Abstract
Auxin regulates the transcription of auxin-responsive genes by the TIR1/AFBs-Aux/IAA-ARF signaling pathway, and in this way facilitates plant growth and development. However, rapid, nontranscriptional responses to auxin that cannot be explained by this pathway have been reported. In this review, we focus on several examples of rapid auxin responses: (1) the triggering of changes in plasma membrane potential in various plant species and tissues, (2) inhibition of root growth, which also correlates with membrane potential changes, cytosolic Ca2+ spikes, and a rise of apoplastic pH, (3) the influence on endomembrane trafficking of PIN proteins and other membrane cargoes, and (4) activation of ROPs (Rho of plants) and their downstream effectors such as the cytoskeleton or vesicle trafficking. In most cases, the signaling pathway triggering the response is poorly understood. A role for the TIR1/AFBs in rapid root growth regulation is emerging, as well as the involvement of transmembrane kinases (TMKs) in the activation of ROPs. We discuss similarities and differences among these rapid responses and focus on their physiological significance, which remains an enigma in most cases.
The phytohormone auxin plays a central role in plant physiology and development. According to our current understanding, it does so by changing gene expression profiles that largely depend on the developmental context and varying auxin concentrations. There are, however, numerous reports from the premolecular auxin era as well as recent publications showing that auxin can initiate rapid responses at the level of organs, cells, and subcellular structures. The rapidity of these responses rules out the involvement of gene transcription and mRNA translation. The eukaryotic transcription rate is ∼1.4 kbp per minute, and the translation rate ∼5 amino acids per second (BioNumbers BNID 111158,104598; Milo et al. 2010). Thus, we can very roughly estimate that an average-length protein of 400 amino acids (Ramírez-Sánchez et al. 2016) needs at least 2 min to be transcribed and translated if all other processes (such as auxin signaling, mRNA processing, nuclear export, and protein maturation) are disregarded. The earliest measurable effect of the canonical auxin transcriptional pathway (the auxin-induced growth) typically shows a 10–20 min lag phase (Senn and Goldsmith 1988; Fendrych et al. 2016). Therefore, we arbitrarily consider responses that happen within a few minutes as rapid, nontranscriptional events that must involve rapid electrochemical responses and/or posttranscriptional modifications.
During canonical auxin transcriptional signaling, when the concentration of auxin drops, the Aux/IAA proteins heterodimerize with the auxin response factors (ARFs) and inhibit auxin-responsive gene transcription (Weijers et al. 2005; Han et al. 2014). Auxin coreceptors TIR1/AFBs are F-box proteins, components of the SKP1–Cullin–F-box (SCF)–E3 ubiquitin ligase complex (Ruegger et al. 1998; Dharmasiri et al. 2005; Kepinski and Leyser 2005; Dezfulian et al. 2016). When auxin concentration increases, TIR1/AFB F-box proteins bind via auxin to Aux/IAA proteins, which results in Aux/IAA ubiquitylation and degradation (Gray et al. 2001; Tan et al. 2007), releasing the inhibition of ARFs. Aux/IAAs interact with auxin and TIR1/AFB via the degron domain (DII), which seals the ligand-binding groove in TIR1/AFB. The degron domain is conserved within the family; the dynamics of degradation mainly depends on the carboxy-terminal domain (Calderón Villalobos et al. 2012; Moss et al. 2015; Winkler et al. 2017). Recently, Niemeyer et al. (2020) and Ramans Harborough et al. (2019) revealed that the KR motif embedded in the intrinsic disorder region (IDR) upstream of the degron confers alternative auxin-independent-binding contacts with TIR1. Therefore, before the actual auxin-induced transcriptional changes, many rapid nontranscriptional events occur, which influence protein localization such as the Aux/IAA relocalization toward the proteasome, protein posttranslational modification, and abundance. This system of auxin-induced Aux/IAA degradation was exploited as a powerful tool to rapidly control protein levels in yeast and in animal cell lines (Nishimura et al. 2009, 2020; Holland et al. 2012). And possibly, some of the nontranscriptional auxin effects that we discuss in this review might be linked to these early phases of the TIR1/AFB-Aux/IAA signaling.
DYNAMICS OF AUXIN AND PROTON TRANSPORT ACROSS MEMBRANES
Polar auxin transport is essential for development (Petrášek and Friml 2009). At the cellular level, auxin fluxes depend on the combination of auxin chemical properties and the controlled activity and localization of influx and efflux auxin carriers (Bennett et al. 1996; Gälweiler et al. 1998; Petrášek et al. 2006). To understand the rapid effects of auxin, we need to discuss how auxin travels across membranes, focusing on associated ion transport processes.
Auxins, as weak acids, are partly undissociated in the acidic apoplast (pH 4.5–5.5) (Barbez et al. 2017), and the uncharged IAA can enter the cell by diffusion. At the more alkaline cytoplasmic pH, IAA releases its H+ and is trapped in the cell. Plasma membrane (PM) permeability for the IAA– anion is ∼100 to 1000 times lower than for IAA (Raven 1975; Delbarre et al. 1996). Uptake of undissociated IAA will continue until the IAA concentration outside and inside equilibrates (Rubery and Sheldrake 1974). Therefore, the existence of H+ gradient (acidic outside) should result in auxin accumulation in the cell even without active transporters (Fig. 1; Lomax et al. 1985).
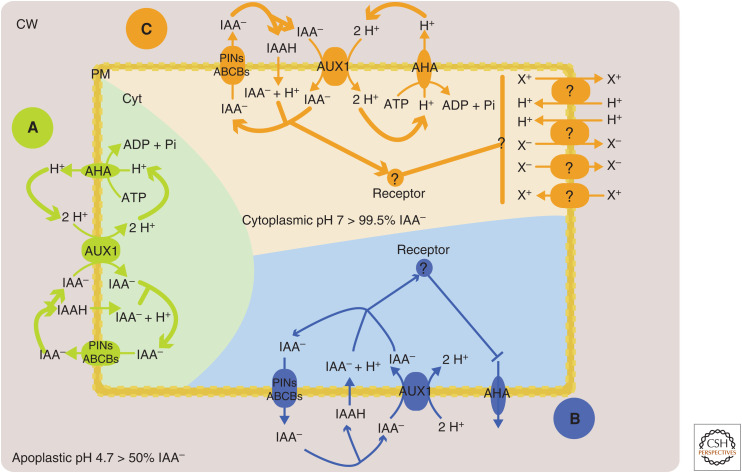
Figure 1.
Possible scenarios that could explain the underlying mechanisms of auxin-induced cell depolarization. (A) Proton influx driven by auxin/proton symport. Import of auxin into the cell participates in the accumulation of 2H+/1IAA– by AUX1/LAX and/or 1H+ by diffusion (dissociation of IAAH at cytoplasmic pH 7). Such accumulation of cytoplasmic-positive charges could increase membrane potential. This could stimulate the proton pump activity, which in turn would fuel auxin import by AUX1/LAX. (B) Inhibition of the proton pump by an intracellular auxin receptor. Similar to A, auxin influx participates in the accumulation of protons in the cytoplasm. In this model, this process is enhanced by the inhibition of the proton pump by active signaling event(s) from auxin receptor(s). (C) Regulation of the activities of ion channels/carriers by intracellular auxin receptor(s). Inhibition of cations efflux or activation of influx and reciprocally for anions could influence membrane potential by accumulation of positive charges in the cytoplasm. The actual mechanism of depolarization is likely to be a combination of these scenarios.
