Abstract
Background:
Human adipose-derived mesenchymal stem cells (AMSCs) are an attractive resource for wound healing because their regenerative capacity improves injury repair. Recently, stem cell-derived exosomes have been shown to play a positive role in stem cell-based therapies. However, the effects of exosomes derived from AMSCs (AEXOs) on wound healing are unclear. In this study, we aimed to examine the role of AEXOs in attenuating inflammation and explore their effects in normal wound healing.
Methods:
We isolated exosomes from AMSCs and established a cellular model of inflammation by treatment with the inflammatory cytokines, interferon gamma and tumor necrosis factor alpha, to determine whether AEXOs can inhibit inflammation. We examined the wound healing effects of AEXOs in in vitro wound healing models and performed a miRNA array to understand the role of AEXOs in inflammation and wound healing.
Results:
A significant difference was observed in wound closure and the expression of anti-inflammatory and wound-healing-related factors between control and AEXO-treated cells.
Conclusion:
Our results showed that besides alleviating the inflammation response, AEXOs also promote wound healing. Thus, AEXOs represent a novel, stem-cell-based, therapeutic strategy for wound healing.
Electronic supplementary material
The online version of this article (10.1007/s13770-020-00316-x) contains supplementary material, which is available to authorized users.
Keywords: Adipose-derived stem cells, Exosomes, Inflammation, Wound healing
Introduction
Wound healing is a complex and dynamic process consisting of molecular and cellular events that regulate homeostasis, inflammation, proliferation, maturation, and tissue remodeling [1]. Although normal wounds heal easily, chronic non-healing wounds can lead to a significant economic burden on the family of affected individuals and society due to ineffective therapies [2]. Therefore, alternative and innovative therapies are urgently needed to treat wounds. Wound healing involves diverse cell types including macrophages, dermal fibroblasts, epidermal keratinocytes, and mesenchymal stem cells [3]. Inflammation is an early stage in the process of wound healing. M2 macrophages promote wound healing by suppressing M1-related inflammatory responses [4]. Fibroblasts play a critical role in wound healing as they produce extracellular matrix (ECM) proteins, including collagen and fibronectin, and release soluble factors such as growth factors and cytokines [5].
Adipose-derived mesenchymal stem cells (AMSCs) are adult stem cells with regenerative capacity that promote wound healing via paracrine signaling, differentiation, exert anti-inflammatory effects, and support resident progenitor cells [6]. In addition, AMSCs can improve diabetic wound healing by enhancing epithelialization, tissue formation, and the synthesis of cytokines that are beneficial for neovascularization and by suppressing inflammation and apoptosis [7]. Exosomes have recently received much attention as paracrine mediators of MSCs for clinical use for cardiovascular disease, acute kidney injury, liver injury, and lung injury because the use of cell-based vesicles is potentially free from tumorigenicity and immune rejection [8–10]. The major mechanism of MSC therapy has been attributed to their exosome-based paracrine action and not to cell replacement or differentiation [11, 12]. MSCs can enhance wound healing without direct contact, thus demonstrating the importance of paracrine signaling [13]. These findings indicate that exosomes could be used a safer therapeutic modality for various diseases [11].
Exosomes, produced in the endosomal compartments of all cells, are small, membrane-bound, extracellular vesicles (30–100 nm diameter) containing biological signaling molecules, such as nucleic acids, proteins, and metabolites, that mediate intercellular communication to cause physiological changes in the recipient cells [14]. In addition to their role as mediators of intercellular interaction, exosomes serve as biomarkers of diseases and vehicles for genes and drug delivery [15]. To date, exosomes have not been reported to have tumorigenicity, and they have a low potential to trigger immune responses [16, 17]. Importantly, exosomes possess homing abilities to migrate to target sites [18], because of which exosome therapy is gaining popularity in immune modulation and regenerative medicine [19]. Recently, many researchers have suggested that exosomes facilitate proliferation, migration, and angiogenesis for wound regeneration [20]. However, it has not been fully elucidated whether exosomes released from AMSCs (AEXOs) are implicated in wound healing, and the mechanism underlying the therapeutic effects of exosomes remains unclear.
Our previous study demonstrated that AEXOs promote the M2 macrophage phenotype by activating Stat6 and MafB transcription factors [21]. The anti-inflammatory properties of M2 macrophages induced by AEXOs are linked to their potent immunosuppressive effects. In this study, we aimed to isolate exosomes from AMSCs and establish a cellular model of inflammation by treatment with the inflammatory cytokines, interferon gamma (IFNγ) and tumor necrosis factor α (TNFα), to determine whether AEXOs can inhibit inflammation. We also aimed to examine the wound healing effects of AEXOs in in vitro wound healing models and perform an miRNA array to understand the role of AEXOs in inflammation and wound healing as miRNA in exosomes are crucial regulators between cells [22]. Our results reveal the potential of exosomes as cell-free therapeutic agents for wound healing.
Materials and methods
Cells
Human adipose tissues were obtained from healthy donors with approval from the Institutional Review Board of Severance Hospital (IRB No. 4-2019-0060), Seoul, the Republic of Korea. The tissues were cut into thin pieces with sterile scissors, and 0.1% of collagenase (Invitrogen, Carlsbad, CA, USA) solution was added to the minced tissues. Next, the mixture was stirred at 37 °C for 1 h. An equal amount of DMEM (low glucose), supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin (all from Invitrogen), was added to stop enzymatic activity. After centrifugation at 1200×g for 5 min, the supernatant was discarded, and the pellet mixture was filtered through a 70-μm cell strainer (BD Biosciences Pharmingen, San Diego, CA, USA). Then, the supernatant was discarded after centrifugation at 1200×g for 5 min. The cells in the pellet were seeded in culture medium (DMEM-low glucose containing 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin) in a 75-cm2 culture flask (Nunc, Roskilde, Denmark) at 37 °C in a 5% CO2-containing atmosphere. The medium was changed every 3 or 4 days with 5 ng/ mL of bFGF (Invitrogen) for expansion of the culture. The cells were maintained until the adherent cells displayed the typical morphology of AMSCs. Human dermal fibroblasts were purchased from Invitrogen.
Exosome isolation and characterization
Exosomes were isolated from cultured AMSC media without FBS by using an exosome isolation kit (System Biosciences, Palo Alto, CA, USA) in accordance with the manufacturer’s protocols. Briefly, cultured medium was transferred to a fresh conical tube. After centrifugation at 1500×g for 5 min, the supernatant was transferred to a fresh conical tube, and ExoQuick-TC reagent was added to the supernatant (1:5 ratio) and mixed by inverting 4 times. The mixture was incubated overnight at 5 °C, and centrifuged at 1500×g for 30 min on the next day. After removal of the supernatant, the exosomes were resuspended in PBS (Invitrogen). The isolated exosomes were quantified by using a BCA protein assay kit (Invitrogen), and the final exosomes were stored at -80 °C for further experiments. To examine their morphology, isolated exosomes were observed using transmission electron microscopy (JEM-1011, JEOL, Tokyo, Japan). The particle size distribution of the exosomes was analyzed with a nanoparticle tracking system according to the manufacturer’s instructions (Nanosight NS300, Malvern Panalytical, Malvern, UK).
