Abstract
Cells and their surrounding microenvironment exist in dynamic reciprocity, where bidirectional feedback and feedforward crosstalk drives essential processes in development, homeostasis, and disease. With the ongoing explosion of customizable biomaterial innovation for dynamic cell culture, an ever-expanding suite of user-programmable scaffolds now exists to probe cell fate in response to spatiotemporally controlled physiochemical cues. Here, we highlight emerging trends in these efforts, emphasizing strategies that offer tunability over complex network mechanics, present biomolecular cues anisotropically, and harness cells as physiochemical actuators of the pericellular niche. Altogether, these material advances will lead to breakthroughs in our basic understanding of how cells interact with, integrate signals from, and influence their surrounding microenvironment.
Keywords: Dynamic Biomaterials, Hydrogel, Microenvironment, Cell Fate
Graphical Abstract
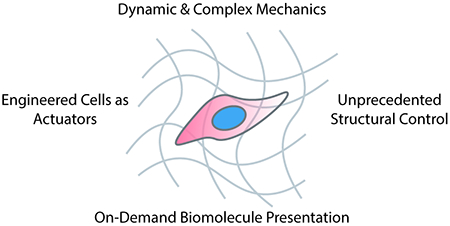
Introduction
It is now widely accepted that the extracellular matrix (ECM) evolves in space and time, harboring persistent recollections of past cellular states. These biological memories are most distinctly present as state-dependent cell-secreted proteins tethered to the ECM and anisotropic variations in matrix mechanics.1,2 Accompanying progression of many diseases, particularly those with a fibrotic element, the ECM binds an altered set of secreted growth factors and develops distinct mechanics from healthy tissue.3,4 With seminal studies highlighting the role of ECM-presented cues in driving significant changes in cell fate,5 researchers now appreciate why seeding healthy cells onto a diseased matrix is often sufficient to induce unhealthy cell phenotypes.6 Therefore, cells exist in dynamic reciprocity with their environment: extracellular cues alter cell behavior, and cells in turn shape their surroundings through secreted bioactive and structural proteins.7
Studies over the past many decades underscore the need to further decouple the ECM’s role in guiding cell behavior throughout development, health, and disease. Engineered microenvironments can provide a user-defined platform in which to precisely tune individual aspects of the ECM to probe and direct encapsulated cell response, increasingly with four-dimensional (4D) and reversible control. To this end, the community has innovated and established a variety of modular hydrogel biomaterial designs that recapitulate critical complexities of the native cellular niche. Here, we highlight recent advances in the synthesis and manipulation of dynamic biomaterials and discuss future strategies to mimic complex biological microenvironments in vitro.
Engineering Tissue Mechanics Beyond the Modulus
Tissue mechanics clearly play an important role in development and disease, and the varied mechanical properties of tissues cannot be fully captured by a single elastic modulus. While most covalently crosslinked polymeric hydrogels are linearly elastic, whole tissues exhibit complex mechanical properties such as strain stiffening/softening and viscoelasticity.8 Though substrate stiffness is often seen as the classic mechanical parameter to characterize and manipulate in an engineered biomaterial, recent efforts have moved to decouple ECM elasticity, viscosity, and fiber thickness/architecture towards elucidating their individual roles on influencing cell function (Figure 1).
Figure 1 –
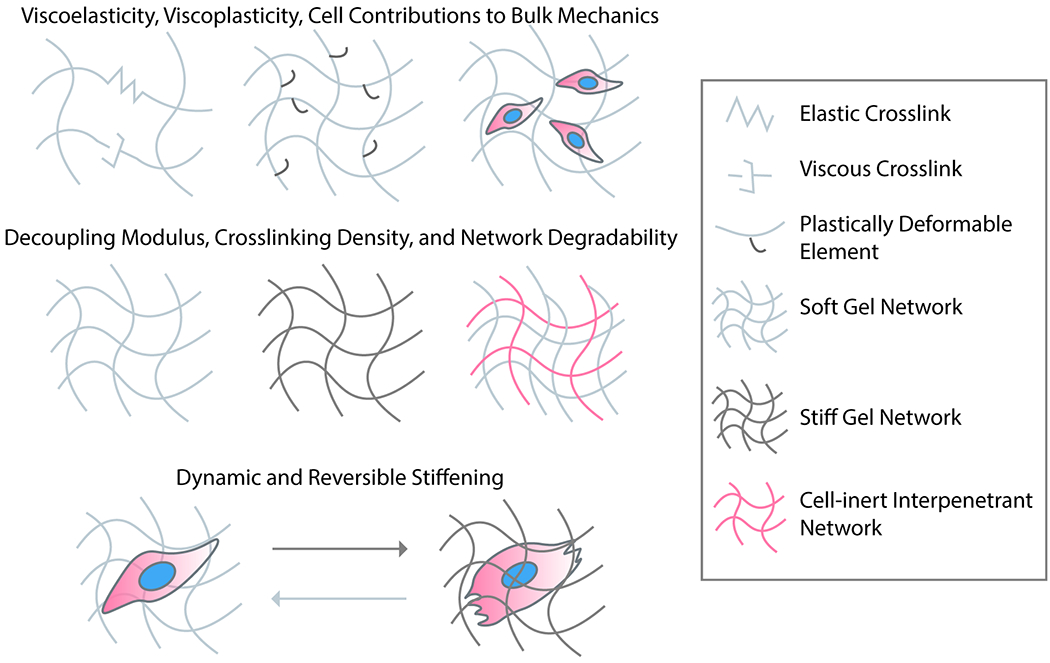
Mimicking complex mechanical aspects of microenvironmental signaling. With the use of modular and synthetic biomaterials, researchers have begun to elucidate the mechanical effects of ECM on cell function beyond its modulus. These properties include the time-dependent components of viscoelasticity and viscoplasticity, as well as dynamic changes to the modulus. Though material modulus and crosslink density are often used interchangeably in the biomaterials community, current efforts seek to decouple material crosslinking, stiffness, degradability, and fibral architecture so as to elucidate their independent effects on cell function.
Underpinning the importance of complex mechanics when engineering cellular microenvironments is the understanding that cellular mechanosensation on and within soft materials is inherently dynamic.9,10 Cells adhere to the matrix through membrane-bound integrins that are clustered into focal adhesions linking the actin cytoskeleton to the ECM. As cells exert spatiotemporally varied forces on their surroundings, time-dependent microenvironmental viscous behaviors complement substrate stiffness in establishing dynamic mechanical reciprocity between intracellular and extracellular tension.
Tuning Viscoelasticity and Viscoplasticity
Non-degradable synthetic polymer hydrogels exhibit dominantly elastic mechanics. This is in stark contrast with the varying stress relaxation responses of soft tissues that can be on the order of several minutes.8 In addition, the viscoelastic properties of tissue have been reported to change throughout the course of disease; patients with cardiomyopathies exhibit increased cardiac muscle viscosity that further contributes to progressive diastolic dysfunction throughout the disease course.11 Conversely, cancerous tissues will also stiffen, but with a lower degree of stress relaxation.12 The viscous behavior of biomaterials has been tuned primarily independent from the storage modulus through encapsulation of non-crosslinked entrapped polymer elements. For example, linear polyacrylamide can be incorporated within a crosslinked polyacrylamide gel to endow this popular and linearly elastic biomaterial platform with tunable viscosity.13 Increasing polyacrylamide viscosity attenuated seeded hepatic myofibroblasts spreading, restoring hallmarks of quiescent hepatic stellate cell phenotype. By incorporating poly(ethylene glycol) (PEG) spacers into an alginate hydrogel, stress relaxation rates can be increased with faster relaxation that drives cell spreading.14 In hyaluronan, the introduction of noncovalent guest-host crosslinks has been used to independently increase the loss modulus of the hydrogel with the same viscosity-dependent effect on cell spreading.15
In addition to the reversible elastic deformation of substrates, cells can also sense the plasticity or irreversible deformation of a biomaterial.16,17 Many natural biomaterials that are noncovalently crosslinked (e.g., gels based on collagen, fibrin, reconstituted basement membrane, agarose, alginate) exhibit some degree of time-dependent plasticity – viscoplasticity.16 Cells encapsulated in these types of materials can plastically remodel their surroundings over time in a manner dependent on integrin-based cellular force transmission and the strength of material crosslinks within the gel. Plasticity can be modulated in a cell adhesion-independent manner through interpenetrating networks of varied molecular weight alginate and ionic crosslinking embedded in a reconstituted basement membrane.17 In this system, highly plastic networks promoted the spreading and invasive behavior of cancer cells independent of matrix modulus or enzymatic degradability. The effects of material plasticity on cell function have also been explored by incorporating PEG spacer sidechains into an alginate hydrogel that are either covalently tethered, dynamically bound and able to rearrange, or free sliding within the network.18 Increased plasticity had profound effects on the transcriptome of mesenchymal stem cells (MSCs) seeded on the gel, especially with respect to pathways regulating focal adhesion remodeling and cell spreading. Gels with intermediate substrate plasticity promoted optimal spreading of MSCs, whereas cell spreading on highly plastic gels could be improved by attenuating cell contraction with the myosin inhibitor blebbistatin.
Whereas many tissues stiffen with compressive strain and soften with extension or shear, natural polymeric hydrogels such as collagen or fibrin do the opposite. These findings highlight the often-overlooked contribution of cells to the overall stiffness and mechanical behaviors of a tissue. Especially at lower strains, the passive stiffness of cells and their cytoskeleton plays a dominant role in dictating tissue stiffness.19 As determined rheologically through progressive decellularization of otherwise intact tissues, cells also contribute to the compressive strain stiffening behavior through both passive stiffness and active contraction.19 These findings offer some explanation as to why many of the most successful hydrogel systems for engineering functional and interconnected constructs are far softer than the tissues they aim to recapitulate.
