Abstract
Background
The recent emergence of a highly infectious and contagious respiratory viral disease known as COVID-19 has vastly impacted human lives and overloaded the health care system. Therefore, it is crucial to develop a fast and accurate diagnostic system for the timely identification of COVID-19 infected patients and thus to help control its spread.
Methods
This work proposes a new deep CNN based technique for COVID-19 classification in X-ray images. In this regard, two novel custom CNN architectures, namely COVID-RENet-1 and COVID-RENet-2, are developed for COVID-19 specific pneumonia analysis. The proposed technique systematically employs Region and Edge-based operations along with convolution operations. The advantage of the proposed idea is validated by performing series of experimentation and comparing results with two baseline CNNs that exploited either a single type of pooling operation or strided convolution down the architecture. Additionally, the discrimination capacity of the proposed technique is assessed by benchmarking it against the state-of-the-art CNNs on radiologist's authenticated chest X-ray dataset. Implementation is available at https://github.com/PRLAB21/Coronavirus-Disease-Analysis-using-Chest-X-Ray-Images.
Results
The proposed classification technique shows good generalization as compared to existing CNNs by achieving promising MCC (0.96), F-score (0.98) and Accuracy (98%). This suggests that the idea of synergistically using Region and Edge-based operations aid in better exploiting the region homogeneity, textural variations, and region boundary-related information in an image, which helps to capture the pneumonia specific pattern.
Conclusions
The encouraging results of the proposed classification technique on the test set with high sensitivity (0.98) and precision (0.98) suggest the effectiveness of the proposed technique. Thus, it suggests the potential use of the proposed technique in other X-ray imagery-based infectious disease analysis.
Keywords: Coronavirus, COVID-19, Chest X-ray, Region homogeneity, Edge, Convolutional neural network, Transfer learning
1. Introduction
Recently, a new member of the coronaviruses (CoV) family named SARS-CoV-2 has appeared, which is highly threatening to human life [1]. This newly emerged member of the CoV family was responsible for the coronavirus disease (COVID-19) epidemic that started in December 2019. The COVID-19 has become a severe public health problem across the globe and was declared a pandemic in 2020 by WHO [2]. The CoV family consists of different human virus subtypes that cause mild to severe respiratory infections, including the common cold and two leading epidemics [3,4].
COVID-19 is a highly transmissible viral respiratory infection that is mostly characterized by lower to upper respiratory tract illness, fever, headache, cough and dyspnea. Besides these, it is accompanied by several other symptoms that vary among individuals. The progression of COVID-19 down to the lower respiratory tract causes inflammation, difficulty in breathing and targets the lungs in most cases. Pneumonia is the most typical clinical manifestation of COVID-19 infected patients, which in severe cases leads to death [5], [6], [7]. COVID-19 transmits primarily through respiratory secretions or droplets, which are discharged during cough, sneezing, talking, and contacting an infected person [8].
The confirmation of COVID-19 is ascertained through molecular diagnostic techniques such as RT-PCR and gene sequencing [9]. However, detection of COVID-19 infection through RT-PCR based testing is time-consuming and requires a duration of at least 4-6 hours from sample collection to results [10,11]. Contrary to this, radiographic imaging-based techniques consisting of X-ray and CT are among the quickest ways of screening.
X-ray is low cost and uses a small dose of radiations for image formation; therefore, it is usually used as an assistive screening tool in addition to RT-PCR for rapid assessment of COVID-19 in symptomatic patients [12,13]. In this regard, X-ray imaging is performed for assessment of the infection spread in lungs, treatment plan, patient care, and follow up. Patients with COVID-19 specific lung infection or pneumonia commonly exhibit patterns of ground-glass opacities and mixed appearance of both ground-glass opacities and consolidation. COVID-19 specific radiographic findings show the predominance of peripheral distribution, bilateral lung involvement and predilection of opacities in lower lobes [14]. Different countries such as China, Spain and Italy used X-ray imaging to administrate patients’ condition and monitor the course of the disease in intensive care patients who are not stable enough for CT scan [15,16].
The rapid transmission rate and critical condition of COVID-19 infected pneumonia patients have significantly burdened the radiologists. Therefore, it is crucial to develop a highly efficient COVID-19 detection technique that can interpret the subtle difference in radiographic patterns to assist the radiologists. Previously, several automated diagnostic systems based on classical Machine Learning (ML) and Deep Learning techniques were developed and successfully deployed due to fast detection speed and good performance [17], [18], [19].
Several researchers exploited the potential of Convolutional Neural Networks (CNNs) to expedite the analysis of COVID-19 infected images. Most of the existing studies adapted state-of-the-art CNN models like ResNet, DenseNet, VGG, MobileNet, Xception, and InceptionNet using Transfer learning (TL) for classification. Recently, a scheme of pre-trained CNN models (ResNet, squezeNet and DensNet) has been utilized to identify COVID-19 in X-ray images. These models are finetuned on the COVID-19 dataset named “COVID-Xray-5k” and achieved an average classification accuracy of 98% [20]. Likewise, [21] finetuned the pre-trained inception model to detect COVID-19 and reported an accuracy of 89.5% [21]. In another study by [22], the ResNet-50 model that was pre-trained on ImageNet data has been employed on small chest X-ray images and reported an accuracy of 98%. Afshar et al. presented COVID-CAPS based on Capsule Net and achieved an accuracy of 98%, sensitivity of 80%, and AUC-ROC of 0.97 [23]. They initially pre-trained the model on X-ray images in comparison to other approaches that adapted CNN models, which are pre-trained on Natural images.
However, due to the unavailability of the consolidated data repository in the early days of the pandemic, these models are evaluated on a small dataset. Moreover, most of the previously conducted studies are based on existing CNN models designed specifically for natural images dataset. These models are adapted for the COVID-19 task without tailoring them by considering the characteristic patterns of COVID-19 pneumonia. Thus, it limits the use of the discussed techniques in real-time diagnostics.
