Abstract
Purpose
COVID-19 pandemic has emerged as a result of infection by the deadly pathogenic severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), causing enormous threats to humans. Coronaviruses are distinguished by a clove-like spike (S) protein, which plays a key role in viral pathogenesis, evolutions, and transmission. The objectives of this study are to investigate the distinctive structural features of SARS-CoV-2 S protein, its essential role in pathogenesis, and its use in the development of potential therapies and vaccines.
Methodology
A literature review was conducted to summarize, analyze, and interpret the available scientific data related to SARS-CoV-2 S protein in terms of characteristics, vaccines development and potential therapies.
Results
The data indicate that S protein subunits and their variable conformational states significantly affect the virus pathogenesis, infectivity, and evolutionary mutation. A considerable number of potential natural and synthetic therapies were proposed based on S protein. Additionally, neutralizing antibodies were recently approved for emergency use. Furthermore, several vaccines utilizing the S protein were developed.
Conclusion
A better understanding of S protein features, structure and mutations facilitate the recognition of the importance of SARS-CoV-2 S protein in viral infection, as well as the development of therapies and vaccines. The efficacy and safety of these therapeutic compounds and vaccines are still controversial. However, they may potentially reduce or prevent SARS-CoV-2 infection, leading to a significant reduction of the global health burden of this pandemic.
Keywords: SARS-CoV-2, Spike protein, Pathogenesis, Mutations, Treatments, Vaccines
Introduction
The severe acute respiratory syndrome coronavirus referred to as SARS-CoV-2 [76] is an enveloped virus possessing a positive-sense single-stranded RNA of approximately 30 Kb length [30, 76]. This virus has emerged in China, particularly in Wuhan, where several severe pneumonia cases were detected in December 2019 [71]. It was initially transmitted from bats through an intermediate host to humans [31]. The virus is made up of several structural and non-structural proteins each responsible for a particular step in viral infectivity and transmission [31, 71]. The spike (S), envelope (E), membrane (M), and nucleocapsid (N) constitute the structural proteins of the virus [31, 71]. People infected with SARS-CoV-2 may experience mild to moderate flu-like symptoms such as fever, cough, diarrhea, and shortness of breath as the most common symptoms [31]. Severe cases that are reported with high-risk individuals include respiratory failure and systemic inflammation, which in turn, can lead to further tragic consequences including multi-organ damage [31]. Unfortunately, asymptomatic patients can contribute to viral transmission [31]. Due to the rapidly increased number of SARS-CoV-2 cases, it was declared by the WHO as a global pandemic. At the time of writing and as per the global statistics, 188,332,972 M are currently confirmed cases, while serious COVID-19 conditions constitute 0.6%, and the remaining 99.4% are relatively mild. The total recovery rates are estimated to be ~ 98% with 2% death rates [105].
Features of S protein
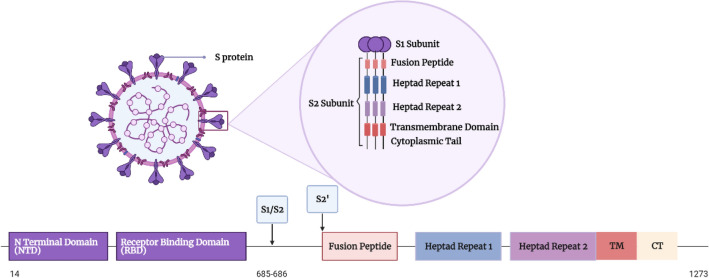
SARS-CoV-2 S protein is a class I fusion transmembrane structural glycoprotein that is composed of S1 and S2 subunits [71]. It is a homotrimer with a size of 180–200 kDa [43], and a total length of between 1273 and 1300 amino acids [113]. The amino acid residues (1–13) constitute a signal peptide [43]. It is a clove-shaped structure [71], protrudes from the surface of the virus (Fig. 1), allowing it to carry the viral binding to the host cell and fusion [30]. It is the basic unit of the virus that recognizes and binds to the host cell receptor ACE-2 [43]; thus, rendering the S protein a main target for neutralizing antibodies.
Fig. 1.
Diagram showing the domain organization of the S protein of SARS-COV-2. NTD N-terminal domain, RBD receptor-binding domain, FP fusion peptide, HR1 heptad repeat 1, HR2 heptad repeat 2, TM transmembrane domain, CT cytoplasmic tail. S1/S2 and S2’: Cleavage sites of the S protein
S1 subunit
S1 is a distal subunit that is composed of 672 amino acids (14–685) residues [30]. These amino acids are organized into four domains: N-terminal domain (NTD), C-terminal domain (CTD), or the receptor-binding domain (RBD), and two subdomains (SD1 and SD2) (Fig. 1) [30]. It is a V-shaped ectotrimer subunit that accommodates one ACE-2 recognition motif per monomer for binding [93]. At the apex of each S1 monomer, there is one RBD [93]. It was revealed that the RBD of SARS-CoV-2 is made up of a core that is built of twisted five-stranded antiparallel β-sheet of β1, β2, β3, β4 and β7 [93]. There is an extended loop located between β4 and β7, which contains short β5 and β6 strands and α4 and α5 helices and loops [93]. This extended loop is known as the receptor-binding motif (RBM), containing most of the amino acids of the RBD that form the interactions with the ACE-2 receptors [30]. The aminopeptidase N region in ACE-2 is responsible for RBD binding [43]. In the RBD, there are nine cysteine residues; eight of them form four pairs of disulfide bonds and three of these pairs are in the core that help in β-sheet structure stabilization, while the remaining pair connects the loops in the RBM distal end [63].
Analysis of the RBD and ACE-2 interface revealed that there are 17 amino acids of RBD in contact with 20 amino acids of ACE-2 [63]. An important feature at the RBD and ACE-2 interface is the hydrophilic interactions network as there are 13 hydrogen bonds and 2 salt bridges [63]. Another feature is the involvement of multiple tyrosine amino acids residues including Tyr449, Tyr 489, and Tyr505 from the RBD that form hydrogen bonds [63]. Additionally, outside the RBM, there is a unique interaction between Lys417 residue in RBD, which forms salt-bridge interactions with Asp30 of ACE-2 [63]. It has been proposed that inhibition of the interaction between RBD and ACE-2 can be useful in the treatment of SARS-CoV-2 infection [63]. However, the RBD is a highly mutable region; thus, it is not an ideal target site for the development of broad-spectrum antiviral [43].
S2 subunit
S2 is a membrane-anchored subunit of 588 amino acid (686–1273) residues (Fig. 1) [30]. It is composed of N-terminal hydrophobic fusion peptide (FP), two heptad-repeat regions (HR1 and HR2), a transmembrane domain (TM), and a cytoplasmic tail (CT) [30]. FP is a short segment of 15–20 amino acids, mainly hydrophobic, such as glycine (G) or alanine (A) [43]. Earlier research has pointed out the vital role of FP in membrane fusion, as it causes disruption and connection to the host cell membrane lipid bilayer [43]. The location of HR1 is at the C-terminus of FP, while HR2 is located at the N-terminus of the TM domain [43]. They are composed of a repetitive heptapeptide HPPHCPC, where H is a hydrophobic or bulky residue, P indicates a hydrophilic residue, and C is a charged amino acid [43]. These regions are critical for viral fusion and entry by forming a six-helical bundle (6-HB) [43]. The TM domain attaches the S protein to the viral membrane [43].
Glycan shield of S protein
S protein is coated with polysaccharides [43]. The S protein gene encodes 22 N-linked glycan sequins per monomer resulting in 66 N-linked glycans in one S glycoprotein [93]. In comparison with S2 monomers, S1 monomers are more decorated with N-linked glycans [93]. This glycan shield has several effects on the S protein folding, its processing by host cell proteases, immune evasion, and the elicitation of humoral immune response as well [93]. Extensive glycan shielding of S protein that blocks the protein surface may thereby, hide specific epitopes from antibody neutralization, thus, masking them and facilitate the immune evasion [93]. Moreover, it was observed that both the glycosylated and de-glycosylated S ectodomains are bound to ACE-2 with almost identical affinities (1.7 nM vs 1.5 nM); hence, it was proposed that S protein glycosylation does not alter the S protein-binding affinity to ACE-2 [30]. However, it is suggested that the glycan shield of SARS-CoV-2 S protein is less dense and less effective compared to glycoproteins of other viruses such as HIV-1, and this might be beneficial for humoral immunity induction and vaccine development [30]. Therefore, there is a huge interest in investigating the potential immunogenicity of the glycan moieties as vaccine candidates [30, 118].
Cleavage site
Cleavage of S protein into S1 and S2 subunits by host proteases is a fundamental step to mediate the fusion process (Fig. 1) [43]. It is usually cleaved by host proteases such as Transmembrane Protease Serine 2 (TMPRSS2), cathepsin L and furin [41], hence, facilitating the viral activation and representing SARS-CoV-2 pathogenicity [72]. In fact, activation of viral spikes could be mediated by other proteases found on host cells such as TMPRSS4, TMPRSS11A, TMPRSS11D and TMPRSS11E1 [72]. A notable feature of the S protein in SARS-CoV-2 is the presence of a unique polybasic furin-like cleavage site (FCS) located at the boundary between the S1 and S2 subunits “S1/S2 site” which increases the rate of its proteolytic processing by host-furin protease during SARS-CoV-2 infection [110]. This, in turn, enhances its infectivity and makes it more contagious [43].
Conformations
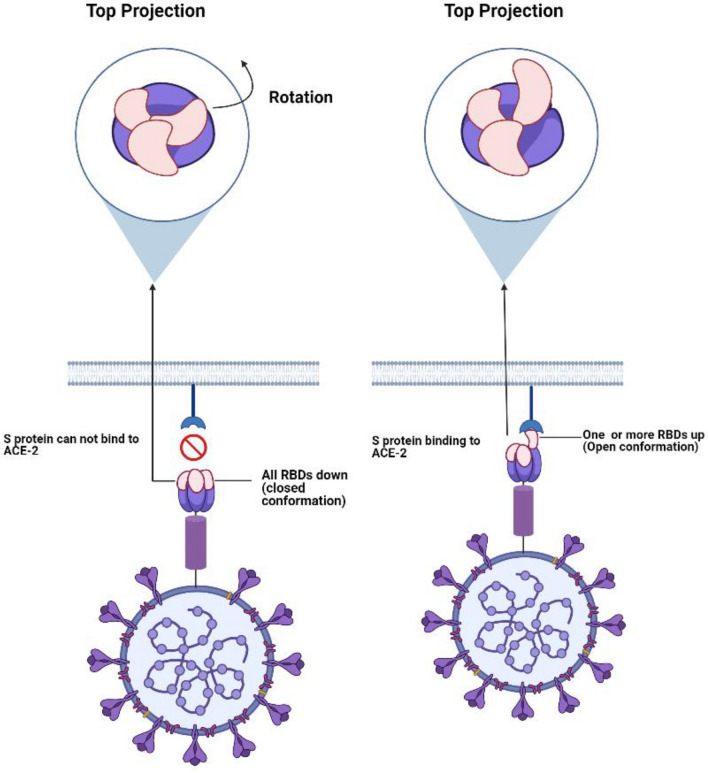
Several structures of S protein are associated with different states, which mainly include the pre-fusion and post-fusion conformations (Fig. 2) [15]. In the metastable pre-fusion conformation, the RBDs may exist in either open or closed forms [30, 31]. In the closed form, all the three RBDs are in the down position representing a (receptor inaccessible) status that hides the receptor-binding region [71]. Due to their flexibility, the RBDs undergo hinge-like movements to a partially open (receptor accessible) state that exposes the receptor-binding region [71]. In this state, one single or more RBDs might be in the “up” position [40]. These forms may co-exist together in varying percentages [101]. However, the closed status is the predominant one [101]. Binding to ACE-2 can stabilize this open dynamic status, thereby undergoing conformational changes through different intermediates, and ultimately, transforming it from pre-fusion to post-fusion conformation [101]. Upon using them as immunogens, these different conformations can induce distinct immune-mediated antibodies constructing a challenge for optimum immunization [101, 111]. Therefore, several research studies are trying to stabilize the S protein conformation [101, 111]. This would have several advantages, as it allows successful vaccine development, improves diagnostics, and additionally, supports the basic research due to its enhanced tolerability for storage conditions on prolonged times [111].
Fig. 2.
Diagram showing the conformational changes of SARS-COV-2 S protein during the binding of ACE-2 receptor
Role of SARS-CoV-2 S protein in viral fusion and host entry
In proximity to the fusion peptide, a structure of the fusion peptide proximal region (FPPR) was found to play a major role in S protein conformational rearrangements [15]. This FPPR is close to the S1/S2 and S2' cleavage sites as well [15]. A crosstalk between the FPPR and the RBDs in the up position causes the latter to clamp down [15]. However, due to their high dynamic energy, the RBDs can flip out reforming the “up” conformation [15]. This partially open conformation allows RBD binding to ACE-2 on the host cell [15]. Consequently, triggering the cleavage at S1/S2 site causing S1-ACE-2 dissociation [51, 101], and creating a flexible FPPR that exposes the S2' cleavage site, leading to the release of fusion peptide and insertion into the host membrane [15, 43]. A subsequent interaction between the HR2 and HR1 takes place exposing the pre-hairpin coiled-coil of the HR1 domain and forming a six-helical bundle (6-HB) [101]. Ultimately, bringing the viral envelope close to the cell membrane for viral fusion and entry [101].
