Abstract
Few studies on mRNA expression of the prolactin receptor (PRLR) isoforms in different tissues of sheep were reported. The objective of this study was to analyze the gene sequence and mRNA expression of PRLR isoforms in the uterus, mammary gland, ovary, spleen and lymph tissue of ewes during the lactation and post-weaning periods. Ten lactating crossbred ewes (Dorper×Hu sheep) with twin lambs were used in this study. Five ewes were chosen randomly and slaughtered at mid-lactation (35 days after lambing). The remaining five ewes were slaughtered on the 5th day after weaning. Samples of uterus, mammary gland, ovary, spleen and lymph tissue were collected from each ewe to determine the mRNA expression of long PRLR (L-PRLR) and short PRLR (S-PRLR) by RT-qPCR. The physical and chemical properties, the similarity of the nucleotides L-PRLR and S-PRLR genes and the secondary and tertiary structure of the L-PRLR and S-PRLR proteins of sheep were analyzed. The results indicated that the predicted protein molecular weights of L-PRLR and S-PRLR are 65235.36 KD and 33847.48 KD, respectively, with isoelectric points of 5.12 and 8.34, respectively. The secondary protein structures of L-PRLR and S-PRLR are different. For L-PRLR these include alpha helix, extended strand and random coils and β-turns for which the content was 16.01%, 21%, 59.55% and 3.44%, respectively, whereas the secondary protein structures of S-PRLR contain only alpha helices, extended strand and random coils, comprising 18.24%, 30.07% and 48.99%, respectively. The L-PRLR and S-PRLR genes of the sheep (Ovis aries) had nucleotide sequences showing much similarity among ruminants. In these sheep, mRNA expression of L-PRLR and S-PRLR was highest in the uterus and differed between the uterus, ovary, mammary gland, spleen and lymph tissue. The mRNA expression of L-PRLR in lymph tissue was higher during lactation than in the post-weaning period (P < 0.01), whereas mRNA expression of S-PRLR in the uterus and the mammary gland was lower during lactation than during the post-weaning period (P < 0.01). In the uterus, mRNA expression of L-PRLR was higher than that of S-PRLR during lactation (P < 0.01) but there were no significant differences (P < 0.05) for the other five tissues. This study that the L-PRLR and S-PRLR proteins in ewes are mainly composed of extended fragments and random coils. The data also indicate that mRNA expression of L-PRLR and S-PRLR genes varies among different tissues in sheep and is higher in the uterus than in the ovary, spleen, mammary gland and lymph tissue throughout lactation and the post-weaning period.
Keywords: Sheep, Prolactin receptor, Lactation, Post-weaning
Introduction
Prolactin (PRL) is a single chain polypeptide hormone synthesized and secreted from the anterior pituitary gland. It belongs to the prolactin/growth hormone family (Goffin et al., 1996) and participates in various physiological processes in mammals such as reproduction, immunity and regulation of metabolism (Freeman et al., 2000). The prolactin receptor, PRLR, has a central role in the PRL signal transduction cascade since PRL exerts its biological functions by binding to PRLR (Bignon et al., 1997). PRLR belongs to the superfamily of cytokine receptors and has been detected in a variety of tissues in many mammals (Motamedi et al., 2020). Ruminants have both a long and a short prolactin receptor (L-PRLR, S-PRLR, respectively) (Bignon et al., 1997). Their genes, L-PRLR and S-PRLR, are expressed by alternative splicing of a single PRLR gene (Chen et al., 2020) and they differ by the lengths of their carboxyl-terminals at the cytoplasmic domains (Viitala et al., 2006).
Many previous studies have reported that PRLR is associated with reproduction (Goffin et al., 1998). In rodent ovaries mRNA expression of L-PRLR is higher than that of S-PRLR during all phases of the estrous cycle and throughout pregnancy (Clarke, Arey & Linzer, 1993; Clarke & Linzer, 1993). PRLR has also been reported to be associated with immunity (Zhou et al., 2020). For instance, the circulating concentration of PRL directly affects the production of CDT cells (Bernichtein, Touraine & Goffin, 2010) and the role of PRL needs to be achieved by the expression of PRLR on immune cells (Zhou et al., 2020). S-PRLR has been cloned in rats (Boutin et al., 1988), and L-PRLR is widely expressed in the muscle, liver, spleen, mammary gland and adipose tissues of dairy goats (Shi et al., 2016). However, few studies on mRNA expression of the PRLR isoforms in different tissues of sheep were reported. We hypothesized that the expression of L-PRLR and S-PRLR were different in uterus, ovary, mammary gland, spleen and lymph tissue of sheep during the different physiological phases. If it does, it will be of great value for further research on the function of L-PRLR and S-PRLR in ruminants. Therefore, the objective of the present study was to analyze the gene sequence of PRLR isoforms and the mRNA expression of L-PRLR and S-PRLR in the ovary, mammary gland, uterus, lymph tissue and spleen in ewes of the Dorper ×Hu breed during the lactation and post-weaning periods.