The natural auxin IAA– is imported with two protons (Rubery and Sheldrake 1974; Hertel et al. 1983; Lomax et al. 1985) by the AUX1/LAX family of transporters (Swarup and Péret 2012). Consequently, the proton gradient or the proton motive force fuels AUX1/LAX activity (Lomax et al. 1985). On the other hand, at higher apoplastic pH, the substrate of AUX1/LAX importer IAA– (Carrier et al. 2008) is more abundant. Finally, AUX1/LAX affinity depends on pH, its optimum being at pH ∼ 5.5 (Carrier et al. 2008). Taken together, these factors explain the nonmonotoniс dependence of IAA influx on pH observed in several studies (Rubery and Sheldrake 1974; Loper and Spanswick 1991; Yang et al. 2006).
Auxin is exported out of cells by the polarly localized Pin Formed (PIN) and apolar ATP-binding cassette subfamily B (ABCBs) transporters (for review, see Petrášek and Friml 2009); some ABCBs can also import auxin (Kamimoto et al. 2012; Kubeš et al. 2012). The structure of PIN proteins and their mechanisms of action remain utterly unknown (Zwiewka et al. 2019). We are not aware of any report that PINs cotransport ions. If we assume that PINs transport the IAA− (at cytoplasmic pH 7.2, 99.7% of IAA is in the IAA− form), then PINs would act as facilitators of IAA− diffusion since, in physiological conditions, the concentration of IAA− in the cytoplasm will always be higher than in the adjacent apoplast. A bile acid–sodium symporter was used as a template for PIN2 modeling (Retzer et al. 2017), indicating a possible relationship. This carrier harnesses the energy of the Na+ gradient (Zhou et al. 2014); however, generalizing this mechanism to PIN proteins would be a wild speculation. Determining the electrical signature of an actively transporting PIN could help resolve the question of what, apart from IAA, PINs actually transport (Zwiewka et al. 2019).
Auxin influx, both by diffusion and carrier-mediated transport, requires a proton gradient across the PM. At the same time, the entry of IAA is accompanied by the entry of H+, one per molecule in the case of diffusion and two per molecule in the case of AUX1/LAX-mediated import. IAA influx therefore reduces the proton motive force and is even electrogenic in the case of AUX1/LAX influx. From what we currently understand, it is impossible to conclude whether auxin efflux directly contributes to ion fluxes across the PM, but the depletion of the cytoplasmic IAA− pool by its efflux promotes the influx carrier activity indirectly and thus contributes to AUX1/LAX-mediated H+ influx.
AUXIN TRIGGERS RAPID CHANGES IN PLASMA MEMBRANE POTENTIAL
Application of auxin to various plant tissues causes very rapid (within seconds) concentration-dependent changes in membrane potential. This potential reflects the difference in cytoplasmic and apoplastic electrical potentials. In resting conditions, in plants, this difference is maintained by active pumping of positively charged protons into the apoplast (Sze 1985). This active process depletes the cytoplasmic potential of positive charges and leads to a membrane potential of approximately −120 to −160 mV (Sze et al. 1999). A negative membrane potential allows secondary active transport of molecules and is essential for cellular energetics. Rapid fluxes of ions in/out of the cell can drastically influence membrane potential causing accumulation of positive charges (depolarization) or the accumulation of negative charges (hyperpolarization) (Weisenseel and Meyer 1997).
Auxin rapidly depolarized PM in coleoptiles of monocots (Goering 1979; Bates and Goldsmith 1983; Felle et al. 1991), in maize (Felle et al. 1991), and white mustard (Tretyn et al. 1991) root hairs. More recently, depolarization was recorded in Arabidopsis thaliana root hairs (Dindas et al. 2018, 2020) and root epidermis (Paponov et al. 2019a). Surprisingly, in white mustard root hairs, picomolar IAA triggered the opposite response—a rapid hyperpolarization (Tretyn et al. 1991). Moreover, in protoplasts (cells with cell walls removed and floating in an isotonic medium), auxin triggered an outward current of positive charge in a rapid manner (∼1 min) (Ruck et al. 1993).
In some tissues, the depolarization was followed by PM hyperpolarization (in 10–20 min); this response was only triggered by active auxins that triggered growth, whereas weak acids with comparable concentrations did not trigger hyperpolarization (Bates and Goldsmith 1983). The slow hyperpolarization response could result from the activation of H+-ATPase, leading to proton extrusion and acid growth of coleoptiles and hypocotyls. This process is governed by the transcriptional activity of the TIR1/AFB1-Aux/IAA coreceptor (Takahashi et al. 2012; Fendrych et al. 2016; Uchida et al. 2018). On the other hand, instantaneous membrane potential responses constitute, without any doubt, a clear nontranscriptional reaction to auxin. What is this response caused by? What is the receptor, if any, triggering it? And, importantly, what is its biological meaning?
PM potential changes by auxin could be achieved by several scenarios (Fig. 1; Tretyn et al. 1991): (1) short-circuiting the primary transport by secondary active transport (cotransport of H+, by AUX1/LAX, for example), (2) inactivating the primary transport (H+-ATPase), and (3) changing the PM permeability for ions by regulating transporter and channel activities (such as Ca2+ and K+). In addition, weak acids themselves, in high concentrations, could lower cytosolic pH, which leads to H+-ATPase stimulation (Bates and Goldsmith 1983; Felle et al. 1991).
Unfortunately, reports vary as to what extent different auxins and weak acids trigger PM depolarization. Both active auxins (IAA, 1-NAA, and 2,4-D) and inactive analogs (2-NAA and 2,3-D) induced rapid PM depolarization (Bates and Goldsmith 1983; Felle et al. 1991; Tretyn et al. 1991; Dindas et al. 2018, 2020; Paponov et al. 2019a). There are indications that rapid depolarization depends on the proton–auxin symport by AUX1/LAX: First, the response is considerably diminished at high pH (Felle et al. 1991; Paponov et al. 2019a), consistent with the above-mentioned AUX1/LAX dependence on proton motive force and pH. Second, the depolarization response in root hairs is drastically diminished in the aux1 mutant (Dindas et al. 2018). However, 1-NAA, which is not a substrate of AUX1/LAX import (Tretyn et al. 1991; Delbarre et al. 1996; Yang et al. 2006), depolarizes PM as well, as does the biologically inactive 2-NAA, at least to some extent (Felle et al. 1991; Tretyn et al. 1991; Paponov et al. 2019a; Dindas et al. 2020). In addition, auxin efflux via PIN2 contributes to PM depolarization (Dindas et al. 2018; Paponov et al. 2019a), possibly by indirectly enhancing AUX1/LAX activity by depleting the intracellular auxin. Auxin transporters are thus important actors in the auxin-induced depolarization.
The fact that picomolar IAA triggered a rapid hyperpolarization in root hairs (Tretyn et al. 1991) could hint to the existence of an auxin receptor rapidly stimulating H+-ATPase. There is recent evidence that, in Arabidopsis root hairs, rapid PM depolarization depends on the TIR1/AFB-Aux/IAA auxin coreceptor. The tir1/afb2/3 triple mutant lacking three of six receptors showed a decreased depolarization response, and the pretreatment with auxinole (a TIR1/AFB-Aux/IAA inhibitor; Hayashi et al. 2012) decreased the depolarization response (Dindas et al. 2018). However, the possibility that auxinole itself triggered depolarization, which would mask the auxin response, cannot be excluded (cf. Supplemental Fig. 4e,f in Dindas et al. 2018). The authors confirmed that the tir1/afb2/3 phenotype was not caused by the lack of AUX1 expression, opening the possibility that the TIR1/AFB coreceptor might indeed trigger signaling that leads to depolarization response and thus be directly involved in rapid, nontranscriptional signaling.
