Abstract
Concepts of evolutionary biology suggest that morphological change may occur by rare punctual but rather large changes, or by more steady and gradual transformations. It can therefore be asked whether genetic changes underlying morphological, physiological, and/or behavioral innovations during evolution occur in a punctual manner, whereby a single mutational event has prominent phenotypic consequences, or if many consecutive alterations in the DNA over longer time periods lead to phenotypic divergence. In the marine teleost, sablefish (Anoplopoma fimbria), complementary genomic and genetic studies led to the identification of a sex locus on the Y Chromosome. Further characterization of this locus resulted in identification of the transforming growth factor, beta receptor 1a (tgfbr1a) gene, gonadal somatic cell derived factor (gsdf), as the main candidate for fulfilling the master sex determining (MSD) function. The presence of different X and Y Chromosome copies of this gene indicated that the male heterogametic (XY) system of sex determination in sablefish arose by allelic diversification. The gsdfY gene has a spatio-temporal expression profile characteristic of a male MSD gene. We provide experimental evidence demonstrating a pivotal role of a transposable element (TE) for the divergent function of gsdfY. By insertion within the gsdfY promoter region, this TE generated allelic diversification by bringing cis-regulatory modules that led to transcriptional rewiring and thus creation of a new MSD gene. This points out, for the first time in the scenario of MSD gene evolution by allelic diversification, a single, punctual molecular event in the appearance of a new trigger for male development.
Concepts of evolutionary biology suggest that morphological change may occur by rare punctual but rather large changes, or by more steady and gradual transformations (Eldredge and Gould 1972; Rhodes 1983; Mayr 1997). Thus, by analogy, the question can be asked whether genetic changes underlying morphological, physiological, or behavioral innovations during evolution occur in a punctual manner, whereby a single mutational event has a prominent phenotypic consequence, or if many consecutive alterations in DNA sequences over longer time periods lead to phenotypic divergence.
To investigate this question, the evolution of sex determination systems offers a very favorable situation because of the peculiarly high turnover rate of its genetic control in certain groups (Charlesworth and Charlesworth 2005; Herpin and Schartl 2008). Among vertebrates, teleost fishes display by far the highest diversity of sex determination systems and sex differentiation mechanisms (Herpin and Schartl 2015; Guiguen et al. 2018). Identification of master sex determining (MSD) genes on sex chromosomes from several fish species confirmed the concept that genetic triggers at the top of the regulatory hierarchy have changed radically as new species have evolved (Pan et al. 2016, 2019, 2021), whereas downstream regulatory networks remained more stable, generally exerting similar functions in driving testicular or ovarian differentiation in different species (Myosho et al. 2012; Kaneko et al. 2015; Zhang et al. 2016). The biological meaning and evolutionary processes of this outstanding molecular diversity to trigger the development of either testes or ovaries remain largely unknown. New MSD genes primarily emanate from one of two evolutionarily conserved processes: (1) sporadic gene duplication and insertion, followed by sub- and/or neo-functionalization; or (2) allelic diversification of a pre-existing locus (Kikuchi and Hamaguchi 2013; Guiguen et al. 2018). However, the molecular changes that allow new MSD genes to exert a novel function are not well understood aside from a few enigmatic studies in model species (for review, Herpin and Schartl 2008, 2015). A much broader knowledge is necessary to conclude about the evolutionary processes at work that bring about the great variety of fundamental steps in development, reproduction, and speciation.
In the marine teleost, sablefish (Anoplopoma fimbria), complementary genomic and genetic studies recently led to the identification of a sex locus on the Y Chromosome (Rondeau et al. 2013). Further characterization of this locus resulted in identification of the transforming growth factor, beta receptor 1a (tgfbr1a) gene, gonadal somatic cell derived factor (gsdf), as the main candidate for fulfilling the MSD function. The presence of different X and Y Chromosome copies of this gene indicated that the male heterogametic (XY) system of sex determination in sablefish arose by allelic diversification.
This study aims to determine what gave rise to diversification of the gsdfX and gsdfY paralogs in sablefish and whether X- and Y-specific DNA inserts in the promoter region upstream of these genes harbor elements that influence their expression. Spatio-temporal expression of the gsdf genes and other key genes associated with sex determination and gonadal differentiation were analyzed to determine if gsdfY exhibits characteristics of a male MSD gene. Moreover, a series of experiments was conducted to examine whether a transposable element (TE) from the hAT family played a critical role in the diversification of gsdfY via introduction of transcription factor-binding sites associated with the initiation and regulation of testicular development. This study is the first in the scenario of MSD gene evolution via allelic diversification to identify a single, punctual molecular event giving rise to a new trigger for male development.
Results
Allelic diversification gave rise to gsdfX and gsdfY with strict gonadal expression
Recent development of genetic tools in sablefish, including polymorphic markers and high-resolution linkage maps, have allowed for the successful mapping of different phenotypes of interest, including sex (Rondeau et al. 2013). Single nucleotide polymorphic (SNP) and microsatellite markers obtained from 35,000 assembled transcript sequences and 360 transcribed polymorphic loci from two families of sablefish permited the production of a map of 24 linkage groups (Rondeau et al. 2013). Although comparative mapping was unsuccessful in linking sex to the female genetic map, this trait clearly mapped to the male linkage group 14 of sablefish, indicating a male heterogametic system (Rondeau et al. 2013), which was subsquently verified by steroid-induced sex reversal and targeted breeding crosses (Luckenbach et al. 2017). The sex-specific regions were located in the vicinity of the gsdf gene. Gsdf has been described as an important component of the male pathway in gonad development in some other fish species (Myosho et al. 2012; Kaneko et al. 2015; Zhang et al. 2016). Several SNPs in the upstream and intronic portions of the sablefish gsdf gene appeared to be linked to sex. Of note, two exonic SNPs were identified within the gsdf coding sequence (CDS), one of which is missense and causes a phenylalanine (F) to leucine (L) change between the X and Y Chromosome copies (Fig. 1A; Rondeau et al. 2013). Otherwise, the gsdfX and gsdfY open reading frames (ORFs) are identical. Determination of the complete gsdfX and gsdfY mRNAs showed that they were each 2182 bp in length and had a 5′ untranslated region (UTR) of 145 and 3′ UTR of 1364 bp. Each mRNA also had an alternate polyadenylation site that reduced the 3′ UTR to 896 bp. Several SNPs were identified in the UTRs, with four being unique to the Y-Chromosomal copy (Fig. 1A). However, the most striking allele-specific features are located upstream of the translated region. The Y-copy has an insertion of 936 bp located 482 bp upstream of the gsdfY start codon, and the X-copy has an insertion of 412 bp located 1298 bp upstream of the gsdfX start codon (Fig. 1B). PCR amplification using primers designed to target these two regions confirmed that these X- and Y-allele-specific insertions segregate in agreement with the expected male and female genotypic sex (Fig. 1B).
Figure 1.
Allelic diversification gave rise to the sablefish (A. fimbria) gsdfX and gsdfY genes with sexually dimorphic expression. (A) Maps of the full-length sablefish gsdfX and gsdfY mRNAs, including polymorphisms and alternative poly-adenylation sites. (CDS) Coding sequence, (UTR) untranslated region, (SNP) single nucleotide polymorphism. (B) PCR amplification of gsdfX and gsdfY on X and Y Chromosomes, respectively, based on specific inserts upstream of the coding sequences. Amplification of an upstream sequence common to the two gsdf paralogs was used as a control. Bands represent amplification from 10 genotypic males (XY) and 10 genotypic females (XX). The depiction shows the general gene structure and placement of assay primers.
gsdfY is expressed during the sex-determining period only in male sablefish and prior to other genes associated with gonadal sex differentiation
Semiquantitative analysis of the tissue distribution of the two gsdf allelic variant mRNAs (PCR targeting both gsdf mRNAs) revealed exclusive gonadal expression in juvenile sablefish (Fig. 2A). To better characterize temporal and spatial expression of the two sex-specific gsdf allelic variants in relation to the molecular and morphological development of male and female gonad primordia, ontogenetic gene expression profiles for gsdfX, gsdfY, and other key markers of gonadal sex differentiation were established (Fig. 2). The gsdfX variant was expressed at very low levels in both XX- and XY-genotype fish from hatching to 40-mm-sized fry (Fig. 2B). However, the gsdfY variant was particularly highly expressed in XY individuals beginning around the time of hatching. During later ontogenetic development, expression of gsdf in XX fish remained low relative to that of XY fish, which exhibited increases during testicular differentiation and development (Fig. 2B). In juvenile gonads, in situ hybridizations (ISHs) using a probe that does not discriminate between the two gsdf allelic variants revealed that, in both testis and ovary, gsdf transcripts are expressed in the somatic supporting cells that surround the germ cells (Fig. 2C–H). Early markers for gonadal sex differentiation and development, such as the transcription factors wt1a, dmrt1, and foxl2, and the steroidogenic enzyme cyp19a1a, start to be expressed at hatching, reaching an initial peak at 5 to 10 mm, at which point none of the genes displayed a sexually dimorphic pattern of expression (Fig. 2I–L), unlike gsdfY as noted above (Fig. 2B). Only during later ontogenetic development, corresponding with the period of gonadal sex differentiation, sexually dimorphic expression was apparent, with dmrt1 being overexpressed in XY fish (Fig. 2J) and cyp19a1a and foxl2 being overexpressed in XX fish (Fig. 2K,L). Transcripts for wt1a, on the other hand, did not exhibit sexually dimorphic expression during ontogeny (Fig. 2I).