Evolutionary pattern of S protein supports the virus infectivity and transmission rates
SARS-CoV-2 has a zoonotic origin [62], where the recent outbreaks have revealed its capability of crossing the inter-species barrier and transmission from animals to humans at any time with unpredictable consequences [61, 113]. In fact, knowing the virus's ability to evolve over time along with its mutation rate would help better to understand the virus transmission, pathogenesis, and infectivity threats [31, 71]. Further, it can help to understand the evasion mechanism of the immune system, the development of drug resistance and hence the development of precise and effective therapy [31, 71].
To better understand SARS-CoV-2 evolution, genome sequencing has been carried out for several strains isolated during the current pandemic [47]. Several genetic evolutionary features were identified and reported, showing that SARS-CoV-2 shares 79.6% of its whole genome sequence with SARS-CoV-1 [109]. Interestingly, SARS-CoV-2 S protein shares ~ 79% amino acid sequence identity with SARS-CoV-1 S protein [31, 48]; hence, the remaining non-conserved nucleotide sequences may explain the different antigenic properties of the new strain [71]. This evolutionary feature is consistent with its major role in viral entry as it confronts the immune system [89]. Based on the large variability in the binding patterns exhibited by RBD and S1-NTD, it can be concluded that both sites are of intense evolutionary pressure [89].
Studies have shown that most evolutionary mutations occurred to both sites represented by a unique pattern of amino acids residues [89]. Such residues with rapid molecular evolution are characterized by an increased flexibility of the protein, being in a closer conformation to ACE-2 interface and important binding sites, and having higher similarity with the host cell surface protein, which in turn increase the potential immune evasion [89]. Moreover, SARS-CoV-2 S protein exhibited a 10–22-fold higher binding affinity to ACE-2 than that of SARS-CoV-1 [101]. This is attributed to the increased contacts in the interface, and the three mutational hotspots in the S protein, which results in a more specific and compact conformation that contributes to the host infectivity range of SARS-CoV-2 [62]. Eventually, being an RNA virus, it is expected that SARS-CoV-2 will continue evolving over time [101].
Notable evolutionary features of SARS-CoV-2 S protein can be summarized as below
Evolutionary mutations of RBD regions responsible for ACE-2 binding
Generally, 20 residues of ACE-2 are responsible for the binding interaction with 16 and 17 amino acid residues of the RBDs of both SARS-CoV-1 and SARS-CoV-2, respectively [63]. Lys417 residue at the SARS-CoV-2 RBD has a positively charged patch that generates an electrostatic potential, which in turn contributes to the higher SARS-CoV-2 RBD-ACE-2-binding affinity when compared to that of SAS-CoV-1 [63]. There are other five residues with different side chains in the two RBDs, yet they share the same biochemical properties [63]. However, Leu472 of SARS-CoV-1 forms interactions with Leu79 and Met82 of ACE-2, while the Phe486 of SARS-CoV-2 forms additional interactions with different amino acid residues of ACE-2 [63]. Furthermore, Asn479 residue of SARS-CoV-1 interacts solely with His34 of ACE-2, while the Gln493 of SARS-CoV-2 forms bonds with Lys31, His34 and particularly, a hydrogen bond with Glu35 of ACE-2 [63]. Additionally, the substitution of Val of SARS-CoV-1 with a lysine residue in SARS-CoV-2 has given it an advantage of a special salt-bridge formation upon the interaction between Lys417 and Asp30 of ACE-2 [63]. A feature that is not permitted by the valine residue of SARS-CoV-1, which explains the stronger interaction between SARS-CoV-2 and ACE-2 receptor and contributes to the higher transmission of SARS-CoV-2 [76, 90]. SARS-CoV-1-receptor-binding motif (RBM) contains a three-residue motif in one of its ridge loops, whereas SARS-CoV-2 contains a four-residue motif [76, 90]. This four-residue motif grants an additional hydrogen bond formation in SARS-CoV-2 RBM causing its loop to move closer to ACE-2, and thus establishing a more compact form [76, 90]. This allows more contact with the N-terminal helix of ACE-2 where a larger binding interface is formed [76, 90].
Virus-binding hotspots on ACE-2 that involve two lysine residues play a significant role in virus binding [90]. A key factor for perfect binding is the neutralization of charges on these lysine residues [90]. This is achieved by specific amino acids, which stabilize these hotspots [90]. This exclusive stability pattern of SARS-CoV-2 may explain its higher infectivity and transmission rates [116].
Polybasic furin cleavage site
SARS-CoV-2 is characterized by a unique polybasic cleavage site at the S1/S2 junction, which is thought to play a critical role in the determination of the viral infectivity, pathogenesis, and transmissibility [116]. It is expected to be responsible for the high virulence of the virus [76]. S protein is pre-activated by furin and furin-like proprotein convertases in the viral cell [48]. This is contrary to SARS-CoV-1, which lacks this furin cleavage site [107].
O-linked glycans
A proline residue has been identified at the S1/S2 junction in SARS-CoV-2 [49], which is predicted to be the reason for the addition of O-linked glycans to S673, T678 and S686 [9]. Its accurate function is unclear; however, it may contribute to the creation of a “mucin-like domain”, which is utilized by the virus as a glycan shield [9]. Yet, additional studies are required to prove their significance in SARS-CoV-2 [9]. Although it has been suggested that this glycan shield may contribute to the virulence of the virus by masking the epitopes, it may flank the cleavage site preventing it from being processed by the activating proteases [49].
SARS-CoV-2 S protein unique mutations
Several nucleotide polymorphisms have been detected in SARS-CoV-2 genomes including 753 missense, 452 silent, 12 nonsense, and four intergenic substitutions [31]. S protein showed a high share of genetic alterations with a mutation rate of ~ 1.21 [31]. These mutations are highly variable as per the geographic distribution, sequencing time and inter-stains recombination [57]. This can also be attributed to its rapid global spread [61]. Nevertheless, other mutations were reported to co-occur [101], thereby hindering the desired complete understanding of its impact on viral transmission, virulence, and disease severity.
D614G mutation of S protein
G614 is a missense mutation in the gene encoding S protein [47]. To highlight, D indicates the aspartic acid residue that is replaced by glycine (G) amino acid at position 614 (D614G) [71]. This is located at the subdomain 2 (SD2) of the S1 subunit outside the RBD [109]. It has been reported that D614G mutation is associated with greater infectivity and transmissibility across different geographic locations [61]. This mutation has increased the viral loads in the respiratory secretions; however, it does not seem to affect the disease severity [104]. Furthermore, the unique multi-basic furin cleavage site has rendered the S protein less stable and triggers the virus to undergo this mutation in order to compensate for this instability [48].
Several studies have demonstrated that the gain of infectivity is correlated to the fact that spike variant D614G favors an open conformational state [38]. Mutation of aspartic acid residue to a smaller glycine amino acid might increase the flexibility, resulting in an easy shift from the closed “down” state to the opened “up” state [61]. Moreover, D614G causes disruption of a significant salt bridge formed between the monomers of S protein by increasing the distance between the amino acids that form this salt-bridge interaction [109]. Therefore, it enhances the accessibility of RBD, promoting the virus binding and fusion into the cell [109]. Of note, limited S1 shedding and higher S protein density were reported in the G614 virion [31, 116]. This, in turn, has raised new hypotheses by which D614G mutation has increased the virus infectivity and enhanced the efficiency of entering ACE-2-expressing cells, and accordingly, increasing the functional S protein incorporation into the virion. This mechanism is referred to as the “density hypothesis” [31, 116]. Another hypothesis “stability hypothesis” proposes that the G614 glycine residue increases the number of binding sites by stabilizing the interaction between S1 and S2 through its loop, which is inserted between NTD and CTD1, and subsequently, reduces the S1 shedding [48, 116, 117]. However, further investigation is required to understand the exact mechanism adopted by this mutation to increase the virus infectivity and transmission [48]. Originally, many SARS-CoV-2 vaccines were designed utilizing the D614 variant of the spike, accordingly, concerns have arisen regarding the G614 mutation consequences on vaccines effectiveness [37]. Furthermore, the majority of data have shown that the G614 variant is more vulnerable to neutralization by RBD-specific monoclonal antibodies and serum samples from people who were previously infected [104]. This can be explained by the higher exposure of the neutralization epitopes in this variant due to the higher percentage of the up conformation of RBD [104].
SARS-CoV-2 S protein as a potential target for therapy and vaccine development
The aforementioned data indicate that in addition to the immunogenic properties of SARS-CoV-2 S protein [45], it is the main responsive for initiation of viral entry, pathogenesis and transmission [79, 87]. Therefore, multiple therapeutic agents have been proposed against S protein. On the other hand, several targets namely, S (including RBD, S1-NTD and S2 subunit), N, M, and E proteins have been studied for possible vaccines development [87]. In fact, E and M proteins are poorly immunogenic, while the N protein, although it is abundant and highly immunogenic and exhibited protection against CoVs in a mouse model, it does not protect against SARS-CoV-2 infection and instead enhanced infection-induced pneumonia. Hence, none of these proteins is a wise choice for the development of SARS-CoV-2 vaccines [25]. Therefore, the optimal target to elicit potent neutralizing antibodies, specific for SARS-CoV-2 is the S protein. Currently, the majority of protective countermeasures are S-based vaccines, and hence they exhibit protective efficacy and an acceptable safety profile [112]. In general, targeting S1-NTD and S2 subunits induce antibodies with neutralizing activities; however, the RBD is considered the major target to elicit highly potent neutralizing antibodies, while providing more selectivity against the current SARS-CoV-2 strain [112].
Potential SARS-CoV-2 S protein-based therapies
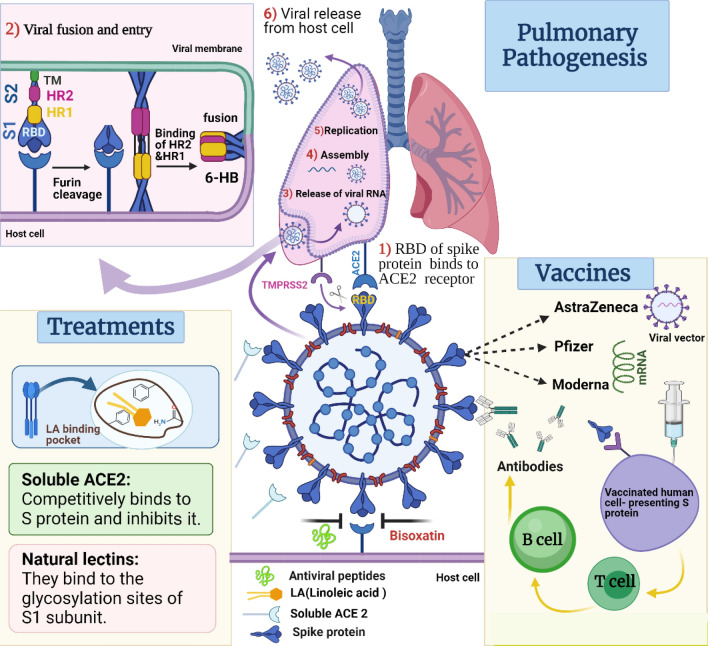
The important features of SARS-CoV-2 S protein make it an excellent target for the development of therapeutic drugs (Table 1; Fig. 3) [9, 97].
Table 1.
Summary of the different potential therapies targeting SARS-CoV-2 S protein
| Treatment | Source | Mechanism of action | Clinical phases (87) |
|---|---|---|---|
| Natural/ Repurposed products | |||
| Linoleic acid | N-6 polyunsaturated fatty acid (PUFA) obtained from vegetable oil | It binds to LA-binding pocket in RBDs of S Protein | NA, Potential drug |
| Emodin | Natural anthraquinon | Potentially inhibits the S protein and ACE-2 receptor interaction, suppresses the inflammation associated with COVID-19 | NA, Potential drug |
| Bisoxatin | Synthetic drug | Significantly binds to RBD of S protein | NA, computational studies, potential lead compound |
| Natural lectins | |||
| GRFT (Griffithsin) | Algae-derived lectin isolated from the red alga Griffithsia sp. | Binds to the glycosylation sites of S1 subunit, preventing the subsequent steps essential for viral entry | In vitro studies, Phase: NA |
| FRIL | Glucose/ mannose lectin isolated from Lablab purpureus plant | Binds to the high-mannose glycans and complex-type N-glycans | In vitro studies |
| Antiviral peptides | |||
| EK1C4 | Synthetic lipopeptide | Targets the HR1 domains of S protein; thus, forming a 6-HB structure similar to the native one, results in inhibiting the viral fusion | Preclinical (In vivo studies) |
| SBP1 | Synthetic peptide | Mimics the residues 21–43 of ACE-2 PD α1 helix sequence, and binds to the RBD, disrupting the ACE-2 and S protein interaction | Potential inhibitors require further testing in animal and human cells |
| Recombinant-based therapy | |||
| Recombinant Soluble ACE-2 | Human recombinant | Competitively binds to S protein and distracts the virus from the membrane-bound ACE-2 receptors | Randomized Interventional (Clinical Trial), Phase: NA |
| Casirivimab, Imdevimab, Bamlanivimab and Etesevimab | Neutralizing monoclonal antibodies (mAbs) | Bind to the SARS-CoV-2 S protein RBD and inhibits its attachment to ACE2 | FDA authorized for emergency emergency use (phase II–III clinical trials) |
Fig. 3.
