***p < 0.001)
CsLHT1 and CsLHT6 are highly expressed in the roots and are localized to the plasma membrane
Tissue-specific expression of genes is generally linked to the biological functions of those genes. For example, AtLHT1 and AtLHT6, functioning in the uptake of amino acids from the soil in Arabidopsis, are highly expressed in roots25,37,47. To assess this relationship for CsLHT1 and CsLHT6, we evaluated their expression in many tea plant tissues, including the leaf buds; first, third and fifth leaves and major veins (MV) (1st leaf and 1st MV; 3rd leaf and 3rd MV; 5th leaf and 5th MV); stems; and vascular bundles extracted from both stem and root tissues (Fig. 7a). Here, CsLHT1 and CsLHT6 were most highly and primarily expressed in the roots, supporting the notion that CsLHT1 and CsLHT6 act in the uptake of amino acids from the soil.
Fig. 7. Tissue-specific expression of CsLHT1 and CsLHT6 and their subcellular localization.
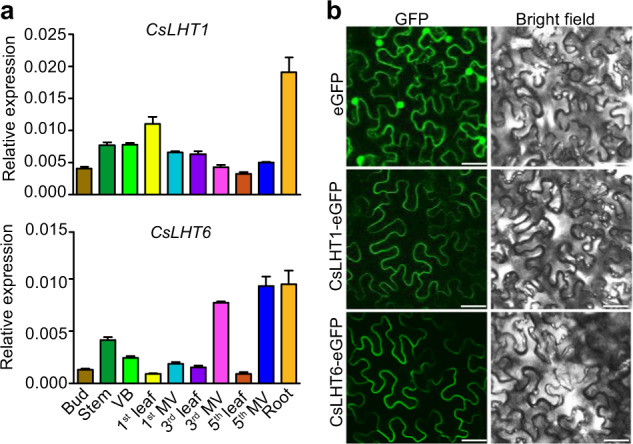
a Expression of CsLHT1/6 in different tea plant tissues. MV major vein, VB vascular bundle, extracted from the indicated stem. The data represent the means ± SDs (n = 3). b CsLHT1-eGFP fusion protein, CsLHT6-eGFP fusion protein and eGFP control transiently expressed in tobacco (N. benthamiana) leaves. Bars = 25 µm
To further explore the mechanism of amino-acid uptake by CsLHT1 and CsLHT6, we examined the subcellular localization of CsLHT1 and CsLHT6. Here, the CaMV 35 S promoter-driven enhanced green fluorescent protein (eGFP)-CsLHT1 or eGFP-CsLHT6 fusion protein and CaMV 35 S promoter-driven eGFP were transiently expressed in epidermal cells of tobacco leaves. Florescent signals of CsLHT1-eGFP and CsLHT6-eGFP were observed at the plasma membrane (Fig. 7b). In contrast, free eGFP florescent signal was distributed within both the cytoplasm and the nucleus. Based on these cellular assays, it appears that CsLHT1 and CsLHT6 are plasma membrane-localized amino-acid transporters, the conclusion of which is also supported by our finding that CsLHT1 and CsLHT6 import amino acids into root cells.
Overexpression of CsLHT1 and CsLHT6 in Arabidopsis increases amino-acid uptake
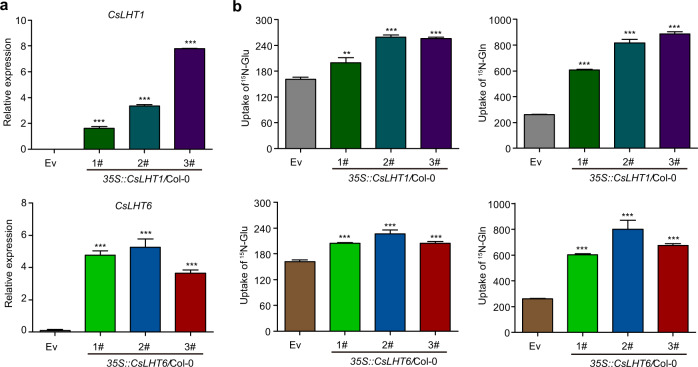
Currently, gene transformation into tea plants remains impractical. Hence, to further verify that CsLHT1 and CsLHT6 participate in amino-acid uptake from the soil, CsLHT1 and CsLHT6 were overexpressed in the model plant species Arabidopsis ecotype Columbia-0 (Col-0) (Fig. 8a). Compared with the empty vector-expressing line, three CsLHT1- or CsLHT6-overexpressing lines presented improved 15N-Glu or 15N-Gln uptake capacity (Fig. 8b). Thus, overexpression of CsLHT1 and CsLHT6 improved amino-acid uptake in planta. Taken together, our findings provide support for a model in which CsLHT1 and CsLHT6 play important roles in amino-acid uptake from the soil into the roots of the tea plant.