Materials and Methods
Animals and experimental design
The study was conducted from August to October 2018 on Weizun Sheep Farm located in the Hebei province of China. All procedures used in this study were approved by the Laboratory Animal Ethics Committee of Hebei Agricultural University (Hebei, P.R. China; permit number 2018082).
Total mixed rations (TMR) were formulated according to NRC (2007). Ewes were fed twice daily, at 0700 h and 1700 h, and had free access to clean water. Feed residuals were 5%–7% of the total offered and these were removed when cleaning was carried out each day after the afternoon feeding.
Ten lactating crossbred ewes (Dorper × Hu sheep, 2.5 years of age) with twin lambs were used. Five ewes were chosen randomly and euthanized in the middle (35 days after lambing) of the lactation period. The remaining five ewes were euthanized on the 5th day after weaning. Samples of tissue from the uterus, mammary gland, ovary, spleen and lymph tissue were collected from each ewe after euthanized.
Data and sample collection
Ten ewes were deprived of feed for 24 h and of water for 16 h then killed humanely at around 0800 h using electro-stunning followed by severance of blood vessels in the neck. Samples of tissue from the uterus, mammary gland, ovary, spleen and lymph tissue were obtained within 20 min of death under sterile conditions by using a sterile scalpel. These were cut into 0.2 cm3 pieces and then immediately frozen in liquid nitrogen for storage until extraction of RNA for the analysis of PRLR expression.
Analytical procedures
Primer design
Conserved regions were found by aligning sheep L-PRLR, S-PRLR and GADPH gene sequences published in GenBank using DNAMAN. Primers were then designed from the conserved region using Primer Premier 5.0 and synthesised by Shanghai Sheng Gong Biotechnology Co., Ltd. Primer sequences and related information are as follows: L-PRLR: 5′-CCCCTTGTTCTCTGCTAAACCC-3′(forward), 5′-CTATCCGTCACCCGAGACACC-3′(reverse) (120 bp); S-PRLR: 5′-AAATACCTTGTGCAGATTCGATG-3′(forward), 5′-AAACACAGACACAAGGCGAGA-3′(reverse) (267 bp); GAPDH: 5′-CTGACCTGCCGCCTGGAGAAA-3′(forward), 5′-GTAGAAGAGTGAGTGTCGCTGTT-3′(reverse) (149 bp).
RNA extraction and quantitative real-time PCR
After thoroughly grinding the collected tissues in liquid nitrogen, total RNA was extracted according to the specification of TRNzol total RNA extraction kit (TIANGEN) and stored at −80 °C. Reverse transcription was carried out using a reagent kit (Takala) according to the manufacturer’s instructions. The reaction mixture, 20 µL, consisted of 5 ×Mix (4 µL), RNA (2 µg), and RNase-free water (16 µL). The reaction was performed at 37 °C for 15 min followed by 85 °C for 5 s; the product was stored at −4 °C.
q-PCR was conducted in strict accordance with the LC-480 PCR system instructions using Ultra SYBR Mixture (with Rox) and the following cycling protocol: 10 min at 95 °C followed by 40 cycles consisting of 15 s at 95 °C and 60 s at 60 °C. The reaction mixtures contained 10 µL Ultra SYBR Mixture (2 ×), 0.4 µL upstream and downstream primers (10 µM) each, 2 µL template and sterile distilled water to a final volume of 20 µL. Relative expression of the target genes was calculated by the 2−ΔΔCT method based on the quantitative real-time PCR results.
Sequencing analysis
EditSeq software was used to predict the physical and chemical properties of L-PRLR and S-PRLR in ewes, which included amino acid composition, isoelectric point and theoretical molecular weight; DNAMAN software was used to analyze the similarity of the nucleotides of L-PRLR and S-PRLR. Online software was used to predict the secondary structure of L-PRLR and S-PRLR proteins (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) and their tertiary structure (http://www.expasy.org/swissmod/swiss-model.html).
Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA) and T-test for comparisons ( T-test was used for mRNA expression in the same tissue of the same isoform at different periods and in the same tissue of different isoforms at the same period. One-way ANOVA was used to analyze the mRNA expression of the same isoform in different tissues at the same period). All calculations were performed with the SAS 9.2 software (SAS Inst., Cary, North Carolina, USA). GraphPad prism 6.0 software was used to make the chart. Differences were considered significant at P <0.05. All data are expressed as the mean ± standard error (S.E.).