Figure 2.
Temporal and spatial expression of gsdfX, gsdfY, and other early gonadal sex marker mRNAs in male and female developing gonads of sablefish. (A) Semiquantitative tissue distribution of gsdf (assay targeting both gsdf variants) in brain (B), pituitary (P), gill (G), heart (H), ovary (O), testis (T), muscle (M), kidney (K), spleen (S), liver (L), intestine (I), and stomach (St). Transcript levels of eef1a1l1 were assessed to verify quality and loading of cDNAs. (B) Ontogenetic gene expression profiles of gsdfX and gsdfY sex-specific allelic variants during early gonadal primordium development and sex differentiation in genotypic female (XX) and male (XY) sablefish. The upper panel in B shows gonadal gsdf expression (nonvariant specific assay) during ontogenetic development. The lower panel (gray shaded) shows gene expression results for assays targeting both X and Y Chromosomal transcripts during early larval development. (C–H) In situ hybridization (ISH) localization of gsdf mRNA in juvenile sablefish gonads. (C,D) ISH, (E) hematoxylin/eosin (HE) staining of sablefish testes. (F,G) ISH, (H) HE staining of sablefish ovaries. (I–L) Ontogenetic expression profiles for wt1a, dmrt1, cyp19a1a, and foxl2, across early gonadal development between genotypic females and males. (B,I–L) Filled circles denote XX-genotype fish and open squares denote XY-genotype fish. See Hayman et al. (2021) for additional detail on gonadal gene expression data. Scale bars: C, F, 200 µm; D, G, E, and H, 20 µm; inserts in E and H, 10 µm. See also Supplemental Data S1 for raw data.
Functional assays reveal no difference in gsdfX and gsdfY biochemical activity/signaling properties
Within their respective CDS, gsdfX and gsdfY allelic variants exhibit a unique missense SNP resulting in a phenylalanine (F) to leucine (L) change at position 5 (pF5L) (see Fig. 1A; Supplemental Fig. S1). Although this residue is not conserved throughout evolution (Gautier et al. 2011) and apparently does not impair the quality of the signal peptide (Supplemental Fig. S1B), we nevertheless explored whether this unique mutation, nested within the signal peptide of the pro-domain of Gsdf, could result in differences in downstream signal transduction of the Gsdf ligand variants after receptor binding. To this end, we used a reporter assay to identify differential activation of Smad effectors (Fig. 3A). A luciferase reporter and transactivator plasmids (for Smad1, 2, 3, 5, or 8) were cotransfected with sablefish GsdfX and GsdfY expression plasmids in medaka fibroblast cells (Fig. 3A) to quantify, after binding to endogenous receptors, the relative differential Smad phosphorylation states using a luciferase activity assay. Although basal phosphorylation states of Smad1, 5, and 8 were not significantly impacted by either GsdfX or GsdfY expression, phosphorylation of Smad2 and 3 significantly increased for both Gsdf variants (Fig. 3B,C). The degree of stimulation of the two Gsdf variants was similar (around three times the basal activity) with respect to activation of Smad2 and 3. In conclusion, the single amino-acid difference between the X- and the Y-encoded proteins does not appear to impact the biochemical signaling function of the sablefish Gsdf proteins with regard to their receptor binding and subsequent Smad activation in the cellular environment tested (i.e., medaka fibroblast cells).
Figure 3.
Differential activation of downstream Gsdf signaling pathway components upon selective GsdfX or GsdfY expression. (A) Medaka fibroblast cells (OLF cell line) were cotransfected with a luciferase reporter construct (UAS-luc) and different combinations of Smad-phosphorylation-dependent transactivating GAL4 constructs (Smad1, 2, 3, 5, or 8 -GAL4). Cells were either stimulated (cotransfection) or not (control) with GsdfX or GsdfY. In the absence of any induced signaling, the fusion proteins Smads-GAL4 remain in the cytoplasm and the luciferase reporter is only activated at a basal level. If activated, the Smads-GAL4 proteins are phosphorylated, translocate into the nucleus, and an increased luciferase expression is recorded. Results are expressed as the relative stimulation of Smad phosphorylation after either GsdfX or GsdfY stimulation compared to control (no stimulation). Irrespective of the Gsdf variants employed, monitoring of Smad1, 2, 3, 5, and 8 phosphorylation states (relative luciferase activity) upon stimulation with either GsdfX (B) or GsdfY (C) revealed that only Smads 2 and 3 were activated in both situations, whereas Smads 1/5/8 always remained unresponsive. See also Supplemental Data S1 for raw data. (n.s.) P > 0.05; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Evolution of the gsdfX/Y promoter sequences
During the process of allelic diversification, the transcriptional context clearly changed between the two gsdf variants (see Fig. 2B). To obtain insights into the sequence evolution of the cis-regulatory regions of the gsdfX and gsdfY genes, their upstream regions were analyzed in detail (Fig. 4A).
Figure 4.
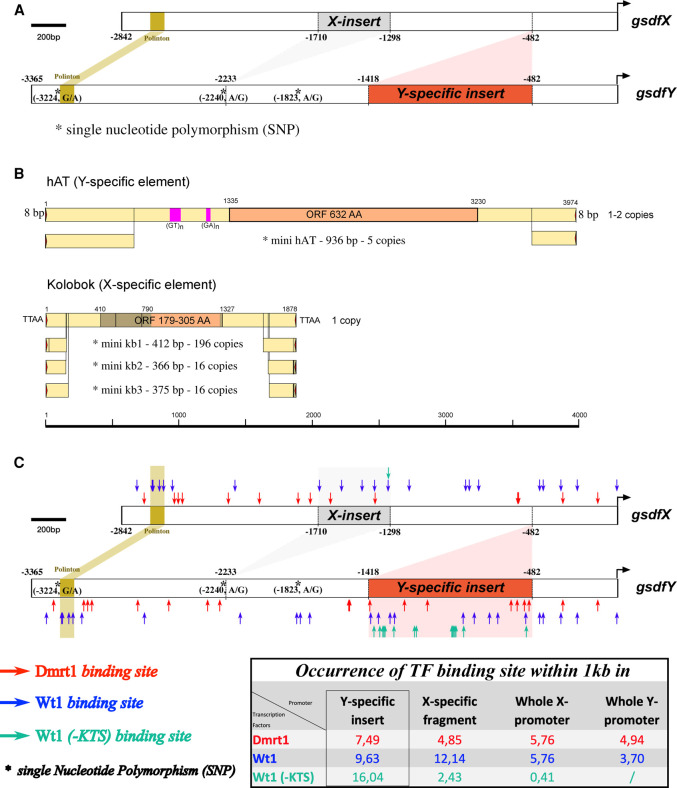
Comparative analysis of the gsdfX and gsdfY promoters and their transcription factor binding sites. (A) The analyzed promoter region of gsdfX in comparison to its gsdfY paralog. Length differences between the two gsdfX and gsdfY promoters are due to Y- and X- specific regions of which unique Y- (936 bp) and X- (412 bp) inserts have been, respectively, added and lost concomitantly during the allelic diversification event. The Y-specific insert is made of a transposable element of the hAT family. (B) Characteristics of Y- and X-specific elements in the gsdf promoter and their relatives. The scale and positions are in nucleotides. For each structure, the copy number identified in the new assembly is indicated. Typical target-site duplication is shown (in size or in sequence) at both ends of the longest elements. For both elements, the copy inserted in the gsdf promoter is an internally deleted version of a longer coding element, present in one or two copies. The ORF is indicated in orange and terminal inverted repeats in pink. For Kolobok, possible extension of the ORF is shown in brown (this would include two frameshift/stop codons). The asterisk indicates the MITE subfamily to which the gsdf-inserted copy belongs. The average number of hits per assembly (with positive hits) is indicated. Please note that the number of detected hits is highly dependent on the assembly quality (N50), so that absence of a hit is not proof that the element is not present in the species. (C) Analysis of the hAT transposable element revealed that its sequence contains an overrepresentation of Dmrt1 and Wt1(-KTS) binding sites compared to the remaining sequences of either the X-specific fragment or the whole X- and Y-promoters.
