ABSTRACT
Background
The essential nutrient choline provides one-carbon units for metabolite synthesis and epigenetic regulation in tissues including brain. Dietary choline intake is often inadequate, and higher intakes are associated with improved cognitive function.
Objective
Choline supplements confer cognitive improvement for those diagnosed with fetal alcohol spectrum disorder (FASD), a common set of neurodevelopmental impairments; however, the effect sizes have been modest. In this retrospective analysis, we report that genetic polymorphisms affecting choline utilization are associated with cognitive improvement following choline intervention.
Methods
Fifty-two children from the upper midwestern United States and diagnosed with FASD, ages 2–5 y, were randomly assigned to receive choline (500 mg/d; n = 26) or placebo (n = 26) for 9 mo, and were genotyped for 384 choline-related single nucleotide polymorphisms (SNPs). Memory and cognition were assessed at enrollment, study terminus, and at 4-y follow-up for a subset.
Results
When stratified by intervention (choline vs. placebo), 14–16 SNPs within the cellular choline transporter gene solute carrier family 44 member 1 (SLC44A1) were significantly associated with performance in an elicited imitation sequential memory task, wherein the effect alleles were associated with the greatest pre-/postintervention improvement. Of these, rs3199966 is a structural variant (S644A) and rs2771040 is a single-nucleotide variant within the 3′ untranslated region of the plasma membrane isoform. An additive genetic model best explained the genotype associations. Lesser associations were observed for cognitive outcome and polymorphisms in flavin monooxygenase-3 (FMO3), methylenetetrahydrofolate dehydrogenase-1 (MTHFD1), fatty acid desaturase-2 (FADS2), and adiponectin receptor 1 (ADIPOR1).
Conclusions
These SLC44A1 variants were previously associated with greater vulnerability to choline deficiency. Our data potentially support the use of choline supplements to improve cognitive function in individuals diagnosed with FASD who carry these effect alleles. Although these findings require replication in both retrospective and prospective confirmatory trials, they emphasize the need to incorporate similar genetic analyses of choline-related polymorphisms in other FASD-choline trials, and to test for similar associations within the general FASD population. This trial was registered at www.clinicaltrials.gov as NCT01149538.
Keywords: fetal alcohol spectrum disorder, choline, SLC44A1, precision nutrition, single nucleotide polymorphism (SNP), rs3199966, rs2771040, nutrigenomics
Introduction
The essential nutrient choline provides one-carbon units for numerous biochemical reactions and DNA and histone methylation (1, 2). In the brain, it contributes to acetylcholine synthesis and cholinergic function, myelination, and the major lipids phosphatidylcholine and sphingomyelin (3). Dietary choline intake is often limiting during the high-demand periods of pregnancy and lactation (4, 5), and higher intakes during pregnancy and early childhood are associated with improved memory, cognition, and attention (5–9) and confer cognitive protection in preclinical models of Down syndrome, Rett syndrome, autism spectrum disorder, and iron deficiency (5, 6, 10). Mechanistically, choline promotes hippocampal and cortical neurogenesis, cholinergic maturation and function, and neuronal survival, in part through effects on DNA methylation and gene expression (5, 6, 11). These actions stimulated interest in choline's potential to remediate the cognitive, learning, and memory deficits that typify fetal alcohol spectrum disorder (FASD), the clinical sequelae of prenatal alcohol exposure (12, 13). Three to five percent of US first-graders meet criteria for an FASD diagnosis (14), and there is high interest in interventions that mitigate alcohol's damage.
Although preclinical models of FASD consistently report that supplemental choline improves cognition, (15), outcomes from clinical intervention trials are more nuanced. In young children diagnosed with FASD, choline (500 mg/d for 9 mo) did not improve pre/post measures of global cognitive functioning (16, 17); however, the youngest children (2.5–4.0 y) had significant improvement in the delayed memory component of a hippocampal-dependent task. In a subset who returned for a 4-y follow-up (18), the choline group had increased nonverbal visual spatial reasoning (P = 0.004) and working memory (P = 0.018), nonverbal IQ (P = 0.03), and working memory (P = 0.01) relative to placebo. In a gestational intervention, infants exposed prenatally to alcohol who received 740 mg choline/d (19) had significantly improved visual memory scores in a habituation-dishabituation test (20) but did not differ from placebo controls on the Bayley Scales of Infant Development (21). Another trial that used a higher choline dose (2 g/d) found that maternal adherence to the choline intervention predicted behavioral improvement (r = 0.63, P < 0.01) (21), and those infants performed better in an eyeblink conditioning task (P < 0.01) at age 6 mo and a visual recognition memory task at age 12 mo.
We hypothesized that genetic differences in choline utilization may partially explain these improvements. Polymorphisms in multiple choline-related genes, including BHMT, CHDH, CHKA, CHDH, flavin monooxygenase-3 (FMO3), methylenetetrahydrofolate dehydrogenase-1 (MTHFD1), PEMT, and SLC44A1, operate as effect alleles that reduce choline synthesis, utilization, or absorption, and thus increase individual choline needs, particularly when intake is limiting (22–30). In this exploratory study, we identify multiple polymorphisms within the choline transporter gene SLC44A1 that associate with cognitive improvement in response to oral choline supplementation, in children diagnosed with FASD.
Methods
Study design and participants
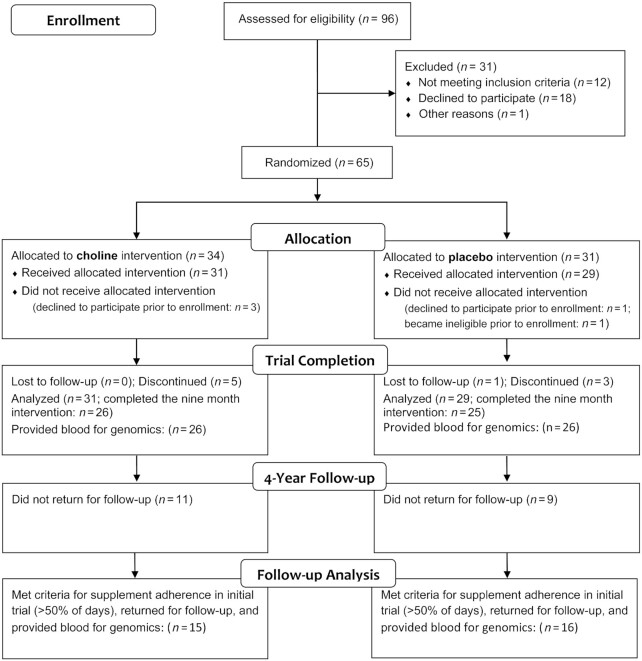
This retrospective analysis used data from a pilot trial wherein 65 individuals diagnosed with FASD, ages 2–5 y and recruited from across the upper midwestern United States, were randomly assigned in a double-blinded trial (NCT01149538) to receive a daily intervention of either 500 mg choline (n = 31), administered as 1.25 g choline bitartrate in a flavored beverage, or the same beverage but lacking choline (placebo; n = 29). The study design (Figure 1) and methods are detailed elsewhere (16–18). This choline dose is twice the Adequate Intake and was selected to be well tolerated and achievable through dietary means (16). Participants received the intervention for 9 mo. The study was completed in 2 phases. The primary outcomes of phase 1 was to assess the side effects or adverse events to the choline intervention; these data were reported in Wozniak et al. (16). For the second phase, the primary outcome was an exploratory assessment of global cognitive functioning, using the Mullen Scales of Early Learning, at baseline and 9 mo (17), and the initial sample size was set to detect an effect size of 0.43 on this measure. The secondary outcome was exploratory assessment of hippocampal-dependent memory function, using an elicited imitation memory paradigm, testing both immediate and delayed recall, at baseline, 6 mo, and 9 mo (17). In a follow-up study 4 y later, a subset of 31 participants (16 placebo, 15 choline) returned for follow-up assessment that included the Stanford-Binet Intelligence Scale, version 5 (SB-5), an age-appropriate elicited imitation memory paradigm; 3 subtests within a developmental NEuroPSYcological Assessment, second edition (NEPSY-II: Memory for Names, Memory for Faces, Narrative Memory); tests of executive function from the NIH Toolbox; and the parent-report Child Behavior Checklist (18). Serum choline, betaine, and phosphatidylcholine were quantified at baseline (0 mo) and at 6 mo and 9 mo 3 h after administering the choline or placebo, using LC/electrospray ionization–isotope dilution MS, as described (17). All procedures were in accordance with the ethical standards of the University of Minnesota's Human Research Protection Program, and in accordance with the Helsinki Declaration of 1975 as revised in 1983.