We can therefore speculate that depolarization is triggered jointly by auxin transport processes, an active signaling response, and in cases of higher concentrations can involve the direct effect of weak acids. However, the most important question remains—what is the biological significance of auxin-induced depolarization? It is hard to imagine a situation in soil when a root hair would suddenly be flushed with micromolar IAA, even though it was shown that strains of Bradyrhizobium japonicum can secrete up to 20 μM IAA into the medium (Minamisawa and Fukai 1991). Root hair growth is indeed regulated by auxin, but this regulation depends on the TIR1/AFB-Aux/IAA-ARF transcriptional signaling (Mangano et al. 2017; Bhosale et al. 2018).
With the boom of molecular biology, many clearly nontranscriptional auxin effects have not been extensively studied (e.g., Marten et al. 1992) and the molecular pathways involved remain unknown. Plant electrophysiology, in contrast to animal electrophysiology, has fallen out of fashion. However, in recent years, efforts have been made to revive those techniques and it is of great importance to now create links between physiology and molecular biology as there are possibly many more rapid auxin responses to be revealed.
ROOT GROWTH INHIBITION BY AUXIN
Whereas the biological significance of auxin-induced depolarization remains a mystery, there is one rapid effect of auxin with an evident function, root growth inhibition by auxin. During root gravitropic response, auxin flows shootward to the lower root side by the coordinate action of PIN2 and AUX1 (Young et al. 1990; Band et al. 2012; Adamowski and Friml 2015) where it inhibits cell elongation. This cellular response is thus the motor of gravitropic root bending. Auxin influences cell expansion of other plant organs as well (for review, see Du et al. 2020); however, outside of the root, the effect is mediated by the canonical TIR1/AFB-Aux/IAA-mediated gene transcription (Fendrych et al. 2016; Uchida et al. 2018; Du et al. 2020).
In nanomolar and higher concentrations, IAA rapidly inhibits root growth (Evans et al. 1994; Monshausen et al. 2011; Scheitz et al. 2013); the detectable inhibition starts within 60 sec of auxin treatment (Shih et al. 2015; Fendrych et al. 2018; Prigge et al. 2020). Such rapidity effectively rules out the involvement of TIR1/AFB-Aux/IAA-ARF-mediated gene transcription. The involvement of transcription has not been formally disproven, but the translation inhibitor, cycloheximide, could not prevent the IAA-triggered growth inhibition (Fendrych et al. 2018). Interestingly and surprisingly, root growth inhibition response is quickly reversible—roots started to resume growth within 3 min after removal of IAA from the medium even after an 80-min inhibition (Fendrych et al. 2018). This indicates that this auxin response is “rapid and nontranscriptional” for a long time, and that roots can slow down and speed up rapidly, depending on the current auxin concentration in the cell.
Indeed, auxin must enter the cell to trigger growth inhibition; Arabidopsis roots lacking the AUX1 auxin influx carrier did not respond to low IAA concentrations, but inhibited growth rapidly when treated with the PM-permeable 1-NAA auxin analog (Fendrych et al. 2018).
The underlying molecular pathway remains unknown, but in agreement with the effect of pH on the cell wall extensibility, inhibition is correlated with alkalinization of the root surface and cell walls. In corn roots, the addition of IAA or 1-NAA led to simultaneous growth inhibition and an increase in the bathing medium pH; 2-NAA had no effect (Evans et al. 1980; Lüthen and Böttger 1988). Imaging, using pH-sensitive dye, revealed that the addition of IAA caused near-instantaneous alkalinization of the root surface in the transition and elongation zones of Arabidopsis root tip. The response of the aux1 mutant to 100 nM IAA was less than the control (Monshausen et al. 2011), which is consistent with the finding that aux1 requires higher IAA doses to trigger growth inhibition (Fendrych et al. 2018). Also, the cell wall pH, visualized by the apopHusion sensor (Gjetting et al. 2012) and a pH-sensitive dye HPTS (Barbez et al. 2017), rose in response to IAA. The alkalinization of root surface and cell wall is probably caused by proton influx into cells, as Monshausen et al. (2011) showed that the auxin-induced alkalinization is accompanied by a “small but significant” decrease of pH inside the cells.
Could the alkalization of the root apoplast be caused directly by the AUX1-mediated IAA-H+ symport? Probably not, because even low IAA concentrations trigger apoplast alkalinization. Also, IAA and PM-permeable 1-NAA can trigger alkalinization in the aux1 mutant. The key protein to understand the connection between apoplastic pH, auxin, and membrane potential changes is the Cyclic Nucleotide-Gated Channel 14 (CNGC14) putative calcium channel. Externally and locally applied IAA induced a rapid cytoplasmic calcium transient, as fast as the pH change along the root axis (Monshausen et al. 2011; Dindas et al. 2018). Moreover, blocking calcium channels by lanthanide impaired the calcium transient and the pH response (Monshausen et al. 2011; Shih et al. 2015). The cngc14 mutant did not accumulate calcium in the cytoplasm after auxin treatment had impaired apoplastic alkalization and delay in rapid growth response (Shih et al. 2015; Dindas et al. 2018). These observations demonstrate the connection between rapid calcium signaling and extracellular alkalization in response to auxin.
Indeed, a massive influx of positively charged calcium into the cytoplasm could create a significant membrane depolarization and, inversely, IAA-triggered proton influx could trigger calcium transient, as pH and calcium spiking are tightly connected (Behera et al. 2018). However, cytosolic calcium concentration, measured by Shih and Dindas, returns to the resting level immediately after reaching the peak value (within 1.5 min). On the other hand, the depolarization is maintained over the course of several minutes (as measured in Dindas et al. 2018; Paponov et al. 2019a) and proton influx lasts much longer (Shih et al. 2015). One could propose that calcium is the secondary messenger in auxin-induced depolarization, and this response is maintained by a nontranscriptional activation of ion channels.
THE NONTRANSCRIPTIONAL FACE OF TIR1/AFB-AUX/IAA AUXIN RECEPTORS
It is well established that the TIR1/AFB-Aux/IAA pathway is involved in root growth inhibition, and the respective mutants show auxin insensitivity and agravitropism (e.g., Ruegger et al. 1998; Knox et al. 2003). But what about the rapid response? Based on a tir1-1/afb1-3/afb2-3/afb3-4 quadruple mutant, Scheitz et al. (2013) showed that TIR1/AFBs are required for growth inhibition, while others claim that auxin-induced rapid pH change is unaffected in tir1 and tir1-1/afb2-3/afb3-4 mutants (Monshausen et al. 2011). Fendrych et al. (2018) showed that the tir1-1/afb2-1/afb3-1 triple mutant has a decreased growth response. Using the modified ccvTIR1 receptor and cvxIAA auxin, this group also showed that the rapid growth response can be triggered directly by the ccvTIR1 protein, even though the response was slightly delayed compared to the control situation (Fendrych et al. 2018). Finally, when analyzing the phenotypes of multiple combinations of TIR1/AFB mutations, Prigge et al. (2020) discovered that each mutant combination lacking the AFB1 protein showed a defect in rapid growth inhibition. The so far enigmatic AFB1 coreceptor thus might be crucial for rapid growth response, while the other paralogs also contribute to the response. In conjunction with the impairment of membrane potential changes in the tir1/afb mutants (see above), this indicates that the TIR1/AFB pathway, so far studied exclusively in connection with transcriptional regulation, has a fast, nontranscriptional signaling branch (Fig. 2).
