Abstract
Genomic instability is a key hallmark of cancer that arises owing to defects in the DNA damage response (DDR) and/or increased replication stress. These alterations promote the clonal evolution of cancer cells via the accumulation of driver aberrations, including gene copy-number changes, rearrangements and mutations; however, these same defects also create vulnerabilities that are relatively specific to cancer cells, which could potentially be exploited to increase the therapeutic index of anticancer treatments and thereby improve patient outcomes. The discovery that BRCA-mutant cancer cells are exquisitely sensitive to inhibition of poly(ADP-ribose) polymerase has ushered in a new era of research on biomarker-driven synthetic lethal treatment strategies for different cancers. The therapeutic landscape of antitumour agents targeting the DDR has rapidly expanded to include inhibitors of other key mediators of DNA repair and replication, such as ATM, ATR, CHK1 and CHK2, DNA-PK and WEE1. Efforts to optimize these therapies are ongoing across a range of cancers, involving the development of predictive biomarker assays of responsiveness (beyond BRCA mutations), assessment of the mechanisms underlying intrinsic and acquired resistance, and evaluation of rational, tolerable combinations with standard-of-care treatments (such as chemotherapeutics and radiation), novel molecularly targeted agents and immune-checkpoint inhibitors. In this Review, we discuss the current status of anticancer therapies targeting the DDR.
DNA damage occurs constantly in cells owing to exogenous and endogenous stressors, and cells have consequently evolved a complex, coordinated DNA damage response (DDR) that encompasses numerous interdependent signalling pathways and machineries. The importance of the DDR in maintaining cell viability and preventing neoplasia is underscored by the additional integral roles of these pathways in regulating the cell cycle, chromatin remodelling, metabolism, immunogenicity and apoptosis1,2. For example, the detection of DNA damage results in the activation of checkpoints that enforce cell cycle arrest to provide the time necessary for DNA repair before cell division; DDR pathways are also closely linked with the apoptotic machinery to enable the elimination of cells with unrepaired DNA damage. Thus, the DDR pathways ultimately enable cell survival in the face of genomic instability and replicative stress, or direct irreparably damaged cells to undergo senescence or programmed death. Genomic instability is a key hallmark of cancer3 and arises as a result of the high rate of cell division and the related rapid accumulation of aberrations on a background of the compromised DDR processes that contribute to cancer initiation and progression. Hence, defects in DDR genes have multiple roles in the promotion of cancer cell growth via accrual of driver mutations, generation of tumour heterogeneity and evasion of apoptosis4. Cells are programmed to constitutively respond to DNA damage, whereby the repair pathways used are dependent on the specific type of damage detected and repair machineries available (FIG. 1). DNA damage most commonly manifests as single-strand breaks (SSBs), although double-strand breaks (DSB) are more lethal to cells and require rapid countermeasures to ensure cell survival. Thus, most contemporary DDR-directed therapies target the signalling and repair mechanisms associated with DSBs, increase replication stress and thereby the frequency of DSBs, or inhibit cell cycle checkpoints that facilitate DSB repair (FIG. 1). Defects in certain high-fidelity DDR machineries, including DSB repair processes involving homologous recombination (HR), increase genomic instability and lead to a greater reliance on compensatory — and often error-prone — DDR and survival pathways2,5,6. These vulnerabilities have been exploited in anticancer therapy through the use of DNA-damaging radiation and chemotherapies and, more recently, with the rapid development of potent and selective molecularly targeted agents against key components of different DDR pathways (herein termed DDR inhibitors) (FIG. 1). However, the development of analytically and clinically validated assays to robustly assess predictive biomarkers of response and/or resistance to DDR inhibitors has lagged behind. In this Review, we provide an overview of the current landscape of DDR-directed therapies, focusing on the respective DDR pathways and replication stress responses pertaining to the most promising targets and emerging therapeutics. We also discuss putative predictive biomarkers of response, mechanisms of resistance and ongoing preclinical and clinical efforts to develop combinatorial strategies to optimize therapeutic targeting of the DDR.
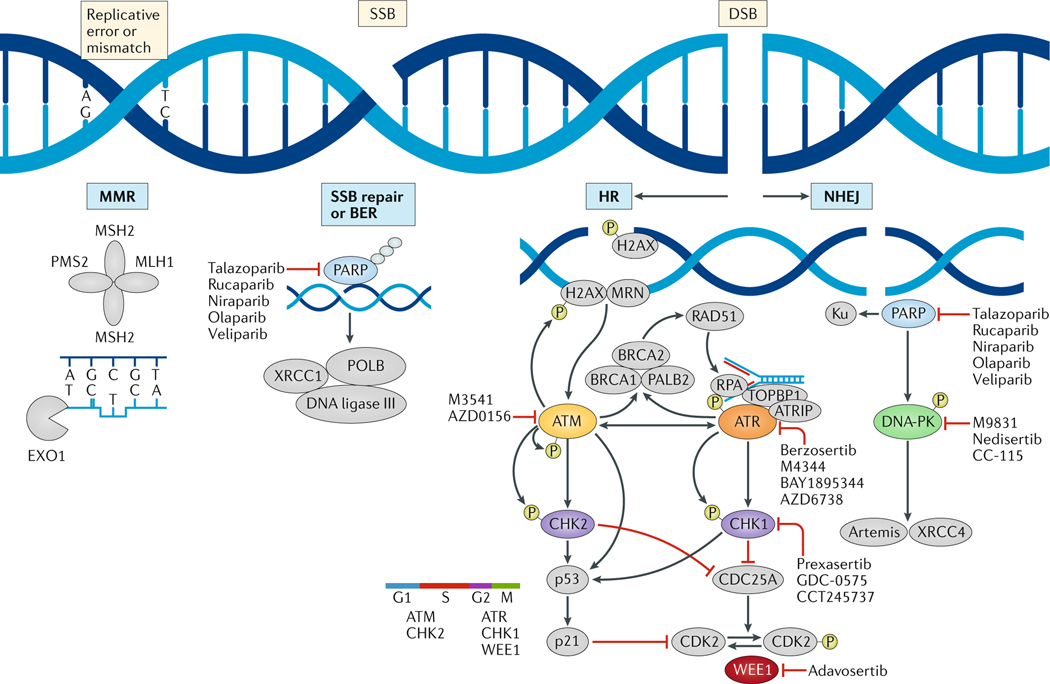
FIG. 1 |. DNA damage response pathways being targeted in the clinic.
Specific types of DNA damage — mismatches due to replication, single-strand DNA breaks (SSBs) or double-strand DNA breaks (DSBs) — result in the activation of specific signalling and repair cascades. DNA damage response (DDR) pathways mitigate replication stress and repair DNA; thus, deficiencies in these pathways result in the accumulation of SSBs and DSBs and increased immunogenicity owing to the generation of neoantigens from mutant proteins. Poly(ADP-ribose) polymerase (PARP) enzymes are key to activating a host of downstream repair mechanisms and are primary proteins involved in SSB repair or base-excision repair (BER). The repair of DSBs occurs predominately through the rapid, error-prone non-homologous end joining (NHEJ) repair pathway in conjunction with the much slower higher-fidelity, error-free homologous recombination (HR) repair pathway. DNA replication is a necessary component of DNA repair and thus cell cycle regulation and replication stress responses are intertwined with DDR pathways. The kinases ATR and ATM have crucial roles in DDR signalling and in maintaining replication fork stability, while also working together via their downstream targets, CHK1 and CHK2, respectively, to regulate cell cycle control checkpoints. The kinase activity of DNA-PK is essential for NHEJ and V(D)J recombination. WEE1 is a distinct nuclear kinase that regulates mitotic entry and nucleotide pools in coordination with DDR. Drugs targeting these key components of the DDR pathways that are undergoing clinical testing are indicated. ATRIP, ATR-interacting protein; EXO1, exonuclease 1; H2AX, histone H2AX; MRN, MRE11, RAD50 and NBS1 complex; POLB, DNA polymerase-β; RPA, replication protein A; TOPBP1, DNA topoisomerase 2-binding protein.
PARP inhibitors
Similarities and differences
The poly(ADP-ribose) polymerase (PARP) family comprises a group of nuclear proteins that are activated upon binding to damaged DNA and have crucial roles in various aspects of the DDR (FIG. 1). The main function of these proteins is to detect SSBs and DSBs, recruit the DNA repair machinery and stabilize replication forks during repair7. The rationale for the antitumour activity of single-agent PARP inhibitors in selected HR-deficient (HRD) tumours, initially those with germline BRCA1 or BRCA2 (BRCA1/2) mutations, is based on the concept of synthetic lethality, whereby the combination of a functional genetic defect in an HR-related gene and pharmacological inhibition of a compensatory DDR-pathway component, such as PARP, leads to insurmountable genomic instability, mitotic catastrophe and cell death7–12. PARP inhibitors are the best-studied class of DDR inhibitors, with robust preclinical and clinical data informing refinements in patient selection and treatment protocols (FIG. 2). With several PARP inhibitors already FDA approved or undergoing testing in late phase clinical trials as single agents in various disease and treatment settings (TABLE 1; Supplementary Table 1), investigators must be cognizant of the mechanisms underlying the anticancer activity of drugs within this class, including any pharmacological similarities and differences, and the respective clinical outcomes and toxicities reported to date. Importantly, a detailed understanding of the toxicities and how to mitigate them will be necessary to optimize the therapeutic index.
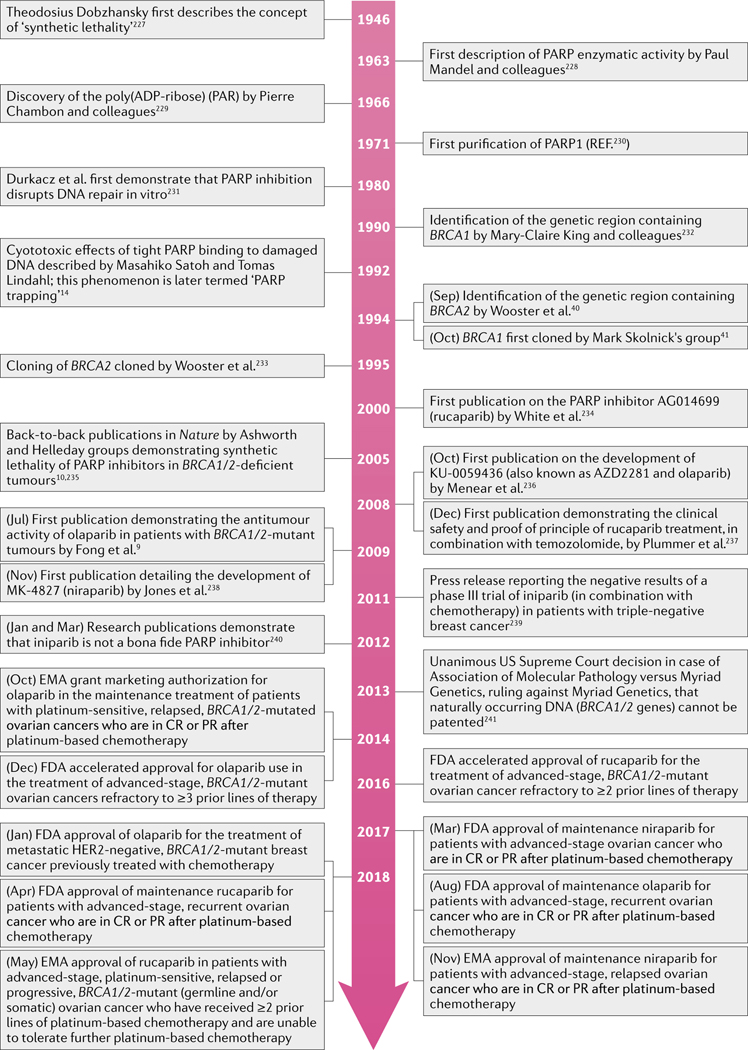
FIG. 2 |. Timeline of key events leading to FDA approvals of PARP inhibitors in cancer medicine.
Landmark discoveries and advances in the development of poly(ADP-ribose) polymerase (PARP) inhibitors are indicated10,14,40,41,227–235, together with the current approved indications for these agents in the USA and the EU. CR, complete remission; PR, partial remission.
Table 1 |.
Registration clinical trials of PARP inhibitors
| Study | Phase | Disease setting | Treatments | Most common grade ≥3 AEs | Efficacy | Reason for notability |
|---|---|---|---|---|---|---|
| Study 19; Ledermann et al. (2012 and 2014)33,34 | II (randomized) | Recurrent ovarian cancer with a PR or CR to most recent line of platinum-based chemotherapy (after ≥2 lines chemotherapy) | Olaparib vs placebo | Fatigue 7% vs 3%; anaemia 5% vs 1%; and diarrhoea 2% vs 2% | • Median PFS 8.4 mo vs 4.8 mo (HR 0.35, 95% CI 0.25–0.49; P < 0.001) • BRCA1/2-mutant: median PFS 11.2 mo vs 4.3 mo (HR 0.18, 95% CI 0.10–0.31; P < 0.0001) • BRCA1/2-wild-type: median PFS 7.4 mo vs 5.5 mo (HR 0.54, 95% CI 0.34–0.85; P = 0.0075) |
• Led to the first approval of a PARP inhibitor, by the EMA in 2014, as a maintenance treatment of patients with relapsed, BRCA1/2-mutated ovarian cancers who are in CR or PR after platinum-based chemotherapy • In conjunction with preliminary results from SOLO2, led to 2017 FDA approval of olaparib maintenance therapy in women with advanced-stage ovarian cancer in CR or PR after platinum-based chemotherapy (irrespective of BRCA1/2 status) |
| Kaufman et al. (2015)11 | II | Advanced-stage and/or recurrent solid tumours with germline BRCA1/2 mutations | Olaparib | Anaemia 17%, fatigue 6% and abdominal pain 6% | • Overall: ORR 78/298 (26.2%) • Ovarian cancers: ORR 60/193 (31.1%) • Breast cancers: ORR 8/62 (12.9%) • Pancreatic cancers: ORR 5/23 (21.7%) • Prostate cancers: ORR 4/8 (50.0%) |
Led to 2014 FDA approval of olaparib for refractory BRCA1/2-mutant ovarian cancer after ≥3 prior lines of chemotherapy |
| ARIEL2 part 1; Swisher et al. (2017)18 | II (non-randomized) | BRCA1/2-mutant, BRCA1/2-wild-type and LOH-high, or BRCA1/2-wild-type and LOH-low recurrent ovarian cancera | Rucaparib | Anaemia 22%, serum ALT and/or AST elevations 12% and fatigue 9% |
• BRCA1/2-mutant: ORR 32/40 (80%); median PFS 12.8 mo • BRCA1/2-wild-type, LOH-high: ORR 24/82 (29%); median PFS 5.7 mo • BRCA1/2-wild-type, LOH-low: ORR 7/70 (10%); median PFS 5.2 mo |
Preliminary results in conjunction with data from Study 10 (REF.39) led to the 2016 FDA and 2017 EMA approvalsb of rucaparib in for BRCA1/2-mutant ovarian cancer refractory to ≥2 prior lines of treatment |
| Study 10; Kristeleit et al. (2016)39 |
I/II | Phase I: advanced-stage ovarian cancer Phase II: germline BRCA1/2-mutant ovarian cancer | • Phase I: escalating rucaparib doses • Phase II: rucaparib 600 mg BID |
Not disclosed separately from ARIEL2 | Phase II: ORR 25/42 (60%) | In conjunction with preliminary results of ARIEL2 (REF.18), led to the 2016 FDA and 2017 EMA approvalsb of rucaparib for BRCA1/2-mutant ovarian cancer refractory to ≥2 prior lines of treatment |
| NOVA; Mirza et al. (2016)37 | III | Platinum-sensitive, recurrent ovarian cancer stratified into two subgroups: germline BRCA1/2 mutant and BRCA1/2 wild type | Niraparib vs placebo | Thrombocytopenia 34% vs 1%; anaemia 25% vs 0%; neutropenia 20% vs 2% | • BRCA1/2-mutant: median 34% vs 1%; PFS 21.0 mo vs 5.5 mo (HR 0.27, 95% CI 0.17–0.41; P < 0.001) • BRCA1/2-wild-type: median PFS 9.3 mo vs3.9 mo (HR 0.45, 95% CI 0.34–0.61; P < 0.001); 12.9 mo vs 3.8 mo in those with HRD (HR 0.38, 95% CI 0.24–0.59; P < 0.001) |
Led to 2017 FDA and EMA approvals of niraparib maintenance therapy for patients with advanced-stage ovarian cancer who are in CR or PR after platinum-based chemotherapy |
| SOLO2/ENGOT-Ov21; Pujade-Lauraine et al. (2017)36 | III | Recurrent BRCA1/2-mutant ovarian cancer with PR or CR to most recent line of platinum-based chemotherapy (after ≥2 lines of chemotherapy) | Olaparib vs placebo | Anaemia 19% vs 2%; fatigue 4% vs 2%; and neutropenia 5% vs 4% | Median PFS 19.1 mo vs 5.5 mo (HR 0.30, 95% CI 0.22–0.41; P > 0.0001) | In conjunction with results from Study 19, led to 2017 FDA approval of olaparib maintenance therapy for patients with advanced-stage ovarian cancer in CR or PR after platinum-based chemotherapy (irrespective of BRCA1/2 status) |
| ARIEL3; Coleman et al. (2017)38 | III | Recurrent ovarian cancer with PR or CR to most recent line of platinum-based chemotherapy (after ≥2 lines chemotherapy) | Rucaparib vs placebo | Anaemia 19% vs 1%; and increased serum ALT and/or AST levels 10% vs 0% | • Median PFS 10.8 mo vs 5.4 mo (HR 0.36, 95% CI 0.30–0.45; P < 0.0001) • BRCA1/2-mutant: 16.6 mo vs 5.4 mo (HR 0.23, 95% CI 0.16–0.34; P < 0.0001). • HRD: 13.6 mo vs 5.4 mo (HR 0.32, 95% CI 0.24–0.42; P < 0.0001) |
Led to 2018 FDA approval of rucaparib maintenance therapy for patients with advanced-stage ovarian cancer in CR or PR after platinum-based chemotherapy (irrespective of BRCA1/2 status) |
| OlympiAD; Robson et al. (2017)31 | III | Metastatic germline BRCA1/2-mutant, HER2-negative breast cancer after ≤2 prior lines of chemotherapy | Olaparib vs physician’s choice of single-agent chemotherapy (capecitabine, eribulin or vinorelbine) | Anaemia 16% vs 4%; neutropenia 9% vs 26%; and decreased white cell count 3% vs 10% | ORR 100 /167 (59.9%) vs 19/66 (28.8%); median PFS 7.0 mo vs 4.2 mo (HR 0.58, 95% CI 0.43–0.80; P < 0.001) | Led to 2018 FDA approval of olaparib for metastatic, HER2-negative breast cancers with BRCA1/2 mutations |
AEs, adverse events; ALT, alanine aminotransferase; AST, alanine aminotransferase; BID, twice daily; CR, complete remission; HRD, homologous recombination deficient; LOH, loss of heterozygosity; ORR, objective response rate; mo, months; PARP, poly(ADP-ribose) polymerase; PFS, progression-free survival; PR, partial remission.
