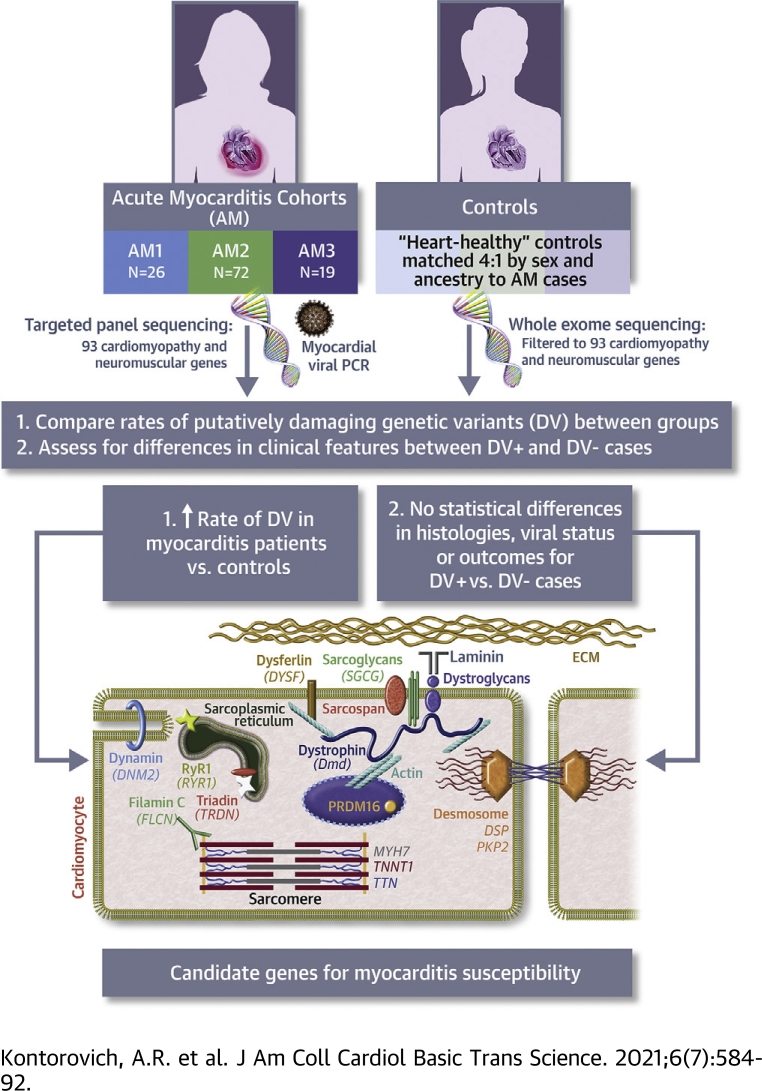
Visual Abstract
Key Words: acute myocarditis, cardiomyopathy, genetics
Abbreviations and Acronyms: AM, acute myocarditis; AM1, acute myocarditis registry 1; ACM, arrhythmogenic cardiomyopathy; CMP, cardiomyopathy; DV, deleterious variant; EF, ejection fraction; ES, exome sequencing; NMD, neuromuscular disorder; OR, odds ratio; TGP, targeted gene panel
Highlights
-
•
AM is an important cause of cardiac dysfunction following some viral infections and has been previously linked with genetic host susceptibilities.
-
•
Sequencing data for genes related to cardiomyocyte structure and function was compared between patients with AM and sex- and ancestry-matched heart-healthy control subjects.
-
•
Putatively DVs among these genes were found in ∼16% of AM cases, contributing to viral susceptibility.
-
•
Implicated genes are ones classically associated with cardiomyopathy or neuromuscular disorders with cardiac involvement, suggesting shared mechanistic pathways.
-
•
Phenotypes, including outcomes, were similar in subjects who have AM with and without DV, therefore genetic testing will be necessary to uncover this subtype and inform risk in relatives.
Summary
Impairments in certain cardiac genes confer risk for myocarditis in children. To determine the extent of this association, we performed genomic sequencing in predominantly adult patients with acute myocarditis and matched control subjects. Putatively deleterious variants in a broad set of cardiac genes were found in 19 of 117 acute myocarditis cases vs 34 of 468 control subjects (P = 0.003). Thirteen genes classically associated with cardiomyopathy or neuromuscular disorders with cardiac involvement were implicated, including >1 associated damaging variant in DYSF, DSP, and TTN. Phenotypes of subjects who have acute myocarditis with or without deleterious variants were similar, indicating that genetic testing is necessary to differentiate them.
The inflammatory disease acute myocarditis (AM) has long been considered an unfortunate and random consequence of infectious, toxic, or autoimmune reactions targeting the myocardium (1). Pathogenic organisms are most frequently implicated, typically involving prevalent viruses (eg, adenoviruses, coxsackieviruses, parvovirus B19, herpesviruses) that cause no more than uncomplicated upper respiratory tract infections in most individuals, yet can lead to sudden cardiac death or permanent heart failure in the subgroup that develop AM (2,3). The outcome of AM from viral infection is, therefore, an outlier, suggesting that genetic factors of the host might influence pathogenesis. The notion of randomness in AM susceptibility is further challenged by the recognition that AM is more common in individuals with genetically mediated derangements in cardiomyocyte cytoskeletal components, including the dilated cardiomyopathy of Duchenne muscular dystrophy and arrhythmogenic cardiomyopathy (ACM) (4, 5, 6). In fact, through exome sequencing (ES) of children with AM, we previously identified enrichment in rare, biallelic nonsynonymous, or splice-site variants among a set of genes related to cardiomyopathies (BAG3, DSP, PKP2, RYR2, SCN5A, and TNNI3) (7), confirming the link between “cardiomyopathic” genes and risk of AM. Brown et al (8) also recently uncovered likely pathogenic and pathogenic variants in an overlapping set of cardiomyopathy (CMP) genes (MYBPC3, TTN, TNNT2, SCN5A) on targeted gene panel (TGP) testing in 5 of 8 children hospitalized for acute heart failure caused by AM. These discoveries cast AM as a human genetic disorder, having genetic overlap with myopathic diseases of the heart.
