Abstract
The development of broad-spectrum antivirals against human coronaviruses (HCoVs) is critical to combat the current coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants, as well as future outbreaks of emerging CoVs. We have previously identified a polyethylene glycol-conjugated (PEGylated) lipopeptide, EK1C4, with potent pan-CoV fusion inhibitory activity. However, PEG linkers in peptide or protein drugs may reduce stability or induce anti-PEG antibodies in vivo. Therefore, we herein report the design and synthesis of a series of dePEGylated lipopeptide-based pan-CoV fusion inhibitors featuring the replacement of the PEG linker with amino acids in the heptad repeat 2 C-terminal fragment (HR2-CF) of HCoV-OC43. Among these lipopeptides, EKL1C showed the most potent inhibitory activity against infection by SARS-CoV-2 and its spike (S) mutants, as well as other HCoVs and some bat SARS-related coronaviruses (SARSr-CoVs) tested. The dePEGylated lipopeptide EKL1C exhibited significantly stronger resistance to proteolytic enzymes, better metabolic stability in mouse serum, higher thermostability than the PEGylated lipopeptide EK1C4, suggesting that EKL1C could be further developed as a candidate prophylactic and therapeutic for COVID-19 and other coronavirus diseases.
Key words: Coronavirus, Lipopeptide, SARS-CoV-2, Polyethylene glycol, Fusion inhibitor
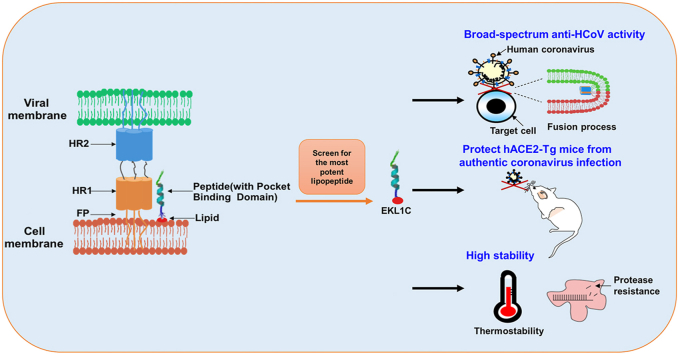
Graphical abstract
The dePEGylated lipopeptides were designed, synthesized, and screened for the most effective one, EKL1C, which was shown to have broad-spectrum anti-coronavirus activity, potent protection of human angiotensin-converting enzyme 2 (hACE2) transgenic mice against authentic coronavirus infection, and high stability.
1. Introduction
Three zoonotic coronaviruses (CoVs) are known to have caused outbreaks of human fatal pneumonia in human and posed a serious threat to public health until now1. Severe acute respiratory syndrome (SARS) caused by SARS-CoV appeared in 2002 and spread to five continents2,3. Middle East respiratory syndrome (MERS) caused by MERS-CoV broke out in 2012 with a high fatality rate4,5. The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to a global pandemic currently. Recent reports have suggested that some bat SARS-related coronaviruses (SARSr-CoVs) may cause novel coronavirus diseases in the near future6,7. These alarming facts call for the development of highly effective pan-CoV inhibitor-based prophylactics and therapeutics to combat both current and future outbreaks of infectious diseases caused by emerging and re-emerging coronaviruses.
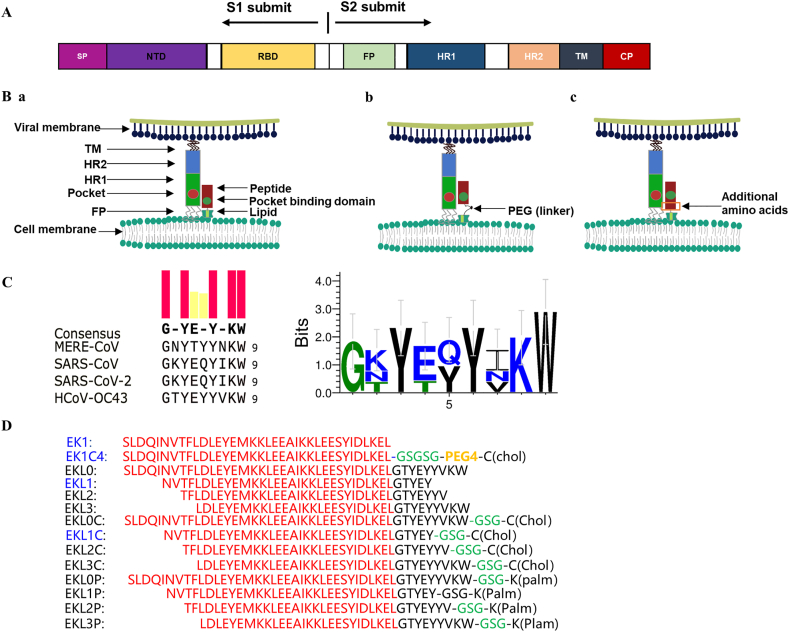
To develop an effective pan-CoV inhibitor-based drug, it is essential to identify a conserved and common target site in the spike (S) proteins of HCoVs, since S protein, which consists of S1 and S2 subunits (Fig. 1A), is responsible for viral attachment with and entry into the host cell. The heptad repeat 1 (HR1) and 2 (HR2) domains in the S2 subunit are highly conserved8,9, and as such, they play an essential role in forming the six-helix bundle (6-HB) core structure during viral fusion. Therefore, the HR1 and HR2 domains are recognized as key targets for the development of pan-CoV fusion and entry inhibitors10.
Figure 1.
Design of the dePEGylated lipopeptide-based pan-CoV fusion inhibitors. (A) Schematic representation of spike (S) protein of a human coronavirus (HCoV). Signal peptide (SP), N-terminal domain (NTD), receptor-binding domain (RBD), fusion peptide (FP), heptad repeat 1 (HR1) domain, HR2 domain, transmembrane domain (TM), and cytoplasmic domain (CP). (B) a: The model of interaction of a lipopeptide-based pan-CoV fusion inhibitor with the HR1 domain in S protein of an HCoV. b: PEG spacer as a flexible linker allowing the lipopeptide to overcome steric hindrance after binding. c: Replacement of the PEG linker with natural amino acids in the HR2 C-terminal fragment (HR2-CF) to facilitate the binding of dePEGylated lipopeptide to the primary binding site in the HR1 domain of viral S protein. (C) Sequence alignment of conserved HR2 sequences of several HCoVs. (D) Amino acid sequences of the dePEGylated lipopeptides and their corresponding peptides without lipid groups, and those of EK1 and EK1C4.