Cell proliferation assay
To identify the effects of exosomes on fibroblast proliferation, cells were seeded at a density of 1 × 103/well in a 96-well plate (BD Falcon, Swedesboro, NJ, USA) and treated with 5 µg/mL of exosomes. After 2 days, proliferation was analyzed using a WST-based assay kit (EZ-Cytox, Daeil Lab, Seoul, Republic of Korea), according to the manufacturer’s guidelines. The absorbance was measured at 450 nm by using a microplate reader (Molecular Devices, San Jose, CA, USA). To create an inflammatory environment, fibroblasts were cultured with recombinant human TNFα (20 ng/mL) and IFNγ (20 ng/mL) (PeproTech, Cranbury, NJ, USA). After 2 days, cell proliferation was determined by using the WST-based assay. Cells cultured without the inflammatory cytokines were used as control. To investigate the antiproliferative activity of exosomes toward T cells stimulated by 10 µg/mL of phytohemagglutinin (Sigma Chemical Co., St. Louis, MO, USA), peripheral blood mononuclear cells (PBMCs) were co-cultured with AMSCs in 12-well plates (Falcon) using transwell inserts (Corning, NY, USA) (PBMCs:AMSCs, 10:1 ratio). Harvested PBMCs were transferred to 96-well plates to measure their growth by adding 10 µL of the WST-based reagent to each well. After incubation for 3 h at 37 °C, the absorbance of the supernatant was measured. Non-stimulated PBMCs were used as control. The absorbance of treated cells was normalized to the absorbance of control cells.
Real-time PCR
To determine gene expression levels, total RNA was isolated by using RiboEx reagent (GeneAll, Seoul, Republic of Korea). The total exosome RNA isolation kit (Invitrogen) was used for RNA isolation from exosomes. The quality and quantity of extracted RNA were analyzed by using a NanoDrop ND-1000 Spectrophotometer (Invitrogen), and the RNA was converted to cDNA using a HiSenScript™ RH[-] cDNA Synthesis Kit (iNtRON, Seongnam, Republic of Korea), according to the manufacturer’s protocols. Quantitative real-time PCR was performed using Light Cycler 480 II SYBR Green I Master mix (Roche Molecular Systems, Pleasanton, CA, USA) under the following conditions: 95 °C for 10 min, 45 cycles of 60 °C for 20 s, and 72 °C for 15 s. miRNA reactions were performed at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and 72 °C for 10 s. All real-time PCR experiments were performed in triplicate using the primer sequences listed in Supplementary table S1. Relative expression levels of control and test groups were determined by advanced relative quantification based on the E-method provided by Roche. The expression levels were normalized to those of GAPDH, and U6 snRNA was used as an internal control gene for miRNA analysis.
Flow cytometry
FACS analysis was used for characterization of cultured AMSCs. Briefly, adherent AMSCs were collected as single cells after treatment with 0.05% trypsin/EDTA (Invitrogen). The harvested cells were stained with FITC-conjugated or PE-conjugated antibodies (CD34, CD45, CD73, CD90, and CD105). The antibodies were purchased from BD Biosciences Pharmingen. FITC- and PE-conjugated isotype IgG were used as negative controls. Data for AMSC characterization were analyzed by using a Cytomics Flow Cytometer (Beckman Coulter, Fullerton, CA, USA).
Western blot
Total protein from exosomes was extracted with RIPA buffer (Biosesang, Seongnam, Republic of Korea), and 20 µg of total protein were separated on precast, 12%-gradient gels by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto PVDF membranes (Bio-Rad Laboratories, Redmond, WA, USA). The membranes were blocked with 5% BSA (Sigma Chemical Co.) in TBST and then incubated overnight with the primary antibodies, CD9 (1:500) and CD63 (1:500) (Abcam, Cambridge, MA, USA) in a 4 °C shaker. The membranes were washed 3 times for 10 min each with TBST and then incubated with HRP-conjugated, goat, anti-mouse IgG (1:1000, Invitrogen) and goat, anti-rabbit IgG (1:1000, Invitrogen) for 1 h at room temperature (RT). The membranes were washed 3 times for 10 min each with TBST and developed with ECL (Bio-Rad) according to the manufacturer’s instructions. The protein bands were detected by using the LAS4000 mini system (GE Healthcare, Uppsala, Sweden).
Carboxyfluorescein succinimidyl ester (CFSE) labeling
To investigate exosome uptake by cells, exosomes were labeled with CFSE (Invitrogen). Briefly, the exosomes were washed twice with warm PBS. Next, 5 µM of a CFSE solution was incubated with the exosomes for 15 min in a 37 °C incubator. Then, 10 mL of cold, serum-free medium was added to stop labeling. The exosomes were ready to use after washing once with cold, serum-free medium. The CFSE-labeled exosomes were co-cultured with PBMCs and fibroblasts for 24 h. The internalization of exosomes into PBMCs and fibroblasts was observed using an Olympus IX71 fluorescent microscope (Olympus, Tokyo, Japan).
Multiplex supernatant cytokine assay
The supernatant was collected and stored at − 80 °C for cytokine assay. The harvested supernatant was analyzed using a Magnetic Luminex Assay-human premixed multi-analyte kit (R&D Systems, Minneapolis, MN, USA). Briefly, to 50 µL of the supernatant or standard in each well were added 50 µL of the diluted microparticle cocktail, followed by a 2-h incubation at RT on a shaker at 800 rpm. After removing the liquid from each well, 100 µL wash buffer was filled and washed 3 times. Then, 50 µL of a diluted biotin-antibody cocktail was added to each well and incubated for 1 h at RT on the shaker at 800 rpm. The wells were washed 4 times. Subsequently, 50 µL of diluted streptavidin-PE was added to each well and incubated for 30 min at RT on the shaker at 800 rpm. After washing 4 times, 100 µL of wash buffer was added to each well and incubated for 2 min at RT on the shaker at 800 rpm. Finally, the resuspended microparticles were analyzed using a Luminex analyzer. All samples were assayed in duplicate, and the concentrations were obtained from an appropriate standard curve.
