Graphical abstract
Keywords: Metabolic models, Drug resistance, Acute Myeloid Leukemia (AML), Metabolic vulnerabilities, Drug targets
Abstract
The development of resistance to chemotherapeutic agents, such as Doxorubicin (DOX) and cytarabine (AraC), is one of the greatest challenges to the successful treatment of Acute Myeloid Leukemia (AML). Such acquisition is often underlined by a metabolic reprogramming that can provide a therapeutic opportunity, as it can lead to the emergence of vulnerabilities and dependencies to be exploited as targets against the resistant cells. In this regard, genome-scale metabolic models (GSMMs) have emerged as powerful tools to integrate multiple layers of data to build cancer-specific models and identify putative metabolic vulnerabilities. Here, we use genome-scale metabolic modelling to reconstruct a GSMM of the THP1 AML cell line and two derivative cell lines, one with acquired resistance to AraC and the second with acquired resistance to DOX. We also explore how, adding to the transcriptomic layer, the metabolomic layer enhances the selectivity of the resulting condition specific reconstructions. The resulting models enabled us to identify and experimentally validate that drug-resistant THP1 cells are sensitive to the FDA-approved antifolate methotrexate. Moreover, we discovered and validated that the resistant cell lines could be selectively targeted by inhibiting squalene synthase, providing a new and promising strategy to directly inhibit cholesterol synthesis in AML drug resistant cells.
1. Introduction
Although great advancement is being made in optimizing existing chemotherapeutics and developing new ones, drug resistance still poses the greatest threat against the successful treatment of cancer. Indeed, over 90% of cancer patients’ deaths can be attributed to drug resistance [1]. Specifically, in Acute Myeloid Leukemia (AML) 35% to 45% of de novo diagnosed patients are bound to develop resistance to the broadly administered induction therapy comprising of an anthracycline drug, e.g., Doxorubicin (DOX), in combination with cytarabine (AraC) [2]. Reportedly, the exact mechanisms of drug resistance are not only complex and elusive but also vary among different subtypes and age groups of AML patients [1], [3], [4].
Metabolism has not only been established as a hallmark of cancer but its involvement in the emergence and instatement of drug resistance in different types of cancer, including AML, has gained increasing interest [5], [6], [7]. In previous studies, we have demonstrated that by targeting glutaminase 1 (GLS1), a metabolic enzyme overexpressed by colon cancer cells in response to palbociclib treatment (an FDA-approved chemotherapeutic that inhibits CDK4/6), it is possible to sensitize cancer cells to CDK4/6 inhibition, thus improving the therapeutic efficacy [8]. Thus, the metabolic adaptations responsible for the therapeutic refractoriness of cancer cells represent, at the same time, vulnerabilities that can be exploited as therapeutic targets to successfully prevent resistance to targeted antineoplastic treatments.
In the last decade, genome-scale metabolic models (GSMMs) have emerged as powerful tools to identify targets against the metabolic reprogramming underlying cancer [9]. A GSMM is a mathematical representation of the entire known network of metabolic reactions of an organism and its functional association with the genome [10]. Human GSMMs, like Recon 2, serve as platforms where multiple layers of data can be integrated to reconstruct cancer-specific GSMMs, which provide a genome-scale representation of the metabolic network supporting cancer cells. Such models allow integrating transcriptomics to identify metabolic vulnerabilities underlying cancer metabolic reprogramming that can be putative drug targets [11], [12]. GSMMs can also integrate metabolomics alongside transcriptomics to be specific enough to identify metabolic vulnerabilities selective for a given subpopulation of cancer cells. Case in point, a GSMM analysis of two isogenic prostate cancer cell lines identified that the cancer cell lines with cancer stem cell like phenotype were vulnerable to the accumulation of long-chain fatty acids with antiproliferative effects and could be selectively killed by blocking fatty acid oxidation [13]. Likewise, in a same patient-derived cell line panel, GSMMs have also enabled the identification that the combined inhibition of cystine uptake and folate metabolism is highly selective against metastatic cells [14].
Here, we reconstructed cell-line specific GSMMs of the THP-1 AML cell line and two derivative cell lines with acquired resistance to AraC and DOX, respectively. The THP-1 cell line was selected for this study since as it is broadly used as a model for AML in drug screenings [15]. Moreover, the THP-1 cell model is part of the Genomic of Drug Resistance in Cancer Project [16], in the Cancer Cell Line Encyclopaedia [17] and in the Cancer Therapeutics Response Portal [18]. We explore how adding the metabolomic layer to the transcriptomic layer enhances the specificity of the resulting condition-specific models. With this layer, we identify and validate folate and cholesterol metabolism as metabolic vulnerabilities that can be used to target the resistant cell lines.
2. Materials and methods
2.1. Chemicals and reagents
Methotrexate, cytarabine and doxorubicin were obtained from Sigma-Aldrich (St. Louis, MO). RPMI-1640 media was obtained from Biowest (Labclinics). Antibiotic (10,000 U/ml penicillin, 10 mg/ml streptomycin), and PBS were obtained from Biological Industries (Kibbutz Beit Haemet, Israel), and fetal bovine serum from Invitrogen (Carlsbad, CA, USA).
2.2. Cell culture
The AML cell line THP-1 was obtained from DSMZ (Braunschweig, Germany). Cytarabine (AraC) and doxorubicin (DOX)-resistant sublines were established by continuous exposure to stepwise increasing drug concentrations as previously described [19] and derived from the Resistant Cancer Cell Line (RCCL) collection (www.kent.ac.uk/stms/cmp/RCCL/RCCLabout.html) [20]. There were no clonal isolation processes involved, thus each subline is expected to be a mixture of clones able to grow optimally under the drug exposure. To ensure AraC and DOX resistance during the entirety of the study, IC50 concentrations against AraC and DOX were determined in the resistant and paired controls each 2–3 weeks. All the cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 4 mM glutamine and 1% penicillin and streptomycin at 37 °C in a humidified incubator with 5% CO2. Particularly, AraC and DOX resistant cells were maintained with 8 µM of AraC or 20 pM of DOX, respectively.