Comparison of the promoter regions of the sablefish gsdf variants upstream of the transcription start sites revealed a clear size difference of about 500 bp in length. This size difference between the gsdfX and gsdfY promoters is due to unique Y- and X-specific insertions (Fig. 4A). Located 482 bp upstream of the transcription start, the Y-specific insertion is 936 bp in length (Fig. 4A). An X-specific insertion of 412 bp is located 1298 bp upstream of the transcription start (Fig. 4A).
Sequence analyses of both inserted sequences revealed that they correspond to repeated TE derivatives (Fig. 4B). Four copies of the Y-specific element were detected at other positions than the gsdfY locus in the A. fimbria genome assembly (NCBI BioProject [https://www.ncbi.nlm.nih.gov/bioproject/] PRJNA656728; GCA_ 000499045.2). The insertion present in gsdfY displays some specific small indels relative to those copies. This element, 927 bp long, does not code for a protein but presents two terminal inverted repeats (TIR) of 12 bp and is inserted in a target site duplication (TSD) of 8 bp. Two other, longer sequences showing homology with this element are present in the genome and contain an internal ORF encoding a putative protein with homology to a hAT Class II element, which is not interrupted by frameshifts or stop codons (Fig. 4B). Hence, this TE family could potentially be active, although the copy number in the assembled genome is very low. The shorter copies correspond to MITEs (miniature inverted transposable elements), resulting from an internal deletion of the longest element, keeping only 595 nt from the 5′ side and 340 nt from the 3′ side.
The X-specific element, on the other hand, has a size of 408 bp and is found in the genome with at least 228 homologous copies shorter than 500 bp. Its TIRs are 12 bp long, and the element is inserted into a TTAA TSD. Eight copies longer than 500 bp were found in the genome, from which two have homology to transposases of the Kolobok superfamily, which is characterized by short TIRs and insertion into TTAA TSD as well (Kapitonov and Jurka 2007). Hence, the X-specific element also corresponds to a MITE. Further detailed analyses of the distribution of the hAT and Kolobok elements in Actinopterygii and phylogenetic analyses are provided in Supplemental Figure S2 and Supplemental Table S2.
Identification of putative transcription factor binding sites within the gsdfX and gsdfY promoters
The sequences of the gsdfX and gsdfY promoters (–2842 and –3365 bp, respectively) (Fig. 4C) were then analyzed for the presence of putative binding sites for transcription factors. The Y-specific insert (hAT transposable element) is characterized by an overrepresentation of putative DNA-binding sites for Wt1 and Dmrt1 (Fig. 4C). Dmrt1 and Wt1, and more specifically the Wt1(-KTS) splice form, are evolutionarily conserved, dose-sensitive transcription factor proteins that are key regulators of male development. Being critical for male sex determination, both dmrt1 and wt1a/b typically exhibit early and male-biased sexually dimorphic gene expression patterns (Suzuki et al. 2002; Klüver et al. 2009; Herpin and Schartl 2011, 2015). We found that the sablefish Y-specific insert upstream of gsdf displays 7, 9, and 16 binding sites for Dmrt1, Wt1, and Wt1(-KTS), respectively (Fig. 4C). Hence, the number of Dmrt1 and Wt1(-KTS) binding sites within the Y-specific insert are up to 39 times higher than in the whole X promoter (Fig. 4C). Nearly all of the Wt1(-KTS) binding sites within the Y promoter, in comparison to the X promoter (15 vs. 1, respectively), were delivered by this Y-specific TE insertion (Fig. 4C).
This, together with the fact that early expression patterns of dmrt1 and wt1 largely overlap with gsdfY expression during the sex determination period (Fig. 2), suggests that the Y-specific region is of primary relevance for controlling gsdfY transcriptional regulation.
Transcriptional activities directed by the gsdfX and gsdfY promoters in different cell lines
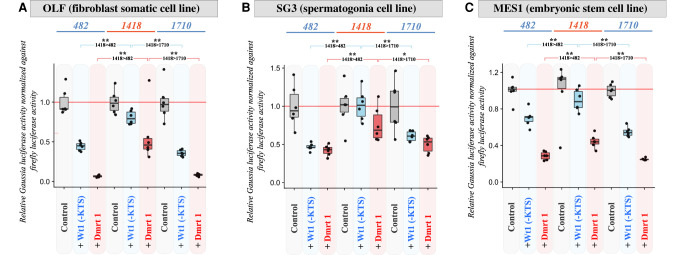
To evaluate the mechanisms possibly regulating differential gsdfX and gsdfY transcription, diverse portions of the gsdfX and gsdfY promoters upstream of the transcriptional start sites were cloned and used for promoter mutagenesis (or promoter bashing) luciferase assays (Fig. 5A–G) using three different medaka cell lines (OLF, MES1, and SG3) (see the Methods section for more information). Basal promoter activity was detectable with the minimal 482-bp proximal region (Fig. 5B–G). Adding more distal portions of the promoter (1298, 2233, 2842, or 3365) (see Fig. 5B–G) to the proximal region resulted in moderate negative modulation of promoter activity. In particular, in all cell lines, a drop in promoter activity was observed when the region encompassing the Y-specific insert was added (Fig. 5B–D, pink shading). Of note, addition of the X-specific region (1298 compared to 1710 in Fig. 5E–G) did not result in further regulation of promoter activity in fibroblast or spermatogonia cells (Fig. 5E,F), whereas a modest repression was recorded in embryonic stem cells (Fig. 5G). Finally, adding the most distal portion of the male promoter (3365) resulted in additional negative regulation of promoter activity in fibroblast cells (Fig. 5B), whereas no further regulation was apparent in either spermatogonia (Fig. 5C) or embryonic stem cells (Fig. 5D). This punctual discrepancy between cell types may point to the importance of the cell's identity (fully differentiated vs. stem cells) for integrating such regulations. Figure 5H recapitulates the observed changes in promoter activity determined by the promoter mutagenesis assays.
Figure 5.
Transient transfection analysis of proximal gsdfX and gsdfY promoter activities. (A) Different deletions of the 5′ gsdf promoters were generated (482, 1298, 1418, 1710, 2233, 2842, and 3365 bp), fused to a luciferase reporter, and analyzed for transcriptional activity after transient transfection in different medaka cell lines (B–G). The data are presented as normalized recorded Gaussia/firefly luciferase activity (see the Methods section for detailed information). For every construct, transfection was repeated six times. Error bars represent the standard deviation of the means. Light-pink shaded areas (B–D) emphasize the hAT- induced modulation of transcriptional activity. (H) Model for the sequential regulation of gsdfX and gsdfY promoter activities. See also Supplemental Data S1 for raw data.