Pulmonary pathogenesis mediated by SARS-CoV-2 S protein upon its fusion and entry, and its potential treatments and current vaccines
Natural products-based therapy
Linoleic acid
Recently, a team of researchers has unexpectedly discovered a key potentially “druggable” binding pocket in the SARS-CoV-2 S protein [51]. Several pharmaceutical agents and fatty acid supplements readily available in the market may show viable affinity to the pocket sites of S trimer; thereby, interfering with S protein accessibility to ACE-2 receptor [9]. A greasy tube-like density buried at the interface between RBDs of the S trimer was observed and interpreted as three free fatty acid-binding pockets [97]. This pocket is composed of hydrophobic amino acids mostly phenylalanine, that can accommodate the hydrocarbons tail of linoleic acid (LA) [97]. Furthermore, the arginine and glutamine residues from the adjacent RBD in the trimer provide hydrophilic positive charges that anchor the negatively charged carboxyl group of LA, allowing LA to properly fit in the binding site [97].
Further investigations indicated that linoleic acid-binding pocket harbors a gating helix at the entrance of the hydrophobic pocket containing two tyrosine residues and a gap between the adjacent RBDs that places arginine and glutamine residues away (~ 10 Å) from the position of the linoleic acid carboxyl group [97, 98]. Therefore, linoleic acid repositions the gating helix by (~ 6 Å) and causes the two tyrosine residues to swing away to avoid a clash with linoleic acid. Subsequently, the pocket will open and the adjacent RBD moves toward its neighbor to lock down the hydrophilic anchor of linoleic acid [97, 98]. Collectively, this results in the formation of rigid compaction in the RBD trimer, producing a closed conformation rendering the S protein inaccessible by the host receptor. Therefore, LA is expected to reduce ACE-2 attachment and, hence, reduce the entry and infectivity [9]. Experimentally, LA supplementation to human epithelial cell culture showed a synergistic effect when administered with remdesivir and markedly suppressed the virus replication [11].
Free fatty acid-binding pocket has been reported in previous pathogenic human coronaviruses (HCoVS) [9]. Structural comparison of SARS-CoV-2 LA-binding pocket with that of SARS-CoV-1 reveals that all the hydrophobic and hydrophilic residues are conserved [97, 98]. In contrast, structural differences in other circulating HCoVs were observed, causing failure in LA binding [97].
It is noteworthy that the discovery of the LA-binding pocket opens the door for the development of new free fatty acid-based therapy [97]. Fatty acids of algae oil such as palmitic acid, oleic acid, stearic acid and linoleic acid have several mechanisms that can prevent COVID-19 infection, including the incorporation into the lipid membrane of SARS-CoV-2 envelopes, thus destabilizing its bilayer or interfering with viral attachment to the host cells [94]. In addition, sesame oil has been proposed to have a potential protective effect against SARS-CoV-2 infection due to its high linoleic acid concentration (41%) [53]. Recent studies suggest the use of oil-based nasal spray containing mainly linoleic acid as a protective method to limit the spread of SARS-CoV-2 infection. However, this proposal needs further clinical and experimental studies [94].
Bisoxatin
Computational studies were performed using drug libraries against the RBD to identify a potent inhibitor of S protein [98]. The selected compounds fulfilled the Lipinski’s rule of five and they were investigated for their significant interactions that may cause disruption of the interaction between the S protein and ACE-2 receptor [98]. These studies indicated that the laxative Bisoxatin (DB09219) can significantly bind to RBD of S protein by forming π–π and hydrophilic interactions with amino acid residues, thus, inhibiting S protein-ACE-2 interaction [98]. Accordingly, Bisoxatin can be utilized as a lead compound for the development of SARS-CoV-2 S protein inhibitors [98].
Emodin
Emodin is a natural anthraquinone that has shown the capability of inhibiting the interaction between S protein and ACE-2 receptor via in vitro analysis [26, 55]. Moreover, it alleviates the asthmatic airway inflammation by inhibiting the activated macrophages polarization, therefore it may suppress the inflammation associated with COVID-19 [108]. However, it has toxic effects and the mechanism of action is not fully understood, which limits its rational use [26].
Antiviral peptides-based therapy
The protein–protein interactions between ACE-2 and S protein represent a potential target for therapeutic interventions [67, 92, 114]. Inhibiting the large interface of RBD by small drug molecules is usually not effective and challenging [92, 114]. Alternatively, peptides can be utilized as they have a larger surface area, allowing more functional groups to interact with the binding site, therefore mimicking and blocking these native protein–protein interactions [92]. Additionally, peptides can be used to inhibit the formation of the six-helical bundle (6-HB) of S protein [69]. Interestingly, antiviral peptides can be formulated as inhalations for local lung delivery [92], thus reducing any possible systemic side effects [51]. Potential anti-SARS-CoV-2 peptides can include:
EK1C4
EK1 is a coronavirus fusion inhibitor composed of 36 amino acids [69], designed to target the heptad repeat-1 domains of S protein in human coronaviruses [51]. Thus, it forms a 6-HB structure similar to the native one [102], and inhibiting the viral fusion [51]. This peptide exhibited potent inhibitory activity against all tested human coronaviruses [51]. Recently, EK1 has been reported as an effective peptide against SARS-CoV-2 [51]. However, the antiviral efficacy of EK1 is less in SARS-CoV-2 due to the high stability of its 6-HB structure [51]. To improve the activity of EK1, its C-terminus has been covalently linked to cholesterol (EK1C) or palmitic acid (EK1P) with a glycine/ serine linker and polyethylene glycol (PEG) spacer. The presence and the length of the linker/ spacer affect the activity of the formed lipopeptide [33].
EK1C4 is a lipopeptide [33] with potent activity against SARS-CoV-2 S protein-mediated membrane fusion [51, 102]. It has been suggested that the EK1 moiety of EK1C4 binds to one of the three hydrophobic grooves of the HR1 trimer and utilizes the cholesterol moiety to anchor to the adjacent hydrophobic groove [51]. In vivo studies confirmed the potency of EK1C4 against coronavirus infection with no or minimal toxicity [51]. Therefore, it can be considered as a promising pan-CoV fusion inhibitor for prophylaxis or treatment of SARS-CoV-2 infection and other human coronaviruses that may emerge in the future [51].
23-Mer peptide (SBP1)
It has been demonstrated that the ACE-2 peptidase domain (PD) α1 helix is critical for the binding with RBD [99]. Accordingly, SBP1 was designed and synthesized to mimic the 21–43 residues of the ACE-2 PD α1 helix sequence [92]; hence, it can bind to the viral RBD, resulting in disruption of the ACE-2 and S protein interaction [33, 114]. SBP1 is not expected to have immunogenic effects since it is derived from human protein [114]. The binding affinity of SBP1 was further improved by minor modifications [92] that led to the newly designed SPB25, which is a 25-mer peptide that was used as a template for further modifications [92]. Accordingly, five peptides were designed, but the most promising one was SPB25F8N [92], mainly due to the involvement of a higher number of predicted hydrogen bonds and the π interactions [92].
Previously, peptides were associated with many challenges including expensive synthesis, limited stability, and bioavailability. However, technological advances and chemical modifications have helped to overcome these challenges [99, 114]. Furthermore, these peptides can be attached to nanoparticles to produce inhibitory effects against several viruses along with enhancement of their binding affinities [99], yet their use in SARS-CoV-2 requires more investigations.
Natural lectins-based therapy
Since S protein is a heavily glycosylated protein, it can be a potential target for carbohydrate-binding agents such as lectins [107].
Griffithsin (GRFT)
GRFT is an algae-derived lectin isolated from the red alga Griffithsia sp., which demonstrates a broad-spectrum antiviral activity [14]. Experimentally, GRFT showed great inhibitory potential against SARS-CoV-2 infection in vitro [14]. It has been reported that GRFT possesses three independent carbohydrate-binding sites [52], which can selectively bind to the glycosylation sites of the S1 subunit, resulting in the formation of a complex that preserves its ability of binding to ACE-2 [52]. However, it prevents the subsequent steps essential for viral entry [52]. Moreover, GRFT has demonstrated immunomodulatory effects, particularly it alters the cytokine response in infected lung cells [52]. Importantly, a previous study has reported a synergistic antiviral activity by combining GFRT with EK1, since both have different binding sites, thus representing a potential cocktail therapy for the prevention and treatment of SARS-CoV-2 infection [14]. Studies have revealed that GFRT at its active antiviral concentration does not exhibit any toxicity and it effectively shows high potency and promising subclinical outcomes [64, 88].
FRIL
FRIL is a glucose/mannose lectin isolated from Lablab purpureus plant [77], and referred to as Flt3 Receptor Interacting Lectin [107]. FRIL showed the ability to neutralize SARS-CoV-2 effectively, resulting in a complete inhibition of viral cytopathic effects [77, 107]. FRIL binds to the complex-type N-glycans, which are the predominant glycans in the S protein N-glycosylation sites. This explains its high binding affinity [107].
Recombinant-based therapy
Soluble ACE-2
ACE-2 has a dual function as it serves as a significant functional receptor for SARS-CoV-2 that permits the viral entry into the host cell, and a protective pathway for multiple organs such as the heart, liver, kidney, and lung [119]. This is attributed to its enzymatic and catalytic function, which regulates the homoeostasis of the renin–angiotensin–aldosterone system (RAAS) [46]. In fact, studies reported that ACE-2 is downregulated following S protein binding, which in turn contributes to severe disease progression and deterioration of pulmonary function [115].
It has been demonstrated that the administration of soluble ACE-2 in excessive amounts can competitively bind to SARS-CoV-2 S protein and distract the virus from the membrane-bound ACE-2 receptors; hence, it can neutralize the virus and inhibit its entry into the host cells [73, 115]. A human recombinant soluble ACE-2 (rhACE2; APN01, GSK2586881) was developed [115] and studies showed that it has ~ 1.2 nM binding affinity to SARS-CoV-2 S protein [119]. APN01 has already passed phase 1 and 2 clinical trials, where it was able to reduce the lung injury in acute respiratory distress syndrome (ARDS), acute lung injury (ALI), and pulmonary arterial hypertension (PAH). In addition, it is well tolerated and safe [54]. Of note, neither an equivalent mouse rsACE-2 nor a murine recombinant soluble ACE-2 (mrsACE2) were able to block SARS-CoV-2 virus entry, revealing the uniqueness of APN01 [73]. Furthermore, data demonstrate the significant ability of APN01 to block the early stages of SARS-CoV-2 infections where it reduces the viral load by 1000–5000. However, its effects in the later stages of the disease are yet to be determined [73, 119]. Although this inhibition was incomplete, possibly due to the presence of other co-receptors [73], a dose-dependent relationship was observed [119].
Importantly, experimental studies indicated the development of IgA and IgG antibodies against SARS-CoV-2 upon treatment with APN01 [8, 119]. Collectively, data suggested that the beneficial effects of APN01 can include slowing down the viral entry, spread and ultimately viral neutralization. It can also preserve cellular ACE-2, hence maintaining its protective activity to vital body organs [8, 56, 115, 119].
Monoclonal neutralizing antibodies
Recently, U.S. FDA has authorized the emergency use of synthetic neutralizing monoclonal antibodies directed against SARS-CoV-2 S protein to inhibit viral attachment and subsequent entry to the host cells [7, 34]. These monoclonal antibodies, namely, Casirivimab, Imdevimab, Bamlanivimab and Etesevimab are indicated for the treatment of mild to moderate COVID-19 in adults and pediatrics over 12 years old or for high-risk individuals who are susceptible to severe disease progression [6, 7]. Bamlanivimab targets the RBD of the S protein of SARS-CoV-2. Casirivimab and Imdevimab are recombinant human monoclonal antibodies that bind to non-overlapping epitopes of the S protein RBD of SARS-CoV-2. Etesevimab binds to a different but overlapping epitope in the RBD of the SARS-CoV-2 S protein [96].
Of note, these drugs should not be used for severe cases, hospitalized patients or who require mechanical ventilation as they may worsen their conditions [6, 7]. Importantly, these therapies should be used in combinations through IV infusions. These combinations include the use of Casirivimab and Imdevimab together [2]. Although Bamlanivimab was authorized for use as a single agent, this authorization was revoked very recently due to lack of efficacy upon the growth of SARS-CoV-2 resistance [2, 3]. Currently, Bamlanivimab is authorized for use in combination with Etesevimab [2, 3]. This emergency authorization is due to the FDA beliefs in the potential benefits of these antibodies that can outweigh the risks since they have shown the ability to decrease the viral loads, and hospitalizations. Thus, it alleviates the health burden of this pandemic [7]. The side effects of these drugs may vary ranging from fever, chills, itching, to infusion-related reactions or anaphylactic shock. However, the safety and efficacy of these mAbs are still under evaluation [6, 7].
A decline in the neutralizing activity of antibodies has been reported in convalescent patients due to the development of recent SARS-CoV-2 variants of concern (VOCs), particularly, those with heavily mutated S protein [23, 42]. A recent study has tested the ability of anti-S protein Casirivimab and Imdevimab monoclonal antibodies combination to neutralize SARS-CoV-2 variants B.1.1.7, B.1.351, mink cluster 5, and COH.20G/677H. The efficacy of this antibody’s combination therapy was declined 9.1-fold on B.1.351 and 16.2-fold on mink cluster 5. These results raised the concerns about the efficacy of using this monoclonal antibodies-based approach in the treatment of new variants, and urge the need to develop new monoclonal antibodies that cover the global SARS-Cov-2 variants [95]. On the other hand, 41 human monoclonal Abs (mAbs) derived from memory B cells have been identified and showed potent neutralizing activity against SARS-CoV-2 S N-terminal domain (NTD), specifically supersite I, suggesting its importance for protective immunity and vaccine design [70].
