Abstract
Background
Artesunate-amodiaquine is a potential therapy for uncomplicated malaria in Cambodia.
Methods
Between September 2016 and January 2017, artesunate-amodiaquine efficacy and safety were evaluated in a prospective, open-label, single-arm observational study at health centers in Mondulkiri, Pursat, and Siem Reap Provinces, Cambodia. Adults and children with microscopically confirmed Plasmodium falciparum malaria received oral artesunate-amodiaquine once daily for 3 days plus single-dose primaquine, with follow-up on days 7, 14, 21, and 28. The primary outcome was day-28 polymerase chain reaction (PCR)-adjusted adequate clinical and parasitological response (ACPR). An amodiaquine parasite survival assay (AQSA) was developed and applied to whole genome sequencing results to evaluate potential amodiaquine resistance molecular markers.
Results
In 63 patients, day-28 PCR-adjusted ACPR was 81.0% (95% confidence interval [CI], 68.9–88.7). Day 3 parasite positivity rate was 44.4% (28/63; 95% CI, 31.9–57.5). All 63 isolates had the K13(C580Y) marker for artemisinin resistance; 79.4% (50/63) had Pfpm2 amplification. The AQSA resistance phenotype (≥45% parasite survival) was expressed in 36.5% (23/63) of isolates and was significantly associated with treatment failure (P = .0020). Pfmdr1 mutant haplotypes were N86/184F/D1246, and Pfcrt was CVIET or CVIDT at positions 72–76. Additional Pfcrt mutations were not associated with amodiaquine resistance, but the G353V mutant allele was associated with ACPR compared to Pfmdr1 haplotypes harboring F1068L or S784L/R945P mutations (P = .030 and P = .0004, respectively).
Conclusions
For uncomplicated falciparum malaria in Cambodia, artesunate-amodiaquine had inadequate efficacy owing to amodiaquine-resistant P. falciparum. Amodiaquine resistance was not associated with previously identified molecular markers.
Keywords: artesunate-amodiaquine, artemisinin, Plasmodium falciparum, Cambodia, drug resistance
Artesunate-amodiaquine has inadequate efficacy in Cambodian patients with uncomplicated falciparum malaria. Amodiaquine resistance is present in Cambodia and not associated with molecular markers reported in Africa and South America.
(See the Editorial Commentary by Petersen and Picot on pages 414–5.)
Artemisinin-based combination therapy (ACT) includes a rapid-acting artemisinin with a longer-acting partner drug. ACTs support effective malaria treatment globally, contributing to recent declines in mortality [1]. In 2006, artemisinin-resistant Plasmodium falciparum was confirmed in Cambodia’s western provinces [2] and subsequently verified in multiple studies [3]. Artemisinin resistance delays parasite killing, but resistance to the partner drug is required before treatment failure rates increase [4, 5]. Unfortunately, P. falciparum resistant to artemisinins and partner drugs (piperaquine, mefloquine) circulate in Cambodia and the Greater Mekong subregion, undermining clinical efficacy and limiting treatment options [1, 6–8].
Artesunate-amodiaquine was not deployed systematically in Cambodia and requires evaluation as a potential replacement for failing ACTs. In Africa, artesunate-amodiaquine is used extensively with 98.5% clinical efficacy [1]. P. falciparum with multidrug resistance 1 (Pfmdr1) alleles N86Y/Y184/D1246Y (YYY haplotype) is associated with amodiaquine treatment failures in Africa [9] but has not been detected in Cambodia [10]. The most prevalent chloroquine resistance transporter gene (Pfcrt) haplotype in Cambodia is CVIET at positions 72–76 (wild-type CVMNT) [10], which is also prevalent in Africa [11, 12], and insufficient to confer amodiaquine resistance in vivo [13, 14]. In Viet Nam, 2 artesunate-amodiaquine clinical trials showed encouraging results with 98% efficacy [15, 16]. However, there are no recent data from Southeast Asia on artesunate-amodiaquine efficacy.
This study investigated artesunate-amodiaquine clinical efficacy for uncomplicated falciparum malaria in Cambodia. In the event of clinical failures, molecular markers associated with amodiaquine resistance were to be investigated.
METHODS
Study Design
This prospective, single-arm, open-label therapeutic efficacy trial of artesunate-amodiaquine plus single-dose primaquine was conducted between September 2016 and January 2017 at 3 health centers in Cambodia: Koh Gnek (Koh Gnek district, Mondulkiri province), Promoy (Veal Veng district, Pursat province), and Khvav (Chi Kraeng district, Siem Reap province) (Supplementary material Figure S1).
The study confirmed to Good Clinical Practice and the Declaration of Helsinki (2000). The protocol followed the standard World Health Organization protocol for the surveillance of antimalarial treatment efficacy [17] and was approved by the National Cambodian Ethical Board and the World Health Organization Regional Office, Western Pacific Region. Owing to an administrative error, this study was registered retrospectively at https://www.anzctr.org.au (identifier ACTRN12619001628134). All patients or their guardians provided written informed consent. Additionally, assent was obtained from children aged ≥12 years.
Patients
Eligible patients were aged 5–60 years with microscopically confirmed P. falciparum monoinfection (1000–250 000 µL−1 blood), fever or history of fever during the past 24 h, who could swallow oral medication. Unmarried girls and women aged 12–18 years were excluded because a pregnancy test would be culturally unacceptable; pregnant and lactating women were also excluded. All other women of child-bearing potential were given a pregnancy test. Exclusion criteria were severe falciparum malaria, severe malnutrition, febrile conditions other than malaria or other underlying chronic illness, medication that might interfere with antimalarial pharmacokinetics, or a history of hypersensitivity to artemisinin or amodiaquine.
