Abstract
A complex assembly of lipids including fatty acids, cholesterol, and ceramides is vital to the integrity of the mammalian epidermal barrier. The formation of this barrier requires oxidation of the substrate fatty acid, linoleic acid (LA), which is initiated by the enzyme 12R-lipoxygenase (LOX). In the epidermis, unoxidized LA is primarily found in long-chain acylceramides termed esterified omega-hydroxy sphingosine (EOS)/phytosphingosine/hydroxysphingosine (collectively EOx). The precise structure and localization of LOX-oxidized EOx in the human epidermis is unknown, as is their regulation in diseases such as psoriasis, one of the most common inflammatory diseases affecting the skin. Here, using precursor LC/MS/MS, we characterized multiple intermediates of EOx, including 9-HODE, 9,10-epoxy-13-HOME, and 9,10,13-TriHOME, in healthy human epidermis likely to be formed via the epidermal LOX pathways. The top layers of the skin contained more LA, 9-HODE, and 9,10,13-TriHOME EOSs, whereas 9,10-epoxy-13-HOME EOS was more prevalent deeper in the stratum corneum. In psoriatic lesions, levels of native EOx and free HODEs and HOMEs were significantly elevated, whereas oxidized species were generally reduced. A transcriptional network analysis of human psoriatic lesions identified significantly elevated expression of the entire biosynthetic/metabolic pathway for oxygenated ceramides, suggesting a regulatory function for EOx lipids in reconstituting epidermal integrity. The role of these new lipids in progression or resolution of psoriasis is currently unknown. We also discovered the central coordinated role of the zinc finger protein transcription factor, ZIC1, in driving the phenotype of this disease. In summary, long-chain oxygenated ceramide metabolism is dysregulated at the lipidomic level in psoriasis, likely driven by the transcriptional differences also observed, and we identified ZIC1 as a potential regulatory target for future therapeutic interventions.
Supplementary key words: ceramides, clinical lipidology, epidermis, lipid metabolism in the skin, lipids/oxidation, lipoxygenase, MS, sphingolipids, EOx, ZIC1
Abbreviations: CID, collision-induced dissociation; EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine; IPA, Ingenuity Pathway Analysis; LA, linoleic acid; LOX, lipoxygenase; NL, neutral loss; NN, healthy control normal; PN, psoriasis normal; PP, psoriasis plaque; SC, stratum corneum; SG, stratum granulosum; SS, stratum spinosum
A complex assembly of lipids including fatty acids, cholesterol, and ceramides is vital to the integrity of the mammalian epidermal barrier. These provide a hydrophobic environment to limit transepidermal water loss and ingress, prevent pathogen invasion, and support a stable microbiome. Correct processing of lipids in keratinocytes deeper in the skin requires several enzymes that include the gene products of ABCA12, transglutaminases (TGM1, TGM3), PLNPLA1, CYP4F22, SDR9C7, and the lipoxygenases ALOX12B (12R-lipoxygenase [LOX]) and ALOXE3 (eLOX3) (1, 2, 3). The importance of the two LOXs is underscored by inactivating mutations in either genes leading to nonbullous ichthyosiform erythroderma, a form of ichthyosis characterized by an increase in transepidermal water loss, compensatory hyperkeratosis, and a general dry, scaly skin (4). Furthermore, 12R-LOX- and eLOX3-deficient mice display a profound skin phenotype, characterized by severe transepidermal water loss leading to death within hours of birth (5, 6, 7). As a corollary to the human phenotype, grafting of 12R-LOX-deficient skin onto nude mice leads to an ichthyosiform phenotype consistent with nonbullous ichthyosiform erythroderma in humans (8). These structural disruptions caused by inactivation of the LOX enzymes highlight the importance of the 12R-LOX pathway and the LOX-derived lipids, but there is still little information on either the products involved or the regulatory role they play in epidermal homeostasis.
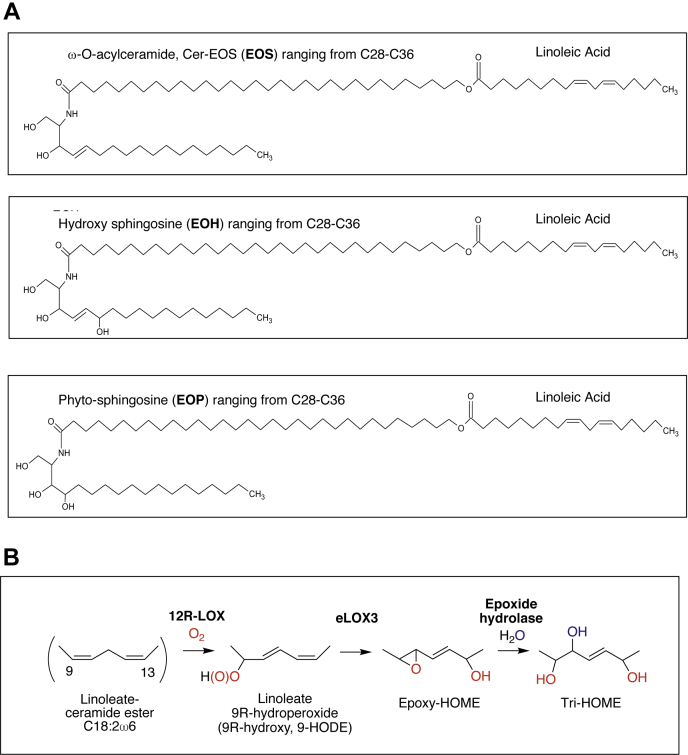
The unsaturated fatty acid substrate of the LOX pathway in the skin is well established as linoleic acid (LA) (18:2), which is esterified to the skin-specific ω-O-acylceramide, Cer-esterified omega-hydroxy sphingosine (EOS) (9). The structure of Cer-EOS comprises an ultra-long N-linked fatty acid (C28 – 36) omega-hydroxylated and ester-linked mainly with LA (Scheme 1A). 12R-LOX and eLOX3 act sequentially on the esterified LA ester in EOS-type ceramides, which comprise both EOS itself and its two common variants, hydroxysphingosine (EOH) and phytosphingosine (EOP), grouped together in this article as EOx (Scheme 1A). Formation of the 9R-hydroperoxide of the LA ester by 12R-LOX is followed by eLOX3-catalyzed isomerization to a 9,10-epoxy-13-hydroxy derivative, the hydrolysis of which yields several trihydroxy products, as demonstrated previously (Scheme 1B) (10). After hydrolysis from the parent ceramides, oxidized LA products have been identified as 9,10,13- and 9,12,13-triHOMEs and biochemical studies using epoxide hydrolases identified the predominance of the 9R,10S,13R-triHOME in human epidermis (11, 12). A recent study showed that triHOME is formed in atopic dermatitis (13). Recently, free acid triHOMEs from eosinophils have also been described. These are generated via 15-LOX, yielding many isomeric species (14). Up to now, triHOMEs have only been characterized as free fatty acids after hydrolysis of epidermal extracts, and the full-length oxidized ceramides were not determined.
Scheme 1.
Representative structures of EOx ceramides including EOS, EOH, and EOP (A) and epidermal lipoxygenase oxidation pathway (B). EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine.
The molecular structures and functions of oxygenated ceramides generated by skin LOXs, from any forms of EOx are not yet known. Indeed, information on the full range of oxidized and substrate ceramide molecular species in human epidermis, their location within the epidermal strata, and their amounts in health and disease is lacking. To address this, we developed new LC/MS/MS methods to enable preliminary identification of large groups of LOX-oxidized EOx ceramides in human epidermis. Over 80 different EOx molecular species were first identified using precursor LC/MS/MS. EOx ceramides were quantified in human epidermis and their localization in human skin in health and disease investigated. A transcriptional analysis of psoriatic skin was then undertaken to examine expression of the genes regulating these lipids.
Materials and methods
Human epidermis isolation and lipid extraction
Full-thickness human breast skin was obtained with informed consent and ethical approval (South East Wales Research Ethics Committee Ref. 08/WSE03/55). Subcutaneous fat was removed by dissection and epidermis isolated via heat separation in a water bath at 60°C for 30 s. The skin was dried, and the epidermis peeled from the underlying tissue. Epidermis was weighed before being homogenized on ice in chloroform:methanol (2:1) until a smooth milky solution was obtained. This was sonicated using a Virsonic sonic probe. After centrifugation (1,500 rpm, 15 min), the organic phase was removed. Chloroform:methanol (2:1) was added to the pellet, followed by a second round of sonication and centrifugation. This was repeated four times and the lipid fractions combined. Internal standards D31-ceramide(N-palmitoyl-d31-D-erythro-sphingosine) (Avanti Polar Lipids) (25 ng) and 12-HETE-d8, 13-HODE-d4, and PGE2-d4 (Cayman Chemical) (10 ng) were added, and the lipids were extracted from the homogenization solution using the Bligh and Dyer method (15). The lipid extract was dried before being loaded onto a pre-equilibrated solid phase silica column in chloroform/hexane (1:1, v/v) (HF Bond Elut SI, Agilent), washed with chloroform/hexane (1:1, v/v), and eluted with chloroform and chloroform/methanol (9:1, v/v). Samples were then taken to dryness, then resuspended in methanol:THF (3:2), and then stored at −80°C before LC/MS/MS analysis as outlined below.
Tape stripping of healthy human volunteer skin and lipid extraction for depth profiling
Tape strip samples were acquired from the volar forearm of healthy volunteers (n = 5) under Cardiff University, School of Medicine, ethical approval (Characterising oxidised lipids in human epidermis–Response REF 13.38). Briefly, tape strips (D-Squame™, 1 cm diameter) were applied to the left volar forearm with gentle pressure and then swiftly removed with sterile tweezers and placed into 3 ml CHCl3: methanol (2:1). In total, 30 strips were taken (sequentially from the same site) and pooled into groups of three, in the same solution. The samples were vortexed thoroughly and placed at −20°C overnight followed by centrifugation at 2000 rpm at 4°C for 10 min. Internal standards were added as above and mixed before the lipid extract was removed from the remaining glue matrix (which remained on surface of liquid) and plastic strip, dried under N2 and resuspended in 1 ml THF:methanol (3:2) for MS analysis as outlined below.
Epidermal analysis of psoriatic patients and healthy controls
Patients with psoriasis were recruited via the Department of Dermatology, University Hospital of Wales, under informed consent and ethical approval from Cardiff and Vale University Health Board and the Wales Research Ethics Committee 1 (reference 15/WA/0285) (n = 9; 5 male and 4 female patients, aged 34–83 years). Healthy controls (differing from those detailed above) were recruited via advert and had no history of psoriasis (n = 5; 3 male and 2 female patients, aged 50–62 years) (supplemental Table S1). Racial composition was predominantly Caucasian, which reflects the demographics of our local area; thus, there was no significant difference in ethnicity between the groups. Data on smoking and comorbidities were not collected, so we are unable to compare specifically between groups, although we note that patients tended to be on a large number of medications including for cardiovascular prevention as well as psoriasis. Gender was evenly spread (55% or 60% male for patients and control groups, respectively), although the patient group had a wider age range. At the time of sampling, patients' lesions had no topical medication applied. We note that our groups are rather small and thus we cannot analyze for confounding factors. Tape strip samples were acquired from both the volar forearm (uninvolved skin in psoriatic patients) and elbow (skin exhibiting psoriatic phenotype in patients). In total, 9 strips were taken and pooled into groups of 3. Samples were taken from healthy participants at the same sites, forearm/elbow (left or right) as controls for psoriatic patients. Internal standards D31-ceramide (25 ng) and 12-HETE-d8, 13-HODE-d4, and PGE2-d4 (10 ng) were added and lipids extracted as described for healthy volunteers above. All human studies abide by the Declaration of Helsinki principles.