Twenty years ago, as gene discovery for Mendelian cardiovascular diseases was in its infancy, Bowles et al (9) proposed that heritable conditions with similar phenotypes and genetic heterogeneity occur because of abnormalities in genes encoding proteins with similar functions or involved in the same pathway. In particular, genetic CMPs were broadly linked to derangements in proteins within sarcomeric and other cytoskeletal structures (10). This “final common pathway” hypothesis has been repeatedly upheld in CMP gene discoveries implicating proteins within cardiomyocyte sarcomere structures, the nuclear membrane, Z-disc, desmosome, spliceosome, and ion channels (11). And whereas recent expert consensus has classified a set of “core genes” for CMP (12), a similar phenotype of cardiac dysfunction also accompanies skeletal myopathy in several genetic neuromuscular disorders (NMDs) (13, 14, 15). We, therefore, hypothesized that vulnerability to AM is caused by genetically mediated impairments in proteins in a range of cardiomyocyte cytoskeletal structures caused by variants in “CMP” genes as well as “NMD” genes that have myocardial expression.
Methods
Subject recruitment, eligibility, and ethics
Subjects with AM were included using deidentified data through 3 separate registries curated by coauthors: AM1 (J.T.), AM2 (K.K.), AM3 (M.B., I.K., K.K.). AM1 cases were adults and children with clinical diagnoses of AM based on historical and diagnostic testing data who were recruited between 1991 and 2000. For these cases, clinical and viral but not genetic data have been previously reported (16, 17, 18). AM2 and AM3 cases were subjects with clinically suspected viral myocarditis, recruited between the 2015-2017 and 1997-2013 periods, respectively, with AM proven by immunohistologic criteria (19). Clinical but not genetic data have been previously reported for AM3 (19, 20, 21). DNAs were provided for all subjects from AM1 and AM2, and samples from AM1 additionally underwent whole genome amplification (Genetic Resources Core Facility DNA services at the Johns Hopkins University) prior to sequencing. For subjects from AM3, genomic DNA extraction was performed from frozen, banked whole blood specimens (Macromolecular Pathology Core, Icahn School of Medicine at Mount Sinai).
Control subjects were identified through previous enrollment in the Icahn School of Medicine’s BioMe Biobank under a protocol approved by the institutional review board (approval 17-02058) and in accordance with institutional guidelines. Broad informed consent for research purposes was secured under the terms of the original BioMe enrollment. To account for the ethnic heterogeneity of the AM cohort, the first 3 principal components of the principal component analysis from sequencing data on AM cases and control cases (Supplemental Methods) were used to generate a subset of genetic ancestry and sex-matched control cases lacking a set of relevant AM, CMP, heart failure, and neuromuscular disorder ICD-9 and ICD-10 codes in the electronic health record at a ratio of 4:1 to each case per cohort (AM1, N = 104; AM2, N = 288; AM3, N = 76).
Genetic analysis
Next-generation sequencing was performed on genomic DNAs from all case subjects using a TGP of 93 genes (Supplemental Table 1) related to cardiomyocyte structure and function, including causative genes for primary CMPs as well as NMDs with established cardiac involvement (Translational Genomics Core, Partners Healthcare). Existing ES data from matched control subjects was available on all BioMe control cases through a prior institutional partnership with Regeneron. TGP and ES methodologies and data analyses are described in the Supplemental Methods. Raw genomic TGP sequencing data from cases were down-sampled to match the ∼30× average read depth of ES. Given the estimated incidence of AM as 1 of 10,000, and the estimated incidence of myocardial involvement in acute viral infections at 1% to 5%, we excluded polymorphisms with a minor allele frequency of >0.1% in the public database of whole genome sequencing from population control subjects, genome Aggregation Database (gnomAD). Nonsynonymous variants were retained as putatively deleterious variants (DVs) if previously interpreted to have a damaging functional effect as determined by designation of “likely pathogenic” or “pathogenic” status without conflicting “likely benign” or “benign” entries among Badge laboratories (22) in the public database ClinVar. Additionally, indel-inframe, indel-frameshift, start-loss, missense, nonsense, stop-loss, synonymous splice-site (within first or last 2 bases of the exon), and essential splice-site (within first or last 2 bases of the intron) variants without any “likely benign” or “benign” designation among Badge laboratories in the public database ClinVar were defined as DVs.
Viral analysis
Viral analyses were performed on myocardial tissues based on previously described methods for subjects in AM1 (23) and AM2 and AM3 (19) and described in more detail in Supplemental Methods.
DV enrichment analysis and statistics
The Student’s t-test (2-tailed, heteroscedastic) was used to compare sequencing depth in AM cases versus control cases. Fisher exact tests (2-tailed) were used to compare DV rates between AM cases and control cases. A value of P < 0.05 was considered statistically significant. Odds ratio (OR) and 95% CIs (24) and associated P values (25) were calculated for each case-control pair. Categorical data are presented as count (percentage) or rates. Continuous data are presented as median (full range).
Pooled cohort analyses and statistics
A logistic regression model of DV outcome (“0” if subject has no DV; “1” if subject has 1 or more DVs) incorporating “study” (AM1, AM2, AM3 cases) and matched control cases within each study was performed using a custom R script (model: DV ∼ Case + Study) to generate a pooled OR and P value for the total combined data set. P < 0.05 was considered statistically significant without adjustment for multiple tests because “study” was a covariate (26). We reported the OR with 95% CI for individual studies and pooled logistic regression model. A P value <0.05 was considered statistically significant. To ensure robustness of our results, we performed a resampling procedure as a secondary statistical analysis. We generated a resampled OR distribution by resampling subjects with replacement within each study and computing an overall OR with the logistic regression model. We repeated this procedure 10,000 times and calculated the overall OR as the mean of simulated ORs, the 95% CI from the simulated ORs occurring between the 2.5th and 97.5th percentiles, and the P value as the proportion of simulations with OR ≤1, considering P < 0.05 to be statistically significant. R version 4.0 (R Foundation) was used for all statistical analyses.
