Abstract
Purpose:
Studies report molecular subtypes within muscle invasive bladder cancer (MIBC) predict clinical outcome. We evaluated whether subtyping by a simplified method and established classifications could predict clinical outcome.
Methods:
Institutional cohort-1 (n=52; MIBC: 39), Oncomine-dataset (MIBC: 151) and The Cancer Genome Atlas (TCGA)-dataset (MIBC: 402) were subtyped by simplified panels (MCG-1; MCG-Ext) that included only transcripts common among published studies, and analyzed for predicting metastasis, cancer-specific survival (CSS), overall-survival (OS), and recurrence–free survival (RFS). TCGA-dataset was further analyzed using Lund-Taxonomy, BC-Molecular Taxonomy Group Consensus (Consensus), and mRNA-subtype (TCGA-2017) classifications.
Results:
MIBC specimens from cohort-1 and Oncomine-dataset showed intra-tumor heterogeneity for transcript/protein expression. MCG-1 subtypes did not predict outcome in univariate or Kaplan-Meier analyses. In multivariate analyses, N-stage (P≤0.007), T-stage (P≤0.04), M-stage (P=0.007) and/or age (P=0.01) predicted metastasis, CSS/OS and/or cisplatin-based adjuvant-chemotherapy response. In the TCGA-dataset, publications report that subtypes risk-stratify patients for OS. Consistently, MCG-1 and MCG-Ext subtypes associated with OS, but not RFS, in univariate and Kaplan-Meier analyses. TCGA-dataset includes low-grade specimens (21/402) and subtypes associated with tumor-grade (P=0.005). However, MIBC is rarely low-grade (<1%). Among only high-grade specimens, MCG-1 and MCG-Ext subtypes could not predict OS. Subtypes by Consensus, TCGA-2017 and Lund-Taxonomy associated with tumor-grade (P<0.0001) and OS (P=0.01-<0.0001) univariately. Regardless of classification, subtypes had ~50%−60% sensitivity and specificity to predict OS/RFS. In multivariate analyses, N-stage and lymphovascular-invasion consistently predicted RFS (P=0.039) and OS (P=0.003).
Conclusion:
Molecular subtypes reflect bladder tumor heterogeneity and associate with tumor-grade. In multiple cohorts/subtyping-classifications, clinical parameters outperform subtypes for predicting outcome.
Keywords: Muscle-invasive bladder cancer, TCGA, Oncomine, molecular subtypes, tumor grade
Introduction
The high morbidity, mortality and healthcare cost associated with muscle-invasive bladder cancer (MIBC) present a need for individualizing patient care1. While the divergent but interrelated two-molecular-pathway model for the development of low-grade and high-grade bladder cancer (BC) is well-established, molecular profiling has revealed heterogeneity in MIBC regarding genetic landscape and clinical outcome2–4. Furthermore, molecular profiling has been proposed in individualizing treatment for MIBC patients5. Retrospective transcriptome profiling studies from different institutions and The Cancer Genome Atlas (TCGA) Research Network have identified distinct molecular subtypes within MIBC and a recent study described them in non-MIBC sepcimens6–9. While different studies report varying MIBC subtypes such as the TCGA clusters, Lund Taxonomy, mRNA expression-based molecular subtypes (TCGA-2017), and the consensus subtypes produced by the Bladder Cancer Molecular Taxonomy Group (Consensus), at the highest level, tumors can be divided into two major subtypes - “basal” (BL) and “luminal” (LU)6–8,10–14. The BL-subtype shares an expression profile with the basal cells of the urothelium and usually associates with a poor prognosis. The LU-subtype shares molecular profiles with differentiated urothelial cells and possibly predicts a better prognosis4,7,8,11,12. In the original Lund-Taxonomy classification from 2012, the best cancer-specific survival (CSS) was associated with “UroA” subtype (now “Urothelial-like”), which represented low-grade non-MIBC with a signature similar to the LU subtype15. Intra-tumor and intra-patient co-existence of basal and luminal tumor regions has been reported in MIBC patients, and many non-MIBC tumors also exhibit MIBC subtypes16,17. Tumor heterogeneity may also be enhanced following neoadjuvant and adjuvant treatment18. Such inter- and intra-tumoral heterogeneity could influence the clinical potential of grouping MIBC into subtypes17.
The primary focus of this study was to develop and validate a subtyping methodology that could be readily applied to any BC cohort for predicting clinical outcome. We initially subtyped our institutional MIBC cohort using a panel of transcripts that are common among published studies4,5,7,8,10,12,15,19–21. We validated the results in an Oncomine-dataset (151 MIBC specimens) and in the commonly used TCGA-dataset (402 MIBC specimens)5,6,10,12,19,20,22. Additionally, we further confirmed the results by analyzing the TCGA-dataset using the subtype identifiers for the specimens according to Lund-Taxonomy, TCGA-2017, and Consensus classifications. In all cohorts and classifications, the subtypes showed strong association with tumor grade, had suboptimal efficacy to predict clinical outcome, and clinical parameters consistently outperformed the subtypes in predicting prognosis.
Materials and methods
Data Acquisition.
All three cohorts are described in Supplementary Table 1. Cohort-1: Our institutional cohort included 52 BC specimens; 39 MIBC. Oncomine-dataset and TCGA-dataset: RNA-Seq data on 151 high-grade MIBC specimens was accessed through Oncomine™23–25 and data on 402 MIBC specimens from the TCGA was accessed through UCSC-Xena Browser26.
Molecular subtyping:
Transcripts included in the subtyping panels (MCG-1, MCG-Ext) are commonly used in subtyping studies4,5,7,8,10,12,15,19–21 (Supplementary Table 2). In cohort-1, transcripts’ expression was measured by reverse transcription quantitative PCR (RT-qPCR) and normalized to β-Actin27. Cohort-1 (transcript levels) and Oncomine-dataset (RNA-Seq) were subtyped by MCG-1. The TCGA-dataset (RNA-Seq) was subtyped by MCG-1, MCG-Ext. Supplementary methods detail the subtyping methodology for MCG-1 and MCG-Ext. The subtype identifiers for specimens in the TCGA-dataset by the Lund-Taxonomy and the Consensus13,14 were kindly provided by Dr. Gottfrid Sjödahl from the Division of Urological Research, Department of Translational Medicine at Lund University. The mRNA subtype (TCGA-2017) identifiers were obtained from a published study12.
Immunohistochemistry (IHC):
IHC was performed on representative patient tissues from cohort-1 to validate RT-qPCR findings (detailed methodology in supplementary materials).
Statistical analysis.
Statistical analyses were performed using SAS9.4, JMP14.0 and GraphPad Prism software (Supplementary Materials). Subtypes’ association with clinical outcome was analyzed by univariate (single parameter logistic regression) analysis. ROC curves were used to compute sensitivity and specificity. Mean, median, and 95% CI of sensitivity and specificity were calculated by Bootstrap modeling with 1000 iterations. Subtypes’ association with time to metastasis, RFS, CSS or OS was determined while adjusting for demographic and clinical covariates described in Supplementary Table 1 using Cox proportional hazard model with stepwise selection procedure. Kaplan-Meier plots with log-rank statistics determined if subtypes classified MIBC patients into risk categories for outcome.