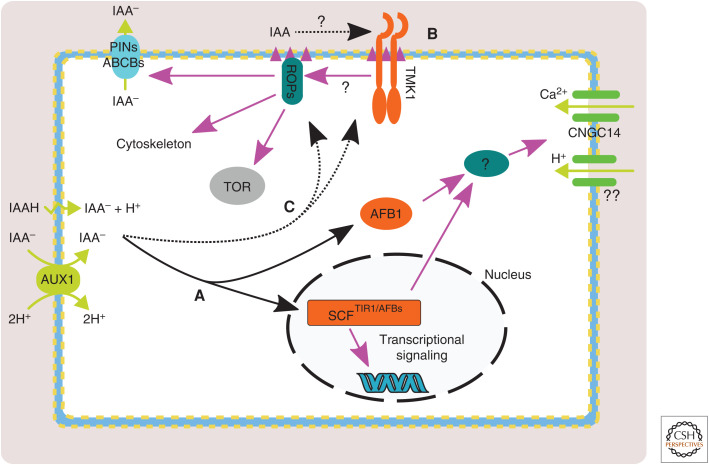
Figure 2.
Schematic model of nontranscriptional auxin signaling at the cellular level. (A) Auxin entering the cell by influx or diffusion activates the TIR1/AFB receptors in the nucleus or possibly in the cytoplasm. Further, these receptors activate Ca2+ and H+ fluxes at the plasma membrane (PM) via an unknown signaling branch and lead to root growth inhibition. (B) Auxin activates ROPs (Rho of plants) by unknown mechanism(s), possibly via the TMK1 receptor. Lipid composition and organization of the membrane play a key role (magenta triangles). Activation by cytoplasmic auxin has not been excluded (C). Activated ROPs trigger downstream events including changes in PIN endomembrane trafficking, cytoskeletal reorganization, and activation of the TOR (target of rapamycin) pathway.
Unlike TIR1, which localizes to the nucleus, some AFBs are both cytoplasmic and nuclear, and this is most prominent in the case of AFB1 (Prigge et al. 2020). Interestingly, the AFB1 protein harbors a polymorphism relative to other members of the family (E8K), which, when introduced into TIR1, untethers TIR1 from the SCF complex, increases its levels, and reduces its ability to respond to auxin (Yu et al. 2015). Is AFB1 associated with the SCF complex to trigger root growth inhibition? Does the cytoplasmic localization of AFB1 matter for the rapid response? These exciting questions should be solved in the near future.
Finally, it remains unclear what the “real life” significance of the rapidity of auxin root growth inhibition is. Unlike organs growing in the air, roots embedded in soil cannot grow back, thus making a turn too late means no return. But regardless of the adaptive value of the response, roots do react quickly to internal auxin during gravitropism and the cngc14 mutant shows a mild delay in gravitropic bending that corresponds to the delay in reaction to auxin added externally (Shih et al. 2015). On the other hand, the single afb1 mutant that showed a much stronger defect in response to external auxin than cngc14 did not have a detectable problem during gravitropism and a gravitropic defect appeared only in the higher-order mutant combinations (Prigge et al. 2020).
AUXIN AND ENDOCYTOSIS
In the last 15 years, the field of rapid auxin responses has been dominated by another nontranscriptional auxin effect: inhibition of endocytosis by auxin. The reason for this interest was that the auxin effect on endocytosis could provide a mechanistic explanation for auxin flux canalization, a symmetry-breaking process that ensures vascular connectivity during auxin-induced vascular tissue differentiation (Ravichandran et al. 2020). Inhibition of endocytosis by auxin postulated extracellular auxin perception and local PIN accumulation due to reduced PIN removal from the PM. As a result, auxin promotes its own efflux because more PINs pump more auxin toward the apoplastic auxin maximum (Paciorek et al. 2005; Robert et al. 2010). Auxin inhibits the intracellular accumulation of PINs, other membrane cargoes, and, importantly, PM-derived vesicles labeled by FM4-64. This means that the entire endocytotic flow is affected (Paciorek et al. 2005) through inhibiting clathrin-mediated endocytosis (Robert et al. 2010). Endocytosis rate was mostly approximated by using Brefeldin A (BFA), which helps visualize cargoes that undergo constitutive recycling (Geldner et al. 2001). BFA bodies accumulate both endocytosed cargoes (e.g., FM4-64) and newly synthesized proteins (Reichardt et al. 2007; Jásik et al. 2016). Consequently, Jásik et al. (2016) used PIN2 tagged with a photoconvertible fluorescent protein to monitor the endocytosis rate of specific protein pools more directly. Whereas auxins decreased PIN2 accumulation in BFA bodies as expected, neither 1-NAA nor IAA reduced PIN2 removal from the PM. Instead, the delivery of PIN2 to the PM was affected. This could be explained by an effect on endomembrane trafficking or PIN2 expression, but not by inhibition of PIN2 endocytosis. On the other hand, in the same PIN2-expressing root cells, 1-NAA triggered the detachment of clathrin light chains from the PM (Robert et al. 2010), indicating that 1-NAA somehow influences the PM pool of clathrin. Clearly, we need detailed and quantitative analysis of auxin effects on endocytosis and endomembrane trafficking to clarify these discrepancies.
The effect of auxin on cargo accumulation in BFA bodies requires high concentrations of the hormone. Importantly, the most potent inhibitor of accumulation of PINs in BFA bodies is 1-NAA, with a near-maximum effect of around 5 µM (Paciorek et al. 2005). The native auxin IAA was effective only in combination with an antioxidant, which should protect the labile IAA molecule. Paponov et al. (2019b) readdressed this discrepancy between IAA and 1-NAA and confirmed that 1-NAA inhibited PIN accumulation in BFA bodies at 10 µM, but IAA did so only at very high concentrations (100 µM). The inactive 2-NAA (with respect to physiological auxin response) triggers a similar cargo accumulation inhibition as 1-NAA, which was also shown independently by Simon et al. (2013). In the original Paciorek et al. (2005) publication, however, 2-NAA did not elicit the inhibitory response. Similarly, in tobacco cells, 1-NAA had a stronger inhibitory effect than IAA (Jelínková et al. 2015). In contrast with these results, there is evidence that genetically increased IAA auxin levels led to reduced BFA accumulation of cargoes (Paciorek et al. 2005). In addition, in a gravistimulated root, the cells on the lower side, which accumulate higher levels of IAA, showed signs of auxin-inhibited PIN internalization (Paciorek et al. 2005). The concentration of IAA in gravistimulated root cells was estimated to be approximately in the range of tens of nanomolars (Band et al. 2012). This means that a long-lasting increase of internal auxin could trigger the changes in PIN internalization. This is in sharp contrast with the transient nature of the effect when 1-NAA is applied externally: after ∼2 h of treatment, the accumulation of membrane cargoes resumes and the clathrin light chain PM localization recovers (Robert et al. 2010). Once again, there are significant disagreements between publications and it is not clear to what extent the effect is relevant for the native IAA auxin and thus for the actual physiology and development of the plant.
Is this phenomenon a nontranscriptional auxin response? The effect is clearly rapid as it can be triggered by a short (<5 min) 1-NAA treatment. Pharmacological inhibition of transcription and translation did not interfere with the 1-NAA effect, which also points to the nontranscriptional nature of the response (Paciorek et al. 2005). In addition, there is evidence that the effect is not mediated by the TIR1/AFB1-Aux/IAA coreceptor or the downstream auxin-regulated gene transcription (Robert et al. 2010). However, there is one publication that claims exactly the opposite—that the response depends on TIR1/AFB-mediated auxin signaling and the downstream sterol composition of membranes (Pan et al. 2009).
