Summary
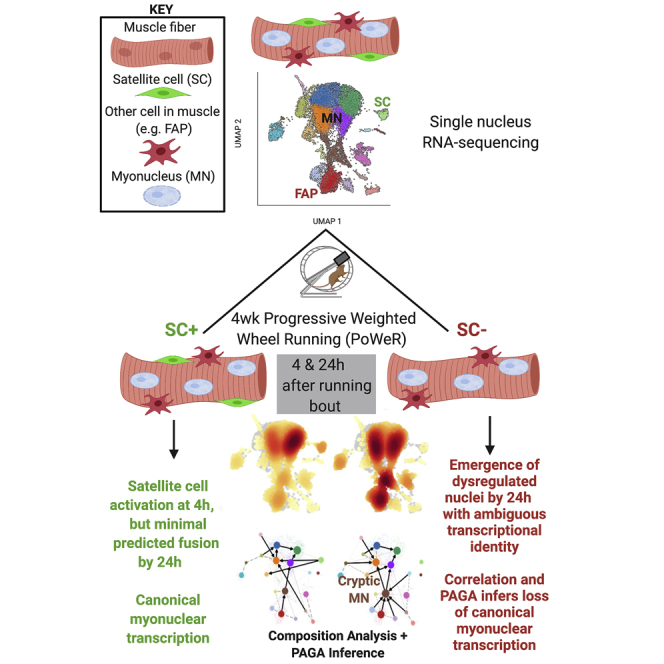
Skeletal muscle is composed of post-mitotic myofibers that form a syncytium containing hundreds of myonuclei. Using a progressive exercise training model in the mouse and single nucleus RNA-sequencing (snRNA-seq) for high-resolution characterization of myonuclear transcription, we show myonuclear functional specialization in muscle. After 4 weeks of exercise training, snRNA-seq reveals that resident muscle stem cells, or satellite cells, are activated with acute exercise but demonstrate limited lineage progression while contributing to muscle adaptation. In the absence of satellite cells, a portion of nuclei demonstrates divergent transcriptional dynamics associated with mixed-fate identities compared with satellite cell replete muscles. These data provide a compendium of information about how satellite cells influence myonuclear transcription in response to exercise.
Subject areas: Biological sciences, Stem cells research, Transcriptomics
Graphical abstract
Highlights
-
•
Single nucleus RNA-sequencing, trajectory inference, and entropy analyses
-
•
Genetic depletion of satellite cells before exercise training in adult mice
-
•
Dysregulated nuclei arise 24 h after last exercise bout when without satellite cells
-
•
Satellite cell depletion alters myonuclear transcription during growth and adaptation
Biological sciences; Stem cells research; Transcriptomics
Introduction
Adult skeletal muscle cells are formed through the fusion of mononucleated precursor cells, forming a myofiber with hundreds of post-mitotic nuclei that share a common cytoplasm spanning several millimeters (Charles et al., 2016). This distinctive cellular structure requires unique adaptive mechanisms such as the involvement of satellite cell activation and myonuclear addition in response to exercise (Masschelein et al., 2020). In addition, satellite cells may affect myonuclear transcription via fusion-independent mechanisms (Kirby et al., 2016; Murach et al., 2020).
Our laboratory developed a translatable model of exercise training (Dungan et al., 2019; Murach et al., 2020) along with genetic models of fluorescent myonuclear labeling (Iwata et al., 2018) and satellite cell depletion (McCarthy et al., 2011). Leveraging these tools in concert with single-nucleus RNA-sequencing (snRNA-seq), we performed state-of-the-art computational analyses comparing thousands of individual nuclear transcriptomes and present findings on how myonuclear transcription is influenced by satellite cells in response to exercise. In our exercise model, myonuclear accretion occurs early during training with few myonuclei added to myofibers beyond 4 weeks (Englund et al., 2020a). After 4 weeks of training, satellite cell activation and transcriptional regulation is apparent within 24 h of an acute exercise bout, and in the absence of satellite cells, transcriptional dynamics are dysregulated in a subset of the nuclear population. These data provide insight into satellite cell-mediated transcriptional coordination among myonuclei within the muscle fiber syncytium and help to explain attenuated adaptation associated with reduced satellite cell content (Englund et al., 2020a; Fry et al., 2014).
Results
Myonuclear transcription in sedentary muscle is coupled with myosin heavy chain gene expression
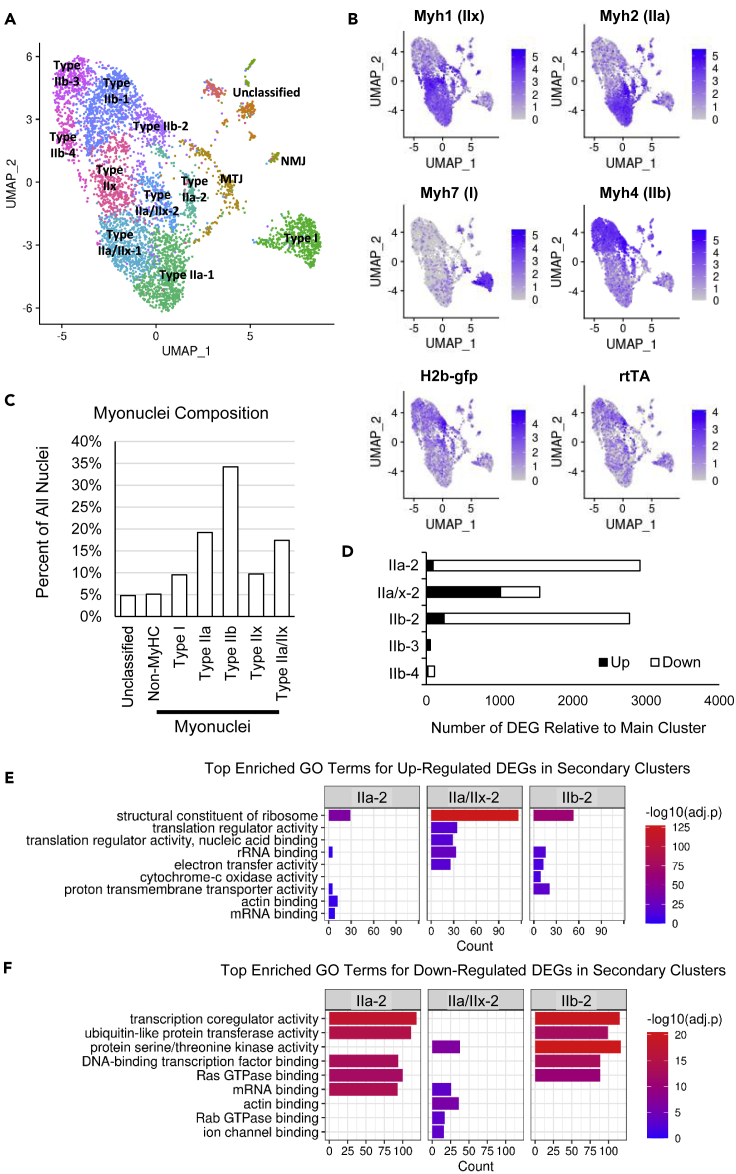
Myonuclei of adult myofibers were specifically GFP labeled using our inducible mouse model (Iwata et al., 2018). We purified labeled myonuclei from sedentary plantaris and soleus muscles via fluorescent activated cell sorting (Figueiredo et al., 2021; Von Walden et al., 2020) and performed snRNA-seq to detect 19,366 expressed genes across 4,900 myonuclei that were grouped into 15 clusters (Figure 1A). Myonuclear clusters segregated by myosin heavy chain (MyHC) gene expression, a determinant of fiber type (Figure 1B). We achieved >95% enrichment of the myonuclei; however, small clusters of “unclassified” nuclei with marker genes representative of non-myonuclei were also identified, totaling 4.8% of the sample (Figure 1C). Two small clusters of myonuclei expressed genes indicative of specialization for the neuromuscular (NMJ) and myo-tendinous (MTJ) junctions (Figure 1A, Data S1). Owing to the diffuse distribution of the MTJ myonuclei, we performed a subset analysis and found four sub-clusters ranging from 32 to 69 nuclei (Data S1 Sheet 2). Although the NMJ and MTJ clusters expressed MyHC, they were grouped as “Non-MyHC” in Figure 1C because they did not express a specific MyHC gene, unlike most of the canonical myonuclei that expressed a single MyHC gene (Figures 1A–1C). A small proportion (17.4%) of myonuclei expressed both Type IIa (Myh2) and IIx (Myh1) MyHCs. We identified a cluster of myonuclei expressing only the type IIx MyHC gene, although most IIx+ myonuclei were also IIa+. More than a third of the myonuclei in the sample were IIb+ (Myh4). Type I MyHC+ (Myh7) myonuclei were separated from the type II MyHC+ myonuclear cloud at a distance comparable with that of non-myonuclei. We observed that very few (<1%) myonuclei expressed developmental MyHCs (Myh3 and Myh8), suggesting minimal satellite cell fusion and/or myogenic developmental program re-activation in soleus and plantaris muscles from sedentary mice.