This study presents a new deep Convolutional Neural Network (CNN) based COVID-19 classification technique for the discrimination of COVID-19 pneumonia patients from healthy individuals based on chest X-Ray images. In this study, we build a large dataset by collecting radiologist's authenticated X-ray images from publicly accessed repositories. We have proposed two novel CNN architectures, namely COVID-RENet-1 and COVID-RENet-2, to classify COVID-19 pneumonia. The idea of the proposed technique is validated by performing series of experimentation and comparing with baseline models. The detection capacity of the proposed technique is evaluated on the seen test set and compared against several state-of-the-art CNNs. The contributions of this work are as follows:
-
•
Two novel CNN architectures, COVID-RENet-1 and COVID-RENet-2, are proposed for the COVID-19 specific pneumonia analysis.
-
•
In the proposed CNN architecture, Region and Edge-based operations are systematically used in combination with convolution operation to better explore the region homogeneity, textural variations, and boundary related information in an image.
-
•
The performance comparison of the proposed COVID-19 classification framework with several existing CNN architectures shows a significant decrease in both the false negatives and false positives.
The rest of the manuscript is arranged as follows: Section 2 presents the details of the proposed COVID-19 classification framework and experimental setup. The results and discussion is made in section 3. The last Section 4 concludes the paper.
2. Materials and methodology
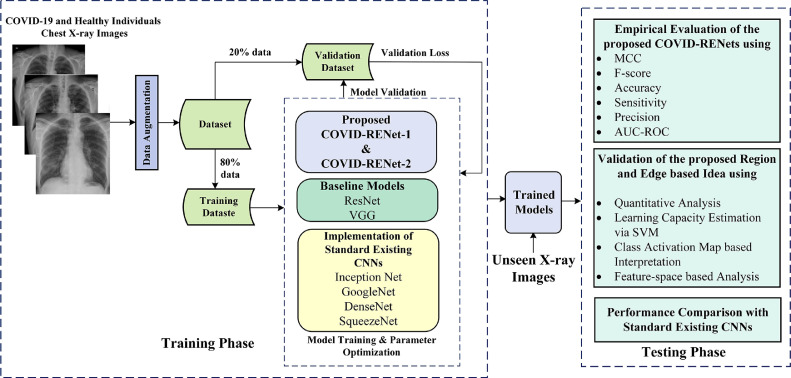
A new classification technique is developed in this work based on two custom CNN architectures for automatic discrimination of COVID-19 pneumonia patients from healthy individuals using chest X-ray images. The discrimination potential of the proposed classification technique is empirically evaluated via several performance metrics and compared with standard state-of-the-art CNNs. In the experimental setup, training samples are augmented to improve the generalization. The overall setup of the proposed COVID-19 classification technique is shown in Fig. 1 .
Fig. 1.
The detailed workflow of the proposed technique for the classification of COVID-19 infected X-ray images.
2.1. Dataset
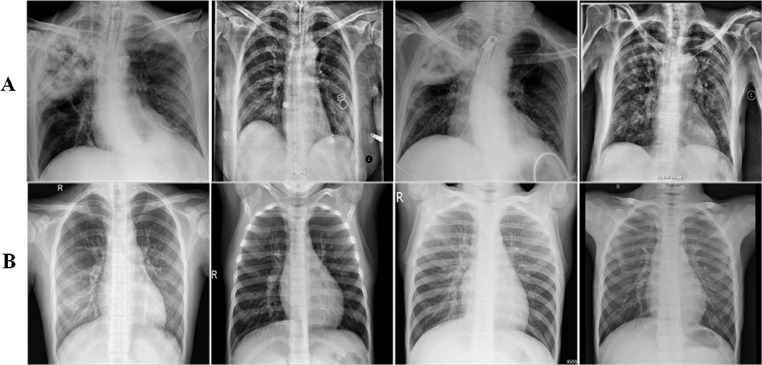
We have built a new dataset consisting of X-ray images of healthy individuals and COVID-19 pneumonia patients in this work. X-ray images were collected from Open Source GitHub repository and Kaggle repository called “pneumonia” [24,25]. Radiologists approved all the images in the aforementioned repositories, and we filtered COVID-19 and healthy samples from these repositories for this experimental setup. Since data is continuously updated by an open-source GitHub repository; therefore, in this experiment, we collected 6448 images consisting of 3224 COVID-19 patients and 3224 healthy individuals; thus, the developed dataset is balanced in nature. Each image in this dataset was resized to 224 × 224 pixels. Some of the COVID-19 infected and healthy images are presented in Fig. 2 .
Fig. 2.
Panel (A) and (B) show COVID-19 infected and healthy images, respectively.
2.2. Data augmentation
Deep learning models largely overfit on an insufficient amount of data. Therefore, a considerable amount of data is required for effective training and achieving good generalization. Data augmentation refers to augmenting the base data to increase data samples [26,27]. In this work, we have augmented the training dataset by performing random rotation (0-360 degree), sharing (±0.05), scaling (0.5-1 range), and image reflection (±1 in left and right direction). These augmentation strategies are used to increase the data as well as generalization of the model to make it effective for different hospitals and lab setups.
2.3. The proposed deep COVID-RENet based classification
In this study, we have exploited the potential of deep CNNs to learn the COVID-19 specific patterns of pneumonia in chest X-ray images. Deep CNNs have been extensively employed for image classification and recognition because of their strong potential in learning salient features and patterns manifested by images. CNNs, on account of their good learning ability, are used for both feature generation and classification [28].
In this work, we have developed two new CNN architectures based on Region and Edge-based operations for COVID-19 specific pneumonia classification in X-ray images and named them as “COVID-RENet-1” (also known as PIEAS Classification-Network-4 (PC-Net-4)) and “COVID-RENet-2” (also known as PIEAS Classification-Network-4 (PC-Net-6)). These models are optimized in an end-to-end manner to capture the pneumonia specific information from the X-ray images. Fully connected layers in the proposed deep CNN models are used for the classification. The architectural details are summarized in the following sections.
2.3.1. Architectural details of the proposed COVID-RENets
The architectural design of the proposed COVID-RENets is motivated by classical image processing techniques [29] and is based on the idea of exploiting the underlying patterns in images. In this regard, we systematically synergized the use of Region and Edge-based operations as well as convolution operation in CNN to learn the COVID-19 specific pneumonia patterns effectively. In this work, we used VGG-16 and ResNet-18 [30,31] as baseline models to validate the advantage of the proposed Region and Edge-based idea for pattern mining in CNNs. VGG-16 is a state-of-the-art CNN that exploits average pooling throughout the architecture for image size regulation and uses convolution operation for feature engineering. ResNet-18 uses strided convolution operation for image downsampling instead of pooling operation and leverage the advantage of convolution operation in combination with the ReLu activation function for feature extraction.
