Abstract
Background
The prognosis for patients with stage II/III non–small cell lung cancer (NSCLC) is unsatisfactory, even after complete tumor resection and adjuvant chemotherapy. Here, we assessed the prognostic and predictive value of immunogenomic signatures for stage II/III NSCLC in Chinese patients.
Methods
A total of 91 paired resected stage II/III NSCLC and normal tissues, including 47 squamous cell lung carcinomas (SCC) and 44 lung adenocarcinomas (ADC), were collected and analyzed using whole exome sequencing (WES) to identify immunogenomic signatures for association with clinicopathological variables and disease‐free survival (DFS).
Results
Higher neoantigen load (NAL, >2 neoantigens/Mb) exhibited better DFS for SCC patients (p = 0.021) but not ADC patients. A benefit from adjuvant chemotherapy was correlated with lower NAL (≤2 neoantigens/Mb) (p = 0.009). However, tumor mutation burden (TMB), mutations of individual gene, oncogene pathways, and antigen presentation machinery genes, and human leukocyte antigen (HLA)‐I number and HLA‐I loss of heterozygosity (LOH) had no prognostic or predictive value for DFS of SCC or ADC patients.
Conclusions
NAL is a useful biomarker for lung SCC prognosis and prediction of chemotherapy responses in Chinese patients. The predictive value of NAL for adjuvant immunotherapy should be further explored in patients with resected NSCLC.
Keywords: biomarker, neoantigen load, NSCLC, prognosis, whole exome sequencing
Kaplan–Meier curves, and log rank test were stratified by neoantigen load (NAL). Prognostic and predictive effect of NAL on DFS of SCC patients. There was an association of low NAL number with poor DFS (months, hazard ratio [HR] = 2.56, 95% CI: 1.15–5.68, p = 0.021) in SCC patients. Furthermore, we also found the benefit of the adjuvant chemotherapy in improvement of DFS in SCCs with a lower NAL number. For patients with SCC, adjuvant chemotherapy, as compared with observation, significantly prolonged DFS among patients with NAL ≤2 (HR = 0.34, 95% CI: 0.11–1.00, p = 0.049) but not among patients with NAL >2 (HR = 0.623, 95% CI: 0.163–2.377, p = 0.483). And the interaction terms between NAL and adjuvant treatment were significant for DFS (p = 0.008).

INTRODUCTION
Lung cancer is still the most significant health burden worldwide and in China, accounting for more than 2.2 million new cases and more than 1.79 million cancer‐related deaths in 2020 globally. 1 According to Chinese national statistics in 2015, approximately 733 000 new cases and 631 000 deaths were reported. 2 Histologically, lung cancer can be divided into small cell lung cancer and non‐small cell lung cancer (NSCLC), while approximately 85% of all lung cases are NSCLC and the latter can be further classified into lung adenocarcinoma (ADC), squamous cell carcinoma (SCC), large cell lung carcinoma, and unclassified carcinoma. 3 Currently, adjuvant chemotherapy in NSCLC patients after complete tumor resection is the standard treatment improving five‐year survival in approximately 5% of patients. 4 However, development and identification of various biomarkers could assist in the appraisal of clinical outcomes of different treatment options and distinguish the benefit of adjuvant chemotherapy in the management of NSCLC patients. 5 For example, both ADAURA and CTONG1104 studies demonstrated that adjuvant therapy of NSCLC with the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (EGFR‐TKI) significantly improved disease‐free survival (DFS) of patients with EGFR‐mutant NSCLC. 6 , 7 The recent development of immune checkpoint inhibitors, such as targeting of programmed death‐1 (PD‐1)/programmed death ligand‐1 (PD‐L1), has revolutionized our control of NSCLC with an undeniable efficacy. 8 Thus, further exploration of immunogenomic signatures in different pathological NSCLC types and evaluation of their impact on clinical outcome by identification of the high‐risk subgroups in disease recurrence or treatment responses could lead to better therapy selections and survival benefit for patients.
Evading immune attack has been determined as a key hallmark of cancer and immune escape has been detected in patients with early stage NSCLC. 9 The absence of immunogenicity is the first step of immune evasion and is strongly associated with clinical outcome in patients. 10 The tumor mutation burden has been characterized as an indicator of immunogenicity given that gene mutations can result in the production of neoantigens which are specifically expressed in tumor rather than normal cells. 11 Several studies have focused on the effect of tumor mutation burden (TMB) on the clinical benefits for patients with resected early‐stage NSCLC. Several studies have shown that a high TMB is associated with a favorable outcome in patients with resected NSCLC 12 , 13 but other researchers have reported that a high TMB is a poor prognostic factor in heterogeneous stages, histology and ethnic groups14, 15 or that TMB is not associated with the overall survival of patients with early‐stage NSCLC, 16 , 17 implying that TMB is not sufficient to predict NSCLC prognosis. However, using computational tools to predict tumor neoantigens based on WES data have been proven to be a meaningful and potential method. 11 McGranahan et al. showed that neoantigen burden was associated with overall survival in ADC but not SCC cohorts of patients from The Cancer Genome Atlas (TCGA). 18 However, the function of immunogenicity‐mediated immune surveillance has so far not been comprehensively studied in patients with resected NSCLC, especially in Chinese NSCLC patients, while the assessment of clinical and survival impact of these distinct mutation context and immunogenic signatures on different pathological types could be clinically useful.
In this study, we assessed gene mutation characteristics using whole exome sequencing (WES) to identify individual oncogenic pathways, DNA repair pathways, and antigen presentation machinery genes, human leukocyte antigen (HLA) ‐I numbers and loss of heterozygosity (LOH), TMB numbers and neoantigen load in stage II/III NSCLC samples and these were then evaluated as prognostic and predictive biomarkers for NSCLC patients. We expected to be able to provide a novel insight into biomarker discovery and identification for the prognosis and prediction of treatment response in Chinese NSCLC patients.