Fig. 8. 15N-Glu and 15N-Gln uptake by Arabidopsis lines overexpressing CsLHT1 and CsLHT6.
a Expression levels of CsLHT1 and CsLHT6 in 35 S::CsLHT1 and 35 S::CsLHT6 transgenic Arabidopsis lines. Arabidopsis transformed with an empty vector (Ev) served as the negative control. The data represent the means ± SDs (n = 3). b Uptake of 15N-Glu and 15N-Gln in Ev-, CsLHT1- and CsLHT6-overexpressing Arabidopsis lines. The data represent the means ± SDs (n = 3). The asterisks represent statistical significance determined by Student’s t test (***p < 0.001). The numbers represent individual transgenic lines
Discussion
N is one of the most important mineral macronutrients essential for plant growth and development. In this regard, it is well known that plants have evolved the capacity to absorb N in the form of amino acid, from aqueous solutions12,48. Furthermore, root uptake of amino acids is known to be energy-dependent and regulated by the concentration of amino acids in solution, indicating that their uptake is an active process mediated by specific transporters16–18,37. In this study, we determined that higher amino-acid levels are present in the soil of an organic tea plantation compared with a conventional tea plantation. Furthermore, 15N-Glu-feeding experiments indicated that tea plants can absorb exogenously applied amino acids that can then be used for N assimilation. In addition, we demonstrated that CsLHT1 and CsLHT6 are involved in the uptake of amino acids from the soil in the tea plant.
Amino acids constitute an important N pool inorganic tea plantations
It has been suggested that tea plants grown inorganic tea plantations are subjected to N-deficient conditions due to the absence of inorganic fertilizer9. Compared with conventional tea, that produced under organic management systems contains higher levels of catechins that are linked to antioxidant effects of tea infusions. However, organic tea contains lower levels of amino acids that are also important compounds in terms of tea quality9. The decay of large amounts of pruned tea shoots may contribute significantly to soil amino-acid levels inorganic tea plantations; the decomposition of such organic matter and nutrient recycling depends largely on soil fungi43,44. Interestingly, the long-term application of high amounts of N fertilizer was found to reduce soil fungal diversity in tea plantations49,50. This likely could account for why we observed higher amino-acid contents in the organic tea plantation compared with the conventional tea plantation (Fig. 1). This implies a more important role for soil amino acids in tea plant grown inorganic tea plantations.
Soil amino acids can be used as sources of N for the tea plant
It has been reported that, in addition to inorganic N, amino acids can support tree growth51,52. As a perennial evergreen tree species, the tea plant can also use organic fertilizer. However, the role of soil amino acids in tea plant growth and metabolism has not yet been investigated. In this study, we observed that the tea plant could take up 15N-Glu, and Glu feeding increased the amino-acid contents in the roots (Figs. 2 and 3a). This revealed that tea plants can take up amino acids from the soil for use in the synthesis of other amino acids. In our study, nine amino acids were detected in the soil of an organic tea plantation, and the utilization of exogenous Glu was analyzed in detail. In future studies, it will be important to test the roles of various mixtures of amino acids for use as fertilizers for the growth and metabolism of the tea plant.