The concentration ranges and spectra of auxin analogs show that the cargo accumulation inhibition is a molecularly distinct process from the pathway that leads to root growth inhibition. Furthermore, these effects do not behave similarly to the rapid membrane potential changes. The effective spectrum of concentrations and auxin analogs resembles the binding preferences of the ABP1 (Badescu and Napier 2006). Using ABP1 knockdown lines, expression of ABP1 variants and the abp1-5 point mutation, Robert et al. (2010) showed that the process is controlled by ABP1. Later, Xu et al. (2014) showed that ABP1 activates ROPs (Rho of plants) via the interaction with the transmembrane kinase 1 (TMK1) membrane receptor, which explained the missing link between apoplastic ABP1 and effects on membrane trafficking. However, in 2015, Gao et al. demonstrated that true ABP1 knockouts do not show any major developmental phenotypes. This was a hard hit for the model because the null mutation of ABP1 was supposedly lethal (Chen et al. 2001), and thus demonstrated the importance of ABP1-mediated regulation of PIN endocytosis. Furthermore, Michalko et al. (2016) demonstrated that the phenotype of the ABP1 knockdown lines was not caused by the down-regulation of ABP1 protein. Finally, Paponov et al. (2019a) showed that in the true ABP1 knockout lines, 1-NAA still inhibits PIN accumulation in BFA bodies. In summary, it is not clear which receptor and signaling pathway participates in this process.
AUXIN AND ROPs
The existence of an apoplastic or PM-localized auxin receptor has been proposed for years, and the receptor-like kinase TMK1 is an excellent candidate. TMK1 seems to regulate both a non-TIR/AFB transcriptional pathway (through noncanonical IAAs; Cao et al. 2019) as well as a nontranscriptional pathway, in which the signal is transduced through ROP GTPases (Xu et al. 2014; Pan et al. 2020).
ROPs belong to a plant-specific, small GTPase superfamily. Like other G-proteins, ROPs act as GTPases that can cycle rapidly between active (GTP-bound) and inactive (GDP-bound) forms based on interaction with their regulatory proteins (for review, see Feiguelman et al. 2018). The first clues that G-proteins may be involved in auxin signaling came from bulk biochemical experiments, where biologically active auxins (IAA, 1-NAA, and 2,4-D) caused a rapid increase in active G-binding protein levels on vesicle membranes of rice coleoptiles (Zaina et al. 1990). On the other hand, the inhibitor of G-protein activation, GDP-β-S, blocked swelling of wheat protoplasts induced by selected hormones, including 1-NAA (Bossen et al. 1991). These data point to possible activation of any class of GTPases present in the cell fractions tested, but so far, only ROPs were reported to be auxin responsive. Tobacco ROP NtRac1 became gradually activated within minutes when seedlings were grown on 1-NAA (Tao et al. 2002), and even finer timescale experiments revealed that Arabidopsis ROP2 and ROP6 are activated in protoplasts after only 30 sec of 1-NAA treatment; this is, beyond doubt, too fast to be transcriptionally regulated (Xu et al. 2010). Activation of these ROPs by the natural auxin IAA was also demonstrated (Xu et al. 2014). In all three studies, the total amount of ROP proteins remained unaffected, clearly indicating a nontranscriptional modulation of their activity.
Auxin perception and signal transduction upstream of ROPs are still enigmatic. Originally, physical interaction of TMK1 with ABP1 upon auxin binding was shown to be responsible for ROP activation (Xu et al. 2014). However, recently, the same group postulated that auxin triggers sterol-dependent nanoclustering of TMK1 that is upstream of ROP6 nanoclustering and activation (Fig. 2; Pan et al. 2020). S-acylation of conserved residues within a G-domain of ROP6 is known to induce its transient activation and association with membrane lipid rafts, while the GTPase activity remains unaffected (Sorek et al. 2010), and this lipid modification is crucial for the ROP6 activation in the auxin-TMK1 pathway (Pan et al. 2020). However, regulation of ROP6 behavior at the PM seems to be more complex. In roots, another phospholipid, phosphatidylserine is required for rapid stabilization of the GTPase into immobile nanoclusters in response to auxin (quantified after 5 min 1-NAA treatment). Abolishing the lipid interaction (but not the GTPase activity) has a direct impact on ROP6 localization and auxin response (Platre et al. 2019). Furthermore, ROP6 binds to phosphatidylglycerol, which activates the GTPase in a dose-dependent manner (Han et al. 2018). Clearly, TMK1 membrane organization and correct lipid composition play a key role in the activation of ROPs. Nevertheless, we do not understand how and where auxin is perceived. Although the direct interaction of ROPs with membrane phospholipids modulate their localization and activity, the spatiotemporal regulation is the function of specific activating (RopGEFs) and inhibiting proteins (RopGAPs, RopGDIs), and we have to bear in mind that these regulatory proteins are also controlled by various interactors including fast-responding kinases (for review, see, e.g., Fehér and Lajkó 2015).
Besides TMK1, mitogen-activated protein kinases (MAPKs) rapidly and transiently respond to auxin. Although relatively high concentrations of auxin were used (20 µM), only IAA, 1-NAA, and 2,4-D were able to trigger the MAPK activation in Arabidopsis roots, but not the inactive auxin analog PCIB (clofibric acid) (Mockaitis and Howell 2000). The auxin-induced MAPK phosphorylation cascade involving MKK3, MPK1, and RBK1 (ROP BINDING PROTEIN KINASE 1) was then shown to regulate cotyledon expansion and root elongation in Arabidopsis seedlings through the activity of ROP4 and ROP6 (Enders et al. 2017).
What is the cellular and physiological role of auxin activation of ROPs? One of the effects is unexpectedly connected to the reinitiation of mRNA translation via the TOR (target of rapamycin) pathway (Fig. 2). After activation by auxin (1-NAA 100 nM), ROP2 directly interacts with TOR and promotes its activation, which becomes profound 60–120 min after the treatment. TOR is a conserved protein kinase that acts as a general growth “sensor,” linked to protein synthesis. In Arabidopsis, auxin enhances its activity by phosphorylation through active ROP2. When activated, TOR promotes translational reinitiation of certain (µORF-containing) mRNAs. These mRNAs can be stored in a cell and, after a certain signal, they assemble into polysomes and are translated. Because of the rather slow activation process, TOR involvement in the rapid auxin response remains questionable (Schepetilnikov et al. 2013, 2017).
In planta, when ROP activity is perturbed, auxin-related phenotypes often emerge. In particular, this research has focused on how ROPs influence PM localization and trafficking of PINs that, in turn, influence auxin fluxes and levels. Polar recruitment of PINs in Arabidopsis seedlings was shown to be regulated by ICR1 (interactor of constitutive active ROP1), and auxin treatment (1-NAA) induced ICR1 expression within 30 min (Hazak et al. 2010). Similarly, PIN2 recycling in roots depends on ROP6 activation by its GEF-protein SPIKE1 (Lin et al. 2012). Whereas the gravitropic root bending of the rop6 mutant is slower (Lin et al. 2012; Han et al. 2018), both overexpression and constitutively active ROP6 bend their roots faster and more than wild-type (WT) plants (Han et al. 2018; Platre et al. 2019). The auxin-ROP-PIN hypothesis has been studied and established in detail on a model system of cotyledon pavement cells, where lobe formation was postulated to be auxin regulated (for review, see Chen et al. 2015). In Arabidopsis, mutants with affected auxin biosynthesis (yuc 1/2/4/6) or ROP signaling (rop2RNAi/rop4-1) share similar features, such as reduced lobe number and length. Addition of exogenous auxin (1-NAA) increased the number of lobes in both WT and the auxin biosynthetic mutant, but not in the ROP signaling mutant. Furthermore, a similar phenotype of pavement cells was shared by abp1-5, pin1, and tmk1 mutants (tmk1234), and none of them could be rescued by application of external auxin (1-NAA). Based on additional experiments, the authors postulated a model of a positive feedback loop where auxin gets recognized by the ABP1-TMK1 complex leading to ROP activation, cytoskeleton remodeling via its effectors and, finally, to PIN1 endocytosis, relocalization, and auxin redistribution to lobe-initiation sites (Xu et al. 2010, 2014; Nagawa et al. 2012). However, independent characterizations of the lobe phenotypes of mutants brought more doubt on the model; pavement cells of abp1 knockouts (represented by two null alleles, abp1-TD1 and abp1-c1) do not differ from WT, and they form more lobes in response to 1-NAA (Gao et al. 2015). Similarly, pavement cells of ric4 (ROP2 effector) or pin1-1 mutants were indistinguishable from WT. In addition, PIN1-GFP protein driven by its native promoter was not expressed in developing pavement cells. Nevertheless, other PINs (PIN3, PIN4, and PIN7) did localize to the PM of expanding pavement cells, but their mutations (either single or multiple) still resulted in reduced cell area but not lobe number, indicating their role in cell expansion but not lobe initiation (Belteton et al. 2018). Clearly, the model requires more experimental attention and deserves revision.