Treatment
Artesunate-amodiaquine (ASAQ Winthrop®, Sanofi, Paris, France) was administered under supervision once-daily for 3 days. Doses were determined by bodyweight to achieve 4 mg/kg/day (range 2–10 mg/kg) artesunate and 10 mg/kg/day (range 7.5–15 mg/kg) amodiaquine. Primaquine was given as a single 15-mg dose (0.25 mg base/kg). All patients were treated as in-patients with out-patient follow-up visits on days 7, 14, 21, and 28. Any recurrence during follow-up was treated with artesunate-mefloquine.
Procedures
At enrollment, a clinical examination was performed and a full medical history taken. Adverse events were recorded at all study visits. Parasitemia and Plasmodium species identification was assessed using Giemsa stained thick and thin blood films obtained at screening, every day following the first treatment dose until samples were parasite negative, at each weekly follow-up visit, and if clinically indicated. Parasite counts were recorded as the average from 2 microscopists using standard methods [17]. Treatment failures were verified as recrudescence using polymerase chain reaction (PCR) genotyping by comparing P. falciparum genes msp1, msp2, and glurp in pretreatment blood samples versus those obtained at recurrence [18].
Molecular Surveillance
Using samples collected on day 0, the Kelch13 (K13) gene was sequenced to identify mutations associated with artemisinin resistance [19], and gene copy numbers for P. falciparum plasmepsin 2/3 (Pfpm2) and Pfmdr1 were determined, as per published methods [20]. The threshold for gene amplification was defined as > 1.5 copies.
Amodiaquine Susceptibility in Vitro
Pretreatment blood samples were collected into acid-citrate-dextrose tubes (Becton-Dickinson, Franklin Lakes, NJ, USA) and processed within 48 h at Institut Pasteur, Cambodia. Clinical isolates were culture adapted using standard methods [4]. P. falciparum reference strains 3D7 (amodiaquine susceptible, from MR4) and 7G8 (amodiaquine resistant, from the European Malaria Reagent Repository) were similarly maintained and used as controls. Molecular markers obtained from day 0 samples were confirmed as identical to those obtained from the corresponding culture-adapted parasites via whole genome sequencing except for 1 isolate that had Pfpm2 amplification at day 0, which reverted to a single gene copy under culture.
In vitro susceptibility to mono-desethyl-amodiaquine (from the WorldWide Antimalarial Resistance Network) was assessed using the [3H]-hypoxanthine assay, according to published methods [4]. Half-maximal inhibitory concentration values (IC50) were determined using ICEstimator software (http://www.antimalarial-icestimator.net).
The amodiaquine survival assay (AQSA) was based on a similar assay for piperaquine [21]. Tightly synchronized ring-stage parasites (0–3 h postinvasion) were exposed to 200 nM mono-desethyl-amodiaquine for 48 h and maintained for a further 24 h in drug-free medium. Live parasites were then enumerated microscopically from Giemsa-stained thin blood films on examination of ≥ 10 000 erythrocytes. Parasite survival following exposure to mono-desethyl-amodiaquine was determined as a percentage relative to untreated controls.
Amodiaquine Resistance and Association With Molecular Markers
Investigation of potential molecular markers associated with amodiaquine resistance used an expanded data set including culture-adapted P. falciparum isolates from this study plus 34 culture-adapted clinical isolates collected from sentinel sites between February 2017 and February 2018 (n = 10 Kampong Speu, n = 13 Mondulkiri, n = 3 Pursat, n = 8 Ratanakiri).
Pfcrt and Pfmdr1 were sequenced using whole genome sequencing with Illumina paired-reads sequencing, according to published protocols [19, 20]. After processing, data were integrated into the Whole-genome Data Manager (version 2.0) [19, 20]. Single-nucleotide polymorphisms (SNPs) were investigated using Phen2gen software [20].
Outcomes
The primary efficacy outcome was day-28 adequate clinical and parasitological response (ACPR) adjusted for reinfection using PCR genotyping. Day-3 parasite positivity rate was the secondary efficacy outcome. Safety outcomes were the frequency of adverse events, serious and severe adverse events.
Statistical Analysis
Data were analyzed with Excel StatX and Graphpad Prism (version 8.3.0). ACPR was evaluated using Kaplan-Meier survival curves and associated 95% confidence intervals (95% CI) and compared using the log-rank test (Mantel-Cox). IC50 and AQSA values versus clinical outcome were compared using the Mann-Whitney test. Resistance thresholds for IC50 and AQSA parasite survival were determined with receiver operating characteristic (ROC) analysis. Kruskal-Wallis tests were used to identify significant differences in Pfcrt–Pfmdr1 haplotype AQSA parasite survival results. Significant P values were <.05.
RESULTS
Patients
Most patients were adult males (87.3%, 55/63) (Table 1). There were no withdrawals or patients lost to follow-up; all 63 patients were included in the analysis. Thirty-one patients were from Mondulkiri, 29 from Pursat, and 3 from Siem Reap.
Table 1.
Baseline Characteristics of the Safety and Efficacy Population
| Characteristic | Study Population (N = 63) |
|---|---|
| Males/females, n | 61/2 |
| Adults aged > 15 years, n | 56 |
| Children aged 5–15 years, n | 7 |
| Mean age (SD) [range], years | 28.2 (11.9) [5–56] |
| Mean weight (SD) [range], kg | 53.7 (10.5) [23–78] |
| Geometric mean parasitemia (range) µL−1 blood | 22 695 (1756–248 000) |
Abbreviation: SD, standard deviation.