Results
Analysis of L-PRLR and S-PRLR and their associated gene sequences
The measured molecular weights of L-PRLR and S-PRLR are 65,235.36 KD and 33,847.48 KD, respectively, and the isoelectric points are 5.12 and 8.34, respectively.
The nucleotide sequence similarity of the CDS region between ovis and other species of L-PRLR and S-PRLR are shown in Table 1. The nucleotide sequences of the L-PRLR and S-PRLR genes are highly conserved among ruminants.
Table 1. The nucleotide sequence similarity of the CDS region of L-PRLR and S-PRLR (%).
| Species | Capra hircus | Bos taurus | Sus scrofa | Mus musculus | Homo sapiens | Gallus gallus |
|---|---|---|---|---|---|---|
| L-PRLR | 97.65 | 94.79 | 77.16 | 65.12 | 71.86 | 39.24 |
| S-PRLR | 98.88 | 96.18 | 85.20 | 16.70 | 65.16 | 53.70 |
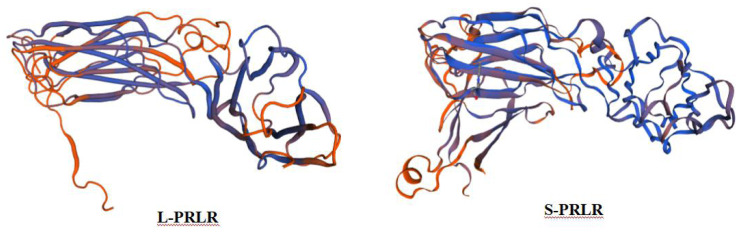
The secondary protein structures of L-PRLR and S-PRLR are different. The secondary protein structure of L-PRLR includes alpha helix, extended strand, random coils and β-turns at proportions of 16.01%, 21%, 59.55% and 3.44%, respectively,whereas the secondary protein structure of S-PRLR contains only alpha helices, extended strand and random coils, at proportions of 18.24%, 30.07% and 48.99%, respectively. The tertiary structures of L-PRLR and S-PRLR encoding proteins are mainly composed of extended fragments and random coils (Fig. 1). The forecast results are consistent with the secondary structures.
Figure 1. Prediction of tertiary structure of L-PRLR and S-PRLR protein in sheep.
Prediction of tertiary structure of L-PRLR and S-PRLR protein in sheep.
mRNA expression of L-PRLR and S-PRLR in different tissues of sheep
All samples were measured by spectrophotometer and had OD260/OD280 values between 1.8 and 2.0, which indicated that they were of good purity.
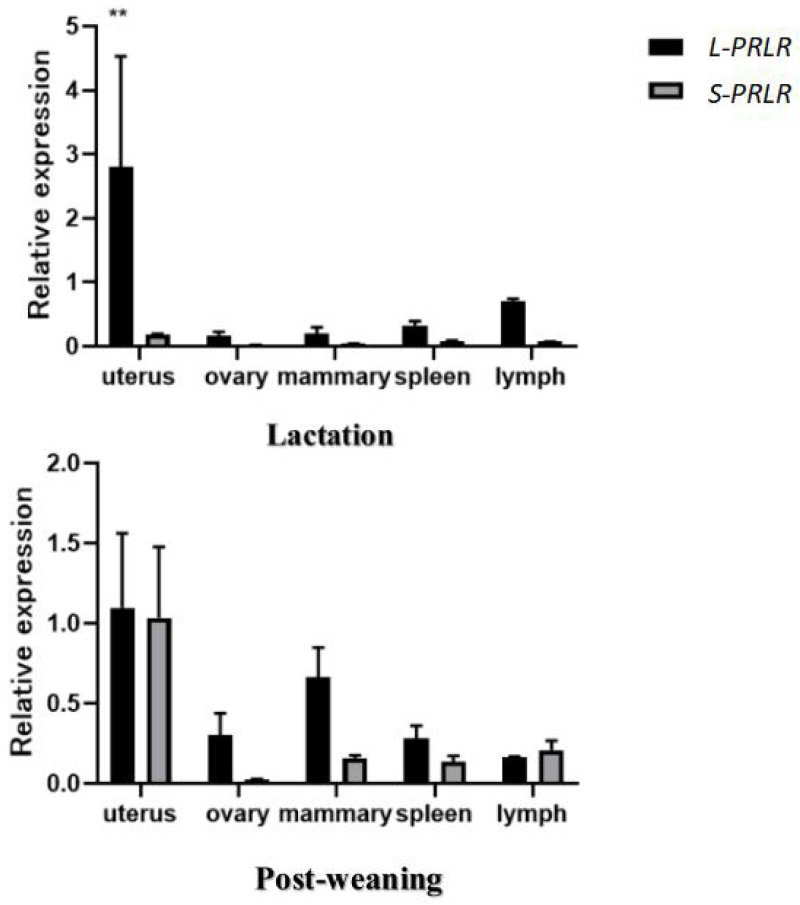
The mRNA expression of L-PRLR and S-PRLR in uterus, mammary gland, ovary, spleen and lymph tissue during lactation and post-weaning periods are shown in Fig. 2 and Table 2.