Oxylipin quantitation using LC/MS/MS
Lipids were separated on a C18 Spherisorb ODS2, 5 μm, 150 × 4.6 mm column (Waters, Hertfordshire, UK) using a gradient of 50%–90% B over 10 min (A, water:acetonitrile:acetic acid, 75:25:0.1; B, methanol:acetonitrile:acetic acid, 60:40:0.1) with a flow rate of 1 ml min−1. Products were quantified by LC/MS/MS ESI on an ABSciex 6500 Q-Trap using precursor-to-product transitions as in supplemental Table S2. Source parameters were declustering potential −20V, collision energy −42V, collision cell exit potential 13V, curtain gas 35, ionSpray voltage −4,500, source temperature 350°C, nebuliser gas 40, and heating gas 30. Lipids were identified and quantified using standard curves for specific positional isomers and appropriate deuterated standards run in parallel under the same conditions (10 ng of 12-HETE-d8, 13-HODE-d4, PGE2-d4). Limit of detection/limit of quantitation was based on signal:noise of at least 5:1 with at least 6 points across a peak.
LC/MS/MS of ceramides
Lipid extracts were separated by reverse-phase HPLC using a Waters Spherisorb ODS2 3 μm C18 (2) 150 × 2-mm column (Waters, Hertfordshire, UK) with a gradient of 20%–0% A over 5 min followed by 50 min at 100% B (A: water, 5 mM ammonium acetate; B: methanol:THF, 80:20, 5 mM ammonium acetate) using a flow rate of 200 μl min−1. Lipids were analyzed using an ABSciex 6500 Q-Trap with a Shimadzu UPLC and ESI. Settings were declustering potential −20 V, collision energy −42 V, collision cell exit potential 13 V, curtain gas 35, ionSpray voltage −4,500, source temperature 350°C, nebuliser gas 40, and heating gas 30. For precursor scanning, spectra were acquired scanning Q1 from m/z 1,000–1,350 over 1.75 s with Q3 set to product ion of interest. Supplemental Table S3 shows all multiple reaction monitoring (MRM) transitions with individual parameters with a scan time of 75 ms per transition. Product ion spectra were obtained at the apex of MRM transitions with the MS operating in the ion trap mode. Scans were acquired at 10,000 Da/s, with a dynamic ion trap fill time. The precursor to product MRM transitions was monitored as in supplemental Table S2.
Native EOS, EOP, and EOH ceramide semiquantitation
Molecular ions corresponding to individual ceramides, differing in the carbon number (C30, C32, or C34) and ceramide species, EOS, EOP, and EOH, were detected as acetate adducts ([M-H+60]−). A C30 EOS standard (N-(30-(9Z,12Z-octadecadienoyloxy)-octacosanoyl)-octasphing-4E-enine), a kind gift from Evonik Industries, was used as a reference standard to quantify all native ceramides. N-palmitoyl-d31-D-erythro-sphingosine (25 ng) was added to each sample as the internal standard, to enable an estimate of nanogram ceramide/milligram epidermis (wet weight) or per tape strip number (16).
Oxidized ceramide semiquantitation and quantification of esterified HODEs and triHOMEs
No oxidized ceramide standards are available. Thus, retention times corresponding to individual C30, C32, and C34 HODE/triHOME-containing ceramides were predicted based on the impact of increasing lipophilicity with increasing chain length, and C30 and C32 structures were subsequently analyzed using MS/MS, generating expected product ions for the oxylipin product. N-palmitoyl-d31-D-erythro-sphingosine (25 ng) was added to each sample as the internal standard, and the amount of oxidized ceramides is displayed as a ratio of analyte area:internal standard area. Next, to directly quantify ceramide-esterified HODEs and triHOMEs, fractions were collected from the LC separation between 16 - 19 min (HODE-ceramides) and 12–17 min (triHOME-ceramides). These were subject to base hydrolysis by solubilizing in 1.5 ml isopropanol and 1.5 ml 1 M NaOH overnight at room temperature. The samples were acidified to pH 5, oxylipin deuterated standards added as above followed by extraction from the isopropanol by 3 ml hexane followed by vortex, centrifugation, and recovery of hexane layer. This process was repeated, and the hexane layers combined. Extracts were dried under N2 and resuspended in methanol before quantitation of free HODEs or triHOMEs using LC/MS/MS as described above. The actual amount of total C30-C34 EOx HODE-or triHOME-ceramide was then estimated using the average MW for these species (average MW C30-C34, EOS, EOH, and EOP = 1,125 (HODE EOx ceramides) and 1,159 (triHOME EOx ceramides)). A molar ratio of 1:1 for EOx to free acid oxylipin was applied. Limit of detection/limit of quantitation was based on signal:noise of at least 5:1 with at least 6 points across a peak.
Expression analysis of gene regulation in psoriatic skin
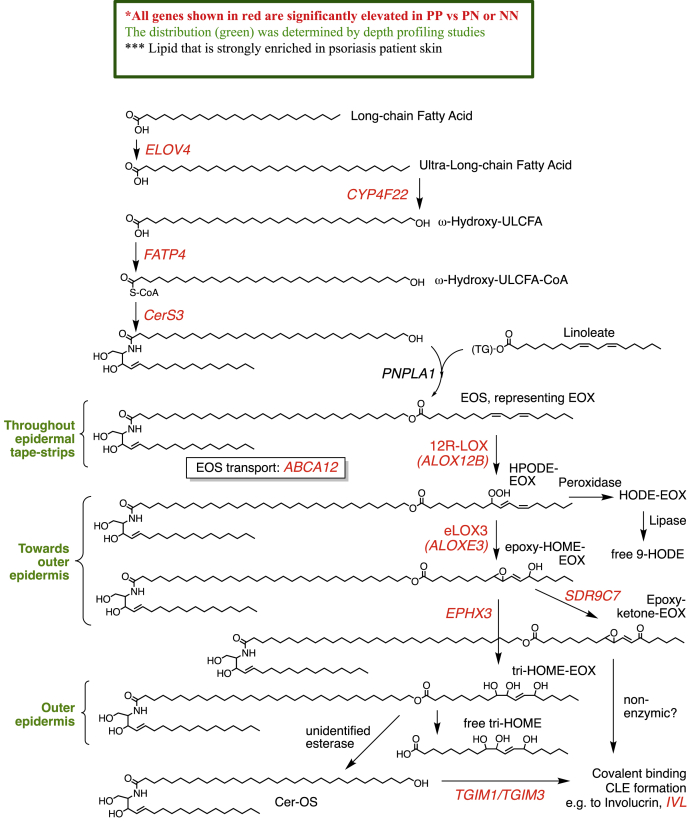
Affymetrix CEL files were downloaded from GEO Database study GSE54456 (17). Study subjects were from the greater Detroit area, and all were Caucasian background. In that study, a set of two biopsies were taken under local anesthesia from each patient as follows; one 6 mm punch biopsy from psoriasis plaque (PP) skin and the other from psoriasis normal (PN) (unaffected) skin, at least 10 cm away from active plaque. Up to 2 biopsies were obtained from separate healthy control normal (NN) skin. The PN and NN skin biopsies were obtained from buttocks or upper thighs in all cases. The full dataset was analyzed in R, using the limma and oligo packages in Bioconductor (18, 19). Genes were corrected using multiple comparison testing (Benjamini-Hochberg). Subsequently, expression data for genes encoding proteins known to be involved in either formation or oxidation of skin ceramides were compared within the patient groups. Pathway analysis was performed in Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, www.ingenuity.com, Redwood City, CA). For network generation, a dataset containing gene identifiers and corresponding expression values was uploaded for significant genes from the ceramide pathway (Scheme 2).
Scheme 2.
Pathway diagram showing the formation and oxidation of long-chain EOS species in human skin. The same enzymes will be responsible for metabolizing EOH and EOP isomers. Gene names in red are all significantly elevated in human psoriatic lesions. EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine.
Statistical analysis
Data are shown as combined experiments, where n represents repeat experiments with combined technical replicates. Human donor skin was cut into 3 sections and each extracted separately. Three separate donors were utilized, resulting in 9 samples; the results from these were averaged and the error reported as ± SEM. For psoriasis tape strips, there were 9 patients and 5 healthy controls. Statistical analysis was performed in GraphPad Prism using a two-tailed Mann-Whitney U test.
Results
Precursor scanning of human epidermal extracts identifies a series of ceramides that contain LA
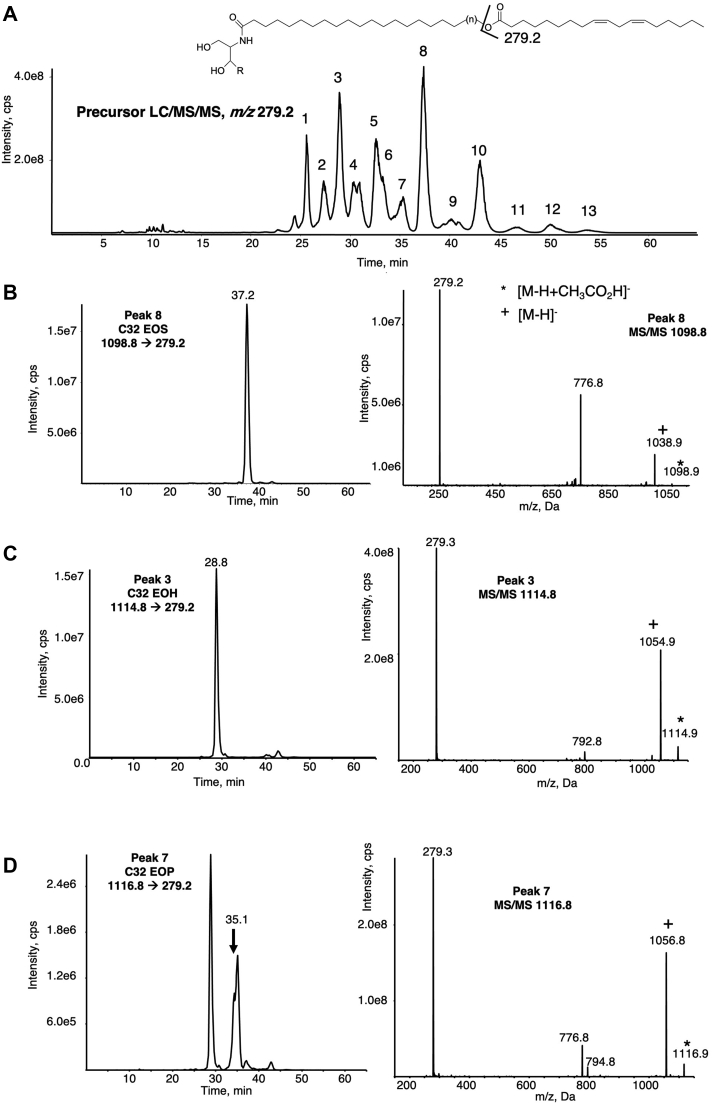
Human epidermal extracts were analyzed using precursor-LC/MS/MS for the LA carboxylate anion (m/z 279.2 [M-H]−). Figure 1A shows a series of ions eluting between 22 and 55 min. MS spectra acquired at the apex of each peak show precursor ions that correspond to putative LA-containing ceramides. This represents multiple species in the range of C28-C36 EOS, EOP, and EOH, with the order of elution corresponding to lipophilicity of the ceramide functional groups and the length of the N-linked fatty acyl (Scheme 1, supplemental Fig. S1). Peak 1 has been putatively assigned (based on parent mass) as C30 EOH at m/z 1086.9 [M-H+CH3CO2H]− (also detected as m/z 1026.9 [M-H]−) (see peak 1 in supplemental Fig. S1). Peak 2 indicates the less polar (more lipophilic) C31 EOH ceramide (m/z 1100.9 [M-H+CH3CO2H]−, m/z 1040.9 [M-H]−). This pattern continues with increasing retention time through Peaks 3–13 (supplemental Fig. S1). Next, lipid extracts were analyzed in the MRM mode for EOS, EOH, and EOP C30, C32, and C34 species, using the product ion for LA [M-H]− at m/z 279.2 and the [M-H+CH3CO2H]− precursor ion. C32 EOS, EOH, and EOP species are shown in Fig. 1B–D, respectively. Here, peaks are seen for each lipid and a representative MS/MS spectrum for each demonstrates the expected product ions including the [M-H+CH3CO2H]− acetate adduct (marked with ∗) and also [M-H]− (marked with +), m/z 279.2 (fatty acyl carboxylate from cleavage of the ω-hydroxylated LA) (Fig. 1B–D). Supplemental Figure S2 illustrates the MS/MS of C30 EOS (panel A), EOH (panel B), and EOP (panel C) ceramides. MRM chromatograms for C34 EOS (panel A), EOH (panel B), and EOP (panel C) are shown in supplemental Fig. S3, with relative retention times inferring the corresponding structures as indicated. Consistent throughout all spectra are product ions corresponding to neutral loss (NL) 262 amu, representing loss of the LA ketene moiety on collision-induced dissociation (CID). Up to now, most studies on skin ceramides have used the positive-ion mode; however, because we focused mainly on oxidized forms, we needed to use the negative-ion mode to detect and characterize the esterified oxylipin groups, especially during precursor scanning which is required for this purpose. However, there is an absence of commercially available standards for these long-chain acyl-esterified ceramides, so we were only able to obtain synthetic LA-C30 EOS for comparison. As expected, this showed an identical retention time and MS/MS spectrum to the putative LA-C30 EOS detected in the skin (supplemental Fig. S4). High-resolution MS/MS for the standard is also shown in the negative-ion mode (supplemental Fig. S4B, C). As already seen for positive MS of skin ceramides, diagnostic ions were seen for loss of fatty acyl (NL 280 amu at m/z 748.7) and its respective ketene (NL 262 amu at m/z 718) (20).