Figure 1.
Myonuclei from sedentary mice demonstrate sub-specialized expression of genes key to metabolic regulation
(A) Unbiased clustering and 2-dimensional uniform manifold approximation and projection (UMAP) representation of myonuclei labeled for myosin heavy chain (MyHC) gene expression in soleus and plantaris muscles. MTJ and NMJ represent myo-tendinous junction and neuromuscular junction myonuclei, respectively.
(B) Expression of key genes in post-mitotic myonuclei and precursor cell nuclei including the transgenes for myonuclear labeling. Parenthesis represent the MyHC isoform encoded by the Myh gene.
(C) Nuclei with non-muscle expression profiles compose less than 5% of all nuclei and myonuclei, whereas MyHC as significant gene markers represent around 90% of all nuclei in the sample.
(D) Number of differentially expressed genes (DEGs) when comparing sub-clusters of each type II MyHC-expressing myonuclei with the main cluster (cluster - 1). Although there are four clusters of IIb+ myonuclei, pairwise comparisons of the IIb+ subpopulations (IIb-2, IIb-3, and IIb-4) against the main population (cluster IIb-1) reveals that the IIb-3 and IIb-4 clusters may be artifacts due to few DEGs with no gene ontology (GO) enrichment.
(E) Top enriched GO terms for up-regulated DEGs (black bars in D) shows commonality among the secondary clusters to specialize for components of the ribosome and mitochondria.
(F) Top enriched GO terms for DEGs that are down-regulated (white bars in D) in the secondary clusters.
A subset of myonuclei show a specialized metabolic gene expression profile
Types I and IIx myonuclei each formed a single cluster, whereas types IIa, IIa/IIx, and IIb myonuclei formed multiple clusters (Figure 1A). For the latter, the main cluster having the most nuclei was designated as “-1” (i.e., IIa-1, IIa/IIx-1, and IIb-1). The genes defining main clusters I, IIa-1, and IIb-1 showed gene ontology (GO) enrichment of moderate significance, whereas IIx and IIa/IIx-1 clusters showed no GO enrichment (Table S1). Of note, we found four clusters of IIb+ myonuclei. Differential gene expression and GO analyses of pairwise comparisons of the IIb+ subpopulations (IIb-2, IIb-3, and IIb-4) against the main population (cluster IIb-1) revealed that the IIb-3 and IIb-4 clusters may be an artifact because there are almost no differentially expressed genes compared with the main IIb-1 cluster (Figure 1D) and no significant GO enrichment. In contrast, comparison between the main fiber type cluster and the respective secondary cluster (IIa-2, IIb-2, and IIax-2) showed a higher abundance of genes that encoded for structural components of the ribosome, rRNA binding, and the electron transport chain in the secondary clusters (Figure 1E, Data S2). These clusters had lower expression of genes enriched for GO terms such as ubiquitination, transcription, and transcription coregulation (Figure 1F).
Integration of snRNA-seq datasets uncovers distinctions between sedentary and exercise trained muscles
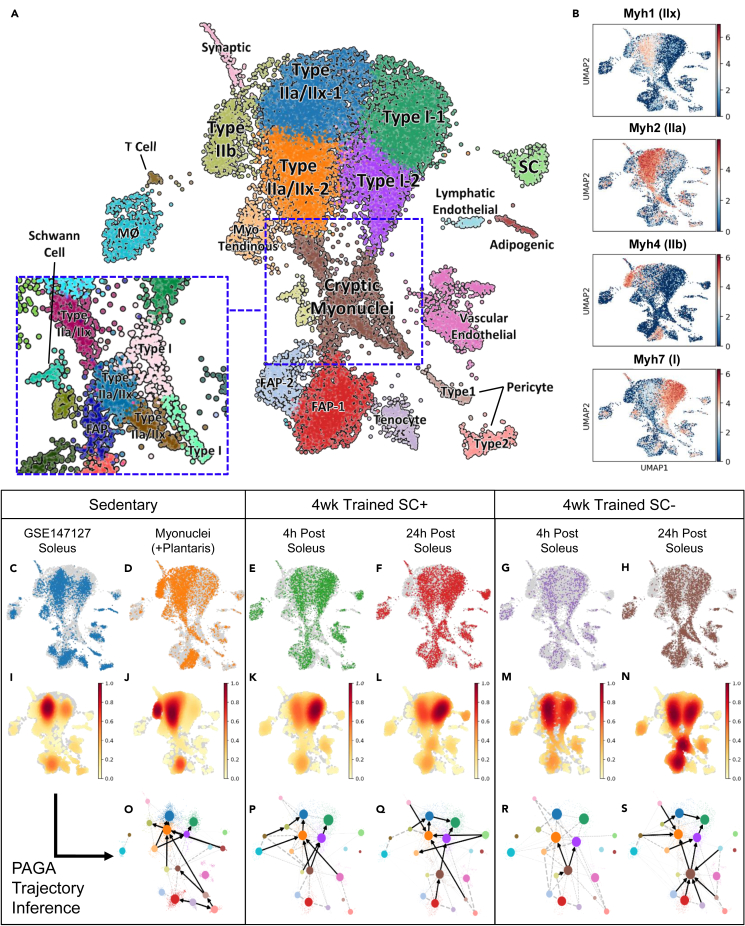
We performed snRNA-seq on soleus muscles from animals subjected to 4 weeks of progressive weighted wheel running in the presence or absence of satellite cells (Englund et al., 2020a). We administered vehicle (SC+) or tamoxifen (SC-) to 4-month-old Pax7CreER/+-R26R Diptheria Toxin A mice (designated Pax7-DTA) to deplete satellite cells prior to exercise training (McCarthy et al., 2011). To analyze and distinguish between the acute and lasting transcriptional responses to exercise training, we collected muscles at 4 and 24 h after the last bout of exercise. We performed an integrated analysis of six snRNA-seq datasets, representing two sedentary (a soleus sample made available through the Gene Expression Omnibus and the combined soleus and plantaris sample described above), two exercise-trained SC+ soleus muscles, and two exercise-trained SC- soleus samples and identified 20 main clusters (Figure 2A). As in the sedentary sample, canonical myonuclei appeared to broadly segregate according to MyHC gene expression (Figure 2B). Furthermore, we identified a cluster of MyHC+ nuclei with ambiguous gene expression that we named cryptic myonuclei (expanded on below). Sub-clustering of the cryptic MyHC+ myonuclear cluster revealed heterogeneous subpopulations expressing genes characteristic of FAPs, and type I and type II myonuclei (Figure 2A, inset). We also identified nuclear populations from satellite cells, macrophages, T cells, FAPs, tenocytes, pericytes, Schwann cells, lymphatic and vascular endothelial cells, and adipogenic cells according to classification by marker gene expression (Data S1).
Figure 2.
Emergence of cryptic myonuclei in exercised soleus muscle in the absence of satellite cells
(A) Unbiased cluster and UMAP representation of integrated datasets of sedentary and exercised muscles with and without satellite cells. Clusters are labeled based on significant gene markers. The clusters enclosed by the blue dotted line can be further subdivided into smaller nuclear clusters with differing expression profiles (inset). SC, FAP, and MØ represent satellite cell, fibro-adipogenic precursor, macrophage, respectively.