Figure 2. MRNA expression of L-PRLR and S-PRLR in different tissues during lactation and post-weaning in sheep.
Asterisks (**) indicate that the difference is extremely significant (P < 0.01).
Table 2. Relative expression of L-PRLR and S-PRLR in different tissues during lactation and post-weaning in sheep.
| Uterus | Ovary | Mammary gland | Spleen | Lymph tissue | ||
|---|---|---|---|---|---|---|
| L-PRLR | Lactation | 2.808 ± 1.725a | 0.164 ± 0.064b | 0.194 ± 0.102b | 0.330 ± 0.065b | 0.700 ± 0.045ab∗∗ |
| Post-weaning | 1.098 ± 0.465a | 0.305 ± 0.134ab | 0.662 ± 0.188ab | 0.282 ± 0.079ab | 0.160 ± 0.009b | |
| S-PRLR | Lactation | 0.186 ± 0.007 | 0.022 ± 0.002 | 0.038 ± 0.004 | 0.076 ± 0.014 | 0.072 ± 0.002 |
| Post-weaning | 1.034 ± 0.444a∗∗ | 0.022 ± 0.006b | 0.156 ± 0.020ab∗∗ | 0.138 ± 0.035b | 0.208 ± 0.060ab |
Notes.
Means with different superscripts within the same row are significantly different (P < 0.05).
Asterisks (**) within the same column represent significant differences (P < 0.01).
The mRNA expression of L-PRLR in uterus, mammary gland, ovary, spleen and lymph tissue during lactation and post-weaning periods in ewes are shown in Table 2. During lactation mRNA expression of L-PRLR was higher in the uterus than in the ovary, mammary gland and spleen (P < 0.05) but was similar to that of lymph tissue. However, during the post-weaning period, mRNA expression of L-PRLR in the uterus was higher than that in lymph tissue (P < 0.05), but it was not different from the values measured in the uterus, ovary, spleen and mammary gland. The expression in lymph tissue was higher during lactation than in the post-weaning period (P < 0.01) but there were no period-related differences in expression in the case of uterus, ovary, mammary gland or spleen.
The mRNA of S-PRLR in uterus, mammary gland, ovary, spleen and lymph tissue during lactation and post-weaning periods in ewes are shown in Table 2. There were no differences in mRNA expression of S-PRLR among the five tissues during the lactation period. However, during the post-weaning period mRNA expression of S-PRLR in uterus was higher than in the ovary and spleen (P < 0.05), although it was similar to that of the mammary gland and lymph tissue. No differences were observed in the mRNA expression of S-PRLR among ovary, mammary gland, spleen and lymph tissue during post-weaning period. Expression of S-PRLR in the uterus and mammary gland was lower during lactation than in the post-weaning period (P < 0.01), but there were no period-related differences in the cases of ovary, spleen or lymph tissue.
The expression of L-PRLR and S-PRLR in 5 tissue during the lactation and post-weaning period are shown in Fig. 2. Expression of L-PRLR in the uterus was higher than that of S-PRLR during the lactation period (P < 0.01). However, there were no differences in expression of the two genes in the ovary, spleen, mammary gland and lymph tissue during the lactation period nor within any of the five tissues during the post-weaning period.
Discussion
Sequence analysis of prolactin receptor isoforms
A long form (L-PRLR), an intermediate form (I-PRLR) and two short forms (S-PRLR) occur in humans (Trott et al., 2003; Abramicheva & Smirnova, 2019). There are at least four PRLR isoforms in mice, including three short and one long, but only two of the short PRLR isoforms are considered to be proteins (Tan et al., 2011) and there are two PRLR isoforms (L-PRLR and S-PRLR) in rats (Jiang et al., 2004). Cloning and genomic analysis of cDNA has revealed that L-PRLR and S-PRLR have arisen from the process of differential alternative splicing of their coding genes (Moore & Oka, 1993). S-PRLR differs from L-PRLR by having a 39 base pair insert at the beginning of the cytoplasmic domain, but it has two contiguous inframe stop codons at its 3′end (Bignon et al., 1997). The present study also reveals differences in the secondary and tertiary structure of the two forms of the receptor protein, which is consistent with there being differences in expression and function of the two protein forms. Our study also showed that the nucleotide sequences of L-PRLR and S-PRLR genes in the sheep were similar to those of other species, indicating that these are highly conserved among ruminants, and could explain why L-PRLR and S-PRLR have similar functions across different species.