Our group had already designed and developed the first pan-CoV fusion inhibitor, EK1, which was derived from the HR2 sequence in S protein of HCoV-OC439. EK1 was effective in inhibiting infection by all HCoVs tested, including SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-229E, and HCoV-NL63, as well as some bat SARSr-CoVs, such as SARSr-CoV-WIV1, SARSr-CoV-Rs3367, and SARSr-CoV-SHC0149,11. Our mechanistic study showed that EK1 bound to the HR1 domain of an HCoV and blocked 6-HB formation between the viral HR1 and HR2 domains, resulting in the inhibition of viral fusion and entry into the host cell9,11.
Then, in 2020, we modified the EK1 peptide by conjugating the peptide with polyethylene glycol (PEG) and cholesterol. The resultant lipopeptide EK1C4 exhibited significantly improved inhibitory activity against infection by all HCoVs tested, including SARS-CoV-2 and SARSr-CoVs11,12. Consistently, several polyethylene glycol-conjugated (PEGylated) lipopeptides targeting the HR1 domain of SARS-CoV-2 also exhibited highly potent inhibitory activity against SARS-CoV-2 infection and its S protein-mediated membrane fusion13,14. However, it is well known that the use of PEG in a peptide- or protein-based drug may elicit anti-PEG antibodies in vivo and thereby diminish the therapeutic efficacy of PEGylated drugs, or even increase adverse risks15, 16, 17. Furthermore, the use of a PEG linker could enhance the sensitivity of the lipopeptides to protease18.
Therefore, this study aimed to design a series of dePEGylated lipopeptide-based pan-CoV fusion inhibitors by replacing the PEG linker in a PEGylated lipopeptide-based pan-CoV fusion inhibitor, such as EK1C4, with some natural amino acids in the HR2 C-terminal fragment (HR2-CF) of HCoV-OC43 S protein (Fig. 1B). In this way, the length of peptides can be modulated to optimize the primary binding site of peptide interact with viral HR1 domains. Subsequently, the resultant dePEGylated lipopeptides were evaluated for their inhibitory activity against infection of SARS-CoV-2 and its variants, as well as other HCoVs, and for their stability.
2. Materials and methods
2.1. Cell lines, viruses, peptides, and animals
Cell lines, including 293T, Vero-E6, RD, A549/ACE2, Caco-2, and Huh-7, were obtained from the American Type Culture Collection (ATCC, USA). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Gibco, USA). The HCoV-OC43 strains obtained from the ATCC were amplified in RD cells. The peptides EK1 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL) and EK1C4 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL-GSG-PEG4-Chol) were synthesized as previously reported9,11. Six-week-old BALB/c mice and eight-week-old specific pathogen free (SPF) female human angiotensin-converting enzyme 2 transgenic (hACE2-Tg) mice were obtained from the Institute of Laboratory Animal Science, Peking Union Medical College, China (Beijing, China). Newborn mice were bred from pregnant mice purchased from Beijing Vital River Laboratory Animal Technology Co. (Beijing, China). All animal experiments were carried out in strict accordance with institutional regulations (approval number 20190221-070; approval date: 21 February 2019; Shanghai Medical College of Fudan University, Shanghai, China).
2.2. Plasmids
The envelope-expressing plasmids pcDNA3.1-SARS-2-S, pcDNA3.1-SARS-S, pcDNA3.1-MERS-S, pcDNA3.1-NL63-S, pcDNA3.1-OC43-S, pcDNA3.1-WIV1-S, pcDNA3.1-Rs3367-S, pNL4-3.Luc.RE (luciferase reporter vector), and pAAV-IRES-EGFP (encodes EGFP), as well as SARS-CoV-2 S mutant envelope-expressing plasmids, were maintained in our laboratory as previously reported9,11.
2.3. Peptide synthesis
Peptides were synthesized by using a standard solid-phase fluorenylmethoxycarbonyl (Fmoc) method with a CEM Liberty Blue (CEM Co., Matthews, NC, USA) automated microwave peptide synthesizer on Rink amide resin (0.44 mmol/g, Nankai Hecheng S&T Co., Ltd., Tianjin, China). Cholesteroylated peptides were prepared by chemoselective thioether conjugation between the peptide precursor that has a C-terminal cysteine residue and the cholesterol derivative, i.e., cholest-5-en-3-yl bromoacetate, as described previously19. For synthesizing fatty acid conjugated peptides, the template peptides containing a N-[1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene) ethyl] (Dde)-protected lysine residue at their C-terminus required a special deprotection step (four 3-min washes of 2% hydrazinehydrate in N,N-dimethylformamide (DMF). This enabled the conjugation of a palmitoyl group (C16), which was performed by the addition of 3 equivalents of palmic acid, 3 equivalents of O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU), and 6 equivalents of N,N-dimethylformamide (DIEA) in DMF to the resin, followed by stirring for 2 h. The crude products were purified by reverse-phase high-performance liquid chromatography (RP-HPLC). The purity of the products was confirmed to be > 95% pure by analytical HPLC. The molecular weight of the peptides was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS, Autoflex III, Bruker Daltonics, Germany).
2.4. Protein information and amino acid sequence alignment and analysis
The protein accession numbers of MERS-CoV (AID55097.1), SARS-CoV (ABD72979.1), SARS-CoV-2 (QIC53213.1), and HCoV-OC43 (CAA83661.1) were downloaded from NCBI GenBank, and sequence alignment was conducted using MEGA and WebLogo 3.
2.5. Assay of cell–cell fusion
S protein-mediated cell–cell fusion was performed as previously described9,11. In brief, 293T effector cells were transfected with plasmid pAAV-IRES-EGFP, encoding the SARS-CoV-2 S protein (293T/SARS-CoV-2/GFP), while Huh-7 cells, expressing various HCoV receptors on the membrane surface, were used as target cells. Huh-7 cells were first seeded in a 96-well plate at 2 × 104 cells/well for 12 h. Then the 293T/SARS-CoV-2/GFP effector cells were added and cocultured with Huh-7 cells for 2 h in DMEM medium containing 10% FBS at 37 °C. After incubation, the number of fused and unfused cells were counted randomly from five selected fields in each well under an inverted fluorescence microscope (Nikon, Tokyo, Japan), and the percent inhibition of cell–cell fusion was calculated as previously reported9,11.