METHODS
Patients and specimens
In this retrospective study, a total of 91 patients with completely resected stage II/III NSCLCs (47 SCCs and 44 ADCs according to the AJCC eighth TNM staging manual) from Zhejiang Cancer Hospital (Hangzhou, China) between August 26, 2006 and September 28, 2014 were included. None of the patients received neoadjuvant chemotherapy. Tissue samples from tumor and matched normal lung were snap‐frozen. All patients were followed up regularly in Zhejiang Cancer Hospital until recurrence or last follow‐up (December 2020). The specimens utilized in this study were comprised of at least 30% tumor cells and each tissue sample was assessed by histological examination. Tumor recurrence was evaluated by two different radiologists for each patient. The detailed clinicopathological features and treatment selections, as well as follow‐up data, were retrieved from patients' medical records and analyzed. This study was approved by our Institutional Review Board of Zhejiang Cancer Hospital and each participant provided a written informed consent form before enrollment into the study.
DNA extraction and whole exome sequencing (WES)
Genomic DNA was extracted from the tumor or normal lung samples using a magnetic genomic DNA kit (Tiangen) according to the manufacturer's protocol and from FFPE of tumor and normal samples using an internally modified magnetic extraction protocol. DNA was randomly broken into 150–200 bp using the ultrasonicator (Covaris). The sheared DNA samples were subjected to the end repair step using the Agilent SureSelectXT Low Input Reagent Kit (Agilent), followed by addition of a base A to the 3′ end to form a sticky end. After that, DNA fragments were ligated with specific barcode adapter sequences and any incompletely ligated fragments using the magnetic beads were removed. The samples were then subjected to a PCR amplification using universal primers complementary with adapter sequences to build a DNA sequencing library. The probes from the Agilent SureSelectXT All Human Exome library were utilized to capture target fragments from the samples and the library concentrations were assessed by using Qubit (Thermo Fisher), and the library size was measured using the Agilent TapeStation. The Illumina HiSeq X Ten system (Illumina, Mingma Technologies Co., Ltd) was used to collect data from 150‐bp pair‐end sequencing.
Assessment of somatic gene mutation and significantly mutated genes (SMG)
After WES of the paired samples, somatic mutation variations using previously reported common algorithms were determined. In particular, the raw reads of WES were filtered using the skewer (v0.2.2) software according to a previous study, 19 while the high‐quality reads were matched to the UCSC human reference genome (hg19) with the Burrows‐Wheeler Aligner (BWA v.0.7.12) accordingly with default parameters. 20 However, all N‐read bases in the reference genome were excluded for calculation of the coverage ratio. Tumor samples were sequenced with an average coverage of 387.6 ×, while normal samples were sequenced with 117.2 ×. We then further performed the deduplication, indel realignment, and base quality recalibration using the Sentieon algorithm (https://www.sentieon.com/). For each sample, we assessed the somatic single nucleotide variations (SNVs) and InDels with the Sentieon TNseq according to a previous study. 21 We then filtered out genomic mutations in a low complexity region, such as the tandem repeat regions or highly homologous regions in the genome, and obtained the repeat regions from the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/) and annotated all high‐confident mutations with ANNOVAR (Version 2016‐02‐01) (http://annovar.openbioinformatics.org/). 22 After that, we assessed the SMGs using MutSigCV (v1.4) software (https://www.genepattern.org/modules/docs/MutSigCV) 23 with the default setting. We defined a gene with the false discovery rate (FDR) <0.05 to be significantly mutated.
Calculation of TMB number
To calculate the TMB number, we first filtered the somatic SNVs and InDels from the WES of our paired samples and matched the variants to one of the following criteria, that is (i) Variant allele frequency < 5%, (ii) inclusion of the 1000 G, ExAC03 and dbSNP151 database with exceptions for the variants with a high count (≥4 observances) in the COSMIC database, (iii) synonymous variants, or (iv) exclusion of the coding sequence of any genes (CDS). The TMB number was calculated using the formula, specifically, TMB = M/L (M represents number of filtered variants, while L represents length of the covered CDS region).
Prediction of HLA and neoantigens
We predicted the neoantigens using a method modified from pVACseq according to a previous study, 21 that is, the HLA type of each paired samples was assessed using the in‐house fq2HLA software analysis of the high quality reads from the normal samples. The unique HLA allele number was then counted as the HLA number, while the LOH of HLA was calculated according to a previous study. 24 Somatic SNVs and InDels were used for the HLA binding affinity prediction using the netMHCpan (v4.0) software (https://services.healthtech.dtu.dk/service.php?NetMHCpan-4.0). Peptides with an affinity <500 nM were considered to be a neoantigen and the number of the NAL of each paired sample was calculated using the formula, that is, NAL = N/L (N represents number of neoantigen, while L represents length of the covered CDS regions).
Statistical analysis
We performed Pearson's χ2 test to associate clinicopathological variables from patients with altered genetic features and then defined the DFS as time from surgery to tumor recurrence, death from any cause, or the last follow‐up of surviving patients. The Kaplan–Meier method and the log‐rank test were performed to associate survival of patients with genomic alterations, such as the TMB number. The Cox regression model was used to predict the role of gene mutations in treatment responses. We then performed univariate and multivariate analyses to assess gene alterations as the prognostic predictors of these NSCLC patients. A p ≤ 0.05 was considered statistically significant.
RESULTS
Clinicopathological characteristics of patients
A total of 91 patients were included in our study and the snap‐frozen tumor tissue samples and matched normal lung tissue were subjected to WES. This study comprised 47 patients with SCCs and 44 with ADCs with a median age of 59 years (ranging between 30 and 71 years old) with 70 male patients (76.7%) and 46 stage II (50.5%) and 45 stage III (49.5%) patients. A total of 43 SCC patients were former or current smokers (93.6%) versus 20 ADC patients (45.5%), while 61.4% (27/44) of patients had an EGFR mutation in ADC, but only 4.26% (2/47) had an EGFR mutation in SCC. In terms of treatment, 73 patients (73/91, 80.2%) received adjuvant chemotherapy after surgery for at least two cycles. A total of 12 (12/47, 25.5%) and six patients (6/44, 13.6%) did not receive adjuvant chemotherapy in the SCC and ADC subgroups separately. The median DFS of SCC and ADC were 69 and 17.5 months, respectively. The clinicopathological data are listed in Table 1.