Results
Case demographics
Table 1 lists the demographics of subjects included in the 3 AM case cohorts. In total there were 13 pediatric (≤20 years of age) cases (11.1%) and 104 adult cases (88.9%). The majority of subjects were of European or Caucasian self-reported ancestry, as most were recruited from sites in Germany.
Table 1.
Patient Demographics
| AM1 (N = 26) | AM2 (N = 72) | AM3 (N = 19) | |
|---|---|---|---|
| Age, y | 38.5 (1.1-78) | 44.5 (18-66) | 40 (17-69) |
| Pediatric | 8 (30.8) | 3 (4.2) | 2 (10.5) |
| Female | 11 (42.3) | 23 (31.9) | 3 (15.8) |
| Self-reported race | |||
| European/Caucasian | 21 (80.8) | 71 (98.6) | 19 (100.0) |
| African/African American | 2 (7.7) | 0 (0.0) | 0 (0.0) |
| Hispanic | 3 (11.5) | 0 (0.0) | 0 (0.0) |
| East Asian | 0 | 0 | 0 |
| South Asian | 0 | 0 | 0 |
| Middle Eastern | 0 (0.0) | 1 (1.4) | 0 (0.0) |
Values are median (range) or n (%).
AM1 = acute myocarditis registry 1.
Genomic characteristics of AM cases and control SUBJECTS
Rates of nonsynonymous and synonymous variants were similar in AM cases and control subjects for the 3 cohorts (AM1, 11.7 vs 13.0 and 12.8 vs 13.0; P = 0.30; AM2, 11.7 vs 12.2 and 12.8 vs 13.2; P = 0.90; AM3, 11.5 vs 13.2 and 11 vs 13.3; P = 0.63), indicating that AM cases and control subjects were appropriately matched within each cohort. Rates of nonsynonymous and synonymous variants were also similar across case cohorts (AM1 vs AM2; P = 1.0; AM1 vs AM3; P = 0.80; AM2 vs AM3; P = 0.70), indicating that global characteristics of sequencing data were similar.
Putatively damaging variants in genes related to cardiomyocyte structure and function are enriched among individuals with AM
DVs were identified in 16.2% of all AM cases (19.2%, 11.1%, and 31.6% of AM1, AM2, and AM3 cases, respectively) (Table 2), compared with 7.2% of all control subjects (5.6%, 7.6%, and 7.9% of the respective matched-control groups). Overall, 3 of 13 pediatric (23.1%) and 16 of 104 adult (15.4%) AM cases were identified as having ≥1 DV (P = 0.44). Monoallelic variants were present in 17 of 19 DV+ cases, whereas 2 cases, both children, bore multiallelic variation. The rates of DV were similar between male and female subjects (18.4% vs 15%; P = 0.80). There was no difference in the median age of the DV+ (40; range 1.5 years-62 years) and DV− (43; range 1.1 years-78.0 years) groups (P = 0.50).
Table 2.
Putatively Damaging Variants in Subjects With AM
| Registry | Case | Age (y) | Sex | Gene | cDNA Change | Function | ClinVar | gnomAD AF | hg38 Coordinates |
|---|---|---|---|---|---|---|---|---|---|
| AM1 | |||||||||
| AM1-1 | 0.6 | F | PRDM16 | c.420delG | Frameshift deletion | – | – | 3244118 | |
| AM1-1 | 0.6 | F | DSP | c.3697dupA | Frameshift insertion | – | – | 7579886 | |
| AM1-1 | 0.6 | F | DNM2 | c.1347dupC | Frameshift insertion | – | – | 10798496 | |
| AM1-1 | 0.6 | F | DMD | c.823dupT | Frameshift insertion | – | – | 31180428 | |
| AM1-2 | 13 | F | TTN | c.76806dupA | Frameshift insertion | – | – | 178533189 | |
| AM1-2 | 13 | F | TTN | c.52867_52868insCA | Frameshift insertion | – | – | 178566645 | |
| AM1-3 | 42 | F | RYR1 | c.G1589A | Nonsynonymous SNV | P/VUS | 5.3 · 10-5 | 38455463 | |
| AM1-4 | 60 | M | DYSF | c.4152dupC | Frameshift insertion | P | 2.8 · 10-5 | 71612666 | |
| AM1-5 | 62 | M | TTN | c.C35680T | Stopgain | – | – | 178589426 | |
| AM2 | |||||||||
| AM2-1 | 19 | F | DSP | c.C1234T | Stopgain | – | – | 7567874 | |
| AM2-2 | 23 | M | PKP2 | c.1771delC | Frameshift deletion | P | – | 32822534 | |
| AM2-3 | 31 | M | DSP | c.C5851T | Stopgain | P | – | 7583113 | |
| AM2-4 | 40 | M | SGCG | c.T581C | Nonsynonymous SNV | LP/P | 1.5 · 10-5 | 23320639 | |
| AM2-5 | 44 | F | MYH7 | c.C2377T | Stopgain | VUS | – | 23425328 | |
| AM2-6 | 48 | M | TTN | c.51250delA | Frameshift deletion | – | – | 178568262 | |
| AM2-7 | 51 | M | FLNC | c.7870delA | Frameshift deletion | – | – | 128858195 | |
| AM2-8 | 54 | F | DYSF | c.C760T | Nonsynonymous SNV | LP/P | 0.0001 | 71515716 | |
| AM3 | |||||||||
| AM3-1 | 34 | M | TTN | c.C22552T | Stopgain | – | – | 178614226 | |
| AM3-2 | 37 | M | TTN | c.50536delT | Frameshift deletion | – | – | 178568976 | |
| AM3-3 | 39 | M | TTN | c.38004delA | Frameshift deletion | – | – | 178585120 | |
| AM3-4 | 46 | M | TTN | c.38004delA | Frameshift deletion | – | – | 178585120 | |
| AM3-5 | 57 | F | TRDN | c.991+2T>A | Splicing | – | – | 123438942 | |
| AM3-6 | 62 | M | TNNT1 | c.G73T | Stopgain | – | 0 | 55146681 |
Dashes indicate that the information is not listed in the database. NM numbers for cDNA variants are as follows: PRDM, NM_022114; DSP, NM_004415; DNM2, NM_001005360; DMD, NM_004015; TTN, NM_133437; RYR1, NM_000540; PLEC, NM_201378; DYSF, NM_001130976; PKP2, NM_001005242; SGCG, NM_000231; MYH7, NM_000257; FLNC, NM_001127487; DYSF, NM_001130455; TRDN, NM_006073; TNNT1, NM_001126133.