ROP signaling has a direct link to actin remodeling (e.g., through SCAR/WAVE, activators of ARP2/3 complex with actin-nucleation activity) (Feiguelman et al. 2018). One important question is whether microfilaments remodeling after ROP activation occurs rapidly enough to act as a messenger. Alternatively, is microfilament remodeling simply a component of the growth response? Arieti and Staiger (2020) followed actin dynamics in vivo in Arabidopsis roots after short-term auxin treatment. At the whole-cell level, after 20–30 min of IAA treatment, the actin network was denser, less bundled, and, interestingly, more organized. Shorter treatment of just 5 min was, however, not sufficient to trigger all these changes. An analysis of individual filaments that can detect finer modifications revealed that a 7-min treatment with 10 nM IAA had a significant impact on actin filament behavior, especially on unbundling throughout both the elongation and transition zone of a root. None of these effects were detectable in the aux1 mutant, indicating that auxin needs to enter the cell (i.e., it cannot trigger actin remodeling from an apoplast located receptor/signal). Together with the timing of actin remodeling, it is plausible that cytoskeleton reacts to the rapid growth inhibition and associated physiological changes.
Actin, the downstream target of ROPs that orchestrates both cell morphogenesis and expansion, also dynamically responds to auxin treatment. Still, too many conclusions, especially in pavement cells studies, are based on transient biolistic experiments that may provide misleading data. We need a detailed characterization of microfilament dynamics within the first minutes following auxin treatment (preferentially IAA) in various tissues/organs to dissect the actin role in the rapid auxin responses. Despite the contradictions in the data and gaps in the models, auxin seems to activate certain ROP GTPases in a transcription-independent way, and ROPs interaction with membrane phospholipids is crucial for their activation and signal transduction (Fig. 2). Interestingly, based on one study (Pan et al. 2009), membrane phospholipid composition depends on auxin signaling via the TIR1/AFB pathway. A putative PM located receptor/receptor-like kinase has been considered so far as an upstream regulator of ROPs, but could the TIR1/AFB nontranscriptional signaling also play a role? And besides ROPs, are there any other GTPases involved?
CONCLUDING REMARKS
Auxin triggers several classes of undoubtedly rapid, nontranscriptional responses. We attempted to discuss the most prominent ones in this review. We think it is critical to clarify which of these are physiologically relevant. Do these responses occur during normal plant growth and development? Although explicitly including the concentration of hormone used in experiments might seem too meticulous, we did so to stress the differences in effectiveness of various auxinic compounds for different auxin rapid responses. Different auxin analogs should not be freely interchanged in interpretation of data and development of models. We believe it is time to shift experimental attention to the effects of lower auxin concentrations that trigger detectable physiological responses.
What do the rapid responses have in common? What are the receptors through which auxin triggers the responses? It is possible that some of the responses do not require a receptor, being triggered directly by the hormone. The weak acid effects discussed in the membrane potential section are one example. Other responses might be activated indirectly by other auxin-triggered processes, for example by the substantial physiological changes that growth inhibition causes in roots. But for others, it is important to unravel the molecular mechanisms that connect auxin perception and the execution of the response. Are there still unknown auxin perception sites in the cell walls, the PM, or the cytoplasm? How is the famous TIR1/AFB-Aux/IAA coreceptor connected to the growth and regulating processes? What activates the ROPs? These and other questions guarantee that the auxin research will not be boring in the future.
ACKNOWLEDGMENTS
The authors thank Maria Akhmanova, Jan Petrášek, and Matouš Glanc for helpful discussions and Jessica L. Barilone for editing the text. The team received support from the European Research Council (Grant No. 803048) and Charles University Primus (Grant No. PRIMUS/19/SCI/09). P.V. was supported by MMVPUK (Grant No. CZ.02.2.69/0.0/0.0/16_027/0008495).
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieti RS, Staiger CJ. 2020. Auxin-induced actin cytoskeleton rearrangements require AUX1. New Phytol 226: 441–459. 10.1111/nph.16382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badescu GO, Napier RM. 2006. Receptors for auxin: will it all end in TIRs? Trends Plant Sci 11: 217–223. 10.1016/j.tplants.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, Sun J, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Peŕet B, et al. 2012. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci 109: 4668–4673. 10.1073/pnas.1201498109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Dünser K, Gaidora A, Lendl T, Busch W. 2017. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc Natl Acad Sci 114: E4884–E4893. 10.1073/pnas.1613499114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates GW, Goldsmith MH. 1983. Rapid response of the plasma-membrane potential in oat coleoptiles to auxin and other weak acids. Planta 159: 231–237. 10.1007/BF00397530 [DOI] [PubMed] [Google Scholar]
- Behera S, Xu Z, Luoni L, Bonza MC, Doccula FG, De Michelis MI, Morris RJ, Schwarzländer M, Costa A. 2018. Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30: 2704–2719. 10.1105/tpc.18.00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteton SA, Sawchuk MG, Donohoe BS, Scarpella E, Szymanski DB. 2018. Reassessing the roles of PIN proteins and anticlinal microtubules during pavement cell morphogenesis. Plant Physiol 176: 432–449. 10.1104/pp.17.01554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950. 10.1126/science.273.5277.948 [DOI] [PubMed] [Google Scholar]
- Bhosale R, Giri J, Pandey BK, Giehl RFH, Hartmann A, Traini R, Truskina J, Leftley N, Hanlon M, Swarup K, et al. 2018. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat Commun 9: 1409. 10.1038/s41467-018-03851-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen ME, Tretyn A, Kendrick RE, Vredenberg WJ. 1991. Comparison between swelling of etiolated wheat (Triticum aestivum L.) protoplasts induced by phytochrome and α-naphthaleneacetic acid, benzylaminopurine, gibberellic acid abscisic acid and acetylcholine. J Plant Physiol 137: 706–710. 10.1016/S0176-1617(11)81226-5 [DOI] [Google Scholar]
- Calderón Villalobos LIA, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Chen R, Li P, Yu Y, Zheng R, Ge D, Zheng W, Wang X, Gu Y, Gelová Z, et al. 2019. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568: 240–243. 10.1038/s41586-019-1069-7 [DOI] [PubMed] [Google Scholar]
- Carrier DJ, Bakar NTA, Swarup R, Callaghan R, Napier RM, Bennett MJ, Kerr ID. 2008. The binding of auxin to the Arabidopsis auxin influx transporter AUX1. Plant Physiol 148: 529–535. 10.1104/pp.108.122044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. 2001. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15: 902–911. 10.1101/gad.866201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wang F, Zheng S, Xu T, Yang Z. 2015. Pavement cells: a model system for non-transcriptional auxin signalling and crosstalks. J Exp Bot 66: 4957–4970. 10.1093/jxb/erv266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J. 1996. Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541. 10.1007/BF00262639 [DOI] [PubMed] [Google Scholar]
- Dezfulian MH, Jalili E, Roberto DKA, Moss BL, Khoo K, Nemhauser JL, Crosby WL. 2016. Oligomerization of SCFTIR1 is essential for Aux/IAA degradation and auxin signaling in Arabidopsis. PLoS Genet 12: e1006301. 10.1371/journal.pgen.1006301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, Von Meyer K, Müller HM, Al-Rasheid KAS, Palme K, Dietrich P, Becker D, Bennett MJ, et al. 2018. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat Commun 9: 1174. 10.1038/s41467-018-03582-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindas J, Becker D, Roelfsema MRG, Scherzer S, Bennett M, Hedrich R. 2020. Pitfalls in auxin pharmacology. New Phytol 227: 286–292. 10.1111/nph.16491 [DOI] [PubMed] [Google Scholar]