Results
Molecular subtyping revealed intra-tumor heterogeneity in MIBC specimens
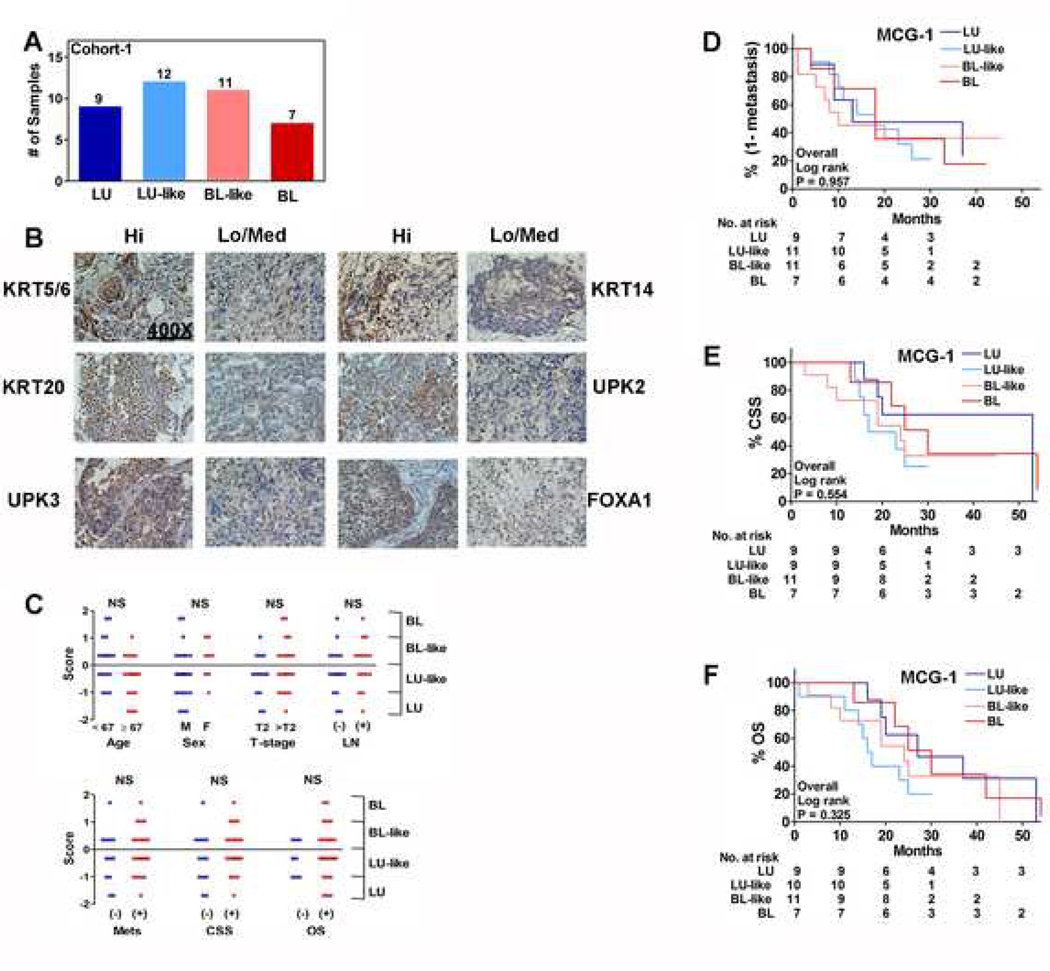
Molecular subtyping studies have used transcriptome data to group MIBC specimens into distinct subtypes6–8,12. To molecularly subtype MIBC specimens in our institutional cohort (cohort-1), we used a subtyping panel, MCG-1, that consisted of only those transcripts that are common in most subtyping studies (Supplementary Table 2)5,7,8,10,12,15,19–22. Since RT-qPCR is a standard technique used to validate transcriptome-based findings, we used RT-qPCR to measure the levels of transcripts in MCG-1 panel. Additionally, if molecular subtypes are to be reduced to clinical practice, RT-qPCR would be a technically easy and relatively inexpensive tool for subtyping patients’ tumors in real-time. In cohort-1 there were 39 high-grade MIBC specimens (Supplementary Table 1). Based on MCG-1, the majority of the specimens in cohort-1 had characteristics of both BL and LU subtypes. Therefore, the subtype scores grouped the specimens into four molecular subtypes: BL, LU, BL-like and LU-like (Figure 1A; Supplementary Methods). BL-like and LU-like subtypes had characteristics of both BL and LU phenotypes, and were the dominant subtypes, suggesting substantial intra-tumor heterogeneity. Based on RT-qPCR data, tissues expressing high- and low-levels of individual BL (KRT5/6, KRT14) and LU (KRT20, UPK2/3, FOXA1) markers were evaluated by IHC. IHC confirmed the expression of markers at the protein level, which also showed intra-tumor heterogeneity in markers’ expression within a single high-power field (Figure 1B).
Figure 1: Association of subtypes to clinical outcome in cohort-1.
A. The distribution of LU, LU-like, BL and BL-like subtypes among specimens in cohort-1. B. IHC of representative clinical specimens. C. Comparison of specimen subtype scores by individual clinical parameters. Each symbol represents an individual specimen/patient in the cohort, categorized with respect to a clinical parameter as shown in the figure. P-values generated by Mann-Whitney U test are two-tailed. D-F. Kaplan-Meier plots for metastasis (D), CSS (E) and OS (F); data stratified by MCG-1 subtypes.
We next evaluated the association of subtype scores generated by MCG-1 to clinical parameters. Unexpectedly, subtype scores did not associate with any clinical or prognostic parameters; although most females had BL-subtype tumors (Figure 1C; Supplementary Figure 1A). Published studies have also reported that bladder tumors from females tend to be of the BL-subtype8. In univariate and multivariate analyses, subtypes by MCG-1 did not significantly predict metastasis, CSS or OS (Tables 1 and 2). Subtypes had 80%−83% sensitivity but 31%−36% specificity for both metastasis and CSS (Table 3). Furthermore, the subtypes did not risk-stratify patients for metastasis, CSS or OS (Figure 1 D-F).
Table 1: Relationship of outcome to clinical parameters and molecular subtypes in cohort-1, the Oncomine-dataset, and TCGA.
Univariate analysis (single parameter logistic regression) was performed to evaluate the ability of clinical parameters and molecular subtypes to associate with metastasis, CSS, OS and RFS. Units odds ratio (OR) and 95% CI (CI) are shown for significant parameters.