TABLE 1.
Clinicopathological and molecular characteristics of non‐small cell lung cancer (NSCLC) patients (n = 91)
| Variables | SCC (n = 47) | ADC (n = 44) |
|---|---|---|
| n (%) | n (%) | |
| Age (years) | ||
| >65 | 5 (10.6) | 7 (16.0) |
| ≤65 | 42 (89.4) | 37 (84.1) |
| Gender | ||
| Male | 45 (95.7) | 25 (56.8) |
| Female | 2 (4.30) | 19 (43.2) |
| Tobacco smoking status | ||
| Never smoker | 3 (6.40) | 24 (54.5) |
| Former/current smoker | 44 (93.6) | 20 (45.5) |
| T stage a | ||
| T1–T2 | 27 (57.4) | 32 (72.7) |
| T3–T4 | 20 (42.6) | 12 (27.3) |
| N stage a | ||
| N0–N1 | 40 (85.1) | 21 (47.7) |
| N2 | 7 (14.9) | 23 (52.3) |
| Stage a | ||
| II | 31 (66.0) | 15(34.1) |
| III | 16 (34.0) | 29 (65.9) |
| EGFR status | ||
| Wild‐type | 45 (95.7) | 17 (38.6) |
| Mutation | 2 (4.26) | 27 (61.4) |
| Treatment | ||
| None | 12 (25.5) | 6 (13.6) |
| Adjuvant chemotherapy | 35 (74.5) | 38 (86.4) |
| Radiotherapy | 4 (8.5) | 14 (31.8) |
| HLA number | ||
| 6 | 32 (68.1) | 34 (77.3) |
| ≤5 | 15 (31.9) | 10 (22.7) |
| HLA LOH | ||
| Yes | 24 (51.1) | 14 (31.8) |
| No | 23 (48.9) | 30 (68.2) |
| TMB | ||
| Median TMB (range) | 4.8 (1.0–29.0) | 2.3 (0.5–32.3) |
| NAL | ||
| Median NAL (range) | 2.4 (0.6–7.4) | 1.0 (0.3–21.8) |
Abbreviations: ADC, adenocarcinoma; LOH, loss of heterozygosity; NAL, tumor neoantigen burden; NSCLC, non‐small cell lung cancer; SCC, squamous cell carcinoma; TMB, tumor mutation burden.
Using the eighth TNM staging classification.
Association of genomic signatures and frequently mutated pathways with ADC and SCC outcomes
Prevalence of the C > T transitions was observed in ADC and the C > A transitions in SCC (Figure 1(a)). An enrichment of the C > A transversions was associated with smoking status that was observed in other cancers for which smoking was a significant risk factor. In SCC, the most frequent mutations were TP53 (41/47, 87.2%), TTN (38/47, 80.8%) and CSMD3 (24/47, 51.1%; Figure 1(b)). EGFR (27/44, 61.4%), TP53 (25/44, 56.8%) and BLLAF1 (16/44, 36.4%) were more frequent in the ADC subgroup (Figure 1(c)). Most patients (44/47, 93.6) were former or current smokers in the SCC subgroup and almost half of patients had smoking histories in the ADC subgroup, thus we detected the genomic characters within smokers and non‐smokers in the ADC subgroup. Frequency of the C > T transition was detected in non‐smokers compared to the enrichment of the C > A transversion in smokers (Figure S1(a)). We also found that TP53 (12/20, 60.0%), BCLAF1 (8/20, 40.0%) and EGFR (8/20, 40.0%) were frequent in tobacco smokers, whereas EGFR (19/24, 79.2%), TP53 (13/24, 54.2%) and BCLAF1 (8/24, 33.3%) were common in patients without smoking history (Figure S1(b)). There were 61.4% of patients with an EGFR mutation (vs. 27% EGFR mutations in Caucasian NSCLC patients) 5 and 4% of ADC patients with a KRAS mutation (vs. 32% of Caucasian patients), 5 suggesting an extraordinary distinct genotypes in different races of patients. 25 We then analyzed the association of individual gene mutations with DFS of patients and found that mutation of USH2A was associated with better DFS in SCC patients (Figure 2(a)). Moreover, mutation of RELN, HMCN1, OR2L8, and NALCN was associated with better DFS in ADC patients, whereas mutated MUC5B was associated with poor DFS in ADC patients (Figure 2(b)). However,there was no association of any mutated genes with DFS after correction of the false discovery rate with the multitests.
FIGURE 1.

Mutation spectra and significantly mutated genes in adenocarcinoma (ADC) and squamous cell carcinoma (SCC). (a) Gene mutation spectra in ADC and SCC. *p ≤ 0.05 and ****p ≤ 0.0001 using Student's t‐test. (b) The top 30 significantly mutated genes in SCC. The samples were aligned according to their somatic nonsynonymous mutation burden (in the top panel) and genes were ranked by mutation frequencies. (c) The top 30 significantly mutated genes in ADC. The samples were made in order based on their somatic nonsynonymous mutation burden (in the top panel) and genes were ranked by mutation frequencies
FIGURE 2.