(B) Expression of Myh genes with the matching protein isoform indicated in parentheses. Color bar represents log normalized expression.
(C–N)(C–H) Nuclear contribution from each individual dataset is plotted in color with the rest of the integrated datasets plotted in gray background to highlight the presence or absence of clusters in A under each condition. This representation cannot differentiate two or more nuclei located at the same UMAP coordinate; thus, multiple nuclei in the same location would appear as a single point. In order to highlight the abundance of nuclei at each UMAP coordinate, I–N show the density estimates of nuclei for each condition, which indicates the level of nuclear overlap; dark red and white indicate the highest and lowest nuclear densities/overlap, respectively.
(O–S) Partition-based graph abstraction trajectory inference using dynamic modeling of RNA velocities for the soleus muscles color coded to match the clusters in A. Arrows indicate potential connections in pseudo time as determined by dynamical modeling of spliced and unspliced transcripts. All data are plotted on the same UMAP projection. 4wk trained samples come from animals subjected to 4 weeks of daily voluntary weighted-wheel running. 4 and 24 h post indicate the amount of time after the wheel was locked at the end of the last bout of running. SC+ and SC- represent satellite cell replete and depleted muscle, respectively.
The nuclei from each dataset were separately plotted in Figures 2C–2H to visualize their respective contributions to the entire nuclear population. By comparing the sedentary datasets, the nuclei from the plantaris muscle became readily apparent, which included a Type IIb cluster, as well as a portion of the main FAP cluster (Figure 2D). The nuclear density heatmaps further confirmed the distinctive pattern of plantaris myonuclear expression (Figure 2J). The sedentary soleus unexpectedly included a small population of IIb+ myonuclei that was absent in exercise-trained muscles, indicative of a training-induced, glycolytic-to-oxidative shift in fiber type distribution (Englund et al., 2020a).
In 4-week trained muscle, trajectory inference shows satellite cells influence myonuclear identity within 24 h after acute exercise
Exercise typically promotes satellite cell activation and, consistent with this scenario, the satellite cell nuclei at both 4 and 24 h after exercise were more transcriptionally active compared with the sedentary soleus sample, indicating a shift from quiescence to cell cycle entry (Data S1). The most significant marker genes in satellite cell nuclei of the exercised samples were ribosomal protein genes, whereas the most significant marker genes in the sedentary satellite cell nuclei included satellite cell markers, Peg3 and Pax7 (Correra et al., 2018; Seale et al., 2000). The number of nuclei in the satellite cell cluster increased from 4 to 24 h post exercise (Figures 2E, 2F, 2K, and 2L), with both time points being higher than the sedentary soleus (Figures 2C and 2I), suggestive of rapid satellite cell expansion. Although the number of nuclei in the remnant satellite cell cluster of the SC- samples also increased modestly from 4 to 24 h post exercise (Figures 2G, 2H, 2M, and 2N), both time points were markedly lower than the sedentary soleus.
Recent advances in trajectory inference allows for prediction of cell fate and commitment, and we employed the partition-based graph abstraction (PAGA) method because of its cluster stability and reproducibility (Trapnell et al., 2014; Wolf et al., 2018, 2019). Incorporating dynamical modeling of RNA velocity using unspliced and spliced transcript counts provide further insights into the transcriptional changes during single-cell lineage progression (Bergen et al., 2020; La Manno et al., 2018; Murach et al., 2021). PAGA trajectory inference with RNA velocity dynamical modeling predicted satellite cell connection to the myonuclear clusters only at 24 h post exercise in the satellite cell replete exercised sample (arrows, Figures 2P and 2Q); however, after 4 weeks of training, our prior work indicates that continued training up to 8 weeks does not result in additional myonuclear accretion (Englund et al., 2020a). No trajectory connection from the satellite cell cluster was present in either the sedentary sample (Figure 2O) or the remnants of the satellite cell cluster in the SC- samples (Figures 2R and 2S). On the other hand, 24 h after the last bout of exercise in the satellite cell deplete muscle, multiple cell types are connected to the cryptic myonuclear cluster (Figure 2S).
In the absence of SCs, trajectory analysis predicts the emergence of cryptic myonuclei that lose their canonical identity after acute exercise
The cryptic myonuclear cluster was expanded in the absence of satellite cells (Figures 2I–2N). The most significant marker genes for this cluster included genes linked to myogenic lineage progression such as Lmod2, Abra, Fhl1, Klhl41, and Csrp3 (Cowling et al., 2008; Londhe and Davie, 2011; Moretti et al., 2016; Ramirez-Martinez et al., 2017; Rashid et al., 2015), in addition to myonuclear genes including Myh1, Myh2, Dmd, and Neb (Data S1). Correlation analysis indicated these cryptic myonuclei had expression patterns that were distinct from and negatively correlated to the canonical myonuclear clusters, highly positively correlated with the FAP clusters, and also slightly correlated with tenocyte, pericyte, and endothelial clusters (Figure S1). This cluster is unlikely to represent aggregate nuclei because these nuclei do not have more genes on average compared with canonical myonuclei (Figure S2A) and their transcriptional dynamics show differential kinetics, which would be expected from nuclei of different cell types (Figure S2B). The transcriptional kinetics differ depending on the gene, which suggests a heterogeneous biological response rather than a homogeneous effect, the latter being indicative of technical artifacts or contaminating RNA species. When satellite cells were ablated, the cluster density of cryptic myonuclei became comparable with the main “canonical” myonuclear clusters at 24 h post exercise, concomitant with increased FAP identity nuclei (Figures 2M and 2N). Trajectory inference suggested these cryptic myonuclei were primarily related to the main FAP nuclei and types I and II myonuclei in the SC+ samples (Figure 2Q). In the SC- sample at 24 h post exercise, multiple non-myonuclear clusters had predicted connections to this cluster, including FAPs, tenocytes, type 1 pericytes, and even lymphatic endothelial cells (Figure 2S). Transcriptional signatures in SC- cryptic myonuclei resembling non-muscle cell types may reflect a dysregulation of resident myonuclei in the absence of satellite cells, leading to a loss of canonical myonuclear identity. An alternative explanation is that loss of satellite cells may exacerbate the ectopic expression of myonuclear genes in non-myonuclei, i.e., loss of fibrogenic and/or endothelial identity. To this point, there is evidence for cardiac fibroblasts adopting a cardiogenic transcription program during development and repair (Furtado et al., 2014), and a more recent study shows that endothelial cells in cardiac tissue can express cardiac-specific genes (Yucel et al., 2020). Only two genes, Neat1 and Txnip, were significantly upregulated in SC- compared with the SC+ 24-h sample, whereas the most significantly downregulated genes included muscle-specific genes such as Acta1, Slc38a2, Cox6a2, Csrp3, Tnnt1, Myl3, Myh7, and Ckm (p < 0.01).