Fig. 1.
Precursor LC/MS/MS identification of native C32-EOx in the healthy human skin. Human epidermal breast tissue was prepared and analyzed using LC/MS/MS as described in Materials and methods. A: Precursor scan for m/z 279.2 ([M-H]− LA) shows a series of ions eluting between 20 and 55 min run, labeled 1–13. These are identified EOS, EOH, and EOP ceramides with varying very-long-chain fatty acid carbon numbers. B–D: Representative chromatograms and MS/MS spectra of C32 EOS, EOH, and EOP detected as the precursor ([M-H+CH3CO2H]−) → m/z 279.2. MS/MS spectra were acquired in the ion trap mode at the apex of elution for each lipid as described in Materials and methods. EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine.
Precursor scanning identifies a series of ceramide lipids that contain oxidized LA in human epidermal extracts
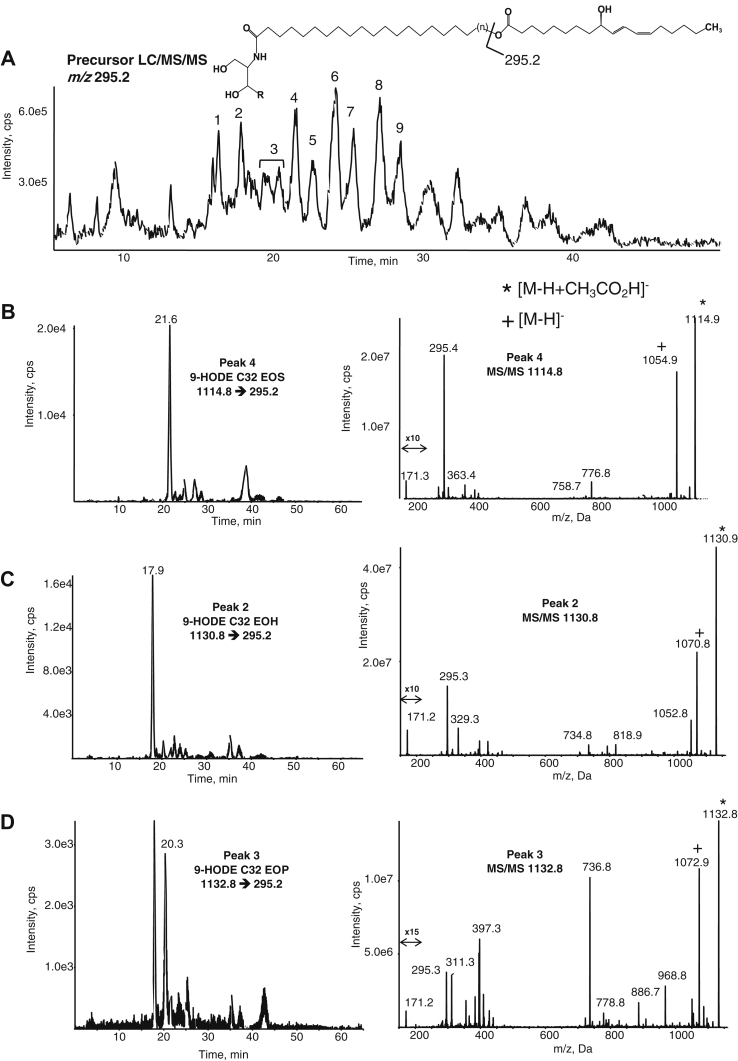
Precursor LC/MS/MS was next used to screen for oxidized EOx putatively generated by 12R-LOX/eLOX3. The product ions used were m/z 311.2 (epoxy-HOME), 295.2 (HODE), and 329.2 (triHOME), [M-H]−. We note that these ions do not specify the position of oxygenation on LA and furthermore do not distinguish the precise oxygenated functional groups at this stage of the analysis.
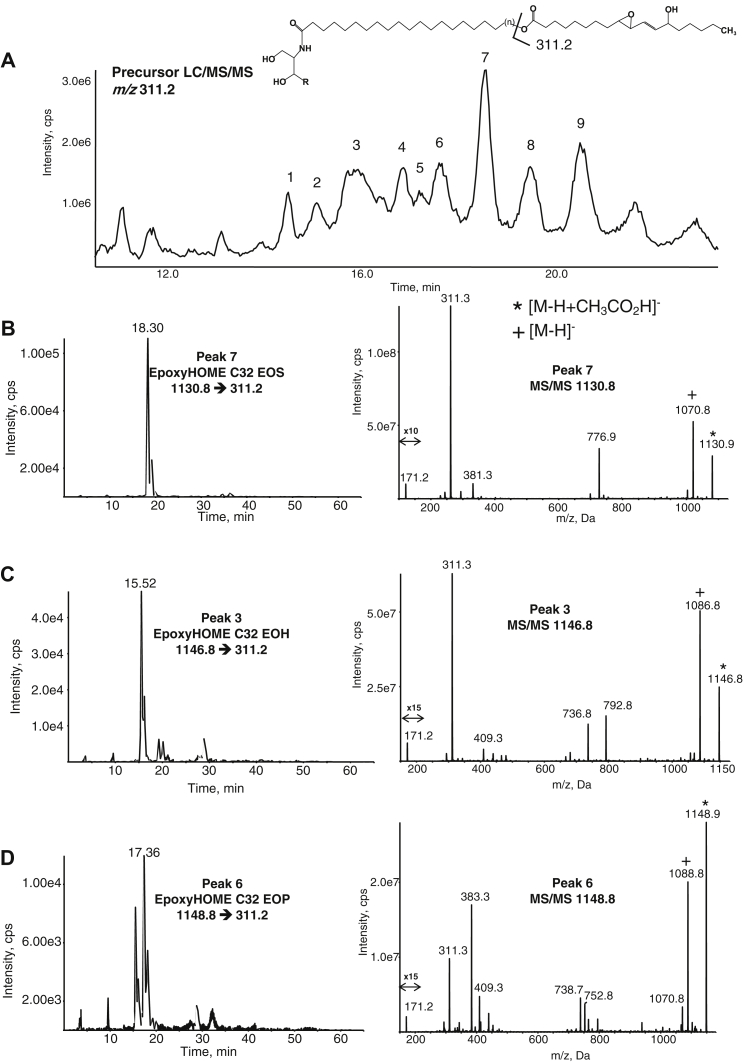
Putative epoxy-HOME-EOS, EOP, and -EOH lipids eluted earlier than native precursors because of reduced lipophilicity from the epoxy-hydroxy functional group, around 14–22 min with nine major peaks (peaks 1–9, Fig. 2A, supplemental Fig. S5). Peak 1 with m/z 1118.9 [M-H+CH3CO2H]− was putatively assigned to the acetate adduct of epoxy-HOME EOH C30 (also seen as m/z 1058.9 [M-H]−) (supplemental Fig. S5). Peak 2, m/z 1132.9 [M-H+CH3CO2H]− was putatively assigned as epoxy-HOME EOH C31 ceramide (supplemental Fig. S5). Peaks 3–9 were then assigned based on parent mass. The chromatograms and product ion spectra for the epoxy-HOME-EOS, EOH, and EOP C32 are shown (Fig. 2B–D, respectively). Similar to the EOx LA substrates, each ceramide species was detected as an [M-H+CH3CO2H]− acetate adduct and also an [M-H]− ion (Fig. 2B–D). The fatty acyl product ion (the ω-hydroxylated esterified epoxy-HOME) is also consistently detected (m/z 311.2 [M-H]−). A m/z 171.2 [M-H]− product ion is also detected for all three indicating the 9,10 epoxide,13 hydroxy positional isomer. Unfortunately, there are currently no available epoxy-HOME standards with which to conduct an MS comparison. However, this finding is consistent with the positional specific oxidation reaction of the 12R-LOX enzyme which ultimately yields the 9,10,13-triHOME (10, 12). Supplemental Figure S6A–C illustrates MS/MS analysis of the epoxy-HOME-EOS (panel A), EOH (panel B), and EOP (panel C) C30 species, and again, these show similar fragmentation. MRM chromatograms for the epoxyHOME-EOS (panel A), EOH (panel B), and EOP (panel C) C34 species are shown in supplemental Fig. S7, with relative retention times inferring the corresponding structures as indicated. For EOH and EOP, the ions were in lower abundance as indicated by the chromatograms. As expected, several epoxyHOME EOx species showed products ions corresponding to NL 294, representing loss of the epoxyHOME ketene moiety on CID.
Fig. 2.
Precursor LC/MS/MS identification of epoxy-HOME EOx in the human skin. Human epidermal breast tissue was prepared and analyzed as described in Materials and methods. A: Precursor scan for m/z 311.2 ([M-H]− for epoxy-HOME) shows a series of ions eluting between 14 and 22 min, labeled 1–9. These are identified as EOS, EOH, and EOP epoxy-HOME ceramides with varying very-long-chain fatty acid carbon numbers. B–D: Representative chromatograms and MS/MS spectra of C32 EOS, EOH, and EOP epoxy-HOME ceramides detected as the precursor ([M-H+CH3CO2H]−) → 311.2. MS/MS spectra were acquired in the ion trap mode at the apex of elution for each lipid as described. EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine.
Precursor scanning LC/MS/MS for m/z 295.2, corresponding to HODE-EOx, was undertaken and identified a series of EOS, EOP, and EOH species (Fig. 3A, supplemental Fig. S8). As before, a consistent pattern in putative assignments is seen in peaks 1–9. The LC/MS/MS and product ion spectra for HODE-EOS, EOH, and EOP C32 are shown (Fig. 3 B–D). A m/z 171.2 [M-H]− daughter ion corresponding to fragmentation of the oxidized LA between C9-C10 is seen (consistent with 9-HODE), supporting C9 oxidation of linoleate via 12R-LOX. Supplemental Fig. S9A–C illustrates MS/MS analysis of the HODE-EOS (panel A), EOH (panel B), and EOP (panel C) C30 species and again, these show daughter ion fragments corresponding to 9-HODE. The MRM chromatograms for the HODE-EOS (panel A), EOH (panel B), and EOP (panel C) C34 species are shown in supplemental Fig. S10, with relative retention times inferring the corresponding structures as indicated. As expected, several HODE EOx species showed product ions corresponding to NL 278 (loss of HODE ketene) and sometimes NL 296 (loss of HODE) on CID.