2.3. Cell characterization and cell viability assay
Cell size was determined using a Scepter TM Handheld Automated Cell Counter (Merck Millipore, Billerica, MA, USA) at different time periods. In addition, protein content differences between the THP-1 cell lines (parental, AraC-resistant and DOX-resistant) were further analyzed by i) collecting in Eppendorf tubes different number of cells (from 250,000 cells/ml to 2x106 cells/ml), ii) counting cell number in each Eppendorf, and iii) lysing cells and measuring the total protein content in all the conditions by Bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Waltham, MA USA).
Cell viability was measured using Cell Titer-Glo® Luminescent Cell Viability Assay. For the cell viability testing after drug incubation, 16,000 cells/well were seeded in 96-well plates using 100 µl suspension cell volume. Fresh media (100 µl) containing the desired concentration of drug, the combination of drugs under study, or vehicle was added, and cells were incubated 72 h. For Cell Titer-Glo® Luminescent Cell Viability Assay, 96-well opaque-walled plates were used and measurements were made according to manufacturer’s instructions. Briefly, plates were removed from the incubator, allowed to equilibrate at room temperature 30 min and 100 µl of Cell Titer-Glo reagent was added directly to the wells. Content was mixed for 2 min on an orbital shaker and plater were allowed to incubate at room temperature for 10 min to stabilize the luminescent signal. Luminescence was determined using a Mithras LB 940 reader (Berthold Technologies, DLReady, Germany), which allows the integration of the signals detected in the short-wavelength filter at 485 nm and the long-wavelength filter at 530 nm. Cell viability was assessed and represented as a percentage of viability relative to untreated control cells. IC50 values were calculated using GraphPad Prism 6 software (La Jolla, CA, USA).
2.4. Measurement of extracellular metabolites
Glucose, lactate, glutamate and glutamine concentrations of THP-1 parental and AraC and DOX resistant cells were determined by spectrophotometry (COBAS Mira Plus, Horiba ABX) from cell culture media by monitoring the production of NAD(P)H in specific reactions for each metabolite at 340 nm wavelength. Glucose concentration was measured using hexokinase (HK) and glucose-6-phosphate dehydrogenase (G6PD) coupled enzymatic reactions (ABX Pentra Glucose HK CP, HORIBA ABX, Montpellier, France). Lactate concentration was determined by lactate dehydrogenase (LDH) reaction at 37 °C by mixing the media samples with 1.55 mg/ml NAD+ and 87.7U/ml LDH (Roche) in 0.2 M hydrazine 12 mM EDTA buffer (pH 9). Glutamate concentration was assessed by its conversion to α-ketoglutarate through glutamate dehydrogenase (GLDH) reaction in the presence of ADP. This reaction was performed at 37 °C by adding media sample to a cuvette containing 2.41 mM ADP, 3.9 mM NAD+ and 39U/ml of GLDH in 0.5 M glycine/0.6 M hydrazine buffer, pH 9. Glutamine was determined by its conversion first to glutamate through glutaminase (GLS) reaction and subsequently quantification of glutamate concentration as described above. GLS reaction was performed by adding media sample to a cuvette containing a mixture consisting of 90 mU/ml GLS in 111 mM acetate buffer, pH 5. The reaction was carried out for 30 min at 37 °C in agitation.
To measure amino acids and biogenic amines we used the Biocrates AbsoluteIDQ p180 kit (UM-P180). In detail, internal standards were applied to the filter inserts of the 96-well kit plate. Standards, quality controls and media samples (10 μl of each) and pellet samples (50 μl each) were added onto the filter inserts and dried for 30 min under a nitrogen stream. Amino acids and biogenic amines were derivatized for 20 min with an excess of 5% phenylisothiocyanate in ethanol/water/pyridine (ratio 1/1/1, v/v/v), and subsequently dried for 60 min under a nitrogen stream. Metabolites and internal standards were then extracted with 300 µl methanol containing 5 mM ammonium acetate by shaking for 30 min, and eluted by centrifugation for 2 min at room temperature and 500 × g. One-half of the eluate was diluted with water (50/50, v/v) was run using tandem mass spectrometry with an MS/MS Sciex Triple Quad 6500 (AB Sciex) coupled to liquid chromatograph UHPLC 1290 Infinity (Agilent).
2.4.1. Normalizing rates of uptake and secretion per cell
Consumption and production rates of glucose, lactate, glutamine and glutamate, as well as essential amino acids in the media, were calculated as follows:
| (1) |
| (2) |
where μ represents the growth rate, N0 and Nf the initial and final number of cells respectively given in [millions of cells] for a time tf given in [h], ΔΜ = Mf-M0 represents the increase or decrease in concentration for a metabolite for the time tf, an initial concentration M0, a final concentration Mf and it is given in [μmol]. Flux rates were normalized by total protein per cell p, given in [mg protein/ million cells] to account for the difference in the size of the cells. Finally, the normalized consumption or production rate of a metabolite kp is calculated in [μmol/ (mg protein∙h)].
2.5. Transcriptomic analysis
2.5.1. RNA extraction
Cell pellets of THP-1 parental and resistant cell lines were collected and frozen. Total RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA integrity was further tested using lab-on-a-chip technology on the BioAnalyzer 2100.
2.5.2. RNA-Seq library preparation, sequencing and generation of FastQ files
High-quality RNA-seq (transcriptome) was performed in the CNIC genomic unit using the Illumina HiSeq 2500 sequencer. There, 200 ng of total RNA were used to generate barcoded RNA-seq libraries using the NEBNext Ultra RNA Library preparation kit (New England Biolabs). Briefly, poly A + RNA was purified using poly-T oligo- attached magnetic beads followed by fragmentation and then first and second cDNA strand synthesis. Next, cDNA ends were repaired and adenylated. The NEBNext adaptor was then ligated, followed by uracil excision from the adaptor and PCR amplification. Finally, the size of the libraries was checked using the Agilent 2100 Bioanalyzer DNA 1000 chip and their concentration was determined using the Qubit® fluorometer (Life Technologies. Libraries were sequenced on a HiSeq2500 (Illumina) to generate 60 bases single reads. Finally, FastQ files for each sample were obtained using CASAVA v1.8 software (Illumina).