| Cohort-1 | ||||||
|---|---|---|---|---|---|---|
| Metastasis | CSS Indicator | |||||
| Parameter | χ2 | P value | OR; 95% CI | χ2 | P value | OR; 95% CI |
| Age | 0.02 | 0.885 | 0.49 | 0.484 | ||
| Sex | 0.54 | 0.462 | 0.29 | 0.590 | ||
| T-Stage | 7.17 | 0.007 | 1.2; 1.1 – 1.4 | 6.76 | 0.009 | 1.2; 1.04 – 1.3 |
| N-stage | 5.15 | 0.023 | 7.5; 1.3 – 42.8 | 6.33 | 0.012 | 9.4; 1.6 – 53.6 |
| CIS | 0 | 0.995 | 0 | 0.994 | ||
| MCG-1 | 0.23 | 0.629 | 0.86 | 0.354 | ||
| Oncomine-dataset | ||||||
| CSS Indicator | OS Indicator | |||||
| Parameter | χ2 | P value | OR; 95% CI | χ2 | P value | OR; 95% CI |
| Age | 3.06 | 0.080 | 1.07 | 0.301 | ||
| Sex | 0.02 | 0.894 | 0.06 | 0.806 | ||
| T-stage | 11.43 | 0.0007 | 1.1; 1.04 – 1.2 | 9.2 | 0.002 | 1.1; 1.04 – 1.2 |
| N-stage | 8.84 | 0.003 | 3.4; 1.5 – 7.5 | 1.55 | 0.213 | |
| M-stage | 0 | 0.998 | 5.38 | 0.020 | 11.9; 1.5 – 96.1 | |
| MCG-1 | 0.06 | 0.808 | 0.98 | 0.323 | ||
| TCGA-dataset | ||||||
| RFS Indicator | OS Indicator | |||||
| Parameter | χ2 | P value | OR; 95% CI | χ2 | P value | OR; 95% CI |
| Age | 0.17 | 0.682 | 15.5 | < 0.0001 | 1.04; 1.02 – 1.06 | |
| Sex | 0 | 0.945 | 0.67 | 0.414 | ||
| Grade | 3.67 | 0.056 | 7.44 | 0.006 | 7.8; 1.8 – 34.1 | |
| T-stage | 4.23 | 0.04 | 1.04; 1.0 – 1.1 | 18.8 | < 0.0001 | 1.08; 1.04 – 1.1 |
| N-stage | 13.95 | 0.0002 | 2.8; 1.6 – 4.9 | 31.1 | < 0.0001 | 3.6; 2.3 – 5.7 |
| LVI | 3.16 | 0.076 | 11.8 | 0.0006 | 2.4; 1.5 – 3.9 | |
| M-stage | 0.98 | 0.323 | 4.44 | 0.035 | 4.3; 1.1 – 16.8 | |
| MCG-1 | 2.28 | 0.131 | 5.5 | 0.019 | 1.3; 1.04 – 1.5 | |
| MCG-Ext | 0.68 | 0.410 | 8.36 | 0.004 | 1.3; 1.1 – 1.6 | |
| Consensus | 4.78 | 0.029 | 1.2; 1.02 – 1.4 | 23.26 | < 0.0001 | 1.3; 1.2 – 1.5 |
| TCGA-2017 | 0.98 | 0.323 | 10.65 | 0.001 | 1.4; 1.1 – 1.6 | |
| Lund-Taxonomy | 1.1 | 0.294 | 8.05 | 0.005 | 1.3; 1.1 – 1.5 | |
Table 2: Relationship of outcome to clinical parameters and molecular subtypes in multivariate analysis in cohort-1 and the Oncomine-dataset.
Cox Proportional Hazards Model with stepwise selection procedure to determine the final model that accurately describes the outcome. In cohort-1, the model included age, sex, T-stage, N-stage, CIS and MCG-1 Subtypes. In the Oncomine-dataset, analyses included age, sex, T-stage, N-stage, M-stage and MCG-1 subtypes. Data are shown only for those parameters that reached significance.
| Cohort 1 Metastasis | |||||
|---|---|---|---|---|---|
| MCG-1 | MCG-1 (with LG) | ||||
| Parameter | P value | HR; 95% CI | P value | HR; 95% CI | |
| N-stage | 0.007 | 3.4; 1.4 – 8.3 | <0.0001 | 9; 3.3 – 24.4 | |
| MCG-1 | 0.006 | 2.9; 2.4 – 32.3 | |||
| Cohort 1 CSS Indicator | |||||
| MCG-1 | MCG-1 (with LG) | ||||
| Parameter | P value | HR; 95% CI | P value | HR; 95% CI | |
| T-stage | 0.027 | 3.9; 1.2 – 12.6 | 0.003 | 5.5; 1.8 – 16.7 | |
| N-stage | 0.0004 | 7.4; 2.5 – 22.2 | |||
| MCG-1 | 0.0192 | 2.3; 0.6 – 9.3 | |||
| Cohort 1 OS Indicator | |||||
| MCG-1 | MCG-1 (with LG) | ||||
| Parameter | P value | HR; 95% CI | P value | HR; 95% CI | |
| T-stage | 0.039 | 3.3; 1.1 – 9.8 | |||
| Oncomine CSS Indicator | |||||
| MCG-1 | |||||
| Parameter | P value | HR; 95% CI | |||
| Age | 0.01 | 1.1; 1.0 – 1.1 | |||
| N-stage | 0.006 | 3.4; 1.4 – 8.2 | |||
| Oncomine OS Indicator | |||||
| MCG-1 | MCG-1 (chemo subgroup) | ||||
| Parameter | P value | HR; 95% CI | P value | HR; 95% CI | |
| T-stage | 0.005 | 37.5; 3.4 – 437 | |||
| M-stage | ≤0.0001 | 7.7; 3.2 – 18.6 | 0.007 | 3.7; 1.4 – 9.8 | |
Table 3: Efficacy analysis of MCG-1 subtypes in cohort-1 and the Oncomine-dataset.
Efficacy was calculated using cut-off limits (Youden’s index) from the ROC curves to calculate metastasis, CSS, and OS. For bootstrap modeling, data were generated using 1000 iterations.
| Cohort-1 | |||||
|---|---|---|---|---|---|
| Metastasis | CSS Indicator (+ = Death due to BC ) | ||||
| Efficacy analysis | Efficacy analysis | ||||
| AUC | % Sensitivity | % Specificity | AUC | % Sensitivity | % Specificity |
| 0.545 | 80.0 | 30.8 | 0.586 | 83.3 | 35.7 |
| Bootstrap modeling | Bootstrap modeling | ||||
| Mean | 63.9 | 57.7 | Mean | 69.5 | 54.7 |
| Median | 68.0 | 56.3 | Median | 78.9 | 50 |
| 95% CI | 17.9 – 95.8 | 23.1 – 100 | 95% CI | 18.2 – 98.3 | 20.0 – 100 |
| Oncomine-dataset | |||||
| CSS Indicator | OS Indicator (+ = Death) | ||||
| Efficacy analysis | Efficacy analysis | ||||
| AUC | % Sensitivity | % Specificity | AUC | % Sensitivity | % Specificity |
| 0.516 | 52.2 | 56.4 | 0.571 | 55.3 | 58.3 |
| Bootstrap modeling | Bootstrap modeling | ||||
| Mean | 67.8 | 46.4 | Mean | 60.2 | 57.3 |
| Median | 66.7 | 49.0 | Median | 58.7 | 60.7 |
| 95% CI | 25.0 – 90.1 | 21.3 – 85.9 | 95% CI | 13.4 – 96.0 | 14.9 – 99.3 |
Oncomine-dataset validated intra-tumor heterogeneity and subtypes’ inability to predict outcome.