Therapeutic Efficacy and Molecular Surveillance
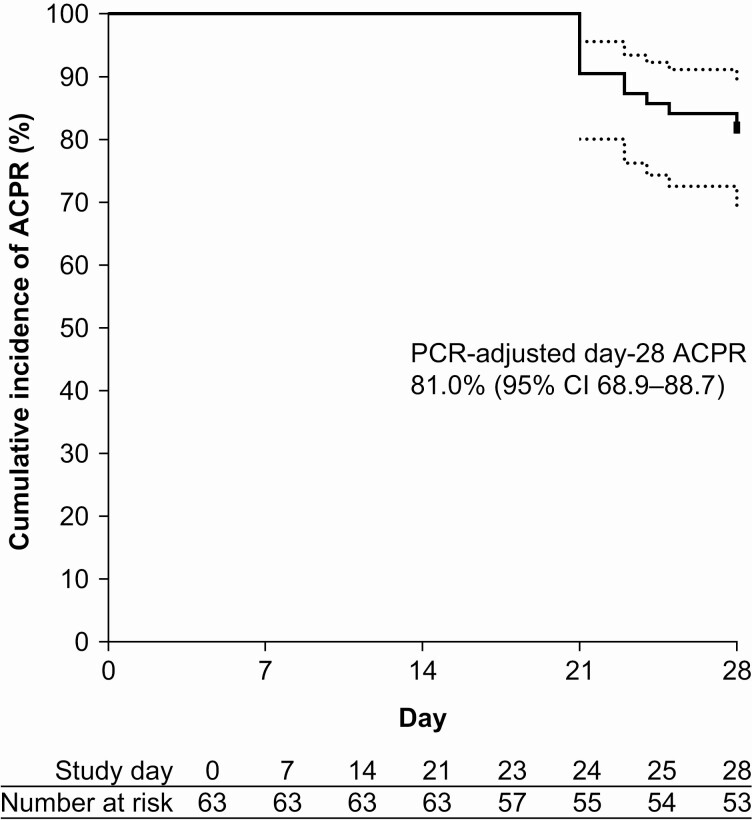
Day-28 ACPR was 81.0% (51/63). All recurrences were late clinical failures (days 21–28) and PCR-confirmed as recrudescence—9 in adults (18−52 years) and 3 in children (8−15 years). The Kaplan-Meier day-28 ACPR estimate was 81.0% (95% CI, 68.9–88.7) (Figure 1); 77.4% (95% CI, 58.4–88.5) for Mondulkiri, 86.2% (95% CI, 67.3–94.6) for Pursat, and 66.7% (95% CI, 5.4–94.5) for Siem Reap. No severe or serious adverse events were reported during the study.
Figure 1.
Kaplan-Meier estimates of ACPR with artesunate-amodiaquine for uncomplicated malaria. There were no reinfections on PCR-adjustment and no patients were censored. Abbreviations: ACPR, adequate clinical and parasitological response; CI, confidence interval; PCR, polymerase chain reaction.
Day-3 parasite positivity rate was 44.4% (28/63; 95% CI, 31.9–57.5); 22.6% (7/31; 95% CI, 9.6–41.1) for Mondulkiri, 69.0% (20/29; 95% CI, 49.2–84.7) for Pursat, and 33.3 (1/3, 95% CI, .8–90.6) for Siem Reap. All 63 isolates had the K13(C580Y) marker for artemisinin resistance. None had increased Pfmdr1 copy number, but 85.7% (54/63) had Pfpm2 amplification.
In Vitro Amodiaquine Resistance
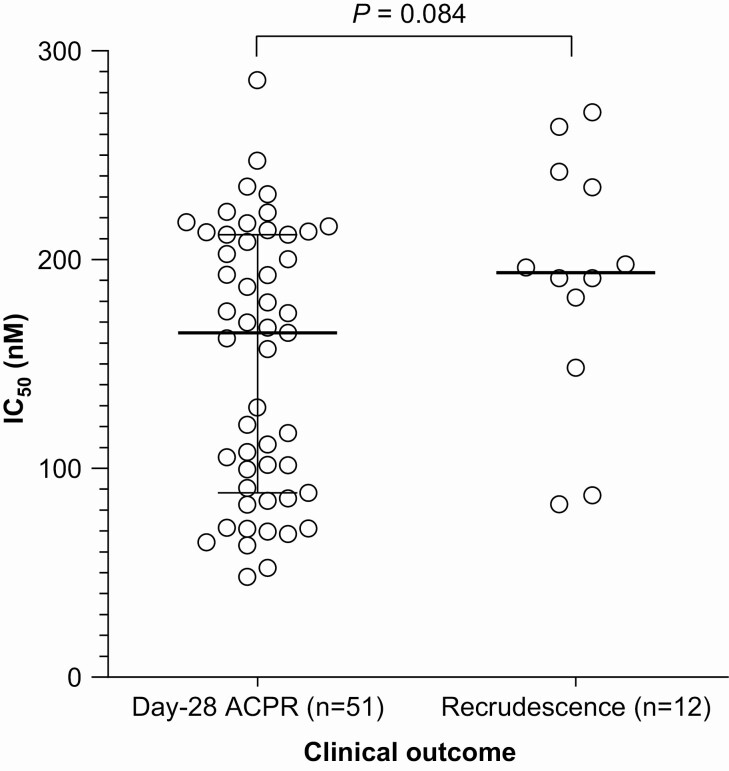
In the [3H]-hypoxanthine assay, the mono-desethyl-amodiaquine median IC50 for the 63 clinical isolates was 174.5 nM (interquartile range [IQR], 90.7–213.1). Isolates from patients with recrudescence (n = 12) had a median IC50 of 193.8 nM (IQR, 156.6–240.3) versus 165.0 nM (IQR, 88.3–212.0) for patients with day-28 ACPR (n = 51) (P = .084, Mann-Whitney) (Figure 2A). ROC analysis indicated an IC50 threshold value most strongly correlated with day-28 ACPR of <181 nM (Supplementary material Figure S2); sensitivity was 61% (95% CI, 47–73) at a specificity of 75% (95% CI, 47–91). Area under the curve (AUC) was 0.66 (95% CI, .49–.83; P = .083). Thus, IC50 had inadequate discriminatory value for predicting clinical outcome.