Convalescent plasma as therapy
Utilization of convalescent plasma as a therapy to prevent or manage COVID-19 infection provides a dual immune and non-immune mechanism of actions, most importantly, through neutralizing antibodies including those against S protein [85]. The peak viremia is observed in most viral diseases within the first week of infection, and the main host immune response usually occurs between the 10th and 14th days of infection, thus, the early stage of infection could be the best time to effectively administer the convalescent plasma [80]. Even though the efficacy of plasma as a therapy in the management of COVID-19 is still controversial, an early administration of convalescent plasma with high-titers to mildly infected patients within 72 h showed a significant reduction by 48% in the risk progression of SARS-CoV-2 infection. Conversely, hospitalized patients who received plasma infusion late during illness has shown no benefit [66, 85].
Immune system antiseptics
Endogenous anti-inflammatory and broad-spectrum anti-pathogenic substances produced by the human body are safe therapy that can be applied in the development of antivirals with respiratory tract infection including in SARS-CoV-2 [59]. N-chlorotaurine (NCT) and hypochlorous acid (HOCl) are examples of endogenous substances produced by the human immune system [12, 59]. N-chlorotaurine (NCT) is a long-lasting oxidant that is produced in the human immune system cells, namely neutrophils, eosinophils, and monocytes. NCT can be synthesized chemically as an antiseptic agent for lower respiratory tract infections to be administered topically via inhalation. Animal studies confirmed its tolerability when being repeatedly inhaled [59]. A study has found that NCT demonstrated a virucidal activity against SARS-CoV-2, and influenza A virus at a therapeutic concentration of 1%. This broad-spectrum activity could eliminate the time consumed in diagnostics as it could be applied instantly regardless of the invading pathogen. Besides, it has activity against bacteria and fungi, which might be greatly useful in preventing secondary and super-infections in COVID-19 patients. Of note, as it is applied topically, it does not cause systemic adverse effects and it does not interact systemically with other medications. However, its clinical efficacy still needs further investigation [59].
HOCL is another endogenous substance that is produced by neutrophils, eosinophils, mononuclear phagocytes, and B lymphocytes in all mammals. It is a powerful oxidizing agent; accordingly, it has a broad-spectrum activity against microorganisms. It destroys the viruses by forming chloramines and radicals that break the viral DNA [12]. It has been widely used topically in the management of multiple infections and inflammatory conditions (Lori Robins 2021). A study showed that cytokine IL-6 failed to bind to IL-6 receptors in vitro after exposure to HOCL and NCT, indicating that they can induce chemical modifications of IL-6. Notably, IL-6 is involved in the pathogenesis of the acute inflammatory conditions associated with SARS-CoV-2 infection. Besides, its levels are typically elevated systemically and locally in pulmonary tissues in COVID-19 patients, suggesting that any measure that reduces its binding with the IL-6 receptor in vivo is expected to relieve the severity and progression of the disease. Using HOCL and NCT to mitigate the cytokine storm could be an alternative approach to monoclonal antibodies. To further highlight, administration of monoclonal antibodies is expensive and requires hospitalization, while HOCL and NCT, when developed in inhalation formulations, can be easily administered by the patients without the need for hospitalization (Lori Robins 2021).
TMPRSS2 targeting therapies
Several studies demonstrated significant higher mortality and hospitalization rates of males than females because of COVID-19. Although a multifactorial reason has been suggested, the difference in the level of androgen between both genders is a major reason, since it modulates the expression of TMPRSS2 [82]. High levels of estrogen increase the expression of TMPRSS2. Therefore, inhibiting the activity of androgen receptors or activating the estrogen pathways can prevent SARS-CoV-2 entry (Wang, 2020). Androgenic antagonists such as darolutamide, apalutamide, and enzalutamide, which are FDA-approved anti-prostate cancer drugs, transcriptionally downregulate TMPRSS2 expression; hence, they can attenuate SARS-CoV-2 infection [82].
SARS-CoV-2 S protein-based vaccines development
Due to its crucial role in controlling the spread of the COVID-19 pandemic, the development of vaccines against S protein has been proposed (Table 2; Fig. 3). As vaccines can decrease disease incidence and increase the overall population immunity; their safety, efficacy, and affordability should be ensured. Indeed, a large number of vaccine candidates are being proposed or tested in clinical trials [100]. These can include:
Table 2.
Summary of the current FDA-approved vaccines designed against SARS-CoV-2 utilizing the S protein
| Vaccine (brand/ generic name) | Manufacturer | Vaccine platform | Antigen | Dose and dosage regimen | Overall efficacy | Clinical phases [50, 108] |
|---|---|---|---|---|---|---|
| Moderna/mRNA-1273 | Moderna (US) | mRNA | Full length S protein stabilized in pre-fusion state by proline substitutions |
100 µg Two doses by IM injection separated by 28 days (~ 4 weeks) |
94.1% |
Phases I–III Authorized by the FDA for emergency use |
| Astrazeneca/ChAdOx1 nCoV-19 | Oxford- Astrazeneca (UK) | Viral vector | Recombinant replication-deficient chimpanzee adenoviral vector utilizing the full-length SARS-CoV-2 S protein gene |
Standard dose: 0.5 mL containing 5 × 1010 viral particles Two doses by IM injection separated by interval of 8–12 weeks |
70·4% |
Phases I–III Recently approved by the WHO for emergency use. Not FDA approved yet |
| Pfizer/BNT162b2 | Pfizer- BioNTech (US) | mRNA | Full length S protein stabilized in pre-fusion state by proline substitutions |
30 μg Two doses regimen separated by 21 days |
95% |
Phases I–III Authorized for emergency use |
| (Janssen/Johnson and Johnson)/Ad26.COV2. S | Janssen/Johnson and Johnson (US) | Recombinant, replication-incompetent human Ad26 vector | Full length S protein stabilized in pre-fusion state by proline substitutions |
One dose (0.5 ml) contains 5 × 1010 viral particles One single dose |
66.3% |
Phases I–III Authorized by the FDA and the WHO for emergency use |
| Novavax/NVX-CoV2373 | Novavax (US) | S protein subunit | Pre-fusion stabilized full-length recombinant S protein |
One dose contains 5 µg protein and 50 µg adjuvant Two doses administered 21 days apart |
90.4% | Phases I–III |
| CureVac/CVnCoV | CureVac AG. (Germany) | mRNA | Pre-fusion stabilized full-length S protein |
12 μg Two doses administered 28 days apart |
48% | Phases I–III |
mRNA-1273 (Moderna vaccine)
mRNA-1273 vaccine, broadly known as Moderna's COVID-19 vaccine is a lipid nanoparticle (LNP)-encapsulated mRNA-based vaccine that encodes a pre-fusion stabilized form of SARS-CoV-2 S protein [10]. It has been developed by the National Institute of Allergy and Infectious Diseases (NIAID) [20, 50]. It belongs to mRNA-based vaccine platforms that are characterized by rapid development, scaling up and quick response compared to traditional platforms. Moreover, they have shown potent elicitation of immunogenic responses with an acceptable safety profile since the early clinical trials of COVID-19 vaccines [20, 50].
Historically, the stabilized pre-fusion conformation of S protein of MERS-CoV, SARS-CoV-1, and HCoV-HKU1 have been found to have 2 proline (2P) substitutions. This was shown to enhance MERS S protein immunogenicity at lower doses than wild-type S protein; in fact, the same concept has been applied to a mRNA-1273 vaccine [22]. These two proline moieties can produce pre-fusion-stabilized SARS-CoV-2 S protein, which was then used for structural evaluation and development of serological assays. Notably, this led to Phase 1 clinical trial on March 16, 2020, phase 2 on May 29, 2020, and, the vaccine is in phase 3 clinical trial [22]. Remarkably, preliminary results from phase 3 showed 94.1% efficacy in preventing COVID-19 infection [10].
Published reports showed the ability of mRNA-1273 to potently induce the release of CD8 T cells and neutralizing antibodies, therefore providing a protection against COVID-19 infection without causing immunopathology [22]. Commensurately, robust neutralizing antibody responses were generated in a dose-dependent fashion at both 50 and 100 µg dose levels [20]. Of note, a mRNA-1273 vaccine is given in two doses by intramuscular injection separated by 28 days [50]. Interestingly, a single dose of mRNA-expressing SARS-CoV-2 S-2P has demonstrated immunogenic and neutralizing activity as well [22].
The safety of mRNA vaccine was investigated including local and systemic adverse effects, where moderate-to-severe systemic side effects have been reported [20]. Notably, the most common systemic adverse effects following the first dose injection were headache, in addition to arthralgia, myalgia, chills, nausea and vomiting [20]. Overall, longitudinal clinical data are critically required to further assess the immunogenicity and safety of the Moderna vaccine [106].
Oxford-AstraZeneca vaccine
COVID-19 Vaccine AstraZeneca vaccine (AZD1222) is a recombinant replication-deficient chimpanzee adenovirus vector, and also known as ChAdOx1 nCoV-19 vaccine that was developed by Oxford University, Astra Zeneca, United Kingdom [35, 100]. The technology utilizes the full-length SARS-CoV-2 immunogenic S glycoprotein gene fused with tissue plasminogen activator (tPA) as a leader sequence [35]. The S protein is stabilized in its expressed trimeric pre-fusion conformation [4]. Oxford vaccine incorporates the antigen-encoding sequences within the DNA carried by Adenovirus. This encapsulation has several advantages as it protects the genetic material since DNA is less fragile than RNA and the adenovirus’s tough protein coat provided additional preservation [17]. In addition, being in the same size scale, allows its entry into the cells, thereby enabling antigens expression from the delivered nucleic acids [21]. Moreover, the immunogenic properties of the chimpanzee adenovirus backbone, for which humans do not have pre-existing immune antibodies, ensures a potent engagement of the immune system, thus enhancing its ability to induce strong cellular and humoral immune response against the S protein without needing an adjuvant [50, 75]. Another advantage is that the vaccine has a natural tropism for the respiratory mucosa, which allows respiratory mucosal vaccination [50, 74].
In clinical trials, the ChAdOx1 nCoV-19 vaccine was found to be highly effective in preventing COVID-19 and has shown efficacy against symptomatic COVID-19 as well [100]. Two dosage regimens were followed, wherein participants who received two standard-dose vaccines containing 5 × 1010 viral particles (standard dose; SD/SD cohort), the efficacy was 62.1% [100]. Whereas in those who received a low dose (2.2 × 1010 viral particles) as their first dose and were boosted with a standard dose as their second dose at least one month apart, (LD/SD cohort), surprisingly, exhibited enhanced efficacy at 90.0% [100]. The combined analysis from both dosing regimens shows overall vaccine efficacy of 70.4% [100]. The reason behind this difference is yet to be understood, however, it was proposed that the increased efficacy with an LD/SD regimen can be attributed to the production of higher levels of neutralizing antibodies [4, 100]. The safety profile indicates that it is well tolerated, without any major safety concerns [4]. Generally, most of the side effects were mild to moderate that would be resolved within few days of vaccination and they were reported to be milder and less frequent after the second dose [4]. Tenderness and pain in the injection site, headache, pyrexia, fatigue, and arthralgia have been noted as the most frequent adverse effects [4].
BNT162b2 (Pfizer vaccine)
BNT162b2 is an mRNA vaccine that has been developed by Pfizer and BioNTech [65]. It is composed of mRNA that encodes the pre-fusion conformation of the full-length S gene which is formulated into a lipid-soluble nanoparticle preparation [65]. It was compared with another vaccine candidate BNT162b1 in which the mRNA encodes the RBD of S protein [65]. The results showed that both BNT162b1 and BNT162b2 produce similar serum neutralizing antibody titers in a dose-dependent manner. However, BNT162b2 produced milder adverse reactions compared to BNT162b1 [65]. It was effective in elderly individuals as well [65]. Additionally, a study has reported that the BNT162b2 vaccine induces the production of SARS-CoV-2 S-specific neutralizing antibodies, CD4 + , CD8 + T cells, and cytokines such as interferon γ (IFNγ) [86].
The 2/ 3 phases of clinical trials were performed to evaluate the efficacy, safety, and immunogenicity of two doses regimen of 30 μg of BNT162b2 in preventing COVID-19 in individuals 16 years old and above [78]. The results have shown that BNT162b2 was 95% effective in preventing COVID-19 infection [78]. Regarding its safety, it was associated with mild to moderate pain at the injection site, headache, fatigue, and a low incidence of serious adverse events [78]. Accordingly, Pfizer submitted an Emergency Use Authorization (EUA) to FDA. On December 11, 2020, the Pfizer-BioNTech COVID-19 vaccine was the first vaccine to be authorized for emergency use by the FDA for the prevention of COVID-19 infection [1].
