Abstract
Introduction:
Derived from genetic alterations, cancer neoantigens are proteins with novel amino acid sequences that can be recognized by the immune system. Recent evidence demonstrates that cancer neoantigens represent important targets of cancer immunotherapy. The goal of cancer neoantigen vaccines is to induce neoantigen-specific immune responses and antitumor immunity, while minimizing the potential for autoimmune toxicity. Advances in sequencing technologies, neoantigen prediction algorithms and other technologies have dramatically improved the ability to identify and prioritize cancer neoantigens. These advances have generated considerable enthusiasm for development of neoantigen vaccines. Several neoantigen vaccine platforms are currently being evaluated in early phase clinical trials including the synthetic long peptide (SLP), RNA, dendritic cell (DC), and DNA vaccine platforms.
Areas covered:
In this review, we describe, evaluate the mechanism(s) of action, compare the advantages and disadvantages, and summarize early clinical experience with each vaccine platform. We provide perspectives on the future directions of the neoantigen vaccine field. All data are derived from Pubmed and ClinicalTrials search updated in October 2020.
Expert opinion:
Although the initial clinical experience is promising, significant challenges to the success of neoantigen vaccines include limitations in neoantigen identification and the need to successfully target the immunosuppressive tumor microenvironment.
Keywords: Neoantigen, cancer vaccine, delivery platforms, clinical trial
1. INTRODUCTION
Genetic alterations are common in cancer. Genetic alterations in cancer driver genes often contribute to the neoplastic phenotype, but genetic alterations are also present in passenger genes. When nonsynonymous genetic alterations are transcribed and translated, novel mRNA and protein sequences are generated [1]. Proteins with novel amino acid sequences can be processed and presented by HLA class I and II molecules, with the potential to induce CD8 and CD4 T cell responses [2]. Mutant proteins that are recognized by the immune system are known as cancer neoantigens. Cancer neoantigens can shape tumor evolution through the process of cancer immunoediting [3,4]. However, the process of cancer immunoediting is complex, and immune regulatory mechanisms such as tumor-associated macrophages, myeloid-derived suppressor cells, inhibitory cytokines and regulatory T cells can restrain antitumor immune responses. Cancer neoantigens often persist in progressing human cancers [5].
CD8 T cells play an important role in the specific recognition of cancer neoantigens, and are considered important effector cells in antitumor immune responses [6,7]. The presence of tumor infiltrating lymphocytes (TILs) in the tumor is an important predictor of prognosis and response to therapy [8–10]. Tumor-infiltrating CD8 T cells can also be inactivated in the tumor microenvironment, a phenomenon associated with expression of exhaustion markers [11]. CD4 T cells also play an important role in antitumor immunity as they help prime and maintain CD8 T cell responses [12–16]. Neoantigen-specific CD4 T cells appear to contribute to antitumor immunity in human cancers. In one of the earliest studies of cancer neoantigens, Linnemann et al. found that cancer neoantigens in melanoma were mainly recognized by CD4 T cells [17]. The role of neoantigen-specific CD4 T cells is not completely understood. There is evidence that CD4 T cells help priming of CD8 T cells by licensing cDC1 via the CD40/CD40L interaction [18]. Of note, help-less CD8 T cells are subject to exhaustion and are unable to control tumor growth [19,20]. CD4 T cells also have effector roles in the tumor microenvironment. These include direct cytotoxicity [21], activation of NK cells and the recruitment of CD8 T cells through the secretion of cytokines such as TNF-α and IFN-γ.
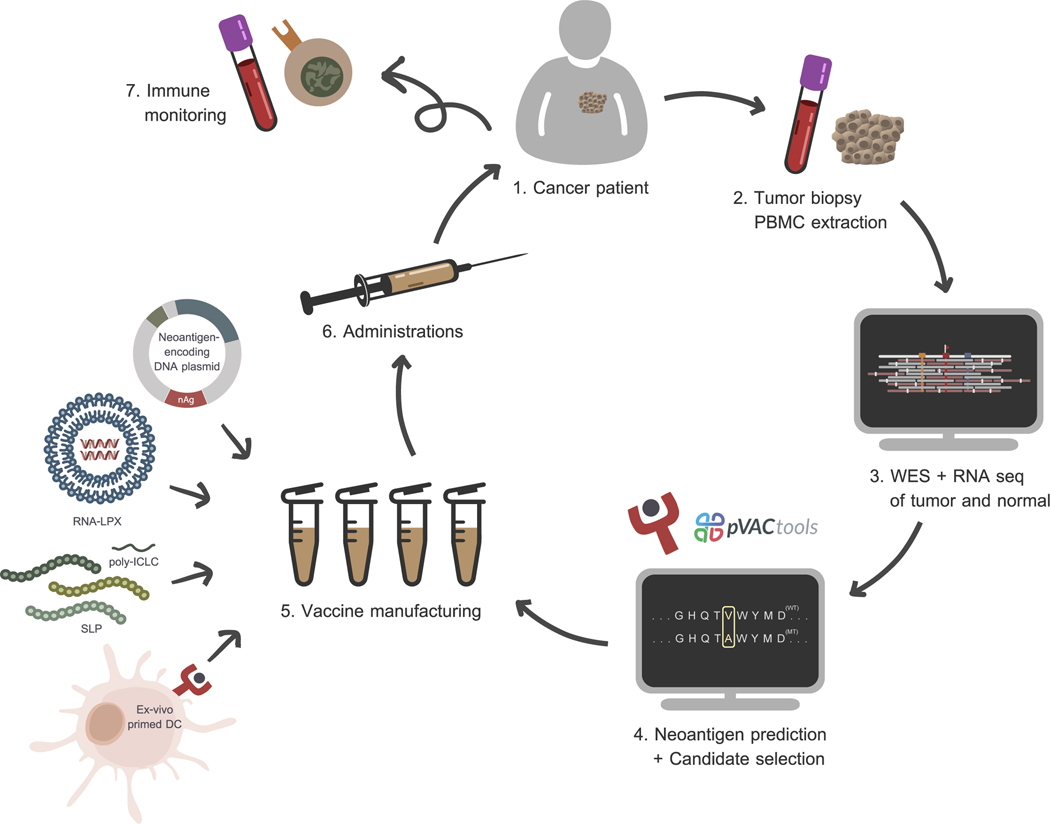
To induce or enhance neoantigen-specific immune responses, cancer neoantigen vaccines have been tested using the synthetic long peptide, RNA, DNA, and dendritic cell vaccine platforms [22,23]. Candidate neoantigens are most commonly identified and prioritized using computational algorithms capable of comparing matched tumor/normal sequencing data, identifying genetic alterations, and then predicting which altered proteins can be recognized by the immune system (Figure 1). Preclinical studies and early phase clinical trials suggest that cancer neoantigens identified by such algorithms may be promising targets for cancer immunotherapy [24–26]. In some types of cancer, such as melanoma, neoantigen-based cancer vaccines have been shown to inhibit tumor growth in mouse models and induce clinical responses in patients [27]. Combining cancer neoantigen vaccines with immune modulators targeting the immunosuppressive tumor environment may enhance vaccine efficacy. This strategy is being tested in several ongoing clinical trials [NCT02950766, NCT03199040].
Figure 1: A personalized cancer vaccine manufacturing workflow.
Inputs of the whole vaccine manufacturing workflow starts from sequencing data of matched tumor-normal samples. Accurate neoepitopes for further vaccine production are derived from computational analysis. Neoantigens can be integrated into the vaccine via several ways such as cloned DNA plasmids, liposome-packed mRNAs, synthetic peptides, and ex-vivo pulsed DCs. Finally, immune responses against delivered epitopes are continuously monitored in the follow-up period.