Threshold for LOH-high versus LOH-low was at a prespecified cut-off of 14%.
EMA approval stipulates that patients must have platinum-sensitive disease but no longer be able to tolerate platinum-based chemotherapy.
AEs, adverse events; BID, twice daily; CLL, chronic lymphocytic leukaemia; DDR, DNA damage response; ORR, objective response rate; PARP, poly(ADP-ribose) polymerase; PD-L1, programmed cell death 1 ligand 1.
Various dosages once a day for days 1 and 2 of 3 consecutive weeks out of every 4-week cycle; maximum tolerated dose (MTD) 175 mg.
Various dosages twice daily for 2.5 days (5 doses) per 21-day cycle; MTD was 200 mg.
Various dosages twice daily for 2.5 days (5 doses) per 21-day cycle; MTD was 225 mg.
All PARP inhibitors currently used in the clinic have a similar capacity to inhibit the catalytic activity of PARP because they share a nicotinamide moiety that competes with NAD+ for binding to this enzyme; however, differences do exist regarding the dose required to inhibit PARP activity and the relative selectivity for different PARP family members. Notwithstanding, the mechanism of cytotoxicity of PARP inhibitors goes beyond simply abrogating catalytic activity. When PARP activity is inhibited, unrepaired SSBs and stalled replication forks accumulate, owing to ‘trapping’ of PARP in a complex with the DNA strand. During S phase of the cell cycle, these unrepaired SSBs convert to DSBs, which are lethal to HRD cancer cells7. Indeed, PARP–DNA complexes are markedly more damaging to the genomic integrity of cells than unbound SSBs alone and activate various pathways outside of HR-dependent DNA repair, including cell cycle checkpoints and post-replication repair13. The concept of PARP trapping was first described more than two decades ago14,15 (FIG. 2), although only within the current decade was DDR demonstrated to be stunted to a greater degree in PARP1-inhibited cells, in which PARP1 remains associated with DNA, than in cells completely lacking PARP1 (REFS14,15). Notably, the PARP-trapping abilities of the five most-studied PARP inhibitors — niraparib, rucaparib, talazoparib, olaparib and veliparib — vary markedly, in contrast with their capacities to inhibit protein poly ADP-ribosylation (PARylation), which have a much narrower range. For example, talazoparib, the PARP inhibitor with the greatest PARP-trapping ability observed preclinically, has cytotoxic potency in the nanomolar range, whereas veliparib results in less PARP trapping and is inactive at 100 mM (REF.13). Furthermore, the maximum tolerated dose (MTD) of these PARP inhibitors mirrors their respective PARP-trapping ability, rather than their capacity to inhibit PARP catalytic activity16,17.
Apart from veliparib, the four other aforementioned PARP inhibitors seem to have generally similar single-agent activity against the advanced-stage cancers evaluated in clinical trials to date9,18–24, although no head-to-head comparisons have been reported. In addition, the varied clinical characteristics of the patient populations studies in the separate trials make cross-study comparisons difficult9,18–24. In clinical studies of PARP inhibitors reported to date, biomarker approaches have focused primarily on single-gene mutations in HR-pathway genes (predominantly BRCA1/2); however, the effects of specific mutations on the HR capacity of cells are not well understood, and thus the development of functional biomarkers of HR capacity is an active area of research. Indeed, while BRCA1/2 mutations decrease HR capacity, cells retain the ability to perform HR, particularly when proteins that push the balance of DNA repair towards the alternative non-homologous end joining (NHEJ) pathway, such as TP53-binding protein 1 (53BP1), are deficient25,26. Better defining the DDR pathways involved in removing PARP–DNA complexes in a cell-specific and mutation-specific manner might improve the identification of predictive biomarkers of optimal benefit and inform treatment strategies to overcome or prevent PARP inhibitor resistance.
PARP inhibitors have shown an overall favourable safety profile as monotherapies; common toxicities shared among these drugs include myelosuppression, gastrointestinal symptoms and fatigue17. Similar to patients with DDR-deficient cancers treated using DNA-damaging chemotherapies, patients treated with PARP inhibitors have a theoretical risk of secondary malignancies owing to DNA damage and genomic instability generating further mutational events. The risk of secondary malignancies, such as myelodysplastic syndrome and acute myeloid leukaemia, in patients treated with PARP inhibitors has generally been low (<1%)27,28, although regular complete blood counts should be undertaken during therapy, with prompt referral of patients with persistent cytopenias to haematology specialists.
FDA approvals in ovarian cancer
The development of PARP inhibitors for the treatment of ovarian cancer has provided clinical proof of concept and paved the way for the development of selective DDR inhibitors in cancer medicine, with three agents — olaparib, rucaparib and niraparib — currently approved in different therapeutic settings (TABLE 1). FDA approvals have typically preceded EMA approvals for similar indications (FIG. 2).
In clinical trials assessing olaparib in patients with ovarian cancer, the BRACAnalysis CDx PCR-based sequencing platform has been used to identify patients with deleterious germline mutations in the protein coding region or the intron–exon boundaries of BRCA1 and BRCA2, and is FDA-approved for this purpose11,29. In clinical studies of rucaparib, investigators used the FoundationFocus CDx platform to perform next-generation sequencing (NGS) for the detection of both somatic and germline BRCA1/2 aberrations18,30; consequently, this test was FDA-approved as a companion diagnostic to select patients with advanced-stage, BRCA1/2-mutated ovarian cancers who have previously received ≥2 lines of chemotherapy to receive rucaparib30. Current National Comprehensive Cancer Network (NCCN) guidelines recommend that deleterious somatic and/or germline BRCA1/2 mutations can be identified using any FDA-approved or other validated test undertaken in a CLIA-approved facility when selecting patients with advanced-stage ovarian cancer to receive olaparib or rucaparib18,29,31,32.
Approvals in the platinum-sensitive, maintenance setting.
In clinical studies to date, PARP inhibitors have provided substantial benefit with tolerable toxicities when used as maintenance therapy following a response to platinum-based chemotherapy (TABLE 1). Indeed, olaparib first gained regulatory approval in 2014 from the EMA for the maintenance treatment of patients with platinum-sensitive, relapsed, germline or somatic BRCA1/2-mutant ovarian cancers who are in complete or partial remission after platinum-based chemotherapy (FIG. 2). This approval was based on data from the randomized phase II Study 19 trial (NCT00753545), which demonstrated improved progression-free survival (PFS) in olaparib capsule-treated versus placebo-treated patients33. A preplanned retrospective analysis of these data demonstrated a PFS benefit34, albeit modest, even for patients lacking BRCA1/2 mutations (TABLE 1), suggesting a need for expanded biomarkers to identify patients with BRCA1/2-wild-type disease who might benefit from PARP inhibitor maintenance therapy. Of note, however, treatment with olaparib capsules was not associated with an improvement in overall survival (OS) in either the BRCA1/2-wild-type or BRCA1/2mutant groups of this study35. The follow-on, placebo-controlled, phase III SOLO2/ENGOT-Ov21 trial involving women with BRCA1/2-mutant ovarian cancers33,36 confirmed that olaparib (tablet formulation) improves PFS in this population, with a tolerable adverse effect profile and no detrimental effects on quality of life (TABLE 1). In 2017, the FDA approved olaparib for the maintenance treatment of ovarian cancer, irrespective of BRCA1/2-mutation status, on the basis of data from these trials (FIG. 2).
In the phase III NOVA study, 553 patients with ovarian cancer who had an objective response to platinum-based chemotherapy were randomly assigned (2:1) to receive either maintenance niraparib or placebo37. The NGS-based myChoice HRD companion diagnostic was used to identify not only BRCA1/2 variants, but also HRD tumours that share molecular hallmarks of BRCA1/2-mutated tumours — that is, loss of heterozygosity (LOH), large-scale translocations (LSTs) and telomeric allelic imbalance (TAI)37. Exploratory analyses revealed that median PFS durations were significantly longer in patients who received niraparib; while the PFS benefit was highest in patients with germline BRCA1/2 mutations and intermediate in patients with BRCA1/2-wild-type HRD tumours, a small but statistically significant PFS benefit was demonstrated in patients without detectable HRD37 (TABLE 1). These findings supported the 2017 FDA and EMA approvals of niraparib for this indication, independent of BRCA1/2 status (FIG. 2).
Similarly, positive data have been reported with maintenance rucaparib in the phase III ARIEL3 trial for patients with high-grade ovarian cancer who had responded to platinum-based chemotherapy in the second-line or third-line settings38. Testing for LOH, somatic BRCA1/2 mutations and other prescribed HR-gene aberrations was conducted using the T5 NGS assay, while germline BRCA1/2 testing was performed using the BRCAnalysis CDx test. Patients who received rucaparib had significantly longer PFS than those who received placebo (TABLE 1), again regardless of BRCA1/2mutation status or the presence of HRD (as defined by high levels of LOH)38, leading to the FDA-approval of rucaparib in this maintenance setting regardless of biomarker status in April 2018 (FIG. 2).
Approvals in the relapsed-disease setting.
PARP inhibitor monotherapy also has demonstrated clinical benefit in selected patients with ovarian cancer that has progressed on prior chemotherapy. For example, on the basis of data from a single-arm phase II trial by Kaufman et al.11 in patients with various advanced-stage, BRCA1/2-mutant cancers (TABLE 1), the FDA in December 2014 granted accelerated approval to olaparib monotherapy for patients with deleterious or suspected deleterious germline BRCA1/2-mutant, advanced-stage, ovarian cancer after ≥3 lines of chemotherapy29 (FIG. 2). In December 2016, the FDA also granted accelerated approval to rucaparib for the treatment of patients with advanced-stage ovarian cancers harbouring deleterious germline and/or somatic BRCA1/2 mutations who have received ≥2 prior lines of chemotherapy30. This approval was based on promising efficacy data from the single-arm, phase II ARIEL2 and Study 10 trials18,39 (TABLE 1). Talazoparib and veliparib are currently in late phase trials in patients with newly diagnosed ovarian cancer (NCT02470585) and in other advanced-stage cancers, mainly in combination with chemotherapies for the latter agent (Supplementary Table 1).
Beyond ovarian cancer
BRCA1 and BRCA2 are the two most studied genes in the HR repair pathway. Germline variants of these genes were initially discovered in patients with hereditary breast or ovarian cancer40–42; however, somatic and germline BRCA1/2 mutations, as well as aberrations affecting other HR genes including ATM, ATR, BARD1, BRIP1, CHK1, CHK2, PALB2, RAD51 and FANC, are increasingly being detected in patients with other tumour types through NGS of paired tumour and non-malignant DNA samples43–47. Antitumour responses have already been reported in a number of phase I/II trials of olaparib monotherapy in patients with various advanced-stage BRCA1/2-mutant cancers, including breast, prostate and pancreatic cancers (Supplementary Table 2).
PARP inhibition for breast cancer.
Data from the phase III OlympiAD trial demonstrated a doubling of the objective response rate (ORR), a significant PFS benefit and a more favourable safety profile for olaparib versus single-agent chemotherapy (not including platinum-based agents) in patients with germline BRCA1/2-mutant, HER2-negative, metastatic breast cancer31 (TABLE 1), leading to FDA approval of olaparib in this patient population (FIG. 2). The efficacy of olaparib in a post-chemotherapy maintenance setting or concurrently with chemotherapy in patients with BRCA1/2-mutant breast cancer is currently being tested in the phase III OlympiA (NCT02032823) and PARTNER (NCT03150576) trials, respectively (Supplementary Table 1).
Encouraging data from the phase III EMBRACA trial investigating single-agent talazoparib for the treatment of advanced-stage, BRCA1/2-mutant, HER2-negative breast cancers indicate a PFS benefit of talazoparib (median 8.6 months versus 5.6 months with chemotherapy; HR 0.54; P < 0.0001), which extended to patients with hormone receptor-positive disease (HR 0.47; 95% CI 0.32–0.71) and those with central nervous system metastases (HR 0.32; 95% CI 0.15–0.88)24. Importantly, the talazoparib-treated patients in this study also had improvements in quality of life and delayed clinically meaningful deterioration compared to patients treated with standard chemotherapies48. A randomized trial of investigator’s choice chemotherapy versus niraparib (NCT01905592) is ongoing.
Substantial efforts are also being applied to early stage clinical testing of PARP inhibitors in the neoadjuvant setting. Veliparib plus carboplatin for triple-negative breast cancer (TNBC) was the first combination to graduate from the I-SPY2 trial, a multicentre, adaptive, platform trial designed to screen multiple experimental compounds on a standard chemotherapy backbone for the treatment of patients with breast cancer49. Data from the phase III BrighTNess trial50, however, did not show a benefit of the addition of veliparib to carboplatin and paclitaxel chemotherapy in terms of pathological complete response (pCR) rate (53% versus 58%; P = 0.36), with carboplatin plus paclitaxel alone providing clinical advantage over single-agent paclitaxel. This lack of benefit could be multifactorial, but, as discussed previously, veliparib is the least potent PARP inhibitor and thus might not be the optimal choice of agent to use in synthetic lethality approaches. By contrast, encouraging data have been reported for the use of single-agent talazoparib in the neoadjuvant setting in patients with BRCA1/2mutant, HER2-negative breast cancer, with 9 of 17 patients (53%) achieving a pCR51,52. Thus, earlier targeting of BRCA1/2-mutant tumours with potent PARP inhibitors, when the accumulation of driver mutations is perhaps more limited, might prove to be particularly effective; however, long-term data are not yet available to discern whether early PARP inhibition has survival benefits.
PARP inhibition for prostate cancer.
Considerable interest surrounds the use of PARP inhibitors in selected patients with metastatic castration-resistant prostate cancer (mCRPC), following positive results from the single-arm phase II TOPARP-A trial of olaparib53 (Supplementary Table 2): the ORR was substantially higher in patients with DDR gene mutations (including BRCA1/2 or ATM mutations) than in the unselected population (88% versus 33%)53. Interestingly, serial analyses of circulating cell-free DNA (cfDNA) revealed that declines in cfDNA concentrations and mutant allele frequencies correlated with better outcomes after olaparib treatment54. The ongoing phase III PROfound (NCT02987543) and TRITON3 (NCT02975934) trials are currently testing olaparib and rucaparib, respectively, versus investigators’ choice therapy for mCRPC with HR gene mutations (Supplementary Table 1).
PARP inhibition for gastrointestinal cancers.
In a subgroup analysis of the phase II trial by Kaufman et al.11, 5 of 23 patients with BRCA1/2-mutant pancreatic cancers (22%) had an objective response and an additional 11 (47%) had stable disease lasting ≥8 weeks (Supplementary Table 2). A phase I/II study of olaparib combined with gemcitabine in patients with advanced-stage pancreatic cancer revealed an ORR of 27% (versus 14% with gemcitabine alone); however, grade ≥3 toxicities were common55 (Supplementary Table 2). In the ongoing phase III POLO trial (NCT02184195; Supplementary Table 1), a switch maintenance approach is being used whereby patients with BRCA1/2-mutated pancreatic cancers who have been on treatment with first-line platinum-based therapy for ≥16 weeks without progression are being randomly assigned to receive either olaparib or placebo.
