ABSTRACT
Terminally differentiated cells are generally thought to have arrived at their final form and function. Many terminally differentiated cell types are polyploid, i.e. they have multiple copies of the normally diploid genome. Mammalian heart muscle cells, termed cardiomyocytes, are one such example of polyploid cells. Terminally differentiated cardiomyocytes are bi- or multi-nucleated, or have polyploid nuclei. Recent mechanistic studies of polyploid cardiomyocytes indicate that they can limit cellular proliferation and, hence, heart regeneration. In this short Spotlight, we present the mechanisms generating bi- and multi-nucleated cardiomyocytes, and the mechanisms generating polyploid nuclei. Our aim is to develop hypotheses about how these mechanisms might relate to cardiomyocyte proliferation and cardiac regeneration. We also discuss how these new findings could be applied to advance cardiac regeneration research, and how they relate to studies of other polyploid cells, such as cancer cells.
KEY WORDS: Cardiomyocytes, Cytokinesis, Differentiation, Karyokinesis, Ploidy, Regeneration
Summary: This Spotlight article discusses the mechanisms that polyploid cardiomyocytes and their relationship with cardiac regeneration.
Introduction
The term ‘terminal differentiation’ has traditionally been used to describe cells that have permanently exited the cell cycle, lost proliferative potential and, consequently, cannot contribute to tissue regeneration after injury. Many terminally differentiated cell types are polyploid (Øvrebø and Edgar, 2018), i.e. they contain multiple copies of the typically diploid genome. In principle, such polyploid cells can contain two or more nuclei, or a nucleus with a higher than normal DNA content. As such, a bi-nucleated cell with two diploid (2N) nuclei is considered to be polyploid, as is a mono-nucleated cell with a tetraploid nucleus.
The heart muscle comprises specialized cells, called cardiomyocytes, that proliferate during development. Most cardiomyocytes in adult mammals are thought to have lost the ability to proliferate (Soonpaa and Field, 1997; Pasumarthi and Field, 2002) and are thus regarded as terminally differentiated. For example, only low levels of cardiomyocytes expressing indicators of cell cycle activity have been reported in adult mice (Walsh et al., 2010; Ali et al., 2014) and in adult humans (Bergmann et al., 2009; Mollova et al., 2013). In addition, although cardiomyocyte DNA synthesis increases in the border zone of a myocardial infarction, adult mammalian hearts do not appear to have sufficient numbers of proliferative cardiomyocytes to allow them to regenerate after injury (Senyo et al., 2013). Thus, although cardiac regeneration research has focused on stimulating cell cycle activity to achieve cardiomyocyte proliferation, a fundamental understanding of terminal differentiation is lacking and has only recently been examined as an approach to understanding the mechanisms that block adult cardiomyocyte proliferation (Tzahor and Poss, 2017; Sadek and Olson, 2020).
Although the presence of multiple and polyploid nuclei in cardiomyocytes has long been known, and is used to identify terminally differentiated cardiomyocytes, the significance of ploidy for proliferation, heart regeneration and heart failure development has only recently begun to be explored. Indeed, recent studies have demonstrated a connection between cardiomyocyte polyploidy and regenerative capacity in vertebrate hearts, suggesting that polyploidy can act as a barrier to regeneration (Patterson et al., 2017; González-Rosa et al., 2018). These reports have underscored the importance of understanding cardiomyocyte terminal differentiation and polyploidy, and have provided new directions for cardiac regeneration research. Moreover, they have highlighted that, although the mechanisms regulating the formation of polyploid cells in other tissues, including trophoblast giant cells in the placenta (Weier et al., 2005; Zybina and Zybina, 2005), hepatocytes of the liver (Duncan et al., 2010; Pandit et al., 2013; Gentric and Desdouets, 2014; Hsu et al., 2016) and platelet-producing megakaryocytes (Corash et al., 1989; Battinelli et al., 2007; Gao et al., 2012), have been characterized, the regulatory mechanisms governing polyploidy in cardiomyocytes are only now beginning to be unraveled.
Here, we review these recent discoveries to develop an integrated view of the mechanisms that direct the formation of polyploid cardiomyocytes. We use the term ‘polyploid’ to refer both to bi- and multi-nucleated cardiomyocytes with 2N DNA content in each nucleus, and to cardiomyocytes with one nucleus of >2N DNA. However, we distinguish the processes forming multi-nucleated cardiomyocytes from those forming polyploid nuclei because they are governed by different mechanisms. Using this approach, we develop a unifying map of cardiomyocyte terminal differentiation, highlighting similarities and differences with other cell types. We hope this map could serve as a framework for future heart regeneration research.
The prevalence of polyploid cardiomyocytes differs between species and changes during development
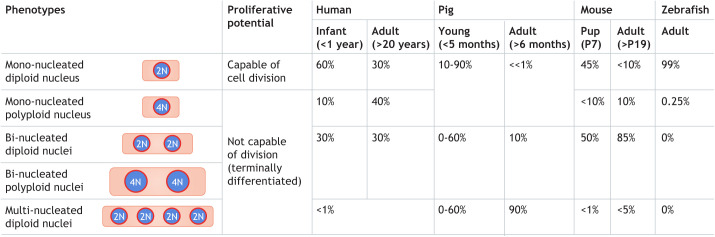
Studies in zebrafish, mice and humans have shown that cardiomyocyte proliferation during development generally involves mono-nucleated cardiomyocytes with diploid nuclei (Wills et al., 2008; Mollova et al., 2013; Patterson et al., 2017; González-Rosa et al., 2018). However, as mammalian cardiomyocytes differentiate, many become bi- and multi-nucleated, and form polyploid nuclei during pre- and postnatal development (Soonpaa and Field, 1997; Mollova et al., 2013; Alkass et al., 2015; Van De Peer et al., 2017; Liu et al., 2019; Han et al., 2020). The prevalence of cardiomyocytes with these different forms of ploidy differs between species and within species between different stages of development (Fig. 1), and could therefore contribute to the differences in regenerative capacity observed between species.
Fig. 1.
Phenotypes of working cardiomyocytes, their proliferative potential and their changing prevalence in common model organisms. Percentages were obtained from studies that used microscopy methods for quantifying cardiomyocyte phenotypes. The prevalence of the different cardiomyocyte phenotypes in humans was taken from Mollova et al. (2013) and Bergmann et al. (2015). In humans with tetralogy of Fallot, a type of congenital heart disease, multi-nucleated cardiomyocytes (>2 nuclei) appear after the first month of life and persist into adulthood (Liu et al., 2019). Although the work of Adler et al. (1996) has served as an important reference for pig data, the results were obtained using an indirect approach. Consequently, recent results from isolated pig cardiomyocytes (Velayutham et al., 2020), using single-cell microscopy, are shown. The values for mouse were taken from Alkass et al. (2015), although nuclear ploidy in mono- and bi-nucleated cardiomyocytes was not clearly distinguished, and from Han et al. (2020). Values for zebrafish were taken from Wills et al. (2008) and González-Rosa et al. (2018).
Cold-blooded organisms, such as zebrafish and newts, are capable of heart regeneration (Poss et al., 2002; Bettencourt-Dias et al., 2003). Zebrafish regenerate their heart muscle via cardiomyocyte proliferation (Poss et al., 2002), raising the following questions: what mechanisms give cold-blooded animals the evolutionary advantage of heart regeneration and how can we leverage this understanding to aid human regenerative medicine? Some research has suggested that low blood pressure in zebrafish and other cold-blooded animals may be responsible for their increased regenerative capacity (Hu et al., 2001). Interestingly, giraffes exhibit one of the highest blood pressures seen in mammals and 88% of their cardiomyocytes have four or more nuclei (Østergaard et al., 2013), whereas all pig cardiomyocytes have 2 or more nuclei (Velayutham et al., 2020). These are the highest known degrees and percentages of bi- and multi-nucleated cardiomyocytes in mammals, although how this relates to regenerative capacity remains unclear. A correlation between metabolic rate, body temperature, serum thyroid hormone levels, percentage of diploid cardiomyocytes and regenerative capability has also been noted among different species, from non-mammals to mammals, including humans (Hirose et al., 2019). However, the trend does not apply to all amphibian species, as medaka (Oryzias latipes), a cold-blooded fish, similar to zebrafish, exhibits almost no cardiomyocyte regeneration following ventricular resection (Ito et al., 2014). Moreover, a comparison between zebrafish and medaka showed that reduced immune responses may prevent regeneration (Lai et al., 2017), which suggests being cold blooded may not be sufficient for heart repair.