2.6. Bioinformatics analysis
Quality Control (QC) of the RNA-seq data was performed using FastQC, a computational Quality control tool for high throughput sequence data in Java developed by Simon Andrews and the Babraham Bioinformatics group (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The alignment of the reads was performed using STAR version 2.5.2a open-source software [21] and the FASTA sequences were generated using the Homo sapiens high coverage GRCh37.75.dna.primary_assembly. Gene counts were analyzed with the DESeq2 package and expressed as fragments per kilobase per million mapped reads (FPKM) [22].
2.7. Modelling
2.7.1. Building condition specific GSMMs
Genome Scale Metabolic Model (GSMM) reconstructions were built for the THP-1 Acute Myeloid Leukaemia (AML) cell-lines under different treatments. All computational analyses for the model reconstructions were performed in Python programming language, using the COBRApy-0.17.1 [23]. The CPLEX 12.6.2 solver was used for the computational simulations.
Recon 2, comprising 5063 metabolites, 7440 reactions and 1734 associated genes, was used as a base model for the reconstructions [24]. Recon 2 includes a biomass reaction that represents the metabolic demands of energy, reductive power and building blocks for growth and proliferation and can be used to simulate proliferation [25].
Transcriptomics and measured rates of metabolite uptake and secretions were integrated to the reconstructions using GIM3E [14]. GIM3E uses flux minimization weighted by transcriptomics Eq. (3) to identify the flux distribution most consistent with transcriptomics and metabolomics measures Eq. (4):
| (3) |
| (4) |
Subject to
where wi is the minimization weight for reaction i. gei is the gene expression value (FPKM) mapped to reaction i using the gene-protein reaction rules of GPR. geth is the gene expression value below which reactions will be penalized. This threshold was set to the 80th percentile for gene expression values of metabolic genes. GIM3Eopt is the optimal value of the GIM3E objective. vi is the flux through reaction i. Sirrev is the stoichiometric matrix of the metabolic network. Prior to applying GIM3E, the network was converted to irreversible by splitting each reversible reaction into a forward and reverse reaction. lb and ub are the lower and upper bound for each reaction. The flux through biomass reaction was also constrained to a lb of 90% of Biomassopt. To integrate the measured consumption or production rate of a metabolite, lb and ub were set to the 95% confidence interval for each measure.
Next, as implemented by Schmidt et al. [26], the GIM3E solution space was identified by maximizing and minimizing each flux within the space defined by 99% optimality of the GIM3E solution (). The obtained flux intervals were added as lower and upper bounds to Recon2, leading to condition specific GSMMs for each cell line.
Then, the OptGpSampler algorithm [27] was applied to obtain 20,000 flux distributions samples in each condition-specific GSMMs. The average of the sampled flux values in each GSMMs was selected as the reference (wild type) flux distribution for each condition.
2.7.2. Identifying putative targets through gene knockouts (KOs)
To identify putative drug targets, the effect of individual gene knockouts (KOs) on growth (i.e., flux through biomass reaction) was simulated in each condition-specific GSMMs [9]. This simulation was performed with the Minimization of metabolic adjustment (MOMA) algorithm [28], which identifies the flux distribution that minimizes the distance between the reference flux distribution for each cell line and the flux distribution after inactivating the reactions associated to the gene KO in each condition specific GSMMs. A gene was considered as essential for growth, and thus a putative target, if it reduced the flux through biomass reaction to less than 20% of the wild type.
2.7.3. Simulating the effect of increasing dosage of Methotrexate
Methotrexate is an anti-folate that inhibits Dihydrofolate Reductase (DHFR). To simulate the effect of increasing dosage of Methotrexate on each cell line, the reactions mapped to the DHFR gene were progressively inhibited (i.e. the upper bound was lowered) relative to the reference flux distribution and the effect of the inhibition on biomass production was evaluated with MOMA. For each condition, inhibitions from 90% to 100% relative to the reference flux distribution were evaluated.
2.8. Venn diagrams
Venn diagrams were generated using the BioVenn version 1.1.1 R package [29], providing the Entrez gene IDs.
3. Results
3.1. Genome scale metabolic reconstructions integrating transcriptomics
We first used the GIM3E algorithm [26] to integrate transcriptomics into the human GSMM Recon2 [24] to build specific models for each condition. The resulting metabolic models for the THP-1 parental untreated, AraC- and DOX-resistant cells incorporating only transcript expression levels were used to perform high throughput simulations of the effects of gene KOs on the capacity to grow and proliferate. These resulted in the identification of 30 putative metabolic vulnerabilities for the THP-1 parental untreated cells and 31 for the AraC-resistant cells, one of which was condition-specific and the remaining 30 were common between the AraC-resistant and the parental untreated models. Finally, 26 essential genes were identified for the DOX-resistant cells, all of which were common targets with the other cell models (Fig 1A).
Fig. 1.
Venn diagram showing the number of condition specific and common essential genes that were identified for the THP-1 parental, AraC- and DOX-resistant cell models only integrating transcriptomic data [A] or integrating both transcriptomics and metabolomics [B].
3.2. Genome-scale metabolic reconstructions integrating transcriptomics and metabolomics
Next, we enhanced the existing THP-1 cell model reconstructions, built using transcriptomic data, with the integration of metabolomics. Specifically, we integrated the consumption rates of the main sources of carbon, i.e., glucose and glutamine, the production rates of their respective products, lactate and glutamate, and the consumption rates of other amino acids as constraints to the models.
The single gene KO simulations performed using these Transcriptomics & Metabolomics models resulted in the identification of 30 putative metabolic vulnerabilities for the THP-1 parental untreated cells, 43 for the AraC-resistant cells and 40 for the DOX-resistant cells (Table 1). No specific metabolic vulnerabilities were found for the THP-1 DOX-resistant cell model (Fig. 1B). One condition-specific essential gene was identified for the parental untreated cells and three metabolic vulnerabilities were found to be common between the THP-1 parental untreated and the AraC-resistant cells.
Table 1.