2.6. Cytotoxicity assay
Cytotoxicity of the peptide EKL1C to the cells (Huh-7, 293T/ACE2, Caco-2) was performed as previously described11. Cells were seeded in a 96-well plate (2 × 104 cells/well) overnight, and EKL1C at graded concentrations was added. After incubation at 37 °C for 48 h, Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) solution was added to measure cell viability, followed by an additional incubation for 4 h. The absorbance was measured at 450 nm with the Multi-Detection Microplate Reader (Tecan, Männedorf, Switzerland). The 50% cytotoxic concentration (CC50) of EKL1C was determined by GraphPad Prism 8.0 software.
2.7. Inhibition of pseudoviruses (PsVs) infection
PsVs were constructed as described previously11,20. Briefly, 293T cells were cotransfected with plasmids pNL4-3.Luc.RE and plasmid pcDNA3.1-HCoV-S (encoding the corresponding HCoV S protein) at mass ratio of 2:1, using VigoFect (Vigorous Biotechnology, Beijing, China). The supernatant containing the PsV was then collected after centrifuging at 1500 ×g for 10 min and maintained at −80 °C until use.
The HCoV PsV inhibition assay was performed as previously described11,21. Briefly, target cells (293T/ACE2 cells for SARS-CoV-2/SARS-CoV/HCoV-NL-63; Caco-2 cells for SARS-CoV-2; A549/ACE2 cells for bat SARSr-CoVs; RD cells for HCoV-OC43 and Huh-7 cells for other CoVs) were seeded at 1 × 104 cells/well into a 96-well plate and incubated overnight at 37 °C. The HCoV PsVs were incubated in the presence or absence of the test peptide which was series diluted with DMEM for 30 min at 37 °C, followed by the addition of target cells. The cells were incubated with or without PsV as virus control and cell control, respectively. And 12 h after infection, the culture was replaced with fresh medium and then incubated for an additional 48 h. Cells were lysed with lysis reagent (Promega, Madison, USA), and the cell lysates were added into a 96-well Costar flat-bottom luminometer plate (Corning Costar, New York, USA), followed by the detection of luminescence with the Multi-Detection Microplate Reader. The SARS-CoV-2 S mutant PsVs were constructed and produced in Huh-7 cells as described previously9,20. An inhibition assay was performed in a manner similar to that described above.
2.8. Inhibition of HCoV replication
The inhibition assay for SARS-CoV-2 was performed in a biosafety level 3 (BSL-3) laboratory as previously described22. Before infection, Vero-E6 cells were seeded at 1 × 104 cells/well in a 96-well plate and incubated overnight at 37 °C. Then peptides with different dilution concentration were mixed with 0.1 multiplicity of infection (MOI) SARS-CoV-2 for 0.5 h. Mixtures were added into the Vero-E6 cells and cocultured for 48 h. The cell supernatants were collected to conduct the quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay. First, the total RNA was extracted using Trizol LS (Thermo, USA). Then the RT-qPCR assay was performed using a One-Step PrimeScrip RT-PCR Kit (Takara, Japan) with primers and probe as follows: SARS-CoV-2-ORF1ab-F: 5′-CCCTGTGGGTTTTACACTTAA-3′, SARS-CoV-2-ORF1ab-R: 5′-ACGATTGTGCATCAGCTGA-3′, SARS-CoV-2-ORF1ab-probe: 5′-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′.
The inhibitory activities of peptides against HoV-OC43 replication on RD cells were assessed as described11. The 102 median tissue culture infective dose (TCID50) of OC43 was mixed with the test peptide at graded concentrations and incubated at 37 °C for 30 min, and then the mixtures were added to the RD cells (1 × 104 cells/well). After culturing for 36 h, CCK-8 solution was added to determine cytopathic effect as described above to measure EKL1C protective effect.
2.9. In vivo protective effect of peptides on mice
To test the protective effect of peptides against SARS-CoV-2 infection in vivo, hACE2-Tg were used22,23. A total of 15 hACE2-Tg mice were randomly assigned to three groups: viral control, prophylactic, and therapeutic groups. In the viral control group, each mouse was challenged intranasally with 105 TCID50 SARS-CoV-2. EKL1C (1.5 mg/kg) was administered intranasally 0.5 h before challenge in the prophylactic group and administered intranasally 0.5 h after challenge in the therapeutic group. Mice were euthanized after 4 days of infection, and lungs were collected and homogenized in Trizol (Takara, Japan). Then total RNA in the lungs were extracted for RT-qPCR assay as described above.
To test the protective effect of peptides against HCoV-OC43 infection in vivo, 18 newborn mice bred from pregnant mice were randomly assigned to three groups: viral control, prophylactic, and therapeutic (each group had six 3-day-old mice). A challenge experiment was performed and evaluated in a manner similar to that described above. Mice in the prevention and treatment groups were intranasally administered with EKL1C (4 mg/kg) 30 min before or after the viral challenge. Each mouse was intranasally challenged with a viral dose of 102 TCID50. Afterwards, the viral titer in mice lungs was collected and assessed9.
2.10. Assay for sensitivity to proteolytic enzymes, proteinase K, and trypsin
Assays for the stability of peptides to proteolytic enzymes, proteinase K, and trypsin were performed as previously described24, 25, 26. First, the concentration of peptide was set according to its 90% inhibition concentration (IC90). For proteinase K tests, peptide was incubated at 37 °C in DMEM containing 1 microunit/mL proteinase K-Acrylic Beads (Sigma–Aldrich, St. Louis, MO, USA). Samples were collected at different times (0, 6, 15, 30, 60, and 240 min) and centrifuged (250 ×g, 5 min, 4 °C) immediately. For trypsin tests, peptide was incubated at 37 °C in DMEM containing 20 μg/mL trypsin (Sigma–Aldrich). Samples were collected at different times (0, 15, 30, 60, 120, 180, and 240 min), 10% FBS was immediately added to neutralize the proteolytic activity of trypsin, followed by inactivating trypsin at 56 °C, 30 min. Then supernatant mixtures were collected and stored at −20 °C before testing. The residual antiviral activity of each sample was detected by PsV inhibition assay as described above.