The expression of prolactin receptor isoforms in different tissues
L-PRLR has been reported as predominantly expressed in the mammary gland, ovary, liver, uterus, skeletal muscle, corpus luteum, and adrenal glands of goats (Shi et al., 2016). This is in keeping with the important role of prolactin receptors as the mediators of prolactin’s actions in processes such as growth, lactation, reproduction and immunity (Posner et al., 1974). Our results confirm the presence of L-PRLR and S-PRLR in tissue from the uterus, mammary gland, ovary, spleen and a lymph tissue in ewes during lactation and the post-weaning period. However, we have found some differences in mRNA expression of L-PRLR and S-PRLR throughout these tissues in sheep. For instance, the high level of expression of PRLR we recorded in the sheep uterus in both periods that were sampled here accords with a similar finding in black Muscovy ducks (Li et al., 2020). Prolactin probably has a vital role in the post-lambing regeneration of the uterine epithelium, which is generally completed within 31 days (O’Shea & Wright, 1984). This process involves reduction of the uterine volume, some tissue degradation, and epithelial repair of the endometrium (Tielgy et al., 1982). The higher mRNA expression of L-PRLR in uterine tissue of the ewes, compared with that of S-PRLR, during lactation and post-weaning indicates that the long isoform of the receptor may be primarily involved in uterine repair and recovery following birth of lambs. The increase in expression of S-PRLR in the uterus from lactation to post-weaning indicates that the short form of the receptor may have a primary role during normal maintenance of the uterus.
L-PRLR and S-PRLR appear to serve different roles in ovary as well. For example, PMSG increased mRNA expression of L-PRLR, suggesting a possible involvement of L-PRLR in folliculogenesis. In contrast, hCG treatment stimulated expression of S-PRLR, indicating a role for the corresponding receptor isoform in formation and maintenance of the corpus luteum (Thompson et al., 2011). Also, reverse-transcription PCR analysis of sheep ovarian tissue showed differences in localization and expression of both S-PRLR and L-PRLR throughout the estrous cycle, with L-PRLR being particularly localized in stromal cells surrounding primordial and primary follicles, whereas genes for both PRLR isoforms were found in granulosa cells of preantral follicles and luteal cells within the corpus luteum (Picazo et al., 2004). The expression of L-PRLR in the sheep ovary is markedly increased around the time of estrus, unlike that of S-PRLR which does not differ throughout the estrous cycle (Picazo et al., 2004). However, during the lactation period follicle development is inhibited and estrus does not occur (Song et al., 2019). In the case of L-PRLR our results show an increased level of expression during the post-weaning period which is consistent with a role for the long form of the receptor during the recovery of ovarian follicular development following the birth of lambs.
In goats, expression of PRLR gradually increases during the dry period after lactation (Song et al., 2019) in a pattern similar to the increases in expression levels of L-PRLR and S-PRLR recorded in the mammary glands of ewes in the present study. At the end of lactation mammary glands enter a degenerative phase, in which the fat pad regenerates in concert with increases in PRLR mRNA content of the adipocytes (Lesueur et al., 1991). This finding and the present results implicate involvement of L-PRLR and S-PRLR in the process of post-lactational mammary remodeling. L-PRLR has a principal role in the induction of milk protein gene transcription (Das & Vonderhaar, 1995) whereas the involvement of S-PRLR is less clear. For instance, L-PRLR could activate the β-casein gene promoter, while S-PRLR did not (Berlanga et al., 1997). Also, over-expression of S-PRLR enabled mammary development and function when PRLR genes were knocked out in heterozygous mice (Zi, Chen & Wang, 2012) and S-PRLR appears to have a negative role in relation to the involvement of PRL in milk protein gene transcription (Berlanga et al., 1997). Together with the present finding of higher expression of L-PRLR compared with that of S-PRLR in the ovine mammary gland, this is evidence for L-PRLR having the predominant role on the maintenance of lactation and in mammary gland repair processes.