Fig. 3.
Precursor LC/MS/MS identification of HODE EOx in the human skin. Human epidermal breast tissue was prepared and analyzed as described in Materials and methods. A: Precursor scan for m/z 295.2 ([M-H]− for HODE) shows a series of ions eluting between 15 and 30 min, labeled 1–9. These are identified as EOS, EOH, and EOP epoxy-HODE ceramides with varying very-long-chain fatty acid carbon numbers. B–D: Representative chromatograms and MS/MS spectra of C32 EOS, EOH, and EOP HODE ceramides detected as precursor ([M-H+CH3CO2H]−) → 295.2. MS/MS spectra were acquired in the ion trap mode at the apex of elution for each lipid as described. EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine.
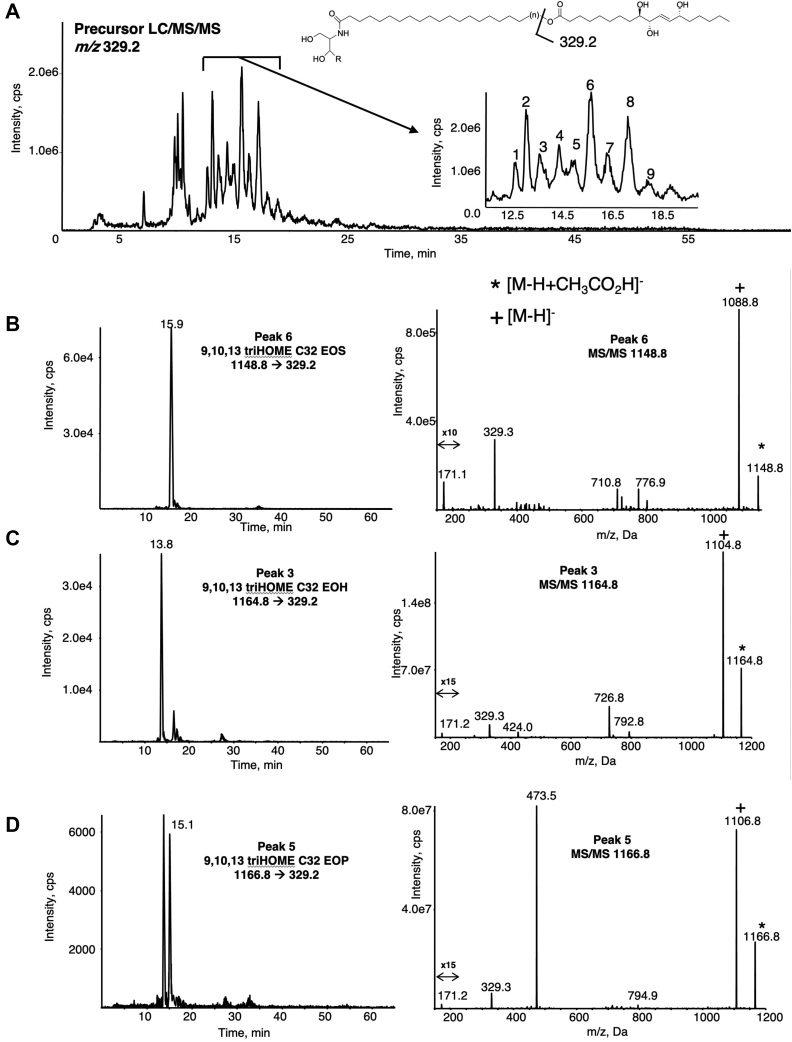
Putative triHOME-EOx lipids eluted earlier than the other oxidized EOx, because of a further reduction in lipophilicity from the triol functional group, around 12–17 min (Fig. 4, supplemental Fig. S11). Peak 1 with m/z 1136.9 [M-H+CH3CO2H]− corresponded to the acetate adduct of triHOME EOH C30 (also seen as m/z 1076.9 [M-H]−) (supplemental Fig. S11). Peak 2 at m/z 1150.9 [M-H+CH3CO2H]− corresponds to the trihydroxy-LA C31 EOH ceramide (supplemental Fig. S11). Overall, the pattern of elution mirrors the ceramides described in Fig. 1, Fig. 2, Fig. 3. LC/MS/MS, monitoring precursor [M-H+CH3CO2H]− to triHOME carboxylate anion corresponding to EOS, EOH, and EOP C32, is shown (Fig. 4B–D). Similar to the EOx LA substrates, each ceramide species was detected as an [M-H+CH3CO2H]− acetate adduct and also an [M-H]− ion (Fig. 4B–D). The fatty acyl product ion (the ω-hydroxylated esterified trihydroxy-LA) is also consistently detected (m/z 329.2 [M-H]−). A m/z 171.2 [M-H]− product ion is seen, suggesting the 9,10,13-triHOME isomer. To further confirm this, MS/MS of 9,10,13- and 9,12,13-triHOME free acids was undertaken for comparison (supplemental Fig. S11A, B). Here, m/z 171 is seen for both positional isomers; however, it is in very low abundance in the 9,12,13-isomer, where instead two ions at m/z 211 and 229 predominate. These ions were not detected in triHOME EOx lipids, which only contained the m/z 171 fragment. This confirms the isomer in the ceramides as the 9,10,13-form (supplemental Fig. S12) and is consistent with the positional specific oxidation reaction of the epidermal 12R-LOX enzyme that has been previously found to yield the 9,10,13 triHOME (10, 12). Finally, many of the triHOME-EOx species yielded characteristic daughter ions for the NL of the ketene (loss of 312 amu). The presence of multiple triHOME-EOx lipids suggests that 12R-LOX and eLOX3 utilize many different epidermal ceramides to yield a large number of oxidized, highly polar products. Supplemental Fig. S13A–C illustrates MS/MS analysis of the triHOME-EOS (panel A), EOH (panel B), and EOP (panel C) C30 species, and again, these show daughter ion fragments (171.2) corresponding to 9,10,13 triHOME. The MRM chromatograms for the triHOME-EOS (panel A), EOH (panel B), and EOP (panel C) C34 species are shown in supplemental Fig. S14.
Fig. 4.
Precursor LC/MS/MS identification of triHOME EOx in the human skin. Human epidermal breast tissue was prepared and analyzed as described in Materials and methods. A: Precursor scan for m/z 329.2 ([M-H]− for triHOME) shows a series of ions eluting between 12 and 18 min, labeled 1–9. These are identified as EOS, EOH, and EOP triHOME ceramides with varying very-long-chain fatty acid carbon numbers. B–D: Representative chromatograms and MS/MS spectra of C32 EOS, EOH, and EOP triHOME ceramides detected as the precursor ([M-H+CH3CO2H]−) → 329.2. MS/MS spectra were acquired in the ion trap mode at the apex of elution for each lipid as described. EOH, hydroxysphingosine; EOP, phytosphingosine; EOS, esterified omega-hydroxy sphingosine; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine.
Semiquantitation of triHOME, HODE fatty acids esterified in EOx ceramides, and free oxylipins
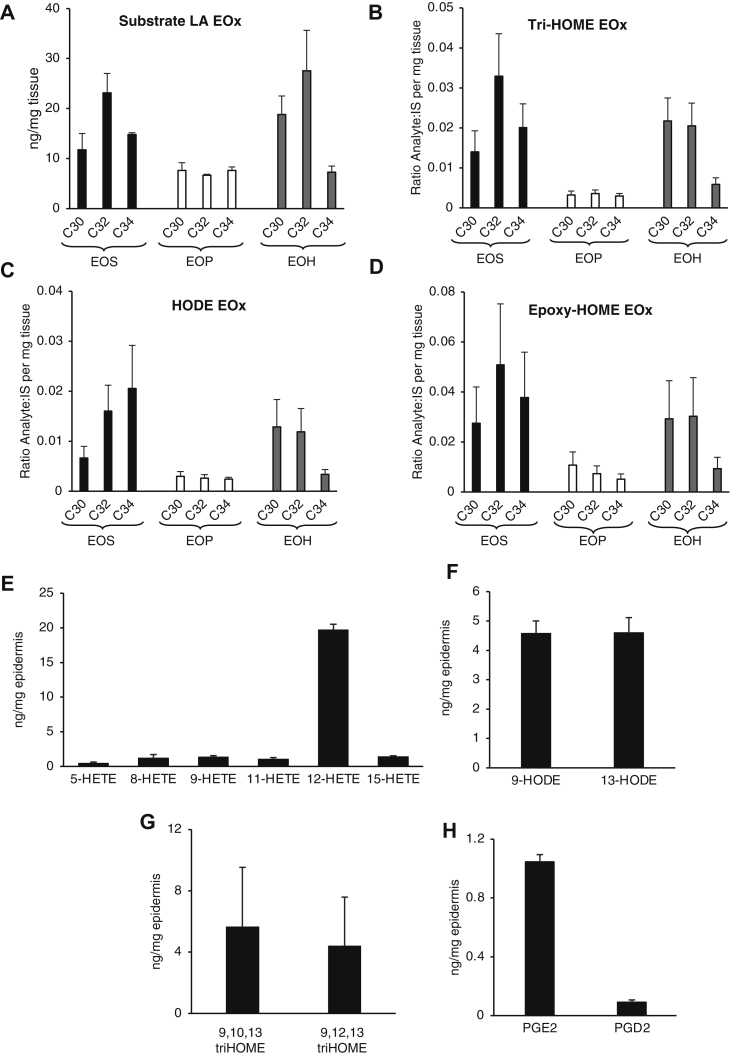
LA-EOS, EOH, and EOP molecular species containing an ultra-long N-linked fatty acid of C30-C34 carbons were semiquantified in healthy human epidermal tissue using LC/MS/MS. Standards for EOP and EOH are not available, and we could only obtain one C32 EOS. Thus, to allow semiquantitation, this lipid was used as the primary standard. We acknowledge some limitations with this approach which include (i) longer and shorter ceramide chains may show differing ionization efficiency and (ii) EOS may ionize differently to EOP and EOH. We note that for all EOx analyzed, the LA carboxylate anion was used as the product ion because we expect fragmentation to generate this to be similar for all lipids analyzed. Of the three ceramide families, EOH appeared the most abundant followed by EOS and then EOP (Fig. 5A). EOS and EOH species showed a similar relative abundance of molecular species in relation to the length of the very long N-linked carbon chain of the ceramide. Generally, the C32 was most abundant, with lower levels of C30 and C34. EOP ceramides showed similar levels of C30, C32, and C34. Because no purified triHOME-containing, HODE-containing, or epoxy-HOME–containing ceramides are available, these are shown as relative to internal standard, normalized per milligram tissue (Fig. 5B–D). The same relative distribution of subspecies based on the carbon chain length is seen for these as for native LA-containing ceramides (Fig. 5A–D). This indicates that the LOX pathway likely metabolizes ceramide substrates based on their availability with no preference for any specific ceramide chain length. As a second approach to quantitation, levels of triHOME and HODEs in EOx ceramides were determined after hydrolysis of HPLC-purified EOx generated from epidermal lipid extracts. These values were next used to generate an estimate of total EOx amounts in the skin containing these oxylipins. Here, we estimate ~118 ± 18.1 and 104 ± 2.1 ng/mg epidermis for total 9,10-13-triHOME- and 9-HODE-EOx, respectively (n = 3 ± SEM). These high levels relative to total LA-EOx detected (173 ng/mg epidermis) (quantified as per Fig. 5) suggest that metabolism of LA-EOx substrate to oxygenated products is highly favored in healthy human skin.
Fig. 5.
Analysis of triHOME, HODE, and epoxy-HOME EOx ceramides and free oxylipins in the healthy human epidermis. All lipids were extracted and analyzed using LC/MS/MS from human epidermal breast tissue as described in Materials and methods (n = 9, mean ± SEM). A: LA-EOx-LA ceramide lipids in the healthy epidermis. B–D: Epidermis levels of EOx oxidized ceramides in the healthy human epidermis. E–H: Free oxylipin quantification in the healthy epidermis. LA, linoleic acid; EOx, omega-hydroxy sphingosine/phytosphingosine/hydroxysphingosine.