Increased diploid mono-nucleated cardiomyocyte content has also been positively correlated with regenerative capacity (Matrone et al., 2017; González-Rosa et al., 2018). Mono-nucleated diploid cardiomyocytes are present in all vertebrate species studied. However, their prevalence varies widely: although almost all zebrafish cardiomyocytes have one diploid nucleus (Wills et al., 2008; González-Rosa et al., 2018), 35% of adult human cardiomyocytes (Mollova et al., 2013; Bergmann et al., 2015) and only 5-10% of adult mice cardiomyocytes (Alkass et al., 2015) have one diploid nucleus. By contrast, pigs develop a high percentage of cardiomyocytes with multiple nuclei within the first 2 months postnatally (Gräbner and Pfitzer, 1974; Adler et al., 1996; Velayutham et al., 2020). Interestingly, the overexpression of microRNA-199a in adult pigs promotes regeneration of the heart following cardiac infarction, indicating that multi-nucleated cardiomyocytes may possess the ability to re-enter the cell cycle and divide (Gabisonia et al., 2019). However, this intervention results in uncontrolled proliferation of poorly differentiated cardiomyocytes and subsequent fatal arrhythmias in pigs, highlighting that the mechanisms of proliferation need to be better understood before any clinical translation studies are pursued. Mice develop mostly bi-nucleated and multi-nucleated cardiomyocytes within 10 days of birth (Soonpaa et al., 1996). However, in mice, the polyploidization of cardiomyocyte nuclei occurs later and does not reach the same proportion as multi-nucleation (Walsh et al., 2010; Alkass et al., 2015) (Fig. 1). Humans also show formation of cardiomyocytes with two diploid nuclei and of cardiomyocytes with one polyploid nucleus at different ages (Brodsky et al., 1994; Mollova et al., 2013; Bergmann et al., 2015), suggesting that the processes that generate these different ploidy types are distinct and may not depend on one another. In the following sections, we review recent reports identifying these distinct processes (Table 1).
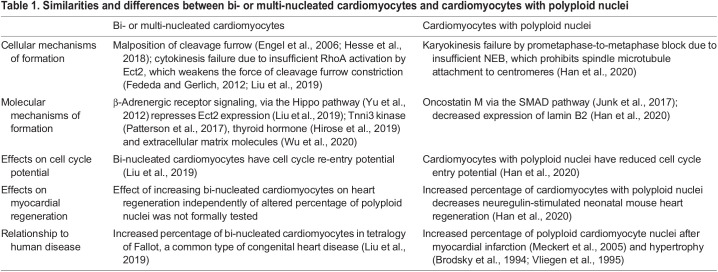
Table 1.
Similarities and differences between bi- or multi-nucleated cardiomyocytes and cardiomyocytes with polyploid nuclei
Cytokinesis failure generates bi- and multi-nucleated cardiomyocytes
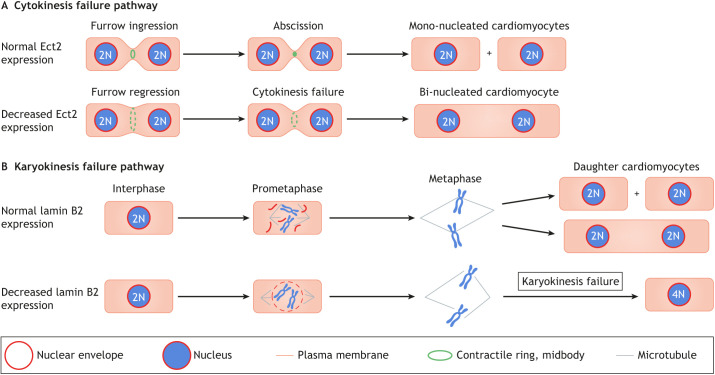
Bi-nucleated cardiomyocytes have two separated nuclei, which means that their mother cardiomyocytes successfully completed nuclear division (karyokinesis) but failed cell body division (cytokinesis). Cytokinesis is the final step in mitosis in which separation of the two daughter cells is mediated by discrete steps (Eggert et al., 2006). The general mechanisms underlying cytokinesis, specifically cleavage furrow constriction and abscission, have been well studied. In short, the GTPase RhoA, guided by the RhoA guanine-nucleotide exchange factor Ect2 (Yüce et al., 2005), recruits anillin (Frenette et al., 2012), a scaffold protein that interacts with non-muscle myosin II (Straight et al., 2005) and F-actin (Oegema et al., 2000) to induce cleavage furrow ingression.
Asymmetric cleavage furrow constriction and close proximity of daughter nuclei during division has been shown during cardiomyocyte cytokinesis failure (Hesse et al., 2018). Cytokinesis failure in cardiomyocytes can be caused by low expression levels of the cytokinesis gene Ect2 (Liu et al., 2019). Ect2 expression is induced in the cell cycle (Liu et al., 2019) and is required for cleavage furrow constriction (Tatsumoto et al., 1999). In cardiomyocytes, Ect2 gene expression is repressed by β-adrenergic receptor (β-AR) signaling via the Hippo tumor suppressor pathway (Liu et al., 2019). The Hippo pathway has been shown to be activated by (β-AR) signaling in the heart (Yu et al., 2012) and is implicated in cardiomyocyte proliferation (Heallen et al., 2011; Von Gise et al., 2012). Ect2 gene repression leads to abscission failure very late in cytokinesis: cardiomyocytes assemble a contractile ring in preparation for abscission but fail to constrict the cleavage furrow sufficiently for abscission (Fig. 2A). Blocking β-AR with propranolol increases Ect2 gene expression, which promotes the division of mitotic cardiomyocytes and increases their total number in mouse hearts (Liu et al., 2019). It has also been shown that cultured cardiomyocytes undergo serum-induced cytokinesis failure, characterized by altered anillin localization and altered cleavage furrow constriction (Engel et al., 2006); while this involves increased p38 mitogen-activated kinase signaling, the responsible serum components are unknown.
Fig. 2.
Cytokinesis and karyokinesis failure mechanisms in cardiomyocytes. (A) Ect2 has a function in mediating cleavage furrow ingression and abscission during cytokinesis. Normal expression of Ect2 during development thus allows for cytokinesis to occur (top). However, a reduction in Ect2 expression (bottom) causes cytokinesis failure, leading to the formation of bi-nucleated (or multi-nucleated) cardiomyocytes. (B) Lamin B2 has a function in karyokinesis, controlling nuclear envelope breakdown (NEB) during prometaphase. If lamin B2 levels are normal, karyokinesis can take place (top). However, insufficient lamin B2 levels (bottom) lead to incomplete NEB. This prevents complete spindle microtubule attachment to centromeres, resulting in prometaphase arrest and formation of polyploid daughter nuclei.
Cytokinesis requires the orchestrated action of multiple proteins (Piekny et al., 2005; D'Avino, 2009; von Dassow, 2009), and the malfunction of any of these could cause cytokinesis failure. Several cytokinesis proteins have been identified to play a role in the formation of bi-nucleated cardiomyocytes. The GTPase activating factor IQGAP3 contributes to the process of actin-myosin ring localization. Its mis-localization results in failure of cleavage furrow formation and mitotic microtubule distribution, thus causing bi-nucleation (Leone et al., 2018). Additional regulators of cardiomyocyte bi-nucleation are troponin i3 kinase (Patterson et al., 2017), thyroid hormone signaling (Hirose et al., 2019), and the extracellular matrix molecules nephronectin and slit homolog 2 protein (Wu et al., 2020), although the cellular mechanisms by which these factors cause cytokinesis failure remain to be investigated. Various interventions that have been reported to stimulate cardiomyocyte proliferation also induce smaller changes in the percentage of bi-nucleated cardiomyocytes, e.g., Meis1 gene inactivation (Mahmoud et al., 2013), telomeric erosion (Aix et al., 2016) and hypoxia (Kimura et al., 2015; Nakada et al., 2017), although their mechanistic connections to cytokinesis failure are unknown.
Cardiomyocyte fusion has also been proposed as a mechanism forming bi- and multi-nucleated cells. However, in zebrafish, cardiomyocyte fusion does not lead to bi-nucleation or polyploidy, but is associated with cell cycle entry, suggesting that fusion could be a trigger for cardiomyocyte re-entry into the cell cycle (Sawamiphak et al., 2017). A paradigm of transient cardiomyocyte fusion has also been suggested in mammals. For example, rat cardiomyocytes fuse with non-cardiomyocyte cell types and re-enter the G2-M phase of the cell cycle; however, these results are limited to cell culture and to neonatal rat cardiomyocytes (Matsuura et al., 2004). Fusion between cardiomyocytes and skeletal muscle cells has also been shown to result in multi-nucleated cells in mice (Reinecke et al., 2004). In addition, a recent report used a confetti Cre-lox system in mice to show putative cardiomyocyte fusion resulting in bi-nucleated cells (Ali et al., 2020). Although, in principle, fusion provides an additional path to generating bi- and multi-nucleated cardiomyocytes, this may not be a dominant mechanism because all of these reports agree that fusion events are rather rare.