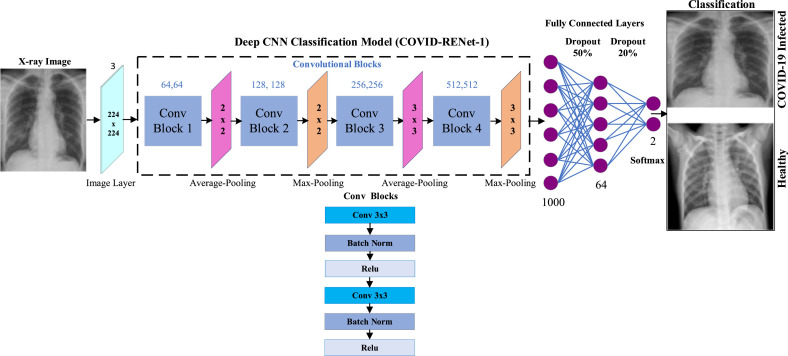
The proposed COVID-RENet-1 consists of four convolutional blocks. Each block consists of a convolutional layer (Eq. (1)), batch normalization, and ReLU as an activation function. After every convolutional block, Region and Edge-based operations are employed using average (Eq. (2)) and max pooling (Eq. (3)), respectively. These operations enhance the region-specific properties and boundary information, whereas convolution operation extracts the pattern defining features from the image. Fully connected layers represented in Eq. (4) are used in the proposed model to obtain target-specific features for classification. Dropout is applied to the fully connected layers to reduce the chances of overfitting.
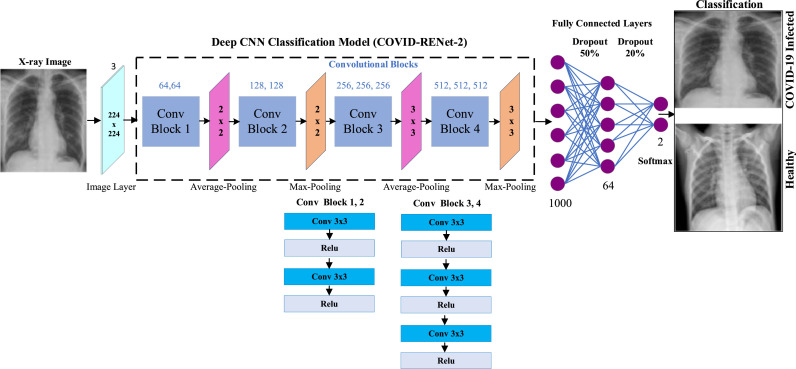
Similarly, the proposed COVID-RENet-2 is based on the same idea with increased depth. COVID-RENet-2 constitutes four convolutional blocks but with a different number of convolutional operations. Fig. 3 illustrates the architecture of the proposed COVID-RENet-1, while Fig. 4 shows the details of the proposed COVID-RENet-2.
| (1) |
| (2) |
| (3) |
| (4) |
Fig. 3.
Architectural details of the proposed COVID-RENet-1.
Fig. 4.
Architectural details of the proposed COVID-RENet-2.
In Eq. (1), the convolutional operation is employed (assuming the filter is symmetric). Input feature map, of size , is represented by f, where kernel, of size p x q, is represented by k. The resultant feature map is represented by f, where x and y run from 1 to X - p + 1 and Y - q + 1, respectively, as depicted in Eq. (1). Eqs. (2) and (3) determine the average and max-pooling operations, and their outputs are represented by and , respectively. In Eqs. (2) and (3), w represents the window size of average and max operation. (Eq. (4)) is an output of a fully connected layer, which applies global operation on the output of the feature extraction stage (consisting of convolution and pooling operation). shows the number of neurons in a fully connected layer.
2.3.2. Advantages of the proposed COVID-RENet in exploring the image content
X-ray imaging of the chest shows several patterns that help display the variation in intensity values of different regions. The underlying structure of these patterns is established on region smoothness, textural variations, and boundaries. X-ray imaging manifests the radiological patterns of COVID-19 pneumonia that are characterized by different types of opacities and obscuration of well-structured marks.
In this work, the systematic combination of convolution operation Eq. (1)), and Region and Edge-based operations (Eqs. (2) and ((3)) enables the proposed architecture in enhancing the region-specific properties. This, in turn, helps distinguish the well-structured healthy regions from the deformed regions in the X-ray images. In contrast to this scheme, most current CNN architectures use different combinations of convolution operations while applying a pooling operation only at the input layer to learn invariant features [31], [32], [33]. Other well-known benchmarked CNNs normally use a single type of pooling down the architecture for down-sampling [30,34]. The advantages of implementing the proposed idea in CNN are as follows:
-
•
The proposed COVID-RENet helps in dynamically emulating image sharpening and smoothing and can automatically adjust the extent of smoothening and sharpening as per the spatial content of the image.
-
•
The systematic use of Region-based operation after each convolutional block helps in enhancing the region homogeneity of different segments. The region operator smoothens the region variations via an average-pooling (Eq. (2)) and thus also acts as a noise suppressant for the distortions acquired during X-ray imaging. On the other hand, the edge operator encourages CNN to learn highly discriminative and local features using max-pooling operation (Eq. (3)).
-
•
Additionally, pooling operations also perform down-sampling, which improves the model robustness against slight variations in the input image.
2.3.3. Validation of the proposed region and edge based idea
The learning capacity of the proposed COVID-RENet-1 and COVID-RENet-2 is evaluated from different perspectives and compare the performance against the baseline models. Initially, consider the proposed models as feature-extractors and transfer their knowledge to linear ML classifier for analyzing discrimination capacity. For this purpose, SVM is used for performing binary classification. Additionally, the class separation ability of the learnt feature-space is analyzed by performing 2-D visualization of the data using principal component analysis. Class activation map based analysis is conducted to gain an insight that the decision-making process of the proposed technique is based on characteristics patterns of COVID-19 infected regions rather than random irrelevant regions.