Independent Control over Gel Mechanics and Network Properties
Another interesting development in the space of engineered tissue mechanics has been the decoupling of stiffness, fiber architecture, and crosslink density in cell-compatible hydrogels. In collagen gels, stiffening the microenvironment by increasing the collagen weight percentage was shown to decrease angiogenic sprouting but stiffening the microenvironment without increasing fiber density through nonenzymatic glycation does the opposite.20 A similar study using pulmonary fibroblasts found that while cells cultured on stiffer gels were more prone to myofibroblastic activation, increased crosslinking density diminished such phenotypic change when cultured in three-dimensional (3D) materials.21 The authors were then able to supplement the hydrogel system with electrospun polymeric fibers in a manner that did not impact bulk storage modulus, demonstrating that increased fiber density promoted fibroblast proliferation and primed for activation.21 Together, these studies reveal the distinct and sometimes opposing effects of fiber density, crosslink density, and substrate stiffness in an engineered biomaterial – three parameters that are often taken for granted as interchangeable in the field. Network crosslink concentration and cell-degradability have also been decoupled in a hydrogel system in which elastin-like polypeptides with varying rates of proteolytic degradation were crosslinked by copper-free click reaction with a suite of non-degradable PEG macromers ranging from 2 to 8 arms.22 To form functional endothelial networks from encapsulated brain microvascular cells, both a low crosslink density and rapid cleavage kinetics were necessary.22
Biomaterials with Dynamic and Reversible Mechanical Control
Temporal evolution of stiffness has been another evolving locus of dynamic biomaterial development. As disease pathophysiology is progressive and chronic, simply lifting cells from a substrate mimicking a healthy mechanical environment and placing them in a diseased environment may not be sufficient to recapitulate the gradual compensation of cells to their changing microenvironment. To overcome this barrier, dynamic materials whose crosslinking density can altered in situ have been the subject of great interest from the field. Many hydrogels that irreversibly stiffen have already been developed, including those based on release of calcium for alginate crosslinking,23 Michael-type addition,24 radical polymerization,25 photoinitiated thiol-ene reaction,26 enzymatic crosslinking,27,28 and anthracene dimerization.29 Similarly, bioorthogonal softening or material degradation can be accomplished through inclusion of a photodegradable moiety (e.g., ortho-nitrobenzyl, allyl sulfide, ruthenium complexes) within a crosslink,30–33 passive crosslinker hydrolysis,34 or even enzymatic transpeptidation in situ.35
With unidirectional control over substrate moduli well established, the field has since turned towards materials capable of reversible stiffening and softening.36 One reversible stiffening cycle can be readily programmed into materials through progressive crosslinking with a subsequently cleavable crosslink, resulting in a system where unidirectional stiffening and softening are controlled orthogonally to one another.37 When true reversibility is desired, the conformational change of a photoresponsive chemical group such as an azobenzene can be exploited for repeat cycling of hydrogel stiffness.38 Through site-specific modification of both the N- and C- termini, full-length proteins may be incorporated as functional crosslinks within a biomaterial. Since the end-to-end translational movement lengths associated with stimuli-sensitive proteins is typically much larger than that obtained by small molecules, systems utilizing photoactivatable proteins can enable cyclic control over stiffness spanning a significantly larger dynamic range.39,40 Using the conformational change of the optogenetic protein pair LOV2-Jα, our group developed one such gel which softens in response to cytocompatible blue light and rapidly recovers its native stiffness in the dark.39 Intriguingly, fibroblast activation was enhanced within these gels when subjected to pulsatile stiffening relative to both persistently soft and stiff controls; these results suggest that cells are not only sensitive towards the static substrate stiffness, but the temporal element of stiffening as well.
Crosslinking gel materials with photoresponsive proteins containing an engineered phytochrome that reversibly dimerize upon red/near-IR light exposure has also provided a route to dynamic mechanical control.40 In this work, human MSCs were seeded onto the gel and subjected to 24 hrs of mechanical priming in the soft configuration, followed by an additional day of either static or dynamic photoswitching of the material. Transcriptomic analysis of cells on the static and dynamic substrates revealed that material stiffness for the first 24 hrs after seeding was more important than any subsequent dynamic alterations to the modulus. Notably, transforming growth factor-β1 (TGF-β1) and yes-associated protein (YAP) pathways were influenced by mechanical priming and less sensitive to more recent modulus switching, underscoring their known roles underpinning longer-term cellular mechanical memory. In contrast, mitogen-activated protein kinase (MAPK) signaling was a key pathway distinguishing slow (160 min) and fast (10 min) cycling of the substrate modulus, supporting its role as a more acute downstream effector of cell mechanotransduction.
With recent efforts to characterize the effects of more complex mechanical and physical properties (e.g., viscoelasticity, material plasticity, strain softening/stiffening, network architecture) on cell fate and function, we anticipate that the field’s next step will be to innovate strategies to reversibly control these material properties as has been done for substrate stiffness. Dynamic control over substrate viscoelasticity poses an interesting challenge, as properties such as stress relaxation are already time-dependent and reliant on non-covalent interactions within the hydrogel network.
Controlling Dynamic Presentation of Bioactive Ligands
Biological tissues are dynamic, not just mechanically, but also biochemically (Figure 2). Cells are exposed to tightly regulated cues in the form of secreted proteins and factors from other cells and the extracellular environment, which in turn influence cell phenotype. Uniform decoration of engineered microenvironment with small molecules, peptides, and whole proteins has become fairly straightforward; several chemical strategies are now in existence to covalently functionalize hydrogels with bioactive elements.41–46 For nonspecific tethering of proteins to a scaffold, custom and commercially available small molecules (e.g., activated esters) can be used to stochastically install functional handles for biomaterial tethering [e.g., azides, alkynes, maleimides, (meth)acrylates] onto a protein under gentle, aqueous conditions. For controlled tethering to a scaffold and minimal impact on protein activity, site-specific modification techniques including through the use of sortase,42 N-myristoyltransferase (NMT),43 and the SpyCatcher/Tag pair44 have proven useful. Further flexibility of chemical group placement within a protein can be achieved with noncanonical amino acid tagging and genetic code expansion.45
Figure 2 –
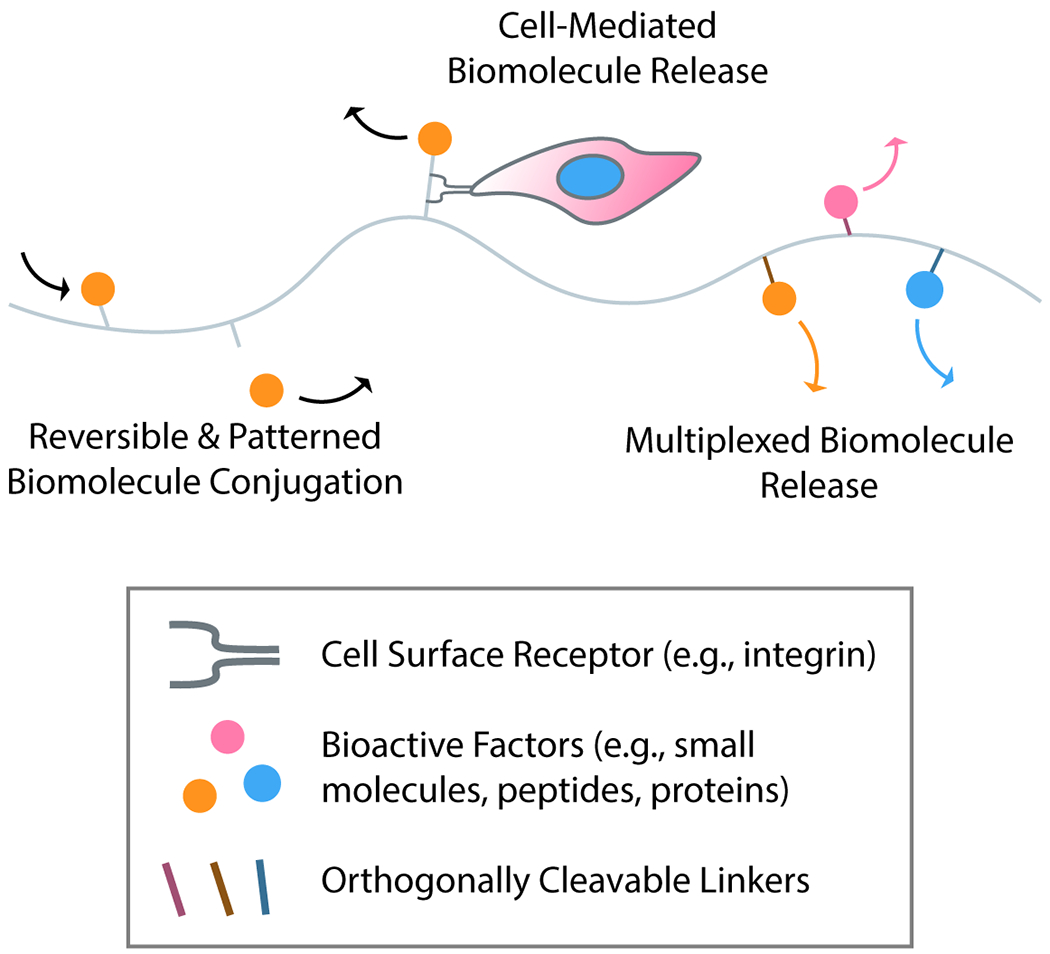
Recapitulating complex biochemical aspects of microenvironmental signaling. Exploiting bioorthogonal chemistries to immobilize or release biomolecules (e.g., small molecules, peptides, proteins) from biomaterials, reversible patterning of synthetic matrices may be achieved. New technologies such as traction-activated payloads (TrAPs) facilitate cell-mediated release of biomolecules from materials. With an ever-increasing suite of triggers for biomolecule release, the field is progressing towards systems capable of highly multiplexed triggers for on-demand and independent biomolecule release.
User-Controlled Presentation of Bioactive Ligands
Heterogenous presentation of biochemical cues within hydrogels has most frequently been achieved using photopatterning, whereby directed light exposure can be used to dictate when and where biomolecules are presented. Photomediated ligations, including those based on acrylates,47,48 thiol-ene,49 oxime,50 and enzymatic chemistries,51 have proven particularly useful for immobilizing small molecules, peptides, and even proteins into hydrogels. Photodegradation reactions, primarily based off of ortho-nitrobenzyl ester,30 coumarin,52 or photocleavable proteins,53 have found benefit for stimulating biomolecule release. These unidirectional material patterning approaches have enabled spatiotemporal control over proliferation, outgrowth, differentiation, and other complex cell fates within 3D gels.