FIGURE 1.
CONSORT flow diagram of the randomized postnatal choline intervention trial. CONSORT, Consolidated Standards of Reporting Trials.
Single nucleotide polymorphism analysis
At the time of the original study, genomic DNA was extracted from blood samples of 52 participants (26 choline, 26 placebo); blood samples were not obtained from 5 participants in the choline group and 3 in the placebo group due to child refusal, difficult veins, etc. Single nucleotide polymorphism (SNP) analysis was performed using an oligo-specific extension-ligation assay on a custom Golden Gate array (Illumina) that contained 384 SNPs in genes relevant to choline and one-carbon metabolism (30), and selected based on published identification of SNPs that altered choline status or affected choline metabolism (24, 25, 29, 31). The data processing and analysis of SNP associations were performed in R (version 3.6.3; R Foundation for Statistical Computing; http://cran.r-project.org/). Using the Entrez global query cross-database search system, 365 SNP identities were mapped into 40 choline-related genes or loci using Rentrez package (version 1.2.2); 19 SNPs could no longer be matched but were retained for further analysis. SNPs removed from analysis were those having a homozygous distribution (n = 60), a frequency of <0.05 (n = 51), or that violated Hardy-Weinberg equilibrium of random mating (n = 24). Thus, the final analysis interrogated 243 SNPs (Supplemental Table 1). Principal components analysis (PCA) from the FactoMineR package (version 2.3; Comprehensive R-Archive Network) was used to examine the genomic separation of participants based on age, sex, race, and treatment group.
The phenotype data set consisted of 230 physical and behavioral characteristics, of which 25 were categorical. The behavioral traits (Table 1) included cognitive performance at baseline and at the end of choline intervention (n = 26 choline, n = 26 placebo) and at the 4-y follow-up (n = 15 choline, n = 16 placebo). Some variables were missing for some participants and, in these instances, participants were nonetheless included for their other existing data. We performed separate association tests between the 243 SNPs and the 230 phenotype categories, and tested for potential interactions under Additive, Dominant, and Recessive genetic models using the SNPassoc package (version 1.9–2) (32). For all 3 genetic models, simple linear modeling was used to apply logistic regression for categorical phenotypes and linear regression for numerical phenotypes; the base R package stats (version 3.6.3) was used to extract P values, coefficients, and effect alleles. P values were adjusted for multiple testing using Bonferroni correction, accounting for SNPs that were in perfect linkage disequilibrium, and a Q value ≤0.05 was considered significant.
TABLE 1.
Definitions of outcome measures1
| Definitions | |
|---|---|
| activity_level.11 | Parent-reported child activity level from the CBCL (short form) at 9 mo; range = 1 to 7, higher scores indicate higher levels of activity |
| blood_pressure_diastolic.1 | Diastolic blood pressure at baseline (0 mo) |
| blood_pressure_diastolic.8 | Diastolic blood pressure at 6 mo |
| bloodlevel_betaine.1 | Plasma betaine concentrations at baseline (0 mo) |
| delta_adjpairs_imm | Change in Elicited Imitation immediate memory task performance (for pairs of items regardless of sequence order) between baseline (0 mo) and 9 mo; higher scores indicate greater positive change (improvement) |
| delta_imt_v1_v13_adjpairs | Change in Elicited Imitation memory task performance (for pairs of adjacent items in the correct sequence order) between baseline (0 mo) and 4-y follow-up; higher scores indicate greater positive change (improvement) |
| delta_pairs_imm | Change in Elicited Imitation immediate memory task performance (pairs) between baseline (0 mo) and 9 mo; higher scores indicate greater positive change (improvement) |
| heart_rate.11 | Resting heart rate at 9 mo |
| height_physical.11 | Height taken during physical examination at 9 mo |
| height_pile_physical.1 | Age-adjusted height percentile at baseline (0 mo) |
| height_pile_physical.11 | Age-adjusted height percentile at 9 mo |
| height_z_physical.1 | z Score for age-adjusted height at 0 mo |
| height_z_physical.11 | z Score for age-adjusted height at 9 mo |
| ibb_tscore21 | CBCL parent-reported “sluggish cognitive tempo” at baseline (0 mo); age-corrected scores with higher scores indicating more problems |
| iib_avg_imt_adjpairs | Elicited Imitation memory task performance (adjacent pairs) at 4-y follow-up; higher scores indicate better memory performance |
| iib_tscore13 | CBCL parent-reported Externalizing problems at baseline (0 mo); age-corrected scores with higher scores indicating more problems |
| iib_tscore21 | CBCL parent-reported Conduct problems at baseline (0 mo); age-corrected scores with higher scores indicating more problems |
| ilb_avg_imt_pairs | Elicited Imitation memory task performance (pairs) at 4-y follow-up; higher scores indicate better memory performance |
| imt_adjpairs_imm_med.11 | Elicited Imitation immediate memory task performance (adjacent pairs) at 9 mo; higher scores indicate better memory performance |
| imt_comp_imm_med.1 | Elicited Imitation immediate memory task performance (components) at baseline (0 mo); higher scores indicate better memory performance |
| imt_comp_sd_avemh.11 | Elicited Imitation short-delay memory task performance (components) at 9 mo; higher scores indicate better memory performance |
| memfaces_delcontrastsc | NEPSY-II Memory for Faces—Immediate vs. Delayed contrast score at 4-y follow-up; age-adjusted scaled score with higher scores indicating better performance |
| memnames_t3total | NEPSY-II Memory for Names Trial 3 total score at 4-y follow-up; age-corrected scaled score with higher scores indicating better performance |
| nihdccs_agecorrstandsc | NIH Toolbox Dimensional Card Sorting test performance at 4-y follow-up; age-corrected standard score with higher scores indicating better performance |
| nihdccs_fullcorrtsc | NIH Toolbox Dimensional Card Sort Test performance at 4-y follow-up; fully corrected (age, education, gender, race/ethnicity) standard score with higher scores indicating better performance |
| nihpsmt_agecorrstandsc | NIH Toolbox Picture Sequence Memory Test performance at 4-y follow-up; age-corrected standard score with higher scores indicating better performance |
| stanfbi_fsiq | Stanford-Binet Intelligence Scale (5th edition) Full Scale Intelligence Quotient at 4-y follow-up; age-corrected scores with higher scores indicating higher ability |
| stanfbi_qr | Stanford-Binet Intelligence Scale (5th edition) Quantitative Reasoning Index at 4-y follow-up; age-corrected scores with higher scores indicating higher ability |
| stanfbi_viq | Stanford-Binet Intelligence Scale (5th edition) Verbal Intelligence Quotient at 4-y follow-up; age-corrected scores with higher scores indicating higher ability |
| stanfbi_wm | Stanford-Binet Intelligence Scale (5th edition) Working Memory index at 4-y follow-up; age-corrected scores with higher scores indicating higher ability |
| t_score_2.11 | CBCL parent-reported social problems at 9 mo; age-corrected scores with higher scores indicating more problems |
| t_score3.1 | CBCL parent-reported school problems at baseline (0 mo); age-corrected scores with higher scores indicating more problems |
| t_score8.11 | CBCL parent-reported thought problems at 9 mo; age-corrected scores with higher scores indicating more problems |
| weight_pile_physical.12 | Age-adjusted weight percentile at 0 mo |
| weight_pile_physical.32 | Age-adjusted weight percentile at 3 mo |
| weight_z_physical.12 | z Score for age-adjusted height at 0 mo |
| weight_z_physical.32 | z Score for age-adjusted height at 3 mo |
| weight_z_physical.82 | z Score for age-adjusted height at 6 mo |
| weight_z_physical.112 | z Score for age-adjusted height at 9 mo |
| weightheight_z_physical.12 | z Score for age-adjusted weight-height at 0 mo |
Child Behavior Checklist; NEPSY-II, NEuroPSYcological Assessment, second edition.