AF = allele frequency; AM = acute myocarditis; cDNA = complementary DNA; gnomAD = genome Aggregation Database; LP = likely pathogenic; NM = neutrophil migration; SNV = single nucleotide variation; P = pathogenic; VUS = variant of uncertain significance.
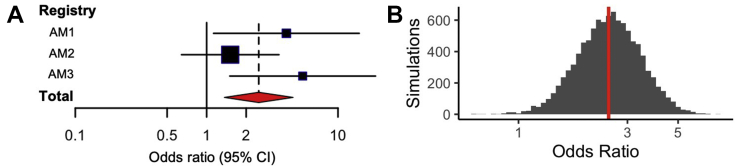
DVs were significantly enriched in AM cases versus control subjects for AM1 (OR: 4.05; 95% CI: 1.13-14.5; P = 0.043) and AM3 (OR: 5.38; 95% CI: 1.5-19.3; P = 0.013) (Table 3). Using a logistic regression model based on subject-level data from all 3 AM cohorts, we observed a statistically significant enrichment of DVs in the pooled AM cohort (OR: 2.50; 95% CI: 1.35-4.54; P = 0.003) (Figure 1A). A resampling procedure was performed as a supporting analysis and confirmed that the results of the pooled analysis were statistically significant (n = 10,000 simulations; OR: 2.48; 95% CI: 1.31-4.54; P = 0.004) (Figure 1B).
Table 3.
Rates of Putatively Damaging Variants in AM Cohorts
| Registry | With DV |
OR (95% CI) | P Value | |
|---|---|---|---|---|
| AM Cases | Control Cases | |||
| AM1 | 5 (19.2) | 6 (5.6) | 4.05 (1.13-14.5) | 0.043 |
| AM2 | 8 (11.1) | 22 (7.6) | 1.51 (0.64-3.55) | 0.344 |
| AM3 | 6 (31.6) | 6 (7.9) | 5.38 (1.5-19.3) | 0.013 |
Figure 1.
Pooled Burden Analysis of Putatively DVs in Patients With AM
(A) A pooled burden analysis of deleterious variant (DV) burden in 3 acute myocarditis (AM) cohorts (AM1, AM2, AM3) identified a statistically significant enrichment of DVs in patients with AM compared with in control subjects (logistic regression odds ratio and 95% CI shown as red diamond; P = 0.003). (B) In a supporting analysis, 10,000 simulations of resampling with replacement within each study based on study sample size (gray) also identified a statistically significant DV burden in patients with AM (odds ratio: 2.48 shown as red line, 95% CI: 1.31-4.54; P = 0.004).
AM-associated genes span associations with multiple myocyte phenotypes
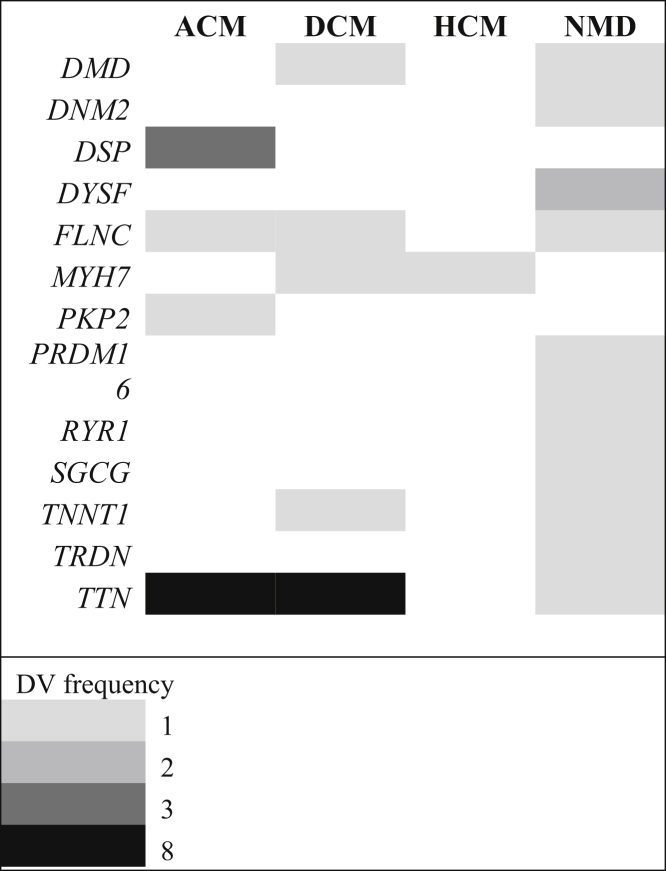
Among subjects with AM, DVs were found in 13 of the 93 genes on the TGP. A single DV was noted in each of 10 of these genes across the 3 AM cohorts. Three genes bore DVs in multiple subjects; relevant variants were found in DSP (n = 3), DYSF (n = 2), and TTN (n = 8) in 3, 2, and 7 subjects with AM, respectively. One TTN allele (c.38004delA) was shared in common between 2 subjects from AM3, who were subsequently determined to be siblings. The Mendelian phenotypes traditionally associated with the 13 AM genes are ACM (n = 2, 15.4%), dilated cardiomyopathy (n = 4, 30.8%), and NMD (n = 8, 61.5%) with several genes known to overlap across these phenotypes (Figure 2).