Amino acid-induced CsLHT1 and CsLHT6 take up amino acids from the soil in the tea plant
Amino-acid transport in plants involves a number of processes, including uptake from the soil solution, cell-to-cell transport across the root cortex and into the stele, xylem loading, root-to-shoot delivery through the transpiration stream, xylem-to-phloem transfer, translocation via the phloem loading to sink regions of the plant, and phloem unloading, and post-vascular movement into sink cells53. These processes are mediated by plasma membrane-localized amino-acid transporters. In plants, there are hundreds of amino-acid transporters belonging to many families. Among these families, the AAPs, LHTs, cation amino-acid transports (CATs), Proline Transporters (ProTs), and Usually Multiple Acids Move In and out Transporters (UMAMITs) have been functionally well characterized54. In Arabidopsis, LHT1, LHT6, AAP1, AAP5, ProT2 mediate amino-acid uptake from the soil; AAP2 and AAP6 act in amino acid xylem and phloem transfer; AAP2, AAP3, AAP5, AAP8, ProT1, CAT1, CAT6, CAT9 are involved in amino acid phloem loading; UmamiT14, UmamiT18, UmamiT28 and UmamiT29 functions in amino acid phloem export into the embryo54.
The molecular mechanism underlying the uptake of amino acids from the soil by trees has not been thoroughly studied. In this study, we identified seven CsLHTs that were grouped into two clusters, which was consistent with LHTs in Arabidopsis (Fig. 3b). CsLHT1 and CsLTH6 in cluster I have amino-acid transport activity (Fig. 5), which is also consistent with AtLHT1 and AtLHT6. Moreover, these two genes were highly expressed in the roots and both encode plasma membrane-localized proteins (Fig. 7). These findings support the hypothesis that CsLHT1 and CsLHT6 play important roles in amino-acid uptake from the soil (Figs. 4–8). However, the members of cluster II, CsLHT2, CsLHT3, CsLHT4, CsLHT5, and CsLHT7, did not display amino-acid transport activity (Fig. 5). Interestingly, except for AtLHT1 and AtLHT6, there are no other AtLHTs being shown to transport amino acids. It is possible that cluster II LHTs are involved in the transport of metabolites other than amino acids. For example, AtLHT2 was recently shown to transport 1-aminocyclopropane-1- carboxylic acid, a biosynthetic precursor of ethylene, in Arabidopsis55.
CsLHT1 and CsLHT6 may play different roles in the uptake of amino acids from the soil
LHT1 has been thoroughly characterized as a high-affinity-amino-acid transporter and has a major role in the uptake of amino acids from the soil in both Arabidopsis and rice27,29,33,37,43,47. In contrast, there is only one report on the function of AtLHT625; it is highly expressed in the roots, and the atlht6 mutant presented reduced amino-acid uptake from media when supplied with a high amount of amino acids. Although the authors did not characterize the amino-acid transport kinetics for AtLHT6, their results are consistent with this protein being a low-affinity-amino-acid transporter.
In the present study, we characterized CsLHT1 to be a high-affinity amino-acid transporter (Km ~40 μM for 15N-Glu), with a capacity to transport a broad spectrum of amino acids (Figs. 4 and 5). By contrast, CsLHT6 exhibited a much lower affinity (Km ~2.5 mM) for 15N-Glu, and it also displayed higher substrate specificity. Considering that amino-acid concentrations in the soil of tea plantations are low (<30 μM) (Fig. 1), CsLHT1 may play a more important function than CsLHT6 in the uptake of amino acids from the soil into tea plants. However, in soils, amino-acid contents could be much higher, locally, particularly in the vicinity of decomposing animal or vegetable matter56. In this situation, CsLHT6 may play an important role in the uptake of amino acids. In addition, CsLHT6 is also highly expressed in the major veins of mature leaves (Fig. 7a), suggesting a role for CsLHT6 in amino-acid transport within these tea leaves.
Given that protocols for the efficient production of transgenic tea cultivars are lacking, CsLHT1 and CsLHT6 expression cannot be modulated by either overexpression or CRISPR/Cas9 gene editing. However, in China, there is an abundance of tea plant germplasm resources. CsLHT1 and CsLHT6 are potential gene markers for selecting germplasms that can efficiently take up amino acids. Moreover, germplasms with high CsLHT1 or CsLHT6 expression can be used as rootstocks for grafting with elite cultivars to improve the ability of these cultivars to take up amino acids from the soil. Alternatively, these germplasms can be utilized through gene introgression. These grafted lines that can efficiently take up amino acids or novel cultivars should be better suited for use inorganic tea plantations than in conventional tea plantations.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31770731 to Z.Z.), the National Key Research and Development Program of China (2018YFD1000601), the Base of Introducing Talents for Tea Plant Biology and Quality Chemistry (D20026), and the Postdoctoral Science Foundation of China (2020M681636).