Height and weight were standardized using normative data (33).
Results
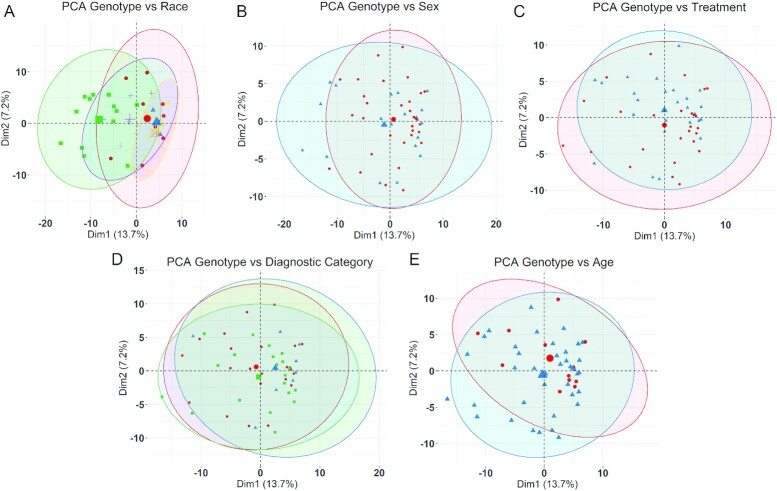
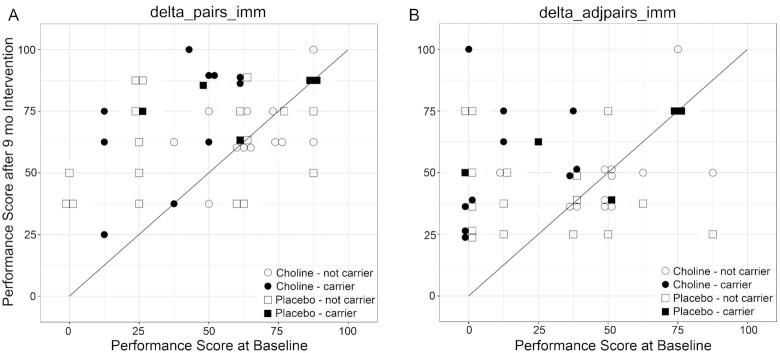
There were no differences in the characteristics of these genotyped participants at baseline and at the 4-y follow-up (Table 2) (16–18). Average daily dietary choline intake did not differ at baseline and averaged 78.7% ± 8.8% of the age-adjusted Adequate Intake (16). Baseline serum values of choline, betaine, and phosphatidylcholine also did not differ (16, 17). PCA of genotype associations against the study population's characteristics also identified no significant associations with the participants’ self-identified race (Figure 2A), sex (Figure 2B), treatment (Figure 2C), diagnostic category (Figure 2D), and age (Figure 2E). In all comparisons, PC1 explained only 13.7% of the association and PC2 explained just 7.2%. We then tested for genetic–phenotypic associations, first evaluating the simplest model in which the polymorphisms acted in an additive manner (AA vs. AB vs. BB). Stratification by intervention (choline vs. placebo) identified genotypes that were significantly associated with cognitive outcome in the choline-treated participants (Table 3). Specifically, the gene variants for 14 SNPs within the choline transporter gene SLC44A1 (listed in Table 3) were significantly associated (P < 0.006 for 13 SNPs; P = 0.04969 for 1 SNP) with the change-score (pre-/postintervention) on an elicited imitation sequential memory task, in which participants were observed to reproduce ordered pairs under immediate recall (delta_pairs_imm). These same 14 gene variants, plus 2 additional variants within SLC44A1, were also associated (P < 0.005 for 15 SNPs, P = 0.023 for 1 SNP) with change scores for adjacent pairs of items from the sequence (delta_adjpairs_imm). Participants having the variant in these SNPs, and also receiving the choline intervention, were more likely to show improvement in the memory task between pre- and postintervention, whereas no such associations were found for those participants in the placebo trial arm. The association between these effect alleles and cognitive performance was also seen in the direct comparison of individual performance in these measures, and choline participants carrying the effect alleles were more likely to show pre-/postintervention improvement compared with those carriers who did not receive choline, or those participants who lacked the effect alleles regardless of intervention (Figure 3). Repeating the association analysis against an assumption that these polymorphisms instead operated under a dominant (AA + AB vs. BB) or recessive (AA vs. AB + BB) genetic model found weaker, largely nonsignificant, associations. The dominant model yielded 8 SNPs within SLC44A1 that were associated with performance in the Elicited Imitation task (delta_adjpairs_imm); these were a subset of those identified in the additive model, and their significance was an order-of-magnitude weaker (P = 0.044; Supplemental Table 2). The recessive model yielded 5 SNPs within SLC44A1 (P = 0.044), all of which were shared with the dominant model. These data indicated that an additive effect of SLC44A1 genotype best modeled the association.
TABLE 2.
Characteristics of participants1
| Participants at baseline | Participants at 4-y follow-up | |||
|---|---|---|---|---|
| Placebo (n = 26) | Choline (n = 26) | Placebo (n = 16) | Choline (n = 15) | |
| Age at enrollment, y | 3.88 ± 0.78 | 3.84 ± 0.78 | 3.81 ± 0.78 | 3.92 ± 0.74 |
| Age at follow-up, y | — | — | 8.44 ± 1.06 | 8.61 ± 0.99 |
| Years since study completion | — | — | 3.96 ± 0.60 | 3.96 ± 0.46 |
| Sex, n (%) | ||||
| Male | 9 (34.6) | 10 (38.5) | 7 (43.8) | 8 (53.3) |
| Female | 17 (65.4) | 16 (61.5) | 9 (56.2) | 7 (46.7) |
| Racial categories,2n (%) | ||||
| White | 7 (26.9) | 12 (46.2) | 4 (25.0) | 7 (46.7) |
| Black or African American | 9 (34.6) | 4 (15.4) | 5 (31.2) | 2 (13.3) |
| American Indian/Alaska Native | 6 (23.0) | 4 (15.4) | 4 (25.0) | 2 (13.3) |
| Asian | 1 (3.8) | 1 (3.8) | 1 (6.3) | 1 (6.7) |
| More than 1 race | 3 (11.5) | 4 (15.4) | 2 (12.5) | 2 (13.3) |
| Not reported | 0 (0) | 1 (3.8) | 0 (0) | 1 (6.7) |
| Ethnic category,2n (%) | ||||
| Hispanic or Latino | 1 (3.8) | 1 (3.8) | 1 (6.3) | 1 (6.7) |
| Not Hispanic or Latino | 24 (92.3) | 24 (92.3) | 14 (87.5) | 13 (86.7) |
| Unknown | 1 (3.8) | 1 (3.8) | 1 (6.3) | 1 (6.7) |
| Dysmorphic facial features, n (%) | ||||
| Lip (score 4 or 5) | 15 (57.7) | 14 (53.8) | 10 (62.5) | 6 (40.0) |
| Philtrum (score 4 or 5) | 18 (69.2) | 13 (50.0) | 12 (75.0) | 6 (40.0) |
| Palpebral fissure (≤10th percentile) | 17 (65.4) | 19 (73.1) | 11 (68.8) | 11 (73.3) |
| ≥2 Facial features present | 15 (57.7) | 15 (57.7) | 10 (62.5) | 6 (40.0) |
| Growth deficiency (≤10th percentile), n (%) | ||||
| Height | 6 (23.0) | 7 (26.9) | 4 (25.0) | 3 (20.0) |
| Weight | 4 (15.4) | 7 (26.9) | 2 (12.5) | 3 (20.0) |
| Deficient brain growth (≤10th percentile), n (%) | ||||
| Occipital-frontal circumference | 10 (38.5) | 6 (23.1) | 5 (31.3) | 5 (33.3) |
| Alcohol exposure, n (%) | ||||
| Alcohol exposure confirmed | 24 (92.3) | 21 (80.8) | 15 (93.7) | 13 (86.7) |
| Alcohol exposure suspected | 1 (3.8) | 3 (11.5) | 1 (6.3) | 1 (6.3) |
| Unknown | 1 (3.8) | 2 (7.7) | 0 (0) | 1 (6.3) |
| Drug exposure, n (%) | ||||
| Drug exposure confirmed | 16 (61.5) | 12 (46.2) | 9 (56.3) | 8 (53.3) |
| Drug exposure suspected | 4 (15.4) | 5 (19.2) | 2 (12.5) | 1 (6.3) |
| Unknown | 6 (23.0) | 9 (34.6) | 5 (31.3) | 5 (33.3) |
| IOM diagnostic category, n (%) | ||||
| Fetal alcohol syndrome | 5 (19.2) | 4 (15.4) | 3 (18.8) | 1 (6.7) |
| Partial fetal alcohol syndrome | 9 (34.6) | 11 (42.3) | 6 (37.5) | 6 (40.0) |
| Alcohol-related neurodevelopmental disorder | 12 (46.2) | 11 (42.3) | 7 (43.8) | 9 (60.0) |
| Baseline Mullen early learning composite | 84.6 ± 20.8 | 84.0 ± 10.9 | 79.8 ± 21.6 | 82.9 ± 11.0 |
Values are n (%) or means ± SDs. No values differed significantly in the comparison between age-matched choline- and placebo-treated participants, using chi-square analysis. IOM, Institute of Medicine.