In vitro wound healing assay
Fibroblasts were seeded into 12-well plates (BD Falcon) at a density of 2 × 105 cells/well. The cells were maintained until they became 100% confluent monolayers. For in vitro injury, the cells in a well were scratched once with a 1,000 µL pipette tip to make an artificial wound. The scratch border was marked with a fine line and photographed using a phase microscope immediately after the scraping. Wounded cells were incubated in the presence of 5 µg/mL exosomes for 24 h; DMEM without exosomes was used as a control. The migration of cells was evaluated as a function of how far the cells had progressed from the scratch line. Wound healing assays were performed in triplicate, and the migration of cells was quantified for depiction in a bar chart.
Library preparation/sequencing and data analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA quality was evaluated by using an Agilent 2100 Bioanalyzer with an RNA 6000 Pico Chip (Agilent Technologies, Amstelveen, The Netherlands). RNA quantification was performed using a NanoDrop 2000 Spectrophotometer system (Thermo Fisher Scientific, Waltham, MA, USA). For AMSCs and exosomal RNAs, an NEBNext Multiplex Small RNA Library Prep kit (New England BioLabs, Inc., Ipswich, MA, USA) was used to construct libraries according to the manufacturer’s instructions. Briefly, 1 µg of total RNA from each sample was used to ligate the adaptors, and cDNA was synthesized using reverse transcriptase with adaptor-specific primers. PCR was performed for library amplification, and libraries were purified using QIAquick PCR Purification Kit (Qiagen, Inc., Hilden, Germany) and AMPure XP beads (Beckman Coulter). The yield and size distribution of the small RNA libraries were assessed by using the Agilent 2100 Bioanalyzer instrument for the High-sensitivity DNA Assay (Agilent Technologies). High-throughput sequences were produced by NextSeq500 system via single-end sequencing (Illumina, San Diego, CA, USA). Sequence reads were mapped by using the software tool, Bowtie2, to obtain a BAM file (alignment file). The mature miRNA sequence was used as a reference for mapping. Read counts mapped on the mature miRNA sequence were extracted from the alignment file using bedtools (v2.25.0) and Bioconductor that uses R (version 3.2.2) statistical programming language (R development Core Team, Vienna, Austria, 2011). Read counts were used to determine the expression levels of miRNAs. Quantile normalization was used for comparison between samples, and miRWalk 2.0 was used for miRNA target study.
Statistical analysis
All data were expressed as means ± Standard Deviation (SD). One-way analysis of variance (ANOVA) for data with > 2 groups with Bonferroni correction and Student’s t test for data of paired groups were performed using SPSS software 18 (SPSS Inc., Chicago, IL, USA). p < 0.05 (*) and < 0.01 (##) were considered significant.
Results
Establishing cell-based models of inflammation with proinflammatory cytokines, IFNγ and TNFα
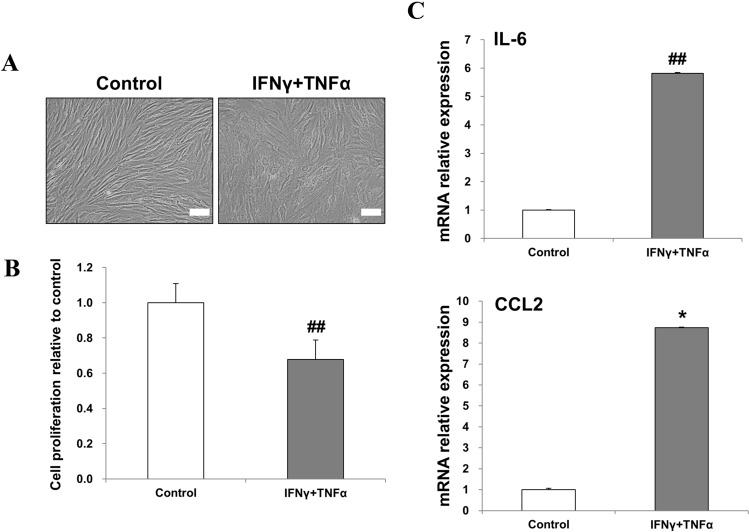
Dermal fibroblasts were used to model inflammation. To determine whether proinflammatory cytokines affect morphology and proliferation of fibroblasts, fibroblasts were incubated with 20 ng/mL of IFNγ and TNFα for 48 h. Untreated cells were used as control. We observed that, in the presence of proinflammatory cytokines, the fibroblasts exhibited change in morphology, appearing as enlarged and flat cells (Fig. 1A). Treatment with proinflammatory cytokines significantly decreased the proliferation of fibroblasts (Fig. 1B). Next, we investigated the effect of proinflammatory stimuli on fibroblasts at the mRNA level. Treatment with proinflammatory cytokines markedly increased the expression of interleukin (IL)-6 and CCL2 (Fig. 1C). Taken together, these results show that a mixture of IFNγ and TNFα can create an inflammatory environment for fibroblasts.
Fig. 1.
Treatment with a mixture of IFNγ and TNFα induced inflammation in fibroblasts. A Representative phase-contrast images of fibroblasts stimulated with 20 ng/mL of IFNγ and TNFα for 48 h (×200 magnification, scale bar = 100 μm). B Cell proliferation was measured by using a EZ-cytox assay kit with WST. C The relative mRNA levels of inflammatory IL-6 and CCL2 were analyzed by real-time PCR. The data are expressed as the mean ± SD of three independent experiments. *Significant difference from untreated control cells, p < 0.05. ##Significant difference from untreated control cells, p < 0.01
Characterization of adipose-derived mesenchymal stem cells (AMSCs) and exosomes produced by AMSCs (AEXOs)
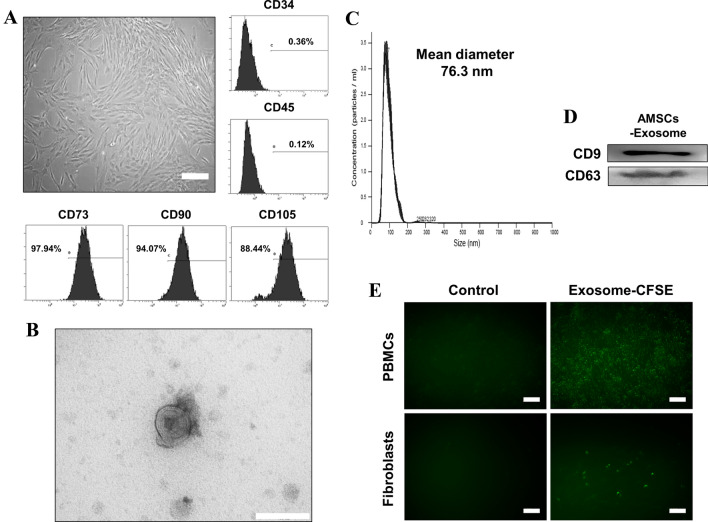
Cells cultured from adipose tissue displayed spindle-shaped and fibroblast-like morphology (Fig. 2A). Flow cytometry data revealed that the cells expressed MSC markers, CD73, CD90, and CD105, but did not express the hematopoietic markers, CD34 and CD45 (Fig. 2A). These results confirm that the cells cultivated from adipose tissue are mesenchymal stem cells. Exosomes isolated from AMSC culture supernatants, and analyzed by TEM, appeared as round, membrane-bound vesicles (Fig. 2B). Nanoparticle tracking analysis revealed that the average diameter of most exosomes was 76.3 nm (Fig. 2C). Western blotting confirmed the expression of the exosomal markers, CD9 and CD63, in AEXOs (Fig. 2D). Fluorescence microscopy revealed green fluorescent cells after internalization of AEXOs by PBMCs and fibroblasts (Fig. 2E).