2.4. Implementation of the standard existing CNNs
For comparison, we implemented different state-of-the-art well-known deep CNN models, including VGG, GoogleNet, Inception, ResNet, SqueezeNet, DenseNet, and Xception [[30], [31], [32],[34], [35], [36], [37]]. These CNNs have been extensively used for a wide range of image classification problems and have been used by several researchers for COVID-19 X-ray classification. These models vary in block design and architecture, but all of them either exploited a single type of pooling operation down the network or replaced the pooling operation with a strided convolution operation for complexity regulation. We implemented these CNNs in an end-to-end manner for classification and added an additional FC and classification layer to tune them for X-ray based COVID-19 pneumonia discrimination.
2.5. Transfer learning based optimization of the proposed technique
The proposed COVID-RENet-1 and COVID-RENet-2 are trained on the X-ray dataset by exploiting TL. CNNs are parameter hungry, and optimal performance requires a large amount of dataset during training, whereas training on a small number of X-ray samples may cause poor convergence [38]. TL is a technique that has shown promising results for CNN models in the non-availability of a large dataset. It allows the reuse of the already trained models’ weight space and prevents highly parameterized models from overfitting by providing a good initial set of weights [39]. Therefore, we customarily exploited the concept of TL in this work to achieve substantial performance. In this regard, we initialized the weights of the proposed COVID-RENets from the pre-trained model's parameter space.
Likewise, for a fair comparison, we adapted the same training strategy for the standard state-of-the-art CNNs. These architectures have been optimized for X-ray images by applying domain-adaptation based TL to fine-tune the ImageNet pre-trained models on the X-ray dataset for classification of COVID-19 specific pneumonia.
2.6. Implementation details
The dataset was divided into two disjoint sets with a ratio of 80:20% for train and test sets, respectively. Further, the training set was partitioned into train and validation datasets for parameter selection. The parameter optimization of the model was carried out using the 5-fold cross-validation. During the training of CNNs, SGD was used as an optimizer with a momentum of 0.95. The model was trained for 10 epochs by initially setting the learning rate to 0.0001 and weight decay as 0.0005. A mini-batch training strategy was employed with a batch size of 16 images per epoch for smooth training. All the deep CNNs were optimized for image classification by minimizing cross-entropy loss, and softmax was used as an activation function. Linear SVM was used for the ML-based classification analysis.
All the simulations were carried out on MATLAB 2019b. Dell Core I i7-7500 CPU with a 2.90 GHz processor and with CUDA-enabled Nvidia® GTX 1060 Tesla was used for MATLAB based simulations. The training of the models approximately took ∼12 hours. The training time for one epoch on Nvidia Tesla K80 was ∼30-60 minutes.
3. Results and discussion
This study proposes a deep CNN-based technique for detecting COVID-19 infected pneumonia patients using chest X-ray images. Two experiments are performed to evaluate the effectiveness of the proposed technique empirically. In the first experiment, we have analyzed the advantages of synchronously using average and max pooling in COVID-RENets for pattern recognition. In the second, a general evaluation for the COVID-19 detection task is performed by comparing performance with popular state-of-the-art techniques.
3.1. Performance measures
The performance of the implemented models is evaluated using various standard performance metrics. These metrics include accuracy, sensitivity, specificity, recall, Mathews Correlation Coefficient (MCC), and F-score. Accuracy is defined in (Eq. (5)), which measures the total number of correct assignments. Similarly, recall (Eq. (6)) and specificity (Eq. (7) measure the proportion of actual COVID-19 patients, and the actual healthy individuals identified, respectively. The precision is specified in (Eq. (8)), F-score in (Eq. (9)), and (Eq. (10)) shows MCC. The mathematical explanation of the different metrics is provided in Table 1 .
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
Table 1.
Explanation of various performance metrics.
| Metric | Symbol | Description |
| Accuracy | Acc | % of correctly predicted samples |
| Recall (Sensitivity) | R | Ratio of correctly predicted COVID-19 samples |
| Specificity | S | Ratio of correctly predicted Healthy samples |
| Precision | P | Ratio of predicted samples close to actual class |
| True Positives | TP | Correctly predicted COVID-19 samples |
| True Negatives | TN | Correctly predicted Healthy examples |
| False Positives | FP | Incorrectly predicted COVID-19 samples |
| False Negatives | FN | Incorrectly predicted Healthy samples |
| Negative Samples | TN+FP | Total Healthy samples in the dataset |
| Positive Samples | TP+FN | Total COVID-19 samples in the dataset |
| Total Samples | TP+TN+FP+FN | Total samples in the dataset |
3.2. Performance analysis of the proposed COVID-RENets
The performance of the proposed COVID-RENet-1 and COVID-RENet-2 is evaluated on an unseen test dataset based on MCC and F-score that are standard measures for medical diagnostic system. Contrary to Accuracy, both MCC and F-score assign weightage to precision along with sensitivity.
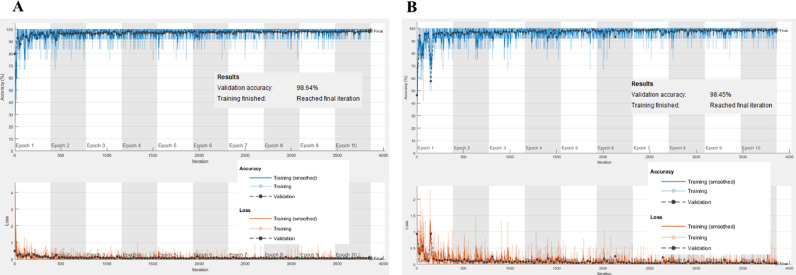
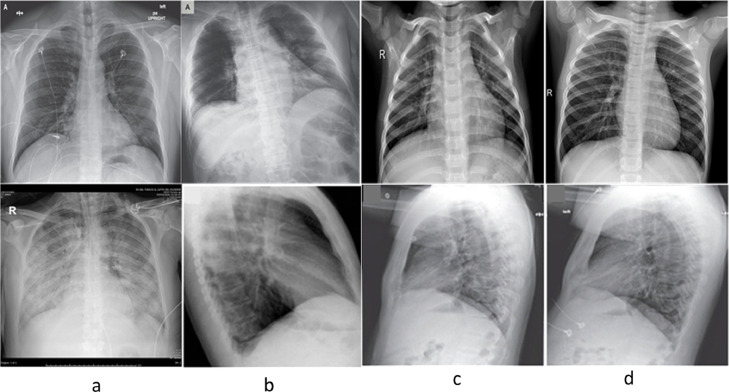
Training based accuracy and loss plots for COVID-RENet-1 and COVID-RENet-2 are shown in Fig. 5 , respectively. Loss and accuracy plots (Fig. 5) show that both the proposed models converge smoothly and reach an optimal value quickly. The proposed COVID-RENet-1 model correctly classified 633 samples of both COVID-19 and healthy individuals. Likewise, the proposed COVID-RENet-2 performs similarly by correctly identifying 628 COVID-19 infected and 637 healthy individuals, respectively. It is observed that an increase in depth improves the detection rate for COVID-19. Fig. 6 shows some of the X-ray images that are misclassified by both COVID-RENet-1 and COVID-RENet-2. Misclassification occurs probably due to illumination variation, low contrast, and intricate pattern of samples. In this regard, we employed several data augmentation strategies during training to maximize the generalization and improve the robustness towards unseen patient samples.