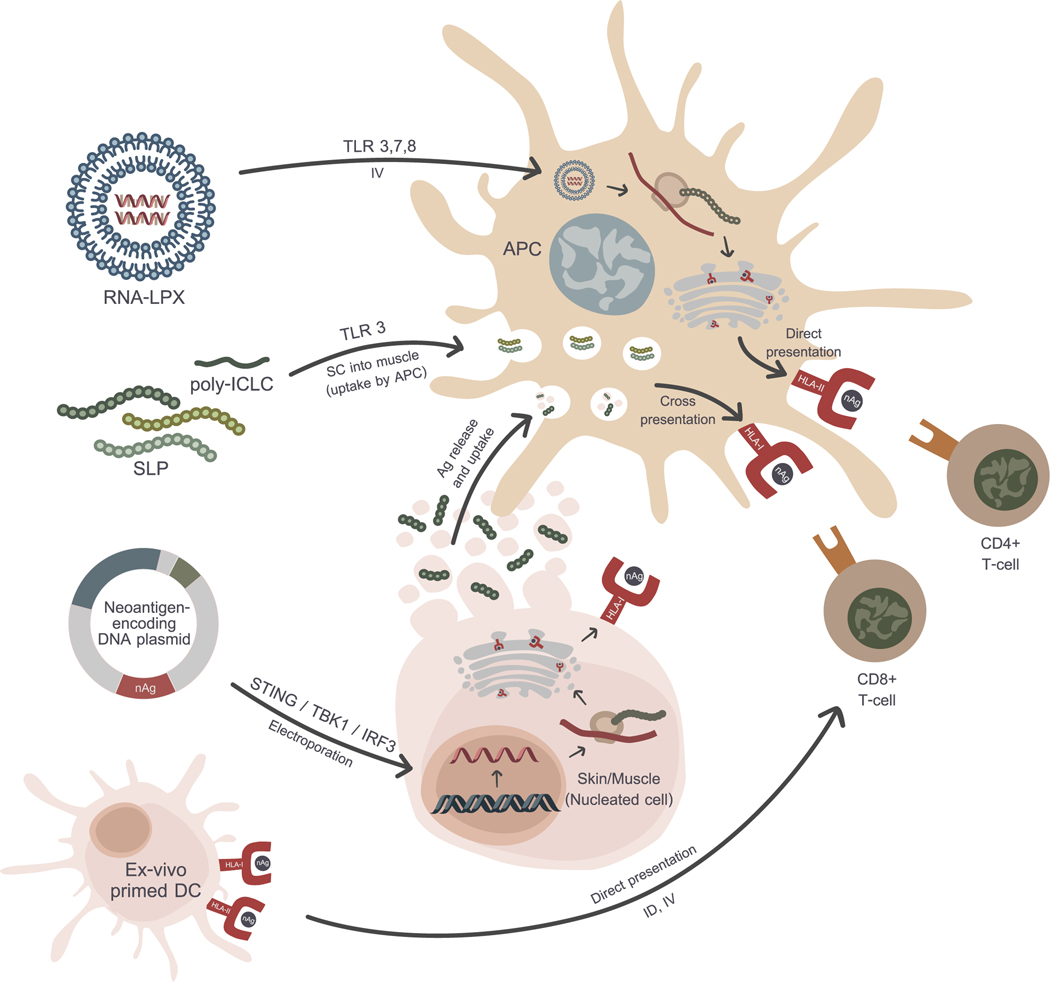
Neoantigen identification and prioritization is just one component of a successful neoantigen vaccine. Vaccine design is also a crucial element in the successful generation of immune responses. Neoantigens must be processed and presented by antigen presenting cells with the appropriate costimulatory signals in order to successfully induce robust CD4 and CD8 neoantigen-specific T cells. Current vaccine platforms that are being tested in ongoing clinical trials include the synthetic long peptide (SLP), RNA, DNA and dendritic cell platforms with or without immune checkpoint inhibitors [28]. Each vaccine platform has a different mechanism of action, but the goal is the successful presentation of cancer neoantigens by antigen presenting cells to T cells with appropriate costimulation so a productive immune response can be induced (Figure 2). Each vaccine platform has intrinsic advantages and disadvantages as summarized in Table 1.
Figure 2: Mechanisms of each delivery platform.
Beside different formulations, each strategy involves different steps in central dogma and antigen presentation pathway. However, their ultimate goal is to activate CD4+ and CD8+ immune responses through neoantigen-primed DCs.
Table 1: Vaccine delivery platform comparison.
Routes of administration, formulations, advantages, and disadvantages among four main delivery platforms are compared in the table. Convincing preclinical studies and ongoing clinical trials of each platform are also provided.
| Platform | Routes | Formulation | Advantages | Disadvantages | Ongoing trials |
|---|---|---|---|---|---|
| Peptide | SC | Synthetic long peptides mixed with poly-ICLC | • Proven safety profile • Stable • Long peptides can stimulate both CD4 and CD8 T cell responses |
• Low immunogenicity unless paired with adjuvant • Unpredictable uptake by DCs • Sequence-specific manufacturing considerations |
NCT03956056
NCT02950766 NCT01970358 NCT03606967 |
| RNA | SC IV | In vitro transcribed RNA encoding cancer neoantigens formulated in liposomes or nanoparticles | • Rapid and inexpensive production • Mimics viral infection • Intrinsic adjuvant properties |
• Relative instability • Susceptible to extracellular degradation by RNAses |
NCT02035956
NCT02316457 NCT03289962 NCT04161755 |
| DC | ID IV | Mature DCs pulsed with peptides or RNA corresponding to cancer neoantigens | • Bypass conventional antigen presentation pathways | • Time and resource intensive manufacturing process • Large-scale production less practical |
NCT03300843
NCT04078269 NCT04105582 NCT01885702 |
| DNA | IM or ID via Electroporation | Plasmid DNA encoding cancer neoantigens | • Rapid and inexpensive production • Mimics viral infection • Flexible platform allows molecular engineering |
• Low immunogenicity • Theoretical risk of host genome integration • Electroporation discomfort |
NCT03122106
NCT03199040 NCT03532217 NCT03914872 |
In this expert opinion, we describe and compare the vaccine platforms, and summarize the results of early clinical trials. To date, there have been relatively few reports of cancer neoantigen vaccines in early stage clinical trials. A total of ten phase 1 clinical trials are summarized in Table 2. Clinical and immune responses observed in these early stage trials is discussed in detail in the sections below. Of note, the safety profile of cancer neoantigen vaccines appears to be excellent regardless of platform. All vaccine platforms were considered to be safe, with the most notable adverse events being injection site reactions, myalgias, fatigue, and chills. Although there are no reports yet from early stage clinical trials evaluating the neoantigen DNA vaccine platform, our group is currently evaluating neoantigen DNA vaccines for the treatment of triple-negative breast cancer (TNBC) and pancreatic cancer with minimal adverse events reported [ NCT03199040, NCT03122106].
Table 2: Published clinical neoantigen studies to date categorized by delivery platform.
From comprehensive Pubmed search with the keyword “(neoantigen OR neoepitope) AND cancer AND vaccin* AND patient*” updated on October 2020, a total of ten clinical neoantigen studies are summarized in the table. Routes of administration, formulations, combined therapies, cancer types, number of patients, number of epitopes vaccinated per patient, clinical, and immunological responses of each study are compared. Case reports and other trials that used tumor-associated antigens are not included.
