Abstract
Increasing transmission of SARS-CoV-2 infection in successive waves may strain the capacity of laboratories performing molecular diagnostic testing. Alternative testing approaches may offer additional diagnostic capacity. A high throughput chemiluminescent antigen assay (Ortho VITROS SARS-CoV-2 antigen test) was evaluated using both an inactivated virus preparation and prospective clinical samples (nasopharyngeal swabs in virus transport medium). The limit of detection of the assay was approximately 0.5 TCID50/ml, equivalent to a Ct value of 33. The assay was linear over a wide range. When 528 clinical samples were tested with the antigen assay, the sensitivity was 84.2% and the specificity was 100% (positive predictive value 100% and negative predictive value 97.7%). High volume antigen tests might be used to supplement molecular diagnostic testing capacity.
Keywords: SARS-CoV-2, antigen detection, COVID-19
1. INTRODUCTION
A cluster of pneumonia cases of unknown aetiology was first reported in Wuhan City, China [1] and this spread rapidly to cause a global pandemic. A novel coronavirus, SARS-CoV-2 [2] was identified as the causative agent of the newly recognized respiratory disease, COVID-19 (COronaVIrus Disease 2019) [3]. Public health agencies and laboratories rapidly developed and implemented nucleic acid-based molecular diagnostic testing. At different stages of the COVID-19 pandemic, rapidly increasing transmission has threatened to overwhelm the capacity of molecular diagnostic testing laboratories, whether locally, regionally or nationally. One approach to mitigate increasing turnaround times for molecular tests has been the development of antigen detection tests for point of care use, almost all of which utilize lateral flow technology. These tests can generate rapid results, within 15-20 minutes of specimen collection [4], [5], [6], [7], [8]. They are simple and straightforward to perform and most do not require the addition of reagents other than a buffer in which the specimen is diluted. This can allow for rapid identification and public health management of cases. However, rapid antigen tests lack sensitivity for detection of SARS-CoV-2, when compared to molecular testing using PCR. Collection and tracking of patient and test data in centralised databases may be difficult to achieve if online access is not available, as in field sites and remote communities. This may delay reporting and public health interventions. Specimen collection devices designed for point of care tests do not generally yield residual sample that can be transported safely and used for further testing.
The limitations of rapid point of care tests can be overcome by using high volume antigen tests that are intended to be run on analyzers in hospital laboratories. Such assays are primarily based upon chemiluminescent detection chemistry, which offers greater sensitivity than the lateral flow assay format. In addition, patient and test data are captured in the LIS and electronic reporting to public health is standard. The specimen collected can also be used for additional testing including PCR, whole genome sequencing or culture. Here we present an evaluation of a SARS-CoV-2 nucleocapsid antigen detection assay (VITROS SARS-CoV-2 antigen test) that has recently received regulatory approval in Europe and in the USA (CE and FDA EUA).
2. METHODS
2.1. Inactivated virus suspension
SARS-CoV-2 was grown on Vero-E6 cells in MEM with 2% foetal bovine serum (Gibco, ThermoFisher Scientific) and the median tissue culture infectious dose (TCID50) was determined. An aliquot was transferred to lysis buffer (MagMAX, ThermoFisher Scientific) for RT-PCR using a laboratory-developed duplex assay that targets RdRP and E genes [9].
The remaining virus was inactivated with β-propiolactone. Briefly, 20 µl of 10% β-propiolactone in Tris-HCl buffer (0.1 M, pH7.8) was added to 1.8 ml viral lysate, followed by 100 µl MEM. The viral suspension was incubated at 4°C for 24 h, and 0.1M NaOH was added dropwise as required to maintain pH near neutrality. The suspension was then incubated at 37°C for 2 h, after which the pH was adjusted again with 0.1M NaOH [10]. The absence of viable virus after inactivation was confirmed by the TCID50 procedure, including inoculation of undiluted virus into cell cultures. PCR was performed on the inactivated virus for comparison with the live virus. Dilutions of inactivated virus were made in MEM before use in the antigen test.