BRCA1/2 mutations are rare in gastric cancers; however, these tumours often have loss of ATM expression, providing a biological rationale for PARP inhibitor therapy56,57. A randomized phase II study has been conducted to compare olaparib plus paclitaxel with placebo plus paclitaxel58. Unsurprisingly, adverse events, including grade ≥3 neutropenia, were more frequently observed with the olaparib combination58. Interestingly, although no difference in PFS was detected, the patients receiving combination therapy with olaparib had longer OS durations (median 13.1 months versus 8.3 months; P = 0.005)58. In addition, a subset of patients with low baseline levels of tumour ATM expression had an even greater OS benefit (median not reached versus 8.2 months; P = 0.002) in a prespecified secondary analysis56, again, without a PFS benefit. Nevertheless, the subsequent placebo-controlled, phase III GOLD trial of the same regimen in Asian patients with gastric cancers did not meet its primary end point of improved OS in the olaparib-treated group, in neither the overall population nor an ATM-low subpopulation59 (Supplementary Table 2). The lack of benefit of olaparib in this setting despite the promising phase II results is likely multifactorial, as discussed elsewhere60. For example, additional factors outside of ATM loss might predict PARP inhibitor sensitivity or resistance in patients with gastric cancer, even in the ATM-low population, highlighting that single-gene or single-protein biomarker approaches might be inadequate60,61.
Other cancer types and chemotherapy combinations.
Large-scale paired tumour and germline sequencing studies have revealed a substantial number of incidental deleterious variants in DDR-related genes, including BRCA1/2 and ATM, across a variety of cancer types, such as prostate cancer, hepatobiliary cancers, sarcomas and bladder cancer43,62,63. Interestingly, prior studies have also revealed a high germline-to-somatic ratio for BRCA1/2 mutations64; therefore, any patient found to have a somatic BRCA1/2 variant in their tumour should be considered for germline testing. Whether context dependency across tumour types will affect antitumour responses remains unclear. In the absence of this knowledge, patients with cancers harbouring pathogenic HR gene mutations should be considered for clinical trials involving PARP inhibitors and other DDR inhibitors, agnostic of tumour type; however, the definition of which genes qualify as ‘HR genes’ is actively evolving65.
In contrast to switch maintenance therapy with chemotherapy followed by PARP inhibition, which has proved successful, simultaneously combining DDR inhibitors, including PARP inhibitors, with chemotherapy has been problematic owing to substantial toxicity. Attempts to combine the first DDR inhibitor, O6-benzylguanine, with alkylating agents were terminated owing to high rates of toxicities. For example, a trial of temozolomide plus O6-benzylguanine for patients with gliomas revealed limited responses, with >45% of patients having grade 4 haematological toxicities66. Multiple subsequent studies of PARP inhibitors, particularly those with a high capacity for PARP trapping, given concurrently with chemotherapeutic agents, have revealed improved response rates across tumour types, compared with chemotherapy alone, but increased toxicity — predominantly myelosuppression — requiring dose reductions or treatment delays in a substantial proportion of patients55,67,68.
Biomarkers beyond BRCA1/2 mutations
Identifying novel predictive biomarkers of benefit from PARP inhibitors is important: although germline or somatic BRCA1/2 mutations can be used to enrich for responders, a substantial number of patients who lack these mutations benefit from PARP inhibitor monotherapy37,38. Furthermore, patients with tumours harbouring BRCA1/2 mutations frequently do not respond to PARP inhibitors (TABLE 1). Multiple studies have been undertaken to investigate whether specific molecular features of BRCA1/2-mutated tumours can serve as biomarkers for selection of patients with BRCA1/2-wild-type disease to receive PARP inhibitors, thereby expanding the potential benefit of these targeted therapies. The term ‘BRCAness’ was originally coined to describe a molecular phenocopy of BRCA1/2-mutated tumours, which can arise through a range of genomic, epigenetic or post-translational alterations69. What BRCAness truly reflects, however, is a HRD phenotype beyond the narrow scope of defects in the BRCA pathway. As such, we propose that the term ‘BRCAness’ should be broadened to ‘HRDness’ to recognize these non-BRCA-related, yet ‘HRD-like’ mechanisms of PARP inhibitor sensitivity (Supplementary Figure 1). Notably, a subset of tumours might also demonstrate ‘PARPness’ — that is, responsiveness to PARP inhibitors in the absence of HRD, potentially owing to PARP trapping or related to abrogation of the activity of PARP in processes other than base-excision repair (BER), such as alternative-NHEJ (alt-N HEJ) or replication-fork protection70,71.
BRCA-like tumours are characterized by frequent genomic structural rearrangements and LSTs resulting in high levels of genomic instability via global LOH and TAI72,73. Within the NOVA37, and ARIEL2 and ARIEL3 trials18,38, different assays designed to quantify HRD via genomic-based analysis of LOH and global genomic alterations were studied as potential companion diagnostics for selecting patients with BRCA1/2-wild-type tumours who are more likely to benefit from treatment with PARP inhibitors74.
The ARIEL2 investigators capped the number of patients with known hereditary BRCA1/2 mutations enrolled in order to test the ability of the T5 NGS LOH assay to predict PARP inhibitor sensitivity in patients with BRCA1/2-wild-type disease18,75. Importantly, the results of this study, and subsequently ARIEL3 (REF.38), showed that maintenance rucaparib improved PFS even in women with BRCA1/2-wild-type, LOH-low ovarian cancer. Similarly, the myChoice HRD assay used in the NOVA trial enabled assessment of BRCAness on the basis of global genomic scarring, including a high degree of LOH, LSTs and TAI; however, PARP inhibition with niraparib improved PFS regardless of biomarker status, albeit with different efficacy in the different patient subsets37. The results of these trials demonstrate that patients with deleterious BRCA1/2 variants achieve the greatest benefit from PARP inhibition, which is biologically in line with preclinical evidence of synthetic lethality, followed by those with molecular genomic features of BRCAness, including high LOH, as defined using companion diagnostic assays37,38 (TABLE 1). However, the observation of clinical benefit from PARP inhibitors in the absence of HRDness defined using the companion diagnostic indicates that genomic scarring assays are either not inclusive enough in defining molecular signatures of HRD tumours or fail to capture mechanisms of PARP inhibitor sensitivity outside of HRDness, or both. Indeed, the inclusion of patients with repeated responses to platinum-based chemotherapy in these trials might have enriched for tumours with HRD, even if this HRD was undetectable in some patients using the companion diagnostic assays, thus skewing the results76,77.
In addition to the aforementioned genomic scarring assays, multiple other mutational signatures of HRDness have been developed through whole-exome or whole-genome sequencing of BRCA1/2-mutant tumours and retrospectively tested in a range of different solid tumour settings. In a seminal study78, all somatic point mutations and larger-scale genomic alterations across 7,042 cancers were catalogued, with subsequently identified mutational signatures serving as avatars of specific aberrant pathways and biological processes. For example, ‘Signature 3’ was strongly associated with BRCA1/2-inactivating mutations across a variety of tumour types78; however, this signature lacks a discreet cut-off to determine BRCA1/2-deficient versus BRCA1/2-proficient tumours. Building on these data, the HRDetect test was subsequently designed and trained by using known BRCA1/2-mutated tumours to generate a unique somatic mutational profile that enabled the identification of BRCA pathway-deficient tumours with 98.7% sensitivity79.
Researchers have also sought to pair data from somatic and germline sequencing of DDR genes with whole-exome or whole-genome mutational signatures80. Whereas Signature 3 and HRDetect have high sensitivity for the detection of BRCA1/2-mutant and so-called BRCA-like tumours, they have failed to enable the identification of tumours with known functional mutations in other HR pathway genes, including ATM, CHEK and ATR, as being HRD79,80. These findings again suggest that some tumours have HRDness through mechanisms unrelated to BRCA pathway function specifically; thus, deleterious variants, epigenetic modifications or post-translational changes affecting other DDR gene products that lead to HRDness might have unique molecular features or signatures that do not entirely align with a BRCA-like or BRCAness signature. Multiple studies assessing PARP inhibitors have reported antitumour responses in patient populations enriched for these non-BRCA HRD mutations or even changes in protein expression levels (for example, loss of ATM)53,57,58,81,82. Knowledge of genes with direct or indirect roles in DDR, cell cycle regulation and chromatin remodelling is constantly expanding, thereby increasing the discovery of aberrations that can lead to HRDness. For example, ARID1A, which encodes a component of the SWI/SNF chromatin remodelling complex, is one of the most frequently mutated genes in cancer and has been shown to facilitate DSB processing, sustain DDR signalling and regulate the cell cycle via CDC25C83. Unsurprisingly, therefore, ARID1A deficiency has been shown to render tumours sensitive to a variety of DDR inhibitors, including PARP inhibitors, in preclinical models84. Aditionally, BAP1, another key SWI/SNF complex unit, has been shown to regulate HR and cellular recovery via its phosphorylation sites and catalytic activity; BAP1 deficiency predicts for PARP inhibitor sensitivity85,86. CDK12 is a transcriptional regulator with roles in maintaining genomic stability by regulating the expression of other DDR genes; loss of CDK12 function leads to HRD and sensitizes ovarian cancer cells to PARP inhibition87. Approximately 5% of mCRPCs harbour deleterious aberrations in CDK12, which have been shown to predict increased T cell infiltration, thus highlighting a new potential biomarker based on genomic instability for selecting patients with prostate cancer to receive immunotherapy88.
Ultimately, the utility of NGS-based biomarkers of sensitivity to PARP inhibitors is limited because they typically provide only a historical, static record of genomic alterations present in the tumour at the time of biopsy sampling or tumour resection and lack the capacity to inform on dynamic, active signalling and, in particular, adaptive processes that occur in the tumour during PARP inhibition. Monitoring cfDNA before and serially during therapy and at the time of disease progression can provide greater insights into tumour genomic evolution as it relates to therapeutic response54. As mentioned previously, decreases in total cfDNA concentration and the allele frequencies of HR gene variants have been independently associated with responsiveness to olaparib in patients with mCRPC, while NGS of cfDNA at the time of progression unveiled somatic reversion mutations in HR genes, thus marrying restoration of DDR gene function to drug resistance54.
Biomarkers of sensitivity to DDR inhibitors that rely on the identification of mutations in cancer-related genes, such as BRCA1/2, are inherently limited by the fact that most variants discovered in these genes are still of undetermined functional significance, and thus their potential for predicting synthetic lethality is unclear. However, the expansion of mutational library screens and collaborative pooling of data from specialist functional laboratories have great potential to differentiate true functional loss from benign exonic point mutations within specific clinical contexts89. For example, the identification of differential gene-expression profiles in known HRD versus HR-proficient cell lines has led to the development and preclinical validation of multiple transcriptomic HRD scores90–93. These RNA profiling-based scores remain in development, but hold great promise as dynamic biomarkers of HR repair function and PARP inhibitor sensitivity90–93. Nevertheless, caution should be exercised in the use of transcriptional signatures that are derived from comparisons of drug-sensitive versus drug-resistant tumour models because results from drug-sensitivity assays in preclinical models have been shown to be highly variable, with low levels of inter-assay concordance94.
A variety of other causes of HRD and/or replication stress have been shown in preclinical models and small cohorts of patients to predict sensitivity to certain DDR inhibitors. For example, a response to platinum-based chemotherapy can be a biomarker in itself, providing insight into the underlying defects in HR and cell cycle control. In a retrospective molecular analysis76, patients with advanced-stage ovarian cancer who responded to multiple lines of platinum-based chemotherapy and, to a lesser extent, long-term responders and/or survivors had tumours enriched for HR defects. Moreover, the co-occurence of HRD and loss of retinoblastoma-associated protein (RB) expression was associated with prolonged survival76. Interestingly, patients with long-term responses to platinum-based chemotherapy and protracted OS also had highly replicative tumours with high Ki67 levels and a high degree of tumour immune infiltration, suggesting increased replication stress and S phase-specific DNA damage76,77.
Other causes of HRDness in cancer cells include the generation of oncometabolites that result from a general shift towards glycolytic metabolism (the Warburg effect), which can result in HRD. For example, hypoxic tumour states and mutations in Krebs cycle genes, such as IDH1 or IDH2, can result in the production of oncometabolites — D-2-hydroxyglutarate in the case of IDH variants — that lead to the downregulation of HR proteins and increased sensitivity to PARP inhibitors in preclinical models95.
In addition, cancers that lack mutations in HR repair genes and do not typically display HRDness, such as small-cell lung cancer (SCLC), have demonstrated sensitivity to platinum-based agents and PARP inhibitors71,96. This increased susceptibility might be a consequence of high levels of replication stress owing to loss of RB1 and TP53 expression combined with MYC overactivation and, therefore, a reliance on the HR and replication stress response pathways for cell viability. High levels of PARP1 expression in the tumour cells, resulting in lethal levels of PARP trapping, is another potential explanation. Thus, even tumours lacking HRDness can be PARP inhibitor sensitive, with these tumours showing features we have termed ‘PARPness’ (Supplementary Figure 1). Potential biomarkers of PARPness include high tumoural levels of PARP1, E-cadherin and/or Schlafen 11 (SLFN11), which predicted sensitivity to platinum-based agents and PARP inhibitors in preclinical models and select clinical trials96–98. High levels of replication stress combined with replication fork instability, as in the context of replication protein A (RPA) exhaustion, can also contribute to PARP and platinum sensitivity99. IDH1 mutations have been shown to confer PARP inhibitor sensitivity via reduced production of NAD+ that is required for PARP1-mediated DNA repair100, in addition to oncometabolite production95. Histological phenotypes, including neuroendocrine differentiation (for example, in SCLC and aggressive-variant prostate cancer), also seem to predict sensitivity to platinum-based agents and PARP inhibitors; studies investigating the underlying mechanisms of responsiveness are ongoing98,101,102. Finally, robust multigene expression signatures based on sensitivity and resistance profiles of cancer cell lines of different lineages have been shown to accurately predict PARP inhibitor sensitivity in preclinical models, as well as benefit from cisplatin chemotherapy in retrospective clinical studies103. These RNA-based expression scores of PARP inhibitor sensitivity reflect tumour PARPness, rather than inherent BRCA1/2 or HR defects103, although the additional mechanisms by which sensitivity to PARP inhibition arise remain under active investigation. To establish the biological rationale for the wider use of DDR inhibitors to treat a broader range of tumours, it is important to move beyond a BRCA1/2-centric view of DDR biomarkers and to consider tumours that demonstrate HRDness and PARPness phenotypes (Supplementary Figure 1).
PARP inhibitor resistance mechanisms
Long-term data from clinical trials of different PARP inhibitors have demonstrated durable responses, mostly in patients with BRCA1/2-mutant cancers, although the majority of patients inevitably develop resistance to platinum-based and/or PARP inhibitor therapy104. Indeed, with the regulatory approval of PARP inhibitors in multiple indications, as well as off-label use of PARP inhibitors, an emerging population of patients with disease that has progressed after PARP inhibition requires new therapeutic options to overcome resistance.
In clinical and preclinical studies, resistance to PARP inhibitors seems to occur by three general mechanisms: acquisition of aberrations that increase HR repair capacity; activation of signalling pathways that decrease cell cycle progression and replication stress; and miscellaneous alterations that cannot currently be assigned to a single DDR pathway-related mechanism (FIG. 3).
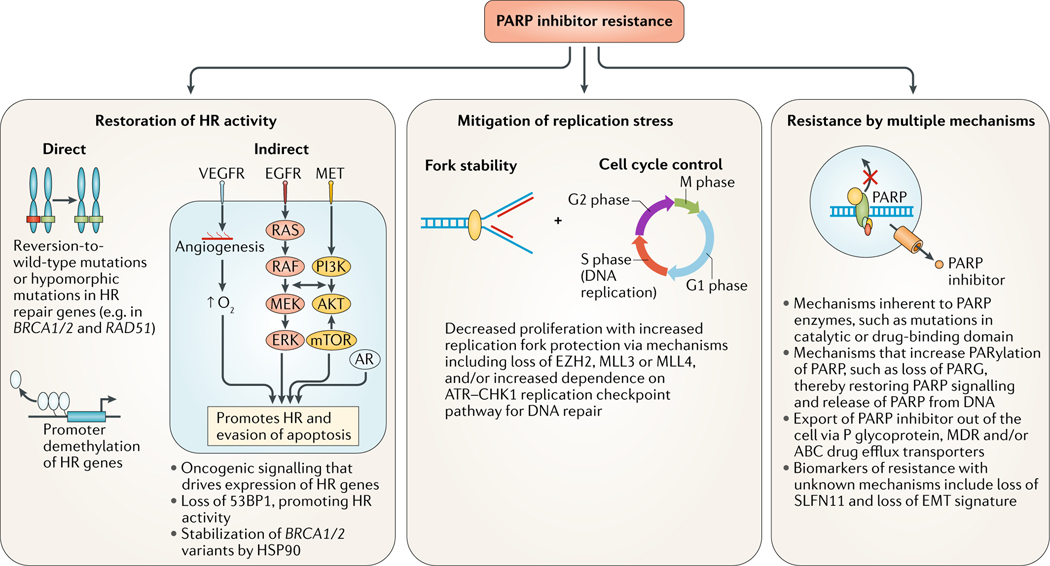
FIG. 3 |. Mechanisms of resistance to PARP inhibitors.