Race and ethnicity are self-identified.
FIGURE 2.
PCA of genotypes and population characteristics. PCA (FactoMineR, v2.3) was used to examine the genomic separation of all 52 participants based on race, sex, treatment, diagnostic category, or age. No significant associations between genotype and any of these population characteristics were observed in the PCA. (A) Self-identified race vs. genotype. Red circles, Native American (n = 10); blue triangles, Asian (n = 2); green squares, Black or African-American (n = 13); purple crosses, multiracial (n = 7); yellow crossed squares, White or Caucasian (n = 19); orange crossed squares, unknown (n = 1). (B) Sex vs. genotype. Red circles, female (n = 33); blue triangles, male (n = 19). (C) Treatment vs. genotype. Red circles, choline-treated (n = 26); blue triangles, placebo-treated (n = 26). (D) Diagnostic category vs. genotype. Red circles, alcohol-related neurodevelopmental disorder (n = 23); blue triangles, FAS (n = 9); green squares, partial FAS (n = 20). (E) Age vs. genotype. Red circles, 3 y or younger (n = 12); blue triangles, older than 3 y (n = 40). The largest symbol for each population characteristic represents the group mean for that characteristic, and the ellipse is the 95% CI surrounding that group mean. Dim1, dimension1; Dim2, dimension 2; FAS, fetal alcohol syndrome; PCA, principal components analysis.
TABLE 3.
SNPs and effect alleles within SLC44A1 associated with improved cognitive outcome in choline-treated participants1
| delta_pairs_imm2 | delta_adjpairs_imm2 | Number w/effect allele5 | ||||
|---|---|---|---|---|---|---|
| SNP | P | Reg. coeff.3 | P | Reg. coeff. | Effect allele4 | |
| rs10123494 | 0.005672 | −1.0505 | 0.000506 | −1.1232 | C (T > C) | 10/22 |
| rs10820801 | 0.005141 | −1.0476 | 0.001469 | −1.0872 | G (A > G) | 9/22 |
| rs10991639 | 0.005141 | −1.0476 | 0.001469 | −1.0872 | A (G > A) | 9/22 |
| rs2417615 | 0.005141 | −1.0476 | 0.001469 | −1.0872 | A (G > A) | 9/22 |
| rs2771040 | 0.005141 | −1.0476 | 0.001469 | −1.0872 | G (A > G) | 9/22 |
| rs3199966 | 0.049690 | −1.0516 | 0.001285 | −1.1981 | G (T > G) | 9/22 |
| rs34750132 | 0.005672 | −1.0505 | 0.000506 | −1.1232 | G (A > G) | 10/22 |
| rs35603631 | 0.005672 | −1.0505 | 0.000506 | −1.1232 | C (T > C) | 10/22 |
| rs4549843 | 0.005141 | −1.0476 | 0.001469 | −1.0872 | A (G > A) | 9/22 |
| rs6479311 | 0.005141 | −1.0476 | 0.001469 | −1.0872 | A (C > A) | 9/22 |
| rs6479313 | 0.005672 | −1.0505 | 0.000506 | −1.1232 | G (C > G) | 10/22 |
| rs7024985 | 0.003171 | −1.0476 | 0.001469 | −1.0872 | T (C > T) | 9/22 |
| rs7029443 | 0.005672 | −1.0505 | 0.000506 | −1.1232 | A (T > A) | 10/22 |
| rs7865985 | 0.005672 | −1.0505 | 0.000506 | −1.1232 | C (T > C) | 10/22 |
| rs12339823 | NS | NS | 0.004567 | −1.1659 | G (A > G) | 7/22 |
| rs16924529 | NS | NS | 0.023036 | −1.3298 | A (G > A) | 5/22 |
Reg. coeff., regression coefficient; SNP, single nucleotide polymorphism.
Associations between SNPs and outcomes were tested under an additive genetic model using the SNPassoc package in R (v.1.9–2). P values were adjusted for multiple testing using Bonferroni correction. Delta_pairs_imm is the change in Elicited Imitation immediate memory task performance (pairs) between baseline (0 mo) and 9 mo; delta_adjpairs_imm is the change in Elicited Imitation immediate memory task performance (adjacent pairs of items) between baseline (0 mo) and 9 mo.
Regression coefficient, extracted from additive model in SNPassoc package in R (v. 1.9–2).
Measured on top strand 5′→ 3′. Effect allele; major > minor allele within dbSNP.
Number of genotyped individuals carrying at least 1 copy of the effect allele within the choline cohort; 4 of the 26 children were missing this behavioral assessment and were excluded from this analysis.
FIGURE 3.
Change in cognitive performance for participants, stratified by intervention and effect allele status. Comparison of individual cognitive performance in the delta_pairs_imm (A) and delta_adjpairs_imm (B) outcomes at baseline (0 mo) and postintervention (9 mo), stratified by whether the participant carried the effect alleles (solid symbols) and received the choline (●) or placebo (▓) intervention, or did not carry the effect alleles (open symbols) and received either the choline (○) or placebo (□) intervention. delta_pairs_imm is the change in Elicited Imitation immediate memory task performance (for pairs of items regardless of sequence order) between baseline (0 mo) and 9 mo, and delta_adjpairs_imm is the change in Elicited Imitation immediate memory task performance (for pairs of adjacent items placed in the correct sequence order) between baseline (0 mo) and 9 mo; for both, higher scores indicate greater positive change (improvement). Carriers receiving choline (solid circles) all reside at or above the regression line that indicates no change in performance (R = 1.00). Because most of the effect alleles were in linkage disequilibrium, all carriers held at least 6 effect alleles. Presented are the 44 of 52 participants who completed this task at baseline and study conclusion; sample sizes are choline carrier, n = 10 of 11 participants: choline not carrier, n = 12 of 15 participants; placebo carrier, n = 5 of 5 participants; placebo not carrier, n = 17 of 21 participants.