2.2. Clinical samples
A panel of 24 samples in VTM (Yocon Biology Technology Company, Beijing, China), previously tested by PCR, were tested using the VITROS SARS-CoV-2 antigen test. This panel included 12 PCR-positive samples (Ct values ranging from 27.14 – 30.08) and 12 PCR-negative samples. Samples were added to lysis buffer within 4 days of collection. A further 528 consecutive clinical samples were tested prospectively. Immediately after sample preparation for PCR testing, samples were added to lysis buffer as described below. The mean time from collection to processing was 28.9 h.
2.3. SARS-CoV-2 Antigen test
Aliquots (400 µl) of inactivated virus or of samples in UTM were added to 100 µl VITROS SARS-CoV-2 antigen extraction buffer (Ortho Clinical Diagnostics, Inc., Rochester, NY) and incubated at room temperature for 2 hours (clinical samples were then held at 4°C overnight before testing). The presence of SARS-CoV-2 antigen was detected using the VITROS SARS-CoV-2 antigen test (Ortho Clinical Diagnostics, Inc., Rochester, NY) on a VITROS XT 7600 analyzer (Ortho Clinical Diagnostics, Inc., Rochester, NY).
3. RESULTS
3.1. Inactivated virus suspension
The titre of the viral lysate was 104.75 TCID50/ml. RT-PCR performed on the viral lysate before inactivation gave Ct values of 12.25 and 12.12 for RdRp and E gene targets respectively. No viable virus was detected after inactivation with β-propiolactone. After inactivation, the Ct values were 12.88 and 13.03 for RdRp and E gene targets respectively.
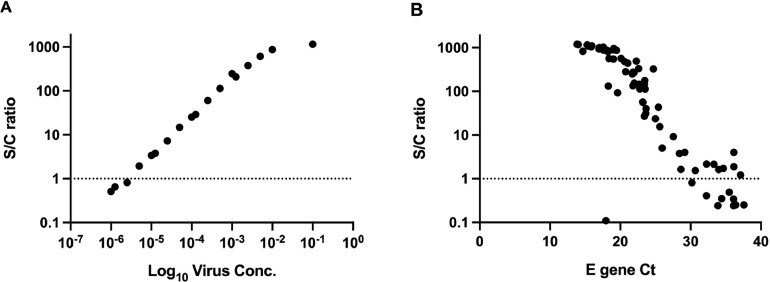
Serial dilutions of inactivated virus from 10−1 – 10−8 were each tested by the VITROS SARS-CoV-2 antigen test on three occasions (see Table 1 ). The coefficient of variation for reactive samples ranged from 0.67-3.22%. The assay repeatedly detected the virus to a 10−5 dilution. The calculated input concentration of virus at the limit of detection was 0.5 TCID50/ml. Additional two-fold dilutions were performed to more closely define the limit of detection, and dilutions close to the cut-off for positivity were tested by PCR to estimate the Ct at the limit of detection. The highest dilution that gave a positive result in the antigen test had Ct values of 33.45 and 33.36 for RdRp and E gene targets respectively (Table 2 ). The assay showed linearity through most of its range (Fig. 1 A).
Table 1.
Determination of sensitivity and precision of the SARS-CoV-2 antigen assay
| Inactivated Virus Dilution | Calculated input conc. (TCID50/ml)a | Calculated input based on Ortho sample (80 µl) | Ortho Vitros S/C ratiob (reactive cutoff ≥1.0)c | CV% | ||
| 10−1 | 4800 | 384 | 1150 | 1180 | 1160 | 1.31 |
| 10−2 | 480 | 38 | 845 | 825 | 826 | 1.35 |
| 10−3 | 48 | 4 | 228 | 226 | 229 | 0.67 |
| 10−4 | 5 | 0.4 | 30.2 | 30.7 | 31.6 | 2.30 |
| 10−5 | 0.5 | 0.04 | 3.14 | 3.35 | 3.3 | 3.32 |
| 10−6 | 0 | 0 | 0.52 | 0.55 | 0.51 | 4.16 |
| 10−7 | 0 | 0 | 0.25 | 0.25 | 0.26 | 2.31 |
| 10−8 | 0 | 0 | 0.25 | 0.24 | 0.23 | 4.55 |
a Based on Ortho buffer dilution 4:1.
b Signal/cutoff ratio.
c Each dilution was tested in triplicate.
Table 2.