The gene essentiality analysis performed in silico resulted in the identification of potential metabolic vulnerabilities for the AML THP-1 parental untreated, AraC-resistant and DOX-resistant cell models. The table presents the potential gene targets grouped according to their metabolic pathway. Genes were considered essential for growth, if their KO reduced the flux through biomass reaction to less than 20% of the wild type. The asterisk (*) denotes the essential genes that were identified only upon the integration of the targeted metabolomics dataset.
| Pathway | Targets | Parental | AraC | DOX |
|---|---|---|---|---|
| Pentose phosphate pathway | G6PD, PGD, PGLS | Essential | Essential | Non-essential |
| RPIA | Essential | Essential | Essential | |
| Cholesterol metabolism | CYP51A1, DHCR7, EBP, SQLS, LSS, MSMO1, NSDHL, SQLE, TM7SF2, HSD17B4, HMGCR, MVD, MVK, PMVK | Non-essential | Essential* | Essential* |
| LIPA | Essential | Essential | Essential | |
| Glycolysis | GPI | Essential | Essential | Essential |
| Folate metabolism | ATIC, DHFR, GART, TYMS | Essential | Essential | Essential |
| Sphingolipid metabolism | KDSR, SGMS1, SPTLC1, SPTLC2, SPTLC3 | Essential | Essential | Essential |
| Nucleotide metabolism | ADSL, CAD, CMPK1, DHODH, PAICS, PFAS, PPAT, TXNRD1, UMPS | Essential | Essential | Essential |
| PNP | Essential | Non-essential | Non-essential | |
| Glycerophospholipid metabolism | CRLS1, LCAT, PGS1, PTPMT1 | Essential | Essential | Essential |
| Alanine, aspartate and glutamate metabolism | ASNS | Essential | Essential | Essential |
In the case of the parental untreated cells, integrating the additional layer of metabolomic data onto the GSMMs that were reconstructed based on the transcriptomics, did not provide any new potential metabolic vulnerabilities. However, further constraining the drug-resistant cell models using targeted metabolomics increased the number of essential genes by 12 for the AraC-resistant and by 14 for the DOX-resistant THP-1 cells (Fig. 2).
Fig. 2.
The integration of targeted metabolomics resulted in the identification of additional and condition-specific putative metabolic vulnerabilities in the case of the drug-resistant THP-1 AML cells models, as opposed to genome-scale metabolic reconstructions built using only transcriptomic data.
Remarkably, the metabolic vulnerabilities shared across all three cell models were also conserved between the reconstructions of different integration levels. Specifically, our analysis revealed that 26 essential genes were shared across the THP-1 parental untreated and the drug-resistant cells and that these genes are involved in glycolysis, the pentose phosphate pathway, folate metabolism, sphingolipid and glycerophospholipid metabolism, nucleotide metabolism and aspartate and asparagine metabolism (Table 1).
3.3. Targeting the folate cycle shows promise against parental and drug resistant THP-1 cells
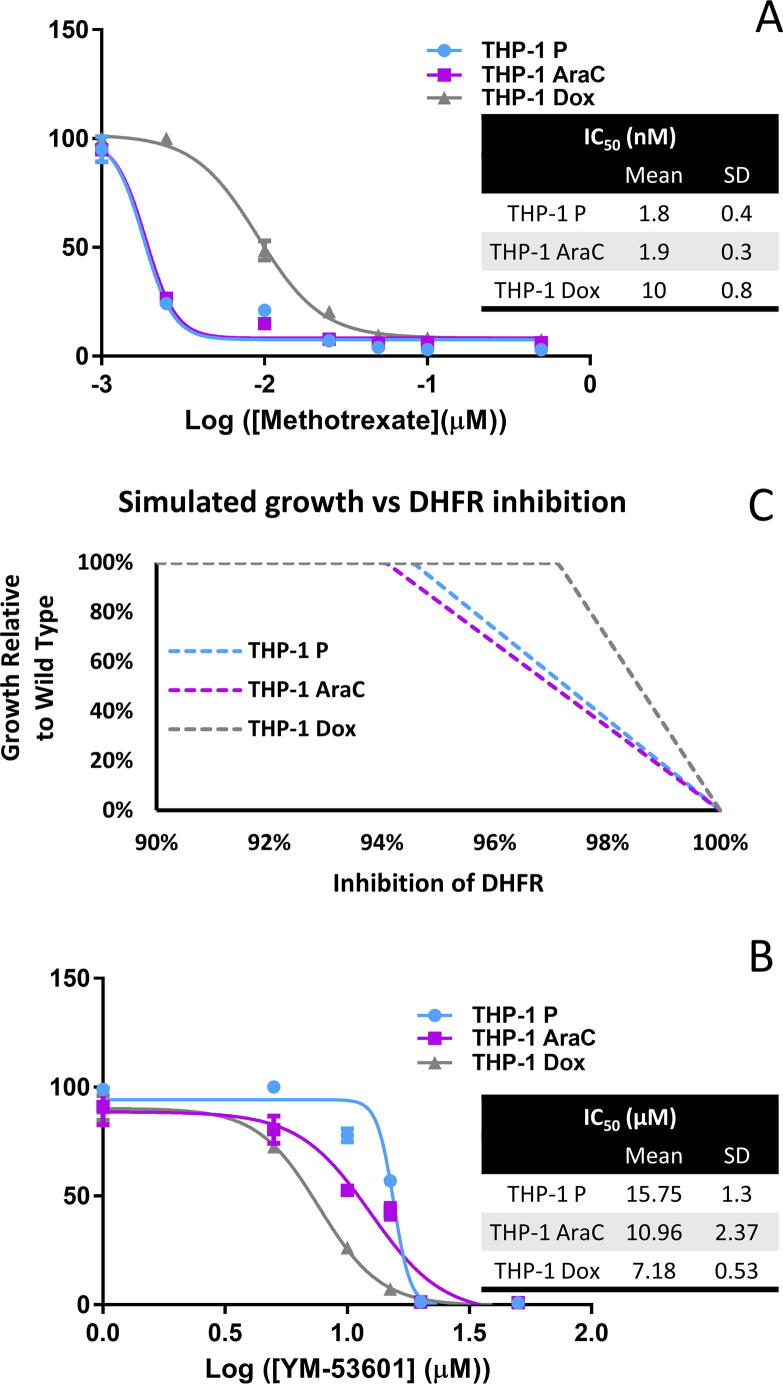
Enzymes involved in Folate metabolism were identified as common metabolic vulnerabilities of the parental untreated THP-1 cells and of both AraC- and DOX-resistant cells, namely thymidylate synthetase (TYMS) and dihydrofolate reductase (DHFR). Both enzymes have a crucial role in DNA biosynthesis [30]. TYMS catalyzes the methylation of 2′-deoxyuridine-5′-monophosphate (dUMP) and 5,10-methylenetetrahydrofolate (CH2THF) to 2′-deoxythymidine-5′-monophosphate (dTMP, thymidylate) and 7,8-dihydrofolate (7,8-DHF) [31]. Then, DHFR catalyses the first of the two reactions needed to transform the reduced 7,8-DHF back to CH2THF [31]. Methotrexate is a DHFR inhibitor that has been approved by the U.S. Food and Drug Administration (FDA) against a variety of pathologies [32], [33]. In our study, we determined that Methotrexate could completely inhibit growth of the parental and resistant cell models at concentrations well under 0.1 μmol. This suggests that Methotrexate could be used to target AraC and DOX resistant populations. Interestingly, the effects of DHFR inhibition were stronger on the THP-1 parental untreated and the AraC-resistant, compared to the DOX-resistant cells (Fig. 3A). This result is supported by the transcriptomic analysis which showed a decrease in DHFR transcript level of 40% in THP-1 DOX-resistant cells when compared to the parental. This could indicate that DOX-resistant cells have a lower dependency on folate cycle for survival than the other cell models. We found that our GSMMs could reproduce this behavior through partial inhibition of DHFR, where it was shown that DOX-resistant cells required a higher degree of inhibition of DHFR to impair cell proliferation (Fig. 3B). Additionally, it has been reported that the inhibition of over 95% DHFR activity is necessary to achieve the therapeutic effect of Methotrexate [34], which matches the growth inhibitory ranges that we identified in silico (Fig. 1, Fig. 3).