Following 4 weeks of training, exercise continues to activate satellite cells without substantial fusion
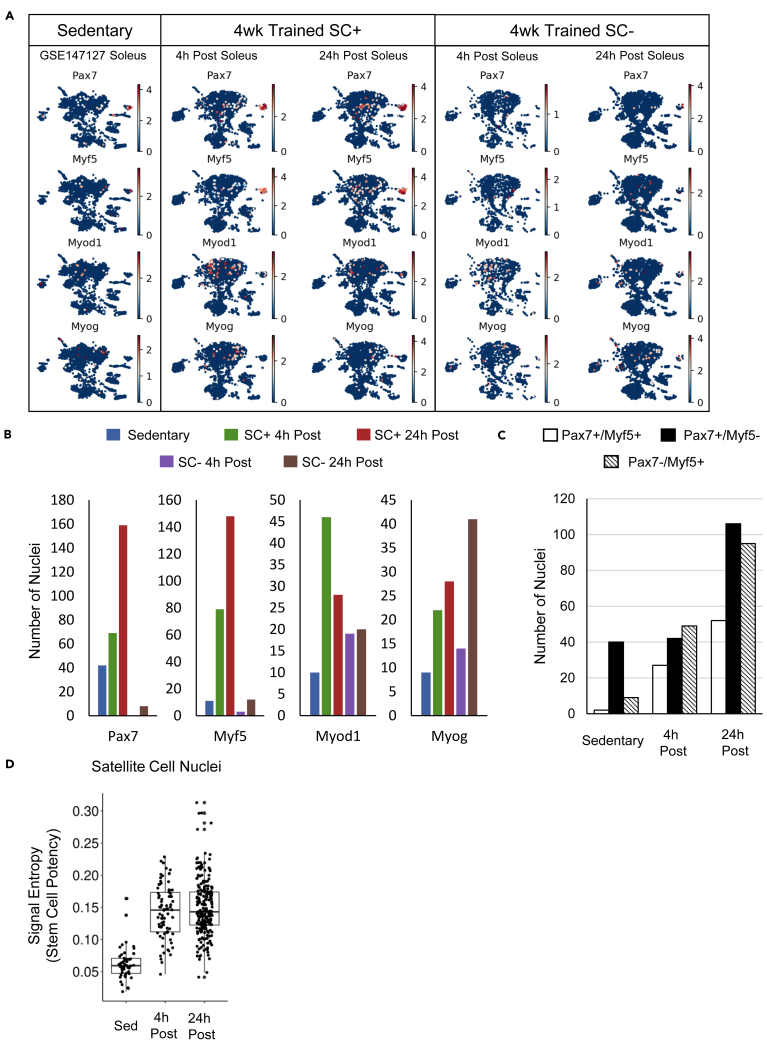
We compared the expression pattern of genes important for myogenesis and lineage progression, including Pax7, and three myogenic regulatory factors, Myf5, Myod1, and Myog (Figures 3A and 3B). We found few Pax7+/Myf5+ nuclei in the sedentary and SC- soleus samples (Figure 3B). In the sedentary soleus, the majority of the Pax7+ nuclei did not express Myf5, indicating a quiescent state. In the 4-week exercise-trained muscles, four hours after the last bout of exercise, there was a higher abundance of Myf5+ nuclei, whereas by 24 h after exercise, both Pax7+/Myf5+ and Pax7+/Myf5- nuclei had a higher abundance (Figure 3C). There were more Myod1+ nuclei 4 h after exercise in SC+ but not SC- soleus compared with sedentary soleus, further supporting activation occurring soon after the end of exercise. In contrast, Myog+ nuclei abundance did not change in SC+ after exercise but were more abundant in SC- soleus 24 h post exercise. Of note, Myod1+ and Myog+ nuclei were found more often in type II and type I MyHC+ myonuclear clusters, respectively, than in satellite cells (Figure 3A). The paucity of nuclei expressing embryonic MyHC (<40) or Myomaker (<4) across all samples suggested that, in both sedentary mice and after 4 weeks of exercise training, there was minimal injury, regeneration, or satellite cell fusion (Millay et al., 2013, 2014, 2016).
Figure 3.
Satellite cells in the exercised muscle activate within the first 24 h of the end of the last bout of exercise
(A) Expression heatmap projected onto UMAP coordinates for satellite cell marker gene, Pax7, and key myogenic regulator factors, Myf5, Myod1, and Myog, in sedentary and exercised soleus muscles in the presence (SC+) and absence (SC-) of satellite cells.
(B) Number of nuclei with expression of key genes in A.
(C) Number of nuclei in sedentary and satellite cell replete exercised soleus muscles with expression of Pax7, Myf5, or both, representing quiescent or activated satellite cells.
(D) Box and whisker plot showing signal entropy estimates for the nuclei in the satellite cell clusters of sedentary and exercised satellite cell replete soleus muscles calculated using discordance between transcript levels in each nucleus and known protein-protein interaction network. Higher signal entropy suggests higher progenitor potential (i.e., stem cell potential). The whiskers indicate 1.5 times the interquartile range above and below the upper and the lower quartiles, respectively. Each dot represents the signal entropy value for a single nucleus. 4wk trained samples come from animals subjected to 4 weeks of daily voluntary weighted-wheel running. 4 and 24 h post indicate the amount of time after the wheel was locked at the end of the last bout of running. SC+ and SC- represent satellite cell replete and depleted muscle, respectively.
The observation that Pax7+/Myf5- nuclei increased at 24 h post but not 4 h post exercise with minimal fusion led us to hypothesize that exercise rejuvenates satellite cells through cycles of self-renewal that improves progenitor cell potential. As a readout for progenitor cell potential we calculated signal entropy by quantifying the discordance between nuclear gene expression and known protein-protein interactions. Higher cellular signal entropy is correlated with higher levels of potency (i.e., precursor/stem cells), and lower entropy indicates lineage commitment (Banerji et al., 2013; Teschendorff and Enver, 2017; Teschendorff and Severini, 2010). We found an elevated level of signal entropy in the satellite cell clusters of the exercised muscle compared with sedentary muscle at both time points post exercise (Figure 3D), indicating that the 4 weeks of exercise training positively affected satellite cell “stemness.”
Discussion
We utilized an exercise training protocol and genetic tools to provide the first information on how satellite cells affect nuclear adaptations to exercise at single nucleus resolution. We focused on the soleus in contrast to previous studies that primarily focused on muscles with high abundance of type IIb MyHC expression, which is not expressed in humans (Chemello et al., 2020; Dos Santos et al., 2020). The mouse soleus contains high numbers of hybrid IIa/IIx+ in addition to type I+ myonuclei, similar to fiber type compositions found in humans (Murach et al., 2019). We found that myonuclei expressing one or more of the adult skeletal muscle MyHC genes (main myonuclear clusters) comprise most of the nuclei and only small populations (<5%) of myonuclei are specialized in the expression of genes necessary for processes in the post-synaptic regions and myo-tendinous junctions. Further sub-clustering of the diffusely clustered myo-tendinous junction myonuclei revealed heterogeneity partially consistent with the previous findings by Kim and colleagues, who recently reported distinct sub-clusters of MTJ myonuclei in the tibialis anterior (TA) muscle with and without injury (Kim et al., 2020). The MTJ-2 sub-cluster in our data shares similar marker genes (Tigd4, Itgb1, and Pdzd2) as the MTJ-A in their report; however, the three other sub-clusters we have found in the soleus and plantaris appear to be distinct from those found in the TA and may represent differences among muscles. In addition, we found three subpopulations of unclassified myonuclei, one of which expresses Meg3 and may be related to one of the four myonuclear subpopulations reported by Kim and colleagues in the TA referred to as the “Rian+” cluster; however, the sizes of the unclassified subpopulations in our dataset were small, totaling less than 5% of all the myonuclei.
Although the main myonuclear clusters mostly express the same genes in sedentary mice, there is a subset of type II myonuclei that express genes encoding components of the ribosome and electron transport chain at a higher level compared with sarcomeric and cytoskeletal components or kinases. This observation raises the possibility that myonuclei in the syncytium may coordinate division of labor by implementing transcriptional programs to achieve specific functional outputs. In addition, GO terms for gene markers of the main type I, IIa, and IIb clusters were enriched but had relatively low significance scores (–log10 of adjusted p value <4). Thus, sedentary myonuclear transcription is relatively homogeneous, which supports coordinated gene expression within the syncytium as previously reported (Dos Santos et al., 2020; Kim et al., 2020; Petrany et al., 2020).