Conflict of interest
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00615-x.
References
- 1.Ruan J, Haerdter R, Gerendas J. Impact of nitrogen supply on carbon/nitrogen allocation: a case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants. Plant Biol. 2010;12:724–734. doi: 10.1111/j.1438-8677.2009.00288.x. [DOI] [PubMed] [Google Scholar]
- 2.Du XH, et al. Inorganic nitrogen fertilizers induce changes in ammonium assimilation and gas exchange in Camellia sinensis L. Turk. J. Agric . 2015;39:28–38. doi: 10.3906/tar-1311-82. [DOI] [Google Scholar]
- 3.Kamau DM, Spiertz JH, Oenema O, Owuor PO. Productivity and nitrogen use of tea plantations in relation to age and genotype. Field Crop Res. 2008;108:60–70. doi: 10.1016/j.fcr.2008.03.003. [DOI] [Google Scholar]
- 4.Ruan JY, Gerendas J, Hardter R, Sattelmacher B. Effect of root zone pH and form and concentration of nitrogen on accumulation of quality-related components in green teat. J. Sci. Food Agric. 2007;87:1505–1516. doi: 10.1002/jsfa.2875. [DOI] [Google Scholar]
- 5.Owuor PO, Ng’etich WK, Obanda M. Quality response of clonal black tea to nitrogen fertiliser, plucking interval and plucking standard. J. Sci. Food Agric. 2000;80:439–446. doi: 10.1002/(SICI)1097-0010(200003)80:4<439::AID-JSFA546>3.0.CO;2-2. [DOI] [Google Scholar]
- 6.Yan P, et al. Tea planting affects soil acidification and nitrogen and phosphorus distribution in soil. Agric Ecosyst. Environ. 2018;254:20–25. doi: 10.1016/j.agee.2017.11.015. [DOI] [Google Scholar]
- 7.Ruan JY, Ma LF, Shi YZ, Han WY. Uptake of fluoride by tea plant (Camellia sinensis L) and the impact of aluminium. J. Sci. Food Agric. 2003;83:1342–1348. doi: 10.1002/jsfa.1546. [DOI] [Google Scholar]
- 8.Seyis F, Yurteri E, Ozcan A, Savsatli Y. Organic tea production and tea breeding in Turkey: challenges and possibilities. Ekin J. 2018;4:60–69. [Google Scholar]
- 9.Han WY, et al. Tea from organic production has higher functional quality characteristics compared with tea from conventional management systems in China. Biol. Agric. Hortic. 2018;34:120–131. doi: 10.1080/01448765.2017.1396497. [DOI] [Google Scholar]
- 10.Williams LE, Miller AJ. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Phys. 2001;52:659. doi: 10.1146/annurev.arplant.52.1.659. [DOI] [PubMed] [Google Scholar]
- 11.Friedel JK, Scheller E. Composition of hydrolysable amino acids in soil organic matter and soil microbial biomass. Soil Biol. Biochem. 2002;34:315–325. doi: 10.1016/S0038-0717(01)00185-7. [DOI] [Google Scholar]
- 12.Yu Z, et al. Contribution of amino compounds to dissolved organic nitrogen in forest soils. Biogeochemistry. 2002;61:173–198. doi: 10.1023/A:1020221528515. [DOI] [Google Scholar]
- 13.Kranabetter JM, Dawson CR, Dunn DE. Indices of dissolved organic nitrogen, ammonium and, nitrate across productivity gradients of boreal forests. Soil Biol. Biochem. 2007;39:3147–3158. doi: 10.1016/j.soilbio.2007.06.026. [DOI] [Google Scholar]
- 14.Kielland K, et al. Rapid cycling of organic nitrogen in taiga forest ecosystems. Ecosystems. 2007;10:360–368. doi: 10.1007/s10021-007-9037-8. [DOI] [Google Scholar]
- 15.Sarwar S, Ahmad F, Waheed A, Zaman Q-U. Study on the determination of nutrient status of NTRI tea gardens soils. Sci. Technol. Dev. 2011;30:39–43. [Google Scholar]
- 16.Fischer WN, et al. Amino acid transport in plants. Trends Plant Sci. 1998;3:188–195. doi: 10.1016/S1360-1385(98)01231-X. [DOI] [Google Scholar]
- 17.Lee YH, et al. AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 2007;50:305–319. doi: 10.1111/j.1365-313X.2007.03045.x. [DOI] [PubMed] [Google Scholar]
- 18.Fischer WN, et al. Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 2002;29:717–731. doi: 10.1046/j.1365-313X.2002.01248.x. [DOI] [PubMed] [Google Scholar]