Of the 32 SNPs within SLC44A1 that were present on the array, 9 of these were in linkage disequilibrium with the 16 significant SNPs. Positional maps revealed that all 26 were distributed across the entire gene structure for SLC44A1, from intron 1 through exon 16 (Table 4). Of particular note, rs3199966 is a coding sequence variant within exon 15, and the variant (T→G) converts serine 446 to alanine (S446A). rs2771040 (A→G) is located in the 3′ untranslated region (3′-UTR) of exon 16, which is unique to the SLC44A1 plasma membrane isoform (34). The remaining SNPs were intronic single nucleotide variants, and none were located within splice junctions or candidate microRNAs. PCA of these 26 SNPs found a significant association with self-identified race (PC1 = 56.6%; Supplemental Figure 1), reflecting that the effect variants are more frequent in populations with African ancestry. There was no association with sex, diagnostic category, or age.
TABLE 4.
SNPs and effect alleles within SLC44A1 showing an association with FASD or are in linkage-disequilibrium with these SNPs1
| SNP | Nucleotide | Location | Effect allele (major > minor allele in dbSNP)2 |
|---|---|---|---|
| rs327959 | 105,246,149 | Intron 1 | Increased with T (C > T) |
| rs327956 | 105,246,635 | Intron 1 | Increased with A (G > A) |
| rs443094 | 105,254,404 | Intron 1 | Increased with C (G > C) |
| rs328006 | 105,277,527 | Intron 1 | Increased with C (G > C) |
| rs327947 | 105,280,309 | Intron 1 | Increased with G (A > G) |
| rs193008 | 105,280,525 | Intron 1 | Increased with C (T > C) |
| rs12335779 | 105,288,137 | Intron 1 | Increased with G (C > G) |
| rs10991611 | 105,301,414 | Intron 2 | Increased with T (C > T) |
| rs9644967 | 105,305,895 | Intron 2 | Increased with A (G > A) |
| rs10991618 | 105,310,238 | Intron 3 | Increased with A (G > A) |
| rs2417615 | 105,325,570 | Intron 3 | Increased with A (G > A) |
| rs6479311 | 105,347,335 | Intron 4 | Increased with A (C > A) |
| rs10991639 | 105,355,290 | Intron 5 | Increased with A (G > A) |
| rs7024985 | 105,358,537 | Intron 7 | Increased with T (C > T) |
| rs7865985 | 105,358,537 | Intron 7 | Increased with C (T > C) |
| rs10123494 | 105,359,416 | Intron 7 | Increased with C (T > C) |
| rs34750132 | 105,360,429 | Intron 7 | Increased with G (A > G) |
| rs6479313 | 105,364,024 | Intron 9 | Increased with G (C > G) |
| rs7029443 | 105,367,465 | Intron 12 | Increased with A (T > A) |
| rs10820801 | 105,367,540 | Intron 12 | Increased with G (A > G) |
| rs4549843 | 105,368,325 | Intron 12 | Increased with A (G > A) |
| rs35603631 | 105,380,371 | Intron 13 | Increased with C (T > C) |
| rs16924529 | 105,382,872 | Intron 13 | Increased with A (G > A) |
| rs3199966 | 105,385,482 | Exon 15 | Increased with G (T > G) |
| rs12339823 | 105,387,394 | Intron E15–E16 | Increased with G (A > G) |
| rs2771040 | 105,389,918 | Exon 16 3′-UTR | Increased with G (A > G) |
E, exon; SNP, single nucleotide polymorphism; 3′-UTR, 3′-untranslated region.
Measured on top strand 5′ → 3′. All of these alleles are single nucleotide variants.
We identified additional associations between the 243 SNP genotypes and the behavioral outcomes and physical characteristics of the choline-supplemented FASD cohort (Table 5). With respect to behavior, improved performance on the working memory measure of the SB-5 (stanfbi_wm) at 4-y follow-up was associated with variants within FMO3 (rs1920149, G, P = 0.0113; rs909531, T, P = 0.00512). Variants in MTHFD1 were associated with improved performance in the Elicited Imitation immediate memory task at baseline (rs3783731, C, P = 0.01122; rs8011839, C, P = 0.01122) and in the NIH Toolbox Dimensional Card Sort Test at the 4-y follow-up (rs8003379, A, P = 0.00595). FMO3 oxidizes the microbial choline metabolite trimethylamine to trimethylamine oxide, and MTHFD1 interconverts the tetrahydrofolate one-carbon unit for purine, pyrimidine, and methionine synthesis. The unassigned SNP rs5899654 is an insertion/deletion (TAA) wherein deletion was associated with improved change-score measures from baseline to 9 mo for the Immediate Memory task in the elicited imitation (delta_pairs_imm, P = 0.00514; delta_adjpairs_imm, P = 0.00147); this SNP was merged with rs3030595 and is located within intron 1 of SLC44A1.
TABLE 5.
Additional SNP associations within choline-treated participants1
| Gene | SNP | P 2 | Reg. coeff. | Outcome3 | Effect allele (major > minor allele in dbSNP)4 | Number w/effect allele5 |
|---|---|---|---|---|---|---|
| ADIPOR1 | rs7539542 | 0.00981 | −1.2034 | nihpsmt_agecorrstandsc | Increased with G (C > G) | 9/15 |
| BHMT | rs567754 | 0.05079 | 0.8758 | height_z_physical.11 | Increased with C (C > T) | 15/23 |
| rs567754 | 0.04701 | 0.8791 | height_pile_physical.11 | Increased with C (C > T) | 15/23 | |
| CHDH | rs881883 | 0.02386 | −1.1318 | ibb_tscore21 | Increased with C (T > C) | 7/15 |
| FADS2 | rs20721146 | 0.04435 | 1.4352 | height_physical.11 | Increased with A (A > G) | 8/23 |
| rs171564426 | 0.00231 | −2.0158 | imt_adjpairs_imm_med.11 | Increased with T (C > T) | 4/23 | |
| FMO3 | rs1920149 | 0.01134 | −1.1290 | stanfbi_wm | Increased with G (A > G) | 11/14 |
| rs909531 | 0.00512 | 1.1902 | stanfbi_wm | Increased with T (T > C) | 7/14 | |
| rs2064074 | 0.04953 | 0.9732 | weight_z_physical.3 | Increased with A (A > G) | 15/21 | |
| rs1736560 | 0.04792 | −0.0892 | bloodlevel_betaine.1 | Increased with G (C > G) | 12/22 | |
| MTHFD1 | rs8003379 | 0.00595 | 1.6801 | nihdccs_agecorrstandsc | Increased with A (A > C) | 7/15 |
| rs8003379 | 0.02407 | 1.6144 | nihdccs_fullcorrtsc | Increased with A (A > C) | 7/15 | |
| rs2295640 | 0.00648 | −2.1758 | blood_pressure_diastolic.8 | Increased with G (C > G) | 3/21 | |
| rs3783731 | 0.01122 | 1.1129 | imt_comp_imm_med.1 | Increased with C (C > T) | 8/25 | |
| rs8011839 | 0.01122 | 1.1129 | imt_comp_imm_med.1 | Increased with C (C > T) | 8/25 | |
| MDR3 | rs31653 | 0.05053 | 2.3077 | memnames_t3total | Increased with G (G > A) | 2/15 |
| PEMT | rs7214988 | 0.04579 | −1.9685 | iib_avg_imt_adjpairs | Increased with G (C > G) | 3/15 |
| rs7224725 | 0.05131 | −2.3063 | iib_avg_imt_adjpairs | Increased with T (C > T) | 2/15 | |
| Unassigned | rs1275103 | 0.01929 | −0.9149 | height_z_physical.11 | Increased with G (C > G) | 17/23 |
| rs1275103 | 0.02179 | −0.9102 | height_pile_physical.11 | Increased with G (C > G) | 17/23 | |
| rs58996547 | 0.00514 | −1.0476 | delta_pairs_imm | Increased with D (I > D) | 9/22 | |
| rs58996547 | 0.00147 | −1.0872 | delta_adjpairs_imm | Increased with D (I > D) | 9/22 | |
| rs998671 | 0.00509 | 1.6972 | activity_level.11 | Increased with G (G > A) | 6/24 |
ADIPOR1, adiponectin receptor 1; D, deletion; BHMT, betaine-homocysteine S-methyltransferase; CHDH, choline dehydrogenase; FADS2, fatty acid desaturase-2; FMO3, flavin monooxygenase-3; I, insertion; MDR3, multidrug resistance protein 3; MTHFD1, methylenetetrahydrofolate dehydrogenase-1; PEMT, Phosphatidylethanolamine N-Methyltransferase; Reg. coeff., regression coefficient [extracted from additive model in SNPassoc package in R (v. 1.9–2)]; SNP, single nucleotide polymorphism.