Fig. 5.
Training plots for the proposed COVID-RENet-2 (panel A) and COVID-RENet-1 (panel B).
Fig. 6.
COVID-19 infected (panel a & b) and healthy (panel c & d) images that are misclassified by both COVID-RENet-1 and COVID-RENet-2.
3.2.1. Performance comparison with baseline techniques
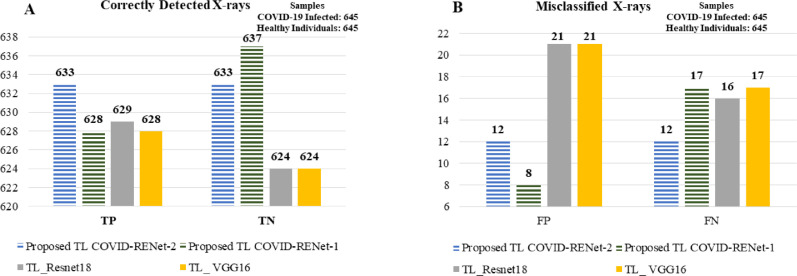
The effectiveness of the proposed idea is evaluated by benchmarking the performance against ResNet and VGG. These two baseline models, ResNet-18 and VGG-16, are approximately as deep as COVID-RENet-1 and COVID-RENet-2. Contrary to the idea of using two opposing pooling operations in COVID-RENets, VGG-16 exploits a single type of pooling operation and ResNet-18 uses strided convolution in place of pooling down the architecture. The comparison is presented in Table 2 . Performance analysis suggests that both the COVID-RENets show gain in performance as compared to ResNet-18 and VGG-16 in terms of MCC (0.96), F-score (0.98), and accuracy (0.98). Fig. 7 shows that the proposed COVID-RENet significantly improves the detection rate for both COVID-19 infected and healthy individuals compared to baseline ResNet-18 and VGG-16.
Table 2.
Performance comparison of the proposed COVID-RENets with baseline models on the test set.
| Models | Depth | MCC | F-score | %Accuracy |
| Proposed TL COVID-RENet-2 | 10 | 0.96 | 0.98 | 98.14 |
| Proposed TL COVID-RENet-1 | 08 | 0.96 | 0.98 | 98.06 |
| TL_Resnet18 | 18 | 0.95 | 0.97 | 97.13 |
| TL_ VGG16 | 16 | 0.94 | 0.97 | 97.05 |
Fig. 7.
Panels (A & B) show the number of correct and misclassified detections, respectively made by COVID-RENet-1, COVID-Net-2 and baseline models.
3.2.2. Learning capacity estimation via ML classifier
Feature engineering plays a significant role in evaluating the learning capacity of deep CNNs. The feature-space of the proposed model is evaluated using the classical ML model to assess its contribution in learning class-specific mappings. The mapping capability of the proposed COVID-RENet is evaluated by extracting features from its penultimate layer and assigning them to the linear ML classifier, SVM. Table 3 shows that the pattern leant by proposed COVID-RENet-1 and COVID-RENet-2 are distinguishable for two classes and can be used to discriminate COVID-19 pneumonia and healthy individuals. Quantitative analysis in terms of MCC (0.97), F-score (0.98), and accuracy (98%) suggest that proposed technique perform better then VGG-16 (MCC:0.94, F-score:0.97, ACC: 97%) and ResNet-18 (MCC:0.95, F-score:0.97, ACC: 97%).
Table 3.
SVM based learning capacity estimation of the proposed COVID-RENets with baseline models on test set.
| Models | MCC | F-score | %Accuracy | TP | FP | FN | TN |
| Proposed TL COVID-RENet-2 | 0.97 | 0.98 | 98.29 | 640 | 18 | 5 | 627 |
| Proposed TL COVID-RENet-1 | 0.96 | 0.98 | 98.14 | 631 | 10 | 14 | 635 |
| TL_Resnet18 | 0.95 | 0.97 | 97.57 | 631 | 18 | 14 | 627 |
| TL_ VGG16 | 0.94 | 0.97 | 97.13 | 629 | 21 | 16 | 624 |
3.2.3. Class activation map based interpretation
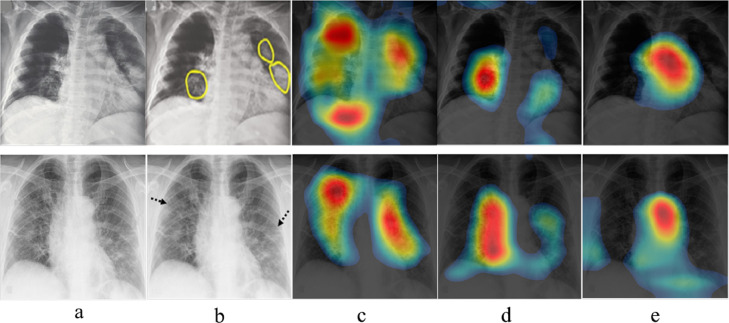
We performed the class activation map based study to empirically investigate the effectiveness of systematically using max and average pooling operations in CNN. Fig. 8 demonstrates the response of the proposed COVID-RENets and best performing baseline model (ResNet-18) towards COVID-19 infected X-rays. Class activation map-based visualization suggests that the synergy of two different pooling operations encourage the model to detect COVID-19 caused pneumonia accurately by focusing on infected regions.