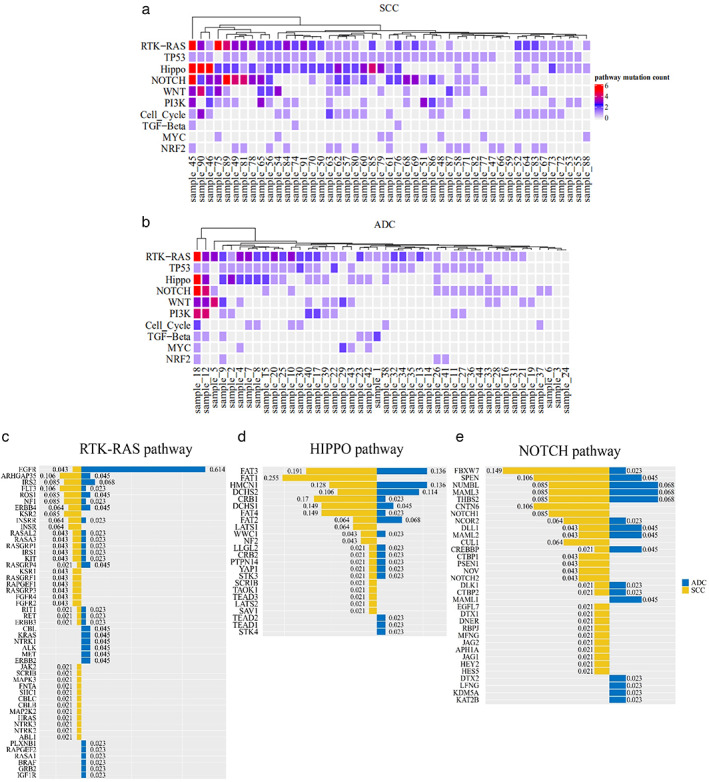
Frequent gene mutations enriched by the oncogenic pathways in squamous cell carcinoma (SCC) and adenocarcinoma (ADC). (a) The mutation status of the top 10 oncogenic pathways in SCC. (b) The mutation status of the top 10 oncogenic pathways in ADC. (c) Gene mutation distribution and types in the RTK‐RAS, HIPPO, and NOTCH signal pathways
Furthermore, we assessed the 10 oncogenic signaling pathways that were genetically altered at high frequency in ADC and SCC. 26 , 27 We found that there were more mutated genes involved in these oncogenic signaling pathways in SCC compared to those of ADC. Among them, the RTK‐RAS (82% in SCC and 84% in ADC) and TP53 pathways (93% in SCC and 59% in ADC) were frequently altered. Moreover, ARHGAP35 (5/47, 10.6%), FLT3 (5/47, 10.6%) and IRS2 (4/47, 8.5%) in the RTK‐RAS signaling were mutated in SCC, whereas alterations of EGFR (27/44, 61.4%), IRS2 (3/44, 6.8%) and ARHGAP35 (2/44, 4.5%) occurred more frequently in ADC. The HIPPO and NOTCH signaling were the third and fourth signaling pathways that were enriched with genetic variants in NSCLC in our study. In the HIPPO pathways, the most frequently mutated genes were FAT1 (12/47, 25.5%), FAT3 (9/47, 19.1%) and CRB1 (8/47, 17.0%) in SCC, whereas mutation of FAT3 (6/44, 13.6%), HMCN1 (6/44, 13.6%) and DCHS2 (5/44, 11.4%) occurred in ADC (Figure 2(c)). In the NOTCH pathways, predominate alterations of FBXW7 (7/47, 14.9%), SPEN (5/47, 10.6%), and CNTN6 (5/47, 10.6%) occurred in SCC, whereas NUMBL (3/44, 6.8%), MAML3 (3/44, 6.8%), and THBS2 (3/44, 6.8%) were mutated in ADC (Figure 2(e)). After that, we analyzed the associations of each pathway gene mutations with survival of patients, but unfortunately, we did not find any association of the altered single oncogenic pathway with DFS in both SCC and ADC (Figure 3(a)–(d)).
FIGURE 3.
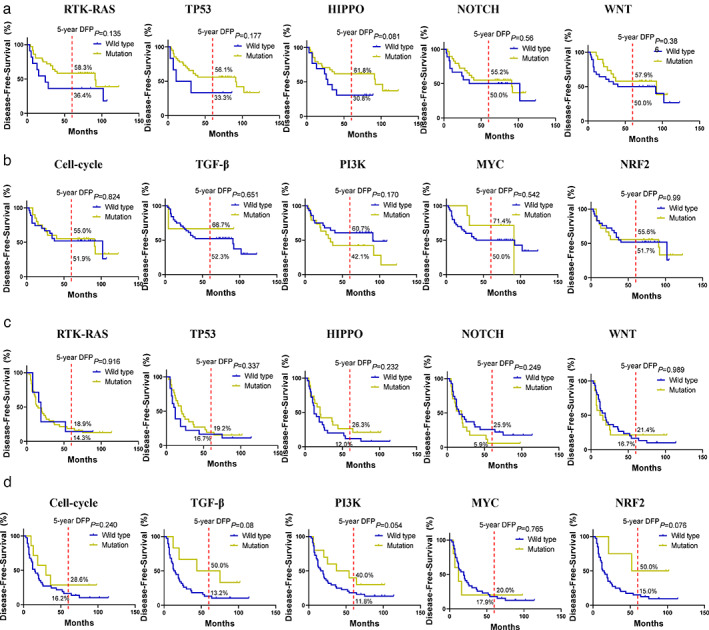
Kaplan–Meier curves of disease‐free survival (DFS) stratified by the 10 oncogenic pathways in squamous cell carcinoma (SCC) and adenocarcinoma (ADC). (a) SCC. (b) ADC
Association of a comprehensive immunogenomic profiling with DFS of ADC and SCC patients
We then performed a comprehensive analysis for the immunogenic profiling using WES data for both ADC and SCC patients, including HLA‐I number, HLA LOH, TMB, DNA repair pathway, and antigen presentation machinery and the calculation to predict the neoantigen to determine the HLA‐binding affinity (<500 nM of peptides derived from somatic SNVs and indels). We found a similar HLA‐I distribution of allele frequency.net of China Jiangsu Han (HLA‐A/B, n = 3238) and China South Han pop 2 (HLA‐C, n = 1098; Figure 4(a)). Most patients had six HLA‐I loci in both SCC (32/47, 68.1%) and ADC (34/44, 77.3%), while almost a half of SCC patients had HLA LOH, but only a third of ADC patients had HLA LOH (Figure 4(b)). Moreover, there was no significant difference in HLA numbers and HLA LOH from tobacco smoking, respectively (Figure S3(a), (b)). Our survival analysis revealed that neither HLA number nor HLA LOH status was associated with DFS of SCC or ADC patients (Figure S3(c), (d) and Table 2).
FIGURE 4.