We identified numerous Myod1+ nuclei in both sedentary and exercise-trained muscles, with a preference for type II MyHC+ clusters, especially the IIb cluster; this is consistent with previous studies demonstrating Myod1 expression bias for type II fibers in adult muscle (Ekmark et al., 2007; Hughes et al., 1993, 1997). Following 4 weeks of daily weighted wheel running in SC+ muscle there was an increase in Myod1+ nuclei 4 h after the last bout of exercise compared with sedentary control that was not seen in the SC- muscle. Myod1 expression points to satellite cell activation, some of which could result in fusion to muscle fibers, although myonuclear number is largely stabilized beyond 4 weeks of exercise training (Englund et al., 2020a). By 24 h, we observed a 4-fold higher number of Pax7+ nuclei in SC+ muscle compared with the sedentary control with a portion being Myf5-, suggesting a lack of myogenic lineage progression and return to quiescence. However, signal entropy for exercised satellite cells is elevated compared with sedentary control, providing some provocative evidence that quiescence in the exercise-trained muscle is different, supporting the idea that exercise rejuvenates satellite cell progenitor potential as speculated by some in the field (Chen et al., 2020). By cycling through activation and back into quiescence, satellite cells may remove cellular waste and repair damage, thereby improving long-term viability (van Velthoven and Rando, 2019). The transient activation of satellite cells may also facilitate their secretory communication to support exercise adaptation (Fry et al., 2017; Murach et al., 2020, 2021). We also observed higher Pax7-/Myf5+ nuclei at 4 h post exercise with further increase at 24 h post exercise, which may be indicative of lineage progression in some satellite cells. However, the low abundance of embryonic MyHC (Myh3)+ nuclei argues against a regenerative response. Very few Mymk+ nuclei in SC+ muscle further suggests that fusion is not the main role for activated satellite cells at this stage of training, consistent with our recent report (Englund et al., 2020a). Myog+ nuclei increased at 24 h in the SC- exercise-trained muscle compared with the sedentary control. Myog+ nuclei are found in the type I myonuclear clusters, consistent with previous reports of its MyHC type affinity. Myogenin enrichment in myonuclei in SC- may represent a compensatory role in promoting oxidative metabolism when myofibers fail to add new myonuclei (Hughes et al., 1999; Meadows et al., 2008; Zhu et al., 2013).
In the absence of SCs, trajectory analysis predicted the emergence of mixed-identity nuclei that became most prominent 24 h after the cessation of exercise. We termed these nuclei “cryptic” owing to their elusive transcription profile. Although these nuclei likely represent myonuclei as indicated by the presence of adult MyHC genes, as well as Neb and Dmd in the list of expressed genes that define this cluster, the origin and function of this subpopulation of nuclei require further experimental verification. Although certain multipotent cells, such as mesoangioblasts and pericytes, have been reported to possess myogenic potential (Ausems et al., 2019; Mavoungou et al., 2019; Messina et al., 2009), we are uncertain how much non-canonical myogenesis occurs in the absence of SCs. We speculate that such contributions to the relatively large myonuclear population is limited considering the lack of evidence for high levels of fusion in our dataset and minimal myonuclear addition in our exercise training model beyond the initial 4 weeks (Englund et al., 2020a). Instead, a subset of the resident myonuclei may lose their canonical transcriptional identity as a compensatory mechanism, part of a dynamic exercise cycle when satellite cells are absent. Anomalous myonuclear transcription could be indicative of myonuclear dysregulation or compensation in the absence of satellite cells that could contribute to impaired long-term muscle adaptation (Englund et al., 2020b; Fry et al., 2014, 2017). We observed an elevated density of nuclei in two FAP clusters of the exercised muscle that is most prominent in the absence of satellite cells 24 h post exercise. This finding is consistent with our previous work showing that activated satellite cells regulate the muscle microenvironment, independent of fusion (Fry et al., 2017; Murach et al., 2018, 2020, 2021). Our data collectively support the idea that activated satellite cells communicate with other cell types in skeletal muscle, including the myofiber, to facilitate an optimal response to exercise.
Limitations of the study
The current study focused solely on nuclear transcription and does not represent changes at the protein level. Therefore, our observations may represent transcriptional steady states or transient adaptations. Owing to the endpoint nature of the analyses, we were unable to determine dynamic transcriptional changes during exercise, and the time points post exercise we have chosen in the study may not be the most optimal. Furthermore, the aberrant nuclear population that emerged after exercise training in the absence of satellite cells appears to be heterogeneous and will require further experimental verification of the RNA-seq data and detailed characterization to elucidate the function of this potentially novel nuclear population.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4′,6-diamidino-2-phenylindole (DAPI) | Molecular Probes | D1306 |
| Spermidine | Sigma | S0266 |
| Spermine tetrahydrochloride | Sigma | S2876 |
| EDTA | Sigma | T9285 |
| EGTA | Sigma | E3889 |
| Magnesium Chloride | Alfa Aesar | J62411 |
| Sucrose | Sigma | S1888 |
| HEPES | Sigma | H3375 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3’ v3 | 10X Genomics | Cat# 1000092 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat# 20020594 |
| Deposited Data | ||
| Single-nucleus RNA-seq of sedentary plantaris and soleus myonuclei and progressive exercise trained soleus nuclei | This paper | GEO: GSE162307 |
| Experimental Models: Organisms/Strains | ||
| Mouse: HSA-rtTA ; TRE-H2B-GFP | (Iwata et al., 2018) | NA |
| Mouse: Pax7CreER/+ ; R26R-DTA | (McCarthy et al., 2011) | NA |
| Software and Algorithms | ||
| Cell Ranger (v4.0.0) | NA | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat (v3.2.2) | (Butler et al., 2018; Hafemeister and Satija, 2019) | https://satijalab.org/seurat/ |
| anndata (v0.7.5.dev13+g77a633c) | (Wolf et al., 2018) | https://github.com/theislab/anndata |
| Scanpy (v1.6.1.dev48+g256f5944) | (Wolf et al., 2018, 2019) | https://github.com/theislab/scanpy |
| Scvelo (v0.2.2) | (Bergen et al., 2020) | https://github.com/theislab/scvelo |
| velocyto (v0.17.17) | (La Manno et al., 2018) | http://velocyto.org/velocyto.py/index.html |
| Harmony | (Korsunsky et al., 2019) | Implemented as harmonypy within scanpy |
| limma (v3.44.3) | (Ritchie et al., 2015) | http://bioconductor.org/packages/release/bioc/html/limma.html |
| Other | ||
| Single-nucleus RNA-seq for sedentary soleus | NA | GEO: GSE147127 |
Resource availability
Lead contact
Charlotte Peterson (cpete4@uky.edu)
Materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact.
Data and code availability
All snRNA-seq datasets are uploaded to the Gene Expression Omnibus (GEO) under the accession number GSE162307 and can be downloaded from the Sequence Read Archive (SRA) under the BioProject number PRJNA681403.
Experimental model details
Mice
All animal procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals as approved by the Animal Care and Use Committee of the University. Mice housed in a humidity- and temperature-controlled facility, maintained on a 14:10 hour light-dark cycle with food and water ad libitum. The adult skeletal muscle myonuclear specific GFP mouse (H2B-GFP) was generated and treated as previously described (Iwata et al., 2018). The reverse tetracycline-controlled transactivator (rtTA) under the control of the human skeletal actin (HSA) promoter was used to activate, in the presence of doxycycline, the expression of a recombinant histone 2b (H2B) attached to an enhanced green fluorescent protein (EGFP) under the control of the tetracycline response element (TRE). The satellite cell specific conditional ablation mouse (Pax7-DTA) was generated and treated as previously described (McCarthy et al., 2011). The Cre recombinase attached to the modified estrogen receptor (CreER(TM)) under the control of the Pax7 promoter was induced in the presence of tamoxifen to translocate into the nucleus. Once in the nucleus, the Cre recombinase targets the loxP sites flanking the stop cassette that prevented the expression of Diphtheria Toxin A (DTA), thereby allowing DTA expression and causes cell death in satellite cells.
Mouse treatment, progressive exercise training, and tissue Collection
H2B-GFP: a 6-month-old female H2B-GFP was administered doxycycline (0.5 mg/mL) in drinking water supplemented with 2% sucrose for 3 weeks. Soleus and plantaris muscles were collected after an overnight fast upon completion of doxycycline treatment and prepared immediately for snRNA-seq.
Pax7-DTA: For snRNA-seq analysis, 4 female mice were randomly assigned to the 4-wk progressive weighted wheel running vehicle treated (SC+) or tamoxifen treated (SC-) groups. Details of the training regimen and outcomes were reported elsewhere (Englund et al., 2020a). Wheels were locked for 4 or 24 hours before sacrifice and mice were fasted during this period. The soleus muscle from one SC+ mouse and one SC- mouse at each time point was excised and prepared immediately for snRNA-seq. Mice from each group (SC+/SC-) were selected based on similarity in body weight and running volume over the 4-wk protocol. All mice in the study were 6 months old at time of sacrifice. Preliminary analyses of the 24 hours post training nuclei were recently reported (Englund et al., 2020a).
