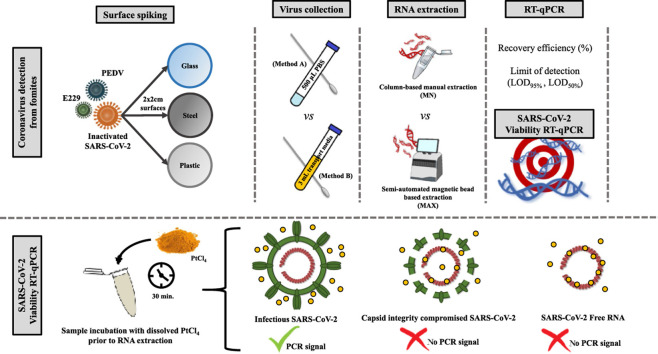
Abstract
The ongoing coronavirus 2019 (COVID-19) pandemic constitutes a concerning global threat to public health and economy. In the midst of this pandemic scenario, the role of environment-to-human COVID-19 spread is still a matter of debate because mixed results have been reported concerning SARS-CoV-2 stability on high-touch surfaces in real-life scenarios. Up to now, no alternative and accessible procedures for cell culture have been applied to evaluate SARS-CoV-2 infectivity on fomites. Several strategies based on viral capsid integrity have latterly been developed using viability markers to selectively remove false-positive qPCR signals resulting from free nucleic acids and damaged viruses. These have finally allowed an estimation of viral infectivity. The present study aims to provide a rapid molecular-based protocol for detection and quantification of viable SARS-CoV-2 from fomites based on the discrimination of non-infectious SARS-CoV-2 particles by platinum chloride (IV) (PtCl4) viability RT-qPCR. An initial assessment compared two different swabbing procedures to recover inactivated SARS-CoV-2 particles from fomites coupled with two RNA extraction methods. Procedures were validated with human (E229) and porcine (PEDV) coronavirus surrogates, and compared with inactivated SARS-CoV-2 suspensions on glass, steel and plastic surfaces. The viability RT-qPCR efficiently removed the PCR amplification signals from heat and gamma-irradiated inactivated SARS-CoV-2 suspensions that had been collected from specified surfaces. This study proposes a rapid viability RT-qPCR that discriminates non-infectious SARS-CoV-2 particles on surfaces thus helping researchers to better understand the risk of contracting COVID-19 through contact with fomites and to develop more efficient epidemiological measures.
Keywords: COVID-19, SARS-CoV-2, Fomites, Viability RT-qPCR, Transmission risk
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the current pandemic of coronavirus 2019 (COVID-19), is an airborne human pathogen transmitted by droplets and aerosols. Detection of infectious SARS-CoV-2 in urine and faecal samples have raised concerns about the possible faecal-oral transmission in COVID-19 spread, however this hypothesis has not been fully elucidated (Patel et al., 2020; Yeo et al., 2020). Furthermore, the question of whether contaminated surfaces (fomites) contribute to COVID-19 transmission remains under debate (Goldman, 2020; Mondelli et al., 2020). For example, inconclusive results have been published regarding the stability of SARS-CoV-2 on fomites. Riddell et al. (2020) reported the persistence of SARS-CoV-2 infectious particles on surfaces for up to 28 days, while other studies described considerably shorter times. Of critical concern is the evaluation of the risk of SARS-CoV-2 transmission through contaminated fomites with infectious respiratory secretions or faecal material. In the context of the current pandemic, environmental fomite screening by molecular techniques has found SARS-CoV-2 RNA contamination on food packaging surfaces (Liu et al., 2020), public transport vehicles (Moreno et al., 2021), hospitals (Liu et al., 2020), cruise ship surfaces (Kampf et al., 2020), and workplaces (Fernández-de-Mera et al., 2020; Harvey et al., 2020). It should be noted however, that most of the fomite samples that tested positive for SARS-CoV-2 RNA showed high Cq values (generally > 33 Cq), making it improbable that they act as environmental reservoirs of infectious viruses (Kampf et al., 2020). Additional attempts have assessed the infectivity of SARS-CoV-2 isolated from hospital fomites by cell culture but the recovered viral particles were found not to be infectious (Mondelli et al., 2020). In contrast, a similar study detected viable SARS-CoV-2 in surface (bedside table, floor, remote control, bed rails, and bedsheets) swabs from the isolation rooms of COVID-19 patients, being the successful cell replication observed for samples with Cq values lower than 34 (Ahn et al., 2020). More recently, infectious SARS-CoV-2 has been isolated from frozen codfish package surfaces (Liu et al., 2020), with no Cq value being reported.
Although the gold standard to assess infectivity relies on viral propagation in susceptible cell cultures, its implementation for SARS-CoV-2 is limited to access to biosafety level 3 laboratories. Given such noteworthy limitation, human coronavirus surrogates (e.g., E229), murine coronavirus or porcine epidemic diarrhoea coronavirus (PEDV), have been used as SARS-CoV-2 surrogates for method development and inactivation studies (Ahmed et al, 2020a, 2020b, ; Pérez-Cataluña et al., 2021). More recently molecular approaches based on viability PCR, either using platinum compounds or photoactivable dyes, have been applied to assess coronavirus’ virion integrity (Blondin-Brosseau et al., 2021; Cuevas-Ferrando et al., 2021; Puente et al., 2020; Wurtzer et al., 2021). Platinum compounds such as platinum chloride (IV) (PtCl4) present some advantages over other photoactivable viability dyes such as propidium monoazide (PMA) and ethidium monoazide (EMA) (Elizaquível et al., 2014; Puente et al., 2020; Randazzo et al., 2018a, 2018b) as they are not sensitive to visible light and are more affordable from an economic perspective (Karami et al., 2014; Soejima and Iwatsuki, 2016). Moreover, photoactivation of azo dyes may be prevented in complex matrices, such as wastewater and molluscs, due to the turbidity, suspended solid and density of concentrated samples (Leifels et al., 2020; Randazzo et al., 2016, 2018a, Polo et al., 2021).