| Route | Formulation | Scheduled timing | Dosing / nAg | Combined therapies | Cancer types | # Pts | # nAg / pt | Predicted HLA | Clinical responses | Immunological responses to nAgs | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide | |||||||||||
| SC | SLP + poly-ICLC | 7 doses • 5 priming (D 1, 4, 8, 15, 22) • 2 boosting (Wk 12, 20) |
300 μg | • Anti PD-1 | Melanoma | 6 | 13–20 | HLA-I | • 4 non-metastatic → no relapses • 2 lung metastatic → recurrences → complete response after pembrolizumab |
ELISPOT All de novo (60% CD4+, 16% CD8+) | Ott et al., 2017 [44] |
| ID | SLP + poly -ICLC + GM-CSF (APVAC2) | At least 8 doses (median =10) • 7 priming (D 1, 2, 4, 8, 15, 22, 36) • At least 1 boosting (q 4 wks) |
400 μg | • Shared TSAs (APVAC1) • Temozolomide |
Glioblastoma | 11 | 2 | HLA- A*02:01, A*24:02 | • 1 complete response • 2 partial response |
ICS 84.7% CD4+ (mostly Th1), (0% CD8+, 38.46% CD4+ and CD8+) | Hilf et al., 2019 [42] |
| SC | SLP + poly -ICLC | 7 doses • 5 priming (D 1, 4, 8, 15, 22) • 2 boosting (Wk 12, 20) |
300 μg | • Anti VEGF (6/8) | Glioblastoma | 8 | 7–20 | HLA-I | • All died from progressive disease • Median progression-free survival = 7.6 mths |
ELISPOT Dexamethasone 0% immunogenic No dexamethasone Mainly de novo CD4+ > CD8+ (N/A %) | Keskin et al., 2019 [43] |
| SC | SLP + GM-CSF | At least 7 doses • 5 priming (D 1, 4, 8, 15, 22) • At least 2 boosting (D 78, 162, andq 2–3 mths) |
• 100 μg (12/22) •300 μg (10/22) |
- | • Melanoma (4/22) • Colon (4/22) • NSCLC (3/22) • Pancreas (2/22) • Biliary tract (2/22) • Ovary (2/22) • Others (5/22) |
22 | 5–20 | HLA-I HLA-II | • 15 stable disease • 6 progressive disease • 1 withdraw |
ELISPOT 79.2% immunogenic | Fang et al., 2020 [41] |
| SC | SLP + poly -ICLC | 7 doses • 5 priming (D 1, 4, q 1 wk x 3) • 2 boosting (Wk 8, 12) |
450 μg | • Anti PD-1 | • Melanoma (27/60) • NSCLC (18/60) • Bladder (15/60) |
60 | Up to 20 | HLA-A HLA-B | Objective response rate • Melanoma 59% • NSCLC 39% • Bladder 27% |
ELISPOTImmunogenicity • Melanoma 52% • NSCLC 47% • Bladder 52% Mainly de novo(42% CD4+, 24% CD8+) |
Ott et al., 2020 [45] |
| RNA | |||||||||||
| IN | Naked | At least 8 doses (median =18) • 8 priming (D 1, 4, 8, 11, 15, 22, 29, 43) • Up to 12 boosting (N/A timing) |
• 500 μg (10/13) • 1,000 μg (3/13) |
• BRAF kinase • inhibitors (3/13) • Anti PD-1 (2/13) • Anti CTLA-4 (2/13) |
Melanoma | 13 | 10 | HLA-I HLA-II | • 8 recurrence-free • 5 relapses (before vaccination) • 2 complete response (1 with anti PD-1) • 1 partial response |
ELISPOT 60% immunogenic: 68% de novo (57% CD4+, 17% CD8+, 26% CD4+ and CD8+) | Sahin et al., 2017 [54] |
| DC | |||||||||||
| IV | DCs pulsed with peptides | 3 doses (D 1, 2, 3) | • First dose: 1.5 × 107 DC • Remaining: • 5 × 106 DC |
• Cyclophosphamide • Anti CTLA-4 |
Melanoma | 3 | 7 | HLA-A*02:01 | • 2 stable disease • 1 complete remission |
CD8+ Dextramer 42.86% immunogenic: 33.33% pre-existing, 66.67% de novo | Carreno et al., 2015 [60] |
| SC | DCs pulsed with peptides | At least 6 doses (q 3 wks) | 5–10 × 106 DC (total) | • Gemcitabine /capecitabine • Aspirin • Anti PD-1 |
Pancreas | 3 | 10 | HLA-I HLA-II | N/A | N/A | Bassani-Sternberg et al., 2019 [61] |
| SC | DCs pulsed with peptides + GM-CSF | At least 2 doses (q 4 wks) | 1 × 107 DC | • Gemcitabine • Cyclophosphamide • nAg-reactive CD8+CD137+ T cells |
• Pancreas (4/6) • Endometrium (1/6) • Thymus (1/6) |
6 | 1 | HLA-I HLA-II | • 1 complete remission • 1 partial response • 4 stable disease |
ELISPOT 16.67% immunogenic All de novo | Chen et al., 2019 [62] |
| IN | DCs pulsed with peptides | At least 6 doses (q 3–4 wks) | 5–10 × 106 DC (total) | • Cyclophosphamide • DCs pulsed with tumor lysate (only for Arm B) |
Ovary | 16 | 10 | HLA-I HLA-II | N/A | N/A | Sarivalasis et al., 2019 [63] |
| DNA | |||||||||||
| No publications | |||||||||||
2. NEOANTIGEN IDENTIFICATION
Neoantigen discovery starts with the identification of tumor-specific nonsynonymous genetic alterations (Figure 1). DNA and RNA are extracted from tumor biopsies and are sequenced using massively parallel next generation sequencing technologies (NGS). DNA is also extracted from peripheral blood mononuclear cells (PBMCs) and used for DNA sequencing and HLA haplotyping. Following alignment of tumor/normal sequencing data to the human reference genome, genetic alterations in the tumor can be detected using variant-calling algorithms. Candidate neoantigens can be prioritized by either computational in silico binding prediction, or less commonly using proteogenomic approaches including MS-based analysis of peptide-HLA (pHLA) immunopeptidomes [29,30]. For proteogenomic approaches, pHLA complexes are first immunoprecipitated following tumor lysis and homogenization. Peptides are then eluted and liquid chromatography with tandem mass spectrometry (LC-MS/MS) is performed to identify the amino acid sequences [31–33].This approach requires considerable amounts of tumor material, ranging from 5 × 107 to 1 × 109 cells per isolation, for HLA-peptide complex precipitation [33]. As tumor samples from patients are usually limited, direct identification of neoantigens by LC-MS/MS seems to be less practical compared to the bioinformatics-based in silico analyses.
Computational algorithms have been developed and are being optimized to allow the prioritization of candidate neoantigens that are likely to generate meaningful immune responses following vaccination. Most neoantigen identification pipelines incorporate algorithms that predict peptide processing, transport and binding affinity to HLA molecules. However, a recent collaborative study involving multiple independent research teams highlights that substantial discrepancies exist in current pipelines, and these discrepancies impact the ability to identify and prioritize immunogenic cancer neoantigens [34]. After systematic analysis of the pipelines, the Consortium identified five parameters that predict epitope immunogenicity. These include so-called presentation features (binding affinity, tumor abundance, and binding stability) and recognition features (agretopicity and foreignness). The term “agretopicity” refers to the difference in predicted epitope-MHC binding affinity between a mutant peptide and its wildtype counterpart. Prioritizing candidate neoantigens based on strong binding affinity, high binding stability and high tumor abundance, coupled with low agretopicity and/or high foreignness were shown to be able to significantly improve the performance of neoantigen prediction algorithms [34].
One such neoantigen prediction algorithm, pVACtools [35], consists of five components including pVACseq, pVACbind, pVACfuse, pVACvector, and pVACviz, has been developed and successfully applied to preclinical and clinical studies. In order to take advantages of neoantigen prediction algorithms like pVACtools, a robust next-generation sequencing pipeline is required to first identify nonsynonymous genetic alterations present in tumors resulting from missense, inframe insertion-deletion, protein-altering, and frameshift mutations. This sequencing pipeline typically includes 1) exome sequencing of tumor and normal DNA; 2) RNA or cDNA-capture sequencing, and 3) data analysis. After identification of genetic alterations, neoantigens are subsequently identified and prioritized using pVACseq [36,37]. Predicted amino acid sequences corresponding to the expressed mutations are pipelined through multiple class I and class II MHC epitope-binding algorithms provided by the Immune Epitope Database and Analysis Resource (http://www.immuneepitope.org) and other publicly available database. These include the class I epitope prediction algorithms (NetMHCpan, NetMHC, NetMHCcons, PickPocket, SMM, SMMPMBEC, MHCflurry, and MHCnuggets) and class II MHC epitope prediction algorithms (NetMHCIIpan, SMMalign, NNalign, and MHCnuggets). Each peptide’s “combined binding score” is defined as the median binding affinity score from all algorithms for each peptide for the best restricting allele. The IEDB consortium [38] has published recommended binding affinity (IC50) cutoffs for the 38 most common HLA-A and HLA-B alleles. For HLA-C alleles and MHC class II, the recommended IC50 cutoff values are 500nM and 1000nM, respectively. The next step is to rank-order the candidate neoantigens based on binding affinity (B), agretopicity (A), mutant allele expression (M) calculated as (geneCPM*MT_allele_RNA_VAF), and DNA VAF (D). The rank-ordered values (1 being the worst) of each criteria are used to generate a final ranking with the formula of Priority Score = B+A+(M*2)+(D/2). Additionally, minimum criteria are set for a candidate neoantigen to be included in the vaccine design. The criteria are: 1) a binding affinity score below the recommended cutoff for that allele; 2) presence in the founding clone or a significant percentage of cells of the tumor; and 3) observed expression of the gene and mutant allele in the RNA. In practice, mutation position may also be considered in the neoantigen selection process. The highest ranked peptides, up to the number that can be accommodated in a vaccine, are selected. In cases where fewer peptides are identified than desired for the vaccine, binding thresholds can be relaxed slightly to include more peptides, provided other criteria are met.