PRL play an crucial role in regulating immunity (Gharbaran et al., 2020), not only by promoting the proliferation of immune cells, but also by stimulating the production of antibodies such as IgG and IgM, and these functions involve expression of the PRL receptor genes (Zhou et al., 2020). For instance, the transcripts encoding both isoforms of PRLR (L-PRLR and S-PRLR) were recorded in all lymphoid tissues examined in mice and rats (Touraine & Kelly, 1995) and in the spleens and lymph tissues of sheep in our study. In the case of thymus and spleen, mRNA expression of L-PRLR was higher than that of S-PRLR (Ouhtit, Kelly & Morel, 1994). In the present study, expression of L-PRLR was higher than that of S-PRLR in lymphatic tissue during lactation, but this pattern reversed during the post-weaning period. These differential findings indicate different specific roles for the two isoforms of the receptor in relation to their involvement in the progression of immune responses during and after lactation that warrant further study.
Conclusions
It was concluded that the mRNA expression of L-PRLR and S-PRLR genes varies among different tissues in sheep. The mRNA expression of L-PRLR and S-PRLR is higher in the uterus than in the ovary, spleen, mammary gland and lymph tissue during both lactation and the post-weaning period. The mRNA expression of L-PRLR is higher than S-PRLR in the uterus, ovary, spleen, mammary gland during both lactation and post-weaning period. The mRNA expression of L-PRLR was higher than S-PRLR in lymph tissue during lactation, but this pattern reversed during the post-weaning period.
Supplemental Information
2−ΔΔCt values of L-PRLR and S-PRLR in uterus, mammary gland, ovary, lymph and spleen of sheep during lactation and post-weaning period. Each group had 5 replicates
Acknowledgments
The authors express their gratitude to Doctor Zhang for laboratory support and to fellow students in the laboratory for assistance.
Funding Statement
This research was supported by the National Modern Agricultural Industry Technology System Construction Project of China [CARS-38] and [CARS-39], the Excellent Youth Program of Hebei Province [8042018-1081034], and the Hebei Province Science Foundation for Youths (C2019204357). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Yingjie Zhang, Email: zhangyingjie66@126.com.
Yueqin Liu, Email: liuyueqin66@126.com.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Ruochen Yang and Chunhui Duan conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Yunxia Guo conceived and designed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Yujing Ma analyzed the data, prepared figures and/or tables, and approved the final draft.
Nazi Niu performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Yingjie Zhang conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Yueqin Liu conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
Hebei, P.R. China; permit number 2018082
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The Prolactin receptor data is available at UniProt: O46561
- L-PRLR(Ovis): identifier: O46561-1
- S-PRLR(Ovis): identifier: O46561-2
The Ovis aries glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mRNA sequence is available at NCBI: NM_001190390.1.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.
References
- Abramicheva & Smirnova (2019).Abramicheva PA, Smirnova OV. Prolactin receptor isoforms as the basis of tissue-specific action of prolactin in the norm and pathology. Biochemistry-Moscow. 2019;84(4):329–345. doi: 10.1134/S0006297919040011. [DOI] [PubMed] [Google Scholar]
- Berlanga et al. (1997).Berlanga JJ, GarciaRuiz JP, PerrotApplanat M, Kelly PA, Edery M. Short form of the prolactin (PRL) receptor silences PRL induction of the beta-casein gene. Molecular Endocrinology. 1997;11(10):1449–1457. doi: 10.1210/me.11.10.1449. [DOI] [PubMed] [Google Scholar]
- Bernichtein, Touraine & Goffin (2010).Bernichtein S, Touraine P, Goffin V. New concepts in prolactin biology. Journal of Endocrinology. 2010;206(1):1–11. doi: 10.1677/JOE-10-0069. [DOI] [PubMed] [Google Scholar]
- Bignon et al. (1997).Bignon C, Binart N, Ormandy C, Schuler LA, Kelly PA, Djiane J. Long and short forms of the ovine prolactin receptor: cDNA cloning and genomic analysis reveal that the two forms arise by different alternative splicing mechanisms in ruminants and in rodents. Journal of Molecular Endocrinology. 1997;19(2):109–120. doi: 10.1677/jme.0.0190109. [DOI] [PubMed] [Google Scholar]