Male-specific transcriptional regulation of gsdfY by Dmrt1 and Wt1(-KTS)
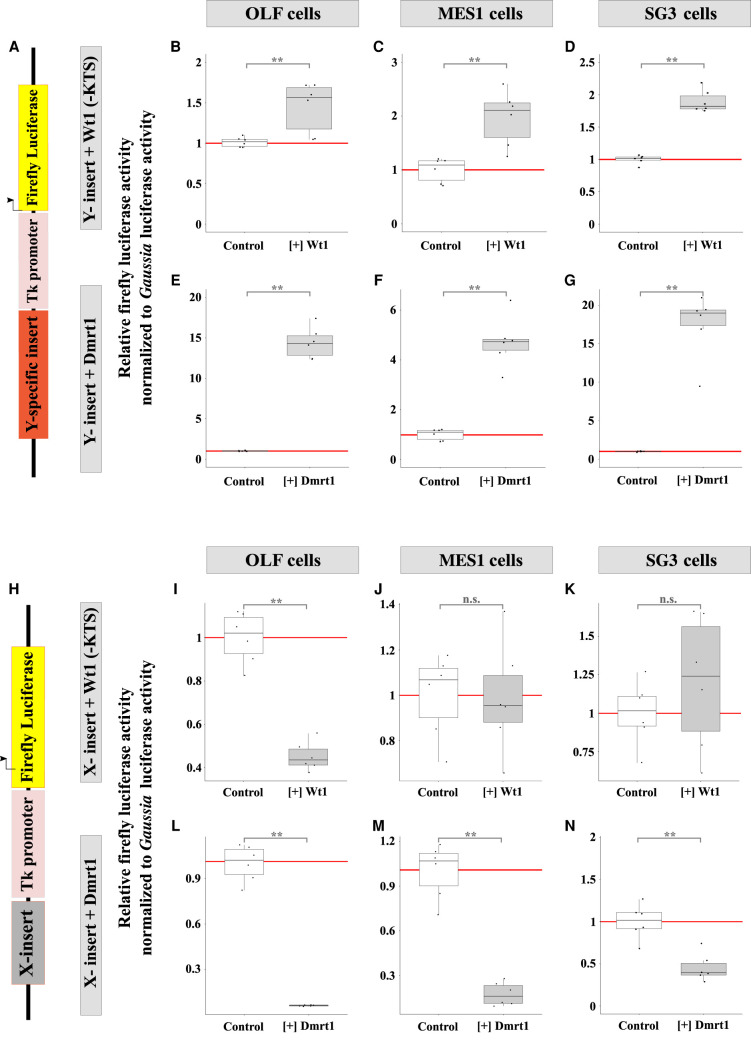
We then sought to determine if the function of the Y- and X- specific inserts in modulating gsdfY and gsdfX transcriptional regulation might be mediated by Dmrt1 and Wt1(-KTS). For this purpose, cotransfections and luciferase assays using either the 482-bp (minimal promoter), the 1418-bp (minimal promoter plus the Y-specific insert), or the 1710-bp (minimal promoter plus the X-specific insert) promoter:luciferase constructs (see Fig. 5A) were cotransfected with either Dmrt1- or Wt1(-KTS)-expressing plasmids in different cell lines (Fig. 6A–C). In the presence of either Dmrt1- or Wt1(-KTS)-expressing constructs, basal transcriptional expression (minimal promoter, 482-bp construct) was reduced in all cell types tested. With the addition of the X-specific insert to the minimal promoter (1710-bp construct), we saw no further effect on the initially observed Dmrt1- and Wt1(-KTS)-induced down-regulation. However, when the Y-specific insert was added to the minimal promoter (1418-bp construct), a clear up-regulation of basal transcriptional activity was apparent in the presence of either Dmrt1 or Wt1(-KTS) in all cell lines tested (Fig. 6A–C).
Figure 6.
hAT-mediated male-specific transcriptional regulation of the gsdfY promoter by Dmrt1 and Wt1(-KTS). (A–C) In vitro quantification of proximal gsdfX and gsdfY promoter activities (luciferase reporters), after Dmrt1 and Wt1(-KTS) transient transfection in OLF fibroblast (A), SG3 spermatogonia (B), and MES1 embryonic stem (C) cell lines from medaka (Oryzias latipes). For every construct, transfection was repeated six times. Error bars represent the standard deviation of the means. See the Methods section for detailed information about the luciferase constructs. See also Supplemental Data S1 for raw data.
To further evaluate the ability of the Y-specific insert to mediate transcriptional regulation on its own and whether it could be modulated by expression of Dmrt1 or Wt1(-KTS), the Y-specific insert alone was fused to the basal thymidine kinase (Tk) promoter, which confers only low transcription of the reporter, and placed in front of a luciferase reporter gene (Fig. 7A). In all cell types tested (Fig. 7B–G), the Y-specific insert conferred strong up-regulation of transcriptional activity—up to 18 times higher—compared to the minimal Tk promoter in the presence of either Wt1(-KTS) (Fig. 7B–D) or Dmrt1 (Fig. 7E–G). When the X-specific insert was fused (Fig. 7H), transcriptional activity of the minimal Tk promoter was either unaffected (Fig. 7J,K) or repressed (Fig. 7I,L–N) when in the presence of Wt1(-KTS) or Dmrt1 (Fig. 7I–N). As noted earlier, differences in the cell type (differentiated vs. stem cells) in the X-specific construct may be responsible for down-regulation in OLF (differentiated) cells when exposed to Wt1(-KTS) compared to MES1 and SG3 cells (stem cells); however, this effect was not apparent when exposed to Dmrt1. Figure 8, A and B, summarizes the above-described regulations.
Figure 7.
hAT- (Y-specific insert) and Kolobok- (X-insert) induced modulation of transcriptional regulation. (A–G) hAT-specific (Y-specific insert) induced modulation of transcriptional activity (luciferase reporter) of a thymidine kinase minimal promoter (A) upon transient transfection of dmrt1 or wt1(-KTS). (H–N) Kolobok-specific (X-insert) induced modulation of transcriptional activity (luciferase reporter) of a thymidine kinase minimal promoter (H) upon transient transfection of dmrt1 or wt1(-KTS). For every construct, transfection was repeated six times. Error bars represent the standard deviation of the means. See the Methods section for detailed information about the luciferase constructs. See also Supplemental Data S1 for raw data.
Figure 8.
Models for gsdfX and gsdfY transcriptional modulation by Dmrt1 and Wt1(-KTS). Due to opposite responsiveness to Dmrt1 and Wt1(-KTS) after the Y-specific hAT element insertion into the gsdfY promoter (A), gsdfX and gsdfY exhibit very early sexually dimorphic expression patterns (B). This example illustrates how sexually dimorphic expression of gsdfY and gsdfX are under the dependence of non-dimorphically expressed regulators (Dmrt1 and Wt1a, for instance). Hence, sexually dimorphic expression of gsdfY and gsdfX could depend on regulators that are not themselves dimorphically expressed.
Discussion
Potent innovations within regulatory networks can occur at the protein sequence level or produce alterations of cis-regulatory sequences defining transcription factor binding sites. After the ubiquitousness of repeated sequences was recognized, a longstanding hypothesis proposed that repeated sequences can take functional roles, for instance, in the 5′ regions of genes, by controlling transcription (Britten and Davidson 1969). This would link them to evolutionary variations and even novelties (Britten and Davidson 1971). Co-option of DNA sequences introduced during the invasion of TEs can generate, ex nihilo, new regulatory units, including enhancers (Glinsky 2015; Lynch et al. 2015; Notwell et al. 2015), repressors (Herpin et al. 2010), insulators (Wang et al. 2015), or even whole alternate gene promoters (Emera et al. 2012; Kapusta et al. 2013), substantially faster than single-point mutations. It is then postulated that co-option of ready-made cis-regulatory motifs nested within TEs facilitated substantial shifts in lineage-specific patterns of gene regulation over short evolutionary timescales (Rebollo et al. 2012).
Although the complex life history of sablefish (e.g., living 100+ yr and reaching sexual maturity at ∼5 yr old and 55 cm long [Mason et al. 1983]) precluded our ability to conduct classical gain- and loss-of-function experiments, our systematic examination of transposon-derived gsdf inserts and their transcriptional regulation provides compelling evidence that gsdfY is the male MSD gene in this species. Comparative mapping of the sablefish genome to that of the three-spined stickleback (Gasterosteus aculeatus) generated linkage maps successfully identifying the locus for sex on the male map (Rondeau et al. 2013). The three-spined stickleback is one of the most closely related species to sablefish (estimated divergence of 150 mya or less) with a fully sequenced and well-annotated genome. These comparative analyses specifically anchored the sablefish MSD region to linkage group 14, corresponding to Chromosome XIV of the three-spined stickleback genome, a region of approximately 2.4 Mbs for the X and Y Chromosomes. However, the sablefish MSD region does not correspond to any of the Y-specific regions of either the three-spined (Chr XIX [Peichel et al. 2004, 2020]) or nine-spined (Chr XII [Ross et al. 2009]) stickleback, which implies that their sex chromosomes evolved independently.