3. SLP VACCINE PLATFORM
The SLP vaccine platform is the most common neoantigen vaccine platform studied to date in preclinical studies and early phase clinical trials. The SLP vaccine platform has significant advantages including a proven safety profile, well characterized GMP manufacturing process, excellent stability, and straightforward administration in human clinical trials. Synthetic long peptides in cancer neoantigen vaccines are typically 20–30 amino acids in length. Peptides of this length may be preferentially processed and presented by antigen presenting cells [39], but they also have the advantage of being able to bind both MHC class I and II molecules with the potential to activate CD8 and/or CD4 T cells. After cleavage by the immunoproteasome and antigen processing, short peptides (usually 9–11 amino acids in length) bind to MHC class I molecules and are presented to CD8 T cells, while long peptides (usually 14–16 amino acids in length) bind to MHC class II molecules and are presented to CD4 T cells [40]. As a result, vaccine-induced CD4 T cells can enhance the priming and function of neoantigens-specific CD8 T cells.
Administration of synthetic peptides without adjuvant does not trigger toll-like receptors (TLR) or activate the innate immune system. Failure to active ate the innate immune system can lead to attenuated or very weak T cell responses. Thus, immune adjuvants need to be co-administered with peptide vaccines in order to induce robust immune responses [41]. Modern adjuvants include ligands for pattern recognition receptors (PRR), which target the APCs and consequently enhance the adaptive immune response by inducing the production of cytokines and chemokines that play a key role in T cell recruitment, priming, expansion and polarization. These adjuvants interact with and signal through specific receptors, providing a danger signal to the immune system which leads to the activation of transcription factors such NF-κB and IRF. One FDA-approved immune adjuvant is polyinosinic-polycytidylic acid-poly-L-lysine (poly-ICLC). As a double-stranded RNA complex, poly-ICLC is similar in structure to RNA viruses, and can be recognized by endosomal receptor TLR3 and cytoplasmic sensors MDA-5 and DHX/DDX RNA helicases [42]. Other immune adjuvants including cyclic dinucleotide (CDN), a potent stimulator of interferon genes (STING) agonist [43], have also been tested. Peptide vaccines are typically delivered via subcutaneous or intramuscular administration near draining lymph nodes [44] with a typical dose range of 300–450 μg per epitope [45–49]. Insights into better SLP vaccine strategies (dosing, frequency, adjuvant, administration, etc) are typically derived from animal studies and further refined in clinical trials. Despite some efforts [50], there is still a lack of systemic comparison of different vaccine regimens for optimal immune response and/or antitumor activity. Furthermore, the potential impact of concomitant therapies (chemotherapy, radiation therapy, and immunotherapy) on the effectiveness of SLP vaccines is not fully understood. The trend in the field is to adopt an SLP vaccine strategy with more frequent but decreased doses of vaccine as supported by computational simulation modeling [51].
Two preclinical studies by Castle et al. and Schreiber et al. demonstrated tumor protection following treatment with SLP-based neoantigen vaccines [24,52]. Castle et al. synthesized two 27-mer peptides (single mutated amino acid at central position flanked by 13 non-mutated amino acids on both sides) and tested these SLP vaccines in B16F10-bearing C57BL/6 mice in prophylactic and therapeutic settings. Vaccines were injected subcutaneously with poly-IC. Neoantigen-specific immune responses were strong enough to inhibit tumor growth in both settings. T cell responses directed at the cancer neoantigens were significantly higher compared to the corresponding wildtype sequences [24]. Gubin et al. conducted similar experiments in an MCA sarcoma cell line. Two H-2Kb-restricted peptides, Lama4 and Alg8, were identified and co-administered with poly-IC. Vaccination was able to elicit antitumor responses in both prophylactic and therapeutic settings [52].
Recent publications have confirmed the therapeutic potential of SLP neoantigen vaccines in several types of human cancer including melanoma, glioblastoma, non-small cell lung cancer (NSCLC), colorectal cancer, and urothelial cancer [45–49]. Melanoma is an attractive target for neoantigen vaccines as it possesses a very high tumor mutational burden and is known to be responsive to cancer immunotherapies. In one study co-led by Wu and Ott [NCT01970358], six treatment-naïve high-risk melanoma patients were vaccinated with 13–20 SLP and poly-ICLC after surgery. The neoantigens were selected based on predicted binding affinity to MHC class I and were administered subcutaneously in prime (5-dose) and boost (2-dose) phases. IFN-gamma ELISPOT assay and intracellular cytokine staining detected polyfunctional CD4 and CD8 T cells targeting 60% and 16%, respectively, of all unique neoantigens across the six patients. Of note, the CD4 T cell response rate was higher than the CD8 T cell response rate despite the fact that the cancer neoantigens were prioritized based on predicted HLA class I binding. This may reflect the fact that MHC class II binding is known to be highly promiscuous. Two years following SLP vaccination, four non-metastatic patients were still free from relapse. The other two patients with metastatic disease encountered disease progression but later experienced clinical responses after receiving four doses of the anti-PD1 checkpoint inhibitor, pembrolizumab [48]. Similar immune responses and clinical outcomes have been observed following vaccination with neoantigen SLP vaccines in other cancer types since this initial report [45,49].
Two important studies of neoantigen SLP vaccines have shown encouraging results in glioblastoma [46,47]. The Glioma Actively Personalized Vaccine Consortium (GAPVAC) in Europe conducted a clinical trial (NCT02149225), in which 15 newly diagnosed glioblastoma patients positive for HLA-A*02:01 or HLA-A*24:02 were vaccinated with conventional tumor associated antigens (APVAC1) followed by cancer neoantigens (APVAC2). All patients underwent surgical resection and received standard adjuvant chemotherapy (temozolomide). The SLP vaccines (APVAC1 and APVAC2) were administered by intradermal injection. Poly-ICLC (s.c.) and GM-CSF (i.d.) were used as adjuvants and applied near the vaccination sites. Although APVAC1 was able to elicit sustained CD8 responses with central memory phenotype, the APVAC2 induced predominantly neoantigen-specific CD4 T cell responses. Out of all neoantigens, 84.7% (11/13) of the vaccinated APVAC2 neoantigens were able to elicit CD4 responses, most of which were polyfunctional with a Th1 phenotype [46]. In the US, neoantigen SLP vaccines were used to treat glioblastoma patients in a phase 1b trial [NCT02287428]. Although all patients eventually died of disease progression, neoantigen SLP vaccines significantly increased the number of TILs and induced strong multifunctional de novo CD4 and CD8 responses against neoantigens in those patients who were not receiving dexamethasone for cerebral edema [47].
4. RNA VACCINE PLATFORM
The RNA vaccine platform has a number of important advantages that make it an attractive platform for cancer neoantigen vaccines [53,54]. For example, RNA vaccine design and manufacture is relatively straightforward and cost effective. RNA vaccines can be produced by in vitro transcription (IVT) using DNA templates derived from synthesized DNA fragments or linearized plasmid DNA [27,55]. RNA vaccines are designed to enter the cytosol where translation of the neoantigen peptides occurs. The RNA does not need to enter the nucleus, minimizing the risk of integration into the host genome [56]. Strategies to enhance stability of RNA vaccines include the use of modified nucleosides, 5’-capping, and formulation into liposomes. RNA vaccines bind directly to TLR7 and have an inherent ability to provide an adjuvant effect, and do not require additional adjuvants [57,58]. RNA vaccines can be administered as naked RNA, but are more often encapsulated into lipid nanoparticles.
RNA vaccines can be administered by various routes of administration (intradermal, intravenous, intramuscular, and intranodal), and RNA uptake appears to be dependent on the route of administration. Intravenous and intranodal administration of RNA vaccines allows direct access of RNA to APCs in lymphoid organs. On the other hand, RNA administered via intramuscular and intradermal routes will require uptake by infiltrating APCs and subsequent transport to draining lymph nodes and other lymphoid organs. Studies have shown that, for naked RNA vaccines, injection into lymph nodes (intranodal) led to the most robust T cell responses [59]. Alternatively, liposomal RNA vaccines delivered intravenously were able to target DCs in the lymphoid tissues and induce robust T cell responses [27]. RNA vaccines preferentially target professional antigen presenting cells (e.g. DCs), resulting in cancer neoantigen presentation in the context of MHC class I and class II complexes. RNA vaccines can induce potent innate type I interferon immune responses through the activation of toll-like receptor signaling pathways (TLR3, 7, and 8) [27]. Type I interferon responses are associated with both inflammation and potentially autoimmunity. Therefore, investigators have been cautious when translating RNA vaccines to clinical practice.