Fig. 3.
[A] Effect of anti-folate analogue Methotrexate on AML THP-1 parental untreated, AraC- and DOX-resistant cells. Cell viability was assessed after 72 h incubation with Methotrexate (nM). Values represent mean ± SD of n = 3. [B] Simulated growth at increasing inhibitions of DHFR, the primary target of Methotrexate. Growth is expressed as the flux through the biomass reaction relative to the wild type. Inhibition of DHFR is relative to the flux through reactions mapped to the DHFR genes in the wild type. [C] Effect of YM-53601 squalene synthase inhibitor on AML THP-1 parental untreated, AraC- and DOX-resistant cells. Cell viability was assessed after 72 h incubation with YM-53601 (μM). Values represent mean ± SD of n = 3.
3.4. Resistant cells are selectively vulnerable to inhibition of cholesterol biosynthesis
Solely upon the integration of targeted metabolomic data, the in silico single gene KO screening highlighted several enzymes that catalyze key reactions in the cholesterol biosynthesis pathway as essential for the viability of the drug-resistant THP-1 cells. Namely, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), mevalonate kinase (MVK), phosphomevalonate kinase (PMVK), mevalonate diphosphate decarboxylase (MVD), squalene synthase (SQLS), squalene epoxidase (SQLE) and lanosterol synthase (LSS) emerged as putative targets against the THP-1 Arac- and DOX-resistant cell models. Previous studies have already established that only the de novo biosynthesis of cholesterol branch of the mevalonate pathway has serious implications in AML and not the branch related with protein isoprenylation [35]. Therefore, we decided to evaluate the effects of direct cholesterol biosynthesis inhibition by using YM-53601, a SQLS specific inhibitor [36]. As predicted by our analysis, the proliferation of THP-1 AraC- and DOX-resistant cells was significantly more compromised compared to the THP-1 parental untreated cells (Fig. 3C). Thus, suggesting that targeting squalene synthase and directly inhibiting cholesterol biosynthesis could be a promising therapeutic strategy against chemoresistant AML populations.
4. Discussion
Most commonly, AML disease diagnosis and subsequent therapeutic planning rely on the identification of specific genomic traits [37] and despite the great efforts put towards designing more effective therapeutic strategies for AML, the fact still remains that drug resistance is the leading cause for treatment failure in this type of cancer. Genomics alone often fails to capture and comprehensively interpret the phenotypic variability that emerges from the complex interactions of the different layers of regulatory events in the cells. The integration of omics data under the framework of GSMMs provides the tools to incorporate a much broader wealth of biological information in the modelling process of different pathologies in a condition-specific manner and therefore reveal not only putative therapeutic drug targets, but also drug suitability or potential side effects [38], [39]. In most mammalian cell models, it has been shown that most transcript levels are correlated to the respective protein abundance [40]. However, in cancer where epigenetic events, such as methylation, contribute to the pathogenesis and the distinct disease subgroup phenotype, not all metabolic flux rates may positively correlate with transcriptomics. Case in point, in previous studies, evidence has been provided, using a breast cancer cell model, that only 50% of metabolic reactions might be regulated directly by transcriptomics [41]. Such cases can be adequately modelled through the integration of other layers such as the metabolome layer into GSMMs.
However, GSMM modelling still has some limitations and further development and improvements are needed. For instance, GSMMs are mainly unable to integrate information regarding enzyme kinetics, allosteric regulation and signal transduction events that might modulate enzyme activity. Data collection is also a limitation to integrate the metabolome layer in genome-scale metabolic modelling in the clinical setting as uptake and secretion rates of metabolites cannot be easily measured in vivo. They can, however, be measured ex vivo using patient-derived primary cell lines.
In this regard, our aim in this study was to evaluate the significance of including the metabolome layer in the reconstruction process of GSMMs. We selected THP-1 parental and AraC and DOX-resistant cells as a case study and we built condition-specific GSMMs to identify metabolic targets against AML cells resistant to the two commonly used chemotherapeutic drugs. Our results showed that adding metabolome constraints, additionally to the transcriptome, leads to the reconstruction of more specific metabolic models of THP-1 cells under AraC and DOX resistance and enhances the identification of selective targets against the different cell models.