Screening a genotyping-by-sequencing (GBS) library allowed narrowing down the sablefish MSD region and isolating a restricted number of SNPs unequivocally linked to sex in the vicinity of the gsdf gene. Gsdf is a growth factor that displays key features of the Gsdf superfamily (Gautier et al. 2011). During vertebrate evolution, it has been lost in tetrapods (Forconi et al. 2013), and its biochemical function is not well studied. Gsdf protein is nevertheless assumed to have a major role in male gonadal development due to its expression in the early differentiating testis of all fish analyzed so far. In medaka, gsdf plays a critical role in testis development (Chakraborty et al. 2016; Zhang et al. 2016). Besides its proposed role in the gonadal downstream regulatory network in Oryzias latipes (Zhang et al. 2016), gsdf has made it to the top of the sex determining regulatory network in Oryzias luzonensis (Myosho et al. 2015), a sister species to medaka, where it serves as the male sex determining gene on the Y Chromosome.
In sablefish, gsdfY, in contrast to its X-linked counterpart, is specifically expressed in male fry earlier than other male or female sex-related genes and prior to both molecular and morphological sexual differentiation of the gonads (Fig. 2; also Hayman et al. 2021). Comparative analysis of the gsdfX and gsdfY expression patterns clearly showed that gsdfY, which is expressed much earlier than gsdfX, experienced transcriptional rewiring during the process of allelic diversification, ultimately giving rise to the X and Y Chromosomes of sablefish. Such acquisition of a new transcriptional context resulting in a different spatio-temporal expression pattern, compatible with a sex-determining function, seems to be the main prerequisite in the process of establishment and fixation of a new MSD gene. In the two sister species, Oryzias latipes and O. dancena, and in mammals, either dmrt1 or sox3 genes, respectively, were subjected to profound transcriptional rewiring for establishing either dmrt1bY (O. latipes; duplication/insertion [Herpin and Schartl 2009; Herpin et al. 2009, 2010]), sox3Y (O. dancena; allelic diversification [Takehana et al. 2014]), or SRY (most mammals; allelic diversification [Sekido and Lovell-Badge 2008]) as MSD genes (for review, see Herpin and Schartl 2015).
In sablefish, because the unique missense mutation between the two gsdf variants does not appear to drastically impact their physiological activity with regard to downstream activation of Smads, any processes of functional divergence of the protein variants after allelic diversification may be reasonably excluded. Uniquely, the newly acquired MSD function of the gsdfY gene seems to be entirely ascribable to its new pattern of expression, indicating a neo-functionalization process.
Here, we report that the high expression of the gsdfY allelic copy during gonadal sex differentiation is largely imputable to a Y-specific insert derived from a TE of the hAT family incorporated into its promoter. Gsdf is an important downstream component of the male sex determination regulatory network, which in sablefish, like in Oryzias luzonensis (Myosho et al. 2012), acquired the role of the MSD gene. Co-option of this TE within the “neo”-gsdfY promoter was likely sufficient for transforming and elevating a protein acting downstream in the sex determination network to a MSD gene being expressed at the right time and the right place. Deciphering the mechanism by which the neo-gsdfY is now transcriptionally controlled and up-regulated by dmrt1 and/or wt1a expression demonstrates the true exaptation of a TE into a regulatory region, thereby creating, de novo, a MSD gene.
Co-option of TEs, or TE-derived enhancers, bringing ready-made regulatory elements in a single step, in contrast to the stepwise accumulation of single mutations, might substantially facilitate immediate shifts in gene regulation over very short timescales. TEs have received particular attention in physiological processes that need rapid adaptation over evolutionary timescales, including reproduction and sex determination (Herpin et al. 2010; Lynch et al. 2011, 2015; Chuong 2013; Schartl et al. 2018). Particularly in medaka, it has been shown that, after local duplication of the dmrt1 gene, a new hierarchy was established following insertion of TEs into the regulatory region of the dmrt1bY gene on the sex chromosome (Herpin et al. 2010; Schartl et al. 2018).
Altogether, our results demonstrate that allelic diversification of the gsdf gene gave rise to the sex determination system in sablefish. Because sablefish is widely considered a panmictic species with no apparent population genetic structure (Jasonowicz et al. 2017), gsdfY is likely conserved as the MSD gene across the entire species. Importantly, the MSD function of gsdfY was not attained by acquisition of a new function of the protein itself but rather through the acquisition of elements in the promotor region. This resulted in a unique expression profile, which relocated gsdfY to the most upstream position in the sex-determining network.
The sablefish Y Chromosome provides an example of how an evolutionary novelty, which is predicted to require transcriptional rewiring of the regulatory network, was brought about after co-option of ready-made cis-regulatory sequences carried by a TE. Bringing another layer of transcriptional modulation via Dmrt1 and Wt1(-KTS) regulation, a unique TE exaptation into the gsdfY promoter appears to have created the MSD gene of sablefish.
Although direct causality between accumulation of Dmrt1 and Wt1(-KTS) binding sites and regulation remains to be investigated in greater detail, the hAT-type TE, in its entirety, confers early up-regulation of gsdfY expression by Dmrt1 and Wt1(-KTS), two key proteins of the canonical gonadal gene regulatory network. Thus, preventing any expression pattern redundancy between the two gsdf allelic copies, such divergent expression regulation might constitute a reasonable evolutionary scenario for the preservation of both gsdf gene copies (X and Y), protecting them from any purification/degeneration processes after allelic diversification. Finally, our data provide strong evidence for an efficient role(s) of TEs in the rewiring of gene regulatory networks in the particular context of establishing new master sex determinants over a very short timescale. An interesting future question regarding the evolutionary timescale of this occurence is whether the rare skilfish (Erilepis zonifer), the only other species with sablefish in the family Anoplopomatidae, also possesses the hAT and Kolobok TEs, particularly in the region upstream of gsdf.
Evolution of new sex determining genes by allelic diversification has often intuitively been associated with gradual processes that occur slowly over evolutionary timescales (Charlesworth 1991; Charlesworth and Charlesworth 2005). We, however, found that in sablefish, allelic diversification of a sex determination gene, initiated by the exaptation of a TE, led to complete transcriptional rewiring of the allele on the proto-Y Chromosome. This provides a unique functional example of a bona fide punctual process as an efficient alternative to the phyletic gradualism model (Sheldon 2001) for the molecular evolution of a master sex determining gene.
Methods
Bioinformatic analyses
Binding sites for Dmrt1, Wt1s, and other transcription factors were identified using MatInspector from the Genomatix portal (http://www.genomatix.de). High molecular weight DNA was extracted using the Qiagen midi prep kit (Qiagen). DNA was used to prepare libraries for three approaches for genome sequence data: (1) overlapping read pair shotgun data generated on the HiSeq X Ten platform; (2) 3-, 8-, and 20-kb mate pair libraries; and (3) Pacific Biosciences (PacBio) long-read data. For the mate pair libraries, the Lucigen NxSeq Clone Free Mate Pair chemistry (Lucigen) was used; each of these libraries was sequenced on the Illumina MiSeq. The data were trimmed for adapters, and then mate read pairs were scanned for the mate pair junction code sequence, and read pairs were split. The HiSeq X Ten data were trimmed and adapters removed using cutadapt v. 1.8.3 (Martin 2011). Data from a prior genome assembly (GCA_000499045.1) were combined with trimmed HiSeq X Ten and mate pair libraries using ALLPATHS-LG 52488 (Gnerre et al. 2011). PacBio data were error-corrected with proovread (Hackl et al. 2014) using the HiSeq X Ten data. The ALLPATHS assembly produced from all Illumina data was then combined with the error-corrected PacBio reads for a hybrid assembly using PBJelly2 (PBSuite v15.8.24 [English et al. 2012]). The completed hybrid assembly was evaluated for completeness using BUSCO (Simão et al. 2015) and the vertebrata_odb9 conserved gene database.