Quantitative analysis of peptides was conducted with liquid chromatography–tandem mass spectrometry (LC–MS). The LC–MS/MS system consisted of a Waters Acquity UPLC system (Waters Corp., Milford, MA, USA) and coupled with an AB SCIEX triple quadrupole mass spectrometer (Sciex Corp., Framingham, MA, USA). Data were acquired and processed using the Analyst 1.6 software supplied by Sciex (Sciex Corp., Framingham, MA, USA). For quantitative analysis, chromatographic separations were performed on a Unitary C8 analytical column [50 mm × 2.1 mm internal diameter (i.d.) 5 μmol/L; ACCHROM, Beijing, China] and column temperature was maintained at 40 °C. Solvent A (0.1% formic acid in water) and solvent B (methanol) were used as mobile phases. The gradient elution program was optimized as follows: 5% phase B at 0.00–1.00 min; from 5% to 70% phase B at 1.00–2.00 min; from 70% to 95% phase B at 2.00–3.00 min; 95% phase B maintaining at 3.00–3.50 min; from 95% to 5% phase B at 3.50–3.60 min, followed by re-equilibration at 5% phase B until 5 min and the flow rate was 0.5 mL/min. Autosampler temperature was kept at 15 °C and the injection volume was set at 4 μL. EKL1C and EK1C4 were dissolved in water to produce 1 mg/mL standard stock solution respectively, and stored at −20 °C. Then the calibrators at 5, 0.5, 0.05 μmol/L were diluted by methanol/water (1/1, v/v). In vitro metabolic study in proteinase K and trypsin were performed according to the reported procedures, the working solutions (10 μmol/L, 0.3 mL) and proteinase K solution (40 μg/mL, 0.3 mL), or trypsin solution (40 μg/mL, 0.3 mL), were mixed in a 1.1 mL microcentrifuge tube. The mixtures were incubated at 37 °C in a water bath for 240 min. The reactions were terminated by drawing 50 μL of the incubation mixtures into tubes containing 50 μL of ice-cold Methanol at 0, 15, 30, 60, 120, 180, and 240 min followed by centrifugation at 5000 ×g for 10 min at 4 °C. The supernatant was transferred to ultra-performance liquid chromatography (UPLC) auto-sampler vials and was analyzed according to the analytical method.
2.11. Assay for stability of peptides stored at different temperatures
To detect the significance of difference in stability between EKL1C and EK1C4, the peptides were stored under the same conditions at 4 °C, room temperature (RT) and 37 °C, respectively. Samples were collected at the same time at different days (15, 30, 60, 100, and 120 days) and stored at −80 °C before testing. The antiviral activity of each sample was detected by PsV inhibition assay as described above.
2.12. Assay for metabolic stability of peptides in mouse serum
To compare significance of difference in metabolic stability of peptides in mouse serum, we collected mouse serum from 6-week BALB/c and mixed the serum with peptides at the concentration that could achieve IC90, estimated according to the result from the previous study. After incubation at 37 °C, samples were collected at different time points (1, 3, 6, and 18 h), followed by inactivating serum at 56 °C for 30 min. Then the mixtures were collected and stored at −20 °C before testing. The residual antiviral activity of each sample was detected by PsV inhibition assay as described above.
2.13. Statistical analysis
All data were presented as the mean ± standard deviation (SD) from at least three experiments. Statistical differences were analyzed with GraphPad Prism software. P < 0.05 was considered significant (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001). One-way ANOVA was used to compare the statistical significance of difference between the pretreatment and viral control groups or between the post-treatment and viral control groups in the inhibition of authentic SARS-CoV-2 and HCoV-OC43 replication in hACE2-Tg mice. Unpaired Student's t-test was used to compare the difference in sensitivity to trypsin, protease K, and serum between EKL1C and EK1C4 and the significance of difference in stability of EKL1C and EK1C4 stored at different temperatures.
3. Results
3.1. Designing dePEGylated lipopeptides and screening for lipopeptides with the most potent inhibitory activity against pseudotyped SARS-CoV-2 infection and S protein-mediated cell–cell fusion
In order to design effective pan-CoV dePEGylated lipopeptides, we first conducted an alignment with the amino acid sequences of the HR2-CF of several HCoVs, including SARS-CoV, MERS-CoV, SARS-CoV-2, and HCoV-OC43. We found that HR2-CF sequences are relatively conserved among these HCoVs (Fig. 1C). Then, we replaced the PEG flexible linker in the EK1C4 lipopeptide with HR2-CF sequence of HCoV-OC43 to design a series of dePEGylated lipopeptides consisting of different length of EK1 and HR2-CF sequences, as well as cholesterol (Chol) or palmitic acid (Palm) (Fig. 1D). The molecular weight of the synthesized peptides was confirmed by MALDI-TOF–MS (Supporting Information Fig. S1).
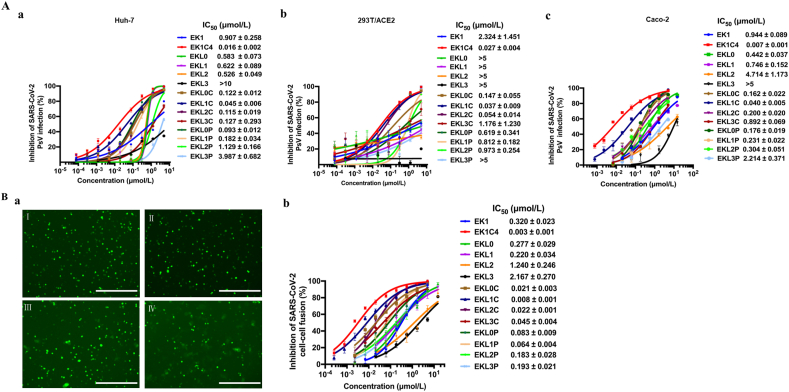
We assessed the antiviral activity of lipopeptides against SARS-CoV-2 PsV infection in Huh-7, 293T/ACE2, and Caco-2 cells, respectively. Among these lipopeptides, EKL1C was found to be the most potent, with IC50s ranging from 0.037 to 0.045 μmol/L, respectively, about 20- to 62-fold more potent than that of EK1 (Fig. 2A), but about 1.3- to 5-fold less potent than that of EK1C4 (IC50 ranging from 0.007 to 0.027 μmol/L).
Figure 2.
The dePEGylated lipopeptide EKL1C showed the most potent inhibitory activity against SARS-CoV-2 PsV infection and S protein-mediated membrane fusion. (A) Inhibition of the peptides against SARS-CoV-2 PsV infection in Huh-7 cells (a), 293T/ACE2 cells (b), and Caco-2 cells (c). Data are presented as mean ± SD (n = 3). (B) Inhibition of the peptides against SARS-CoV-2 S protein-mediated cell–cell fusion. a: Images of SARS-CoV-2 S protein-mediated cell–cell fusion in the presence of EK1 (I), EKL1C (II), EKL1 (III), and PBS (IV). Scale bars: 200 μm. b: Inhibitory activity of the peptides on SARS-CoV-2 S protein-mediated cell–cell fusion. Data are presented as mean ± SD (n = 3).