Fig. 2.
Characterization of AMSCs and exosomes derived from AMSCs. A The morphology and expression of cell surface markers of AMSCs. Cells cultured from adipose tissue displayed the typical fibroblast-like morphology of MSCs (×100 magnification, scale bar = 200 μm). Flow cytometry analysis revealed that cells were positive for CD73, CD90, and CD105, all MSC markers, but negative for hematopoietic markers, CD34 and CD45. B Transmission electron microscopy image of AEXOs (scale bar = 200 nm). C Exosome particle size was analyzed by using Nanosight; the mean diameter was 76.3 nm. D CD9 and CD63, both exosomal markers, were detected by western blotting. E Fluorescence images were obtained after treating PBMCs and fibroblasts with CFSE-labeled (green) exosomes for 12 h. Unlabeled exosomes were used as negative control (×200 magnification, scale bar = 100 μm)
AEXOs induced M2 macrophage polarization and attenuated inflammation with immune modulation
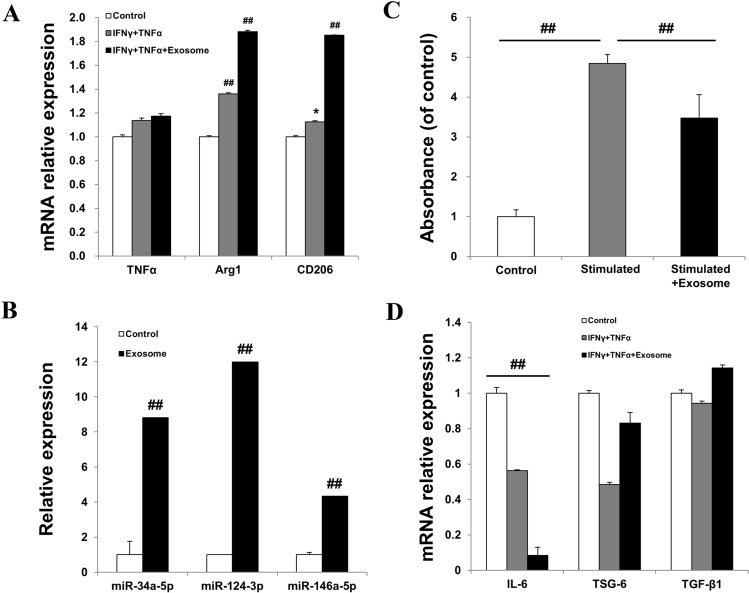
To further confirm M2 macrophage polarization by AEXOs, we used a cell-based model of inflammation as shown in Fig. 1. In a co-culture system of PBMCs and IFNγ-and-TNFα-treated fibroblasts, exosome-treated cells showed the most significant increase in expression of arginase (Arg)1 and CD206, markers of M2 polarized macrophages, compared with untreated fibroblasts, demonstrating that AEXOs induced polarization to the M2 macrophage phenotype (Fig. 3A). Studies have shown that miR-34a-5p, miR-124-3p, and miR-146a-5p induce M2 polarization in macrophages by targeting various transcription factors and proteins [23]. Hence, we analyzed miRNA levels using real-time PCR, and found that the miRNAs known to mediate M2 macrophage polarization were highly expressed in the exosomes when compared to the AMSCs (Fig. 3B). Thus, miR-34a-5p, miR-124-3p, and miR-146a-5p expressed in these exosomes were probably responsible for the polarization to the M2 macrophage phenotype. M2 polarization of macrophages is linked to wound repair and suppression of the immune response and of inflammation [23]. To determine the effects of AEXOs on immunosuppression, PBMCs were stimulated by phytohemagglutinin (PHA) and treated with AEXOs. As shown in Fig. 3C, AEXOs suppressed the proliferation of PBMCs. In our cell-based model of inflammation, we observed that exposure of PBMCs to exosomes significantly decreased the expression levels of the IL-6 gene (Fig. 3D). Under the same treatment conditions, AEXO-treated PBMCs showed upregulation of anti-inflammatory genes, tumor necrosis factor-inducible gene 6 (TSG-6), compared to their expression in IFNγ-and-TNFα-treated PBMCs (Fig. 3D). Collectively, these results suggest that AEXOs induce M2-phenotype macrophage polarization via expression of miR-34a-5p, miR-124-3p, and miR-146a-5p. Moreover, our data indicate that AEXOs can attenuate immune response and inflammation.
Fig. 3.
Exosomes derived from AMSCs induced an anti-inflammatory M2 phenotype in PBMCs. A Relative expression levels of TNFα, Arg1, and CD206 were determined by real-time PCR. PBMCs were co-cultured with IFNγ-and-TNFα-treated fibroblasts. Untreated fibroblasts were used as control. B Expression of miR-34a-5p, miR-124-3p, and miR-146a-5p was upregulated in exosomes compared to that in AMSCs. Exosomes derived from AMSCs exerted immunomodulatory effects and suppress inflammation. C The suppression of the proliferation of PHA-stimulated PBMCs was evaluated by using a proliferation assay kit. Unstimulated PBMCs were used as control. D The mRNA levels of IL-6, TSG-6, and TGF-β1 in PBMCs co-cultured with IFNγ-and-TNFα-stimulated fibroblasts were analyzed by real-time PCR. PBMCs co-cultured with unstimulated fibroblasts were used as control. The data are expressed as the mean ± SD of three independent experiments. *Significant difference from untreated control cells, p < 0.05. ##Significant difference from untreated control cells, p < 0.01
Effects of AEXOs on IFNγ-and-TNFα-treated fibroblasts
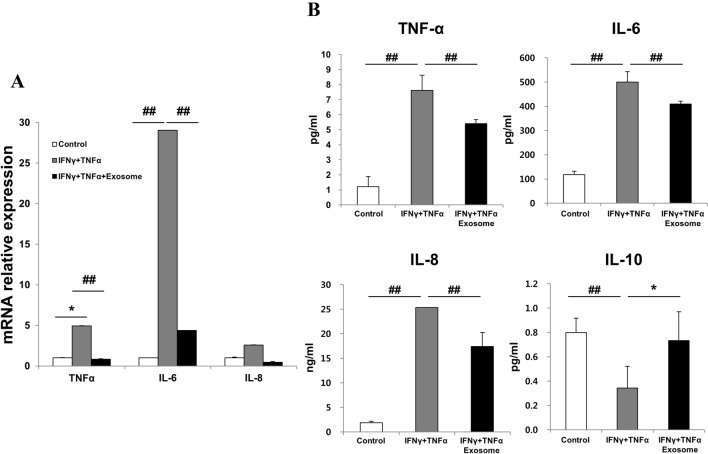
We investigated the effects of AEXOs in inflammatory-cytokine-stimulated fibroblasts. Treatment with IFNγ and TNFα increased the expression of inflammatory markers, TNFα, IL-6, and IL-8 in fibroblasts, which was significantly reduced after treatment with AEXOs (Fig. 4A). To evaluate changes in inflammatory cytokine levels after AEXO treatment, the culture medium was subjected to a magnetic beads-based multiplex assay. We found that treatment with IFNγ and TNFα increased the concentrations of the TNFα, IL-6, and IL-8 in the culture medium, whereas the level of IL-10 was significantly reduced (Fig. 4B). Of note, decrease in protein levels of TNFα, IL-6, and IL-8 and increase in IL-10 concentrations were significant after treatment with AEXOs (Fig. 4B). These results indicate that AEXOs play a role in controlling the cellular response to proinflammatory environments.