Resistance of cancers to poly(ADP-ribose) polymerase (PARP) inhibitors can be inherent or acquired. The potential mechanisms of resistance are varied and can be multifactorial but centre around three main categories: restoration of homologous recombination (HR) repair activity through direct (genomic, epigenetic or post-translational changes in the HR machinery itself) or indirect mechanisms (signalling that increases the activity and/or expression of the HR machinery); replication stress mitigation, whereby the cancer cell slows the cell cycle and stabilizes replication forks; and mechanisms not currently assigned to a single DNA repair pathway-related process but still alter the response to PARP inhibition, such as mutations in PARP itself, genomic events that alter protein poly ADP-ribosylation (PARylation) and/or PARP trapping, upregulation of drug efflux pumps and loss of biomarkers of sensitivity to PARP inhibition, such as expression of Schlaffen 11 (SLFN11) and/or a epithelial-to-mesenchymal (EMT) signature. 53BP1, TP53-binding protein 1, AR, androgen receptor; HSP90, heat-shock protein 90; MDR, multidrug resistance protein; MLL3, histone-lysine N-methyltransferase 2C; MLL4, histone-lysine N-methyltransferase 2B; PARG, poly(ADP-ribose) glycohydrolase
Mechanisms of acquired resistance to PARP inhibition in patients with BRCA1/2-mutant tumours have been determined through genomic analyses of sequential tumour biopsy and cfDNA samples. Across multiple cancer types, secondary mutations have been discovered that cause a reversion to the wild-type sequence and/or function of the DDR gene or protein, including BRCA1/2, RAD51C, RAD51D and PALB2, thereby restoring HR repair capacity and negating synthetic lethality54,105,106 (FIG. 3). These types of mutations have also been implicated in resistance to platinum-based chemotherapy107. However, these reversion mutations are identified in only a small subset (approximately 20–25%) of patients with PARP inhibitor resistance108. Moving forward, larger clinical data sets incorporating liquid or tumour biopsy sampling before and during treatment and at the time of disease progression are needed to better understand the prevalence and influence of these reversion mutations in the context of PARP inhibition and to better define changes in the function of DDR pathways during therapy108.
Observations from preclinical models have revealed other mechanisms of resistance to PARP inhibitors that rely on the restoration of HR repair function. For example, HSP90 can stabilize and prevent the degradation of a subset of BRCA1 variants, resulting in retention of some HR repair function109 (FIG. 3). In addition, changes in the promoter regions of DDR genes, in the form of gene fusions or loss of promoter hypermethylation, can restore the activity of HR genes, such as BRCA1 (REF.110). Loss of 53BP1 expression has been shown to influence responsiveness and resistance to PARP inhibitors, probably by shifting the balance of DNA repair from NHEJ to HR25,26,111. Mitigation of replication stress via replication fork stabilization, by a variety of potential pathways, often combined with slowing of cell cycle progression, might also have an important role in PARP inhibitor resistance, distinct from restoration of HR repair112,113 (FIG. 3).
Multiple biomarkers of PARP inhibitor resistance do not coalesce on a single DDR pathway-related mechanism (FIG. 3). For example, SLFN11 expression predicts PARP inhibitor sensitivity96–98 and unsurprisingly, therefore, SLFN11 inactivation can confer PARP inhibitor resistance114. SLFN11 responds to replication stress by binding chromatin at stressed replication foci and stalling replication, making cancer cells with overexpression of SLFN11 susceptible to synthetic lethal therapies; thus, loss of SLFN11 expression, which is often achieved through promoter hypermethylation in a variety of cancers, leads to resistance to such treatments, probably through increased reliance on the ATR–CHK1 axis97,114–116. Overexpression of the multidrug efflux transporter P glycoprotein has also been implicated in resistance to both PARP inhibitors and chemotherapy117. Inherent resistance to PARP inhibition might also be attributable to deficient PAR glycohydrolase (PARG) activity, which increases PARP1 auto-PARylation, thereby restoring PARP signalling, releasing PARP1 from DNA and ultimately decreasing PARP inhibitor-induced DNA damage118. Additionally, resistance can arise through mutations or loss of PARP1 that decrease PARP inhibitor binding and/or PARP trapping119.
PARP inhibitor resistance is probably multifactorial and research on strategies to overcome these various forms of acquired resistance is ongoing114,120,121. Interestingly, certain markers of resistance to PARP inhibitors, such as loss of SLFN11 expression, might predict for sensitivity to other DDR inhibitors. Finally, genomic, transcriptomic and proteomic profiling of sensitive and resistant cell lines has revealed that the tissue-specific activation of certain oncogenic pathways, such as RAS, PI3K or androgen receptor (AR) signalling, can promote HR repair activity and PARP inhibitor resistance in certain cancer contexts, providing the rationale for targeted combination strategies90,122–125 (FIG. 3).
Moving beyond PARP in targeting the DDR
Inhibitors of ATR and ATM
ATR inhibition.
Unlike PARP inhibitors, the use of other antitumour agents targeting key components of the DDR is currently limited to early phase clinical trials (TABLE 2; Supplementary Table 3). ATR and ATM are prime targets of DDR inhibitors, given their central regulatory function in activating the response to both SSBs and DSBs126. Both of these proteins work through distinct but overlapping pathways to halt the cell cycle and initiate DDR pathways (FIG. 1). Four ATR inhibitors are currently undergoing clinical trial testing: M6620 (VX-970 or berzosertib), M4344 (VX-803), AZD6738 and BAY1895344 (Supplementary Table 3).
Table 2 |.
Trials of DDR inhibitors other than PARP inhibitors
| Study | Phase | Disease setting | Treatments | Most common grade ≥3 AEs | ORR | Reason for notability |
|---|---|---|---|---|---|---|
| Yap et al. (2015)128 | I | Advanced-stage and/or recurrent solid tumours | Escalating doses of M6620 (VX-970 or berzosertib; ATR inhibitor) alone or in combination with carboplatin | M6620 alone: noneM6620 plus carboplatin: neutropenia 38%, hypersensitivity 8% and thrombocytopenia 8% | • M6620 alone: 1/11 (9%) • M6620 plus carboplatin: 1/15 (7%) |
First-in-human study of an ATR inhibitor alone and with chemotherapy |
| Do et al. (2015)172 | I | Advanced-stage and/or recurrent solid tumours | Escalating doses of adavosertib (AZD1775; WEE1 inhibitor) | Leukopenia 8%, lymphopenia 8% and neutropenia 8% | 2/21 (10%); both responders had BRCA1/2 mutations | First-in-human trial of a WEE1 inhibitor |
| Leijen et al. (2016)171 | I | Advanced-stage and/or recurrent solid tumours | Escalating doses of adavosertib alone or combined with gemcitabine, cisplatin or carboplatin | Overall: thrombocytopenia 23%, neutropenia 22%, anaemia 12%, leukopenia 10% and diarrhoea 7% | • Adavosertib (multiple dosesa) plus gemcitabine: 3/67 (4%) • Adavosertib (multiple dosesb) plus cisplatin: 7/45 (16%) • Adavosertib (multiple dosesc) plus carboplatin: 2/46 (4%) |
First-in-human trial combining WEE1 inhibitor with chemotherapy |
| Hong et al. (2016)161 | I | Advanced-stage and/or recurrent solid tumours and CLL | Escalating doses of prexasertib (LY2606368; CHK1 inhibitor) | Neutropenia 89%, leukopenia 71% and thrombocytopenia 53% | 2/45 (4%) | First-in-human trial of a CHK1 inhibitor |
| Munster et al. (2016)148 | I | Advanced-stage and/or recurrent solid and haematological cancers | Escalating doses of CC-115 (dual mTOR and DNA-PK inhibitor) | Not reported | 2/44 (4%) | First-in-human trial of a DNA-PK inhibitor |
| Yap et al. (2017)132 | I | Advanced-stage and/or recurrent solid tumours | Escalating doses AZD6738 (ATR inhibitor) plus carboplatin, olaparib (PARP inhibitor) or durvalumab (anti-PD-L1 antibody) | • Carboplatin plus AZD6738: anaemia 19%, neutropenia 19% and thrombocytopenia 19% • Olaparib plus AZD6738: anaemia 7%, low white blood cell count 7% and neutropenia 7% • Durvalumab plus AZD6738: anaemia 8%, lung infection 8% and hyponatraemia 8% |
• AZD6738 plus carboplatin: 3/37 (8%) • AZD6738 plus olaparib: 3/28 (11%) • AZD6738 plus durvalumab: 2/12 (17%) |
First-in-human study of ATR inhibitor in combination with a PARP inhibitor or a immune-checkpoint inhibitor |
AEs, adverse events; BID, twice daily; CLL, chronic lymphocytic leukaemia; DDR, DNA damage response; ORR, objective response rate; PARP, poly(ADP-ribose) polymerase; PD-L1, programmed cell death 1 ligand 1.
Various dosages once a day for days 1 and 2 of 3 consecutive weeks out of every 4-week cycle; maximum tolerated dose (MTD) 175 mg.
Various dosages twice daily for 2.5 days (5 doses) per 21-day cycle; MTD was 200 mg.
Various dosages twice daily for 2.5 days (5 doses) per 21-day cycle; MTD was 225 mg.
M6620 is the first-in-class ATR inhibitor and has been tested as monotherapy and in combination with different chemotherapies, including topotecan, carboplatin, gemcitabine and cisplatin127–130. M6620 monotherapy was well tolerated, with no dose-limiting toxicities (DLTs) observed; a durable complete response (CR) lasting >19 months was observed in a patient with advanced-stage colorectal cancer and 100% ATM loss on immunohistochemistry127 (TABLE 2). Preliminary data from phase I trials have also now been reported for the chemotherapy combinations; while signals of antitumour activity were observed, unlike monotherapy, the chemotherapy combinations were associated with higher rates of bone marrow toxicities, requiring frequent dose delays and reductions127–130. Unsurprisingly, the MTDs of M6620 in combination with chemotherapy were lower than the recommended phase II dose (RP2D) of M6620 monotherapy127. Together with the aforementioned studies combining PARP inhibition with chemotherapy55,67,68, these findings emphasize that DDR inhibitor–chemotherapy combinations have problematic toxicities, particularly bone marrow suppression, without any clear clinical benefits over the use of either treatment alone (although direct comparisons are lacking).
The safety and efficacy of AZD6738 monotherapy in patients with advanced-stage solid tumours have been investigated in the phase I PATRIOT study; two partial responses (PRs) were observed, although one was unconfirmed131. Owing to bone marrow suppression observed beyond cycle 1 with continuous dosing, the investigators are exploring whether different dosing schedules improve long-term tolerability131. Patients with Ki67-high and/or HRD tumours will be enrolled in future expansion cohorts131. In a parallel phase of the study, AZD6738 treatment is being combined with palliative radiotherapy131. Early data on AZD6738 in combination with carboplatin, olaparib or the anti-programmed cell death 1 ligand 1 (PD-L1) antibody durvalumab have also been reported132 (TABLE 2); drug-related toxicities were substantially more common with the AZD6738–carboplatin combination, including grade ≥3 thrombocytopenia, neutropenia and anaemia requiring dose delays and modifications132. With the AZD6738–carboplatin combination, three PRs (one unconfirmed) were reported, two of which involved patients with ATM-aberrant tumours (rectal and clear cell ovarian cancers)132. In comparison with the carboplatin regimen, the AZD6738–olaparib combination seemed to be well tolerated, although myelosuppression remained the predominant reported toxicity132. PRs were observed in two patients with BRCA1-mutant TNBC and in a patient with BRCA2-mutant oestrogen receptor-positive breast cancer132. Testing of AZD6738–durvalumab revealed that this combination was well tolerated, with no grade ≥3 or DLTs reported; PRs were reported in a patient with HNSCC and in another with non-small-cell lung cancer (NSCLC)132. While objective responses were observed with each combination, it remains unclear whether these are truly biologically active combinations because the antitumour activity could be attributable to any of the individual agents. However, the non-overlapping toxicities of DDR inhibitors and immune-checkpoint inhibitors makes combinations of these agents attractive. The oral ATR inhibitors BAY1895344 and M4344 are currently in phase I testing in combination with different chemotherapies and/or as single agents in patients with advanced-stage solid tumours (NCT03188965 and NCT02278250, respectively; Supplementary Table 3).
Predictive biomarkers of ATR inhibitor sensitivity.
Preclinical screens for predictive biomarkers have revealed that HR defects, including deleterious ATM and BRCA1/2 mutations, confer sensitivity to ATR inhibition133–135. This observation suggests that many biomarkers of HRDness might predict benefit from ATR inhibitors, as well as PARP inhibitors (Supplementary Figure 1). Indeed, anecdotal responses of patients with ATM-aberrant tumours to regimens containing ATR or PARP inhibitors have been reported in early phase clinical trials53,56,58,132, although the optimal method of assessing ATM status — assays for deleterious mutations or loss of protein expression — remains unclear126. Of note, the failure of ATM loss (defined as nuclear staining of ATM in <25% of tumour cells) to predict benefit from olaparib in the aforementioned phase III GOLD trial59 might in part reflect limitations of the immunohistochemistry assay and scoring system used. Moving forward, it might be prudent to define only tumours with 100% loss of ATM expression as ATM deficient. Notwithstanding, ATM levels should be better quantified to help determine whether a more precise threshold would enable more accurate prediction of ATR inhibitor sensitivity. In addition, p53 deficiency, as an indication of compromised ATM signalling and DNA damage checkpoints, has been shown in preclinical studies to confer cancer cells with sensitivity to ATR inhibition; this association has not been recapitulated in early phase clinical studies, but warrants further investigation126,136. As with PARP inhibitors, mutations in ARID1A have also been shown to predict for ATR inhibitor responsiveness, which is related to the reliance of ARID1A-deficient cells on ATR checkpoint activity to prevent premature mitotic entry and subsequent apoptosis137.
Alternative lengthening of telomeres (ALT), a telomerase-independent mechanism of maintaining telomere length and overcoming replicative senescence, is a biological process not directly related to HRD that has been investigated as a predictor of ATR inhibitor sensitivity. ALT is detected across a variety of tumour types and arises predominantly owing to deleterious mutations in the ATRX gene and less commonly owing to mutations in DAXX138,139. ATR inhibition in ALT-positive cancer cells has been shown to disrupt ALT, leading to chromosome fragmentation and ultimately apoptosis140. Fluorescence in situ hybridization (FISH)-based assays that enable the identification and quantification of ALT in tumour specimens have been developed138. However, preclinical data on ALT positivity as a predictor of ATR inhibitor sensitivity remain conflicting139–141, and clinical investigations of the predictive value of ALT biomarkers are warranted.
HER2 signalling has been shown in preclinical models to promote activation of the ATM–ATR signalling cascades in response to DNA damage, and selected HER2-positive breast cancer cell lines display sensitivity to ATR inhibition with AZD6738 (REFS142,143). However, the mechanism of this sensitivity is not well understood and clinical trials of ATR inhibitors are warranted in this setting.
ATM inhibition.
ATM is another logical therapeutic target for DDR inhibitors, given its close association with ATR and its crucial function as a central regulator of the DDR, especially in DSB repair144. Two ATM inhibitors are currently being tested in phase I trials: M3541, combined with fractionated palliative radiotherapy, in patients with solid tumours (NCT03225105); and AZD0156, as a monotherapy and in combination with olaparib or 5-fluorouracil, folinic acid and irinotecan, in patients with advanced-stage solid cancers (NCT02588105).
Inhibitors of DNA-P K
DNA-P K, a member of the PI3K–mTOR enzyme family, is a critical enzyme involved in the NHEJ pathway of DNA repair145 (FIG. 1). The specific targeting of NHEJ makes this class of drug especially attractive for combination with radiation because NHEJ is the predominant mechanism for repair of traditional (non-heavy ion) radiation therapy146. Three DNA-PK inhibitors are currently being investigated in phase I/II trials (Supplementary Table 3): M9831 (VX-984), nedisertib (M3814; MSC2490484A) and CC-115 (FIG. 1). CC-115 is a small-molecule inhibitor of both DNA-P K and mTOR that was developed through optimization of a novel series of triazole-containing mTOR inhibitors147. CC-115 monotherapy has been evaluated in a phase I study (NCT01353625) with an initial 44 patients treated across 10 dose-escalation cohorts148. Preliminary antitumour activity was reported, although whether these responses are attributable to activity against DNA-PK or mTOR is unclear — especially considering that CC-115 led to hyperglycaemia, which is consistent with mTOR inhibition, and that associated pharmacodynamic studies provided evidence of mTOR complex 1 (mTORC1) and mTORC2 inhibition148. Planned phase Ib/II trials include combination studies of CC-115 with androgen-deprivation therapy (ADT) in patients with CRPC (NCT02833883) or with radiation in those with glioblastoma (NCT02977780).
The DNA-PK inhibitor nedisertib has been tested in combination with palliative radiation therapy in a phase I trial involving patients with tumours or metastases in the head and neck or thoracic regions; 2 of 7 patients had grade 3 mucositis, and 2 patients had local disease control lasting >300 days149. Multiple trials of nedisertib alone or with definitive chemotherapy and/or radiotherapy are underway (Supplementary Table 3). M9831 is being assessed with and without pegylated doxorubucin in an ongoing phase I study initially in patients with advanced-stage solid tumours, followed by an expansion cohort comprising patients with metastatic endometrial cancer that has progressed after platinum-based chemotherapy (NCT02644278). Biomarkers of responsiveness to DNA-PK inhibition remain in preclinical development, but HRDness could theorectically predict sensitivity to DNA-PK inhibitors given the increased reliance of HRD cells on NHEJ.