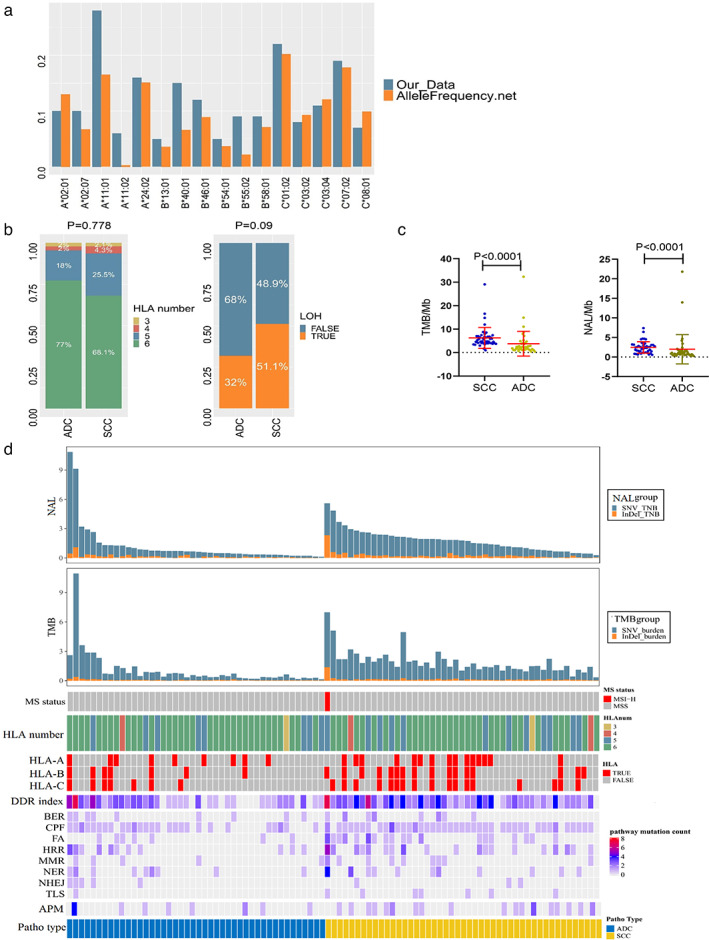
Immunogenomic profiling of altered genes in squamous cell carcinoma (SCC) and adenocarcinoma (ADC). (a) Comparison of human leukocyte antigen (HLA‐I) type in our cohort of patients versus the online database. Our data are consistent with Allelefrequency.net of China Jiangsu Han (HLA‐A/B, n = 3238) and China South Han pop 2 (HLA‐C, n = 1098). (b) Comparison of HLA‐I number and loss of heterozygosity (LOH) between our ADC and SCC samples. (c) Tumor mutation burden (TMB) and neoantigen load (NAL). Our data showed that TMB and NAL were higher in SCC than in ADC. (d) Illustration of immunogenomic features between ADC and SCC. The HLA number, LOH, MS status, DNA damage repair pathway and antigen presentation machinery pathway were profiled stratified by non‐small cell lung cancer (NSCLC) histological types
TABLE 2.
Univariate analysis of DFS predicators
| Variables | SCC (n = 47) | ADC (n = 44) | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |
| Age (years) | ||||||
| >65 vs. ≤65 | 1.56 | 0.53–4.57 | 0.42 | 1.58 | 0.69–3.65 | 0.28 |
| Gender | ||||||
| Male vs. female | 0.07 | 0.01–0.37 | 0.002 | 0.77 | 0.40–1.46 | 0.42 |
| Smoking status | ||||||
| Former/current smoker vs. never smoker | 0.38 | 0.09–1.65 | 0.2 | 0.62 | 0.32–1.20 | 0.16 |
| T stage a | ||||||
| T1–T2 vs. T3–T4 | 0.67 | 0.31–1.48 | 0.33 | 2.29 | 1.02–5.11 | 0.044 |
| N stage a | ||||||
| N2 vs. N0‐N1 | 2.12 | 0.78–5.81 | 0.14 | 1.98 | 1.00–3.91 | 0.049 |
| Stage a | ||||||
| II vs. III | 0.50 | 0.22–1.1 | 0.085 | 0.91 | 0.45–1.79 | 0.79 |
| HLA number | ||||||
| 6 vs. ≤5 | 0.95 | 0.42–2.14 | 0.98 | 1.80 | 0.82–3.95 | 0.14 |
| HLA LOH | ||||||
| Yes vs. No | 0.52 | 0.23–1.17 | 0.11 | 1.14 | 0.57–2.26 | 0.71 |
| Treatment | ||||||
| Adjuvant chemotherapy vs. none | 0.44 | 0.19–1.00 | 0.051 | 1.16 | 0.45–2.98 | 0.76 |
| Radiotherapy vs. none | 0.27 | 0.04–1.96 | 0.19 | 0.96 | 0.49–1.90 | 0.91 |
| TMB | ||||||
| ≤4 vs. >4 | 1.28 | 0.52–3.20 | 0.6 | 2.12 | 0.96–4.84 | 0.075 |
| NAL | ||||||
| ≤2 vs. >2 | 2.56 | 1.15–5.68 | 0.021 | 1.67 | 0.65–4.29 | 0.29 |
| DDR index | ||||||
| Low vs. high | 2.79 | 1.15–6.78 | 0.024 | 0.72 | 0.38–1.4 | 0.31 |
| APM status | ||||||
| Mutation vs. wild‐type | 0.85 | 0.38–1.90 | 0.69 | 1.67 | 0.69–4.00 | 0.26 |
Abbreviations: ADC, adenocarcinoma; LOH, loss of heterozygosity; NAL, neoantigen load; NSCLC, non‐small cell lung cancer; SCC, squamous cell carcinoma; TMB, tumor mutation burden.
Using the eighth TNM staging classification.