Figure 2.
AM Genes and Traditional Associated Phenotypes
The numbers1, 2, 3, and 8 indicate the DV frequencies. ACM = arrhythmogenic cardiomyopathy; AM = acute myocarditis; DCM = dilated cardiomyopathy; DV = deleterious variant; HCM = hypertrophic cardiomyopathy; NMD = neuromuscular disease.
Although truncating variants in TTN have been identified in 1% to 2.9% of control populations (27), the rate of TTN DV in our AM cohort (7 of 117 subjects, 6%) was significantly higher than this background prevalence (P = 0.019).
Genotype-phenotype associations in the AM cohort
Detailed histologic data were available for 116 of 117 subjects, enabling classification (Table 4, Supplemental Table 2). DV+ status was more frequent in lymphocytic (16 of 87, 18.4%) versus nonlymphocytic (eosinophilic, giant cell, or granulomatous; 2 of 29, 6.9%) cases, but this association did not achieve significance (P = 0.23). Notably, in AM2, the only cohort without statistical enrichment of DVs, >40% of cases were non-lymphocytic.
Table 4.
Histologic AM Type
| Registry | AM Type | Lymphocytic | Giant Cell | Eosinophilic | Granulomatous |
|---|---|---|---|---|---|
| AM1 | DV+ | 5 (19.2) | – | – | – |
| DV− | 21 (80.8) | – | – | – | |
| Total | 26 (100.0) | – | – | – | |
| AM2a | DV+ | 5 (7.0) | 1 (1.4) | – | 1 (1.4) |
| DV− | 37 (52.1) | 11 (15.5) | 13 (18.3) | 3 (4.2) | |
| Total | 42 (59.2) | 12 (16.9) | 13 (18.3) | 4 (5.6) | |
| AM3 | DV+ | 6 (31.6) | – | – | – |
| DV− | 13 (68.4) | – | – | – | |
| Total | 19 (100.0) | – | – | – |
Viral etiologies were proven by polymerase chain reaction of myocardial tissues for 8 of 19 subjects who were DV+ (42.1%) and 34 of 99 subjects who were DV− (34.3%; P = 0.60) without statistical predominance of any specific pathogen between groups (Table 5), which is similar to rates of viral genomes found in other published AM cohorts (26%-38%) (17,23).
Table 5.
Results of Viral PCR on Myocardial Specimens From Subjects Who Were DV+ vs. DV− in AM1, AM2, and AM3
| Adenoviruses | HHV6 | Enteroviruses | Parvovirus B19 | Not Found | |
|---|---|---|---|---|---|
| DV+ | 0 | 1 | 2 | 5 | 11 |
| DV− | 10 | 5 | 6 | 13 | 65 |
| Total | 10 | 6 | 8 | 18 | 76 |
Values are n. One subject had both parvovirus B19 and HHV6 DNA in myocardium.
HHV6 = human herpesvirus 6; PCR = polymerase chain reaction; other abbreviations as in Table 1.
Outcomes data were available for AM1 and AM3 but not AM2. There were 5 of 11 subjects who were DV+ (45.5%) from these 2 cohorts who either died or required transplant or left ventricular assist device versus 14 of 35 subjects who were DV− (40%; P = 1.00). In AM3 (Supplemental Table 3), where ejection fractions (EFs) during acute illness and at follow-up were available on all subjects, 2 of 6 subjects who were DV+ experienced persistence of cardiomyopathy versus normalization of left ventricular systolic function; 11 of 14 subjects who were DV− (78.6%) had chronic cardiomyopathy versus recovery of function (P = 0.12). Among all subjects for whom acute EF data was available (n = 83, from AM2 and AM3 only), DVs were found in 12 of 55 with EF <40% versus 2 of 28 with EF ≥40% (P = 0.12).
Of the 2 children (ages 18 months and 9 years) with multiallelic DVs, histology revealed lymphocytic AM and myocardial viral RNA/DNA was not found in both, and outcomes were transplant and ventricular assist device, respectively.
Discussion
Risk for the development of AM is likely multifactorial, caused by both environmental exposure and underlying genetic vulnerability of the substrate. Modifiers may include sex (28), gut microbiome (29), vitamins, and trace elements (30), as seen in animal models. The particular interplay between viruses and host genetics is highlighted by previous rodent studies demonstrating that genetic dystrophin deficiency increases enterovirus-induced CMP (31) and that coxsackievirus B3 targets the cardiomyocyte dystrophin-glycoprotein complex to impair sarcolemmal integrity (32).
Shifts toward genomic paradigms have recently occurred for several myocardial diseases that were previously attributed to environmental, toxic, or immunologic factors. Pregnancy-associated and peripartum CMP were both long without clear delineation of risk factors, although parity, gravidity, and history of preeclampsia were suggested. In recent years, however, several familial and registry studies have linked these conditions with pathogenic variants in CMP genes (26,33,34). Ware et al (35) found that 15% of women with peripartum cardiomyopathy harbored rare truncating variants among 8 CMP genes, similar to rates seen in patients with dilated CMP (17%) and significantly higher than in a reference population (4.7%); the most frequently implicated gene was TTN. Heart failure guidelines have recently been updated to acknowledge these results and recommend genetic testing in women with peripartum CMP (12). As a second example, chemotherapy-induced CMP occurs as a complication in 9% of patients treated with anthracyclines, and clinical factors do not fully account for susceptibility (36). Several recent case studies and series have illuminated the role of genetics in this condition, identifying pathogenic variants in the dilated CMP genes TTN (37,38) and MYH7 (39) among affected individuals. These data suggest that genetic substrate plays a critical role in cardiomyopathic outcome among patients exposed to certain environmental or toxic factors. This “2-hit” hypothesis may extend to AM (with infectious exposure as the second hit) given similar associations and patterns of genetic enrichment.