Reversible biochemical control uniquely enables researchers to probe feedback loops between cells and their environment, which may be informative to identify tipping points in disease pathophysiology.36 The first path to reversible payload tethering and release from biomaterial is simply combining an additive chemistry with an orthogonal subtractive one, and this strategy has been successfully used to sequentially tether and release whole proteins to create complex and temporally evolving patterns capable of directing cell fate.50,54,55 However, this approach only allows one cycle of reversion, and current efforts seek to identify fully reversibly chemistries. One of the most promising approaches to date employs an allyl sulfide chain transfer, in which active radicals can help to trade one network-bound thiolated biomolecule for another.56 While this reversible chemistry offers some repeatability, nonspecific reactions associated with free-radicals limit reversibility and may be undesirable in the presence of cells. The reversible association of protein binding pairs has also been exploited through the optogenetic LOVTRAP system in hydrogels, which also enabled repetitive cycles of protein patterning and release.57 While fully reversible, this strategy relies on non-covalent protein association with the gel that is comparatively unstable. Ongoing efforts seek to identify truly reversible and covalent strategies for biomolecule patterning.
Cell-Dictated Release of Bioactive Factors
Beyond user-directed ligand presentation, an emergent line of materials development focuses on systems that present biochemical cues in a cell-directed manner. The extracellular matrix acts not just as a structural scaffold but also as a reservoir for sequestered growth factors that become available to the cell upon matrix strain or remodeling.58,59 One well-characterized example is the sequestration of TGF-β1 in the form of a large latent complex in the ECM, which when activated by strain or ECM degradation, causes fibroblast activation in the initiation of tissue repair.60 Disrupting this sequestration capacity leads to dysregulated TGF-β1 signaling, in turn causing developmental defects, cardiac disease, and cancers.61 The context of growth factor presentation is also incredibly important; when cells release sequestered growth factors from the ECM, receptor clustering with integrins can alter the nature of downstream intracellular signaling cascades. Therefore, mimicking cell-mediated release of bioactive factors from the ECM has been of great interest both for researchers wishing to deliver these factors therapeutically, as well as for those studying growth factor signaling through disease.
Growth factor sequestration within an engineered matrix for subsequent cell-mediated release can be accomplished through the inclusion of natural ECM components with growth factor affinity (e.g., fibronectin, heparin) or engineered components (e.g., antibodies, binding peptides).58 More recently, synthetic aptamers have been designed to bind growth factors with high affinity and tether them to the matrix.62 By conjugating a cell-adhesive peptide to the free end of the aptamer, growth factors may be released from their bind by cell-mediated traction forces. The aptamer design of these Traction-Activated Payloads (TrAPs) could be modified to accommodate nearly any protein payload, whereas cell-selective release can also be achieved by modifying the cell-adhesive ligand through which the TrAP unravels. TrAP delivery of platelet-derived growth factor-BB (PDGF-BB) promoted denser cell growth in serum-free conditions that persisted for two weeks, suggesting that the growth factor could be stabilized by the aptamer in a manner similar to that of native ECM sequestration.
One of the most important aspects in designing a dynamic microenvironment for cells is the selection of an exogenous or cell-mediated trigger to induce material dynamics.63 The number of triggers available to a researcher is ever-increasing and now includes bioorthogonal mechanisms including remote fields (e.g., light, ultrasound, magnetism)64 and engineered enzymes35. Utilization of many of these exogenous triggers frequently requires specialty chemistries that are synthetic inaccessible and are limited in their capacity for multiplexing. Technologies for gene editing, specifically Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas, are now easily adaptable to target nearly any DNA sequence of interest, allowing researchers to quickly customize the DNA sequence which Cas targets by changing the sequence of the guide RNA. This complete customizability is highly desired for programming the stimulus response of a biomaterial, and for this reason the CRISPR/Cas12a system has been recently adapted to the materials space.65 Cas12a specifically recognizes a double-stranded DNA (dsDNA) trigger, but upon sequence recognition collaterally cleaves nearby single-stranded DNA (ssDNA) nonspecifically.65 By incorporating a payload tethered to a hydrogel backbone by ssDNA or incorporating ssDNA into the material crosslink, stimulus-responsive payload release or bulk material degradation was induced upon introduction of a specific dsDNA trigger. By customizing the guide RNA and dsDNA trigger sequences, this strategy is easily adapted to any oligonucleotide trigger; since the mechanism for degradation is the nonspecific collateral cleavage of ssDNA downstream to dsDNA recognition, payload release is not entirely specific. Exploring material dynamics reliant only on specific dsDNA cleavage could open the door to near limitless multiplexing potential based on distinct target sequences and gRNAs.
Fabricating Complex Tissue Structures
The heterogeneous and hierarchical structures of biological tissues provide both an inspiration and a challenge to engineers looking to recapitulate structurally facilitated tissue functions. The human body is full of branching structures scaling many orders of magnitude in size for gas, liquid, and nutrient transport, anisotropic tissues optimized for the generation of force, and gradients of stiffness or biochemical factors. Appropriate structures are often necessary for the function of bioengineered tissues; as one example, engineered heart muscles must align to mature and generate appreciable force, and large tissues are limited by nutrient diffusion and require perfusable vasculature. Recent advances in spatially controlling material structural properties have opened the door to tissues with greater functionality (Figure 3).
Figure 3 –
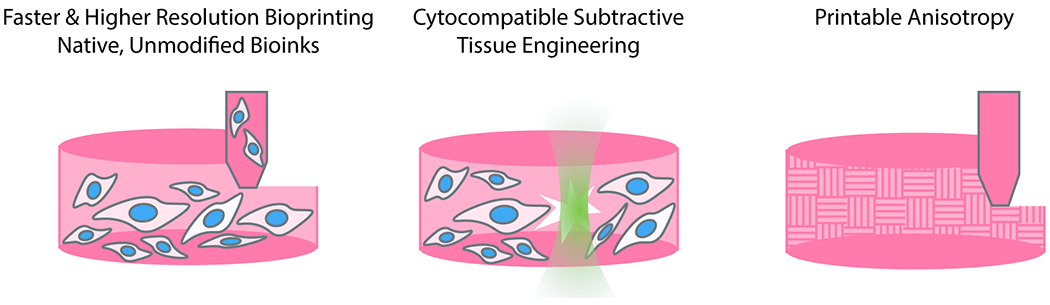
Advances in engineering complex tissue structures. Recent efforts in bioprinting has boosted resolution and speed, yielding faster, parallelizable, and more generalizable techniques for additive tissue construction. Photolabile hydrogel crosslinks now permit creation of intricate voids and microvascular structures through subtractive engineering. Bioprinting dynamic materials with switchable anisotropy also has opened the door to local control over material microstructure.
Additive Manufacturing of Engineered Tissues
Additive manufacturing of complex architectures with native and specialty biomaterials has hit maturity, and several advances to the field over the past few years now enable facile printing of exquisitely complex geometries. Specialty bioinks are no longer required to print high resolution structures – with the development of freeform reversible embedding of suspended hydrogels (FRESH), native collagen can be sculpted down to a resolution of 20 micrometers through extrusion into a buffered gelatin slurry that is washed away upon incubation at 37 °C.66 This technique has enabled perfusable vessels, a contractile ventricular model, and an at-scale heart to be printed using unmodified collagen.66,67 Though extrusion is perhaps the simplest method for bioprinting, stereolithographic photopolymerization of materials enables highly parallelized fabrication of structures. While this approach is much faster and offers fewer architectural constraints, cytotoxic photoinitiators/photoabsorbers have limited its use for bioprinting.68 Recently, the cytocompatible food dye tartrazine was identified as an alternative photoabsorber for stereolithography, leading to the development of a technique termed stereolithography apparatus for tissue engineering (SLATE). By minimizing unwanted out-of-plane photopolymerization, SLATE has proven useful for the fabrication of complex and interlocking void spaces within hydrogels to recapitulate vascular geometries, and can be used to rapidly construct engineered tissues containing a wide variety of cell types, including primary human stem cells.69
While stereolithographic polymerization of tissues enables precise and parallelized deposition of voxels of a single material by layer, it does not readily enable voxel-by-voxel control of material composition. To this end, multimaterial multinozzle arrays have been reported, capable of switching between up to 8 materials that can be switched at the nozzle head level at a frequency of up to 50Hz.70 Applied to bioprinting, this approach could provide unprecedented control over the composition of a biomaterial in 3D, with the benefits of parallelized material deposition. To further expand the palette of bioinks that can be 3D printed, eliminating requirements of photopolymerization and material extrudability, open-microfluidic well-plate inserts can be exploited to sculpt gel precursors by capillary action, facilitating controlled deposition of 3D structures with nearly any hydrogel chemistry.71
Another limitation of conventional stereolithography for additive manufacture is its resolution (typically tens to hundreds of microns), which may not be small enough for applications exploring subcellular patterning of topographical and biological cues.72 An alternative means for additive manufacture is multiphoton polymerization, a versatile strategy in which a femtosecond-pulsed laser can initiate polymerization on a voxel-by-voxel basis, enabling a much finer resolution (hundreds of nanometers).73 Many of the conventionally used bioinks used for stereolithography also have multiphoton absorbance, and thus can be readily used with this technique. Though these methods offer unmatched spatial patterning resolution, one critical limitation lies in fabrication speed; as polymerization occurs one voxel at a time, these techniques are largely reserved for small-featured structures.74
Just as the biomaterials field has successfully coopted multiphoton laser scanning from the photonics community for additive manufacturing, light sheet microscopy is now also poised for adoption to rapidly generate relatively high-resolution structures. In light sheet microscopy, the sample is illuminated through one axis while its fluorescence signal is detected through one perpendicular, enabling very fast scanning of large volumes that would be prohibitive to image by conventional laser scanning microscopy.75 One early and very recent adaptation of this technique for additive manufacturing – xolography – polymerizes structures by illuminating the resin with a projected image and an intersecting light sheet at different wavelengths and along orthogonal axes.76 Photopolymerization is achieved in a dual-color photoinitiator (DCPI) system, in which the activity of a benzophenone type II photoinitiator is optically regulated with a ultraviolet light-responsive spiropyran photoswitch. This approach improves upon the resolution of stereolithography by an order of magnitude, while generating large-scale objects at four-to-five orders of magnitude faster than multiphoton lithography. For biological applications, we anticipate that the first-generation DCPI will be limited by cytotoxicity and carcinogenicity similar to its benzophenone precursor, but look forward to the development of biocompatible initiators for this uniquely enabling additive manufacturing technique.77
Subtractive Manufacturing of Engineered Tissues
Subtractive manufacturing, whereby patterned removal of a subset of bulk starting material, has also found utility for tissue fabrication; micron-scale resolution over complex void volumes have made these the strategy of choice for creating well-defined microvascular networks. Utilizing high-intensity femtosecond-pulsed lasers to induce nonspecific photoablation of hydrogel materials (e.g., PEG, collagen), early efforts demonstrated the feasibility of laser-based subtractive manufacturing as a technique to generate perfusable microvascular arrays within a biomaterial.78,79 Though nonspecific photoablation can be used to fabricate vessels down to the size of a human capillary,80 the process is slow and requires high illumination intensities which are generally not cytocompatible. This process can be sped up dramatically through employment of photolabile moieties within the gel backbone, enabling material degradation and capillary-sized vessel patterning in the presence of living cells.30,81 Further improvements have been made by employing small molecule photosensitizers in conjunction with degradable gels.82 Despite these successes, there remains substantial room for improvements on the speed and throughput of subtractive tissue engineering, calling for the same ingenuity which catalyzed the additive manufacturing improvements reviewed above.