BLAST searches in the Anoplopoma fimbria assembled genome
Sequences for the X-specific and the Y-specific elements were used in a megaBLASTN search (BLAST 2.6.0+ [Camacho et al. 2009], default parameters) against the A. fimbria genome assembly (GCA_000499045.2). Hits present on the same scaffold or contig with a distance of <2000 bp were reassembled into a single copy. We further discarded incomplete or truncated copies (missing one end), or copies that possessed more than 100 Ns within the sequence. Copies longer than 1000 bp were used in a BLASTX search (evalue -1E-10) against the Repbase protein database (Repbase20.05_REPET edition, https://www.girinst.org/) in order to identify the elements and extract the ORF sequences.
Homology searches in the Perciformes and Actinopterygii genomes
For both elements, MITE and ORF sequences were used as queries in BLASTN searches (evalue -1E-10) against all Actinopterygii genomes present in the NCBI genome databases (932 assemblies, 672 species, https://www.ncbi.nlm.nih.gov/genome/), including Perciformes (38 species). Only hits longer than 200 bp (for search with ORFs) or longer than 100 bp (for search with MITE sequences) were kept. Distributions in the different Actinopterygii orders are shown in Supplemental Figure S2 and Supplemental Table S2.
Phylogenetic analysis
Sequences presenting homology with coding sequences were then used in a BLASTX search against the Repbase protein database (-evalue 1E-10, -max_targets_seqs 1). For each assembly, we then selected the sequence presenting the highest homology. Sequences were aligned according to codon sequences and then translated and aligned along the Repbase protein sequences using MAFFTv7.2 (Katoh and Standley 2013). We further removed close sequences from the same species and sequences presenting long internal deletions or insertions. Amino-acid alignments were trimmed in order to keep only conserved regions. Phylogenies (Supplemental Fig. S2) were reconstructed using FastTree v2.1.7 (Price et al. 2009). Robustness was assessed by the Shimodaira-Hasegawa (SH) test implemented in FastTree.
Cell lines and cell transfections
Medaka fibroblast-like (OLF), spermatogonia (SG3), and embryonic stem (MES1) cells were cultured as previously described (Hong et al. 2004; Thoma et al. 2011; Zhang et al. 2016). For transfection, cells were grown to 80% confluency in six-well plates and subsequently transfected with 5 µg expression vectors using either Lipofectamine (Invitrogen) or FuGENE (Roche) reagents as described by the manufacturers.
Luciferase assays
For promoter analyses (Figs. 5, 6A–C), 482-, 1298-, 1418-, 1710-, 2233-, 2842-, and 3365-bp fragments upstream of gsdfX and gsdfY promoters (see Fig. 5A for the map) were isolated by PCR from genomic DNA of sablefish, sequenced, and cloned into the pGLuc-basic plasmid (Gaussia luciferase; New England Biolabs). Differential Gaussia luciferase activities were then quantified using the Dual Luciferase Reporter Assay System from Promega and normalized against the cotransfected firefly luciferase-expressing control plasmid ptkLUC+ (accession number AF027128). For Figure 6D–H, the hAT Y-specific insert was cloned into the ptkLUC+ plasmid (firefly luciferase), upstream of the thymidine kinase minimal promoter, and (firefly) luciferase activities normalized using the Gaussia luciferase expressing pCMV-GLuc plasmid (New England Biolabs). Experiments for which error bars are shown resulted from at least six replicates and represent the standard deviation of the mean. Statistical significance was assessed by means of the Mann-Whitney U test.
UAS-GAL4-Smad-AD assay
For monitoring differential activation of Smads upon selective GsdfX or GsdfY expression, medaka fibroblast cells were seeded in six-well plates and cotransfected with a combination of four kinds of plasmids (see also Fig. 3): (1) an expression plasmid which encodes a fusion protein (Smad1-AD-GAL4-DBD, Smad2-AD-GAL4-DBD, Smad3-AD-GAL4-DBD, Smad5-AD-GAL4-DBD, or Smad8-AD-GAL4-DBD; 300 ng per well) that will translocate to the nucleus upon phosphorylation by the different Gsdfs and in return transactivate the UAS-4 promoter through its GAL4 DNA-binding domain; (2) a reporter plasmid, which codes for luciferase under the control of a minimal promoter, which contains UAS sequences (UAS-luc, firefly luciferase; 300 ng per well); (3) plasmids coding for the different Gsdf ligands to be tested for signaling activity (either pCMV-gsdf-X, pCMV-gsdf-Y, or a control plasmid; 400 ng per well); and (4) a Gaussia luciferase expression plasmid for normalization (pCMV-Gluc; 5 ng per well). After 24 h, cells were washed with 2× phosphate-buffered saline solution (PBS) and lysed with 75 µL of passive lysis buffer (Dual Luciferase Reporter Kit Assay; Promega), and then subjected to luciferase assay. Firefly luciferase activity (UAS-luc reporter constructs) was quantified using the Dual Luciferase Reporter Assay System (Promega) and normalized against cotransfected Gaussia luciferase-expressing plasmid. Data sets are the result of at least four independent cell transfections and luciferase measurements. Statistical significance was assessed by means of the Mann-Whitney U test (n = 4 or 8).
Nucleotide isolation and cDNA synthesis
Sablefish tissues used for various analyses (gsdf mRNA cloning, PCR, tissue distribution, and ontogenetic expression) were collected from animals cultured at the Northwest Fisheries Science Center, Manchester Research Station. All fish sampled were produced by in vitro fertilization using wild sablefish broodstock captured off the coast of Washington State, USA (Cook et al. 2015). Genomic DNA was isolated from fin clips using the DNeasy Blood and Tissue kit (Qiagen) or gonadal tissue following the Tri-Reagent (Molecular Research Center) DNA isolation protocol. RNA isolation and cDNA synthesis generally followed previous work (Luckenbach et al. 2011; Hayman et al. 2021). In brief, tissues were preserved in RNAlater (Ambion) or quickly frozen in liquid nitrogen and total RNA later extracted using 1 mL Tri-Reagent following the manufacturer's protocol including the optional spin step after homogenization. Total RNA was then DNase-treated using the TURBO DNA-free kit (Invitrogen) and quality assessed by spectrophotometry (NanoDrop). Reverse transcription (RT) of 500 ng (tissue distribution) or 250 ng (gene expression) of RNA per 10 µL reaction was done using SuperScript II (Invitrogen).
Full-length gsdfX and gsdfY mRNA sequences
To obtain full-length gsdfX and gsdfY mRNA sequences, DNase-treated RNA samples from immature testis and ovary were used to generate RACE-ready first-strand cDNA (GeneRacer kit; Invitrogen) following the manufacturer's protocol. GeneRacer and gene-specific primers were used for RACE PCR (Supplemental Table S1). Subsequent 5′ and 3′ products were run on agarose gels, bands were punched and purified (Qiagen Gel Extraction Kit) and then cloned using the Zero Blunt TOPO PCR Cloning kit (Invitrogen) with at least three clones per amplicon sequenced in both directions (GenScript). Full-length sequences for gsdfX and gsdfY were aligned using MacVector software version 12.6 (Accelrys) and deposited in the NCBI GenBank database (see Data access).
Genotypic sexing assays
For determination of genotypic sex in sablefish, PCR assays were conducted that targeted the X-insert in the gsdf promoter (Rondeau et al. 2013; Luckenbach et al. 2017). To demonstrate that the gsdfX and gsdfY alleles were in accordance with phenotypic sex, PCRs were developed targeting chromosome-specific inserts in the regions upstream of gsdf (Supplemental Table S1). Primer pairs amplified sequences upstream of and into target X and Y inserts. As a positive control, an upstream sequence common to both gsdf paralogs was also amplified. PCR products were resolved on a 1.5% agarose gel with GelRed stain (Biotium).
Tissue distribution and ontogenetic expression
For tissue distribution and ontogenetic gene expression analysis, some data from a previous study characterizing sablefish sex differentiation (Hayman et al. 2021) were incorporated. In addition, an early developmental series of samples was collected that targeted the period of sex determination. For this, cultured sablefish embryos were collected beginning 5 d prior to hatching, and whole larvae were collected at hatch and then sampled weekly through ∼40 mm fork length (FL).