Here, we report an optimized protocol for the detection and quantification of potentially infectious SARS-CoV-2 particles on fomites based on a viability RT-qPCR technique. By discriminating non-infectious SARS-CoV-2 particles on fomites, the method could help to better understand the risk of contracting COVID-19 through contact with fomites and to develop more efficient mitigation measures.
2. Material and methods
2.1. Coronavirus stocks
This study used inactivated preparations with SARS-CoV-2 viral particles obtained from USA-WA1/2020 isolate and provided by BEI Resources (VA, US) after being either gamma-irradiated (5 × 106 RADs) (NRC-52287) or heat treated (65 °C for 30 min) (NR-52286). For comparison purposes, the E229 strain of human coronavirus (ATCC-VR740) and the CV777 strain of Porcine Epidemic Diarrhoea Coronavirus (PEDV provided by Friedrich-Loeffler-Institut ,Greifswald, Germany) were assayed as SARS-CoV-2 surrogates. The E229 and the PEDV are enveloped virus member of the Coronaviridae family, genus Alphacoronavirus, being the former responsible for the common cold in humans and bats (Gaunt et al., 2010) and the latter the etiological agent of porcine epidemic diarrhoea, a highly contagious intestinal disease causing a severe diarrhoea in pigs of all ages (Pensaert and de Bouck, 1978).
Both E229 and PEDV were assayed as semipurified in-house preparations of infected cell lysates from MRC-5 (ATCC- CCL171) and Vero cells, respectively. Semipurified virus stocks were obtained by centrifugation of infected cell lysates at 660×g for 30 min at 2–3 days post infection for both viruses. MRC-5 and Vero cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Biowest, Nuaillé, France) supplemented with 10% heat-inactivated fetal bovine serum (GE Healthcare Bio-Sciences, Austria), 100 units/ml of penicillin, 100 mg/mL of streptomycin, and 0.25 mg/mL of Fungizone® (Antibiotic-Antimycotic 100X, Gibco, Spain) at 37 °C and 5% CO2.
2.2. Swab sampling of coronavirus contaminated surfaces
Two different swabbing methods were compared by artificially inoculating clean and disinfected steel, plastic and glass surfaces (2 × 2 cm) with a total volume of 15 μL of coronavirus E229, PEDV, and gamma-inactivated-SARS-CoV-2 suspensions prepared in PBS and containing approximately 104 genomic copies (gc) per each virus. Contaminated surfaces were dried at room temperature for 30 min, time needed for inoculum to visually dry. Swabbing method A followed the recommendations of the ISO 15216:1 standard norm. In brief, the surface under test was intensively swabbed using a sterile, cotton-headed, plastic-shaft swab moistened in phosphate buffer solution (PBS), applying a little pressure to detach the viruses. Then, the swab was immediately immersed in a tube containing 500 μL PBS, vortexed and then pressed against the side of the tube to release the liquid. The immersion and pressing cycle was repeated three or four times to ensure maximum yield of virus collection. Swabbing method B adapted the previously described procedure to EUROTUBO®, a commercial specimen collection swab with a cotton head and plastic-shaft (Deltalab, Spain) containing 3 mL of transport media whose formulation is confidential. Recapitulating, the swabbing procedures differed on the type and volume of the elution media (500 μL PBS vs. 3 mL transport media).
Regardless of the method, for each experiment, a recovery control was prepared by inoculating the same amount of viruses to a single tube containing 1 mL of either PBS (Method A) or transport media (Deltalab, Spain, Method B).
2.3. Viral RNA extraction and amplification
Two different viral RNA extraction procedures were compared for protocol optimization: a) nucleic acid extraction with the bench top Maxwell® RSC 16 Instrument (Promega, Spain) (referred to as MAX) using the Maxwell RSC Pure Food GMO and authentication kit (Promega). The Manufacturer's recommended protocol was modified according to previous laboratory sample-based method optimization assays (Pérez-Cataluña et al., 2021). Specifically, 400 μL of cetyltrimethyl ammonium bromide (CTAB) and 40 μL of proteinase K solution were added to 300 μL of selected samples, the mix was subsequently incubated at 60 °C and transferred to the loading cartridge along with 300 μL of lysis buffer. The cartridge was then placed in the Maxwell® RSC Instrument and the extraction performed by selecting the “Maxwell RSC Viral total Nucleic Acid” program incorporated in the instrument software; b) RNA purification with the NucleoSpin RNA virus kit (Macherey-Nagel GmbH & Co.) (referred to as MN) which was conducted according to the manufacturer's instructions using an initial sample volume of 150 μL. SARS-CoV-2 amplification was carried out by targeting the N1 region of the nucleocapsid gene (CDC, 2020), PEDV's primers targeted a 140 bp sequence within the highly conserved M gene (Zhou et al., 2017). A target region of 70 bp for the membrane protein HCoV was selected for E229 amplification (Vijgen et al., 2005). All RT-qPCRs were performed using the One Step PrimeScript™ RT-PCR Kit (Takara Bio Inc.) on a LightCycler 480 instrument (Roche Diagnostics, Germany). Thermal cycling conditions and sequences for primers and probes are shown in Table 1 . All reactions were performed in duplicate and included with nuclease-free water as negative control. Amplification curve showing Ct values < 40 were considered as positive and quantified as follow. For SARS-CoV-2 quantification, a calibration curve was produced using a quantified SARS-CoV-2 genomic RNA suspension (VR-1986D™, ATCC, VA, US). Both PEDV and E229 quantification curves were generated by amplifying serial end-point tenfold dilutions of viral suspensions in quintuplicates and calculating the numbers of PCR units (PCRU). Detailed information on calibration curves and R2 is provided elsewhere (Bäuerl et al., 2021; Pérez-Cataluña et al., 2021). Final quantifications were adjusted for the extraction volume of the swabbing method.