Fig. 4.
Effects of exosomes derived from AMSCs on proinflammatory-cytokine-treated fibroblasts. A Changes in gene expression of TNFα, IL-6, and IL-8 were detected by real-time PCR after IFNγ-and-TNFα treatment. Untreated fibroblasts were used as control. B A multiplex assay was performed to determine the changes in inflammation-related factors at protein levels in conditioned media. The conditioned media of unstimulated fibroblasts were used as control. The data are expressed as the mean ± SD of three independent experiments. *Significant difference from untreated control cells, p < 0.05. ##Significant difference from untreated control cells, p < 0.01
AEXOs accelerate the proliferation and migration of fibroblasts
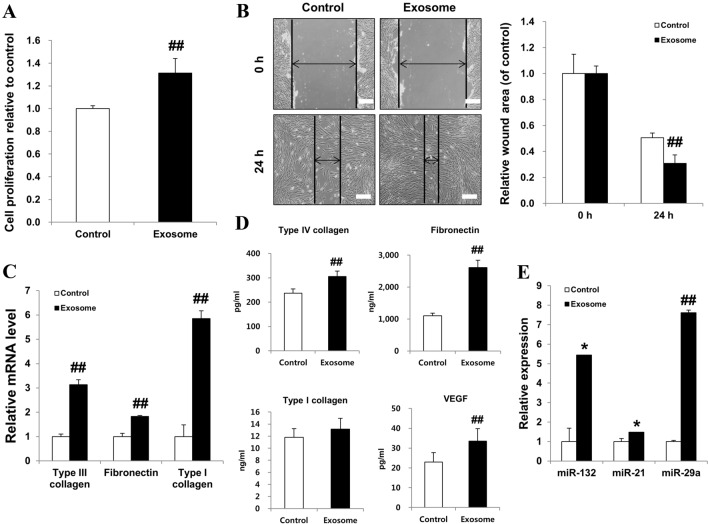
We investigated the proliferation of fibroblasts cultured with AEXOs and found that the proliferation and expansion of fibroblasts treated with AEXOs were higher than in the control cells (Fig. 5A). Additionally, we explored whether AEXOs could influence the migration of fibroblasts in an in vitro scratch assay. Quantification of the migrated cells revealed significantly faster migration of fibroblasts treated with AEXOs compared with untreated cells in an artificial wound model (Fig. 5B). Moreover, real-time PCR results revealed that the expression of extracellular matrix (ECM) molecules involved in cell migration, such as type III collagen, fibronectin, and type I collagen, was elevated in AEXO-treated fibroblasts compared to that in control cells (Fig. 5C). Compared to control fibroblasts, we observed significantly higher concentrations of fibronectin, collagen, and VEGF proteins in the supernatants of AEXO-treated fibroblasts (Fig. 5D). Numerous studies have demonstrated the important roles of miRNAs such as miR-132, miR-21, and miR-29a in wound healing [24]. Figure 5E shows that miR-132, miR-21, and miR-29a were expressed in AEXOs, which indicates that AEXOs enhance wound healing, confirmed by proliferation and migration of fibroblasts as well as ECM production.
Fig. 5.
Exosomes derived from AMSCs promoted proliferation and migration of fibroblasts. A Proliferation rates of fibroblasts treated with exosomes were analyzed by using a proliferation assay kit. B Migration of fibroblasts treated with exosomes was evaluated by an in vitro scratch wound healing assay. A representative of three independent experiments is shown (×100 magnification, scale bar = 200 μm). Fibroblast migration was quantified. C Real-time PCR analysis of the relative mRNA levels of type III collagen, fibronectin, and type I collagen genes was performed in fibroblasts after treatment with exosomes and compared to the respective mRNA levels in untreated cells. D Fibronectin, collagen, and VEGF concentrations were measured in conditioned media by multiplex assay. E The expression of miR-132, miR-21, and miR-29a was analyzed in exosomes produced by AMSCs by real-time PCR. The data are expressed as the mean ± SD of three independent experiments. *Significant difference from untreated control cells, p < 0.05. ##Significant difference from untreated control cells, p < 0.01
AEXOs contained miRNAs involved in inflammation and wound healing
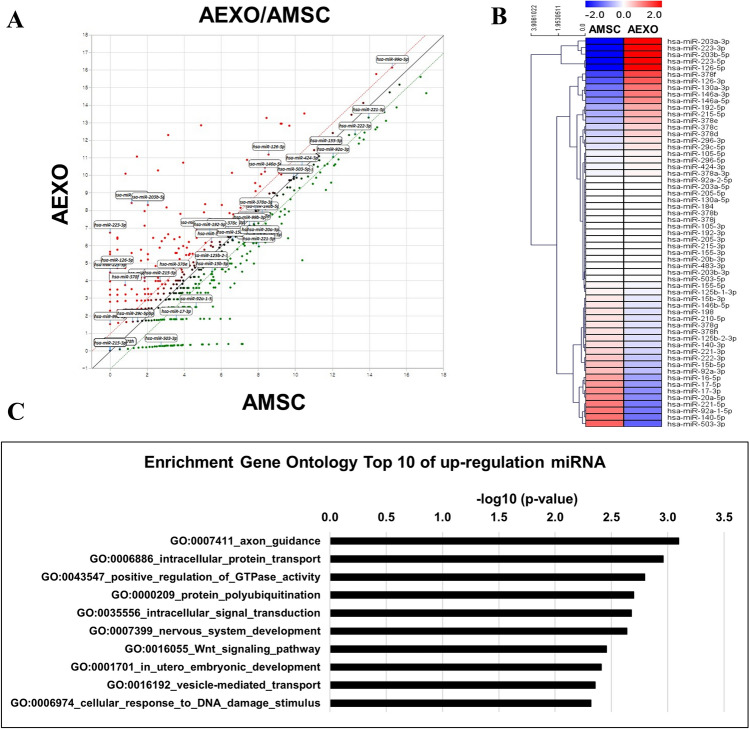
We used microRNA sequencing to profile miRNAs associated with inflammation and wound healing with AMSCs as control samples and exosomes released by the same AMSCs as test samples. The fold change was calculated by dividing the normalized expression profile of the exosome sample by that of the corresponding AMSC sample. Interestingly, scatter plots of the global miRNA expression showed that the AEXO profile was different from that of the AMSCs (Fig. 6). To reduce the complexity of analysis, we focused on five well-characterized functional categories involved in wound healing: anti-inflammatory, pro-angiogenic, remodeling, proinflammatory, and anti-angiogenic [25]. As illustrated in Fig. 6, we identified miRNAs associated with wound healing. Upregulated miRNAs included miR-223, miR-203, and miR-146a in the anti-inflammatory category; miR-126, miR-130a, miR-296, and miR-378 in the pro-angiogenic category; and miR-29c, miR192, miR-215 in the remodeling category. Downregulated miRNAs included miR-140 in the proinflammatory category; miR-92a, miR-17, miR-15b, miR-16, miR-20a, miR-221, miR-222, and miR-503 in the anti-angiogenic category [25]. Loss and gain of miRNAs in exosomes produced by AMSCs are listed in Supplementary tables S2 and S3.