The DNA damage repair (DDR) signaling includes eight pathways, namely, the check point factors (CPF), homologous recombination repair (HRR), Fanconi anemia (FA), nucleotide excision repair (NER), mismatch repair (MMR), base excision repair (BER), nonhomologous end‐joining (NHEJ) and DNA translation synthesis (TLS). 28 Genomic variants in CPF (93% in SCC and 61% in ADC) and the HRR pathway (43% in SCC and 25% in ADC) were more obvious in both SCC and ADC. Indeed, SCC patients showed a higher prevalence of gene alterations in the DNA repair pathway and the antigen presentation machines compared to those of ADC patients (Figure 4(d)). In this study, we explored the mutations of the antigen presentation machinery (APM)‐related genes for association with prognosis of Chinese lung cancer patients, but there was no positive or useful finding (Figure S4(c), (d)). In line with this, the mutated status of each of DDR signaling was not associated with the clinical outcomes (Figure S4(a), (b)). When tumors were stratified into dichotomy (low DDR index with gene mutations in <3 pathways versus high DDR index with gene mutations in ≥3 pathways), the low DDR index was associated with poor DFS of SCC patients (Figure 5(a) and Table 2), although the DDR index did not have a predictable value for chemotherapy of SCC patients (Figure 5(a)). However, interestingly, the DDR index was significantly associated with neoantigen load, although there was no correlation observed between the DDR index and TMB in SCC patients, suggesting a key role of the DDR index in neoantigen production in SCC (Figure S5(a)). However, the HLA number and LOH were no differences between the low and high DDR index groups (Figure S6(b)). Furthermore, we observed a significant difference in TMB and NAL between low and high DDR index groups in ADC patients (Figure S5(c)), although there was no association of the DDR index in prediction of DFS in ADC (Figure S5(d)). There was also no significant difference in HLA number and LOH between low and high DDR indexes in ADC (Figure S5(e)).
FIGURE 5.

Neoantigen load (NAL) as a prognostic and predictive indicator for lung squamous cell carcinoma (SCC). (a) Kaplan–Meier curves, and log rank test stratified by the DNA damage repair (DDR) index. Prognostic and predictive effect of the low versus high DDR index on disease‐free survival (DFS) of SCC patients. (b) Kaplan–Meier curves, and log rank test stratified by the NAL. Prognostic and predictive effect of NAL on DFS of SCC patients. (c) Comparison of NAL with the DDR index in SCC patients. (d) Comparison of the different gene mutation types and characters between NAL and tumor mutation burden (TMB) in adenocarcinoma (ADC) and SCC
Specifically, SCC had a higher level of TMB and NAL than those of ADC (Figure 4(c)). A high TMB number per Mb (i.e., 4 mutations per Mb) was associated with better DFS of these 91 NSCLC patients (Figure S6(a)). However, there was no association observed between TMB number and DFS of ADC (Figure S6(b)) or SCC patients (Figure S6(c)). We then used NAL number (>2 neoantigens per Mb as a cutoff point) and further analyzed these data and still not find any statistical significance in ADC (Figure S6(d)). Interestingly, there was an association of low NAL number with poor DFS (months, hazard ratio [HR] = 2.56, 95% CI: 1.15–5.68, p = 0.021) in SCC patients (Figure 5(b) and Table 2). Our multivariate analysis also showed that NAL number was an independent prognostic predicator for SCC patients, while the DDR index was not included in the multivariate analysis considering the strong association between the DDR index and NAL (see Table S1). Furthermore, we also found a benefit of adjuvant chemotherapy in improvement of DFS in SCCs with a lower NAL number (Figure 5(b)). For patients with SCC, adjuvant chemotherapy, as compared with observation, significantly prolonged DFS among patients with NAL ≤2 (HR = 0.34, 95% CI: 0.11–1.00, p = 0.049) but not among patients with NAL >2 (HR = 0.623, 95% CI: 0.163–2.377, p = 0.483). The interaction terms between NAL and adjuvant treatment were significant for DFS (HR = 0.41, 95% CI: 0.18–0.95, p = 0.038). These results allowed us to explore the detailed association of DDR pathway and NAL numbers in SCC. As shown in Figure 5(c), patients with high NAL numbers were enriched in high DDR index group and CPF, FA, and HRR pathways were the most frequently mutated among the high DDR index and NAL groups in SCC, indicating that the DNA damage repair pathway contributed to the neoantigen productions and neoantigen‐directed immune surveillance favored SCC patients. However, the immune escape may be adapted by other immune escape mechanisms in ADC patients (Figure 5(c)). In addition, we analyzed the different pattern of TMB and NAL number in NSCLC and found that one half of oncogenic mutations did not create neoantigen, and indels variants created 1.75‐fold neoantigens compared to SNV in our study among ADC and SCC, suggesting TMB number was not a good surrogate marker of the immunogenic neoantigen (Figure 5(d)).
In summary, predicted neoantigen load acted as a more useful indicator of immunogenicity than TMB, and provided effective stratification variable in prognosticating disease outcome and benefits from adjuvant chemotherapy for patients with lung squamous cell carcinoma.
DISCUSSION
Lung cancer is still a significant global health problem and adjuvant chemotherapy, a standard therapeutic strategy for NSCLC, has only been reported to improve the five‐year survival by 5%. 4 Recent utilization of treatment with immune checkpoint inhibitors has been reported to promote the overall survival of patients with advanced NSCLC. 8 Identification of comprehensively genomic signatures and gene alterations could be useful biomarkers to predict NSCLC survival and treatment responses. The results from this study found a higher NAL number was able to better predict favorable DFS and successful adjuvant chemotherapy in SCC patients (but not for ADC patients). In addition, the high DNA damage repair (DDR) index also predicted a high NAL number and favorable DFS of SCC patients (also not for ADC patients). However, we failed to find any predictive values of mutations of individual, oncogene pathway, and antigen presentation machinery genes, and HLA‐I number and LOH for DFS of SCC or ADC patients or treatment responses. In conclusion, our current data revealed analysis of NAL number as a biomarker for lung SCC prognosis and prediction of chemotherapy responses.
Several studies have previously focused on TMB for the prediction of clinical outcomes of patients with resected early stage NSCLC; for example, Siddhartha et al. showed that a high TMB (>8 mutations per Mb) was associated with a favorable outcome (overall survival [OS], disease‐free survival [DFS], and lung cancer‐specific survival [LCSS]) of patients with resected NSCLC after adjuvant chemotherapy. 12 However, controversially, Owada‐Ozaki et al. reported that a high TMB number (>62 mutations per Mb) was associated with poor NSCLC prognosis in Japanese patients. 14 Chun et al. showed that lung adenocarcinoma patients with low TMB numbers had better DFS than those with high TMB in Korean patients. 15 Our current data also confirmed that use of TMB is not sufficient to predict NSCLC prognosis.