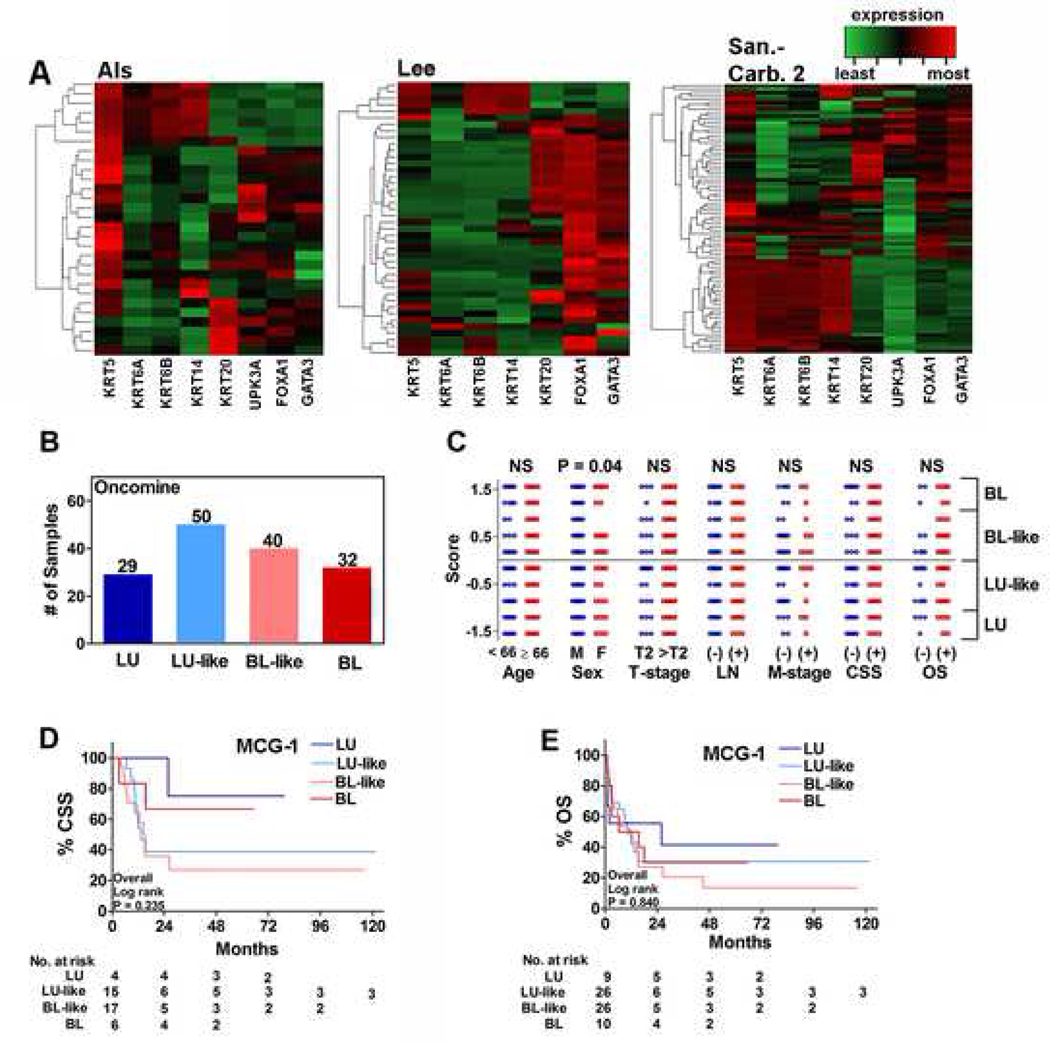
Since cohort-1 has a relatively small sample size and RT-qPCR was used for subtyping as opposed to RNA-Seq, we accessed three independent transcriptome datasets through Oncomine™ (Oncomine-dataset; Supplementary Table 1)23–25. Hierarchical cluster analysis of the high-grade MIBC specimens by MCG-1 showed some grouping of MIBC specimens, but the groups were heterogeneous among datasets (Figure 2A). MCG-1 again grouped the 151 specimens into four subtypes (Figure 2B). Among all clinical parameters, subtype scores significantly associated only with sex; females mostly had basal tumors (BL+BL-like; Supplementary Figure 1A; Figure 2C). In univariate analysis, TNM-staging, but not subtype scores, significantly associated with CSS or OS (Table 1). In multivariate analysis, age and TNM-staging significantly predicted CSS or OS (Table 2). Subtypes failed to risk stratify patients by Kaplan-Meier plots and had 52%−55% sensitivity and 56%−58% specificity to predict CSS and OS (Figure 2D, E; Table 3).
Figure 2: Association of subtypes to clinical outcome in the Oncomine-dataset.
A. Hierarchical clustering of high-grade MIBC specimens in Als (a), Lee (b) and Sanchez-Carbayo 2 (c) datasets deposited in Oncomine™. Note: data on UPK3B was not available and hence could not be included. B. The distribution of LU, LU-like, BL and BL-like subtypes among specimens in the Oncomine-dataset of 151 high-grade MIBC specimens. C. Comparison of specimen subtype scores by clinical parameters. Each symbol represents an individual specimen/patient, categorized with respect to a clinical parameter as shown in the figure. P-values generated by Mann-Whitney U tests are two-tailed. D, E. Kaplan-Meier plots for CSS (D) and (OS) stratified by MCG-1 subtypes using the specimens on which follow-up data were available.
Molecular subtypes associated with OS in the TCGA-dataset.
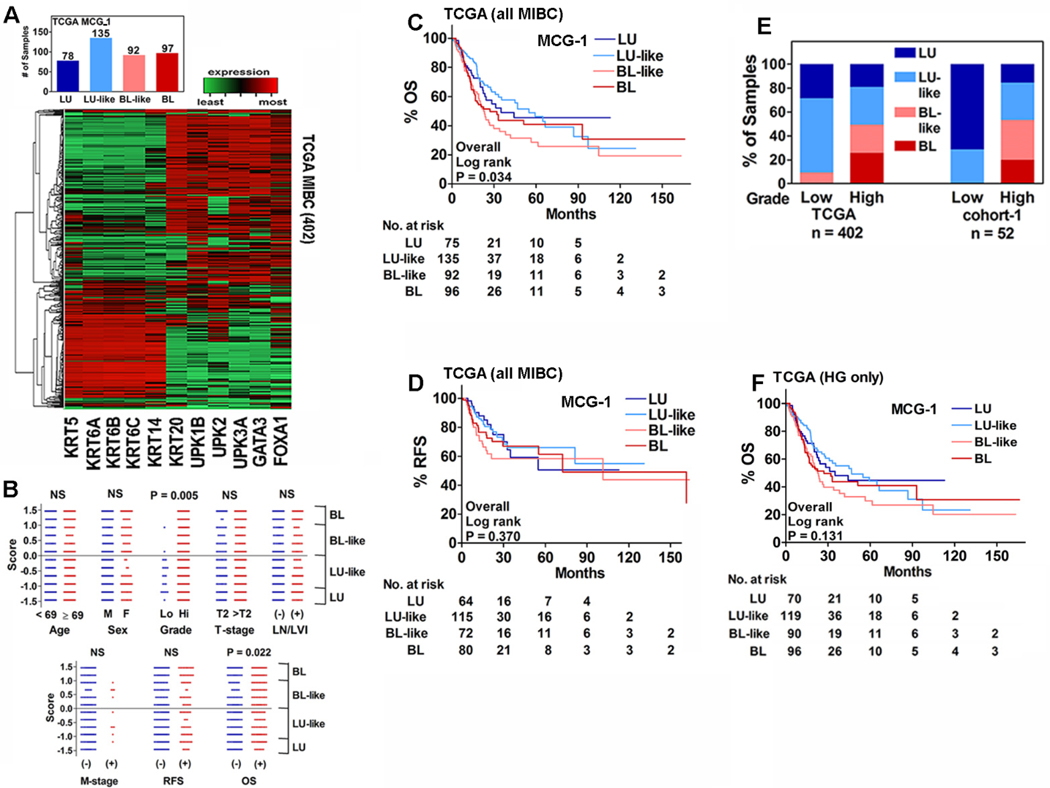
Since results of the Oncomine-dataset and cohort-1 were in agreement, the inability of the subtypes to associate with clinical outcome was likely not due to sample size or methodology. Many groups have reported an association of the molecular subtypes to OS using the TCGA-dataset5,10,12,19. Therefore, we sought to reproduce the published results by MCG-1 subtyping on all 402 MIBC specimens in the TCGA-dataset (Supplementary Table 1).