Karyokinesis failure generates polyploid cardiomyocyte nuclei
Karyokinesis involves a series of steps that lead to the division of the nucleus of the cell, resulting in two daughter nuclei (Fig. 2B). Specifically, the process begins with nuclear envelope breakdown (NEB), followed by the alignment and segregation of the chromosomes. Karyokinesis concludes with nuclear envelope reassembly around the two sets of daughter chromosomes. Karyokinesis failure involves the uncoupling of DNA replication from the completion of nuclear division (Edgar et al., 2014). Several studies of cardiomyocytes have shown that the mechanisms generating polyploid nuclei are distinct from the aforementioned cytokinesis failure mechanisms that form bi- and multi-nucleated cardiomyocytes (Table 1).
Depletion of survivin, an inhibitor of apoptosis protein, significantly increases DNA content in cardiomyocyte nuclei without cell division (Levkau et al., 2008). Deletion of glycogen synthase kinase (GSK)-3α also promotes polyploidization, specifically an increase in 4N nuclei, owing to incomplete karyokinesis (Zhou et al., 2016). In addition, inducible transgenic expression of the transcription factor Myc increases the number of polyploid cardiomyocyte nuclei in adult mice (Xiao et al., 2001). However, more recent results suggest that increasing the expression of Myc together with that of the positive transcription elongation factor (P-TEFb) increases cardiomyocyte proliferation without changing the percentages of bi- and multi-nucleated cardiomyocytes (Bywater et al., 2020). Decreased expression of Lamin B2, an intermediate filament of the nuclear lamina, is also responsible for karyokinesis failure in cardiomyocytes. Lamin B2 gene inactivation in mice inhibits progression of prometaphase to metaphase, due to failure of proper NEB. Incomplete NEB prevents effective attachment of mitotic spindle microtubules to centromeres in prometaphase (Fig. 2B), creating a prometaphase to metaphase block and resulting in the formation of polyploid nuclei (Han et al., 2020). Overall, these findings indicate that the mechanisms forming polyploid nuclei are different from those forming bi- and multi-nucleated cardiomyocytes. This is also in agreement with the observation that bi-nucleated cardiomyocytes and polyploid cardiomyocyte nuclei are formed over different timescales (discussed below) in both mice and humans (Soonpaa et al., 1996; Mollova et al., 2013; Bergmann et al., 2015).
The relationship between karyokinesis failure and cytokinesis failure in cardiomyocytes
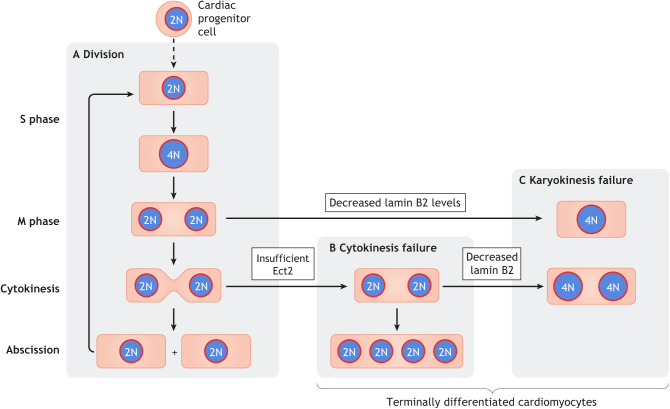
In mice, 80-95% of cardiomyocytes become bi-nucleated within 7-10 days of birth (Soonpaa et al., 1996). The formation of polyploid nuclei occurs around 3 weeks after birth, i.e. after the surge of bi-nucleation (Walsh et al., 2010; Alkass et al., 2015). In humans, a surge of polyploidization occurs in cardiomyocytes at around 10 years after birth to establish a level of 35% mono-nucleated cardiomyocytes with polyploid nuclei. The percentage of cardiomyocytes with polyploid nuclei then steadily increases until 20 years (Herget et al., 1997; Mollova et al., 2013; Bergmann et al., 2015). A further small increase in the percentage of cardiomyocytes with polyploid nuclei has been reported in humans over the age of 20 years (Mollova et al., 2013), which is consistent with the observation that some cardiomyocytes are seen to exist in S phase and M phase at this age (Bergmann et al., 2009; Mollova et al., 2013). Thus, although in mice the formation of bi- and multi-nucleated cardiomyocytes with polyploid nuclei occurs within 3 weeks of birth, this process takes more than a decade in humans. This suggests that karyokinesis failure likely occurs after cytokinesis failure, implying that cardiomyocytes with polyploid nuclei could represent a more advanced stage of differentiation. However, cytokinesis failure may not be required for karyokinesis failure to occur.
Repression of the cytokinesis gene Ect2 is linked to the formation of bi-nucleated cardiomyocytes in mice and in human infants with congenital heart disease (Liu et al., 2019). However, it is important to emphasize that Ect2 gene inactivation does not alter S-phase or M-phase entry in cardiomyocytes (Liu et al., 2019). As such, successive cytokinesis failure could generate multi-nucleated cardiomyocytes, which are prevalent in pigs (Adler et al., 1996; Velayutham et al., 2020) and found at raised levels in human infants with tetralogy of Fallot (Liu et al., 2019) (Fig. 3). The cell cycle potential of bi-nucleated cardiomyocytes also allows subsequent cell cycle entry, thus providing a mechanism for the generation of polyploid nuclei in bi-nucleated cardiomyocytes (Fig. 3). Although it has been suggested that bi-nucleated cardiomyocytes can divide in mice (Naqvi et al., 2014), live cell imaging of cardiomyocytes in culture shows that the division of bi-nucleated cardiomyocytes is rare (Bersell et al., 2009; Leone and Engel, 2019).
Fig. 3.
The generation of terminally differentiated cardiomyocytes involves hierarchical karyokinesis and cytokinesis failure. The dashed vertical arrow indicates the multiple maturation steps leading from cardiac progenitor cells to cardiomyocytes. Cell cycle phases are indicated on the left. (A) In a normal cell division cycle, cardiomyocytes express the complete set of cell cycle genes required for transition of the complete cell cycle. (B) Insufficient levels of the cytokinesis gene Ect2 cause cytokinesis failure, leading to the formation of bi-nucleated cardiomyocytes. These cells can re-enter the cell cycle and progress to cytokinesis such that consecutive cytokinesis failure generates multi-nucleated (≥4 nuclei) cardiomyocytes. (C) Insufficient levels of lamin B2 can cause karyokinesis failure, resulting in the formation of cardiomyocytes with polyploid nuclei. These cells have a very low potential to re-enter the cell cycle.
Cardiomyocytes with polyploid nuclei have a lower cell cycle activity than those with diploid nuclei (Han et al., 2020), suggesting that karyokinesis failure could be the final process in the formation of terminally differentiated cardiomyocytes. It is known that Lamin B2 protein levels in mice decrease after birth, thereby reducing NEB and preventing spindle microtubule attachment to centromeres, which prevents karyokinesis and, consequently, cytokinesis. Accordingly, experimental gene inactivation of Lamin B2 increases the percentage of cardiomyocytes with polyploid nuclei. As these cells have decreased cell cycle potential, there are fewer cell cycle re-entries that end with cytokinesis failure, resulting in a lower percentage of bi-nucleated cardiomyocytes, which further supports the notion that karyokinesis failure could be the final process in the formation of terminally differentiated cardiomyocytes.
Some inbred mouse strains have different proportions of mono- versus bi-nucleated diploid cardiomyocytes, suggesting a genetic influence (Patterson et al., 2017). This has been linked to polymorphisms in the gene encoding the cardiomyocyte-specific kinase Tnni3k. Tnni3k gene inactivation increases the percentage of diploid mono-nucleated cardiomyocytes (Patterson et al., 2017). However, the function of Tnni3k in cytokinesis and karyokinesis is unclear, and it is possible that Tnni3k participates in the formation of both bi-nucleated cardiomyocytes and polyploid nuclei via unknown mechanisms.
The significance of cardiomyocyte polyploidy for heart regeneration
Although it has been commonly thought that hearts with a high percentage of mono-nucleated diploid (2N) cardiomyocytes can regenerate, functional support for this notion was lacking until recently. Studies in zebrafish have shown that overexpression of a C-terminally truncated mouse Ect2, which lacks the guanine-nucleotide exchange factor (GEF) domain but retains RhoA-GDP-binding activity, thus exhibiting dominant-negative function by interfering with the activation of RhoA by wild-type Ect2, increases not only the percentage of bi-nucleated cardiomyocytes but also, surprisingly, the percentage of cardiomyocytes with polyploid nuclei (González-Rosa et al., 2018). This mutant was used to show that zebrafish hearts, in which the combined percentages of bi-nucleated cardiomyocytes and cardiomyocytes with polyploid nuclei was greater than 45%, exhibit decreased cardiomyocyte proliferation and heart regeneration. This study established a functional connection between a lower percentage of mono-nucleated diploid cardiomyocytes and a decrease in heart regeneration. However, it remained unclear whether polyploid nuclei, bi-nucleated cardiomyocytes, or a combination of both establish a barrier to cell cycle entry and division.