Indeed, other recent studies using WES and computational analysis were able to utilize NAL as a biomarker to predict and assess cancer immunity and immunotherapy in human cancers. 11 , 29 Specifically, tumor neoantigens have been defined as peptides derived from somatic gene mutations and expressed in tumor cells but not in normal cells that can be recognized by tumor‐infiltrating lymphocytes (TILs) 29 ; thus, NAL is an ideal surrogate of immunogenicity. Moreover, various mutation types have been found to contribute to different neoantigen productions; for example, the probability of the indels that alter a given gene open‐reading frame could generate a neoantigen has been found to be three‐fold higher than that of nonsynonymous SNV 30 ; thus, use of NAL number to predict DFS could be much better than TMB numbers. Our study further showed that patients with a low NAL SCC benefited from adjuvant chemotherapy. Indeed, neoantigen load acted as a useful indicator of immunogenicity compared with TMB and could provide a stratification variable for prognosis or outcome of patients with lung SCC cancer after adjuvant chemotherapy.
However, our current study does have some limitations. This study was retrospective with a relatively small sample size. Considering the possibility for clinical decision‐making, future validation is needed in an independent larger cohort of patients. Moreover, NAL used in our current study was only based on the HLA‐I binding prediction; however, other processes involved in neoantigen production, such as the processing and presentation of antigens, stability of the MHC–peptide complex and immune recognition have not been calculated in our current prediction algorithms. Last, but not least, our current study did not evaluate OS.
In conclusion, our current study demonstrated the use of NAL number to predict DFS and treatment responses of SCC patients, which could be a more useful indicative marker for immunogenicity than TMB to provide an effective stratification as a prognostic marker for SCC outcome and treatment response. Further studies with a larger cohort from multiple institutions are needed to confirm our current data.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Figure S1. Gene mutation spectra and significantly mutated genes in ADC stratified by tobacco smoking status and EGFR mutations. (a) Gene mutation spectra in ADC with or without tobacco smoking history. *p ≤ 0.05 and ***p ≤ 0.001 using Student's t‐test. (b) The top 30 significantly mutated genes in ADC stratified by tobacco smoking history. The samples were made in order according to their somatic non‐synonymous mutation burden (in the top panel) and genes were ranked by mutation frequencies.
Figure S2. Association of mutated individual genes with DFS of SCC and ADC patients. (a), SCC. (b) ADC.
Figure S3. Illustration of HLA‐I number and LOH and EGFR mutations in NSCLC. (a) the distribution of HLA‐I number in ADC stratified by tobacco smoking status and EGFR mutations. (b) the distribution of HLA‐I LOH in ADC stratified by tobacco smoking status and EGFR mutations. (c) Kaplan–Meier curves. The data showed a prognostic significance of HLA number and LOH in prediction of DFS in SCC. (d) Kaplan–Meier curves. The data showed a prognostic significance of HLA number and LOH in prediction of DFS in ADC.
Figure S4. Kaplan–Meier plots. The data showed the prognostic significance of the DDR and APM pathways in prediction of DFS in SCC (a and c) and ADC (b and d).
Figure S5. Association of the DDR index with other immunogenic variants in NSCLC. (a) Association of the DDR index with TMB and NAL in SCC patients. (b) Association of the DDR index with HLA number and LOH in SCC patients. (c) Association of the DDR index with TMB and NAL in ADC patients. (d), Kaplan–Meier plots. The data showed a prognostic significance of the DDR index in prediction of DFS in ADC. E, Association of the DDR index and HLA number and LOH in ADC patients.
Figure S6. Association of different clinicopathological data with DFS of NSCLC patients. (a) Comparison of TMB with NAL between ADC patients with or without tobacco smoking history. (b) Comparison of TMB with NAL between ADC patients with or without EGFR mutations. (c) Kaplan–Meier plots. The data showed a prognostic significance of TMB in prediction of DFS in all NSCLC patients. (d) Kaplan–Meier plots. The data showed a prognostic significance of TMB in prediction of DFS in ADC. (e) Kaplan–Meier plots. The data showed a prognostic significance of TMB in prediction of DFS in SCC. F, Kaplan–Meier plots. The data showed a prognostic significance of NAL in prediction of DFS in ADC.
Table S1. Multivariate analysis of prognostic predictors of DFS
Table S2. Association of clinicopathological and molecular characteristics with tumor neoantigen burden (NAL) level
Gong L, He R, Xu Y, et al. Neoantigen load as a prognostic and predictive marker for stage II/III non‐small cell lung cancer in Chinese patients. Thorac Cancer. 2021;12:2170–2181. 10.1111/1759-7714.14046
Lei Gong and Ronghui He contributed equally to this work.
Funding information the Natural Science Foundation of Zhejiang Province, Grant/Award Number: LQ21H160004; Zhejiang Medical technology program, Grant/Award Number: 202143913
Contributor Information
Zhiguo Zheng, Email: zhengzg@zjcc.org.cn.