Our previous work and that of others supports the 2-hit theory through identification of deleterious variation in cardiomyocyte genes in pediatric AM cases and cohorts. The current investigation extends this in the largest genomic investigation of patients with AM to date studying a primarily adult population. We found significant enrichment in putatively damaging variants in genes related to cardiomyocyte sarcolemmal, desmosomal, and cytoskeletal structures and functions among subjects with AM compared with in their age-, sex-, and ancestry-matched control subjects. The rates of these variants were not statistically different between pediatric and adult cases, nor between male or female subjects. Most of the relevant variation was heterozygous, but 2 pediatric cases harbored multiple DVs; although our prior ES study of children with AM identified homozygosity and compound heterozygosity, double heterozygosity may also confer a higher risk of AM in younger individuals. Three genes (DSP, DYSF, TTN) were implicated in multiple subjects, raising the possibility that the encoded proteins and downstream pathways are of particular importance for AM susceptibility. Indeed, putatively damaging variants in DSP, along with PKP2 (another closely related desmosomal ACM gene) were also identified among subjects in our prior pediatric AM cohort. DSP has also been implicated in several other recent case reports of myocarditis (6,40,41). The AM genes observed in this study are traditionally associated with both CMP (arrhythmogenic, dilated, hypertrophic) and NMD phenotypes. Within the “final common pathway” framework, these findings suggest that impairments in the structural integrity of cardiomyocytes render them susceptible to injury by infectious pathogens.
Although the subgroups in this study were small, we did not observe any statistically significant associations between DV status and either presence or absence of myocardial viral genomes or AM outcome, although there was a trend toward higher rates of DVs in lymphocytic histologic subtypes. Interestingly, in the only cohort in which DVs were not statistically enriched, almost one-half of subjects had nonlymphocytic histology and 5 of 7 DV+ cases were in subjects with lymphocytic histology; the other 2 cases were a subject with giant cell histology and a stopgain MYH7 variant and a subject with granulomatous histology and a missense DYSF variant. The trend of DVs seen less frequently in giant cell, granulomatous, and eosinophilic histologic cases suggests that mechanisms for susceptibility to such forms of AM may fall outside of the final common pathway. Overall, although this study was not powered to assess clinical outcome by DV status, our findings suggest that the presence of DVs may not accelerate or worsen AM course. Future studies including larger numbers of patients with AM with uniform clinical data will be needed to clarify whether DV+ status is merely 1 hit in the 2-hit mechanism, or rather it alters outcomes (ie, brisker inflammatory response).
Recognizing the genetic underpinnings in AM not only contributes to mechanistic insight, it may also be useful for clinical assessments. The lack of association with any specific clinical features suggests that only genetic testing can uncover DV+ cases. For both autosomal dominant and recessive forms of genetic AM, family screening may be warranted to identify and counsel potential at-risk relatives. A recent study of genetic testing in clinically suspected AM revealed 56% of cases harboring pathogenic variants in CMP genes (predominantly ACM, frequently PKP2 but also DSP) (42). In the present study, a 39-year-old man (AM3-3) developed AM caused by parvovirus B19, with an acute EF of 14% and required transplantation. Four years later, his brother (AM3-4), age 46 years, developed AM with acute EF of 30%, improving only to 42% at follow-up; no myocardial viral RNA/DNA was found in that case. These 2 individuals shared the same presumed genetic AM risk factor, a pathogenic variant in TTN. Indeed, review of the medical records revealed a physician letter documenting a history of dilated cardiomyopathy after parvovirus B19 infection in their mother. These cases further highlight the value of genetic testing and family screening, as the second brother could have been counseled on antiviral precautions or managed differently at the early stages of AM had his genotype been known. As with peripartum CMP, clinical genetic testing for AM may one day be recommended within society guidelines. Based on our findings, a panel comprising both CMP and NMD genes may be of the highest yield.
Study limitations
The present analyses were limited by differences in the extent of data available within the 3 AM cohorts. Detailed histologic data were only available for 2 of the cohorts. Furthermore, all 3 cohorts comprised individuals predominantly from European ancestry groups. Genomic findings derived from this study may not be applicable in more diverse populations.
CONCLUSIONS
Putatively damaging variants in genes related to cardiomyocyte structure and function are enriched in patients with AM compared to sex- and ancestry-matched controls individuals. Clinical outcomes were similar between patients with or without genetically-mediated AM, highlighting the value of genetic testing to identify genotype-positive cases and perform downstream cascade testing in families.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE 1: AM is an important cause of heart failure and CMP in adults and children. Although risk factors are not fully understood, some cases of myocarditis have been linked to suspected pathogenic variants in cardiac genes traditionally associated with CMPs.
COMPETENCY IN MEDICAL KNOWLEDGE 2: In this study, putatively damaging variants among 93 cardiac genes were enriched in subjects with myocarditis compared with in their age-, sex- and ancestry-matched control subjects.
COMPETENCY IN MEDICAL KNOWLEDGE 3: Genes associated with myocarditis included DMD, DNM2, DSP, DYSF, FLNC, MYH7, PKP2, PRDM16, RYR1, SGCG, TNNT1, TRDN, and TNN, which encode proteins spanning a range of sarcolemmal, desmosomal, sarcomeric, and other cytoskeletal cardiomyocyte constituents. Impairments in these proteins and related pathways may therefore confer vulnerability to viral infection.
COMPETENCY IN MEDICAL KNOWLEDGE 4: Because myocarditis phenotypes of subjects with and without putatively damaging variants were similar, genetic testing is necessary to identify those at increased genetic risk.