Table 1.
Primers and probes used for the detection of SARS-CoV-2, Porcine Epidemic Diarrhoea Coronavirus (PEDV) and human coronavirus E229.
| Virus | Primers and probe | Sequence | RT-qPCR conditions | Reference |
|---|---|---|---|---|
| SARS-CoV-2 | 2019-nCoV_N1–F | GACCCCAAAATCAGCGAAAT | (CDC, 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel For Emergency Use Only Instructions for Use, n.d.) | |
| PCR (45 cycles) | ||||
| 95 °C for 03 s | ||||
| 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | 55 °C for 30 s | ||
| 2019-nCoV_N1–P | [FAM]-TCCTGAGGTCAATGCA- ZEN™/3′ Iowa Black™ | |||
| PEDV | PEDV_forward | CAGGACACATTCTTGGTGGTCTT | RT: 45 °C for 15 min, Preheating: 95 °C for 2 min | (Zhou et al., 2017) |
| PCR (45 cycles) | ||||
| 95 °C for 15 s | ||||
| PEDV_reverse | CAAGCAATGTACCACTAAGGAGTGTT | 60 °C for 60 s | ||
| PEDV_probe | [FAM]-ACGCGCTTCTCACTAC-NFQ-[MGB] | |||
| E229 | 229 E-FP | TTCCGACGTGCTCGAACTTT | RT: 45 °C for 15 min, Preheating: 95 °C for 2 min | Vijgen et al. (2005) |
| PCR (45 cycles) | ||||
| 95 °C for 15 s | ||||
| 229 E-RP | CCAACACGGTTGTGACAGTGA | 60 °C for 60 s | ||
| 229 E-TP | [FAM]-TCCTGAGGTCAATGCA-NFQ-[MGB] |
2.4. SARS-CoV-2 and surrogates detection limit on fomites
The limits of detection at 95% and 50% confidence intervals (LoD95% and LoD50%, respectively) were obtained by detecting ten-fold serially diluted gamma-irradiated SARS-CoV-2 (5.10 × 104 to 5.1 gc/4 cm2), PEDV (1.42 × 104 to 1.42 gc/4 cm2) and E229 (2.28 × 105 to 22.8 gc/4 cm2) seeded on glass, steel and plastic surfaces following the selected sampling Method A coupled with MAX RNA extraction procedure. All data were compiled from three independent experiments with three technical replicates for each inoculation level. LoD95% and LoD50% were calculated according to (Wilrich and Wilrich, 2009).
2.5. Rapid molecular assessment of SARS-CoV-2 viability
A viral viability molecular assay was carried out on heat-inactivated and gamma-irradiated SARS-CoV-2 recovered from plastic, glass and steel surfaces following the swabbing method A (based on ISO 15216:1 standard norm). The viability RT-qPCR assay was assessed using both heat-inactivated and gamma-irradiated SARS-CoV-2 because the mode of viral inactivation may influence the performance of this technique (Leifels et al., 2020; Randazzo et al., 2018). Platinum chloride (IV) (PtCl4) was dissolved in DMSO at 1 M and then diluted in water to a final 50 mM ready-to-use stock. For each recovered virus suspension, 300 μL of sample were incubated at room temperature for 30 min with 0.5 mM–2.5 mM concentrations of PtCl4. RNA was immediately extracted using the MAX RNA extraction procedure as previously described. A PtCl4 non-treated sample was included as control in each experiment.
2.6. Statistical analyses
All data were compiled from three independent experiments with three technical replicates for each variable.
Viral recoveries of samples (r) were calculated as a percentage (%) with respect to the control (virus directly spiked into the buffer) following the formula:
To test the impact of each variable on viral recovery results, data were subjected to the analysis of variance (ANOVA) followed by Tukey's HSD as a post hoc test to obtain homogenous groups. Differences in means were considered significant when the p-value was <0.05. Statistical data processing and graphic constructions were done by STATISTICA version 7 (StatSoft Inc., Tulsa, OK, USA) and GraphPad Prism version 8.0.2 (GraphPad Software, USA) software.