BioNTech and Moderna are biotechnology firms that have pioneered the RNA vaccine platform. In a preclinical study, Kreiter et al. demonstrated the efficacy of RNA vaccines in three murine cancer models (B16F10 melanoma, CT 26 colon carcinoma, and 4T1 breast carcinoma). RNA vaccines encoding 27-mer neoantigens were formulated in cationic liposomes and delivered intravenously. RNA vaccines were able to induce robust neoantigen-specific CD4 and CD8 T cell responses, with CD4 T cell responses being predominant. The strong immune responses were associated with antitumor immunity and increased survival of tumor-bearing mice [27]. Recently, Sahin et al. reported favorable outcomes of a phase 1 clinical trial treating patients with metastatic melanoma with a nanoparticle RNA vaccine expressing four melanoma tumor associated antigens (TAA) [60]. The RNA vaccine was formulated in cationic liposomes and was administered intravenously to patients who were previously treated with anti-PD1 immune checkpoint inhibitors. The RNA vaccine was able to generate TAA-specific immune responses in the majority of patients (39/50 or 78%). Among 17 patients treated with the vaccine in combination with anti-PD1 blockade, six (35%) developed a partial response. As with many early phase clinical trials, this study used a dose-escalating regimen. Patients were treated with eight infusions of RNA vaccine within 64 days with RNA doses ranging from14.4 μg to 400 μg. Some patients also received optional continued vaccinations. The development of an optimal RNA vaccine platform for the treatment of cancer is still under investigation. No consensus has been reached in terms of dosing, formulation and administration. Of note, two SARS-CoV2 (COVID-19) vaccines recently approved for emergency use in the US are based on the same mRNA vaccine platform using LNP as a nucleic acid carrier [61,62]. These vaccines are the BNT162b2 vaccine by Pfizer-BioNTech and the mRNA-1273 vaccine by Moderna.
Although clinical studies of RNA vaccines to date have focused on targeting TAA, there is an increasing interest in testing RNA vaccines targeting cancer neoantigens. For example, Sahin et al. treated 13 stage III-IV melanoma patients with an RNA vaccine targeting cancer neoantigens in combination with anti PD-1, anti CTLA-4, and BRAF kinase inhibitors [63]. Ten neoantigens per patient were selected based on predicted binding affinity to HLA class I and HLA class II. Each RNA vaccine encoded five 27-mer neoantigens which were connected via linkers. All patients were vaccinated with 0.5 or 1 mg mRNA per vaccination with a maximum of 20 vaccine doses by ultrasound-guided intranodal injection into both inguinal lymph nodes. Of note, during the vaccine production period, patients with tumors expressing NY-ESO-1 or tyrosinase also received the mRNA-based vaccine encoding these TAAs. IFN-γ ELISPOT data indicated that 60% of all targeted neoantigens were immunogenic. Neoantigen-specific immune responses were detectable in all patients, and were mostly de novo (68%). CD4 T cell responses were predominant (57%), compared to CD8 T cell responses. RNA vaccination was associated with prolonged progression-free survival (PFS). Several patients who did progress developed clinical responses after receiving mRNA vaccines alone or in combination with immune checkpoint inhibitors [63].
5. DC VACCINE PLATFORM
Dendritic cells are the most potent APCs and autologous DCs generated ex vivo have been studied as cell-based cancer vaccines. Monocyte-derived DC can be expanded and matured from leukapheresis specimens using a cocktail of IL-4, GM-CSF and TNF-alpha cytokines [64]. However, ex vivo generation of DCs is complicated and patient variability can impact reproducibility [25]. It is also costly and time-consuming to generate the large number of DC required for vaccination [65]. Cancer neoantigens can be loaded onto DC by pulsing DC with synthetic peptides or with tumor lysate. Alternatively, DC can be transfected with RNA encoding cancer neoantigens by electroporation [64,66]. Synthetic RNA expressing molecules present on activated DC such as TLR4, CD40L, and CD70 can also be transfected into DC [67].
The ex vivo expanded, matured and neoantigen-loaded DC can then be administered to the patient. Routes of administration for DC-based vaccines include intranodal, intradermal, intravenous, or intralymphatic injection [65]. While timing and dosing are being investigated, most clinical trials applied 5–10 × 106 DCs per dose every 3–4 weeks [68–71]. Once administered, DC must travel to lymphoid tissues in order to stimulate antigen-specific T cells. Although the mechanism of DC trafficking following injection remains poorly understood, studies indicate that pre-conditioning of the vaccination site with recall antigens such as tetanus toxoid appeared to facilitate DC homing to lymph nodes in both human and mice, a chemokine CCL3-dependent process [72]. In addition to directly stimulating CD4 and CD8 T cells, injected DCs can interact with endogenous lymph node-resident DCs by transferring antigens to and stimulating IL-12 production in resident XCR1+ cDC1 [73]. Animal studies have documented the requirement of IL-12 produced by host DC for DC vaccine-induced Th1 response [73]. This may be of particular relevance for DC-based vaccination for cancer as cancer patients may have impaired endogenous DC function due to prior or concomitant treatments.
Carreno et al. were the first to treat cancer patients with cancer neoantigen DC vaccines [NCT00683670] [69]. Immature DCs were generated in culture containing GM-CSF and IL-4 from three stage 3C cutaneous melanoma patients positive for HLA-A*02:01. DC maturation was induced by IFN-γ, poly-IC, R848 and CD40L. Two hours prior to intravenous administration, mature DCs were pulsed with neoantigens identified for each patient. Cyclophosphamide was administered prior to the first dose of DC in an effort to deplete Treg. HLA-A*02:01-peptide dextramer staining of pre- and post- vaccine PBMC revealed that DC vaccine can augment pre-existing as well as induce de novo neoantigen-specific CD8 T cell responses. TCR-β sequencing data showed an increase in both frequency and repertoire for dominant and subdominant neoantigens. Unfortunately, tumor regression was not monitored in this study because tumors were resected before vaccination. This study, together with a few others [68,70,71], has documented safety profile of DC-based neoantigen cancer vaccine. Neoantigen vaccines based on DC platform are currently under investigation to treat patients with TNBC [NCT04105582], hepatocellular carcinoma [NCT03674073], and NSCLC [NCT04078269].
6. DNA VACCINE PLATFORM
Plasmid DNA vaccines are relatively easy and cost effective to manufacture compared to other conventional vaccines [74], making the DNA vaccine platform attractive for neoantigen vaccines. Similar to RNA vaccines, plasmid DNA can be readily engineered to encode multiple neoantigens. Additional immune modulators can also be integrated into the vaccine to augment immune responses. In order to elicit maximal immune responses, DNA vaccines integrate potent eukaryote promoters, a strong polyadenylation/transcriptional termination signal, and codon-optimized gene sequences. Despite initial concerns that plasmid DNA might integrate into patients’ genomes, the DNA vaccine platform has an extraordinary safety profile in clinical translation to date. Initial studies confirmed that the probability of human genome integration is extremely low for DNA vaccines, at or even lower than that of spontaneous mutations [75]. DNA vaccine manufacture involves purification of plasmid DNA from bacterial cultures. Plasmid DNA is most commonly administered intramuscularly or intradermally. Electroporation devices are commonly used to improve DNA uptake and antigen expression by nucleated skin or muscle cells. DNA vaccines can also be formulated in liposomes for mucosal delivery or intravenous administration. Successful translation of DNA vaccines requires optimal design of the vector and vaccine insert, effective delivery to target APCs, and maximal antigen expression, processing and presentation. Although the DNA vaccine platform has demonstrated its potential in preventing infection and treating tumors in preclinical models, the efficacy of DNA vaccine in generating immune response in human is currently under investigation.