Our study highlighted several metabolic vulnerabilities shared by the parental and resistant cells that have gained scientific interest in AML and are being tested as potential drug targets in clinical trials. For instance, we identified some members of the de novo pyrimidine biosynthesis pathway as putative targets for all cell line models (i.e., THP-1 parental untreated, AraC- and DOX-resistant), including dihydroorotate dehydrogenase (DHODH). There are reports in the literature about the use of different DOHDH inhibitors in AML with very promising results in several clinical and preclinical studies [42], [43], [44], [45]. In the latest of them, Cao et al., 2019, presented a novel inhibitory compound that displayed increased efficacy and low cytotoxicity in preclinical AML trials [46].
Our analysis also predicted that inhibition of DHFR with the anti-folate Methotrexate could target both resistant and parental cell lines. Folate metabolism is often over-activated in cancer cells and plays a key role in supporting de novo nucleotide synthesis, epigenetic regulation, and energy and redox balance, thus making it a prominent therapeutic target against cancer. Methotrexate is approved for clinical use against several autoimmune diseases and cancer types, including Acute Lymphoblastic Leukemia (ALL) where it is administered alone or in combination therapies [47], [48]. Studies have shown that in vitro AML cell lines respond to Methotrexate and that in vivo acute monocytic leukemia seems to be particularly sensitive [48], [49]. However, its clinical potential against AML resistant cells has not been explored yet. In fact, Methotrexate in AML has only been studied in a clinical setting against childhood acute monocytic leukemia and has only been proposed in combination therapy with Asparaginase [50]. Remarkably, our simulation showed that it would be effective against both parental and resistant cells and this was confirmed in vitro with both parental and resistant cell lines having high sensitivity to Methotrexate. In addition, we determined that Methotrexate does not exhibit cross-resistance with AraC in THP-1 cells and only moderate cross-resistance with DOX. Worth noting, even in DOX-resistant cells, the measured IC50s for Methotrexate were significantly under the range of concentrations (>0.1–10 μM) that cause methotrexate‐associated toxicity [51]. Thus, the use of Methotrexate could be explored in a clinical setting to improve the therapeutic outcome of AraC- or DOX-resistant patients.
Additionally, taking full advantage of the metabolomic layer, we identified cholesterol metabolism as a selective target against AraC- and DOX-resistant THP-1 cells. Indeed, the experimental validation of targeting the de novo cholesterol biosynthesis pathway as putative targets using YM-5360, which inhibits squalene synthase, provided very promising results against the resistant cells. Remarkably, the reconstructed GSMMs were able to predict the enhanced sensitivity of resistant THP-1 to YM-53601 compared to the parental cells. Indeed, many of the targets predicted to be selective against the resistant cell lines were associated with cholesterol metabolism, confirming that this pathway plays a key role in the metabolic reprogramming underlying the acquisition of chemoresistance in our cell model. In this regard, it has been demonstrated that exposing AML cell lines to radiotherapy and chemotherapy increases cholesterol levels and blocking cholesterol synthesis increases the susceptibility of the cell lines to therapy [35]. Even more, statins and specific inhibitors of the mevalonate pathway have been evaluated in combination therapy with chemotherapeutics, including AraC [52], [53]. The response rate to this combinatorial approach has been estimated at around 75% for relapse patients [54]. Interestingly, to our knowledge, our work is the first instance where squalene synthase inhibitors have been used against AML or chemoresistance, paving the way for new therapeutic combinations against AML. Notably, squalene synthase and cholesterol metabolism only emerged as a target when metabolomics data were integrated. Therefore, our results suggest that transcriptomic data alone can fail to capture the shift in metabolic requirements and dependencies that accompanied the acquisition of drug resistance in the THP-1 cells.
Our results provide a proof of concept that when condition-specific GSMMs are constructed, adding the metabolomic enhances the specificity of the condition-specific GSMM. Indeed, we show that additional metabolic vulnerabilities against the resistant cells that were not identifiable using only transcriptomic data are unveiled following the proposed workflow. The transcriptomics-metabolomics integration in GSMMs presented in this paper clearly contributes towards improving the predictive capacity of the flux distribution beyond transcriptomic regulation and sets the basis for clinical omics-data integration. Mapping metabolic regulation in AML at a patient-specific level could be the key to select the best treatment for AML patients and can be analogously applied in additional cancer types.
5. Conclusions
In this study, we use a workflow for the reconstruction of condition-specific GSMMs to build metabolic models of the AML cell line THP-1 and two derivatives with resistance to AraC and DOX. The resulting models enabled us to identify that drug-resistant cell lines could be targeted with the FDA-approved anti-folate Methotrexate. Moreover, we determined that the resistant cell lines could also be selectively targeted with YM-53601, an inhibitor of squalene synthase, providing a new and promising strategy to directly inhibit cholesterol synthesis in AML drug-resistant cells. Future works should continue to evaluate the therapeutic potential of using squalene synthase combined with chemotherapeutic agents in AML and other cancer types. Important to mention, our work demonstrated the importance of integrating the metabolomics layer into GSMMs, as it enabled a clearer distinction of putative metabolic vulnerabilities between the parental untreated and the drug-resistant AML THP-1 cells and led to the identification of squalene synthase as a putative target. Henceforth, the integration of such layer could be key to building condition-specific GSMMS of in vitro models of drug resistance, not only in the case of AML but also for other cancer types, and enable the identification of novel putative targets against chemoresistance. Indeed, metabolism has been recognized as an important driver in the emergence and instatement of drug resistance in different cancers. Therefore, our methodology could be applied not only for AML but also to other hematologic and non-hematologic cancers to enhance the identification of metabolic vulnerabilities specific to the different malignancies significantly. Given how straightforward the methodology described is, it could also be applied on patient-derived cell lines to build accurate patient-specific GSMMS incorporating the particular metabolomic and transcriptomic alterations for applications-oriented at patient risk stratification and personalized medicine.