Tissue distribution and gene expression methods were reported previously or as generally described (Luckenbach et al. 2017; Hayman et al. 2021). In brief, tissue distribution semiquantitative PCRs were conducted across 11 tissues from an immature female sablefish, with the exception of testis tissue, which was collected from an immature male (both fish were 251 mm FL). Equal amounts of cDNA (2 ng) were amplified over 32 cycles and resolved on a 1.5% agarose gel. For gene expression analyses, RT-quantitative PCR (RT-qPCR) assays were conducted in 384-well plates run on a 7900HT Fast Real-Time PCR machine (Applied Biosystems). Reactions contained 1× Power SYBR Green Master Mix (Applied Biosystems), 150 nM of each gene-specific primer (Supplemental Table S1), and 2 ng of cDNA template in 12.5-μL volumes. Standards were serially diluted and run in triplicate with pooled cDNA amounts of 5, 1, 0.25, or 0.05 ng per reaction. No amplification and no template controls (NACs and NTCs) were included in all assays and had no amplification. To confirm amplification of a single PCR and correct target in experimental samples, melt curves were included and PCR products were directly sequenced for each targeted gene. All RT-qPCR data were normalized to the geometric mean of three established reference genes (actb1, btf3, and rpl4) (Hayman et al. 2021).
In situ hybridization
Testis and ovary samples collected from juvenile sablefish (male: 235 mm FL, 125g body weight; female: 235 mm FL, 130g) were fixed in Bouin's fixative at 4°C. After fixation, the tissues were dehydrated, embedded in paraffin, and sectioned at a depth of 4 μm. A subset of the paraffin sections was stained with hematoxylin and eosin (HE) stains. The localization of sablefish gsdf mRNA was assessed by in situ hibridization. The full gsdf CDS (648 bp) was amplified using SF-GSDF-ISH-F: 5′-ATGTCCTTTACCCTCGTTGTCACGACGATG-3′ and SF-GSDF-ISH-R: 5′-TTACTCTCTGCTGGGTGGCTGGAGGTT-3′ primers (GenBank Accession # MT900066) and subcloned into the pGEM T-easy vector (Promega). Sense- and antisense-RNA probes were transcribed in vitro using digoxigenin-labeled uridine triphosphate (UTP; Roche) and SP6 or T7 RNA polymerase (Promega). ISH was performed as described by Octavera and Yoshizaki (2019).
Ethics statement
All sablefish were handled by NOAA Northwest Fisheries Science Center staff in accordance with National Research Council guidelines for aquatic animals (National Research Council 2011) and the American Veterinary Medical Association (https://olaw.nih.gov/sites/default/files/Euthanasia2007.pdf). For sampling, fish were first euthanized using a lethal dose of Tricaine-S (200 mL/L; Western Chemical) and then decapitated prior to tissue collections.
Data access
The original sablefish genome assembly (GCA_000499045.1) presented in Rondeau et al. (2013) was updated with the collection of additional sequence data from the same individual (a male) and was submitted to the NCBI Assembly database (https://www.ncbi.nlm.nih.gov/assembly) under accession number GCA_000499045.2. The updated Whole Genome Shotgun project was submitted to the Assembly database under accession number AWGY00000000.2. Full-length mRNA sequences for gsdfX and gsdfY were submitted to the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MT900065 and MT900066, respectively. All relevant data are within the paper or as Supplemental Material (Supplemental Data S1).
Supplementary Material
Acknowledgments
The authors thank the staff of the Marine Fish and Shellfish Biology Program at NOAA's Northwest Fisheries Science Center for producing cultured sablefish used in this project. Marian R. Fairgrieve, Elizabeth K. Smith, and William T. Fairgrieve provided valuable assistance with fish care and/or tissue sampling. Ben F. Koop and Eric B. Rondeau shared tissue for the updated sablefish genome assembly. This project was made possible via support provided by National Oceanic and Atmospheric Administration (NOAA) Fisheries International Science Grants from the Office of Science and Technology to J.A.L., K.M.N., and other NOAA colleagues for research with INRAE. Funding for additional sablefish genome sequencing was obtained through the USDA NRSP8 program. This work was also supported by a grant (SCHA 408/12-1; HE 7135/2-1) from the Deutsche Forschungsgemeinschaft to A.H. and M.S. and Crédits Incitatifs Phase 2015/Emergence to A.H. A.H. was also funded by the AquaCRISPR (ANR-16-COFA-0004-01), TUNESAL (Research Project- HAVBRUK2, PN: 294971), Higher Education Discipline Innovation project: 111 Project (China, Grant No. D20007), and AquaExcel3.0 (Grant Agreement No. 871108) projects.
Author contributions: A.H., J.A.L., and K.M.N. acquired the funding; A.H., J.A.L., K.M.N., M.S., A.H.-V., and G.W.G. conceived and planned the study; A.H., A.D., Y.G., J.B., A.H.-V., E.S.H., A.O., and G.Y. performed the experiments; A.H., M.S., J.A.L., J.B., K.M.N., A.H.-V., and G.W.G. analyzed the data and discussed the experiments; A.H., M.S., and J.A.L. wrote the original draft of the article.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.274266.120.
Competing interest statement
The authors declare no competing interests.
References
- Britten RJ, Davidson EH. 1969. Gene regulation for higher cells: a theory. Science 165: 349–357. 10.1126/science.165.3891.349 [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. 1971. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol 46: 111–138. 10.1086/406830 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, Zhou LY, Chaudhari A, Iguchi T, Nagahama Y. 2016. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci Rep 6: 19480. 10.1038/srep19480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science 251: 1030–1033. 10.1126/science.1998119 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. 2005. Sex chromosomes: evolution of the weird and wonderful. Curr Biol 15: R129–R131. 10.1016/j.cub.2005.02.011 [DOI] [PubMed] [Google Scholar]
- Chuong EB. 2013. Retroviruses facilitate the rapid evolution of the mammalian placenta. Bioessays 35: 853–861. 10.1002/bies.201300059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MA, Massee KC, Wade TH, Oden SM, Jensen C, Jasonowicz A, Immerman DA, Goetz FW. 2015. Culture of sablefish (Anoplopoma fimbria) larvae in four experimental tank designs. Aquacult Eng 69: 43–49. 10.1016/j.aquaeng.2015.09.003 [DOI] [Google Scholar]
- Eldredge N, Gould SJ. 1972. Punctuated equilibria: an alternative to phyletic gradualism. Freeman, Cooper and Company, San Francisco. [Google Scholar]
- Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP. 2012. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol Biol Evol 29: 239–247. 10.1093/molbev/msr189 [DOI] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7: e47768. 10.1371/journal.pone.0047768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forconi M, Canapa A, Barucca M, Biscotti MA, Capriglione T, Buonocore F, Fausto AM, Makapedua DM, Pallavicini A, Gerdol M, et al. 2013. Characterization of sex determination and sex differentiation genes in Latimeria. PLoS One 8: e56006. 10.1371/journal.pone.0056006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Le Gac F, Lareyre J-J. 2011. The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 472: 7–17. 10.1016/j.gene.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Glinsky GV. 2015. Transposable elements and DNA methylation create in embryonic stem cells human-specific regulatory sequences associated with distal enhancers and noncoding RNAs. Genome Biol Evol 7: 1432–1454. 10.1093/gbe/evv081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci 108: 1513–1518. 10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiguen Y, Fostier A, Herpin A. 2018. Sex determination and differentiation in fish. In Sex control in aquaculture (ed. Wang H.-P., et al. ), pp. 35–63. J. Wiley, New York. https://onlinelibrary.wiley.com/doi/abs/10.1002/9781119127291.ch2 [Google Scholar]
- Hackl T, Hedrich R, Schultz J, Förster F. 2014. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30: 3004–3011. 10.1093/bioinformatics/btu392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman ES, Fairgrieve WT, Luckenbach JA. 2021. Molecular and morphological sex differentiation in sablefish (Anoplopoma fimbria), a marine teleost with XX/XY sex determination. Gene 764: 145093. 10.1016/j.gene.2020.145093 [DOI] [PubMed] [Google Scholar]