- Boutin et al. (1988).Boutin JM, Jolicoeur C, Okamura H, Gagnon J, Edery M, Shirota M, Banville D, Dusanter-Fourt I, Djiane J, Kelly PA. Cloning and expression of the rat prolactin receptor, a member of the growth hormone/prolactin receptor gene family. Cell. 1988;53(1):69–77. doi: 10.1016/0092-8674(88)90488-6. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2020).Chen YX, Moutal A, Navratilova E, Kopruszinski C, Yue X, Ikegami M, Chow M, Kanazawa I, Bellampalli SS, Xie J, Patwardhan A, Rice K, Fields H, Akopian A, Neugebauer V, Dodick D, Khanna R, Porreca F. The prolactin receptor long isoform regulates nociceptor sensitization and opioid-induced hyperalgesia selectively in females. Science Translational Medicine. 2020;12:529. doi: 10.1126/scitranslmed.aay7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, Arey & Linzer (1993).Clarke DL, Arey BJ, Linzer DI. Prolactin receptor messenger ribonucleic acid expression in the ovary during the rat estrous cycle. Endocrinology. 1993;133(6):2594–2603. doi: 10.1210/en.133.6.2594. [DOI] [PubMed] [Google Scholar]
- Clarke & Linzer (1993).Clarke DL, Linzer DIH. Changes in prolactin receptor expression during pregnancy in the mouse ovary. Endocrinology. 1993;133(1):224–232. doi: 10.1210/en.133.1.224. [DOI] [PubMed] [Google Scholar]
- Das & Vonderhaar (1995).Das R, Vonderhaar BK. Transduction of prolactin (PRL) growth signal through both long and short forms of the PRL receptor. Molecular Endocrinology. 1995;9(12):1750–1759. doi: 10.1210/me.9.12.1750. [DOI] [PubMed] [Google Scholar]
- Freeman et al. (2000).Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiological Reviews. 2000;80(4):1523–1631. doi: 10.1152/physrev.2000.80.4.1523. [DOI] [PubMed] [Google Scholar]
- Gharbaran et al. (2020).Gharbaran R, Onwumere O, Codrington N, Somenarain L, Redenti S. Immunohistochemical localization of prolactin receptor (PRLR) to Hodgkin’s and Reed-Sternberg cells of Hodgkin’s lymphoma. Acta Histochemica. 2020;123(1):151657–151657. doi: 10.1016/j.acthis.2020.151657. [DOI] [PubMed] [Google Scholar]
- Goffin et al. (1998).Goffin V, Bouchard B, Ormandy CJ, Weimann E, Ferrag F, Touraine P, Bole-Feysot C, Maaskant RA, Clement-Lacroix P, Edery M, Binart N, Kelly PA. Prolactin: a hormone at the crossroads of neuroimmunoendocrinology. Annals of the New York Academy of Sciences. 1998;840(1):498–509. doi: 10.1111/j.1749-6632.1998.tb09588. [DOI] [PubMed] [Google Scholar]
- Goffin et al. (1996).Goffin V, Shiverick KT, Kelly PA, Martial JA. Sequence-function relationships within the expanding family of prolactin, growth hormone, placental lactogen, and related proteins in mammals. Endocrine Reviews. 1996;17(4):385–410. doi: 10.1210/er.17.4.385. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2004).Jiang R, Li J, Qu L, Li H, Yang N. A new single nucleotide polymorphism in the chicken pituitary-specific transcription factor (POU1F1) gene associated with growth rate. Animal Genetics. 2004;35(4):344–346. doi: 10.1111/j.1365-2052.2004.01164.x. [DOI] [PubMed] [Google Scholar]
- Lesueur et al. (1991).Lesueur L, Edery M, Ali S, Paly J, Kelly PA, Djiane J. Comparison of long and short forms of the prolactin receptor on prolactin-induced milk protein gene transcription. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(3):824–828. doi: 10.1073/pnas.88.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2020).Li X, Ji W, Sun G, Xiao W, Bian Y, Qing H. Cloning and expression analysis of PRL and PRLR genes in black Muscovy duck. British Poultry Science. 2020;61(1):92–96. doi: 10.1080/00071668.2019.1680800. [DOI] [PubMed] [Google Scholar]
- Moore & Oka (1993).Moore RC, Oka T. Cloning and sequencing of the cDNA encoding the murine mammary gland long-form prolactin receptor. Gene. 1993;134(2):263–265. doi: 10.1016/0378-1119(93)90104. [DOI] [PubMed] [Google Scholar]
- Motamedi et al. (2020).Motamedi B, Rafiee-Pour HA, Khosravi MR, Kefayat A, BaradaranN A, Amjadi E, Goli P. Prolactin receptor expression as a novel prognostic biomarker for triple negative breast cancer patients. Annals of Diagnostic Pathology. 2020;46:151507. doi: 10.1016/j.anndiagpath.2020.151507. [DOI] [PubMed] [Google Scholar]