3. Results
3.1. Recovery efficiency and limit of RT-qPCR detection of SARS-cov-2 and surrogates on glass, steel and plastic surfaces
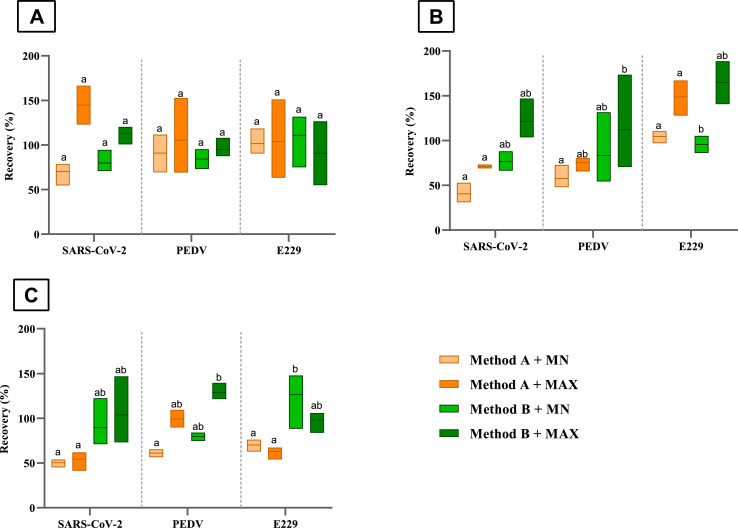
Two swabbing protocols (referred to as Method A and Method B) and two RNA extraction procedures (referred to as MN and MAX) were assessed in this study to provide a suitable method to efficiently detect SARS-CoV-2 RNA on fomites. We determined SARS-CoV-2, PEDV and E229 recovery yields and the limit of detection at 95 and 50% confidence interval (LOD95% and LOD50%) for each method combination, as the analytic characteristics for viral detection on spiked glass, steel and plastic surfaces (Fig. 1 ; Table 2 ). Overall, all coronaviruses were efficiently recovered from all surfaces (mean recovery value = 95.77%) (Fig. 1). Specifically, mean recovery values for gamma-inactivated SARS-CoV-2 on glass surfaces ranged from 70.28 ± 13.58% (Method A combined with MN extraction), to 112.63 ± 10.57% (Method B combined with MAX extraction), and these values were those with the widest recovery variability among all tested coronavirus. On steel surfaces, mean recovery values ranged from 40.62 ± 11.19% for SARS-CoV-2 recovered by Method A combined with MN extraction, to 164.87 ± 23.94% for E229 recovered by Method B combined with MAX extraction. On plastic surfaces, mean recovery values ranged from 50.32 ± 4.55% for SARS-CoV-2 recovered by Method A combined with MN extraction, to 133.99 ± 94.58% for PEDV recovered by Method B combined with MN extraction. Univariate statistics showed that the type of virus, the swabbing procedures and extraction methods significantly affected the recoveries (p < 0.05), but not the type of surfaces (p = 0.062). Interestingly, multivariate analyses showed no statistically significant differences for the combination of type of virus, swabbing procedures and extraction methods (p = 0.83), thus Method A coupled with MAX was the selected combination for the further assessment of the limits of detection and SARS-CoV-2 viability assay.
Fig. 1.
Recovery yields of SARS-CoV-2, PEDV, and E229 on artificially contaminated glass (panel A), steel (panel B) and plastic (panel C) surfaces (4 cm2). Tested procedures compared two swabbing methods (Method A based on ISO 15216:1 standard norm vs Method B based on commercial specimen collection swab) and two RNA extraction techniques (MN based on manual column assisted extraction vs MAX semi-automated extraction). For each surface, boxes with the same letter show differences not statistically significant (p-value < 0.05, Tukey HSD test).
Table 2.
Limit of detection at 95 and 50% confidence interval (LOD95% and LOD50%) for SARS-CoV-2 and surrogates PEDV and E229 on glass, steel and plastic surfaces following the selected sampling Method A coupled with MAX RNA extraction procedure.
| Inoculated virus | Glass |
Steel |
Plastic |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positivesa | LOD95%b | LOD50%b | Positives | LOD95% | LOD50% | Positives | LOD95% | LOD50% | ||
| SARS-CoV-2 (gc/4 cm2) | 5.10 × 104 | 6/6 | 3.31 × 102 | 7.65 × 101 | 6/6 | 2.18 × 102 | 5.03 × 101 | 6/6 | 6.37 × 102 | 1.47 × 102 |
| 5.10 × 103 | 6/6 | 6/6 | 6/6 | |||||||
| 5.10 × 102 | 6/6 | 6/6 | 6/6 | |||||||
| 5.10 × 101 | 2/6 | 3/6 | 0/6 | |||||||
| 5.10 × 100 | 0/6 | 0/6 | 0/6 | |||||||
| E229 (PCRU/4 cm2) | 2.28 × 105 | 6/6 | 1.60 × 103 | 3.70 × 102 | 6/6 | 4.92 × 103 | 1.14 × 103 | 6/6 | 4.65 × 103 | 1.08 × 103 |
| 2.28 × 104 | 6/6 | 6/6 | 6/6 | |||||||
| 2.28 × 103 | 6/6 | 5/6 | 4/6 | |||||||
| 2.28 × 102 | 2/6 | 0/6 | 2/6 | |||||||
| 2.28 × 101 | 0/6 | 0/6 | 0/6 | |||||||
| PEDV (PCRU/4 cm2) | 1.42 × 104 | 6/6 | 3.60 × 102 | 8.34 × 101 | 6/6 | 6.94 × 102 | 1.61 × 102 | 6/6 | 1.01 × 103 | 2.33 × 102 |
| 1.42 × 103 | 6/6 | 6/6 | 6/6 | |||||||
| 1.42 × 102 | 4/6 | 3/6 | 2/6 | |||||||
| 1.42 × 101 | 1/6 | 0/6 | 0/6 | |||||||
| 1.42 × 100 | 0/6 | 0/6 | 0/6 | |||||||
Virus positive/total number of samples.
Calculated according to Wilrich and Wilrich (2009).
Detection limits of SARS-CoV-2 and its surrogates on different surfaces were evaluated through the analysis of serial diluted spiked samples. The results showed that the combination of swabbing Method A with MAX semi-automated RNA extraction rendered SARS-CoV-2 LOD95% values of 3.31 × 102, 2.18 × 102, and 6.37 × 102 gc/4 cm2 for glass, steel and plastic surfaces, respectively. The same procedure yielded mean SARS-CoV-2 LOD50% values of 7.65 × 101, 5.03 × 101, and 1.47 × 101 gc/4 cm2 for glass, steel and plastic surfaces, respectively. The same assay on SARS-CoV-2 surrogates E229 and PEDV resulted in higher LOD95% and LOD50% values. Extended data are presented in Table 2.