Fig. 6.
Differential expression of miRNAs. The distribution of differentially expressed miRNAs in exosomes released by AMSCs compared with control AMSCs. A Scatter plots of miRNA expression in AEXOs versus AMSCs. B Hierarchical clustering analysis of miRNA expression related to wound regeneration in AEXOs and AMSCs. C Gene ontology analysis of top 10 upregulated miRNAs AEXOs versus AMSCs
Discussion
The original concept of stem-cell therapy was based on the hypothesis that stem cells would home and migrate to injured sites and then differentiate to replace damaged cells. However, numerous studies have shown that therapeutic effects of MSCs are mediated by paracrine signaling and anti-inflammatory, anti-fibrotic, and pro-angiogenic activities instead of direct cell replacement or differentiation [26–28]. Recent studies of exosomes have helped us gain a better understanding of the underlying mechanism of the paracrine effects of MSCs [29]. Although a number of experiments have shown positive effects of AMSCs in various wound healing models, the effects of exosomes from AMSCs on wound healing have not been studied to the same extent [30]. Furthermore, little is known about the contents (proteins, lipids, and nucleic acids) of the exosomes that mediate wound healing. Hence, this study aimed to shed light on the beneficial effects and molecular mechanisms of exosomes from AMSCs in wound healing and provide directions for future stem-cell-based research. To this end, we successfully isolated exosomes from AMSCs and verified exosomal markers, demonstrated the internalization of fluorescently labeled AEXOs by PBMCs and fibroblasts, and confirmed the biological effects of AEXOs on the target cells by AEXOs; our results were consistent with previous findings [31, 32].
Wound healing is a sophisticated process that is associated with three phases: inflammation, proliferation, and remodeling [33]. Inflammation plays an important role in the pathogenesis of wounds. Furthermore, multiple cellular events involving platelets, neutrophils, macrophages, and fibroblasts contribute to wound repair [34]. In particular, macrophages resolve inflammation and support cell proliferation and tissue restoration in wounds. Dynamic changes occur in response to environmental influence in the PBMC population. As wounds heal, the macrophage population transforms from the proinflammatory M1 macrophage phenotype to the anti-inflammatory M2 macrophage phenotype [35]. We have previously shown that AMSCs induce the M2 macrophage phenotype via exosomes [21]. Fibroblasts are the major cell type involved in wound healing [36]. Hence, we constructed a cell-based model of inflammation by treating fibroblasts with proinflammatory cytokines, IFNγ and TNFα, to explore the role of AEXOs in inflammation. Interestingly, we found that AEXOs increased the induction of the M2 macrophage phenotype in PBMCs, indicating the active role of AEXOs in M2 macrophage polarization. Accumulating evidence has shown that the miRNA-mediated pathway plays an important role in macrophage polarization [23]. Herein, we confirmed that AEXOs contained the important miRNAs (miR-34a-5p, miR-124-3p, miR-146a-5p) related to the M2 phenotype. The proinflammatory factors, CCL2 and IL-6, are known to induce M2 macrophage polarization [37]. In our experiments, we found that the expression of Arg1 and CD206, both M2 macrophage markers, increased after co-culturing PBMCs with IFNγ-and-TNFα-treated fibroblasts, despite the absence of AEXO treatment. We assumed that CCL2 and IL-6 released by the fibroblasts treated with IFNγ and TNFα may have stimulated M2 polarization in PBMCs. Thus, besides AEXOs, CCL2 and IL-6 also played a role in inducing M2 type macrophage polarization.
M2 macrophages have dual effects in creating an immunosuppressive and anti-inflammatory microenvironment [38]. Hence, we examined the immunosuppressive effects of AEXOs by using PBMCs stimulated by PHA and observed that AEXOs reduced the proliferation of PBMCs. Exosomes harbor cytokines and growth factors, which contribute to immunoregulation [39]. In our study, we found that AEXOs downregulated IL-6 expression and upregulated expression of TSG-6 and transforming growth factor beta 1 (TGF-β1) in the inflammatory environment. These findings are consistent with previous reports that AEXOs can promote M2 macrophage polarization and exert immunomodulatory effects [21]. Indeed, we found that the expression of inflammatory molecules such as TNFα, IL-6, and IL-8 at both mRNA and protein levels was reduced in AEXO-treated fibroblasts. Domenis et al. suggested that the ability of AEXOs to modulate immune cells is activated by signals derived from a proinflammatory microenvironment [40]. In agreement with these previous findings, educated AEXOs primed with proinflammatory molecules may have exerted immunomodulatory effects in our study [29]. Taken together, these results suggest that AEXOs suppress an inflammatory environment.