Although in the TCGA-dataset hierarchical clustering showed an overall grouping of specimens, based on the subtype scores, the majority were BL-like and LU-like (Figure 3A). Contrary to our findings in cohort-1 and the Oncomine-dataset, but consistent with published studies5,10,12,19, subtype scores associated with OS in the TCGA-dataset (Figure 3B). In univariate analysis, subtypes by MCG-1, along with all clinical parameters except sex, significantly predicted OS; the subtypes did not predict RFS (Table 1). Kaplan-Meier plots showed that subtypes significantly stratified patients for risk of death (OS; Figure 3C; P = 0.034). However, the subtypes could not stratify patients regarding risk of RFS (Figure 3D). In multivariate analysis LVI and N-stage were significant for predicting OS and RFS, respectively (Table 4). Subtypes had 55.7% sensitivity and 59.5% specificity for OS, and 54% sensitivity and 57% specificity for RFS (Table 5).
Figure 3: Association of subtypes to clinical parameters, OS and RFS in the TCGA-dataset and the influence of grade.
A. Hierarchical clustering of 402 MIBC specimens in the TCGA-dataset and the distribution of LU, LU-like, BL and BL-like subtypes. B. Comparison of specimen subtype scores by clinical parameters. Each symbol represents an individual specimen. P-values generated by Mann-Whitney U test are two-tailed. C and D. Kaplan-Meier plots for OS (C) and RFS (D) of MIBC specimens in the TCGA-dataset stratified by MCG-1 subtypes. E. Distribution of the molecular subtypes between low- and high-grade tumors in the TCGA and cohort-1 datasets. Note: There were only high-grade MIBC specimens in the Oncomine-dataset. F. Kaplan-Meier plot for OS including only high-grade MIBC specimens in the TCGA-dataset.
Table 4: Multivariate analysis to determine relationship of outcome to clinical parameters and molecular subtype classifications in the TCGA-dataset.
In all analyses we performed Cox Proportional Hazards Model with stepwise selection procedure to determine the final model that accurately describes the outcome. The parameters included in each analysis were: Age, sex, T-stage, N-stage, M-stage, LVI, and subtype classification. Only significant parameters based on Wald’s test with overall hazard ratio (HR) and 95% CI are shown.
| TCGA dataset OS Indicator | ||||||||
|---|---|---|---|---|---|---|---|---|
| MCG-1 | Consensus | TCGA-2017 | Lund-Taxonomy | |||||
| Parameter | P value | HR; 95% CI | P value | HR; 95% CI | P value | HR; 95% CI | P value | HR; 95% CI |
| LVI | 0.003 | 2.7; 1.4 – 5.3 | 0.003 | 2.7; 1.4 – 5.3 | 0.016 | 2.3; 1.2 – 4.7 | 0.003 | 2.7; 1.4 – 5.3 |
| Subtype Classification | 0.045 | 4.4; 1.1 – 16.5 | ||||||
| TCGA dataset RFS Indicator | ||||||||
| MCG-1 | Consensus | TCGA-2017 | Lund-Taxonomy | |||||
| Parameter | P value | HR; 95% CI | P value | HR; 95% CI | P value | HR; 95% CI | P value | HR; 95% CI |
| N-stage | 0.039 | 2.1; 1.04 – 4.2 | 0.039 | 2.1; 1.04 – 4.2 | 0.039 | 2.1; 1.04 – 4.2 | 0.039 | 2.1; 1.04 – 4.2 |
Note: Analysis of only HG specimens yields the same values.
Table 5: Efficacy analysis of molecular subtypes in the TCGA-dataset.
Efficacy of the subtypes was calculated using cut-off limits (Youden’s index) from the ROC curves to calculate OS and RFS. For bootstrap modeling, data were generated using 1000 iterations.
| TCGA dataset OS Indicator | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MCG-1 | Consensus | TCGA 2017 | Lund-Taxonomy | ||||||||
| Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | ||||||||
| AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. |
| 0.568 | 55.7 | 59.5 | 0.636 | 63.1 | 59.9 | 0.593 | 68.2 | 53.2 | 0.578 | 59.1 | 56.3 |
| Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | ||||||||
| % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | ||||
| Mean | 55.7 | 59.6 | Mean | 72.3 | 52.0 | Mean | 67.7 | 53.5 | Mean | 57.4 | 58.0 |
| Med. | 55.9 | 59.6 | Med. | 70.6 | 54.9 | Med. | 67.9 | 53.1 | Med. | 58.7 | 56.8 |
| 95% CI | 46.7 - 63.5 |
52.5 – 67.3 | 95% CI | 56.5 – 86.1 | 36.4 – 67.6 | 95% CI | 59.7 – 74.6 | 46.5 – 61.0 | 95% CI | 40.3 – 66.2 | 49.2 – 73.4 |
| MCG-1 (HG only) | Consensus (HG only) | TCGA 2017 (HG only) | Lund-Taxonomy (HG only) | ||||||||
| Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | ||||||||
| AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | ROC AUC | % Sens. | % Spec. |
| 0.548 | 55.2 | 55.2 | 0.611 | 63.2 | 56.2 | 0.574 | 68.4 | 48.8 | 0.564 | 59.8 | 52.7 |
| Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | ||||||||
| % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | ||||
| Mean | 56.6 | 54.3 | Mean | 70.3 | 50.0 | Mean | 66.6 | 50.6 | Mean | 55.7 | 57.0 |
| Med. | 56.0 | 55.8 | Med. | 66.7 | 53.8 | Med. | 68.2 | 49.2 | Med. | 59.2 | 54.1 |
| 95% CI | 25.7 – 86.3 | 21.2 – 81.3 | 95% CI | 51.8 – 86.5 | 32.2 – 67.0 | 95% CI | 42.3 – 75.7 | 42.2 – 74.7 | 95% CI | 8.0 – 67.4 | 46.8 – 98.0 |
| TCGA dataset RFS Indicator | |||||||||||
| MCG-1 | Consensus | TCGA 2017 | Lund-Taxonomy | ||||||||
| Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | ||||||||
| AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. |
| 0.554 | 54.0 | 57.0 | 0.578 | 48.3 | 65.2 | 0.536 | 60.9 | 48.4 | 0.535 | 40.2 | 66.4 |
| Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | ||||||||
| % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | ||||
| Mean | 56.0 | 55.8 | Mean | 57.4 | 57.9 | Mean | 51.2 | 60.4 | Mean | 51.2 | 58.0 |
| Med. | 55.6 | 57.4 | Med. | 55.6 | 60.8 | Med. | 56.6 | 53.9 | Med. | 51.0 | 59.9 |
| 95% CI | 24.7 – 88.0 | 20.8 – 81.7 | 95% CI | 41.2 – 79.2 | 34.2 – 71.2 | 95% CI | 14.3 – 71.4 | 42.7 – 92.9 | 95% CI | 7.4 – 94.5 | 11.0 – 97.6 |
| MCG-1 (HG only) | Consensus (HG only) | TCGA 2017 (HG only) | Lund-Taxonomy (HG only) | ||||||||
| Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | Efficacy Analysis | ||||||||
| AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. | AUC | % Sens. | % Spec. |
| 0.538 | 54.1 | 53.4 | 0.559 | 49.4 | 62.3 | 0.522 | 43.5 | 62.8 | 0.512 | 40.0 | 63.2 |
| Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | Bootstrap Modeling | ||||||||
| % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | % Sens. | % Spec. | ||||
| Mean | 58.3 | 51.0 | Mean | 47.3 | 62.3 | Mean | 47.3 | 62.3 | Mean | 53.8 | 53.7 |
| Med. | 56.8 | 53.6 | Med. | 48.2 | 62.7 | Med. | 48.2 | 62.7 | Med. | 52.5 | 54.6 |
| 95% CI | 23.7 – 89.5 | 18.9 – 81.7 | 95% CI | 12.8 – 93.7 | 10.9 – 93.5 | 95% CI | 12.8 – 93.7 | 10.9 – 93.5 | 95% CI | 6.3 – 95.1 | 10.3 – 98.2 |
In the TCGA-dataset Prognostic capability of subtypes depended on low-grade MIBC specimens.