3.2. Application of SARS-CoV-2 viability RT-qPCR on fomites
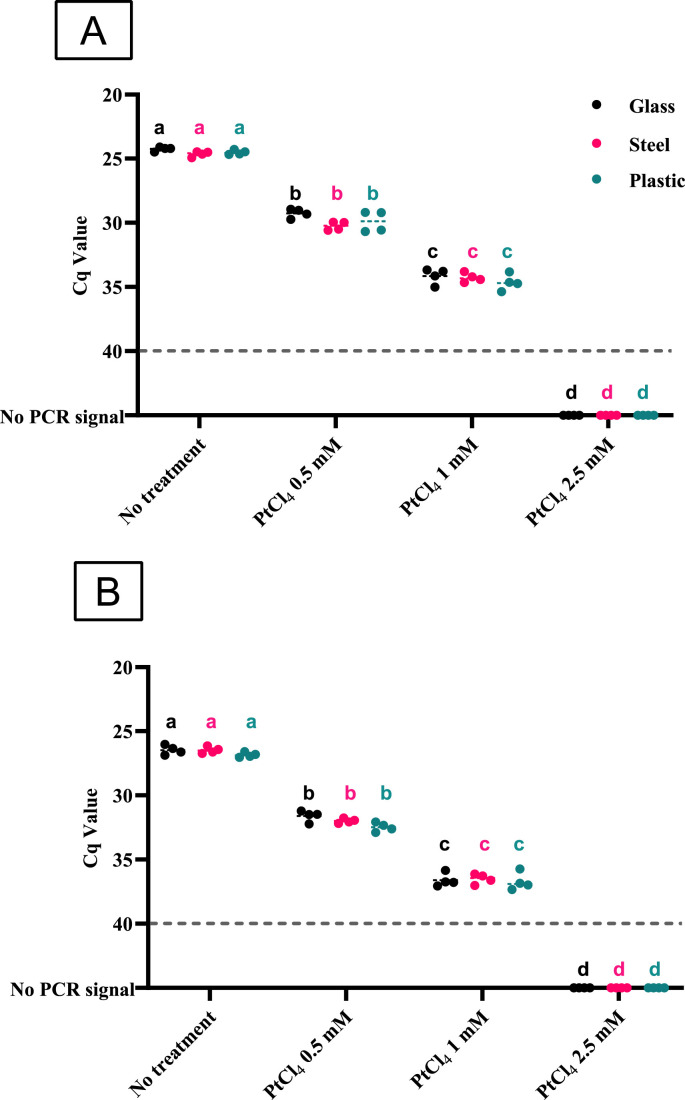
A viability RT-qPCR assay was optimized testing 0.5–2.5 M PtCl4 concentrations in gamma-irradiated and heat-inactivated SARS-CoV-2 suspensions recovered from the artificially contaminated glass, steel, and plastic surfaces. Compared to the PtCl4 untreated control, significant differences were observed for all tested PtCl4 concentrations (0.5, 1.0, and 2.5 mM) (Fig. 2 ). A sharp difference of above one logarithm of gc (ΔCqs ≈ 3.3) was observed with 0.5 mM PtCl4, the lowest viability marker concentration tested. Viability treatment with 1 mM PtCl4 resulted in 2–3 log gc reductions (ΔCqs ≈ 9–10). The complete suppression of the RT-qPCR amplification signal was accomplished when pretreating the suspensions with 2.5 mM PtCl4.
Fig. 2.
Platinum chloride (PtCl4) viability RT-qPCR on gamma-inactivated (panel A) and heat-inactivated (panel B) SARS-CoV-2 suspensions recovered from glass, steel and plastic surfaces artificially contaminated with approximately 6 log10 gc/4 cm2. Dashed grey line represents the RT-qPCR limit of detection. The same letter shows differences not statistically significant for each surface (p < 0.05, Tukey HSD test).
4. Discussion
The global health crisis caused by the COVID-19 pandemic began in late 2019, and since SARS-CoV-2 has been confirmed to be excreted in faeces, urine and nasopharyngeal secretions, surface-mediated transmission of this pathogen has been suggested. Thus, research has been rapidly oriented to understand the distribution, exposure, and persistence of the virus on fomites as crucial factors for implementing environmental surveillance and finally mitigating disease risk.
High-touch fomites in community settings and clinical facilities have been confirmed to be contaminated by SARS-CoV-2 RNA demonstrating that the molecular detection on fomites can be used to track contamination, to identify targets for intervention, and finally to guide any public health response (Ahn et al., 2020; Ben-Shmuel et al., 2020; Elbadawy et al., 2021; Fernández-de-Mera et al., 2020; Harvey et al., 2021; Moreno et al., 2021). However, the risk of infection from touching a contaminated surface has been determined using different environmental models with surprisingly divergent outcomes ranging from low to substantial risk estimates (Harvey et al., 2020; Lopman et al., 2011; Mondelli et al., 2020). Such a discrepancy could be explained by the bias introduced by data on viral exposure and persistence generated in simulated laboratory conditions and those observed in naturally contaminated real-life scenarios (Goldman, 2020). For example, a number of studies reported that infectious SARS-CoV-2 spiked on fomites of different materials persist from a few up to 21 days (Ben-Shmuel et al., 2020; Chin et al., 2020; Kasloff et al., 2021; Riddell et al., 2020; van Doremalen et al., 2020), while attempts to recover infectious virus from naturally contaminated fomites were unsuccessful (Ben-Shmuel et al., 2020; Mondelli et al., 2020) with the only exception of a specific highly contaminated indoor setting, such as the isolation room of a COVID-19 patient requiring ventilation (7 positive culture results for SARS-CoV-2 out 16 samples tested) (Ahn et al., 2020).