Most DNA-based cancer vaccine studies have targeted tumor-associated antigens rather than cancer neoantigens, such as HPV E6 and E7 in cervical cancer, mammaglobin-A in breast cancer, and HER2/CEA in solid cancer [76–79]. Duperret et al. studied polyepitope DNA vaccines targeting cancer neoantigens in three murine cancer cell lines, TC1, LLC, and ID8. Polyepitope inserts encoding 33-mer neoantigens separated by furin cleavage sites were cloned into the pVAX plasmid backbone. DNA vaccines were able to induce robust neoantigen-specific CD8 T cell responses and antitumor immunity as measured by ELISPOT assay and tumor challenge [80]. Li et al. have also tested an optimized polyepitope DNA vaccine targeting cancer neoantigens identified in murine breast cancer E0771 and 4T1 models. Vaccination with DNA vaccine was able to induce robust neoantigen-specific T cell responses to some but not all candidate neoantigens. In combination with anti-PD-L1, polyepitope DNA vaccines were able to suppress tumor growth [81]. We and others are conducting clinical trials using neoantigen DNA vaccines to treat cancer patients with pancreatic carcinoma [NCT03122106], TNBC [NCT03199040], and glioblastoma [NCT04015700]. In patients with TNBC, vaccination with neoantigen DNA vaccines is associated with robust immune responses and prolongation of progression-free survival.
7. OTHER VACCINE PLATFORMS
In addition to the four major vaccine platforms discussed above, other platforms are being explored for targeting neoantigens. Viral vectors can be modified to express neoantigens. Among viral vectors, replication-incompetent adenoviruses (Ad2 and Ad5) have been popular in delivering tumor antigens in clinical trials with or without co-stimulatory molecules. Because most adults have pre-existing immunity to adenovirus, new vectors derived from chimpanzee and gorilla adenovirus are being developed. Another viral platform is the attenuated poxvirus MVA (Modified Vaccinia Ankara). MVA has been extensively studied (e.g. heterologous prime-boost strategy) in animal models and in patients, and has demonstrated safety. In addition to viral vectors, bacteria such as Salmonella typhimurium and Listeria monocytogenes have also been tested as vectors to express tumor antigens. These platforms are in the early stages of clinical development and their potential in targeting neoantigens has not been studied.
The use of liposomes as carriers has been investigated for decades. Recent studies have demonstrated that RNA-LPX [27,60] and SLP-LPX [82] vaccines preferentially deliver the RNA/SLP cargos to lymphoid organs and induce potent CD8 and CD4 T cell responses and antitumor activity in both preclinical and clinical settings. Therefore, nanoparticle liposomes represent a novel and promising new neoantigen vaccine platform that warrants further investigation..
8. CONCLUSION
Current neoantigen vaccine platforms have distinct mechanisms of action, advantages and disadvantages. Although multiple platforms are currently being investigated, it remains unclear which strategy will prove superior in terms of feasibility in clinical translation, and ability to induce robust immune and clinical responses. Data from ongoing early stage clinical trials are very promising. Severe immune-related adverse events (irAEs) have not been observed, and mild irAEs are generally reversible and are typically associated with combination therapies such as immune checkpoint inhibitors [83]. Additional study is required to determine the optimal platform, formulation and dosing that will allow successful integration of neoantigen vaccines into broader clinical practice. Finally, even optimized neoantigen vaccines alone may not be adequate as monotherapy for cancer treatment. The success of neoantigen vaccines may depend on combination with other cancer immunotherapies such as immune checkpoint inhibition, and/or immune modulatory agents targeting the tumor microenvironment.
9. EXPERT OPINION
The evolution of sequencing technologies has made it possible to identify cancer neoantigens and study their biology. This has led to the important recognition that cancer neoantigens play a critical role in the response to cancer immunotherapies. This has generated significant interest in the development and clinical translation of neoantigen vaccines. Initial clinical experience is promising across multiple different neoantigen vaccine platforms. All appear capable of generating immune responses.
Significant challenges remain to the successful translation of neoantigen vaccines into broader clinical practice. Challenges include limitations in neoantigen identification and prioritization, integration of neoantigen vaccines into an appropriate clinical context, and defining how to best overcome regulatory networks in the tumor microenvironment restraining antitumor immune responses. Cancer sequencing is a critical first step in neoantigen identification and prioritization. Clinical-grade cancer sequencing is now much more widely available and decreasing in price. It is likely that cancer sequencing will be increasingly integrated into clinical treatment paradigms. Still, the quality of cancer sequencing remains highly dependent on the quality of the biopsy, tumor purity, and even tumor heterogeneity [84]. Variant calling algorithms are excellent for identification of single nucleotide variants, but are less refined for identification of gene fusions, indels and other more complex genetic alterations. As highlighted in the early stage clinical trials above, current neoantigen prediction algorithms need to continue to be refined so the most immunogenic cancer neoantigens can be prioritized for inclusion in cancer vaccines. This is true for algorithms predicting immunogenic cancer neoantigens restricted by both MHC class I and II alleles. Even with improved neoantigen identification algorithms, there may not be sufficient neoantigens in cancers with low mutational burden to target effectively.
Design and manufacture of neoantigen vaccines is a complex, time consuming, and resource-intensive process. Steps in neoantigen vaccine design and manufacture include obtaining archival tumor tissue or performing a dedicated tumor biopsy, nucleic acid isolation, tumor/normal sequencing, variant calling, neoantigen prediction, vaccine design, vaccine manufacture, and product release tests. This entire process can take 3–6 months [48,63], even if all steps are expedited. Patient selection is critical. Ideally, appropriate clinical contexts can be identified such that patients can be identified early, and vaccine treatment can reliably begin after vaccine manufacture has been successfully completed. Many early stage clinical trials have been performed in the adjuvant setting. Typically, patients are treated with standard of care therapies and have no evidence of disease following treatment but are at high risk of disease recurrence. These patients are candidates for neoantigen vaccine therapy given the high risk of disease recurrence. Alternatively, patients with metastatic disease may be candidates for vaccine therapy. These patients would typically need to be treated with chemotherapy or other systemic therapy to control disease, providing a window of opportunity to complete the steps required for vaccine design and manufacture. Unfortunately, patients with metastatic disease and significant comorbidities and/or decreased performance status related to disease may not be realistic candidates for neoantigen vaccine therapy.
Neoantigen vaccine therapy appears to be extremely well tolerated with minimal side effects and/or adverse events. As such, neoantigen vaccines can be used alone (for example in the adjuvant setting), or can be readily combined with other treatments. Based on mechanism of action, it is likely that neoantigen vaccines will be particularly effective in combination with cancer immunotherapies [85–88]. The tumor microenvironment is rarely hospitable to tumor-specific T cells. It is likely that cancer immunotherapies designed to abrogate immune checkpoint pathways and/or target the immunosuppressive tumor microenvironment will be particularly effective. Early stage clinical trials are ongoing combining neoantigen vaccines and immune checkpoint inhibitors. Targeting CTLA4 may facilitate neoantigen-specific T cell priming, while targeting PD-1/L1 may prevent exhaustion once T cells arrive in the tumor. Strategies targeting tumor associated macrophages, and/or myeloid-derived suppressor cells are also likely to be important.
Although a single vaccine platform may ultimately prove to be superior, our Expert Opinion is that addressing some of the challenges highlighted above may move the field forward more quickly rather than focusing exclusively on optimizing a single vaccine platform. In our opinion two of the most significant challenges that need to be addressed include limitations in neoantigen prediction algorithms, and the need to successfully target the immunosuppressive tumor microenvironment. For example, independent of vaccine platform, most neoantigens prioritized by current neoantigen prediction algorithms fail to induce immune responses. Similarly, neoantigen-specific T cell responses may not be effective in mediating antitumor immunity if the neoantigen-specific T cells are suppressed in the tumor microenvironment. Ultimately, the most effective vaccine platform may be dependent on the specific cancer and/or clinical context that is being investigated.
ARTICLE HIGHLIGHTS.
The goal of cancer neoantigen vaccines is to induce neoantigen-specific immune responses and antitumor immunity, while minimizing the potential for autoimmune toxicity.
For a successful neoantigen cancer vaccine, neoantigen identification, prioritization, and delivery platform are important components in a vaccine manufacturing workflow.
Several neoantigen vaccine platforms are currently being evaluated in early phase clinical trials including the synthetic long peptide (SLP), RNA, dendritic cell (DC), and DNA vaccine platforms, and it remains unclear if one platform is superior.