Fig. 8.
Panel (a) shows the original chest X-ray image. Panel (b) shows the radiologist defined COVID-19 infected regions highlighted by yellow circle or black arrow. The resulted class activation map of the proposed COVID-RENet-2 and COVID-RENet-1 is shown in panels (c & d), respectively. Panel (e) shows the class activation map of the best performing baseline CNN, ResNet.
3.2.4. Feature space based analysis
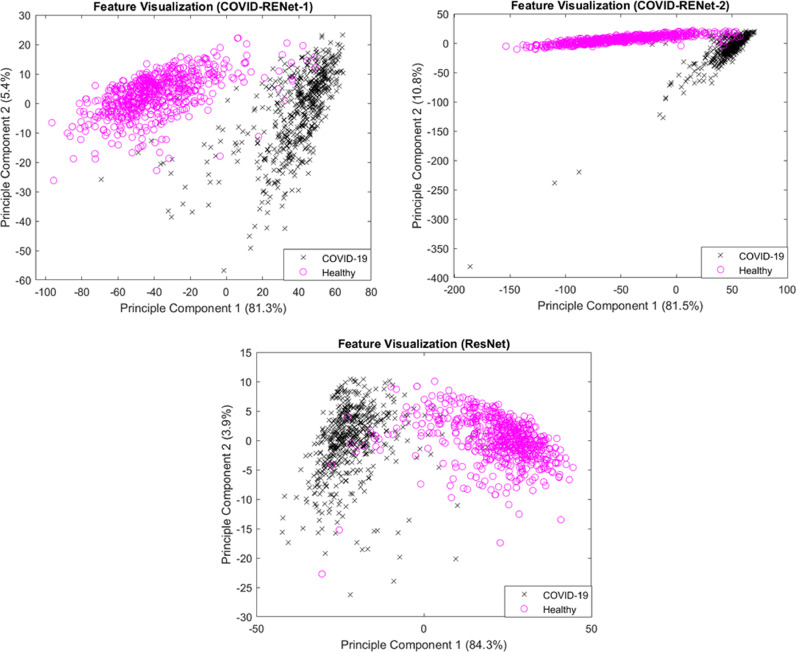
Feature space learnt by the proposed COVID-RENets and best performing baseline model, ResNet, is analyzed to understand the decision-making behaviour in a better way. The good discrimination ability of a classifier is generally associated with the characteristics of the feature space. Class distinguishable features improve the learning and lower the model's variance on the diverse set of examples. Visualization of feature space is performed by plotting principal components of the data. Fig. 9 shows the 2-D plots of principal component 1 and principal component 2 and their percentage variance for the proposed COVID-RENet-1, COVID-RENet-2 and ResNet-18 for the test set. Data plotting shows that exploitation of both max and average pooling operation considerably improves the feature space diversity and thus resulted in improved classification performance.
Fig. 9.
Feature visualization for the proposed COVID-RENets and the best performing baseline CNN (ResNet) on the test set.
3.3. Discrimination capability of the proposed COVID-RENets
We compared the performance of the proposed technique with popular CNNs on unseen chest X-ray images for comprehensive empirical evaluation. The results are evaluated in terms of MCC, F-score, Accuracy, AUC-ROC, sensitivity and precision.
3.3.1. Performance comparison with existing CNNs
The performance of the proposed COVID-RENet-1 and COVID-RENet-2 is compared with GoogleNet, InceptionV3, ResNet-50, SqueezeNet, Xception and DenseNet-201. Table 4 shows that the performance comparison suggests that proposed models COVID-RENet-1 and COVID-RENet-2 can better recognize the COVID-19 specific pneumonia patterns from X-ray images in terms of standard metrics MCC, F-score, and accuracy. This improvement in the performance is because of the systematic use of average and max pooling operators in the proposed CNN architecture. In essence, this systematic use of these two opposite pooling operators encourages the model to learn both fine-grained details and highly discriminative features from the raw X-ray images.
Table 4.
Performance comparison of the proposed COVID-RENets with standard existing CNNs on test set.
| Models | MCC | F-score | %Accuracy | TP | FP | FN | TN |
| Proposed TL COVID-RENet-2 | 0.96 | 0.98 | 98.14 | 633 | 12 | 12 | 633 |
| Proposed TL COVID-RENet-1 | 0.96 | 0.98 | 98.06 | 628 | 8 | 17 | 637 |
| TL_Inceptionv3 | 0.93 | 0.96 | 96.51 | 620 | 20 | 25 | 625 |
| TL_DenseNet201 | 0.93 | 0.96 | 96.51 | 626 | 26 | 19 | 619 |
| TL_Google Net | 0.93 | 0.96 | 96.51 | 621 | 21 | 24 | 624 |
| TL_Xception | 0.93 | 0.96 | 96.43 | 625 | 26 | 20 | 619 |
| TL_Resnet50 | 0.94 | 0.97 | 97.05 | 629 | 22 | 16 | 623 |
| TL_Squeeze Net | 0.93 | 0.96 | 96.51 | 625 | 25 | 20 | 620 |
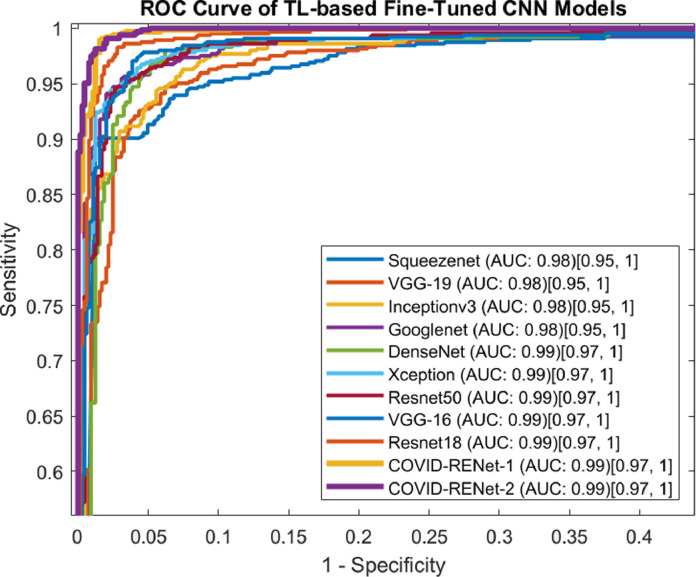
3.3.2. ROC-AUC based analysis
ROC curve has an essential role in achieving the optimal analytical threshold for the classifier. ROC curve graphically illustrates the classifier segregation ability at possible threshold values. Fig. 10 shows that our proposed COVID-RENet-1 and COVID-RENet-2 achieved an AUC of (0.99) on COVID-19 chest X-ray datasets. It is evident from ROC based quantitative analysis that the proposed technique upholds high sensitivity with a low False-positive rate. This suggests that the proposed COVID-19 classification technique has a significant potential to deploy for COVID19 patients’ analysis.