Free acid oxylipins were next quantified using LC/MS/MS, so that quantitative comparisons with EOx forms could be made. Only a few eicosanoids and prostaglandins were detected (Fig. 5E–H). 12-HETE (19.7 ng/mg) and 13-HODE (4.6 ng/mg) are likely originating from 12R-LOX and/or 15-LOX-2 in the epidermis (21). Levels of 9-HODE and 13-HODE are equivalent (4.6 ng/mg); however, 9-HODE would be expected to be generated from hydrolysis from ceramides as human epidermal 12R-LOX prefers esterified linoleate as the substrate (22). Standards used for analysis were all R enantiomers as the products were expected to be derived from the 12R-LOX pathway. However, the oxylipins quantified here could be a combination of S/R enantiomers as there would be no discernible difference in the retention time during the HPLC analysis. Indeed, it has been shown that 12S-HETE is also present in the epidermal tissue (23). 9,12,13-triHOME (4.4 ng/mg) and 9,10,13-triHOME (5.6 ng/mg) were detected in far lower amounts than their corresponding ceramide esters (calculated as 118 ng/mg tissue). Free 9,10-epoxy-13-hydroxy linoleate was not analyzed as there is no internal standard available. The cyclooxygenase pathway products PGE2 and PGD2 were also detected at low levels, as previously reported (24). Overall, this indicates that triHOMEs and HODEs predominate in the skin as esterified forms in EOx.
TriHOME-EOx are concentrated in the upper epidermis, whereas all other EOx show enrichment in deeper layers
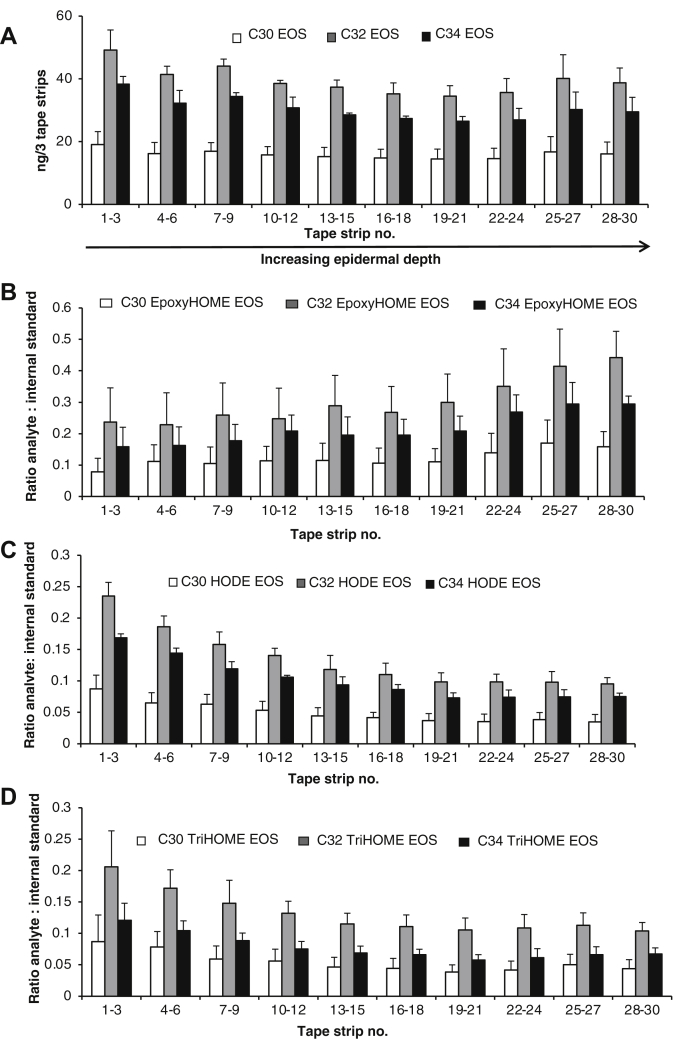
12R-LOX is primarily found in the lowest layers of the epidermis, including the stratum basale, stratum spinosum (SS), and lower stratum granulosum (SG), with the stratum basale being the lowest followed by the SS and so forth. eLOX3 is expressed throughout the epidermis (25). The location of oxidized EOx generation in the skin, however, is currently unknown (6, 9, 25). To determine this, we applied 30 tape strips to the same area of the forearm to repeatedly remove one layer of skin cells at a time. Because the uppermost layer of the skin, the stratum corneum (SC), is approximately 15 layers thick, the use of 30 strips should achieve penetration into the deeper SG and possibly the SS of the epidermis. We first determined that native LA-EOx ceramides are fairly evenly localized throughout the epidermal depth (Fig. 6A, supplemental Figs. S15A, S16A). In contrast, epoxy-HOME-ceramides (generated by 12R-LOX/eLOX3, Scheme 1) increase in abundance deeper into the skin (strips 22–30) (Fig. 6B, supplemental Fig. S16B). Of note, epoxy-HOME EOP products were below the limit of detection. Conversely, HODE-EOx and triHOME-EOx were more abundant closer to the skin surface, strips 1–18, and therefore likely to be in the upper SG and the SC (Fig. 6C, D, supplemental Figs. S15B, C, S16C, D). This is consistent with the location of the 12R-LOX being in the lower layers of the epidermis as the initial product is generated before rapid eLOX3 generation of the epoxy-HOME and implies that the enzyme responsible for the hydrolysis of the epoxide (recently identified as EH3) to yield the triHOME is expressed higher in the epidermis (likely the upper SG and SC) (26). Of relevance, this coincides with keratinocyte differentiation and markers associated with CLE formation, including involucrin, transglutaminase, loricrin, and filaggrin supporting a role for these ceramides in these events (27).
Fig. 6.
Human epidermis shows higher levels of triHOME and HODE-EOS and reduced levels of epoxyHOME-EOS in upper layers. Tape strips were acquired from the volar forearm of healthy volunteers (n = 5 subjects, 30 strips per person, mean ± SEM). Strips were combined into groups of 3 and lipids extracted and analyzed using LC/MS/MS as described in Materials and methods for total levels of lipid present. A: LA-EOS is evenly distributed throughout the skin layers. B: Epoxy-HOME EOS increases with greater epidermal depth and is the highest in the stratum granulosum (SG)/stratum spinosum (SS) layers. C and D: TriHOME EOS and HODE EOS are increased in the upper layers of the epidermis and appear the highest in the stratum corneum (SC). EOS, esterified omega-hydroxy sphingosine; LA, linoleic acid.
Human psoriatic patients have altered levels of oxidized and native ceramides both in lesions and nonlesion healthy skin
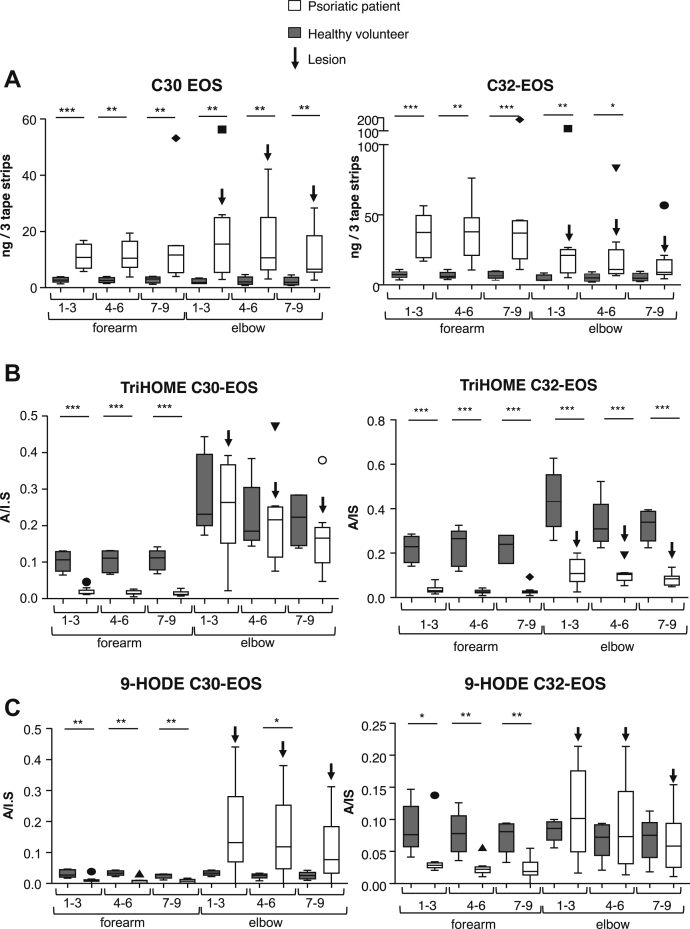
Elevated levels of free and esterified 9-HODE and 13-HODE have been previously detected in psoriatic lesions with 9R and 13S enantiomers predominating (28). To extend this to the study of specific ceramides, we determined the relative abundance of oxidized EOx identified herein. Two different comparisons were used. First, psoriatic plaques (left or right outer elbow) and uninvolved lesion-free skin (volar forearm) were tape stripped from the same patients. This allowed comparison within the same person for diseased versus nonlesion skin. Next, outer elbows (left or right) from healthy volunteers were tape stripped. These were then compared directly with psoriatic lesions that come from the same site in patients because it is recognized that the skin at different sites may show a distinct lipid profile. Data for LA, TriHOME, epoxyHOME, and HODE isomers of C30, C32, and C34-EOX (EOS, EOP, and EOH) are shown (Fig. 7, Supplemental Figs. S17–S20). Epoxy-HOME-EOx were below the limit of detection. LA containing C30, 32, and 34-EOx ceramides were significantly higher in the psoriatic patient skin, both for the forearm (no lesion, ~10-fold elevated) and elbow (plaque lesion, ~5-fold elevated) (Fig. 7A, Supplemental Figs. S17–S20). Thus, substrate ceramides are elevated in psoriatic patient skin regardless of whether a psoriasis plaque is present. Hyperproliferation of the SG and lack of terminal differentiation, providing increased abundance of EOS-producing cells in psoriasis plaques is known, providing a potential explanation (29). However, this was unexpected in unaffected skin in psoriasis where there is a lack of concomitant hyperproliferation. In contrast, triHOME-C32-EOS was significantly reduced in psoriatic patient skin (lesions or nonlesional skin) versus healthy controls (Fig. 7B). This contrast is also seen with triHOME-C34-EOS, triHOME-C30/C32/C34-EOH, and triHOME-C34-EOP (Supplemental Figs. S17–S20). Finally, HODE-C32-EOS were similar across forearm and elbow in healthy subjects, but for psoriatic patients, they were low in the unaffected forearm skin but far higher and more variable in involved elbow skin (Fig. 7C). This variability is also evident with the C30/C34-EOS/EOP and EOH species with no clear significance between the elbow and forearm (Supplemental Figs. S18–S20). There is a significant difference in HODE-C34-EOS species with this lipid being significantly higher in healthy volunteers at both sample sites (Supplemental Fig. S16C). The overall pattern is that substrate EOS was significantly elevated in the patient skin versus healthy controls, regardless of whether there was a lesion, whereas oxidized products were either significantly lower (nonlesion) or similar (lesions) in the patient skin.
Fig. 7.
Lipidomics analysis of psoriasis lesions shows elevated native EOS species and significantly altered levels of oxidized species. Tape strips were acquired from the volar forearm (uninvolved skin) and elbow (location of psoriatic lesion), from patients with psoriasis (n = 9) and from healthy controls (n = 5). From each site, 9 tape strips were obtained and pooled into groups of 3. Lipids were extracted and analyzed as described in Materials and methods. Increasing the tape strip number corresponds to increasing epidermal depth. Psoriatic patients (empty bars), healthy controls (gray bars), mean ± SEM.∗P<0.05, ∗∗P<0.01, ∗∗∗P<0.001. The bold down arrow (↓) corresponds to skin lesion samples. Geometric shapes show outliers. A: Epidermal tape strip profile of C30 and C32 EOS. B: Epidermal tape strip profile of triHOME EOS. C: Epidermal tape strip profile of 9-HODE EOS in psoriatic versus healthy control. EOS, esterified omega-hydroxy sphingosine.