- Herpin A, Schartl M. 2008. Regulatory putsches create new ways of determining sexual development. EMBO Rep 9: 966–968. 10.1038/embor.2008.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M. 2009. Molecular mechanisms of sex determination and evolution of the Y-chromosome: insights from the medakafish (Oryzias latipes). Mol Cell Endocrinol 306: 51–58. 10.1016/j.mce.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Herpin A, Schartl M. 2011. Dmrt1 genes at the crossroads: a widespread and central class of sexual development factors in fish. FEBS J 278: 1010–1019. 10.1111/j.1742-4658.2011.08030.x [DOI] [PubMed] [Google Scholar]
- Herpin A, Schartl M. 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep 16: 1260–1274. 10.15252/embr.201540667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Nakamura S, Wagner TU, Tanaka M, Schartl M. 2009. A highly conserved cis-regulatory motif directs differential gonadal synexpression of Dmrt1 transcripts during gonad development. Nucleic Acids Res 37: 1510–1520. 10.1093/nar/gkn1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Braasch I, Kraeussling M, Schmidt C, Thoma EC, Nakamura S, Tanaka M, Schartl M. 2010. Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet 6: e1000844. 10.1371/journal.pgen.1000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Liu T, Zhao H, Xu H, Wang W, Liu R, Chen T, Deng J, Gui J. 2004. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci 101: 8011–8016. 10.1073/pnas.0308668101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasonowicz AJ, Goetz FW, Goetz GW, Nichols KM. 2017. Love the one you're with: genomic evidence of panmixia in the sablefish (Anoplopoma fimbria). Can J Fish Aquat Sci 74: 377–387. 10.1139/cjfas-2016-0012 [DOI] [Google Scholar]
- Kaneko H, Ijiri S, Kobayashi T, Izumi H, Kuramochi Y, Wang D-S, Mizuno S, Nagahama Y. 2015. Gonadal soma-derived factor (gsdf), a TGF-β superfamily gene, induces testis differentiation in the teleost fish Oreochromis niloticus. Mol Cell Endocrinol 415: 87–99. 10.1016/j.mce.2015.08.008 [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2007. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet 23: 521–529. 10.1016/j.tig.2007.08.004 [DOI] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Hamaguchi S. 2013. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn 242: 339–353. 10.1002/dvdy.23927 [DOI] [PubMed] [Google Scholar]
- Klüver N, Herpin A, Braasch I, Drieβle J, Schartl M. 2009. Regulatory back-up circuit of medaka Wt1 co-orthologs ensures PGC maintenance. Dev Biol 325: 179–188. 10.1016/j.ydbio.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Luckenbach JA, Dickey JT, Swanson P. 2011. Follicle-stimulating hormone regulation of ovarian transcripts for steroidogenesis-related proteins and cell survival, growth and differentiation factors in vitro during early secondary oocyte growth in coho salmon. Gen Comp Endocrinol 171: 52–63. 10.1016/j.ygcen.2010.12.016 [DOI] [PubMed] [Google Scholar]
- Luckenbach JA, Fairgrieve WT, Hayman ES. 2017. Establishment of monosex female production of sablefish (Anoplopoma fimbria) through direct and indirect sex control. Aquaculture 479: 285–296. 10.1016/j.aquaculture.2017.05.037 [DOI] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1159. 10.1038/ng.917 [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grützner F, Bauersachs S, et al. 2015. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep 10: 551–561. 10.1016/j.celrep.2014.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mason JC, Beamish RJ, McFarlane GA. 1983. Sexual maturity, fecundity, spawning, and early life history of sablefish (Anoplopoma fimbria) off the Pacific coast of Canada. Can J Fish Aquat Sci 40: 2126–2134. 10.1139/f83-247 [DOI] [Google Scholar]
- Mayr E. 1997. Evolution and the diversity of life: selected essays. Harvard University Press, Cambridge, MA. [Google Scholar]
- Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, Naruse K, Hamaguchi S, Sakaizumi M. 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. 10.1534/genetics.111.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M. 2015. Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda) 5: 2685–2691. 10.1534/g3.115.021543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. 2011. Aquatic animals. In Guide for the care and use of laboratory animals, 8th ed., pp. 77−103. The National Academies Press, Washington, DC. 10.17226/12910 [DOI] [Google Scholar]
- Notwell JH, Chung T, Heavner W, Bejerano G. 2015. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nat Commun 6: 6644. 10.1038/ncomms7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octavera A, Yoshizaki G. 2019. Production of donor-derived offspring by allogeneic transplantation of spermatogonia in Chinese rosy bitterling. Biol Reprod 100: 1108–1117. 10.1093/biolre/ioy236 [DOI] [PubMed] [Google Scholar]
- Pan Q, Anderson J, Bertho S, Herpin A, Wilson C, Postlethwait JH, Schartl M, Guiguen Y. 2016. Vertebrate sex-determining genes play musical chairs. C R Biol 339: 258–262. 10.1016/j.crvi.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Feron R, Yano A, Guyomard R, Jouanno E, Vigouroux E, Wen M, Busnel J-M, Bobe J, Concordet J-P, et al. 2019. Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet 15: e1008013. 10.1371/journal.pgen.1008013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Feron R, Jouanno E, Darras H, Herpin A, Koop B, Rondeau E, Goetz FW, Larson WA, Bernatchez L, et al. 2021. The rise and fall of the ancient northern pike master sex-determining gene. eLife 10: e62858. 10.7554/eLife.62858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol 14: 1416–1424. 10.1016/j.cub.2004.08.030 [DOI] [PubMed] [Google Scholar]
- Peichel CL, McCann SR, Ross JA, Naftaly AFS, Urton JR, Cech JN, Grimwood J, Schmutz J, Myers RM, Kingsley DM, et al. 2020. Assembly of the threespine stickleback Y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biol 21: 177. 10.1186/s13059-020-02097-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42. 10.1146/annurev-genet-110711-155621 [DOI] [PubMed] [Google Scholar]
- Rhodes FHT. 1983. Gradualism, punctuated equilibrium and the Origin of Species. Nature 305: 269–272. 10.1038/305269a0 [DOI] [PubMed] [Google Scholar]
- Rondeau EB, Messmer AM, Sanderson DS, Jantzen SG, von Schalburg KR, Minkley DR, Leong JS, Macdonald GM, Davidsen AE, Parker WA, et al. 2013. Genomics of sablefish (Anoplopoma fimbria): expressed genes, mitochondrial phylogeny, linkage map and identification of a putative sex gene. BMC Genomics 14: 452. 10.1186/1471-2164-14-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. 2009. Turnover of sex chromosomes in the stickleback fishes (gasterosteidae). PLoS Genet 5: e1000391. 10.1371/journal.pgen.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Schories S, Wakamatsu Y, Nagao Y, Hashimoto H, Bertin C, Mourot B, Schmidt C, Wilhelm D, Centanin L, et al. 2018. Sox5 is involved in germ-cell regulation and sex determination in medaka following co-option of nested transposable elements. BMC Biol 16: 16. 10.1186/s12915-018-0485-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. 2008. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453: 930–934. 10.1038/nature06944 [DOI] [PubMed] [Google Scholar]
- Sheldon PR. 2001. Punctuated equilibrium and phyletic gradualism. In Encyclopedia of life sciences. Wiley. https://onlinelibrary.wiley.com/doi/abs/10.1038/npg.els.0001774 [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Mizusaki H, Kawabe K, Kasahara M, Yoshioka H, Morohashi K. 2002. Concerted regulation of gonad differentiation by transcription factors and growth factors. Novartis Found Symp 244: 68–77. [PubMed] [Google Scholar]
- Takehana Y, Matsuda M, Myosho T, Suster ML, Kawakami K, Shin-I T, Kohara Y, Kuroki Y, Toyoda A, Fujiyama A, et al. 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat Commun 5: 4157. 10.1038/ncomms5157 [DOI] [PubMed] [Google Scholar]
- Thoma EC, Wagner TU, Weber IP, Herpin A, Fischer A, Schartl M. 2011. Ectopic expression of single transcription factors directs differentiation of a medaka spermatogonial cell line. Stem Cells Dev 20: 1425–1438. 10.1089/scd.2010.0290 [DOI] [PubMed] [Google Scholar]
- Wang J, Vicente-García C, Seruggia D, Moltó E, Fernandez-Miñán A, Neto A, Lee E, Gómez-Skarmeta JL, Montoliu L, Lunyak VV, et al. 2015. MIR retrotransposon sequences provide insulators to the human genome. Proc Natl Acad Sci 112: E4428–E4437. 10.1073/pnas.1507253112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guan G, Li M, Zhu F, Liu Q, Naruse K, Herpin A, Nagahama Y, Li J, Hong Y. 2016. Autosomal gsdf acts as a male sex initiator in the fish medaka. Sci Rep 6: 19738. 10.1038/srep19738 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.