Fomite swabbing represents a critical component in environmental SARS-CoV-2 monitoring, and the analytical performance of the method significantly impacts the accuracy of downstream detection with either molecular or cell culture approaches. To shed light on this critical matter, our work aimed to tackle the first of these hurdles which is the characterization of the viral recovery rate and the limit of detection of coronaviruses on fomites following a swabbing procedure and RNA amplification by RT-qPCR. We have demonstrated the efficient recovery of coronavirus, including SARS-CoV-2, PEDV and E229, from glass, steel and plastic surfaces with swabs (mean recovery value = 95.77%) and their sensitive detection by RT-qPCR (overall LOD95% = 1.60 × 103 gc/4 cm2, and LOD50% = 4.55 × 102 gc/4 cm2).
Moreover, PBS swabbing (Method A) and semiautomatic extraction (MAX) was revealed to be a sensitive method to recover SARS-CoV-2 from fomites given 3.95 × 102 and 9.13 × 101, gc/4 cm2 as LOD95% and LOD50%, respectively. CDC-approved (SYN), consumer-grade (CGp), and bulk (TMI) swabs were recently assessed as an alternative to clinical-grade hospital tests for SARS-CoV-2 recovery from fomites. The resulting LOD values ranged from 3.62 × 102 (SYN) to 1.45 × 103 (CGp and TMI) gc on an approx. 26 cm2 area (Minich et al., 2021), a figure in the same order of magnitude as our findings.
Our multivariate results showed no statistical differences for the combination of type of virus, swabbing procedures and extraction methods, finally demonstrating E229 and PEDV as suitable surrogates for SARS-CoV-2 to define viral recovery from surfaces. Similarly, Guillier and colleagues analysing data of Alphacoronavirus and Betacoronavirus, concluded that the persistence of different coronaviruses on fomites is comparable (2020). However, when defining the LoD, we observed some variability being PEDV more similar to SARS-CoV-2 than E229. Multiple mechanisms may explain the slight different LoD of surrogates compared to SARS-CoV-2 on surfaces, including the effect of desiccation on the viruses (Cox, 1993). Considering the use of our procedure in terms of routine fomite monitoring, the combination of PBS with semiautomatic extraction represents additional advantages such as overcoming the shortage of lab consumables and curtailing the turnaround time for results. However, the efficiency of coronavirus recovery from hard surfaces other than stainless steel, glass and plastic may be lower and requires further evaluation. As well, our analytical determinations are based on contaminated surfaces with viral suspensions prepared in cell culture medium. Even beyond the scope of this work, additional experiments should confirm the performance of the proposed swabbing and detection workflow for coronavirus in human secretions (e.g. saliva, urine, nasal mucus).
Molecular detection does not reveal viral infectivity and viral replication in permissive cell line is not sensitive enough for environmental samples. Thus, alternative approaches need to be explored to characterize the health risk from exposure and persistence of SARS-CoV-2 on fomites. We therefore suggest a basic protocol to apply a viability RT-qPCR method for SARS-CoV-2 in environmental swabbing with the aim of differentiating intact and damaged viral particles, and free RNA. Based on findings recently published in a study by our group (Cuevas-Ferrando et al., 2021), we demonstrated that a viability RT-qPCR using 2.5 mM PtCl4 prevented PCR amplification (>4 log of genome copies/4 cm2) of inactivated SARS-CoV-2 recovered from fomites.
Given the much longer stability of RNA compared to infectious particles demonstrated for SARS- CoV-2 on fomites (Kasloff et al., 2021), the viability RT-qPCR allows the elimination of the PCR signals of free RNA and damaged particles (at least partly) thus finally providing more robust data on potential viral infectivity to be used for viral exposure and persistence estimates.
Moreover, the viability RT-qPCR provides results in a significantly shorter turnaround time (a few hours vs days) and benefits from a better analytical sensitivity than conventional cell-culture procedure. This is an aspect of extreme value for the timely implementation of epidemiological control measures, and also considerably expands the number of laboratory facilities where these investigations can be carried out given the BSL-3 requirements needed to cultivate SARS-CoV-2.
In summary, our findings primarily suggest detection of SARS-CoV-2 RNA in the environment could be performed using less expensive and PBS-based storage solutions, without loss of analytical sensitivity. Additional studies would confirm the use of viability RT-qPCR to infer potential infectivity of SARS-CoV-2 samples recovered from fomites, greatly supporting the environmental surveillance of COVID-19 worldwide, particularly in resource-limited communities.
Author contributions
Conceptualization: APC, RA, WR, and GS; Methodology: ECF, IGG, IF, and ADR; Formal analysis: ECF, and WR; Resources: RA, WR, and GS; Writing – original draft: ECF and WR; Writing – review & editing: ECF, IGG, APC, IF, ADR, RA, WR and GS. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The study was supported by EIT-Food (COVID-19 BAEMitup), CSIC (202070E101), Generalitat Valenciana (Covid_19-SCI), MICINN co-founded by AEI/FEDER, UE (AGL2017-82909), and MICINN/AEI (PID2019-105509RJ-I00). EC-F is recipient of a predoctoral contract from the MICINN, Call 2018. IGG is recipient of a Collaboration Grant 2020–2021 from the MEFP.