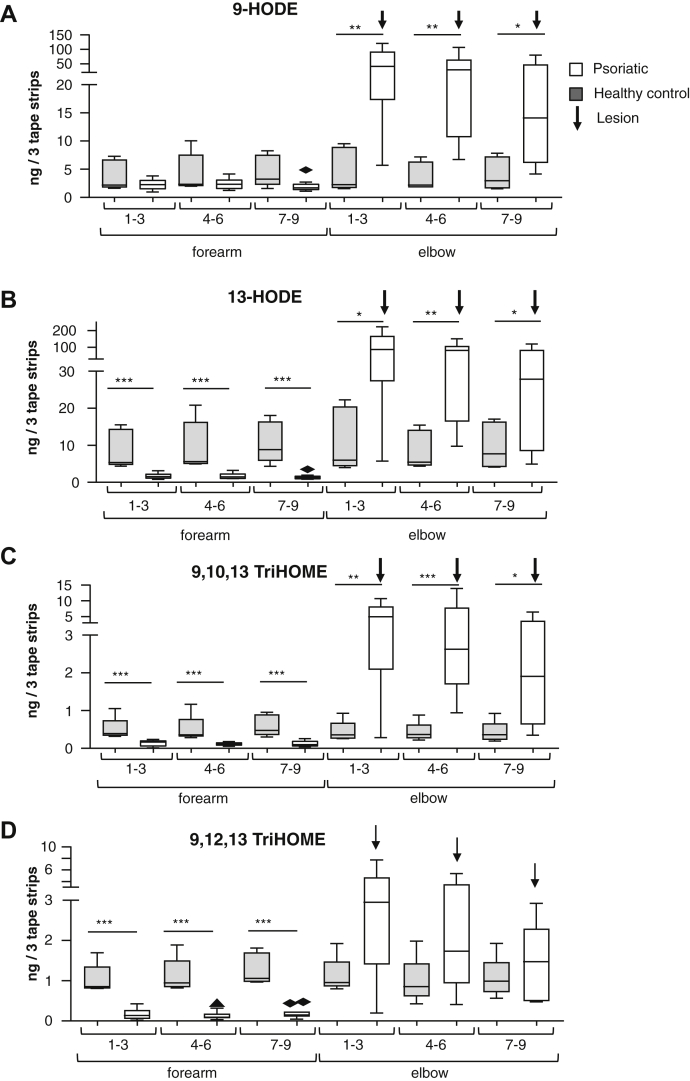
Free oxylipins were analyzed in psoriatic and healthy skin, to directly compare with EOS. 9- or 13-HODE and 9,10,13- or 9,12,13-triHOMEs were significantly increased in lesions of psoriatic patients versus healthy volunteer elbows (Fig. 8A–D). Owing to unavailability of standards, we were unable to quantify epoxyHOMEs. 9-HODE and 13-HODE were significantly higher in psoriatic lesions than the matched forearm skin matched from the same patient, as well as being significantly higher than uninvolved patient skin (30) (Fig. 8A, B). The abundant 13-HODE (which has been shown to be primarily 13S (28)) is partly generated by a 15-LOX, either 15-LOX-1 or 15-LOX-2 (21, 31). 9,10,13-triHOME and 9,12,13-triHOME showed the same trend as HODEs (Fig. 8C, D). In the skin, it is well known that 12R-LOX has very poor activity toward free LA; thus, these oxidation products most likely arise after hydrolysis of the oxidized fatty acyl from the LOX-oxidized EOx (32). In summary, free oxylipins are present at significantly higher levels in lesional epidermis than both nonlesional epidermis from the same patient and epidermis of healthy controls.
Fig. 8.
Free triHOMEs and HODEs are elevated in human psoriatic lesions. Tape strips were acquired from the volar forearm (uninvolved skin) and elbow (location of psoriatic lesion), from patients with psoriasis (n = 9) and healthy controls (n = 5). From each site, nine tape strips were obtained and pooled into groups of 3. Lipids were extracted and analyzed as described in Materials and methods. An increasing tape strip number corresponds to increasing epidermal depth. Psoriatic patients (empty bars), healthy controls (gray bars), mean ± SEM. ∗ P < 0.05, ∗∗ P < 0.01, ∗∗∗ P < 0.001. The bold down arrow (↓) corresponds to skin lesion samples. Geometric shapes show outliers. A: 9-HODE. B: 13-HODE. C: 9,10,13 triHOME. D: 9,12,13 triHOME.
The biosynthetic and metabolic pathway for oxidized EOx is highly upregulated in psoriatic lesions
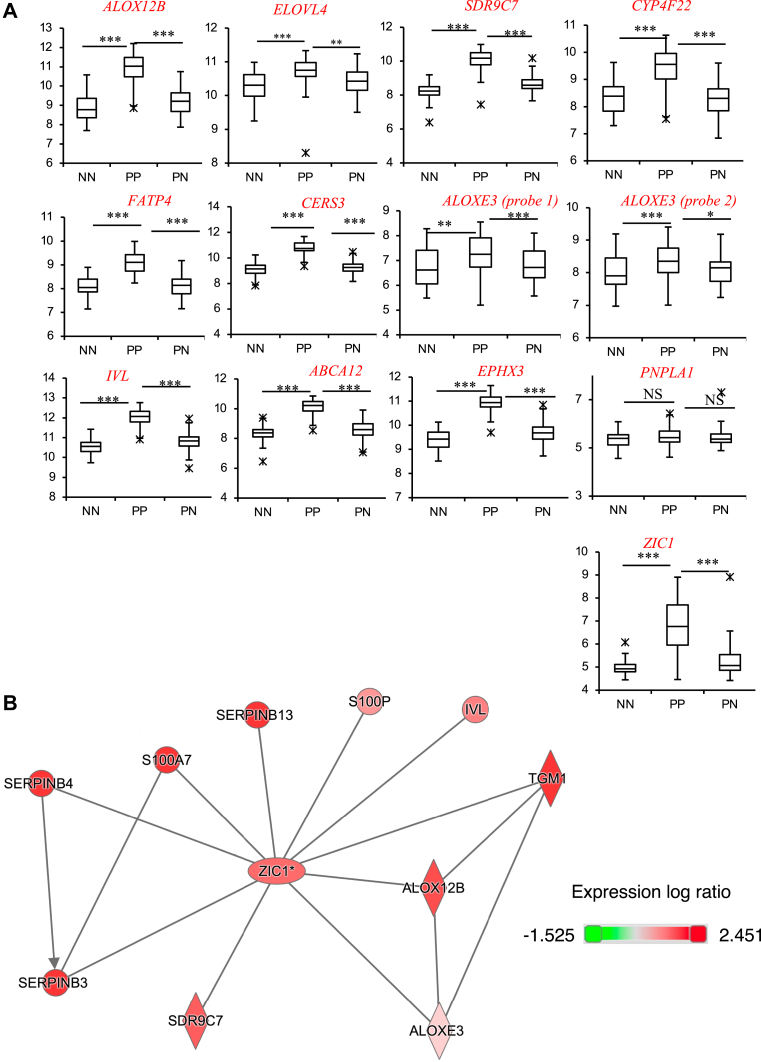
Lipidomics of psoriasis lesions demonstrated distinctive native and oxidized EOx profiles. This could result from altered expression and/or activity of the genes involved in their biosynthesis or degradation. To investigate this, a transcriptomic study on psoriatic lesions (PP) compared with healthy skin from the patient (PN) or healthy donors (NN) was downloaded and analyzed for differentially expressed genes relevant to EOx metabolism (17). Genes included in the analysis are labeled on Scheme 2. Notably, almost all genes that regulate the biosynthesis, metabolism, and coupling of EOx to form the CLE were significantly upregulated in this dataset when psoriatic lesions are compared with either uninvolved skin in the same patients, or with control skin from healthy volunteers (Scheme 2, Fig. 9A, Supplemental Fig. S21). This indicates that the oxidized EOx gene network that is required for forming an epidermal barrier is highly upregulated in psoriasis.
Fig. 9.
Several ceramide pathway genes are upregulated in psoriasis and functionally linked with the transcription factor ZIC1. Affymetrix CEL files from a psoriasis study were downloaded and genes in the ceramide pathway (Scheme 2) analyzed (17). Robust multiarray averaging-normalized gene expression level was corrected for multiple comparisons on a total of 50,683 probes (Benjamini-Hochberg test). Psoriasis plaque (PP), n = 64; psoriasis normal skin (PN), n = 58; healthy control normal skin (NN), n = 58. A: Box and whisker plots showing gene expression level for genes in the ceramide pathway which are significantly different. ∗ P < 0.05, ∗∗ P < 0.01, ∗∗∗ P < 0.005. B: IPA identifies many significantly different genes in the ceramide pathway link to ZIC1. Gene identifiers and expression values were uploaded for significant genes and network analysis performed (adjusted P value < 0.01). IPA, Ingenuity Pathway Analysis.
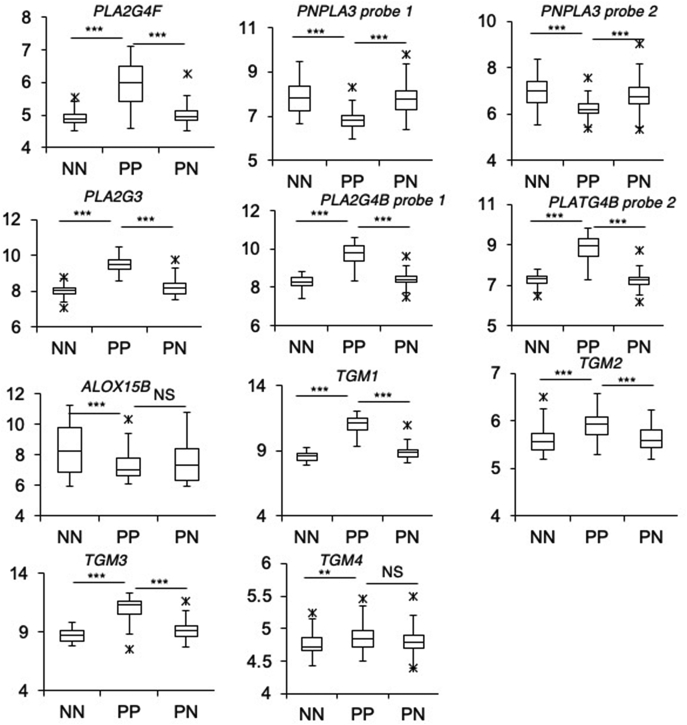
Next, the psoriasis dataset was analyzed using the Ingenuity Pathway Analysis (comparing PP with NN). Here, ALOX12B was found to link with two other highly upregulated genes in the pathway that forms and couples these lipids to form the CLE, specifically TGM1 and ALOXE3, as shown (Fig. 9B). These three genes linked directly through the IPA network to ZIC1, a zinc finger protein transcription factor (Fig. 9A, B) (33). Importantly, IPA directly linked ZIC1 directly with other relevant genes including IVL, and SDR9C7 (Fig. 9B). Furthermore, the ZIC1 gene was strongly upregulated in PP (Fig. 9A, B). Several of the upregulated genes (TGM1, ALOX12B, ALOXE3, SDR9C7, IVL, S100P, S100A7, SERPINB4, SERPINB3, SERPINB13) are included in a ZIC1 interactome network in HEK293 cells, which was identified using affinity purification-MS methodology (33). Increased expression of several phospholipase A2 isoforms was also noted in psoriatic skin, specifically PLA2G4B, PLA2G3, and PLA2G2F, whereas PNPLA3 was downregulated (Fig. 10).
Fig. 10.