COMPETENCY IN MEDICAL KNOWLEDGE 5: As with other intermediate cardiac phenotypes in which genetic background interacts with host environmental factors (eg, peripartum CMP), clinical genetic testing may one day be included in society guidelines and should include a broad set of genes, including those that have traditionally been associated with neuromuscular disorders having cardiac involvement.
COMPETENCY IN MEDICAL KNOWLEDGE 6: It is important to counsel patients with myocarditis about the possibility of genetic associations so family members may be screened. Because the “myocarditis genes” overlap with other disorders, relatives may also be at risk for CMP and/or neuromuscular disease.
TRANSLATIONAL OUTLOOK: Understanding the role of host cardiomyocyte genetics on viral susceptibility should improve mechanistic understanding of the pathways leading to myocarditis and potentially inform new therapies. In vitro and animal models of myocarditis should incorporate deleterious variants in cardiac genes such as those identified in this study.
Funding Support and Author Disclosures
This study is supported in part by the National Heart Lung and Blood Institute of the National Institutes of Health (HL140083 [to Dr Kontorovich]) and (HL135742 [to Dr Gelb]), a grant from the Children’s Cardiomyopathy Foundation (to Dr Kontorovich), and the German Research Foundation (KL593 2-3 [to Dr Klingel]). Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health (S10OD026880). Funders had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors acknowledge the Charles Bronfman Institute for Personalized Medicine at the Icahn School of Medicine at Mount Sinai for use of BioMe resources.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental methods and tables, please see the online version of this paper.
Appendix
References
- 1.Pollack A., Kontorovich A.R., Fuster V., Dec G.W. Viral myocarditis—diagnosis, treatment options, and current controversies. Nat Rev Cardiol. 2015;12(11):670–680. doi: 10.1038/nrcardio.2015.108. [DOI] [PubMed] [Google Scholar]
- 2.Fabre A., Sheppard M.N. Sudden adult death syndrome and other non-ischaemic causes of sudden cardiac death. Heart. 2006;92(3):316–320. doi: 10.1136/hrt.2004.045518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felker G.M., Hu W., Hare J.M., Hruban R.H., Baughman K.L., Kasper E.K. The spectrum of dilated cardiomyopathy: the Johns Hopkins experience with 1,278 patients. Medicine (Baltimore) 1999;78(4):270–283. doi: 10.1097/00005792-199907000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Mavrogeni S., Papavasiliou A., Spargias K. Myocardial inflammation in Duchenne Muscular Dystrophy as a precipitating factor for heart failure: a prospective study. BMC Neurol. 2010;10:33. doi: 10.1186/1471-2377-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles N.E., Ni J., Marcus F., Towbin J.A. The detection of cardiotropic viruses in the myocardium of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol. 2002;39(5):892–895. doi: 10.1016/s0735-1097(02)01688-1. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Ayala J.M., Pastor-Quirante F., Gonzalez-Carrillo J. Genetics of myocarditis in arrhythmogenic right ventricular dysplasia. Heart Rhythm. 2015;12(4):766–773. doi: 10.1016/j.hrthm.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Belkaya S., Kontorovich A.R., Byun M. Autosomal recessive cardiomyopathy presenting as acute myocarditis. J Am Coll Cardiol. 2017;69(13):1653–1665. doi: 10.1016/j.jacc.2017.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown E.E., McMilllan K.N., Halushka M.K. Genetic aetiologies should be considered in paediatric cases of acute heart failure presumed to be myocarditis. Cardiol Young. 2019;29(7):917–921. doi: 10.1017/S1047951119001124. [DOI] [PubMed] [Google Scholar]
- 9.Bowles N.E., Bowles K.R., Towbin J.A. The “final common pathway” hypothesis and inherited cardiovascular disease: the role of cytoskeletal proteins in dilated cardiomyopathy. Herz. 2000;25(3):168–175. doi: 10.1007/s000590050003. [DOI] [PubMed] [Google Scholar]
- 10.Towbin J.A. The role of cytoskeletal proteins in cardiomyopathies. Curr Opin Cell Biol. 1998;10(1):131–139. doi: 10.1016/s0955-0674(98)80096-3. [DOI] [PubMed] [Google Scholar]
- 11.Rosenbaum A.N., Agre K.E., Pereira N.L. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020;17(5):286–297. doi: 10.1038/s41569-019-0284-0. [DOI] [PubMed] [Google Scholar]
- 12.Hershberger R.E., Givertz M.M., Ho C.Y. Genetic evaluation of cardiomyopathy: a Heart Failure Society of America practice guideline. J Card Fail. 2018;24(5):281–302. doi: 10.1016/j.cardfail.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldfarb L.G., Park K.Y., Cervenakova L. Missense mutations in desmin associated with familial cardiac and skeletal myopathy. Nat Genet. 1998;19(4):402–403. doi: 10.1038/1300. [DOI] [PubMed] [Google Scholar]
- 14.Savarese M., Sarparanta J., Vihola A., Udd B., Hackman P. Increasing role of titin mutations in neuromuscular disorders. J Neuromuscul Dis. 2016;3(3):293–308. doi: 10.3233/JND-160158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNally E.M., Sparano D. Mechanisms and management of the heart in myotonic dystrophy. Heart. 2011;97(13):1094–1100. doi: 10.1136/hrt.2010.214197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina K.M., Garcia X., Denfield S.W. Parvovirus B19 myocarditis causes significant morbidity and mortality in children. Pediatr Cardiol. 2013;34(2):390–397. doi: 10.1007/s00246-012-0468-4. [DOI] [PubMed] [Google Scholar]
- 17.Bowles N.E., Ni J., Kearney D.L. Detection of viruses in myocardial tissues by polymerase chain reaction: evidence of adenovirus as a common cause of myocarditis in children and adults. J Am Coll Cardiol. 2003;4293:466–472. doi: 10.1016/s0735-1097(03)00648-x. [DOI] [PubMed] [Google Scholar]
- 18.Griffin L.D., Kearney D., Ni J. Analysis of formalin-fixed and frozen myocardial autopsy samples for viral genome in childhood myocarditis and dilated cardiomyopathy with endocardial fibroelastosis using polymerase chain reaction (PCR) Cardiovasc Pathol. 1995;4(1):3–11. doi: 10.1016/1054-8807(94)00025-m. [DOI] [PubMed] [Google Scholar]
- 19.Kindermann I., Kindermann M., Kandolf R. Predictors of outcome in patients with suspected myocarditis. Circulation. 2008;118(6):639–648. doi: 10.1161/CIRCULATIONAHA.108.769489. [DOI] [PubMed] [Google Scholar]
- 20.Ukena C., Kindermann M., Mahfoud F. Diagnostic and prognostic validity of different biomarkers in patients with suspected myocarditis. Clin Res Cardiol. 2014;103(9):743–751. doi: 10.1007/s00392-014-0709-z. [DOI] [PubMed] [Google Scholar]
- 21.Pavlicek V., Kindermann I., Wintrich J. Ventricular arrhythmias and myocardial inflammation: long-term follow-up of patients with suspected myocarditis. Int J Cardiol. 2019;274:132–137. doi: 10.1016/j.ijcard.2018.07.142. [DOI] [PubMed] [Google Scholar]
- 22.ClinGen Clinical Laboratories Meeting Minimum Requirements for Data Sharing to Support Quality Assurance. https://www.clinicalgenome.org/tools/clinical-lab-data-sharing-list/ Accessed March 13, 2019.