Engineered Control of Tissue Anisotropy
The microstructural alignment, or anisotropy, of an engineered biomaterial can impact a variety of tissue and cellular responses, including neurite outgrowth along aligned surfaces, fibroblast activation following myocardial infarction, and the coordinated contraction of muscle for force generation.83–85 Anisotropic biomaterials are traditionally fabricated through electrospinning or by directional freezing prior to lyophilization.86,87 Alternatively, as collagen in solution has negative diamagnetic anisotropy, aligned collagen hydrogels can be formed under a supermagnetic field.83 Until recently, however, the magnetic alignment of collagen gels could only be accomplished in bulk and was limited by the specialized instrumentation required.88
Even more recently, local spatial control of material anisotropy has been achieved on a bioprinter with a regular magnet by embedding streptavidin-conjugated iron nanoparticles into a collagen/agarose bioink such that magnetic field-induced particle movement during gelation promoted collagen fiber alignment.89 By equipping the 3D bioprinter with a magnet, anisotropy could be induced upon ink deposition through pulsed magnetic fields.89
In another powerful approach complementary to directed material alterations, tissue anisotropy can be introduced directly through acoustic patterning of cells.90 In this technique, cells are forced into the pressure nodes of a standing ultrasound wave as a hydrogel is polymerized around them, thus generating repeating lines or points of cells within the gel as the basis for microstructural anisotropy. This approach has been successfully employed to pattern myoblasts into aligned engineered muscle.
Cell-Guided Construction of Engineered Tissues
All of the above fabrication techniques offer a high degree of user control over guiding biological structure. Yet, complex biological structures may also be fabricated by offering control over self-assembly to encapsulated cells and providing minimal cues to guide self-emergent tissue architecture. This approach to tissue construction comes from the philosophy of organoid biology, which harnesses stem-cell aggregation and self-guided differentiation into complex biological structures.91 To take organoid biology beyond the millimeter scale, the same cells or cellular aggregates used to form organoids can also be extruded through a syringe at high density, and allowed to self-aggregate in a workflow termed bioprinting-assisted tissue emergence (BATE).92 This approach derives all complexity from organoid self-assembly, and as a result requires no specialized equipment for the syringe extrusion printing. The future of bioprinting holds larger and even more complex structures than ever, yet these advances may very well be delivered in simple and streamlined workflows.
Engineering Cells as Mechanical and Biochemical Actuators of the Microenvironment
Genetic engineering and synthetic biological approaches have become more accessible than ever, leading to several recent studies that use engineered cells themselves to pattern phenotype and the microenvironment (Figure 4). Both biochemical and mechanical actuation through synthetic gene circuits have been explored to spatially control tissue structure and function.
Figure 4 –
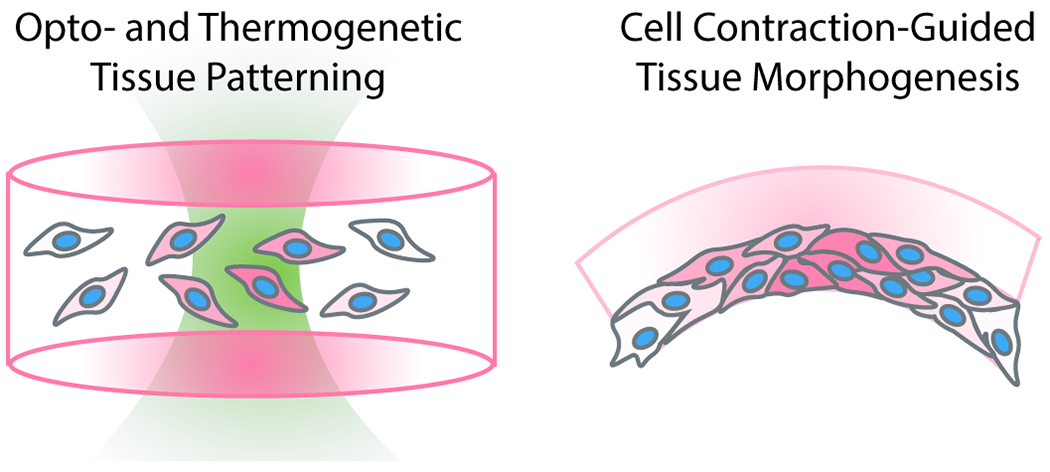
Employing cells as direct biochemical and mechanical actuators of the microenvironment. Borrowing tools from optogenetics, genetic engineering, and synthetic biology, an expanded toolbox exists to direct cell function within engineered materials. Opto- and thermogenetic cell patterning approaches provide a powerful route to directly modulate cell state within a material. Cells may also be used to guide macroscale tissue structure through controlled contraction.
The most extensive suite of remote triggers for cells may be borrowed from the field of optogenetics, which focuses on the use of light to precisely direct gene regulation and cell function. Optogenetic triggers, which come in the form of proteins that may dimerize, conformationally change, aggregate, or open/close a channel, have been developed to manipulate nearly every level of biological signaling.93 Though we have already introduced studies which borrow optogenetic proteins to impart dynamic biomaterial properties, this rich toolkit can also affords control over cell adhesion to the matrix and neighboring cells,94,95 migration,96 protein expression,97–99 and ion flux,100 all of which could be used to control the extracellular microenvironment of an engineered tissue from the inside out. Paralleling development of orthogonal biomaterial triggers, orthogonal optogenetic triggers have also been developed and enable multiplexed control over cell function.101,102 Optogenetic control over cell migration can be combined with light-sensitive material chemistries.103 For example, stem cells transfected with a photoactivatable Rac kinase were rendered susceptible to migration in the direction of a 458 nm light pulse, while hydrogel channels could be dynamically ablated through cleavage of an ortho-nitrobenzyl moiety as a PEG hydrogel crosslink.103 Simultaneously controlling cell behavior from inside-out and outside-in offers unique opportunities to engineer the dynamic reciprocity between cell and environment that can be ubiquitously found throughout development and disease.
Using the human heat shock promoter HSPA7, several past studies have successfully hijacked the heat stress response to induce transgene expression in response to mild heating.104,105 Recent work adopts this strategy for tissue engineering, through the incorporation of perfusable channels through which hot fluid may be pumped, thus creating well-defined temperature profiles throughout the material that governs patterned gene expression of encapsulated cells.106 This method was then used to induce spatial expression of liver enzymes through patterned expression of a Wnt signaling regulator.106
Most biological tissues are not uniform slabs, and instead exhibit complex, curved geometries that spontaneously emerge throughout development and dictate tissue-specific functions. Just as bulk mechanical properties of a tissue can influence cell state, local topological and mechanical cues also dictate the behavior of tissues and the cells within them.107,108 Both as a means of replicating developmental emergence of tissue shapes and as a strategy for creating appropriately shaped tissues for regenerative therapies, the actuation of cell contraction has been harnessed as a mechanism for generating curved structures from biomaterials.109,110 This can be accomplished by either patterning a contractile cell type or modulating that cell’s ability to compact zones of a material. DNA-programmed assembly of cells has been used to localize seed mesenchymal cells onto collagen hydrogels and rationally induce folding of these gels into a user-specified shape by cellular contraction.109 Through entirely synthetic cell patterning, this technique was used to recapitulate the incredibly complex tessellated curvature of embryonic chick gut lumen. During the dynamic folding process, actively contractile mesenchymal cells were found able to guide the migration of “passenger” vascular endothelial cells into the nascent folds. Alternatively, by spatially controlling the incorporation of an peptide which inhibits cell contraction in an evenly seeded gel, flat hydrogels can be made curved by inducing regions of high contraction by cells in the material.110 These curved gels were fabricated to recreate the geometry of the human cornea, and were able to guide the differentiation pattern of human epithelial stem cells into corneal epithelium. Though in this case contraction was indirectly induced by providing fetal bovine serum in the medium, logical “AND” gate crosslinkers susceptible to one exogenous stimulus and one cell-secreted factor could be harnessed for tighter control over cell remodeling and contraction of a material.111
In the future, we envision that instead of producing a protein in bulk and biochemically patterning it within gels, proteins may instead be produced in situ by encapsulated cells under tight 4D transcriptional control by optogenetic or thermogenetic means. This concept has only recently been preliminarily explored, whereby bacteria transformed with an optogenetic protein plasmid (pDawn) and encapsulated within a gel expressed and secreted Red Fluorescent Protein in response to blue light.112 With several strategies to genetically install chemical groups which facilitate gel-protein conjugation, secreted proteins could be sequestered by the hydrogel in a manner mimicking the role of the ECM.
Engineering Simplicity
The options available to a researcher wishing to engineer a biological microenvironment – material platforms, conjugation chemistries, mechanical and biochemical factors to consider – are nearly limitless. Contrary as it may sound, simplicity is also a key factor to consider when engineering complex microenvironments and is a crucial to the utility of any tool towards the study of disease. Many life-sciences labs are not equipped with the instrumentation or personnel for organic synthesis of the precursors and reagents used in many dynamic biomaterials. It is for this reason that Matrigel remains the one of the most widely-used 3D matrices, despite limitations in batch variability and a lack of tunability.113 Conversely, biomaterials labs depend on expert collaborators to provide impactful applications for the uniquely enabling materials that they develop. In many ways, biomaterials development to study the microenvironment have outpaced the utilization of such materials for studying biology.