Figure 2.
Relationship between mono-desethyl-amodiaquine IC50 determined in the [3H]-hypoxanthine uptake inhibition assay and clinical outcome at day 28 following treatment with artesunate-amodiaquine for 63 P. falciparum clinical isolates. Open circles represent P. falciparum isolates and black horizontal bars and Ι bars indicate the median and interquartile range, statistical comparison used the Mann-Whitney test. Abbreviations: ACPR, adequate clinical and parasitological response; IC50, half-maximal inhibitory concentration.
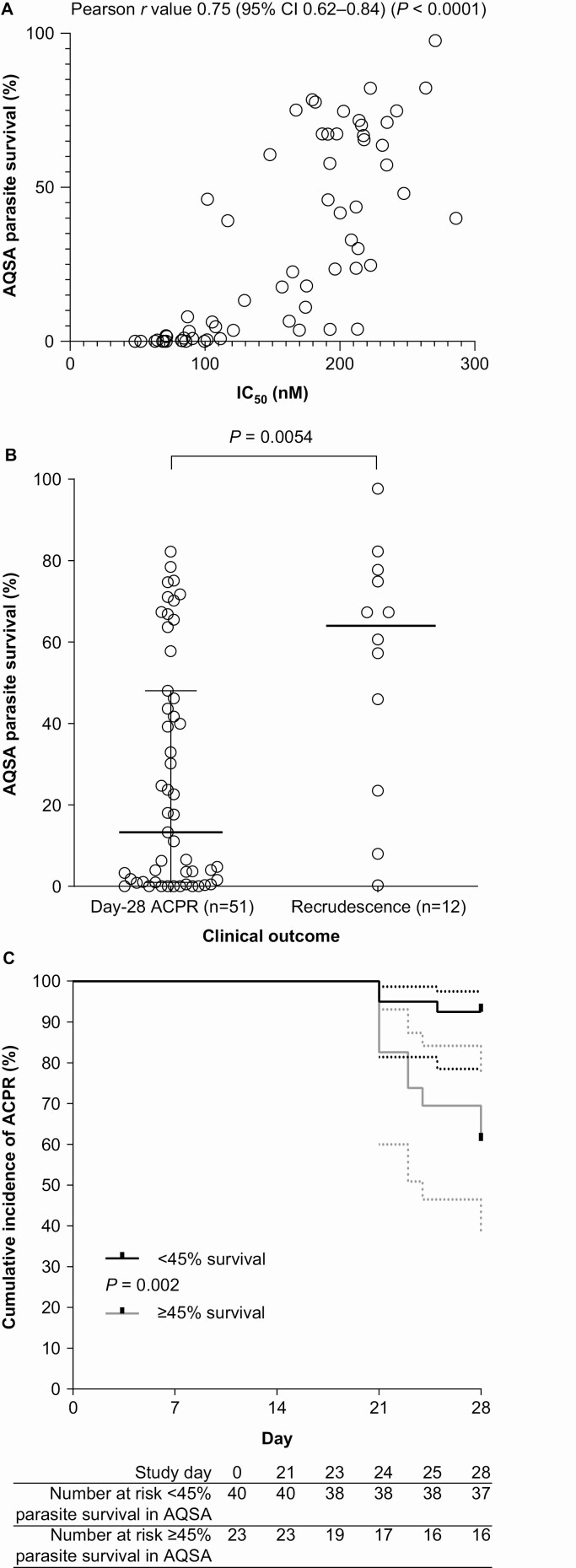
Amodiaquine Survival Assay (AQSA)
In the 63 clinical isolates, AQSA median parasite survival was 23.5% (IQR, 1.6–65.5). Quality control values were 72.8% parasite survival for the amodiaquine-resistant 7G8 strain and 0% for the susceptible 3D7 strain. There was a strong positive correlation between IC50 and AQSA parasite survival (Pearson r value 0.75 [95% CI, .62–.84] P < .0001) (Figure 3A). Recrudescent isolates had a significantly higher median survival (64.0% [IQR, 29.1–77.0]) versus those from patients with day-28 ACPR (13.3% [IQR, 0.9–48.1]) (P = .0054, Mann-Whitney) (Figure 3B). ROC analysis indicated an AQSA threshold predictive of day-28 ACPR of <45% survival, with 75% sensitivity (95% CI, 47–91) and 73% specificity (95% CI, 59–83). AUC was 0.75 (95% CI, .60–.91; P = .0063) (Supplementary material Figure S3). ACPR occurred with 92.5% (37/40) of isolates with <45% AQSA parasite survival but decreased significantly to 60.9% (14/23) for those with ≥45% survival (P = .0020, log-rank Mantel-Cox) (Figure 3C). Therefore, ≥45% parasite survival in the AQSA was a clinically relevant resistance phenotype.
Figure 3.