- 19.Ma HL, et al. Genome-wide survey and expression analysis of the amino acid transporter superfamily in potato (Solanum tuberosum L.) Plant Physiol. Biochem. 2016;107:164–177. doi: 10.1016/j.plaphy.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Wu M, et al. Genome-wide survey and expression analysis of the amino acid transporter gene family in poplar. Tree Genet. Genomes. 2015;11:83. doi: 10.1007/s11295-015-0908-4. [DOI] [Google Scholar]
- 21.Zhao HM, Ma HL, Yu L, Wang X, Zhao J. Genome-wide survey and expression analysis of amino acid transporter gene family in Rice (Oryza sativa L.) PLoS ONE. 2012;7:e49210. doi: 10.1371/journal.pone.0049210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng L, et al. Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in Glycine Max. Front. Plant Sci. 2016;7:515. doi: 10.3389/fpls.2016.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia JX, et al. Genome-wide identification and expression analysis of amino acid transporters in the Whitefly, Bemisia tabaci (Gennadius) Int J. Biol. Sci. 2017;13:735–747. doi: 10.7150/ijbs.18153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tegeder M, Offler CE, Frommer WB, Patrick JW. Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol. 2000;122:319–325. doi: 10.1104/pp.122.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perchlik M, Foster J, Tegeder M. Different and overlapping functions of Arabidopsis LHT6 and AAP1 transporters in root amino acid uptake. J. Exp. Bot. 2014;65:5193–5204. doi: 10.1093/jxb/eru278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svennerstam H, Ganeteg U, Nasholm T. Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease. N. Phytol. 2008;180:620–630. doi: 10.1111/j.1469-8137.2008.02589.x. [DOI] [PubMed] [Google Scholar]
- 27.Svennerstam H, et al. Transporters in Arabidopsis roots mediating uptake of amino acids at naturally occurring concentrations. N. Phytol. 2011;191:459–467. doi: 10.1111/j.1469-8137.2011.03699.x. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann S, et al. In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 2011;62:787–796. doi: 10.1093/jxb/erq320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganeteg U, et al. Amino acid transporter mutants of Arabidopsis provides evidence that a non-mycorrhizal plant acquires organic nitrogen from agricultural soil. Plant Cell Environ. 2017;40:413–423. doi: 10.1111/pce.12881. [DOI] [PubMed] [Google Scholar]
- 30.Su YH, Frommer WB, Ludewig U. Molecular and functional characterization of a family of amino acid transporters from. Arabidopsis. Plant Physiol. 2004;136:3104–3113. doi: 10.1104/pp.104.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Bush DR. Expression and transcriptional regulation of amino acid transporters in plants. Amino Acids. 2006;30:113–120. doi: 10.1007/s00726-005-0248-z. [DOI] [PubMed] [Google Scholar]
- 32.Tegeder M, Tan Q, Grennan AK, Patrick JW. Amino acid transporter expression and localisation studies in pea (Pisum sativum) Funct. Plant Biol. 2007;34:1019–1028. doi: 10.1071/FP07107. [DOI] [PubMed] [Google Scholar]
- 33.Wang XH, et al. Disruption of an amino acid transporter LHT1 leads to growth inhibition and low yields in rice. BMC Plant Biol. 2019;19:268. doi: 10.1186/s12870-019-1885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang T, et al. Transcriptional regulation of amino acid metabolism in response to nitrogen deficiency and nitrogen forms in tea plant root (Camellia sinensis L.) Sci. Rep. 2020;10:6868. doi: 10.1038/s41598-020-63835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, et al. Seasonal theanine accumulation and related gene expression in the roots and leaf buds of tea plants (Camellia Sinensis L.) Front. Plant Sci. 2019;10:1397. doi: 10.3389/fpls.2019.01397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using RT-PCR and the 2 (-Delta C (T)) methods. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Hirner A, et al. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong C, et al. Theanine transporters identified in tea plants (Camellia sinensis L.) Plant J. 2019;101:57–70. doi: 10.1111/tpj.14517. [DOI] [PubMed] [Google Scholar]