Limit of detection of the SARS-CoV-2 antigen assay
| Inactivated Virus Dilution | Ortho S/C Ratioa | RdRP/E gene Ct values |
| 0.1 | 1160 | ndb |
| 0.01 | 874 | nd |
| 0.005 | 612 | nd |
| 0.0025 | 380 | nd |
| 0.00125 | 210 | nd |
| 0.001 | 247 | nd |
| 0.0005 | 114 | nd |
| 0.00025 | 60.4 | nd |
| 0.000125 | 29.1 | nd |
| 0.0001 | 25.3 | nd |
| 0.00005 | 14.8 | nd |
| 0.000025 | 7.27 | 31.46/31.56 |
| 0.0000125 | 3.8 | 32.1/32.47 |
| 0.00001 | 3.38 | 32.27/32.07 |
| 0.000005 | 1.94 | 33.45/33.36 |
| 0.0000025 | 0.82 | 33.56/34.82 |
| 0.00000125 | 0.65 | 35.66/35.54 |
| 0.000001 | 0.51 | nd |
Fig. 1.
Linearity of the SARS-CoV-2 antigen assay. (A) S/C ratio displays linearity over wide range of inactivated virus concentration. (B) Distribution of E gene Ct values for antigen-positive samples. Dotted line indicates positive cut-off for antigen assay.
3.2. Clinical samples
A panel of 24 samples in UTM, previously tested by PCR, were tested using the antigen test. This panel included 12 PCR-positive samples (Ct values ranging from 27.14 – 30.08) and 12 PCR-negative samples. Antigen was detected in 10/12 (83%) PCR-positive samples (Ct from 27.14-30.00), but not in two samples with Ct of 29.5 and 30. Antigen was not detected in 12 PCR-negative samples.
Of the 528 samples tested prospectively, 70 were positive by PCR, of which 59 were also antigen-positive (sensitivity 84.2%). Ct values in antigen-positive samples ranged from 13.84-37.09; most antigen-positive samples had Ct values below 25 (n=44, 74%). The distribution of S/C ratio values was linear over a range of Ct values in the 20-30 range (Fig. 1B).
In the PCR positive, antigen-negative samples, Ct ranged from 17.95-37.57. The sample with a Ct of 17.95 was not available for repeat antigen testing. If this sample was excluded, the remaining antigen-negative, PCR-positive samples ranged from Ct 30.18-37.57.
Antigen was not detected in PCR-negative samples (specificity 100%). The positive predictive value of the antigen assay was 100% and the negative predictive value was 97.7%.
4. DISCUSSION
Antigen tests do not reach the same level of diagnostic sensitivity as PCR assays [8], but a correlation with excretion of infectious, cultivable virus has been reported [11, 12]. We evaluated the performance of a high throughput SARS-CoV-2 antigen assay (VITROS SARS-CoV-2 antigen test), using an inactivated virus suspension to determine the limit of detection of the assay. The use of β-propiolactone to inactivate the virus allowed us to compare the antigen test results with PCR Ct values. The limit of detection was approximately 0.5 TCID50/ml, and a Ct value of 33. However, when clinical samples were tested, the Ct cut-off for reliable antigen detection was approximately 30. The sensitivity of the assay was 84%. Interpretation of sensitivity estimates requires caution; the prospective clinical samples were tested during a period of rapid growth in case numbers, with a test-positivity rate of 13%. At this time, most positive samples had relatively low Ct values. The specificity of the assay was 100% when tested on prospective clinical samples.
The antigen assay showed linearity over a range of virus concentrations (Fig. 1). This suggests that, with appropriate standardization, this assay could be applied to estimate the viral burden that might be considered infectious, as has been proposed for rapid antigen tests [12].
The sensitivity of this assay was significantly higher than that expected of rapid antigen tests [13], but high volume antigen tests also have relative disadvantages, which include the need for transport to a central testing laboratory, potential concerns about the stability of viral antigens during transport, the requirement for safe handling of the specimen in a biosafety cabinet while preparing for analysis and the additional time required for virus inactivation. After inactivation of sample, the VITROS SARS-CoV-2 antigen test requires 48 minutes for completion and is intended to run on VITROS 3600 immunodiagnostic systems and VITROS 5600 or XT 7600 integrated systems, with a throughput of up to 130 samples per hour [14]. The ability to run large numbers of tests through analyzers installed in core laboratories potentially increases the testing capacity in all hospital laboratory sites.