Parasite survival in the amodiaquine survival assay (AQSA). A, Correlation between AQSA parasite survival and IC50 determined in the [3H]-hypoxanthine uptake inhibition assay. B, Relationship between AQSA parasite survival and clinical outcome at day 28 following treatment with artesunate-amodiaquine. Open circles represent P. falciparum isolates and black horizontal bars and Ι bars indicate the median and interquartile range, statistical comparison used the Mann-Whitney U test. C, Kaplan-Meier estimates of ACPR for parasites with the AQSA resistance phenotype (≥45% survival) versus those with the susceptible phenotype (<45% survival), statistical comparison used the log-rank test (Mantel-Cox). Abbreviations: ACPR, adequate clinical and parasitological response; IC50, half-maximal inhibitory concentration.
Molecular Signature Associated With Amodiaquine Resistance
In the expanded data set of 97 clinical isolates, median AQSA parasite survival was 8.0% (IQR, 1.0−50.2), and 28.9% (28/97) had ≥45% parasite survival. Ninety-six isolates were K13(C580Y) and 1 was K13(Y493H). One isolate had Pfmdr1 amplification, whereas 78.4% (76/97) had Pfpm2 amplification. There was no significant difference in the median AQSA value of strains with single-copy Pfpm2 (6.3% [IQR, 1.1–56.4]) versus those with multiple copies (12.3% [IQR, 0.91–47.6]) (P = .74, Mann-Whitney).
The frequency of SNPs for the key P. falciparum resistance genes Pfcrt and Pfmdr1 are shown in Supplementary material Figure S4. For isolates with complete Pfcrt haplotypes (N = 94), 12 different haplotypes were identified, of which 97.9% (92/94) were either Dd2 or on the Dd2 background (Table 2). For Pfmdr1, 6 different haplotypes were found, with 51.1% (48/94) having Y148F plus at least 1 other mutation (Table 2).
Table 2.
Pfcrt or Pfmdr1 Haplotype and Parasite Survival in the Amodiaquine Survival Assay (AQSA)
| Haplotype | n (%) | Median Parasite Survival, % (IQR) |
|---|---|---|
| Pfcrt | N = 94 | |
| Dd2a | 13 (13.8) | 48.1 (1.5–77.2) |
| Dd2 F145I | 7 (7.4) | 0.0 (0–1.8) |
| Dd2 G353V | 13 (13.8) | 0.4 (0–1.0) |
| Dd2 G367C | 1 (1.1) | 0.4 |
| Dd2 H97Y | 14 (14.9) | 15.9 (4.5–46.8) |
| Dd2 I210F | 13 (13.8) | 8.0 (3.8–31.5) |
| Dd2 I218F | 1 (1.1) | 0.0 |
| Dd2 M343I | 2 (2.1) | 1.6 (0.2–3.0) |
| Dd2 N88K | 1 (1.1) | 0.7 |
| Dd2 T93S | 27 (28.7) | 57.8 (22.6–71.7) |
| Cam734a | 1 (1.1) | 11.1 |
| GB4a | 1 (1.1) | 0.0 |
| Pfmdr1 | N = 94 | |
| Wild type | 3 (3.2) | 4.8 (1.4–11.1) |
| Y184F | 43 (45.7) | 1.3 (0–6.3) |
| P72S/Y184F | 3 (3.2) | 24.7 (0.6–32.9) |
| Y184F/F1068L | 19 (20.2) | 64.2 (46.0–74.7) |
| Y184F/G1314D | 18 (19.1) | 13.7 (3.6–42.8) |
| Y184F/S784L/R945P | 8 (8.5) | 67.8 (48.1–81.6) |
Abbreviations: IQR, interquartile range; Pfcrt, P. falciparum chloroquine resistance transporter; Pfmdr1, P. falciparum multidrug resistance 1.
aMutations for Pfcrt D2d: M74I/N75E/K76T/A220S/Q271E/N326S/I356T/R371I; Pfcrt Cam734: M74I/N75D/K76T/A220S/Q271E/T333S; Pfcrt GB4: 74I/75E/76T/A220S/Q271E/R371I.
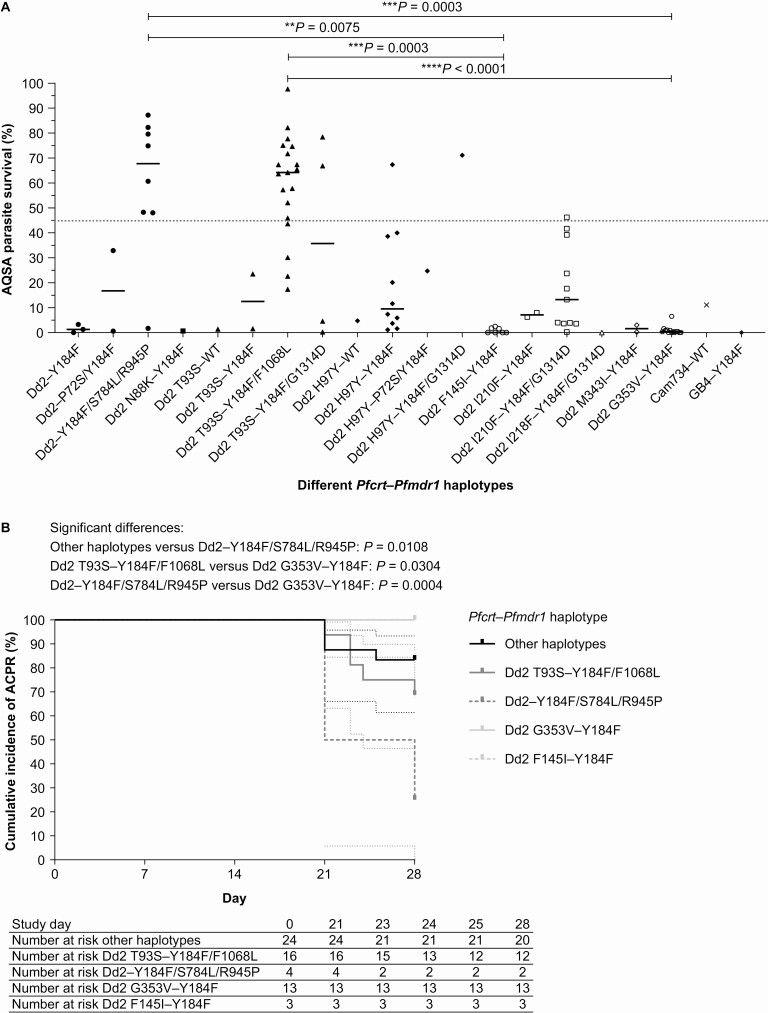
AQSA results for isolates with complete Pfcrt–Pfmdr1 sequences (N = 92) indicated significant differences in survival between 4 haplotypes: Dd2–Y184F/S784L/R945P versus Dd2 F145I–Y184F and versus Dd2 G353V–Y184F (P = .0075 and P = .0003, respectively; Kruskal-Wallis); and between Dd2 T93S–Y184F/F1068L versus Dd2 F145I–Y184F and versus Dd2 G353V–Y184F (P = .0003 and P < .0001, respectively; Kruskal-Wallis) (Figure 4A). When examining clinical outcomes, although the data set was smaller (N = 60), there was still a significant difference between Dd2 G353V–Y184F versus Dd2–Y184F/S784L/R945P or Dd2 T93S–Y184F/F1068L (P = .0004 and P = .0304, respectively; log-rank Mantel-Cox) (Figure 4B).