Fig. 10.
ROC curve based analysis of the proposed COVID-RENet-1 and COVID-RENet-2 with exisitng CNNs. The values in square bracket show a standard error at the 95% confidence interval.
3.3.3. Diagnostic significance of the proposed technique
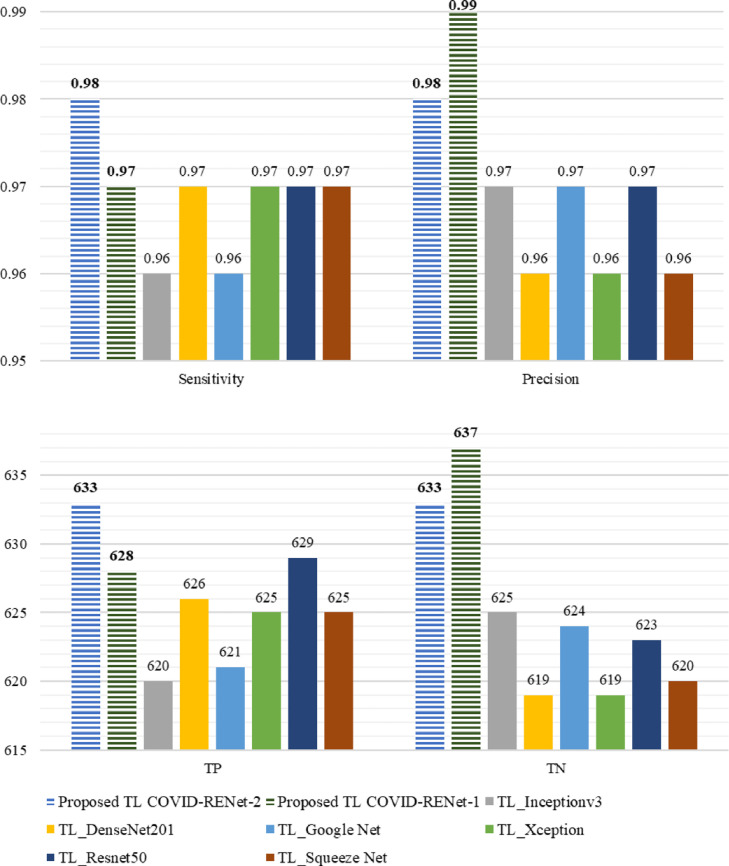
The effectiveness of a medical diagnostic system is mostly assessed through detection rate (sensitivity) and precision. An accurate detection rate is important for the COVID-19 detection system in controlling infection spread. Therefore, the proposed technique's detection rate and precision are explored for COVID-19 X-ray images, as shown in Fig. 11 and Table 4. The quantitative analysis shows (Fig. 11) that the COVID-RENet-1 (Sen: 0.97, Pre: 0.99) and COVID-RENet-2 (Sen: 0.98, Pre: 0.98) improves the precision of the classification system along with a high detection rate. Thus, it is likely to assist the radiologist with high accuracy and can be used to increase the throughput by reducing the burden on expert radiologists.
Fig. 11.
Performance comparison of the proposed COVID-RENet-1 and COVID-RENet-2 with state-of-the-art CNNs.
4. Conclusions
Early diagnosis of COVID-19 patients is essential in preventing the spread of disease. Therefore, two novel custom, deep CNN models are proposed to discriminate the X-ray images of COVID-19 pneumonia patients from healthy individuals. The performance analysis of the proposed COVID-19 classification technique is carried out with the standard existing CNN models. Experimental results demonstrated that the proposed COVID-RENet-1 and COVID-RENet-2 perform better than baseline and existing CNN models by improving Accuracy, F-score, and MCC. The proposed technique achieved an MCC of 0.97 for discriminating COVID-19 samples from healthy individuals with a sensitivity and precision of 0.98. The proposed technique is expected to help medical practitioners with the diagnosis of COVID-19 infected patients. Moreover, it has a strong potential to be used for the analysis of different types of chest X-ray image abnormalities.
Availability of data and material
Publicly available dataset is used in this work that is accessible at https://www.kaggle.com/khoongweihao/covid19-xray-dataset-train-test-sets and https://github.com/ieee8023/covid-chestxray-dataset. Whereas all the data generated during analysis is accessable from corresponding author on reseasonable request.
Code availability
All the scripts that are developed for the simulations are available from the corresponding author on reasonable request.
Declaration of Competing Interest
Authors declare no conflict of interest.