Many systems highlighted herein gain utility from simplicity. SLATE bioprinting uses Food and Drug Administration (FDA)-approved food additive dyes as photoabsorbers.69 Recent studies pushing forward the culture of organoids in synthetic matrices have used hydrolysis as a trigger for the dynamic substrate softening required to support organoid formation.34 BATE relies upon biology to provide emergent complexity in printed organoids, but only requires simple extrusion printing using a syringe and a manually controlled microscope stage.92 These types of systems are easy for a non-engineer to adopt and exploit to catalyze impactful and translatable findings with respect to any disease of interest.
Our group and others have made a turn towards genetically encoded approaches for hydrogel formation and modulation, circumventing many of the insurmountable barriers that synthetic organic chemistry have imposed on biology labs interest in using biomaterial tools.53,114–116 By co-expressing pre-existing enzymes for site-specific protein modification (e.g., NMT, sortase) alongside a protein of interest, bioorthogonal handles can be installed in situ for direct incorporation into a hydrogel network with no post-synthetic modification.53 Spontaneous protein-protein binding (e.g., SpyCatcher/Tag ligation) can be exploited for hydrogel crosslinking with no synthetic elements or catalysts, distilling the field of synthetic hydrogel matrices into a format accessible to biologists.116,117
Future Directions
As the field of dynamic biomaterials develops increasingly modular biomaterials platforms and flexible bioconjugation chemistries, it is now possible to take many popular biomaterials platforms off the shelf and simultaneously specify an expansive set of biochemical and physical properties in tandem. Future work will certainly continue to push the limits of stimulus responsiveness towards improved multiplexing, utilization of triggers with in vivo relevance, scaffolding elements which integrate biological signals and generate feedback, and full spatiotemporal regulation that matches all biological scales. With such expanded levels of customization, it will be increasingly important to seek out the simplest mechanisms of control required for any given experimental question.
We believe that the field still has much to borrow from the emergent and neighboring spheres of optogenetics and protein engineering. Current approaches typically use a naturally derived protein or peptide as is to impart a biomaterial with a biological function. Yet, with modern-day protein engineering tools, it is possible to optimize proteins for sustained bioactivity and tunable release from a biomaterial scaffold.118 Furthermore, as de novo protein design continues to reach maturity, we envision a future in which protein elements may be rationally designed from grounds up as desired components of biomaterials.119 Already there is an ever-expanding toolkit of structural components120–122 and protein logic gates123–125 which – in combination with site-specific modification strategies to incorporate these elements into a material – provide ripe grounds for exploration.
Lastly, while the philosophy of engineering complexity from the outside in has yielded unprecedented control over the biological microenvironment, approaches that harness self-emergent complexity such as BATE and the broader field of organoid biology have demonstrated that engineered tissues may also be constructed from naïve cell types; as complex 3D folds may be generated from relatively simple 2D cell patterns, these strategies promise to yield relatively mature engineered tissues with very few exogenous cues in a largely unsupervised manner. Such emergent strategies may prove more effective for some tissue engineering applications than traditional approaches involving patterned materials. Certainly, both philosophies should be considered by biologists, biomaterial scientists, and tissue engineers alike, and we turn towards many exciting future studies to identify which approach is best suited for a given biological application.
Funding Sources
This work was supported by a Graduate Research Fellowship (2018261576, R.C.B.) and a CAREER Award (DMR 1652141, C.A.D.) from the National Science Foundation, as well as a Maximizing Investigators’ Research Award (R35GM138036, C.A.D.) from the National Institutes of Health.
ABBREVIATIONS
- 3D
three-dimensional
- 4D
four-dimensional
- BATE
bioprinting-assisted tissue emergence
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- DCPI
dual-color photoinitiator
- dsDNA
Double-stranded deoxyribonucleic acid
- ECM
extracellular matrix
- FDA
United States Food and Drug Administration
- FRESH
freeform reversible embedding of suspended hydrogels
- MAPK
mitogen-activated protein kinase
- MSC
mesenchymal stem cell
- NMT
N-myristoyltransferase
- PEG
Poly(ethylene glycol)
- PDGF-BB
platelet-derived growth factor-BB
- SLATE
stereolithographic apparatus for tissue engineering
- ssDNA
single-stranded deoxyribonucleic acid
- TrAP
traction-activated payload
- YAP
yes-associated protein
REFERENCES
- (1).Muncie JM; Weaver VM The Physical and Biochemical Properties of the Extracellular Matrix Regulate Cell Fate. In Current Topics in Developmental Biology; Academic Press Inc., 2018; Vol. 130, pp 1–37. 10.1016/bs.ctdb.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wang Y; Hu G; Hill RC; Dzieciatkowska M; Hansen KC; Zhang XB; Yan Z; Pei M Matrix Reverses Immortalization-Mediated Stem Cell Fate Determination. Biomaterials 2021, 265, 120387. 10.1016/j.biomaterials.2020.120387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lampi MC; Reinhart-King CA Targeting Extracellular Matrix Stiffness to Attenuate Disease: From Molecular Mechanisms to Clinical Trials. Science Translational Medicine. American Association for the Advancement of ScienceJanuary3, 2018, p 475. 10.1126/scitranslmed.aao0475. [DOI] [PubMed] [Google Scholar]
- (4).Dalby MJ; García AJ; Salmeron-Sanchez M Receptor Control in Mesenchymal Stem Cell Engineering. Nature Reviews Materials. Nature Publishing GroupJanuary31, 2018, pp 1–14. 10.1038/natrevmats.2017.91. [DOI] [Google Scholar]
- (5).Engler AJ; Sen S; Sweeney HL; Discher DE Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126 (4), 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- (6).Sewanan LR; Schwan J; Kluger J; Park J; Jacoby DL; Qyang Y; Campbell SG Extracellular Matrix From Hypertrophic Myocardium Provokes Impaired Twitch Dynamics in Healthy Cardiomyocytes. JACC Basic to Transl. Sci 2019, 4 (4), 495–505. 10.1016/j.jacbts.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Xu R; Boudreau A; Bissell MJ Tissue Architecture and Function: Dynamic Reciprocity via Extra- and Intra-Cellular Matrices. Cancer and Metastasis Reviews. NIH Public Access2009, pp 167–176. 10.1007/s10555-008-9178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chaudhuri O; Cooper-White J; Janmey PA; Mooney DJ; Shenoy VB Effects of Extracellular Matrix Viscoelasticity on Cellular Behaviour. Nature. Nature ResearchAugust27, 2020, pp 535–546. 10.1038/s41586-020-2612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Sun Z; Guo SS; Fässler R Integrin-Mediated Mechanotransduction. Journal of Cell Biology. Rockefeller University PressNovember21, 2016, pp 445–456. 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Elosegui-Artola A; Trepat X; Roca-Cusachs P Control of Mechanotransduction by Molecular Clutch Dynamics. Trends in Cell Biology. Elsevier LtdMay1, 2018, pp 356–367. 10.1016/j.tcb.2018.01.008. [DOI] [PubMed] [Google Scholar]
- (11).Caporizzo MA; Chen CY; Bedi K; Margulies KB; Prosser BL Microtubules Increase Diastolic Stiffness in Failing Human Cardiomyocytes and Myocardium. Circulation 2020, 141 (11), 902–915. 10.1161/CIRCULATIONAHA.119.043930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).A Simple Indentation Device for Measuring Micrometer-Scale Tissue Stiffness. 2010. 10.1088/0953-8984/22/19/194120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Charrier EE; Pogoda K; Wells RG; Janmey PA Control of Cell Morphology and Differentiation by Substrates with Independently Tunable Elasticity and Viscous Dissipation. Nat. Commun 2018, 9 (1), 1–13. 10.1038/s41467-018-02906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chaudhuri O; Gu L; Darnell M; Klumpers D; Bencherif SA; Weaver JC; Huebsch N; Mooney DJ Substrate Stress Relaxation Regulates Cell Spreading. Nat. Commun 2015, 6 (1), 6365. 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Loebel C; Mauck RL; Burdick JA Local Nascent Protein Deposition and Remodelling Guide Mesenchymal Stromal Cell Mechanosensing and Fate in Three-Dimensional Hydrogels. Nat. Mater 2019, 18 (8), 883–891. 10.1038/s41563-019-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nam S; Lee J; Brownfield DG; Chaudhuri O Viscoplasticity Enables Mechanical Remodeling of Matrix by Cells. 2016. 10.1016/j.bpj.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wisdom KM; Adebowale K; Chang J; Lee JY; Nam S; Desai R; Rossen NS; Rafat M; West RB; Hodgson L; Chaudhuri O Matrix Mechanical Plasticity Regulates Cancer Cell Migration through Confining Microenvironments. Nat. Commun 2018, 9 (1), 1–13. 10.1038/s41467-018-06641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Grolman JM; Weinand P; Mooney DJ Extracellular Matrix Plasticity as a Driver of Cell Spreading. Proc. Natl. Acad. Sci 2020, 202008801. 10.1073/pnas.2008801117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).van Oosten ASG; Chen X; Chin LK; Cruz K; Patteson AE; Pogoda K; Shenoy VB; Janmey PA Emergence of Tissue-like Mechanics from Fibrous Networks Confined by Close-Packed Cells. Nature 2019, 573 (7772), 96–101. 10.1038/s41586-019-1516-5. [DOI] [PubMed] [Google Scholar]