References
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J.Y., An S., Sohn Y., Cho Y., Hyun J.H., Baek Y.J., Kim M.H., Jeong S.J., Kim J.H., Ku N.S., Yeom J.S., Smith D.M., Lee H., Yong D., Lee Y.J., Kim J.W., Kim H.R., Hwang J., Choi J.Y. Environmental contamination in the isolation rooms of COVID-19 patients with severe pneumonia requiring mechanical ventilation or high-flow oxygen therapy. J. Hosp. Infect. 2020;106:570–576. doi: 10.1016/j.jhin.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäuerl C., Randazzo W., Sánchez G., Selma-Royo M., Garcia-Verdevio E., Martínez-Rodríguez L., Parra-Llorca A., Lerin C., Fumadó V., Crovetto F., Crispi F., Pérez-Cano F.J., Rodríguez G., Ruíz-Redondo G., Campoy C., Martínez-Costa C., Collado M.C. SARS-CoV-2 RNA and antibody detection in human milk from a prospective multicenter study in Spain. medRxiv. 2021 doi: 10.1101/2021.05.06.21256766. 2021. 05.06.21256766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shmuel A., Brosh-Nissimov T., Glinert I., Bar-David E., Sittner A., Poni R., Cohen R., Achdout H., Tamir H., Yahalom-Ronen Y., Politi B., Melamed S., Vitner E., Cherry L., Israeli O., Beth-Din A., Paran N., Israely T., Yitzhaki S., Levy H., Weiss S. Detection and infectivity potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clin. Microbiol. Infect. 2020;26:1658–1662. doi: 10.1016/j.cmi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondin-Brosseau M., Harlow J., Doctor T., Nasheri N. Examining the persistence of human Coronavirus 229E on fresh produce. Food Microbiol. 2021;98 doi: 10.1016/j.fm.2021.103780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2019. Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel for Emergency Use Only Instructions for Use. (n.d) [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020;1:e10. doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C.S. Roles of water molecules in bacteria and viruses. Orig. life Evol. Biosph. J. Int. Soc. Study Orig. Life. 1993;23:29–36. doi: 10.1007/BF01581988. [DOI] [PubMed] [Google Scholar]
- Cuevas-Ferrando E., Randazzo W., Pérez-Cataluña A., Navarro D., Martin-Latin S., Díaz-Reolid A., Girón-Guzmán I., Allende A., Sánchez G. Viability RT-PCR for SARS-CoV-2: a step forward to solve the infectivity quandary 1 2. medRxiv 2021. 2021;3:22. doi: 10.1101/2021.03.22.21253818. 21253818. [DOI] [Google Scholar]
- Elbadawy H.M., Khattab A., Alalawi A., Dakilallah Aljohani F., Sundogji H., Mahmoud A.S., Abouzied M., Eltahir H.M., Alahmadey Z., Bahashwan S., Suliman B.A. The detection of SARS‐CoV‐2 in outpatient clinics and public facilities during the COVID‐19 pandemic. J. Med. Virol. jmv. 2021;26819 doi: 10.1002/jmv.26819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizaquível P., Aznar R., Sánchez G. Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 2014;116:1–13. doi: 10.1111/jam.12365. [DOI] [PubMed] [Google Scholar]
- Fernández‐de‐Mera I.G., Rodríguez del‐Río F.J., Fuente J., Pérez‐Sancho M., Hervás D., Moreno I., Domínguez M., Domínguez L., Gortázar C. Detection of environmental SARS‐CoV‐2 RNA in a high prevalence setting in Spain. Transbound. Emerg. Dis. tbed. 2020;13817 doi: 10.1111/tbed.13817. [DOI] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman E. Exaggerated risk of transmission of COVID-19 by fomites. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier L., Martin-Latil S., Chaix E., Thébault A., Pavio N., Le Poder S., Batéjat C., Biot F., Koch L., Schaffner D.W., Sanaa M. Modeling the inactivation of viruses from the Coronaviridae family in response to temperature and relative humidity in suspensions or on surfaces. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.01244-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A.P., Fuhrmeister E.R., Cantrell M., Pitol A.K., Swarthout J.M., Powers J.E., Nadimpalli M.L., Julian T.R., Pickering A.J. Longitudinal monitoring of SARS-CoV-2 RNA on high-touch surfaces in a community setting. medRxiv. 2020 doi: 10.1101/2020.10.27.20220905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A.P., Fuhrmeister E.R., Cantrell M.E., Pitol A.K., Swarthout J.M., Powers J.E., Nadimpalli M.L., Julian T.R., Pickering A.J. Longitudinal monitoring of SARS-CoV-2 RNA on high-touch surfaces in a community setting. Environ. Sci. Technol. Lett. 2021;8:168–175. doi: 10.1021/acs.estlett.0c00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020 Mar;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. Epub 2020 Feb 6. Erratum in: J Hosp Infect. 2020 Jun 17;: PMID: 32035997; PMCID: PMC7132493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami K., Hosseini-Kharat M., Sadeghi-Aliabadi H., Lipkowski J., Mirian M. In vitro cytotoxicity studies of palladacyclic complexes containing the symmetric diphosphine bridging ligand. Studies of their interactions with DNA and BSA. Eur. J. Med. Chemi. 2014;73:8–17. doi: 10.1016/j.ejmech.2013.11.042. [DOI] [PubMed] [Google Scholar]