Analysis of the clinical data in all three cohorts revealed that while neither cohort-1 nor the Oncomine-dataset included low-grade MIBC specimens, in the TCGA-dataset 5.2% (21/402) of the specimens were low-grade MIBC (Supplementary Table 1). Nineteen of these 21 specimens (90.5%) were either the LU or LU-like subtype and subtypes associated with tumor grade (P=0.005; Figure 3B, E). Molecular subtyping studies that used the TCGA-dataset and reported prognostic significance included these low-grade specimens in their analyses5,10,12,19. However, low-grade MIBC is a rare occurrence; only 0.988% (55/5,564) MIBC cases in the SEER database are low-grade. When we re-performed MCG-1 subtype analyses of the TCGA-dataset using only the high-grade MIBC specimens, the subtypes neither associated with OS in univariate analysis, nor could they stratify patients for OS (Table 6, Figure 3F). In multivariate analysis, LVI and N-stage were still the only significant parameters for OS and RFS, respectively (Table 4). In efficacy analysis of only the high-grade specimens, subtypes by MCG-1 had suboptimal sensitivity and specificity for OS and RFS (Table 5).
Table 6: Influence of grade on subtypes’ association to clinical outcome.
Univariate analysis for evaluating subtype scores’ association with clinical outcome when 7 low-grade specimens were added in cohort-1 and 21 low-grade specimens were removed from the TCGA-dataset.
| TCGA (HG only) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| RFS Indicator | OS Indicator | ||||||||
| Parameter | χ2 | P value | OR; 95% CI | χ2 | P value | OR; 95% CI | |||
| MCG-1 | 1.09 | 0.296 | 2.61 | 0.11 | |||||
| Cohort-1 (HG + LG specimens) | |||||||||
| Metastasis | CSS Indicator | OS Indicator | |||||||
| Parameter | χ2 | P value | OR; 95% CI | χ2 | P value | OR; 95% CI | χ2 | P value | OR; 95% CI |
| MCG-1 | 4.28 | 0.039 | 1.91; 1.9 – 3.5 | 4.65 | 0.031 | 2; 1.1 – 3.7 | 3.88 | 0.049 | 1.9; 1.0 – 3.7 |
MCG-Ext subtypes validates results of MCG-1 subtypes in the TCGA-dataset.
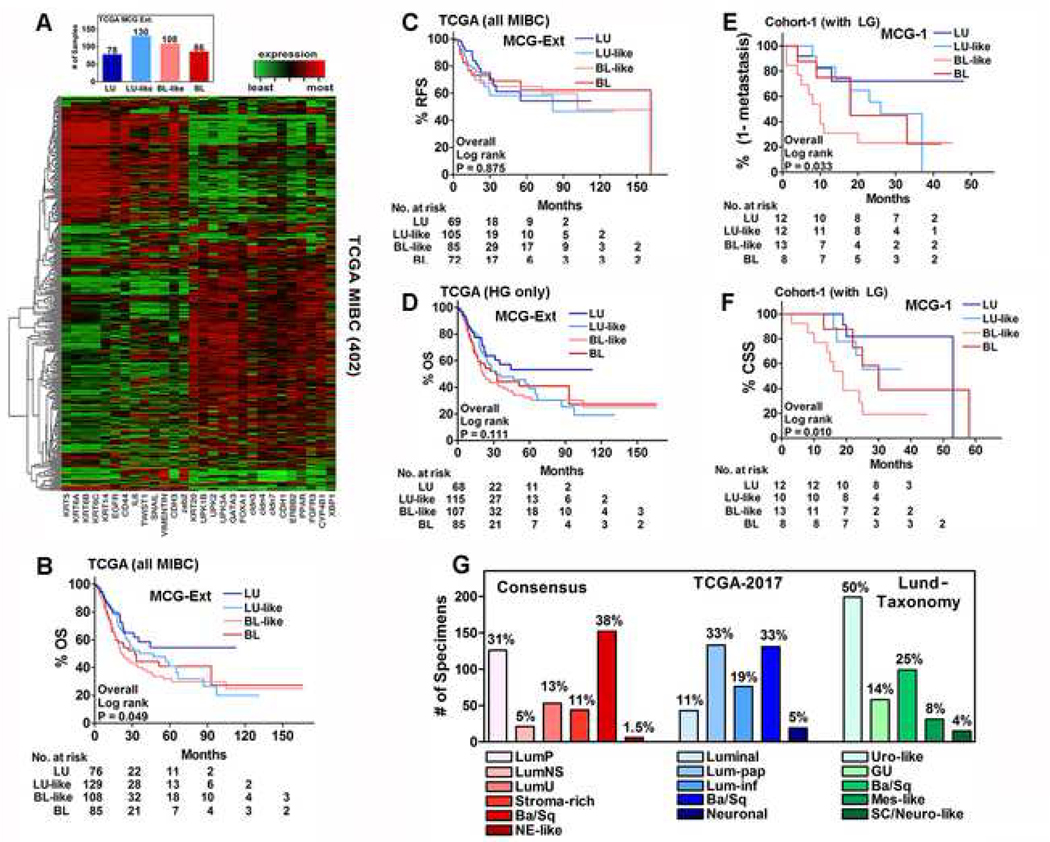
To validate results from MCG-1, we expanded the panel by including additional genes reported in at least two subtyping studies (MCG-Ext; Supplementary Table 2)7,8,10,12,19–22. Hierarchical clustering by MCG-Ext showed more obvious heterogeneity than MCG-1. Nevertheless, subtype scores grouped the majority of specimens into BL-like and LU-like subtypes (Figure 4A). Out of 21 low-grade tumors, MCG-Ext classified 20 (95.2%) as LU or LU-like and in univariate analysis, subtypes showed strong association with tumor-grade (P=0.0004). In univariate analysis, subtypes by MCG-Ext, significantly associated with OS, but not RFS (Table 1). Kaplan-Meier plots showed that subtypes significantly stratified patients for OS, but not RFS (Figure P=0.049; Figure 4B, C). In multivariate analysis LVI and N-stage were significant for predicting OS and RFS, respectively (Table 4). Consistent with MCG-1 panel, the subtypes did not stratify patients for OS when only high-grade specimens were analyzed (Figure 4D).
Figure 4: Analysis of MCG-Ext subtypes and distribution of subtypes among established classifications in the TCGA-dataset.