References
- 1.Pang L., Liu S., Zhang X., Tian T., Zhao Z. Transmission dynamics and control strategies of COVID-19 in Wuhan, China. J. Biol. Syst. 2020 doi: 10.1142/S0218339020500096. [DOI] [Google Scholar]
- 2.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svyatchenko V.A., Nikonov S.D., Mayorov A.P., Gelfond M.L., Loktev V.B. Antiviral photodynamic therapy: inactivation and inhibition of SARS-CoV-2 in vitro using methylene blue and Radachlorin. Photodiagn. Photodyn. Ther. 2021 doi: 10.1016/j.pdpdt.2020.102112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G., Revel M.P. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagn. Interv. Imaging. 2020 doi: 10.1016/j.diii.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and multiorgan response. Curr. Probl. Cardiol. 2020 doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias L.D., Blanco K.C., Bagnato V.S. COVID-19: beyond the virus. The use of photodynamic therapy for the treatment of infections in the respiratory tract. Photodiagnosis Photodyn. Ther. 2020 doi: 10.1016/j.pdpdt.2020.101804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewnard J.A., Lo N.C. Scientific and ethical basis for social-distancing interventions against COVID-19. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Kang H., Liu X., Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020;92:538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020 doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei X., Lee H.C., yue Diao K., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiaffino S., Tritella S., Cozzi A., Carriero S., Blandi L., Ferraris L., Sardanelli F. Diagnostic performance of Chest X-ray for COVID-19 pneumonia during the SARS-CoV-2 pandemic in Lombardy, Italy. J. Thorac. Imaging. 2020 doi: 10.1097/RTI.0000000000000533. [DOI] [PubMed] [Google Scholar]
- 13.Carey S., Kandel S., Farrell C., Kavanagh J., Chung T., Hamilton W., Rogalla P. Comparison of conventional chest X-ray with a novel projection technique for ultra-low dose CT. Med. Phys. 2020;1 doi: 10.1002/mp.14142. [DOI] [PubMed] [Google Scholar]
- 14.Rousan L.A., Elobeid E., Karrar M., Khader Y. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020 doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borghesi A., Maroldi R. COVID-19 outbreak in Italy: experimental chest X-ray scoring system for quantifying and monitoring disease progression. Radiol. Med. 2020 doi: 10.1007/s11547-020-01200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue F., Chen C., Yan Z., Chen C., Guo Z., Zhang Z., Chen Z., Zhang F., Lv X. Fourier transform infrared spectroscopy combined with deep learning and data enhancement for quick diagnosis of abnormal thyroid function. Photodiagn. Photodyn. Ther. 2020 doi: 10.1016/j.pdpdt.2020.101923. [DOI] [PubMed] [Google Scholar]
- 18.Maroof N., Khan A., Qureshi S.A., ul Rehman A., Khalil R.K., Shim S.O. Mitosis detection in breast cancer histopathology images using hybrid feature space. Photodiagn. Photodyn. Ther. 2020 doi: 10.1016/j.pdpdt.2020.101885. [DOI] [PubMed] [Google Scholar]
- 19.Sohail A., Khan A., Wahab N., Zameer A., Khan S. A multi-phase deep CNN based mitosis detection framework for breast cancer histopathological images. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-85652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-COVID: predicting COVID-19 FROM CHEst X-ray images using deep transfer learning. Med. Image Anal. 2020 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.S. Wang, B. Kang, J. Ma, X. Zeng, M. Xiao, J. Guo, M. Cai, J. Yang, Y. Li, X. Meng, B. Xu, A deep learning algorithm using CT images to screen for corona virus disease (COVID-19), MedRxiv. (2020) 2020.02.14.20023028. https://doi.org/10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- 22.A. Narin, C. Kaya, Z. Pamuk, Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks, (2020). http://arxiv.org/abs/2003.10849. [DOI] [PMC free article] [PubMed]
- 23.P. Afshar, S. Heidarian, F. Naderkhani, A. Oikonomou, K.N. Plataniotis, A. Mohammadi, COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images, (2020) 1–5. http://arxiv.org/abs/2004.02696. [DOI] [PMC free article] [PubMed]
- 24.COVID-19 Xray Dataset (Train & Test Sets) | Kaggle, (n.d.). https://www.kaggle.com/khoongweihao/covid19-xray-dataset-train-test-sets (accessed November 26, 2020).
- 25.J.P. Cohen, P. Morrison, L. Dao, COVID-19 image data collection, (2020). http://arxiv.org/abs/2003.11597 (accessed May 2, 2020).
- 26.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J. Big Data. 2019;6 doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez L., Wang J. 2017. The Effectiveness of Data Augmentation in Image Classification using Deep Learning.http://arxiv.org/abs/1712.04621 [Google Scholar]
- 28.Khan A., Sohail A., Zahoora U., Qureshi A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020;53:1–68. doi: 10.1007/s10462-020-09825-6. [DOI] [Google Scholar]
- 29.Mallick A., Roy S., Chaudhuri S.S., Roy S. Optimization of Laplace of Gaussian (LoG) filter for enhanced edge detection: a new approach. Int. Conf. Control. Instrum. Energy Commun. CIEC. 2014:2014. doi: 10.1109/CIEC.2014.6959172. [DOI] [Google Scholar]
- 30.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings, arXiv Prepr. ArXiv 1409.1556, vol. 493, no. 6, pp. 405–10, Sep. 2014. http://arxiv.org/abs/1409.1556.
- 31.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conf. Comput. Vis. Pattern Recognit. IEEE; 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 32.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proc. - 30th IEEE Conf. Comput. Vis. Pattern Recognit. CVPR 2017; 2017. Densely connected convolutional networks; pp. 2261–2269. 2017-Janua. [DOI] [Google Scholar]
- 33.Zagoruyko S., Komodakis N. Procedings Br. Mach. Vis. Conf. 2016. Vol. 12. 2016. Wide residual networks. 87.1-87. [DOI] [Google Scholar]
- 34.F.N. Iandola, M.W. Moskewicz, K. Ashraf, S. Han, W.J. Dally, K. Keutzer, SqueezeNet, ArXiv. (2016).
- 35.Szegedy C., Liu Wei, Jia Yangqing, Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. 2015 IEEE Conf. Comput. Vis. Pattern Recognit. IEEE; 2015. Going deeper with convolutions; pp. 1–9. [DOI] [Google Scholar]
- 36.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the inception architecture for computer vision. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2016:2818–2826. doi: 10.1109/CVPR.2016.308. IEEE. [DOI] [Google Scholar]
- 37.F. Chollet, Xception: Deep learning with depthwise separable convolutions, ArXiv Prepr. (2017) 1610–2357.
- 38.Chougrad H., Zouaki H., Alheyane O. Multi-label transfer learning for the early diagnosis of breast cancer. Neurocomputing. 2020 doi: 10.1016/j.neucom.2019.01.112. [DOI] [Google Scholar]
- 39.Wahab N., Khan A., Lee Y.S. Transfer learning based deep CNN for segmentation and detection of mitoses in breast cancer histopathological images. Microscopy. 2019;68:216–233. doi: 10.1093/jmicro/dfz002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available dataset is used in this work that is accessible at https://www.kaggle.com/khoongweihao/covid19-xray-dataset-train-test-sets and https://github.com/ieee8023/covid-chestxray-dataset. Whereas all the data generated during analysis is accessable from corresponding author on reseasonable request.