- Du M, Spalding EP, Gray WM. 2020. Rapid auxin-mediated cell expansion. Annu Rev Plant Biol 71: 379–402. 10.1146/annurev-arplant-073019-025907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders TA, Frick EM, Strader LC. 2017. An Arabidopsis kinase cascade influences auxin-responsive cell expansion. Plant J 92: 68–81. 10.1111/tpj.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ML, Mulkey TJ, Vesper MJ. 1980. Auxin action on proton influx in corn roots and its correlation with growth. Planta 148: 510–512. 10.1007/BF02395322 [DOI] [PubMed] [Google Scholar]
- Evans ML, Ishikawa H, Estelle MA. 1994. Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194: 215–222. 10.1007/BF01101680 [DOI] [Google Scholar]
- Fehér A, Lajkó DB. 2015. Signals fly when kinases meet Rho-of-plants (ROP) small G-proteins. Plant Sci 237: 93–107. 10.1016/j.plantsci.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Feiguelman G, Fu Y, Yalovsky S. 2018. ROP GTPases structure–function and signaling pathways. Plant Physiol 176: 57–79. 10.1104/pp.17.01415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle H, Peters W, Palme K. 1991. The electrical response of maize to auxins. Biochim Biophys Acta 1064: 199–204. 10.1016/0005-2736(91)90302-O [DOI] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J. 2016. Tir1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. 10.7554/eLife.19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230. 10.1126/science.282.5397.2226 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. 2015. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci 112: 2275–2280. 10.1073/pnas.1500365112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. 2001. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- Gjetting KSK, Ytting CK, Schulz A, Fuglsang AT. 2012. Live imaging of intra-and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J Exp Bot 63: 3207–3218. 10.1093/jxb/ers040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering H. 1979. Depolarization of transmembrane potential of corn and wheat coleoptiles under reduced water potential and after IAA application. Plant Cell Physiol 20: 649–656. [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Han M, Park Y, Kim I, Kim EH, Yu TK, Rhee S, Suh JY. 2014. Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci 111: 18613–18618. 10.1073/pnas.1419525112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Shi Y, Liu G, Guo Y, Yang Y. 2018. Activation of ROP6 GTPase by phosphatidylglycerol in Arabidopsis. Front Plant Sci 9: 347. 10.3389/fpls.2018.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi KI, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H. 2012. Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem Biol 7: 590–598. 10.1021/cb200404c [DOI] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S. 2010. A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8: e1000282. 10.1371/journal.pbio.1000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel R, Lomax TL, Briggs WR. 1983. Auxin transport in membrane vesicles from Cucurbita pepo L. Planta 157: 193–201. 10.1007/BF00405182 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci 109: E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jásik J, Bokor B, Stuchlík S, Mičieta K, Turňa J, Schmelzer E. 2016. Effects of auxins on PIN-FORMED2 (PIN2) dynamics are not mediated by inhibiting PIN2 endocytosis. Plant Physiol 172: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelínková A, Müller K, Fílová-Pařezová M, Petrášek J. 2015. NtGNL1a ARF-GEF acts in endocytosis in tobacco cells. BMC Plant Biol 15: 272. 10.1186/s12870-015-0621-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimoto Y, Terasaka K, Hamamoto M, Takanashi K, Fukuda S, Shitan N, Sugiyama A, Suzuki H, Shibata D, Wang B, et al. 2012. Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol 53: 2090–2100. 10.1093/pcp/pcs149 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. 2003. AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777. 10.1242/dev.00659 [DOI] [PubMed] [Google Scholar]
- Kubeš M, Yang H, Richter GL, Cheng Y, Młodzińska E, Wang X, Blakeslee JJ, Carraro N, Petrášek J, Zažímalová E, et al. 2012. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J 69: 640–654. 10.1111/j.1365-313X.2011.04818.x [DOI] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. 2012. A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325. 10.1016/j.cub.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax TL, Mehlhorn RJ, Briggs WR. 1985. Active auxin uptake by zucchini membrane vesicles: quantitation using ESR volume and delta pH determinations. Proc Natl Acad Sci 82: 6541–6545. 10.1073/pnas.82.19.6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper MT, Spanswick RM. 1991. Auxin transport in suspension-cultured soybean root cells. II: Anion effects on carrier-mediated uptake. Plant Physiol 96: 192–197. 10.1104/pp.96.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthen H, Böttger M. 1988. Kinetics of proton secretion and growth in maize roots: action of various plant growth effectors. Plant Sci 54: 37–43. 10.1016/0168-9452(88)90053-2 [DOI] [Google Scholar]
- Mangano S, Denita-Juarez SP, Choi HS, Marzol E, Hwang Y, Ranocha P, Velasquez SM, Borassi C, Barberini ML, Aptekmann AA, et al. 2017. Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci 114: 5289–5294. 10.1073/pnas.1701536114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten I, Zeilinger C, Redhead C, Landry DW, Al-Awqati Q, Hedrich R. 1992. Identification and modulation of a voltage-dependent anion channel in the plasma membrane of guard cells by high-affinity ligands. EMBO J 11: 3569–3575. 10.1002/j.1460-2075.1992.tb05440.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Glanc M, Perrot-Rechenmann C, Friml J. 2016. Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000Res 5: 86. 10.12688/f1000research.7654.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Jorgensen P, Moran U, Weber G, Springer M. 2010. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res 38: D750–D753. 10.1093/nar/gkp889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamisawa K, Fukai K. 1991. Production of indole-3-acetic acid by Bradyrhizobium japonicum: a correlation with genotype grouping and rhizobitoxine production. Plant Cell Physiol 32: 1–9. [Google Scholar]
- Mockaitis K, Howell SH. 2000. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J 24: 785–796. 10.1046/j.1365-313x.2000.00921.x [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S. 2011. Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J 65: 309–318. 10.1111/j.1365-313X.2010.04423.x [DOI] [PubMed] [Google Scholar]
- Moss BL, Mao H, Guseman JM, Hinds TR, Hellmuth A, Kovenock M, Noorassa A, Lanctot A, Calderón Villalobos LIA, Zheng N, et al. 2015. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol 169: 803–813. 10.1104/pp.15.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z. 2012. ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10: e1001299. 10.1371/journal.pbio.1001299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer M, Moreno Castillo E, Ihling CH, Iacobucci C, Wilde V, Hellmuth A, Hoehenwarter W, Samodelov SL, Zurbriggen MD, Kastritis PL, et al. 2020. Flexibility of intrinsically disordered degrons in AUX/IAA proteins reinforces auxin co-receptor assemblies. Nat Commun 11: 2277. 10.1038/s41467-020-16147-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Yamada R, Hagihara S, Iwasaki R, Uchida N, Kamura T, Takahashi K, Torii KU, Fukagawa T. 2020. A super-sensitive auxin-inducible degron system with an engineered auxin-TIR1 pair. Nucleic Acids Res 48: e108. 10.1093/nar/gkaa748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, Petrášek J, Stierhof Y-D, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256. 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- Pan J, Fujioka S, Peng J, Chen J, Li G, Chen R. 2009. The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580. 10.1105/tpc.108.061465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Fang L, Liu J, Senay-Aras B, Lin W, Zheng S, Zhang T, Guo J, Manor U, Van Norman J, et al. 2020. Auxin-induced signaling protein nanoclustering contributes to cell polarity formation. Nat Commun 11: 3914. 10.1038/s41467-020-17602-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Dindas J, Król E, Friz T, Budnyk V, Teale W, Paponov M, Hedrich R, Palme K. 2019a. Auxin-induced plasma membrane depolarization is regulated by auxin transport and not by AUXIN BINDING PROTEIN1. Front Plant Sci 9: 1953. 10.3389/fpls.2018.01953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Friz T, Budnyk V, Teale W, Wüst F, Paponov M, Al-Babili S, Palme K. 2019b. Natural auxin does not inhibit Brefeldin A induced PIN1 and PIN2 internalization in root cells. Front Plant Sci 10: 574. 10.3389/fpls.2019.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Friml J. 2009. Auxin transport routes in plant development. Development 136: 2675–2688. 10.1242/dev.030353 [DOI] [PubMed] [Google Scholar]