Associations between SNPs and outcomes were tested under an additive genetic model using the SNPassoc package in R (v.1.9–2). P values are adjusted for multiple testing using Bonferroni correction.
Outcomes are defined in Table 1.
Measured on top strand, 5′ → 3′. All other alleles are single nucleotide variants.
Number of genotyped individuals carrying the effect allele within the choline cohort; some outcomes were missing for some children and thus were excluded from that analysis.
Also observed in analysis of all participants (Supplemental Table 3); P = 0.03062 for rs2072114, P = 0.03616 for rs17156442.
Merged with rs3030595 (10 May 2012); housed in intron 1 of SLC44A1 (TAA > deletion).
Additional, novel associations were found for physical characteristics of those receiving choline (Table 5). An allelic variant in BHMT, rs567754 (C→T), was weakly associated with greater height gain (P = 0.051) and height percentile at 9 mo (P = 0.047), as was a variant in the unassigned SNP (rs1275103, G; P = 0.0193 and P = 0.022, respectively) located on chromosome 5. A variant in FMO3 (rs2064074, A, P = 0.0495) weakly correlated with greater weight gain. At baseline, there were no treatment-group differences in serum concentrations for choline, phosphatidylcholine, and betaine (17), and we observed no associations with these apart from a weak, positive association between baseline serum betaine and a variant in FMO3 (rs1736560, G, P = 0.048). There were no genotype interactions with serum choline and its metabolites during the study intervention period.
Finally, several additional associations emerged in 2 genes involved in lipid metabolism (Table 5). A variant in the adiponectin receptor 1 (ADIPOR1; rs7539542, G, P = 0.0098) positively correlated with improved performance on the picture sequence memory task in the NIH Toolbox (nihpsmt_agecorrstandsc). For fatty acid desaturase-2 (FADS2), the variant rs17156442 (C→T, P = 0.0023) was associated with improved performance on the immediate recall adjacent pairs task at the 4-y follow-up, and the variant rs2072114 correlated with greater height (P = 0.044). To explore this latter finding further, analysis across the entire set of study participants revealed novel associations between physical characteristics (height, weight) and polymorphisms in BHMT (P < 0.012), FADS2 (P = 0.0306), and SLC44A1 (P < 0.034; Supplemental Table 3). Similar novel height and weight associations were present in the placebo controls for FADS2 (P < 0.025), MDR3 (P < 0.046), and SLC44A1 (P = 0.0074; Supplemental Table 3), and the MTHFR variant rs17421511 G (P ≤ 0.040) was associated with improved cognitive measures on the SB-5 assessment.
Discussion
Data herein may help explain the variable impact of choline supplementation upon cognitive functioning in children diagnosed with FASD. Ongoing clinical trials report lesser cognitive improvements (17, 18, 20, 21, 35) compared with the large effect sizes obtained in animal models of FASD (15). We find that, for this cohort, the benefits are masked when cognitive performance was pooled across genotypes. Stratification by genotype in SLC44A1, the gene coding for low-affinity choline transport across the plasma and mitochondrial membranes, revealed that choline supplementation was associated with increased sequential memory task scores for those participants who carried the minor allelic variants. Importantly, this same cognitive measure, the elicited imitation immediate performance, showed a nonsignificant trend for improvement in the original, nongenotypic analysis (17); our analysis shows that those carrying the effect variants (Table 2) in SLC44A1 account for this trend. There is a significant a priori rationale for such a genotype interaction, as polymorphisms that modulate individual choline need have been described for multiple choline-related genes, including SLC44A1, PEMT, CHDH, and BHMT, among others (23–31). Such a gene–nutrient interaction has not been previously identified for FASD. A prior choline intervention trial in heavy-drinking pregnant women found no interactions between outcomes and the rs12325817 polymorphism in PEMT (35); however, potential interactions may have been difficult to identify due to the participants’ low protocol adherence. It will be important to replicate these preliminary findings in a prospective a priori trial of choline versus genotype, complemented by confirmatory post hoc analysis of other FASD-choline intervention trials (20, 21, 35), and within the general population of those diagnosed with FASD (6, 7, 9, 36).
The association between SLC44A1 and cognitive improvement under choline supplementation is consistent with this protein's known function. SLC44A1, also known as the choline transporter-like protein 1 (CTL-1), is ubiquitous and mediates the bulk of choline transport for most tissues, including brain, where it is expressed by neurons, oligodendrocytes, and endothelial cells (37, 38). SLC44A1 is essential for brain development, and in humans its functional loss is associated with cognitive decline, ataxia, tremor, white matter deficits, hearing loss, and cerebral and cerebellar atrophy (38–40). These deficits may reflect, in part, its role in myelin compaction through interactions with nectin-like 4, whereas its loss-of-function impairs neurite myelination (41). The predicted protein structure of SLC44A1 features 7 transmembrane domains, and alternate splicing at its carboxy-terminus directs the protein to either the cell membrane (SLC44A1a; exon 16) or the mitochondria [SLC44A1b; exon 17; (38)].
Although most of the 25 SNPs implicating SLC44A1 in FASD cognitive outcomes were intronic, 2 may have functional relevance. rs3199966 (T > G) is a structural variant (S644A) in the carboxy-terminus of both cell membrane and mitochondrial isoforms, and is predicted to reside in an extracellular domain (34). The rs3199966 GG genotype is associated with greater vulnerability to skeletal muscle damage (P-adjusted = 0.0493) under a low-choline diet (26). Under high choline intake (930 mg/d), women carrying the rs3199966 variants TG or GG convert more deuterated choline into betaine and methionine synthesis, an effect not observed under adequate choline intake (430 mg) (34). rs2771040(A > G) is 4.4 kb downstream of rs3199966 and is located in the 3′-UTR of the SLC44A1a plasma membrane isoform, where it could potentially impact, for example, translation efficiency or structural motifs that modulate transcript stability. rs2771040 also positively correlates with more severe skeletal muscle damage (P-adjusted = 0.0024) when people are fed low-choline diets (26). Thus, both SNPs are associated with increased vulnerability to choline insufficiency. Baseline dietary choline intake in these choline-treated participants averaged 68.6% of their age-adjusted Adequate Intake (16); thus, it is possible that some individuals carrying these SNPs had an inadequate choline intake.