CRediT authorship contribution statement
Effrosyni Karakitsou: Writing - original draft, Writing - review & editing, Conceptualization, Formal analysis, Data curation, Investigation, Visualization, Methodology. Carles Foguet: Writing - original draft, Writing - review & editing, Conceptualization, Formal analysis, Data curation, Investigation, Visualization, Methodology. Miriam G. Contreras Mostazo: Writing - original draft, Writing - review & editing, Validation, Investigation, Visualization. Nina Kurrle: Supervision, Writing - review & editing, Resources, Funding acquisition. Frank Schnütgen: Supervision, Writing - review & editing, Resources, Funding acquisition. Martin Michaelis: Writing - review & editing, Resources. Jindrich Cinatl: Writing - review & editing, Resources. Silvia Marin: Writing - original draft, Writing - review & editing, Supervision, Data curation, Formal analysis, Resources, Funding acquisition. Marta Cascante: Writing - original draft, Writing - review & editing, Supervision, Data curation, Formal analysis, Conceptualization, Resources, Visualization, Funding acquisition.
Funding
E. Karakitsou and M.G. Contreras Mostazo were supported by the EU grant HaemMetabolome H2020-MSCA-ITN-2015-675790. F. Schnütgen, S. Marin, N. Kurrle and M. Cascante acknowledge support from the European Commission (HaemMetabolome [EC-675790]). M. Cascante, C. Foguet and S. Marin acknowledge the Spanish Ministerio de Economia y Competitividad (MINECO) and Ministerio de Ciencia e Innovación -European Commission FEDER funds—“Una manera de hacer Europa” (SAF2017-89673-R and PID2020-115051RB-I00), the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) Generalitat de Catalunya (2017SGR1033) and CIBERehd (CB17/04/00023) (ISCIII, Spain). M. Cascante also received support through the prize “ICREA Academia” for excellence in research, funded by ICREA foundation–Generalitat de Catalunya. F. Schnütgen was supported by the Wilhelm Sander-Foundation (2015.138.1) and the Deutsche Forschungsgemeinschaft (SCHN1166/4-1). N. Kurrle and H. Serve acknowledge Deutsche Forschungsgemeinschaft (SFB 815, TP A08, TP A10).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Si W., Shen J., Zheng H., Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11(1) doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thol F. Can we forecast induction failure in acute myeloid leukemia? Haematologica. 2018;103(3):375–377. doi: 10.3324/haematol.2018.187575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. [DOI] [PMC free article] [PubMed]
- 4.Gurnari C., Pagliuca S., Visconte V. Deciphering the therapeutic resistance in acute myeloid leukemia. Int J Mol Sci. 2020;21(22):8505. doi: 10.3390/ijms21228505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaal E.A., Berkers C.R. The influence of metabolism on drug response in cancer. Front Oncol. 2018;8(500) doi: 10.3389/fonc.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, Y., E.B. Butler, and M. Tan, Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis, 2013. 4(3): p. e532. [DOI] [PMC free article] [PubMed]
- 7.Rahman M., Hasan M.R. Cancer metabolism and drug resistance. Metabolites. 2015;5(4):571–600. doi: 10.3390/metabo5040571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarrado-Castellarnau M. De novo MYC addiction as an adaptive response of cancer cells to CDK4/6 inhibition. Mol Syst Biol. 2017;13(10):940. doi: 10.15252/msb.20167321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson A., Nielsen J. Genome scale metabolic modeling of cancer. Metab Eng. 2017;43:103–112. doi: 10.1016/j.ymben.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Aguilar E., Marin de Mas I., Zodda E., Marin S., Morrish F., Selivanov V. Metabolic reprogramming and dependencies associated with epithelial cancer stem cells independent of the epithelial-mesenchymal transition program. Stem Cells. 2016;34(5):1163–1176. doi: 10.1002/stem.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folger O. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M., Nielsen J. identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10(3):721. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marín de Mas I., Aguilar E., Zodda E., Balcells C., Marin S., Dallmann G. Model-driven discovery of long-chain fatty acid metabolic reprogramming in heterogeneous prostate cancer cells. PLoS Comput Biol. 2018;14(1):e1005914. doi: 10.1371/journal.pcbi.1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarragó-Celada J., Foguet C., Tarrado-Castellarnau M., Marin S., Hernández-Alias X., Perarnau J. Cysteine and folate metabolism are targetable vulnerabilities of metastatic colorectal cancer. Cancers. 2021;13(3):425. doi: 10.3390/cancers13030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26(2):171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 16.Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu A., Bodycombe N., Cheah J., Price E., Liu K., Schaefer G. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaelis M, et al., Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis, 2011. 2(12): p. e243. [DOI] [PMC free article] [PubMed]
- 20.Michaelis M, Wass MN, Cinatl J. Drug-adapted cancer cell lines as preclinical models of acquired resistance. [DOI] [PMC free article] [PubMed]
- 21.Dobin A. et al., STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 2012. 29(1): p. 15–21. [DOI] [PMC free article] [PubMed]
- 22.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst Biol. 2013;7(1):74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swainston N. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orth J.D., Thiele I., Palsson B. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt BJ, et al., GIM3E: condition-specific models of cellular metabolism developed from metabolomics and expression data. Bioinformatics, 2013. 29(22): p. 2900–2908. [DOI] [PMC free article] [PubMed]
- 27.Megchelenbrink W., Huynen M., Marchiori E., Rogers S. optGpSampler: an improved tool for uniformly sampling the solution-space of genome-scale metabolic networks. PLoS ONE. 2014;9(2):e86587. doi: 10.1371/journal.pone.0086587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segre D., Vitkup D., Church G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99(23):15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulsen T., de Vlieg J., Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9(1):488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu E., Callender M.A., Farrell M.P., Schmitz J.C. Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother Pharmacol. 2003;52(0):80–89. doi: 10.1007/s00280-003-0625-9. [DOI] [PubMed] [Google Scholar]
- 31.Gangjee A. Dual inhibitors of thymidylate synthase and dihydrofolate reductase as antitumor agents: design, synthesis, and biological evaluation of classical and nonclassical pyrrolo[2,3-d]pyrimidine anti-folates (1) J Med Chem. 2006;49(3):1055–1065. doi: 10.1021/jm058276a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakura T, et al., High-dose methotrexate therapy significantly improved survival of adult acute lymphoblastic leukemia: a phase III study by JALSG. [DOI] [PubMed]
- 33.Gonen N., Assaraf Y.G. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat. 2012;15(4):183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Zhao R., Goldman I.D. resistance to anti-folates. Oncogene. 2003;22(47):7431–7457. doi: 10.1038/sj.onc.1206946. [DOI] [PubMed] [Google Scholar]
- 35.Li HY, et al., Cholesterol-modulating agents kill acute myeloid leukemia cells and sensitize them to therapeutics by blocking adaptive cholesterol responses. Blood, 2003. 101(9): p. 3628–3634. [DOI] [PubMed]
- 36.UgawaT, et al., YM-53601, a novel squalene synthase inhibitor, suppresses lipogenic biosynthesis and lipid secretion in rodents. Br J Pharmacol, 2003. 139(1): p. 140–6. [DOI] [PMC free article] [PubMed]
- 37.Zaidi S.Z., Owaidah T., Al Sharif F., Ahmed S.Y., Chaudhri N., Aljurf M. The challenge of risk stratification in acute myeloid leukemia with normal karyotype. Hematol Oncol Stem Cell Ther. 2008;1(3):141–158. doi: 10.1016/s1658-3876(08)50023-9. [DOI] [PubMed] [Google Scholar]
- 38.Dougherty BV, Moutinho TJ Jr, Papin J. Accelerating the drug development pipeline with genome-scale metabolic network reconstructions. Systems Biology, 2017. 6.