Figure 4.
Effect of P. falciparum Pfcrt–Pfmdr1 haplotype on resistance phenotype. A, Parasite survival in the amodiaquine survival assay (AQSA) for Cambodian clinical isolates (N = 92), the horizontal gray dotted line indicates the AQSA resistance phenotype (≥45% parasite survival). Symbols represent P. falciparum isolates, and black horizontal bars and Ι bars indicate the median and interquartile range; statistical comparison used the Kruskal-Wallis test. B, Kaplan-Meier plots for day-28 ACPR in 60 patients with P. falciparum malaria treated with artesunate-amodiaquine for amodiaquine-resistant and -susceptible Pfcrt–Pfmdr1 haplotypes. Abbreviation: ACPR, adequate clinical and parasitological response.
DISCUSSION
Antimalarial drug resistance curtails ACT efficacy for uncomplicated malaria in Cambodia, with the emergence of triple mutants (artemisinin, piperaquine, and mefloquine resistant) underlining the need for new therapeutic options [22–24]. High artesunate-amodiaquine efficacy in Africa and Viet Nam, and the absence of known amodiaquine resistance markers in Cambodia, suggested that this ACT would be efficacious. Surprisingly, 19.0% of patients had recrudescence, sufficient to exclude artesunate-amodiaquine as an uncomplicated malaria treatment in Cambodia.
The K13(C580Y) artemisinin resistance marker was ubiquitous in this study and is the predominant K13 mutant in Cambodia [25]. Consistent with this, the day-3 parasite positivity rate was 44.4%. However, artemisinin resistance increases recrudescence probability only if there is also partner drug resistance [4, 5]. Thus, the high treatment failure rate is most likely explained by amodiaquine resistance.
Previous studies in Africa indicated a mono-desethyl-amodiaquine IC50 resistance threshold of >60 nM with amodiaquine monotherapy [26], compared with >180 nM for artesunate-amodiaquine in the current study. This could be because artemisinin resistance is partial, and a higher degree of amodiaquine resistance is necessary to support parasite recrudescence following artesunate-amodiaquine versus amodiaquine monotherapy.
In the current study, IC50 lacked sufficient discriminatory power to differentiate between ACPR and recrudescence. This lack of correlation between IC50 and clinical outcome was observed previously for artemisinin and piperaquine, promoting the development of parasite survival assays measuring cytocidal activity [4, 21, 27]. This study validated the AQSA, a novel amodiaquine parasite survival assay, which correlated well with clinical outcome, and was sufficiently sensitive and specific to use as a resistance phenotype to investigate potential amodiaquine resistance molecular markers. Consequently, we propose AQSA ≥45% parasite survival as a novel definition for amodiaquine resistance.
In Africa, Pfmdr1 N86 and D1246 selection following artemether-lumefantrine improves clinical outcomes with artesunate-amodiaquine in P. falciparum malaria [28]. This suggests that an artemether-lumefantrine-amodiaquine triple therapy could counterselect for resistance, with ongoing initiatives to develop the combination [29]. However, the rationale for artemether-lumefantrine-amodiaquine in Cambodia has not been demonstrated. Our data show that amodiaquine resistance in Cambodia exists in the absence of the African amodiaquine-resistant haplotype Pfmdr1 86Y/Y184/1246Y [10], as all mutant Pfmdr1 haplotypes were N86/184F/D1246. Also, mutations common in South American P. falciparum Pfmdr1 (S1034C, N1042D and D1246Y) were absent. Molecular markers for lumefantrine resistance are not validated for Southeast Asia, and our data do not support counter selection for amodiaquine susceptibility. Rather, resistance to both amodiaquine and lumefantrine appears possible in Cambodian P. falciparum.
In the expanded parasite data set, all except 2 Pfcrt haplotypes were on the Dd2 background (ie, C72/V73/74I/75E/76T/220S/271E/326S/356T/371I). The amodiaquine-resistant 72–76 SVMNT haplotype reported in South American parasites was absent, consistent with previous data [10].