Although each vaccine platform has different formulations, routes of administration, and mechanism(s) of action, they share a common goal of presentation of cancer neoantigens to T cells in the context of appropriate stimulatory signals.
The success of neoantigen vaccines may depend on combination with other cancer immunotherapies such as immune checkpoint inhibition, and/or immune modulatory agents targeting the tumor microenvironment.
Acknowledgments
Funding
This project was supported by grants from Susan G. Komen for the Cure (KG111025), the Alvin J. Siteman Cancer Center (Siteman Investment Program grant 4035), the National Cancer Institute at the National Institute of Health (Cancer Center Support Grant P30-CA091842 and SPORE in Pancreatic Cancer P50-CA196510), the Foundation for Barnes-Jewish Hospital (to SPG), and Prince Mahidol Award Youth Program of Prince Mahidol Award Foundation under the Royal Patronage of HM the king of Thailand (to SS).
Footnotes
Declaration of Interests
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial relationships or otherwise to disclose.
REFERENCES
Papers of particular interest are identified using one or two asterisk symbols(* = of interest, ** = of considerable interest).
- 1.Jiang T, Shi T, Zhang H, et al. Tumor neoantigens: from basic research to clinical applications. J Hematol Oncol. 2019. September 6;12(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Efremova M, Finotello F, Rieder D, et al. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front Immunol. 2017;8:1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelova M, Mlecnik B, Vasaturo A, et al. Evolution of Metastases in Space and Time under Immune Selection. Cell. 2018. October 18;175(3):751–765 e16. [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019. March;16(3):151–167. [DOI] [PubMed] [Google Scholar]
- 5.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015. April 3;348(6230):74–80. [DOI] [PubMed] [Google Scholar]
- 6.Coulie PG, Van den Eynde BJ, van der Bruggen P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014. February;14(2):135–46. [DOI] [PubMed] [Google Scholar]
- 7.Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol. 2019. June;234(6):8509–8521. [DOI] [PubMed] [Google Scholar]
- 8.Feldman SA, Assadipour Y, Kriley I, et al. Adoptive Cell Therapy--Tumor-Infiltrating Lymphocytes, T-Cell Receptors, and Chimeric Antigen Receptors. Semin Oncol. 2015. August;42(4):626–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasetto A, Gros A, Robbins PF, et al. Tumor- and Neoantigen-Reactive T-cell Receptors Can Be Identified Based on Their Frequency in Fresh Tumor. Cancer Immunol Res. 2016. September 2;4(9):734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto TN, Kishton RJ, Restifo NP. Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat Med. 2019. October;25(10):1488–1499. [DOI] [PubMed] [Google Scholar]
- 11.Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017. November 2;171(4):934–949 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogen B, Fauskanger M, Haabeth OA, et al. CD4(+) T cells indirectly kill tumor cells via induction of cytotoxic macrophages in mouse models. Cancer Immunol Immunother. 2019. November;68(11):1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015. April 30;520(7549):692–6.** preclinical study using RNA vaccine targeting MHC class II epitopes
- 14.Ostroumov D, Fekete-Drimusz N, Saborowski M, et al. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell Mol Life Sci. 2018. February;75(4):689–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2020. May 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veatch JR, Jesernig BL, Kargl J, et al. Endogenous CD4(+) T Cells Recognize Neoantigens in Lung Cancer Patients, Including Recurrent Oncogenic KRAS and ERBB2 (Her2) Driver Mutations. Cancer Immunol Res. 2019. June;7(6):910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linnemann C, van Buuren MM, Bies L, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015. January;21(1):81–5. [DOI] [PubMed] [Google Scholar]
- 18.Ferris ST, Durai V, Wu R, et al. cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature. 2020. August;584(7822):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alspach E, Lussier DM, Miceli AP, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 2019. October;574(7780):696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busselaar J, Tian S, van Eenennaam H, et al. Helpless Priming Sends CD8(+) T Cells on the Road to Exhaustion. Front Immunol. 2020;11:592569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quezada SA, Simpson TR, Peggs KS, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010. March 15;207(3):637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodge K, Supabphol S, Kumar P, et al. Recent developments in neoantigen-based cancer vaccines. Asian Pac J Allergy Immunol. 2020. June;38(2):91–101. [DOI] [PubMed] [Google Scholar]
- 23.Peng M, Mo Y, Wang Y, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. 2019. August 23;18(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castle JC, Kreiter S, Diekmann J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012. March 1;72(5):1081–91.* demonstrates antitumor activity of SLP neoantigen vaccines in preclinical models
- 25.Li L, Goedegebuure SP, Gillanders WE. Preclinical and clinical development of neoantigen vaccines. Ann Oncol. 2017. December 1;28(suppl_12):xii11–xii17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014. November 27;515(7528):572–6.* combines both computational and proteomic approaches to design SLP neoantigen vaccines
- 27.Kranz LM, Diken M, Haas H, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016. June 16;534(7607):396–401.** despcibes liposomal RNA vaccines targeting TAA in preclinical and clinical settings
- 28.Fennemann FL, de Vries IJM, Figdor CG, et al. Attacking Tumors From All Sides: Personalized Multiplex Vaccines to Tackle Intratumor Heterogeneity. Front Immunol. 2019;10:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roudko V, Greenbaum B, Bhardwaj N. Computational Prediction and Validation of Tumor-Associated Neoantigens. Front Immunol. 2020;11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Qi Y, Zhang Q, et al. Application of mass spectrometry-based MHC immunopeptidome profiling in neoantigen identification for tumor immunotherapy. Biomed Pharmacother. 2019. December;120:109542. [DOI] [PubMed] [Google Scholar]
- 31.Abelin JG, Keskin DB, Sarkizova S, et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity. 2017. February 21;46(2):315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassani-Sternberg M, Braunlein E, Klar R, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun. 2016. November 21;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell AW, Ramarathinam SH, Ternette N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc. 2019. June;14(6):1687–1707. [DOI] [PubMed] [Google Scholar]
- 34.Wells DK, van Buuren MM, Dang KK, et al. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell. 2020. October 29;183(3):818–834 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hundal J, Kiwala S, McMichael J, et al. pVACtools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunol Res. 2020. March;8(3):409–420.* describes the development of the pVACtools
- 36.Hundal J, Carreno BM, Petti AA, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016. January 29;8(1):11.* descibes the pVAC-seq algorithm
- 37.Hundal J, Kiwala S, Feng YY, et al. Accounting for proximal variants improves neoantigen prediction. Nat Genet. 2019. January;51(1):175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleri W. Selecting thresholds (cut-offs) for MHC class I and II binding predictions [Web Page]. Immune Epitope Database and Analysis Resource. Available from: https://help.iedb.org/hc/en-us/articles/114094151811-Selecting-thresholds-cut-offs-for-MHC-class-I-and-II-binding-predictions [Google Scholar]
- 39.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz FM, Colbert JD, Merino E, et al. The Biology and Underlying Mechanisms of Cross-Presentation of Exogenous Antigens on MHC-I Molecules. Annu Rev Immunol. 2017. April 26;35:149–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azmi F, Ahmad Fuaad AA, Skwarczynski M, et al. Recent progress in adjuvant discovery for peptide-based subunit vaccines. Hum Vaccin Immunother. 2014;10(3):778–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena M, Sabado RL, La Mar M, et al. Poly-ICLC, a TLR3 Agonist, Induces Transient Innate Immune Responses in Patients With Treated HIV-Infection: A Randomized Double-Blinded Placebo Controlled Trial. Front Immunol. 2019;10:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinkead HL, Hopkins A, Lutz E, et al. Combining STING-based neoantigen-targeted vaccine with checkpoint modulators enhances antitumor immunity in murine pancreatic cancer. JCI Insight. 2018. October 18;3(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneble E, Clifton GT, Hale DF, et al. Peptide-Based Cancer Vaccine Strategies and Clinical Results. Methods Mol Biol. 2016;1403:797–817. [DOI] [PubMed] [Google Scholar]