- (20).Bordeleau F; Mason BN; Lollis EM; Mazzola M; Zanotelli MR; Somasegar S; Califano JP; Montague C; LaValley DJ; Huynh J; Mencia-Trinchant N; Abril YLN; Hassane DC; Bonassar LJ; Butcher JT; Weiss RS; Reinhart-King CA Matrix Stiffening Promotes a Tumor Vasculature Phenotype. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (3), 492–497. 10.1073/pnas.1613855114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Matera DL; DiLillo KM; Smith MR; Davidson CD; Parikh R; Said M; Wilke CA; Lombaert IM; Arnold KB; Moore BB; Baker BM Microengineered 3D Pulmonary Interstitial Mimetics Highlight a Critical Role for Matrix Degradation in Myofibroblast Differentiation. Sci. Adv 2020, 6 (37), eabb5069. 10.1126/sciadv.abb5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Madl CM; Katz LM; Heilshorn SC Tuning Bulk Hydrogel Degradation by Simultaneous Control of Proteolytic Cleavage Kinetics and Hydrogel Network Architecture. ACS Macro Lett. 2018, 7 (11), 1302–1307. 10.1021/acsmacrolett.8b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Stowers RS; Allen SC; Suggs LJ; Anseth KS Dynamic Phototuning of 3D Hydrogel Stiffness. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (7), 1953–1958. 10.1073/pnas.1421897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Young JL; Engler AJ Hydrogels with Time-Dependent Material Properties Enhance Cardiomyocyte Differentiation in Vitro. Biomaterials 2011, 32 (4), 1002–1009. 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Guvendiren M; Burdick JA Stiffening Hydrogels to Probe Short- and Long-Term Cellular Responses to Dynamic Mechanics. Nat. Commun 2012, 3 (1), 1–9. 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- (26).Mabry KM; Lawrence RL; Anseth KS Dynamic Stiffening of Poly(Ethylene Glycol)-Based Hydrogels to Direct Valvular Interstitial Cell Phenotype in a Three-Dimensional Environment. Biomaterials 2015, 49, 47–56. 10.1016/J.BIOMATERIALS.2015.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu H-Y; Greene T; Lin T-Y; Dawes CS; Korc M; Lin C-C Enzyme-Mediated Stiffening Hydrogels for Probing Activation of Pancreatic Stellate Cells. Acta Biomater. 2017, 48, 258–269. 10.1016/j.actbio.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Stoppel WLWL; Gao AEAE; Greaney AMAMAM; Partlow BPBPBP; Bretherton RCRC; Kaplan DLDLDL; Black LDLD Elastic, Silk-Cardiac Extracellular Matrix Hydrogels Exhibit Time-Dependent Stiffening That Modulates Cardiac Fibroblast Response. J. Biomed. Mater. Res. - Part A 2016, 104 (12), 3058–3072. 10.1002/jbm.a.35850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Günay KA; Ceccato TL; Silver JS; Bannister KL; Bednarski OJ; Leinwand LA; Anseth KS PEG–Anthracene Hydrogels as an On-Demand Stiffening Matrix To Study Mechanobiology. Angew. Chemie - Int. Ed 2019, 58 (29), 9912–9916. 10.1002/anie.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kloxin AM; Kasko AM; Salinas CN; Anseth KS Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science 2009, 324 (5923), 59–63. 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).DeForest CA; Anseth KS Cytocompatible Click-Based Hydrogels with Dynamically Tunable Properties through Orthogonal Photoconjugation and Photocleavage Reactions. Nat. Chem 2011, 3 (12), 925–931. 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Brown TE; Marozas IA; Anseth KS Amplified Photodegradation of Cell-Laden Hydrogels via an Addition-Fragmentation Chain Transfer Reaction. Adv. Mater 2017, 29 (11), 1605001. 10.1002/adma.201605001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rapp TL; Highley CB; Manor BC; Burdick JA; Dmochowski IJ Ruthenium-Crosslinked Hydrogels with Rapid, Visible-Light Degradation. Chem. - A Eur. J 2018, 24 (10), 2328–2333. 10.1002/chem.201704580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gjorevski N; Sachs N; Manfrin A; Giger S; Bragina ME; Ordóñez-Morán P; Clevers H; Lutolf MP Designer Matrices for Intestinal Stem Cell and Organoid Culture. Nature 2016, 539 (7630), 560–564. 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- (35).Valdez J; Cook CD; Ahrens CC; Wang AJ; Brown A; Kumar M; Stockdale L; Rothenberg D; Renggli K; Gordon E; Lauffenburger D; White F; Griffith L On-Demand Dissolution of Modular, Synthetic Extracellular Matrix Reveals Local Epithelial-Stromal Communication Networks. Biomaterials 2017, 130, 90–103. 10.1016/J.BIOMATERIALS.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Rosales AM; Anseth KS The Design of Reversible Hydrogels to Capture Extracellular Matrix Dynamics. Nat. Rev. Mater 2016, 1. 10.1038/natrevmats.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rosales AM; Vega SL; DelRio FW; Burdick JA; Anseth KS Hydrogels with Reversible Mechanics to Probe Dynamic Cell Microenvironments. Angew. Chemie - Int. Ed 2017, 56 (40), 12132–12136. 10.1002/anie.201705684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Rosales AM; Mabry KM; Nehls EM; Anseth KS Photoresponsive Elastic Properties of Azobenzene-Containing Poly(Ethylene-Glycol)-Based Hydrogels. Biomacromolecules 2015, 16 (3), 798–806. 10.1021/bm501710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liu L; Shadish JA; Arakawa CK; Shi K; Davis J; DeForest CA Cyclic Stiffness Modulation of Cell□Laden Protein–Polymer Hydrogels in Response to User□Specified Stimuli Including Light. Adv. Biosyst 2018, 2 (12), 1800240. 10.1002/adbi.201800240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hörner M; Raute K; Hummel B; Madl J; Creusen G; Thomas OS; Christen EH; Hotz N; Gübeli RJ; Engesser R; Rebmann B; Lauer J; Rolauffs B; Timmer J; Schamel WWA; Pruszak J; Römer W; Zurbriggen MD; Friedrich C; Walther A; Minguet S; Sawarkar R; Weber W Phytochrome□Based Extracellular Matrix with Reversibly Tunable Mechanical Properties. Adv. Mater 2019, 31 (12), 1806727. 10.1002/adma.201806727. [DOI] [PubMed] [Google Scholar]
- (41).Shadish JA; DeForest CA Site-Selective Protein Modification: From Functionalized Proteins to Functional Biomaterials. Matter. Cell PressJanuary8, 2020, pp 50–77. 10.1016/j.matt.2019.11.011. [DOI] [Google Scholar]
- (42).Guimaraes CP; Witte MD; Theile CS; Bozkurt G; Kundrat L; Blom AEM; Ploegh HL Site-Specific C-Terminal and Internal Loop Labeling of Proteins Using Sortase-Mediated Reactions. Nat. Protoc 2013, 8 (9), 1787–1799. 10.1038/nprot.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Heal WP; Wright MH; Thinon E; Tate EW Multifunctional Protein Labeling via Enzymatic N-Terminal Tagging and Elaboration by Click Chemistry. Nat. Protoc 2012, 7 (1), 105–117. 10.1038/nprot.2011.425. [DOI] [PubMed] [Google Scholar]
- (44).Zakeri B; Fierer JO; Celik E; Chittock EC; Schwarz-Linek U; Moy VT; Howarth M Peptide Tag Forming a Rapid Covalent Bond to a Protein, through Engineering a Bacterial Adhesin. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (12), E690–E697. 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Krall N; Da Cruz FP; Boutureira O; Bernardes GJL Site-Selective Protein-Modification Chemistry for Basic Biology and Drug Development. Nature Chemistry. Nature Publishing GroupFebruary1, 2016, pp 103–113. 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- (46).Hermanson GT Bioconjugate Techniques: Third Edition; Elsevier Inc., 2013. 10.1016/C2009-0-64240-9. [DOI] [Google Scholar]
- (47).Hahn MS; Miller JS; West JL Three-Dimensional Biochemical and Biomechanical Patterning of Hydrogels for Guiding Cell Behavior. Adv. Mater 2006, 18 (20), 2679–2684. 10.1002/adma.200600647. [DOI] [Google Scholar]
- (48).Luo Y; Shoichet MS A Photolabile Hydrogel for Guided Three-Dimensional Cell Growth and Migration. Nat. Mater 2004, 3 (4), 249–254. 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- (49).DeForest CA; Polizzotti BD; Anseth KS Sequential Click Reactions for Synthesizing and Patterning Three-Dimensional Cell Microenvironments. Nat. Mater 2009, 8 (8), 659–664. 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).DeForest CA; Tirrell DA A Photoreversible Protein-Patterning Approach for Guiding Stem Cell Fate in Three-Dimensional Gels. Nat. Mater 2015, 14 (5), 523–531. 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- (51).Broguiere N; Lüchtefeld I; Trachsel L; Mazunin D; Rizzo R; Bode JW; Lutolf MP; Zenobi□Wong M Morphogenesis Guided by 3D Patterning of Growth Factors in Biological Matrices. Adv. Mater 2020, 32 (25), 1908299. 10.1002/adma.201908299. [DOI] [PubMed] [Google Scholar]