Inhibitors of CHK1/2
The cell cycle checkpoint kinases CHK1 and CHK2 act in coordination with DDR pathways and are immediate targets of ATR and ATM, respectively (FIG. 1). Inhibitors of CHK1 and/or CHK2 have a long development history with numerous compounds being discontinued before phase III testing, in most cases owing to toxicity150, such as UCN-01 (7-hydroxystaurosporine)151,152. Many of these inhibitors, including UCN-01 are nonspecific, inhibiting both CHK1 and CHK2, as well as other targets150. AZD7762, an inhibitor of CHK1 with equal potency against CHK2, has been tested alone or in combination with gemcitabine or irinotecan in patients with advanced-stage solid tumours153–155. In addition to low efficacy, severe cardiac toxicities, including myocardial infarction, ventricular dysfunction and troponin elevations, were observed153–155. Interestingly, one patient with RAD50-mutant, ATM-deficient SCLC achieved a CR with the combination of irinotecan and AZD7762, suggesting that biomarkers of sensitivity to CHK1 inhibition will likely overlap with those of ATR inhibitors156. The relatively selective CHK1 inhibitor rabusertib (LY2603618), has also been assessed in combination with different chemotherapies, but these trials revealed limited antitumour activity and a high incidence of serious thromboembolic events157,158. Another selective CHK1 inhibitor, MK-8776 (SCH 900776) has undergone phase I testing as a monotherapy and in combination with gemcitabine and cytarabine for solid tumours and acute myeloid leukaemia, respectively; a high frequency of QTc prolongation with both MK-8776 monotherapy and combination regimens was a concern159,160. Neither AZD7726, rabusertib, nor MK-8776 are being tested in active clinical trials.
Currently, clinical evaluation of three selective CHK1 inhibitors is ongoing: prexasertib (LY2606368), GDC-575 (ARRY-575; RG7741) and CCT245737 (SRA737). A high frequency of grade 4 neutropenia (~73%) was observed in patients treated on the phase I and phase Ib trials with single-agent prexasertib, a second-generation CHK1-selective inhibitor, which was generally manageable and transient (lasting <5 days)161,162. Preliminary data from a phase II trial in 22 patients with advanced-stage, high-grade serous ovarian cancers demonstrated 5 PRs in patients without detectable BRCA1/2 mutations; again, grade 3–4 prexasertib-related neutropenia was common163. This drug is undergoing further testing in a number of phase I–II trials across various treatment and disease settings (Supplementary Table 3). GDC-575 (NCT01564251) and CCT245737 (NCT02797964 and NCT02797977) are in phase I testing as single agents or in combination with gemcitabine-based chemotherapy.
Inhibitors of WEE1
WEE1 is a protein kinase that inhibits cyclin-dependent kinase 1 and 2 (CDK1/2), thereby activating the G2/M cell cycle checkpoint, causing cell cycle arrest and providing time for DNA damage repair (FIG. 1). Thus, inhibition of WEE1 prevents G2 checkpoint initiation, leading to unscheduled mitotic entry, increased replication stress via uncontrolled firing of replication origins, subsequent nucleotide starvation and loss of genomic integrity164,165. Given these effects, a strong biological rationale supports the targeting of p53-deficient cells with WEE1 inhibitors, given the key role of p53 in the regulation of the G1 checkpoint and, therefore, the increased reliance on the G2 checkpoint in p53-deficient cells164,165. Indeed, preclinical studies with adavosertib (AZD1775; MK-1775) have shown that this WEE1 inhibitor abrogates the G2 checkpoint and sensitizes p53-deficient cells to DNA-damaging chemotherapies and radiation owing to mitotic lethality164–166. As such, current developmental strategies have focused on using WEE1 inhibitors in combination with other DNA-damaging treatments (including PARP inhibitors, chemotherapy or radiation therapy) in patients with tumours harbouring TP53 mutations. In addition, preclinical data also suggest increased sensitivity to WEE1 inhibition through mechanisms outside of cell cycle checkpoint defects, such as DDR aberrations and nucleotide resource starvation, with single-agent activity observed even in TP53-wild-type cancer cells167–170.
Adavosertib is the first-in-class WEE1 inhibitor and the only WEE1 inhibitor currently in clinical development (TABLE 2; Supplementary Table 3). Establishing an optimal dose and schedule of adavosertib that achieves an acceptable therapeutic index at active drug doses seems to be a major challenge. The phase I study of adavosertib either as monotherapy or in combination with DNA-damaging chemotherapy in patients with advanced-stage solid tumours resulted in a somewhat unconventional dose and schedule171 (TABLE 2). The rationale for the MTDs of adavosertib in combination with chemotherapeutic agents was based on pharmacokinetic data indicating that, when given in combination with chemotherapy, serum adavosertib concentrations after the fifth dose exceeded the threshold established in preclinical models to be safe171. Nevertheless, grade 3 adverse events were common (55% in all evaluable patients), including haematological and gastrointestinal toxicities171 (TABLE 2). Promisingly, in another phase I study172, adavosertib monotherapy led to 2 PRs in 25 evaluable patients, both in patients with refractory BRCA1-mutant solid tumours. Retrospective tissue analysis showed 5 patients had tumours with TP53 mutations, but none had a response (despite the strong preclinical rationale for WEE1 inhibition in this context)172. While the sample set was limited, pharmacodynamic analyses of paired skin biopsy samples showed a reduction in the levels of phosphorylated CDK1/2, with phosphorylated histone H2AX staining providing evidence of DSBs172.
Adavosertib has been tested in combination with gemcitabine, cisplatin or carboplatin in the aforementioned phase I study171. When adavosertib was administered in multiple doses with chemotherapy, 47–67% of patients had grade ≥3 adverse events, primarily haematological toxicities (frequencies of 29–57%). TP53 mutations were only weakly associated with an antitumour response (across treatments), with an ORR of 21% in 19 evaluable patients with TP53-mutant tumours versus 12% in 33 patients with TP53-wild-type disease171. In a phase II study of adavosertib plus carboplatin in patients with TP53-mutant refractory ovarian cancer, the ORR was 43%, with one patient having a prolonged CR (lasting >30 months); however, toxicities were again frequent, particularly fatigue, nausea and thrombocytopenia173. In a subsequent phase II study, 121 patients with platinum-sensitive, TP53-mutant ovarian cancers were randomly assigned to receive carboplatin plus paclitaxel with or without adavosertib, with preliminary data at 57% maturity revealing improved PFS with the addition of adavosertib (median 42.9 weeks versus 34.9 weeks; HR 0.55, 95% CI 0.32–0.95; P = 0.03)174. Clinical studies to evaluate adavosertib combined with cisplatin and docetaxel as neoadjuvant therapy before resection (NCT02508246) or with definitive chemoradiation and concurrent cisplatin175 (NCT02585973) in patients with locoregionally advanced HNSCC, as well as in other settings, are ongoing (Supplementary Table 3).
In addition to p53 deficiency164,165, HR repair defects owing to mutations in BRCA1/2 and/or the FANC genes have been shown to predict a response to WEE1 inhibitors176, similar to PARP inhibitors. Other predictive biomarkers being explored for WEE1 inhibition include high levels of EZH2 and mitotic cyclins165. Moreover, deficiencies in trimethylation of histone H3K36 (H3K36me3), which are observed in a range of cancers and specifically in SETD2-mutant cancers168, have been shown to predict for sensitivity to WEE1 inhibition in preclinical models via a mechanism of nucleotide resource depletion and cancer cell starvation167,168. Mechanistically, H3K36me3 deficiency leads to reduced expression of the ribunucleotide-diphosphate reductase subunit M2 (RRM2) and depletion of deoxynucleoside triphosphate pools resulting in apoptosis; in this context, WEE1 inhibition induces synthetic lethality by promoting degradation of RRM2 via dysregulated CDK1/2 activity167. H3K36me3 deficiency and SETD2 mutations might thus serve as potential biomarkers of response to WEE1 inhibitors across different cancer types, and a clinical trial of single-agent adavosertib for patients with SETD2-deficient cancers is underway (NCT03284385).
DR inhibitor combinations
The lack of regular and prolonged responses to DDR inhibitors, even in biomarker-selected populations, points to inherent or acquired mechanisms of resistance to single-agent therapy. In general, tumour sensitivity and resistance to DDR inhibitors will largely be dependent on the remaining proficiency of the underlying SSB and DSB response and repair, cell cycle regulation and chromatin remodelling pathways, as well as the active oncogenic pathways, which can also influence DDR and the availability and/or utilization of cellular resources. Understanding the system-wide biology of these sensitivity and resistance patterns can directly inform combination treatment strategies in order to overcome or prevent resistance and to expand the potential patient populations that might benefit from DDR inhibitors (FIG. 4).
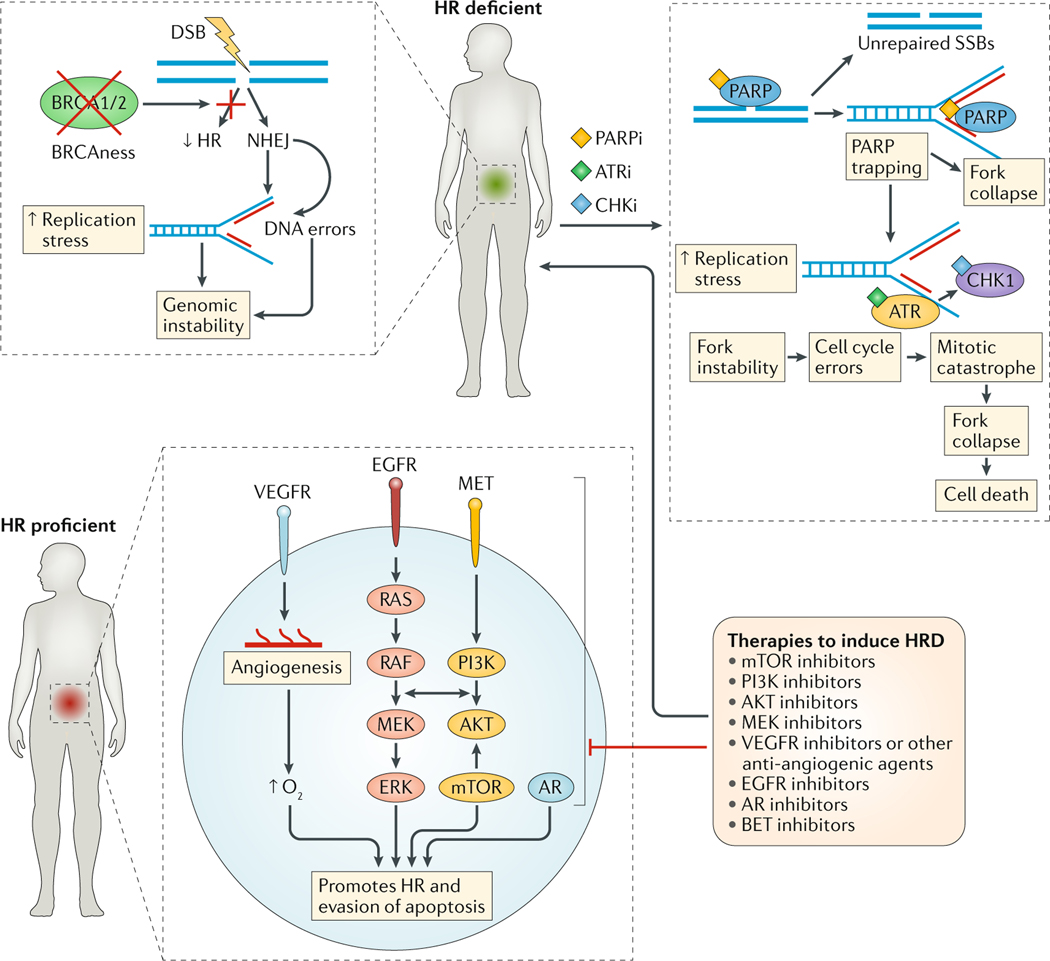
FIG. 4 |. Biomarker-driven combination strategies to augment PARP inhibitor responses.
Cancers that are inherently homologous recombination deficient (HRD) or display ‘BRCAness’/‘HRDness’ are susceptible to poly(ADP-ribose) polymerase (PARP) inhibition (PARPi). Acquired PARP inhibitor resistance arises owing to the phenotypic rescue of homologous recombination (HR) or by mitigation of replication stress, and could potentially be overcome through combination of PARP inhibitors with ATR inhibition (ATRi) and/or inhibition of cell cycle checkpoint kinases (CHKi), such as CHK1. Selected oncogenic drivers and metabolic pathways specific to certain tumour types can drive HR activity to enable cancer cell survival and PARP inhibitor resistance. Therefore, HR-proficient cancer cells can be induced to become HRD through the concept of chemical HRDness by targeting these pro-survival pathways with different molecularly targeted agents, thereby engendering PARP inhibitor sensitivity. Given the rapidly growing number of rational combinations, functional biomarkers of HR, replication stress and PARP trapping are now urgently needed to provide guidance as to which combination should be used for which tumour type and at what time point. AR, androgen receptor; BET, bromodomain and extraterminal motif; DSB, double-strand DNA break; NHEJ, non-homologous end joining; SSBs, single-strand DNA breaks.
Combinations with DNA-damaging agents
Given the ability of radiation to cause anatomically targeted DNA damage, combination of DDR inhibitors with radiotherapy has been the subject of considerable ongoing clinical research. However, early data, mostly with PARP inhibitors, have not demonstrated compelling and consistent evidence of synergy, although nor have they shown unexpected toxic effects177–179. This lack of synergy might be attributable to the fact that DSB repair in response to conventional radiation damage occurs predominately via the NHEJ pathway, rather than by the BER pathway that is primarily mediated by PARP (FIG. 1). Moreover, most DDR inhibitors, including PARP and ATR inhibitors, exploit aberrations in HR repair pathway components, rather than defects in the NHEJ pathway; DNA-PK inhibitors are a notable exception and are promising combination partners for radiotherapy. Furthermore, preclinical studies have revealed that, in comparison with conventional photon-based radiation, more densely ionizing DNA damage induced by irradiation with heavier ions (carbon and iron) or high energy protons results in greater engagement of HR repair and might, therefore, be more attractive for combination with DDR inhibitors146,180.
As discussed, multiple studies of concurrent treatment with DDR inhibitors and chemotherapy have been performed, with the majority of these combinations resulting in additive toxicities with only modest clinical benefit; in most cases, studies comparing the use of DDR inhibitor alone versus in combination with chemotherapy have not been performed55,67,68. By contrast, approaches using sequential administration of chemotherapy followed by DDR inhibitor have proven to be more tolerable and clinically successful, as discussed previously34,36,37,104.
DDR inhibitor–DDR inhibitor combinations
Preclinical studies combining agents targeting key components of the DDR have been shown to overcome acquired resistance to single-agent DDR inhibitors and to induce synthetic lethality. Multiple drug combinations have been evaluated in preclinical studies with the aim of overcoming acquired resistance to DDR inhibitors, predominantly PARP inhibitors (for which the mechanisms of resistance are best understood). For example, PARP inhibitor resistance due to SLFN11 inactivation can be overcome using ATR inhibition, owing to the reliance of SLFN11-deficient cells on the ATR pathway for survival under PARP inhibitor treatment114. Similarly, in BRCA-deficient cancer cells with resistance to PARP inhibition, ATR inhibition further disrupts HR repair leading to replication fork collapse, supporting the development of PARP–ATR inhibitor combinations120 (FIG. 4).
Preclinical studies combining adavosertib with olaparib revealed that WEE1 inhibition enhances PARP inhibitor-mediated radiosensitization of pancreatic cancer cells, apparently by reducing HR repair capacity and abrogating the G2 cell cycle checkpoint181. Similarly, depletion of DNA topoisomerase 2-binding protein 1 (TOPBP1) has been shown to induce sensitivity to PARP inhibition by reducing HR efficiency. The bromodomain and extraterminal (BET) protein BRD4 promotes global gene transcription by RNA polymerase II; BET inhibition has also been shown to suppress the transcription of key DDR genes, including CTIP, BRCA1, RAD51, TOPBP1 and WEE1, thereby increasing the sensitivity of HR-proficient cell lines to PARP inhibition182–184. Furthermore, BET inhibition has been shown to synergize with PARP inhibition in multiple mouse models of HR-proficient tumours182–184.
Early phase clinical trials of PARP inhibitors combined with ATR or WEE1 inhibitors are currently underway (NCT02723864, NCT02576444 and NCT02511795; Supplementary Table 3). Given the vital roles of CHK1, DNA-PK and ATM, as well as ATR and WEE1, at the interface between DDR and cell cycle checkpoint signalling, combination of one or more of these agents with PARP inhibition to induce replication fork collapse and/or mitotic lethality is biologically feasible, even in HR-proficient cells185. In addition, HRD cancers have an increased reliance on alternative repair pathways, including microhomology-mediated end joining (MMEJ) for DSB repair. Thus, a biological rationale exists for combining PARP inhibitors with inhibitors of DNA polymerase-θ (POLQ), a key protein mediating MMEJ repair, to achieve synthetic lethality186. However, these agents all have overlapping toxicity profiles, most notably bone marrow suppression, which will pose a challenge to such combination strategies; both dosing and scheduling of the agents will be crucial to optimizing their application in the clinic.