Next, we tested the potential inhibitory activity of the lipopeptides against SARS-CoV-2 S protein-mediated cell–cell fusion. As shown in Fig. 2Ba, EKL1C could completely inhibit cell–cell fusion at the concentration of 2.5 μmol/L and showed the most potent inhibitory activity among the lipopeptides tested, consistent with the results from the SARS-CoV-2 PsV infection experiments. The IC50 value of EKL1C for inhibiting SARS-CoV-2 S protein-mediated cell–cell fusion was 0.008 μmol/L, about 40-fold more potent than that of EK1 (IC50 = 0.320 μmol/L), but about 2.6-fold less potent than that of EK1C4 (IC50 = 0.003 μmol/L).
We then assessed the cytotoxicity of EKL1C and found that its CC50 was 10, 13.81, and 8.49 μmol/L on Huh-7, Caco-2, and 293T/ACE2 cells, respectively (Supporting Information Fig. S2), and all the selectivity indexes (SI=CC50/IC50) were in a range of 222–345, justifying the further study of EKL1C.
3.2. The dePEGylated lipopeptide EKL1C potently inhibited infection by various pseudotyped SARS-CoV-2 S variants and multiple HCoV PsVs
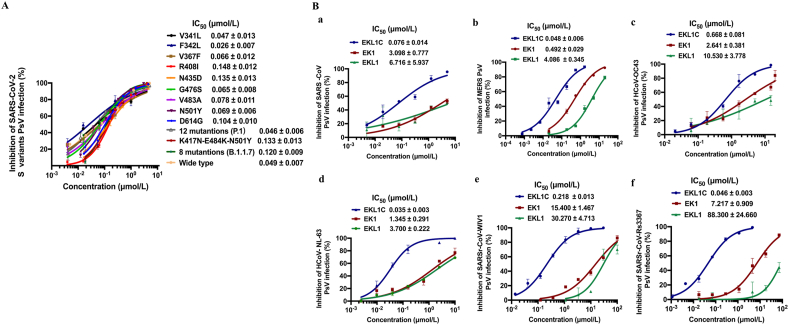
Mutations in the spike protein of SARS-CoV-2 are well known, e.g., D614G and N501Y, detected in some SARS-CoV-2 variants isolated from different regions at different times. It is also suspected that such variants have the potential to escape from effective immune responses against the wild-type SARS-CoV-227,28. Here we assessed the inhibitory activity of EKL1C peptide against infection of Huh-7 cells by pseudotyped SARS-CoV-2 strains with single or multiple key mutations in S protein of some SARS-CoV-2 variants, including 9 mutants with a single mutation in S protein (V341L, F342L, V367F, R408I, N435D, G476S, V483A, N501Y, D614G), P.1 variant with mutations in S protein (L18F/T20N/P26S/D138Y/R190S/K417T/E484K/N501Y/D614G/H655Y/T1027I/V1176F)29, B.1.351 variant with mutations in receptor-binding domain (RBD) (N501Y/E484K/K417N)30, and B.1.1.7 variant with mutations in S protein (ΔH69/V70/ΔY144/N501Y/A570D/D614G/P681H/T716I/S982A/D1118H)31. We found that EKL1C could effectively inhibit infection of all 12 mutants with IC50 below 0.15 μmol/L (Fig. 3A).
Figure 3.
The dePEGylated lipopeptide EKL1C exhibited broad-spectrum anti-CoV activity. (A) Inhibitory activity of EKL1C against infection of pseudotyped SARS-CoV-2, as well as its S protein carrying single or multiple key mutations, in Huh-7 cells. Data are presented as mean ± SD (n = 3). (B) Inhibitory activity of EKL1C against infection of pseudotyped SARS-CoV (a), MERS-CoV (b), HCoV-OC43 (c), HCoV-NL63 (d), WIV1 (e), and Rs3367 (f). Data are presented as mean ± SD (n = 3).
We then compared the inhibitory activity of the cholesterol-conjugated EKL1 peptide, EKL1C, with that of EK1 and EKL1 peptides (Fig. 1D) against infection by different pseudotyped HCoVs. We found that EKL1C was highly effective in inhibiting infection by the pseudotyped β-coronaviruses, including SARS-CoV (Fig. 3Ba), MERS-CoV (Fig. 3Bb), and HCoV-OC43 (Fig. 3Bc) with IC50 of 0.076, 0.048, and 0.668 μmol/L, respectively, showing higher potency than that of either EK1 (IC50: 0.49–3.09 μmol/L) or EKL1 (IC50: 4.08–10.53 μmol/L). EKL1C could also effectively block infection of the pseudotyped α-HCoV, HCoV-NL63 with IC50 of 0.035 μmol/L, about 37- and 105-fold more effective than either EK1 (IC50 = 1.345 μmol/L) or EKL1 (IC50 = 3.7 μmol/L), respectively (Fig. 3Bd). EKL1C was also highly effective against infection of pseudotyped SARSr-CoVs, including WIV1 and Rs3367, with IC50s of 0.218 and 0.046 μmol/L, respectively, about 70- and 156-fold more potent than that of EK1, and 138- and 1919-fold more potent than that of EKL1 (Fig. 3Be and f).