Besides suppression of inflammation, optimization of fibroblasts can accelerate wound closure by generating beneficial ECM proteins. Our results suggest that AEXOs had an enhancing effect on the proliferation and migration of fibroblasts by stimulating ECM protein synthesis. Analysis using the multiplex assay further confirmed significantly higher ECM protein levels than those of the control group, implying that AEXOs promoted ECM protein synthesis for wound healing. Previous studies have proposed that miRNAs, including miR-132, miR-21, and miR-29a, are key components that play critical roles in wound healing [41]. We detected the expression of miR-132, miR-21, and miR-29a in AEXOs, which indicates that miR-132, miR-21, and miR-29a inside the AEXOs may be vital mediators of wound repair.
Recent studies have demonstrated that small RNAs within exosomes play a crucial role in enhancing the curative effects of exosomes [42]. To elucidate the molecular mechanisms underlying wound healing, we analyzed miRNAs associated with inflammation and wound repair using a comprehensive microarray approach. miR-223 involved in suppressing inflammation induces M2 polarization in macrophages by targeting various factors such as Mef2c [23]. miR-203 also potentiates the differentiation of monocytes to M2 macrophages by targeting TNF-α and IL-24 [43]. miR-146a modulates macrophage polarization by suppressing the Notch1 pathway [44]. Our results showed that miRNAs regulating the M2 phenotype (miR-223, miR-203, miR-146a) may participate in anti-inflammatory effects. Of note, it has been proposed that miR-29c, miR-192, and miR-215 promote remodeling through the activation of SMADs, β-catenin, E-cadherin, and SIP1 [45–47]. In line with our results, it has been hypothesized that the expression of miRNAs associated with wound healing regardless of the most significant and highest expression in AMSCs-derived exosomes, could reduce inflammation and enhance wound healing. The therapeutic effects on injury are likely to result from multiple factors such as miRNA, proteins, mRNA, lipids, and various molecules. In the present study, our primary focus was on identifying candidate miRNAs that are involved in inflammation and tissue regeneration. By comparing our results with previous data, we determined that the majority of identified miRNAs were known exosomal miRNAs. These results indicate the reliability of our analysis. There are some limitations of our data. Validation of the RNA sequencing data and evaluation of the up- or downstream effects of miRNA expression related to wound repair need to be further explored.
Taken together, our data suggest that AEXOs may exert equivalent or more potent effects on wound healing. In this study, we compared and analyzed AMSCs and AEXOs derived from the same donor at the miRNA level. We have shown that AEXOs could be used as therapeutic agents instead of AMSCs as the combination of miR-34a-5p, miR-124-3p, miR-146a-5p, miR-132, miR-21, and miR-29a, which are highly expressed in AEXOs, reduces inflammation and enhance wound healing. Of note, besides these miRNAs, other molecules carried by exosomes and crosstalk with cellular pathways, could also play a role in wound repair. In conclusion, our results suggest that AEXOs containing miRNAs that suppress inflammation and enhance remodeling may be used as a next-generation, stem-cell-based, cell-free strategy for wound healing. Further investigation with chronic wound and in vivo wound models and the effects of AEXOs on wound angiogenesis will be undertaken in our laboratory.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgement
This research was funded by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (No. 2019R1C1C1007036).
Compliance with ethical standards
Conflict of interest
The authors declare that they no conflict of interest.
Ethical statement
The study was approved by the Institutional Review Board at Severance Hospital (IRB No. 4-2019-0060).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Seung Yong Song, Email: PSSYSONG@yuhs.ac.
Hyun Ok Kim, Email: HYUNOK1019@yuhs.ac.
References
- 1.Rodriguez-Menocal L, Salgado M, Ford D, Van Badiavas E. Stimulation of skin and wound fibroblast migration by mesenchymal stem cells derived from normal donors and chronic wound patients. Stem Cells Transl Med. 2012;1:221–229. doi: 10.5966/sctm.2011-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao P, Sui BD, Liu N, Lv YJ, Zheng CX, Lu YB, et al. Anti-aging pharmacology in cutaneous wound healing: effects of metformin, resveratrol, and rapamycin by local application. Aging Cell. 2017;16:1083–1093. doi: 10.1111/acel.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin P. Wound healing–aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 4.Kim SY, Nair MG. Macrophages in wound healing: activation and plasticity. Immunol Cell Biol. 2019;97:258–267. doi: 10.1111/imcb.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon D, Yoon D, Sim H, Hwang I, Lee JS, Chun W. Accelerated wound healing by fibroblasts differentiated from human embryonic stem cell-derived mesenchymal stem cells in a pressure ulcer animal model. Stem Cells Int. 2018;2018:4789568. doi: 10.1155/2018/4789568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kariminekoo S, Movassaghpour A, Rahimzadeh A, Talebi M, Shamsasenjan K, Akbarzadeh A. Implications of mesenchymal stem cells in regenerative medicine. Artif Cells Nanomed Biotechnol. 2016;44:749–757. doi: 10.3109/21691401.2015.1129620. [DOI] [PubMed] [Google Scholar]
- 7.Gadelkarim M, Abushouk AI, Ghanem E, Hamaad AM, Saad AM, Abdel-Daim MM. Adipose-derived stem cells: effectiveness and advances in delivery in diabetic wound healing. Biomed Pharmacother. 2018;107:625–633. doi: 10.1016/j.biopha.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Wang S, Wu S, Hao Q, Li Y, Guo Z, et al. Exosomes secreted by adipose-derived mesenchymal stem cells regulate type I collagen metabolism in fibroblasts from women with stress urinary incontinence. Stem Cell Res Ther. 2018;9:159. doi: 10.1186/s13287-018-0899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendt M, Rezvani K, Shpall E. Mesenchymal stem cell-derived exosomes for clinical use. Bone Marrow Transplant. 2019;54:789–792. doi: 10.1038/s41409-019-0616-z. [DOI] [PubMed] [Google Scholar]
- 10.Rani S, Ryan AE, Griffin MD, Ritter T. Mesenchymal stem cell-derived extracellular vesicles: toward cell-free therapeutic applications. Mol Ther. 2015;23:812–823. doi: 10.1038/mt.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011;6:481–492. doi: 10.2217/rme.11.35. [DOI] [PubMed] [Google Scholar]