A positive correlation between a higher diploid mono-nuclear cardiomyocyte content and proliferation capacity after injury has also been observed in adult mice (Patterson et al., 2017). In this case, genetic differences between inbred mouse strains influenced the diploid mono-nuclear cardiomyocyte content (Gan et al., 2020). A molecular connection between decreased thyroid hormone receptor signaling and a lower percentage of polyploid cardiomyocytes has also been demonstrated pharmacologically and genetically in mice, and has been linked to improved heart regeneration (Hirose et al., 2019). These results are surprising as thyroid hormone is beneficial for tissue repair in numerous organs (Mourouzis et al., 2013), and as thyroid hormone improves both cardiac contractility and cardiovascular function after injury (Chowdhury et al., 2001; Pingitore et al., 2019; von Hafe et al., 2019).
Relatively small increases in the percentage of mono-nucleated diploid cardiomyocytes in mice (Patterson et al., 2017; González-Rosa et al., 2018; Han et al., 2020) appear to have positive effects on myocardial regeneration. This could be explained by the existence of privileged subpopulations of cardiomyocytes that are more capable of proliferation. Molecular interventions that alter the size of this subpopulation could thus cause a disproportionate improvement of heart regeneration. However, human infants, despite 70% of their cardiomyocytes being mono-nucleated diploid (2N), develop scars after myocardial infarction and permanent myocardial dysfunction after accidental coronary artery obstruction during neonatal congenital heart disease surgery (Franciosi and Blanc, 1968; Kirklin et al., 1992). This suggests that the presence of a large percentage of mono-nucleated diploid cardiomyocytes alone may not be sufficient for heart regeneration.
Cytokinesis failure and karyokinesis failure in cancer cells and other cell types
Cytokinesis failure is one of the possible intermediate steps in the formation of tetraploid or octaploid cancer cells, which can become senescent or undergo further aberrant divisions that may result in chromosomal instability and subsequent aneuploidy (Ganem et al., 2007; Vitale et al., 2011). Cancer cells can also form micronuclei from lagging chromosomes during aberrant mitosis (Crasta et al., 2012). These micronuclei have abnormal composition of nuclear lamins (Vitale et al., 2011), and it has been shown that B-type lamins are involved in micronuclei stability in cancer cells (Hatch et al., 2013). However, micronuclei have not been reported in cardiomyocytes, likely due to the rare occurrence of mitotic events. Moreover, although depletion of Lamin B2 reduces NEB in cardiomyocytes and acts as a roadblock for karyokinesis and cardiomyocyte proliferation, decreased expression of Lamin B2 in human cancer cells can result in chromosome instability and mitotic spindle misalignment (Kuga et al., 2014). Although cardiomyocytes do not undergo uncontrolled proliferation, loss of glycogen synthase kinase (GSK) 3 isoforms has been shown to induce cardiomyocyte mitotic catastrophe (Zhou et al., 2016). Interestingly, cardiomyocytes become polyploid before undergoing mitotic catastrophe (Zhou et al., 2016), although the details of this process are unknown.
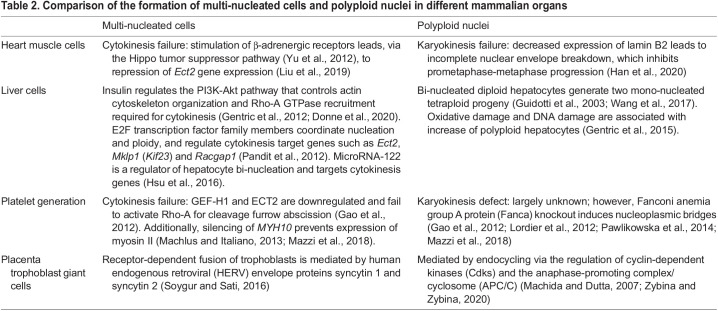
Cytokinesis failure also contributes to the formation of diverse polyploid cell types in animals (Van De Peer et al., 2017; Fox et al., 2020). For example, cytokinesis is the primary mechanism involved in the formation of polyploid megakaryocytes that are responsible for platelet production (Gao et al., 2012; Machlus and Italiano, 2013; Mazzi et al., 2018). Interestingly, downregulation of Ect2 expression is essential for polyploidization in megakaryocytes (Gao et al., 2012). Bi-nucleated hepatocytes are also generated by cytokinesis failure, which is also mediated by decreased Ect2 expression (Pandit et al., 2012; Hsu et al., 2016). Thus, decreased Ect2 expression may be a general mechanism of programmed cytokinesis failure in animal cells. In contrast, cancer cell proliferation is associated with overexpression of Ect2 and constitutively active Ect2 mutants (Salhia et al., 2008; Hirata et al., 2009; Justilien and Fields, 2009; Fields and Justilien, 2010) (Table 2). In summary, cytokinesis and karyokinesis failure are programmed in different cell types as part of development and differentiation.
Table 2.
Comparison of the formation of multi-nucleated cells and polyploid nuclei in different mammalian organs
Although polyploid cells are found in diverse organs and tissues, they may have distinct functions. As highlighted above, polyploid cells in the adult heart appear to act as a barrier to regeneration. However, epicardial cells, which make up the outer layer of the heart and support cardiomyocyte proliferation (Cao and Poss, 2018), become polyploid during regeneration. Specifically, in zebrafish, epicardial cells at the injury border become polyploid and undergo hypertrophy in the process of regeneration (Cao et al., 2017). In mammals, polyploid hepatocytes contribute to liver regeneration (Pandit et al., 2012; Matsumoto et al., 2020). The cell cycle transcription factors E2F7 and E2F8 regulate hepatocyte polyploidization (Pandit et al., 2012), although whether they have a similar function in cardiomyocytes is unclear. The formation of polyploid cells is also a mechanism of tissue regeneration in Drosophila (Losick et al., 2013, 2016; Xiang et al., 2017; Cohen et al., 2018; Grendler et al., 2019), indicating that polyploid cells and their formation can also be part of regeneration.
Conclusions and perspectives
Failures in karyokinesis and cytokinesis appear to be regulated processes that can generate polyploid nuclei and multi-nucleated cardiomyocytes, respectively, via distinct molecular mechanisms. There are distinguishing features between karyokinesis and cytokinesis failure in cardiomyocytes vis-à-vis abnormal mitoses in cancer: the programmed nature, as opposed to dysregulated cancer cell mitosis; and the resulting decrease of the proliferative potential of the polyploid daughter cardiomyocytes, as opposed to the increased malignant potential in aneuploid cancer cells.
Differences in nucleation and ploidy raise questions about the molecular differences between diploid and polyploid cardiomyocytes at the transcriptional level. Although one report showed that all adult mouse cardiomyocytes have a largely uniform transcriptional profile (Yekelchyk et al., 2019), another group reported differences as a result of Ect2 gene inactivation-induced cytokinesis failure (Windmueller et al., 2020). A recently published study identified that the transcriptional profile of cardiomyocytes is independent of ploidy or nucleation but only differs in response to injury (Hesse et al., 2021). It is possible that many currently used techniques for single-cell transcriptional profiling do not capture the genome-wide transcriptome of single cells and thus do not have the sufficient depth to reveal all differences at the single cell level, highlighting that further studies are needed. Likewise, the functional differences between diploid and polyploid cardiomyocytes deserves future examination.
Knowledge of the mechanisms generating bi-nucleated cardiomyocytes and cardiomyocytes with polyploid nuclei could help to advance our understanding of ploidy changes in heart development and regeneration. Importantly, several issues relating to reported molecular interventions for stimulating cardiomyocyte regeneration in adult mice could be examined. For example, although it has been shown that neuregulin induces cardiomyocyte cell cycle entry, which promotes heart repair, this was associated with a 50% division and a 50% cytokinesis failure in adult mice (Bersell et al., 2009; Polizzotti et al., 2015); it would therefore be of interest to study how other interventions affect karyokinesis failure and cytokinesis failure. These include manipulation of Fstl1 (Wei et al., 2015), Meis1 (Mahmoud et al., 2013), the Hippo pathway (Heallen et al., 2011; Von Gise et al., 2012), agrin (Bassat et al., 2017), cyclin A2 (Shapiro et al., 2014), a combination of four transcription factors (Mohamed et al., 2018) and hypoxia (Kimura et al., 2015; Nakada et al., 2017). This new knowledge could also help advance the differentiation of embryonic stem cells and induced pluripotent stem cells (iPSCs) into terminally differentiated cardiomyocytes, e.g. by pushing them towards polyploidy. In this regard, a polyploidy-induced proliferative block in iPSC-derived cardiomyocytes could be a safety feature for transplantation, where unwanted proliferation of the transplanted cells needs to be avoided. Finally, a tantalizing opportunity for applying this new fundamental knowledge to improve human health exists in individuals with tetralogy of Fallot, the most common form of cyanotic congenital heart disease, in which the myocardial wall of the right ventricle is exposed to increased mechanical strain due to right ventricular hypertension (van der Ven et al., 2019). This increases β-AR signaling, which drives cytokinesis failure via of Ect2 gene repression, resulting in a predicted 30% reduction in the number of cardiomyocytes in the right ventricle. However, this could be prevented with β-blocker administration (Liu et al., 2019), highlighting the fundamental importance of understanding the basic mechanisms of cardiomyocyte terminal differentiation and how they relate to human health and disease.