- 39.Rolletschek H, et al. Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 2005;137:1236–1249. doi: 10.1104/pp.104.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santiago JP, Tegeder M. Connecting source with sink: the role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol. 2016;171:508–521. doi: 10.1104/pp.16.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, et al. Theanine transporters are involved in nitrogen deficiency response in tea plant (Camellia sinensis L.) Plant Signal. Behav. 2020;15:1728109. doi: 10.1080/15592324.2020.1728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, et al. Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in. Arabidopsis. Plant Cell. 2010;22:3603–3620. doi: 10.1105/tpc.110.073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo N, et al. Oryza sativa Lysine-Histidine-type transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. 2020;103:395–411. doi: 10.1111/tpj.14742. [DOI] [PubMed] [Google Scholar]
- 44.Kan C, et al. Exogenous glutamate rapidly induces the expression of genes involved in metabolism and defense responses in rice roots. BMC Genomics. 2017;18:186. doi: 10.1186/s12864-017-3588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia EH, et al. Tea Plant Information Archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 2019;17:1938–1953. doi: 10.1111/pbi.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Besnard J, et al. UMAMIT14 is an amino acid exporter involved in phloem unloading in Arabidopsis roots. J. Exp. Bot. 2016;67:6385–6397. doi: 10.1093/jxb/erw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svennerstam H, Ganeteg U, Bellini C, Nasholm T. Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol. 2007;143:1853–1860. doi: 10.1104/pp.106.092205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipson D, Nasholm T. The unexpected versatility of plants: organic nitrogen use and availability in terrestrial ecosystems. Oecologia. 2001;128:305–316. doi: 10.1007/s004420100693. [DOI] [PubMed] [Google Scholar]
- 49.Cobb AB, et al. The role of arbuscular mycorrhizal fungi in grain production and nutrition of sorghum genotypes: enhancing sustainability through plant-microbial partnership. Agric. Ecosyst. Environ. 2016;233:432–440. doi: 10.1016/j.agee.2016.09.024. [DOI] [Google Scholar]
- 50.Klaubauf S, et al. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fungal Divers. 2010;44:65–75. doi: 10.1007/s13225-010-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang XD, et al. Long-term nitrogen fertilization indirectly affects soil fungi community structure by changing soil and pruned litter in a subtropical tea (Camellia sinensis L.) plantation in China. Plant Soil. 2019;444:409–426. doi: 10.1007/s11104-019-04291-8. [DOI] [Google Scholar]
- 52.Ohlund J, Nasholm T. Growth of conifer seedlings on organic and inorganic nitrogen sources. Tree Physiol. 2001;21:1319–1326. doi: 10.1093/treephys/21.18.1319. [DOI] [PubMed] [Google Scholar]
- 53.Tegeder M. Transporters for amino acids in plant cells: some functions and many unknowns. Curr. Opin. Plant Biol. 2012;15:315–321. doi: 10.1016/j.pbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Dinkeloo K, Boyd S, Pilot G. Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Semin Cell Dev. Biol. 2018;74:105–113. doi: 10.1016/j.semcdb.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 55.Choi J, et al. Identification of lysine histidine transporter 2 as an 1-aminocyclopropane carboxylic acid transporter in Arabidopsis thaliana by transgenic complementation approach. Front. Plant Sci. 2019;10:1092. doi: 10.3389/fpls.2019.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walch-Liu P, et al. Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:1045–1057. doi: 10.1093/pcp/pcj075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.