A. Hierarchical clustering of 402 MIBC specimens in the TCGA dataset and the distribution of LU, LU-like, BL and BL-like subtypes according to MCG-Ext. B, C. Kaplan-Meier plots including all MIBC specimens in the TCGA-dataset for OS (B) and RFS (C) based on MCG-Ext. D. Kaplan-Meier plot for OS including only HG MIBC specimens in the TCGA-dataset based on MCG-Ext. E, F. Kaplan-Meier plots including LG specimens (all non-MIBC) in cohort-1 for metastasis (E) and CSS (F) based on MCG-1. G. Distribution of subtypes in each established classification in the TCGA-dataset.
Abbreviations: BC: bladder cancer; BL: basal; CSS: cancer-specific survival; KRT: Cytokeratin(s); LN: lymph node; LU: luminal; LVI: lymphovascular invasion; MIBC: muscle invasive bladder cancer; OS: overall survival; RFS: recurrence-free survival; TCGA: The Caner Genome Atlas; TNM: Tumor-stage, lymph node, metastasis; UPK: Uroplakin(s)
Subtypes predicted clinical outcome in cohort-1 with inclusion of low-grade specimens
We also evaluated whether addition of low-grade specimens in cohort-1 would enable the subtypes to predict clinical outcome. Therefore, we re-analyzed cohort-1 including low-grade specimens (all non-MIBC). Similar to the TCGA-dataset, all seven low-grade specimens were the LU or LU-like subtype (Figure 3E). Kaplan-Meier plots showed that the subtypes could now significantly risk-stratify patients for metastasis and CSS (Fig 4E, F); similar results were obtained for OS (P = 0.023). Univariate analysis confirmed these results (Table 6). In multivariate analysis, subtypes, along with N-stage and T-stage were significant predictors of metastasis and CSS (Table 2). Analysis using high-grade non-MIBC and MIBC specimens showed roughly equal distribution of the subtypes between these groups (Supplementary Figure 1B). Among this high-grade subgroup (MIBC and non-MIBC), subtypes did not stratify patients for risk of metastasis, CSS, or OS (Supplementary Figure 1 C-E).
Subtypes do not predict response to cisplatin-based chemotherapy
In the TCGA-dataset there are 10 specimens from patients who underwent neoadjuvant chemotherapy. Although the response data are not provided, all 10 patients had high-grade MIBC (pathologic stage: II: n=3; ≥ III: n=7), suggesting that they did not respond to NAC. Five specimens were the BL/BL-like subtype, and five were the LU/LU-like subtype. In the Oncomine-dataset, 43 patients received adjuvant cisplatin-based chemotherapy and had follow-up data (Supplementary Table 1)23,25. Subtypes by MCG-1 could not risk-stratify these patients with respect to response, i.e. OS, and in multivariate analysis only M-stage was an independent predictor of OS (Supplementary Figure 1F, Table 2).
Three published subtype classifications validate results of MCG-1 and MCG-Ext in the TCGA-dataset
We also analyzed the TCGA-dataset using three established subtyping classifications: Lund-Taxonomy, TCGA-2017, and Consensus12–14. Although each classification system divides the TCGA-dataset into five or six subtypes, the majority (66%–75%) of specimens belong to two subtypes, while the remaining subtypes are underrepresented (Figure 4G). In each classification the NE-like/Neuronal subtype accounts for only six (1.5%) to 19 (4.7%) specimens in the dataset (Figure 4G). In univariate analysis, subtypes by these classifications associated with OS, and only Consensus subtypes modestly associated with RFS (Table 1). Of the 21 low-grade tumors, 20 (95.2%) were Luminal Papillary according to the TCGA-2017 and Consensus classifications, and all 21 were Urothelial-like according to Lund-Taxonomy. Furthermore, as observed for MCG-1 and MCG-Ext, tumor grade significantly associated with subtypes in these classifications (P<0.0001), univariately. In multivariate analysis, the TCGA-2017 subtypes predicted OS (P=0.045); however, subtypes by neither the Lund-Taxonomy nor Consensus were able to predict OS; no classification predicted RFS (Table 4). In each classification, LVI and N-stage significantly predicted OS and RFS, respectively (Table 4). Subtypes by these classifications had 59.1%−68.2% sensitivity and 53.2%−59.9% specificity for OS and 40.2%−60.9% sensitivity and 48.4%−66.4% specificity for RFS (Table 5).
It is noteworthy that despite association of subtypes to tumor grade, in multivariate analyses, subtypes by Consensus, TCGA-2017, and Lund-Taxonomy did not reach significance whether low-grade tumors were included or not. In the high-grade only subgroup, efficacy values were similar - OS: sensitivity, 59.8%−68.4%; specificity, 48.8%−56.2%; RFS: sensitivity, 40%−49.4%; specificity: 62.3%−63.2% (Table 5).
Discussion
Historically, molecular classification of bladder tumors has identified two divergent pathways with distinct genetic hallmarks characterizing low-grade and high-grade tumors2,3. Recent studies, including those by the TCGA Research Network, identified molecular subtypes that could potentially predict outcome and individualize care of MIBC patients4–8,10,19,20. The salient findings of our study are: 1. Development of a RT-qPCR based simplified MIBC subtyping methodology that can be applied to any cohort. Prognostic predictions by this methodology are consistent with those obtained using established classification systems. 2. MCG-1, MCG-Ext, Lund-Taxonomy, Consensus and TCGA-2017 classifications consistently showed an association of subtypes with tumor grade. 4. Regardless of sample size, cohort/dataset, or subtyping classification, the subtypes were consistently outperformed by clinical parameters (mainly N-Stage and LVI) for predicting BC-specific outcomes and OS. 5. Subtypes could not predict response to cisplatin-based adjuvant chemotherapy.
Molecular subtyping classifications have provided insight into the biology of bladder tumors. For example, the existence of multiple subtypes demonstrates heterogeneity among tumors. Our study reveals that both BL and LU markers exist within one tumor, reflecting intra-tumor heterogeneity. Furthermore, different cells within a tumor express different levels of a single marker. Such heterogeneity has also been previously reported within a bladder tumor and among clonally related bladder tumors in a single patient16. These observations indicate two possibilities; first, tumor cells may express BL or LU markers relative to their temporal and spatial positioning, and this expression may be altered by the tumor microenvironment, disease progression and/or treatment. Second, tumors contain tumor cell clones representing either “BL” or “LU” phenotype arising from different cells of origin. Our IHC data support both possibilities and are consistent with the reported differences in genetic landscapes between primary tumor biopsies and multiple metastatic sites18. Tumor sampling technique could also have an impact on subtyping. A single tumor could plausibly be classified as different subtypes depending on location and/or size of the sample. Therefore, intra-tumor heterogeneity associated with molecular subtypes could be a reflection of differences in etiology, cell of origin, tumor sampling, tumor microenvironment and/or therapeutic response.
It was reported that MIBC subtypes may predict patient response to chemotherapy and could be used to develop targeted therapies5,10,12,19,20. In the Oncomine-dataset, subtypes could not stratify patients regarding response to cisplatin-based adjuvant chemotherapy. Together with the heterogeneity introduced by sampling and tumor microenvironment is the complexity that marker profiles are a reflection of gene expression. Since gene expression patterns change, subtype drifting or switching due to tumor evolution and reaction to therapy are reported. Therefore, there is a need for systematic biological studies to better stratify BC based on genetic drivers of treatment response rather than subtyping based on expression profiling7,28.