Method details
Nuclear isolation
Hindlimb muscles were harvested following euthanasia and nuclei were isolated using the biochemical protocol developed by (Cutler et al., 2017). Briefly, muscles were minced with scissors in homogenization buffer (500 μl HEPES (1 M) 3 ml KCl (1 M) 250 μl spermidine (100 mM) 750 μl spermine tetrahydrochloride (10 mM) 10 ml EDTA (10 mM) 250 μl EGTA (100 mM) 2.5 ml MgCl (100 mM) 5.13 g sucrose, Dounce homogenized on ice in homogenization buffer and passed through a 40 μm filter into sorting buffer. For the Pax7-DTA soleus muscle samples, DAPI was added to label nuclei. Fluorescently labeled nuclei (DAPI+ or GFP+) were purified via Fluorescence-activated cell sorting (FACS) and collected in reverse transcription (RT) buffer.
Library preparation and single-nucleus RNA sequencing
Nuclei were loaded into the 10X Chromium system using the Single Cell 3’ Reagent Kit v3 according to the manufacturer’s protocol. Following library construction, libraries were sequenced on the Illumina NextSeq 500 System at the University of Florida.
Bioinformatic analysis pipeline
Sequenced reads were converted to fastq format after demultiplexed using the “cellranger mkfastq” function and aligned to the reference mouse genome using Cell Ranger (v4.0.0) developed and maintained by 10X Genomics. The reference genome for the mouse was downloaded from Ensembl (GRCm38.p6, release 101, repeat masked, RefSeq: GCA_000001635.8), which included the matching set of genome sequence (FASTA) and gene annotations (GTF). A custom transcriptome reference was created by appending the sequences and annotations of the transgenes of interest to the downloaded FASTA and GTF files, respectively. The sequence for rtTA (Tet-On 3G) was obtained from Cloning vector pDXTR (GenBank: MN_044710.1). The sequences for H2B-GFP, CreER(TM), and DTA were obtained from plasmid sequences for pBABE-H2BGFP (Addgene: # 26790, 15028, 22677) (Coschi et al., 2010; Nam and Benezra, 2009), respectively. In order to accommodate the high levels of unspliced transcripts in the nucleus, a “pre-mRNA” transcriptome reference was created through a two-step process. First, the previously generated custom GTF file was modified by converting gene labels from “transcript” to “exon” and appending the “_premrna” tag to the original “transcript_id”. Then, the modified GTF was used with the previously generated custom FASTA file with the added transgenes to generate the “pre-mRNA” reference using the “cellranger mkref” function. Reads from the Cell Ranger FASTQ files were aligned to the custom “pre-mRNA: reference using the “cellranger count” function.
The Cell Ranger alignment results in the form of filtered count matrices labeled with gene symbols and nuclei barcodes were imported into R (v4.0.2) programming environment using Seurat (v3.2.2). A summary table of quality control metrics are provided in Data S2. Only genes with non-zero expression in at least three nuclei were retained for each sample. Quality control was performed by calculating the number of unique genes per nucleus and the percent of extra-nuclear transcripts (i.e., mitochondrial-encoded genes) and filtering out nuclei with less than 200 or greater than 3000 unique detected genes or with greater than 5% extra-nuclear genes. Due to variability in the percent of mitochondrial-encoded genes and number of unique genes per nucleus, these two variables along with the read depth were used as covariates in the SCTransform function, which applied the regularized negative binomial regression on 3000 variable genes per dataset to normalize and scale the UMI count data (Hafemeister and Satija, 2019). Dimensionality reduction was performed using principal component analysis, and the first 50 principal components were used to construct the Shared Nearest Neighbor (SNN) graph (Butler et al., 2018). Unbiased clustering was performed with a resolution set to 1 using the FindClusters function which employs the Louvain algorithm. Non-linear dimension reduction using the first 50 principal components was achieved by the Uniform Manifold Approximation and Projection (UMAP) technique to visualize nuclear gene expression in 2-dimentional space.
Integration of six datasets (sedentary soleus (GEO: GSE147127), sedentary soleus and plantaris, exercised SC+ and SC- soleus at 4 and 24 hours after the last bout of exercise) was achieved using Harmony (Korsunsky et al., 2019). Briefly, spliced and unspliced RNA reads were extracted from the Cell Ranger alignment results using the velocyto (v0.17.17) command line python tool (La Manno et al., 2018) with inputs being the custom FASTA and GFT files that included the transgenes. The “.loom” output of from velocyto was analyzed using scvelo (v0.2.2), scanpy (v1.6.1.dev48+g256f5944), and anndata (v0.7.5.dev13+g77a633c) python packages (Bergen et al., 2020; Wolf et al., 2018). Quality control and normalization were performed on each dataset the same way as described above for Seurat with the exception that SCTransform was not available in scanpy. Instead, a simple linear regression similar to a previous version of Seurat’s “regressOut” function was applied. Harmony integration using the “harmonypy” implementation was performed with a maximum of 25 iterations, achieving convergence after 7 iterations. Neighbors graph was constructed using the harmony correction with local neighborhood size set to 30 and using the first 50 principal components. Unbiased clustering was performed on the combined and corrected dataset after integration using the Leiden algorithm (Traag et al., 2019). Partition-based graph abstraction (PAGA) positions were computed after Leiden clustering and UMAP embedding of the neighborhood graph was initialized using the PAGA positions (Wolf et al., 2019). For RNA velocity calculations, the first- and second-order moments were first computed using the Harmony corrected neighbor graph followed by recovery of transcriptional dynamics and finally, RNA velocities were computed using the “dynamical” mode and subsequently adjusted for differences in transcriptional kinetics among various cell types. Importantly, for snRNA-seq data, cellular RNA degradation was assumed to be replaced by nuclear export. RNA velocity pseudotime was subsequently used as priors to correct PAGA connectivity for each dataset.
Signaling entropy was estimated using the “CompCCAT” (Correlation of Connectome and Transcriptome) function from the SCENT (v1.0.2) R package using the protein-protein interaction network “net17Jan16.m” provided by the package. We used CCAT instead of the actual signaling entropy rates because the computation of CCAT is much more efficient and thus is amenable to be used on the large datasets such as snRNA-seq from multiple experiments. The signaling entropy rate has been used as a proxy to determine the differential potential of single cells (Teschendorff and Enver, 2017; Teschendorff and Severini, 2010).
Quantification and statistical analysis
Gene expression signatures for each nuclear cluster was determined using the “FindAllMarkers” function in Seurat or the “tl.rank_genes_groups” function in scanpy, both of which performs the Wilcoxon Rank Sum test to determine significantly (adjusted p < 0.05) differentially expressed gene computed using the log normalized read counts. Seurat default parameters included additional filters for a minimum natural log fold difference of 0.25 (~28% difference) and a minimum expression in 25% of nuclei in that cluster. Clusters with marker genes that include Myh7, Myh2, Myh4, or Myh1 were classified as Type I, IIa, IIb, or IIx myonuclei. Clusters with markers genes that include a combination of one or more of these MyHC genes were classified accordingly as hybrid myonuclear clusters. Clusters without one of the MyHC genes as a marker were deemed as “Non-MyHC”.
Differential gene expression analyses among datasets and among subsets within datasets were performed using the limma (v3.44.3) R package (Ritchie et al., 2015). We chose this package because previous studies report consistent performance in terms of sensitivity, specificity, and rediscovery rates for limma compared to numerous other available packages (Mou et al., 2019; Soneson and Robinson, 2018). Read counts were extracted from Seurat objects using the “GetAssayData” function and lowly expressed genes (total expression across all nuclei < 50 or having non-zero expression in fewer than 10 nuclei) were removed from analysis improve specificity because previous studies have found that too many lowly expressed genes contribute to increased false discovery (Soneson and Robinson, 2018). In the sedentary plantaris and soleus dataset, we filtered out lowly expressed genes and tested for statistical significance using an empirical Bayes prior trend model.