Hui Li, Email: 11118029@zju.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 3. Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer. 1995;75:191–202. [DOI] [PubMed] [Google Scholar]
- 4. Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–9. [DOI] [PubMed] [Google Scholar]
- 5. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446–54. [DOI] [PubMed] [Google Scholar]
- 6. Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in resected EGFR‐mutated non‐small‐cell lung cancer. N Engl J Med. 2020;383:1711–23. [DOI] [PubMed] [Google Scholar]
- 7. Zhong WZ, Wang Q, Mao WM, Xu ST, Wu L, Shen Y, et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II‐IIIA (N1‐N2) EGFR‐mutant NSCLC (ADJUVANT/CTONG1104): a randomised, open‐label, phase 3 study. Lancet Oncol. 2018;19:139–48. [DOI] [PubMed] [Google Scholar]
- 8. Wang L, Ma Q, Yao R, Liu J. Current status and development of anti‐PD‐1/PD‐L1 immunotherapy for lung cancer. Int Immunopharmacol. 2020;79:106088. [DOI] [PubMed] [Google Scholar]
- 9. Mascaux C, Angelova M, Vasaturo A, Beane J, Hijazi K, Anthoine G, et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature. 2019;571:570–5. [DOI] [PubMed] [Google Scholar]
- 10. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. [DOI] [PubMed] [Google Scholar]
- 11. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. [DOI] [PubMed] [Google Scholar]
- 12. Devarakonda S, Rotolo F, Tsao MS, Lanc I, Brambilla E, Masood A, et al. Tumor mutation burden as a biomarker in resected non‐small‐cell lung cancer. J Clin Oncol. 2018;36:2995–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Talvitie EM, Vilhonen H, Kurki S, Karlsson A, Orte K, Almangush A, et al. High tumor mutation burden predicts favorable outcome among patients with aggressive histological subtypes of lung adenocarcinoma: a population‐based single‐institution study. Neoplasia. 2020;22:333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Owada‐Ozaki Y, Muto S, Takagi H, Inoue T, Watanabe Y, Fukuhara M, et al. Prognostic impact of tumor mutation burden in patients with completely resected non‐small cell lung cancer: brief report. J Thorac Oncol. 2018;13:1217–21. [DOI] [PubMed] [Google Scholar]
- 15. Chun YJ, Choi JW, Hong MH, Jung D, Son H, Cho EK, et al. Molecular characterization of lung adenocarcinoma from Korean patients using next generation sequencing. PLoS One. 2019;14:e0224379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang T, Shi J, Dong Z, Hou L, Zhao C, Li X, et al. Genomic landscape and its correlations with tumor mutational burden, PD‐L1 expression, and immune cells infiltration in Chinese lung squamous cell carcinoma. J Hematol Oncol. 2019;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu H, Chen Z, Ballman KV, Watson MA, Govindan R, Lanc I, et al. Correlation of PD‐L1 expression with tumor mutation burden and gene signatures for prognosis in early‐stage squamous cell lung carcinoma. J Thorac Oncol. 2019;14:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next‐generation sequencing paired‐end reads. BMC Bioinformatics. 2014;15:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H, Durbin R. Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC‐Seq: a genome‐guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high‐throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer‐associated genes. Nature. 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele‐specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sekine I, Yamamoto N, Nishio K, Saijo N. Emerging ethnic differences in lung cancer therapy. Br J Cancer. 2008;99:1757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52:177–86. [DOI] [PubMed] [Google Scholar]
- 27. Morgensztern D, Devarakonda S, Govindan R. Genomic landscape of squamous cell carcinoma of the lung. Am Soc Clin Oncol Educ Book. 2013;15:348–53. [DOI] [PubMed] [Google Scholar]
- 28. Scarbrough PM, Weber RP, Iversen ES, Brhane Y, Amos CI, Kraft P, et al. A cross‐cancer genetic association analysis of the DNA repair and DNA damage signaling pathways for lung, ovary, prostate, breast, and colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Efremova M, Finotello F, Rieder D, Trajanoski Z. Neoantigens generated by individual mutations and their role in cancer immunity and immunotherapy. Front Immunol. 2017;8:1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion‐and‐deletion‐derived tumour‐specific neoantigens and the immunogenic phenotype: a pan‐cancer analysis. Lancet Oncol. 2017;18:1009–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gene mutation spectra and significantly mutated genes in ADC stratified by tobacco smoking status and EGFR mutations. (a) Gene mutation spectra in ADC with or without tobacco smoking history. *p ≤ 0.05 and ***p ≤ 0.001 using Student's t‐test. (b) The top 30 significantly mutated genes in ADC stratified by tobacco smoking history. The samples were made in order according to their somatic non‐synonymous mutation burden (in the top panel) and genes were ranked by mutation frequencies.
Figure S2. Association of mutated individual genes with DFS of SCC and ADC patients. (a), SCC. (b) ADC.
Figure S3. Illustration of HLA‐I number and LOH and EGFR mutations in NSCLC. (a) the distribution of HLA‐I number in ADC stratified by tobacco smoking status and EGFR mutations. (b) the distribution of HLA‐I LOH in ADC stratified by tobacco smoking status and EGFR mutations. (c) Kaplan–Meier curves. The data showed a prognostic significance of HLA number and LOH in prediction of DFS in SCC. (d) Kaplan–Meier curves. The data showed a prognostic significance of HLA number and LOH in prediction of DFS in ADC.
Figure S4. Kaplan–Meier plots. The data showed the prognostic significance of the DDR and APM pathways in prediction of DFS in SCC (a and c) and ADC (b and d).
Figure S5. Association of the DDR index with other immunogenic variants in NSCLC. (a) Association of the DDR index with TMB and NAL in SCC patients. (b) Association of the DDR index with HLA number and LOH in SCC patients. (c) Association of the DDR index with TMB and NAL in ADC patients. (d), Kaplan–Meier plots. The data showed a prognostic significance of the DDR index in prediction of DFS in ADC. E, Association of the DDR index and HLA number and LOH in ADC patients.
Figure S6. Association of different clinicopathological data with DFS of NSCLC patients. (a) Comparison of TMB with NAL between ADC patients with or without tobacco smoking history. (b) Comparison of TMB with NAL between ADC patients with or without EGFR mutations. (c) Kaplan–Meier plots. The data showed a prognostic significance of TMB in prediction of DFS in all NSCLC patients. (d) Kaplan–Meier plots. The data showed a prognostic significance of TMB in prediction of DFS in ADC. (e) Kaplan–Meier plots. The data showed a prognostic significance of TMB in prediction of DFS in SCC. F, Kaplan–Meier plots. The data showed a prognostic significance of NAL in prediction of DFS in ADC.
Table S1. Multivariate analysis of prognostic predictors of DFS
Table S2. Association of clinicopathological and molecular characteristics with tumor neoantigen burden (NAL) level