Acknowledgements
We are thankful to members of the Kühn lab for critical discussions. We thank Michael Tsang and Andrew W. Duncan (University of Pittsburgh) and Vicki Losick (Boston College) for critical reading and commenting on this manuscript. We apologize to our colleagues whose work we could not discuss due to space limitations.
Footnotes
Competing interests
B.K. and H.L. are inventors on a patent application (PCT/US20/41808; 62/873,483) filed by the University Pittsburgh that covers the use of β-blockers for preventing increased cardiomyocyte cytokinesis failure in pediatric patients.
Funding
We acknowledge research support from the Richard King Mellon Foundation Institute for Pediatric Research (UPMC Children's Hospital of Pittsburgh), from a Transatlantic Network of Excellence grant from the Fondation Leducq (15CVD03) and from the National Heart, Lung, and Blood Institute (R01HL151386, R01HL155597 and R01HL151415 to B.K.). L.H. received support from the American Heart Association (18CDA34110053). A.K. received support from the National Institute of General Medical Sciences (T32GM008208). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Deposited in PMC for release after 12 months.
References
- Adler, C.-P., Friedburg, H., Herget, G. W., Neuburger, M. and Schwalb, H. (1996). Variability of cardiomyocyte DNA content, ploidy level and nuclear number in mammalian hearts. Virchows Arch. 429, 159-164. 10.1007/BF00192438 [DOI] [PubMed] [Google Scholar]
- Aix, E., Gutiérrez-Gutiérrez, Ó., Sánchez-Ferrer, C., Aguado, T. and Flores, I. (2016). Postnatal telomere dysfunction induces cardiomyocyte cell-cycle arrest through p21 activation. J. Cell Biol. 213, 571-583. 10.1083/jcb.201510091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, S. R., Hippenmeyer, S., Saadat, L. V., Luo, L., Weissman, I. L. and Ardehali, R. (2014). Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc. Natl. Acad. Sci. USA 111, 8850-8855. 10.1073/pnas.1408233111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, S. R., Menendez-Montes, I., Warshaw, J., Xiao, F. and Sadek, H. A. (2020). Homotypic fusion generates multinucleated cardiomyocytes in the murine heart. Circulation, 141, 1940-1942. 10.1161/CIRCULATIONAHA.119.043530 [DOI] [PubMed] [Google Scholar]
- Alkass, K., Panula, J., Westman, M., Wu, T.-D., Guerquin-Kern, J.-L. and Bergmann, O. (2015). No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 163, 1026-1036. 10.1016/j.cell.2015.10.035 [DOI] [PubMed] [Google Scholar]
- Bassat, E., Mutlak, Y. E., Genzelinakh, A., Shadrin, I. Y., Baruch Umansky, K., Yifa, O., Kain, D., Rajchman, D., Leach, J., Riabov Bassat, D.et al. (2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547 179-184. 10.1038/nature22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battinelli, E. M., Hartwig, J. H. and Italiano, J. E. (2007). Delivering new insight into the biology of megakaryopoiesis and thrombopoiesis. Curr. Opin Hematol. 14, 419-426. 10.1097/MOH.0b013e3282bad151 [DOI] [PubMed] [Google Scholar]
- Bergmann, O., Bhardwaj, R. D., Bernard, S., Zdunek, S., Barnabé-Heider, F., Walsh, S., Zupicich, J., Alkass, K., Buchholz, B. A., Druid, H.et al. (2009). Evidence for cardiomyocyte renewal in humans. Science 324, 98-102. 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, O., Zdunek, S., Felker, A., Salehpour, M., Alkass, K., Bernard, S., Sjostrom, S. L., Szewczykowska, M., Jackowska, T., dos Remedios, C.et al. (2015). Dynamics of cell generation and turnover in the human heart. Cell 161, 1566-1575. 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- Bersell, K., Arab, S., Haring, B. and Kühn, B. (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257-270. 10.1016/j.cell.2009.04.060 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., Mittnacht, S. and Brockes, J. P. (2003). Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. J. Cell Sci. 116, 4001-4009. 10.1242/jcs.00698 [DOI] [PubMed] [Google Scholar]
- Brodsky, V. Y., Arefyeva, A. M., Gvasava, I. G., Sarkisov, D. S. and Panova, N. W. (1994). Polyploidy in cardiac myocytes of normal and hypertrophic human hearts; range of values. Virchows Arch. 424, 429-435. 10.1007/BF00190566 [DOI] [PubMed] [Google Scholar]
- Bywater, M. J., Burkhart, D. L., Straube, J., Sabò, A., Pendino, V., Hudson, J. E., Quaife-Ryan, G. A., Porrello, E. R., Rae, J., Parton, R. G.et al. (2020). Reactivation of Myc transcription in the mouse heart unlocks its proliferative capacity. Nat. Commun. 11, 1827. 10.1038/s41467-020-15552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. and Poss, K. D. (2018). The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol. 15, 631-647. 10.1038/s41569-018-0046-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Wang, J., Jackman, C. P., Cox, A. H., Trembley, M. A., Balowski, J. J., Cox, B. D., De Simone, A., Dickson, A. L., Di Talia, S.et al. (2017). Tension creates an endoreplication wavefront that leads regeneration of epicardial tissue. Dev. Cell 42, 600-615.e4. 10.1016/j.devcel.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, D., Ojamaa, K., Parnell, V. A., Mcmahon, C., Sison, C. P. and Klein, I. (2001). A prospective randomized clinical study of thyroid hormone treatment after operations for complex congenital heart disease. J. Thoracic. Cardiovasc. Surg. 122, 1023-1025. 10.1067/mtc.2001.116192 [DOI] [PubMed] [Google Scholar]
- Cohen, E., Allen, S. R., Sawyer, J. K. and Fox, D. T. (2018). Fizzy-Related dictates A cell cycle switch during organ repair and tissue growth responses in the Drosophila hindgut. eLife, 7, e38327. 10.7554/eLife.38327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corash, L., Levin, J., Mok, Y., Baker, G. and McDowell, J. (1989). Measurement of megakaryocyte frequency and ploidy distribution in unfractionated murine bone marrow. Exp. Hematol. 17, 278-286. Available at: http://europepmc.org/abstract/MED/2465169. [PubMed] [Google Scholar]
- Crasta, K., Ganem, N. J., Dagher, R., Lantermann, A. B., Ivanova, E. V., Pan, Y., Nezi, L., Protopopov, A., Chowdhury, D. and Pellman, D. (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53-58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avino, P. P. (2009). How to scaffold the contractile ring for a safe cytokinesis - Lessons from Anillin-related proteins. J. Cell Sci. 122, 1071-1079. 10.1242/jcs.034785 [DOI] [PubMed] [Google Scholar]
- Donne, R., Saroul-Aïnama, M., Cordier, P., Celton-Morizur, S. and Desdouets, C. (2020). Polyploidy in liver development, homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 17, 391-405. 10.1038/s41575-020-0284-x [DOI] [PubMed] [Google Scholar]
- Duncan, A. W., Taylor, M. H., Hickey, R. D., Hanlon Newell, A. E., Lenzi, M. L., Olson, S. B., Finegold, M. J. and Grompe, M. (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707-710. 10.1038/nature09414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B. A., Zielke, N. and Gutierrez, C. (2014). Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15, 197-210. 10.1038/nrm3756 [DOI] [PubMed] [Google Scholar]
- Eggert, U. S., Mitchison, T. J. and Field, C. M. (2006). Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75, 543-566. 10.1146/annurev.biochem.74.082803.133425 [DOI] [PubMed] [Google Scholar]
- Engel, F. B., Schebesta, M. and Keating, M. T. (2006). Anillin localization defect in cardiomyocyte binucleation. J. Mol. Cell. Cardiol. 41, 601-612. 10.1016/j.yjmcc.2006.06.012 [DOI] [PubMed] [Google Scholar]
- Fededa, J. P. and Gerlich, D. W. (2012). Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14, 440-447. 10.1038/ncb2482 [DOI] [PubMed] [Google Scholar]
- Fields, A. P. and Justilien, V. (2010). The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv. Enzyme Regul. 50, 190-200. 10.1016/j.advenzreg.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, D. T., Soltis, D. E., Soltis, P. S., Ashman, T.-L. and Van De Peer, Y. (2020). Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol. 30, 688-694. 10.1016/j.tcb.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciosi, R. A. and Blanc, W. A. (1968). Myocardial infarcts in infants and children. I. A necropsy study in congenital heart disease. J. Pediatr. 73, 309-319. 10.1016/S0022-3476(68)80106-4 [DOI] [PubMed] [Google Scholar]