- (52).Azagarsamy MA; Anseth KS Wavelength-Controlled Photocleavage for the Orthogonal and Sequential Release of Multiple Proteins. Angew. Chemie - Int. Ed 2013, 52 (51), 13803–13807. 10.1002/anie.201308174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Shadish JA; Strange AC; DeForest CA Genetically Encoded Photocleavable Linkers for Patterned Protein Release from Biomaterials. J. Am. Chem. Soc 2019, 141 (39), 15619–15625. 10.1021/jacs.9b07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Shadish JA; Benuska GM; DeForest CA Bioactive Site-Specifically Modified Proteins for 4D Patterning of Gel Biomaterials. Nat. Mater 2019, 1. 10.1038/s41563-019-0367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).DeForest CA; Anseth KS Photoreversible Patterning of Biomolecules within Click-Based Hydrogels. Angew. Chemie - Int. Ed 2012, 51 (8), 1816–1819. 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Grim JC; Brown TE; Aguado BA; Chapnick DA; Viert AL; Liu X; Anseth KS A Reversible and Repeatable Thiol–Ene Bioconjugation for Dynamic Patterning of Signaling Proteins in Hydrogels. ACS Cent. Sci 2018, 4 (7), 909–916. 10.1021/acscentsci.8b00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Hammer JA; Ruta A; West JL Using Tools from Optogenetics to Create Light-Responsive Biomaterials: LOVTRAP-PEG Hydrogels for Dynamic Peptide Immobilization. Ann. Biomed. Eng 2020, 48 (7), 1885–1894. 10.1007/s10439-019-02407-w. [DOI] [PubMed] [Google Scholar]
- (58).Teixeira SPB; Domingues RMA; Shevchuk M; Gomes ME; Peppas NA; Reis RL Biomaterials for Sequestration of Growth Factors and Modulation of Cell Behavior. Adv. Funct. Mater 2020, 1909011. 10.1002/adfm.201909011. [DOI] [Google Scholar]
- (59).Belair DG; Le NN; Murphy WL Design of Growth Factor Sequestering Biomaterials. Chem. Commun 2014, 50 (99), 15651–15668. 10.1039/c4cc04317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hinz B The Extracellular Matrix and Transforming Growth Factor-B1: Tale of a Strained Relationship. Matrix Biol. 2015, 47, 54–65. 10.1016/j.matbio.2015.05.006. [DOI] [PubMed] [Google Scholar]
- (61).Sterner-Kock A; Thorey IS; Koli K; Wempe F; Otte J; Bangsow T; Kuhlmeier K; Kirchner T; Jin S; Keski-Oja J; Von Melchner H Disruption of the Gene Encoding the Latent Transforming Growth Factor-β Binding Protein 4 (LTBP-4) Causes Abnormal Lung Development, Cardiomyopathy, and Colorectal Cancer. Genes Dev. 2002, 16 (17), 2264–2273. 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Stejskalová A; Oliva N; England FJ; Almquist BD Biologically Inspired, Cell-Selective Release of Aptamer-Trapped Growth Factors by Traction Forces. Adv. Mater 2019, 31 (7), 1806380. 10.1002/adma.201806380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Badeau BA; DeForest CA Programming Stimuli-Responsive Behavior into Biomaterials. Annu. Rev. Biomed. Eng 2019, 21 (1), 241–265. 10.1146/annurev-bioeng-060418-052324. [DOI] [PubMed] [Google Scholar]
- (64).Armstrong JPK; Stevens MM Using Remote Fields for Complex Tissue Engineering. Trends in Biotechnology. Elsevier Ltd; March1, 2020, pp 254–263. 10.1016/j.tibtech.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).English MA; Soenksen LR; Gayet RV; de Puig H; Angenent-Mari NM; Mao AS; Nguyen PQ; Collins JJ Programmable CRISPR-Responsive Smart Materials. Science 2019, 365 (6455), 780–785. 10.1126/science.aaw5122. [DOI] [PubMed] [Google Scholar]
- (66).Lee A; Hudson AR; Shiwarski DJ; Tashman JW; Hinton TJ; Yerneni S; Bliley JM; Campbell PG; Feinberg AW 3D Bioprinting of Collagen to Rebuild Components of the Human Heart. Science 2019, 365 (6452), 482–487. 10.1126/science.aav9051. [DOI] [PubMed] [Google Scholar]
- (67).Mirdamadi E; Tashman JW; Shiwarski DJ; Palchesko RN; Feinberg AW FRESH 3D Bioprinting a Full-Size Model of the Human Heart. ACS Biomater. Sci. Eng 2020, acsbiomaterials.0c01133. 10.1021/acsbiomaterials.0c01133. [DOI] [PubMed] [Google Scholar]
- (68).Mondschein RJ; Kanitkar A; Williams CB; Verbridge SS; Long TE Polymer Structure-Property Requirements for Stereolithographic 3D Printing of Soft Tissue Engineering Scaffolds. Biomaterials. Elsevier LtdSeptember1, 2017, pp 170–188. 10.1016/j.biomaterials.2017.06.005. [DOI] [PubMed] [Google Scholar]
- (69).Grigoryan B; Paulsen SJ; Corbett DC; Sazer DW; Fortin CL; Zaita AJ; Greenfield PT; Calafat NJ; Gounley JP; Ta AH; Johansson F; Randles A; Rosenkrantz JE; Louis-Rosenberg JD; Galie PA; Stevens KR; Miller JS Multivascular Networks and Functional Intravascular Topologies within Biocompatible Hydrogels. Science 2019, 364 (6439), 458–464. 10.1126/science.aav9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Skylar-Scott MA; Mueller J; Visser CW; Lewis JA Voxelated Soft Matter via Multimaterial Multinozzle 3D Printing. Nature 2019, 575 (7782), 330–335. 10.1038/s41586-019-1736-8. [DOI] [PubMed] [Google Scholar]
- (71).Lee UN; Day JH; Haack AJ; Bretherton RC; Lu W; DeForest CA; Theberge AB; Berthier E Layer-by-Layer Fabrication of 3D Hydrogel Structures Using Open Microfluidics. Lab Chip 2020, 20 (3), 525–536. 10.1039/c9lc00621d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Ligon SC; Liska R; Stampfl J; Gurr M; Mülhaupt R Polymers for 3D Printing and Customized Additive Manufacturing. Chemical Reviews. American Chemical SocietyAugust9, 2017, pp 10212–10290. 10.1021/acs.chemrev.7b00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Ovsianikov A; Mironov V; Stampf J; Liska R Engineering 3D Cell-Culture Matrices: Multiphoton Processing Technologies for Biological and Tissue Engineering Applications. Expert Review of Medical Devices. November9, 2012, pp 613–633. 10.1586/erd.12.48. [DOI] [PubMed] [Google Scholar]
- (74).Saha SK; Wang D; Nguyen VH; Chang Y; Oakdale JS; Chen SC Scalable Submicrometer Additive Manufacturing. Science 2019, 366 (6461), 105–109. 10.1126/science.aax8760. [DOI] [PubMed] [Google Scholar]
- (75).Reynaud EG; Peychl J; Huisken J; Tomancak P Guide to Light-Sheet Microscopy for Adventurous Biologists; 2014. 10.1038/nmeth.3222. [DOI] [PubMed] [Google Scholar]
- (76).Regehly M; Garmshausen Y; Reuter M; König NF; Israel E; Kelly DP; Chou CY; Koch K; Asfari B; Hecht S Xolography for Linear Volumetric 3D Printing. Nature 2020, 588 (7839), 620–624. 10.1038/s41586-020-3029-7. [DOI] [PubMed] [Google Scholar]
- (77).Taschner R; Gauss P; Knaack P; Liska R Biocompatible Photoinitiators Based on Poly□α□ketoesters. J. Polym. Sci 2020, 58 (2), 242–253. 10.1002/pol.20199929. [DOI] [Google Scholar]
- (78).Brandenberg N; Lutolf MP In Situ Patterning of Microfluidic Networks in 3D Cell-Laden Hydrogels. Adv. Mater 2016, 28 (34), 7450–7456. 10.1002/adma.201601099. [DOI] [PubMed] [Google Scholar]
- (79).Heintz KA; Bregenzer ME; Mantle JL; Lee KH; West JL; Slater JH Fabrication of 3D Biomimetic Microfluidic Networks in Hydrogels. Adv. Healthc. Mater 2016, 5 (17), 2153–2160. 10.1002/adhm.201600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Arakawa C; Gunnarsson C; Howard C; Bernabeu M; Phong K; Yang E; DeForest CA; Smith JD; Zheng Y Biophysical and Biomolecular Interactions of Malaria-Infected Erythrocytes in Engineered Human Capillaries. Sci. Adv 2020, 6 (3), eaay7243. 10.1126/sciadv.aay7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Arakawa CK; Badeau BA; Zheng Y; DeForest CA Multicellular Vascularized Engineered Tissues through User□Programmable Biomaterial Photodegradation. Adv. Mater 2017, 29 (37), 1703156. 10.1002/adma.201703156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Lunzer M; Shi L; Andriotis OG; Gruber P; Markovic M; Thurner PJ; Ossipov D; Liska R; Ovsianikov A A Modular Approach to Sensitized Two□Photon Patterning of Photodegradable Hydrogels. Angew. Chemie Int. Ed 2018, 57 (46), 15122–15127. 10.1002/anie.201808908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Morgan JR; Yarmush ML; Girton TS; Dubey N; Tranquillo RT Magnetic-Induced Alignment of Collagen Fibrils in Tissue Equivalents. In Tissue Engineering; Humana Press, 2003; Vol. 18, pp 67–74. 10.1385/0-89603-516-6:67. [DOI] [PubMed] [Google Scholar]
- (84).Bugg D; Bretherton RC; Kim P; Olszewski E; Nagle A; Schumacher AE; Chu N; Gunaje J; DeForest CA; Stevens K; Kim D-H; Davis JM Infarct Collagen Topography Regulates Fibroblast Fate via P38-Yes-Associated Protein Transcriptional Enhanced Associate Domain Signals. Circ. Res 2020, CIRCRESAHA.119.316162. 10.1161/CIRCRESAHA.119.316162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Jana S; Levengood SKL; Zhang M Anisotropic Materials for Skeletal-Muscle-Tissue Engineering. Advanced Materials. Wiley-VCH VerlagDecember28, 2016, pp 10588–10612. 10.1002/adma.201600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Wegst UGK; Schecter M; Donius AE; Hunger PM Biomaterials by Freeze Casting. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci 2010, 368 (1917), 2099–2121. 10.1098/rsta.2010.0014. [DOI] [PubMed] [Google Scholar]
- (87).Sill TJ; von Recum HA Electrospinning: Applications in Drug Delivery and Tissue Engineering. Biomaterials. ElsevierMay1, 2008, pp 1989–2006. 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- (88).Eguchi Y; Ohtori S; Sekino M; Ueno S Effectiveness of Magnetically Aligned Collagen for Neural Regeneration in Vitro and in Vivo. Bioelectromagnetics 2015, 36 (3), 233–243. 10.1002/bem.21896. [DOI] [PubMed] [Google Scholar]
- (89).Betsch M; Cristian C; Lin Y-Y; Blaeser A; Schöneberg J; Vogt M; Buhl EM; Fischer H; Duarte Campos DF Incorporating 4D into Bioprinting: Real-Time Magnetically Directed Collagen Fiber Alignment for Generating Complex Multilayered Tissues. Adv. Healthc. Mater 2018, 7 (21), 1800894. 10.1002/adhm.201800894. [DOI] [PubMed] [Google Scholar]