JNJ-78436735/Ad26.COV2.S (Johnson vaccine)
JNJ-78436735/Ad26.COV2.S vaccine is an adenoviral vector-based SARS-CoV-2 vaccine, that has already received an EUA. It is composed of recombinant, replication-incompetent naturally occurring, low-prevalence human adenovirus type 26 (Ad26) vector that expresses the S protein of SARS-CoV-2 in its pre-fusion-stabilized conformation [83]. Utilizing this rare Ad26 serotype overcomes the obstacle of pre-existing immunity exhibited with other viral vector vaccines, therefore, impairing the magnitude of elicited immune responses is not expected to be a major concern [24, 58]. This vaccine has been developed by the Janssen Pharmaceutical Company of Johnson & Johnson, USA for the immunization of individuals aged 18 and above [32]. The vaccine shows strong immunogenic properties following a single vaccination dose, which is an obvious advantage over a two-dose vaccine, especially during a pandemic to meet the global demands at an affordable cost and limited time [58],Laffoley et al.). Ad26.COV2.S vaccine has an acceptable safety, reactogenicity, and immunogenicity profile [83]. Moreover, recently published data have demonstrated its efficiency in eliciting a strong, persistent immune response against the rapidly spreading Delta variant of SARS-CoV-2 and other prevalent viral variants (July 1, 2021). In comparison to Pfizer and Moderna vaccines, Johnson & Johnson’s vaccine requires easier storage conditions at refrigerator temperature, which facilitates its global distribution and administration [29].
NVX-CoV2373 (Novavax vaccine)
NVX-CoV2373, Novavax’ COVID-19 is a protein subunit vaccine that consists of a stable, pre-fusion S protein antigen derived from SARS-CoV-2. NVX-CoV2373 was created using Novavax’ recombinant nanoparticle technology and is formulated with Novavax’ patented saponin-based Matrix-M™ adjuvant to enhance the immune response and stimulate the production of neutralizing antibodies [58, 84]. The vaccine exhibits an overall 90.4% efficacy in Phase 3 clinical trial and maintains consistent, high efficacy among the newly circulating viral variants. It also demonstrates reassuring tolerability and safety profiles and the robust capability of elicitation of antibody response [27, 84].
CVnCoV (CureVac’s vaccine)
CureVac’s CVnCoV vaccine is a COVID-19 vaccine candidate developed by a German biotechnology company that utilizes novel mRNA-based technology. Similar to Pfizer-BioNTech and Moderna, the vaccine encodes the SARS-CoV-2 S protein gene that is encapsulated by a lipid-soluble nanoparticle [19]. However, CureVac’s vaccine keeps the natural non-chemically modified mRNA nucleotides sequence rather than substituting Uridine with pseudouridine in the other vaccines [13, 19], Dolgin). This vaccine has been expected to owe some advantages over its mRNA vaccine competitors since it can be stored at standard refrigerator temperature, and requires a lower dosage of 12 μg, thus, providing a faster and cheaper mass production. Accordingly, it can expand the distribution of mRNA-based vaccines in lower-income or warmer countries, in addition to minimizing the gap of vaccines shortage in Europe [19],Dolgin). However, the vaccine showed inadequate results since it shows a 48% efficacy in its final analysis of Phase III trials, which is below the acceptance threshold set by WHO. Nevertheless, some arguments attribute this low efficacy to the timing of clinical trials, while newer variants are dominant, the use of RNA, and the dose that is thought to be suboptimal (Dolgin). Currently, CureVac is collaborating with GlaxoSmithKline (GSK) to produce a second-generation vaccine that has been modulated to boost its capability of eliciting neutralizing antibodies [19],Dolgin).
Future perspectives of SARS-CoV-2 S protein mutations and predictive measures
A deep understanding of SARS-CoV-2 evolution and its ongoing mutagenesis can facilitate future planning to identify and restrict the spread of potential new variants [36, 81]. Tracking and analyzing the mutagenic pattern adopted by the SARS-CoV-2 S protein can reveal that the ongoing increase in population immunity against the virus, either due to getting infected, vaccinated or via neutralizing antibodies can be a source of strong selection pressure on the virus (Fig. 4A) [14, 81]. As a result, the general tendency of the newly emerging variants is towards increasing the viral fitness and binding affinity to the ACE-2 receptor through increasing its binding free energy [14, 81]. These can serve as escape mechanisms that allow immune evasion and more efficient host infection [14, 81].
Fig. 4.
Future perspectives of SARS-CoV-2 S protein mutations and planning to identify and restrict the spread of potential new variants. A Tracking and analyzing the mutagenic pattern adopted by the SARS-CoV-2 S protein. B Factors that significantly contribute to infection range and protection
In general, predicting the future mutations of S protein can be achieved by extensive, large-scale tracking and analysis of viral dynamics, evolutionary adaptions, and genetic sequencing over time (Fig. 4B) [36, 62, 81]. Other factors that should be considered can include the possible variant combinations, the influence of population immunity, structural interactions analysis to recognize key residues contributing to binding affinity, free energy, and stability taking into account both individual and concurrent mutations as they can potentially show additive effects (Fig. 4B) [36, 62, 81]. Building structural models that incorporate demographics and climate data, which can be an evolutionary driving force, can improve the predictive outcomes as well [36, 62, 81]. Additionally, since receptor binding mediated by the S protein is a major contributor to the viral host range, it should be expected that being a multi-host pathogen that has many possible animal reservoirs may significantly impact and guide its genomic surveillance and future mutations prediction [39, 62, 91]. Of note, analysis of its phylogenetic patterns indicates that co-infection and genetic recombination events across multiple host species and geographic regions are possible [39, 62, 91].
These extensive studies could be performed utilizing a multi-approach that combines computational and experimental in silico mutagenesis methods, and molecular dynamics simulation programs [18, 44, 77]. Using bioinformatics tools can speed up and simplify the research process as well [18, 44, 77]. This process can be facilitated by the available international websites and databases that allow following and mapping the mutations and tracing their transmission, thus assisting more efficient predictions [18, 44, 77]. In this context, it is worth mentioning that a simple, freely accessible, computational model was developed by some researchers. It is referred to as “SpikePro”, which is a useful instrument that predicts, describes and interprets the effect of new S protein variants in its future evolution concerning the viral fitness, transmissibility, infectivity, and host’s immune system evasion capability [81]. Practically, some studies were able to predict a total of 3686 possible mutations in the S protein upon Codon analysis of RBD amino acids residues and identifying the hotspots [14]. These mutations were categorized as most likely, likely, and unlikely mutations [14]. The impacts of these mutations were analyzed as well [14]. In general, the more likely to happen mutations had higher free energy and are predicted to be infectivity-strengthening ones [14].
Conclusion
In summary, there is well-established evidence that SARS-CoV-2 S protein plays vital role in virus pathogenesis and transmission, making it an important target for the development of therapeutics and vaccines. This interest was further enhanced by the deep understanding of S protein including its structural features, binding interactions, and mutations. This promotes the discovery of novel potential applications in diagnosis and immunotherapy. However, most of the drug candidates are still in the pre-clinical phases. Thus, further studies are necessary to ensure their safety and efficacy. As for the current emergency-used vaccines, it has been believed that performing a continuous comprehensive evaluation of post-vaccination immune response is crucial. The findings described in this review will guide rational drug and vaccine designs that prove effective in mitigating the morbidity and mortality of the ongoing pandemic as a global priority.
Acknowledgements
The authors acknowledge the generous support from Sandooq Al-Watan and University of Sharjah to SS.
Abbreviations
- ACE-2
Angiotensin-converting enzyme 2
- ALI
Acute lung injury
- ARDS
Acute respiratory distress syndrome
- COVID-19
Coronavirus Disease 2019
- CTD
C-terminal domain
- EUA
Emergency use authorization
- FCS
Furin-like cleavage site
- FP
Fusion peptide
- FDA
Food and Drug Administration
- FPPR
Fusion peptide proximal region
- GRFT
Griffithsin
- 6-HB
Six-helical bundle
- HCoVs
Human coronaviruses
- HCoV-HKU1
Human coronavirus HKU1
- HIV-1
Human immunodeficiency virus
- HPPHCPC
Repetitive heptapeptide
- HR1
Heptad-repeat region1
- HR2
Heptad-repeat region2
- IgA
Immunoglobulin A
- IgG
Immunoglobulin G
- LA
Linoleic acid
- LNP
Lipid nanoparticle
- MERS-CoV
Middle East respiratory syndrome coronavirus
- NIAID
National Institute of Allergy and Infectious Diseases
- NTD
N-terminal domain
- PAH
Pulmonary arterial hypertension
- PEG
Polyethylene glycol
- PD
Peptidase domain
- PUFA
Polyunsaturated fatty acid
- RAAS
Renin–angiotensin–aldosterone system
- RBD
Receptor-binding domain
- RBM
Receptor-binding motif
- RhACE2
Recombinant human angiotensin-converting enzyme 2
- SD1
Subdomain 1
- SD2
Subdomain 2
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- S protein
Spike protein
- TMPRSS2
Transmembrane protease serine 2
- tPA
Tissue plasminogen activator
Declarations
Conflict of interest
All authors declare there is no conflict of interest.
References
- 1.U.S. Food and Drug Administration (FDA). Pfizer-BioNTech COVID-19 vaccine. (2021).
- 2.U.S. Centers for Medicare & Medicaid Services (CMS.gov). Monoclonal antibody COVID-19 infusion. (2021).
- 3.U.S. Food and Drug Administration (FDA). Coronavirus (COVID-19) update: FDA revokes emergency use authorization for monoclonal antibody bamlanivimab. (2021).
- 4.World Health Organization (WHO). AZD1222 vaccine against COVID-19 developed by Oxford University and Astra Zeneca: Background paper (2021)
- 5.Johnson & Johnson. Positive new data for Johnson & Johnson single-shot COVID-19 vaccine on activity against delta variant and long-lasting durability of response. (2021) July 1, 2021.
- 6.U.S. Food and Drug Administration (FDA). Coronavirus (COVID-19) Update: FDA authorizes monoclonal antibody for treatment of COVID-19. (2020).
- 7.U.S. Food and Drug Administration (FDA). Coronavirus (COVID-19) Update: FDA authorizes monoclonal antibodies for treatment of COVID-19. (2020).
- 8.Abd El-Aziz TM, Al-Sabi A, Stockand JD. Human recombinant soluble ACE2 (hrsACE2) shows promise for treating severe COVID19. Signal Transduct Target Ther. 2020;5:258. doi: 10.1038/s41392-020-00374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger I, Schaffitzel C. The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res. 2020;30:1059–1060. doi: 10.1038/s41422-020-00430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block MS, Rowan BG. Hypochlorous acid: a review. J Oral Maxillofac Surg. 2020;78:1461–1466. doi: 10.1016/j.joms.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borah P, Deb PK, Al-Shari NA, Dahabiyeh LA, Venugopala KN, Singh V, Shinu P, Hussain S, Deka S, Chandrasekaran B, Jaradat DSMM. Perspectives on RNA vaccine candidates for COVID-19. Front Mol Biosci. 2021;8:30. doi: 10.3389/fmolb.2021.635245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Xu W, Gu C, Cai X, Qu D, Lu L, Xie Y, Jiang S. Griffithsin with a broad-spectrum antiviral activity by binding glycans in viral glycoprotein exhibits strong synergistic effect in combination with a pan-coronavirus fusion inhibitor targeting SARS-CoV-2 spike S2 subunit. Virol Sin. 2020;35:857–860. doi: 10.1007/s12250-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM, Rawson S, Rits-Volloch S, Chen B. Distinct conformational states of SARS-CoV-2 spike protein. bioRxiv.2020b; 2020.2005.2016.099317 [DOI] [PMC free article] [PubMed]
- 16.Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM, Rawson S, Rits-Volloch S, Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaitanya KV. Structure and organization of virus genomes. Genome Genomics 2019;1–30
- 18.Chen C, Boorla VS, Banerjee D, Chowdhury R, Cavener VS, Nissly RH, Gontu A, Boyle NR, Vandergrift K, Nair MS, Kuchipudi SV, Maranas CD. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike protein and the human ACE2 receptor. bioRxiv. 2021;2021.2003.2024.436885. [DOI] [PMC free article] [PubMed]
- 19.Cheung CW, Gibbons N, Johnson DW, Nicol DL. Silibinin–a promising new treatment for cancer. Anticancer Agents Med Chem. 2010;10:186–195. doi: 10.2174/1871520611009030186. [DOI] [PubMed] [Google Scholar]
- 20.Chu L, McPhee R, Huang W, Bennett H, Pajon R, Nestorova B, Leav B. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine. 2021. [DOI] [PMC free article] [PubMed]
- 21.Chung YH, Beiss V, Fiering SN, Steinmetz NF. COVID-19 vaccine frontrunners and their nanotechnology design. ACS Nano. 2020;14:12522–12537. doi: 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- 22.Corbett KS, Edwards D, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, KongW-P, Schmidt SD, Wang L, Zhang Y, Stevens LJ, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Nason MC, Ruckwardt TJ, McLellan JS, Denison MR, Chappell JD, Moore IN, Morabito KM, Mascola JR, Baric RS, Carfi A, Graham BS. SARS-CoV-2 mRNA Vaccine development enabled by prototype pathogen preparedness. bioRxiv. 2020; 2020.2006.2011.145920.