- O’Shea & Wright (1984).O’Shea JD, Wright PJ. Involution and regeneration of the endometrium following parturition in the ewe. Cell and Tissue Research. 1984;236(2):477–485. doi: 10.1007/BF00214253. [DOI] [PubMed] [Google Scholar]
- Ouhtit, Kelly & Morel (1994).Ouhtit A, Kelly PA, Morel G. Visualization of gene expression of short and long forms of prolactin receptor in rat digestive tissues. The American Journal of Physiology. 1994;266(5):G807–G815. doi: 10.1152/ajpgi.1994.266.5.G807. [DOI] [PubMed] [Google Scholar]
- Picazo et al. (2004).Picazo RA, Ruiz JPG, Moreno JS, de Bulnes AG, Munoz J, Silvan G, Lorenzo PL, Illera JC. Cellular localization and changes in expression of prolactin receptor isoforms in sheep ovary throughout the estrous cycle. Reproduction. 2004;128(5):545–553. doi: 10.1530/rep.1.00343. [DOI] [PubMed] [Google Scholar]
- Posner et al. (1974).Posner BI, Kelly PA, Shiu RP, Friesen HG. Studies of insulin, growth hormone and prolactin binding: tissue distribution, species variation and characterization. Endocrinology. 1974;95(2):521–531. doi: 10.1210/endo-95-2-521. [DOI] [PubMed] [Google Scholar]
- Shi et al. (2016).Shi HP, Zhang TY, Yi YQ, Wang H, Luo J. Long form PRLR (lPRLR) regulates genes involved in the triacylglycerol synthesis in goat mammary gland epithelial cells. Small Ruminant Research. 2016;139:7–14. doi: 10.1016/j.smallrumres.2016.04.008. [DOI] [Google Scholar]
- Song et al. (2019).Song YX, Hu P, Bai YL, Zhao C, Xia C, Xu C. Plasma metabolic characterisation of dairy cows with inactive ovaries and oestrus during the peak of lactation. Journal of Veterinary Research. 2019;63(3):359–367. doi: 10.2478/jvetres-2019-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan et al. (2011).Tan DY, Chen KHE, Khoo T, Walker AM. Prolactin increases survival and migration of ovarian cancer cells: Importance of prolactin receptor type and therapeutic potential of S179D and G129R receptor antagonists. Cancer Letters. 2011;310(1):101–108. doi: 10.1016/j.canlet.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Thompson et al. (2011).Thompson IM, Ozawa M, Bubolz JW, Yang Q, Dahl GE. Bovine luteal prolactin receptor expression: potential involvement in regulation of progesterone during the estrous cycle and pregnancy. Journal of Animal Science. 2011;89(5):1338–1346. doi: 10.2527/jas.2010-3559. [DOI] [PubMed] [Google Scholar]
- Tielgy et al. (1982).Tielgy AH, Fathalla M, Omar MA, Al-Dahash S. The clinical and morphological characteristics of the uterus of the goat during the period of involution. The Canadian Veterinary Journal. 1982;23(4):138–140. [PMC free article] [PubMed] [Google Scholar]
- Touraine & Kelly (1995).Touraine P, Kelly PA. Expression of the short and long forms of the prolactin receptor in Murine lymphoid-tissues. Recent Progress in Hormone Research. 1995;50:423–428. doi: 10.1016/b978-0-12-571150-0.50031-7. [DOI] [PubMed] [Google Scholar]
- Trott et al. (2003).Trott JF, Hovey RC, Koduri S, Vonderhaar BK. Alternative splicing to exon 11 of human prolactin receptor gene results in multiple isoforms including a secreted prolactin-binding protein. Journal of Molecular Endocrinology. 2003;30(1):31–47. doi: 10.1677/jme.0.0300031. [DOI] [PubMed] [Google Scholar]
- Viitala et al. (2006).Viitala S, Szyda J, Blott S, Schulman N, Lidauer M, Maki-Tanila A, George M, Vilkki J. The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics. 2006;173(4):2151–2164. doi: 10.1534/genetics.105.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2020).Zhou YX, Zong HF, Han L, YQ XIE, Jiang H, Gilly J, Zhang BH, Lu HL, Chen J, Sun R, Pan ZD, Zhu JW. A novel bispecific antibody targeting CD3 and prolactin receptor (PRLR) against PRLR-expression breast cancer. Journal of Experimental & Clinical Cancer Research. 2020;39(7528):667–674. doi: 10.1186/s13046-020-01564-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi, Chen & Wang (2012).Zi XD, Chen DW, Wang HM. Molecular characterization, mRNA expression of prolactin receptor (PRLR) gene during pregnancy, nonpregnancy in the yak (Bos grunniens) General and Comparative Endocrinology. 2012;175(3):384–388. doi: 10.1016/j.ygcen.2011.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2−ΔΔCt values of L-PRLR and S-PRLR in uterus, mammary gland, ovary, lymph and spleen of sheep during lactation and post-weaning period. Each group had 5 replicates
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.