Acknowledgments
The authors thank Dr. Eric T. Wang and Lance Denes at the University of Florida for sequencing expertise, Jennifer Strange of the University of Kentucky Flow Cytometry Core and Dr. Doug Harrison of the University of Kentucky Biology Department/Genetics and Genomics Imaging Center for their technical expertise with fluorescent activated cell sorting and single cell RNA sequencing, respectively. Partial computational support was provided by The University of Kentucky High Performance Computing complex. This work was supported by NIH grants from the National Institutes of Arthritis and Musculoskeletal and Skin Diseases (AR060701 to C.A.P. and J.J.M., AR071753 to K.A.M, and AR075364 to D.A.E.) and National Institute on Aging (AG049086 to C.A.P. and J.J.M. and AG063994 to K.A.M). The graphical abstract was generated using BioRender.
Author contributions
Y.W., D.A.E., J.J.M., and C.A.P. conceived the project and designed study approach. Y.W. performed the transcriptomic and computational analysis. D.A.E. and K.A.M. oversaw mouse experimentation. D.A.E., B.D.P., and K.A.M. performed nuclear isolation and processed samples for snRNA-seq. J.J.M. and C.A.P. supervised and coordinated all aspects of the study, computational analysis, and manuscript writing. The manuscript was written by Y.W. with input from D.A.E. and K.A.M. Figures were created by Y.W. and K.A.M. All authors have critically revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102838.
Supplemental information
Cluster marker genes. Sheet 1: Marker genes for each cluster in sedentary soleus and plantaris myonuclei. Sheet 2: Sub-clustering of the MTJ cluster and marker genes for each sub-cluster. Sheet 3: Top 200 markers genes (based on p values) for each cluster after integration of sedentary and exercised datasets. Sheet 4: Differentially expressed genes in the cryptic myonuclear cluster comparing SC- 24hrs post with SC+ 24hrs post last bout of exercise.
Differential gene expression data for the sedentary myonuclei from soleus and plantaris muscles. Sheet 1: Statistical results comparing genes expressed in the secondary type IIa cluster compared with the primary type IIa cluster shown in Figure 1. Sheet 2: Statistical results comparing genes expressed in the secondary type IIa/IIx cluster compared to the primary type IIa/IIx cluster shown in Figure 1 Sheet 3: Statistical results comparing genes expressed in the secondary type IIb cluster compared with the primary type IIb cluster shown in Figure 1. Sheet 4: Summary table of quality control metrics for all the analyzed datasets.
References
- Ausems C.R.M., Raaijmakers R.H.L., van den Broek W., Willemse M., van Engelen B.G.M., Wansink D.G., van Bokhoven H. Intrinsic myogenic potential of skeletal muscle-derived pericytes from patients with myotonic dystrophy type 1. Mol. Ther. Methods Clin. Dev. 2019;15:120–132. doi: 10.1016/j.omtm.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji C.R., Miranda-Saavedra D., Severini S., Widschwendter M., Enver T., Zhou J.X., Teschendorff A.E. Cellular network entropy as the energy potential in Waddington's differentiation landscape. Sci. Rep. 2013;3:3039. doi: 10.1038/srep03039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles J.P., Cappellari O., Spence A.J., Hutchinson J.R., Wells D.J. Musculoskeletal geometry, muscle architecture and functional specialisations of the mouse hindlimb. PLoS One. 2016;11:e0147669. doi: 10.1371/journal.pone.0147669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemello F., Wang Z., Li H., McAnally J.R., Liu N., Bassel-Duby R., Olson E.N. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. U S A. 2020;117:29691–29701. doi: 10.1073/pnas.2018391117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Datzkiw D., Rudnicki M.A. Satellite cells in ageing: use it or lose it. Open Biol. 2020;10:200048. doi: 10.1098/rsob.200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correra R.M., Ollitrault D., Valente M., Mazzola A., Adalsteinsson B.T., Ferguson-Smith A.C., Marazzi G., Sassoon D.A. The imprinted gene Pw1/Peg3 regulates skeletal muscle growth, satellite cell metabolic state, and self-renewal. Sci. Rep. 2018;8:14649. doi: 10.1038/s41598-018-32941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi C.H., Martens A.L., Ritchie K., Francis S.M., Chakrabarti S., Berube N.G., Dick F.A. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling B.S., McGrath M.J., Nguyen M.A., Cottle D.L., Kee A.J., Brown S., Schessl J., Zou Y., Joya J., Bonnemann C.G. Identification of FHL1 as a regulator of skeletal muscle mass: implications for human myopathy. J. Cell Biol. 2008;183:1033–1048. doi: 10.1083/jcb.200804077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler A.A., Corbett A.H., Pavlath G.K. Biochemical isolation of myonuclei from mouse skeletal muscle tissue. Bio Protoc. 2017;7:e2654. doi: 10.21769/BioProtoc.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos M., Backer S., Saintpierre B., Izac B., Andrieu M., Letourneur F., Relaix F., Sotiropoulos A., Maire P. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat. Commun. 2020;11:5102. doi: 10.1038/s41467-020-18789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungan C.M., Murach K.A., Frick K.K., Jones S.R., Crow S.E., Englund D.A., Vechetti I.J., Jr., Figueiredo V.C., Levitan B.M., Satin J. Elevated myonuclear density during skeletal muscle hypertrophy in response to training is reversed during detraining. Am. J. Physiol. Cell Physiol. 2019;316:C649–C654. doi: 10.1152/ajpcell.00050.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmark M., Rana Z.A., Stewart G., Hardie D.G., Gundersen K. De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. J. Physiol. 2007;584:637–650. doi: 10.1113/jphysiol.2007.141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund D.A., Figueiredo V.C., Dungan C.M., Murach K.A., Peck B.D., Petrosino J.M., Brightwell C.R., Dupont A.M., Neal A.C., Fry C.S. Satellite cell depletion disrupts transcriptional coordination and muscle adaptation to exercise. Function. 2020;2:zqaa033. doi: 10.1093/function/zqaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund D.A., Murach K.A., Dungan C.M., Figueiredo V.C., Vechetti I.J., Jr., Dupont-Versteegden E.E., McCarthy J.J., Peterson C.A. Depletion of resident muscle stem cells negatively impacts running volume, physical function, and muscle fiber hypertrophy in response to lifelong physical activity. Am. J. Physiol. Cell Physiol. 2020;318:C1178–C1188. doi: 10.1152/ajpcell.00090.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo V.C., Wen Y., Alkner B., Fernandez-Gonzalo R., Norrbom J., Vechetti I.J., Jr., Valentino T., Mobley C.B., Zentner G.E., Peterson C.A. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J. Physiol. 2021;599:3363–3384. doi: 10.1113/JP281244. [DOI] [PubMed] [Google Scholar]
- Fry C.S., Kirby T.J., Kosmac K., McCarthy J.J., Peterson C.A. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell. 2017;20:56–69. doi: 10.1016/j.stem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry C.S., Lee J.D., Jackson J.R., Kirby T.J., Stasko S.A., Liu H., Dupont-Versteegden E.E., McCarthy J.J., Peterson C.A. Regulation of the muscle fiber microenvironment by activated satellite cells during hypertrophy. FASEB J. 2014;28:1654–1665. doi: 10.1096/fj.13-239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado M.B., Costa M.W., Pranoto E.A., Salimova E., Pinto A.R., Lam N.T., Park A., Snider P., Chandran A., Harvey R.P. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ. Res. 2014;114:1422–1434. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.M., Chi M.M., Lowry O.H., Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J. Cell Biol. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.M., Koishi K., Rudnicki M., Maggs A.M. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mech. Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Hughes S.M., Taylor J.M., Tapscott S.J., Gurley C.M., Carter W.J., Peterson C.A. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Iwata M., Englund D.A., Wen Y., Dungan C.M., Murach K.A., Vechetti I.J., Jr., Mobley C.B., Peterson C.A., McCarthy J.J. A novel tetracycline-responsive transgenic mouse strain for skeletal muscle-specific gene expression. Skelet. Muscle. 