- 23.Corti D, Purcell LA, Snell G, Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:3086–3108. doi: 10.1016/j.cell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 Vaccines. JAMA. 2021;325:1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 25.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dellafiora L, Dorne JL, Galaverna G, Dall'Asta C. Preventing the interaction between coronaviruses spike protein and angiotensin I converting enzyme 2: an in silico mechanistic case study on emodin as a potential model compound. Appl Sci. 2020;10:6358. doi: 10.3390/app10186358. [DOI] [Google Scholar]
- 27.Diaz-Marrero AR, Austin P, Van Soest R, Matainaho T, Roskelley CD, Roberge M, Andersen RJ. Avinosol, a meroterpenoid-nucleoside conjugate with antiinvasion activity isolated from the marine sponge Dysidea sp. Org Lett. 2006;8:3749–3752. doi: 10.1021/ol061333p. [DOI] [PubMed] [Google Scholar]
- 28.Dolgin E. CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature. 2021; 594:483. [DOI] [PubMed]
- 29.Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020a;11. [DOI] [PMC free article] [PubMed]
- 31.Durmaz B, Abdulmajed O, Durmaz R. Mutations observed in the SARS-CoV-2 spike glycoprotein and their effects in the interaction of virus with ACE-2 receptor. Medeni Med J. 2020;35:253–260. doi: 10.5222/MMJ.2020.98048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, Hall MD. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faheem Kumar BK, Sekhar KVGC, Kunjiappan S, Jamalis J, Balaña-Fouce R, Tekwani BL, Sankaranarayanan M. Druggable targets of SARS-CoV-2 and treatment opportunities for COVID-19. Bioorg Chem. 2020;104:104269. doi: 10.1016/j.bioorg.2020.104269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.U. S. Food and Drug Administration (FDA). Coronavirus (COVID-19) update: FDA authorizes monoclonal antibodies for treatment of COVID-19. 2021.
- 35.Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, Bellamy D, Bibi S, Bittaye M, Clutterbuck EA, Dold C, Faust SN, Finn A, Flaxman AL, Hallis B, Heath P, Jenkin D, Lazarus R, Makinson R, Minassian AM, Pollock KM, Ramasamy M, Robinson H, Snape M, Tarrant R, Voysey M, Green C, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ, Aboagye J, Adams K, Ali A, Allen E, Allison JL, Anslow R, Arbe-Barnes EH, Babbage G, Baillie K, Baker M, Baker N, Baker P, Baleanu I, Ballaminut J, Barnes E, Barrett J, Bates L, Batten A, Beadon K, Beckley R, Berrie E, Berry L, Beveridge A, Bewley KR, Bijker EM, Bingham T, Blackwell L, Blundell CL, Bolam E, Boland E, Borthwick N, Bower T, Boyd A, Brenner T, Bright PD, Brown-O'Sullivan C, Brunt E, Burbage J, Burge S, Buttigieg KR, Byard N, Cabera Puig I, Calvert A, Camara S, Cao M, Cappuccini F, Carr M, Carroll MW, Carter V, Cathie K, Challis RJ, Charlton S, Chelysheva I, Cho J-S, Cicconi P, Cifuentes L, Clark H, Clark E, Cole T, Colin-Jones R, Conlon CP, Cook A, Coombes NS, Cooper R, Cosgrove CA, Coy K, Crocker WEM, Cunningham CJ, Damratoski BE, Dando L, Datoo MS, Davies H, De Graaf H, Demissie T, Di Maso C, Dietrich I, Dong T, Donnellan, F.R, Douglas N, Downing C, Drake J, Drake-Brockman R, Drury R.E, Dunachie, S.J, Edwards, N.J, Edwards, F.D.L, Edwards, C.J, Elias, S.C, Elmore, M.J, Emary, K.R.W, English, M.R, Fagerbrink S, Felle S, Feng S, Field S, Fixmer C, Fletcher C, Ford, K.J, Fowler J, Fox P, Francis E, Frater J, Furze J, Fuskova M, Galiza E, Gbesemete D, Gilbride C, Godwin K, Gorini G, Goulston L, Grabau C, Gracie L, Gray Z, Guthrie, L.B, Hackett M, Halwe S, Hamilton E, Hamlyn J, Hanumunthadu B, Harding I, Harris, S.A, Harris A, Harrison D, Harrison C, Hart, T.C, Haskell L, Hawkins S, Head I, Henry, J.A, Hill J, Hodgson, S.H.C, Hou, M.M, Howe E, Howell N, Hutlin C, Ikram S, Isitt C, Iveson P, Jackson S, Jackson F, James, S.W, Jenkins M, Jones E, Jones K, Jones, C.E, Jones B, Kailath R, Karampatsas K, Keen J, Kelly S, Kelly D, Kerr D, Kerridge S, Khan L, Khan U, Killen A, Kinch J, King TB, King L, King J, Kingham-Page L, Klenerman P, Knapper F, Knight JC, Knott D, Koleva S, Kupke A, Larkworthy, C.W, Larwood, J.P.J, Laskey A, Lawrie, A.M, Lee A, Ngan Lee KY, Lees EA, Legge H, Lelliott A, Lemm N-M, Lias, A.M, Linder A, Lipworth S, Liu X, Liu S, Lopez Ramon R, Lwin M, Mabesa F, Madhavan M, Mallett G, Mansatta K, Marcal I, Marinou S, Marlow E, Marshall, J.L, Martin J, McEwan J, McInroy L, Meddaugh G, Mentzer AJ, Mirtorabi N, Moore M, Moran E, Morey E, Morgan V, Morris SJ, Morrison H, Morshead G, Morter R, Mujadidi YF, Muller J, Munera-Huertas T, Munro C, Munro A, Murphy S, Munster VJ, Mweu P, Noé A, Nugent FL, Nuthall E, O'Brien K, O'Connor D, Oguti B, Oliver, J.L, Oliveira C, O'Reilly, P.J, Osborn M, Osborne P, Owen C, Owens D, Owino N, Pacurar M, Parker K, Parracho H, Patrick-Smith M, Payne, V, Pearce J, Peng Y, Peralta Alvarez, M.P, Perring J, Pfafferott K, Pipini D, Plested E, Pluess-Hall H, Pollock K, Poulton I, Presland L, Provstgaard-Morys S, Pulido D, Radia K, Ramos Lopez F, Rand J, Ratcliffe H, Rawlinson T, Rhead S, Riddell A, Ritchie, A.J, Roberts H, Robson J, Roche S, Rohde C, Rollier, C.S, Romani R, Rudiansyah I, Saich S, Sajjad S, Salvador S, Sanchez Riera L, Sanders H, Sanders K, Sapaun S, Sayce C, Schofield E, Screaton G, Selby B, Semple C, Sharpe, H.R, Shaik I, Shea A, Shelton H, Silk S, Silva-Reyes L, Skelly, D.T, Smee H, Smith, C.C, Smith, D.J, Song R, Spencer, A.J, Stafford E, Steele A, Stefanova E, Stockdale L, Szigeti A, Tahiri-Alaoui A, Tait M, Talbot H, Tanner R, Taylor, I.J, Taylor, V, Te Water Naude R, Thakur N, Themistocleous Y, Themistocleous A, Thomas M, Thomas, T.M, Thompson A, Thomson-Hill S, Tomlins J, Tonks S, Towner J, Tran N, Tree, J.A, Truby A, Turkentine K, Turner C, Turner N, Turner S, Tuthill T, Ulaszewska M, Varughese R, Van Doremalen N, Veighey K, Verheul, M.K, Vichos I, Vitale E, Walker L, Watson, M.E.E, Welham B, Wheat J, White C, White R, Worth, A.T, Wright D, Wright S, Yao, X.L, Yau Y. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. The Lancet 2020;396:467–478 [DOI] [PMC free article] [PubMed]
- 36.Garvin MR, Prates TE, Pavicic M, Jones P, Amos BK, Geiger A, Shah MB, Streich J, Felipe Machado Gazolla JG, Kainer D, Cliff A, Romero J, Keith N, Brown JB, Jacobson D. Potentially adaptive SARS-CoV-2 mutations discovered with novel spatiotemporal and explainable AI models. Genome Biol. 2020;21:304. doi: 10.1186/s13059-020-02191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobeil SMC, Janowska K, McDowell S, Mansouri K, Parks R, Manne K, Stalls V, Kopp MF, Henderson R, Edwards RJ, Haynes BF, Acharya P. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34:108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groves DC, Rowland-Jones SL, Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2020. [DOI] [PMC free article] [PubMed]
- 39.Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci. 2020;117:15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V, Gobeil SMC, Kopp M, Li D, Parks R, Hsu AL, Borgnia MJ, Haynes BF, Acharya P. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Peng P, Wang K, Fang L, Luo F-Y, Jin A-S, Liu B-Z, Tang N, Huang A-L. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, Yang C, Xu X-F, Xu W, Liu S-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hufsky F, Lamkiewicz K, Almeida A, Aouacheria A, Arighi C, Bateman A, Baumbach J, Beerenwinkel N, Brandt C, Cacciabue M, Chuguransky S, Drechsel O, Finn RD, Fritz A, Fuchs S, Hattab G, Hauschild A-C, Heider D, Hoffmann M, Hölzer M, Hoops S, Kaderali L, Kalvari I, von Kleist M, Kmiecinski R, Kühnert D, Lasso G, Libin P, List M, Löchel HF, Martin MJ, Martin R, Matschinske J, McHardy AC, Mendes P, Mistry J, Navratil V, Nawrocki EP, O'Toole ÁN, Ontiveros-Palacios N, Petrov AI, Rangel-Pineros G, Redaschi N, Reimering S, Reinert K, Reyes A, Richardson L, Robertson DL, Sadegh S, Singer JB, Theys K, Upton C, Welzel M, Williams L, Marz M. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief Bioinform. 2021;22:642–663. doi: 10.1093/bib/bbaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iacob S, Iacob DG. SARS-CoV-2 Treatment approaches: numerous options, no certainty for a versatile virus. Front Pharmacol 2020;11. [DOI] [PMC free article] [PubMed]
- 46.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui C-C, Hein L, Uhlig S, Slutsky AS, Jiang C, Penninger JM. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isabel S, Graña-Miraglia L, Gutierrez JM, Bundalovic-Torma C, Groves HE, Isabel MR, Eshaghi A, Patel SN, Gubbay JB, Poutanen T, Guttman DS, Poutanen SM. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci Rep. 2020;10:14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson CB, Zhang L, Farzan, M., Choe, H., 2020. Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem Biophys Res Commun. 2021;538:108-115. [DOI] [PMC free article] [PubMed]
- 49.Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 2020;23:101212. [DOI] [PMC free article] [PubMed]
- 50.Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, Lu JM, Peukes J, Xiong X, Kräusslich H-G, Scheres SHW, Bartenschlager R, Briggs JAG. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keefe BR, Giomarelli B, Barnard DL, Shenoy SR, Chan PKS, McMahon JB, Palmer KE, Barnett BW, Meyerholz DK, Wohlford-Lenane CL, McCray PB. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol. 2010;84:2511. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khalil MI, Salih MA, Mustafa AA. Broad beans (Vicia faba) and the potential to protect from COVID-19 coronavirus infection. Sudan J Paediatr. 2020;20:10–12. doi: 10.24911/SJP.1061585398078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, Hardes K, Powley WM, Wright TJ, Siederer SK, Fairman DA, Lipson DA, Bayliffe AI, Lazaar AL. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Critical Care 2017;21:234. [DOI] [PMC free article] [PubMed]
- 55.Khare P, Sahu U, Pandey SC, Samant M. Current approaches for target-specific drug discovery using natural compounds against SARS-CoV-2 infection. Virus Res. 2020;290:198169. doi: 10.1016/j.virusres.2020.198169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khodarahmi R, Sayad B, Sobhani M. The ACE2 as a “rescue protein” or “suspect enzyme” in COVID-19: possible application of the “engineered inactive hrsACE2” as a safer therapeutic agent in the treatment of SARS-CoV-2 infection. J Iran Chem Soc. 2021;18:495–502. doi: 10.1007/s13738-020-02049-z. [DOI] [Google Scholar]
- 57.Korber B, Fischer W, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Foley B, Giorgi E, Bhattacharya T, Parker M, Partridge D, Evans C, Freeman T, de Silva T, LaBranche C, Montefiori D. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020;2020.2004.2029.069054.