- Frenette, P., Haines, E., Loloyan, M., Kinal, M., Pakarian, P. and Piekny, A. (2012). An anillin-ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS ONE 7, e34888. 10.1371/journal.pone.0034888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabisonia, K., Prosdocimo, G., Aquaro, G. D., Carlucci, L., Zentilin, L., Secco, I., Ali, H., Braga, L., Gorgodze, N., Bernini, F.et al. (2019). MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 569, 418-422. 10.1038/s41586-019-1191-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, P., Patterson, M., Watanabe, H., Wang, K., Edmonds, R. A., Reinholdt, L. G. and Sucov, H. M. (2020). Allelic variants between mouse substrains BALB/cJ and BALB/cByJ influence mononuclear cardiomyocyte composition and cardiomyocyte nuclear ploidy. Sci. Rep. 10, 7605. 10.1038/s41598-020-64621-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N. J., Storchova, Z. and Pellman, D. (2007). Tetraploidy, aneuploidy and cancer. Curr. Opin Genet. Dev. 17, 157-162. 10.1016/j.gde.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Gao, Y., Smith, E., Ker, E., Campbell, P., Cheng, E.-c., Zou, S., Lin, S., Wang, L., Halene, S. and Krause, D. S. (2012). Role of RhoA-specific guanine exchange factors in regulation of endomitosis in megakaryocytes. Dev. Cell 22, 573-584. 10.1016/j.devcel.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric, G. and Desdouets, C. (2014). Polyploidization in liver tissue. Am. J. Pathol. 184, 322-331. 10.1016/j.ajpath.2013.06.035 [DOI] [PubMed] [Google Scholar]
- Gentric, G., Desdouets, C. and Celton-Morizur, S. (2012). Hepatocytes polyploidization and cell cycle control in liver physiopathology. Int. J. Hepatol. 2012, 282430. 10.1155/2012/282430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric, G., Maillet, V., Paradis, V., Couton, D., L'Hermitte, A., Panasyuk, G., Fromenty, B., Celton-Morizur, S. and Desdouets, C. (2015). Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J. Clin. Investig. 125, 981-992. 10.1172/JCI73957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Rosa, J. M., Sharpe, M., Field, D., Soonpaa, M. H., Field, L. J., Burns, C. E. and Burns, C. G. (2018). Myocardial polyploidization creates a barrier to heart regeneration in zebrafish. Dev. Cell 44, 433-446.e7. 10.1016/j.devcel.2018.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräbner, W. and Pfitzer, P. (1974). Number of nuclei in isolated myocardial cells of pigs. Virchows Archiv. B Cell Pathol. 15, 279-294. 10.1007/BF02889344 [DOI] [PubMed] [Google Scholar]
- Grendler, J., Lowgren, Sara, Mills, Monique and Losick, Vicki P. (2019). Wound-induced polyploidization is driven by Myc and supports tissue repair in the presence of DNA damage. Development 146, dev173005. 10.1242/dev.173005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti, J.-E., Brégerie, O., Robert, A., Debey, P., Brechot, C. and Desdouets, C. (2003). Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J. Biol. Chem. 278, 19095-19101. 10.1074/jbc.M300982200 [DOI] [PubMed] [Google Scholar]
- Han, L., Choudhury, S., Mich-Basso, J. D., Ammanamanchi, N., Ganapathy, B., Suresh, S., Khaladkar, M., Singh, J., Maehr, R., Zuppo, D. A.et al. (2020). Lamin B2 levels regulate polyploidization of cardiomyocyte nuclei and myocardial regeneration. Dev. Cell 53, 42-59.e11. 10.1016/j.devcel.2020.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, E. M., Fischer, A. H., Deerinck, T. J. and Hetzer, M. W. (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47-60. 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen, T., Zhang, M., Wang, J., Bonilla-Claudio, M., Klysik, E., Johnson, R. L. and Martin, J. F. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458-461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herget, G. W., Neuburger, M., Plagwitz, R. and Adler, C. P. (1997). DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovasc. Res. 36, 45-51. 10.1016/S0008-6363(97)00140-5 [DOI] [PubMed] [Google Scholar]
- Hesse, M., Doengi, M., Becker, A., Kimura, K., Voeltz, N., Stein, V. and Fleischmann, B. K. (2018). Midbody positioning and distance between daughter nuclei enable unequivocal identification of cardiomyocyte cell division in mice. Circ. Res. 123, 1039-1052. 10.1161/CIRCRESAHA.118.312792 [DOI] [PubMed] [Google Scholar]
- Hesse, M., Bednarz, R., Carls, E., Becker, C., Bondareva, O., Lother, A., Geisen, C., Dreßen, M., Krane, M., Roell, W.et al. (2021). Proximity to injury, but neither number of nuclei nor ploidy define pathological adaptation and plasticity in cardiomyocytes. J. Mol. Cell. Cardiol. 152, 95-104. 10.1016/j.yjmcc.2020.11.012 [DOI] [PubMed] [Google Scholar]
- Hirata, D., Yamabuki, T., Miki, D., Ito, T., Tsuchiya, E., Fujita, M., Hosokawa, M., Chayama, K., Nakamura, Y. and Daigo, Y. (2009). Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal Cancer progression. Clin. Cancer Res. 15, 256-266. 10.1158/1078-0432.CCR-08-1672 [DOI] [PubMed] [Google Scholar]
- Hirose, K., Payumo, A. Y., Cutie, S., Hoang, A., Zhang, H., Guyot, R., Lunn, D., Bigley, R. B., Yu, H., Wang, J.et al. (2019). Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364, 184-188. 10.1126/science.aar2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, S.-H., Delgado, E. R., Otero, P. A., Teng, K.-y., Kutay, H., Meehan, K. M., Moroney, J. B., Monga, J. K., Hand, N. J., Friedman, J. R.et al. (2016). MicroRNA-122 regulates polyploidization in the murine liver. Hepatology 64, 599-615. 10.1002/hep.28573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, N., Yost, H. J. and Clark, E. B. (2001). Cardiac morphology and blood pressure in the adult zebrafish. Anat. Rec. 264, 1-12. 10.1002/ar.1111 [DOI] [PubMed] [Google Scholar]
- Ito, K., Morioka, M., Kimura, S., Tasaki, M., Inohaya, K. and Kudo, A. (2014). Differential reparative phenotypes between zebrafish and medaka after cardiac injury. Dev. Dyn. 243, 1106-1115. 10.1002/dvdy.24154 [DOI] [PubMed] [Google Scholar]
- Junk, D. J., Bryson, B. L., Smigiel, J. M., Parameswaran, N., Bartel, C. A. and Jackson, M. W. (2017). Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 36, 4001-4013. 10.1038/onc.2017.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien, V. and Fields, A. P. (2009). Ect2 links the PKCι–Par6α complex to Rac1 activation and cellular transformation. Oncogene 28, 3597-3607. 10.1038/onc.2009.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, W., Xiao, F., Canseco, D. C., Muralidhar, S., Thet, S. W., Zhang, H. M., Abderrahman, Y., Chen, R., Garcia, J. A., Shelton, J. M.et al. (2015). Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature 523, 226-230. 10.1038/nature14582 [DOI] [PubMed] [Google Scholar]
- Kirklin, J. W., Blackstone, E. H., Tchervenkov, C. I. and Castaneda, A. R. (1992). Clinical outcomes after the arterial switch operation for transposition. Patient, support, procedural, and institutional risk factors. Congenital heart surgeons society. Circulation 86, 1501-1515. 10.1161/01.CIR.86.5.1501 [DOI] [PubMed] [Google Scholar]
- Kuga, T., Nie, H., Kazami, T., Satoh, M., Matsushita, K., Nomura, F., Maeshima, K., Nakayama, Y. and Tomonaga, T. (2014). Lamin B2 prevents chromosome instability by ensuring proper mitotic chromosome segregation. Oncogenesis 3, e94. 10.1038/oncsis.2014.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, S.-L., Marín-Juez, R., Moura, P. L., Kuenne, C., Lai, J. K. H., Tsedeke, A. T., Guenther, S., Looso, M. and Stainier, D. Y. R. (2017). Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. eLife 6, e25605. 10.7554/eLife.25605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone, M. and Engel, F. B. (2019). Pseudo-bipolar spindle formation and cell division in postnatal binucleated cardiomyocytes. J. Mol. Cell. Cardiol. 134, 69-73. 10.1016/j.yjmcc.2019.07.005 [DOI] [PubMed] [Google Scholar]
- Leone, M., Musa, G. and Engel, F. B. (2018). Cardiomyocyte binucleation is associated with aberrant mitotic microtubule distribution, mislocalization of RhoA and IQGAP3, as well as defective actomyosin ring anchorage and cleavage furrow ingression. Cardiovasc. Res. 114, 1115-1131. 10.1093/cvr/cvy056 [DOI] [PubMed] [Google Scholar]