2018;8:33. doi: 10.1186/s13395-018-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Franke V., Brandt B., Lowenstein E.D., Schowel V., Spuler S., Akalin A., Birchmeier C. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat. Commun. 2020;11:6375. doi: 10.1038/s41467-020-20064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T.J., Patel R.M., McClintock T.S., Dupont-Versteegden E.E., Peterson C.A., McCarthy J.J. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Mol. Biol. Cell. 2016;27:788–798. doi: 10.1091/mbc.E15-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lonnerberg P., Furlan A. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londhe P., Davie J.K. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet. Muscle. 2011;1:14. doi: 10.1186/2044-5040-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masschelein E., D'Hulst G., Zvick J., Hinte L., Soro-Arnaiz I., Gorski T., von Meyenn F., Bar-Nur O., De Bock K. Exercise promotes satellite cell contribution to myofibers in a load-dependent manner. Skelet. Muscle. 2020;10:21. doi: 10.1186/s13395-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavoungou L.O., Neuenschwander S., Pham U., Iyer P.S., Mermod N. Characterization of mesoangioblast cell fate and improved promyogenic potential of a satellite cell-like subpopulation upon transplantation in dystrophic murine muscles. Stem Cell Res. 2019;41:101619. doi: 10.1016/j.scr.2019.101619. [DOI] [PubMed] [Google Scholar]
- McCarthy J.J., Mula J., Miyazaki M., Erfani R., Garrison K., Farooqui A.B., Srikuea R., Lawson B.A., Grimes B., Keller C. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138:3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows E., Cho J.H., Flynn J.M., Klein W.H. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev. Biol. 2008;322:406–414. doi: 10.1016/j.ydbio.2008.07.024. [DOI] [PubMed] [Google Scholar]
- Messina G., Sirabella D., Monteverde S., Galvez B.G., Tonlorenzi R., Schnapp E., De Angelis L., Brunelli S., Relaix F., Buckingham M. Skeletal muscle differentiation of embryonic mesoangioblasts requires pax3 activity. Stem Cells. 2009;27:157–164. doi: 10.1634/stemcells.2008-0503. [DOI] [PubMed] [Google Scholar]
- Millay D.P., Gamage D.G., Quinn M.E., Min Y.L., Mitani Y., Bassel-Duby R., Olson E.N. Structure-function analysis of myomaker domains required for myoblast fusion. Proc. Natl. Acad. Sci. U S A. 2016;113:2116–2121. doi: 10.1073/pnas.1600101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay D.P., O'Rourke J.R., Sutherland L.B., Bezprozvannaya S., Shelton J.M., Bassel-Duby R., Olson E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay D.P., Sutherland L.B., Bassel-Duby R., Olson E.N. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28:1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti I., Ciciliot S., Dyar K.A., Abraham R., Murgia M., Agatea L., Akimoto T., Bicciato S., Forcato M., Pierre P. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nat. Commun. 2016;7:12397. doi: 10.1038/ncomms12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou T., Deng W., Gu F., Pawitan Y., Vu T.N. Reproducibility of methods to detect differentially expressed genes from single-cell RNA sequencing. Front. Genet. 2019;10:1331. doi: 10.3389/fgene.2019.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach K.A., Dungan C.M., Kosmac K., Voigt T.B., Tourville T.W., Miller M.S., Bamman M.M., Peterson C.A., Toth M.J. Fiber typing human skeletal muscle with fluorescent immunohistochemistry. J. Appl. Physiol. (1985) 2019;127:1632–1639. doi: 10.1152/japplphysiol.00624.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach K.A., Fry C.S., Kirby T.J., Jackson J.R., Lee J.D., White S.H., Dupont-Versteegden E.E., McCarthy J.J., Peterson C.A. Starring or supporting role? Satellite cells and skeletal muscle fiber size regulation. Physiology (Bethesda) 2018;33:26–38. doi: 10.1152/physiol.00019.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach K.A., Peck B.D., Policastro R.A., Vechetti I.J., Van Pelt D.W., Dungan C.M., Denes L.T., Fu X., Brightwell C.R., Zentner G.E. Early satellite cell communication creates a permissive environment for long-term muscle growth. iScience. 2021;24:102372. doi: 10.1016/j.isci.2021.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach K.A., Vechetti I.J., Jr., Van Pelt D.W., Crow S.E., Dungan C.M., Figueiredo V.C., Kosmac K., Fu X., Richards C.I., Fry C.S. Fusion-independent satellite cell communication to muscle fibers during load-induced hypertrophy. Function (Oxf) 2020;1:zqaa009. doi: 10.1093/function/zqaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H.S., Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrany M.J., Swoboda C.O., Sun C., Chetal K., Chen X., Weirauch M.T., Salomonis N., Millay D.P. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 2020;11:6374. doi: 10.1038/s41467-020-20063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Martinez A., Cenik B.K., Bezprozvannaya S., Chen B., Bassel-Duby R., Liu N., Olson E.N. KLHL41 stabilizes skeletal muscle sarcomeres by nonproteolytic ubiquitination. Elife. 2017;6:e26439. doi: 10.7554/eLife.26439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M.M., Runci A., Polletta L., Carnevale I., Morgante E., Foglio E., Arcangeli T., Sansone L., Russo M.A., Tafani M. Muscle LIM protein/CSRP3: a mechanosensor with a role in autophagy. Cell Death Discov. 2015;1:15014. doi: 10.1038/cddiscovery.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- Teschendorff A.E., Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell's transcriptome. Nat. Commun. 2017;8:15599. doi: 10.1038/ncomms15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E., Severini S. Increased entropy of signal transduction in the cancer metastasis phenotype. BMC Syst. Biol. 2010;4:104. doi: 10.1186/1752-0509-4-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velthoven C.T.J., Rando T.A. Stem cell quiescence: dynamism, restraint, and cellular idling. Cell Stem Cell. 2019;24:213–225. doi: 10.1016/j.stem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Walden F., Rea M., Mobley C.B., Fondufe-Mittendorf Y., McCarthy J.J., Peterson C.A., Murach K.A. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics. 2020;15:1151–1162. doi: 10.1080/15592294.2020.1755581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Gottgens B., Rajewsky N., Simon L., Theis F.J. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel N., Axsom J., Yang Y., Li L., Rhoades J.H., Arany Z. Cardiac endothelial cells maintain open chromatin and expression of cardiomyocyte myofibrillar genes. Elife. 2020;9:e55730. doi: 10.7554/eLife.55730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.N., Ren Y., Chen J.Q., Wang Y.Z. Effects of myogenin on muscle fiber types and key metabolic enzymes in gene transfer mice and C2C12 myoblasts. Gene. 2013;532:246–252. doi: 10.1016/j.gene.2013.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cluster marker genes. Sheet 1: Marker genes for each cluster in sedentary soleus and plantaris myonuclei. Sheet 2: Sub-clustering of the MTJ cluster and marker genes for each sub-cluster. Sheet 3: Top 200 markers genes (based on p values) for each cluster after integration of sedentary and exercised datasets. Sheet 4: Differentially expressed genes in the cryptic myonuclear cluster comparing SC- 24hrs post with SC+ 24hrs post last bout of exercise.
Differential gene expression data for the sedentary myonuclei from soleus and plantaris muscles. Sheet 1: Statistical results comparing genes expressed in the secondary type IIa cluster compared with the primary type IIa cluster shown in Figure 1. Sheet 2: Statistical results comparing genes expressed in the secondary type IIa/IIx cluster compared to the primary type IIa/IIx cluster shown in Figure 1 Sheet 3: Statistical results comparing genes expressed in the secondary type IIb cluster compared with the primary type IIb cluster shown in Figure 1. Sheet 4: Summary table of quality control metrics for all the analyzed datasets.
Data Availability Statement
All snRNA-seq datasets are uploaded to the Gene Expression Omnibus (GEO) under the accession number GSE162307 and can be downloaded from the Sequence Read Archive (SRA) under the BioProject number PRJNA681403.