Inducing HRDness with targeted agents
Preclinical studies have shown that actionable oncoproteins can directly or indirectly regulate DDR and cell cycle checkpoint pathways by driving HR gene expression. For example, prior studies have demonstrated that the RAS, PI3K and AR signalling pathways can all promote HR repair; thus, similar to inhibition of BET proteins182–184, targeting of these oncoproteins together with DDR pathway components has the potential to pharmacologically induce an HRDness phenotype and lead to synthetic lethality in otherwise DDR inhibitor-resistant cells123–125,187, which we have termed ‘chemical HRDness’ (FIG. 4). Preclinical experiments providing evidence of such a chemical HRDness concept have paved the way for a diverse array of combination strategies using a variety of molecularly targeted agents in combination with inhibitors of PARP and potentially other DDR proteins (Supplementary Figure 2), which hold great potential for the treatment of a wide range of tumour types122–125,187–190. In addition, DDR pathways overlap considerably with chromatin remodelling mediated by histone modifications, providing the rationale for combining DDR inhibitors with epigenetic agents, beyond those targeting BRD4 (REF.191). For example, inhibition of PARP attenuates the PARP1-mediated downregulation of EZH2 induced by alkylating DNA damage, and the addition of an EZH2 inhibitor further sensitizes BRCA1/2-mutant cells to PARP inhibition in preclinical models192. Histone deacetylase inhibitors have also been reported to synergize with PARP inhibitors in preclinical studies, a finding associated with downregulation of HR repair genes, suggesting induction of HRDness191,193. Furthermore, DNA methyltransferase (DNMT) inhibitors can improve PARP inhibitor efficacy in BRCA1/2-wild-type cells and leukaemia models by increasing tight binding of PARP1 to chromatin at sites of DNA damage194.
In the preclinical setting, genetically induced PARP knockout models have been shown to inhibit in vivo angiogenesis; although, paradoxically, PARP inhibition has been shown to selectively target hypoxic tumour cells and increase tumour blood vessel perfusion as a potential mechanism of engendering chemosensitivity and radioresponsiveness195–197. Despite the lack of a complete understanding of the effects of PARP inhibition on hypoxia and angiogenesis, promising preclinical findings have led to multiple clinical trials assessing the combination of anti-angiogenic agents with PARP inhibitors. The synergy of such combinations might partially be explained by hypoxia induced by anti-angiogenic therapy leading to HRD through the downregulation of key repair proteins, thereby enhancing sensitivity to PARP inhibitors95,198. In the clinical setting, a phase I trial to assess olaparib combined with the anti-VEGFA antibody bevacizumab showed that the regimen was safe and tolerated, with no DLTs199. In a proof-of-concept trial200, the small-molecule VEGFR inhibitor cediranib was also combined with olaparib in patients with recurrent ovarian cancer (n = 20) or metastatic TNBC (n = 8), resulting in an ORR of 44% in 18 evaluable patients with ovarian cancer; no objective responses were observed in 7 evaluable patients with TNBC. Notably, 75% of the patients had grade ≥3 toxicities, predominantly bone marrow suppression, hypotension and fatigue200. Preliminary data from the ongoing phase II study of this same combination (NCT01116648) in 90 patients with recurrent platinum-sensitive ovarian cancer demonstrated a PFS benefit compared with single-agent olaparib across the entire study population (HR 0.50, 95% CI 0.30–0.83, P = 0.007), with a trend towards improved OS, although not statistically significant201. Grade 3–4 toxicities, including fatigue, diarrhoea and hypertension, were more common with the combination188,201. Interestingly, the PFS benefit of the combination was not seen in subgroup analysis of patients with known germline BRCA1/2 mutations201. These findings are perhaps unsurprising considering that this combination is likely to benefit patients with BRCA1/2-wild-type disease to a greater extent than those with pre-existing HRD due to germline BRCA1/2 mutations, who will already gain benefit from olaparib monotherapy. These data potentially justify the increased risk of toxicities with the combination therapy in the former group. Nevetheless, biomarker-driven trials are needed to determine the optimal subgroup of patients to target. A phase III trial of the olaparib–cediranib combination in patients with BRCA1/2-mutant ovarian cancers is underway (Supplementary Table 1).
Findings from preclinical models have provided a biological rationale and robust evidence of antitumour activity that support combined inhibition of PARP and the PI3K–AKT–mTOR pathway122,187,189 (FIG. 4). Genetic or pharmacological inhibition of PI3K in BRCA1/2-wild-type TNBC cells was shown to induce HRDness via suppression of BRCA1/2 expression, increased DNA damage and compensatory PARP activation, and subsequently sensitized tumours to PARP inhibition187. Downregulation of BRCA1/2 expression upon PI3K inhibition is hypothesized to be mediated via a compensatory increase in MEK–ERK signalling resulting in binding of the transcriptional repressor ETS1 to the BRCA1/2 promoter regions187. By contrast, marked synergy between PARP and MEK inhibitors via reduced HR gene expression, HR repair capacity and cell cycle checkpoint activity and increased PARP1 expression and PARP inhibitor-mediated DNA damage has been reported in other preclinical studies, at least in RAS-mutant or RAF-mutant tumours124. Not surprisingly, in vivo synergy between PI3K and PARP inhibitors has been shown in mouse models of BRCA1-mutant breast cancer, PIK3CA-mutant ovarian cancer and PTEN-mutant endometrial cancers, among others189,202,203. Transcriptomic profiling and pathway analysis has led to the identification of mTOR inhibitors as top candidates for inducing HRDness, even in BRCA1/2-wild-type breast cancer cell lines and xenografts90,123. Indeed, the combination of the mTOR inhibitor everolimus and talazoparib had greater efficacy than either agent alone in preclinical models123. Interestingly, mTOR inhibition did not alter the expression of key HR genes, such as BRCA1/2 or RAD51, but instead induced HRD via the suppression of SUV39H1, a histone methyltransferase involved in DSB repair123.
In the clinical setting, a phase I trial of the pan-class I PI3K inhibitor buparlisib (BKM120) in combination with olaparib (NCT01623349) was undertaken in 70 patients with advanced-stage, high-grade serous ovarian or breast cancers204,205. The ORRs were 29% in those with ovarian cancer, irrespective of platinum sensitivity, and 28% in those with breast cancer204,205 (Supplementary Table 2). Nausea and fatigue were the most common toxicities, whereas DLTs included transaminitis and depression204,205. In view of multiple buparlisib dose reductions needed owing to drug-related toxicities, olaparib was subsequently combined with the PI3Kα-specific inhibitor alpelisib (BYL719)205,206. The MTD was lower than the monotherapy RP2D; DLTs included hyperglycaemia, rash and fever with decreased neutrophil counts. The most common toxicities were those expected with PI3K inhibition, including gastrointestinal symptoms, hyperglycaemia and fatigue. Impressively, among 28 patients with advanced-stage ovarian cancer, 26 (93%) of whom had platinum-resistant disease, the ORR was 36%, with a median duration of response of 167 days (range 16–398 days). The ORR was similar between patients with or without germline BRCA1/2 mutations in both the buparlisib and alpelisib cohorts204–206.
A phase I trial of AKT inhibitor capivasertib (AZD5363) in combination with olaparib in patients with advanced-stage solid tumours (NCT02338622) revealed that the combination was well tolerated; antitumour activity was observed independent of BRCA1/2 status or previous exposure to platinum-based chemotherapy, PARP inhibitors or PI3K pathway inhibitors207.
Supported by the aforementioned preclinical findings124, the combination of the MEK inhibitor selumetinib and olaparib is currently being assessed in a phase I trial involving patients with endometrial, ovarian or other solid tumours with RAS pathway alterations, as well as those with difficult to treat PARP inhibitor-resistant ovarian cancers (NCT03162627). This combination could potentially be of great importance, given the difficulties associated with targeting RAS-mutant tumours (and considering that RAS pathway alterations are among the most common drivers of tumorigenesis). Furthermore, the approval of PARP inhibitors for multiple indications and the almost inevitable emergence of resistance is rapidly leading to a population of patients for whom resensitization to PARP inhibitors is an urgent need. In PARP inhibitor-resistant preclinical models, the combination of PARP and MEK or other MAPK inhibitors is highly active124,208.
Induction of oncogenic MYC in myeloma cell lines induces replication and oxidative stresses, which cause DSBs requiring the activation of HR repair pathways209. Similarly, amplification of MYC correlates with increased RAD51 expression in both germline BRCA1/2-mutant and sporadic TNBCs, and women with tumours overexpressing both MYC and RAD51 have an unfavourable prognosis190. Treatment with the pan-CDK inhibitor dinaciclib (SCH-727965) downregulates MYC expression in TNBC cells, although studies have shown that MYC loss alone is insufficiently lethal, thus providing the rationale for combination therapy190. The combination of dinaciclib with niraparib results in increased DNA damage and downregulated HR activity, resulting in suppression of epithelial-to-mesenchymal transition, and importantly resensitizes PARP inhibitor-resistant TNBC cells to niraparib190. This synthetic lethal treatment strategy is active across ovarian, prostate, pancreatic, colon and lung cancer cell lines190. The precise mechanism of dinaciclib-induced MYC downregulation remains unclear, although several CDKs are implicated in regulating DNA repair190. A phase I study of dinaciclib in combination with veliparib (NCT01434316) has been undertaken in patients with HR-proficient advanced-stage solid tumours. Overall, the combination was poorly tolerated, probably owing to the toxicities associated with dinaciclib monotherapy, and limited antitumour activity was observed210. Combinations involving more-selective CDK inhibitors and more-potent PARP inhibitors might be a better approach to testing this hypothesis in patients.
Emerging data also support a synthetic lethal relationship between AR signalling and the PARP and ATR–CHK1 axes in prostate cancers. AR signalling has a key role in regulating HR repair, whereas AR inhibition activates the PARP pathway in mouse models of this disease125,211,212. Moreover, ADT has been shown to functionally impair HR repair before the development of castration resistance in prostate cancer180, providing the rationale for the use of PARP inhibitors in combination with ADT upfront in patients with advanced-stage or high-risk prostate cancers. Preclinical models have revealed that the combined or sequential targeting of the AR with PARP and/or ATR inhibition leads to synthetic lethality, regardless of DDR gene mutation status125,211,212. Correspondingly, data from a phase II trial of abiraterone combined with olaparib in patients with mCRPC showed improved radiographic PFS compared with that observed with abiraterone alone, including in patients without DDR gene mutations213. Another randomized phase II trial is currently underway to compare the AR antagonist enzalutamide, olaparib and combination of both agents specifically in patients with mCRPC harbouring DNA repair defects (NCT03012321).
Thus, multiple combination or sequential treatment strategies involving DDR inhibitors are now being tested. In order to optimize their development, efforts should be focused on using adaptive trial designs with dynamic, functional biomarkers of DDR and replication stress in order to pursue strategies to maintain the cancer cells in a state of perpetual DDR deficiency and minimize issues of acquired resistance.
Combinations with immunotherapies
Multiple lines of evidence have provided a biological rationale, and demonstrated synergistic benefit, for combining immune-checkpoint inhibitors (ICIs) with DDR inhibitors, while defects in DDR pathways might potentially serve as predictive biomarkers of responsiveness to ICIs. DDR proteins maintain the integrity of the genome and, therefore, the use of DDR inhibitors might increase the tumour mutational burden (TMB), which in turn could lead to neoantigen production and enhanced anticancer T cell activity. Studies have supported the association of a high TMB and dynamic neoantigen renewal with neoepitope-specific T cell responses against mismatch repair (MMR)-deficient tumours treated with ICIs214–216. Thus, the high ORRs (40–71%) are observed in patients with MMR-deficient tumours treated with anti-programmed cell death protein 1 (PD-1) antibodies are unsurprising, given the mutator phenotype engendered by defects in this DNA repair pathway214,215. These impressive clinical findings led to the histology-agnostic FDA approval of the anti-PD-1 antibody pembrolizumab for patients with MMR-deficient cancers. However, direct evidence that targeting of DSB repair proteins with DDR inhibitors causes an increased TMB is only beginning to emerge217. The mechanisms underlying the potential benefit of immunotherapy in patients with HRD tumours and/or the rationale for combining ICIs with DDR inhibitors are multifactorial.
First, S phase-specific DSBs in cells with DDR gene aberrations result in the accumulation of cytosolic DNA, which activates stimulator of interferon genes (STING); STING subsequently mediates the release of type I interferons, which activate T cells and facilitate the innate immune recognition of immunogenic tumours, but is associated with upregulation of immunosuppressive PD-L1 expression5,180,218. Preclinical studies have also revealed that PARP inhibition upregulates PD-L1 expression via inactivation of GSK3β and consequently attenuates antitumour immunity219. Moreover, BRCA2 or Ku70/80 depletion has been shown to enhance CHK1-dependent and interferon signalling-dependent upregulation of PD-L1 on cancer cell lines following PARP inhibition and/or irradiation220.
Second, a high burden of indel mutations in tumours predicts for higher levels of neoantigens and better responses to ICIs in preclinical models221. Given that HRD typically results in a higher number of indels, neoantigen generation might contribute to the improved immunotherapy responses of these tumours. In HRD tumours, DSBs might also contribute to genomic instability and copy-number aberrations, which seem to be associated with responsiveness to both PARP inhibitors and ICIs222. Thus, the neoantigen burden and profile of a tumour is probably partially dependent on the specific defect in the DDR, resulting in substantial heterogeneity.
Together, these findings suggest that HRD and/or DDR inhibitors might create immunological vulnerabilities in tumours, while simultaneously activating immunosuppressive pathways. Thus, targeting HRD or even HR-proficient tumours with anti-PD-1 or anti-PD-L1 antibodies in combination with PARP inhibitors or other DDR inhibitors is attractive, particularly given that these agents have distinct, mostly non-overlapping toxicities. Indeed, combinations of ICIs and DDR inhibitors are currently undergoing clinical testing.
In a small cohort of patients with advanced-stage gynaecological or breast cancers (n = 12), Lee and colleagues223 established the combination RP2D at full monotherapy doses of both olaparib and durvalumab, with no DLTs observed (Supplementary Table 2). Overall, two PRs (lasting ≥15 months and ≥11 months) and eight stable disease responses with a median duration of 8 months (range 4 months to 14.5 months) were observed, equating to a disease control rate (DCR) of 83%223. Preliminary data from the phase II MEDIOLA trial of the same drug combination indicated a similarly impressive DCR of 80% for patients with advanced-stage germline BRCA1/2-mutant breast cancers; the ORR (all PRs) was 52%224.
Early data from a phase II trial of the olaparib–durvalumab combination in patients with mCRPC (NCT02484404) demonstrated an acceptable toxicity profile and serum prostate-specific antigen responses in 12 of 17 patients (71%), with a median PFS of 16.1 months for all patients who received the combination225. Importantly, biochemical responses were observed independently of known mutations in DNA repair genes225. Interestingly, T cell activation was shown to be associated with improved PFS225. At this early stage of clinical development, although apparently well tolerated, the extent to which anti-PD-1 or anti-PD-L1 antibodies will enhance the effects of PARP inhibition in patients with germline BRCA1/2-mutant cancers remains unclear. Other active areas of research include the determination of other tumours or molecular subtypes of cancers in which this combination will ultimately have biological and clinical activity.
Outside of PARP inhibitors, the ATR inhibitor AZD6738 is currently being tested in combination with durvalumab in a phase Ib trial involving patients with advanced-stage NSCLC or HNSCC (NCT02264678; Supplementary Table 3); patients will receive AZD6738 daily during a run-in period before each dose of durvalumab in 28-day cycles. Initial data from this trial indicate that the combination is well tolerated, with no DLTs observed, with preliminary signals of activity132 (TABLE 2). In the phase Ib, randomized, multi-drug, biomarker-directed BISCAY study in patients with metastatic muscle-invasive bladder cancer (NCT02546661), patients with any HR repair defect detected will receive durvalumab with or without olaparib; those with detectable CDKN2A or RB1 deficiency and/or amplifications of CCNE1, MYC, MYCL or MYCN will receive durvalumab with or without the WEE1 inhibitor adavosertib, owing to the increased replication stress and dysfunction in cell cycle response associated with these genomic aberrations226. Adavosertib is also being combined with durvalumab in a phase I trial to test different treatment schedules in patients with advanced-stage solid tumours (NCT02617277).
Conclusions
Great strides have been made in successfully targeting DNA repair in cancer medicine, with multiple selective and potent DDR inhibitors emerging in the clinic. The regulatory approvals of different PARP inhibitors in the management of patients with BRCA1/2-mutant ovarian and breast cancers and in the maintenance setting for those with platinum-sensitive ovarian cancer, regardless of mutation status, are major breakthroughs. Indeed, ovarian cancer has served as the poster child for the use of PARP inhibitors, although clear signals of acceptable safety and antitumour activity have been observed across multiple other cancer types and molecular subtypes, including in patients who are wild type for canonical DDR genes. The oncology community’s clinical research focus for the development of PARP and other DDR inhibitors should now shift to transforming more nonresponders into responders and turning more responders into ‘super-responders’ with a markedly increased depth and duration of response.