3.3. The dePEGylated lipopeptide EKL1C effectively inhibited authentic SARS-CoV-2 and HCoV-OC43 infection in vitro and in vivo
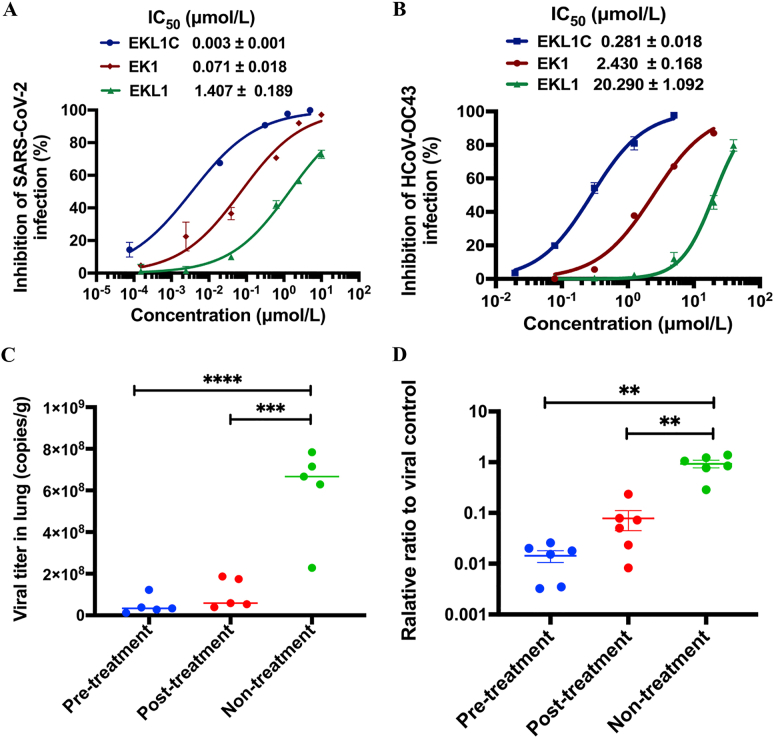
We further assessed the inhibitory activity of EKL1C against authentic SARS-CoV-2 and HCoV-OC43 infection. As expected, EKL1C blocked SARS-CoV-2 infection in Vero-E6 cells in a dose-dependent manner with IC50 of 0.003 μmol/L, about 23- and 469-fold more potent than that of EK1 (IC50 = 0.071 μmol/L) and EKL1 (IC50 = 1.407 μmol/L), respectively, which is consistent with the results from the PsV infection assay as described above (Fig. 4A). Again, EKL1C could effectively inhibit authentic HCoV-OC43 infection in RD cells in a dose-dependent manner with IC50 of 0.281 μmol/L, about 8- and 71-fold more effective than that of EK1 (IC50 = 2.43 μmol/L) and EKL1 (IC50 = 20.29 μmol/L) (Fig. 4B).
Figure 4.
EKL1C effectively inhibits authentic SARS-CoV-2 and HCoV-OC43 infection in vitro and in vivo. (A) EKL1C inhibited authentic SARS-CoV-2 infection in Vero E6 cells in a dose-dependent manner. Data are presented as mean ± SD (n = 3). (B) EKL1C inhibited authentic HCoV-OC43 infection in RD cells in a dose-dependent manner. Data are presented as mean ± SD (n = 3). (C) Reduction of SARS-CoV-2 titer in the lungs of hACE2-Tg mice in the EKL1C pre- and post-treatment groups, compared with the non-treatment control group. ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (D) Reduction of HCoV-OC43 titer in the lungs of newborn mice in the EKL1C pre- and post-treatment groups, compared with the non-treatment control group. ∗∗P < 0.01.
As HCoVs transmit and infect humans mainly though the respiratory tract, hACE2-Tg mice were administered with EKL1C via an intranasal route at 0.5 h before or after intranasal challenge with 105 TCID50 SARS-CoV-2 in order to assess the prophylactic and therapeutic potential of EKL1C. Four days post-challenge, lungs of mice in the three groups were collected for the quantification of viral titers in lung tissues. Compared with the nontreatment groups, viral titer in the lungs of mice in the prophylactic and therapeutic groups decreased significantly (∗∗∗P < 0.001, ∗∗∗∗P < 0.0001, Fig. 4C). To assess the inhibitory activity of EKL1C peptides against authentic HCoV-OC43 infection in vivo, HCoV-OC43-infected newborn mice were treated with EKL1C at 0.5 h before or after intranasal challenge with 102 TCID50 HCoV-OC43. As shown in Fig. 4D, the viral titers detected in the lungs of mice in the prophylactic and therapeutic groups were decreased about 100- and 10-fold, compared with those in the lungs of mice in the nontreatment control group. These results confirmed that EKL1C is effective against authentic SARS-CoV-2 and HCoV-OC43 infection in vivo.
3.4. The dePEGylated lipopeptide EKL1C showed stronger resistance to proteinase K and trypsin, better metabolic stability in mouse serum and higher thermostability than the PEGylated lipopeptide EK1C4
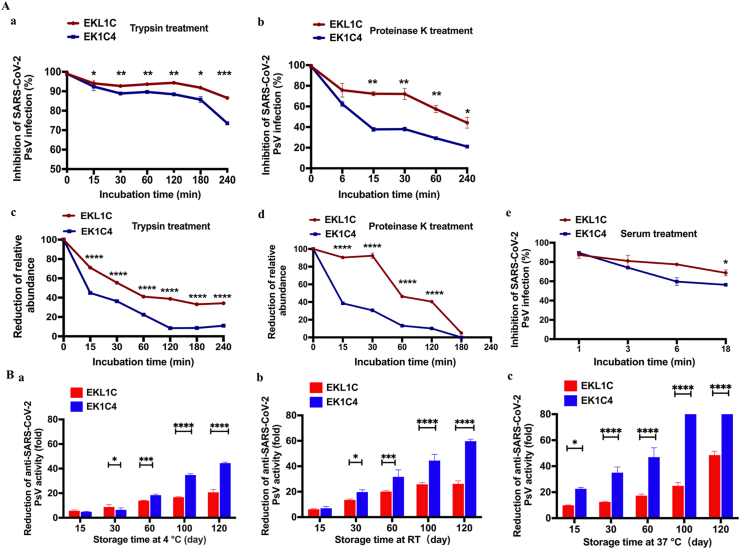
To compare the sensitivity of the dePEGylated lipopeptide EKL1C with the PEGylated lipopeptide EK1C4 to proteolytic enzymes, both peptides were treated with trypsin or proteinase K at 37 °C, followed by collection of samples at different time points post-treatment for detection of their inhibitory activity against SARS-CoV-2 PsV infection. As shown in Fig. 5Aa and b, the inhibitory activity of EK1C4 against SARS-CoV-2 PsV infection gradually decreased more significantly than that of EKL1C under the same treatment condition (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). These samples were also analyzed by LC–MS quantitative analysis and similar trends were obtained (Fig. 5Ac and d). These results suggest that the dePEGylated lipopeptide EKL1C is significantly more resistant than the PEGylated lipopeptide EK1C4 to the proteolytic enzymes trypsin and proteinase K.
Figure 5.