In this report we describe the performance characteristics of a high volume antigen assay. The analytical sensitivity is high, and the assay appears to be sensitive and specific in clinical use.
Author Statements
The authors declare that there are no conflicts of interest. This work received no specific grant from any funding agency.
UBC Clinical Research Ethics Board review was not required under article 2.5 of the TCPS2, the overarching ethical framework for research involving human participants in Canada.
CRediT authorship contribution statement
Paul N. Levett: Conceptualization, Methodology, Data curation, Writing – original draft, Writing – review & editing, Visualization. Branco Cheung: Investigation, Writing – review & editing. Jesse Kustra: Investigation, Writing – review & editing. Tamara Pidduck: Validation, Data curation. Annie Mak: Validation, Writing – review & editing. Frankie Tsang: Resources. Martin Petric: Investigation, Resources, Writing – review & editing. Mel Krajden: Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy D, Sidorov IA, Sola I, Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization . 2020. Naming the coronavirus disease (COVID-19) and the virus that causes it.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Accessed March 30, 2021. [Google Scholar]
- 4.Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes MA, Martínez M, Poujois S, Forqué L, Valdivia A, Solano de la Asunción C, Ferrer J, Colomina J, Navarro D. Field evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2021;27 doi: 10.1016/j.cmi.2020.11.004. 472 e7-472 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toptan T, Eckermann L, Pfeiffer AE, Hoehl S, Ciesek S, Drosten C, Corman VM. Evaluation of a SARS-CoV-2 rapid antigen test: Potential to help reduce community spread? J Clin Virol. 2020;135 doi: 10.1016/j.jcv.2020.104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jääskeläinen AE, Ahava MJ, Jokela P, Szirovicza L, Pohjala S, Vapalahti O, Lappalainen M, Hepojoki J, Kurkela S. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Virol. 2021;137 doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, Griego-Fullbright C, Burgard C, Fernandez C, Eckert K, Andrews JC, Ren H, Allen J, Ackerman R, Cooper CK. Clinical Evaluation of BD Veritor SARS-CoV-2 Point-of-Care Test Performance Compared to PCR-Based Testing and versus the Sofia 2 SARS Antigen Point-of-Care Test. J Clin Microbiol. 2020;59 doi: 10.1128/JCM.02338-20. e02338-e02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blairon L, Wilmet A, Beukinga I, Tré-Hardy M. Implementation of rapid SARS-CoV-2 antigenic testing in a laboratory without access to molecular methods: Experiences of a general hospital. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeBlanc JJ, Gubbay JB, Li Y, Needle R, Arneson SR, Marcino D, Charest H, Desnoyers G, Dust K, Fattouh R, Garceau R, German G, Hatchette TF, Kozak RA, Krajden M, Kuschak T, Lang ALS, Levett P, Mazzulli T, McDonald R, Mubareka S, Prystajecky N, Rutherford C, Smieja M, Yu Y, Zahariadis G, Zelyas N, Bastien N, Covid- Pandemic Diagnostics Investigation Team of the Canadian Public Health Laboratory Network Respiratory Virus Working Group Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zakhartchouk AN, Liu Q, Petric M, Babiuk LA. Augmentation of immune responses to SARS coronavirus by a combination of DNA and whole killed virus vaccines. Vaccine. 2005;23:4385–4391. doi: 10.1016/j.vaccine.2005.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekosz A, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S, Gary DS, Roger-Dalbert C, Leitch J, Cooper CK. Antigen-Based Testing but Not Real-Time Polymerase Chain Reaction Correlates With Severe Acute Respiratory Syndrome Coronavirus 2 Viral Culture. Clin Infect Dis. 2021 doi: 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C, Laffineur K, Rosseels C, Van Eeckhoudt S, Nicolas JB, Morimont L, Dogné JM, Douxfils J. Head-to-Head Comparison of Rapid and Automated Antigen Detection Tests for the Diagnosis of SARS-CoV-2 Infection. J Clin Med. 2021;10 doi: 10.3390/jcm10020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortho Clinical Diagnostics. 2021. VITROS Immunodiagnostic Products SARS-CoV-2 Antigen Reagent Pack - Instructions for Use. https://www.fda.gov/media/145073/download. Accessed March 31st 2021.