To produce a large step change and improvements in patient outcomes, clear priority should now be given to the development of more inclusive and functionally based predictive biomarker assays. Moreover, an improved understanding of resistance mechanisms to different DDR inhibitors and their interactions with one another is needed to facilitate the development of novel combination strategies. While combinations designed to induce tumour HRDness are a rational approach and have demonstrated clinical potential, the appropriate dosing and scheduling of each agent to minimize adverse events while maximizing benefit will also be key to optimizing outcomes. The clear and mounting evidence that genomic instability strongly influences the tumour immune microenvironment is paving the way for biomarker development as well as treatment strategies to target this hallmark of cancer with DDR inhibitors in conjunction with immunotherapy.
Preclinical and clinical data discussed herein emphasize the complex and far-reaching nature of DDR pathways. The ability to maximize the benefit of DDR inhibitors requires a better system-wide understanding of the dynamic mechanisms that tissue-specific cancer cells use to mitigate genomic instability and how this instability influences innate and acquired immunity. In the clinic, treatment strategies using DDR inhibitors should be optimized through the use of adaptive, longitudinal trial designs with integrated functional biomarkers to truly personalize antitumour DDR therapies and increase patient benefit.
Supplementary Material
Key points.
The DNA damage response (DDR) involves a complex network of genes responsible for sensing and responding to specific types of DNA damage, encompassing specific machineries mediating DNA repair, cell cycle regulation, replication stress responses and apoptosis.
Defects in the DDR give rise to genomic instability in cells, aiding in cancer initiation and progression via mutation accumulation, but also providing targetable vulnerabilities relatively specific to cancer cells that can be exploited for clinical benefit with the use of DDR inhibitors.
Targeting BRCA1/2-deficient cancers using poly(ADP-ribose) polymerase (PARP) inhibitors is the archetype of synthetic lethality, but now the therapeutic landscape of DDR inhibitors is rapidly expanding; bridging preclinical data on each of these agents to the clinical setting is vital to inform appropriate biomarkers and timing for their use.
Preclinical and clinical data on DDR inhibitors indicate that biomarkers of response and resistance extend beyond BRCA1/2 to provide a more inclusive and functionally informed approach to patient selection.
Preclinical and clinical research with PARP inhibition has revealed multiple resistance mechanisms across a variety of cancer subtypes, highlighting the need for functional biomarkers and sequential or combination treatment strategies.
While impressive clinical responses can be seen rarely with the use of single-agent DDR inhibitors, a multitude of biologically informed combination treatment strategies using a backbone of DDR inhibitors are under development to extend their use to larger populations, while minimizing overlapping toxicities.
Acknowledgements
The University of Texas MD Anderson Cancer Center (Houston, TX, USA) is supported by the NIH Cancer Center Support Grant CA016672.
G.B.M. has received research support from AstraZeneca, Pfizer and Tesaro, and has been a consultant for AstraZeneca, Pfizer and Tesaro. T.A.Y. has received research support from AstraZeneca, Bayer, Pfizer, Tesaro and Vertex Pharmaceuticals; has served on advisory boards of Aduro, Almac, AstraZeneca, Atrin, Bristol-Myers Squibb, Clovis, EMD Serono, Ignyta, Jansen, Merck, Pfizer, Roche, Seattle Genetics and Vertex Pharmaceuticals; and has received speaker bureau from AstraZeneca, Merck and Tesaro.
Footnotes
Competing interests
The other authors declare no competing interests.
Reviewer information
Nature Reviews Clinical Oncology thanks T. Laetsch and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41571–018-0114-z.
References
- 1.Roos WP, Thomas AD & Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 16, 20–33 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Xu Y. DNA damage: a trigger of innate immunity but a requirement for adaptive immune homeostasis. Nat. Rev. Immunol. 6, 261–270 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Tubbs A & Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkes EE et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J. Natl. Cancer Inst. 109, djw199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson LR et al. Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin. Cancer Biol. 35 (Suppl.), 5–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lord CJ & Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong PC et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 28, 2512–2519 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Fong PC et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Bryant HE et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kaufman B. et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 33, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashworth A & Lord CJ Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 15, 564–576 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Murai J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satoh MS & Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Kedar PS, Stefanick DF, Horton JK & Wilson SH Increased PARP-1 association with DNA in alkylation damaged, PARP-inhibited mouse fibroblasts. Mol. Cancer Res. 10, 360–368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carney B. et al. Target engagement imaging of PARP inhibitors in small-cell lung cancer. Nat. Commun. 9, 176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown JS, Kaye SB & Yap TA PARP inhibitors: the race is on. Br. J. Cancer 114, 713–715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swisher EM et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 18, 75–87 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Gelmon KA et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 12, 852–861 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Kaye SB et al. Phase II, open-label, randomized, multicenter study comparing the efficacy and safety of olaparib, a poly (ADP-ribose) polymerase inhibitor, and pegylated liposomal doxorubicin in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer. J. Clin. Oncol. 30, 372–379 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Oza AM et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: integrated analysis of data from Study 10 and ARIEL2. Gynecol. Oncol. 147, 267–275 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Sandhu SK et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 14, 882–892 (2013). [DOI] [PubMed] [Google Scholar]
- 23.de Bono J. et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 7, 620–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litton JK et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 379, 753–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong R. et al. 53BP1 depletion causes PARP inhibitor resistance in ATM-deficient breast cancer cells. BMC Cancer 16, 725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaspers JE et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 3, 68–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenblick A, de Azambuja E, Azim HA & Piccart M. An update on PARP inhibitors — moving to the adjuvant setting. Nat. Rev. Clin. Oncol. 12, 27–41 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Moore KN, Mirza MR & Matulonis UA The poly (ADP ribose) polymerase inhibitor niraparib: management of toxicities. Gynecol. Oncol. 149, 214–220 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Kim G. et al. FDA Approval Summary: Olaparib monotherapy in patients with deleterious germline BRCA-m utated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 21, 4257–4261 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Balasubramaniam S. et al. FDA Approval Summary: Rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin. Cancer Res. 23, 7165–7170 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Robson M. et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 377, 523–533 (2017). [DOI] [PubMed] [Google Scholar]
- 32.National Comprehensive Cancer Network. NCCN guidelines. NCCN https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf (2017). [Google Scholar]
- 33.Ledermann J. et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 366, 1382–1392 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Ledermann J. et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 15, 852–861 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Ledermann JA et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 17, 1579–1589 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Pujade-Lauraine E. et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-O v21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1274–1284 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Mirza MR et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 375, 2154–2164 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Coleman RL et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 1949–1961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristeleit RS et al. Clinical activity of the poly(ADP-ribose) polymerase (PARP) inhibitor rucaparib in patients (pts) with high-grade ovarian carcinoma (HGOC) and a BRCA mutation (BRCAmut): Analysis of pooled data from Study 10 (parts 1, 2a, and 3) and ARIEL2 (parts 1 and 2). Ann. Oncol. 27, 8560–8560 (2016). [Google Scholar]
- 40.Wooster R. et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science 265, 2088–2090 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Miki Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994). [DOI] [PubMed] [Google Scholar]
- 42.King M-C, Marks JH & Mandell JB, New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Mandelker D. et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA versus guideline-based germline testing. JAMA 318, 825–835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritchard CC et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 375, 443–453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrader KA et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2, 104–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meric-Bernstam F. et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann. Oncol. 27, 795–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shindo K. et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J. Clin. Oncol. 35, 3382–3390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettl J. et al. Quality of life with talazoparib versus physician’s choice of chemotherapy in patients with advanced breast cancer and germline BRCA1/2 mutation: patient-reported outcomes from the EMBRACA phase III trial. Ann. Oncol. 10.1093/annonc/mdy257 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Rugo HS et al. Adaptive randomization of veliparib-carboplatin treatment in breast cancer. N. Engl. J. Med. 375, 23–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loibl S. et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol. 19, 497–509 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Litton JK, Scoggins M, Ramirez D & Arun B. A pilot study of neoadjuvant talazoparib for early-stage breast cancer patients with a BRCA mutation. Ann. Oncol. 27, 43–67 (2016). [Google Scholar]
- 52.Litton JK et al. Neoadjuvant talazoparib (TALA) for operable breast cancer patients with a BRCA mutation (BRCA+). J. Clin. Oncol. 36, 508–508 (2018). [Google Scholar]
- 53.Mateo J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodall J. et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 7, 1006–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bendell J. et al. Phase I study of olaparib plus gemcitabine in patients with advanced solid tumours and comparison with gemcitabine alone in patients with locally advanced/metastatic pancreatic cancer. Ann. Oncol. 26, 804–811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim HS et al. Concordance of ATM (ataxia telangiectasia mutated) immunohistochemistry between biopsy or metastatic tumor samples and primary tumors in gastric cancer patients. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 80, 127–137 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Kubota E. et al. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle 13, 2129–2137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bang Y-J et al. Randomized, double-blind phase II trial with prospective classification by ATM protein level to evaluate the efficacy and tolerability of olaparib plus paclitaxel in patients with recurrent or metastatic gastric cancer. J. Clin. Oncol. 33, 3858–3865 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Bang Y-J et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1637–1651 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Smyth E. Missing a GOLDen opportunity in gastric cancer. Lancet Oncol. 18, 1561–1563 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Mo M, Yang J, Zhu X & Zhu J. Use of olaparib in patients with advanced gastric cancer. Lancet Oncol. 19, e75 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Ballinger ML et al. Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol. 17, 1261–1271 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Felsenstein KM & Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat. Rev. Urol. 15, 92–111 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Daly MB et al. NCCN Guidelines Insights: Genetic/familial high-risk assessment: breast and ovarian, version 2.2017. J. Natl Compr. Cancer Netw. 15, 9–20 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Knijnenburg TA et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 23, 239–254.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinn JA et al. Phase II trial of temozolomide plus o6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J. Clin. Oncol. 27, 1262–1267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajan A. et al. A phase I combination study of olaparib with cisplatin and gemcitabine in adults with solid tumors. Clin. Cancer Res. 18, 2344–2351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dréan A, Lord CJ & Ashworth A. PARP inhibitor combination therapy. Crit. Rev. Oncol. Hematol. 108, 73–85 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Turner N, Tutt A & Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer 4, 814–819 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Metzger MJ, Stoddard BL & Monnat RJ PARP-mediated repair, homologous recombination, and back-up non-homologous end joining-like repair of single-strand nicks. DNA Repair 12, 529–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sen T, Gay CM & Byers LA Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl Lung Cancer Res. 7, 50–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abkevich V. et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 107, 1776–1782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watkins JA, Irshad S, Grigoriadis A & Tutt ANJ Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 16, 211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang ZC et al. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin. Cancer Res. 18, 5806–5815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.González Martín A. Progress in PARP inhibitors beyond BRCA mutant recurrent ovarian cancer? Lancet Oncol. 18, 8–9 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Garsed DW et al. Homologous recombination DNA repair pathway disruption and retinoblastoma protein loss are associated with exceptional survival in high-grade serous ovarian cancer. Clin. Cancer Res. 24, 569–580 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Peng G & Mills GB Surviving ovarian cancer: an affair between defective DNA repair and RB1. Clin. Cancer Res. 24, 508–510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davies H. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polak P. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 49, 1476–1486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lord CJ & Ashworth A. BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Gilardini Montani MS et al. ATM-depletion in breast cancer cells confers sensitivity to PARP inhibition. J. Exp. Clin. Cancer Res. CR 32, 95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu C. et al. Targeting AURKA-CDC25C axis to induce synthetic lethality in ARID1A-deficient colorectal cancer cells. Nat. Commun. 9, 3212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen J. et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 5, 752–767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu H. et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Natl Acad. Sci. USA 111, 285–290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parrotta R. et al. A novel BRCA1-associated protein-1 isoform affects response of mesothelioma cells to drugs impairing BRCA1-mediated DNA repair. J. Thorac. Oncol. 12, 1309–1319 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Bajrami I. et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 74, 287–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Y-M et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yi S. et al. Functional variomics and network perturbation: connecting genotype to phenotype in cancer. Nat. Rev. Genet. 18, 395–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peng G. et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun. 5, 3361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Konstantinopoulos PA et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 28, 3555–3561 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Severson TM et al. The BRCA1ness signature is associated significantly with response to PARP inhibitor treatment versus control in the I-SPY 2 randomized neoadjuvant setting. Breast Cancer Res. 19, 99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolf DM et al. DNA repair deficiency biomarkers and the 70-gene ultra-high risk signature as predictors of veliparib/carboplatin response in the I-SPY 2 breast cancer trial. NPJ Breast Cancer 3, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weinstein JN & Lorenzi PL Cancer: discrepancies in drug sensitivity. Nature 504, 381–383 (2013). [DOI] [PubMed] [Google Scholar]
- 95.Sulkowski PL et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl Med. 9, eaal2463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pietanza MC et al. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386–2394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Allison Stewart C. et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8, 28575–28587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Byers LA et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2, 798–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bélanger F. et al. Replication protein a availability during DNA replication stress is a major determinant of cisplatin resistance in ovarian cancer cells. Cancer Res. 10.1158/0008-5472.CAN-180618 (2018). [DOI] [PubMed] [Google Scholar]
- 100.Lu Y. et al. Chemosensitivity of IDH1-mutated gliomas due to an impairment in PARP1-mediated DNA repair. Cancer Res. 77, 1709–1718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aparicio AM et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin. Cancer Res. 22, 1520–1530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aparicio AM et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin. Cancer Res. 19, 3621–3630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McGrail DJ et al. Improved prediction of PARP inhibitor response and identification of synergizing agents through use of a novel gene expression signature generation algorithm. NPJ Syst. Biol. Appl. 3, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lheureux S. et al. Long-term responders on olaparib maintenance in high-grade serous ovarian cancer: clinical and molecular characterization. Clin. Cancer Res. 23, 4086–4094 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Kondrashova O. et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 7, 984–998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Quigley D. et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 7, 999–1005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patch A-M et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015). [DOI] [PubMed] [Google Scholar]
- 108.Domchek SM Reversion mutations with clinical use of PARP inhibitors: many genes, many versions. Cancer Discov. 7, 937–939 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Johnson N. et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl Acad. Sci. USA 110, 17041–17046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ter Brugge P. et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1deficient breast cancer. J. Natl. Cancer Inst. 108, djw148 (2016). [DOI] [PubMed] [Google Scholar]
- 111.Fojo T & Bates S. Mechanisms of resistance to PARP inhibitors — three and counting. Cancer Discov. 3, 20–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ray Chaudhuri A. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S-B et al. Tousled-like kinases stabilize replication forks and show synthetic lethality with checkpoint and PARP inhibitors. Sci. Adv. 4, eaat4985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murai J. et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 7, 76534–76550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lok BH et al. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin. Cancer Res. 23, 523–535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Murai J. et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol. Cell 69, 371–384. e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Henneman L. et al. Selective resistance to the PARP inhibitor olaparib in a mouse model for BRCA1deficient metaplastic breast cancer. Proc. Natl Acad. Sci. USA 112, 8409–8414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gogola E. et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 33, 1078–1093.e12 (2018). [DOI] [PubMed] [Google Scholar]