- Levkau, B., Schäfers, M., Wohlschlaeger, J., Von Wnuck Lipinski, K., Keul, P., Hermann, S., Kawaguchi, N., Kirchhof, P., Fabritz, L., Stypmann, J.et al. (2008). Survivin determines cardiac function by controlling total cardiomyocyte number. Circulation 117, 1583-1593. 10.1161/CIRCULATIONAHA.107.734160 [DOI] [PubMed] [Google Scholar]
- Liu, H., Zhang, C.-H., Ammanamanchi, N., Suresh, S., Lewarchik, C., Rao, K., Uys, G. M., Han, L., Abrial, M., Yimlamai, D.et al. (2019). Control of cytokinesis by β-adrenergic receptors indicates an approach for regulating cardiomyocyte endowment. Sci. Transl. Med. 11, eaaw6419. 10.1126/scitranslmed.aaw6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lordier, L., Bluteau, D., Jalil, A., Legrand, C., Pan, J., Rameau, P., Jouni, D., Bluteau, O., Mercher, T., Leon, C.et al. (2012). RUNX1-induced silencing of non-muscle myosin heavy chain IIB contributes to megakaryocyte polyploidization. Nat. Commun. 3, 717. 10.1038/ncomms1704 [DOI] [PubMed] [Google Scholar]
- Losick, V. P., Fox, D. T. and Spradling, A. C. (2013). Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr. Biol. 23, 2224-2232. 10.1016/j.cub.2013.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick, V. P., Jun, A. S. and Spradling, A. C. (2016). Wound-induced polyploidization: regulation by hippo and JNK signaling and conservation in Mammals. PLoS ONE 11, e0151251. 10.1371/journal.pone.0151251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, Y. J. and Dutta, A. (2007). The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 21, 184-194. 10.1101/gad.1495007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlus, K. R. and Italiano, J. E. (2013). The incredible journey: from megakaryocyte development to platelet formation. J. Cell Biol. 201, 785-796. 10.1083/jcb.201304054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud, A. I., Kocabas, F., Muralidhar, S. A., Kimura, W., Koura, A. S., Thet, S., Porrello, E. R. and Sadek, H. A. (2013). Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature, 497, 249-253. 10.1038/nature12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone, G., Tucker, C. S. and Denvir, M. A. (2017). Cardiomyocyte proliferation in zebrafish and mammals: lessons for human disease. Cell. Mol. Life Sci. 74, 1367-1378. 10.1007/s00018-016-2404-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, T., Wakefield, L., Tarlow, B. D. and Grompe, M. (2020). In vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell 26, 34-47.e3. 10.1016/j.stem.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, K., Wada, H., Nagai, T., Iijima, Y., Minamino, T., Sano, M., Akazawa, H., Molkentin, J. D., Kasanuki, H. and Komuro, I. (2004). Cardiomyocytes fuse with surrounding noncardiomyocytes and reenter the cell cycle. J. Cell Biol. 167, 351-363. 10.1083/jcb.200312111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzi, S., Lordier, L., Debili, N., Raslova, H. and Vainchenker, W. (2018). Megakaryocyte and polyploidization. Exp. Hematol. 57, 1-13. 10.1016/j.exphem.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Meckert, P. C., Rivello, H., Vigliano, C., Gonzalez, P., Favaloro, R. and Laguens, R. (2005). Endomitosis and polyploidization of myocardial cells in the periphery of human acute myocardial infarction. Cardiovasc. Res. 67, 116-123. 10.1016/j.cardiores.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Mohamed, T. M. A., Ang, Y.-S., Radzinsky, E., Zhou, P., Huang, Y., Elfenbein, A., Foley, A., Magnitsky, S. and Srivastava, D. (2018). Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104-116.e12. 10.1016/j.cell.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova, M., Bersell, K., Walsh, S., Savla, J., Das, L. T., Park, S.-Y., Silberstein, L. E., Dos Remedios, C. G., Graham, D., Colan, S.et al. (2013). Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA 110, 1446-1451. 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourouzis, I., Politi, E. and Pantos, C. (2013). Thyroid hormone and tissue repair: new tricks for an old hormone ? J. Thyroid. Res. 2013, 312104. 10.1155/2013/312104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, Y., Canseco, D. C., Thet, S. W., Abdisalaam, S., Asaithamby, A., Santos, C. X., Shah, A. M., Zhang, H., Faber, J. E., Kinter, M. T.et al. (2017). Hypoxia induces heart regeneration in adult mice. Nature 541, 222-227. 10.1038/nature20173 [DOI] [PubMed] [Google Scholar]
- Naqvi, N., Li, M., Calvert, J. W., Tejada, T., Lambert, J. P., Wu, J., Kesteven, S. H., Holman, S. R., Matsuda, T., Lovelock, J. D.et al. (2014). A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell 157, 795-807. 10.1016/j.cell.2014.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Savoian, M. S., Mitchison, T. J. and Field, C. M. (2000). Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 150, 539-552. 10.1083/jcb.150.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard, K. H., Baandrup, U. T., Wang, T., Bertelsen, M. F., Andersen, J. B., Smerup, M. and Nyengaard, J. R. (2013). Left ventricular morphology of the giraffe heart examined by stereological methods. Anat. Rec. 296, 611-621. 10.1002/ar.22672 [DOI] [PubMed] [Google Scholar]
- Øvrebø, J. I. and Edgar, B. A. (2018). Polyploidy in tissue homeostasis and regeneration. Development 145(14), dev156034. 10.1242/dev.156034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, S. K., Westendorp, B., Nantasanti, S., Van Liere, E., Tooten, P. C. J., Cornelissen, P. W. A., Toussaint, M. J. M., Lamers, W. H. and De Bruin, A. (2012). E2F8 is essential for polyploidization in mammalian cells. Nat. Cell Biol. 14, 1181-1191. 10.1038/ncb2585 [DOI] [PubMed] [Google Scholar]
- Pandit, S. K., Westendorp, B. and De Bruin, A. (2013). Physiological significance of polyploidization in mammalian cells. Trends Cell Biol. 23, 556-566. 10.1016/j.tcb.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Pasumarthi, K. B. S. and Field, L. J. (2002). Cardiomyocyte cell cycle regulation. Circ. Res. 90, 1044-1054. 10.1161/01.RES.0000020201.44772.67 [DOI] [PubMed] [Google Scholar]
- Patterson, M., Barske, L., Van Handel, B., Rau, C. D., Gan, P., Sharma, A., Parikh, S., Denholtz, M., Huang, Y., Yamaguchi, Y.et al. (2017). Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 49, 1346-1353 10.1038/ng.3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska, P., Fouchet, P., Vainchenker, W., Rosselli, F. and Naim, V. (2014). Defective endomitosis during megakaryopoiesis leads to thrombocytopenia in Fanca−/− mice. Blood 124, 3613-3623. 10.1182/blood-2014-01-551457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny, A., Werner, M. and Glotzer, M. (2005). Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 15, 651-658. 10.1016/j.tcb.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Pingitore, A., Mastorci, F., Piaggi, P., Aquaro, G. D., Molinaro, S., Ravani, M., De Caterina, A., Trianni, G., Ndreu, R., Berti, S.et al. (2019). Usefulness of Triiodothyronine replacement therapy in patients with ST elevation myocardial infarction and borderline/reduced Triiodothyronine Levels (from the THIRST study). Am. J. Cardiol. 123, 905-912. 10.1016/j.amjcard.2018.12.020 [DOI] [PubMed] [Google Scholar]
- Polizzotti, B. D., Ganapathy, B., Walsh, S., Choudhury, S., Ammanamanchi, N., Bennett, D. G., Dos Remedios, C. G., Haubner, B. J., Penninger, J. M. and Kühn, B. (2015). Neuregulin stimulation of cardiomyocyte regeneration in mice and human myocardium reveals a therapeutic window. Sci. Transl. Med. 7, 281ra45. 10.1126/scitranslmed.aaa5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss, K. D., Wilson, L. G. and Keating, M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Reinecke, H., Minami, E., Poppa, V. and Murry, C. E. (2004). Evidence for fusion between cardiac and skeletal muscle cells. Circ. Res. 94, e56-e60. 10.1161/01.RES.0000125294.04612.81 [DOI] [PubMed] [Google Scholar]