- Kasloff S.B., Leung A., Strong J.E., Funk D., Cutts T. Stability of SARS-CoV-2 on critical personal protective equipment. Sci. Rep. 2021;11:984. doi: 10.1038/s41598-020-80098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifels M., Dan C., Sozzi E., Shoults D.C., Wuertz S., Mongkolsuk S., Sirikanchana K. Capsid integrity quantitative PCR to determine virus infectivity in environmental and food applications – a systematic review. Water Res. X. 2020;11:100080. doi: 10.1016/j.wroa.2020.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Yang M., Zhao X., Guo Y., Wang L., Zhang J., Lei W., Han W., Jiang F., Liu W.J., Gao G.F., Wu G. Cold-chain transportation in the frozen food industry may have caused a recurrence of COVID-19 cases in destination: successful isolation of SARS-CoV-2 virus from the imported frozen cod package surface. Biosaf. Heal. 2020;2:199–201. doi: 10.1016/j.bsheal.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman B.A., Hall A.J., Curns A.T., Parashar U.D. Increasing rates of gastroenteritis hospital discharges in US adults and the contribution of norovirus, 1996-2007. Clin. Infect. Dis. 2011;52:466–474. doi: 10.1093/cid/ciq163. [DOI] [PubMed] [Google Scholar]
- Minich J.J., Ali F., Marotz C., Belda-Ferre P., Chiang L., Shaffer J.P., Carpenter C.S., McDonald D., Gilbert J., Allard S.M., Allen E.E., Knight R., Sweeney D.A., Swafford A.D. Feasibility of using alternative swabs and storage solutions for paired SARS-CoV-2 detection and microbiome analysis in the hospital environment. Microbiome. 2021;9:1–13. doi: 10.1186/s40168-020-00960-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondelli M.U., Colaneri M., Seminari E.M., Baldanti F., Bruno R. Low risk of SARS-CoV-2 transmission by fomites in real-life conditions. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno T., Pintó R.M., Bosch A., Moreno N., Alastuey A., Minguillón M.C., Anfruns-Estrada E., Guix S., Fuentes C., Buonanno G., Stabile L., Morawska L., Querol X. Tracing surface and airborne SARS-CoV-2 RNA inside public buses and subway trains. Environ. Int. 2021;147:106326. doi: 10.1016/j.envint.2020.106326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KP, Vunnam SR, Patel PA, Krill KL, Korbitz PM, Gallagher JP, Suh JE, Vunnam RR. Transmission of SARS-CoV-2: an update of current literature. Eur. J. Clin. Microbiol. Infect. Dis. 2020 Nov;39(11):2005–2011. doi: 10.1007/s10096-020-03961-1. Epub 2020 Jul 7. PMID: 32638221; PMCID: PMC7339796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758:143870. doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D, Lois M, Fernández-Núñez M.T., Romalde J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021;786:147534. doi: 10.1016/j.scitotenv.2021.147534. ISSN 0048-9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente H., Randazzo W., Falcó I., Carvajal A., Sánchez G. Rapid selective detection of potentially infectious porcine epidemic diarrhea coronavirus exposed to heat treatments using viability RT-qPCR. Front. Microbiol. 2020;11:1911. doi: 10.3389/fmicb.2020.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Khezri M., Ollivier J., Le Guyader F.S., Rodríguez-Díaz J., Aznar R., Sánchez G. Optimization of PMAxx pretreatment to distinguish between human norovirus with intact and altered capsids in shellfish and sewage samples. Int. J. Food Microbiol. 2018;266 doi: 10.1016/j.ijfoodmicro.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Randazzo W., López-Gálvez F., Allende A., Aznar R., Sánchez G. Evaluation of viability PCR performance for assessing norovirus infectivity in fresh-cut vegetables and irrigation water. Int. J. Food Microbiol. 2016;229:1–6. doi: 10.1016/j.ijfoodmicro.2016.04.010. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Piqueras J., Rodríguez-Díaz J., Aznar R., Sánchez G. Improving efficiency of viability-qPCR for selective detection of infectious HAV in food and water samples. J. Appl. Microbiol. 2018;124 doi: 10.1111/jam.13519. [DOI] [PubMed] [Google Scholar]
- Randazzo Walter, Vasquez-García A., Aznar R., Sánchez G. Viability RT-qPCR to distinguish between HEV and HAV with intact and altered capsids. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell S., Goldie S., Hill A., Eagles D., Drew T.W. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol. J. 2020;17:145. doi: 10.1186/s12985-020-01418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima T., Iwatsuki K.J. Innovative use of palladium compounds to selectively detect live Enterobacteriaceae in milk by PCR. Appl. Environ. Microbiol. 2016;82(23):6930–6941. doi: 10.1128/AEM.01613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/nejmc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Maes P., Duson G., Van Ranst M. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 2005;43:5452–5456. doi: 10.1128/JCM.43.11.5452-5456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilrich C., Wilrich P.-T. Estimation of the pod function and the LOD of a qualitative microbiological measurement method. J. AOAC Int. 2009;92:1763–1772. doi: 10.1093/jaoac/92.6.1763. [DOI] [PubMed] [Google Scholar]
- Wurtzer S, Waldman P, Ferrier-Rembert A, Frenois-Veyrat G, Mouche J.M., et al. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Research. 2021;198:117183. doi: 10.1016/j.watres.2021.117183. https://www.sciencedirect.com/science/article/pii/S004313542100381X ISSN 0043-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet. Gastroenterol. Hepatol. 2020 Apr;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. Epub 2020 Feb 20. PMID: 32087098; PMCID: PMC7130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhang T., Song D., Huang T., Peng Q., Chen Y., Li A., Zhang F., Wu Q., Ye Y., Tang Y. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes. 2017;33:36–41. doi: 10.1016/j.mcp.2017.02.002. [DOI] [PubMed] [Google Scholar]