Biochemical studies of SLC44A1 offer additional insight into how it could modulate differential sensitivity to choline status. Choline intake modulates SLC44A1 activity (42–44), and under low choline SLC44A1 can be internalized via phosphorylation at serine 12 and serine 13 (43). A reduction in SLC44A1 membrane abundance has been hypothesized to favor choline uptake by the high-affinity transporter SLC5A7, which is enriched on cholinergic neurons and with the outcome of prioritizing acetylcholine synthesis when choline is limiting (37). This concentration-dependent activity of SLC44A1 may explain why cognitive enhancement was only associated with those carrying the effect variants in SLC44A1 (Table 2). As shown by da Costa et al. (26), those carrying these variants have greater vulnerability for poor choline status under conditions of low choline intake and would therefore derive greater benefit from choline supplementation. Functional studies are necessary to define the underlying mechanism; for example, the effect variant might be associated with reduced transporter activity, or with a greater reduction in membrane abundance when choline intake is low, and thus causing a greater reduction in cellular choline content. Alternately, the effect allele might confer greater membrane abundance under choline supplementation, and we note that, under supraphysiological choline intake, carriers of the effect allele in rs319996 direct greater quantities of choline into methylation via PEMT, rather than into the direct synthesis of phosphatidylcholine (23); this observation may offer mechanistic insight into how choline improves cognition in this FASD subpopulation. Conclusions regarding these putative mechanisms await functional testing of SLC44A1 and its allelic variants.
This study has multiple limitations. A genetic association analysis was not a primary outcome of the original study, and thus our analysis was likely underpowered to detect choline–gene associations for many of the study's measures. A significant benefit was only identified for SLC44A1, and the size effects were an order-of-magnitude smaller for polymorphisms in other choline-related genes known to affect choline requirements, including FMO3, BHMT, and MTHFD; their lower significance may be a limitation of the small cohort size. Our failure to identify parallel genetic-cognitive associations at the 4-y follow-up may similarly reflect the underpowered sample size. We did not identify associations with additional cognitive measures within the choline intervention cohort; the benefit was observed for the easiest task, and perhaps supplementation for a longer duration, or at younger ages, will confer greater benefit. Supporting this is the demonstration that alcohol-abusing women who were most compliant with a gestational choline supplement gave birth to children with the greatest choline-related cognitive performance (35). Finally, there was largely no association between these polymorphisms and choline-related metabolites, and thus plasma values did not reflect who gained the greatest benefit from the choline supplement.
In summary, we have identified genotypes affecting choline metabolism in individuals diagnosed with FASD and that are associated with significant cognitive improvement in an early childhood intervention trial with choline. SLC44A1 also mediates placental choline transport (38), and we predict that a similar association will likely emerge from the gestational choline intervention trials. Polymorphisms that affect the activity of additional choline-related genes will likely have similar influences, and lacked a robust association here, perhaps because the cohort was underpowered. It is challenging to prevent FASD due to the complex social issues that surround binge drinking, and we have few interventions to improve the daily function of those who are affected. Our data support the continued use of supraphysiological choline intake to improve cognitive function in those who are diagnosed with FASD, and the need to incorporate similar genetic analyses in future choline intervention trials for FASD and for other cognitive disorders.
Supplementary Material
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—JRW, SHZ, MKG, and JKE: designed the research; JRW, CJB, KES, JKE, and SHZ: conducted the research; MSV: analyzed the data; SMS, MSV, JRW, and SHZ: wrote the manuscript; SMS and JRW: have primary responsibility for final content; and all authors: read and approved the final manuscript. SHZ is the founder of SNP Therapeutics, he is on advisory boards for Ingenuity Brands and ByHeart, and has received grant funding from Balchem; all of these companies have an interest in choline. The other authors report no conflicts of interest.
Notes
Supported by NIH awards R01AA011085 and R01AA022999 to SMS; R01AA024123, R21AA019580, and R33AA019580 to JRW; R01DK115380 and R01DK056350 to SHZ; and internal funding from the UNC (University of North Carolina) Nutrition Research Institute.
Supplemental Figure 1 and Supplemental Tables 1–3 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: CBCL, Child Behavior Checklist; FADS2, fatty acid desaturase-2; FASD, fetal alcohol spectrum disorder; FMO3, flavin monooxygenase-3; MTHFD1, methylenetetrahydrofolate dehydrogenase-1; PCA, principal components analysis; SB-5, Stanford-Binet Intelligence Scale, version 5; SNP, single nucleotide polymorphism; SLC44A1, solute carrier family 44 member 1; 3′-UTR, 3′-untranslated region.
Contributor Information
Susan M Smith, Nutrition Research Institute, University of North Carolina at Chapel Hill, Kannapolis, NC, USA; Department of Nutrition, University of North Carolina at Chapel Hill, Kannapolis, NC, USA.
Manjot S Virdee, Nutrition Research Institute, University of North Carolina at Chapel Hill, Kannapolis, NC, USA.
Judith K Eckerle, Department of Pediatrics, University of Minnesota Twin Cities, Minneapolis, MN, USA.
Kristin E Sandness, Department of Pediatrics, University of Minnesota Twin Cities, Minneapolis, MN, USA.
Michael K Georgieff, Department of Pediatrics, University of Minnesota Twin Cities, Minneapolis, MN, USA.
Christopher J Boys, Department of Pediatrics, University of Minnesota Twin Cities, Minneapolis, MN, USA.
Steven H Zeisel, Nutrition Research Institute, University of North Carolina at Chapel Hill, Kannapolis, NC, USA; Department of Nutrition, University of North Carolina at Chapel Hill, Kannapolis, NC, USA.
Jeffrey R Wozniak, Department of Psychiatry and Behavioral Sciences, University of Minnesota Twin Cities, Minneapolis, MN, USA.
Data Availability
Deidentified data described in the manuscript, code book, and analytic code will be made available upon request.
References
- 1. Caudill MA, Obeid R, Derbyshire E, Bernhard W, Lapid K, Walker SJ, Zeisel SH. Building better babies: should choline supplementation be recommended for pregnant and lactating mothers? Literature overview and expert panel consensus. Eur Gyn Obstet. 2020;2:149–61. [Google Scholar]
- 2. Zeisel SH, Klatt KC, Caudill MA. Choline. Adv Nutr. 2018;9:58–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dawson G. Measuring brain lipids. Biochim Biophys Acta. 2015;1851:1026–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chester DN, Goldman JD, Ahuga JK, Moshfegh AJ. Dietary intakes of choline: What We Eat in America, NHANES 2007–2008. Dietary Data Brief. 2011;9:1–4. [PubMed] [Google Scholar]
- 5. Wallace TC, Blusztajn JK, Caudill MA, Klatt KC, Zeisel SH. Choline: the neurocognitive essential nutrient of interest to obstetricians and gynecologists. J Diet Suppl. 2020;17:733–52. [DOI] [PubMed] [Google Scholar]