- Sadek, H. and Olson, E. N. (2020). Toward the goal of human heart regeneration. Cell Stem Cell, 26, 7-16. 10.1016/j.stem.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhia, B., Tran, N. L., Chan, A., Wolf, A., Nakada, M., Rutka, F., Ennis, M., Mcdonough, W. S., Berens, M. E., Symons, M.et al. (2008). The guanine nucleotide exchange factors Trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am. J. Pathol. 173, 1828-1838. 10.2353/ajpath.2008.080043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamiphak, S., Kontarakis, Z., Filosa, A., Reischauer, S. and Stainier, D. Y. R. (2017). Transient cardiomyocyte fusion regulates cardiac development in zebrafish. Nat. Commun. 8(1), 1525. 10.1038/s41467-017-01555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo, S. E., Steinhauser, M. L., Pizzimenti, C. L., Yang, V. K., Cai, L., Wang, M., Wu, T.-D., Guerquin-Kern, J.-L., Lechene, C. P. and Lee, R. T. (2013). Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433-436. 10.1038/nature11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, S. D., Ranjan, A. K., Kawase, Y., Cheng, R. K., Kara, R. J., Bhattacharya, R., Guzman-Martinez, G., Sanz, J., Garcia, M. J. and Chaudhry, H. W. (2014). Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci. Transl. Med. 6, 224ra27. 10.1126/scitranslmed.3007668 [DOI] [PubMed] [Google Scholar]
- Soonpaa, M. H. and Field, L. J. (1997). Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am. J. Physiol. Heart Circ. Physiol. 272, H220-H226. 10.1152/ajpheart.1997.272.1.h220 [DOI] [PubMed] [Google Scholar]
- Soonpaa, M. H., Kim, K. K., Pajak, L., Franklin, M. and Field, L. J. (1996). Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiology Heart Circ. Physiol. 271, H2183-H2189. 10.1152/ajpheart.1996.271.5.h2183 [DOI] [PubMed] [Google Scholar]
- Soygur, B. and Sati, L. (2016). The role of syncytins in human reproduction and reproductive organ cancers. Reproduction 152, R167-R178. 10.1530/REP-16-0031 [DOI] [PubMed] [Google Scholar]
- Straight, A. F., Field, C. M. and Mitchison, T. J. (2005). Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell 16, 193-201. 10.1091/mbc.e04-08-0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumoto, T., Xie, X., Blumenthal, R., Okamoto, I. and Miki, T. (1999). Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 147, 921-928. 10.1083/jcb.147.5.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor, E. and Poss, K. D. (2017). Cardiac regeneration strategies: staying young at heart. Science 356, 1035-1039. 10.1126/science.aam5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Peer, Y., Mizrachi, E. and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411-424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Van Der Ven, J. P. G., Van Den Bosch, E., Bogers, A. J. C. C. and Helbing, W. A. (2019). Current outcomes and treatment of tetralogy of fallot. F1000Research 8, 1530. 10.12688/f1000research.17174.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayutham, N., Alfieri, C. M., Agnew, E. J., Riggs, K. W., Baker, R. S., Ponny, S. R., Zafar, F. and Yutzey, K. E. (2020). Cardiomyocyte cell cycling, maturation, and growth by multinucleation in postnatal swine. J. Mol. Cell. Cardiol. 146 95-108. 10.1016/j.yjmcc.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale, I., Galluzzi, L., Castedo, M. and Kroemer, G. (2011). Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12, 385-392. 10.1038/nrm3115 [DOI] [PubMed] [Google Scholar]
- Vliegen, H. W., Eulderink, F., Bruschke, A. V., van der Laarse, A. and Cornelisse, C. J. (1995). Polyploidy of myocyte nuclei in pressure overloaded human hearts: a flow cytometric study in left and right ventricular myocardium. Am. J. Cardiovasc. Pathol. 5, 27-31 [PubMed] [Google Scholar]
- Von Dassow, G. (2009). Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 19, 165-173. 10.1016/j.tcb.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Von Gise, A., Lin, Z., Schlegelmilch, K., Honor, L. B., Pan, G. M., Buck, J. N., Ma, Q., Ishiwata, T., Zhou, B., Camargo, F. D.et al. (2012). YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 109, 2394-2399. 10.1073/pnas.1116136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hafe, M., Neves, J. S., Vale, C., Borges-Canha, M. and Leite-Moreira, A. (2019). The impact of thyroid hormone dysfunction on ischemic heart disease. Endocr. Connections 8, R76-R90. 10.1530/EC-19-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, S., Pontén, A., Fleischmann, B. K. and Jovinge, S. (2010). Cardiomyocyte cell cycle control and growth estimation in vivo—an analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 86, 365-373. 10.1093/cvr/cvq005 [DOI] [PubMed] [Google Scholar]
- Wang, M.-J., Chen, F., Lau, J. T. Y. and Hu, Y.-P. (2017). Hepatocyte polyploidization and its association with pathophysiological processes. Cell Death Dis. 8, e2805. 10.1038/cddis.2017.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, K., Serpooshan, V., Hurtado, C., Diez-Cuñado, M., Zhao, M., Maruyama, S., Zhu, W., Fajardo, G., Noseda, M., Nakamura, K.et al. (2015). Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 525, 479-485. 10.1038/nature15372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier, J. F., Weier, H.-U. G., Jung, C. J., Gormley, M., Zhou, Y., Chu, L. W., Genbacev, O., Wright, A. A. and Fisher, S. J. (2005). Human cytotrophoblasts acquire aneuploidies as they differentiate to an invasive phenotype. Dev. Biol. 279, 420-432. 10.1016/j.ydbio.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Wills, A. A., Holdway, J. E., Major, R. J. and Poss, K. D. (2008). Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 135, 183-192. 10.1242/dev.010363 [DOI] [PubMed] [Google Scholar]
- Windmueller, R., Leach, J. P., Babu, A., Zhou, S., Morley, M. P., Wakabayashi, A., Petrenko, N. B., Viatour, P. and Morrisey, E. E. (2020). Direct comparison of mononucleated and binucleated cardiomyocytes reveals molecular mechanisms underlying distinct proliferative competencies. Cell Rep. 30, 3105-3116.e4. 10.1016/j.celrep.2020.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.-C., Jeratsch, S., Graumann, J. and Stainier, D. Y. R. (2020). Modulation of mammalian cardiomyocyte cytokinesis by the extracellular matrix. Circ. Res. 127, 896-907. 10.1161/CIRCRESAHA.119.316303 [DOI] [PubMed] [Google Scholar]
- Xiang, J., Bandura, J., Zhang, P., Jin, Y., Reuter, H. and Edgar, B. A. (2017). EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nat. Commun. 8, 15125. 10.1038/ncomms15125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, G., Mao, S., Baumgarten, G., Serrano, J., Jordan, M. C., Roos, K. P., Fishbein, M. C. and Maclellan, W. R. (2001). Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ. Res. 89, 1122-1129. 10.1161/hh2401.100742 [DOI] [PubMed] [Google Scholar]
- Yekelchyk, M., Guenther, S., Preussner, J. and Braun, T. (2019). Mono- and multi-nucleated ventricular cardiomyocytes constitute a transcriptionally homogenous cell population. Basic Res. Cardiol. 114, 36. 10.1007/s00395-019-0744-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.-X., Zhao, B., Panupinthu, N., Jewell, J. L., Lian, I., Wang, L. H., Zhao, J., Yuan, H., Tumaneng, K., Li, H.et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780-791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce, Ö., Piekny, A. and Glotzer, M. (2005). An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170, 571-582. 10.1083/jcb.200501097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Ahmad, F., Parikh, S., Hoffman, N. E., Rajan, S., Verma, V. K., Song, J., Yuan, A., Shanmughapriya, S., Guo, Y.et al. (2016). Loss of adult cardiac myocyte GSK-3 leads to mitotic catastrophe resulting in fatal dilated cardiomyopathy. Circ. Res. 118, 1208-1222. 10.1161/CIRCRESAHA.116.308544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybina, T. G. and Zybina, E. V. (2005). Cell reproduction and genome multiplication in the proliferative and invasive trophoblast cell populations of mammalian placenta. Cell Biol. Int. 29, 1071-1083. 10.1016/j.cellbi.2005.10.015 [DOI] [PubMed] [Google Scholar]
- Zybina, T. G. and Zybina, E. V. (2020). Role of cell cycling and polyploidy in placental trophoblast of different mammalian species. Reprod. Domest. Anim. 55, 895-904. 10.1111/rda.13732 [DOI] [PubMed] [Google Scholar]