Comparison of the sensitivity to proteolytic enzymes (trypsin and proteinase K), metabolic stability in mouse serum, and thermostability between EKL1C and EK1C4. (A) The sensitivity of EKL1C and EK1C4 to trypsin (a, c) and proteinase K (b, d) after the peptides were treated with trypsin or proteinase K at 37 °C for different time as determined by SARS-CoV-2 PsV inhibition assay (a, b) and LC–MS quantitative analysis (c, d), respectively. The metabolic stability of EKL1C and EK1C4 in mouse serum was assessed by SARS-CoV-2 PsV inhibition assay (e). Data are presented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. EK1C4 groups. (B) Thermostability of EKL1C and EK1C4 stored at 4 °C for 120 days (a), RT for 120 days (b), and at 37 °C for 120 days (c). The inhibitory activity of the peptides against SARS-CoV-2 PsV infection was detected. Data are presented as mean ± SD (n = 3). ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
To compare the metabolic stability of the dePEGylated lipopeptide EKL1C with that of the PEGylated lipopeptide EK1C4 in mouse serum, we added the peptides into mouse serum. After incubation at 37 °C for 18 h, the inhibitory activity of EKL1C on SARS-CoV-2 PsV infection decreased about 20%, while that of EK1C4 decreased about 40% (∗P < 0.05), indicating that EKL1C has better metabolic stability than EK1C4 (Fig. 5Ae).
To detect the thermostability of lipopeptides, we tested the antiviral activities of EKL1C and EK1C4 stored at different temperatures and different times. As shown in Fig. 5B, after being stored at 4 °C, RT, and 37 °C for 120 days, the inhibitory activity of EK1C4 against SARS-CoV-2 PsV infection was reduced by about 44-, 58-, and 92-fold, respectively, while that of EKL1C decreased about 20-, 26-, and 48-fold, respectively, confirming that dePEGylated lipopeptide EKL1C is significantly more thermostable than the PEGylated lipopeptide EK1C4 (∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001).
4. Discussion
The current COVID-19 pandemic caused by SARS-CoV-2 and its variants has posed a serious threat to the global heath and economy27,32, 33, 34, 35, while future outbreaks of highly pathogenic infectious diseases may be caused by emerging and re-emerging coronaviruses36. These current and potential threats call for the development of potent pan-CoV inhibitor-based antivirals targeting the conserved sites in the functional proteins in HCoVs10.
Our recent studies have shown that peptides derived from the HR2 domain in the S2 subunit of HCoV-OC43 S protein, such as OC43-HR2P and its mutant peptide EK1, which target the conserved HR1 domain, exhibit effective and broad-spectrum inhibitory activities against infection of SARS-CoV-2 and its variants, as well as all HCoVs and some SARSr-CoVs tested, and their S protein-mediated membrane fusion9,37. To improve upon the anti-HCoV activity of EK1, we conjugated cholesterol to the C-terminus of EK1 under the assistance of a PEG linker because PEG, as a flexible linker, could facilitate overcoming the steric hindrance against the binding of lipopeptide to the target site38 (Fig. 1B). Indeed, the resultant PEGylated lipopeptide, EK1C4 exhibited much more potent anti-CoV activity than EK111. A number of other reports also show that PEGylated lipopeptides possess much improved antiviral activity over that of unmodified peptides11,13,39, possibly because the lipopeptide, through cholesterol linkage at its C-terminus, can anchor to the host cell membrane, thus raising the local concentration of the lipopeptide at the fusion site (Fig. 1B).
However, recent studies have shown that some PEG-modified therapeutics could induce PEG-specific antibodies in vivo, resulting in decreased therapeutic efficacy40, 41, 42, 43. It was also reported that PEG conjugation at the C-terminus of a lipopeptide could enhance its sensitivity to proteolytic enzymes18,44. Most recently, it was reported that at least eight health volunteers who received the mRNA COVID-19 vaccine produced by Pfizer and BioNTech suffered severe allergy-like reactions, possibly because these people may have had high levels of the preexisting antibodies against PEG, which was used in the nanoparticles for mRNA packaging of the vaccine45.
Therefore, to circumvent these limitations, we designed and synthesized eight dePEGylated lipopeptides by replacing the PEG linker with some amino acids at the HR2-CF of HCoV-OC43 as a linker with an optimal length between the pan-CoV fusion inhibitory peptide and the cholesterol or palmitic acid, and four corresponding peptides without lipid groups as controls (Fig. 1D). Among these dePEGylated lipopeptides, EKL1C showed the most effective anti-SARS-CoV-2 activity with IC50 at nmol/L level, confirming its high level of anti-SARS-CoV-2 activity.
Based on our comprehensive results, EKL1C could maintain its potent pan-CoV fusion inhibitory activity by flexibly binding to the HR1 domains in the S proteins of all above HCoVs and SARSr-CoVs tested.
Most importantly, the dePEGylated lipopeptide EKL1C exhibited significantly stronger resistance to the proteolytic enzymes, trypsin, and proteinase, better metabolic stability in mouse sera, and higher thermostability than the PEGylated lipopeptide EK1C4, suggesting that our strategy offers a possible solution to the limitations of PEG linkers in PEGylated drugs. Since the dePEGylated lipopeptide EKL1C contains no PEG, it is unlikely to cause severe allergy-like reactions in people who have high levels of the preexisting anti-PEG antibodies.
5. Conclusions
We have designed and developed a highly stable dePEGylated lipopeptide, EKL1C, with highly potent pan-CoV fusion inhibitory activity. First, EKL1C targets a highly conserved target site in HR1 domains in S protein of HCoVs, thus possessing broad-spectrum anti-HCoV activity and high genetic barrier for drug resistance, successfully addressing the shortcomings of antibody drugs. Second, EKL1C could be used in an intranasal formulation to prevent HCoV infection in the respiratory system, thus reducing viral loads and pathological damage in lungs caused by HCoV infection. Third, EKL1C has shown relatively stronger resistance to proteolytic enzymes and higher thermostability, thereby avoiding the limitations of a PEGylated lipopeptide drug candidate like EK1C4. Collectively, these findings suggest that EKL1C is a promising candidate for further development as a potent and stable pan-HCoV fusion inhibitor that can be used alone or in combination with other broad-spectrum anti-HCoV drugs for the treatment and prevention of infection by SARS-CoV-2 and its variants, other HCoVs, and any future emerging and re-emerging coronaviruses.
Acknowledgments
We thank members of the Core Facility of Microbiology and Parasitology (SHMC) and the Biosafety Level 3 Laboratory at Shanghai Medical College of Fudan University (China), especially Drs. Di Qu, Xia Cai, Chengjian Gu, and Gaowei Hu for their technical assistance in this study. This work was supported by the National Natural Science Foundation of China (81822045 and 82041036 to Lu Lu; 82041025 to Shibo Jiang; 21877127 to Chao Wang) and the Program of Shanghai Academic/Technology Research Leader (20XD1420300 to Lu Lu, China).