- 58.Kyriakidis NC, López-Cortés A, González EV, Grimaldos AB, Prado EO. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lackner M, Rössler A, Volland A, Stadtmüller M, Müllauer B, Banki Z, Ströhle J, Luttick A, Fenner J, Stoiber H, Laer D, Wolff T, Schwarz C, Nagl M. N-chlorotaurine, a novel inhaled virucidal antiseptic is highly active against respiratory viruses including SARS-CoV-2 (COVID-19). 2020. [DOI] [PMC free article] [PubMed]
- 60.Laffoley D, Baxter JM, Amon DJ, Claudet J, Hall-Spencer JM, Grorud-Colvert K, Levin LA, Reid PC, Rogers AD, Taylor ML, Woodall LC, Andersen NF. Evolving the narrative for protecting a rapidly changing ocean, post-COVID-19. Aquatic Conserv: Mar FreshW Ecosyst. 2021;31:1512-1534. [DOI] [PMC free article] [PubMed]
- 61.Laha S, Chakraborty J, Das S, Manna SK, Biswas S, Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lam SD, Bordin N, Waman VP, Scholes HM, Ashford P, Sen N, van Dorp L, Rauer C, Dawson NL, Pang CSM, Abbasian M, Sillitoe I, Edwards SJL, Fraternali F, Lees JG, Santini JM, Orengo CA. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci Rep. 2020;10:16471. doi: 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 64.Lee C. Griffithsin, a highly potent broad-spectrum antiviral lectin from red algae: from discovery to clinical application. Mar Drugs. 2019;17:567. doi: 10.3390/md17100567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li D-D, Li Q-H. SARS-CoV-2: vaccines in the pandemic era. Mil Med Res. 2021;8:1. doi: 10.1186/s40779-020-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Libster R, Pérez Marc G, Wappner D, Coviello S, Bianchi A, Braem V, Esteban I, Caballero MT, Wood C, Berrueta M, Rondan A, Lescano G, Cruz P, Ritou Y, Fernández Viña V, Álvarez Paggi D, Esperante S, Ferreti A, Ofman G, Ciganda Á, Rodriguez R, Lantos J, Valentini R, Itcovici N, Hintze A, Oyarvide ML, Etchegaray C, Neira A, Name I, Alfonso J, López Castelo R, Caruso G, Rapelius S, Alvez F, Etchenique F, Dimase F, Alvarez D, Aranda SS, Sánchez Yanotti C, De Luca J, Jares Baglivo S, Laudanno S, Nowogrodzki F, Larrea R, Silveyra M, Leberzstein G, Debonis A, Molinos J, González M, Perez E, Kreplak N, Pastor Argüello S, Gibbons L, Althabe F, Bergel E, Polack FP. Early high-titer plasma therapy to prevent severe Covid-19 in older adults. N Engl J Med. 2021;384:610–618. doi: 10.1056/NEJMoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ling R, Dai Y, Huang B, Huang W, Yu J, Lu X, Jiang Y. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130:170328. doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lori Robins EK, Robins D, Edgar S, Meschke J, Gafken P, Williams J. Modification of IL-6 by hypochlorous acid: effects on receptor binding and possible role in treatment of COVID-19. chemrxiv. 2021 [DOI] [PMC free article] [PubMed]
- 69.Mahendran ASK, Lim YS, Fang C-M, Loh H-S, Le CF. The potential of antiviral peptides as COVID-19 therapeutics. Front Pharmacol. 2020;11. [DOI] [PMC free article] [PubMed]
- 70.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo S-YC, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e2316. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mittal A, Manjunath K, Ranjan RK, Kaushik S, Kumar S, Verma V. COVID-19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020;16:1008762. doi: 10.1371/journal.ppat.1008762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mollica V, Rizzo A, Massari F. The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Fut Oncol. 2020;16:2029–2033. doi: 10.2217/fon-2020-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mudgal R, Nehul S, Tomar S. Prospects for mucosal vaccine: shutting the door on SARS-CoV-2. Hum Vaccin Immunother. 2020;16:2921–2931. doi: 10.1080/21645515.2020.1805992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mullard A. COVID-19 vaccines start moving into advanced trials. Nat Rev Drug Discov. 2020;19:435. doi: 10.1038/d41573-020-00107-y. [DOI] [PubMed] [Google Scholar]
- 76.Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, Atif SM, Hariprasad G, Hasan GM, Hassan MI. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878–165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nascimento da Silva LC, Mendonça JSP, de Oliveira WF, Batista KLR, Zagmignan A, Viana IFT, dos Santos Correia MT. Exploring lectin–glycan interactions to combat COVID-19: lessons acquired from other enveloped viruses. Glycobiology. 2020;31:358-371. [DOI] [PMC free article] [PubMed]
- 78.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, Kumar S, Bhattacharyya A, Kumar H, Bansal S, Medhi B. Drug targets for corona virus: a systematic review. Indian J Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Psaltopoulou T, Sergentanis TN, Pappa V, Politou M, Terpos E, Tsiodras S, Pavlakis GN, Dimopoulos MA. The emerging role of convalescent plasma in the treatment of COVID-19. Hemasphere. 2020;4:e409–e409. doi: 10.1097/HS9.0000000000000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pucci F, Rooman M. bioRPrediction and evolution of the molecular fitness of SARS-CoV-2 variants: introducing SpikePro. Viruses. 2021;13:935. [DOI] [PMC free article] [PubMed]
- 82.Qiao Y, Wang X-M, Mannan R, Pitchiaya S, Zhang Y, Wotring JW, Xiao L, Robinson DR, Wu Y-M, Tien JCY, Cao X, Simko SA, Apel IJ, Bawa P, Kregel S, Narayanan SP, Raskind G, Ellison SJ, Parolia A, Zelenka-Wang S, McMurry L, Su F, Wang R, Cheng Y, Delekta AD, Mei Z, Pretto CD, Wang S, Mehra R, Sexton JZ, Chinnaiyan AM. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc Natl Acad Sci. 2021;118:118. doi: 10.1073/pnas.2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B, Goepfert PA, Truyers C, Fennema H, Spiessens B, Offergeld K, Scheper G, Taylor KL, Robb ML, Treanor J, Barouch DH, Stoddard J, Ryser MF, Marovich MA, Neuzil KM, Corey L, Cauwenberghs N, Tanner T, Hardt K, Ruiz-Guiñazú J, Le Gars M, Schuitemaker H, Van Hoof J, Struyf F, Douoguih M. Safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sáez-Álvarez Y, Arias A, del Águila C, Agudo R. Development of a fluorescence-based method for the rapid determination of Zika virus polymerase activity and the screening of antiviral drugs. Sci Rep. 2019;9:5397. doi: 10.1038/s41598-019-41998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saha S, Kadam S. Convalescent plasma therapy—a silver lining for COVID-19 management? Hematol Transfus Cell Therapy. 2021;43:201–211. doi: 10.1016/j.htct.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz, L.M, Vormehr M, Quandt J, Bidmon N, Ulges A, Baum A, Pascal, K., Maurus D, Brachtendorf S, Lörks, V, Sikorski J, Koch P, Hilker R, Becker D, Eller, A.-K, Grützner J, Tonigold M, Boesler C, Rosenbaum C, Heesen L, Kühnle, M.-C, Poran A, Dong, J.Z, Luxemburger U, Kemmer-Brück A, Langer D, Bexon M, Bolte S, Palanche T, Schultz A, Baumann S, Mahiny, A.J, Boros G, Reinholz J, Szabó, G.T, Karikó K, Shi, P.-Y, Fontes-Garfias C, Perez, J.L, Cutler M, Cooper D, Kyratsous, C.A, Dormitzer, P.R, Jansen, K.U, Türeci, Ö. BNT162b2 induces SARS-CoV-2-neutralising antibodies and poly-specific T cells in humans. Nature. 2021;595:572-577 [DOI] [PubMed]
- 87.Salvatori G, Luberto L, Maffei M, Aurisicchio L, Roscilli G, Palombo F, Marra E. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J Transl Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Santos IDA, Grosche VR, Bergamini FRG, Sabino-Silva R, Jardim ACG. Antivirals against coronaviruses: candidate drugs for SARS-CoV-2 treatment? Front Microbiol. 2020;11. [DOI] [PMC free article] [PubMed]
- 89.Saputri DS, Li S, van Eerden FJ, Rozewicki J, Xu Z, Ismanto HS, Davila A, Teraguchi S, Katoh K, Standley DM. Flexible, functional, and familiar: characteristics of SARS-CoV-2 spike protein evolution. Front Microbiol 2020;11. [DOI] [PMC free article] [PubMed]
- 90.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh D, Yi SV. On the origin and evolution of SARS-CoV-2. Exp Mol Med. 2021;53:537-547. [DOI] [PMC free article] [PubMed]
- 92.Sitthiyotha T, Chunsrivirot S. Computational design of 25-mer peptide binders of SARS-CoV-2. J Phys Chem B. 2020;124:10930–10942. doi: 10.1021/acs.jpcb.0c07890. [DOI] [PubMed] [Google Scholar]
- 93.Sternberg A, Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;257:118056. doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subhash DGV, Kumar G, Sapre A, Dasgupta S. Possible Prevention of COVID 19 by using linoleic acid (C18) rich algae oil. AIJR Preprints. 2020.
- 95.Tada T, Dcosta BM, Zhou H, Vaill A, Kazmierski W, Landau NR. Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. bioRxiv. 2021;2021.2002.2018.431897.
- 96.Taylor PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev Immunol. 2021;21:382-393 [DOI] [PMC free article] [PubMed]
- 97.Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD, Kavanagh Williamson M, Shoemark DK, Garzoni F, Staufer O, Milligan R, Capin J, Mulholland AJ, Spatz J, Fitzgerald D, Berger I, Schaffitzel C. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Unni S, Aouti S, Thiyagarajan S, Padmanabhan B. Identification of a repurposed drug as an inhibitor of Spike protein of human coronavirus SARS-CoV-2 by computational methods. J Biosci. 2020;45:130. doi: 10.1007/s12038-020-00102-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.VanPatten S, He M, Altiti A, Cheng K, Ghanem MH, Al-Abed Y. Evidence supporting the use of peptides and peptidomimetics as potential SARS-CoV-2 (COVID-19) therapeutics. Future Med Chem. 2020;12:1647–1656. doi: 10.4155/fmc-2020-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KsJ, Fairlie L, Faust, S.N, Feng S, Ferreira, D.M, Finn A, Goodman, A.L, Green, C.M, Green, C.A, Heath, P.T, Hill C, Hill H, Hirsch I, Hodgson, S.H.C, Izu A, Jackson S, Jenkin D, Joe, C.C.D, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie, A.M, Lelliott A, Libri V, Lillie, P.J, Mallory R, Mendes, A.V.A, Milan, E.P, Minassian, A.M, McGregor A, Morrison H, Mujadidi, Y.F, Nana A, O’Reilly, P.J, Padayachee, S.D, Pittella A, Plested E, Pollock, K.M, Ramasamy, M.N, Rhead S, Schwarzbold, A.V, Singh N, Smith A, Song R, Snape, M.D, Sprinz E, Sutherland, R.K, Tarrant R, Thomson, E.C, Török, M.E, Toshner M, Turner, D.P.J, Vekemans J, Villafana, T.L, Watson, M.E.E, Williams, C.J, Douglas, A.D, Hill, A.V.S, Lambe T, Gilbert, S.C, Pollard, A.J, Aban M, Abayomi F, Abeyskera K, Aboagye J, Adam M, Adams K, Adamson J, Adelaja, Y.A, Adewetan G, Adlou S, Ahmed K, Akhalwaya Y, Akhalwaya S, Alcock A, Ali A, Allen, E.R, Allen L, Almeida, T.C.D.S.C, Alves, M.P.S, Amorim F, Andritsou F, Anslow R, Appleby M, Arbe-Barnes, E.H, Ariaans, M.P, Arns B, Arruda L, Azi P, Azi L, Babbage G, Bailey C, Baker, K.F, Baker M, Baker N, Baker P, Baldwin L, Baleanu I, Bandeira D, Bara A, Barbosa, M.A.S, Barker D, Barlow, G.D, Barnes E, Barr, A.S, Barrett, J.R, Barrett J, Bates L, Batten A, Beadon K, Beales E, Beckley R, Belij-Rammerstorfer S, Bell J, Bellamy D, Bellei N, Belton S, Berg A, Bermejo L, Berrie E, Berry L, Berzenyi D, Beveridge A, Bewley, K.R, Bexhell H, Bhikha S, Bhorat, A.E, Bhorat, Z.E, Bijker E, Birch G, Birch S, Bird A, Bird O, Bisnauthsing K, Bittaye M, Blackstone K, Blackwell L, Bletchly H, Blundell, C.L, Blundell, S.R, Bodalia P, Boettger, B.C, Bolam E, Boland E, Bormans D, Borthwick N, Bowring F, Boyd A, Bradley P, Brenner T, Brown P, Brown C, Brown-O'Sullivan C, Bruce S, Brunt E, Buchan R, Budd W, Bulbulia, Y.A, Bull M, Burbage J, Burhan H, Burn A, Buttigieg, K.R, Byard N, Cabera Puig I, Calderon G, Calvert A, Camara S, Cao M, Cappuccini F, Cardoso, J.R, Carr M, Carroll, M.W, Carson-Stevens A, Carvalho, Y.d.M, Carvalho, J.A.M, Casey, H.R, Cashen P, Castro T, Castro, L.C, Cathie K, Cavey A, Cerbino-Neto J, Chadwick J, Chapman D, Charlton S, Chelysheva I, Chester O, Chita S, Cho, J.