In Asia, novel Pfcrt mutations have emerged on the Dd2 chloroquine-resistant allelic background, in contrast to Africa where 3D7, GB4, and Cam783 haplotypes predominate [30]. In clinical isolates and gene edited parasites, Pfcrt Dd2 decreases in vitro susceptibility to amodiaquine relative to Pfcrt 3D7 and thus can be considered an amodiaquine-tolerant background [30, 31]. In the current study, 85.9% (79/92) of the Dd2-based haplotypes had additional mutations. The Pfcrt mutations G353V and F145I were significantly associated with amodiaquine sensitivity in the AQSA, and G353V was associated with ACPR. These findings are consistent with data in gene-edited parasites showing that these mutations confer resistance to piperaquine but sensitize parasites to amodiaquine, chloroquine, and quinine [32, 33]. Notably, in gene-edited parasites neither G353V nor F145I had any impact on lumefantrine susceptibility, and counterselection between lumefantrine and amodiaquine appears unlikely [32]. In previous studies, Pfcrt T93S has been associated with piperaquine resistance, and its prevalence has been increasing in Cambodia [33–35]. In the current study, Pfcrt T93S was the most common Pfcrt haplotype and was associated with both amodiaquine-susceptible and amodiaquine-resistant haplotypes. Thus, elevated AQSA values were not associated with an identified polymorphism in Pfcrt. Rather, key mutations in this gene appear to be associated with sensitization to amodiaquine.
Pfmdr1 showed 6 haplotypes in this study, and 51.1% (48/94) were Y184F plus another mutation. Of these, S784L/R945P and F1068L were associated with clinical and in vitro resistance. Pfmdr1 S784L has previously been noted from several locations in Cambodia at frequencies between 0.5 and 29.8% but was not associated with R945P [10]. Pfmdr1 F1068L was reported from Pailin at a low frequency (4.2%) [10], versus 20.2% (19/94) in the current study. This study is the first report to our knowledge associating these Pfmdr1 haplotypes with amodiaquine resistance. However, our data set is insufficient to show causality or to perform multivariate analysis. Thus, extended genome-wide association studies and genome editing are required to validate our findings. The single amodiaquine-susceptible Pfmdr1 S784L/R945P mutant in the AQSA was the only isolate with Pfmdr1 amplification. Conclusions cannot be drawn from one isolate, but additional investigations may be valuable.
The origin of amodiaquine resistance in Cambodia is unclear. This drug was not used recently and saw only limited implementation in the 1990s. Piperaquine resistance selection is unlikely to be associated with amodiaquine resistance emergence, as the associated genotype, that is, Pfpm2 amplification and Pfcrt mutations have either no effect or sensitize parasites to amodiaquine [32]. We could thus hypothesize that amodiaquine resistance emerged in Cambodia in the past consecutively to extensive chloroquine use and since then has been perpetuated by an unidentified mechanism.
While confirming artesunate-amodiaquine resistance in Cambodia, this was a small study conducted in a limited geographical region. Although significant relationships between SNPs in key resistance genes and amodiaquine resistance phenotypes were observed, causality cannot be determined. For example, the associations could result from the close relatedness of parasites in this study. Further investigations are required to confirm the putative amodiaquine resistance markers and assess their relevance to other malaria endemic areas. In the absence of molecular markers, the AQSA provides a novel methodology to assess clinically relevant amodiaquine resistance. However, AQSA specificity and sensitivity were determined according to the limited study size and location, and the ≥ 45% parasite survival AQSA resistance phenotype may require revision with additional data.
This study highlights the need for careful assessment of therapeutic outcomes and molecular markers before introducing a new antimalarial treatment in Cambodia. Resistance to amodiaquine was unexpected and was not associated with any known resistance genotype from other malaria endemic areas. Our findings indicate that clinical resistance was linked to the acquisition of high-level resistance against an amodiaquine-tolerant background. Thus, any amodiaquine-based combination would place partner compounds under a high selective pressure and be inappropriate in Cambodia.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. R. L., P. R., F. A., and B. W. contributed to the concept and design of the study. M. M.-K., C. M., N. K., S. K., S. Ke, C. K., N. Kl, R. E., S. C., and B. I. were involved in data acquisition and M. M.-K., R. L., C. M., N. K., D. M. B., M. D. B., P. R., F. A., and B. W. in data analysis and interpretation. All authors critically reviewed the manuscript, approved the final version, and take full responsibility for the publication.
Acknowledgments. Naomi Richardson of Magenta Communications Ltd developed a first draft of this article from the statistical output, collated author contributions, provided graphic services, and was funded by the World Health Organization.
Disclaimer. The funding source was not involved in the design and conduct of the study, the interpretation of the results, and the development of this publication. The funders of this study had no role in study design, data collection, data analysis, data interpretation, writing of the report, and the decision to submit. The corresponding author had full access to all data in the study and final responsibility for the decision to submit for publication.
Financial support. This work was supported by the Bill & Melinda Gates Foundation and United State Agency for International development-President’s Malaria Initiative through the World Health Organization. M. M.-K. and B. W. were supported by 5% initiative (MIVS-ACT, grant number 15SANIN211).
Potential conflicts of interest. M. D. B., D. M. B., and P. R. are staff members of the World Health Organization. The authors alone are responsible for the views expressed in this publication and they do not necessarily represent the decisions, policy, or views of the World Health Organization. All other authors have no conflicts of interest to report. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. World Malaria Report. 2018. Available at: https://www.who.int/malaria/publications/world-malaria-report-2018/en/. Accessed 18 September 2019.