The LU subtype is reported to predict a better prognosis4,7,8,11,12,20. In the TCGA-dataset and cohort-1, 91% and 100% of the low-grade tumors were LU phenotype (LU/LU-like) by MCG-1, respectively. Using the established classifications (Consensus, TCGA-2017, and Lund-Taxonomy), 95.2%−100% of low-grade tumors were LU-related subtypes. In all cohorts, subtypes by all classifications associated with tumor grade. The predictive ability of MCG-1 and MCG-Ext subtypes for OS was lost when low-grade specimens were excluded. In agreement, the subtypes’ gained predictive ability when low-grade specimens were included in analysis of cohort-1. A recent meta-cohort study describing six molecular subtypes showed that subtypes did not associate with OS in the TCGA-dataset of 383 specimens (P=0.077)9. This sample size is only plausible if all or most of the 21 low-grade specimens were excluded. The study also analyzed 418 specimens from the “UROMOL” non-MIBC cohort (n=476; Array Express for UROMOL E-MATB-4321). In this cohort, >50% of specimens are low-grade and subtypes were reported as predicting disease-free survival9. Another group applied their own subtyping classification to the UROMOL-cohort and reported subtypes’ predicted progression-free survival29. However, the UROMOL-cohort has only 31 patients progressing to MIBC, and 22 had high-grade tumors. This made grade a better predictor for progression (log-rank: P<0.0001) than the subtypes reported in both studies9,29.
Our study demonstrates that subtyping by a simplified RT-qPCR-based method yields results comparable to established RNA-Seq based classification systems. This is demonstrated by subtypes by any classification system correlating with histopathologic grade, and low-grade tumors being of luminal lineage. Furthermore, regardless of subtyping classification, only clinical parameters such as N-stage and LVI were independent predictors of prognosis in multivariate analyses. In any cohort, subtypes by any classification had suboptimal efficacy to predict clinical outcome.
Conclusions
Molecular subtyping classifications have provided insight into the biology of bladder tumors, especially regarding tumor heterogeneity. However, results from multiple cohorts and classification systems reveal that subtypes strongly associate with histopathologic grade and are consistently outperformed by clinical parameters to predict BC patient outcome. Further investigation is needed into the clinical applicability of molecular subtypes before their incorporation into the personalized care of MIBC patients.
Supplementary Material
Acknowledgments:
We are indebted to Dr. Gottfrid Sjödahl from Division of Urological Research, Department of Translational Medicine, Lund University, Skåne University Hospital, Malmö, Sweden for graciously providing the subtype identifiers for specimens in the TCGA-dataset according to the Lund-Taxonomy and the Consensus classifications. We thank Mrs. Cynthia Soloway and Dr. Bal Lokeshwar for their input and critical reading and editing of this manuscript.
Funding: This work was supported by the National Cancer Institute at the National Institutes of Health (grant number 1R01CA227277-01A1 to VBL, 5R01CA176691-05 to VBL, 1F31 CA210612-01 to ARJ, 1F31 CA236437-01 to DSM); and The Department of Defense Peer Reviewed Cancer Research Program (grant number W81XWH1810277 (CA170470; PRCRP) to VBL).
Abbreviations:
- BC
bladder cancer
- BL
basal
- CSS
cancer-specific survival
- KRT
Cytokeratin(s)
- LN
lymph node
- LU
luminal
- LVI
lymphovascular invasion
- MIBC
muscle invasive bladder cancer
- OS
overall survival
- RFS
recurrence-free survival
- TCGA
The Caner Genome Atlas
- TNM
Tumor-stage, lymph node, metastasis
- UPK
Uroplakin(s)
References
- 1.Milowsky MI, Rumble RB, Booth CM, et al. Guideline on Muscle-Invasive and Metastatic Bladder Cancer (European Association of Urology Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34(16):1945–1952. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Droller MJ. Pathways of development and progression in bladder cancer: new correlations between clinical observations and molecular mechanisms. Seminars in urology. 1993;11(4):177–192. [PubMed] [Google Scholar]
- 3.Droller MJ. Bladder cancer. Current problems in surgery. 1981;18(4):205–279. [DOI] [PubMed] [Google Scholar]
- 4.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41. [DOI] [PubMed] [Google Scholar]
- 5.Seiler R, Ashab HAD, Erho N, et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. European urology. 2017;72(4):544–554. [DOI] [PubMed] [Google Scholar]
- 6.TCGA Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25(2):152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. Vol 1112014:3110–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan TZ, Rouanne M, Tan KT, Huang RY, Thiery JP. Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-cohort Analysis of 2411 Tumors. European urology. 2019;75(3):423–432. [DOI] [PubMed] [Google Scholar]
- 10.Dadhania V, Zhang M, Zhang L, et al. Meta-Analysis of the Luminal and Basal Subtypes of Bladder Cancer and the Identification of Signature Immunohistochemical Markers for Clinical Use. EBioMedicine. Vol 122016:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConkey DJ, Choi W. Molecular Subtypes of Bladder Cancer. Curr Oncol Rep. 2018;20(10):77. [DOI] [PubMed] [Google Scholar]
- 12.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171(3):540–556.e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzouka NA, Eriksson P, Rovira C, Liedberg F, Sjodahl G, Hoglund M. A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Scientific reports. 2018;8(1):3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamoun A, de Reynies A, Allory Y, et al. The Bladder Cancer Molecular Taxonomy Group. The consensus molecular classification of muscle-invasive bladder cancer. BioRxiv 488460; doi: 10.1101/488460 [DOI] [Google Scholar]
- 15.Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. [DOI] [PubMed] [Google Scholar]
- 16.Thomsen MBH, Nordentoft I, Lamy P, et al. Comprehensive multiregional analysis of molecular heterogeneity in bladder cancer. Scientific reports. 2017;7(1):11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyrskjot L Molecular Subtypes of Bladder Cancer: Academic Exercise or Clinical Relevance? European urology. 2019;75(3):433–434. [DOI] [PubMed] [Google Scholar]
- 18.Faltas BM, Prandi D, Tagawa ST, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet. 2016;48(12):1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochoa AE, Choi W, Su X, et al. Specific micro-RNA expression patterns distinguish the basal and luminal subtypes of muscle-invasive bladder cancer. Oncotarget. 2016;7(49):80164–80174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinaldetti S, Rempel E, Worst TS, et al. Subclassification, survival prediction and drug target analyses of chemotherapy-naive muscle-invasive bladder cancer with a molecular screening. Oncotarget. 2018;9(40):25935–25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seiler R, Gibb EA, Wang NQ, et al. Divergent Biological Response to Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-18-1106. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 22.Choi W, Ochoa A, McConkey DJ, et al. Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. European urology. 2017;72(3):354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Als AB, Dyrskjot L, von der Maase H, et al. Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer. Clin Cancer Res. 2007;13(15 Pt 1):4407–4414. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24(5):778–789. [DOI] [PubMed] [Google Scholar]
- 25.Kim WJ, Kim EJ, Kim SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haussler MG, Brian C, Mim H, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. 2019. [Google Scholar]
- 27.Yates TJ, Knapp J, Gosalbez M, et al. C-X-C chemokine receptor 7: a functionally associated molecular marker for bladder cancer. Cancer. 2013;119(1):61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjodahl G, Eriksson P, Lovgren K, et al. Discordant molecular subtype classification in the basal-squamous subtype of bladder tumors and matched lymph-node metastases. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2018. [DOI] [PubMed] [Google Scholar]
- 29.Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell. 2016;30(1):27–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.