Expression of phospholipases, ALOX15B, and TGM genes in psoriasis shows candidate PLA2 genes for oxylipin hydrolysis from ceramides and significant elevation in TGMs in psoriasis lesions. Affymetrix CEL files from a psoriasis study were downloaded and genes in the ceramide pathway analyzed (17). RMA-normalized gene expression level was corrected for multiple comparisons on a total of 50,683 probes (Benjamini-Hochberg test). Psoriasis plaque (PP), n = 64; psoriasis normal skin (PN), n = 58; healthy control normal skin (NN) n = 58. Box and whisker plots show gene expression level for genes in the ceramide pathway, which are significantly different. ∗∗P < 0.01, ∗∗∗P < 0.005.
Discussion
Little data exist on the regulation of 12R-LOX and its associated genes in healthy skin or in disease despite the fact that this pathway is essential for normal barrier function. Here, we present the first analysis of epidermal oxidized ceramides in the healthy human skin and in a skin with inflammatory disease characterized by impaired barrier function. To identify the lipids, we used a “fishing” approach termed precursor scanning in the negative-ion mode. Because most MS/MS analysis of long-chain ceramides in the skin used the positive-ion mode, we first analyzed the native EOS, EOH, and EOP with the most abundant long chains (C28-C36) with this method (20, 34, 35). The expected species were found, in particular abundant ions for the C30-C34 EOS, EOH, and EOP esterified to LA. Next, using precursor scanning LC/MS/MS, large numbers of oxidized forms of LA were detected esterified to long-chain EOx species, including epoxy-HOME, 9-HODE, and 9,10,13-triHOME. Their enzymatic generation was indicated by the relative abundance of the positional isomers of HODEs and triHOMEs (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5).
Depth profiling of the oxygenated ceramides suggested that eLOX3 and 12R-LOX are predominantly localized to the lower epidermis with epoxide hydrolase activity (recently identified as EPHX3 (26)) that forms the trihydroxy-LA ceramides more active in the upper epidermis (Fig. 6). The existence of polar oxidized ceramides in the upper layers may have a significant effect on CLE lipid organization in the SC, influencing the ability of other ceramides, cholesterol, and fatty acids to spatially arrange and form an adequate lipid barrier. Bouwstra et al. mathematically described the molecular composition of the SC to have an optimal mix of FAs, ceramides, and cholesterol to mimic what is known about in vivo epidermis (36, 37, 38). These models can now be updated by inclusion of oxidized ceramides as described herein.
Psoriatic lesions showed a complex altered phenotype of ceramides and free oxylipins when compared with either uninvolved skin or skin from healthy subjects (Figs. 7, 8); however, because these measurements only reflect steady state levels of lipids, they do not provide any information on flux through the metabolic pathway. Thus, they do not inform on whether the pathway is upregulated or downregulated because the level of a lipid is a function of its rate of formation and removal. To interrogate this, we analyzed the expression of all known relevant genes using a transcriptomic dataset. Almost all the known genes involved in generation of either long-chain ceramides or their oxygenation were significantly upregulated in psoriatic lesions, indicating that this pathway is highly active (Fig. 9). This significantly expands on previous observations that 12R-LOX is upregulated in both lesional and nonlesional skin in psoriasis, showing the entire pathway is affected (21, 22, 39, 40).
Significantly increased oxidized free fatty acids were seen in psoriatic elbow versus the healthy controls, along with reduced levels of oxidized EOS (Fig. 8). This may result from altered lipase activities in the skin, and our gene expression analysis identified three potential candidates, PLA2G3, PLA2G2F, and PLA2G4B (Fig. 10). This is backed up by previous studies that show an abundance of free arachidonic acid in psoriatic lesions compared with the healthy skin, where an upregulation of a PLA2 type activity was also suggested (39). Indeed, mice overexpressing epidermal PLA2G2F were reported previously as exhibiting a psoriasis phenotype, suggesting this as a likely candidate (41). It is important to clarify that as yet no enzyme has been identified to cleave the oxidized LA from the ω-hydroxy ceramide and thus further studies may look to express these enzymes to characterize their activity with an oxidized EOx substrate. Two other lipases were also identified in our gene expression analysis, PNPLA1 and PNPLA3, and both have been linked to epidermal barrier formation (42, 43). Substrates for these enzymes are yet to be identified, and they may therefore be a possible candidate for the enzymatic cleavage of the oxidized EOx. Previously, LIPN was proposed as a candidate because it is associated with the development of skin barrier impairment (44), albeit not as a classical ichthyosis gene (45). However, LIPN was not listed in the array. In summary, several additional phospholipases are identified that represent additional potential candidates and can be tested in future studies.
Our network analysis identified ZIC1 as a potential master regulator of many genes that are represented in the oxidized ceramide pathway (33). This includes several enzymes that directly oxygenate ceramides (ALOX12B, ALOXE3, SDR9C7), as well as downstream genes such as TGM1, which encodes the transglutaminase that couples the hydrolyzed omega hydroxy ceramide to involucrin (IVL) and other CLE proteins (46). Some of these genes are already known to be upregulated in psoriasis, for example, the antimicrobial proteins, SERPINs and S100s (47, 48). This phenotype could represent an attempt to repair barrier function in a situation where it is compromised and to dampen infection. ZIC1 has been found to be overexpressed in a variety of cancers modulating multiple pathways involved in tumor cell proliferation (49, 50, 51, 52). Here, it induces proliferation in some tumors while inhibiting in others (50). Another zinc finger protein, ZNF450, has been shown to act as a keratinocyte proliferation regulator, and mutations in ZNF450 have been implicated in the development of psoriasis (53, 54). However, to our knowledge, this article is the first to indicate a role of ZIC1 in keratinocyte proliferation in psoriasis via possible regulation of oxidized ceramide formation.
In summary, here we structurally characterized the major oxygenated ceramide lipids present in the epidermis of the human skin using a new LC/MS/MS method capable of analyzing > 80 molecular species. Combined with a transcriptional analysis, we identify a new regulatory network involving the zinc finger transcription factor, ZIC1, that may play a central role in upregulating their metabolism in psoriasis. Inhibition of ZIC1 may provide a potential therapeutic target in the treatment of psoriasis via downregulation of the oxidized fatty acids seen in abundance in this disease. The role of the oxidized free acid in the epidermis is yet to be determined, but data presented here make it apparent that the ability of the skin to retain the oxidized moiety as long-chain ceramides appears to be vital in maintaining a homeostatic epidermal barrier. The role of these new lipids in progression or resolution of psoriasis is currently unknown.
Data availability
All data concerned with this study are presented within this article.
Supplemental data
This article contains supplemental data.
Conflict of interest
J. R. I. is a consultant for UCB Pharma, Boehringer Ingelheim, Novartis, and ChemoCentryx and has served on advisory boards for Viela Bio and Kymera Therapeutics, all in the field of hidradenitis suppurativa. He is the Editor-in-Chief of the British Journal of Dermatology. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
V. J. T., F. A., W. E. B., V. B. O., and C. P. T. investigation; V. J. T., A. R. B., V. B. O., and C. P. T. visualization; V. J. T., J. C. B., J. R. I., V. P., A. R. B., and V. B. O. writing-review and editing; R. A. and J. B. formal analysis; J. C. B., J. R. I., V. P., A. R. B., V. B. O., and C. P. T. supervision; J. R. I., R. C. M., V. P., and V. B. O. resources; A. R. B., V. B. O., and C. P. T. conceptualization; A. R. B., V. B. O., and C. P. T. methodology; V. B. O. and C. P. T. funding acquisition; C. P. T. writing-original draft; C. P. T. project administration.
Funding and additional information
Funding is gratefully acknowledged from Medical Research Council MR/M011445/1 (V. B. O., C. P. T., V. J. T., J. B., J. C. B., and V. P.), Marie Curie International Outgoing Fellowship (FP7) (C. P. T.), and the NIH Grant GM134548 (A. R. B.). V. B. O. holds a Royal Society Wolfson Merit Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplemental data
References
- 1.Takeichi T., Akiyama M. Inherited ichthyosis: Non-syndromic forms. J. Dermatol. 2016;43:242–251. doi: 10.1111/1346-8138.13243. [DOI] [PubMed] [Google Scholar]
- 2.Feingold K.R. The importance of lipids in cutaneous function. J. Lipid Res. 2007;48:2529–2530. doi: 10.1194/jlr.E700004-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Vahlquist A., Fischer J., Torma H. Inherited nonsyndromic ichthyoses: an update on pathophysiology, diagnosis and treatment. Am. J. Clin. Dermatol. 2018;19:51–66. doi: 10.1007/s40257-017-0313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jobard F., Lefevre C., Karaduman A., Blanchet-Bardon C., Emre S., Weissenbach J., Ozgüc M., Lathrop M., Prud'homme J.F., Fischer J. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum. Mol. Genet. 2002;11:107–113. doi: 10.1093/hmg/11.1.107. [DOI] [PubMed] [Google Scholar]
- 5.Krieg P., Rosenberger S., de Juanes S., Latzko S., Hou J., Dick A., Kloz U., van der Hoeven F., Hausser I., Esposito I., Rauh M., Schneider H. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J. Invest. Dermatol. 2013;133:172–180. doi: 10.1038/jid.2012.250. [DOI] [PubMed] [Google Scholar]
- 6.Epp N., Furstenberger G., Muller K., de Juanes S., Leitges M., Hausser I., Thieme F., Liebisch G., Schmitz G., Krieg P. 12R-lipoxygenase deficiency disrupts epidermal barrier function. J. Cell Biol. 2007;177:173–182. doi: 10.1083/jcb.200612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran J.L., Qiu H., Turbe-Doan A., Yun Y., Boeglin W.E., Brash A.R., Beier D.R. A mouse mutation in the 12R-lipoxygenase, Alox12b, disrupts formation of the epidermal permeability barrier. J. Invest. Dermatol. 2007;127:1893–1897. doi: 10.1038/sj.jid.5700825. [DOI] [PubMed] [Google Scholar]
- 8.de Juanes S., Epp N., Latzko S., Neumann M., Furstenberger G., Hausser I., Stark H.J., Krieg P. Development of an ichthyosiform phenotype in Alox12b-deficient mouse skin transplants. J. Invest. Dermatol. 2009;129:1429–1436. doi: 10.1038/jid.2008.410. [DOI] [PubMed] [Google Scholar]
- 9.Hansen H.S., Jensen B., von Wettstein-Knowles P. Apparent in vivo retroconversion of dietary arachidonic to linoleic acid in essential fatty acid-deficient rats. Biochim. Biophys. Acta. 1986;878:284–287. doi: 10.1016/0005-2760(86)90158-x. [DOI] [PubMed] [Google Scholar]