Author contributions
Qian Wang, Shibo Jiang, Lu Lu, and Youhua Xie designed the study. Jie Zhou, Wei Xu, Zezhong Liu, Chao Wang, Shuai Xia, Qiaoshuai Lan, Yanxing Cai, Shan Su, Jing Pu, and Lixiao Xing performed the biochemical and viral experiments and carried out data analysis. Jie Zhou and Chao Wang wrote the manuscript. Qian Wang, Shibo Jiang, Lu Lu, and Youhua Xie revised the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.026.
Contributor Information
Youhua Xie, Email: yhxie@fudan.edu.cn.
Lu Lu, Email: lul@fudan.edu.cn.
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
Qian Wang, Email: wang_qian@fudan.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 4.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Gallagher T., Perlman S. Public health: broad reception for coronavirus. Nature. 2013;495:176–177. doi: 10.1038/495176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Xia S., Wang Q., Xu W., Li W., Lu L., et al. Broad-spectrum coronavirus fusion inhibitors to combat COVID-19 and other emerging coronavirus diseases. Int J Mol Sci. 2020;21:3843. doi: 10.3390/ijms21113843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.K., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L., Su S., Yang H., Jiang S. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184:1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries R.D., Schmitz K.S., Bovier F.T., Predella C., Khao J., Noack D., et al. Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science. 2021;371:1379–1382. doi: 10.1126/science.abf4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J Virol. 2020;94 doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garay R.P., El-Gewely R., Armstrong J.K., Garratty G., Richette P. Antibodies against polyethylene glycol in healthy subjects and in patients treated with PEG-conjugated agents. Expet Opin Drug Deliv. 2012;9:1319–1323. doi: 10.1517/17425247.2012.720969. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q., Jacobs T.M., McCallen J.D., Moore D.T., Huckaby J.T., Edelstein J.N., et al. Analysis of pre-existing IgG and IgM antibodies against polyethylene glycol (PEG) in the general population. Anal Chem. 2016;88:11804–11812. doi: 10.1021/acs.analchem.6b03437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Povsic T.J., Lawrence M.G., Lincoff A.M., Mehran R., Rusconi C.P., Zelenkofske S.L., et al. Pre-existing anti-PEG antibodies are associated with severe immediate allergic reactions to pegnivacogin, a PEGylated aptamer. J Allergy Clin Immunol. 2016;138:1712–1715. doi: 10.1016/j.jaci.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 18.Mathieu C., Augusto M.T., Niewiesk S., Horvat B., Palermo L.M., Sanna G., et al. Broad spectrum antiviral activity for paramyxoviruses is modulated by biophysical properties of fusion inhibitory peptides. Sci Rep. 2017;7:43610. doi: 10.1038/srep43610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingallinella P., Bianchi E., Ladwa N.A., Wang Y.J., Hrin R., Veneziano M., et al. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc Natl Acad Sci U S A. 2009;106:5801–5806. doi: 10.1073/pnas.0901007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z., Xia S., Wang X., Lan Q., Xu W., Wang Q., et al. Inefficiency of sera from mice treated with pseudotyped SARS-CoV to neutralize 2019-nCoV infection. Virol Sin. 2020;35:340–343. doi: 10.1007/s12250-020-00214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct Target Ther. 2020;5:282. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 24.Tong P., Lu Z., Chen X., Wang Q., Yu F., Zou P., et al. An engineered HIV-1 gp41 trimeric coiled coil with increased stability and anti-HIV-1 activity: implication for developing anti-HIV microbicides. J Antimicrob Chemother. 2013;68:2533–2544. doi: 10.1093/jac/dkt230. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Lu L., Qi Z., Lu H., Wang J., Yu X., et al. Novel recombinant engineered gp41 N-terminal heptad repeat trimers and their potential as anti-HIV-1 therapeutics or microbicides. J Biol Chem. 2010;285:25506–25515. doi: 10.1074/jbc.M110.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pang W., Wang R.R., Yang L.M., Liu C.M., Tien P., Zheng Y.T. Recombinant protein of heptad-repeat HR212, a stable fusion inhibitor with potent anti-HIV action in vitro. Virology. 2008;377:80–87. doi: 10.1016/j.virol.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182 doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 29.Hirotsu Y., Omata M. Discovery of a SARS-CoV-2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J Infect. 2021;82 doi: 10.1016/j.jinf.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 31.Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., et al. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., et al. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J Virol. 2019;93 doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Augusto M.T., Hollmann A., Castanho M.A., Porotto M., Pessi A., Santos N.C. Improvement of HIV fusion inhibitor C34 efficacy by membrane anchoring and enhanced exposure. J Antimicrob Chemother. 2014;69:1286–1297. doi: 10.1093/jac/dkt529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Augusto M.T., Hollmann A., Porotto M., Moscona A., Santos N.C. Antiviral lipopeptide–cell membrane interaction is influenced by PEG linker length. Molecules. 2017;22:1190. doi: 10.3390/molecules22071190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q., Lai S.K. Anti-PEG immunity: emergence, characteristics, and unaddressed questions. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2015;7:655–677. doi: 10.1002/wnan.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abu Lila A.S., Kiwada H., Ishida T. The accelerated blood clearance (ABC) phenomenon: clinical challenge and approaches to manage. J Control Release. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu T., Abu Lila A.S., Fujita R., Awata M., Kawanishi M., Hashimoto Y., et al. A hydroxyl PEG version of PEGylated liposomes and its impact on anti-PEG IgM induction and on the accelerated clearance of PEGylated liposomes. Eur J Pharm Biopharm. 2018;127:142–149. doi: 10.1016/j.ejpb.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 43.McSweeney M.D., Versfeld Z.C., Carpenter D.M., Lai S.K. Physician awareness of immune responses to polyethylene glycol–drug conjugates. Clin Transl Sci. 2018;11:162–165. doi: 10.1111/cts.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vigen M., Ceccarelli J., Putnam A.J. Protease-sensitive PEG hydrogels regulate vascularization in vitro and in vivo. Macromol Biosci. 2014;14:1368–1379. doi: 10.1002/mabi.201400161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vrieze Jd. March. 8 2021. Suspicions grow that nanoparticles in Pfizer's COVID-19 vaccine trigger rare allergic reactions.https://www.sciencemag.org/news/2020/12/suspicions-grow-nanoparticles-pfizer-s-covid-19-vaccine-trigger-rare-allergic-reactions Available from: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.