- 12.Ko KW, Yoo YI, Kim JY, Choi B, Park SB, Park W, et al. Attenuation of tumor necrosis factor-α induced inflammation by umbilical cord-mesenchymal stem cell derived exosome-mimetic nanovesicles in endothelial cells. Tissue Eng Regen Med. 2020;17:155–163. doi: 10.1007/s13770-019-00234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shabbir A, Cox A, Rodriguez-Menocal L, Salgado M, Van Badiavas E. Mesenchymal stem cell exosomes induce proliferation and migration of normal and chronic wound fibroblasts, and enhance angiogenesis in vitro. Stem Cells Dev. 2015;24:1635–1647. doi: 10.1089/scd.2014.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Jia L, Zheng Y, Li W. Salivary exosomes: emerging roles in systemic disease. Int J Biol Sci. 2018;14:633–643. doi: 10.7150/ijbs.25018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 16.Ankrum JA, Ong JF, Karp JM. Mesenchymal stem cells: immune evasive, not immune privileged. Nat Biotechnol. 2014;32:252–260. doi: 10.1038/nbt.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou YF, Bosch-Marce M, Okuyama H, Krishnamachary B, Kimura H, Zhang L, et al. Spontaneous transformation of cultured mouse bone marrow-derived stromal cells. Cancer Res. 2006;66:10849–10854. doi: 10.1158/0008-5472.CAN-06-2146. [DOI] [PubMed] [Google Scholar]
- 18.Jiang L, Zhang S, Hu H, Yang J, Wang X, Ma Y, et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate acute liver failure by reducing the activity of the NLRP3 inflammasome in macrophages. Biochem Biophys Res Commun. 2019;508:735–741. doi: 10.1016/j.bbrc.2018.11.189. [DOI] [PubMed] [Google Scholar]
- 19.Bjørge IM, Kim SY, Mano JF, Kalionis B, Chrzanowski W. Extracellular vesicles, exosomes and shedding vesicles in regenerative medicine—a new paradigm for tissue repair. Biomater Sci. 2017;6:60–78. doi: 10.1039/C7BM00479F. [DOI] [PubMed] [Google Scholar]
- 20.Goodarzi P, Larijani B, Alavi-Moghadam S, Tayanloo-Beik A, Mohamadi-Jahani F, Ranjbaran N, et al. Mesenchymal stem cells-derived exosomes for wound regeneration. Adv Exp Med Biol. 2018;1119:119–131. doi: 10.1007/5584_2018_251. [DOI] [PubMed] [Google Scholar]
- 21.Heo JS, Choi Y, Kim HO. Adipose-derived mesenchymal stem cells promote M2 macrophage phenotype through exosomes. Stem Cells Int. 2019;2019:7921760. doi: 10.1155/2019/7921760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Li D, Wikstrom JD, Pivarcsi A, Sonkoly E, Stahle M, et al. MicroRNA-132 promotes fibroblast migration via regulating RAS p21 protein activator 1 in skin wound healing. Sci Rep. 2017;7:7797. doi: 10.1038/s41598-017-07513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herter EK, Xu Landén N. Non-coding RNAs: new players in skin wound healing. Adv Wound Care (New Rochelle) 2017;6:93–107. doi: 10.1089/wound.2016.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulholland EJ, Dunne N, McCarthy HO. MicroRNA as therapeutic targets for chronic wound healing. Mol Ther Nucleic Acids. 2017;8:46–55. doi: 10.1016/j.omtn.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Dong Z, Cao Y, Wang H, Liu S, Liao L, et al. MSC-derived exosome promotes M2 polarization and enhances cutaneous wound healing. Stem Cells Int. 2019;2019:7132708. doi: 10.1155/2019/7132708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roşca AM, Ţuţuianu R, Titorencu ID. Mesenchymal stromal cells derived exosomes as tools for chronic wound healing therapy. Rom J Morphol Embryol. 2018;59:655–662. [PubMed] [Google Scholar]
- 28.Zhong S, He X, Li Y, Lou X. Conditioned medium enhances osteogenic differentiation of induced pluripotent stem cell-derived mesenchymal stem cells. Tissue Eng Regen Med. 2019;16:141–150. doi: 10.1007/s13770-018-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blazquez R, Sanchez-Margallo FM, de la Rosa O, Dalemans W, Alvarez V, Tarazona R, et al. Immunomodulatory potential of human adipose mesenchymal stem cells derived exosomes on in vitro stimulated T cells. Front Immunol. 2014;5:556. doi: 10.3389/fimmu.2014.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Guo X. A review: therapeutic potential of adipose-derived stem cells in cutaneous wound healing and regeneration. Stem Cell Res Ther. 2018;9:302. doi: 10.1186/s13287-018-1044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Che Y, Shi X, Shi Y, Jiang X, Ai Q, Shi Y, et al. Exosomes derived from miR-143-overexpressing MSCs inhibit cell migration and invasion in human prostate cancer by downregulating TFF3. Mol Ther Nucleic Acids. 2019;18:232–244. doi: 10.1016/j.omtn.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep. 2017;7:1878. doi: 10.1038/s41598-017-01731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra MB, Barroso WA, da Silva NN, Silva SDN, Borges ACR, Abreu IC, et al. From inflammation to current and alternative therapies involved in wound healing. Int J Inflam. 2017;2017:3406215. doi: 10.1155/2017/3406215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah JM, Omar E, Pai DR, Sood S. Cellular events and biomarkers of wound healing. Indian J Plast Surg. 2012;45:220–228. doi: 10.4103/0970-0358.101282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzyszczyk P, Schloss R, Palmer A, Berthiaume F. The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front Physiol. 2018;9:419. doi: 10.3389/fphys.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darby IA, Laverdet B, Bonté F, Desmoulière A. Fibroblasts and myofibroblasts in wound healing. Clin Cosmet Investig Dermatol. 2014;7:301–311. doi: 10.2147/CCID.S50046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J Biol Chem. 2009;284:34342–34354. doi: 10.1074/jbc.M109.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26:78. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrello J, Monticone S, Gai C, Gomez Y, Kholia S, Camussi G. Stem cell-derived extracellular vesicles and immune-modulation. Front Cell Dev Biol. 2016;4:83. doi: 10.3389/fcell.2016.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domenis R, Cifù A, Quaglia S, Pistis C, Moretti M, Vicario A, et al. Pro inflammatory stimuli enhance the immunosuppressive functions of adipose mesenchymal stem cells-derived exosomes. Sci Rep. 2018;8:13325. doi: 10.1038/s41598-018-31707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee J, Sen CK. microRNA and wound healing. Adv Exp Med Biol. 2015;888:291–305. doi: 10.1007/978-3-319-22671-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ti D, Hao H, Fu X, Han W. Mesenchymal stem cells-derived exosomal microRNAs contribute to wound inflammation. Sci China Life Sci. 2016;59:1305–1312. doi: 10.1007/s11427-016-0240-4. [DOI] [PubMed] [Google Scholar]
- 43.Primo MN, Bak RO, Schibler B, Mikkelsen JG. Regulation of pro-inflammatory cytokines TNFalpha and IL24 by microRNA-203 in primary keratinocytes. Cytokine. 2012;60:741–748. doi: 10.1016/j.cyto.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 44.Huang C, Liu XJ, QunZhou XJ, Xie J, Ma TT, Meng XM, et al. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int Immunopharmacol. 2016;32:46–54. doi: 10.1016/j.intimp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Herman-Edelstein M, Koh P, Burns W, Jandeleit-Dahm K, Watson A, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794–1802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.