- (90).Armstrong JPK; Puetzer JL; Serio A; Guex AG; Kapnisi M; Breant A; Zong Y; Assal V; Skaalure SC; King O; Murty T; Meinert C; Franklin AC; Bassindale PG; Nichols MK; Terracciano CM; Hutmacher DW; Drinkwater BW; Klein TJ; Perriman AW; Stevens MM Engineering Anisotropic Muscle Tissue Using Acoustic Cell Patterning. Adv. Mater 2018, 30 (43), 1802649. 10.1002/adma.201802649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Simian M; Bissell MJ Organoids: A Historical Perspective of Thinking in Three Dimensions. J. Cell Biol 2017, 216 (1), 31–40. 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Brassard JA; Nikolaev M; Hübscher T; Hofer M; Lutolf MP Recapitulating Macro-Scale Tissue Self-Organization through Organoid Bioprinting. Nat. Mater 2020, 1–8. 10.1038/s41563-020-00803-5. [DOI] [PubMed] [Google Scholar]
- (93).Mumford TR; Roth L; Bugaj LJ Reverse and Forward Engineering Multicellular Structures with Optogenetics. Curr. Opin. Biomed. Eng 2020, 100250. 10.1016/j.cobme.2020.100250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Ollech D; Pflästerer T; Shellard A; Zambarda C; Spatz JP; Marcq P; Mayor R; Wombacher R; Cavalcanti-Adam EA An Optochemical Tool for Light-Induced Dissociation of Adherens Junctions to Control Mechanical Coupling between Cells. Nat. Commun 2020, 11 (1), 1–13. 10.1038/s41467-020-14390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Baaske J; Mühlhäuser WWD; Yousefi OS; Zanner S; Radziwill G; Hörner M; Schamel WWA; Weber W Optogenetic Control of Integrin-Matrix Interaction. Commun. Biol 2019, 2 (1), 1–8. 10.1038/s42003-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Zhang J; Luo Y; Poh CL Blue Light-Directed Cell Migration, Aggregation, and Patterning. J. Mol. Biol 2020, 432 (10), 3137–3148. 10.1016/j.jmb.2020.03.029. [DOI] [PubMed] [Google Scholar]
- (97).Kim NY; Lee S; Yu J; Kim N; Won SS; Park H; Heo W Do. Optogenetic Control of MRNA Localization and Translation in Live Cells. Nat. Cell Biol 2020, 22 (3), 341–352. 10.1038/s41556-020-0468-1. [DOI] [PubMed] [Google Scholar]
- (98).Konermann S; Brigham MD; Trevino AE; Hsu PD; Heidenreich M; Cong L; Platt RJ; Scott DA; Church GM; Zhang F Optical Control of Mammalian Endogenous Transcription and Epigenetic States. Nature 2013, 500 (7463), 472–476. 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Nguyen NT; He L; Martinez-Moczygemba M; Huang Y; Zhou Y Rewiring Calcium Signaling for Precise Transcriptional Reprogramming. ACS Synth. Biol 2018, 7 (3), 814–821. 10.1021/acssynbio.7b00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Deisseroth K; Hegemann P The Form and Function of Channelrhodopsin. Science. American Association for the Advancement of ScienceSeptember15, 2017. 10.1126/science.aan5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Kramer MM; Mühlhäuser WWD; Weber W; Radziwill G Multichromatic Control of Signaling Pathways in Mammalian Cells. Adv. Biosyst 2020, 2000196. 10.1002/adbi.202000196. [DOI] [PubMed] [Google Scholar]
- (102).Rasoulinejad S; Mueller M; Nzigou Mombo B; Wegner SV Orthogonal Blue and Red Light Controlled Cell-Cell Adhesions Enable Sorting-out in Multicellular Structures. ACS Synth. Biol 2020, 9 (8), 2076–2086. 10.1021/acssynbio.0c00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Guo Q; Wang X; Tibbitt MW; Anseth KS; Montell DJ; Elisseeff JH Light Activated Cell Migration in Synthetic Extracellular Matrices. Biomaterials 2012, 33 (32), 8040–8046. 10.1016/j.biomaterials.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Smith RC; Machluf M; Bromley P; Atala A; Walsh K Spatial and Temporal Control of Transgene Expression through Ultrasound-Mediated Induction of the Heat Shock Protein 70B Promoter in Vivo. Hum. Gene Ther 2002, 13 (6), 697–706. 10.1089/104303402317322267. [DOI] [PubMed] [Google Scholar]
- (105).Miller IC; Gamboa Castro M; Maenza J; Weis JP; Kwong GA Remote Control of Mammalian Cells with Heat-Triggered Gene Switches and Photothermal Pulse Trains. ACS Synth. Biol 2018, 7 (4), 1167–1173. 10.1021/acssynbio.7b00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Corbett DC; Fabyan WB; Grigoryan B; O’Connor CE; Johansson F; Batalov I; Regier MC; DeForest CA; Miller JS; Stevens KR Thermofluidic Heat Exchangers for Actuation of Transcription in Artificial Tissues. Sci. Adv 2020, 6 (40), eabb9062. 10.1126/sciadv.abb9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Iskratsch T; Wolfenson H; Sheetz MP Appreciating Force and Shape-the Rise of Mechanotransduction in Cell Biology. Nature Reviews Molecular Cell Biology. Nature Publishing GroupDecember11, 2014, pp 825–833. 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Callens SJP; Uyttendaele RJC; Fratila-Apachitei LE; Zadpoor AA Substrate Curvature as a Cue to Guide Spatiotemporal Cell and Tissue Organization. Biomaterials. Elsevier LtdFebruary1, 2020, p 119739. 10.1016/j.biomaterials.2019.119739. [DOI] [PubMed] [Google Scholar]
- (109).Hughes AJ; Miyazaki H; Coyle MC; Zhang J; Laurie MT; Chu D; Vavrušová Z; Schneider RA; Klein OD; Gartner ZJ Engineered Tissue Folding by Mechanical Compaction of the Mesenchyme. Dev. Cell 2018, 44 (2), 165–178.e6. 10.1016/j.devcel.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Miotto M; Gouveia RM; Ionescu AM; Figueiredo F; Hamley IW; Connon CJ 4D Corneal Tissue Engineering: Achieving Time-Dependent Tissue Self-Curvature through Localized Control of Cell Actuators. Adv. Funct. Mater 2019, 29 (8), 1807334. 10.1002/adfm.201807334. [DOI] [Google Scholar]
- (111).Badeau BA; Comerford MP; Arakawa CK; Shadish JA; DeForest CA Engineered Modular Biomaterial Logic Gates for Environmentally Triggered Therapeutic Delivery. Nat. Chem 2018, 10 (3), 251–258. 10.1038/nchem.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Sankaran S; del Campo A Optoregulated Protein Release from an Engineered Living Material. Adv. Biosyst 2018, 3 (2), 1800312. 10.1002/adbi.201800312. [DOI] [PubMed] [Google Scholar]
- (113).Aisenbrey EA; Murphy WL Synthetic Alternatives to Matrigel. Nature Reviews Materials. Nature ResearchJuly1, 2020, pp 539–551. 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Xiang D; Wu X; Cao W; Xue B; Qin M; Cao Y; Wang W Hydrogels With Tunable Mechanical Properties Based on Photocleavable Proteins. Front. Chem 2020, 8, 7. 10.3389/fchem.2020.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Yang Z; Yang Y; Wang M; Deng X; Zhang W-B; Sun F; Wang T; Kiu Francis Fok H; Jiang B; Xiao W; Kou S; Guo Y; Yan Y Dynamically Tunable, Macroscopic Molecular Networks Enabled by Cellular Synthesis of 4-Arm Star-like Proteins HIGHLIGHTS The Integration of Protein Topology Engineering and Materials Science Cellular Synthesis of 4-Arm Star-like Proteins Enabled by Split. Cell Matter 2019. 10.1016/j.matt.2019.09.013. [DOI] [Google Scholar]
- (116).Sun F; Zhang W. Bin; Mahdavi A; Arnold FH; Tirrell DA Synthesis of Bioactive Protein Hydrogels by Genetically Encoded SpyTag-SpyCatcher Chemistry. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (31), 11269–11274. 10.1073/pnas.1401291111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Yang Z; Yang Y; Wang M; Wang T; Fok HKF; Jiang B; Xiao W; Kou S; Guo Y; Yan Y; Deng X; Zhang W-B; Sun F Dynamically Tunable, Macroscopic Molecular Networks Enabled by Cellular Synthesis of 4-Arm Star-like Proteins. Matter 2019, 0 (0). 10.1016/j.matt.2019.09.013. [DOI] [Google Scholar]
- (118).Hettiaratchi MH; O’meara MJ; O’meara TR; Pickering AJ; Letko-Khait N; Shoichet MS Reengineering Biocatalysts: Computational Redesign of Chondroitinase ABC Improves Efficacy and Stability; 2020; Vol. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Huang P-S; Boyken SE; Baker D The Coming of Age of de Novo Protein Design. 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- (120).Hsia Y; Mout R; Sheffler W; Edman NI; Vulovic I; Park Y-J; Redler RL; Bick MJ; Bera AK; Courbet A; Kang A; Brunette TJ; Nattermann U; Tsai E; Saleem A; Chow CM; Ekiert D; Bhabha G; Veesler D; Baker D Hierarchical Design of Multi-Scale Protein Complexes by Combinatorial Assembly of Oligomeric Helical Bundle and Repeat Protein Building Blocks. bioRxiv 2020, 2020.07.27.221333. 10.1101/2020.07.27.221333. [DOI] [Google Scholar]
- (121).hsia Y; Bale JB; Gonen S; Shi D; Sheffler W; Fong KK; nattermann U; Xu C; huang P-S; Ravichandran R; Yi S; Davis trisha; Gonen tamir; King neil P.; Baker D Design of a Hyperstable 60-Subunit Protein Icosahedron. Nature 2016. 10.1038/nature18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Bale JB; Gonen S; Liu Y; Sheffler W; Ellis D; Thomas C; Cascio D; Yeates TO; Gonen T; King NP; Baker D Accurate Design of Megadalton-Scale Two-Component Icosahedral Protein Complexes. Science 2016, 353 (6297), 389–394. 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Foight GW; Wang Z; Wei CT; Greisen P; Warner KM; Cunningham-Bryant D; Park K; Brunette TJ; Sheffler W; Baker D; Maly DJ Multi-Input Chemical Control of Protein Dimerization for Programming Graded Cellular Responses. Nat. Biotechnol 2019, 37 (10), 1209–1216. 10.1038/s41587-019-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Chen Z; Kibler RD; Hunt A; Busch F; Pearl J; Jia M; VanAernum ZL; Wicky BIM; Dods G; Liao H; Wilken MS; Ciarlo C; Green S; El-Samad H; Stamatoyannopoulos J; Wysocki VH; Jewett MC; Boyken SE; Baker D De Novo Design of Protein Logic Gates. Science 2020, 368 (6486), 78–84. 10.1126/science.aay2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Lajoie MJ; Boyken SE; Salter AI; Bruffey J; Rajan A; Langan RA; Olshefsky A; Muhunthan V; Bick MJ; Gewe M; Quijano-Rubio A; Johnson J; Lenz G; Nguyen A; Pun S; Correnti CE; Riddell SR; Baker D Designed Protein Logic to Target Cells with Precise Combinations of Surface Antigens. Science 2020, 369 (6511), eaba6527. 10.1126/science.aba6527. [DOI] [PMC free article] [PubMed] [Google Scholar]