-S, Cifuentes L, Clark E, Clark M, Clarke A, Clutterbuck, E.A, Collins, S.L.K, Conlon, C.P, Connarty S, Coombes N, Cooper C, Cooper R, Cornelissen, L., Corrah, T., Cosgrove, C., Cox, T., Crocker, W.E.M., Crosbie, S., Cullen, L., Cullen, D., Cunha, D.R.M.F., Cunningham, C., Cuthbertson, F.C., Da Guarda, S.N.F., da Silva, L.P., Damratoski, B.E., Danos, Z., Dantas, M.T.D.C., Darroch, P., Datoo, M.S., Datta, C., Davids, M., Davies, S.L., Davies, H., Davis, E., Davis, J., Davis, J., De Nobrega, M.M.D, De Oliveira Kalid, L.M, Dearlove D, Demissie T, Desai A, Di Marco S, Di Maso C, Dinelli, M.I.S, Dinesh T, Docksey C, Dold C, Dong T, Donnellan, F.R, Dos Santos T, dos Santos, T.G, Dos Santos, E.P, Douglas N, Downing C, Drake J, Drake-Brockman R, Driver K, Drury R, Dunachie, S.J, Durham, B.S, Dutra L, Easom, N.J.W, van Eck S, Edwards M, Edwards, N.J, El Muhanna, O.M, Elias, S.C, Elmore M, English M, Esmail A, Essack, Y.M, Farmer E, Farooq M, Farrar M, Farrugia L, Faulkner B, Fedosyuk S, Felle S, Feng S, Ferreira Da Silva C, Field S, Fisher R, Flaxman A, Fletcher J, Fofie H, Fok H, Ford, K.J, Fowler J, Fraiman, P.H.A, Francis E, Franco, M.M, Frater J, Freire, M.S.M, Fry, S.H, Fudge S, Furze J, Fuskova M, Galian-Rubio P, Galiza E, Garlant H, Gavrila M, Geddes A, Gibbons, K.A, Gilbride C, Gill H, Glynn S, Godwin K, Gokani K, Goldoni, U.C, Goncalves M, Gonzalez, I.G.S, Goodwin J, Goondiwala A, Gordon-Quayle K, Gorini G, Grab J, Gracie L, Greenland M, Greenwood N, Greffrath J, Groenewald, M.M, Grossi L, Gupta G, Hackett M, Hallis B, Hamaluba M, Hamilton E, Hamlyn J, Hammersley D, Hanrath, A.T, Hanumunthadu B, Harris, S.A, Harris C, Harris T, Harrison, T.D, Harrison D, Hart, T.C, Hartnell B, Hassan S, Haughney J, Hawkins S, Hay J, Head I, Henry J, Hermosin Herrera M, Hettle, D.B, Hill J, Hodges G, Horne E, Hou, M.M, Houlihan C, Howe E, Howell N, Humphreys J, Humphries, H.E, Hurley K, Huson C, Hyder-Wright A, Hyams C, Ikram S, Ishwarbhai A, Ivan M, Iveson P, Iyer V, Jackson F, De Jager J, Jaumdally S, Jeffers H, Jesudason N, Jones B, Jones K, Jones E, Jones C, Jorge, M.R, Jose A, Joshi A, Júnior, E.A.M.S, Kadziola J, Kailath R, Kana F, Karampatsas K, Kasanyinga M, Keen J, Kelly, E.J, Kelly, D.M, Kelly D, Kelly S, Kerr D, Kfouri, R.d.Á, Khan L, Khozoee B, Kidd S, Killen A, Kinch J, Kinch P, King, L.D.W, King, T.B, Kingham L, Klenerman P, Knapper F, Knight, J.C, Knott D, Koleva S, Lang M, Lang G, Larkworthy, C.W, Larwood, J.P.J, Law R, Lazarus, E.M, Leach A, Lees, E.A, Lemm, N.-M, Lessa A, Leung S, Li Y, Lias, A.M, Liatsikos K, Linder A, Lipworth S, Liu S, Liu X, Lloyd A, Lloyd S, Loew L, Lopez Ramon R, Lora L, Lowthorpe V, Luz K, MacDonald, J.C, MacGregor G, Madhavan M, Mainwaring, D.O, Makambwa E, Makinson R, Malahleha M, Malamatsho R, Mallett G, Mansatta K, Maoko T, Mapetla K, Marchevsky, N.G, Marinou S, Marlow E, Marques, G.N, Marriott P, Marshall, R.P, Marshall, J.L, Martins, F.J, Masenya M, Masilela M, Masters, S.K, Mathew M, Matlebjane H, Matshidiso K, Mazur O, Mazzella A, McCaughan H, McEwan J, McGlashan J, McInroy L, McIntyre Z, McLenaghan D, McRobert N, McSwiggan S, Megson C, Mehdipour S, Meijs W, Mendonça RNÁ, Mentzer AJ, Mirtorabi N, Mitton C, Mnyakeni S, Moghaddas F, Molapo K, Moloi M, Moore M, Moraes-Pinto MI, Moran M, Morey E, Morgans R, Morris S, Morris S, Morris HC, Morselli F, Morshead G, Morter R, Mottal L, Moultrie A, Moya N, Mpelembue M, Msomi S, Mugodi Y, Mukhopadhyay E, Muller J, Munro A, Munro C, Murphy S, Mweu P, Myasaki CH, Naik G, Naker K, Nastouli E, Nazir A, Ndlovu B, Neffa F, Njenga C, Noal H, Noé A, Novaes G, Nugent FL, Nunes G, O'Brien K, O'Connor D, Odam M, Oelofse S, Oguti B, Olchawski V, Oldfield NJ, Oliveira MG, Oliveira C, Oosthuizen A, O'Reilly P, Osborne P, Owen D.R.J, Owen L, Owens D, Owino N, Pacurar M, Paiva, B.V.B, Palhares, E.M.F, Palmer S, Parkinson S, Parracho, H.M.R.T, Parsons K, Patel D, Patel B, Patel F, Patel K, Patrick-Smith M, Payne, R.O, Peng Y, Penn, E.J, Pennington A, Peralta Alvarez, M.P, Perring J, Perry N, Perumal R, Petkar S, Philip T, Phillips, D.J, Phillips J, Phohu, M.K, Pickup L, Pieterse S, Piper J, Pipini D, Plank M, Du Plessis J, Pollard S, Pooley J, Pooran A, Poulton I, Powers C, Presa, F.B, Price, D.A, Price V, Primeira M, Proud, P.C, Provstgaard-Morys S, Pueschel S, Pulido D, Quaid S, Rabara R, Radford A, Radia K, Rajapaska D, Rajeswaran T, Ramos, A.S.F, Ramos Lopez F, Rampling T, Rand J, Ratcliffe H, Rawlinson T, Rea D, Rees B, Reiné J, Resuello-Dauti M, Reyes Pabon E, Ribiero, C.M, Ricamara M, Richter A, Ritchie N, Ritchie, A.J, Robbins, A.J, Roberts H, Robinson, R.E, Robinson H, Rocchetti, T.T, Rocha, B.P, Roche S, Rollier C, Rose L, Ross Russell, A.L, Rossouw L, Royal S, Rudiansyah I, Ruiz S, Saich S, Sala C, Sale J, Salman, A.M, Salvador N, Salvador S, Sampaio M, Samson, A.D, Sanchez-Gonzalez A, Sanders H, Sanders K, Santos E, Santos Guerra, M.F.S, Satti I, Saunders, J.E, Saunders C, Sayed A, Schim van der Loeff I, Schmid, A.B, Schofield E, Screaton G, Seddiqi S, Segireddy, R.R, Senger R, Serrano S, Shah R, Shaik I, Sharpe, H.E, Sharrocks K, Shaw R, Shea A, Shepherd A, Shepherd, J.G, Shiham F, Sidhom E, Silk, S.E, da Silva Moraes, A.C, Silva-Junior G, Silva-Reyes L, Silveira, A.D, Silveira, M.B.V, Sinha J, Skelly, D.T, Smith, D.C, Smith N, Smith, H.E, Smith, D.J, Smith, C.C, Soares A, Soares T, Solórzano C, Sorio, G.L, Sorley K, Sosa-Rodriguez T, Souza, C.M.C.D.L, Souza, B.S.D.F, Souza, A.R, Spencer, A.J, Spina F, Spoors L, Stafford L, Stamford I, Starinskij I, Stein R, Steven J, Stockdale L, Stockwell, L.V, Strickland, L.H, Stuart, A.C, Sturdy A, Sutton N, Szigeti A, Tahiri-Alaoui A, Tanner R, Taoushanis C, Tarr, A.W, Taylor K, Taylor U, Taylor, I.J, Taylor J, te Water Naude R, Themistocleous Y, Themistocleous A, Thomas M, Thomas K, Thomas, T.M, Thombrayil A, Thompson F, Thompson A, Thompson K, Thompson A, Thomson J, Thornton-Jones V, Tighe, P.J, Tinoco, L.A, Tiongson G, Tladinyane B, Tomasicchio M, Tomic A, Tonks S, Towner J, Tran N, Tree J, Trillana G, Trinham C, Trivett R, Truby A, Tsheko, B.L, Turabi A, Turner R, Turner C, Ulaszewska M, Underwood, B.R, Varughese R, Verbart D, Verheul M, Vichos I, Vieira T, Waddington, C.S, Walker L, Wallis E, Wand M, Warbick D, Wardell T, Warimwe G, Warren, S.C, Watkins B, Watson E, Webb S, Webb-Bridges A, Webster A, Welch J, Wells J, West A, White C, White R, Williams P, Williams, R.L, Winslow R, Woodyer M, Worth, A.T, Wright D, Wroblewska M, Yao A, Zimmer R, Zizi D, Zuidewind, P. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet. 2021;397:99-111 [DOI] [PMC free article] [PubMed]
- 101.Wang L, Xiang Y. Spike glycoprotein-mediated entry of SARS coronaviruses. Viruses. 2020;12:1289. doi: 10.3390/v12111289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X, Xia S, Zhu Y, Lu L, Jiang S. Pan-coronavirus fusion inhibitors as the hope for today and tomorrow. Protein Cell. 2021;12:84–88. doi: 10.1007/s13238-020-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang XDR, Povysil G, Zoghbi A, Motelow J, Hostyk J, Nickols N, Rettig M, Goldstein DB. TMPRSS2 transcriptional inhibition as a therapeutic strategy for COVID-19. 2020 10.20944/preprints202003.0360.v2
- 104.Weissman D, Alameh M-G, de Silva T, Collini P, Hornsby H, Brown R, LaBranche CC, Edwards RJ, Sutherland L, Santra S, Mansouri K, Gobeil S, McDanal C, Pardi N, Hengartner N, Lin PJC, Tam Y, Shaw PA, Lewis MG, Boesler C, Şahin U, Acharya P, Haynes BF, Korber B, Montefiori DC. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e24. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.World Health Organization (WHO). WHO coronavirus (COVID-19) dashboard, 04/22/2021 ed. 2021.
- 106.Widge AT, Rouphael NG, Jackson LA, Anderson EJ, Roberts PC, Makhene M, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott AB, Flach B, Lin BC, Doria-Rose NA, O’Dell S, Schmidt SD, Neuzil KM, Bennett H, Leav B, Makowski M, Albert J, Cross K, Edara V-V, Floyd K, Suthar MS, Buchanan W, Luke CJ, Ledgerwood JE, Mascola JR, Graham BS, Beigel JH. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N Engl J Med. 2020;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, Zhu Y, Wang Q, Lu L, Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xian Y, Zhang J, Bian Z, Zhou H, Zhang Z, Lin Z, Xu H. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharmaceut Sin B. 2020;10:1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xiao-Yong Z, Ying Z, Xuefu Z, Ke H, Yichao Q, Yang L, Leping Y, Bihui H, Yulong H. Molecular Evolution of SARS-CoV-2 Structural Genes: Evidence of Positive Selection in Spike Glycoprotein. BioRixv. 2020. 10.1101/2020.06.25.170688
- 110.Xing Y, Li X, Gao X, Dong Q. Natural polymorphisms are present in the furin cleavage site of the SARS-CoV-2 spike glycoprotein. Front Genet. 2020;11:783. doi: 10.3389/fgene.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiong X, Qu K, Ciazynska KA, Hosmillo M, Carter AP, Ebrahimi S, Ke Z, Scheres SHW, Bergamaschi L, Grice GL, Zhang Y, Bradley J, Lyons PA, Smith KGC, Toshner M, Elmer A, Ribeiro C, Kourampa J, Jose S, Kennet J, Rowlands J, Meadows A, O’Brien C, Rastall R, Crucusio C, Hewitt S, Price J, Calder J, Canna L, Bucke A, Tordesillas H, Harris J, Ruffolo V, Domingo J, Graves B, Butcher H, Caputo D, Le Gresley E, Dunmore BJ, Martin J, Legchenko E, Treacy C, Huang C, Wood J, Sutcliffe R, Hodgson J, Shih J, Graf S, Tong Z, Mescia F, Tilly T, O’Donnell C, Hunter K, Pointon L, Pond N, Wylot M, Jones E, Fawke S, Bullman B, Bergamaschi L, Turner L, Jarvis I, Omarjee O, De Sa A, Marsden J, Betancourt A, Perera M, Epping M, Richoz N, Bower G, Sharma R, Nice F, Huhn O, Stark H, Walker N, Stirrups K, Ovington N, Dewhust E, Li E, Papadia S, Nathan JA, Baker S, James LC, Baxendale HE, Goodfellow I, Doffinger R, Briggs JAG, The C-NC-BC. A thermostable, closed SARS-CoV-2 spike protein trimer. Nat Struct Mol Biol. 2020;27:934–941. doi: 10.1038/s41594-020-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Y, Du L. SARS-CoV-2 spike protein: a key target for eliciting persistent neutralizing antibodies. Signal Transduct Target Ther. 2021;6:95. doi: 10.1038/s41392-021-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yesudhas D, Srivastava A, Gromiha MM. COVID-19 outbreak: history, mechanism, transmission, structural studies and therapeutics. Infection. 2021;49:199–213. doi: 10.1007/s15010-020-01516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang G, Pomplun S, LoftisAR, Tan X, Loas A, Pentelute BL. Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. bioRxiv. 2020a;2020.2003.2019.999318.
- 115.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang J, Cai Y, Xiao T, Lu J, Peng H, Sterling SM, Walsh RM, Rits-Volloch S, Sliz P, Chen B. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021;372:525-530. [DOI] [PMC free article] [PubMed]
- 117.Zhang L, JacksonCB, Mou H, Ojha A, Rangarajan ES, Izard T, Farzan M, Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020d;2020.2006.2012.148726.
- 118.Zhao X, Chen H, Wang H. Glycans of SARS-CoV-2 spike protein in virus infection and antibody production. Front Mol Biosci. 2021;8:53. doi: 10.3389/fmolb.2021.629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zoufaly A, Poglitsch M, Aberle JH, Hoepler W, Seitz T, Traugott M, Grieb A, Pawelka E, Laferl H, Wenisch C, Neuhold S, Haider D, Stiasny K, Bergthaler A, Puchhammer-Stoeckl E, Mirazimi A, Montserrat N, Zhang H, Slutsky AS, Penninger JM. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]