- 23.Pauschinger M., Bowles N.E., Fuentes-Garcia F.J. Detection of adenoviral genome in the myocardium of adult patients with idiopathic left ventricular dysfunction. Circulation. 1999;99:1348–1354. doi: 10.1161/01.cir.99.10.1348. [DOI] [PubMed] [Google Scholar]
- 24.Altman D.G. Chapman and Hall/CRC; 1999. Practical Statistics for Medical Research. [Google Scholar]
- 25.Sheskin D. 3rd ed. Chapman and Hall/CRC; 2004. Handbook of Parametric and Nonparametric Statistical Procedures. [Google Scholar]
- 26.Morales A., Painter T., Li R. Rare variant mutations in pregnancy-associated or peripartum cardiomyopathy. Circulation. 2010;121(20):2176–2782. doi: 10.1161/CIRCULATIONAHA.109.931220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts A.M., Ware J.S., Herman D.S. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7(270):270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Florio D.N., Sin J., Coronado M.J., Atwal P.S., Fairweather D. Sex differences in inflammation, redox biology, mitochondria and autoimmunity. Redox Biol. 2020;31:101482. doi: 10.1016/j.redox.2020.101482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil-Cruz C., Perez-Shibayama C., De Martin A. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science. 2019;366(6467):881–886. doi: 10.1126/science.aav3487. [DOI] [PubMed] [Google Scholar]
- 30.Beck M.A. Selenium and vitamin E status: impact on viral pathogenicity. J Nutr. 2007;137(5):1338–1340. doi: 10.1093/jn/137.5.1338. [DOI] [PubMed] [Google Scholar]
- 31.Xiong D., Lee G.H., Badorff C. Dystrophin deficiency markedly increases enterovirus-induced cardiomyopathy: a genetic predisposition to viral heart disease. Nat Med. 2002;8(8):872–877. doi: 10.1038/nm737. [DOI] [PubMed] [Google Scholar]
- 32.Badorff C., Lee G.H., Lamphear B.J. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat Med. 1999;5(3):320–326. doi: 10.1038/6543. [DOI] [PubMed] [Google Scholar]
- 33.van Spaendonck-Zwarts K.Y., Posafalvi A., van den Berg M.P. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur Heart J. 2014;35(32):2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 34.van Spaendonck-Zwarts K.Y., van Tintelen J.P., van Veldhuisen D.J. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation. 2010;121(20):2169–2175. doi: 10.1161/CIRCULATIONAHA.109.929646. [DOI] [PubMed] [Google Scholar]
- 35.Ware J.S., Li J., Mazaika E. IMAC-2 and IPAC Investigators. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374(3):233–241. doi: 10.1056/NEJMoa1505517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardinale D., Colombo A., Bacchiani G. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation. 2015;131(22):1981–1988. doi: 10.1161/CIRCULATIONAHA.114.013777. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Pavia P., Kim Y., Restrepo-Cordoba M.A. Genetic variants associated with cancer therapy-induced cardiomyopathy. Circulation. 2019;140(1):31–41. doi: 10.1161/CIRCULATIONAHA.118.037934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linschoten M., Teske A.J., Baas A.F. Truncating titin (TTN) variants in chemotherapy-induced cardiomyopathy. J Card Fail. 2017;23(6):476–479. doi: 10.1016/j.cardfail.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Wasielewski M., van Spaendonck-Zwarts K.Y., Westerink N.D. Potential genetic predisposition for anthracycline-associated cardiomyopathy in families with dilated cardiomyopathy. Open Heart. 2014;1(1):e000116. doi: 10.1136/openhrt-2014-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith E.D., Lakdawala N.K., Papoutsidakis N. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. 2020;141(23):1872–1884. doi: 10.1161/CIRCULATIONAHA.119.044934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kissopoulou A., Fernlund E., Holmgren C. Monozygotic twins with myocarditis and a novel likely pathogenic desmoplakin gene variant. ESC Heart Fail. 2020;7(3):1210–1216. doi: 10.1002/ehf2.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ader F., Surget E., Charron P. Inherited cardiomyopathies revealed by clinically suspected myocarditis: highlights from genetic testing. Circ Genom Precis Med. 2020;13(4):e002744. doi: 10.1161/CIRCGEN.119.002744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.