- Petrášek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. 2006. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, del Mar Marquès-Bueno M, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, et al. 2019. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62. 10.1126/science.aav9959 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: 1–28. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramans Harborough S, Kalverda A, Thompson G, Kieffer M, Kubes M, Quareshy M, Uzunova V, Prusinska J, Hayashi K, Napier R, et al. 2019. A fuzzy encounter complex precedes formation of the fully-engaged TIR1-Aux/IAA auxin co-receptor system. bioRxiv 10.1101/781922 [DOI] [Google Scholar]
- Ramírez-Sánchez O, Pérez-Rodríguez P, Delaye L, Tiessen A. 2016. Plant proteins are smaller because they are encoded by fewer exons than animal proteins. Genome Proteom Bioinf 14: 357–370. 10.1016/j.gpb.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. 1975. Transport of indoleacetic acid in plant cells in relation to pH and electrical potential gradients, and its significance for polar IAA transport. New Phytol 74: 163–172. 10.1111/j.1469-8137.1975.tb02602.x [DOI] [Google Scholar]
- Ravichandran SJ, Linh NM, Scarpella E. 2020. The canalization hypothesis—challenges and alternatives. New Phytol 227: 1051–1059. 10.1111/nph.16605 [DOI] [PubMed] [Google Scholar]
- Reichardt I, Stierhof Y-D, Mayer U, Richter S, Schwarz H, Schumacher K, Jürgens G. 2007. Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr Biol 17: 2047–2053. 10.1016/j.cub.2007.10.040 [DOI] [PubMed] [Google Scholar]
- Retzer K, Lacek J, Skokan R, Del Genio CI, Vosolsobě S, Laňková M, Malínská K, Konstantinova N, Zažímalová E, Napier RM, et al. 2017. Evolutionary conserved cysteines function as cis-acting regulators of Arabidopsis PIN-FORMED 2 distribution. Int J Mol Sci 18: 2274. 10.3390/ijms18112274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. 2010. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121. 10.1016/j.cell.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubery PH, Sheldrake AR. 1974. Carrier-mediated auxin transport. Planta 118: 101–121. 10.1007/BF00388387 [DOI] [PubMed] [Google Scholar]
- Ruck A, Palme K, Venis MA, Napier RM, Felle HH. 1993. Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J 4: 41–46. 10.1046/j.1365-313X.1993.04010041.x [DOI] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12: 198–207. 10.1101/gad.12.2.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheitz K, Lüthen H, Schenck D. 2013. Rapid auxin-induced root growth inhibition requires the TIR and AFB auxin receptors. Planta 238: 1171–1176. 10.1007/s00425-013-1941-x [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102. 10.1038/emboj.2013.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA. 2017. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J 36: 886–903. 10.15252/embj.201694816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn AP, Goldsmith MH. 1988. Regulation of electrogenic proton pumping by auxin and fusicoccin as related to the growth of Avena coleoptiles. Plant Physiol 88: 131–138. 10.1104/pp.88.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HW, Depew CL, Miller ND, Monshausen GB. 2015. The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr Biol 25: 3119–3125. 10.1016/j.cub.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Simon S, Kubeš M, Baster P, Robert S, Dobrev PI, Friml J, Petrášek J, Zažímalová E. 2013. Defining the selectivity of processes along the auxin response chain: a study using auxin analogues. New Phytol 200: 1034–1048. 10.1111/nph.12437 [DOI] [PubMed] [Google Scholar]
- Sorek N, Segev O, Gutman O, Bar E, Richter S, Poraty L, Hirsch JA, Henis YI, Lewinsohn E, Jürgens G, et al. 2010. An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr Biol 20: 914–920. 10.1016/j.cub.2010.03.057 [DOI] [PubMed] [Google Scholar]
- Swarup R, Péret B. 2012. AUX/LAX family of auxin influx carriers—an overview. Front Plant Sci 3: 225. 10.3389/fpls.2012.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H. 1985. H+-translocating ATPases: advances using membrane vesicles. Annu Rev Plant Physiol 36: 175–208. 10.1146/annurev.pp.36.060185.001135 [DOI] [Google Scholar]
- Sze H, Li X, Palmgren MG. 1999. Energization of plant cell membranes by H+-pumping ATPases: regulation and biosynthesis. Plant Cell 11: 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. 2012. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159: 632–641. 10.1104/pp.112.196428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Tao L, Cheung AY, Wu H. 2002. Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14: 2745–2760. 10.1105/tpc.006320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyn A, Wagner G, Felle HH. 1991. Signal transduction in Sinapis alba root hairs: auxins as external messengers. J Plant Physiol 139: 187–193. 10.1016/S0176-1617(11)80606-1 [DOI] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, Endo TA, Kimura S, Zhang H, Nomoto M, Tada Y, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat Chem Biol 14: 299–305. 10.1038/nchembio.2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. 2005. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885. 10.1038/sj.emboj.7600659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenseel MH, Meyer AJ. 1997. Bioelectricity, gravity and plants. Planta 203: S98–S106. 10.1007/PL00008122 [DOI] [PubMed] [Google Scholar]
- Winkler M, Niemeyer M, Hellmuth A, Janitza P, Christ G, Samodelov SL, Wilde V, Majovsky P, Trujillo M, Zurbriggen MD, et al. 2017. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat Commun 8: 15706. 10.1038/ncomms15706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. 2010. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110. 10.1016/j.cell.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, et al. 2014. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028. 10.1126/science.1245125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. 2006. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127. 10.1016/j.cub.2006.04.029 [DOI] [PubMed] [Google Scholar]
- Young LM, Evans ML, Hertel R. 1990. Correlations between gravitropic curvature and auxin movement across gravistimulated roots of Zea mays. Plant Physiol 92: 792–796. 10.1104/pp.92.3.792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhang Y, Moss BL, Bargmann BOR, Wang R, Prigge M, Nemhauser JL, Estelle M. 2015. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants 1: 14030. 10.1038/nplants.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaina S, Reggiani R, Bertani A. 1990. Preliminary evidence for involvement of GTP-binding protein(s) in auxin signal transduction in rice (Oryza sativa L.) coleoptile. J Plant Physiol 136: 653–658. 10.1016/S0176-1617(11)81339-8 [DOI] [Google Scholar]
- Zhou X, Levin EJ, Pan Y, McCoy JG, Sharma R, Kloss B, Bruni R, Quick M, Zhou M. 2014. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 505: 569–573. 10.1038/nature12811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M, Bilanovičová V, Seifu YW, Nodzyński T. 2019. The nuts and bolts of PIN auxin efflux carriers. Front Plant Sci 10: 985. 10.3389/fpls.2019.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]