- 10.Thomas C.P., Boeglin W.E., Garcia-Diaz Y., O'Donnell V.B., Brash A.R. Steric analysis of epoxyalcohol and trihydroxy derivatives of 9-hydroperoxy-linoleic acid from hematin and enzymatic synthesis. Chem. Phys. Lipids. 2013;167-168:21–32. doi: 10.1016/j.chemphyslip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamanashi H., Boeglin W.E., Morisseau C., Davis R.W., Sulikowski G.A., Hammock B.D., Brash A.R. Catalytic activities of mammalian epoxide hydrolases with cis and trans fatty acid epoxides relevant to skin barrier function. J. Lipid Res. 2018;59:684–695. doi: 10.1194/jlr.M082701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba T., Thomas C.P., Calcutt M.W., Boeglin W.E., O'Donnell V.B., Brash A.R. The precise structures and stereochemistry of trihydroxy-linoleates esterified in human and porcine epidermis and their significance in skin barrier function: implication of an epoxide hydrolase in the transformations of linoleate. J. Biol. Chem. 2016;291:14540–14554. doi: 10.1074/jbc.M115.711267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiba T., Nakahara T., Kohda F., Ichiki T., Manabe M., Furue M. Measurement of trihydroxy-linoleic acids in stratum corneum by tape-stripping: Possible biomarker of barrier function in atopic dermatitis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs D., Tang X., Johnsson A.K., Dahlen S.E., Hamberg M., Wheelock C.E. Eosinophils synthesize trihydroxyoctadecenoic acids (TriHOMEs) via a 15-lipoxygenase dependent process. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158611. doi: 10.1016/j.bbalip.2020.158611. [DOI] [PubMed] [Google Scholar]
- 15.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 16.Thomas C.P., Clark S.R., Hammond V.J., Aldrovandi M., Collins P.W., O'Donnell V.B. Identification and quantification of aminophospholipid molecular species on the surface of apoptotic and activated cells. Nat. Protoc. 2014;9:51–63. doi: 10.1038/nprot.2013.163. [DOI] [PubMed] [Google Scholar]
- 17.Gudjonsson J.E., Ding J., Li X., Nair R.P., Tejasvi T., Qin Z.S., Ghosh D., Aphale A., Gumucio D.L., Voorhees J.J., Abecasis G.R., Elder J.T. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J. Invest. Dermatol. 2009;129:2795–2804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Smeden J., Hoppel L., van der Heijden R., Hankemeier T., Vreeken R.J., Bouwstra J.A. LC/MS analysis of stratum corneum lipids: ceramide profiling and discovery. J. Lipid Res. 2011;52:1211–1221. doi: 10.1194/jlr.M014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nugteren D.H., Kivits G.A. Conversion of linoleic acid and arachidonic acid by skin epidermal lipoxygenases. Biochim. Biophys. Acta. 1987;921:135–141. doi: 10.1016/0005-2760(87)90179-2. [DOI] [PubMed] [Google Scholar]
- 22.Boeglin W.E., Kim R.B., Brash A.R. A 12R-lipoxygenase in human skin: mechanistic evidence, molecular cloning, and expression. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6744–6749. doi: 10.1073/pnas.95.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arenberger P., Kemeny L., Ruzicka T. Characterization of high-affinity 12(S)-hydroxyeicosatetraenoic acid (12(S)-HETE) binding sites on normal human keratinocytes. Epithelial Cell Biol. 1993;2:1–6. [PubMed] [Google Scholar]
- 24.Leong J., Hughes-Fulford M., Rakhlin N., Habib A., Maclouf J., Goldyne M.E. Cyclooxygenases in human and mouse skin and cultured human keratinocytes: association of COX-2 expression with human keratinocyte differentiation. Exp. Cell Res. 1996;224:79–87. doi: 10.1006/excr.1996.0113. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Lorie E.P., Fischer J., Vahlquist A., Torma H. The expression of epidermal lipoxygenases and transglutaminase-1 is perturbed by NIPAL4 mutations: indications of a common metabolic pathway essential for skin barrier homeostasis. J. Invest. Dermatol. 2012;132:2368–2375. doi: 10.1038/jid.2012.160. [DOI] [PubMed] [Google Scholar]
- 26.Edin M.L., Yamanashi H., Boeglin W.E., Graves J.P., DeGraff L.M., Lih F.B., Zeldin D.C., Brash A.R. Epoxide hydrolase 3 (Ephx3) gene disruption reduces ceramide linoleate epoxide hydrolysis and impairs skin barrier function. J. Biol. Chem. 2020;296 doi: 10.1074/jbc.RA120.016570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckert R.L., Sturniolo M.T., Broome A.M., Ruse M., Rorke E.A. Transglutaminase function in epidermis. J. Invest. Dermatol. 2005;124:481–492. doi: 10.1111/j.0022-202X.2005.23627.x. [DOI] [PubMed] [Google Scholar]
- 28.Baer A.N., Costello P.B., Green F.A. Stereospecificity of the products of the fatty acid oxygenases derived from psoriatic scales. J. Lipid Res. 1991;32:341–347. [PubMed] [Google Scholar]
- 29.Wraight C.J., White P.J., McKean S.C., Fogarty R.D., Venables D.J., Liepe I.J., Edmondson S.R., Werther G.A. Reversal of epidermal hyperproliferation in psoriasis by insulin-like growth factor I receptor antisense oligonucleotides. Nat. Biotechnol. 2000;18:521–526. doi: 10.1038/75382. [DOI] [PubMed] [Google Scholar]
- 30.Sorokin A.V., Domenichiello A.F., Dey A.K., Yuan Z.X., Goyal A., Rose S.M., Playford M.P., Ramsden C.E., Mehta N.N. Bioactive lipid mediator profiles in human psoriasis skin and blood. J. Invest. Dermatol. 2018;138:1518–1528. doi: 10.1016/j.jid.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baer A.N., Costello P.B., Green F.A. Free and esterified 13(R,S)-hydroxyoctadecadienoic acids: principal oxygenase products in psoriatic skin scales. J. Lipid Res. 1990;31:125–130. [PubMed] [Google Scholar]
- 32.Yu Z., Schneider C., Boeglin W.E., Brash A.R. Human and mouse eLOX3 have distinct substrate specificities: implications for their linkage with lipoxygenases in skin. Arch. Biochem. Biophys. 2006;455:188–196. doi: 10.1016/j.abb.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H., Szpyt J., Tam S., Zarraga G., Pontano-Vaites L., Swarup S. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.t'Kindt R., Jorge L., Dumont E., Couturon P., David F., Sandra P., Sandra K. Profiling and characterizing skin ceramides using reversed-phase liquid chromatography-quadrupole time-of-flight mass spectrometry. Anal. Chem. 2012;84:403–411. doi: 10.1021/ac202646v. [DOI] [PubMed] [Google Scholar]
- 35.Kawana M., Miyamoto M., Ohno Y., Kihara A. Comparative profiling and comprehensive quantification of stratum corneum ceramides in humans and mice by LC/MS/MS. J. Lipid Res. 2020;61:884–895. doi: 10.1194/jlr.RA120000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Smeden J., Janssens M., Gooris G.S., Bouwstra J.A. The important role of stratum corneum lipids for the cutaneous barrier function. Biochim. Biophys. Acta. 2014;1841:295–313. doi: 10.1016/j.bbalip.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Mojumdar E.H., Gooris G.S., Barlow D.J., Lawrence M.J., Deme B., Bouwstra J.A. Skin lipids: localization of ceramide and fatty acid in the unit cell of the long periodicity phase. Biophys. J. 2015;108:2670–2679. doi: 10.1016/j.bpj.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danso M., Boiten W., van Drongelen V., Gmelig Meijling K., Gooris G., El Ghalbzouri A., Absalah S., Vreeken R., Kezic S., van Smeden J., Lavrijsen S., Bouwstra J. Altered expression of epidermal lipid bio-synthesis enzymes in atopic dermatitis skin is accompanied by changes in stratum corneum lipid composition. J. Dermatol. Sci. 2017;88:57–66. doi: 10.1016/j.jdermsci.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Hammarstrom S., Hamberg M., Samuelsson B., Duell E.A., Stawiski M., Voorhees J.J. Increased concentrations of nonesterified arachidonic acid, 12L-hydroxy-5,8,10,14-eicosatetraenoic acid, prostaglandin E2, and prostaglandin F2alpha in epidermis of psoriasis. Proc. Natl. Acad. Sci. U.S.A. 1975;72:5130–5134. doi: 10.1073/pnas.72.12.5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeichi T., Kinoshita F., Tanaka H., Fujita S., Kobayashi Y., Nakatochi M., Sugiura K., Akiyama M. The lipoxygenase-hepoxilin pathway is activated in cutaneous plaque lesions of psoriasis. J. Cutan. Immunol. Allergy. 2019;2:15–24. [Google Scholar]
- 41.Yamamoto K., Miki Y., Sato M., Taketomi Y., Nishito Y., Taya C., Muramatsu K., Ikeda K., Nakanishi H., Taguchi R., Kambe N., Kabashima K., Lambeau G., Gelb M.H., Murakami M. The role of group IIF-secreted phospholipase A2 in epidermal homeostasis and hyperplasia. J. Exp. Med. 2015;212:1901–1919. doi: 10.1084/jem.20141904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toulza E., Mattiuzzo N.R., Galliano M.F., Jonca N., Dossat C., Jacob D., de Daruvar A., Wincker P., Serre G., Guerrin M. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007;8:R107. doi: 10.1186/gb-2007-8-6-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crumrine D., Khnykin D., Krieg P., Man M.Q., Celli A., Mauro T.M., Wakefield J.S., Menon G., Mauldin E., Miner J.H., Lin M.H., Brash A.R., Sprecher E., Radner F.P.W., Choate K. Mutations in recessive congenital ichthyoses illuminate the origin and functions of the corneocyte lipid envelope. J. Invest. Dermatol. 2019;139:760–768. doi: 10.1016/j.jid.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Israeli S., Goldberg I., Fuchs-Telem D., Bergman R., Indelman M., Bitterman-Deutsch O., Harel A., Mashiach Y., Sarig O., Sprecher E. Non-syndromic autosomal recessive congenital ichthyosis in the Israeli population. Clin. Exp. Dermatol. 2013;38:911–916. doi: 10.1111/ced.12148. [DOI] [PubMed] [Google Scholar]
- 45.Hotz A., Bourrat E., Kusel J., Oji V., Alter S., Hake L., Korbi M., Ott H., Hausser I., Zimmer A.D., Fischer J. Mutation update for CYP4F22 variants associated with autosomal recessive congenital ichthyosis. Hum. Mutat. 2018;39:1305–1313. doi: 10.1002/humu.23594. [DOI] [PubMed] [Google Scholar]
- 46.Nemes Z., Marekov L.N., Steinert P.M. Involucrin cross-linking by transglutaminase 1. Binding to membranes directs residue specificity. J. Biol. Chem. 1999;274:11013–11021. doi: 10.1074/jbc.274.16.11013. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y., Sheshadri N., Zong W.X. SERPINB3 and B4: From biochemistry to biology. Semin. Cell Dev. Biol. 2017;62:170–177. doi: 10.1016/j.semcdb.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schonthaler H.B., Guinea-Viniegra J., Wculek S.K., Ruppen I., Ximenez-Embun P., Guio-Carrion A., Navarro R., Hogg N., Ashman K., Wagner E.F. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity. 2013;39:1171–1181. doi: 10.1016/j.immuni.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Ma G., Dai W., Sang A., Yang X., Li Q. Roles of ZIC family genes in human gastric cancer. Int. J. Mol. Med. 2016;38:259–266. doi: 10.3892/ijmm.2016.2587. [DOI] [PubMed] [Google Scholar]
- 50.Zhong J., Chen S., Xue M., Du Q., Cai J., Jin H., Si J., Wang L. ZIC1 modulates cell-cycle distributions and cell migration through regulation of sonic hedgehog, PI(3)K and MAPK signaling pathways in gastric cancer. BMC Cancer. 2012;12:290. doi: 10.1186/1471-2407-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gan L.H., Pan J., Chen S.J., Zhong J., Wang L.J. [DNA methylation of ZIC1 and KLOTHO gene promoters in colorectal carcinomas and its clinicopathological significance] Zhejiang Da Xue Xue Bao Yi Xue Ban. 2011;40:309–314. doi: 10.3785/j.issn.1008-9292.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 52.Brill E., Gobble R., Angeles C., Lagos-Quintana M., Crago A., Laxa B., Decarolis P., Zhang L., Antonescu C., Socci N.D., Taylor B.S., Sander C., Koff A., Singer S. ZIC1 overexpression is oncogenic in liposarcoma. Cancer Res. 2010;70:6891–6901. doi: 10.1158/0008-5472.CAN-10-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cassandri M., Smirnov A., Novelli F., Pitolli C., Agostini M., Malewicz M., Melino G., Raschellà G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3:17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birnbaum R.Y., Hayashi G., Cohen I., Poon A., Chen H., Lam E.T., Kwok P.Y., Birk O.S., Liao W. Association analysis identifies ZNF750 regulatory variants in psoriasis. BMC Med. Genet. 2011;12:167. doi: 10.1186/1471-2350-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data concerned with this study are presented within this article.