- 119.Pettitt SJ et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yazinski SA et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bitler BG, Watson ZL, Wheeler LJ & Behbakht K. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol. Oncol. 147, 695–704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cardnell RJ et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin. Cancer Res. 19, 6322–6328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mo W. et al. mTOR inhibitors suppress homologous recombination repair and synergize with PARP inhibitors via regulating SUV39H1 in BRCA-proficient triple-negative breast cancer. Clin. Cancer Res. 22, 1699–1712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun C. et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci. Transl Med. 9, eaal5148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li L. et al. Androgen receptor inhibitor-induced ‘BRCAness’ and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal. 10, eaam7479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sundar R, Brown J, Ingles Russo A & Yap TA Targeting ATR in cancer medicine. Curr. Probl. Cancer 41, 302–315 (2017). [DOI] [PubMed] [Google Scholar]
- 127.O’Carrigan B. et al. Phase I trial of a first-in-class ATR inhibitor VX-970 as monotherapy (mono) or in combination (combo) with carboplatin (CP) incorporating pharmacodynamics (PD) studies [abstract]. J. Clin. Oncol. 34, 2504 (2016). [Google Scholar]
- 128.Yap TA et al. Phase I trial of first-in-class ataxia telangiectasia-mutated and Rad3-related (ATR) inhibitor VX-970 as monotherapy (mono) or in combination with carboplatin (CP) in advanced cancer patients (pts) with preliminary evidence of target modulation and antitumor activity [abstract]. Mol. Cancer Ther. 14, R14 (2015). [Google Scholar]
- 129.Shapiro G. et al. Phase 1 trial of first-in-class ATR inhibitor VX-970 in combination with cisplatin (Cis) in patients (pts) with advanced solid tumors (NCT02157792) [abstract]. Cancer Res. 76, CT012 (2016). [Google Scholar]
- 130.Plummer ER et al. Phase I trial of first-in-class ATR inhibitor VX-970 in combination with gemcitabine (Gem) in advanced solid tumors (NCT02157792) [abstract]. J. Clin. Oncol. 34, 2513 (2016). [Google Scholar]
- 131.Dillon MT et al. A Phase I dose-escalation study of ATR inhibitor monotherapy with AZD6738 in advanced solid tumors (PATRIOT Part A) [abstract]. Cancer Res. 77, CT084 (2017). [Google Scholar]
- 132.Yap TA et al. Phase I modular study of AZD6738, a novel oral, potent and selective ataxia telangiectasia Rad3-related (ATR) inhibitor in combination (combo) with carboplatin, olaparib or durvalumab in patients (pts) with advanced cancers [abstract 1LBA]. Eur. J. Cancer 69, S2 (2016). [Google Scholar]
- 133.Middleton FK et al. Common cancer-associated imbalances in the DNA damage response confer sensitivity to single agent ATR inhibition. Oncotarget 6, 32396–32409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mohni KN et al. A synthetic lethal screen identifies DNA repair pathways that sensitize cancer cells to combined ATR inhibition and cisplatin treatments. PLOS ONE 10, e0125482 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim H. et al. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin. Cancer Res. 23, 3097–3108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reaper PM et al. Selective killing of ATM-or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 7, 428–430 (2011). [DOI] [PubMed] [Google Scholar]
- 137.Williamson CT et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat. Commun. 7, 13837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.VandenBussche CJ et al. Alternative lengthening of telomeres and ATRX/DAXX loss can be reliably detected in FNAs of pancreatic neuroendocrine tumors. Cancer 125, 544–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cesare AJ & Reddel RR Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 (2010). [DOI] [PubMed] [Google Scholar]
- 140.Flynn RL et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 347, 273–277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deeg KI, Chung I, Bauer C & Rippe K. Cancer cells with alternative lengthening of telomeres do not display a general hypersensitivity to ATR inhibition. Front. Oncol. 6, 186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim H-J et al. Anti-tumor activity of the ATR inhibitor AZD6738 in HER2 positive breast cancer cells. Int. J. Cancer 140, 109–119 (2017). [DOI] [PubMed] [Google Scholar]
- 143.Yan Y. et al. A novel function of HER2/Neu in the activation of G2/M checkpoint in response to γ-irradiation. Oncogene 34, 2215–2226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Weber AM & Ryan AJ ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Brown JS, O’Carrigan B, Jackson SP & Yap TA Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 7, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Willers H, Dahm-Daphi J & Powell SN Repair of radiation damage to DNA. Br. J. Cancer 90, 1297–1301 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mortensen DS et al. Optimization of a series of triazole containing mammalian target of rapamycin (mTOR) kinase inhibitors and the discovery of CC-115. J. Med. Chem. 58, 5599–5608 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Munster PN et al. Phase I trial of a dual TOR kinase and DNA-PK inhibitor (CC-115) in advanced solid and hematologic cancers. J. Clin. Oncol. 34, 2505 (2016). [Google Scholar]
- 149.Van Triest B. et al. A phase Ia/Ib trial of the DNA-dependent protein kinase inhibitor (DNA-PKi) M3814 in combination with radiotherapy in patients with advanced solid tumors. J. Clin. Oncol. 35, e14048 (2016). [Google Scholar]
- 150.Matthews TP, Jones AM & Collins I. Structure-based design, discovery and development of checkpoint kinase inhibitors as potential anticancer therapies. Expert Opin. Drug Discov. 8, 621–640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Welch S. et al. UCN-01 in combination with topotecan in patients with advanced recurrent ovarian cancer: a study of the Princess Margaret Hospital Phase II consortium. Gynecol. Oncol. 106, 305–310 (2007). [DOI] [PubMed] [Google Scholar]
- 152.Ma CX et al. A phase II study of UCN-01 in combination with irinotecan in patients with metastatic triple negative breast cancer. Breast Cancer Res. Treat. 137, 483–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sausville E. et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother. Pharmacol. 73, 539–549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seto T. et al. Phase I, dose-escalation study of AZD7762 alone and in combination with gemcitabine in Japanese patients with advanced solid tumours. Cancer Chemother. Pharmacol. 72, 619–627 (2013). [DOI] [PubMed] [Google Scholar]
- 155.Ho AL et al. Phase I, open-label, dose-escalation study of AZD7762 in combination with irinotecan (irino) in patients (pts) with advanced solid tumors. J. Clin. Oncol. 29, 3033–3033 (2011). [Google Scholar]
- 156.Al-Ahmadie H. et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov. 4, 1014–1021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wehler T. et al. A randomized, phase 2 evaluation of the CHK1 inhibitor, LY2603618, administered in combination with pemetrexed and cisplatin in patients with advanced nonsquamous non-small cell lung cancer. Lung Cancer 108, 212–216 (2017). [DOI] [PubMed] [Google Scholar]
- 158.Laquente B. et al. A phase II study to evaluate LY2603618 in combination with gemcitabine in pancreatic cancer patients. BMC Cancer 17, 137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Daud AI et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J. Clin. Oncol. 33, 1060–1066 (2015). [DOI] [PubMed] [Google Scholar]
- 160.Karp JE et al. Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin. Cancer Res. 18, 6723–6731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hong D. et al. Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J. Clin. Oncol. 34, 1764–1771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hong DS et al. Evaluation of prexasertib, a checkpoint kinase 1 inhibitor, in a phase Ib study of patients with squamous cell carcinoma. Clin. Cancer Res. 24, 3263–3272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee JM et al. A phase II study of the cell cycle checkpoint kinases 1 and 2 inhibitor (LY2606368; Prexasertib monomesylate monohydrate) in sporadic high-grade serous ovarian cancer (HGSOC) and germline BRCA mutation-associated ovarian cancer (gBRCAm+ OvCa). Ann. Oncol. 27, 8550 (2016). [Google Scholar]
- 164.Hirai H. et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 8, 2992–3000 (2009). [DOI] [PubMed] [Google Scholar]
- 165.Aarts M. et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2, 524–539 (2012). [DOI] [PubMed] [Google Scholar]
- 166.Bridges KA et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin. Cancer Res. 17, 5638–5648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pfister SX. et al. Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell 28, 557–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pfister SX et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Guertin AD et al. Preclinical evaluation of the WEE1 inhibitor MK-1775 as single-agent anticancer therapy. Mol. Cancer Ther. 12, 1442–1452 (2013). [DOI] [PubMed] [Google Scholar]
- 170.Mizuarai S. et al. Discovery of gene expression-based pharmacodynamic biomarker for a p53 context-specific anti-tumor drug Wee1 inhibitor. Mol. Cancer 8, 34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leijen S. et al. Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J. Clin. Oncol. 34, 4371–4380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Do K. et al. Phase I study of single-agent AZD1775 (MK-1775), a Wee1 kinase inhibitor, in patients with refractory solid tumors. J. Clin. Oncol. 33, 3409–3415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leijen S. et al. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J. Clin. Oncol. 34, 4354–4361 (2016). [DOI] [PubMed] [Google Scholar]
- 174.Oza AM et al. An international, biomarker-directed, randomized, phase II trial of AZD1775 plus paclitaxel and carboplatin (P/C) for the treatment of women with platinum-sensitive, TP53-mutant ovarian cancer. J. Clin. Oncol. 33, 5506–5506 (2015). [DOI] [PubMed] [Google Scholar]
- 175.Chera BS et al. Phase Ib trial of dose-escalating AZD1775 in combination with concurrent radiation and cisplatin for intermediate and high risk head and neck squamous cell carcinoma. J. Clin. Oncol. 34, TPS6106 (2016). [DOI] [PubMed] [Google Scholar]
- 176.Lal S. et al. WEE1 inhibition in pancreatic cancer cells is dependent on DNA repair status in a context dependent manner. Sci. Rep. 6, 33323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chabot P. et al. Veliparib in combination with whole-brain radiation therapy for patients with brain metastases from non-small cell lung cancer: results of a randomized, global, placebo-controlled study. J. Neurooncol. 131, 105–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reiss KA et al. A final report of a phase I study of veliparib (ABT-888) in combination with low-dose fractionated whole abdominal radiation therapy (LDFWAR) in patients with advanced solid malignancies and peritoneal carcinomatosis with a dose escalation in ovarian and fallopian tube cancers. Gynecol. Oncol. 144, 486–490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Blumenthal DT et al. A Phase III study of radiation therapy (RT) and O6-benzylguanine + BCNU versus RT and BCNU alone and methylation status in newly diagnosed glioblastoma and gliosarcoma: Southwest Oncology Group (SWOG) study S0001. Int. J. Clin. Oncol. 20, 650–658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bhattacharya S. et al. RAD51 interconnects between DNA replication, DNA repair and immunity. Nucleic Acids Res. 45, 4590–4605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Karnak D. et al. Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin. Cancer Res. 20, 5085–5096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Karakashev S. et al. BET Bromodomain inhibition synergizes with PARP inhibitor in epithelial ovarian cancer. Cell Rep. 21, 3398–3405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yang L. et al. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci. Transl Med. 9, eaal1645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sun C. et al. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell 33, 401–416. e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.O’Connor MJ Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560 (2015). [DOI] [PubMed] [Google Scholar]
- 186.Ceccaldi R. et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ibrahim YH et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2, 1036–1047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu JF et al. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol. 15, 1207–1214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Juvekar A. et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2, 1048–1063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Carey JP et al. Synthetic lethality of PARP inhibitors in combination with MYC blockade is independent of BRCA status in triple negative breast cancer. Cancer Res. 78, 742–757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Min A. et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 17, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yamaguchi H. et al. EZH2 contributes to the response to PARP inhibitors through its PARP-mediated poly-ADP ribosylation in breast cancer. Oncogene 37, 208–217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chao OS & Goodman OB Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Mol. Cancer Res. 12, 1755–1766 (2014). [DOI] [PubMed] [Google Scholar]
- 194.Muvarak NE et al. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents — a potential therapy for cancer. Cancer Cell 30, 637–650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tentori L. et al. Poly(ADP-ribose) polymerase (PARP) inhibition or PARP-1 gene deletion reduces angiogenesis. Eur. J. Cancer 43, 2124–2133 (2007). [DOI] [PubMed] [Google Scholar]
- 196.Borst GR et al. Neoadjuvant olaparib targets hypoxia to improve radioresponse in a homologous recombination-proficient breast cancer model. Oncotarget 8, 87638–87646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ali M. et al. Vasoactivity of AG014699, a clinically active small molecule inhibitor of poly(ADP-ribose) polymerase: a contributory factor to chemopotentiation in vivo? Clin. Cancer Res. 15, 6106–6112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bindra RS et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell. Biol. 24, 8504–8518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dean E. et al. Phase I study to assess the safety and tolerability of olaparib in combination with bevacizumab in patients with advanced solid tumours. Br. J. Cancer 106, 468–474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liu JF et al. A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur. J. Cancer 49, 2972–2978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Liu JF et al. Overall survival and updated progression-free survival results from a randomized phase 2 trial comparing the combination of olaparib and cediranib against olaparib alone in recurrent platinum-sensitive ovarian cancer. J. Clin. Oncol. 35, 5535–5535 (2017). [Google Scholar]
- 202.Philip C-A et al. Inhibition of PI3K-AKT-mTOR pathway sensitizes endometrial cancer cell lines to PARP inhibitors. BMC Cancer 17, 638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang D. et al. Effective use of PI3K inhibitor BKM120 and PARP inhibitor Olaparib to treat PIK3CA mutant ovarian cancer. Oncotarget 7, 13153–13166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Matulonis UA et al. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high-grade serous ovarian and breast cancer. Ann. Oncol. 28, 512–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Matulonis UA et al. Phase I of oral BKM120 or BYL719 and olaparib for high-grade serous ovarian cancer or triple-negative breast cancer: final results of the BKM120 plus olaparib cohort [abstract]. Cancer Res. 75, CT324 (2015). [Google Scholar]
- 206.Konstantinopoulos PA et al. Phase I study of the alpha specific PI3-Kinase inhibitor BYL719 and the poly (ADP-Ribose) polymerase (PARP) inhibitor olaparib in recurrent ovarian and breast cancer: analysis of the dose escalation and ovarian cancer expansion cohort [abstract]. Cancer Res. 77, CT008 (2017). [Google Scholar]
- 207.Michalarea V. et al. Phase I trial combining the PARP inhibitor olaparib (Ola) and AKT inhibitor AZD5363 (AZD) in germline (g)BRCA and non-BRCA mutant (m) advanced cancer patients (pts) incorporating noninvasive monitoring of cancer mutations [abstract]. Cancer Res. 76, CT010 (2016). [Google Scholar]
- 208.Du Y. et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat. Med. 22, 194–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cottini F. et al. Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 5, 972–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shapiro GI et al. Phase 1 dose-escalation study of the CDK inhibitor dinaciclib in combination with the PARP inhibitor veliparib in patients with advanced solid tumors [abstract]. Cancer Res. 77, CT047 (2016). [Google Scholar]
- 211.Karanika S. et al. Targeting DNA damage response in prostate cancer by inhibiting androgen receptor-CDC6-ATR-Chk1 signaling. Cell Rep. 18, 1970–1981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Asim M. et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat. Commun. 8, 374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Clarke N. et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 19, 975–986 (2018). [DOI] [PubMed] [Google Scholar]
- 214.Le DT et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Germano G. et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017). [DOI] [PubMed] [Google Scholar]
- 217.Brown JS, Sundar R & Lopez J. Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. Br. J. Cancer 118, 312–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Härtlova A. et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015). [DOI] [PubMed] [Google Scholar]
- 219.Jiao S. et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin. Cancer Res. 23, 3711–3720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sato H. et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 8, 1751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Turajlic S. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 222.McGrail DJ et al. Multi-omics analysis reveals neoantigen-independent immune cell infiltration in copy-number driven cancers. Nat. Commun. 9, 1317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lee J-M et al. Safety and clinical activity of the programmed death-ligand 1 inhibitor durvalumab in combination with poly (ADP-ribose) polymerase inhibitor olaparib or vascular endothelial growth factor receptor 1–3 inhibitor cediranib in women’s cancers: a dose-escalation, phase i study. J. Clin. Oncol. 35, 2193–2202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Domchek S. et al. An open-label, multitumor, phase II basket study of olaparib and durvalumab (MEDIOLA): results in germline BRCA-mutated (gBRCAm) HER2negative metastatic breast cancer (MBC) [abstract]. Cancer Res. 78, PD6–11 (2018). [Google Scholar]
- 225.Karzai F. et al. A phase 2 study of olaparib and durvalumab in metastatic castrate-resistant prostate cancer (mCRPC) in an unselected population. J. Clin. Oncol. 36, 163 (2018). [Google Scholar]
- 226.Powles T. et al. BISCAY, a phase Ib, biomarker-directed multidrug umbrella study in patients with metastatic bladder cancer. J. Clin. Oncol. 34, TPS4577 (2017). [Google Scholar]
- 227.Dobzhansky T. Genetics of natural populations; rec ombi nation and variability in populations of Drosophila pseudoobscura. Genetics 31, 269–290 (1946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chambon P, Weill JD & Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 11, 39–43 (1963). [DOI] [PubMed] [Google Scholar]
- 229.Chambon P, Weill JD, Doly J, Strosser M & Mandell P. On the formation of a novel adelylic compound by enzymatic extracts of liver nuclei. Biochem. Biophys. Res. Commun. 25, 638–643 (1966). [Google Scholar]
- 230.Yamada M, Miwa M & Sugimura T. Studies on poly (adenosine diphosphate-ribose). X. Properties of a partially purified poly (adenosine diphosphate-ribose) polymerase. Arch. Biochem. Biophys. 146, 579–586 (1971). [DOI] [PubMed] [Google Scholar]
- 231.Durkacz BW, Omidiji O, Gray DA & Shall S. (ADP-ribose)n participates in DNA excision repair. Nature 283, 593 (1980). [DOI] [PubMed] [Google Scholar]
- 232.Hall JM et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689 (1990). [DOI] [PubMed] [Google Scholar]
- 233.Wooster R. et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789–792 (1995). [DOI] [PubMed] [Google Scholar]
- 234.White AW et al. Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase. J. Med. Chem. 43, 4084–4097 (2000). [DOI] [PubMed] [Google Scholar]
- 235.Farmer H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 236.Menear KA et al. 4-[3-(4-cyclopropane-carbonylpiperazine-1-carbonyl)-4-fluorobenzyl]2H-phthalazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J. Med. Chem. 51, 6581–6591 (2008). [DOI] [PubMed] [Google Scholar]
- 237.Plummer R. et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 14, 7917–7923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jones P. et al. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}−2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and −2 mutant tumors. J. Med. Chem. 52, 7170–7185 (2009). [DOI] [PubMed] [Google Scholar]
- 239.Sanofi-aventis. Sanofi-aventis reports top-line results from phase III study with iniparib (BSI-201) in metastatic triple-negative breast cancer. Sanofi http://www.news.sanofi.us/press-releases?item=118547 (2011). [Google Scholar]
- 240.Patel AG, De Lorenzo SB, Flatten KS, Poirier GG & Kaufmann SH Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin. Cancer Res. 18, 1655–1662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Supreme Court of the United States. Association for Molecular Pathology et al. petitioners v. Myriad Genetics, INC. et al. Supreme Court of the United States https://www.supremecourt.gov/opinions/12pdf/12-398_1b7d.pdf (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.