- 39.Zielinski D.C., Filipp F.V., Bordbar A., Jensen K., Smith J.W., Herrgard M.J. Pharmacogenomic and clinical data link non-pharmacokinetic metabolic dysregulation to drug side effect pathogenesis. Nat Commun. 2015;6(1) doi: 10.1038/ncomms8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machado D., Herrgård M., Maranas C.D. Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLoS Comput Biol. 2014;10(4):e1003580. doi: 10.1371/journal.pcbi.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katzir R., Polat I.H., Harel M., Katz S., Foguet C., Selivanov V.A. The landscape of tiered regulation of breast cancer cell metabolism. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-54221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christian S., Merz C., Evans L., Gradl S., Seidel H., Friberg A. The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia. 2019;33(10):2403–2415. doi: 10.1038/s41375-019-0461-5. [DOI] [PubMed] [Google Scholar]
- 43.Sykes D.B. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell. 2016;167(1):171–186.e15. doi: 10.1016/j.cell.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ladds M.J.G.W. A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage. Nat Commun. 2018;9(1):1107. doi: 10.1038/s41467-018-03441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu D., Wang W., Chen W., Lian F., Lang L., Huang Y. Pharmacological inhibition of dihydroorotate dehydrogenase induces apoptosis and differentiation in acute myeloid leukemia cells. Haematologica. 2018;103(9):1472–1483. doi: 10.3324/haematol.2018.188185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao L., Weetall M., Trotta C., Cintron K., Ma J., Kim M.J. Targeting of hematologic malignancies with PTC299, a novel potent inhibitor of dihydroorotate dehydrogenase with favorable pharmaceutical properties. Mol Cancer Ther. 2019;18(1):3–16. doi: 10.1158/1535-7163.MCT-18-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakura T., Hayakawa F., Sugiura I., Murayama T., Imai K., Usui N. High-dose methotrexate therapy significantly improved survival of adult acute lymphoblastic leukemia: a phase III study by JALSG. Leukemia. 2018;32(3):626–632. doi: 10.1038/leu.2017.283. [DOI] [PubMed] [Google Scholar]
- 48.Zarou M.M., Vazquez A., Vignir Helgason G. Folate metabolism: a re-emerging therapeutic target in haematological cancers. Leukemia. 2021;35(6):1539–1551. doi: 10.1038/s41375-021-01189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rots M.G., Pieters R., Jansen G., Kaspers G.J.L., Van Zantwijk C.H., Noordhuis P. A possible role for Methotrexate in the treatment of childhood acute myeloid leukaemia, in particular for acute monocytic leukaemia. Eur J Cancer. 2001;37(4):492–498. doi: 10.1016/s0959-8049(00)00433-0. [DOI] [PubMed] [Google Scholar]
- 50.Buaboonnam J., Cao X., Pauley J.L., Pui C.-H., Ribeiro R.C., Rubnitz J.E. Sequential administration of Methotrexate and asparaginase in relapsed or refractory pediatric acute myeloid leukemia. Pediatr Blood Cancer. 2013;60(7):1161–1164. doi: 10.1002/pbc.24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widemann B.C., Adamson P.C. Understanding and managing methotrexate nephrotoxicity. Oncologist. 2006;11(6):694–703. doi: 10.1634/theoncologist.11-6-694. [DOI] [PubMed] [Google Scholar]
- 52.Stirewalt D.L., Appelbaum F.R., Willman C.L., Zager R.A., Banker D.E. Mevastatin can increase toxicity in primary AMLs exposed to standard therapeutic agents, but statin efficacy is not simply associated with ras hotspot mutations or overexpression. Leuk Res. 2003;27(2):133–145. doi: 10.1016/s0145-2126(02)00085-1. [DOI] [PubMed] [Google Scholar]
- 53.Lee J.S., Roberts A., Juarez D., Vo T.-T., Bhatt S., Herzog L.-o. Statins enhance efficacy of venetoclax in blood cancers. Sci Transl Med. 2018;10(445):eaaq1240. doi: 10.1126/scitranslmed.aaq1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shadman M., Mawad R., Dean C., Chen T.L., Shannon-Dorcy K., Sandhu V. Idarubicin, cytarabine, and pravastatin as induction therapy for untreated acute myeloid leukemia and high-risk myelodysplastic syndrome. Am J Hematol. 2015;90(6):483–486. doi: 10.1002/ajh.23981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.