- 45.Fang Y, Mo F, Shou J, et al. A Pan-cancer Clinical Study of Personalized Neoantigen Vaccine Monotherapy in Treating Patients with Various Types of Advanced Solid Tumors. Clin Cancer Res. 2020. September 1;26(17):4511–4520.** clinical study of SLP-based neoantigen cancer vaccine
- 46.Hilf N, Kuttruff-Coqui S, Frenzel K, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019. January;565(7738):240–245.** clinical study of SLP-based neoantigen cancer vaccines
- 47.Keskin DB, Anandappa AJ, Sun J, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019. January;565(7738):234–239.** clinical study of SLP-based neoantigen cancer vaccine
- 48.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017. July 13;547(7662):217–221.** clinical study of SLP-based neoantigen cancer vaccine
- 49.Ott PA, Hu-Lieskovan S, Chmielowski B, et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell Lung Cancer, or Bladder Cancer. Cell. 2020. October 15;183(2):347–362 e24.** clinical study of SLP-based neoantigen cancer vaccine
- 50.Wages NA, Slingluff CL Jr., Bullock TN, et al. Tailoring early-phase clinical trial design to address multiple research objectives. Cancer Immunol Immunother. 2020. January;69(1):95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumbhari A, Kim PS, Lee PP. Optimisation of anti-cancer peptide vaccines to preferentially elicit high-avidity T cells. J Theor Biol. 2020. February 7;486:110067. [DOI] [PubMed] [Google Scholar]
- 52.Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014. November 27;515(7528):577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson NAC, Kester KE, Casimiro D, et al. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNamara MA, Nair SK, Holl EK. RNA-Based Vaccines in Cancer Immunotherapy. J Immunol Res. 2015;2015:794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez-Aguado I, Rodriguez-Castejon J, Vicente-Pascual M, et al. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials (Basel). 2020. February 20;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinle H, Behring A, Schlensak C, et al. Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges. Stem Cells. 2017. January;35(1):68–79. [DOI] [PubMed] [Google Scholar]
- 57.Kowalczyk A, Doener F, Zanzinger K, et al. Self-adjuvanted mRNA vaccines induce local innate immune responses that lead to a potent and boostable adaptive immunity. Vaccine. 2016. July 19;34(33):3882–93. [DOI] [PubMed] [Google Scholar]
- 58.Schlake T, Thess A, Fotin-Mleczek M, et al. Developing mRNA-vaccine technologies. RNA Biol. 2012. November;9(11):1319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreiter S, Selmi A, Diken M, et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res. 2010. November 15;70(22):9031–40. [DOI] [PubMed] [Google Scholar]
- 60.Sahin U, Oehm P, Derhovanessian E, et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020. July 29.** clinical study of liposomal RNA vaccines targeting TAA in melanoma
- 61.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med. 2020. November 12;383(20):1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polack FP, Thomas SJ, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020. December 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017. July 13;547(7662):222–226.** clinical study of RNA-based neoantigen cancer vaccine
- 64.Nair S, Archer GE, Tedder TF. Isolation and generation of human dendritic cells. Curr Protoc Immunol. 2012. November;Chapter 7:Unit 7 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santos PM, Butterfield LH. Dendritic Cell-Based Cancer Vaccines. J Immunol. 2018. January 15;200(2):443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tran E, Turcotte S, Gros A, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014. May 9;344(6184):641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Keersmaecker B, Claerhout S, Carrasco J, et al. TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: link between T-cell activation and clinical responses in advanced melanoma. J Immunother Cancer. 2020. February;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bassani-Sternberg M, Digklia A, Huber F, et al. A Phase Ib Study of the Combination of Personalized Autologous Dendritic Cell Vaccine, Aspirin, and Standard of Care Adjuvant Chemotherapy Followed by Nivolumab for Resected Pancreatic Adenocarcinoma-A Proof of Antigen Discovery Feasibility in Three Patients. Front Immunol. 2019;10:1832.** clinical study of DC-based neoantigen vaccine
- 69.Carreno BM, Magrini V, Becker-Hapak M, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015. May 15;348(6236):803–8.** first clinical study using DC-based neoantigen vaccine
- 70.Chen F, Zou Z, Du J, et al. Neoantigen identification strategies enable personalized immunotherapy in refractory solid tumors. J Clin Invest. 2019. May 1;129(5):2056–2070.** clinical study of DC-based neoantigen vaccine
- 71.Sarivalasis A, Boudousquie C, Balint K, et al. A Phase I/II trial comparing autologous dendritic cell vaccine pulsed either with personalized peptides (PEP-DC) or with tumor lysate (OC-DC) in patients with advanced high-grade ovarian serous carcinoma. J Transl Med. 2019. November 26;17(1):391.** clinical study of DC-based neoantigen vaccine
- 72.Mitchell DA, Batich KA, Gunn MD, et al. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature. 2015. March 19;519(7543):366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ashour D, Arampatzi P, Pavlovic V, et al. IL-12 from endogenous cDC1, and not vaccine DC, is required for Th1 induction. JCI Insight. 2020. May 21;5(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopes A, Vandermeulen G, Preat V. Cancer DNA vaccines: current preclinical and clinical developments and future perspectives. J Exp Clin Cancer Res. 2019. April 5;38(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faurez F, Dory D, Le Moigne V, et al. Biosafety of DNA vaccines: New generation of DNA vectors and current knowledge on the fate of plasmids after injection. Vaccine. 2010. May 21;28(23):3888–95. [DOI] [PubMed] [Google Scholar]
- 76.Diaz CM, Chiappori A, Aurisicchio L, et al. Phase 1 studies of the safety and immunogenicity of electroporated HER2/CEA DNA vaccine followed by adenoviral boost immunization in patients with solid tumors. J Transl Med. 2013. March 8;11:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tiriveedhi V, Fleming TP, Goedegebuure PS, et al. Mammaglobin-A cDNA vaccination of breast cancer patients induces antigen-specific cytotoxic CD4+ICOShi T cells. Breast Cancer Res Treat. 2013. February;138(1):109–18.* clinical study of a DNA vaccine targeting TAA in breast cancer
- 78.Tiriveedhi V, Tucker N, Herndon J, et al. Safety and preliminary evidence of biologic efficacy of a mammaglobin-a DNA vaccine in patients with stable metastatic breast cancer. Clin Cancer Res. 2014. December 1;20(23):5964–75.* clinical study of a DNA vaccine targeting TAA in breast cancer
- 79.Trimble CL, Morrow MP, Kraynyak KA, et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015. November 21;386(10008):2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duperret EK, Perales-Puchalt A, Stoltz R, et al. A Synthetic DNA, Multi-Neoantigen Vaccine Drives Predominately MHC Class I CD8(+) T-cell Responses, Impacting Tumor Challenge. Cancer Immunol Res. 2019. February;7(2):174–182.* preclinical study of a polyepitope DNA vaccine
- 81.Li L, Zhang X, Wang X, et al. Optimized polyepitope neoantigen DNA vaccines elicit neoantigen-specific immune responses in preclinical models and in clinical translation. Genome Med. 2021, April 21;13(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arbelaez CA, Estrada J, Gessner MA, et al. A nanoparticle vaccine that targets neoantigen peptides to lymphoid tissues elicits robust antitumor T cell responses. NPJ Vaccines. 2020. November 12;5(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lopez JS, Camidge R, Iafolla M, et al. Abstract CT301: A phase Ib study to evaluate RO7198457, an individualized Neoantigen Specific immunoTherapy (iNeST), in combination with atezolizumab in patients with locally advanced or metastatic solid tumors. Cancer Research. 2020;80(16 Supplement):CT301–CT301. [Google Scholar]
- 84.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012. March 8;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Miguel-Luken MJ, Mansinho A, Boni V, et al. Immunotherapy-based combinations: current status and perspectives. Curr Opin Oncol. 2017. September;29(5):382–394. [DOI] [PubMed] [Google Scholar]
- 86.Liu CJ, Schaettler M, Blaha DT, et al. Treatment of an aggressive orthotopic murine glioblastoma model with combination checkpoint blockade and a multivalent neoantigen vaccine. Neuro Oncol. 2020. September 29;22(9):1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Narayanan JSS, Ray P, Hayashi T, et al. Irreversible Electroporation Combined with Checkpoint Blockade and TLR7 Stimulation Induces Antitumor Immunity in a Murine Pancreatic Cancer Model. Cancer Immunol Res. 2019. October;7(10):1714–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao J, Chen Y, Ding ZY, et al. Safety and Efficacy of Therapeutic Cancer Vaccines Alone or in Combination With Immune Checkpoint Inhibitors in Cancer Treatment. Front Pharmacol. 2019;10:1184. [DOI] [PMC free article] [PubMed] [Google Scholar]