- 2. Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM; Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium . Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 2008; 359:2619–20. [DOI] [PubMed] [Google Scholar]
- 3. Miotto O, Almagro-Garcia J, Manske M, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 2013; 45:648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Witkowski B, Amaratunga C, Khim N, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 2013; 13:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Witkowski B, Khim N, Chim P, et al. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother 2013; 57:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duru V, Witkowski B, Ménard D. Plasmodium falciparum resistance to artemisinin derivatives and piperaquine: a major challenge for malaria elimination in Cambodia. Am J Trop Med Hyg 2016; 95:1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rogers WO, Sem R, Tero T, et al. Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar J 2009; 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denis MB, Tsuyuoka R, Lim P, et al. Efficacy of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in northwest Cambodia. Trop Med Int Health 2006; 11:1800–7. [DOI] [PubMed] [Google Scholar]
- 9. Humphreys GS, Merinopoulos I, Ahmed J, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 2007; 51:991–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Srimuang K, Miotto O, Lim P, et al. ; Tracking Resistance to Artemisinin Collaboration . Analysis of anti-malarial resistance markers in pfmdr1 and pfcrt across Southeast Asia in the tracking resistance to artemisinin collaboration. Malar J 2016; 15:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oladipo OO, Wellington OA, Sutherland CJ. Persistence of chloroquine-resistant haplotypes of Plasmodium falciparum in children with uncomplicated Malaria in Lagos, Nigeria, four years after change of chloroquine as first-line antimalarial medicine. Diagn Pathol 2015; 10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veiga MI, Dhingra SK, Henrich PP, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 2016; 7:11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 2002; 298:210–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beshir K, Sutherland CJ, Merinopoulos I, et al. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother 2010; 54:3714–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thanh NX, Trung TN, Phong NC, et al. Open label randomized comparison of dihydroartemisinin-piperaquine and artesunate-amodiaquine for the treatment of uncomplicated Plasmodium falciparum malaria in central Vietnam. Trop Med Int Health 2009; 14:504–11. [DOI] [PubMed] [Google Scholar]
- 16. Thanh NX, Trung TN, Phong NC, et al. The efficacy and tolerability of artemisinin-piperaquine (Artequick®) versus artesunate-amodiaquine (Coarsucam™) for the treatment of uncomplicated Plasmodium falciparum malaria in south-central Vietnam. Malar J 2012; 11:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. World Health Organization. Methods for surveillance of antimalarial drug efficacy. 2009. Available at: https://apps.who.int/iris/bitstream/handle/10665/44048/9789241597531_eng.pdf;jsessionid=BF00D558415E9CB80B884CCBD6269BF7?sequence=1. Accessed 27 February 2019.
- 18. World Health Organization. Methods and techniques for clinical trials on antimalarial drug efficacy: genotyping to identify parasite populations. 2008. Available at: http://www.who.int/malaria/publications/atoz/9789241596305/en/. Accessed 26 February 2019.
- 19. Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014; 505:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Witkowski B, Duru V, Khim N, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 2017; 17:174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duru V, Khim N, Leang R, et al. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med 2015; 13:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rossi G, De Smet M, Khim N, Kindermans JM, Menard D. Emergence of Plasmodium falciparum triple mutant in Cambodia. Lancet Infect Dis 2017; 17:1233. [DOI] [PubMed] [Google Scholar]
- 23. Leang R, Khim N, Chea H, et al. Pyronaridine-artesunate plus single-dose primaquine efficacy and safety for the treatment of malaria in western Cambodia. Antimicrob Agents Chemother 2019; 63:e01273–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bui Quang P, Huynh Hong Q, Tran Thanh D, et al. Pyronaridine-artesunate efficacy and safety in uncomplicated Plasmodium falciparum malaria in areas of artemisinin-resistant falciparum in Viet Nam (2017–2018). Clin Infect Dis 2019; 70:2187–95. [DOI] [PubMed] [Google Scholar]
- 25. Kheang ST, Sovannaroth S, Ek S, et al. Prevalence of K13 mutation and Day-3 positive parasitaemia in artemisinin-resistant malaria endemic area of Cambodia: a cross-sectional study. Malar J 2017; 16:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basco LK, Ringwald P. Molecular epidemiology of malaria in Cameroon. XXIV. Trends of in vitro antimalarial drug responses in Yaounde, Cameroon. Am J Trop Med Hyg 2007; 76:20–6. [PubMed] [Google Scholar]
- 27. Paguio MF, Bogle KL, Roepe PD. Plasmodium falciparum resistance to cytocidal versus cytostatic effects of chloroquine. Mol Biochem Parasitol 2011; 178:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okell LC, Reiter LM, Ebbe LS, et al. Emerging implications of policies on malaria treatment: genetic changes in the Pfmdr-1 gene affecting susceptibility to artemether-lumefantrine and artesunate-amodiaquine in Africa. BMJ Glob Health 2018; 3:e000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. University of Oxford. A trial to compare the efficacy, safety and tolerability of combinations of 3 anti-malarial drugs against combinations of 2 anti-malarial drugs (Asia) (DeTACT-ASIA). 2019. Available at: https://clinicaltrials.gov/ct2/show/NCT03939104. Accessed 5 December 2019.
- 30. Dhingra SK, Gabryszewski SJ, Small-Saunders JL, et al. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. MBio 2019; 10:e02731–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venkatesan M, Gadalla NB, Stepniewska K, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 2014; 91:833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ross LS, Dhingra SK, Mok S, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 2018; 9:3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dhingra SK, Small-Saunders JL, Ménard D, Fidock DA. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect Dis 2019; 19:1168–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Pluijm RW, Imwong M, Chau NH, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis 2019; 19:952–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamilton WL, Amato R, van der Pluijm RW, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis 2019; 19:943–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.