- 6. Blusztajn JK, Slack BE, Mellott TJ. Neuroprotective actions of dietary choline. Nutrients. 2017;9:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boeke CE, Gillman MW, Hughes MD, Rifas-Shiman SL, Villamor E, Oken E. Choline intake during pregnancy and child cognition at age 7 years. Am J Epidemiol. 2013;177:1338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caudill MA, Strupp BJ, Muscalu L, Nevins JEH, Canfield RL. Maternal choline supplementation during the third trimester of pregnancy improves infant information processing speed: a randomized, double-blind, controlled feeding study. FASEB J. 2018;32:2172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu BT, Dyer RA, King DJ, Richardson KJ, Innis SM. Early second trimester maternal plasma choline and betaine are related to measures of early cognitive development in term infants. PLoS One. 2012;7:e43448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kennedy BC, Tran PV, Kohli M, Maertens JJ, Gewirtz JC, Georgieff MK. Beneficial effects of postnatal choline supplementation on long-term neurocognitive deficit resulting from fetal-neonatal iron deficiency. Behav Brain Res. 2018;336:40–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeisel S. Choline, other methyl-donors and epigenetics. Nutrients. 2017;9:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoyme HE, Kalberg WO, Elliott AJ, Blankenship J, Buckley D, Marais AS, Manning MA, Robinson LK, Adam MP, Abdul-Rahman Oet al. . Updated clinical guidelines for diagnosing fetal alcohol spectrum disorders. Pediatrics. 2016;138:e20154256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mattson SN, Roesch SC, Glass L, Deweese BN, Coles CD, Kable JA, May PA, Kalberg WO, Sowell ER, Adnams CMet al. . Further development of a neurobehavioral profile of fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2013;37:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. May PA, Chambers CD, Kalberg WO, Zellner J, Feldman H, Buckley D, Kopald D, Hasken JM, Xu R, Honerkamp-Smith Get al. . Prevalence of fetal alcohol spectrum disorders in 4 US communities. JAMA. 2018;319:474–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akison LK, Kuo J, Reid N, Boyd RN, Moritz KM. Effect of choline supplementation on neurological, cognitive, and behavioral outcomes in offspring arising from alcohol exposure during development: a quantitative systematic review of clinical and preclinical studies. Alcohol Clin Exp Res. 2018;42:1591. [DOI] [PubMed] [Google Scholar]
- 16. Wozniak JR, Fuglestad AJ, Eckerle JK, Kroupina MG, Miller NC, Boys CJ, Brearley AM, Fink BA, Hoecker HL, Zeisel SHet al. . Choline supplementation in children with fetal alcohol spectrum disorders has high feasibility and tolerability. Nutr Res. 2013;33:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wozniak JR, Fuglestad AJ, Eckerle JK, Fink BA, Hoecker HL, Boys CJ, Radke JP, Kroupina MG, Miller NC, Brearley AMet al. . Choline supplementation in children with fetal alcohol spectrum disorders: a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr. 2015;102:1113–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wozniak JR, Fink BA, Fuglestad AJ, Eckerle JK, Boys CJ, Sandness KE, Radke JP, Miller NC, Lindgren C, Brearley AMet al. . Four-year follow-up of a randomized controlled trial of choline for neurodevelopment in fetal alcohol spectrum disorder. J Neurodevelop Disord. 2020;12(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chambers CD, Yevtushok L, Zymak-Zakutnya N, Korzhynskyy Y, Ostapchuk L, Akhmedzhanova D, Chan PH, Xu R, Wertelecki W. Prevalence and predictors of maternal alcohol consumption in 2 regions of Ukraine. Alcohol Clin Exp Res. 2014;38:1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kable JA, Coles CD, Keen CL, Uriu-Adams JY, Jones KL, Yevtushok L, Kulikovsky Y, Wertelecki W, Pedersen TL, Chambers CDet al. . The impact of micronutrient supplementation in alcohol-exposed pregnancies on information processing skills in Ukrainian infants. Alcohol. 2015;49:647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coles CD, Kable JA, Keen CL, Jones KL, Wertelecki W, Granovska IV, Pashtepa AO, Chambers CD; Collaborative Initiative on FASD . Dose and timing of prenatal alcohol exposure and maternal nutritional supplements: developmental effects on 6-month-old infants. Matern Child Health J. 2015;19:2605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corbin KD, Zeisel SH. The nutrigenetics and nutrigenomics of the dietary requirement for choline. Prog Mol Biol Transl Sci. 2012;108:159–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ganz AB, Klatt KC, Caudill MA. Common genetic variants alter metabolism and influence dietary choline requirements. Nutrients. 2017;9(8):E837.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fischer LM, da Costa KA, Kwock L, Galanko J, Zeisel SH. Dietary choline requirements of women: effects of estrogen and genetic variation. Am J Clin Nutr. 2010;92:1113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. da Costa KA, Kozyreva OG, Song J, Galanko JA, Fischer LM, Zeisel SH. Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 2006;20:1336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. da Costa KA, Corbin KD, Niculescu MD, Galanko JA, Zeisel SH. Identification of new genetic polymorphisms that alter the dietary requirement for choline and vary in their distribution across ethnic and racial groups. FASEB J. 2014;28:2970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ganz AB, Shields K, Fomin VG, Lopez YS, Mohan S, Lovesky J, Chuang JC, Ganti A, Carrier B, Yan Jet al. . Genetic impairments in folate enzymes increase dependence on dietary choline for phosphatidylcholine production at the expense of betaine synthesis. FASEB J. 2016;30:3321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ganz AB, Cohen VV, Swersky CC, Stover J, Vitiello GA, Lovesky J, Chuang JC, Shields K, Fomin VG, Lopez YSet al. . Genetic variation in choline-metabolizing enzymes alters choline metabolism in young women consuming choline intakes meeting current recommendations. IJMS. 2017;18:E252.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kohlmeier M, da Costa KA, Fischer LM, Zeisel SH. Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc Natl Acad Sci. 2005;102:16025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silver MJ, Corbin KD, Hellenthal G, da Costa KA, Dominguez-Salas P, Moore SE, Owen J, Prentice AM, Hennig BJ, Zeisel SH. Evidence for negative selection of gene variants that increase dependence on dietary choline in a Gambian cohort. FASEB J. 2015;29:3426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resseguie ME, da Costa KA, Galanko JA, Patel M, Davis IJ, Zeisel SH. Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction. J Biol Chem. 2011;286:1649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao F, Song M, Wang Y, Wang W. Genetic model. J Cell Mol Med. 2016;20:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, Flegal KM, Guo SS, Wei R, Curtin LR, Roche AF, Johnson CL. CDC growth charts: United States. Adv Data. 2000;314:1–27. [PubMed] [Google Scholar]
- 34. Michel V, Bakovic M. The ubiquitous choline transporter SLC44A1. Cent Nerv Syst Agents Med Chem. 2012;12:70–81. [DOI] [PubMed] [Google Scholar]
- 35. Jacobson SW, Carter RC, Molteno CD, Stanton ME, Herbert JS, Lindinger NM, Lewis CE, Dodge NC, Hoyme HE, Zeisel SHet al. . Efficacy of maternal choline supplementation during pregnancy in mitigating adverse effects of prenatal alcohol exposure on growth and cognitive function: a randomized, double-blind, placebo-controlled clinical trial. Alcohol Clin Exp Res. 2018;42:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Signore C, Ueland PM, Troendle J, Mills JL. Choline concentrations in human maternal and cord blood and intelligence at 5 y of age. Am J Clin Nutr. 2008;87:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Inazu M. Functional expression of choline transporters in the blood-brain barrier. Nutrients. 2019;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Traiffort E, O'Regan S, Ruat M. The choline transporter-like family SLC44: properties and roles in human diseases. Mol Aspects Med. 2013;34:646–54. [DOI] [PubMed] [Google Scholar]
- 39. Fagerberg CR, Taylor A, Distelmaier F, Schrøder HD, Kibæk M, Wieczorek D, Tarnopolsky M, Brady L, Larsen MJ, Jamra RAet al. . Choline transporter-like 1 deficiency causes a new type of childhood-onset neurodegeneration. Brain. 2020;143:94–111. [DOI] [PubMed] [Google Scholar]
- 40. Reuter MS, Tawamie H, Buchert R, Hosny Gebril O, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel Met al. . Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry. 2017;74:293–9. [DOI] [PubMed] [Google Scholar]
- 41. Heffernan C, Jain MR, Liu T, Kim H, Barretto K, Li H, Maurel P. Nectin-like 4 complexes with choline transporter-like protein-1 and regulates Schwann cell choline homeostasis and lipid biogenesis in vitro. J Biol Chem. 2017;292:4484–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fullerton MD, Wagner L, Yuan Z, Bakovic M. Impaired trafficking of choline transporter-like protein-1 at plasma membrane and inhibition of choline transport in THP-1 monocyte-derived macrophages. Am J Physiol Cell Physiol. 2006;290:C1230–8. [DOI] [PubMed] [Google Scholar]
- 43. Ishikawa T, Suwanai H, Shikuma J, Suzuki R, Yamanaka T, Odawara M, Inazu M. Protein kinase C promotes choline transporter–like protein 1 function via improved cell surface expression in immortalized human hepatic cells. Mol Med Rep. 2020;21:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michel V, Singh RK, Bakovic M. The impact of choline availability on muscle lipid metabolism. Food Funct. 2011;2:53–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified data described in the manuscript, code book, and analytic code will be made available upon request.